Abstract
Polymorphisms in antioxidant enzymes and innate immune receptors have been implicated in the development of various types of cancer. The present study aimed to investigate whether polymorphisms of glutathione S-transferase π 1 (GSTP1) and toll-like receptors (TLRs) 2 and 9 are associated with susceptibility to breast cancer among females. The study was conducted on 72 Egyptian female patients with breast cancer, along with 100 healthy volunteers. Polymorphisms of GSTP1 (codon 105 Ile/Val) and TLR9 rs187084 (1237T/C) genes were assessed by polymerase chain reaction (PCR)-restriction fragment length polymorphism, while the −196 to −174 deletion/insertion (del/ins) polymorphism of TLR2 was detected by PCR. The results indicated a decrease in GSTP1 Val allele frequency in breast cancer patients compared with healthy controls, at rates of 22.9 vs. 32.5%, respectively. In addition, the breast cancer group demonstrated a decreased TLR9 C allele frequency compared with the control group, at rates of 36.1 vs. 51.5%, respectively (P=0.0047). A non-significant difference was detected in the frequency of the TLR2 −196 to −174 del allele in breast cancer patients when compared to normal controls. In conclusion, these results suggested that the GSTP1 Val and TLR9 1237C alleles, but not TLR2 −196 to −174 del, are likely to be associated with breast cancer development among females.
Keywords: breast cancer, polymorphisms, toll-like receptor 2, toll-like receptor 9, glutathione S-transferase π 1
Introduction
Breast cancer is one of the most commonly occurring cancers in females, with ~1.7 million new cases diagnosed and 0.5 million mortalities reported annually, worldwide (1). The main treatments for breast cancer include hormonal therapy, chemotherapy, radiotherapy and surgery (1,2). The survival rates for breast cancer are dependent on the stage at which the tumor is diagnosed. According to the National Cancer Institute's Surveillance, Epidemiology, and End Results 2015 database, the relative 5-year survival rates of breast cancer stages I, II, III and IV, are 100, 93, 72 and 22%, respectively (3). If breast cancer is not appropriately diagnosed and treated, numerous complications may develop, such as psychological, nervous, lymph and circulatory disorders. Furthermore, advanced breast cancer may lead to life-threatening metastasis of the tumor to numerous organs, including the liver, kidney, bone, skin and lung, ultimately leading to patient mortality (4). Variations in host genetic factors are emerging as key determinants for breast cancer risk and responsiveness to chemotherapeutic agents (5). Among these variations, polymorphisms in antioxidant enzymes and innate immune receptors have been demonstrated to play major roles in the development of malignancies (6).
Glutathione S-transferase is a phase II detoxifying enzyme that catalyzes the conjunction of reduced glutathione with a wide variety of electrophilic substrates and reactive oxygen species (7). Polymorphisms consisting of single-nucleotide substitutions in the coding sequence of the glutathione S-transferase π 1 gene (GSTP1; 1578A>G) give rise to the Ile105Val amino acid substitution; this lies within the substrate-binding site of GSTP1 (8). Patients with homozygous isoleucine (Ile/Ile) have the highest level of GSTP1 activity. The GSTP1 105Val variant is associated with a lower thermal stability and altered catalytic activity in response to a variety of substrates compared with GSTP1 105Ile (9). GSTP1 activity is somewhat reduced in heterozygotes (Ile/Val) and further diminished for those with two copies of valine (Val/Val) (10).
Toll-like receptors (TLRs) are important members of the host innate immune response and their genes have been found to be polymorphic (11). TLR2 stimulation on the surface of breast cancer cells has been demonstrated to increase the invasive potential of the disease through NF-κB signaling (12). Furthermore, emerging evidence suggests that TLR2 signaling may aid tumor cells in overcoming immune surveillance and avoiding attack by the host immune system (13). Genetic studies on the TLR2 gene have identified a polymorphism that causes a 22-bp nucleotide deletion (−196 to −174 del) in its promoter. This substitution may significantly alter the function of the promoter, likely leading to decreased transcription of TLR2 (14). This polymorphism is reported to be associated with increased susceptibility to hepatocellular carcinoma and gastric cancer (15,16).
TLR9 is a well-known mediator of innate immunity that is capable of detecting DNA from microbial and endogenous sources (17,18). In addition to its role in innate immunity, TLR9 has been demonstrated to be widely expressed in breast cancers, and its stimulation has been implicated in breast cancer cell invasion (19). However, the complete role of TLR9 in breast pathophysiology has not been clearly established. Recent data indicate that polymorphisms in the TLR9 gene (rs352144, rs187084, rs352139, rs352140 and rs445676) may cause an imbalance between pro- and anti-inflammatory cytokines, resulting in chronic inflammation and cancer development (20–22). The majority of studies have focused on three common single nucleotide polymorphisms (SNPs): rs352140, rs5743836 and rs187084. Nevertheless, data regarding the rs187084 genetic variant of TLR9 and its link to breast cancer development are still lacking.
