Abstract
Hepatocellular carcinoma (HCC) is a fatal disease, primarily due to the limited effective therapies available for patients with advanced or recurrent stages of the disease. Therefore, in order to improve patient prognosis, it is important to identify an informative biomarker for HCC progression, as well as a molecular target for therapy. Neurotrophin receptor-interacting melanoma antigen-encoding protein (NRAGE), a member of the type II melanoma-associated antigen family, mediates apoptosis and cell death through interactions with a wide range of proteins, and is implicated as a tumor suppressor or oncoprotein depending on cell type. However, the role of NRAGE in HCC is currently unknown, therefore, the present study aimed to identify the underlying function of NRAGE in HCC tumorigenesis. Resected tumor and non-cancerous liver tissues from 151 patients with HCC, alongside HCC cell lines, were analyzed by polymerase chain reaction and immunohistochemical techniques to determine NRAGE expression levels, as well as the expression levels of potential genes encoding interacting proteins. It was demonstrated that the expression levels of NRAGE mRNA correlated significantly with those of apoptosis-antagonizing transcription factor (AATF), and were not affected by cirrhosis in non-cancerous liver tissues when compared to elevated levels in HCC tissues. The expression patterns of NRAGE protein and mRNA were consistent among 30 representative specimen pairs. Furthermore, increased NRAGE expression in patients with HCC correlated significantly with a shorter disease-specific survival time, and was identified as an independent prognostic factor via multivariate analysis (hazard ratio, 2.23; 95% confidence interval, 1.06–3.83; P=0.020). Therefore, the results of the present study indicated that increased NRAGE expression affects HCC progression via its interaction with AATF, and may represent a novel biomarker and molecular target for the treatment of HCC.
Keywords: hepatocellular carcinoma, neurotrophin receptor-interacting melanoma antigen-encoding protein, expression, progression, apoptosis-antagonizing transcription factor
Introduction
Hepatocellular carcinoma (HCC) is one of the most prevalent malignant tumors, with high rates of mortality and morbidity (1,2). The worldwide incidence of HCC is ~1,000,000 cases per year and is equivalent to its mortality rate (3). Furthermore, HCC ranks as the third highest cause of cancer-associated mortality (4). Such statistics are largely explained by the difficulties faced when diagnosing HCC, with diagnosis often only confirmed once the disease is at a late stage and palliative care is the only treatment option available. An important strategy to improve patient outcome involves the identification of disease risk factors and the maintenance of vigilant surveillance of high-risk individuals to allow for recognition of the disease at a stage that is responsive to treatment with curative intent. Therefore, it is fundamental that high-risk groups are identified, followed by successful implementation of a surveillance program and recall protocol following any abnormal findings (5–8). Progress has been made in identifying molecular markers for the initiation and progression of HCC that will likely increase the rate of early and potentially life-saving diagnoses (9–12). The expanding amount of knowledge regarding the molecular foundations of HCC makes it increasingly clear that successful therapy requires treatment tailored to the individual patient (8).
Melanoma-associated antigens (MAGEs), members of the cancer/testis antigen family that consists of >50 proteins, are divided into two types, MAGE-I and MAGE-II, dependent on varying gene structures and tissue-specific expression patterns (13,14). The MAGE-I subgroup consists of products yielded by numerous × chromosome clustered genes, including MAGE-A, -B and -C, which are typically expressed in cancer cells of various origins, but not in adult tissues, with the exception of germ-line cells in the placenta, ovaries and testes (15,16). By contrast, the MAGE-II subgroup, including MAGE-D variants, do not have defined chromosome clustering and are cancer non-specific, with near universal expression in normal adult tissues and germ-line cells (14,16,17). MAGE proteins, as tumor-associated antigens, have attracted increasing attention regarding the development of vaccine-based immunotherapy for the treatment of cancer (18). Our strategy for addressing this issue involves detailed analysis of the literature to identify potential markers and targets for therapy of HCC (15). For example, a previous meta-analysis indicated that glypican-3, des-γ-carboxyprothrombin, α-L-fucosidase and vascular endothelial growth factor may serve as suitable serological markers for HCC (19). Meta-analyses are incisive and may potentially be more informative compared with omic surveys that are expensive, technology-intensive and arguably more time-consuming. This lead the current study to focus on the 86-kDa NRAGE protein, also known as MAGE-D1, which is encoded by the NRAGE gene located on the × chromosome (20–22). Cells of diverse embryonic and adult tissues, particularly those of the nervous system, express NRAGE, which subsequently interacts with proteins that regulate cell adhesion and migration, the cell cycle, cell differentiation, apoptosis and gene transcription (22–24). However, little is understood regarding the physiological relevance of these interactions.
