Abstract
Osteopontin (OPN) is a chemokine-like glycoprotein that has a prominent role in regulating inflammation and immunity. OPN polymorphisms and elevated OPN levels are associated with systemic lupus erythematosus (SLE) in several populations. The aim of present study was to evaluate the association between the OPN rs1126616 polymorphism and OPN level with SLE susceptibility. A total of 163 SLE patients and 180 age-, gender- and ethnically matched controls were genotyped for the rs1126616 polymorphism by the polymerase chain reaction-restriction fragment length polymorphism method. Serum OPN levels were assayed by the enzyme-linked immunosorbent assay. There was no association between the OPN rs1126616 C/T polymorphism and SLE. The frequency of the OPN rs1126616 CT genotype was significantly higher in SLE patients with nephritis compared to SLE patients without nephritis and controls. Additionally, the frequency of TT genotypes was higher in SLE patients with nephritis compared to controls. The serum OPN levels were significantly higher in SLE patients compared to controls (50.6±22 vs. 35.6±15.8 ng/ml, P<0.001). Increased serum OPN levels were observed in SLE patients with lupus nephritis and joint symptoms. There was no correlation between OPN levels and the OPN rs1126616 polymorphism. The present data suggest that the CT and TT genotypes of the OPN rs1126616 polymorphism could be a risk factor for lupus nephritis. The OPN level is associated with SLE and certain SLE manifestations. However, there was no association between the OPN rs1126616 C/T polymorphism and SLE susceptibility.
Keywords: systemic lupus erythematosus, osteopontin, gene, polymorphism, lupus nephritis
Introduction
Systemic lupus erythematosus (SLE) is a human chronic inflammatory disorder encompassing multisystem organs with a broad spectrum. Overproduction of auto-antibodies in the formation of immune complexes may lead to hyperactivation of the immune system. These immune complexes, accumulated in several tissues and organs, have key roles in certain clinical manifestations of SLE. The prevalence of SLE in the general population is 20–150 cases/100,000 people worldwide and it is more common in women; almost 10-fold higher than men (1). Although genetic and environmental factors appear to have a significant role in the pathogenesis of SLE, the etiology of this disease remains to be elucidated. Familial studies have revealed that 10–12% of SLE patients that have an affected first-degree relative show a 20 to 50-fold occurrence increment when compared to the general population. Evidence in monozygotic and dizygotic twins has indicated that the disease occurs in 24–69% of the cases in monozygotic twins, but only 2–9% of the cases in dizygotic twins (2,3).
Osteopontin (OPN) is a chemokine-like glycoprotein, rich in aspartate and sialic acid residues, which has a key function in bone biology, and was recently found as prominent in regulating inflammation and immunity. As OPN is produced upon activation of cell, this glycoprotein was termed early T-lymphocyte activation protein-1 (Eta-1), which increases the Th1 response as well as inhibits the Th2 response (4). There are at least two OPN isoforms, including secreted OPN (sOPN) and intracellular OPN (iOPN), which are produced by translation from alternative initiation sites. The OPN protein is exposed to various post-translational modifications including glycosylation, phosphorylation and tyrosine sulfation, which are specific for cell type. These modifications could depend on physiological and pathophysiological factors and affect the OPN structure and function (5).
OPN activates immune cells, such as T cells, natural killer (NK) cells, macrophages and Kupffer cells (6). There are multiple receptors for OPN, and amongst them, cluster of differentiation 44 (CD44) is the most known receptor and mediates the cell chemotaxis and attachment (7). A number of studies have reported that serum OPN levels are increased in several autoimmune diseases, such as multiple sclerosis and SLE (8–10), therefore, OPN could contribute to the SLE pathogenesis or its manifestation. The association between OPN polymorphisms and SLE susceptibility has been studied in different populations; however, their results have been controversial (11–13).
As there are few studies regarding the effect of the OPN rs1126616 polymorphism on SLE and its association with serum OPN levels, the present study was performed to investigate the association between the OPN rs1126616 polymorphism and serum level of OPN with SLE susceptibility and its manifestations in southeast Iran.
