Abstract
To develop widely-applicable diagnostic and potentially therapeutic approaches overcoming protein heterogeneity in human cancer, we have developed a technology to unbiasedly select high specificity compound(s) that bind any bio-molecule (e.g., proteins, lipids, carbohydrates) presented on the cancer cell surface but not on normal cells. We utilized a peptidomimetic based on-bead two-color (OBTC) combinatorial cell screen that can detect differences between two cell surfaces at high accuracy by looking for beads (where each bead in the library had one peptide-peptoid hybrid on the surface) that only bound cancer but not normal cells. We screened a library of 393,216 compounds targeting HCC4017 lung adenocarcinoma cells (labeled in red) in the presence of HBEC30KT normal bronchial epithelial cells (labeled in green) derived from the same tissue of the same patient. This screen identified a peptide-peptoid hybrid called PPS1 which displayed high specific binding for HCC4017 cancer cells over HBEC30KT cells. Specificity was validated through: on-bead, ELISA-like and magnetic bead pulldown studies; while a scrambled version of PPS1 did not show any binding. Of interest, the simple dimeric version (PPS1D1) displayed cytotoxic activity on HCC4017 cells, but not on normal HBEC30KT cells. PPS1D1 also strongly accumulated in HCC4017 lung cancer xenografts in mice over control constructs. We conclude that such combinatorial screens using tumor and normal cells from the same patient have significant potential to develop new reagents for cancer biology, diagnosis, and potentially therapy.
INTRODUCTION
A standard approach in drug development is to target bio-molecules that have known functions related to a given disease state. The majority of these bio-molecules are proteins such as enzymes, hormones, receptors, or signaling molecules. While this approach has been successful in some cases, targeting immensely diverse pathological states such as cancer through this conventional approach is challenging.1-5 The expression level of target proteins is highly variable between cancer types6, and even between different cancer cells within a single tumor.7, 8 Additionally, cross-talk between signaling networks often results in compensation that blunts the efficacy of targeting a single pathway.3 For cancer therapy, combination of drugs is often used to improve efficacy; however, an increase in therapy-related toxicity is a frequent consequence of this approach. Therefore, new approaches that are not constrained by limitations of protein targets are needed.
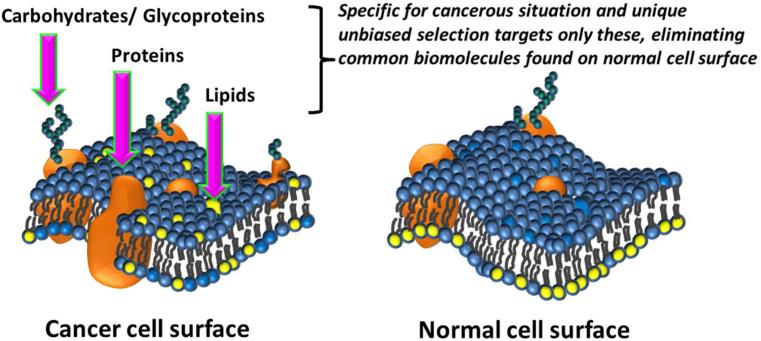
Anionic phospholipids, sialic acid residues and heparin sulfates are examples of molecular classes that are overexpressed on the cancer cell surface that have not been exploited extensively for anti-cancer drug development. These targets are often absent on normal cells and present universally in cancer cells.9-13 Therefore, targeting such non-protein biomolecules may provide a unique opportunity to address some of the challenges associated with drugs that target heterogeneously expressed proteins in cancer. Current technologies to develop drug leads are extensively based on macromolecular structural characteristics of proteins and cannot easily be applied to directly target lipids or carbohydrates. We set out to develop screening strategies that can directly target lipid, carbohydrates or proteoglycans as well as proteins on the surface of tumor cells. Here we propose to consider cellular differences by directly targeting cancer cells over normal cells derived from same person using a suitable combinatorial high throughput screening approach. The goal would be to develop an unbiased selection method that could recognize a biomolecule on the cancer cell surface which is most importantly not found on the normal cell surface, comparing cancer cell vs normal cells simultaneously (Figure 1). This biomolecule could still be a protein, but if we apply this selection criterion carefully, it will give an equal chance to recognize a lipid or a carbohydrate specifically found on the cancer cell surface (Figure 1). This approach may even find compounds that may target combinations of biomolecules (e.g. glycoproteins) or higher order structural arrangements of those biomolecules that are unique for cancerous situation as they present naturally on the cancer cell surface. The key point here is to apply a method that can eliminate compounds targeting all the biomolecules on the normal cell surface and pick a compound that target any additionally expressed biomolecule on cancer cell surface. This approach does not need any prior knowledge of the biomolecule we target and that can be identified later.
Figure 1.
Schematic comparison of bimolecular asymmetry in cancer and normal cell membranes. Cancer cell surface may display specific protein, lipid, carbohydrates and glycoproteins that are expressed under cancerous situation that may be absent or minimal on normal cell surface under healthy biological conditions. These specific biomolecules will be targeted in the unbiased selection.
There are several reported methods for unbiased selection of cell surface targeted compounds. Phage display, introduced about two decades ago14, has proven to be an excellent method to identify high specificity peptides as cell surface markers.15-17 Unbiased phage display peptide selection has been used to target a particular cell surface without prior knowledge of the target.18 But, the methodology is time consuming and limited to natural peptides, which have limited serum stability and can be immunogenic. Later, live cell screening methods using large combinatorial libraries of natural and unnatural amino acids containing synthetic peptides has been reported.19 Even though some of these methods contain a secondary screening step to eliminate compounds that bind to control cells20, this requires a subsequent step and additional time and resources. To address these concerns, we previously reported a rapid and convenient on-bead two-color (OBTC) cell screen to directly identify high specificity ligands for known cell surface receptors. The assay is based on real time comparison of two identical cell groups that differ only by a single receptor type on the cell surface and we identified high specificity peptoid ligands for VEGFR2.21 This assay was subsequently used to select high specificity peptoid ligands for EAE (experimental autoimmune encephalomyelitis) responsive T-cell receptors that are elevated as compared to normal cells T-cell populations.22 Herein we report a complete unbiased selection application of our OBTC cell screen to identify peptide-peptoid hybrids targeting lung cancer cells over normal bronchial epithelial cells derived from the same patient. Peptoids are emerging as a novel class of biologically compliant compounds with rapid and cost effective synthesis and optimization.21, 23-28 They are protease insensitive, cell permeable, highly diverse, and non-immunogenic and have been reported as effective antagonists for various bio-molecules.29-33 The minimum pharmacophore of these peptoids can be easily identified24 and that knowledge can be used to rapidly optimize activities.34 We and others have shown that peptoids can be applied in therapeutic21, 35-37 and diagnostic38 applications in vivo.