NRAGE serves a role in the process of apoptosis through interactions with the p75 neurotrophin receptor (p75NTR) (25) and apoptosis-antagonizing transcription factor (AATF) (26,27). As NRAGE interacts with proteins with diverse functions, it is not unexpected that its effects are cell-type specific. For example, the downregulation of NRAGE transcription serves an important function in apoptosis, and when expressed ectopically, NRAGE inhibits the proliferation of breast cancer cells (27). Furthermore, downregulation of NRAGE in colorectal cancer is associated with a negative clinical course (24,28,29). By contrast, NRAGE functions as an oncogene in esophageal and lung cancers (22,30). Yang et al (22) demonstrated that the overexpression of NRAGE exerts tumor-promoting effects by interacting with the DNA polymerase III domain of proliferating cell nuclear antigen (PCNA) and consequently inhibits K48-polyubiquitin chain-mediated proteasome degradation of PCNA in esophageal cancer. However, to the best of our knowledge, there are currently no studies concerning NRAGE expression in HCC or its role in the pathogenesis of this disease. Therefore, the aim of the present study was to assess the clinical significance of NRAGE expression in HCC, as well as its relevance as a novel biomarker for tumor progression.
Materials and methods
Sample collection
The HCC cell lines (Hep3B, HepG2, HLE, HLF, HuH1, HuH2, HuH7, PLC/PRF/5 and SK-Hep1) were obtained from the American Type Culture Collection (Manassas, VA, USA). All cell lines were cultured in Dulbecco's modified Eagle's Medium (Sigma-Aldrich, St. Louis, MO, USA), supplemented with 10% fetal bovine serum at 37°C in an atmosphere containing 5% CO2 (6). The primary HCC tissues and the corresponding non-cancerous tissues were collected from 151 patients with HCC who had undergone liver resection at the Nagoya University Hospital (Nagoya, Japan) between January 1998 and July 2012. The specimens were classified histologically according to the criteria published in the Classification of Malignant Tumours, Union for International Cancer Control (UICC) (31). Clinicopathological data were collected from medical records, and written informed consent for the use of clinical samples and data was obtained from all patients, as required by the Institutional Review Board of Nagoya University, Japan (32).
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Expression levels of mRNA in all samples were analyzed using RT-qPCR, which was performed using an Applied Biosystems StepOne Plus (Thermo Fisher Scientific, Inc., Waltham, MA, USA) in triplicate. Total RNA (10 µg) isolated from the HCC cell lines, and the 151 primary HCC specimens and corresponding adjacent non-cancerous tissues, was used as the template for the synthesis of complementary DNA. RT-qPCR was performed using the SYBR® Green PCR Core Reagents kit (Applied Biosystems; Thermo Fisher Scientific, Inc.) and specific primers for NRAGE (Hokkaido System Science Co., Ltd., Tokyo, Japan) as follows: One cycle at 95°C for 10 min, 40 cycles at 95°C for 5 sec and 60°C for 30 sec. For standardization, glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA (TaqMan GAPDH Control Reagents; Applied Biosystems; Thermo Fisher Scientific, Inc.) was quantified in each sample. Expression levels are presented as the value of NRAGE mRNA divided by the value of GAPDH mRNA (33,34). Expression levels of mRNAs were normalized by the serially diluted standards (35). AATF, p75NTR and PCNA encode proteins that may interact with NRAGE, and the specific primers (Hokkaido System Science Co., Ltd.) used for each of these genes are listed in Table I.
Table I.
Primers and annealing temperatures.
