Abstract
Circulating cluster of differentiation (CD)14+ human leukocyte antigen (HLA)-DRlow/- monocytes, those with a lower HLA-DR expression or are negative for HLA-DR, are considered to be involved in systemic immunosuppression in patients with several malignant tumors. However, few studies have investigated in detail the gene expression profile of CD14+HLA-DRlow/- monocytes. In the present study, the mRNA expression levels of immune-associated molecules in CD14+ monocytes isolated from healthy donors and patients with renal cell carcinoma (RCC) were analyzed. Consistent with previous studies, the percentage of HLA-DRlow/- cells in CD14+ monocytes was significantly increased in patients with RCC compared with healthy donors. In 3 of the 4 patients who underwent surgical resection of the primary tumor, the percentage of CD14+HLA-DRlow/- cells was significantly decreased following surgery. The mRNA expression levels of cyclooxygenase 2, transforming growth factor β, interleukin 6R, chemokine (C-C motif) ligand 2 (CCL2), chemokine (C-X-C motif) ligand 10 (CXCL10), oncostatin M, and vascular endothelial growth factor-A in CD14+ monocytes were quantified using reverse transcription-quantitative polymerase chain reaction. The results of the present study revealed that increased expression levels of CCL2 and CXCL10 were inversely correlated with the percentage of CD14+HLA-DRlow/- monocytes. This suggested that monocytes in RCC patients were immunologically suppressed, and that immunosuppression in RCC patients may be due, in part, to the dysfunction of circulating monocytes.
Keywords: myeloid-derived suppressor cells, chemokine (C-C motif) ligand 2, chemokine (C-X-C motif) ligand 10, renal cell carcinoma, cluster of differentiation 14
Introduction
Systemic immunosuppression in cancer patients is considered to affect the progression of cancer, and therefore the treatment outcome (1,2). Renal cell carcinoma (RCC) is known to be resistant to conventional chemotherapeutic agents and is known to induce an immunosuppressive environment. Although tyrosine kinase inhibitors (TKIs), including sorafenib and sunitinib, are widely utilized for the treatment of patients with metastatic RCC and TKIs are expected to act as adjuvants for immunotherapeutic effects (3,4), the anticancer effects of TKIs may be unable to overcome the immunosuppressive microenvironment of RCC hosts (5).
Previous studies have indicated that myeloid lineage cells, including tumor-associated macrophages, inflammatory monocytes and myeloid-derived suppressor cells (MDSCs), have a significant role in cancer-induced immunosuppression (6,7). MDSCs were initially described in murine cancer models. However, it remains to be elucidated which cell populations in humans are comparable to murine MDSCs (6). Several studies have revealed that cancer patients exhibit an increase in the number of cluster of differentiation (CD)14+ human leukocyte antigen (HLA)-DRlow/− cells, those with a lower HLA-DR expression or are negative for HLA-DR, circulating in the blood (8–10). CD14+HLA-DRlow/− cells that were isolated from cancer patients were identified to suppress T-cell activation in culture, and thus these CD14+HLA-DRlow/− monocyte populations were considered to act as MDSCs (6,8–10).
In the present study, in order to investigate the immunological characteristics of human monocyte populations, HLA-DR and the gene expression of immune-associated molecules in circulating CD14+ myeloid cells from RCC patients were evaluated.
Materials and methods
Blood samples
The protocol of the present study was approved by the Kumamoto University Review Board (ethical permit no. 509; Kumamoto University Hospital, Kumamoto, Japan). All healthy donors and patients reviewed the study objectives and agreed to provide a blood sample based on consent in accordance with the Declaration of Helsinki. The clinical data of all patients and healthy donors is summarized in Table I. Additional blood samples were collected from 4 of the patients 2–3 months after surgery. All patients had not received treatment with TKIs or immunotherapy prior to sample collection. Patients with chronic renal failure and diabetes mellitus were excluded from the present study.
Table I.