Our overall strategy for unbiased selection of high specificity ligands that target any type of bio-molecules on the cancer cell surface involved the following steps: (I) Design and synthesis of peptide-peptoid combinatorial library. Here we introduce amino acids to a limited number of positions in a mainly peptoid-based 8-mer library, increasing the structural diversity. (II) A suitable cancer and normal cell pair were selected. We carefully selected cancer (test) and normal (control) cells derived from the same patient to increase the likelihood of targeting cancer specific bio-molecules. (III) We exploited a rapid, reliable, and economical way to unbiasedly select ligands using our unique OBTC cell screen. (IV) Robust validation methods to confirm the binding, specificity and activity of the compounds were performed.
RESULTS AND DISCUSSION
Design and synthesis of peptide-peptoid hybrid library
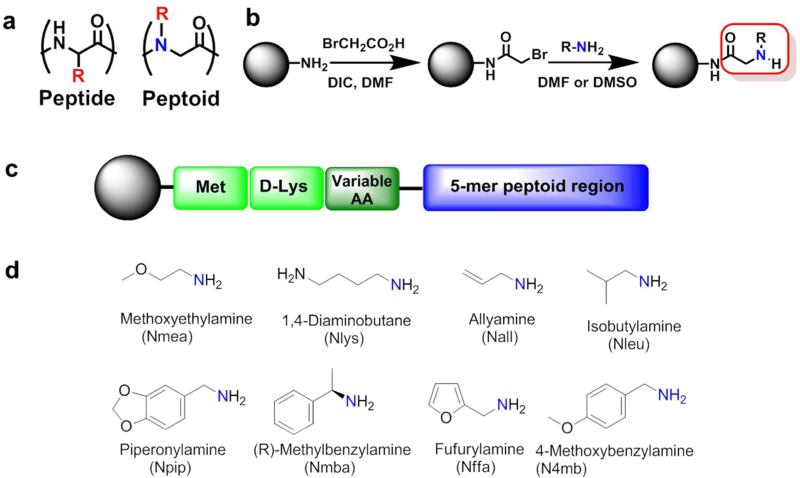
As mentioned, peptoids have a great potential to rapidly move from the “bench to bedside”, thus, they were chosen as the most suitable molecular class for this study. Peptoids are oligo-N-substituted glycines and closely resemble peptides except that the side chains extend from the main chain nitrogen rather than the α-carbon (Figure 2a). Peptoid synthesis is straightforward; bromoacetic acid coupling brings the 2 carbon unit and the Br can be replaced by any amine group (Figure 2b), completing each of these reactions in less than one minute using microwave assisted protocol. First we developed a unique one-bead-one-compound combinatorial library with theoretical diversity of 393,216 permutations. Each of those sequences contains three amino acids followed by a 5-mer highly diversified peptoid region (Figure 2c). Methionine at the first position supports Cyanogen bromide (CNBr) cleavage from tentagel beads for mass spectroscopic sequencing and D-Lysine at the second position acts as a linker. In addition, this positively charged Lys at the base of the sequence reduces aggregation of library molecules and facilitates proper display of peptoids on the bead surface. The third position was randomly filled with one of the 12 different amino acids (Supplementary Figure S1) to improve diversity. All three amino acid positions were carefully designed to avoid vulnerability towards serum proteases and should be stable in biological systems. The next five positions were completely randomized and contain peptoid units developed using eight highly diverse organic amines (Figure 2d). We hypothesized that the peptide-peptoid sequence scaffold may bring additional structural features leading to interesting biological activities.
Figure 2.
The peptide-peptoid hybrid library development. (a) Peptide vs. Peptoid comparison. (b) Two step peptoid synthesis. (c) Library composition; three C-terminal amino acid residues, followed by 5-mer peptoid region. (d) Different amines employed in the 5-mer peptoid region.
Unbiased selection of peptide-peptoid hybrid compounds for cancer cells over normal cells
Our main aim was to identify peptide-peptoid hybrid compounds that specifically distinguish the surface of cancer cells from normal cells. We decided to target HCC4017 lung cancer cells using our OBTC assay,21, 39 as the paired normal HBEC30KT normal epithelial cells from the same patient were available. HCC4017 cells were stained with Qtracker 655 quantum dots (red) and HBEC30KT cells with Qtracker 565 quantum dots (green). Cells were mixed in a 1:1 ratio and exposed to approximately 100,000 library beads (Figure 3a). After 30 minutes incubation with shaking at room temperature, unbound cells were washed off and beads bound only with red-labeled cells (HCC4017) were selected (Figure 3b) as candidates that have high specificity towards HCC4017 cancer cells. The assay was performed 4 times, each time using approximately 100,000 beads to roughly cover the total theoretical diversity of the entire library. Out of the 4 panning attempts only 3 beads that were bound exclusively by HCC4017 cells were identified. Single bead Edman sequencing was used to determine the sequences of the peptide-peptoid hybrids on those 3 beads (Supplementary Figure S2) and the structure of one specific peptide-peptoid hybrid, PPS1 is shown in Figure 3d.
Figure 3.
The On-Bead Two-Color (OBTC) combinatorial cell screen. (a) Schematic representation of the assay. One-bead one-compound library beads (large blue circles) were treated with 1:1 mixture of red and green quantum dot stained HCC4017 cells and HBEC30KT cells respectively. A bead bound only by red cells indicates that the compound on that bead recognized a biomolecule uniquely present on HCC4017 cell surface that is absent (or negligible) in HBEC30KT cell surface. (b & c) Fluorescent microscopic images of beads; (b) One of the three beads found with only red stained HCC4017 cells bound. (c) A bead bound to both red and green stained cells (non-specifics). (d) The structure of PPS1 identified from the screen.