| Gene | Oligo sequence (5′–3′) | Product size, bp | Annealing temperature, °C |
|---|---|---|---|
| NRAGE | |||
| Forward | GATTCCCTCAGACCTTTGC | 170 | 60 |
| Reverse | GAAGGAATCTGAGGCTTCAG | ||
| AATF | |||
| Forward | ACAAAGGTGGCCCAGAATTT | 103 | 62 |
| Reverse | TGGAAAAGCAACTCTTCCTGA | ||
| p75NTR | |||
| Forward | CTGCTGCTGTTGCTGCTTCT | 98 | 60 |
| Reverse | CAGGCTTTGCAGCACTCAC | ||
| PCNA | |||
| Forward | TGCAAGTGGAGAACTTGGAA | 128 | 58 |
| Reverse | TCAGGTACCTCAGTGCAAAAG | ||
| GAPDH | |||
| Forward | GAAGGTGAAGGTCGGAGTC | 226 | 60 |
| Probe | CAAGCTTCCCGTTCTCAGCC | ||
| Reverse | GAAGATGGTGATGGGATTTC |
NRAGE, neurotrophin receptor-interacting melanoma antigen-encoding protein; AATF, apoptosis-antagonizing transcription factor; p75NTR, p75 neurotrophin receptor; PCNA, proliferating cell nuclear antigen; GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
Immunohistochemistry (IHC)
IHC was performed to determine the expression and localization of NRAGE in 30 representative well-preserved HCC samples. Formalin-fixed, paraffin-embedded tissue samples were dewaxed twice in xylene for 5 min, rehydrated sequentially using a graded series of alcohol concentrations (100, 90 and 70%) for 2 min each and treated with 3% H2O2 to inhibit endogenous peroxidases. Antigen retrieval was performed by incubating the sections 5 times in citrate buffer (10 mM) at 95°C for 5 min. The samples were then washed with phosphate-buffered saline, followed by a 10-min incubation with biotinylated goat anti-rabbit IgG secondary antibody (Histofine SAB PO(R) kit; Code 424032; Nichirei Corporation, Tokyo, Japan) for 5 min to limit non-specific reactivity, and incubated for 1 h with a rabbit polyclonal anti-human NRAGE antibody (catalog no., LS-C100414; LifeSpan BioSciences, Inc., Seattle, WA, USA) in a 1:200 dilution with ChemMateT antibody diluent (Dako Japan Co., Ltd., Tokyo, Japan). The samples were then washed with phosphate-buffered saline, followed by a 10-min incubation with biotinylated goat anti-rabbit IgG secondary antibody (Histofine SAB-PO(R) kit; Nichirei Corporation) in a 1:1,000 dilution with ChemMateT antibody dilutent. Subsequently, the sections were incubated for 1 min with liquid 3,3′-diaminobenzidine (Nichirei Corporation) to detect antigen-antibody complexes. Staining of NRAGE was evaluated using vessels as internal controls. To avoid bias when interpreting data, specimens were randomized and coded prior to analysis by two independent observers who were uninformed of the status of the samples. Each observer evaluated all specimens at least twice within a given time interval to minimize intraobserver variation (36,37).
Statistical analysis
The significance of the association between the levels of NRAGE mRNA and the clinicopathological features was evaluated using the χ2 test, and differences between groups were evaluated using the Mann-Whitney U test. Correlations between the level of NRAGE mRNA and encoding AATF, p75NTR and PCNA, as well as those of pre-operative serum tumor markers, were analyzed using Spearman's rank correlation coefficient. Disease-specific survival rates were calculated using the Kaplan-Meier method, and the differences in survival curves were evaluated using the generalized Wilcoxon rank-sum test. P<0.05 was considered to indicate a statistically significant difference. All statistical analyses were performed using JMP® software, version 10 (SAS Institute Inc., Cary, NC, USA).
Results
Analysis of NRAGE, AATF, p75NTR and PCNA mRNA expression in HCC cell lines
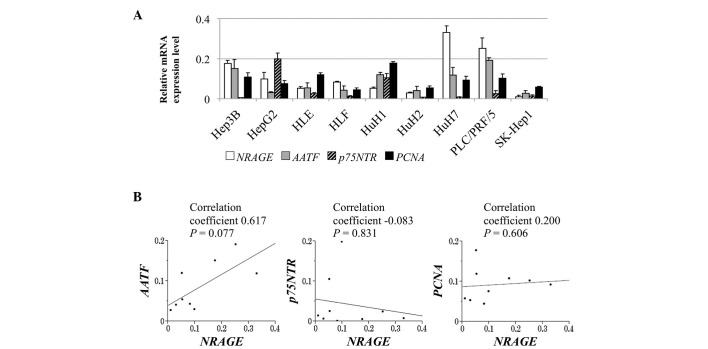
The relative levels of NRAGE mRNA and those of its putative interacting partners, AATF, p75NTR and PNCA, in the HCC cell lines are presented in Fig. 1A. Heterogeneity was observed in the NRAGE mRNA levels between 9 HCC cell lines, which correlated significantly with those of AATF (correlation coefficient, 0.617), whereas no significant correlation was observed between the expression of NRAGE and p75NTR or PCNA (Fig. 1B).
Figure 1.
Analysis of NRAGE mRNA expression in the HCC cell lines. (A) Levels of NRAGE mRNA and its putative interacting partners in the HCC cell lines. Error bars indicate the standard deviation of three biological replicates. (B) Analysis of the correlation between NRAGE mRNA level and those of its putative interacting partners AATF, p75NTR and PCNA. NRAGE, neurotrophin receptor-interacting melanoma antigen-encoding protein; HCC, hepatocellular carcinoma; AATF, apoptosis-antagonizing transcription factor; p75NTR, p75 neurotrophin receptor; PCNA, proliferating cell nuclear antigen.
