Abstract
Glucocorticoids have been administered to mothers at risk of premature delivery to induce maturation of preterm fetal lungs and prevent the development of respiratory distress syndrome. Micro (mi)RNAs serve various crucial functions in cell proliferation, differentiation and organ development; however, few studies have demonstrated an association between miRNAs and lung development. The aim of the present study was to investigate alterations in the miRNA profiles of rat lung tissue following prenatal glucocorticoid therapy for fetal lung development. The differences in miRNA expression profiles were compared between postnatal days 7 (D7) and 120 (D120) rat lung tissues, followed by validation using reverse transcription-quantitative polymerase chain reaction. The miRNA profiles of rat lung tissues following prenatal dexamethasone (DEX) therapy were also investigated. miRNAs with 2-fold changes were selected for further analysis. At D120, 6 upregulated and 6 downregulated miRNAs were detected, compared with D7. Among these differentially expressed miRNAs, miR-101-3p and miR-99b-5p were associated with the lowest and highest expressions of miRNA at D7, respectively. A limited impact on the miRNA profiles of rat lung tissues was observed following prenatal DEX treatment, which may help to further clarify the mechanisms underlying normal lung development. However, the results of the present study cannot entirely elucidate the effects of prenatal DEX treatment on the lung development of premature infants, and further studies investigating the impact of prenatal corticosteroids on fetal lung miRNA profiles are required.
Keywords: prenatal glucocorticoid, rat, lung, microRNA
Introduction
Pulmonary immaturity is a major cause of morbidity and mortality in premature infants. For >30 years glucocorticoids have been clinically administered to mothers at risk of premature delivery in order to induce maturation of preterm fetal lungs and prevent the development of respiratory distress syndrome (1,2). However, glucocorticoids have been reported to affect the stimulation of surfactant synthesis (3,4), structural lung growth (5–9), and the maturation of pulmonary epithelial cells and differentiation of type 2 cells (10,11) during lung development.
Drake et al (12) have demonstrated the intergenerational consequences of fetal programming in utero by exposure to glucocorticoids in rats. Furthermore, increasing evidence in humans suggests that prenatal overexposure to glucocorticoids may result in adverse adult cardiovascular, metabolic, neuroendocrine and behavioral phenotypes, and these effects appear to be transmitted across generations (13). This transmission across generations without further exposure to glucocorticoids suggests an epigenetic mechanism (13). The authors of the present study have previously demonstrated that high dose prenatal dexamethasone (DEX) treatment increases the expression levels of TNF-α and decreased the levels of histone deacetylase 2 protein in rat lungs in the acute stage (14).
In addition to methylation and histone acetylation, epigenetic alterations include regulation of micro (mi)RNA. It is hypothesized that miRNAs are involved in the regulation of almost every cellular and physiological process; however, the specific biological and physiological functions of many miRNAs remain unknown (15). Certain miRNAs have been reported to modulate the development of the lungs, surfactant secretion and PPAR-γ expression during rat lung development (15–17). Furthermore, Williams et al (18) have demonstrated that the overall miRNA expression profile is similar for mouse and human lung tissue, which suggests that miRNA expression is evolutionarily conserved during lung development. Various miRNAs, including miR-150, miR-375 and miR-26a, have been reported to be involved in the regulation of pulmonary surfactant secretion (15,19,20). Since miRNA expression profiling is important for the investigation of potential differences between physiological and pathophysiological status, the aim of the present study was to investigate alterations in miRNA profiles following prenatal glucocorticoid treatment for fetal lung development.
Materials and methods
Animals
This study was conducted in strict accordance with the recommendations outlined in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. The protocol was approved by the Institutional Animal Care and Use Committee of the Kaohsiung Chang Gung Memorial Hospital (Kaohsiung, Taiwan). Virgin Sprague-Dawley (SD) rats (12–16 weeks old) were obtained (BioLASCO Taiwan Co., Ltd., Taipei, Taiwan), and housed and maintained in a facility accredited by the Association for Assessment and Accreditation of Laboratory Animal Care International. Virgin SD female rats were allowed to mate with male rats for 24 h, and were separated from the male rats and housed individually in a standard plastic home cage. Following confirmation of pregnancy on day 14 following mating, pregnant females were randomly assigned into prenatal steroid treatment or untreated until delivery groups. Pregnant rats were checked for litters daily at 10 a.m. The day of birth was designated postnatal day 0 (D0) and rat pups were weaned on postnatal day 21 (D21), with ad libitum access to standard chow and water. Only male rat pups were used in the present study. In the short-term prenatal steroid group, pregnant rats were administered i.p. DEX (0.1 mg/kg/day; 026804; Taiwan Biotech Co., Ltd., Taoyuan, Taiwan) at gestational day 19–20 (DEX2 group); whereas pregnant rats in the long-term antenatal steroid group were administered i.p. DEX (0.1 mg/kg/day) at gestational day 14–20 (DEX7 group). The early effects of prenatal programming using glucocorticoids were assessed on postnatal day 7 (D7), whereas the late programming effects were assessed on postnatal day 120 (D120). The vehicle group comprised of pregnant SD rats at gestational day 14–20 that received i.p. normal saline.
Histological examination
Rats were anesthetized by an intramuscular injection of a 1:1 mixture (100 mg/kg) of ketamin (#003542; United Biomedical, Inc., Hsinchu, Taiwan) and rompun (#06713; Bayer Taiwan Co., Ltd., Taipei, Taiwan), followed by cardiac puncture and perfusion. Immediately after the rats were sacrificed as described previously (14), the lungs were harvested and stored in saline on ice and were subsequently dissected from the surrounding tissues. Lung tissue was then fixed in 10% formalin neutral buffer solution, pH 7.4 (Wako Pure Chemical Industries, Ltd., Osaka, Japan). Subsequently, the tissues were cut into 4-µm sections and stained with hematoxylin (Merck, Darmstadt, Germany) and eosin Y (HT110180; Sigma-Aldrich, St. Louis, MO, USA) for morphometric analysis. Images were captured using a digital camera (CTR 5000; Leica) and a Nikon Eclipse E600 microscope (magnification, ×10).
