Abstract
More than 100,000 genetic variants are reported to cause Mendelian disease in humans, but the penetrance - the probability that a carrier of the purported disease-causing genotype will indeed develop the disease - is generally unknown. Here we assess the impact of variants in the prion protein gene (PRNP) on the risk of prion disease by analyzing 16,025 prion disease cases, 60,706 population control exomes, and 531,575 individuals genotyped by 23andMe, Inc. We show that missense variants in PRNP previously reported to be pathogenic are at least 30× more common in the population than expected based on genetic prion disease prevalence. While some of this excess can be attributed to benign variants falsely assigned as pathogenic, other variants have genuine effects on disease susceptibility but confer lifetime risks ranging from <0.1% to ~100%. We also show that truncating variants in PRNP have position-dependent effects, with true loss-of-function alleles found in healthy older individuals, supporting the safety of therapeutic suppression of prion protein expression.
INTRODUCTION
The study of pedigrees with Mendelian disease has been tremendously successful in identifying variants that contribute to severe inherited disorders (1–3). Causal variant discovery is enabled by selective ascertainment of affected individuals, and especially of multiplex families. Although efficient from a gene discovery perspective, the resulting ascertainment bias confounds efforts to accurately estimate the penetrance of disease-causing variants, with profound implications for genetic counseling (4–7). The development of large-scale genotyping and sequencing methods has recently made it tractable to perform unbiased assessments of penetrance in population controls. In several instances, such studies have suggested that previously reported Mendelian variants, as a class, are substantially less penetrant than had been believed (8–11). To date, however, all of these studies have been limited to relatively prevalent (>0.1%) diseases, and point estimates of the penetrance of individual variants have been limited to large copy number variations (8, 11).
Here we demonstrate the use of large-scale population data to infer the penetrance of variants in rare, dominant, monogenic disease, using the example of prion diseases. These invariably fatal neurodegenerative disorders are caused by misfolding of the prion protein (PrP, the product of PRNP) (12) and have an annual incidence of 1 to 2 cases per 1 million population (13). A small, albeit infamous, minority of cases (<1% in recent years (14, 15)) are acquired through dietary or iatrogenic routes. The majority (~85%) of cases are defined as sporadic, occurring in individuals with two wild-type PRNP alleles and no known environmental exposures. Finally, ~15% of cases occur in individuals with rare, typically heterozygous, coding variants in PRNP, including missense variants, truncating variants, and octapeptide repeat insertions or deletions (Table S1). Centralized ascertainment of cases by national surveillance centers (Materials and Methods) makes prion disease a good test case for using reference datasets to assess the penetrance of these variants.
PRNP was conclusively established as a dominant disease gene due to clear Mendelian segregation of a few variants with disease (16–18). Yet ascertainment bias (19), low rates of predictive genetic testing (20), and frequent lack of family history (21, 22) confound attempts to estimate penetrance by survival analysis (19, 23–26). Meanwhile, the existence of non-genetic etiologies leaves doubt as to whether novel variants are causal or coincidental.
A fully penetrant disease genotype should be no more common in the population than the disease that it causes. This observation allows us to leverage two large population control datasets to re-evaluate the penetrance of reported disease variants in PRNP. The recently reported Exome Aggregation Consortium (ExAC) dataset (27) contains variant calls on 60,706 people ascertained for various common diseases, without any ascertainment on neurodegenerative disease. 23andMe’s database contains genotypes on 531,575 customers of its direct-to-consumer genotyping service who have opted in to participate in research, pruned to remove related individuals (first cousins or closer; Materials and Methods), preventing enrichment due to large families with prion disease.
RESULTS
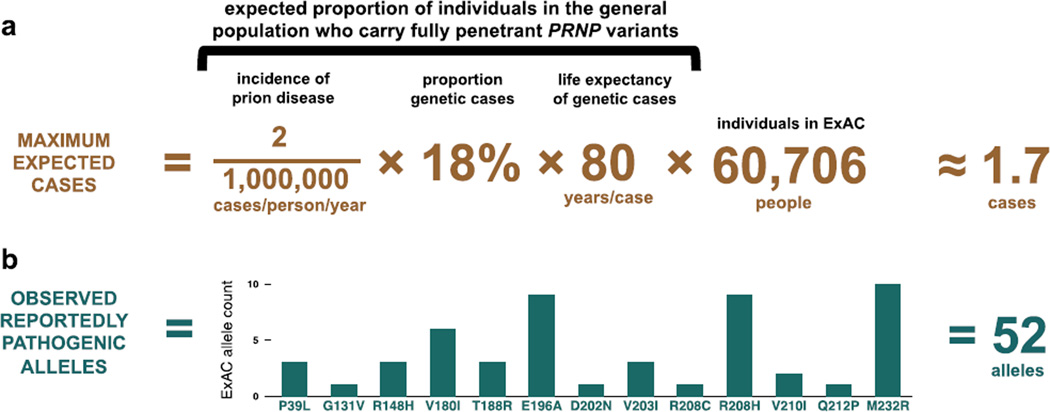
We began by asking whether reportedly pathogenic variants are as rare as expected in these population control datasets. The proportion of people alive in the population today who harbor completely penetrant variants causal for prion disease can be approximated by the product of three numbers: the annual incidence of prion disease, the proportion of cases with such a genetic variant, and the life expectancy of individuals harboring these variants. Based on upper bounds of these numbers (Figure 1A), and assuming ascertainment is neutral with respect to neurodegenerative disease, we would expect no more than ~1.7 such individuals in the 60,706 exomes in the ExAC dataset (27), and ~15 such individuals among the ~530,000 genotyped 23andMe customers who opted to participate in research.
Figure 1. Reportedly pathogenic PRNP variants are >30 times more common in controls than expected based on disease incidence.
Reported prion disease incidence varies with the intensity of surveillance efforts (13), with an apparent upper bound of ~2 cases per million population per year (Materials and Methods). In our surveillance cohorts, 65% of cases underwent PRNP open reading frame sequencing, with 12% of all cases, or 18% of sequenced cases, possessing a rare variant (Table S1), consistent with an oft-cited estimate that 15% of cases of Creutzfeldt-Jakob disease are familial (31). Genetic prion diseases typically strike in midlife, with mean age of onset for different variants ranging from 28 to 77 (22, 32) (Table S10); we accepted 80, a typical human life expectancy, as an upper bound for mean age of onset, and to be additionally conservative, we assumed that all individuals in ExAC and 23andMe were below any age of onset, even though both contain elderly individuals (33) (Figure S1). Thus, no more than ~29 people per million in the general population should harbor high-penetrance prion disease-causing variants. Therefore at most ~1.7 people in ExAC (A) and ~15 people in 23andMe would be expected to harbor such variants. In fact, reportedly pathogenic variants are seen in 52 ExAC individuals (B) and on 141 alleles in the 23andMe database.
Through reviews (28–30) and PubMed searches, we identified 63 rare genetic variants reported to cause prion disease (Table S2). We reviewed ExAC read-level evidence for every rare (<0.1% allele frequency) variant call in PRNP (Materials and Methods; Table S3–4) and found that 52 individuals in ExAC harbor reportedly pathogenic missense variants (Figure 1B), at least a 30-fold excess over expectation if all such variants were fully penetrant. Similarly, in the 23andMe database we observed a total of 141 alleles of 16 reportedly pathogenic variants genotyped on their platform (Table S5).
Individuals with reportedly pathogenic PRNP variants did not cluster within any one cohort within ExAC (Table S6), arguing against enrichment due to comorbidity with a common disease ascertained for exome sequencing. ExAC does include populations, such as South Asians, in which prion disease is not closely surveilled and we cannot rule out a higher incidence than that reported in developed countries, yet the individuals with reportedly pathogenic variants in either ExAC or 23andMe were of diverse inferred ancestry (Table S7–9). These individuals’ ages were consistent with the overall ExAC age distribution (Figure S1), rather than being enriched below some age of disease onset. ExAC genotypes at the prion disease modifier polymorphism M129V (34) were consistent with population allele frequencies (Table S7), rather than enriched for the lower-risk heterozygous genotype. Certain PRNP variants are associated with highly atypical phenotypes (35, 36), which are mistakable for other dementias and may not be well ascertained by current surveillance efforts. Most of the variants found in our population control cohorts, however, have been reported in individuals with a classic, sporadic Creutzfeldt-Jakob disease phenotype (22, 28, 30, 37–39), arguing that the discrepancy between observed and expected allele counts does not result primarily from an underappreciated prevalence of atypical prion disease.
Having observed a large excess of reportedly pathogenic variants over expectation in two datasets, and having excluded the most obvious confounders, we hypothesized that the unexpectedly high frequency of these variants in controls might arise from benign and/or low-risk variants.