Patient characteristics
The ages of the 151 patients ranged from 34–84 years (mean ± standard deviation, 64.7±9.8 years), and the male to female ratio was 5:1. A total of 37 and 84 patients were infected with hepatitis B and C virus, respectively. The numbers of patients with a normal liver, chronic hepatitis or cirrhosis were 10, 87 and 54, respectively. When classified according to the UICC's tumor-node-metastasis classification, 94, 39 and 18 patients were in stages I, II and III, respectively.
Analysis of NRAGE and AATF mRNA levels in surgically resected liver tissues
There were no significant differences observed in the NRAGE mRNA levels in the non-cancerous tissue samples between patients with or without cirrhosis (Fig. 2A). However, the mean level of NRAGE mRNA in the HCC tissues was significantly higher compared with that in the corresponding normal tissues (P=0.006; Fig. 2B).
Figure 2.
Analysis of NRAGE expression in the liver tissues. (A) NRAGE mRNA levels did not differ significantly between non-cancerous liver tissues of patients with and without cirrhosis. (B) NRAGE mRNA levels were significantly elevated in the HCC tissues compared with the corresponding non-cancerous liver tissues. (C) Two representative cases demonstrating strong immunoreactivity of NRAGE specific to the HCC tissues (magnification: Left, ×200; right, ×100; and inset, ×400). (D) Analysis of the correlation between NRAGE and AATF mRNA levels in the HCC tissues. HCC, hepatocellular carcinoma; NRAGE, neurotrophin receptor-interacting melanoma antigen-encoding protein; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; N, normal; T, tumor; AATF, apoptosis-antagonizing transcription factor.
IHC analysis was conducted to determine whether NRAGE expression correlated with NRAGE mRNA levels in the tumor and non-cancerous tissues. Representative images showing strong staining intensities of NRAGE in cancer tissues are presented in Fig. 2C. The expression patterns of NRAGE protein and mRNA in the HCC and non-cancerous tissues were consistent among 30 representative pairs of specimens. It was observed that the level of NRAGE and AATF mRNA was directly correlated, consistent with results of the analysis of the HCC cell lines (Fig. 2C).
Clinical significance of NRAGE mRNA expression
Tissue sections were assigned to two groups (high and low expression) according to their level of NRAGE mRNA. The high and low expression groups included 93 and 58 patients, respectively. The high expression group was associated with increased age and an α-fetoprotein level of >20 ng/ml (P=0.014 and P=0.020, respectively; Table II). The disease-specific survival time of the patients was significantly shorter in the high expression group compared with the low expression group (5-year survival rate, 60 vs. 75%; P=0.008; Fig. 3A). Subgroup analysis according to UICC staging indicated that the disease-specific survival curves showed a similar tendency of differences between high and low NRAGE expression groups in each disease stage (Fig. 3B). Multivariate analysis identified vascular invasion and increased NRAGE expression as independent prognostic factors (hazard ratios, 2.24 and 2.23, respectively; Table III).
Table II.
Association between NRAGE mRNA levels and clinicopathological parameters of 151 hepatocellular carcinoma patients.
| Clinicopathological parameters | Increased expression of NRAGE mRNA, n | Others, n | P-value |
|---|---|---|---|
| Age, years | 0.014a | ||
| <65 | 34 | 33 | |
| ≥65 | 59 | 25 | |
| Gender | 0.096 | ||
| Male | 74 | 52 | |
| Female | 19 | 6 | |
| Background of liver | 0.628 | ||
| Normal | 6 | 4 | |
| Chronic hepatitis | 51 | 36 | |
| Cirrhosis | 36 | 18 | |
| Child-Pugh classification | 0.621 | ||
| A | 87 | 53 | |
| B | 6 | 5 | |
| Hepatitis virus | 0.728 | ||
| Absent | 17 | 13 | |
| HBV | 22 | 15 | |
| HCV | 54 | 30 | |
| AFP, ng/ml | 0.020a | ||
| ≤20 | 43 | 38 | |
| >20 | 50 | 20 | |
| PIVKA II, mAU/ml | 0.350 | ||
| ≤40 | 33 | 25 | |
| >40 | 60 | 33 | |
| Tumor multiplicity | 0.707 | ||
| Solitary | 73 | 44 | |
| Multiple | 20 | 14 | |
| Tumor size, cm | 0.733 | ||
| <3.0 | 28 | 19 | |
| ≥3.0 | 65 | 39 | |
| Differentiation | 0.314 | ||
| Well | 19 | 16 | |
| Moderate to poor | 74 | 42 | |
| Growth type | 0.208 | ||
| Expansive | 81 | 46 | |
| Invasive | 12 | 12 | |
| Serosal infiltration | 0.389 | ||
| Absent | 68 | 46 | |
| Present | 25 | 12 | |
| Formation of capsule | 0.289 | ||
| Absent | 26 | 21 | |
| Present | 67 | 37 | |
| Infiltration to capsule | 0.101 | ||
| Absent | 37 | 31 | |
| Present | 56 | 27 | |
| Septum formation | 0.407 | ||
| Absent | 35 | 18 | |
| Present | 58 | 40 | |
| Vascular invasion | 0.386 | ||
| Absent | 68 | 46 | |
| Present | 25 | 12 | |
| UICC pathological stage | 0.919 | ||
| I | 59 | 35 | |
| II | 23 | 16 | |
| III | 11 | 7 |
P<0.05. HCC, hepatocellular carcinoma; NRAGE, neurotrophin receptor-interacting melanoma antigen-encoding protein; HBV, hepatitis B virus; HCV, hepatitis C virus; AFP, α-fetoprotein; PIVKA, protein induced by vitamin K antagonists; UICC, Union for International Cancer Control.
