Abstract
Small secondary metabolites, including glucosinolates and the major phytoalexin camalexin, play important roles in immunity in Arabidopsis thaliana. We isolated an Arabidopsis mutant with increased resistance to the powdery mildew fungus Golovinomyces cichoracearum and identified a mutation in the gene encoding cytochrome P450 83A1 monooxygenase (CYP83A1), which functions in glucosinolate biosynthesis. The cyp83a1-3 mutant exhibited enhanced defense responses to G. cichoracearum and double mutant analysis showed that this enhanced resistance requires NPR1, EDS1, and PAD4, but not SID2 or EDS5. In cyp83a1-3 mutants, the expression of genes related to camalexin synthesis increased upon G. cichoracearum infection. Significantly, the cyp83a1-3 mutant also accumulated higher levels of camalexin. Decreasing camalexin levels by mutation of the camalexin synthetase gene PAD3 or the camalexin synthesis regulator AtWRKY33 compromised the powdery mildew resistance in these mutants. Consistent with these observations, overexpression of PAD3 increased camalexin levels and enhanced resistance to G. cichoracearum. Taken together, our data indicate that accumulation of higher levels of camalexin contributes to increased resistance to powdery mildew.
Keywords: camalexin, powdery mildew, CYP83A1, plant immunity, Arabidopsis thaliana
Introduction
To protect themselves against pathogens, plants have evolved intricate immune responses that include accumulation of reactive oxygen species, deposition of callose, enhanced expression of pathogenesis-related (PR) genes, and biosynthesis of phytoalexins. Phytoalexins are low molecular mass secondary metabolites that are induced by both biotic and abiotic stress. During pathogen infection, plants synthesize a wide variety of structurally different phytoalexins to defend against pathogen invasion (Hammerschmidt, 1999; Pedras et al., 2011). Camalexin, 3-thiazol-2′-yl-indole, is one of the major phytoalexins of Arabidopsis thaliana and was long considered to be the only phytoalexin of Arabidopsis, until the discovery of rapalexin A (Pedras and Adio, 2008).
The biosynthesis and regulation of camalexin in Arabidopsis remain only partially understood, and the full scope of camalexin functions also remains to be defined. Camalexin is derived from tryptophan and requires many cytochrome P450s, including CYP79B2, CYP71A13, and CYP71B15 [which corresponds to the camalexin-deficient mutant PHYTOALEXIN DEFICIENT 3 (PAD3); Nafisi et al., 2007; Schuhegger et al., 2007a,b]. The Arabidopsis transcription factors WRKY33, WRKY18, and WRKY40 appear to be involved in the regulation of camalexin biosynthesis (Qiu et al., 2008; Pandey et al., 2010; Mao et al., 2011). Camalexin plays an important role in the response to necrotrophic pathogens Alternaria brassicicola and Botrytis cinerea (Thomma et al., 1999; Ferrari et al., 2003; Kliebenstein et al., 2005; Nafisi et al., 2007), and the oomycete Phytophthora brassicae (Schlaeppi et al., 2010), as well as the biotrophic fungus Golovinomyces orontii (Consonni et al., 2010; Pandey et al., 2010). Although, camalexin produces broad-spectrum resistance to many species of plant pathogens, how it functions remains unclear.
In addition to camalexin, plants synthesize other, related secondary metabolites, such as glucosinolates, that participate in the defense response. Plant cells usually store glucosinolates in stable forms; during insect and/or pathogen attack, myrosinases hydrolyze these stable forms into active compounds (Halkier and Gershenzon, 2006). According to their side-chain radical, glucosinolates can be divided into aliphatic glucosinolates, indole glucosinolates, and aromatic glucosinolates. Many cytochrome P450s function in glucosinolate synthesis, including CYP83A1 and CYP83B1. The Arabidopsis cytochrome P450 monooxygenase CYP83A1 participates in the biosynthesis of aliphatic glucosinolates from aliphatic oximes, whereas CYP83B1, the Arabidopsis protein most similar to CYP83A1, functions in the biosynthesis of indole glucosinolates.
The biosynthetic pathways of alkylglucosinolates and indole glucosinolates affect each other; for instance, the cyp83a1-2 (also called ref2-1) mutant produces lower levels of aliphatic glucosinolates, but accumulates higher levels of indole-derived glucosinolates compared with wild-type (Hemm et al., 2003; Naur et al., 2003; Sonderby et al., 2010). The biosynthesis of glucosinolates, especially indole glucosinolates, shares the intermediate product indole-3-acetaldoxime (IAOx) with biosynthetic pathways that produce many other secondary metabolites or hormones like camalexin and indole-3-acetic acid (IAA), respectively (Hemm et al., 2003; Grubb and Abel, 2006; Nafisi et al., 2007). Although the regulation of callose biosynthesis, in response to bacterial elicitors of Arabidopsis immunity, requires 4-methoxy-indol-3-ylmethylglucosinolate (4MI3G; Bednarek et al., 2009; Clay et al., 2009), how 4MI3G and related metabolites participate in the immune response is not well-understood.
Powdery mildew fungi, as biotrophic pathogens, infect many plant species and cause huge agricultural losses worldwide. The plant hormone salicylic acid (SA) plays an important role in resistance to powdery mildew in Arabidopsis, and many mutants showing enhanced resistance to powdery mildew require SA signaling for their resistance phenotype; these mutants include edr1 (enhanced disease resistance 1), edr2 and edr4 (Frye and Innes, 1998; Frye et al., 2001; Tang et al., 2005a,b; Zhao et al., 2014; Wu et al., 2015).
To further study resistance to powdery mildew in Arabidopsis, we characterized an Arabidopsis mutant that exhibits enhanced resistance to a variety of powdery mildew species. Here, we report that a mutation in the gene encoding cytochrome P450 monooxygenase CYP83A1, a component of the glucosinolate pathway, leads to higher accumulation of camalexin and enhanced resistance to the powdery mildew fungus Golovinomyces cichoracearum, which is consistent with the previous finding that cyp83a1 exhibits increased resistance to powdery mildew fungus Erysiphe cruciferarum (Weis et al., 2013). We show that cyp83a1-3 accumulates higher levels of camalexin. We also show that mutations in genes affecting camalexin production suppress the resistance of cyp83a1-3, indicating that higher accumulation of camalexin in cyp83a1-3 mutants contributes to their enhanced powdery mildew resistance.
Materials and Methods
Isolation of the cyp83a1-3 Mutant
The cyp83a1-3 mutant was identified in a population of transgenic Arabidopsis Col-0 plants that expressed a PR2::GUS transgene and had been mutagenized with ethyl methanesulfonate as described (Cao et al., 1994). Two leaves of each of 3850 M2 plants were infiltrated with Pseudomonas syringae pv. maculicola strain ES4326 at a dose of 105 cells per cm2 leaf area. Four putative mutants reproducibly exhibited reduced disease symptoms 3 days after infection, and two of these exhibited significantly reduced growth (about 10-fold less) of P. syringae ES4326. One of these latter two mutants, which was eventually named cyp83a1-3, also exhibited reduced symptom development when infected with the G. orontii strain MGH. The cyp83a1-3 mutant was backcrossed twice to the parental line carrying the PR2::GUS reporter. Genetic analysis showed that the resistance phenotype of cyp83a1-3 was recessive and segregated 1:3 as expected for a single recessive Mendelian gene.
Plant Materials and Growth Conditions
pad3-1 (Zhou et al., 1999), cyp83a1-1 (Salk_123405), cyp83a1-2 (ref2-1; Hemm et al., 2003) and wrky33-2 (GABI_324B11; Zheng et al., 2006) were described previously. Arabidopsis plants were grown in a growth room at 20–22°C under a 9-h-light/15-h-dark cycle for phenotyping or a 16-h-light/8-h-dark cycle for seed setting, as described previously (Nie et al., 2011).
Pathogen Infection and Microscopy
Powdery mildew pathogens G. orontii strain MGH (Plotnikova et al., 1998) and G. cichoracearum strain UCSC1 were maintained on pad4-1 plants (Jirage et al., 1999) as described previously (Frye et al., 2001). Four-weeks-old plants were inoculated with powdery mildew using a settling tower to achieve an even distribution of conidia (Wang et al., 2011). To quantify fungal growth and conidiation, the number of conidiophores per colony was counted at 5 dpi (Consonni et al., 2006). At least 30 colonies were counted for each genotype in each experiment. Trypan blue staining was used to visualize fungal hyphae and dead cells (Frye and Innes, 1998), and H2O2 accumulation was detected with 3,3′-diaminobenzidine hydrochloride (DAB) staining (Koch and Slusarenko, 1990). Samples were photographed with an Olympus BX53 microscope.