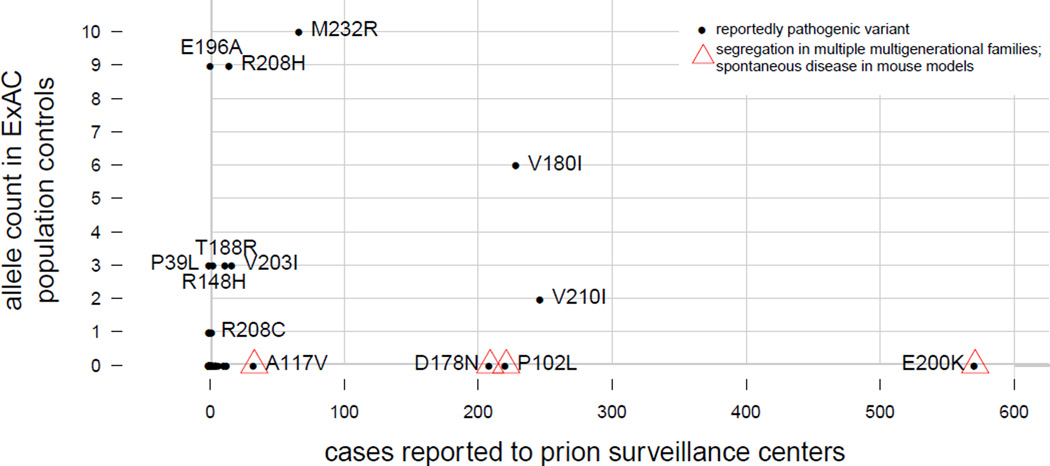
We investigated which variants were responsible for the observed excess (Figure 2). Variants with the strongest prior evidence of pathogenicity are absent from ExAC and cumulatively account for ≤5 alleles in 23andMe, consistent with the known rarity of genetic prion disease. Much of the excess allele frequency in population controls is due, instead, to variants with very weak prior evidence of pathogenicity (Figure 2 and Supplementary Discussion). For four variants observed in controls (V180I, R208H, V210I, and M232R), pathogenicity is controversial (40, 41) or reduced penetrance has been suggested (42, 43), but quantitative estimates of penetrance have never been produced, and the variants remain categorized as causes of genetic Creutzfeldt-Jakob disease (21, 22). Although we cannot prove that any one of the variants we observe in population controls is completely neutral, the list of reported pathogenic variants likely includes false positives. Indeed, the observation that 0.4% (236 / 60,706) of ExAC individuals harbor a rare (<0.1%) missense variant (Table S4) suggests that ~4 of every 1000 sporadic prion disease cases will, by chance, harbor such a variant, which in many cases will be interpreted and reported as causal given the long-standing classification of PRNP as a Mendelian disease gene.
Figure 2. Reportedly pathogenic PRNP variants include Mendelian, benign, and intermediate variants.
Prior evidence of pathogenicity is extremely strong for four missense variants - P102L, A117V, D178N and E200K - each of which has been observed to segregate with disease in multiple multigenerational families (16–18, 44–48) and to cause spontaneous disease in mouse models (49–54). These account for >50% of genetic prion disease cases (Table S1), yet are absent from ExAC (Table S3), and collectively appear on ≤5 alleles in 23andMe’s cohort (Table S5), indicating allele frequencies sufficiently low to be consistent with the prevalence of genetic prion disease (Figure 1). Conversely, the variants most common in controls and rare in cases had categorically weak prior evidence for pathogenicity. R208C (8 alleles in 23andMe) and P39L were observed in patients presenting clinically with other dementias, with prion disease suggested as an alternative diagnosis solely on the basis of finding a novel PRNP variant (55, 56). E196A was originally reported in a single patient, with a sporadic Creutzfeldt-Jakob disease phenotype and no family history (37), and appeared in only 2 of 790 Chinese prion disease patients in a recent case series (57), consistent with the ~0.1% allele frequency among Chinese individuals in ExAC (Tables S5 and S8). At least three variants (M232R, V180I, and V210I) occupy a space inconsistent with either neutrality or with complete penetrance (see main text and Figure 3). R148H, T188R, V203I, R208H and additional variants are discussed in Supplementary Discussion.
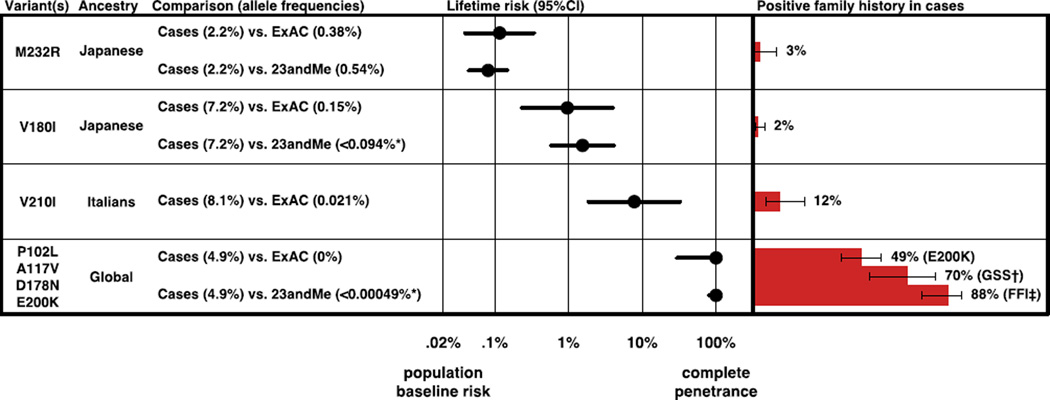
At least three variants, however (V180I, V210I, and M232R) fail to cluster with either the likely benign or likely Mendelian variants (Figure 2). Because each of these three appears primarily in one population in both cases and controls (Tables S1, S5, S7), we compared allele frequencies in matched population groups. Each has an allele frequency in controls that is too high for a fully penetrant, dominant prion disease-causing variant, and yet far lower than the corresponding allele frequency in cases (Figure 3).
Figure 3. Certain variants confer intermediate amounts of lifetime risk.
M232R, V180I, and V210I show varying degrees of enrichment in cases over controls, indicating a weak to moderate increase in risk. Best estimates of lifetime risk in heterozygotes (Materials and Methods) range from ~0.08% for M232R to ~7.8% for V210I, and correlate with the proportion of patients with a positive family history. Allele frequencies for P102L, A117V, D178N and E200K are consistent with up to 100% penetrance, with confidence intervals including all reported estimates of E200K penetrance based on survival analysis, which range from ~60% to ~90% (19, 23–26). Rates of family history of neurodegenerative disease in Japanese cases are from (Table S10) and in European populations are from Kovacs et al (21), with Wilson binomial 95% confidence intervals shown. *Based on allele counts rounded for privacy (Materials and Methods). †GSS, Gerstmann Straussler Scheinker disease associated with variants P102L, A117V and G131V. ‡FFI: fatal familial insomnia associated with a D178N cis 129M haplotype.
Because we lack genome-wide SNP data on cases we are unable to directly correct for population stratification, which thus may contribute to the observed differences in allele frequencies. Geographic clusters of genetic prion disease have been recognized for decades (26, 31, 58). For example, nearly half of Italian prion disease cases with the V210I variant are concentrated within two regions of Italy (59), so any non-uniform geographic sampling in cases versus controls would add some uncertainty to our penetrance estimates.
Nonetheless, the magnitude of the enrichment of certain variants in cases over controls in our datasets makes substructure an implausible explanation for the entire difference. In order for V210I to be neutral and yet appear with an allele frequency of 8.1% in Italian cases despite an apparent allele frequency of 0.02% in Italian controls, it would need to be fixed in a subpopulation comprising 8% of Italy’s populace. Under this scenario, this subpopulation would need to be virtually unsampled in any of our control cohorts, and V210I cases would contain many homozygotes. In reality, no cases have been reported homozygous for this variant. Conversely, if V210I were fully penetrant, family history would be positive in most cases, and the variant’s appearance on 13 alleles in 23andMe (Table S5) would indicate that this variant alone accounts for three times the known prevalence of genetic prion disease (Figure 1A). Finally, if the low family history rate were due to many de novo mutations, then V210I cases would be more uniformly distributed across populations (Table S1). Similar arguments rule out V180I being either benign or Mendelian. M232R, though clearly not Mendelian, could still be benign as it exhibits only 4- to 6-fold enrichment in cases, an amount that might conceivably be explained by Japanese population substructure alone. However, because even common variants in PRNP affect prion disease risk with odds ratios of 3 or greater (60–62), it is not implausible that M232R has a similar effect size, and our data suggest this a more likely scenario than it being neutral.
Satisfied that these three variants are likely neither benign nor Mendelian, we estimated lifetime risk in heterozygotes (Materials and Methods). The ~2 in 1 million annual incidence of prion disease translates into a baseline lifetime risk of ~1 in 5,000 in the general population (Materials and Methods). Because prion diseases are so rare, even the massive enrichment of heterozygotes in cases (Figure 3), implying odds ratios on the order of 10 to 1,000, corresponds to only low penetrance, with lifetime risk for M232R, V180I and V210I estimated near 0.1%, 1%, and 10%, respectively. Although our estimates are imperfect due to population stratification, they accord well with family history rates (Figure 3) and explain the unique space that these variants occupy in the plot of case versus control allele count (Figure 2). These data indicate that PRNP missense variants occupy a risk continuum rather than a dichotomy of causal versus benign.
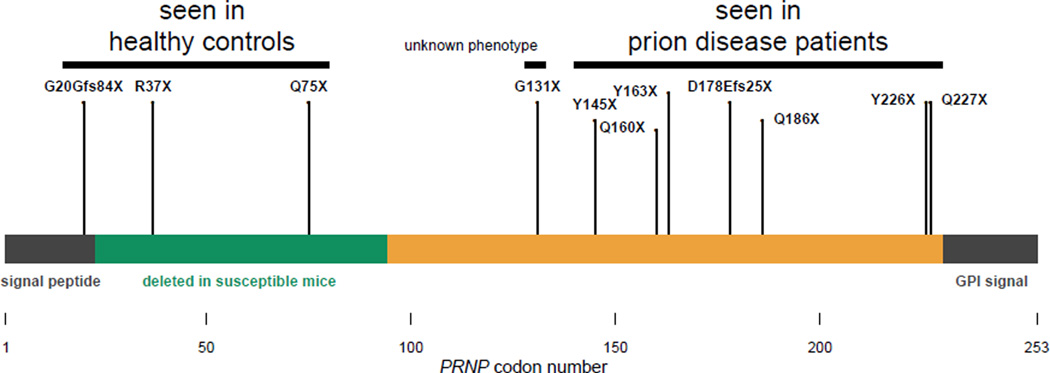
We asked whether the same was true of protein-truncating variants. PRNP possesses only one protein-coding exon, so premature stop codons are expected to result in truncated polypeptides rather than in nonsense-mediated decay. Prion diseases are known to arise from a gain of function, as neurodegeneration is not seen in mice, cows, or goats lacking PrP (63–66), and the rate of prion disease progression is tightly correlated with PrP expression level (67). Yet heterozygous C-terminal (residue ≥145) truncating variants are known to cause prion disease, sometimes with peripheral amyloidosis (35). Some of these patients also experience sensorimotor neuropathy phenotypically similar to that present in homozygous, but not heterozygous, PrP knockout mice (68), but attributed to amyloid infiltration of peripheral nerves, rather than loss of PrP function (35).