Figure 3.
Patient survival analysis. (A) Patients with increased expression of NRAGE mRNA experienced significantly shorter disease-specific survival times. (B) Subgroup analysis as a function of Union for International Cancer Control tumor-node-metastasis staging. Increased levels of NRAGE mRNA were associated with shorter survival times in patients with stages I or II–III HCC. HCC, hepatocellular carcinoma; NRAGE, neurotrophin receptor-interacting melanoma antigen-encoding protein.
Table III.
Prognostic factors of 151 patients with hepatocellular carcinoma.
| Univariate analysis | Multivariate analysis | ||||||
|---|---|---|---|---|---|---|---|
| Variable | n | Hazard ratio | 95% CI | P-value | Hazard ratio | 95% CI | P-value |
| Age ≥65 years | 84 | 1.92 | 1.07–3.57 | 0.030a | 1.39 | 0.75–2.65 | 0.301 |
| Male gender | 126 | 1.27 | 0.60–3.13 | 0.553 | |||
| Cirrhosis of liver | 4 | 1.58 | 0.88–2.81 | 0.123 | |||
| Child-Pugh classification B | 11 | 0.93 | 0.28–2.32 | 0.889 | |||
| AFP >20 ng/ml | 70 | 1.90 | 1.07–3.42 | 0.029a | 1.30 | 0.70–2.44 | 0.790 |
| PIVKA II >40 mAU/ml | 93 | 2.10 | 1.14–4.07 | 0.016a | 1.12 | 0.55–2.40 | 0.770 |
| Tumor multiplicity | 34 | 2.09 | 1.11–3.76 | 0.023a | 1.32 | 0.67–2.53 | 0.416 |
| Tumor size ≥3.0 cm | 104 | 2.20 | 1.13–4.71 | 0.020a | 1.87 | 0.82–4.64 | 0.140 |
| Well-differentiated tumor | 35 | 0.55 | 0.25–1.10 | 0.095 | |||
| Invasive growth | 24 | 1.44 | 0.69–2.76 | 0.318 | |||
| Serosal infiltration | 37 | 2.51 | 1.32–4.61 | 0.006a | 1.53 | 0.76–2.95 | 0.225 |
| Formation of capsule | 104 | 1.05 | 0.57–2.02 | 0.884 | |||
| Infiltration to capsule | 83 | 1.20 | 0.67–2.18 | 0.537 | |||
| Septum formation | 98 | 0.87 | 0.49–1.60 | 0.651 | |||
| Vascular invasion | 37 | 3.40 | 1.87–6.07 | <0.001a | 2.24 | 1.12–4.41 | 0.022a |
| Increased NRAGE expression | 93 | 2.42 | 1.27–4.99 | 0.006a | 2.23 | 1.06–3.83 | 0.020a |
Univariate analysis was performed using the log-rank test. Multivariate analysis was performed using the Cox proportional hazards model.
P<0.05. CI, confidence interval; AFP, α-fetoprotein; PIVKA, protein induced by vitamin K antagonists; NRAGE, neurotrophin receptor-interacting melanoma antigen-encoding protein.
Discussion
Originally identified by Salehi et al (25) in a two-hybrid screen utilizing p75NTR as bait, NRAGE is a signaling cascade component that mediates apoptosis by interacting with p75NTR to antagonize its association with the nerve growth factor receptor tropomyosin receptor kinase A. Studies have indicated that NRAGE promotes apoptosis through the ubiquitination of AATF (26,27), therefore suggesting that NRAGE functions as a tumor suppressor by inducing tumor cell apoptosis. However, overexpression of NRAGE accelerates the proliferation and migration of esophageal cancer cells via interactions with PCNA. A genome-wide association study demonstrated that the initiation of NRAGE signals through the JNK pathway is associated with non-small cell lung cancer (22,30). Xue et al (38) identified an association between elevated NRAGE expression and the increased radioresistance of esophageal carcinoma cells. Such studies indicate that NRAGE functions to inhibit or promote oncogenesis depending on cell type, leading to the rationale behind the present study.