Map-Based Cloning and Complementation
For map-based cloning, we crossed the cyp83a1-3 mutant with Landsberg erecta and the mutation was mapped using a variety of molecular markers to a 128-kb region on chromosome 4 spanned by two BAC clones, T6G15 and F18A5 (https://www.arabidopsis.org). This region contains 24 predicted genes. Using a candidate gene approach, genes in the region were amplified by PCR and sequenced until the mutation was identified as a single nucleotide change in At4g13770, which encodes the cytochrome P450 CYP83A1. The mutation results in the substitution of glutamic acid for a conserved glycine at amino acid position 346 in the heme-binding site of the enzyme.
The genomic DNA sequence of CYP83A1 including 1.1 kb upstream of the ATG start codon and 0.4 kb downstream of the stop codon of At4g13770 was cloned into binary vector pCambia1300 for complementation analysis. The derived genomic construct was verified by sequencing and introduced into Agrobacterium tumefaciens strain GV3101, then transformed into cyp83a1-3 using the floral dip method (Clough and Bent, 1998). The transgenic plants were selected on 1/2 MS medium with 50 mg/L hygromycin.
Construction of Double Mutants
The following mutants were crossed with cyp83a1-3 to construct double mutants: sid2-2 (Wildermuth et al., 2001), pad4-1 (Jirage et al., 1999), eds1-2 (Bartsch et al., 2006), eds5-1 (Nawrath et al., 2002), npr1-63 (Alonso et al., 2003), pad3-1 (Zhou et al., 1999), and wrky33-2 (Zheng et al., 2006). Double mutants were identified by PCR, except for the pad3-1 mutation in pad3-1 cyp83a1-3, which was identified by PCR followed by sequencing.
Quantitative Real-Time RT-PCR
Real-time quantitative PCR was performed as described previously (Nie et al., 2012).
SA Extraction and Measurement
Salicylic acid extraction and measurement were performed as described previously (Gou et al., 2009).
Vector Construction
The full-length CYP83A1 coding sequence (CDS) without the stop codon was amplified by PCR from Col-0 cDNA and inserted into the Gateway vector pDONR207 using a BP Clonase kit (Invitrogen) to create a pDONR207-CYP83A1 CDS entry clone. Then an LR Clonase kit (Invitrogen) was used to introduce the inserts into the pEarleyGate 101/103 destination vector containing a 35S promoter and C-terminal HA/GFP fusion (Earley et al., 2006). The same methods were used to construct PAD3 overexpression constructs.
Camalexin Measurement
Camalexin content was determined using a previously described fluorometric method (Glazebrook and Ausubel, 1994) with excitation at 315 nm and emission at 385 nm using a HITACHI F4500 spectrofluorometer. The concentration of camalexin was determined by comparison with a camalexin standard curve using purified camalexin kindly provided by Dr. Shuqun Zhang (University of Missouri).
The Statistical Analysis
Statistical comparison of counts in genotypes in each of three independent experiments was performed using a mixed effects model for nested ANOVA, implemented in R. The genotypes were treated as fixed effects, whereas different experiments were treated as random effects. The resulting ANOVA P-values were used as estimates of statistical significance of the difference between genotypes.
Gene ID Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative databases under the following gene ID numbers: Arabidopsis CYP83A1 (At4g13770), PAD3 (At3g26 830), CYP71A13 (At2g30770), WRKY33 (At2g38470), PR1 (At2g14610), PR2 (At3g57260), FRK1 (At2g19190), and ACTIN2 (At3g18780).
Primers used for genotyping and gene expression analysis are listed in Supplementary Table S1.
Results
The cyp83a1-3 Mutant Displays Reduced Susceptibility to the Powdery Mildew Fungi G. orontii and G. cichoracearum
To study the molecular mechanism of plant resistance to powdery mildew, we initially screened an ethyl methanesulfonate-mutagenized Arabidopsis ecotype Col-0 population for mutants with enhanced resistance to P. syringae pv. maculicola strain ES4326 and then subsequently for resistance to the powdery mildew fungi G. orontii and G. cichoracearum. In this screen (see Materials and Methods), we identified two mutants with enhanced resistance to P. syringae, one of which also showed increased resistance to powdery mildew. We designated this latter mutant cyp83a1-3 based on subsequent characterization, as described below.
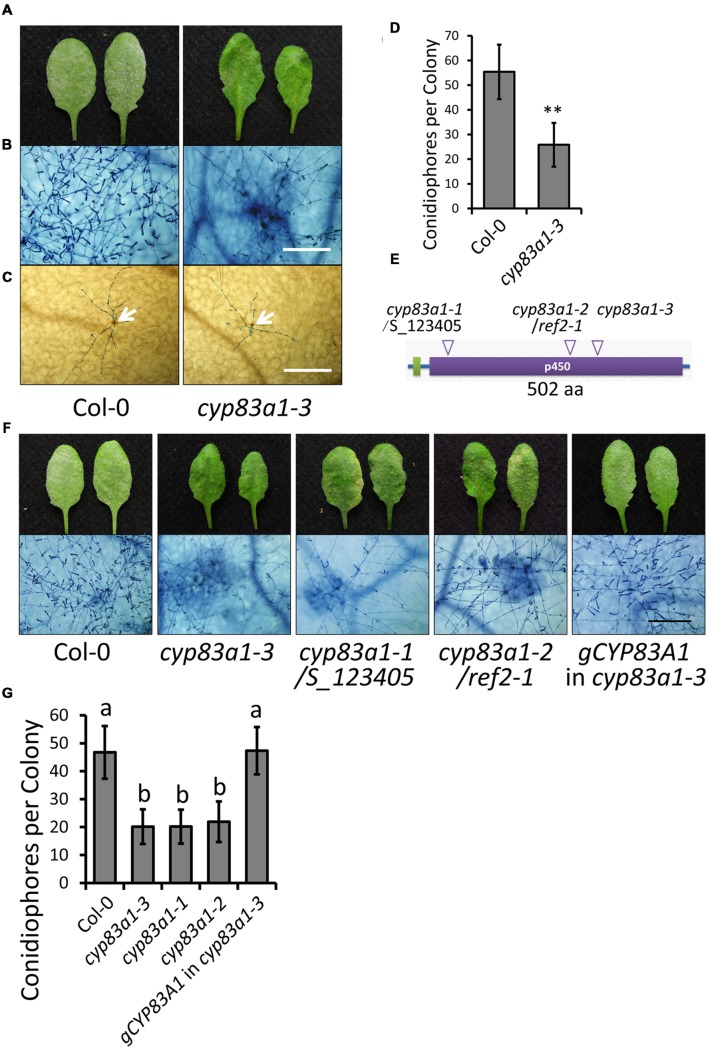
In the absence of pathogen, the growth of the cyp83a1-3 mutant was similar to the wild-type under standard short-day conditions (Supplementary Figure S1). However, cyp83a1-3 mutants showed significantly fewer conidia with minor G. cichoracearum-induced lesions in comparison to wild-type plants at 8 days post-infection (dpi; Figures 1A,B). Powdery mildew infection often causes accumulation of H2O2 in resistant plants (Shi et al., 2013; Wu et al., 2015; Zhao et al., 2015). We used DAB staining to measure H2O2 accumulation at 2 dpi, but we found that the cyp83a1-3 and wild-type plants had similar levels of H2O2 (Figure 1C), suggesting that an enhanced oxidative burst is not responsible for the resistance phenotype of the cyp83a1-3 mutant. To quantitate the level of enhanced resistance of cyp83a1-3 to G. cichoracearum, we quantified fungal growth by counting the number of conidiophores per colony and found that the cyp83a1-3 mutant showed significantly fewer conidiophores per colony than the wild-type at 5 dpi (Figure 1D). These results are consistent with previous results showing that plants deficient in CYP83A1 are more resistant to a different powdery mildew species, E. cruciferarum (Weis et al., 2013).
FIGURE 1.
The cyp83a1-3 mutant displays enhanced resistance to Golovinomyces cichoracearum. (A) Four-weeks-old Arabidopsis wild-type and cyp83a1-3 mutant plants were infected with G. cichoracearum, and representative leaves were removed and photographed at 8 dpi. (B) Leaves were stained with trypan blue at 8 dpi, bar = 200 μm. (C) Leaves were stained with DAB and trypan blue at 2 dpi; arrows indicate H2O2 accumulation, bar = 200 μm. (D) Quantification of fungal growth in plants at 5 dpi by counting the number of conidiophores per colony. Results represent the mean and standard deviation in three independents experiments (n = 30). Asterisk represents statistically significant differences from wild-type (P < 0.01, nested ANOVA). (E) Schematic representation of the CYP83A1 protein, arrows indicate mutation sites of three cyp83a1 mutant alleles. (F) Four-weeks-old wild-type, cyp83a1-3, cyp83a1-1, cyp83a1-2, and transgenic cyp83a1-3 mutant plants complemented with the wild-type CYP83A1 gene (gCYP83A1) were infected with G. cichoracearum and representative leaves were stained with trypan blue at 8 dpi, bar = 200 μm. (G) Quantification of fungal growth in plants at 5 dpi by counting the number of conidiophores per colony. Results represent the mean and standard deviation in three independent experiments (n = 30; P < 0.01, nested ANOVA).