To the best of our knowledge, there is extremely little data in the literature regarding TLR2 and TLR9 polymorphisms in patients with breast cancer. Therefore, the aim of the present study was to assess whether an association exists between the development of breast cancer in Egyptian females and genetic polymorphisms of GSTP1, TLR2 and TLR9.
Subjects and methods
Subjects
The present study was conducted on 72 patients diagnosed with breast cancer at the Oncology Center of Mansoura University (Mansoura, Egypt) between September 2013 and December 2013. Prior to commencing treatment, relevant information was obtained from each patient regarding age, menopausal status, number of children, lactation history and family history of breast cancer. Patients with chronic diseases (such as diabetes mellitus, liver dysfunction or rheumatoid arthritis) were excluded from the study. All pathological data were recorded following surgery (Table I). The control group included 100 healthy female volunteers. Each study participant provided written informed consent. The study was approved by the ethics committee of the Faculty of Medicine of Mansoura University.
Table I.
Clinicopathological characteristics of the studied patients (n=72).
| Patient characteristics | n | % |
|---|---|---|
| Age, years (range, 27–76) | ||
| ≤45 | 19 | 26.4 |
| 46–55 | 31 | 43.1 |
| >55 | 22 | 30.6 |
| Menopausal status | ||
| Premenopausal | 28 | 38.9 |
| Postmenopausal | 44 | 61.1 |
| Number of children | ||
| 0 | 2 | 2.8 |
| 1 | 2 | 2.8 |
| 2 | 10 | 13.9 |
| 3 | 30 | 41.7 |
| 4 | 23 | 31.9 |
| 5 | 1 | 1.4 |
| 6 | 3 | 4.2 |
| 7 | 1 | 1.4 |
| Lactation history | ||
| Positive | 69 | 95.8 |
| Negative | 3 | 4.2 |
| Family history of breast cancer | ||
| Positive | 6 | 8.3 |
| Negative | 66 | 91.7 |
| Pathology | ||
| Infiltrating ductal carcinoma | 64 | 88.9 |
| Infiltrating lobular carcinoma | 6 | 8.3 |
| Paget disease | 2 | 2.8 |
| Grade | ||
| I | 6 | 8.3 |
| II | 47 | 65.3 |
| III | 19 | 26.4 |
| Number of infiltrated lymph nodes | ||
| 0–10 | 61 | 84.7 |
| 11–20 | 8 | 11.1 |
| 21–30 | 3 | 4.2 |
| Lymphovascular invasion | ||
| Positive | 52 | 72.2 |
| Negative | 20 | 27.8 |
| T stage | ||
| 1 | 11 | 15.3 |
| 2 | 42 | 58.3 |
| 3 | 15 | 20.8 |
| 4 | 4 | 5.6 |
| N stage | ||
| 0 | 20 | 27.8 |
| 1 | 20 | 27.8 |
| 2 | 19 | 26.4 |
| 3 | 13 | 18.1 |
| M stage | ||
| 0 | 57 | 79.2 |
| 1 | 15 | 20.8 |
| ER-positive cases, %a | ||
| 0 | 9 | 12.5 |
| 10–20 | 12 | 16.7 |
| 30–40 | 17 | 23.6 |
| 50–60 | 12 | 16.7 |
| 70–80 | 17 | 23.6 |
| 90–100 | 5 | 6.9 |
| PR-positive cases, %a | ||
| 0 | 6 | 8.3 |
| 10–20 | 13 | 18.1 |
| 30–40 | 21 | 29.2 |
| 50–60 | 15 | 20.8 |
| 70–80 | 13 | 18.1 |
| 90–100 | 4 | 5.6 |
| HER2 expression | ||
| Positive | 17 | 23.6 |
| Negative | 55 | 76.4 |
% of positively-stained cells in the tissue samples. T, tumor size; N, lymph node status; M, metastatic status; ER, estrogen receptor; PR, progesterone receptor; HER2, human epidermal growth factor receptor 2.
Specimen collection
For the assessment of GSTP1, TLR2 and TLR9 genetic polymorphisms, 5 ml blood was drawn from each subject enrolled in the study following an overnight fast, and was collected in EDTA tubes (BD Vacutainer Systems, Plymouth, UK) for DNA extraction. DNA was extracted from EDTA-anticoagulated blood using a Gentra Puregene Blood Kit for DNA purification (Qiagen, Inc., Valencia, CA, USA).