In our OBTC assay, a bead bound with only red stained cells (Figure 3b) indicates that the compound on this bead binds to a biomolecule that is present only on the red stained cancer cell surface and that biomolecule is not found or is much less abundant on the green stained normal cell surface. If binding to any common cell surface biomolecule occurs, it will register both red and green cells (Figure 3c). Therefore, highly cancer specific ligands can be rapidly and easily selected. We applied this unique capability of recognizing differences between two cell surfaces of our OBTC assay as our main hypothesis in this study. More importantly, the identified compound may binds to a protein, lipid or carbohydrate unique to this cancer cell surface. One critically important factor here is to have both cancer and normal cells derived from same individual. Otherwise, the differences picked here may be due to the genetic differences between individuals, and not exactly cancerous vs normal differences. After very careful search and considerations, we decided to use the HCC4017 lung cancer cell line as our target, since the HBEC30KT normal immortalized bronchial epithelial cell line that is originated from the lungs of the same patient is available. We have obtained the genetic analysis of both cell lines to make sure both cell lines are derived from the same patient (Supplementary Figure S3).
The selected peptoid-peptide hybrid, PPS1, consists of four hydrophobic residues towards the N-terminus and three positively charged residues towards the C-terminus (Figure 3d). All four hydrophobic residues are peptoid residues and interestingly contain bulkier aromatic rings on each side chain. Two of them contain Oxygen as heteroatoms. From the three positive charges, one was the fixed D-lysine at the 2nd position of the library. The next position was the variable amino acid region and lysine was selected for this 3rd position during the screen. The remaining positive charge at the 4th position from the C-terminus is a peptoid residue with a lysine-like side chain. Very interestingly, one of the other two ‘hits’ identified had almost the same structure as PPS1 – different only by a single residue. These two compounds are currently under investigation and will be published elsewhere.
Binding and specificity validation of PPS1
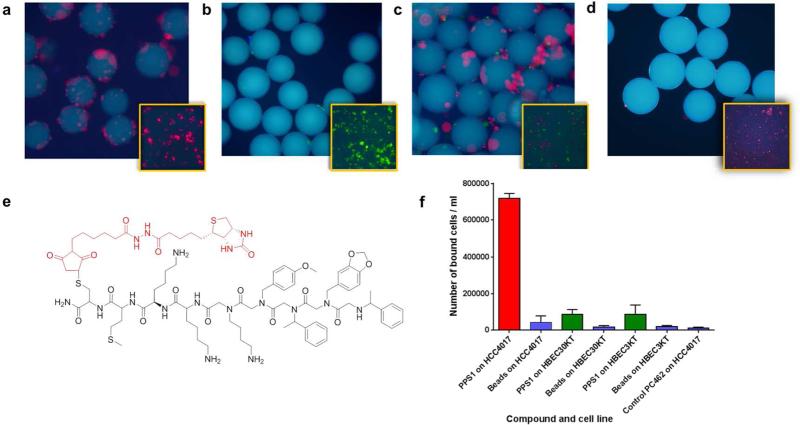
After sequence determination, both qualitative and quantitative methods were used to characterize the binding of PPS1 to cells. First the PPS1 compound was re-synthesized on Tentagel beads and exposed to red quantum dot labeled HCC4017 cells alone (Figure 4a), green quantum dot labeled HBEC30KT cells alone (Figure 4b), and a 1:1 mixture of red and green labeled cells (Figure 4c). PPS1 bearing beads readily bound to HCC4017 lung cancer cells (Figure 4a & c) but rarely bound to green labeled HBEC30KT normal lung cells (Figure 4b & c), validating the high specificity of PPS1 for to HCC4017 lung cancer cells. The scrambled version of PPS1, PC2 (Supplementary Figure S11) bearing beads did not show any binding to HCC4017, indicating the sequence specificity (Figure 4d). We also devised a semi-quantitative magnetic bead pulldown assay to further confirm the specificity of PPS1. Here, PPS1 (Figure 4e) and another non-binding control compound PC462 (Supplementary Figure S4) were synthesized with biotin tag at the C-terminus. Biotin-PPS1 and biotin-PC462 were linked to streptavidin-magnetic beads and then equilibrated with 1 million cells of HCC4017 and HBEC30KT cell types. These magnetic bead bound cells were precipitated with a magnet and counted. As shown in Figure 4f, PPS1 coated magnetic beads readily pulled down about 70-75% of the HCC4017 cells, while only less than 10% of the normal HBEC30KT and HBEC3KT cells (another normal bronchial epithelial cell line) were pulled down. PC462 coated magnetic beads were unable to pulldown HCC4017 cells.
Figure 4.
Qualitative binding and specificity of the PPS1. PPS1 containing tentagel beads were exposed to: (a) Red stained HCC4017 cells alone, (b) Green stained HBEC30KT cells alone, (c) Red and green stained cells at 1:1 mixture. PPS1 predominantly bound to red stained HCC4017 cells over HBEC30KT cells. (d) Red stained HCC4017 cells did not bind to tentagel beads carrying scrambled version PC2. (E) Structure of the c-terminal biotinylated PPS1. (e) Streptavidin-magnetic beads coated with biotinylated PPS1 pulled down only HCC4017, but not HBEC30KT or HBEC3KT cells. The magnetic beads alone or control PC462 compound coated magnetic beads fail to pulldown HCC4017 cells.