Data from healthy donors and renal cell carcinoma patients.
| Patient | Age, years | Gender | Tumor classification | Surgery | Histology | Size, cm | White blood cells, cells/µl | Lymphocytes, % | Monocytes, % | CD14+HLADRlow/−, % |
|---|---|---|---|---|---|---|---|---|---|---|
| HD1 | 37 | M | – | – | – | – | – | – | – | 14 |
| HD2 | 58 | F | – | – | – | – | – | – | – | 16 |
| HD3 | 49 | F | – | – | – | – | – | – | – | 13 |
| HD4 | 58 | M | – | – | – | – | – | – | – | 10 |
| HD5 | 57 | M | – | – | – | – | – | – | – | 10 |
| P1 | 64 | M | T3a(rt),cT1a(lt) | Yes | Clear cell | 6.5 (rt), 2.5 (lt) | 3700 | 34 | 5.9 | 38 |
| P2 | 62 | F | T1a | Yes | Clear cell | 3.1 | 3200 | 45 | 6.0 | 29 |
| P3 | 61 | M | T2a | Yes | Clear cell | 7.1 | 4600 | 54 | 4.5 | 24 |
| P4 | 64 | F | T1a | Yes | Papillary | 2.5 | 6100 | 53 | 6.3 | 24 |
| P5 | 51 | M | T1a | Yes | Clear cell | 3.0 | 4100 | 70 | 5.4 | 18 |
| P6 | 75 | F | T1b | Yes | Clear cell | 6.5 | 6300 | 68 | 8.6 | 35 |
| P7 | 65 | M | – | Yes | – | – | 7000 | 65 | 5.1 | 31 |
| P8 | 71 | M | T1a(rt),T1b(lt) | Yes | Papillary | 2.0 (rt), 4.8 (lt) | 6500 | 51 | 4.8 | 12 |
| P9 | 73 | F | T1a | Yes | Clear cell | 4.0 | 6800 | 47 | 3.9 | 34 |
| P10 | 81 | F | T1a | Yes | Clear cell | 3.3 | 2900 | 44 | 5.5 | 10 |
| P11 | 66 | M | T1a | Yes | Papillary | 3.0 | 6600 | 63 | 4.1 | 17 |
| P12 | 47 | M | multiple(NA) | No | Papillary | – | 2500 | 66 | 5.7 | 12 |
| P13 | 42 | M | T1a | Yes | Clear cell | 1.8 | 4000 | 49 | 4.5 | 14 |
| P14 | 82 | F | T1b | Yes | Clear cell | 4.7 | 7400 | 56 | 4.3 | 20 |
| P15 | 77 | F | T1b(rt),T1a(lt) | Yes | Clear cell | 4.7 (rt), 1.8 (lt) | 3700 | 44 | 7.5 | 28 |
| P16 | 57 | F | T1b | Yes | Clear cell | 5.0 | 4200 | 44 | 5.5 | 31 |
| P17 | 79 | M | T3a | Yes | Clear cell | 7.0 | 5600 | 67 | 6.4 | 8 |
| P18 | 69 | M | T3a | Yes | Clear cell | – | 5900 | 65 | 6.8 | 23 |
| P19 | 51 | M | T1b | Yes | Clear cell | 5.0 | 7100 | 66 | 3.7 | 10 |
| P20 | 66 | F | T3a | Yes | Clear cell | 7.0 | 7200 | 81 | 5.2 | 45 |
| P21 | 75 | M | T4 | No | Clear cell | – | 5100 | 66 | 8.9 | 40 |
| P22 | 37 | M | T4 | Yes | Clear cell | 10.0 | 3900 | 61 | 10.6 | 23 |
HD, healthy donor; P, patient; rt, right; lt, left; -, not applicable; CD, cluster of differentiation; HLA-DRlow, human leukocyte antigen-DR lower fraction.