Materials and methods
Patients and sample collection
The present study was approved by the Zahedan University of Medical Sciences Ethics Committee (Zahedan, Iran). This case-control investigation was conducted on all SLE patients (163 patients: 13 men and 150 women) who were referred to the Rheumatology Clinics of Ali-Ebne-Abitaleb Hospital in Zahedan between 2011 and 2013.
Individuals with malignant tumors, infectious disease and other rheumatic diseases were excluded from the study. All SLE patients fulfilled at least four items of SLE according to the ACR 1997 criteria (American College of Rheumatology) (14).
The control group consisted of 180 gender-, age- and ethnically matched volunteers (14 men and 166 women) with a negative antinuclear antibodies test that was randomly selected from healthy individuals who were referred to the Internal Medicine Clinic for check-up examinations.
The control group had no systemic disease and family association with lupus patients. Written informed consent according to the declaration of Helsinki was provided from all participants.
Genotyping of the OPN gene rs1126616 polymorphism
Genomic DNA was isolated from whole blood leukocytes by the salting-out procedure. OPN polymorphisms were identified by the polymerase chain reaction (PCR)-restriction fragment length polymorphism technique. Two oligonucleotide primers: Forward, 5′-TACCCTGATGCTACAGACGAGG-3′ and reverse, 5′-CTGACTATCAATCACATCGGAATG-3′; based on the flanking sequences of the OPN rs1126616 polymorphism were used to amplify the corresponding DNA. PCR was carried out in a 25 µl total final volume containing 25 pmol of each primer, 0.1 mM dNTP, 0.5–1 µg genomic DNA, 1.5 mM MgCl2, 2.5 µl of 10X PCR buffer and 1.5 unit TaqDNA polymerase (both from Fermentas, Vilnius, Lithuania). Thermal cycling conditions were as follows: Initial denaturation at 94°C for 5 min; 35 cycles of denaturation at 94°C for 40 sec, annealing for 30 sec at 60°C, and extension at 72°C for 45 sec; and final extension at 72°C for 6 min. The 252-base pair (bp) PCR fragment was digested with the AluI restriction enzyme. The C allele had one AluI cleavage site and was digested to 147 and 105 bp fragments, whereas for the T allele, the 105 bp fragment was cleaved to 61 and 44 bp fragments.
Digested products were separated by electrophoresis on a 3% agarose gel and visualized by ethidium bromide staining.
OPN measurement
Blood samples were collected after 12 h of fasting. Following this, samples were centrifuged at 3,000 × g for 10 min after clotting for 30 min at room temperature and were stored at −70°C until analysis. Serum OPN levels were measured using human OPN kit (R&D Systems, Minneapolis, MN, USA) according to the manufacturer's protocol.
Statistical analysis
Data were analyzed using the statistical software SPSS version 18 (SPSS, Inc., Chicago, IL, USA). The differences between the groups were analyzed by independent sample t-test, χ2 test or Fisher's exact test, as appropriate. For determining the frequency of the allele, the direct gene counting method was used. The genotype and allele frequencies were compared between SLE patients and controls by χ2 test and Fisher's exact test. The odds ratio (OR) and 95% confidence intervals (CI) were also estimated. Logistic regression analysis was used to assess the independent effect of each risk polymorphism on SLE. P<0.05 was considered to indicate a statistically significant difference.
Results
Demographic and clinical characteristics
Demographic and clinical data of the SLE patients and control group are shown in Table I. There were no significant differences in age, gender and ethnicity between SLE and control groups (Table I). Dermomucus manifestations were identified in 83% of SLE patients (54% with malar rash). Joint symptoms were improved in 87% of patients and neuropsychiatric manifestations were observed in 24% of patients. Lupus nephritis (LN) was developed with raised serum creatinine in 22% of patients.
Table I.