We identified, for the first time, heterozygous N-terminal (residue ≤131) truncating variants in four ExAC individuals and were able to obtain Sanger validation (Figure S1) and limited phenotype data (Table S11) for three. These individuals are free of overt neurological disease at ages 79, 73, and 52, and report no personal or family history of neurodegeneration nor of peripheral neuropathy. Therefore, the pathogenicity of protein-truncating variants appears to be dictated by position within PrP’s amino acid sequence (Figure 4). Observing three PRNP nonsense variants in ExAC is consistent with the expected number (~3.9) once we adjust our model (69) to exclude codons ≥145, where truncations cause a dominant gain-of-function disease. Thus, we see no evidence that PRNP is constrained against truncation in its N terminus. This, combined with the lack of any obvious phenotype in individuals with N-terminal truncating variants, suggests that heterozygous loss of PrP function is tolerated.
Figure 4. Effects of truncating variants in the human prion protein are position-dependent.
Truncating variants reported in prion disease cases in the literature (Table S2) and in our cohorts (Table S1) cluster exclusively in the C-terminal region (residue ≥145), while truncating variants in ExAC are more N-terminal (residue ≤131). The ortholog of each residue from 23–94 is deleted in at least one prion-susceptible transgenic mouse line (70). C-terminal truncations abolish PrP’s glycosylphosphatidylinositol anchor but leave most of the protein intact, a combination that mediates gain of function through mislocalization, causing this normally cell-surface-anchored protein to be secreted. Consistent with this model of pathogenicity, mice expressing full-length secreted PrP develop fatal and transmissible prion disease (71, 72). By contrast, the N-terminal truncating variants that we observe retain only residues dispensable for prion propagation, and are likely to cause a total loss of protein function.
DISCUSSION
Over 100,000 genetic variants have been reported to cause Mendelian disease in humans (73, 74). Many such reports do not meet current standards for assertions of pathogenicity (75, 76), and if all such reports were believed, the cumulative frequency of these variants in the population would imply that most people have a genetic disease (27). It is generally unclear how much of the excess burden of purported disease variants in the population is due to benign variants falsely associated, and how much is due to variants with genuine association but incomplete penetrance.
Here we leverage newly available large genomic reference datasets to re-evaluate reported disease associations in a dominant disease gene, PRNP. We identify some missense variants as likely benign while showing that others span a spectrum from <0.1% to ~100% penetrance. Our analyses provide quantitative estimates of lifetime risk for hundreds of asymptomatic individuals who have inherited incompletely penetrant PRNP variants.
Available datasets are only now approaching the size and quality required for such analyses, resulting in limitations for our study. The confidence intervals on our lifetime risk estimates span more than an order of magnitude, and our inability to perfectly control for population stratification injects additional uncertainty. We have been unable to reclassify those PRNP variants that are very rare both in cases and in controls (Supplementary Discussion). We have avoided analysis of large insertions that are poorly called with short sequencing reads, though we note that existing literature on these insertions is consistent with a spectrum of penetrance similar to that which we observe for missense variants (28, 77). Penetrance estimation in Mendelian disease will be improved by the collection of larger case series, particularly with genome-wide SNP data to allow more accurate population matching. This, coupled with continued large-scale population control sequencing and genotyping efforts, should reveal whether the dramatic variation in penetrance that we observe here is a more general feature of dominant disease genes.
Because PrP is required for prion pathogenesis and reduction in gene dosage slows disease progression (67, 78–80), several groups have sought to therapeutically reduce PrP expression using RNA interference (81–83), antisense oligonucleotides (84), or small molecules (85, 86). Our discovery of heterozygous loss-of-function variants in three healthy older humans provides the first human genetic data regarding the effects of a 50% reduction in gene dosage for PRNP. Both the number of individuals and the depth of available phenotype data are limited, and lifelong heterozygous inactivation of a gene is an imperfect model of the effects of pharmacological depletion of the gene product. With those limitations, our data provide preliminary evidence that a reduction in PRNP dosage, if achievable in patients, is likely to be tolerated. Increasingly large control sequencing datasets will soon enable testing whether the same is true of other genes currently being targeted in substrate reduction therapeutic approaches for other protein-folding disorders.
Together, our findings highlight the value of large reference datasets of human genetic variation for informing both genetic counseling and therapeutic strategy.
MATERIALS AND METHODS
Prion disease case series
Prion disease is considered a notifiable diagnosis in most developed countries, with mandatory reporting of all suspect cases to a centralized surveillance center. Surveillance was carried out broadly according to established guidelines (87, 88), with specifics as described previously for Australia (89), France (90), Germany (91–93), Italy (94), Japan (22), and the Netherlands (95). Sanger sequencing of the PRNP open reading frame was performed as described (96). We included only prion disease cases classified as definite (autopsy-confirmed) or probable according to published guidelines (88). Criteria for genetic testing vary between countries and over the years of data collection, with testing offered only on indication of family history in some times and places, and testing of all suspect cases with tissue available in other instances. Summary statistics on the total number and proportion of cases sequenced are presented in Table S1.
Exome sequencing and analysis
The ascertainment, sequencing, and joint calling of the ExAC dataset have been described previously (97). We extracted all rare (<0.1%) coding variant calls in PRNP with genotype quality (GQ) ≥10, alternate allele depth (AD) ≥3 and alternate allele balance (AB) ≥20%. Read-level evidence was visualized using Integrative Genomics Viewer (IGV) (98) for manual review. Because most ExAC exomes were sequenced with 76bp reads and the PRNP octapeptide repeat region (codons 50–90 inclusive) is 123bp long, it was impossible to determine whether genotype calls in this region were correct, and they were not considered further. After review of IGV screenshots, 87% of genotype calls were judged to be correct and were included in Table S3. Of the genotype calls judged to be correct, 99% had genotype quality (GQ) ≥95, 99% had allelic balance (AB) between 30% and 70%, and 97% had ≥10 reads supporting the alternate allele. All participants provided informed consent for exome sequencing and analysis. The Exome Aggregation Consortium’s aggregation and release of exome data have been approved by the Partners Healthcare Institutional Research Board (2013P001339). ExAC data have been publicly released at http://exac.broadinstitute.org/ and IGV screenshots of the rare PRNP variants deemed to be genuine and included in this study are available at https://github.com/ericminikel/prnp_penetrance/tree/master/supplement/igv
23andMe research participants and genotyping
Participants were drawn from the customer base of 23andMe, Inc., a personal genetics company (accessed February 6, 2015). All participants provided informed consent under a protocol approved by an external AAHRPP-accredited IRB, Ethical & Independent Review Services (E&I Review). DNA extraction and genotyping were performed on saliva samples by National Genetics Institute (NGI), a CLIA-licensed clinical laboratory and a subsidiary of Laboratory Corporation of America. Samples were genotyped on one of four Illumina platforms (V1-V4) as described previously (99). Of the PRNP SNPs considered, two (P105L and E200K) were genotyped on all four platforms while the other 14 were genotyped only on V3 and V4, resulting in differing numbers of total samples genotyped (Table S5). Genotypes were called with Illumina GenomeStudio. A 98.5% call rate were required for all samples. As with all 23andMe research participants, individuals whose genotyping analyses failed to reach the desired call rate repeatedly were recontacted to provide additional samples. A maximal set of unrelated individuals was chosen based on segmental identity-by-descent (IBD) estimation(100). Individuals were defined as related if they shared more than 700 cM IBD (approximately the minimal expected sharing between first cousins). Allele counts between 1 and 5 were rounded up to 5 to protect individual privacy (Table S5). Rounding down to 1 instead would raise our estimates of penetrance for V180I to 7.7% (95%CI, 1.2% – 50%) and for P102L, A117V, D178N and E200K collectively to 100% (95%CI, 100% – 100%), but the confidence intervals would still overlap those based on ExAC allele frequencies, and the overall conclusions of our study would remain unchanged.
23andMe ancestry composition
Ancestral origins of chromosomal segments were assigned on a continental level (European, Latino, African, and East Asian) and a country level (Japanese) as described by Durand et al (101). Briefly, after phasing genotypes using an out-of-sample implementation of the Beagle algorithm (102), a string kernel support vector machine classifier assigns tentative ancestry labels to local genomic regions. Then an autoregressive pair hidden Markov model was used to simultaneously correct phasing errors and produce reconciled local ancestry estimates and confidence scores based on the initial assignment. Finally, isotonic regression models were used to recalibrate the confidence estimates.
Europeans and East Asians were defined as individuals with more than 97% of chromosomal segments predicted as being from the respective ancestries. Because African Americans and Latinos are highly admixed, no single threshold of genome-wide ancestry is sufficient to distinguish them. However, segment length distributions of European, African, and Native American ancestries are different between African Americans and Latinos, due to distinct admixture timing in the two ethnic groups. Thus, a logistic classifier based on segment length of European, African, and Native American ancestries was used to distinguish between African Americans and Latinos.