Isolation of peripheral blood mononuclear cells and CD14+ monocytes
Peripheral blood mononuclear cells (PBMCs) were obtained from 30-ml blood samples using Lymphoprep™ (Axis-Shield Density Gradient Media; Alere Technologies GmbH, Jena, Germany) according to the manufacturer's protocols. Half of the PBMCs were suspended in CELLBANKER® medium (Nippon Zenyaku Kogyo Co., Ltd., Fukushima, Japan) and were stored in liquid nitrogen. The remaining half of the PBMCs were used for isolation of CD14+ monocytes using CD14 MicroBeads (Miltenyi Biotec, Inc., Auburn, CA, USA) according to the manufacturer's protocols.
Flow cytometry
PBMCs (5×105/tube) were treated with Fc Receptor Blocking Solution and subsequently stained with mouse monoclonal fluorescein isothiocyanate-labeled anti-human CD14 (catalog no., 325604; clone, HCD14; 1:20) and phycoerythrin-labeled anti-human HLA-DR (catalog no., 307606; clone, L243; 1:20) antibodies. Fc receptor Blocking Solution and all antibodies, including mouse monoclonal isotype-matched control antibodies (clones, MOPC-173 and MOPC-21; catalog no's., 400212 and 400110, respectively; 1:20), were obtained from BioLegend, Inc. (San Diego, CA, USA). The stained cell samples were analyzed using a FACSverse™ and FACSuite software (BD Biosciences, San Jose, CA, USA).
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was extracted with RNAiso Plus (catalog no., 6110A; Takara Bio Inc., Otsu, Japan). RNA was reverse-transcribed using the PrimeScript RT Reagent kit and DNase (catalog no., 2270A) from Takara Bio, Inc. The complementary DNA product (25 µl) was amplified using qPCR at 94°C for 5 min, then 40 cycles of 94°C for 30 sec and 60°C for 30 sec. qPCR was performed using TaqMan polymerase with SYBR Green fluorescence (Takara Bio, Inc.) with an ABI PRISM® 7300 Sequence Detection System (Applied Biosystems; Thermo Fisher Scientific, Waltham, MA, USA). The relative expression level was determined using the 2−∆∆Cq normalization method (11). The sequences of the primers were designed using the Primer3 website (version 0.4.0; avaliable from http://bioinfo.ut.ee/primer3–0.4.0/) and were synthesised by Hokkaido System Science Co., Ltd. (Tokyo, Japan). The primer seqeunces are shown in Table II. The internal control gene used was β-actin, and 3 parallel wells were set up for each DNA sample (25 µl/well). The data was representative of ≥2 independent experiments.
Table II.
List of primers used for reverse transcription-quantitative polymerase chain reaction.
| Gene | Primer sequence (5′-3′) |
|---|---|
| TGF-β | F: TTGCTTCAGCTCCACGGAGAA |
| R: ACGTAGTACACGATGGGCAGC | |
| CXCL10 | F: CGCTGTACCTGCATCAGCATTAG |
| R: CTGGATTCAGACATCTCTTCTCACC | |
| PTGS2 (COX2) | F: ACTATGGCTACAAAAGCTGGGAAG |
| R: ATCATCAGGCACAGGAGGAAG | |
| VEGFA | F: CAGGAGTACCCTGATGAGATCG |
| R: TCTGCATGGTGATGTTGGAC | |
| IL-6R | F: CACGACTCTGGAAACTATTCATGC |
| R: AGGACCCCACTCACAAACAAC | |
| CCL2 | F: GTGTCCCAAAGAAGCTGTGATCT |
| R: TGTCCAGGTGGTCCATGGA | |
| OSM | F: GCCCAGGATTTGGAGAGGTCTGG |
| R: GCGATGGTAGCCATGCAGGAACCT | |
| β-actin | F: ATTCCTATGTGGGCGACGAG |
| R: AAGGTGTGGTGCCAGATTTTC |
F, forward; R, reverse; TGF, tumor growth factor; CXCL10, chemokine (C-X-C motif) ligand 10; PTGS, prostaglandin-endoperoxide synthase; COX; cyclooxygenase; VEGF, vascular endothelial growth factor; IL; interleukin; CCL2, chemokine (C-C motif) ligand 2; OSM, oncostatin M.