Demographic and clinical characteristics of SLE patients and controls.
| Parameter | SLE patients, n=163 | Controls, n=180 | P-value |
|---|---|---|---|
| Age, year | 32.6±8.6 | 32.1±11.7 | 0.68 |
| Gender (male/female), n | 13/150 | 14/166 | 0.55 |
| Ethnicity, n (%) | |||
| Persian | 82 (50) | 86 (48) | 0.36 |
| Balouch | 81 (50) | 94 (52) | |
| Joint symptoms, n (%) | 141 (87) | − | |
| Renal diseases, n (%) | 36 (22) | − | |
| Dermomucus disorder, n (%) | 135 (83) | − | |
| Neurological disorder, n (%) | 23 (14) | − | |
| Hematological disorder, n (%) | 98 (60) | − | |
| Oral ulcer, n (%) | 45 (28) | ||
| Antinuclear antibodies, n (%) | 147 (90) | − | |
| Anti-dsDNA antibodies, n (%) | 122 (75) | − |
SLE, systemic lupus erythematosus.
Polymorphism frequency
The frequency of the OPN rs1126616 polymorphism genotypes and alleles between the two groups is summarized in Table II. The frequency of the OPN rs1126616 polymorphism genotypes was in Hardy-Weinberg equilibrium for cases and controls. Although the frequency of the OPN rs1126616 T allele was higher in SLE patients compared to the controls (17.2 vs. 12.2%), the difference was not statistically significant. There were no significant differences in the CT and TT genotypes between the SLE patients and control group. However, a higher frequency of the CT genotype was observed in patients with LN compared to patients without LN and healthy controls (Table III), therefore this genotype may increase the LN risk (OR, 3.1; 95% CI, 1.4–6.9; P=0.004 and OR, 3.5; 95% CI, 1.6–7.4; P=0.001, respectively). The frequency of the TT genotype was significantly higher in patients with LN compared to the controls (OR, 8.1; 95% CI, 1.1–62.5). There was no association between the OPN rs1126616 polymorphism and other SLE manifestations.
Table II.
Genotypes and alleles frequency of OPN rs1126616 polymorphism in SLE patients and controls.
| OPN rs1126616 | SLE patients, n=163 | Controls, n=180 | P-value | OR (95% CI) |
|---|---|---|---|---|
| Genotypes, n (%) | ||||
| CC | 112 (69) | 138 (77) | 1.0 | |
| CT | 46 (28) | 40 (22) | 0.17 | 1.4 (0.9–2.3) |
| TT | 5 (3) | 2 (1) | 0.18 | 1.8 (0.8–4.0) |
| CT + TT | 51(31) | 42 (23) | 0.10 | 1.5 (0.9–2.4) |
| Alleles, n (%) | ||||
| C | 270 (82.8) | 316 (87.8) | 1.0 | |
| T | 56 (17.2) | 44 (12.2) | 0.07 | 1.5 (1–2.3) |
SLE, systemic lupus erythematosus; OPN, osteopontin; OR, odds ratio; CI, confidence interval.
Table III.
Genotypes frequency of OPN rs1126616 polymorphism in SLE patients with and without LN.
| OPN rs1126616 | SLE patients with LN n=36 | SLE patients without LN n=127 | Controls n=180 | P-valuea | OR (95% CI)a | P-valueb | OR (95% CI)b |
|---|---|---|---|---|---|---|---|
| Genotypes, n (%) | |||||||
| CC | 17 (47) | 94 (74) | 138 (77) | 1 | 1 | ||
| CT | 17 (47) | 30 (24) | 40 (22) | 0.004 | 3.1 (1.4–6.9) | 0.001 | 3.5 (1.6–7.4) |
| TT | 2 (6) | 3 (2) | 2 (1) | 0.170 | 3.7 (0.6–23.7) | 0.043 | 8.1 (1.1–62.5) |
Between SLE patients with LN and without LN
between SLE patients with LN and controls. SLE, systemic lupus erythematosus; OPN, osteopontin; OR, odds ratio; CI, confidence interval; LN, lupus nephritis.