At the country level, individuals were classified as Japanese based on the fraction of the respective local ancestry using a threshold of 90% for classifying Japanese ancestry. This threshold is based on the average fraction of local ancestry in the reference population (23andMe research participants with all four grandparents from the reference country): 94% (5% SD, N=533) for Japanese. Using the same approach, we were unable to obtain a confident set of Italian individuals for analysis of V210I due to extensive admixture. 23andMe research participants with all four grandparents from Italy only have 66% (18% SD, N=2090) Italian ancestry, and only ~60 participants have >90% Italian ancestry.
ExAC ancestry inference
We computed ten principal components based on ~5,800 common SNPs as described (27, 103). A centroid in eigenvalue-weighted principal component space was generated for each HapMap population based on 1000 Genomes individuals in ExAC. The remaining individuals in ExAC were assigned to the HapMap population with the nearest centroid according to eigenvalue-weighted Euclidean distance. Ancestries of all individuals, including those with reportedly pathogenic variants, are summarized in (Tables S7, S8).
Prion disease incidence and baseline risk
The reported incidence of prion disease varies between countries and between years, with much of the variability explained by the intensity of surveillance, as measured by the number of cases referred to national surveillance centers (13). Rates of ~1 case per million population per year have been reported, for instance in the U.S. (104) and in Japan (22), however, the countries with the most intense surveillance (greatest number of referrals per capita), such as France and Austria, observe incidence figures as high as 2 cases per million population per year (13). Only in small countries where the statistics are dominated by a particular genetic prion disease founder mutation, such as Israel and Slovakia (23, 26), has an incidence higher than 2 per million been consistently observed (105). We therefore accepted 2 cases per million as an upper bound for the true incidence of prion disease. Assuming an all-causes death rate of ~10 per 1,000 annually (106), this incidence corresponds to prion disease accounting for ~0.02% of all deaths, which we accepted as the baseline disease risk in the general population.
Lifetime risk estimation
By Bayes' theorem, the probability of disease given a genotype (penetrance or lifetime risk, P(D|G)) is equal to the proportion of individuals with the disease who have the genotype (genotype frequency in cases, P(G|D)) times the prevalence of the disease (baseline lifetime risk in the general population, P(D)), divided by the frequency of the genotype in the general population (here, population control allele frequency, P(G)). The use of this formula to estimate disease risk dates back at least to Cornfield's estimation of the probability of lung cancer in smokers (107), with later contributions by Woolf (108) and a synthesis by C.C. Li with application to genetics (109).
We used an allelic rather than genotypic model, such that lifetime risk in an individual with one allele is equal to case allele frequency (based on the number of prion disease cases that underwent PRNP sequencing) times baseline risk divided by population control allele frequency, P(D|A) = P(A|D)×P(D)/P(A). Note that we assume that our population control datasets include individuals who will later die of prion disease, thus enabling direct use of the ExAC and 23andMe allele frequencies as the denominator P(A). Following Kirov (11), we compute Wilson 95% confidence intervals on the binomial proportions P(A|D) and P(A), and calculate the upper bound of the 95% confidence interval for penetrance using the upper bound on case allele frequency and the lower bound on population control allele frequency, and vice versa for the lower bound on penetrance.
Supplementary Material
Acknowledgments
Funding
Research reported in this publication was partially supported by the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) and the National Institute of General Medical Sciences (NIGMS) of the National Institutes of Health, under awards U54DK105566 and R01GM104371, and by Broad Institute NextGen funds. Sonia Vallabh is supported by the National Science Foundation (NSF) Graduate Research Fellowship Program (GRFP) grant number 2015214731. U.S. prion surveillance work was conducted under Centers for Disease Control and Prevention (CDC) contract UR8/CCU515004. Japanese prion surveillance work was supported by a grant-in-aid from the Research Committee of Prion Disease and Slow Virus Infection, the Ministry of Health, Labour and Welfare of Japan, and from the Research Committee of Surveillance and Infection Control of Prion Disease, the Ministry of Health, Labour and Welfare of Japan. The French surveillance network is supported by the Institut National de veille Sanitaire. German prion surveillance work was supported by Robert Koch-Institute / Federal Ministry of Health grant 1369-341. The UK National CJD Research and Surveillance Unit is supported by the Department of Health and the Scottish Executive. The Australian National Creutzfeldt-Jakob Disease Registry is funded by the Commonwealth Department of Health. SJC is supported by a NHMRC Practitioner Fellowship: identification #APP1005816. Contributions at Erasmus MC were supported by Netherlands Genomics Initiative (NGI)/Netherlands Organisation for Scientific Research (NWO) sponsored Netherlands Consortium for Healthy Aging (NCHA; project 050-060-810), by the Genetic Laboratory of the Department of Internal Medicine, Erasmus MC, by a Complementation Project of the Biobanking and Biomolecular Research Infrastructure Netherlands (BBMRI-NL; www.bbmri.nl; project number CP2010-41), by Erasmus Medical Center and Erasmus University, Rotterdam, Netherlands Organization for the Health Research and Development (ZonMw), the Research Institute for Diseases in the Elderly (RIDE), the Ministry of Education, Culture and Science, the Ministry for Health, Welfare and Sports, the European Commission (DG XII), and the Municipality of Rotterdam. We thank the customers of 23andMe who participated in this research.
Footnotes
Source code availability
Data processing, analysis, and figure generation utilized custom scripts written in Python 2.7.6 and R 3.1.2. These scripts, along with vector graphics of all figures and tab-delimited text versions of all supplementary tables, are available online at https://github.com/ericminikel/prnp_penetrance
Author contributions
E.V.M., S.M.V. and D.G.M. conceived and designed the study. E.V.M analyzed data, generated figures, and wrote the manuscript. S.M.V. and E.V.M. reviewed literature and IGV screenshots. K.E.S. performed constraint analyses. M. Lek, K.E., K.E.S., K.J.K., A.H.O.-L., M.J.D., and D.G.M. consulted on data analysis and interpretation. J.F.S., C.Y.M., J.Y.C., and L.P.C.Y. prepared and consulted on analysis of 23andMe data. P.G., J.B., S.Z., Y.C., W.C., M.Y., T.H., N.S., H.M., Y.N., T.K., S.J.C., A.B., R.G.W., R. Knight, C.P., I.Z., T.F.J.K., S.E., A.G., M.C., J.d.P.C., S.H., J.-L.L., E.B.-A., J.-P.B., S.C, P.P., A.L., A.P., R. Kraaij., J.G.J.v.R., S.J.v.d.L., R.M., and C.v.D. prepared and consulted on analysis of prion surveillance data. E.V.M., J.L.M., M.B., M. Laakso, K.M., A.K., K.C., S.A.M., P.S., C.M.H., S.M.P., P.S., C.v.D., F.R.R., A.H., A.I., S.J.v.d.L., J.M.V.-D., and A.G.U. prepared and consulted on analysis of data regarding protein-truncating variants. ExAC provided exome sequence data.