The current study demonstrated that NRAGE mRNA levels positively correlated with those of AATF, but not those of p75NTR or PCNA. The presence of cirrhosis had a minimal affect on NRAGE expression, and NRAGE mRNA levels were significantly higher in the HCC tissues compared with the corresponding non-cancerous tissues; this suggests that NRAGE is involved in hepatocarcinogenesis or the subsequent progression of HCC.
Multivariate analysis of disease-specific survival time following curative hepatectomy and subgroup analysis, according to UICC staging, identified that increased NRAGE mRNA expression in the HCC tissues functioned as an independent prognostic factor. These results indicate that NRAGE serves as a tumor promoting factor, and that the levels of NRAGE mRNA in resected primary lesions may function as a biomarker for the progression of HCC. Furthermore, the results of IHC analysis demonstrated that NRAGE expression correlated with mRNA level, subsequently indicating the physiological significance of the latter. Therefore, future investigation into the function of NRAGE in HCC may be facilitated using RT-qPCR analysis of NRAGE mRNA levels.
In conclusion, NRAGE mediates the progression of HCC and may serve as a novel biomarker for the malignant phenotype of the disease. The present study indicated that the analysis of NRAGE expression may enhance the clinical management of HCC. For example, the levels of NRAGE mRNA in liver biopsies or surgically resected tissues may facilitate risk stratification of patients with HCC, and may also serve as a criterion for determining the most appropriate therapy tailored to individual patients. Furthermore, the findings of the current study demonstrate promise for the development of novel therapies for HCC that employ small molecules or antibodies that target NRAGE and its interacting partners, including AATF. Research to further investigate the signaling pathways that are regulated by or function through NRAGE may expose alternative targets for the treatment of HCC.
References
- 1.Shiraha H, Yamamoto K, Namba M. Human hepatocyte carcinogenesis (review) Int J Oncol. 2013;42:1133–1138. doi: 10.3892/ijo.2013.1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bruix J, Gores GJ, Mazzaferro V. Hepatocellular carcinoma: Clinical frontiers and perspectives. Gut. 2014;63:844–855. doi: 10.1136/gutjnl-2013-306627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yang JD, Roberts LR. Hepatocellular carcinoma: A global view. Nat Rev Gastroenterol Hepatol. 2010;7:448–458. doi: 10.1038/nrgastro.2010.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.GLOBOCAN Estimated Cancer Incidence, Mortality and Prevalence Worldwide in 2012. Stomach Cancer. 2012 [Google Scholar]
- 5.Zhao YJ, Ju Q, Li GC. Tumor markers for hepatocellular carcinoma. Mol Clin Oncol. 2013;1:593–598. doi: 10.3892/mco.2013.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kanda M, Sugimoto H, Nomoto S, Oya H, Hibino S, Shimizu D, Takami H, Hashimoto R, Okamura Y, Yamada S, et al. B-cell translocation gene 1 serves as a novel prognostic indicator of hepatocellular carcinoma. Int J Oncol. 2015;46:641–648. doi: 10.3892/ijo.2014.2762. [DOI] [PubMed] [Google Scholar]
- 7.Flores A, Marrero JA. Emerging trends in hepatocellular carcinoma: Focus on diagnosis and therapeutics. Clin Med Insights Oncol. 2014;8:71–76. doi: 10.4137/CMO.S9926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Giannelli G, Rani B, Dituri F, Cao Y, Palasciano G. Moving towards personalised therapy in patients with hepatocellular carcinoma: The role of the microenvironment. Gut. 2014;63:1668–1676. doi: 10.1136/gutjnl-2014-307323. [DOI] [PubMed] [Google Scholar]
- 9.Kanda M, Nomoto S, Okamura Y, et al. Detection of metallothionein 1G as a methylated tumor suppressor gene in human hepatocellular carcinoma using a novel method of double combination array analysis. Int J Oncol. 2009;35:477–483. doi: 10.3892/ijo_00000359. [DOI] [PubMed] [Google Scholar]
