Abstract
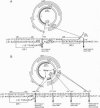
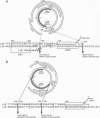
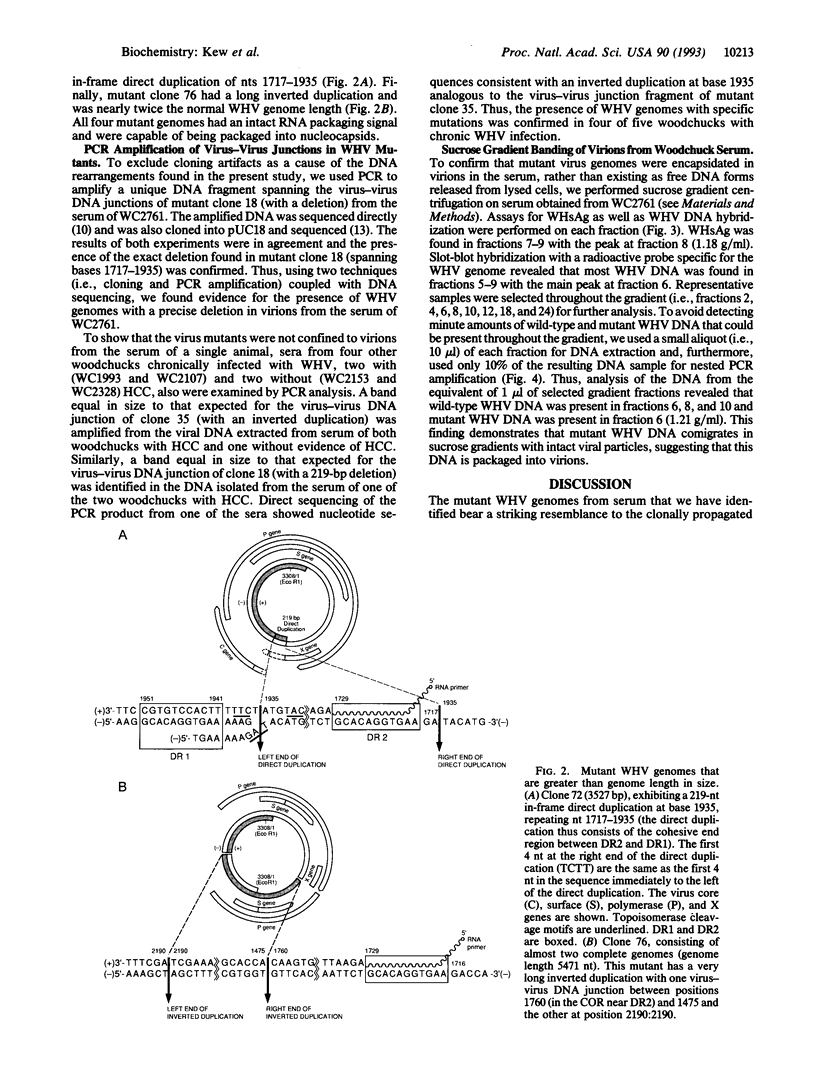
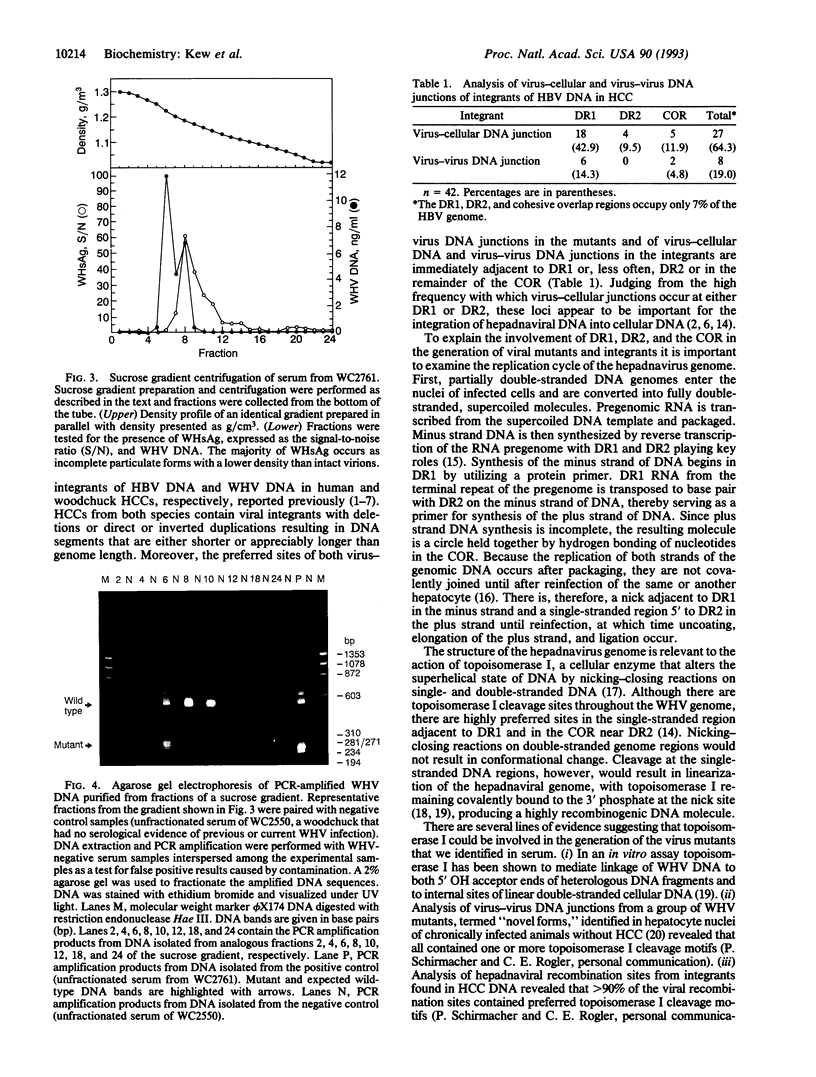
Although hepadnaviruses are implicated in the etiology of hepatocellular carcinoma, the pathogenic mechanisms involved remain uncertain. Clonally propagated integrations of hepadnaviral DNA into cellular DNA can be demonstrated in most virally induced hepatocellular carcinomas. Integration occurs at random sites in cellular DNA, but the highly preferred sites in viral DNA are adjacent to the directly repeated sequence DR1, less often DR2, or in the cohesive overlap region. Integrants invariably contain simple deletions or complex rearrangements that have been thought to occur after integration. We report here the detection of mutant woodchuck hepatitis virus (WHV) genomes cloned from virions in serum that are strikingly similar to the rearranged hepadnaviral genomes found previously as integrated sequences in cellular DNA. Of 102 cloned genomes studied, 2 had large inverted duplications, 1 a 219-nucleotide direct duplication, and 1 a 219-nucleotide deletion. Virus-virus DNA junctions occurred either adjacent to DR1 or DR2 or in the cohesive overlap region at preferred topoisomerase I cleavage sites. Since these sites are located in the single-stranded regions of the genome, cleavage by topoisomerase I would produce linear molecules that would be expected to be highly recombinogenic since this enzyme, possessing nicking and ligating activities, would remain covalently attached. Sucrose density gradient centrifugation coupled with polymerase chain reaction studies confirmed that the mutant WHV DNA forms resided in virions and did not represent free viral DNA released from infected cells or were unlikely to be an artifact of the cloning process. Thus, the finding in virions of mutant WHV DNA similar to WHV DNA integrated into cellular DNA suggests that the processes of mutation and integration are linked in some instances. Furthermore, the mutant genomes that are preferentially integrated into cellular DNA may have an etiologic role in hepatocarcinogenesis.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Botchan M., Stringer J., Mitchison T., Sambrook J. Integration and excision of SV40 DNA from the chromosome of a transformed cell. Cell. 1980 May;20(1):143–152. doi: 10.1016/0092-8674(80)90242-1. [DOI] [PubMed] [Google Scholar]
- Bullock P., Champoux J. J., Botchan M. Association of crossover points with topoisomerase I cleavage sites: a model for nonhomologous recombination. Science. 1985 Nov 22;230(4728):954–958. doi: 10.1126/science.2997924. [DOI] [PubMed] [Google Scholar]