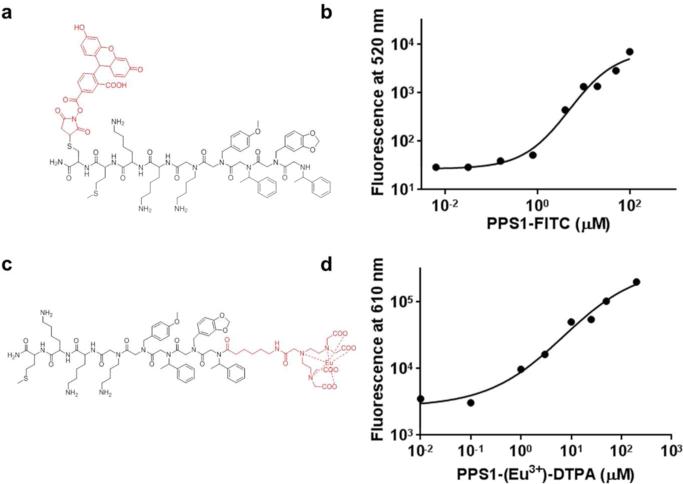
To quantitate binding of PPS1 to HCC4017 cells, an ELISA-like assay using fluorescein isothiocyanate (FITC) labelled PPS1 (Figure 5a) was used. First PPS1 was synthesized with C-terminal cysteine and the thiol group was used to attach FITC through standard maleimide chemistry. HCC4017 cells were grown in 96 well plates and blocked for nonspecific binding. PPS1-FITC was added in serial dilution to the cells, incubated for 1 hr, washed and fluorescence was detected at 520 nm. As shown in Figure 5b this assay indicated that PPS1-FITC binds to HCC4017 cells with a KD ~ 5 μM. We also used a recently published Europium (Eu3+) labelled diethylenetriaminepentaacetic acid (DTPA) based cell surface binding detection assay40. Lanthanide-based (e.g. Eu3+) luminescent ligand binding assays are superior to traditional radio-labelled and FITC-labelled assays due to improved sensitivity and also the capability of eliminating auto-fluorescence challenges. We synthesized DTPA labelled PPS1 and chelated DTPA with Eu3+ (Figure 5c). The binding assay was conducted in standard ELISA-like approach as described above. The binding curve obtained (Figure 5d) using this assay and apparent KD (~7 μM) was similar to what we observed using FITC-PPS1.
Figure 5.
ELISA-like quantitative binding and specificity validation of the PPS1. (a) Chemical structure of c-terminus fluorescein isothiocyanate (FITC) labelled PPS1 (FITC-PPS1). (b) Binding curve of HCC4017 cells with PPS1-FITC indicates KD around 5μM. (c) Chemical structure of N-terminus modified Eu3+-chelated DTPA labelled PPS1. (d) Binding curve of HCC4017 cells with PPS1-(Eu3+)-DTPA indicates KD around 5-7μM.
Further improvements and in vitro activity validation of PPS1
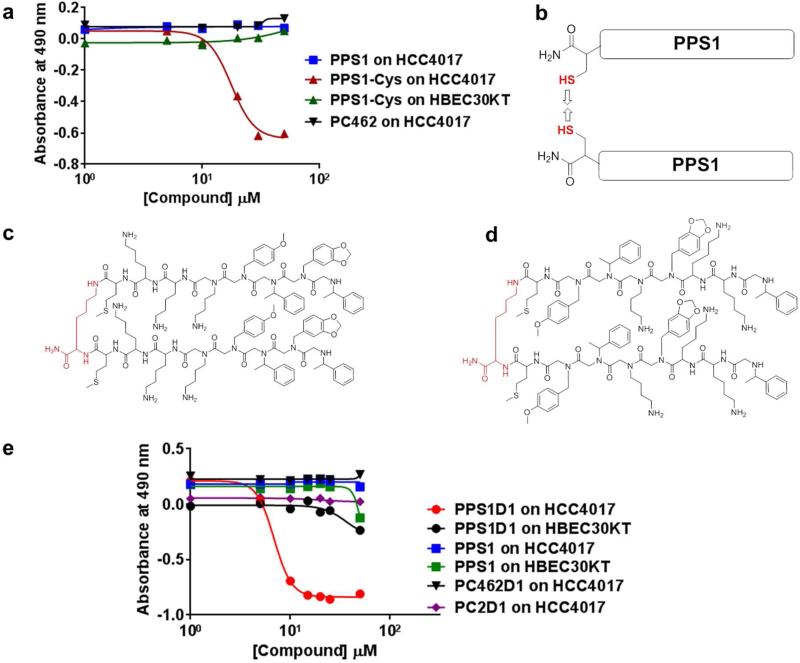
After confirming the binding and specificity of PPS1 for HCC4017 cells, we investigated the biological effects of PPS1 on HCC4017 and HBEC cells. The effect of PPS1 on cell viability was determined using the standard MTS assay. PPS1 or the control compound C462 at concentrations of up to 150 μM had no effect on viability of HCC4017, HBEC30KT and HBEC3KT cells (Figure 6a & e). However when the Cysteine-PPS1 version was used (the intermediate PPS1 synthesized with c-terminal Cysteine, used in developing FITC coupled PPS1), showed moderate cell killing activity on HCC4017 cells (Figure 6a). Toxicity was specific to HCC4017 cells as HBEC30KT and HBEC3KT cells were unaffected. The only difference between Cysteine-PPS1 and PPS1 was a single Cysteine amino acid at the C-terminus. Since it is very difficult to rationalize having a single amino acid residue at the C-terminal of the sequence could bring such a large difference of activity, we thought of other possibilities that might have occurred. Cysteine has a sulfur (-SH) group on the side chain which can easily form disulfide bonds resulting in a dimeric structure of PPS1 (Figure 6b). As reported in the literature, multimerization can improve the binding and activity of compounds.41, 42 For instance, we have previously observed over 90-fold activity improvements through homo-dimerization of the VEGF receptor-2 targeted peptoid we identified using the same OBTC assay.21 To explore this possibility, we synthesized a simple homo-dimeric version of PPS1 by covalently conjugating two monomeric units through a lysine residue using the method we described previously.43 The resulting dimeric version of PPS1 compound, PPS1D1 (Figure 6c), the analogous dimeric versions of scrambled PC2, PC2D1 (Figure 6d) and control compound C462, PC462D1 (Supplementary Figure S5) were then evaluated for activity. In contrast to PPS1, PPS1D1 showed clear cytotoxic activity against HCC4017 cells with an IC50 of ~10 μM (Figure 6e). Importantly, PPS1D1 did not have any activity on either of the normal HBEC cells lines tested (Figure 6e). Additionally, PC2D1 or PC462D1 had no effect on cell viability (Figure 6e).
Figure 6.