Western blot analysis
A 50-mg lung tissue sample was homogenized using 500 µl PRO-PREP Protein Extraction Solution (#17081; iNtRON Biotechnology, Inc., Seongnam, Korea). Cell lysis was induced by incubation for 30 min on ice and the samples was centrifuged at 14,000 × g for 20 min at 4°C. Protein concentrations were determined using a Bio-Rad Protein Assay kit II (#5000002; Bio-Rad Laboratories, Inc., Hercules, CA, USA). Protein samples (50 µg) were boiled with gel-loading buffer (10 mM Tris-HCl, 1% sodium dodecyl sulfate (SDS), 25% glycerol, 0.1 mM β-mercaptoethanol and 0.03% bromophenol blue; pH 6.8) for 5 min and separated by 10% SDS-polyacrylamide gel electrophoresis. Following transfer to a polyvinylidene fluoride membrane (Roche Diagnostics, Basel, Switzerland) and blocking with phosphate-buffered saline-Tween containing 5% dry milk, the membranes were incubated for 2 h with anti-rat lamin A/C antibody (#4777; Cell Signaling Technology, Inc., Danvers, MA, USA) diluted 1:200 in Tris-buffered saline (TBS) containing 1.21 g Tris (#640310, Merck) and 8.76 g NaCl (#106404; Merck) in 1 L ddH2O (pH 7.6), supplemented with 1% skimmed milk. Following five washes with 0.1% Tween-TBS (T-TBS), the membranes were incubated for 1 h with horseradish peroxidase-labeled secondary antibody (#7076; Cell Signaling Technology, Inc.) diluted 1:1,000 in T-TBS. Bands of interest were visualized using Western Lightning Plus enhanced chemiluminescence reagents (NEL105001EA; PerkinElmer, Inc., Waltham, MA, USA) and quantified using densitometry (Quantity One Analysis software, version 4.6.7; Bio-Rad Laboratories, Inc.) as integrated optical density after subtraction of background. The integrated optical density was factored for Ponceau S staining (PonS; P7170; Sigma-Aldrich) to correct for any variations in total protein loading. The protein abundance was represented as integrated optical density/PonS.
RNA isolation
Total RNA was extracted using TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA) according to the manufacturer's protocol. Purified RNA was quantified at an optical density of 260 nm using a ND-1000 spectrophotometer (NanoDrop Technologies; Thermo Fisher Scientific, Inc., Wilmington, DE, USA) and measured using a Bioanalyzer 2100 with an RNA 6000 LabChip kit (both Agilent Technologies, Inc., Santa Clara, CA, USA).
Library preparation and sequencing
Small RNA library construction and deep sequencing were performed by Welgene Biotech, Co., Ltd. (Taipei, Taiwan). Samples were prepared using an Illumina Sample Preparation kit according to the TruSeq® Small RNA Sample Preparation Guide (Illumina, Inc., San Diego, CA, USA). Subsequently, 5′ and 3′ adaptors (Illumina, Inc.) were ligated to total RNA, followed by reverse transcription-quantitative polymerase chain reaction (RT-qPCR) amplification (described below). Enriched cDNA constructs were size-fractionated and purified by electrophoresis in 6% polyacrylamide gel with 30% acrylamide/Bis solution (#1610156) at a ratio of 29:1, and using a Mini-PROTEAN Tetra Vertical Electrophoresis System (Bio-Rad Laboratories, Inc.). Bands containing 18–40 nucleotide RNA fragments, 140–155 nucleotides in length with both adapters, were harvested. Libraries were sequenced using an Genome Analyzer IIx (50 cycle single read; Illumina, Inc.).
Next-generation sequencing (NGS) data analysis
According to the manufacturer's protocol, the generated NGS data were analyzed using the miRSeq software package (21), which enables the acquisition of miRNA expression profiles and the evaluation of the overall quality of the NGS libraries. Using the readPro tool in miRSeq, a 15–27 length parameter was specified. Each sample used 1 ng RNA in the NGS experiment.
Prediction of target genes of differentially-expressed miRNAs
Targets of miRNAs were predicted using an online target prediction tool; TargetScan 6.2 (http:www.targetscan.org/). A prediction score of >0.5 was selected as the criterion for target genes with each miRNA.
RT-qPCR analysis
In order to validate the miRNA profiling results, RT-qPCR was conducted on all samples using a SYBR® Green miRNA-based assay including the Universal cDNA Synthesis Kit (#203301) and an ExiLENT SYBR® Green Master Mix Kit (#203420; Exiqon, Inc., Woburn, MA, USA). Total RNA was extracted from the lung tissue using TRIzol® reagent. A Universal cDNA Synthesis kit (#203301; Exiqon, Inc.) was used to reverse transcribe 20 ng RNA from the tissues using the 2X ExiLENT SYBR® Green Master Mix, according to the manufacturer's protocol. A total of 50 ng RNA was reverse transcribed to cDNA in qPCR experiment. The mixture was incubated at 42°C for 60 min, followed by 95°C for 5 min. RT-qPCR reactions were subsequently performed using a total reaction volume of 10 µl containing 4 µl cDNA, 1 µl PCR primer mix and 5 µl ExiLENT SYBR® Green master mix (203420; Exiqon, Inc.). The PCR cycling was conducted as follows: Pre-incubation at 95°C for 10 min; then amplification for 45 cycles of 95°C for 10 sec and 60°C for 10 min; then melting cycle of 95°C for 5 sec, 65°C for 1 min and 97°C with continuous per 5°C acquisition of fluorescence; and then cooling at 40°C for 10 sec. All RT-qPCR reactions, including the controls, were performed in triplicate using a LightCycler® 480 system (Roche Diagnostics) (14). Relative miRNA expression levels were analyzed using the Cq method and normalized against U6 snRNA (203907; Exiqon, Inc.) expression levels. Target miRNA primer sequences were as follows: Rno-let-7b-5p, 5′-UGAGGUAGUAGGUUGUGUGGUU-3′; rno-miR-26a-5p, 5′-UUCAAGUAAUCCAGGAUAGGCU-3′; rno-miR-27a-3p, 5′-UUCACAGUGGCUAAGUUCCGC-3′; rno-miR-101a-3p, 5′-UACAGUACUGUGAUAACUGAA-3′; rno-miR-375–3p, 5′-UUUGUUCGUUCGGCUCGCGUGA-3′. Data were analyzed as previously described (22).
Statistical analysis
Data are expressed as mean ± standard error of the mean. Mann-Whitney U test was used when two groups were analyzed. All statistical tests were performed using SPSS software, version 19.0 (IBM SPSS, Armonk, NY, USA). P<0.05 was considered to indicate a statistical significant difference.
Results
Prenatal DEX treatment induces alveolar tissue dysplasia and lamin A/C reduction
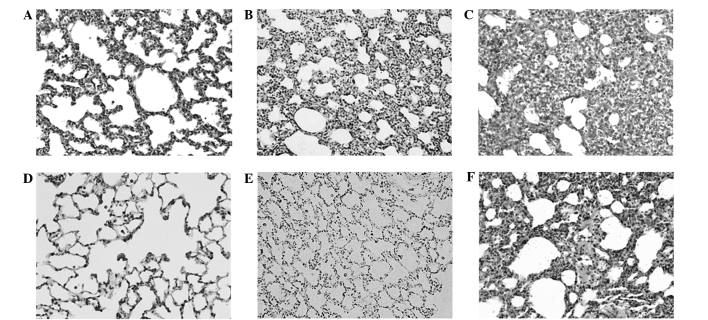
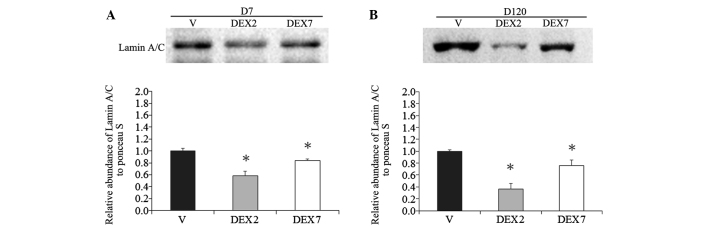
Histological examination of the prenatal DEX treatment group using hematoxylin and eosin staining demonstrated obvious dysplasia of the lung alveolar tissue at D7 and D120, as compared with the vehicle group (Fig. 1). The greatest dysplastic change was detected in the rat lungs of the DEX7 group, which were administered prenatal DEX for 7 days. Since lamin A/C level has been demonstrated to be an important determinant for lineage-specific differentiation during embryonic development (23), the abundance of lamin A/C was determined using western blotting. A reduction in lamin A/C was detected in the lung alveolar tissue of the prenatal DEX group at D7 and D120, as compared with the vehicle group (Fig. 2). The lowest lamin A/C expression levels were detected in the rat lungs of the DEX2 group, which were administered prenatal DEX for 2 days.