- 10.Miki D, Ochi H, Hayes CN, Aikata H, Chayama K. Hepatocellular carcinoma: Towards personalized medicine. Cancer Sci. 2012;103:846–850. doi: 10.1111/j.1349-7006.2012.02242.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Khare S, Zhang Q, Ibdah JA. Epigenetics of hepatocellular carcinoma: Role of microRNA. World J Gastroenterol. 2013;19:5439–5445. doi: 10.3748/wjg.v19.i33.5439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kanda M, Nomoto S, Oya H, Takami H, Hibino S, Hishida M, Suenaga M, Yamada S, Inokawa Y, Nishikawa Y, et al. Downregulation of DENND2D by promoter hypermethylation is associated with early recurrence of hepatocellular carcinoma. Int J Oncol. 2014;44:44–52. doi: 10.3892/ijo.2013.2165. [DOI] [PubMed] [Google Scholar]
- 13.Chomez P, De Backer O, Bertrand M, De Plaen E, Boon T, Lucas S. An overview of the MAGE gene family with the identification of all human members of the family. Cancer Res. 2001;61:5544–5551. [PubMed] [Google Scholar]
- 14.Tseng HY, Chen LH, Ye Y, Tay KH, Jiang CC, Guo ST, Jin L, Hersey P, Zhang XD. The melanoma-associated antigen MAGE-D2 suppresses TRAIL receptor 2 and protects against TRAIL-induced apoptosis in human melanoma cells. Carcinogenesis. 2012;33:1871–1881. doi: 10.1093/carcin/bgs236. [DOI] [PubMed] [Google Scholar]
- 15.Takami H, Kanda M, Oya H, Hibino S, Sugimoto H, Suenaga M, Yamada S, Nishikawa Y, Asai M, Fujii T, et al. Evaluation of MAGE-D4 expression in hepatocellular carcinoma in Japanese patients. J Surg Oncol. 2013;108:557–562. doi: 10.1002/jso.23440. [DOI] [PubMed] [Google Scholar]
- 16.Sang M, Wang L, Ding C, Zhou X, Wang B, Wang L, Lian Y, Shan B. Melanoma-associated antigen genes - an update. Cancer Lett. 2011;302:85–90. doi: 10.1016/j.canlet.2010.10.021. [DOI] [PubMed] [Google Scholar]
- 17.Oya H, Kanda M, Takami H, Hibino S, Shimizu D, Niwa Y, Koike M, Nomoto S, Yamada S, Nishikawa Y, et al. Overexpression of melanoma-associated antigen D4 is an independent prognostic factor in squamous cell carcinoma of the esophagus. Dis Esophagus. 2015;28:188–195. doi: 10.1111/dote.12156. [DOI] [PubMed] [Google Scholar]
- 18.Chang CC, Campoli M, Luo W, Zhao W, Zaenker KS, Ferrone S. Immunotherapy of melanoma targeting human high molecular weight melanoma-associated antigen: Potential role of nonimmunological mechanisms. Ann NY Acad Sci. 2004;1028:340–350. doi: 10.1196/annals.1322.040. [DOI] [PubMed] [Google Scholar]
- 19.Hussein TD. Serological tumor markers of hepatocellular carcinoma: A meta-analysis. Int J Biol Markers. 2015;30:e32–e42. doi: 10.5301/jbm.5000119. [DOI] [PubMed] [Google Scholar]
- 20.Jordan BW, Dinev D, LeMellay V, Troppmair J, Gotz R, Wixler L, Sendtner M, Ludwig S, Rapp UR. Neurotrophin receptor-interacting mage homologue is an inducible inhibitor of apoptosis protein-interacting protein that augments cell death. J Biol Chem. 2001;276:39985–39989. doi: 10.1074/jbc.C100171200. [DOI] [PubMed] [Google Scholar]
- 21.Wang X, Gao X, Xu Y. MAGED1: Molecular insights and clinical implications. Ann Med. 2011;43:347–355. doi: 10.3109/07853890.2011.573806. [DOI] [PubMed] [Google Scholar]
- 22.Yang Q, Ou C, Liu M, Xiao W, Wen C, Sun F. NRAGE promotes cell proliferation by stabilizing PCNA in a ubiquitin-proteasome pathway in esophageal carcinomas. Carcinogenesis. 2014;35:1643–1651. doi: 10.1093/carcin/bgu084. [DOI] [PubMed] [Google Scholar]
- 23.Barker PA, Salehi A. The MAGE proteins: Emerging roles in cell cycle progression, apoptosis, and neurogenetic disease. J Neurosci Res. 2002;67:705–712. doi: 10.1002/jnr.10160. [DOI] [PubMed] [Google Scholar]
- 24.Du Q, Zhang Y, Tian XX, Li Y, Fang WG. MAGE-D1 inhibits proliferation, migration and invasion of human breast cancer cells. Oncol Rep. 2009;22:659–665. doi: 10.3892/or_00000486. [DOI] [PubMed] [Google Scholar]