Dimerization of PPS1 triggers the activity of PPS1. (a) MTS cell viability assay results on HCC4017 with the treatment of PPS1, Cystein-PPS1 and PC462 and Cystein-PPS1 treated on HBEC30KT. Only Cystein-PPS1 treated with HCC4017 displayed cell killing activity. (b) Cartoon depicting the suspected disulfide bond formation between two c-terminal Cysteine labelled PPS1. (c) Structure of PPS1 homo-dimer PPS1D1. (d) Scrambled version of PPS1D1, PC2D1. (e) MTS cell viability assay results on HCC4017 and HBEC30KT treated with PPS1D1, PPS1, PC2D1, PC462D1. Only PPS1D1 treated with HCC4017 displayed cell killing activity.
These data demonstrate that PPS1 has a high specificity binding towards a biomolecule presented on HCC4017 cells, which is not found or is substantially less abundant in normal HBEC cells. Moreover, upon dimerization PPS1D1 displays considerable cell killing activity on HCC4017 cancer cells and has no effect on HBEC normal cells within the concentration range tested.
In vivo validation of PPS1D1
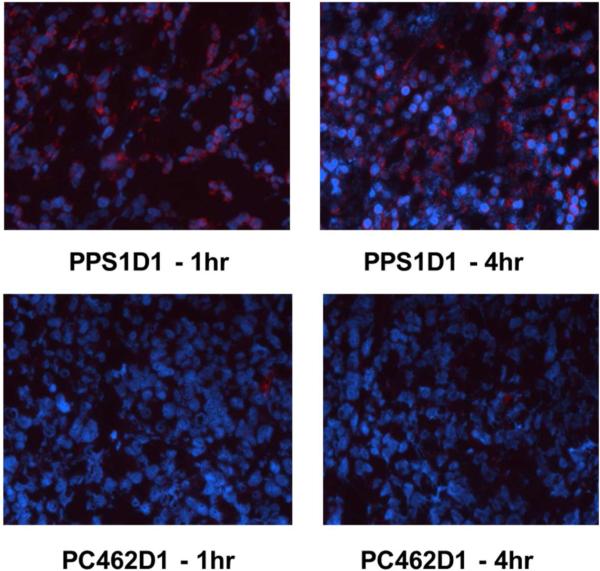
To determine if the specificity of PPS1D1 was maintained in the in vivo setting, we evaluated the localization of PPS1D1 in mice bearing HCC4017 xenografts. Initial Maximum tolerated dose (MTD) studies indicated that the MTD for non-tumor bearing animals was 5 mg/kg and 1 mg/kg for mice bearing subcutaneous HCC4017 xenografts. In vivo localization studies were performed with biotinylated versions of PPS1D1 and PC462D1. Animals were injected iv with 100 μl of biotinylated PPS1D1 or PC462D1 (500 μg/ml) and sacrificed at 1 hr and 4 hr post injection. Tissue was collected; snap frozen, sectioned and the presence of the PPS1D1 and PC462D1 determined by streptavidin-Cy3. Biotinylated PPS1D1 accumulated in the tumor microenvironment at 1 hr and with increased signal intensity at 4hr (Figure 7). There was no detectable signal from the control compound at either time point. This indicates the specificity of PPS1D1 towards HCC4017 cells was promptly maintained in vivo as well.
Figure 7.
Tumor accumulation studies of PPS1D1 and control P462D1 compounds on HCC4017 xenografts in NOD/SCID mice. PPS1D1 strongly accumulated in the tumor in both at 1 hr and 4 hr time points, while the control PC462D1 was not detected.
In conclusion, we applied our OBTC combinatorial cell screening technology to identify a highly specific peptide-peptoid hybrid PPS1 for HCC4017 lung cancer cells. The screen was unique in that it exploited the use of lung cancer cells and normal human bronchial epithelial cells (HBEC30KT) derived from the same patient. Our hypothesis was to apply this assay to identify compounds that can recognize any type of bio-molecule (e.g. protein, lipid or carbohydrate) only present on the HCC4017 lung cancer cell surface. We found that the monomeric version of PPS1 while specific had little biologic activity; however the dimer, PPS1D1 displayed cell killing activity of HCC4017 lung cancer cells, but not on normal cells. Further we demonstrated that PPS1D1 specifically accumulated in HCC4017 xenografts. These data highlight the potential of the OBTC unbiased selection approach to effectively bypass the time and resource consuming conventional drug lead development approach, where knowledge of the targeted bio-molecule is a prerequisite. The next major goal of this study is to identify the target of PPS1 on HCC4017 cells. This technology can be applied to other cancer types providing a platform for unbiased selection of high specificity ligands for various disease specific biological targets.
METHODS
Cell lines
Lung cancer cell line HCC4017 and normal HBEC30KT and HBEC3KT cell lines were obtained from the cell collection of Dr. John Minna's research group at UT Southwestern Medical Center.44 HCC4017 was grown in RPMI supplemented with 5% FBS. Normal lung cell line HBEC30KT and HBEC3KT were grown with keratinocyte serum free media (KSFM) supplemented with human recombinant epidermal growth factor and bovine pituitary extract.
Library synthesis
The library consists of three amino acids followed by 5-mer diversified peptoid region. Detailed procedure is listed in the Supplementary Material (S11). Briefly, TentaGel macrobeads were swelled in dimethylformamide (DMF) and treated with premixed 0.4M Fmoc-Met-OH, 0.4M HBTU and 0.8M N-methyl morphaline overnight. Fmoc group was removed by piperidine treatment for 2×10 minutes and Fmoc-D-Lys(Boc)-OH was added as described previously. After removing Fmoc group, the split-pool synthesis protocol was followed to build the diversified region of the library sequence.45 Briefly, the beads were equally distributed into 12 reaction vessels and 12 different amino acids were added. Then, the beads were pooled together, Fmoc group was removed and divided equally into 8 reaction vessels for adding 8 different amines (peptoid residues) in split-pool fashion. Each of the reaction vessels was twice treated with 2M Bromoacetic acid and 2M DIC in anhydrous DMF for 30s and microwaved for 2×15s with the power set at 10%. After washing with DMF, each reaction vessel was twice treated with 1M solution of the primary amine and microwaved for 2×15s. The beads were washed, pooled and divided equally into 8 reaction vessels for the addition of next peptoid residue. This procedure was repeated until 5-mer peptoid region is completed. Finally beads were washed with Dichloromethane (DCM) and side chain protection groups were cleaved with TFA cleavage cocktail.