- Bullock P., Forrester W., Botchan M. DNA sequence studies of simian virus 40 chromosomal excision and integration in rat cells. J Mol Biol. 1984 Mar 25;174(1):55–84. doi: 10.1016/0022-2836(84)90365-6. [DOI] [PubMed] [Google Scholar]
- Clayton C. E., Rigby P. W. Cloning and characterization of the integrated viral DNA from three lines of SV40-transformed mouse cells. Cell. 1981 Aug;25(2):547–559. doi: 10.1016/0092-8674(81)90073-8. [DOI] [PubMed] [Google Scholar]
- Cohen J. I., Miller R. H., Rosenblum B., Denniston K., Gerin J. L., Purcell R. H. Sequence comparison of woodchuck hepatitis virus replicative forms shows conservation of the genome. Virology. 1988 Jan;162(1):12–20. doi: 10.1016/0042-6822(88)90389-3. [DOI] [PubMed] [Google Scholar]
- Dejean A., Sonigo P., Wain-Hobson S., Tiollais P. Specific hepatitis B virus integration in hepatocellular carcinoma DNA through a viral 11-base-pair direct repeat. Proc Natl Acad Sci U S A. 1984 Sep;81(17):5350–5354. doi: 10.1073/pnas.81.17.5350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiNardo S., Voelkel K., Sternglanz R. DNA topoisomerase II mutant of Saccharomyces cerevisiae: topoisomerase II is required for segregation of daughter molecules at the termination of DNA replication. Proc Natl Acad Sci U S A. 1984 May;81(9):2616–2620. doi: 10.1073/pnas.81.9.2616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganem D., Varmus H. E. The molecular biology of the hepatitis B viruses. Annu Rev Biochem. 1987;56:651–693. doi: 10.1146/annurev.bi.56.070187.003251. [DOI] [PubMed] [Google Scholar]
- Goto T., Wang J. C. Cloning of yeast TOP1, the gene encoding DNA topoisomerase I, and construction of mutants defective in both DNA topoisomerase I and DNA topoisomerase II. Proc Natl Acad Sci U S A. 1985 Nov;82(21):7178–7182. doi: 10.1073/pnas.82.21.7178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hino O., Ohtake K., Rogler C. E. Features of two hepatitis B virus (HBV) DNA integrations suggest mechanisms of HBV integration. J Virol. 1989 Jun;63(6):2638–2643. doi: 10.1128/jvi.63.6.2638-2643.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hino O., Shows T. B., Rogler C. E. Hepatitis B virus integration site in hepatocellular carcinoma at chromosome 17;18 translocation. Proc Natl Acad Sci U S A. 1986 Nov;83(21):8338–8342. doi: 10.1073/pnas.83.21.8338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko S., Feinstone S. M., Miller R. H. Rapid and sensitive method for the detection of serum hepatitis B virus DNA using the polymerase chain reaction technique. J Clin Microbiol. 