To identify the mutation in cyp83a1-3 responsible for the enhanced resistance phenotype, we carried out standard map-based cloning as described in Materials and Methods and identified a mutation (GA) in At4g13770, which encodes CYP83A1 and causes an amino acid substitution (G346E). The cytochrome P450 monooxygenase CYP83A1 functions in the biosynthesis of aliphatic glucosinolates from aliphatic oximes. Previous studies also identified mutations in the CYP83A1 gene. The cyp83a1-1 mutant (SALK_123405) contains a T-DNA insertion in the open reading frame of CYP83A1 (Weis et al., 2013), and cyp83a1-2/ref2-1 contains a loss-of-function point mutation leading to a premature stop codon (W58stop) in the CYP83A1 gene (Hemm et al., 2003; Figure 1E).
To correlate the mutation in cyp83a1-3 with the powdery mildew resistance phenotype, we cloned the wild-type CYP83A1 gene driven by its native promoter and transformed this construct into the cyp83a1-3 mutant. Arabidopsis wild-type Col-0 is susceptible to G. cichoracearum. This construct reversed the powdery mildew resistance phenotype in the cyp83a1-3 mutant. In addition, the allelic mutants cyp83a1-1 and cyp83a1-2 showed similar G. cichoracearum resistant phenotypes as cyp83a1-3 (Figures 1F,G). Taken together, these results indicate that the mutation in CYP83A1 in cyp83a1-3 causes the powdery mildew resistance phenotype.
Resistance of cyp83a1-3 to G. cichoracearum Does Not Require SA Signaling
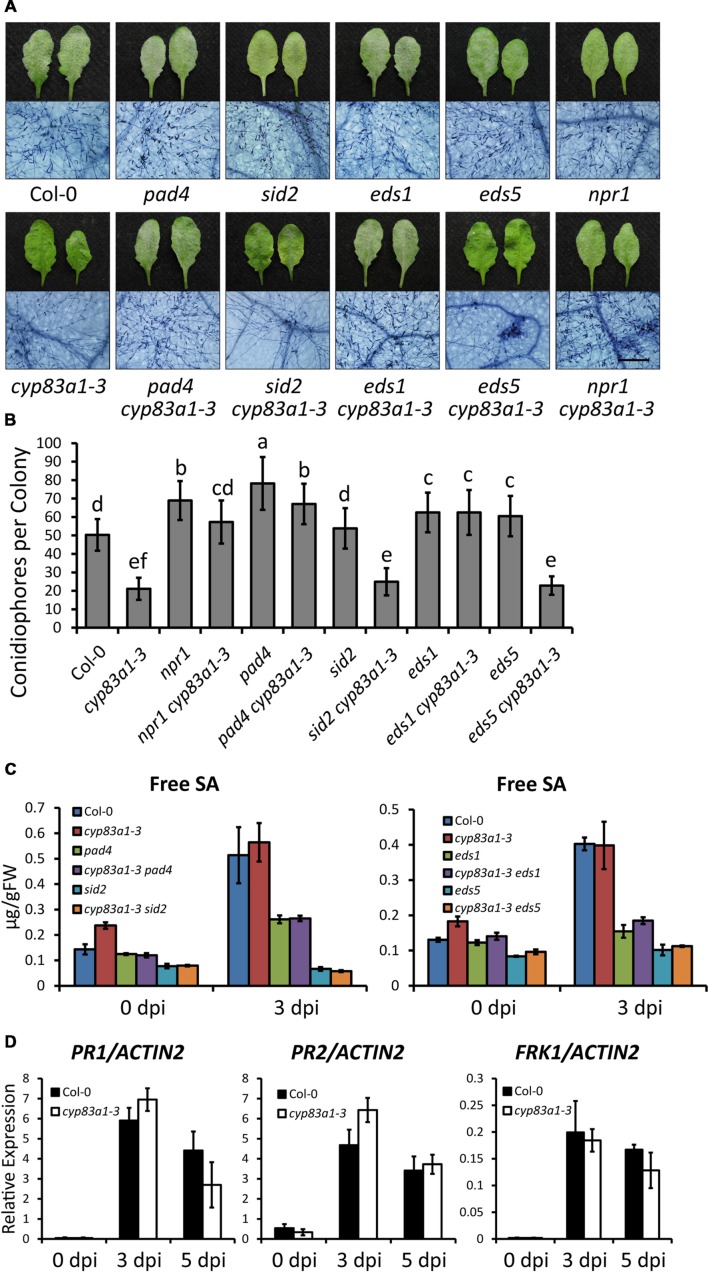
To investigate the cause of the resistance to G. cichoracearum in cyp83a1-3, we first examined whether the phytohormone SA, which plays an important role in resistance to biotrophic pathogens, is involved. We constructed double mutants by crossing cyp83a1-3 with sid2, eds5, npr1, pad4, and eds1, well-characterized mutants with defects in SA accumulation or signaling. Double mutants were identified by PCR amplification (Supplementary Figure S2). We then inoculated the wild-type, single, and double mutants with G. cichoracearum, and performed trypan blue staining at 8 dpi. As shown in Figure 2A, the resistance of cyp83a1-3 requires PAD4, EDS1, and NPR1, but not SID2 or EDS5. We also counted the number of conidiophores per colony in these plants, and the results were consistent with the staining assay (Figure 2B).
FIGURE 2.
The resistance in cyp83a1-3 is SA-independent. (A) Four-weeks-old Arabidopsis wild-type, cyp83a1-3 mutant, and double mutant plants were infected with G. cichoracearum, and representative leaves were removed and stained with trypan blue at 8 dpi, bar = 200 μm. (B) Quantification of fungal growth of the plants in (a) at 5 dpi by counting the number of conidiophores per colony. Results represent the mean and standard deviation in three independent experiments (n = 30). Different letters represent statistically significant differences (P < 0.01, nested ANOVA). (C) Free SA levels were measured in the uninfected and infected (3 dpi) leaves after inoculation with G. cichoracearum. FW, fresh weight. (D) Accumulation of PR1, PR2, and FRK1 transcripts in 4-weeks-old plants infected by G. cichoracearum examined by quantitative real-time PCR. Results represent the mean and standard deviation in three independent experiments (n = 4).
To further assess the role of SA in powdery mildew resistance in cyp83a1-3, we measured the accumulation of SA before and after G. cichoracearum infection. The cyp83a1-3 mutant accumulated similar levels of SA as the wild-type at 3 dpi, and the levels of SA induced by powdery mildew were suppressed by mutations in PAD4, SID2, EDS1, and EDS5 (Figure 2C). Mutation of SID2 or EDS5 suppressed SA accumulation in cyp83a1-3, but it did not suppress the resistance phenotype, indicating that the cyp83a1-3 powdery mildew resistance phenotype may not require SA.
In addition, we measured the expression of defense-related genes, including PR1 (PATHOGENESIS-RELATED GENE1), PR2, and FRK1 (FLG22-INDUCED RECEPTOR-LIKE KINASE1), and found that the cyp83a1-3 mutant accumulated similar levels of PR1, PR2, and FRK1 transcripts as the wild-type at both 3 and 5 days after infection with G. cichoracearum (Figure 2D). Taken together, these results indicate that the resistance of cyp83a1-3 to G. cichoracearum is dependent on PAD4, EDS1, and NPR1, but does not appear to be due to the increased accumulation of SA.
The Expression of Genes Related to Camalexin Synthesis Increase upon G. cichoracearum Infection
To understand what causes powdery mildew resistance in cyp83a1-3 mutants, we analyzed the secondary metabolic network that involves CYP83A1 (Supplementary Figure S3). Since the biosynthetic pathways for production of aliphatic glucosinolates and many indole-derived compounds are closely related, defects in CYP83A1 could change other indole-derived pathways and in turn affect powdery mildew resistance.