Statistical analysis
The Mann-Whitney U and Spearman's rank correlation tests were performed for statistical analysis using StatMate (GraphPad Software, Inc., La Jolla, CA, USA). P<0.05 was considered to indicate a statistically significant difference.
Results
CD14+HLA-DRlow/− monocyte percentage is increased in patients with RCC
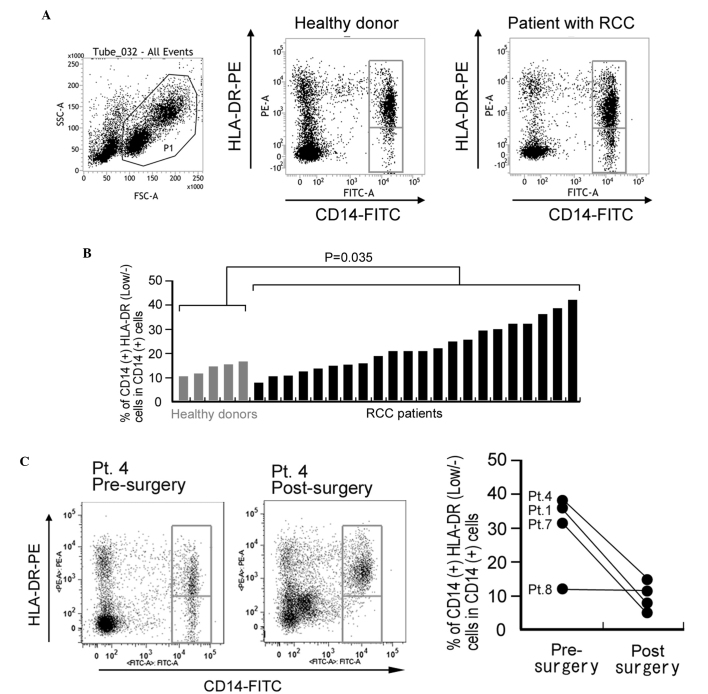
In total, 22 patients with RCC and 5 age-matched healthy donors were enrolled in the present study (Table I). The mean percentage of CD14+HLA-DRlow/- cells in CD14+ monocytes isolated from PBMCs was 23.9% in the RCC patients and 12.6% in the healthy donors. This difference was statistically significant (P=0.035; Fig. 1A and B). For 4 patients, the percentage of CD14+HLA-DRlow/- cells was compared pre- and post-surgery (2–3 months later). A significant reduction in the percentage of CD14+HLA-DRlow/- cells was observed following surgical resection in 3 of the 4 patients (Fig. 1C). By contrast, the percentage of CD14+HLA-DRlow/- monocytes demonstrated no correlation with age, gender, tumor classification or other laboratory data (Table I).
Figure 1.
Percentage of CD14+HLA-DRlow/− monocytes in circulating blood is increased in patients with RCC. (A) Representative FACS analysis of CD14+HLA-DRlow/− monocytes in the peripheral blood mononuclear cells of a healthy control donor and a patient with RCC. (B) The percentage of CD14+HLA-DRlow/− monocytes in circulating CD14+ monocytes of healthy donors and patients was analyzed. The difference between the groups was statistically analyzed using the Mann-Whitney U test. (C) The percentage of CD14+HLA-DRlow/− monocytes in blood samples of 4 RCC patients pre-surgery and post-surgery. Left panel: Representative FACS analysis of pt.4. Right panel: Quantification of all patients. CD, cluster of differentiation; HLA, human leukocyte antigen; RCC, renal cell carcinoma; HLA-DRlow/−, cells with a lower HLA-DR expression or are negative for HLA-DR; FSC, forward scatter; SSC, side scatter; PE, phycoerythrin; FITC, fluorescein isothiocyanate; Pt, patient; FACS, fluorescence-activated cell sorting.