Serum OPN levels
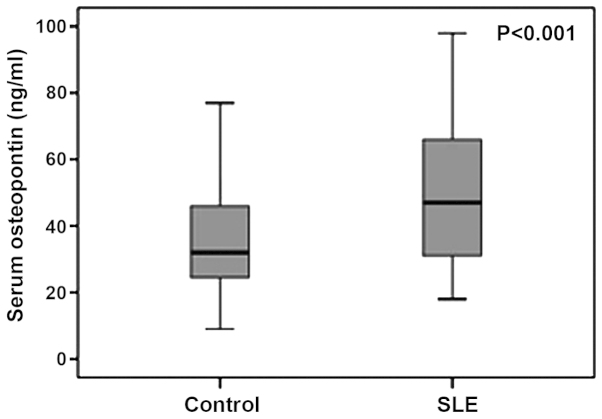
The serum OPN levels were 50.6±22 ng/ml in SLE patients (n=46) and 35.6±15.8 ng/ml in the control group (n=43), which indicated a significant elevation in SLE patients (P<0.001) (Fig. 1).
Figure 1.
Comparison of median serum osteopontin levels between systemic lupus erythematosus (SLE) patients and controls.
The serum OPN levels were 48.7±21.5 ng/ml in individuals with the T allele (CT+TT genotypes) and 52.2±22.7 ng/ml in those without the T allele (CC genotype) in SLE patients, which were not statistically different (P=0.13) (Fig. 2A). In addition, the serum OPN levels were not different between individuals with the T allele (39.7±16.1 ng/ml) and individuals without the T allele (32.4±15 ng/ml) in the control group (P=0.6) (Fig. 2B).
Figure 2.
Comparison of the median serum osteopontin (OPN) levels between individuals with CT + TT genotypes and CC genotype in (A) systemic lupus erythematosus (SLE) patients and (B) the control group.
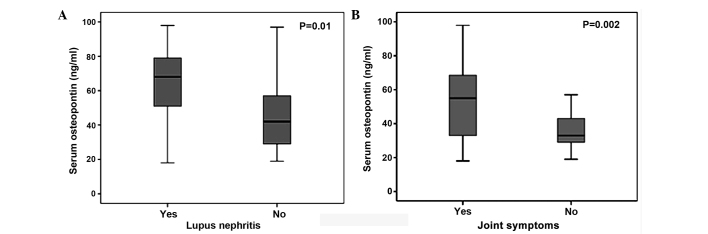
Higher serum OPN levels were observed in SLE patients with LN compared to SLE patients without this manifestation (63.9±19.7 vs. 45.3±20.9 mg/ml, respectively, P=0.01) (Fig. 3A). The serum OPN levels were significantly higher in SLE patients with joint symptoms compared to those without joint symptoms (54.4±22 vs. 36.6±12 mg/ml, respectively, P=0.002) (Fig. 3B).
Figure 3.
Comparison of the median serum osteopontin levels between systemic lupus erythematosus patients (A) with lupus nephritis and without lupus nephritis and (B) with joint symptoms and without joint symptoms.
Discussion
In the present study no association between the allelic frequency of the OPN rs1126616 polymorphism and SLE susceptibility was observed. Although the frequency of CT and TT genotypes was higher in SLE patients, the differences were not significant. However the frequency of the OPN rs1126616CT genotype was significantly higher in patients with LN compared to patients without LN and controls. Furthermore, the frequency of OPN rs1126616TT was higher in the LN group compared to the control. There was no correlation between the rs1126616 polymorphism and other SLE manifestations. Additionally, an elevated serum OPN level was identified only in SLE patients. A higher serum OPN concentration was observed in SLE patients with LN and joint symptoms compared to SLE patients without these manifestations.