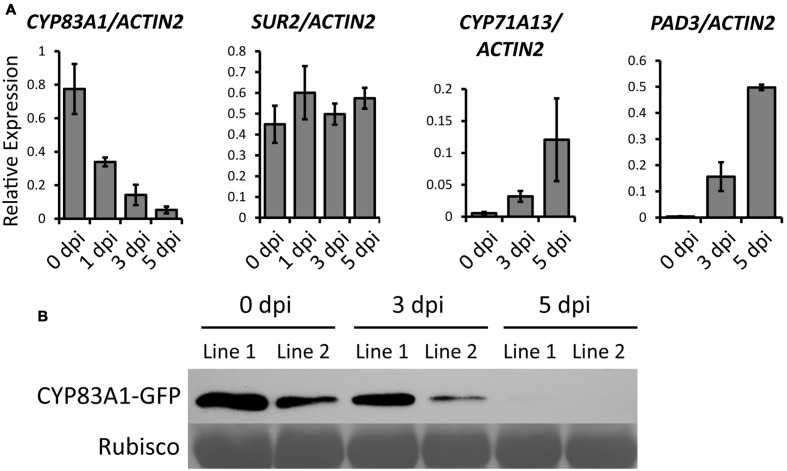
To investigate which alkylglucosinolate synthesis-related pathway contributes to the powdery mildew resistance, we first analyzed the expression of genes in three pathways, including the alkylglucosinolate pathway, indole glucosinolate pathway, and camalexin pathway, upon powdery mildew infection in Col-0 wild-type plants. We chose CYP83A1 to represent the alkylglucosinolate pathway, CYP83B1/SUR2 for the indole glucosinolate pathway, and CYP71A13 and CYP71B15/PAD3 for the camalexin pathway. We examined the expression of these genes using quantitative RT-PCR before and after G. cichoracearum infection. As shown in Figure 3A, the transcript levels of CYP83A1 decreased, the transcript level of SUR2 remained the same, but transcript levels of the camalexin synthetase genes CYP71A13 and PAD3 increased upon infection. These results suggested that camalexin may play an important role in the resistance to G. cichoracearum in Arabidopsis.
FIGURE 3.
Transcript accumulation of genes related to glucosinolate or camalexin biosynthesis upon G. cichoracearum infection. (A) Four-weeks-old wild-type plants were infected with G. cichoracearum, and the accumulation of CYP83A1, SUR2, CYP71A13, and PAD3 transcripts was examined by quantitative real-time PCR. Results represent the mean and standard deviation in three independent experiments (n = 4). (B) Four-weeks-old CYP83A1-GFP transgenic plants were infected with G. cichoracearum and immunoblot analysis was performed using an anti-GFP antibody. The large subunit of Rubisco was used as a protein loading control.
To examine whether CYP83A1 protein levels also decrease during powdery mildew infection, we constructed a plasmid to express a CYP83A1-GFP chimeric protein by fusing the genomic CYP83A1 sequence to a C-terminal GFP sequence driven by the native CYP83A1 promoter, and transformed it into the cyp83a1-3 mutant. The transgene rescued the mutant phenotype (Supplementary Figures S4A,B), indicating that CYP83A1-GFP was functional. We inoculated the CYP83A1-GFP transgenic plants with powdery mildew and examined the accumulation of CYP83A1-GFP by immunoblot with a GFP antibody. As shown in Figure 3B, the CYP83A1 protein level decreased during infection, consistent with the observation that the CYP83A1 mRNA level also decreased.
The cyp83a1-3 Mutant Accumulates High Levels of Camalexin and Resistance Requires the Camalexin Synthetase PAD3
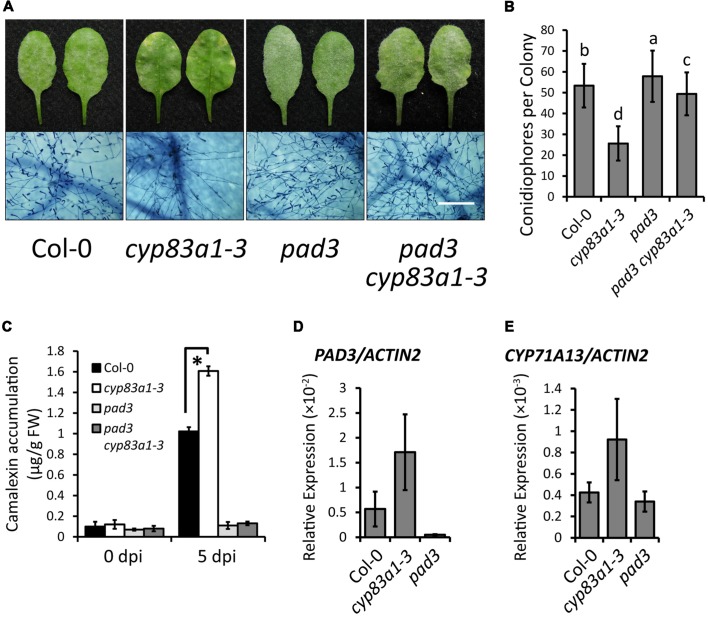
Since the biosynthetic pathways of glucosinolates and camalexin involve the same intermediates, and the expression of genes related to camalexin synthesis increase upon G. cichoracearum infection (Figure 3A), we hypothesized that increased levels of camalexin in the cyp83a1-3 mutant compared to wild-type plants may be the reason that cyp83a1-3 displays enhanced powdery mildew resistance. To test this hypothesis, we first constructed double mutants of cyp83a1-3 with pad3, which accumulates lower levels of camalexin, compared with wild-type (Glazebrook and Ausubel, 1994). We then infected the double mutants with G. cichoracearum. As shown in Figures 4A,B, the pad3 mutation suppressed the resistance phenotype of cyp83a1-3, indicating that the cyp83a1-3 phenotype requires the PAD3 camalexin synthetase (CYP71B15).
FIGURE 4.
The cyp83a1-3 mutant accumulates high levels of camalexin upon G. cichoracearum infection, which is suppressed by mutation of PAD3. (A) Four-weeks-old wild-type, cyp83a1-3, pad3 mutant, and double-mutant plants were infected with G. cichoracearum. Representative leaves were removed and stained with trypan blue at 8 dpi, bar = 200 μm. (B) Quantification of fungal growth of the plants in (a) at 5 dpi by counting the number of conidiophores per colony. Results represent the mean and standard deviation in three independent experiments (n = 30; P < 0.01, nested ANOVA). (C) Four-weeks-old plants were infected with G. cichoracearum. Camalexin accumulation was determined at 0 and 5 dpi. Results represent the mean and standard deviation in three experiments (n = 3). Asterisk represents statistically significant difference from wild-type (P < 0.01, nested ANOVA). (D) The transcript accumulation of PAD3 was examined by quantitative real-time PCR on samples from 4-weeks-old wild-type, cyp83a1-3 and pad3 mutant plants. (E) The transcript accumulation of CYP71A13 examined by quantitative real-time PCR. Results represent the mean and standard deviation in three independent experiments (n = 4).
To further assess the role of camalexin in cyp83a1-3 resistance, we also measured the camalexin levels in cyp83a1-3 before infection and at 5 dpi with G. cichoracearum. As shown in Figure 4C, after infection, the cyp83a1-3 mutant accumulated significantly more camalexin than the wild-type, and this increase was suppressed by a mutation in PAD3. In addition, we found that the transcript levels of the camalexin synthesis genes PAD3 and CYP71A13 were higher in cyp83a1-3 than in the wild-type in the absence of pathogen (Figures 4D,E).
Taken together, these results demonstrate that cyp83a1-3 accumulates more camalexin compared with wild-type plants, and that the camalexin synthetase PAD3 is required for cyp83a1-3-mediated resistance, thus suggesting that the phytoalexin camalexin contributes to powdery mildew resistance.