mRNA expression of chemokine (C-C motif) ligand 2 (CCL2) and chemokine (C-X-C motif) ligand 10 (CXCL10) is negatively associated with the percentage of CD14+HLA-DRlow/− monocytes
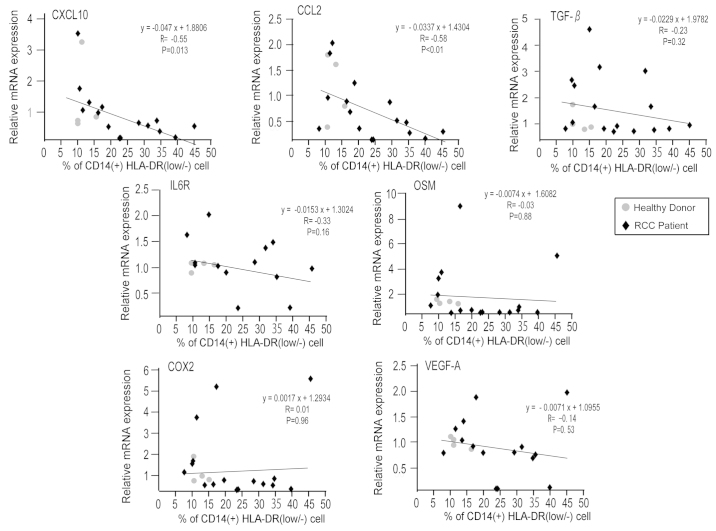
The present study analyzed the mRNA expression of immune-associated molecules in CD14+ monocytes, and subsequently evaluated the association between mRNA expression levels and frequency of CD14+HLA-DRlow/- monocytes. This method of analysis was used due to the fact that the mRNA expression of CD14+HLA-DRlow/- monocytes could not be directly measured, as the total number of CD14+HLA-DRlow/- monocytes was too low to undergo mRNA extraction. mRNA was extracted from 16 patient and 4 healthy donor samples. A total of 6 patient samples and 1 healthy donor sample were excluded due to the low quality of the mRNA. Extremely low levels of gene expression of interleukin (IL)-6, IL-10, programmed death-1 (PD-1), PD-1 ligand (PD-L1), PD-L2, nitric oxide synthase 2, indoleamine 2,3-dioxygenase 1 and arginase 1 were observed in the preliminary analysis of microarray data (data not shown). The gene expression levels of cyclooxygenase 2 (COX2), transforming growth factor β (TGF-β), IL-6R, CCL2, CXCL10, oncostatin M (OSM) and vascular endothelial growth factor A (VEGF-A) were quantified using RT-qPCR (Fig. 2). Increased expression levels of CCL2 and CXCL10 were significantly correlated with a reduced percentage of CD14+HLA-DRlow/- monocytes (P<0.010; R=−0.58; and P=0.013; R=−0.55; respectively).
Figure 2.
mRNA expression of immune-associated genes is negatively associated with a low percentage of CD14+HLA-DRlow/− monocytes. The mRNA expression of COX2, TGF-β, IL-6R, CCL2, CXCL10, OSM and VEGF-A in CD14+ monocytes of healthy donors and RCC patients was examined by reverse transcription-quantitative polymerase chain reaction. The correlation between indicated gene expression and the percentage of CD14+HLA-DRlow/− monocytes in CD14+ monocytes was evaluated using Spearman's rank correlation test. CD, cluster of differentiation; HLA, human leukocyte antigen; HLA-DRlow/−, cells with a lower HLA-DR expression or are negative for HLA-DR; COX2, cyclooxygenase 2; TGF-β, transforming growth factor β; IL, interleukin; CCL2, chemokine (C-C motif) ligand 2; CXCL10, chemokine (C-X-C motif) ligand 10; OSM, oncostatin M; VEGF, vascular endothelial growth factor; RCC, renal cell carcinoma.