- 25.Salehi AH, Roux PP, Kubu CJ, Zeindler C, Bhakar A, Tannis LL, Verdi JM, Barker PA. NRAGE, a novel MAGE protein, interacts with the p75 neurotrophin receptor and facilitates nerve growth factor-dependent apoptosis. Neuron. 2000;27:279–288. doi: 10.1016/S0896-6273(00)00036-2. [DOI] [PubMed] [Google Scholar]
- 26.Di Certo MG, Corbi N, Bruno T, Iezzi S, De Nicola F, Desantis A, Ciotti MT, Mattei E, Floridi A, Fanciulli M, Passananti C. NRAGE associates with the anti-apoptotic factor Che-1 and regulates its degradation to induce cell death. J Cell Sci. 2007;120:1852–1858. doi: 10.1242/jcs.03454. [DOI] [PubMed] [Google Scholar]
- 27.Passananti C, Fanciulli M. The anti-apoptotic factor Che-1/AATF links transcriptional regulation, cell cycle control, and DNA damage response. Cell Div. 2007;2:21. doi: 10.1186/1747-1028-2-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Krämer BF, Schoor O, Krüger T, Reichle C, Müller M, Weinschenk T, Hennenlotter J, Stenzl A, Rammensee HG, Stevanovic S. MAGED4-expression in renal cell carcinoma and identification of an HLA-A*25-restricted MHC class I ligand from solid tumor tissue. Cancer Biol Ther. 2005;4:943–948. doi: 10.4161/cbt.4.9.1907. [DOI] [PubMed] [Google Scholar]
- 29.Zeng ZL, Wu WJ, Yang J, Tang ZJ, Chen DL, Qiu MZ, Luo HY, Wang ZQ, Jin Y, Wang DS, Xu RH. Prognostic relevance of melanoma antigen D1 expression in colorectal carcinoma. J Transl Med. 2012;10:181. doi: 10.1186/1479-5876-10-181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee D, Lee GK, Yoon KA, Lee JS. Pathway-based analysis using genome-wide association data from a Korean non-small cell lung cancer study. PLoS One. 2013;8:e65396. doi: 10.1371/journal.pone.0065396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sobin LH, Gospodarowicz MK, Wittekind C, editors. TNM Classification of Malignant Tumors. 7th. New York: Wiley-Blackwell; 2009. International Union Against Cancer. [Google Scholar]
- 32.Kanda M, Shimizu D, Nomoto S, Hibino S, Oya H, Takami H, Kobayashi D, Yamada S, Inokawa Y, Tanaka C, et al. Clinical significance of expression and epigenetic profiling of TUSC1 in gastric cancer. J Surg Oncol. 2014;110:136–144. doi: 10.1002/jso.23614. [DOI] [PubMed] [Google Scholar]
- 33.Kanda M, Nomoto S, Oya H, Shimizu D, Takami H, Hibino S, Hashimoto R, Kobayashi D, Tanaka C, Yamada S, et al. Dihydropyrimidinase-like 3 facilitates malignant behavior of gastric cancer. J Exp Clin Cancer Res. 2014;33:66. doi: 10.1186/s13046-014-0066-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kanda M, Shimizu D, Nomoto S, Takami H, Hibino S, Oya H, Hashimoto R, Suenaga M, Inokawa Y, Kobayashi D, et al. Prognostic impact of expression and methylation status of DENN/MADD domain-containing protein 2D in gastric cancer. Gastric Cancer. 2015;18:288–296. doi: 10.1007/s10120-014-0372-0. [DOI] [PubMed] [Google Scholar]
- 35.Kubista M, Andrade JM, Bengtsson M, Forootan A, Jonák J, Lind K, Sindelka R, Sjöback R, Sjögreen B, Strömbom L, et al. The real-time polymerase chain reaction. Mol Aspects Med. 2006;27:95–125. doi: 10.1016/j.mam.2005.12.007. [DOI] [PubMed] [Google Scholar]
- 36.Kanda M, Nomoto S, Oya H, Hashimoto R, Takami H, Shimizu D, Sonohara F, Kobayashi D, Tanaka C, Yamada S, et al. Decreased expression of prenyl diphosphate synthase subunit 2 correlates with reduced survival of patients with gastric cancer. J Exp Clin Cancer Res. 2014;33:88. doi: 10.1186/s13046-014-0088-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kanda M, Nomoto S, Okamura Y, Hayashi M, Hishida M, Fujii T, Nishikawa Y, Sugimoto H, Takeda S, Nakao A. Promoter hypermethylation of fibulin 1 gene is associated with tumor progression in hepatocellular carcinoma. Mol Carcinog. 2011;50:571–579. doi: 10.1002/mc.20735. [DOI] [PubMed] [Google Scholar]
- 38.Xue XY, Liu ZH, Jing FM, Li YG, Liu HZ, Gao XS. Relationship between NRAGE and the radioresistance of esophageal carcinoma cell line TE13R120. Chin J Cancer. 2010;29:900–906. doi: 10.5732/cjc.010.10141. [DOI] [PubMed] [Google Scholar]