The Resistance in cyp83a1-3 Mutants Requires the Camalexin Synthesis Regulator AtWRKY33
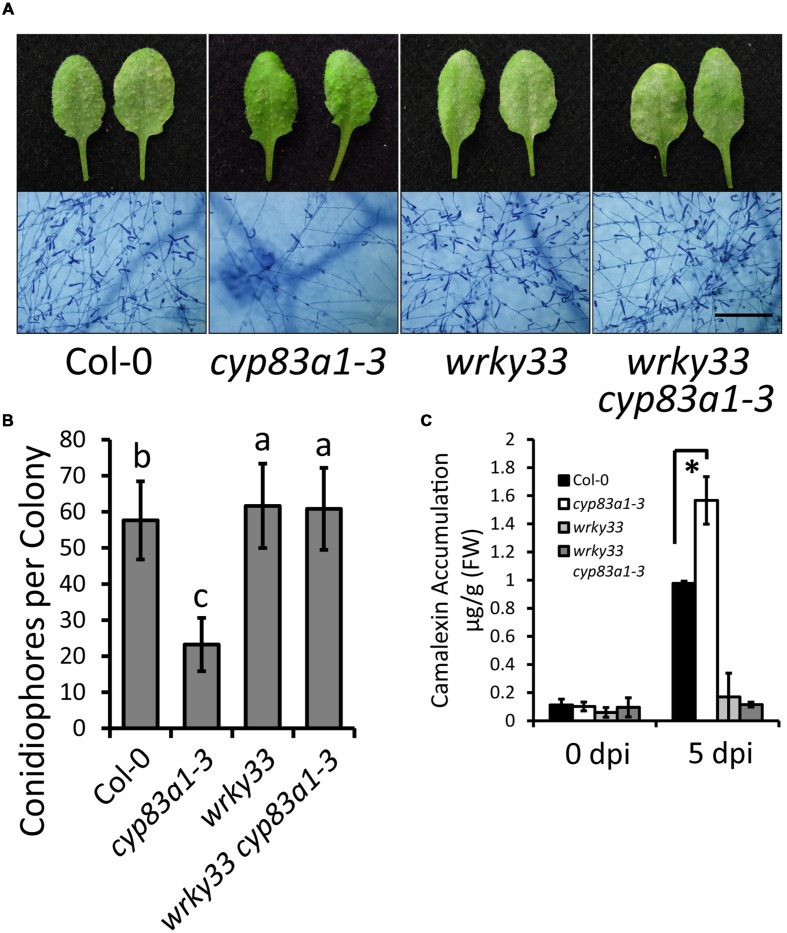
To further confirm the role of camalexin accumulation in cyp83a1-3 resistance, we tested whether the resistance in cyp83a1-3 mutants requires AtWRKY33, a transcription factor that positively regulates the expression of many camalexin synthetase genes. We constructed a cyp83a1-3 wrky33 double mutant and examined the powdery mildew responses and camalexin levels in the single and double mutants. As shown in Figures 5A–C, the wrky33 mutation not only suppressed powdery mildew resistance, but it also decreased camalexin accumulation in the cyp83a1-3 mutant.
FIGURE 5.
The resistance phenotype and high levels of camalexin in cyp83a1-3 is suppressed by mutation of WRKY33. (A) Four-weeks-old wild-type, cyp83a1-3, wrky33, and wrky33 cyp83a1-3 double-mutant plants were infected with G. cichoracearum. Representative leaves were removed and stained with trypan blue at 8 dpi, bar = 200 μm. (B) Quantification of fungal growth of the plants in (a) at 5 dpi by counting the number of conidiophores per colony. Results represent the mean and standard deviation in three independent experiments (n = 30; P < 0.01, nested ANOVA). (C) Camalexin accumulation of the plants in (A) was determined at 0 and 5 dpi. Results represent the mean and standard deviation in three independent experiments (n = 3). Asterisk represents statistically significant difference from wild-type (P < 0.01, nested ANOVA).
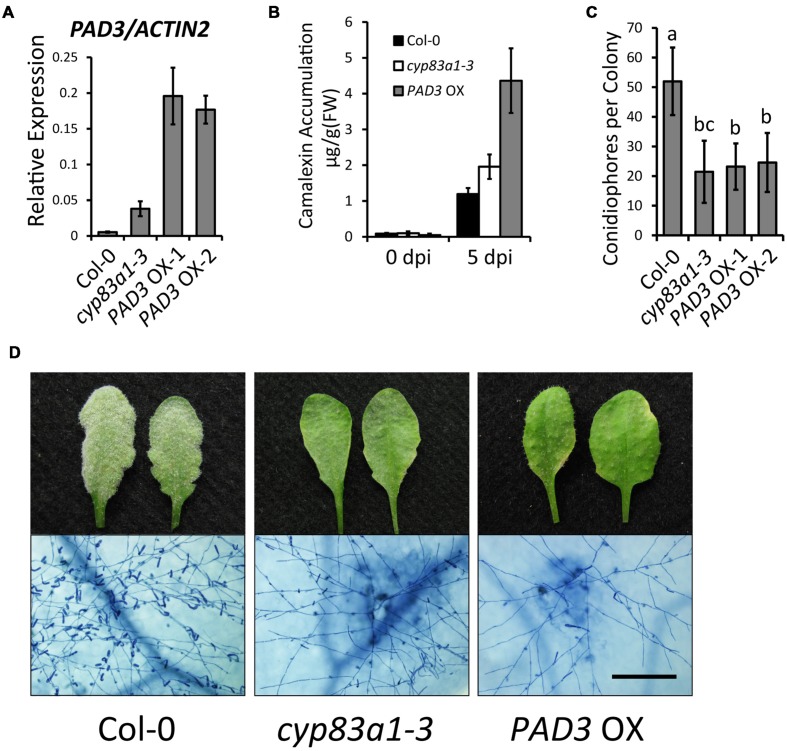
Overexpression of PAD3 Leads to Increased Camalexin Accumulation and Enhanced Resistance to G. cichoracearum
To further confirm the role of camalexin in powdery mildew resistance, we constructed transgenic plants that overexpress PAD3 (PAD3-OX). For this, we made a construct with the PAD3 coding sequence driven by the 35S promoter and transformed this construct into wild-type plants. We used quantitative RT-PCR to measure the PAD3 expression levels of two independent lines of T3 generation PAD3-OX plants (Figure 6A). After G. cichoracearum infection, the PAD3-OX plants showed higher camalexin accumulation at 5 dpi, compared with wild-type (Figure 6B). The PAD3-OX plants also displayed enhanced resistance to powdery mildew compared with the wild-type (Figures 6C,D), similar to the cyp83a1-3 mutant. These results provide further evidence that higher levels of camalexin cause enhanced resistance to G. cichoracearum. Taken together, our findings revealed that the resistance to G. cichoracearum in cyp83a1-3 is at least partially due to higher levels of camalexin accumulation.
FIGURE 6.
PAD3-overexpressing plants accumulate higher levels of camalexin and display a cyp83a1-3-like resistance phenotype. (A) The accumulation of PAD3 transcript in 4-weeks-old plants was examined by quantitative real-time PCR. Results represent the mean and standard deviation in three independent experiments (n = 4). (B) Four-weeks-old plants were infected with G. cichoracearum and camalexin accumulation was determined at 0 and 5 dpi. Three PAD3-OX independent lines were tested, and similar results were obtained in these lines. One representative PAD3-OX line (PAD3-OX-1) is shown. Results represent the mean and standard deviation in three experiments (n = 3). (C) Quantification of fungal growth of wild-type, cyp83a1-3, and PAD3-OX plants at 5 dpi by counting the number of conidiophores per colony. Results represent the mean and standard deviation in three independent experiments (n = 30). Different letters represent statistically significant differences (P < 0.01, nested ANOVA). (D) Four-weeks-old wild-type, cyp83a1-3, and PAD3-OX-1 plants were infected with G. cichoracearum. Three PAD3-OX independent lines were examined, and similar results were obtained in these three lines. Representative leaves were removed and stained with trypan blue at 8 dpi, bar = 200 μm.
Discussion
The cyp83a1-3 mutant displays enhanced resistance to powdery mildews, including G. cichoracearum. The levels of SA accumulation and PR gene expression in cyp83a1-3 mutants were similar to the wild-type, and double mutant analysis showed that resistance in cyp83a1-3 was dependent on PAD4, EDS1, and NPR1 but independent of SID2 and EDS5. Mutations in SID2 or EDS5 suppressed SA accumulation induced by powdery mildew but did not suppress the disease resistance, indicating that resistance in cyp83a1-3 is not caused by enhanced SA signaling. These observations indicate that cyp83a1-3-mediated resistance differs from that in the edr1 and edr2 mutants (Frye et al., 2001; Tang et al., 2005b). Moreover, the cyp83a1-3 mutant accumulates more camalexin upon powdery mildew infection, and mutations in PAD3 or WRKY33 suppressed both the disease resistance and the high accumulation of camalexin, indicating a link between camalexin levels and responses to powdery mildew. Consistent with a role of camalexin in powdery mildew resistance, the PAD3-overexpressing plants accumulated more camalexin and mimicked the resistance phenotype of the cyp83a1-3 mutant. Taken together, those findings indicate that the higher level of camalexin contributes to the enhanced resistance to G. cichoracearum observed in the cyp83a1-3 mutant. Our finding that eds1 and pad4 mutations suppressed powdery mildew resistance in cyp83a1 is consistent with previous work showing that eds1 and pad4 have reduced levels of camalexin (Glazebrook et al., 1997; Mert-Türk et al., 2003). It would be interesting to examine whether NPR1 contributes to camalexin accumulation since NPR1 is also required for powdery mildew resistance in cyp83a1.