REFERENCES
- 1.Brunham LR, Hayden MR. Hunting human disease genes: lessons from the past, challenges for the future. Hum. Genet. 2013;132:603–617. doi: 10.1007/s00439-013-1286-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amberger J, Bocchini C, Hamosh A. A new face and new challenges for Online Mendelian Inheritance in Man (OMIM®) Hum. Mutat. 2011;32:564–567. doi: 10.1002/humu.21466. [DOI] [PubMed] [Google Scholar]
- 3.Chong JX, Buckingham KJ, Jhangiani SN, Boehm C, Sobreira N, Smith JD, Harrell TM, McMillin MJ, Wiszniewski W, Gambin T, Coban Akdemir ZH, Doheny K, Scott AF, Avramopoulos D, Chakravarti A, Hoover-Fong J, Mathews D, Witmer PD, Ling H, Hetrick K, Watkins L, Patterson KE, Reinier F, Blue E, Muzny D, Kircher M, Bilguvar K, López-Giráldez F, Sutton VR, Tabor HK, Leal SM, Gunel M, Mane S, Gibbs RA, Boerwinkle E, Hamosh A, Shendure J, Lupski JR, Lifton RP, Valle D, Nickerson DA, Centers for Mendelian Genomics. Bamshad MJ. The Genetic Basis of Mendelian Phenotypes: Discoveries, Challenges, and Opportunities. Am. J. Hum. Genet. 2015;97:199–215. doi: 10.1016/j.ajhg.2015.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Crow JF. Hardy, Weinberg and language impediments. Genetics. 1999;152:821–825. doi: 10.1093/genetics/152.3.821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Begg CB. On the use of familial aggregation in population-based case probands for calculating penetrance. J. Natl. Cancer Inst. 2002;94:1221–1226. doi: 10.1093/jnci/94.16.1221. [DOI] [PubMed] [Google Scholar]
- 6.Goldwurm S, Zini M, Mariani L, Tesei S, Miceli R, Sironi F, Clementi M, Bonifati V, Pezzoli G. Evaluation of LRRK2 G2019S penetrance: relevance for genetic counseling in Parkinson disease. Neurology. 2007;68:1141–1143. doi: 10.1212/01.wnl.0000254483.19854.ef. [DOI] [PubMed] [Google Scholar]
- 7.Cooper DN, Krawczak M, Polychronakos C, Tyler-Smith C, Kehrer-Sawatzki H. Where genotype is not predictive of phenotype: towards an understanding of the molecular basis of reduced penetrance in human inherited disease. Hum. Genet. 2013;132:1077–1130. doi: 10.1007/s00439-013-1331-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cooper GM, Coe BP, Girirajan S, Rosenfeld JA, Vu TH, Baker C, Williams C, Stalker H, Hamid R, Hannig V, Abdel-Hamid H, Bader P, McCracken E, Niyazov D, Leppig K, Thiese H, Hummel M, Alexander N, Gorski J, Kussmann J, Shashi V, Johnson K, Rehder C, Ballif BC, Shaffer LG, Eichler EE. A copy number variation morbidity map of developmental delay. Nat. Genet. 2011;43:838–846. doi: 10.1038/ng.909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bick AG, Flannick J, Ito K, Cheng S, Vasan RS, Parfenov MG, Herman DS, DePalma SR, Gupta N, Gabriel SB, Funke BH, Rehm HL, Benjamin EJ, Aragam J, Taylor HA, Fox ER, Newton-Cheh C, Kathiresan S, O’Donnell CJ, Wilson JG, Altshuler DM, Hirschhorn JN, Seidman JG, Seidman C. Burden of rare sarcomere gene variants in the Framingham and Jackson Heart Study cohorts. Am. J. Hum. Genet. 2012;91:513–519. doi: 10.1016/j.ajhg.2012.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Flannick J, Beer NL, Bick AG, Agarwala V, Molnes J, Gupta N, Burtt NP, Florez JC, Meigs JB, Taylor H, Lyssenko V, Irgens H, Fox E, Burslem F, Johansson S, Brosnan MJ, Trimmer JK, Newton-Cheh C, Tuomi T, Molven A, Wilson JG, O’Donnell CJ, Kathiresan S, Hirschhorn JN, Njølstad PR, Rolph T, Seidman JG, Gabriel S, Cox DR, Seidman CE, Groop L, Altshuler D. Assessing the phenotypic effects in the general population of rare variants in genes for a dominant Mendelian form of diabetes. Nat. Genet. 2013;45:1380–1385. doi: 10.1038/ng.2794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kirov G, Rees E, Walters JTR, Escott-Price V, Georgieva L, Richards AL, Chambert KD, Davies G, Legge SE, Moran JL, McCarroll SA, O’Donovan MC, Owen MJ. The penetrance of copy number variations for schizophrenia and developmental delay. Biol. Psychiatry. 2014;75:378–385. doi: 10.1016/j.biopsych.2013.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Prusiner SB. Prions. Proc. Natl. Acad. Sci. U. S. A. 1998;95:13363–13383. doi: 10.1073/pnas.95.23.13363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Klug GMJA, Wand H, Simpson M, Boyd A, Law M, Masters CL, Matěj R, Howley R, Farrell M, Breithaupt M, Zerr I, van Duijn C, Ibrahim-Verbaas C, Mackenzie J, Will RG, Brandel J-P, Alperovitch A, Budka H, Kovacs GG, Jansen GH, Coulthard M, Collins SJ. Intensity of human prion disease surveillance predicts observed disease incidence. J. Neurol. Neurosurg. Psychiatry. 2013;84:1372–1377. doi: 10.1136/jnnp-2012-304820. [DOI] [PubMed] [Google Scholar]
- 14.U. S. National Prion Disease Pathology Surveillance Center. CDC - Creutzfeldt-Jakob Disease, Classic (CJD) available at http://web.archive.org/web/20150202162606/ http://www.cdc.gov/ncidod/dvrd/cjd/
- 15.U. K. National Creutzfeldt-Jakob Disease Research and Surveillance Unit. CREUTZFELDT-JAKOB DISEASE IN THE UK - figs.pdf. 2015 available at http://web.archive.org/web/20150330211505/ http://www.cjd.ed.ac.uk/documents/figs.pdf. [Google Scholar]
- 16.Hsiao K, Baker HF, Crow TJ, Poulter M, Owen F, Terwilliger JD, Westaway D, Ott J, Prusiner SB. Linkage of a prion protein missense variant to Gerstmann-Sträussler syndrome. Nature. 1989;338:342–345. doi: 10.1038/338342a0. [DOI] [PubMed] [Google Scholar]
- 17.Hsiao K, Meiner Z, Kahana E, Cass C, Kahana I, Avrahami D, Scarlato G, Abramsky O, Prusiner SB, Gabizon R. Mutation of the prion protein in Libyan Jews with Creutzfeldt-Jakob disease. N. Engl. J. Med. 1991;324:1091–1097. doi: 10.1056/NEJM199104183241604. [DOI] [PubMed] [Google Scholar]
- 18.Medori R, Tritschler HJ, LeBlanc A, Villare F, Manetto V, Chen HY, Xue R, Leal S, Montagna P, Cortelli P. Fatal familial insomnia, a prion disease with a mutation at codon 178 of the prion protein gene. N. Engl. J. Med. 1992;326:444–449. doi: 10.1056/NEJM199202133260704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Minikel EV, Zerr I, Collins SJ, Ponto C, Boyd A, Klug G, Karch A, Kenny J, Collinge J, Takada LT, Forner S, Fong JC, Mead S, Geschwind MD. Ascertainment bias causes false signal of anticipation in genetic prion disease. Am. J. Hum. Genet. 2014;95:371–382. doi: 10.1016/j.ajhg.2014.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Owen J, Beck J, Campbell T, Adamson G, Gorham M, Thompson A, Smithson S, Rosser E, Rudge P, Collinge J, Mead S. Predictive testing for inherited prion disease: report of 22 years experience. Eur. J. Hum. Genet. EJHG. 2014 doi: 10.1038/ejhg.2014.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kovács GG, Puopolo M, Ladogana A, Pocchiari M, Budka H, van Duijn C, Collins SJ, Boyd A, Giulivi A, Coulthart M, Delasnerie-Laupretre N, Brandel JP, Zerr I, Kretzschmar HA, de Pedro-Cuesta J, Calero-Lara M, Glatzel M, Aguzzi A, Bishop M, Knight R, Belay G, Will R, Mitrova E. EUROCJD, Genetic prion disease: the EUROCJD experience. Hum. Genet. 2005;118:166–174. doi: 10.1007/s00439-005-0020-1. [DOI] [PubMed] [Google Scholar]
- 22.Nozaki I, Hamaguchi T, Sanjo N, Noguchi-Shinohara M, Sakai K, Nakamura Y, Sato T, Kitamoto T, Mizusawa H, Moriwaka F, Shiga Y, Kuroiwa Y, Nishizawa M, Kuzuhara S, Inuzuka T, Takeda M, Kuroda S, Abe K, Murai H, Murayama S, Tateishi J, Takumi I, Shirabe S, Harada M, Sadakane A, Yamada M. Prospective 10-year surveillance of human prion diseases in Japan. Brain J. Neurol. 2010;133:3043–3057. doi: 10.1093/brain/awq216. [DOI] [PubMed] [Google Scholar]
- 23.Chapman J, Ben-Israel J, Goldhammer Y, Korczyn AD. The risk of developing Creutzfeldt-Jakob disease in subjects with the PRNP gene codon 200 point mutation. Neurology. 1994;44:1683–1686. doi: 10.1212/wnl.44.9.1683. [DOI] [PubMed] [Google Scholar]
- 24.Spudich S, Mastrianni JA, Wrensch M, Gabizon R, Meiner Z, Kahana I, Rosenmann H, Kahana E, Prusiner SB. Complete penetrance of Creutzfeldt-Jakob disease in Libyan Jews carrying the E200K mutation in the prion protein gene. Mol. Med. Camb. Mass. 1995;1:607–613. [PMC free article] [PubMed] [Google Scholar]
- 25.D’Alessandro M, Petraroli R, Ladogana A, Pocchiari M. High incidence of Creutzfeldt-Jakob disease in rural Calabria, Italy. Lancet. 1998;352:1989–1990. doi: 10.1016/S0140-6736(05)61335-9. [DOI] [PubMed] [Google Scholar]
- 26.Mitrová E, Belay G. Creutzfeldt-Jakob disease with E200K mutation in Slovakia: characterization and development. Acta Virol. 2002;46:31–39. [PubMed] [Google Scholar]
- 27.E.A. Consortium. Lek M, Karczewski K, Minikel E, Samocha K, Banks E, Fennell T, O’Donnell-Luria A, Ware J, Hill A, Cummings B, Tukiainen T, Birnbaum D, Kosmicki J, Duncan L, Estrada K, Zhao F, Zou J, Pierce-Hoffman E, Cooper D, DePristo M, Do R, Flannick J, Fromer M, Gauthier L, Goldstein J, Gupta N, Howrigan D, Kiezun A, Kurki M, Moonshine AL, Natarajan P, Orozco L, Peloso G, Poplin R, Rivas M, Ruano-Rubio V, Ruderfer D, Shakir K, Stenson P, Stevens C, Thomas B, Tiao G, Tusie-Luna M, Weisburd B, Won H-H, Yu D, Altshuler D, Ardissino D, Boehnke M, Danesh J, Roberto E, Florez J, Gabriel S, Getz G, Hultman C, Kathiresan S, Laakso M, McCarroll S, McCarthy M, McGovern D, McPherson R, Neale B, Palotie A, Purcell S, Saleheen D, Scharf J, Sklar P, Patrick S, Tuomilehto J, Watkins H, Wilson J, Daly M, MacArthur D. Analysis of protein-coding genetic variation in 60,706 humans. bioRxiv. 2015:030338. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kong Q, Surewicz WK, Petersen RB, Chen SG, Gambetti P, Parchi P, Capellari S, Goldfarb L, Montagna P, Lugaresi E, Piccardo P, Ghetti B. Prion Biology and Diseases. Cold Spring Harbor Laboratory Press; 2004. [Google Scholar]