On bead two color (OBTC) combinatorial cell binding assay
Detailed procedure is listed in the Supplementary Material (S12).21, 39 Briefly, about 100,000 peptoid library beads were pre-blocked with RPMI medium with 5% FBS and 2% BSA for 1 hour. HCC4017 and HBEC30KT cells were removed from culture plates using GIBCO enzyme free cell dissociation buffer, washed, counted and labelled with red Qtracker 655 (HCC4017) and green Qtracker 565 (HBEC30KT) quantum dots (Invitrogen). After washing, each cell line was re-suspended in RPMI medium with 5% FBS and 2% BSA and mixed thoroughly at 1:1 ratio. Then, this cell mixture was added to polypropylene tubes containing library beads and incubated at room temperature for 30 minutes. The beads were washed and visualized under the fluorescent microscope using DAPI filter. Single positive beads bound only with red stained cells were identified, removed manually and processed for Edman sequencing to identify the sequence of the peptide-peptoid hybrid.
Synthesis of PPS1 compound
Synthesis of PPS1 compound was done on: (I) TentaGel resin (for on-bead cell binding assay), and (II) NovaSyn TGR resin (for all the other assays). First three amino acids, Fmoc-Met-OH, Fmoc–D-Lys(Boc)-OH and Fmoc-Lys(Boc)-OH were loaded to the resin after Fmoc removal each time. Then 5-mer peptoid region containing Boc-Diaminobutane, 4-methoxybenzylamine, (R)-Methylbenzylamine, Piperonylamine and (R)-Methylbenzylamine was completed using microwave assisted peptoid synthesis protocol. At the end, beads were washed with DCM and cleaved off with TFA cleavage cocktail (Please refer to Supporting Information for more synthesis procedures).
Magnetic bead binding assay
The assay was done using Dynabeads M-280 Streptavidin (Invitrogen). Nearly 9×106 beads were transferred, re-suspended in PBS with 0.1% BSA. Biotinylated PPS1 and control PC462 were added to each vial and incubated for 30 minutes at RT. The beads were washed 3 times and 1 million of each HCC4017, HBEC30KT and HBEC3KT cells were added to each tube and incubated for 30 minutes at RT with gentle shaking. The bead bound cells were isolated by placing the vial on the magnet and after removing supernatant, cells were counted with hemocytometer.
ELISA-like binding assay
HCC4017 cells were grown in clear bottom 96 well plates and blocked with 100 μl of 5% BSA in PBS for 15 minutes. After removing blocking solution, each well was treated with concentration gradients of 50μl of FITC-PPS1 or PPS1-(Eu3+)-DTPA compounds and incubated for 45 minutes at room temperature. Wells were washed with PBS and fluorescence was measured at 520 nm (for FITC) and 610 nm [for (Eu3+)-DTPA] using the plate reader.40
Cell viability assay
HCC4017 and HBEC30KT cells were grown in clear bottom 96 well plates. On second day, cells were treated with PPS1, PPS1D1, PPS1-Cys and control PC462D1 in RPMI medium with 5% FBS containing 3% BSA. For HBEC30KT, Keratinocyte-SFM with 3% BSA media was used. On day 4, the treatment was repeated. On day 5, 20 μl of CellTiter 96® AQueous One Solution (Promega) was added to each well and absorbance was measured at 490 nm.
In vivo studies
All animals were housed in a pathogen-free facility with continuous access to food and water. Experiments were approved by and performed in accordance with the Institutional Animal Care and Use Committee at the University of Texas Southwestern. Mice were purchased from the core breeding facility at UT Southwestern. For MTD studies non-tumor bearing or tumor bearing animals were injected iv with the following doses: 0.2 mg/kg, 1 mg/kg, or 5 mg/kg. Animals were subsequently monitored immediately afterwards and hourly for 8 hrs. Animals were sacrificed if signs of distress were significant (labored breathing, lethargy, loss of mobility). For tumor localization studies six- to eight-week-old female NOD/SCID mice were injected with 2.5×106 HCC4017 cells subcutaneously. Tumors were used for localization when they reached ~500 mm3 in volume. Biotinylated PPS1D1 or PC462D1 (control) were diluted in saline and injected iv (100 μl at 500 μg/ml, n=3/compound/time point). Animals were sacrificed at 1 and 4 hrs post injection. Tissues including the tumors were harvested, snap frozen and sectioned. Detection of biotinylated peptoid-peptide hybrid was conducted by streptavidin-Cy3 and visualized by fluorescence microscopy.
Supplementary Material
ACKNOWLEDGEMENT
We thank Drs. M. Peyton and J. Larsen at University of Texas Southwestern Medical Center (UTSW) for their initial help in establishing cancer and normal cell lines as well as important discussions. We also thank Dr. L. Girard at UTSW for confirming the genetic background of HCC4017 and HBEC30KT cell lines and Proteomics Core Facility at UTSW for conducting Edman Sequencing. We also thank Dr. K. Brown at UTSW for providing access to plater reader and MALDI mass spectrometer. This work was supported by National Cancer Institute (NCI) at National Institute of Health (NIH) grant R01CA175779, Cancer Prevention and Research Institute of Texas Grant RP130258, Lung Cancer SPORE P50CA70907 grant and funding from University of Texas Southwestern Medical Center.
Footnotes
ASSOCIATED CONTENT
Supporting Information Available:
Detailed procedures, Edman sequencing data, HCC4017 and HBEC30KT genetic analysis data, all the compound structures and characterizations (MALDI-TOF and HPLC spectra) are available as Supporting Information free of charge via the Internet.
The authors declare competing financial interests in the form of a pending patent application.