OPN as a secretory ECM protein, is involved in different physiological as well as pathophysiological processes, such as cell attachment, migration, proliferation, bone formation, cytokine expression, acute and chronic inflammation, leukocyte recruitment and immunological activity regulation (15–18). OPN can exert pro-inflammatory and anti-inflammatory functions, and its mere effect depends on the nature of scenario (4). OPN has a role in the production of type I interferon (IFN) and can be involved in autoimmune disease pathogenesis (19). Several studies have shown the association between OPN and SLE pathogenesis, as well as other autoimmune diseases (8,9).
A higher OPN expression has been observed in macrophages and CD4−/CD8− T cells in MRL/lpr lupus murine, which stimulates polyclonal antibodies expression (20). In addition, Li et al (21) reported that OPN and OPN mRNA expression was elevated in peripheral blood mononuclear cells of SLE patients.
In an experimental study performed by Iizuka (22), it was shown that OPN may have an important role in proliferation and differentiation of B1 cells and autoantibody production.
Wong et al (10) revealed that the plasma OPN level was significantly higher in SLE patients and the increased OPN level correlated with the SLE disease activity index score in all SLE patients. Additionally, elevated plasma OPN concentrations were identified in SLE patients with renal impairment compared to controls and they suggested that OPN production was correlated with the inflammatory process.
A recent study by Briggs (9) confirmed the correlation between raised baseline plasma OPN and IFN levels. Furthermore, the study reported the elevated level of OPN prior to the onset of increased cumulative disease and end-organ damage. These associations were particularly observed in pediatric SLE, which signified that OPN may be a useful biomarker of disease activity in childhood lupus.
Although the OPN gene has not been introduced as a SLE susceptibility gene, various polymorphisms of the OPN gene have been associated with elevated IFN-α induction in young patients with SLE and specific clinical phenotypes (23,24).
Therefore, several studies have been performed to assess the correlation between the OPN gene polymorphisms and predisposition to SLE and its manifestations. In 2002, Forton et al (12) showed the association between the C707T polymorphism of the OPN gene with SLE susceptibility. In addition, the study suggested the association between this polymorphism with renal disease and opportunistic infections. D'Alfonso et al (11) scanned the coding 5′ and 3′ flanking regions of the OPN gene in SLE patients and revealed the correlation between the −156G allele in the promoter and +1239C allele in the 3′-untranslated region of the OPN gene and SLE susceptibility. In addition, an association between lymphadenopathy and −156 genotypes was observed.
Although Xu et al (13) observed a lower frequency of the TT genotype and higher frequency of the TC genotype of the OPN polymorphism in SLE patients compared to controls, no association between this polymorphism and SLE was observed in the present study. Similar to the results of the present study, the correlation between the OPN polymorphism and LN has been reported in the southern Chinese Han population (25).
Han et al (26) performed a large cohort study regarding the effect of A7385G, A1239C and C750T polymorphisms of the OPN gene in SLE susceptibility in European-American and African-American populations. The study concluded that the frequency of the 750T and 1239C minor alleles were associated with a higher risk of SLE in males only. Additionally, a significant association between the 750T-7385A-1239C haplotype with SLE was observed, particularly in males.
Similarly, Kariuki et al (23) reported the association between the OPN 1239C allele with higher serum OPN and IFN-α levels in men. This association in women was restricted to younger subjects. Trivedi et al (24) showed a significant association between the 1239C allele and photosensitivity in SLE patients. Additionally, an association between the C allele with thrombocytopenia and hemolytic anemia in patients was identified.
Although the majority of the available SLE patients in southeast Iran were included in the present study, the study is limited by the small sample size. In addition, as the number of males (13 men) that were included in the SLE group was low, the effect of gene-gender interaction could not be analyzed. Therefore, further studies using higher sample size is suggested for assessing the effect of the gene-gender interaction.
In conclusion, there was no association between the OPN rs1126616 polymorphism and predisposition to SLE. The OPN rs1126616 polymorphism was correlated with LN. Additionally, higher serum OPN levels were observed in SLE patients and SLE patients with LN and joint symptoms.
Acknowledgements
The authors would like to thank the Deputy of Research at Zahedan University of Medical Sciences for the financial support of this research.