Our finding that camalexin plays an important role in powdery mildew resistance is also consistent with previous work showing that loss-of-function mutations in two transcription factors, WRKY18 and WRKY40, results in the accumulation of higher levels of camalexin as well as both preinvasive and post-invasive resistance against the powdery mildew fungus G. orontii (Pandey et al., 2010; Schön et al., 2013). Similar to our results, these authors also found that PAD3 (a key enzyme in camalexin biosynthesis) is required for the preinvasive resistance (but surprisingly not for the post-invasive resistance) against G. orontii in a wrky18 wrky40 background (Schön et al., 2013). Moreover, Schön et al. (2013) also report that wrky18 wrky40 plants do not show increased resistance against two other powdery mildews, G. cichoracearum and G. cruciferarum. The reason why we observed enhanced resistance to G. cichoracearum in cyp83a1-3 whereas Schön et al. (2013) did not observe enhanced resistance in wrky18 wrky40 is not clear, but may be related to differences in camalexin levels in the two mutants or to differences in susceptibility of different G. cichoracearum isolates to camalexin.
The function of CYP83A1 has been studied previously. An earlier study showed that overexpression of CYP83A1 could rescue the auxin-excess phenotype of cyp83b1/rnt1/sur2 mutants (Bak and Feyereisen, 2001). Further studies showed that levels of many phenylpropanoid pathway-derived products were reduced in the cyp83a1-2/ref2-1 mutant, indicating crosstalk between the pathways producing aliphatic glucosinolates and indole glucosinolates (Hemm et al., 2003; Naur et al., 2003). Recent work showed that CYP83A1 interacts with BAX INHIBITOR-1, a cell death suppressor in plants and animals. The loss-of-function mutants cyp83a1-1 and cyp83a1-2 displayed enhanced resistance to the powdery mildew fungus E. cruciferarum (Weis et al., 2013). A more recent study measured the levels of several glucosinolates in cyp83a1-1 mutants, but found only marginally increased amounts of indole-derived glucosinolates. The cyp83a1 mutants lack very-long-chain aldehydes and accumulate more 5-methylthiopentanaldoxime (5-MPTO), a potentially toxic substrate of CYP83A1 (Weis et al., 2014). As very-long-chain aldehydes promote germination and appressorium formation of E. cruciferarum, it was proposed that lack of very-long-chain aldehydes causes the resistance phenotypes in cyp83a1 mutants (Weis et al., 2014).
Here, we showed that the high level of camalexin contributes to resistance to the powdery mildew fungus G. cichoracearum. It is worth noting that different species of powdery mildew, G. cichoracearum and E. cruciferarum, were used in our study and in the Weis et al. (2014) study, respectively. Although cyp83a1 mutants displayed enhanced resistance to both powdery mildew strains, the mechanisms could differ. Consistent with this notion, several studies observed differences in infection phenotypes between different powdery mildew species in Arabidopsis. For example, G. cichoracearum and G. orontii have different host ranges/responses (Plotnikova et al., 1998) and many Arabidopsis accessions show different responses to the powdery mildew species E. cruciferarum UEA1 and G. cichoracearum UCSC1 (Adam et al., 1999). Here, we showed that pad3 and wrky33 suppressed the accumulation of camalexin and the enhanced resistance in cyp83a1-3 mutants, indicating a role of camalexin in cyp83a1-3-mediated resistance. It would be interesting to examine the responses of pad3 cyp83a1-3 and wrky33 cyp83a1-3 mutants to E. cruciferarum, and to measure the levels of very-long-chain aldehydes and 5-MPTO in those mutants. It is also possible that both very-long-chain aldehydes and camalexin contribute to resistance against to G. cichoracearum and E. cruciferarum.
CYP83A1 functions in the biosynthesis of aliphatic glucosinolates, so one interesting question is how the mutation of the glucosinolate synthetase gene CYP83A1 affects the accumulation of camalexin. One explanation is that it may cause crosstalk within the complicated metabolic network. In this scenario, the biosynthetic pathway of aliphatic glucosinolates, which involves CYP83A1, and indole glucosinolates, which share the IAOx intermediate with camalexin (Nafisi et al., 2007; Schuhegger et al., 2007a; Bottcher et al., 2009), can affect each other. So when the aliphatic glucosinolates pathway is blocked in the cyp83a1 mutant, the pathway for indole-derived products, including indole glucosinolates and camalexin, is enhanced. Indole glucosinolates are known to contribute to plant immunity (Bednarek, 2012). Arabidopsis PENETRATION2 (PEN2), which initiates indole glucosinolate metabolism, plays an important role in penetration resistance (Bednarek et al., 2009; Clay et al., 2009). In addition to increased levels of camalexin, the cyp83a1 mutant has several other aberrant phenotypes including decreased levels of very-long-chain aldehydes and alterations in many metabolites that could also contribute to resistance, so the exact mechanism of powdery mildew resistance in cyp83a1 needs to be further studied. Consistent with this notion, PAD3-overexpression plants accumulate much higher levels of camalexin than the cyp83a1-3 mutant, but no further increase in powdery mildew resistance was observed. It would be interesting to measure very-long-chain aldehydes and indole-derived products in pad3 cyp83a1 double mutants to further examine whether altered camalexin levels or differences in other metabolites are responsible for the powdery mildew resistance phenotype in cyp83a1.
Conclusion
We showed that camalexin plays an important role in resistance to the powdery mildew pathogen G. cichoracearum. Our study provides new insights into the role of camalexin in plant immunity against powdery mildew.
Author Contributions
DT, SL, and FA designed the project. SL, LB, and SV performed experiments. SL, LB, SV, FA, and DT discussed and interpreted results. SL and DT wrote the manuscript. All authors carefully read and edited the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Shuqun Zhang for purified camalexin and Julia Dewdney and Carine Denoux for identifying the mutation in CYP83A1 in the cyp83a1-3 mutant, Ruslan Sadreyev for assistance with the statistical analysis, and Zhenyu Cheng for reading the manuscript.
Footnotes
Funding. The work was supported by grants from the Strategic Priority Research Program of the Chinese Academy of Sciences (XDB11020100), National Science Fund for Distinguished Young Scholars of China (31525019), the Ministry of Science and Technology of China (2014DFA31540) to DT, and by U.S. National Institutes of Health grant R37 GM48707 and U.S. National Science Foundation grants MCB-0519898 and IOS 0929226 to FA.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2016.00227
FIGURE S1 | The growth phenotype of the cyp83a1-3 mutant is similar to wild-type in the absence of G. cichoracearum. Wild-type and cyp83a1-3 mutants were grown under standard short day conditions. Five-weeks-old plants were photographed.
FIGURE S2 | Identification of double mutants by PCR amplification. The pad4-1 cyp83a1-3, sid2-2 cyp83a1-3, eds1-2 cyp83a1-3 eds5-1 cyp83a1-3, npr1-63 cyp83a1-3, and wrky33-2 cyp83a1-3 mutants were identified by PCR amplification.
FIGURE S3 | The biosynthetic network of glucosinolates and related compounds in Arabidopsis. Schematic representation of the biosynthesis pathway derived from tryptophan and methionine.
FIGURE S4 | CYP83A1-GFP rescued the powdery mildew-resistant phenotype of cyp83a1-3. (A) Four-weeks-old wild-type, cyp83a1-3, and cyp83a1-3 transgenic plants expressing the CYP83A1-GFP fusion protein driven by native promoter were infected with G. cichoracearum. Representative leaves were removed (upper panel) and stained with trypan blue (lower panel) at 8 dpi, bar = 200 μm. (B) Quantification of fungal growth of the plants in (a) at 5 dpi by counting the number of conidiophores per colony. Results represent the mean and standard deviation of three independent experiments (n = 30). Different letters represent statistically significant differences (P < 0.01, nested ANOVA).