Figure 1.
Hematoxylin and eosin staining of rat lungs. Representative fields at postnatal day 7 in the groups treated with (A) vehicle, (B) dexamethasone (DEX) for 2 days and (C) 7 days groups. Representative fields at postnatal day 120 in the groups treated with (D) vehicle, (E) DEX for 2 days and (F) 7 days.
Figure 2.
Western blot analysis of lamin A/C in the V and prenatal DEX groups of lung tissue at D7 and D120. Representative blots of the V, DEX2 and DEX7 groups at (A) D7 and (B) D120. Lung tissue of prenatal DEX treatment showed decreased lamin A/C expression, particularly in the DEX7 group, as compared with the V group. Data were calculated from six replicated experiments. *P<0.05 vs. V group. D7, postnatal day 7; V, vehicle; DEX2, dexamethasone treatment for 2 days; DEX7, dexamethasone treatment for 2 days DEX7, dexamethasone treatment for 7 days; D120, postnatal day 120.
Summaries of NGS libraries
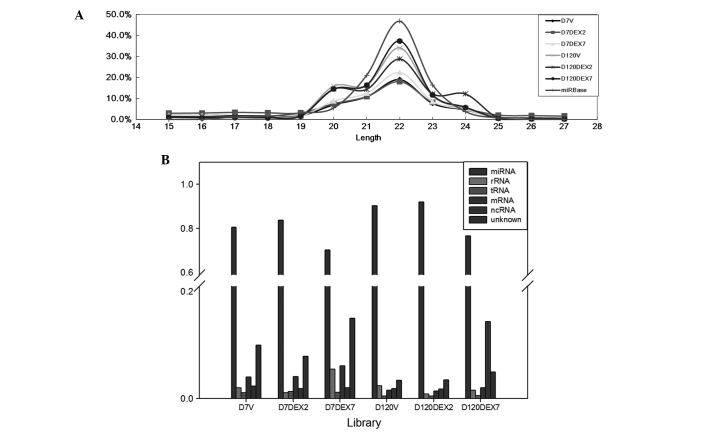
In the present study, six libraries were used from rat lung tissues sequenced using the NGS platform and the generated NGS data were analyzed using the miRSeq software package. The results demonstrated that the six libraries had similar performances regarding self-ligation reads, which are the reads generated from 5′ and 3′ adaptor self-ligation at the sample preparation step, and clean reads, which are the reads with the 3′ adaptor identified and trimmed. However, further investigation demonstrated that the clean reads of the D7 libraries tended to be less enriched in 22-nt length (Fig. 3A). The length distribution patterns of the D120 libraries were more homologous to the rat miRNAs in miRBase 20; therefore, these had a higher likelihood of being miRNAs (Fig. 2B). The D120 libraries accounted for a significantly higher proportion of the miRNA reads (P=0.015 pair-wise t-test) (Fig. 3B). Furthermore, among the three D120 libraries, a peak at 24 nt was detected at D120 in the DEX7 group, resulting in a higher proportion of non-coding RNA reads. Therefore, the sample preparation steps performed better in the D120 libraries, among which the D120 V and D120 DEX2 groups were particularly successful.
Figure 3.
Next generation sequencing data. (A) Length distributions of the clean reads in the libraries, which were defined as reads with the 3′ adaptor identified and trimmed. (B) The read category of the next generation sequencing reads. Length distribution patterns suggested that miRNA reads accounted for a higher proportion of the reads in the D120 libraries, as compared with the D7 libraries, which was confirmed by the read category. D7, postnatal day 7; D120, postnatal day 120; DEX2, prenatal dexamethasone treatment for 2 days; DEX7, prenatal dexamethasone treatment for 7 days; miRNA, micro RNA; rRNA, ribosomal RNA; tRNA, transfer RNA;mRNA, messenger RNA; ncRNA, non-coding RNA.
miRNA expression profiles varied between D7 and D120
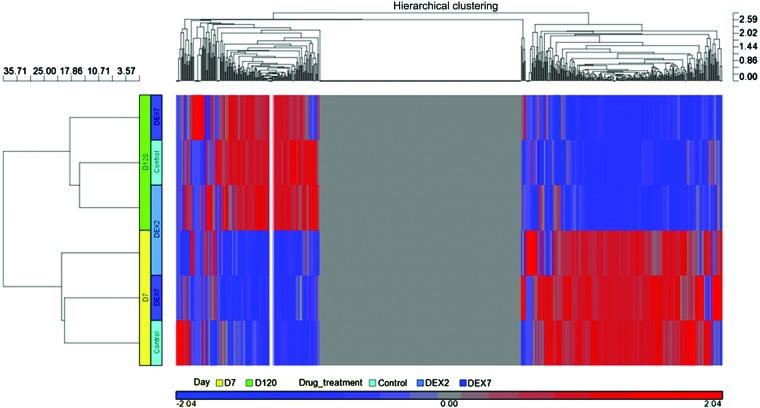
Following miRSeq analysis, miRNA expression profiles in six libraries were acquired. In order to compare the overall miRNA expression profiles among the six libraries, respective heat maps were plotted using the Partek® analysis toolkit (Partek, Inc., St. Louis, MO, USA). The six libraries were first classified into two clusters belonging to D7 and D120, respectively (Fig. 4). Therefore, age was the predominant factor for the observed differences in the miRNA expression profiles. In the D7 libraries, the vehicle and DEX7 groups demonstrated more homologous patterns; whereas, in the D120 libraries, the vehicle group was most homologous to the DEX2 group. Therefore, the results of the present study suggest that DEX treatment produced inconsistent effects on the miRNA expression profiles between the D7 and D120 libraries.
Figure 4.
Heat map of microRNA expression preference. The heat map demonstrated that majority of the differences detected in the expression levels were associated with age rather than the prenatal DEX steroid treatment. D7, postnatal day 7; D120, postnatal day 120; DEX2, prenatal dexamethasone treatment for 2 days; DEX7, prenatal dexamethasone treatment for 7 days.