Discussion
The present study evaluated the mRNA expression of immune-associated molecules in circulating CD14+ monocytes. The number of CD14+HLA-DRlow/− monocytes is considered to be a prognostic factor; however, an evaluation of the association with the clinical prognosis of patients was not performed by the present study. A high number (>40%) of CD14+HLA-DRlow/− monocytes was significantly associated with a poorer clinical prognosis in patients with chronic lymphocytic leukemia (12). In addition, an increased number of CD14+HLA-DRlow/− monocytes was demonstrated to be an independent prognostic factor in patients with non-small lung cancer (13). The present study revealed that the number of CD14+HLA-DRlow/− monocytes was associated with CXCL10 and CCL2 expression, and in a previous study a high CXCL10 expression in cancer tissues was demonstrated to be associated with a favorable survival rate in patients with esophageal cancer (14). Therefore, it may be of interest to investigate whether the serum concentration of CXCL10 or CCL2 is a useful prognostic marker for patients with cancer. A higher frequency of CD14+HLA-DRlow/− monocytes in RCC patients was observed compared with healthy donors, confirming observations made in previous studies (8–10). Notably, he present study showed that the frequency of CD14+HLA-DRlow/− monocytes reverted to normal following surgical resection of the primary tumor. This observation indicated that unknown molecules derived from RCC tissues may affect the frequency of CD14+HLA-DRlow/− monocytes. The combined observations of the present study suggested that modification of circulating monocytes by cancer-derived factors may be involved in immunosuppression in patients with RCC. However, to the best of our knowledge, there has been no evidence (such as mRNA expression levels of immune-associated molecules) that has been able to directly support the possibility that immunosuppressive molecules are expressed by CD14+HLA-DRlow/− monocytes. The present study speculated that increased expression levels of immunosuppressive molecules would be observed in circulating monocytes along with the high percentage of CD14+HLA-DRlow/− monocytes. However, the results of the present study revealed that the expression of major immunosuppressive molecules, including COX2, OSM, TGF-β and VEGF-A, was not correlated with the frequency of CD14+HLA-DRlow/− monocytes. By contrast, increased expression levels of CXCL10, which is associated with immune activation, and CCL2, which is known to be secreted from activated monocytes and macrophages, were significantly associated with a reduced percentage of CD14+HLA-DRlow/− monocytes (15). The results of the present study suggested that the expression of inflammatory cytokines is typically decreased in CD14+HLA-DRlow/− monocytes.
In conclusion, the present study did not observe an increase in the expression of any immune-associated molecules investigated in CD14+ monocytes isolated from patients with RCC. However, increased mRNA expression of CCL2 and CXCL10 was inversely correlated with the frequency of CD14+HLA-DRlow/− monocytes, suggesting that cytokine expression may be suppressed in these cells. As the dysfunction of monocytes appears to be attributable to immunosuppression in RCC patients, we are currently in the process of identifying the molecules that are derived from cancer tissue and are able to suppress monocytes. Additional investigation regarding the strategy required to overcome the dysfunction of CD14+HLA-DRlow/− monocytes and to generate immune stimulatory myeloid lineages is required in order to achieve an improved outcome for cancer patients.
Acknowledgements
The authors would like to thank Mr. Takenobu Nakagawa, Mr. Osamu Nakamura, Ms. Emi Kiyota and Ms. Yui Hayashida (Department of Cell Pathology, Graduate School of Medical Sciences, Kumamoto University, Kumamoto, Japan) for providing technical assistance. The present study was supported by a research grant from Ono Pharmaceutical Corporation (Osaka, Japan; grant no. #ONO-2013-Kumamoto1).