References
- 1.Crispín JC, Liossis SN, Kis-Toth K, Lieberman LA, Kyttaris VC, Juang YT, Tsokos GC. Pathogenesis of human systemic lupus erythematosus: Recent advances. Trends Mol Med. 2010;16:47–57. doi: 10.1016/j.molmed.2009.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Harley JB, Kelly JA, Kaufman KM. Unraveling the genetics of systemic lupus erythematosus. Springer Semin Immunopathol. 2006;28:119–130. doi: 10.1007/s00281-006-0040-5. [DOI] [PubMed] [Google Scholar]
- 3.Deapen D, Escalante A, Weinrib L, Horwitz D, Bachman B, Roy-Burman P, Walker A, Mack TM. A revised estimate of twin concordance in systemic lupus erythematosus. Arthritis Rheum. 1992;35:311–318. doi: 10.1002/art.1780350310. [DOI] [PubMed] [Google Scholar]
- 4.Mazzali M, Kipari T, Ophascharoensuk V, Wesson JA, Johnson R, Hughes J. Osteopontin - a molecule for all seasons. QJM. 2002;95:3–13. doi: 10.1093/qjmed/95.1.3. [DOI] [PubMed] [Google Scholar]
- 5.Christensen B, Kazanecki CC, Petersen TE, Rittling SR, Denhardt DT, Sorensen ES. Cell type-specific post-translational modifications of mouse osteopontin are associated with different adhesive properties. J Biol Chem. 2007;282:19463–19472. doi: 10.1074/jbc.M703055200. [DOI] [PubMed] [Google Scholar]
- 6.Scatena M, Liaw L, Giachelli CM. Osteopontin: a multifunctional molecule regulating chronic inflammation and vascular disease. Arterioscler Thromb Vasc Biol. 2007;27:2302–2309. doi: 10.1161/ATVBAHA.107.144824. [DOI] [PubMed] [Google Scholar]
- 7.Weber GF, Ashkar S, Glimcher MJ, Cantor H. Receptor-ligand interaction between CD44 and osteopontin (Eta-1) Science. 1996;271:509–512. doi: 10.1126/science.271.5248.509. [DOI] [PubMed] [Google Scholar]
- 8.Murugaiyan G, Mittal A, Weiner HL. Increased osteopontin expression in dendritic cells amplifies IL-17 production by CD4+ T cells in experimental autoimmune encephalomyelitis and in multiple sclerosis. J Immunol. 2008;181:7480–7488. doi: 10.4049/jimmunol.181.11.7480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Briggs TA. Osteopontin - a biomarker for organ damage in paediatric lupus? Arthritis Res Ther. 2013;15:110. doi: 10.1186/ar4172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wong CK, Lit LC, Tam LS, Li EK, Lam CW. Elevation of plasma osteopontin concentration is correlated with disease activity in patients with systemic lupus erythematosus. Rheumatology (Oxford) 2005;44:602–606. doi: 10.1093/rheumatology/keh558. [DOI] [PubMed] [Google Scholar]
- 11.D'Alfonso S, Barizzone N, Giordano M, Chiocchetti A, Magnani C, Castelli L, Indelicato M, Giacopelli F, Marchini M, Scorza R, et al. Two single-nucleotide polymorphisms in the 5′ and 3 ends of the osteopontin gene contribute to susceptibility to systemic lupus erythematosus. Arthritis Rheum. 2005;52:539–547. doi: 10.1002/art.20808. [DOI] [PubMed] [Google Scholar]
- 12.Forton AC, Petri MA, Goldman D, Sullivan KE. An osteopontin (SPP1) polymorphism is associated with systemic lupus erythematosus. Hum Mutat. 2002;19:459. doi: 10.1002/humu.9025. [DOI] [PubMed] [Google Scholar]
- 13.Xu AP, Bai J, Lü J, Liang YY, Li JG, Lai DY, Wan X, Huang HH. Osteopontin gene polymorphism in association with systemic lupus erythematosus in Chinese patients. Chin Med J (Engl) 2007;120:2124–2128. [PubMed] [Google Scholar]