Since age was associated with notable differences, alterations in the miRNA expression profiles of rat lung tissues were compared between D7 and D120. miRNA profiles were grouped according to D7 or D120, regardless of prenatal DEX treatment. In this NGS profile, 167 differentially expressed miRNAs were detected between the D7 and D120 groups, and 12 of these miRNAs demonstrated a >2-fold change, including six upregulated miRNAs: miR-101a-3p; let-7b-5p; let-7c-5p, miR-23b-3p; miR-27a-3p; and miR30b-5p, and six downregulated miRNAs: miR-99b-5p; miR-146b-5p; miR-181b-5p; miR-182; miR-199a-3p; and mir-351-5p. (Table I). When the fold-change cutoff was set to 1.5–2.0, seven upregulated miRNAs and nine downregulated miRNAs were detected at D120 (Table II).
Table I.
Differentially expressed miRNAs with >2-fold change between D7 and D120 (transcripts per million).
| D7 | D120 | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| miR_ID | V | DEX2 | DEX7 | Mean ± SE | V | DEX2 | DEX7 | Mean ± SE | D120/D7 ratio |
| rno-miR-101a-3p | 1,944.1 | 2,008.1 | 1,735.1 | 1,896±82 | 8,774.1 | 9,926.1 | 10,070.1 | 9,590±410 | 5.06 |
| rno-miR-30b-5p | 1,872.1 | 1,583.1 | 1,487.1 | 1,647±116 | 3,959.1 | 4,839.1 | 4,319.1 | 4,372±255 | 2.65 |
| rno-miR-27a-3p | 2,278.1 | 1,866.1 | 2,149.1 | 2,098±122 | 4,409.1 | 5,952.1 | 3,981.1 | 4,781±599 | 2.28 |
| rno-miR-23b-3p | 1,665.1 | 1,677.1 | 1,755.1 | 1,699±28 | 3,747.1 | 3,861.1 | 3,966.1 | 3,858±63 | 2.27 |
| rno-let-7c-5p | 6,885.2 | 8,289.2 | 7,993.2 | 7,723±427 | 15,630.2 | 16,148.2 | 16,302.2 | 16,027±203 | 2.08 |
| rno-let-7b-5p | 1,907.1 | 2,133.1 | 2,355.1 | 2,132±129 | 4,463.1 | 4,397.1 | 4,405.1 | 4,422±21 | 2.07 |
| rno-miR-146b-5p | 5,756.1 | 6,051.1 | 6,828.1 | 6,212±320 | 3,845.1 | 2,190.1 | 3,173.1 | 3,069±481 | 0.49 |
| rno-miR-181b-5p | 3,553.2 | 3,839.2 | 4,497.2 | 3,963±279 | 1,895.2 | 1,890.2 | 1,968.2 | 1,918±25 | 0.48 |
| rno-miR-182 | 11,330.1 | 9,066.1 | 9,610.1 | 10,002±682 | 4,050.1 | 5,689.1 | 4,013.1 | 4,584±553 | 0.46 |
| rno-miR-199a-3p | 11,301.1 | 13,021.1 | 11,249.1 | 11,857±582 | 4,014.1 | 4,244.1 | 4,002.1 | 4,087±79 | 0.34 |
| rno-miR-351–5p | 9,872.1 | 8,745.1 | 10,055.1 | 9,557±410 | 1,829.1 | 2,052.1 | 1,649.1 | 1,843±117 | 0.19 |
| rno-miR-99b-5p | 38,160.1 | 34,937.1 | 37,580.1 | 36,892±992 | 5,815.1 | 6,246.1 | 6,643.1 | 6,235±239 | 0.17 |
Differences between D7 and D120 were P<0.05. miRNA, micro RNA; D7, postnatal day 7; D120, postnatal day 120; V, vehicle; DEX2, prenatal dexamethasone treatment for 2 days; DEX7, prenatal dexamethasone treatment for 7 days; SE, standard error of the mean.
Table II.
Differentially expressed miRNAs with a fold change of 1.5–2-fold between D7 and D120 (transcripts per million).
| D7 | D120 | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| miR_ID | V | DEX2 | DEX7 | Mean ± SE | V | DEX2 | DEX7 | Mean ± SE | D120/D7 ratio |
| rno-miR-24–3p | 1,478.2 | 1,652.2 | 1,588.2 | 1,573±51 | 3,586.2 | 2,718.2 | 3,068.2 | 3,124±252 | 1.99 |
| rno-miR-22–3p | 34,445.1 | 31,739.1 | 32,412.1 | 32,865±813 | 62,815.1 | 72,094.1 | 59,302.1 | 64,737±3,816 | 1.97 |
| rno-miR-30a-5p | 70,582.1 | 62,221.1 | 57,131.1 | 63,311±3,921 | 112,501.1 | 112,489.1 | 124,269.1 | 116,420±3,925 | 1.84 |
| rno-miR-26b-5p | 4,819.1 | 3,493.1 | 4,496.1 | 4,269±399 | 8,013.1 | 8,604.1 | 6,330.1 | 7,649±681 | 1.79 |
| rno-miR-126a-3p | 11,682.1 | 10,692.1 | 12,468.1 | 11,614±514 | 19,567.1 | 21,152.1 | 18,153.1 | 19,624±866 | 1.69 |
| rno-miR-34c-5p | 2,917.1 | 3,165.1 | 3,207.1 | 3,096±90 | 4,733.1 | 5,210.1 | 5,355.1 | 5,099±188 | 1.65 |
| rno-miR-375–3p | 2,200.1 | 1,742.1 | 2,183.1 | 2,042±150 | 3,145.1 | 3,007.1 | 3,116.1 | 3,089±42 | 1.51 |
| rno-miR-16–5p | 19,969.1 | 17,390.1 | 21,315.1 | 19,558±1,152 | 13,711.1 | 12,966.1 | 12,399.1 | 13,025±380 | 0.67 |
| rno-miR-151–5p | 5,036.1 | 5,846.1 | 4,952.1 | 5,278±285 | 3,568.1 | 3,745.1 | 3,210.1 | 3,508±157 | 0.66 |
| rno-miR-450a-5p | 3,921.1 | 3,304.1 | 3,100.1 | 3,442±247 | 2,560.1 | 2,426.1 | 1,716.1 | 2,234±262 | 0.65 |
| rno-miR-92a-3p | 13,189.2 | 11,645.2 | 14,568.2 | 13,134±844 | 7,602.2 | 8,403.2 | 9,040.2 | 8,349±416 | 0.64 |
| rno-miR-25–3p | 3,437.1 | 3,411.1 | 3,578.1 | 3,475±52 | 2,060.1 | 2,220.1 | 2,338.1 | 2,206±81 | 0.63 |
| rno-miR-151–3p | 7,984.1 | 7,373.1 | 8,401.1 | 7,919±299 | 4,718.1 | 4,124.1 | 5,128.1 | 4,657±291 | 0.59 |
| rno-miR-125a-5p | 7,931.1 | 7,749.1 | 7,260.1 | 7,647±200 | 4,633.1 | 4,911.1 | 3,730.1 | 4,425±356 | 0.58 |
| rno-miR-92b-3p | 2,728.1 | 2,191.1 | 3,271.1 | 2,730±312 | 1,450.1 | 1,435.1 | 1,571.1 | 1,485±43 | 0.54 |
| rno-miR-181c-5p | 6,190.1 | 6,712.1 | 6,829.1 | 6,577±196 | 3,264.1 | 3,235.1 | 3,586.1 | 3,362±112 | 0.51 |
Differences between D7 and D120 were P<0.05. miRNA, microRNA; D7, postnatal day 7; D120, postnatal day 120; V, vehicle; DEX2, dexamethasone treatment for 2 days; DEX7, prenatal dexamethasone treatment for 7 days; SE, standard error of the mean.
miRNA expression profile of rat lungs in the vehicle and prenatal DEX treatment groups
miRNA expression profiles of the rat lungs were compared between the vehicle and prenatal DEX treatment groups, including day 7 in the vehicle group (D7 V), prenatal DEX treatment for 2 days (D7 DEX2) and prenatal DEX treatment for 7 days (D7 DEX7). Since prenatal DEX treatment did not cause a > 2-fold increase in the miRNA expression levels, as compared with the vehicle, the cutoff value was set as a 1.2-fold change between the D7 DEX2 and D7 V groups or the D7 DEX7 and D7 V groups. Following the implementation of the 1.2-fold change cutoff, 12 upregulated and 20 downregulated miRNAs were detected following prenatal DEX treatment at D7 (Table III).