- 29.Beck JA, Poulter M, Campbell TA, Adamson G, Uphill JB, Guerreiro R, Jackson GS, Stevens JC, Manji H, Collinge J, Mead S. PRNP allelic series from 19 years of prion protein gene sequencing at the MRC Prion Unit. Hum. Mutat. 2010;31:E1551–E1563. doi: 10.1002/humu.21281. [DOI] [PubMed] [Google Scholar]
- 30.Mastrianni JA. The genetics of prion diseases. Genet. Med. Off. J. Am. Coll. Med. Genet. 2010;12:187–195. doi: 10.1097/GIM.0b013e3181cd7374. [DOI] [PubMed] [Google Scholar]
- 31.Masters CL, Harris JO, Gajdusek DC, Gibbs CJ, Bernoulli C, Asher DM. Creutzfeldt-Jakob disease: patterns of worldwide occurrence and the significance of familial and sporadic clustering. Ann. Neurol. 1979;5:177–188. doi: 10.1002/ana.410050212. [DOI] [PubMed] [Google Scholar]
- 32.Laplanche JL, Hachimi KH, Durieux I, Thuillet P, Defebvre L, Delasnerie-Lauprêtre N, Peoc’h K, Foncin JF, Destée A. Prominent psychiatric features and early onset in an inherited prion disease with a new insertional mutation in the prion protein gene. Brain J. Neurol. 1999;122(Pt 12):2375–2386. doi: 10.1093/brain/122.12.2375. [DOI] [PubMed] [Google Scholar]
- 33.Servick K. Can 23andMe have it all? Science. 2015;349:1472–1474. 1476–1477. doi: 10.1126/science.349.6255.1472. [DOI] [PubMed] [Google Scholar]
- 34.Capellari S, Strammiello R, Saverioni D, Kretzschmar H, Parchi P. Genetic Creutzfeldt-Jakob disease and fatal familial insomnia: insights into phenotypic variability and disease pathogenesis. Acta Neuropathol. (Berl.) 2011;121:21–37. doi: 10.1007/s00401-010-0760-4. [DOI] [PubMed] [Google Scholar]
- 35.Mead S, Reilly MM. A new prion disease: relationship with central and peripheral amyloidoses. Nat. Rev. Neurol. 2015 doi: 10.1038/nrneurol.2014.263. [DOI] [PubMed] [Google Scholar]
- 36.Moore RC, Xiang F, Monaghan J, Han D, Zhang Z, Edström L, Anvret M, Prusiner SB. Huntington disease phenocopy is a familial prion disease. Am. J. Hum. Genet. 2001;69:1385–1388. doi: 10.1086/324414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang H, Wang M, Wu L, Zhang H, Jin T, Wu J, Sun L. Novel prion protein gene mutation at codon 196 (E196A) in a septuagenarian with Creutzfeldt-Jakob disease. J. Clin. Neurosci. Off. J. Neurosurg. Soc. Australas. 2014;21:175–178. doi: 10.1016/j.jocn.2013.03.016. [DOI] [PubMed] [Google Scholar]
- 38.Tartaglia MC, Thai JN, See T, Kuo A, Harbaugh R, Raudabaugh B, Cali I, Sattavat M, Sanchez H, DeArmond SJ, Geschwind MD. Pathologic evidence that the T188R mutation in PRNP is associated with prion disease. J. Neuropathol. Exp. Neurol. 2010;69:1220–1227. doi: 10.1097/NEN.0b013e3181ffc39c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Peoc’h K, Manivet P, Beaudry P, Attane F, Besson G, Hannequin D, Delasnerie-Lauprêtre N, Laplanche JL. Identification of three novel mutations (E196K, V203I, E211Q) in the prion protein gene (PRNP) in inherited prion diseases with Creutzfeldt-Jakob disease phenotype. Hum. Mutat. 2000;15:482. doi: 10.1002/(SICI)1098-1004(200005)15:5<482::AID-HUMU16>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 40.Beck J, Collinge J, Mead S. Prion protein gene M232R variation is probably an uncommon polymorphism rather than a pathogenic mutation. Brain J. Neurol. 2012;135:e209. doi: 10.1093/brain/awr294. author reply e210. [DOI] [PubMed] [Google Scholar]
- 41.Nozaki I, Sakai K, Kitamoto T, Yamada M. Reply: Prion protein gene M232R variation is probably an uncommon polymorphism rather than a pathogenic mutation. Brain. 2012;135:e210–e210. doi: 10.1093/brain/awr294. [DOI] [PubMed] [Google Scholar]
- 42.Capellari S, Cardone F, Notari S, Schininà ME, Maras B, Sità D, Baruzzi A, Pocchiari M, Parchi P. Creutzfeldt-Jakob disease associated with the R208H mutation in the prion protein gene. Neurology. 2005;64:905–907. doi: 10.1212/01.WNL.0000152837.82388.DE. [DOI] [PubMed] [Google Scholar]
- 43.Ripoll L, Laplanche JL, Salzmann M, Jouvet A, Planques B, Dussaucy M, Chatelain J, Beaudry P, Launay JM. A new point mutation in the prion protein gene at codon 210 in Creutzfeldt-Jakob disease. Neurology. 1993;43:1934–1938. doi: 10.1212/wnl.43.10.1934. [DOI] [PubMed] [Google Scholar]
- 44.Goldfarb LG, Korczyn AD, Brown P, Chapman J, Gajdusek DC. Mutation in codon 200 of scrapie amyloid precursor gene linked to Creutzfeldt-Jakob disease in Sephardic Jews of Libyan and non-Libyan origin. Lancet. 1990;336:637–638. doi: 10.1016/0140-6736(90)93443-s. [DOI] [PubMed] [Google Scholar]
- 45.Hsiao KK, Cass C, Schellenberg GD, Bird T, Devine-Gage E, Wisniewski H, Prusiner SB. A prion protein variant in a family with the telencephalic form of Gerstmann-Sträussler-Scheinker syndrome. Neurology. 1991;41:681–684. doi: 10.1212/wnl.41.5.681. [DOI] [PubMed] [Google Scholar]
- 46.Medori R, Montagna P, Tritschler HJ, LeBlanc A, Cortelli P, Tinuper P, Lugaresi E, Gambetti P. Fatal familial insomnia: a second kindred with mutation of prion protein gene at codon 178. Neurology. 1992;42:669–670. doi: 10.1212/wnl.42.3.669. [DOI] [PubMed] [Google Scholar]
- 47.Mastrianni JA, Curtis MT, Oberholtzer JC, Da Costa MM, DeArmond S, Prusiner SB, Garbern JY. Prion disease (PrP-A117V) presenting with ataxia instead of dementia. Neurology. 1995;45:2042–2050. doi: 10.1212/wnl.45.11.2042. [DOI] [PubMed] [Google Scholar]
- 48.Webb TEF, Poulter M, Beck J, Uphill J, Adamson G, Campbell T, Linehan J, Powell C, Brandner S, Pal S, Siddique D, Wadsworth JD, Joiner S, Alner K, Petersen C, Hampson S, Rhymes C, Treacy C, Storey E, Geschwind MD, Nemeth AH, Wroe S, Collinge J, Mead S. Phenotypic heterogeneity and genetic modification of P102L inherited prion disease in an international series. Brain J. Neurol. 2008;131:2632–2646. doi: 10.1093/brain/awn202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hsiao KK, Scott M, Foster D, Groth DF, DeArmond SJ, Prusiner SB. Spontaneous neurodegeneration in transgenic mice with mutant prion protein. Science. 1990;250:1587–1590. doi: 10.1126/science.1980379. [DOI] [PubMed] [Google Scholar]
- 50.Jackson WS, Borkowski AW, Faas H, Steele AD, King OD, Watson N, Jasanoff A, Lindquist S. Spontaneous generation of prion infectivity in fatal familial insomnia knockin mice. Neuron. 2009;63:438–450. doi: 10.1016/j.neuron.2009.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yang W, Cook J, Rassbach B, Lemus A, DeArmond SJ, Mastrianni JA. A New Transgenic Mouse Model of Gerstmann-Straussler-Scheinker Syndrome Caused by the A117V Mutation of PRNP. J. Neurosci. Off. J. Soc. Neurosci. 2009;29:10072–10080. doi: 10.1523/JNEUROSCI.2542-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jackson WS, Borkowski AW, Watson NE, King OD, Faas H, Jasanoff A, Lindquist S. Profoundly different prion diseases in knock-in mice carrying single PrP codon substitutions associated with human diseases. Proc. Natl. Acad. Sci. U. S. A. 2013;110:14759–14764. doi: 10.1073/pnas.1312006110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dossena S, Imeri L, Mangieri M, Garofoli A, Ferrari L, Senatore A, Restelli E, Balducci C, Fiordaliso F, Salio M, Bianchi S, Fioriti L, Morbin M, Pincherle A, Marcon G, Villani F, Carli M, Tagliavini F, Forloni G, Chiesa R. Mutant prion protein expression causes motor and memory deficits and abnormal sleep patterns in a transgenic mouse model. Neuron. 2008;60:598–609. doi: 10.1016/j.neuron.2008.09.008. [DOI] [PubMed] [Google Scholar]
- 54.Bouybayoune I, Mantovani S, Del Gallo F, Bertani I, Restelli E, Comerio L, Tapella L, Baracchi F, Fernández-Borges N, Mangieri M, Bisighini C, Beznoussenko GV, Paladini A, Balducci C, Micotti E, Forloni G, Castilla J, Fiordaliso F, Tagliavini F, Imeri L, Chiesa R. Transgenic fatal familial insomnia mice indicate prion infectivity-independent mechanisms of pathogenesis and phenotypic expression of disease. PLoS Pathog. 2015;11:e1004796. doi: 10.1371/journal.ppat.1004796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bernardi L, Cupidi C, Frangipane F, Anfossi M, Gallo M, Conidi ME, Vasso F, Colao R, Puccio G, Curcio SAM, Mirabelli M, Clodomiro A, Di Lorenzo R, Smirne N, Maletta R, Bruni AC. Novel N-terminal domain mutation in prion protein detected in 2 patients diagnosed with frontotemporal lobar degeneration syndrome. Neurobiol. Aging. 2014;35(2657):e7–e11. doi: 10.1016/j.neurobiolaging.2014.06.006. [DOI] [PubMed] [Google Scholar]
- 56.Zheng L, Longfei J, Jing Y, Xinqing Z, Haiqing S, Haiyan L, Fen W, Xiumin D, Jianping J. PRNP mutations in a series of apparently sporadic neurodegenerative dementias in China. Am. J. Med. Genet. Part B Neuropsychiatr. Genet. Off. Publ. Int. Soc. Psychiatr. Genet. 2008;147B:938–944. doi: 10.1002/ajmg.b.30761. [DOI] [PubMed] [Google Scholar]