References
- Adam L., Ellwood S., Wilson I., Saenz G., Xiao S., Oliver R. P., et al. (1999). Comparison of Erysiphe cichoracearum and E. cruciferarum and a survey of 360 Arabidopsis thaliana accessions for resistance to these two powdery mildew pathogens. Mol. Plant-Microbe Interact. 12 1031–1043. 10.1094/MPMI.1999.12.12.1031 [DOI] [PubMed] [Google Scholar]
- Alonso J. M., Stepanova A. N., Leisse T. J., Kim C. J., Chen H., Shinn P., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657. 10.1126/science.1086391 [DOI] [PubMed] [Google Scholar]
- Bak S., Feyereisen R. (2001). The involvement of two p450 enzymes, CYP83B1 and CYP83A1, in auxin homeostasis and glucosinolate biosynthesis. Plant Physiol. 127 108–118. 10.1104/pp.127.1.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartsch M., Gobbato E., Bednarek P., Debey S., Schultze J. L., Bautor J., et al. (2006). Salicylic acid-independent ENHANCED DISEASE SUSCEPTIBILITY1 signaling in Arabidopsis immunity and cell death is regulated by the monooxygenase FMO1 and the nudix hydrolase NUDT7. Plant Cell 18 1038–1051. 10.1105/tpc.105.039982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bednarek P. (2012). Chemical warfare or modulators of defence responses - the function of secondary metabolites in plant immunity. Curr. Opin. Plant Biol. 15 407–414. 10.1016/j.pbi.2012.03.002 [DOI] [PubMed] [Google Scholar]
- Bednarek P., Pislewska-Bednarek M., Svatos A., Schneider B., Doubsky J., Mansurova M., et al. (2009). A glucosinolate metabolism pathway in living plant cells mediates broad-spectrum antifungal defense. Science 323 101–106. 10.1126/science.1163732 [DOI] [PubMed] [Google Scholar]
- Bottcher C., Westphal L., Schmotz C., Prade E., Scheel D., Glawischnig E. (2009). The multifunctional enzyme CYP71B15 (PHYTOALEXIN DEFICIENT3) converts cysteine-indole-3-acetonitrile to camalexin in the indole-3-acetonitrile metabolic network of Arabidopsis thaliana. Plant Cell 21 1830–1845. 10.1105/tpc.109.066670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao H., Bowling S. A., Gordon A. S., Dong X. N. (1994). Characterization of an Arabidopsis mutant that is nonresponsive to inducers of systemic acquired-resistance. Plant Cell 6 1583–1592. 10.1105/tpc.6.11.1583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clay N. K., Adio A. M., Denoux C., Jander G., Ausubel F. M. (2009). Glucosinolate metabolites required for an Arabidopsis innate immune response. Science 323 95–101. 10.1126/science.1164627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S. J., Bent A. F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16 735–743. 10.1046/j.1365-313x.1998.00343.x [DOI] [PubMed] [Google Scholar]
- Consonni C., Bednarek P., Humphry M., Francocci F., Ferrari S., Harzen A., et al. (2010). Tryptophan-derived metabolites are required for antifungal defense in the Arabidopsis mlo2 mutant. Plant Physiol. 152 1544–1561. 10.1104/pp.109.147660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consonni C., Humphry M. E., Hartmann H. A., Livaja M., Durner J., Westphal L., et al. (2006). Conserved requirement for a plant host cell protein in powdery mildew pathogenesis. Nat. Genet. 38 716–720. 10.1038/ng1806 [DOI] [PubMed] [Google Scholar]
- Earley K. W., Haag J. R., Pontes O., Opper K., Juehne T., Song K. M., et al. (2006). Gateway-compatible vectors for plant functional genomics and proteomics. Plant J. 45 616–629. 10.1111/j.1365-313X.2005.02617.x [DOI] [PubMed] [Google Scholar]
- Ferrari S., Plotnikova J. M., De Lorenzo G., Ausubel F. M. (2003). Arabidopsis local resistance to Botrytis cinerea involves salicylic acid and camalexin and requires EDS4 and PAD2, but not SID2, EDS5 or PAD4. Plant J. 35 193–205. 10.1046/j.1365-313X.2003.01794.x [DOI] [PubMed] [Google Scholar]
- Frye C. A., Innes R. W. (1998). An Arabidopsis mutant with enhanced resistance to powdery mildew. Plant Cell 10 947–956. 10.1105/tpc.10.6.947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frye C. A., Tang D., Innes R. W. (2001). Negative regulation of defense responses in plants by a conserved MAPKK kinase. Proc. Natl. Acad. Sci. U.S.A. 98 373–378. 10.1073/pnas.011405198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glazebrook J., Ausubel F. M. (1994). Isolation of phytoalexin-deficient mutants of Arabidopsis thaliana and characterization of their interactions with bacterial pathogens. Proc. Natl. Acad. Sci. U.S.A. 91 8955–8959. 10.1073/pnas.91.19.8955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glazebrook J., Zook M., Mert F., Kagan I., Rogers E. E., Crute I. R., et al. (1997). Phytoalexin-deficient mutants of Arabidopsis reveal that PAD4 encodes a regulatory factor and that four PAD genes contribute to downy mildew resistance. Genetics 146 381–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gou M. Y., Su N., Zheng J., Huai J. L., Wu G. H., Zhao J. F., et al. (2009). An F-box gene, CPR30, functions as a negative regulator of the defense response in Arabidopsis. Plant J. 60 757–770. 10.1111/j.1365-313X.2009.03995.x [DOI] [PubMed] [Google Scholar]
- Grubb C. D., Abel S. (2006). Glucosinolate metabolism and its control. Trends Plant Sci. 11 89–100. 10.1016/j.tplants.2005.12.006 [DOI] [PubMed] [Google Scholar]
- Halkier B. A., Gershenzon J. (2006). Biology and biochemistry of glucosinolates. Annu. Rev. Plant Biol. 57 303–333. 10.1146/annurev.arplant.57.032905.105228 [DOI] [PubMed] [Google Scholar]
- Hammerschmidt R. (1999). Phytoalexins: what have we learned after 60 years? Annu. Rev. Phytopathol. 37 285–306. 10.1146/annurev.phyto.37.1.285 [DOI] [PubMed] [Google Scholar]
- Hemm M. R., Ruegger M. O., Chapple C. (2003). The Arabidopsis ref2 mutant is defective in the gene encoding CYP83A1 and shows both phenylpropanoid and glucosinolate phenotypes. Plant Cell 15 179–194. 10.1105/tpc.006544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jirage D., Tootle T. L., Reuber T. L., Frost L. N., Feys B. J., Parker J. E., et al. (1999). Arabidopsis thaliana PAD4 encodes a lipase-like gene that is important for salicylic acid signaling. Proc. Natl. Acad. Sci. U.S.A. 96 13583–13588. 10.1073/pnas.96.23.13583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kliebenstein D. J., Rowe H. C., Denby K. J. (2005). Secondary metabolites influence Arabidopsis/Botrytis interactions: variation in host production and pathogen sensitivity. Plant J. 44 25–36. 10.1111/j.1365-313X.2005.02508.x [DOI] [PubMed] [Google Scholar]
- Koch E., Slusarenko A. (1990). Arabidopsis is susceptible to infection by a downy mildew fungus. Plant Cell 2 437–445. 10.1105/tpc.2.5.437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao G., Meng X., Liu Y., Zheng Z., Chen Z., Zhang S. (2011). Phosphorylation of a WRKY transcription factor by two pathogen-responsive MAPKs drives phytoalexin biosynthesis in Arabidopsis. Plant Cell 23 1639–1653. 10.1105/tpc.111.084996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mert-Türk F., Bennett M. H., Mansfield J. W., Holub E. B. (2003). Camalexin accumulation in Arabidopsis thaliana following abiotic elicitation or inoculation with virulent or avirulent Hyaloperonospora parasitica. Physiol. Mol. Plant Pathol. 62 137–145. 10.1016/s0885-5765(03)00047-x [DOI] [Google Scholar]
- Nafisi M., Goregaoker S., Botanga C. J., Glawischnig E., Olsen C. E., Halkier B. A., et al. (2007). Arabidopsis cytochrome P450 monooxygenase 71A13 catalyzes the conversion of indole-3-acetaldoxime in camalexin synthesis. Plant Cell 19 2039–2052. 10.1105/tpc.107.051383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naur P., Petersen B. L., Mikkelsen M. D., Bak S., Rasmussen H., Olsen C. E., et al. (2003). CYP83A1 and CYP83B1, two nonredundant cytochrome P450 enzymes metabolizing oximes in the biosynthesis of glucosinolates in Arabidopsis. Plant Physiol. 133 63–72. 10.1104/pp.102.019240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawrath C., Heck S., Parinthawong N., Metraux J. P. (2002). EDS5, an essential component of salicylic acid-dependent signaling for disease resistance in Arabidopsis, is a member of the MATE transporter family. Plant Cell 14 275–286. 10.1105/tpc.010376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie H., Wu Y., Yao C., Tang D. (2011). Suppression of edr2-mediated powdery mildew resistance, cell death and ethylene-induced senescence by mutations in ALD1 in Arabidopsis. J. Genet. Genomics 38 137–148. 10.1016/j.jgg.2011.03.001 [DOI] [PubMed] [Google Scholar]