Table III.
Differences in miRNA expression profiles at D7 (transcripts per million).
| miR_ID | V | DEX2 | DEX7 | DEX2/V ratio | DEX7/V ratio |
|---|---|---|---|---|---|
| rno-let-7b-5p | 1,907.1 | 2,133.1 | 2,355.1 | 1.12 | 1.23 |
| rno-let-7c-5p | 6,885.2 | 8,289.2 | 7,993.2 | 1.20 | 1.16 |
| rno-miR-10a-5p | 54,083.1 | 64,522.1 | 55,017.1 | 1.19 | 1.02 |
| rno-miR-21–5p | 10,869.1 | 13,347.1 | 11,374.1 | 1.23 | 1.05 |
| rno-miR-26a-5p | 70,422.1 | 56,575.1 | 58,489.1 | 0.80 | 0.83 |
| rno-miR-26b-5p | 4,819.1 | 3,493.1 | 4,496.1 | 0.72 | 0.93 |
| rno-miR-27a-3p | 2,278.1 | 1,866.1 | 2,149.1 | 0.82 | 0.94 |
| rno-miR-28–3p | 3,702.1 | 2,921.1 | 3,449.1 | 0.79 | 0.93 |
| rno-miR-30a-5p | 70,582.1 | 62,221.1 | 57,131.1 | 0.88 | 0.81 |
| rno-miR-30b-5p | 1,872.1 | 1,583.1 | 1,487.1 | 0.85 | 0.79 |
| rno-miR-30c-5p | 7,164.2 | 6,454.2 | 5,816.2 | 0.90 | 0.81 |
| rno-miR-30d-5p | 23,793.1 | 19,892.1 | 22,603.1 | 0.84 | 0.95 |
| rno-miR-92b-3p | 2,728.1 | 2,191.1 | 3,271.1 | 0.80 | 1.20 |
| rno-miR-100-5p | 1,970.1 | 2,304.1 | 2,399.1 | 1.17 | 1.22 |
| rno-miR-126a-5p | 39,461.1 | 33,312.1 | 36,780.1 | 0.84 | 0.93 |
| rno-miR-127–3p | 19,146.1 | 14,062.1 | 17,011.1 | 0.73 | 0.89 |
| rno-miR-143–3p | 205,095.1 | 245,257.1 | 215,878.1 | 1.20 | 1.05 |
| rno-miR-146a-5p | 1,401.1 | 1,523.1 | 2,035.1 | 1.09 | 1.45 |
| rno-miR-146b-5p | 5,756.1 | 6,051.1 | 6,828.1 | 1.05 | 1.19 |
| rno-miR-148b-3p | 1,184.1 | 1,659.1 | 1,333.1 | 1.40 | 1.13 |
| rno-miR-181a-5p | 61,870.2 | 71,522.2 | 79,498.2 | 1.16 | 1.28 |
| rno-miR-181b-5p | 3,553.2 | 3,839.2 | 4,497.2 | 1.08 | 1.27 |
| rno-miR-182 | 11,330.1 | 9,066.1 | 9,610.1 | 0.80 | 0.85 |
| rno-miR-183–5p | 1,652.1 | 1,318.1 | 1,536.1 | 0.80 | 0.93 |
| rno-miR-186–5p | 2,128.1 | 1,372.1 | 1,653.1 | 0.64 | 0.78 |
| rno-miR-200b-3p | 1,791.1 | 1,428.1 | 1,658.1 | 0.80 | 0.93 |
| rno-miR-322–5p | 2,786.1 | 2,097.1 | 2,181.1 | 0.75 | 0.78 |
| rno-miR-375–3p | 2,200.1 | 1,742.1 | 2,183.1 | 0.79 | 0.99 |
| rno-miR-411–5p | 3,302.1 | 2,481.1 | 2,920.1 | 0.75 | 0.88 |
| rno-miR-429 | 1,646.1 | 1,258.1 | 1,313.1 | 0.76 | 0.80 |
| rno-miR-434–3p | 1,903.1 | 1,044.1 | 1,393.1 | 0.55 | 0.73 |
| rno-miR-449a-5p | 2,708.1 | 2,806.1 | 3,497.1 | 1.04 | 1.29 |
| rno-miR-450a-5p | 3,921.1 | 3,304.1 | 3,100.1 | 0.84 | 0.79 |
miRNA, microRNA; D7, postnatal day 7; V, vehicle; DEX2, prenatal dexamethasone treatment for 2 days; DEX7, prenatal dexamethasone treatment for 7 days.
Differences in the miRNA expression profiles of the rat lungs at D120 were compared among the vehicle (D120 V), prenatal DEX treatment for 2 days (D120 DEX2) and D120 DEX7) groups. Following the implementation of the 1.2-fold change cutoff, four upregulated and five downregulated miRNAs were detected following prenatal DEX treatment at D120 (Table IV). These results suggest that the efficacy of prenatal treatment with DEX on the miRNA profiles in rat lung tissues is markedly decreased in the chronic stage.
Table IV.
Differences in miRNA expression profiles at D120 (transcripts per million).