REFERENCES
- 1.Sawyers C. Targeted cancer therapy. Nature. 2004;432:294–297. doi: 10.1038/nature03095. [DOI] [PubMed] [Google Scholar]
- 2.Petrelli A, Giordano S. From single- to multi-target drugs in cancer therapy: when aspecificity becomes an advantage. Current medicinal chemistry. 2008;15:422–432. doi: 10.2174/092986708783503212. [DOI] [PubMed] [Google Scholar]
- 3.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 4.Singh A, Settleman J. EMT, cancer stem cells and drug resistance: an emerging axis of evil in the war on cancer. Oncogene. 2010;29:4741–4751. doi: 10.1038/onc.2010.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clevers H. The cancer stem cell: premises, promises and challenges. Nature medicine. 2011;17:313–319. doi: 10.1038/nm.2304. [DOI] [PubMed] [Google Scholar]
- 6.Ross DT, Scherf U, Eisen MB, Perou CM, Rees C, Spellman P, Iyer V, Jeffrey SS, Van de Rijn M, Waltham M, Pergamenschikov A, Lee JC, Lashkari D, Shalon D, Myers TG, Weinstein JN, Botstein D, Brown PO. Systematic variation in gene expression patterns in human cancer cell lines. Nature genetics. 2000;24:227–235. doi: 10.1038/73432. [DOI] [PubMed] [Google Scholar]
- 7.Marusyk A, Polyak K. Tumor heterogeneity: causes and consequences. Biochimica et biophysica acta. 2010;1805:105–117. doi: 10.1016/j.bbcan.2009.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Meacham CE, Morrison SJ. Tumour heterogeneity and cancer cell plasticity. Nature. 2013;501:328–337. doi: 10.1038/nature12624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Riedl S, Rinner B, Asslaber M, Schaider H, Walzer S, Novak A, Lohner K, Zweytick D. In search of a novel target - phosphatidylserine exposed by non-apoptotic tumor cells and metastases of malignancies with poor treatment efficacy. Biochimica et biophysica acta. 2011;1808:2638–2645. doi: 10.1016/j.bbamem.2011.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ran S, He J, Huang X, Soares M, Scothorn D, Thorpe PE. Antitumor effects of a monoclonal antibody that binds anionic phospholipids on the surface of tumor blood vessels in mice. Clinical cancer research : an official journal of the American Association for Cancer Research. 2005;11:1551–1562. doi: 10.1158/1078-0432.CCR-04-1645. [DOI] [PubMed] [Google Scholar]
- 11.Balasubramanian K, Schroit AJ. Aminophospholipid asymmetry: A matter of life and death. Annual review of physiology. 2003;65:701–734. doi: 10.1146/annurev.physiol.65.092101.142459. [DOI] [PubMed] [Google Scholar]
- 12.Sasisekharan R, Shriver Z, Venkataraman G, Narayanasami U. Roles of heparan-sulphate glycosaminoglycans in cancer. Nature reviews. Cancer. 2002;2:521–528. doi: 10.1038/nrc842. [DOI] [PubMed] [Google Scholar]
- 13.Plucinsky MC, Riley WM, Prorok JJ, Alhadeff JA. Total and lipid-associated serum sialic acid levels in cancer patients with different primary sites and differing degrees of metastatic involvement. Cancer. 1986;58:2680–2685. doi: 10.1002/1097-0142(19861215)58:12<2680::aid-cncr2820581222>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 14.Scott JK, Smith GP. Searching for peptide ligands with an epitope library. Science. 1990;249:386–390. doi: 10.1126/science.1696028. [DOI] [PubMed] [Google Scholar]
- 15.Brown KC. New approaches for cell-specific targeting: identification of cell-selective peptides from combinatorial libraries. Current opinion in chemical biology. 2000;4:16–21. doi: 10.1016/s1367-5931(99)00045-9. [DOI] [PubMed] [Google Scholar]
- 16.Landon LA, Deutscher SL. Combinatorial discovery of tumor targeting peptides using phage display. Journal of cellular biochemistry. 2003;90:509–517. doi: 10.1002/jcb.10634. [DOI] [PubMed] [Google Scholar]
- 17.Shadidi M, Sioud M. Selection of peptides for specific delivery of oligonucleotides into cancer cells. Methods Mol Biol. 2004;252:569–580. doi: 10.1385/1-59259-746-7:569. [DOI] [PubMed] [Google Scholar]
- 18.Elayadi AN, Samli KN, Prudkin L, Liu YH, Bian A, Xie XJ, Wistuba, Roth JA, McGuire MJ, Brown KC. A peptide selected by biopanning identifies the integrin alphavbeta6 as a prognostic biomarker for nonsmall cell lung cancer. Cancer research. 2007;67:5889–5895. doi: 10.1158/0008-5472.CAN-07-0245. [DOI] [PubMed] [Google Scholar]
- 19.Aina OH, Liu R, Sutcliffe JL, Marik J, Pan CX, Lam KS. From combinatorial chemistry to cancer-targeting peptides. Molecular pharmaceutics. 2007;4:631–651. doi: 10.1021/mp700073y. [DOI] [PubMed] [Google Scholar]
- 20.Peng L, Liu R, Marik J, Wang X, Takada Y, Lam KS. Combinatorial chemistry identifies high-affinity peptidomimetics against alpha4beta1 integrin for in vivo tumor imaging. Nature chemical biology. 2006;2:381–389. doi: 10.1038/nchembio798. [DOI] [PubMed] [Google Scholar]
- 21.Udugamasooriya DG, Dineen SP, Brekken RA, Kodadek T. A peptoid “antibody surrogate” that antagonizes VEGF receptor 2 activity. Journal of the American Chemical Society. 2008;130:5744–5752. doi: 10.1021/ja711193x. [DOI] [PubMed] [Google Scholar]
- 22.Gocke AR, Udugamasooriya DG, Archer CT, Lee J, Kodadek T. Isolation of antagonists of antigen-specific autoimmune T cell proliferation. Chemistry & biology. 2009;16:1133–1139. doi: 10.1016/j.chembiol.2009.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Simon RJ, Kania RS, Zuckermann RN, Huebner VD, Jewell DA, Banville S, Ng S, Wang L, Rosenberg S, Marlowe CK, et al. Peptoids: a modular approach to drug discovery. Proceedings of the National Academy of Sciences of the United States of America. 1992;89:9367–9371. doi: 10.1073/pnas.89.20.9367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Udugamasooriya DG, Dunham G, Ritchie C, Brekken RA, Kodadek T. The pharmacophore of a peptoid VEGF receptor 2 antagonist includes both side chain and main chain residues. Bioorganic & medicinal chemistry letters. 2008;18:5892–5894. doi: 10.1016/j.bmcl.2008.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zuckermann RN, Kerr JM, Kent SBH, Moos WH. Efficient method for the preparation of peptoids [oligo(N-substituted glycines)] by submonomer solid-phase synthesis. In J. Am. Chem. Soc. 1992:10646–10647. [Google Scholar]