- 14.Hochberg MC. Updating the American College of Rheumatology revised criteria for the classification of systemic lupus erythematosus. Arthritis Rheum. 1997;40:1725. doi: 10.1002/art.1780400928. [DOI] [PubMed] [Google Scholar]
- 15.Ashkar S, Weber GF, Panoutsakopoulou V, Sanchirico ME, Jansson M, Zawaideh S, Rittling SR, Denhardt DT, Glimcher MJ, Cantor H. Eta-1 (osteopontin): An early component of type-1 (cell-mediated) immunity. Science. 2000;287:860–864. doi: 10.1126/science.287.5454.860. [DOI] [PubMed] [Google Scholar]
- 16.El-Tanani MK. Role of osteopontin in cellular signaling and metastatic phenotype. Front Biosci. 2008;13:4276–4284. doi: 10.2741/3004. [DOI] [PubMed] [Google Scholar]
- 17.Weber GF, Cantor H. The immunology of Eta-1/osteopontin. Cytokine Growth Factor Rev. 1996;7:241–248. doi: 10.1016/S1359-6101(96)00030-5. [DOI] [PubMed] [Google Scholar]
- 18.Weber GF, Zawaideh S, Hikita S, Kumar VA, Cantor H, Ashkar S. Phosphorylation-dependent interaction of osteopontin with its receptors regulates macrophage migration and activation. J Leukoc Biol. 2002;72:752–761. [PubMed] [Google Scholar]
- 19.Shinohara ML, Kim JH, Garcia VA, Cantor H. Engagement of the type I interferon receptor on dendritic cells inhibits T helper 17 cell development: Role of intracellular osteopontin. Immunity. 2008;29:68–78. doi: 10.1016/j.immuni.2008.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lampe MA, Patarca R, Iregui MV, Cantor H. Polyclonal B cell activation by the Eta-1 cytokine and the development of systemic autoimmune disease. J Immunol. 1991;147:2902–2906. [PubMed] [Google Scholar]
- 21.Li L, Yu XQ, Yin PD, Chen YX, Liu YG, Xu HS. OPN expression of peripheral blood mononuclear cells in systemic lupus erythematosus (SLE) and its clinical significance. Chin J Rheumatol Chin. 1999;3:84–86. (In Chinese) [Google Scholar]
- 22.Iizuka J. Relationship between osteopontin expression and autoimmune disease - analysis of osteopontin expressed in transgenic mice. Hokkaido Igaku Zasshi. 1998;73:487–495. (In Japanese) [PubMed] [Google Scholar]
- 23.Kariuki SN, Moore JG, Kirou KA, Crow MK, Utset TO, Niewold TB. Age- and gender-specific modulation of serum osteopontin and interferon-alpha by osteopontin genotype in systemic lupus erythematosus. Genes Immun. 2009;10:487–494. doi: 10.1038/gene.2009.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Trivedi T, Franek BS, Green SL, Kariuki SN, Kumabe M, Mikolaitis RA, Jolly M, Utset TO, Niewold TB. Osteopontin alleles are associated with clinical characteristics in systemic lupus erythematosus. J Biomed Biotechnol. 2011;2011:802581. doi: 10.1155/2011/802581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xu AP, Liang YY, Lü J, Li JG, Wang Z. Association of osteopontin gene polymorphism with lupus nephritis in Chinese Han population. Nan Fang Yi Ke Da Xue Xue Bao. 2007;27:1348–1351. (In Chinese) [PubMed] [Google Scholar]
- 26.Han S, Guthridge JM, Harley IT, Sestak AL, Kim-Howard X, Kaufman KM, Namjou B, Deshmukh H, Bruner G, Espinoza LR, et al. Osteopontin and systemic lupus erythematosus association: A probable gene-gender interaction. PLoS One. 2008;3:e0001757. doi: 10.1371/journal.pone.0001757. [DOI] [PMC free article] [PubMed] [Google Scholar]