- 57.Shi Q, Zhou W, Chen C, Zhang B-Y, Xiao K, Zhang X-C, Shen X-J, Li Q, Deng L-Q, Dong J-H, Lin W-Q, Huang P, Jiang W-J, Lv J, Han J, Dong X-P. The Features of Genetic Prion Diseases Based on Chinese Surveillance Program. PloS One. 2015;10:e0139552. doi: 10.1371/journal.pone.0139552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lee HS, Sambuughin N, Cervenakova L, Chapman J, Pocchiari M, Litvak S, Qi HY, Budka H, del Ser T, Furukawa H, Brown P, Gajdusek DC, Long JC, Korczyn AD, Goldfarb LG. Ancestral origins and worldwide distribution of the PRNP 200K mutation causing familial Creutzfeldt-Jakob disease. Am. J. Hum. Genet. 1999;64:1063–1070. doi: 10.1086/302340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ladogana A, Puopolo M, Poleggi A, Almonti S, Mellina V, Equestre M, Pocchiari M. High incidence of genetic human transmissible spongiform encephalopathies in Italy. Neurology. 2005;64:1592–1597. doi: 10.1212/01.WNL.0000160118.26865.11. [DOI] [PubMed] [Google Scholar]
- 60.Shibuya S, Higuchi J, Shin RW, Tateishi J, Kitamoto T. Codon 219 Lys allele of PRNP is not found in sporadic Creutzfeldt-Jakob disease. Ann. Neurol. 1998;43:826–828. doi: 10.1002/ana.410430618. [DOI] [PubMed] [Google Scholar]
- 61.Bishop MT, Pennington C, Heath CA, Will RG, Knight RSG. PRNP variation in UK sporadic and variant Creutzfeldt Jakob disease highlights genetic risk factors and a novel non-synonymous polymorphism. BMC Med. Genet. 2009;10:146. doi: 10.1186/1471-2350-10-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mead S, Uphill J, Beck J, Poulter M, Campbell T, Lowe J, Adamson G, Hummerich H, Klopp N, Rückert I-M, Wichmann H-E, Azazi D, Plagnol V, Pako WH, Whitfield J, Alpers MP, Whittaker J, Balding DJ, Zerr I, Kretzschmar H, Collinge J. Genome-wide association study in multiple human prion diseases suggests genetic risk factors additional to PRNP. Hum. Mol. Genet. 2012;21:1897–1906. doi: 10.1093/hmg/ddr607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Büeler H, Fischer M, Lang Y, Bluethmann H, Lipp HP, DeArmond SJ, Prusiner SB, Aguet M, Weissmann C. Normal development and behaviour of mice lacking the neuronal cell-surface PrP protein. Nature. 1992;356:577–582. doi: 10.1038/356577a0. [DOI] [PubMed] [Google Scholar]
- 64.Richt JA, Kasinathan P, Hamir AN, Castilla J, Sathiyaseelan T, Vargas F, Sathiyaseelan J, Wu H, Matsushita H, Koster J, Kato S, Ishida I, Soto C, Robl JM, Kuroiwa Y. Production of cattle lacking prion protein. Nat. Biotechnol. 2007;25:132–138. doi: 10.1038/nbt1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yu G, Chen J, Xu Y, Zhu C, Yu H, Liu S, Sha H, Chen J, Xu X, Wu Y, Zhang A, Ma J, Cheng G. Generation of goats lacking prion protein. Mol. Reprod. Dev. 2009;76:3. doi: 10.1002/mrd.20960. [DOI] [PubMed] [Google Scholar]
- 66.Benestad SL, Austbø L, Tranulis MA, Espenes A, Olsaker I. Healthy goats naturally devoid of prion protein. Vet. Res. 2012;43:87. doi: 10.1186/1297-9716-43-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Fischer M, Rülicke T, Raeber A, Sailer A, Moser M, Oesch B, Brandner S, Aguzzi A, Weissmann C. Prion protein (PrP) with amino-proximal deletions restoring susceptibility of PrP knockout mice to scrapie. EMBO J. 1996;15:1255–1264. [PMC free article] [PubMed] [Google Scholar]
- 68.Bremer J, Baumann F, Tiberi C, Wessig C, Fischer H, Schwarz P, Steele AD, Toyka KV, Nave K-A, Weis J, Aguzzi A. Axonal prion protein is required for peripheral myelin maintenance. Nat. Neurosci. 2010;13:310–318. doi: 10.1038/nn.2483. [DOI] [PubMed] [Google Scholar]
- 69.Samocha KE, Robinson EB, Sanders SJ, Stevens C, Sabo A, McGrath LM, Kosmicki JA, Rehnström K, Mallick S, Kirby A, Wall DP, MacArthur DG, Gabriel SB, DePristo M, Purcell SM, Palotie A, Boerwinkle E, Buxbaum JD, Cook EH, Gibbs RA, Schellenberg GD, Sutcliffe JS, Devlin B, Roeder K, Neale BM, Daly MJ. A framework for the interpretation of de novo mutation in human disease. Nat. Genet. 2014;46:944–950. doi: 10.1038/ng.3050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Aguzzi A, Baumann F, Bremer J. The prion’s elusive reason for being. Annu. Rev. Neurosci. 2008;31:439–477. doi: 10.1146/annurev.neuro.31.060407.125620. [DOI] [PubMed] [Google Scholar]
- 71.Chesebro B, Race B, Meade-White K, Lacasse R, Race R, Klingeborn M, Striebel J, Dorward D, McGovern G, Jeffrey M. Fatal transmissible amyloid encephalopathy: a new type of prion disease associated with lack of prion protein membrane anchoring. PLoS Pathog. 2010;6:e1000800. doi: 10.1371/journal.ppat.1000800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Stöhr J, Watts JC, Legname G, Oehler A, Lemus A, Nguyen H-OB, Sussman J, Wille H, DeArmond SJ, Prusiner SB, Giles K. Spontaneous generation of anchorless prions in transgenic mice. Proc. Natl. Acad. Sci. U. S. A. 2011;108:21223–21228. doi: 10.1073/pnas.1117827108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Landrum MJ, Lee JM, Riley GR, Jang W, Rubinstein WS, Church DM, Maglott DR. ClinVar: public archive of relationships among sequence variation and human phenotype. Nucleic Acids Res. 2014;42:D980–D985. doi: 10.1093/nar/gkt1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Stenson PD, Mort M, Ball EV, Shaw K, Phillips A, Cooper DN. The Human Gene Mutation Database: building a comprehensive mutation repository for clinical and molecular genetics, diagnostic testing and personalized genomic medicine. Hum. Genet. 2014;133:1–9. doi: 10.1007/s00439-013-1358-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.MacArthur DG, Manolio TA, Dimmock DP, Rehm HL, Shendure J, Abecasis GR, Adams DR, Altman RB, Antonarakis SE, Ashley EA, Barrett JC, Biesecker LG, Conrad DF, Cooper GM, Cox NJ, Daly MJ, Gerstein MB, Goldstein DB, Hirschhorn JN, Leal SM, Pennacchio LA, Stamatoyannopoulos JA, Sunyaev SR, Valle D, Voight BF, Winckler W, Gunter C. Guidelines for investigating causality of sequence variants in human disease. Nature. 2014;508:469–476. doi: 10.1038/nature13127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, Grody WW, Hegde M, Lyon E, Spector E, Voelkerding K, Rehm HL ACMG Laboratory Quality Assurance Committee. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. Off. J. Am. Coll. Med. Genet. 2015;17:405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mead S. Prion disease genetics. Eur. J. Hum. Genet. EJHG. 2006;14:273–281. doi: 10.1038/sj.ejhg.5201544. [DOI] [PubMed] [Google Scholar]
- 78.Büeler H, Aguzzi A, Sailer A, Greiner RA, Autenried P, Aguet M, Weissmann C. Mice devoid of PrP are resistant to scrapie. Cell. 1993;73:1339–1347. doi: 10.1016/0092-8674(93)90360-3. [DOI] [PubMed] [Google Scholar]
- 79.Mallucci G, Dickinson A, Linehan J, Klöhn P-C, Brandner S, Collinge J. Depleting neuronal PrP in prion infection prevents disease and reverses spongiosis. Science. 2003;302:871–874. doi: 10.1126/science.1090187. [DOI] [PubMed] [Google Scholar]
- 80.Safar JG, DeArmond SJ, Kociuba K, Deering C, Didorenko S, Bouzamondo-Bernstein E, Prusiner SB, Tremblay P. Prion clearance in bigenic mice. J. Gen. Virol. 2005;86:2913–2923. doi: 10.1099/vir.0.80947-0. [DOI] [PubMed] [Google Scholar]
- 81.White MD, Farmer M, Mirabile I, Brandner S, Collinge J, Mallucci GR. Single treatment with RNAi against prion protein rescues early neuronal dysfunction and prolongs survival in mice with prion disease. Proc. Natl. Acad. Sci. U. S. A. 2008;105:10238–10243. doi: 10.1073/pnas.0802759105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Pulford B, Reim N, Bell A, Veatch J, Forster G, Bender H, Meyerett C, Hafeman S, Michel B, Johnson T, Wyckoff AC, Miele G, Julius C, Kranich J, Schenkel A, Dow S, Zabel MD. Liposome-siRNA-peptide complexes cross the blood-brain barrier and significantly decrease PrP on neuronal cells and PrP in infected cell cultures. PloS One. 2010;5:e11085. doi: 10.1371/journal.pone.0011085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ahn M, Bajsarowicz K, Oehler A, Lemus A, Bankiewicz K, DeArmond SJ. Convection-enhanced delivery of AAV2-PrPshRNA in prion-infected mice. PloS One. 2014;9:e98496. doi: 10.1371/journal.pone.0098496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Nazor Friberg K, Hung G, Wancewicz E, Giles K, Black C, Freier S, Bennett F, Dearmond SJ, Freyman Y, Lessard P, Ghaemmaghami S, Prusiner SB. Intracerebral Infusion of Antisense Oligonucleotides Into Prion-infected Mice. Mol. Ther. Nucleic Acids. 2012;1:e9. doi: 10.1038/mtna.2011.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Karapetyan YE, Sferrazza GF, Zhou M, Ottenberg G, Spicer T, Chase P, Fallahi M, Hodder P, Weissmann C, Lasmézas CI. Unique drug screening approach for prion diseases identifies tacrolimus and astemizole as antiprion agents. Proc. Natl. Acad. Sci. U. S. A. 2013;110:7044–7049. doi: 10.1073/pnas.1303510110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Silber BM, Gever JR, Rao S, Li Z, Renslo AR, Widjaja K, Wong C, Giles K, Freyman Y, Elepano M, Irwin JJ, Jacobson MP, Prusiner SB. Novel compounds lowering the cellular isoform of the human prion protein in cultured human cells. Bioorg. Med. Chem. 2014;22:1960–1972. doi: 10.1016/j.bmc.2014.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.World Health Organization. Global Surveillance, Diagnosis and Therapy of Human Transmissible Spongiform Encephalopathies: Report of a WHO Consultation Geneva, Switzerland, 9–11 February 1998. 1998 http://www.who.int/csr/resources/publications/bse/whoemczdi989.pdf.