- 26.Zuckermann RN, Kodadek T. Peptoids as potential therapeutics. Curr Opin Mol Ther. 2009;11:299–307. [PubMed] [Google Scholar]
- 27.Fowler SA, Blackwell HE. Structure-function relationships in peptoids: recent advances toward deciphering the structural requirements for biological function. Org Biomol Chem. 2009;7:1508–1524. doi: 10.1039/b817980h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yoo B, Kirshenbaum K. Peptoid architectures: elaboration, actuation, and application. Current opinion in chemical biology. 2008;12:714–721. doi: 10.1016/j.cbpa.2008.08.015. [DOI] [PubMed] [Google Scholar]
- 29.Reddy MM, Kodadek T. Protein “fingerprinting” in complex mixtures with peptoid microarrays. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:12672–12677. doi: 10.1073/pnas.0501208102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Simpson LS, Burdine L, Dutta AK, Feranchak AP, Kodadek T. Selective toxin sequestrants for the treatment of bacterial infections. Journal of the American Chemical Society. 2009;131:5760–5762. doi: 10.1021/ja900852k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Alluri P, Liu B, Yu P, Xiao X, Kodadek T. Isolation and characterization of coactivator-binding peptoids from a combinatorial library. Molecular bioSystems. 2006;2:568–579. doi: 10.1039/b608924k. [DOI] [PubMed] [Google Scholar]
- 32.Alluri PG, Reddy MM, Bachhawat-Sikder K, Olivos HJ, Kodadek T. Isolation of protein ligands from large peptoid libraries. Journal of the American Chemical Society. 2003;125:13995–14004. doi: 10.1021/ja036417x. [DOI] [PubMed] [Google Scholar]
- 33.Astle JM, Udugamasooriya DG, Smallshaw JE, Kodadek T. A VEGFR2 antagonist and other peptoids evade immune recognition. Int J Pept Res Ther. 2008;14:223–227. [Google Scholar]
- 34.Lynn KD, Udugamasooriya DG, Roland CL, Castrillon DH, Kodadek TJ, Brekken RA. GU81, a VEGFR2 antagonist peptoid, enhances the anti-tumor activity of doxorubicin in the murine MMTV-PyMT transgenic model of breast cancer. BMC cancer. 2010;10:397. doi: 10.1186/1471-2407-10-397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kapoor R, Eimerman PR, Hardy JW, Cirillo JD, Contag CH, Barron AE. Efficacy of Antimicrobial Peptoids against Mycobacterium tuberculosis. Antimicrob Agents Ch. 2011;55:3058–3062. doi: 10.1128/AAC.01667-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Levine PM, Lee E, Greenfield A, Bonneau R, Logan SK, Garabedian MJ, Kirshenbaum K. Androgen Receptor Antagonism by Divalent Ethisterone Conjugates in Castrate-Resistant Prostate Cancer Cells. ACS chemical biology. 2012;7:1693–1701. doi: 10.1021/cb300332w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee J, Udugamasooriya DG, Lim HS, Kodadek T. Potent and selective photo-inactivation of proteins with peptoid-ruthenium conjugates. Nature chemical biology. 2010;6:258–260. doi: 10.1038/nchembio.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.De Leon-Rodriguez LM, Lubag A, Udugamasooriya DG, Proneth B, Brekken RA, Sun X, Kodadek T, Dean Sherry A. MRI Detection of VEGFR2 in Vivo Using a Low Molecular Weight Peptoid-(Gd)(8)-Dendron for Targeting. Journal of the American Chemical Society. 2010 doi: 10.1021/ja105563a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Udugamasooriya DG, Kodadek T. On-Bead Two-Color (OBTC) Cell Screen for Direct Identification of Highly Selective Cell Surface Receptor Ligands. Current protocols in chemical biology. 2012;4:35–48. doi: 10.1002/9780470559277.ch110199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.De Silva CR, Vagner J, Lynch R, Gillies RJ, Hruby VJ. Optimization of time-resolved fluorescence assay for detection of europium-tetraazacyclododecyltetraacetic acid-labeled ligand-receptor interactions. Analytical biochemistry. 2010;398:15–23. doi: 10.1016/j.ab.2009.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kiessling LL, Gestwicki JE, Strong LE. Synthetic multivalent ligands in the exploration of cell-surface interactions. Current opinion in chemical biology. 2000;4:696–703. doi: 10.1016/s1367-5931(00)00153-8. [DOI] [PubMed] [Google Scholar]
- 42.Kiessling LL, Gestwicki JE, Strong LE. Synthetic multivalent ligands as probes of signal transduction. Angewandte Chemie. 2006;45:2348–2368. doi: 10.1002/anie.200502794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hooks JC, Matharage JP, Udugamasooriya DG. Development of homomultimers and heteromultimers of lung cancer-specific peptoids. Biopolymers. 2011;96:567–577. doi: 10.1002/bip.21596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim HS, Mendiratta S, Kim J, Pecot CV, Larsen JE, Zubovych I, Seo BY, Kim J, Eskiocak B, Chung H, McMillan E, Wu S, De Brabander J, Komurov K, Toombs JE, Wei S, Peyton M, Williams N, Gazdar AF, Posner BA, Brekken RA, Sood AK, Deberardinis RJ, Roth MG, Minna JD, White MA. Systematic identification of molecular subtype-selective vulnerabilities in non-small-cell lung cancer. Cell. 2013;155:552–566. doi: 10.1016/j.cell.2013.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lam KS, Salmon SE, Hersh EM, Hruby VJ, Kazmierski WM, Knapp RJ. A new type of synthetic peptide library for identifying ligand-binding activity. Nature. 1991;354:82–84. doi: 10.1038/354082a0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.