References
- 1.Vieweg J, Su Z, Dahm P, Kusmartsev S. Reversal of tumor-mediated immunosuppression. Clin Cancer Res. 2007;13(Suppl):S727–S732. doi: 10.1158/1078-0432.CCR-06-1924. [DOI] [PubMed] [Google Scholar]
- 2.Dunn GP, Koebel CM, Schreiber RD. Interferons, immunity and cancer immunoediting. Nat Rev Immunol. 2006;6:836–848. doi: 10.1038/nri1961. [DOI] [PubMed] [Google Scholar]
- 3.Motzer RJ, Bukowski RM. Targeted therapy for metastatic renal cell carcinoma. J Clin Oncol. 2006;24:5601–5608. doi: 10.1200/JCO.2006.08.5415. [DOI] [PubMed] [Google Scholar]
- 4.Tanigawa G, Kawashima A, Yamaguchi S, Nishimura K, Miyoshi S, Kajikawa J, Meguro N, Yosioka T, Oka T, Hara T, et al. Clinical outcome and prognostic factors of sorafenib in Japanese patients with advanced renal cell carcinoma in general clinical practice. Jap J Clin Oncol. 2011;41:1265–1270. doi: 10.1093/jjco/hyr137. [DOI] [PubMed] [Google Scholar]
- 5.Finke JH, Rayman PA, Ko JS, Bradley JM, Gendler SJ, Cohen PA. Modification of the tumor microenvironment as a novel target of renal cell carcinoma therapeutics. Cancer J. 2013;19:353–364. doi: 10.1097/PPO.0b013e31829da0ae. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Greten TF, Manns MP, Korangy F. Myeloid derived suppressor cells in human diseases. Int Immunopharmacol. 2011;11:802–807. doi: 10.1016/j.intimp.2011.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ochoa AC, Zea AH, Hernandez C, Rodriguez PC. Arginase, prostaglandins, and myeloid-derived suppressor cells in renal cell carcinoma. Clin Cancer Res. 2007;13(Suppl):S721–S726. doi: 10.1158/1078-0432.CCR-06-2197. [DOI] [PubMed] [Google Scholar]
- 8.Lin Y, Gustafson MP, Bulur PA, Gastineau DA, Witzig TE, Dietz AB. Immunosuppressive CD14+HLA-DR(low)/- monocytes in B-cell non-Hodgkin lymphoma. Blood. 2011;117:872–881. doi: 10.1182/blood-2010-05-283820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chikamatsu K, Sakakura K, Toyoda M, Takahashi K, Yamamoto T, Masuyama K. Immunosuppressive activity of CD14+ HLA-DR- cells in squamous cell carcinoma of the head and neck. Cancer Sci. 2012;103:976–983. doi: 10.1111/j.1349-7006.2012.02248.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hoechst B, Ormandy LA, Ballmaier M, Lehner F, Krüger C, Manns MP, Greten TF, Korangy F. A new population of myeloid-derived suppressor cells in hepatocellular carcinoma patients induces CD4(+)CD25(+)Foxp3(+) T cells. Gastroenterology. 2008;135:234–243. doi: 10.1053/j.gastro.2008.03.020. [DOI] [PubMed] [Google Scholar]
- 11.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 12.Liu J, Zhou Y, Huang Q, Qiu L. CD14+HLA-DRlow/− expression: A novel prognostic factor in chronic lymphocytic leukemia. Oncol Lett. 2015;9:1167–1172. doi: 10.3892/ol.2014.2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vetsika EK, Koinis F, Gioulbasani M, Aggouraki D, Koutoulaki A, Skalidaki E, Mavroudis D, Georgoulias V, Kotsakis A. A circulating subpopulation of monocytic myeloid-derived suppressor cells as an independent prognostic/predictive factor in untreated non-small lung cancer patients. J Immunol Res. 2014;2014:659294. doi: 10.1155/2014/659294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sato Y, Motoyama S, Nanjo H, Wakita A, Yoshino K, Sasaki T, Nagaki Y, Liu J, Imai K, Saito H, Minamiya Y. CXCL10 expression status is prognostic in patients with advanced thoracic esophageal squamous cell carcinoma. Ann Surg Oncol: Oct. 2015;13 doi: 10.1245/s10434-015-4909-1. (Epub ahead of print) [DOI] [PubMed] [Google Scholar]
- 15.Fujita M, Kohanbash G, Fellows-Mayle W, Hamilton RL, Komohara Y, Decker SA, Ohlfest JR, Okada H. COX-2 blockade suppresses gliomagenesis by inhibiting myeloid-derived suppressor cells. Cancer Res. 2011;71:2664–2674. doi: 10.1158/0008-5472.CAN-10-3055. [DOI] [PMC free article] [PubMed] [Google Scholar]