1989 Sep;27(9):1930–1933. doi: 10.1128/jcm.27.9.1930-1933.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kew M. C., Chestnut T., Baldwin B. H., Hornbuckle W. E., Tennant B. C., Purcell R. H., Miller R. H. Heterogeneity of the woodchuck hepatitis virus genome in a chronically infected woodchuck. Virus Res. 1993 Mar;27(3):229–237. doi: 10.1016/0168-1702(93)90035-l. [DOI] [PubMed] [Google Scholar]
- Konopka A. K. Compilation of DNA strand exchange sites for non-homologous recombination in somatic cells. Nucleic Acids Res. 1988 Mar 11;16(5):1739–1758. doi: 10.1093/nar/16.5.1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogata N., Tokino T., Kamimura T., Asakura H. A comparison of the molecular structure of integrated hepatitis B virus genomes in hepatocellular carcinoma cells and hepatocytes derived from the same patient. Hepatology. 1990 Jun;11(6):1017–1023. doi: 10.1002/hep.1840110617. [DOI] [PubMed] [Google Scholar]
- Ogston C. W., Jonak G. J., Rogler C. E., Astrin S. M., Summers J. Cloning and structural analysis of integrated woodchuck hepatitis virus sequences from hepatocellular carcinomas of woodchucks. Cell. 1982 Jun;29(2):385–394. doi: 10.1016/0092-8674(82)90155-6. [DOI] [PubMed] [Google Scholar]
- Sambrook J., Greene R., Stringer J., Mitchison T., Hu S. L., Botchan M. Analysis of the sites of integration of viral DNA sequences in rat cells transformed by adenovirus 2 or SV40. Cold Spring Harb Symp Quant Biol. 1980;44(Pt 1):569–584. doi: 10.1101/sqb.1980.044.01.059. [DOI] [PubMed] [Google Scholar]
- Shih C., Burke K., Chou M. J., Zeldis J. B., Yang C. S., Lee C. S., Isselbacher K. J., Wands J. R., Goodman H. M. Tight clustering of human hepatitis B virus integration sites in hepatomas near a triple-stranded region. J Virol. 1987 Nov;61(11):3491–3498. doi: 10.1128/jvi.61.11.3491-3498.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong D. C., Shih J. W., Purcell R. H., Gerin J. L., London W. T. Natural and experimental infection of woodchucks with woodchuck hepatitis virus, as measured by new, specific assays for woodchuck surface antigen and antibody. J Clin Microbiol. 1982 Mar;15(3):484–490. doi: 10.1128/jcm.15.3.484-490.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaginuma K., Kobayashi M., Yoshida E., Koike K. Hepatitis B virus integration in hepatocellular carcinoma DNA: duplication of cellular flanking sequences at the integration site. Proc Natl Acad Sci U S A. 1985 Jul;82(13):4458–4462. doi: 10.1073/pnas.82.13.4458. [DOI] [PMC free article] [PubMed] [Google Scholar]