| miR_ID | V | DEX2 | DEX7 | DEX2/V ratio | DEX7/V ratio |
|---|---|---|---|---|---|
| rno-miR-24–3p | 3,586.2 | 2,718.2 | 3,068.2 | 0.76 | 0.86 |
| rno-miR-26a-5p | 83,002.1 | 99,966.1 | 71,257.1 | 1.20 | 0.86 |
| rno-miR-26b-5p | 8,013.1 | 8,604.1 | 6,330.1 | 1.07 | 0.79 |
| rno-miR-27a-3p | 4,409.1 | 5,952.1 | 3,981.1 | 1.35 | 0.90 |
| rno-miR-28–3p | 1,883.1 | 2,265.1 | 1,961.1 | 1.20 | 1.04 |
| rno-miR-30b-5p | 3,959.1 | 4,839.1 | 4,319.1 | 1.22 | 1.09 |
| rno-miR-92a-3p | 7,602.2 | 8,403.2 | 9,040.2 | 1.11 | 1.19 |
| rno-miR-125a-5p | 4,633.1 | 4,911.1 | 3,730.1 | 1.06 | 0.81 |
| rno-miR-126a-5p | 40,074.1 | 30,726.1 | 38,860.1 | 0.77 | 0.97 |
| rno-miR-145–5p | 1,204.1 | 1,897.1 | 1,417.1 | 1.58 | 1.18 |
| rno-miR-146b-5p | 3,845.1 | 2,190.1 | 3,173.1 | 0.57 | 0.83 |
| rno-miR-150–5p | 1,623.1 | 1,810.1 | 1,341.1 | 1.12 | 0.83 |
| rno-miR-181a-5p | 66,412.2 | 54,932.2 | 63,020.2 | 0.83 | 0.95 |
| rno-miR-182 | 4,050.1 | 5,689.1 | 4,013.1 | 1.40 | 0.99 |
| rno-miR-192–5p | 1,683.1 | 1,760.1 | 2,144.1 | 1.05 | 1.27 |
| rno-miR-450a-5p | 2,560.1 | 2,426.1 | 1,716.1 | 0.95 | 0.67 |
miRNA, microRNA; V, vehicle; D120, postnatal day 120; DEX2, prenatal dexamethasone treatment for 2 days; DEX7, prenatal dexamethasone treatment for 7 days at postnatal day 7.
RT-qPCR validation
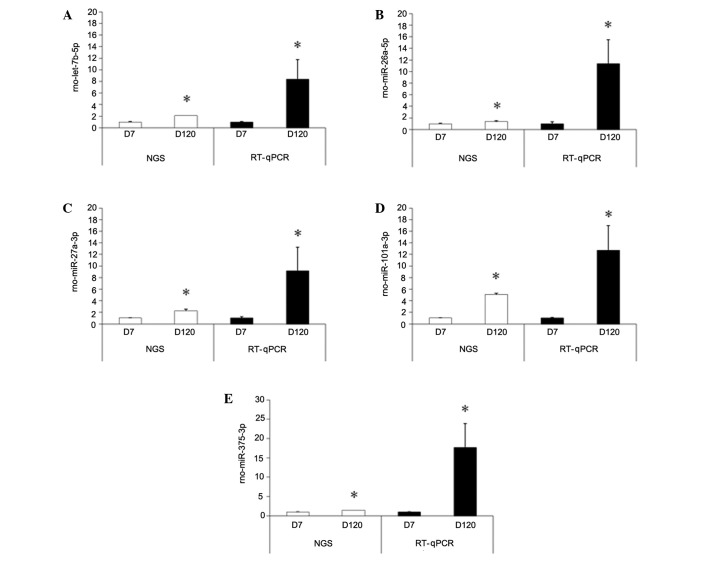
The following differentially expressed miRNAs were selected to perform RT-qPCR, based on the NGS data: let-7b-5p, miR-26a-5p, miR-27a-3p, miR-101a-3p, and miR-375. The relative expression of these five miRNAs are shown in Fig. 5. These five miRNAs were selected for further study as, among the consistently upregulated miRNAs, let-7b-5p, miR-27a-3p, and miR-101a-3p demonstrated the greatest fold changes between the D7 and D120 groups. Furthermore, let-7b-5p, miR-27a-3p, and miR-101a-3p are reportedly associated with lung tumor development (24), and miR-26a and miR-375 have been associated with the regulation of pulmonary surfactant secretion. The alterations in miRNAs following prenatal DEX treatment were not validated by RT-qPCR as the effects of prenatal DEX treatment appear to be limited.
Figure 5.
Reverse transcription-quantitative polymerase chain reaction analysis (RT-qPCR) was conducted to validate the miRNA profiles of five miRNAs (A-E) determined using next generation sequencing (NGS). The results demonstrated that the expression patterns of these five microRNAs were consistent with those determined by NGS. *P<0.05 vs. D7. Significant differences between the data determined by NGS and RT-qPCR were also detected. D7, day 7; D120, day 120.
Discussion
In the present study, miRSeq was used to analyze six libraries from rat lung tissues. The D7 libraries were less enriched as they contained unknown reads with nucleotides of 31, 32 and 33 bp in length (data not shown).
miRNAs serve a crucial function in cellular proliferation, differentiation and organ development; however, few previous miRNA studies have investigated their connection with diseases of lung development (25). In the present study, differences in miRNA expression profiles were compared between the D7 and D120 groups, and the miRNAs with a >2-fold change were selected. Six upregulated and six downregulated miRNAs were detected at D120, as compared with D7. Among these differentially expressed miRNAs, miR-101-3p and miR-99b-5p were responsible for the lowest and highest expressions of miRNA at D7, respectively.
It has previously been demonstrated that miR-101-3p is downregulated following infection with hepatitis B virus (26); however, the role of miR-101-3p in lung development remains unclear. Li et al (27) demonstrated that the altered serum levels of miR-21, miR-155 and miR-101-3p were associated with the degree of forced vital capacity and radiographic features in idiopathic pulmonary fibrosis. miR-101 and miR-144 have also been demonstrated to regulate the expression levels of the cystic fibrosis transmembrane conductance regulator in the lungs (28). Furthermore, previous studies have demonstrated that mir-101 exerts a tumor suppressive function in certain lung cancer conditions (29,30), and the overexpression of miR-101-3p has been reported to inhibit cellular proliferation, migration and reduce apoptosis (31). Low expression levels of miR-101-3p at D7 in the present study suggested its immaturity.
The mature form of let-7 and its family members are highly conserved. The key functions of let-7 genes are to promote terminal differentiation in development and tumor suppression (32,33). The let-7 family has previously been identified as a tumor marker and a potential therapeutic regimen for the treatment of certain tumors (32,34–36). Furthermore, previous studies have demonstrated that mir-27a may be associated with malignant changes in bronchial epithelial cells and signal interactions in lung cancer (37,38). However, to the best of our knowledge, the associations between mir-23b-3p and mir-30b-5p, and lung development is yet to be investigated.
It has previously been suggested that miR-99b is capable of potentiating endothelial cell differentiation from stem cells (39). Enhanced miR-99b levels following lentiviral-mediated transfer were demonstrated to potentiate the mRNA and protein expression of endothelial cell specific markers, increase nitric oxide production and improve neovascularization in vivo (39). The high expression levels of miR-99b detected at D7 in the present study are consistent with growth and vascularization. Furthermore, among the downregulated miRNAs following lung maturation (Table I), miR-99b-5p was responsible for the least miRNA in D120 rat lungs. In order to elucidate the underlying mechanisms of its regulatory role, the predicted target genes of miR-99b-5p were collected from TargetScan 6.2 and the pathways in which the target genes were enriched were derived. Among the enriched pathways, the Hippo signaling pathway (Kyoto Encyclopedia of Genes and Genomes, map04390) was significantly associated with organ growth (P=0.0003) (40). In animals, the Hippo signaling pathway controls organ size through the regulation of cell proliferation and apoptosis (40); therefore, it is reasonable that the Hippo signaling pathway is activated during lung tissue maturation in order to restrict organ size overgrowth by suppressing miR-99b-5p.