- 88.World Health Organization. WHO manual for surveillance of human transmissible spongiform encephalopathies including variant Creutzfeldt-Jakob disease. 2003 http://www.who.int/bloodproducts/TSE-manual2003.pdf.
- 89.Collins S, Boyd A, Lee JS, Lewis V, Fletcher A, McLean CA, Law M, Kaldor J, Smith MJ, Masters CL. Creutzfeldt-Jakob disease in Australia 1970–1999. Neurology. 2002;59:1365–1371. doi: 10.1212/01.wnl.0000031793.11602.8c. [DOI] [PubMed] [Google Scholar]
- 90.Brandel J-P, Welaratne A, Salomon D, Capek I, Vaillant V, Aouba A, Aouaba A, Haïk S, Alpérovitch A. Can mortality data provide reliable indicators for Creutzfeldt-Jakob disease surveillance? A study in France from 2000 to 2008. Neuroepidemiology. 2011;37:188–192. doi: 10.1159/000332764. [DOI] [PubMed] [Google Scholar]
- 91.Windl O, Giese A, Schulz-Schaeffer W, Zerr I, Skworc K, Arendt S, Oberdieck C, Bodemer M, Poser S, Kretzschmar HA. Molecular genetics of human prion diseases in Germany. Hum. Genet. 1999;105:244–252. doi: 10.1007/s004399900124. [DOI] [PubMed] [Google Scholar]
- 92.Grasbon-Frodl E, Lorenz H, Mann U, Nitsch RM, Windl O, Kretzschmar HA. Loss of glycosylation associated with the T183A mutation in human prion disease. Acta Neuropathol. (Berl.) 2004;108:476–484. doi: 10.1007/s00401-004-0913-4. [DOI] [PubMed] [Google Scholar]
- 93.Zerr I, Kallenberg K, Summers DM, Romero C, Taratuto A, Heinemann U, Breithaupt M, Varges D, Meissner B, Ladogana A, Schuur M, Haik S, Collins SJ, Jansen GH, Stokin GB, Pimentel J, Hewer E, Collie D, Smith P, Roberts H, Brandel JP, van Duijn C, Pocchiari M, Begue C, Cras P, Will RG, Sanchez-Juan P. Updated clinical diagnostic criteria for sporadic Creutzfeldt-Jakob disease. Brain J. Neurol. 2009;132:2659–2668. doi: 10.1093/brain/awp191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Puopolo M, Ladogana A, Almonti S, Daude N, Bevivino S, Petraroli R, Poleggi A, Quanguo L, Pocchiari M. Mortality trend from sporadic Creutzfeldt-Jakob disease (CJD) in Italy, 1993–2000. J. Clin. Epidemiol. 2003;56:494–499. doi: 10.1016/s0895-4356(02)00606-6. [DOI] [PubMed] [Google Scholar]
- 95.Jansen C, Parchi P, Capellari S, Ibrahim-Verbaas CA, Schuur M, Strammiello R, Corrado P, Bishop MT, van Gool WA, Verbeek MM, Baas F, van Saane W, Spliet WGM, Jansen GH, van Duijn CM, Rozemuller AJM. Human prion diseases in the Netherlands (1998–2009): clinical, genetic and molecular aspects. PloS One. 2012;7:e36333. doi: 10.1371/journal.pone.0036333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Parchi P, Giese A, Capellari S, Brown P, Schulz-Schaeffer W, Windl O, Zerr I, Budka H, Kopp N, Piccardo P, Poser S, Rojiani A, Streichemberger N, Julien J, Vital C, Ghetti B, Gambetti P, Kretzschmar H. Classification of sporadic Creutzfeldt-Jakob disease based on molecular and phenotypic analysis of 300 subjects. Ann. Neurol. 1999;46:224–233. [PubMed] [Google Scholar]
- 97.Exome Aggregation Consortium. Combined analysis of protein-coding genetic variation in 60,706 humans. doi: 10.1038/nature19057. Prep. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP. Integrative genomics viewer. Nat. Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Bryc K, Durand EY, Macpherson JM, Reich D, Mountain JL. The genetic ancestry of African Americans, Latinos, and European Americans across the United States. Am. J. Hum. Genet. 2015;96:37–53. doi: 10.1016/j.ajhg.2014.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Durand EY, Eriksson N, McLean CY. Reducing pervasive false-positive identical-by-descent segments detected by large-scale pedigree analysis. Mol. Biol. Evol. 2014;31:2212–2222. doi: 10.1093/molbev/msu151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Durand EY, Do CB, Mountain JL, Macpherson JM. Ancestry Composition: A Novel, Efficient Pipeline for Ancestry Deconvolution. bioRxiv. 2014:010512. [Google Scholar]
- 102.Browning SR, Browning BL. Rapid and accurate haplotype phasing and missing-data inference for whole-genome association studies by use of localized haplotype clustering. Am. J. Hum. Genet. 2007;81:1084–1097. doi: 10.1086/521987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Purcell SM, Moran JL, Fromer M, Ruderfer D, Solovieff N, Roussos P, O’Dushlaine C, Chambert K, Bergen SE, Kähler A, Duncan L, Stahl E, Genovese G, Fernández E, Collins MO, Komiyama NH, Choudhary JS, Magnusson PKE, Banks E, Shakir K, Garimella K, Fennell T, DePristo M, Grant SGN, Haggarty SJ, Gabriel S, Scolnick EM, Lander ES, Hultman CM, Sullivan PF, McCarroll SA, Sklar P. A polygenic burden of rare disruptive mutations in schizophrenia. Nature. 2014;506:185–190. doi: 10.1038/nature12975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Holman RC, Belay ED, Christensen KY, Maddox RA, Minino AM, Folkema AM, Haberling DL, Hammett TA, Kochanek KD, Sejvar JJ, Schonberger LB. Human prion diseases in the United States. PloS One. 2010;5:e8521. doi: 10.1371/journal.pone.0008521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Surveillance Data - CJD International Surveillance Network. 2015 available at http://web.archive.org/web/20151102144718/ http://www.eurocjd.ed.ac.uk/surveillance%20data%201.html. [Google Scholar]
- 106.United Nations Statistics Division - Demographic and Social Statistics. available at http://unstats.un.org/unsd/demographic/products/vitstats/ [Google Scholar]
- 107.Cornfield J. A method of estimating comparative rates from clinical data; applications to cancer of the lung, breast, and cervix. J. Natl. Cancer Inst. 1951;11:1269–1275. [PubMed] [Google Scholar]
- 108.Woolf B. On Estimating the Relation Between Blood Group and Disease. Ann. Hum. Genet. 1955;19:251–253. doi: 10.1111/j.1469-1809.1955.tb01348.x. [DOI] [PubMed] [Google Scholar]
- 109.Li C-C. Human genetics: principles and methods. New York: The Blakiston Division, McGraw-Hill Book Company Inc; 1961. http://catalog.hathitrust.org/Record/001496005. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.