- Nie H., Zhao C., Wu G., Wu Y., Chen Y., Tang D. (2012). SR1, a calmodulin-binding transcription factor, modulates plant defense and ethylene-induced senescence by directly regulating NDR1 and EIN3. Plant Physiol. 158 1847–1859. 10.1104/pp.111.192310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey S. P., Roccaro M., Schon M., Logemann E., Somssich I. E. (2010). Transcriptional reprogramming regulated by WRKY18 and WRKY40 facilitates powdery mildew infection of Arabidopsis. Plant J. 64 912–923. 10.1111/j.1365-313X.2010.04387.x [DOI] [PubMed] [Google Scholar]
- Pedras M. S. C., Adio A. M. (2008). Phytoalexins and phytoanticipins from the wild crucifers Thellungiella halophila and Arabidopsis thaliana: rapalexin A, wasalexins and camalexin. Phytochemistry 69 889–893. 10.1016/j.phytochem.2007.10.032 [DOI] [PubMed] [Google Scholar]
- Pedras M. S. C., Yaya E. E., Glawischnig E. (2011). The phytoalexins from cultivated and wild crucifers: chemistry and biology. Nat. Prod. Rep. 28 1381–1405. 10.1039/C1np00020a [DOI] [PubMed] [Google Scholar]
- Plotnikova J. M., Reuber T. L., Ausubel F. M. (1998). Powdery mildew pathogenesis of Arabidopsis thaliana. Mycologia 90 1009–1016. 10.2307/3761274 [DOI] [Google Scholar]
- Qiu J. L., Fiil B. K., Petersen K., Nielsen H. B., Botanga C. J., Thorgrimsen S., et al. (2008). Arabidopsis MAP kinase 4 regulates gene expression through transcription factor release in the nucleus. EMBO J. 27 2214–2221. 10.1038/emboj.2008.147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlaeppi K., Abou-Mansour E., Buchala A., Mauch F. (2010). Disease resistance of Arabidopsis to Phytophthora brassicae is established by the sequential action of indole glucosinolates and camalexin. Plant J. 62 840–851. 10.1111/j.1365-313X.2010.04197.x [DOI] [PubMed] [Google Scholar]
- Schön M., Töller A., Diezel C., Roth C., Westphal L., Wiermer M., et al. (2013). Analyses of wrky18 wrky40 plants reveal critical roles of SA/EDS1 signaling and indole-glucosinolate biosynthesis for Golovinomyces orontii resistance and a loss-of resistance towards Pseudomonas syringae pv. tomato AvrRPS4. Mol. Plant-Microbe Interact. 26 758–767. 10.1094/MPMI-11-12-0265-R [DOI] [PubMed] [Google Scholar]
- Schuhegger R., Nafisi M., Mansourova M., Petersen B. L., Olsen C. E., Svatos A., et al. (2007a). CYP71B15 (PAD3) catalyzes the final step in camalexin biosynthesis - Correction. Plant Physiol. 145 1086–1086. 10.1104/pp.104.900240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuhegger R., Rauhut T., Glawischnig E. (2007b). Regulatory variability of camalexin biosynthesis. J. Plant Physiol. 164 636–644. 10.1016/j.jplph.2006.04.012 [DOI] [PubMed] [Google Scholar]
- Shi H., Shen Q., Qi Y., Yan H., Nie H., Chen Y., et al. (2013). BR-SIGNALING KINASE1 physically associates with FLAGELLIN SENSING2 and regulates plant innate immunity in Arabidopsis. Plant Cell 25 1143–1157. 10.1105/tpc.112.107904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonderby I. E., Geu-Flores F., Halkier B. A. (2010). Biosynthesis of glucosinolates–gene discovery and beyond. Trends Plant Sci. 15 283–290. 10.1016/j.tplants.2010.02.005 [DOI] [PubMed] [Google Scholar]
- Tang D., Ade J., Frye C. A., Innes R. W. (2005a). Regulation of plant defense responses in Arabidopsis by EDR2, a PH and START domain-containing protein. Plant J. 44 245–257. 10.1111/j.1365-313X.2005.02523.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang D., Christiansen K. M., Innes R. W. (2005b). Regulation of plant disease resistance, stress responses, cell death, and ethylene signaling in Arabidopsis by the EDR1 protein kinase. Plant Physiol. 138 1018–1026. 10.1104/pp.105.060400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomma B. P. H. J., Nelissen I., Eggermont K., Broekaert W. F. (1999). Deficiency in phytoalexin production causes enhanced susceptibility of Arabidopsis thaliana to the fungus Alternaria brassicicola. Plant J. 19 163–171. 10.1046/j.1365-313X.1999.00513.x [DOI] [PubMed] [Google Scholar]
- Wang Y., Nishimura M. T., Zhao T., Tang D. (2011). ATG2, an autophagy-related protein, negatively affects powdery mildew resistance and mildew-induced cell death in Arabidopsis. Plant J. 68 74–87. 10.1111/j.1365-313X.2011.04669.x [DOI] [PubMed] [Google Scholar]
- Weis C., Hildebrandt U., Hoffmann T., Hemetsberger C., Pfeilmeier S., Konig C., et al. (2014). CYP83A1 is required for metabolic compatibility of Arabidopsis with the adapted powdery mildew fungus Erysiphe cruciferarum. New Phytol. 202 1310–1319. 10.1111/nph.12759 [DOI] [PubMed] [Google Scholar]
- Weis C., Pfeilmeier S., Glawischnig E., Isono E., Pachl F., Hahne H., et al. (2013). Co-immunoprecipitation-based identification of putative BAX INHIBITOR-1-interacting proteins involved in cell death regulation and plant-powdery mildew interactions. Mol. Plant Pathol. 14 791–802. 10.1111/mpp.12050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wildermuth M. C., Dewdney J., Wu G., Ausubel F. M. (2001). Isochorismate synthase is required to synthesize salicylic acid for plant defence. Nature 414 562–565. 10.1038/35107108 [DOI] [PubMed] [Google Scholar]
- Wu G., Liu S., Zhao Y., Wang W., Kong Z., Tang D. (2015). ENHANCED DISEASE RESISTANCE4 associates with CLATHRIN HEAVY CHAIN2 and modulates plant immunity by regulating relocation of EDR1 in Arabidopsis. Plant Cell 27 857–873. 10.1105/tpc.114.134668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C., Nie H., Shen Q., Zhang S., Lukowitz W., Tang D. (2014). EDR1 physically interacts with MKK4/MKK5 and negatively regulates a MAP kinase cascade to modulate plant innate immunity. PLoS Genet. 10:e1004389 10.1371/journal.pgen.1004389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao T., Rui L., Li J., Nishimura M. T., Vogel J. P., Liu N., et al. (2015). A truncated NLR protein, TIR-NBS2, is required for activated defense responses in the exo70B1 mutant. PLoS Genet. 11:e1004945 10.1371/journal.pgen.1004945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Z., Qamar S. A., Chen Z., Mengiste T. (2006). Arabidopsis WRKY33 transcription factor is required for resistance to necrotrophic fungal pathogens. Plant J. 48 592–605. 10.1111/j.1365-313X.2006.02901.x [DOI] [PubMed] [Google Scholar]
- Zhou N., Tootle T. L., Glazebrook J. (1999). Arabidopsis PAD3, a gene required for camalexin biosynthesis, encodes a putative cytochrome P450 monooxygenase. Plant Cell 11 2419–2428. 10.1105/tpc.11.12.2419 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
FIGURE S1 | The growth phenotype of the cyp83a1-3 mutant is similar to wild-type in the absence of G. cichoracearum. Wild-type and cyp83a1-3 mutants were grown under standard short day conditions. Five-weeks-old plants were photographed.
FIGURE S2 | Identification of double mutants by PCR amplification. The pad4-1 cyp83a1-3, sid2-2 cyp83a1-3, eds1-2 cyp83a1-3 eds5-1 cyp83a1-3, npr1-63 cyp83a1-3, and wrky33-2 cyp83a1-3 mutants were identified by PCR amplification.
FIGURE S3 | The biosynthetic network of glucosinolates and related compounds in Arabidopsis. Schematic representation of the biosynthesis pathway derived from tryptophan and methionine.
FIGURE S4 | CYP83A1-GFP rescued the powdery mildew-resistant phenotype of cyp83a1-3. (A) Four-weeks-old wild-type, cyp83a1-3, and cyp83a1-3 transgenic plants expressing the CYP83A1-GFP fusion protein driven by native promoter were infected with G. cichoracearum. Representative leaves were removed (upper panel) and stained with trypan blue (lower panel) at 8 dpi, bar = 200 μm. (B) Quantification of fungal growth of the plants in (a) at 5 dpi by counting the number of conidiophores per colony. Results represent the mean and standard deviation of three independent experiments (n = 30). Different letters represent statistically significant differences (P < 0.01, nested ANOVA).