In the present study, the differences in the miRNA expression profiles of rat lung tissues were compared between vehicle and prenatal DEX treatment groups. Prenatal DEX treatment had a limited impact on the miRNA profiles of lung tissue at D7 and D120. Therefore, the fold-change cutoff value was set at 1.2, which facilitated the detection of 12 upregulated and 20 downregulated miRNAs at D7 following prenatal DEX treatment (Table III). Among these 20 downregulated miRNAs, miR-434–3p was demonstrated to be the most suppressive. Various miRNAs, including miR-150, miR-375, and miR-26a have been associated with the regulation of pulmonary surfactant secretion (15,19,20). The results of the present study agree in part with these previous studies, and downregulated miR-375 and miR-26a were compatible with prenatal glucocorticosteroid treatment. However, further miRNAs that are downregulated following prenatal DEX treatment have been detected in our study. The authors of the present study hypothesize that these newly identified miRNAs may be crucially involved in the development of fetal lungs, and the results of the present study may aid further clarification of the mechanisms of normal lung development. However, the results of the present study cannot fully represent the effects of prenatal DEX treatment on the lung development of premature babies since significant prenatal DEX-induced changes in the miRNA profiles may appear earlier; therefore more intensive time course studies are required.
Acknowledgements
The present study was supported in part by grants from the National Science Council, Taiwan (grant nos. CMRPG8B0141, CMRPG8B0142, CMRPG8C0171 and NSC 102-2314-B-182A-042-MY3). The authors thank the Genomics and Proteomics Core Laboratory, Department of Medical Research at Kaohsiung Chang Gung Memorial Hospital for their technical support.
References
- 1.Crowley P, Chalmers I, Keirse MJ. The effects of corticosteroid administration before preterm delivery: An overview of the evidence from controlled trials. Br J Obstet Gynaecol. 1990;97:11–25. doi: 10.1111/j.1471-0528.1990.tb01711.x. [DOI] [PubMed] [Google Scholar]
- 2.Roberts D, Dalziel S. Antenatal corticosteroids for accelerating fetal lung maturation for women at risk of preterm birth. Cochrane Database Syst Rev. 2006;19:CD004454. doi: 10.1002/14651858.CD004454.pub2. [DOI] [PubMed] [Google Scholar]
- 3.Liggins GC. The role of cortisol in preparing the fetus for birth. Reprod Fertil Dev. 1994;6:141–150. doi: 10.1071/RD9940141. [DOI] [PubMed] [Google Scholar]
- 4.Gross I. Regulation of fetal lung maturation. Am J Physiol. 1990;259:L337–L344. doi: 10.1152/ajplung.1990.259.6.L337. [DOI] [PubMed] [Google Scholar]
- 5.Schittny JC, Djonov V, Fine A, Burri PH. Programmed cell death contributes to postnatal lung development. Am J Respir Cell Mol Biol. 1998;18:786–793. doi: 10.1165/ajrcmb.18.6.3031. [DOI] [PubMed] [Google Scholar]
- 6.Whitsett JA, Stahlman MT. Impact of advances in physiology, biochemistry and molecular biology on pulmonary disease in neonates. Am J Respir Crit Care Med. 1998;157:S67–S71. doi: 10.1164/ajrccm.157.4.nhlb1-2. [DOI] [PubMed] [Google Scholar]
- 7.Walther FJ, Ikegami M, Warburton D, Polk DH. Corticosteroids, thyrotropin-releasing hormone and antioxidant enzymes in preterm lamb lungs. Pediatr Res. 1991;30:518–521. doi: 10.1203/00006450-199112000-00004. [DOI] [PubMed] [Google Scholar]
- 8.Pinkerton KE, Willet KE, Peake JL, Sly PD, Jobe AH, Ikegami M. Prenatal glucocorticoid and T4 effects on lung morphology in preterm lambs. Am J Respir Crit Care Med. 1997;156:624–630. doi: 10.1164/ajrccm.156.2.9701018. [DOI] [PubMed] [Google Scholar]
- 9.Beck JC, Mitzner W, Johnson JW, Hutchins GM, Foidart JM, London WT, Palmer AE, Scott R. Betamethasone and the rhesus fetus: Effect on lung morphometry and connective tissue. Pediatr Res. 1981;15:235–240. doi: 10.1203/00006450-198103000-00007. [DOI] [PubMed] [Google Scholar]
- 10.Vyas J, Kotecha S. Effects of antenatal and postnatal corticosteroids on the preterm lung. Arch Dis Child Fetal Neonatal Ed. 1997;77:F147–F150. doi: 10.1136/fn.77.2.F147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang J, Kuliszewski M, Yee W, Sedlackova L, Xu J, Tseu I, Post M. Cloning and expression of glucocorticoid-induced genes in fetal rat lung fibroblasts. Transforming growth factor-beta 3. J Biol Chem. 1995;270:2722–2728. doi: 10.1074/jbc.270.6.2722. [DOI] [PubMed] [Google Scholar]
- 12.Drake AJ, Walker BR, Seckl JR. Intergenerational consequences of fetal programming by in utero exposure to glucocorticoids in rats. Am J Physiol Regul Integr Comp Physiol. 2005;288:R34–R38. doi: 10.1152/ajpregu.00106.2004. [DOI] [PubMed] [Google Scholar]
- 13.Drake AJ, Tang JI, Nyirenda MJ. Mechanisms underlying the role of glucocorticoids in the early life programming of adult disease. Clin Sci (Lond) 2007;113:219–232. doi: 10.1042/CS20070107. [DOI] [PubMed] [Google Scholar]
- 14.Yu HR, Kuo HC, Chen CC, Sheen JM, Tiao MM, Chen YC, Chang KA, Tain YL, Huang LT. Prenatal dexamethasone exposure in rats results in long-term epigenetic histone modifications and tumor necrosis factor-α production decrease. Immunology. 2014;143:651–660. doi: 10.1111/imm.12346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang H, Mishra A, Chintagari NR, Gou D, Liu L. Micro-RNA-375 inhibits lung surfactant secretion by altering cytoskeleton reorganization. IUBMB Life. 2010;62:78–83. doi: 10.1002/iub.286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee EK, Lee MJ, Abdelmohsen K, Kim W, Kim MM, Srikantan S, Martindale JL, Hutchison ER, Kim HH, Marasa BS, et al. MiR-130 suppresses adipogenesis by inhibiting peroxisome proliferator-activated receptor gamma expression. Mol Cell Biol. 2011;31:626–638. doi: 10.1128/MCB.00894-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bhaskaran M, Wang Y, Zhang H, Weng T, Baviskar P, Guo Y, Gou D, Liu L. MicroRNA-127 modulates fetal lung development. Physiol Genomics. 2009;37:268–278. doi: 10.1152/physiolgenomics.90268.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Williams AE, Moschos SA, Perry MM, Barnes PJ, Lindsay MA. Maternally imprinted microRNAs are differentially expressed during mouse and human lung development. Dev Dyn. 2007;236:572–580. doi: 10.1002/dvdy.21047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weng T, Mishra A, Guo Y, Wang Y, Su L, Huang C, Zhao C, Xiao X, Liu L. Regulation of lung surfactant secretion by microRNA-150. Biochem Biophys Res Commun. 2012;422:586–589. doi: 10.1016/j.bbrc.2012.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang XQ, Zhang P, Yang Y, Qiu J, Kan Q, Liang HL, Zhou XY, Zhou XG. Regulation of pulmonary surfactant synthesis in fetal rat type II alveolar epithelial cells by microRNA-26a. Pediatr Pulmonol. 2014;49:863–872. doi: 10.1002/ppul.22975. [DOI] [PubMed] [Google Scholar]
- 21.Pan CT, Tsai KW, Hung TM, Lin WC, Pan CY, Yu HR, Li SC. MiRSeq: A user-friendly standalone toolkit for sequencing quality evaluation and miRNA profiling. Biomed Res Int. 2014;2014:462135. doi: 10.1155/2014/462135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yu HR, Chang JC, Chen RF, Chuang H, Hong KC, Wang L, Yang KD. Different antigens trigger different Th1/Th2 reactions in neonatal mononuclear cells (MNCs) relating to T-bet/GATA-3 expression. J Leukoc Biol. 2003;74:952–958. doi: 10.1189/jlb.0902474. [DOI] [PubMed] [Google Scholar]
- 23.Sehgal P, Chaturvedi P, Kumaran RI, Kumar S, Parnaik VK. Lamin A/C haploinsufficiency modulates the differentiation potential of mouse embryonic stem cells. PLoS One. 2013;8:e57891. doi: 10.1371/journal.pone.0057891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang QZ, Xu W, Habib N, Xu R. Potential uses of microRNA in lung cancer diagnosis, prognosis, and therapy. Curr Cancer Drug Targets. 2009;9:572–594. doi: 10.2174/156800909788486731. [DOI] [PubMed] [Google Scholar]
- 25.Sessa R, Hata A. Role of microRNAs in lung development and pulmonary diseases. Pulm Circ. 2013;3:315–328. doi: 10.4103/2045-8932.114758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sheng Y, Li J, Zou C, Wang S, Cao Y, Zhang J, Huang A, Tang H. Downregulation of miR-101-3p by hepatitis B virus promotes proliferation and migration of hepatocellular carcinoma cells by targeting Rab5a. Arch Virol. 2014;159:2397–2410. doi: 10.1007/s00705-014-2084-5. [DOI] [PubMed] [Google Scholar]
- 27.Li P, Li J, Chen T, Wang H, Chu H, Chang J, Zang W, Wang Y, Ma Y, Du Y, et al. Expression analysis of serum microRNAs in idiopathic pulmonary fibrosis. Int J Mol Med. 2014;33:1554–1562. doi: 10.3892/ijmm.2014.1712. [DOI] [PubMed] [Google Scholar]
- 28.Hassan F, Nuovo GJ, Crawford M, Boyaka PN, Kirkby S, Nana-Sinkam SP, Cormet-Boyaka E. MiR-101 and miR-144 regulate the expression of the CFTR chloride channel in the lung. PLoS One. 2012;7:e50837. doi: 10.1371/journal.pone.0050837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cho HM, Jeon HS, Lee SY, Jeong KJ, Park SY, Lee HY, Lee JU, Kim JH, Kwon SJ, Choi E, et al. MicroRNA-101 inhibits lung cancer invasion through the regulation of enhancer of zeste homolog 2. Exp Ther Med. 2011;2:963–967. doi: 10.3892/etm.2011.284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang JG, Guo JF, Liu DL, Liu Q, Wang JJ. MicroRNA-101 exerts tumor-suppressive functions in non-small cell lung cancer through directly targeting enhancer of zeste homolog 2. J Thorac Oncol. 2011;6:671–678. doi: 10.1097/JTO.0b013e318208eb35. [DOI] [PubMed] [Google Scholar]
- 31.Sheng Y, Ding S, Chen K, Chen J, Wang S, Zou C, Zhang J, Cao Y, Huang A, Tang H. Functional analysis of miR-101-3p and Rap1b involved in hepatitis B virus-related hepatocellular carcinoma pathogenesis. Biochem Cell Biol. 2014;92:152–162. doi: 10.1139/bcb-2013-0128. [DOI] [PubMed] [Google Scholar]
- 32.Barh D, Malhotra R, Ravi B, Sindhurani P. MicroRNA let-7: An emerging next-generation cancer therapeutic. Curr Oncol. 2010;17:70–80. doi: 10.3747/co.v17i1.356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Johnson SM, Grosshans H, Shingara J, Byrom M, Jarvis R, Cheng A, Labourier E, Reinert KL, Brown D, Slack FJ. RAS is regulated by the let-7 microRNA family. Cell. 2005;120:635–647. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 34.Childs G, Fazzari M, Kung G, Kawachi N, Brandwein-Gensler M, McLemore M, Chen Q, Burk RD, Smith RV, Prystowsky MB, et al. Low-level expression of microRNAs let-7d and miR-205 are prognostic markers of head and neck squamous cell carcinoma. Am J Pathol. 2009;174:736–745. doi: 10.2353/ajpath.2009.080731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schubert M, Spahn M, Kneitz S, Scholz CJ, Joniau S, Stroebel P, Riedmiller H, Kneitz B. Distinct microRNA expression profile in prostate cancer patients with early clinical failure and the impact of let-7 as prognostic marker in high-risk prostate cancer. PLoS One. 2013;8:e65064. doi: 10.1371/journal.pone.0065064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Takamizawa J, Konishi H, Yanagisawa K, Tomida S, Osada H, Endoh H, Harano T, Yatabe Y, Nagino M, Nimura Y, et al. Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival. Cancer Res. 2004;64:3753–3756. doi: 10.1158/0008-5472.CAN-04-0637. [DOI] [PubMed] [Google Scholar]
- 37.Acunzo M, Romano G, Palmieri D, Laganá A, Garofalo M, Balatti V, Drusco A, Chiariello M, Nana-Sinkam P, Croce CM. Cross-talk between MET and EGFR in non-small cell lung cancer involves miR-27a and Sprouty2. Proc Natl Acad Sci USA. 2013;110:8573–8578. doi: 10.1073/pnas.1302107110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang Q, Li DC, Li ZF, Liu CX, Xiao YM, Zhang B, Li XD, Zhao J, Chen LP, Xing XM, et al. Upregulation of miR-27a contributes to the malignant transformation of human bronchial epithelial cells induced by SV40 small T antigen. Oncogene. 2011;30:3875–3886. doi: 10.1038/onc.2011.103. [DOI] [PubMed] [Google Scholar]
- 39.Kane NM, Howard L, Descamps B, Meloni M, McClure J, Lu R, McCahill A, Breen C, Mackenzie RM, Delles C, et al. Role of microRNAs 99b, 181a and 181b in the differentiation of human embryonic stem cells to vascular endothelial cells. Stem cells. 2012;30:643–654. doi: 10.1002/stem.1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pan D. Hippo signaling in organ size control. Genes Dev. 2007;21:886–897. doi: 10.1101/gad.1536007. [DOI] [PubMed] [Google Scholar]