Histopathological and immunohistochemical examination
Tumor evaluation and grading were assessed histopathologically, as previously described (23). Tumor size, regional lymph nodes and distant metastasis were also evaluated and staged according to the American Joint Committee on Cancer Staging Manual (6th edition) (24). For immunohistochemical staining, samples were fixed with 10% neutral-buffered formalin and routinely processed to paraffin blocks. Next, 4-µm sections were cut from the blocks and mounted on poly-L-lysine pre-coated glass slides. Antigen retrieval was performed in a pressure cooker for 20 min at 120°C followed by incubation with blocking 6% H2O2 for 3 min to block endogenous peroxidase activity. The sections were then incubated with the Lab Vision™ Avidin Biotin Blocking Solution (cat. no. TA-015-BB (Thermo Fisher Scientific, Pittsburgh, PA, USA) to block nonspecific protein binding. Following antigen retrieval, the sections were incubated for 1 h at room temperature with primary antibodies, followed by horseradish peroxidase-conjugated secondary antibodies (Dako, Glostrup, Denmark) for 30 min at room temperature. The primary antibodies used were as follows: Monoclonal rabbit anti-human estrogen receptor (ER) antibody (clone SP1; cat. no. MA1-39540; 1:200; Thermo Fisher Scientific, Waltham, MA, USA), monoclonal mouse anti-human progesterone receptor (PR) antibody (clone PgR 636; 1:50; cat. no. M3569; Dako) and monoclonal mouse anti-human human epidermal growth factor receptor 2 (HER2) antibody (clone CB11; 1:10; cat. no. 18-7107; Thermo Fisher Scientific). The secondary antibodies used were as follows: Polyclonal goat anti-rabbit IgG antibody (1:200; cat. no. P0448; Dako) or polyclonal rabbit anti-mouse IgG antibody (1:200; cat. no. P0260; Dako). 3,3′-diaminobenzidine (Thermo Fisher Scientific) was then added as a chromogen for color development. The expression of the ER and PR were assessed immunohistochemically, as described previously (25). Scoring for both ER and PR expression was based on the proportion of cells in a given tumor specimen exhibiting distinct nuclear immunopositivity, as well as the intensity of staining (0, 5, 10, 20, 30, 40, 50, 60, 70, 80, 90 or 100%). Positive immunostaining for ER/PR expression was defined as >10% of cells with positively stained nuclei of any intensity. Samples exhibiting >10% immunoexpression were considered positive. HER 2-positivity was defined immunohistochemically, according to the Food and Drug Administration-approved criteria, as uniform and intense circumferential membrane staining in >10% of invasive tumor cells (26).
Detection of GSTP1 polymorphism
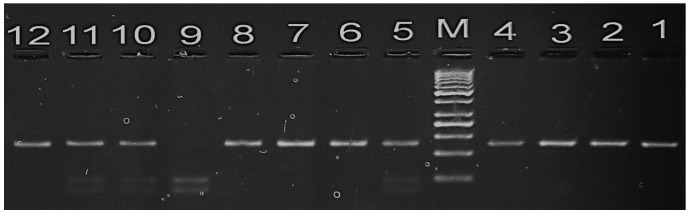
The GSTP1 105 Ile/Val polymorphism was investigated using a polymerase chain reaction (PCR)-restriction fragment length polymorphism (RFLP) method with the following primers (27): p105 forward, 5′-ACCCCAGGGCTCTATGGGAA-3′; and p105 reverse, 5′-TGAGGGCACAAGAAGCCCCT-3′ (Biosearch Technologies, Petaluma, CA, USA). PCR was performed using DreamTaq Green PCR Master Mix (Thermo Fisher Scientific). The procedure was conducted in a total reaction volume of 40 µl. containing 1.5 mM MgCl2, 1 unit of Taq DNA polymerase, 200 ng of each primer and 10 µl DNA as template. PCR cycling conditions were as follows: Initial denaturation at 95°C for 5 min; 30 cycles of denaturation at 95°C for 30 sec, primer annealing at 55°C for 30 sec and extension at 72°C for 1 min; and a final polymerization step of 72°C for 5 min to complete the elongation processes. The PCR products (20 µl) were then digested using 5 units of Alw261 restriction enzyme (Thermo Fisher Scientific) in a total volume of 25 µl, and subsequently separated on a 3.5% agarose gel (Bio Basic Inc., Markham, ON, Canada) prior to staining with ethidium bromide (10 mg/ml; Bio Basic Inc.) to visualize the bands (Fig. 1). Individuals with the wild genotype (Ile/Ile) exhibit a single band (176 bp), whereas those with the mutant homozygote (Val/Val) exhibit two bands (85 and 91 bp). Individuals with the variant allele (Ile/Val) exhibit three bands (176 bp corresponding to Ile; and 91 and 85 bp corresponding to Val).
Figure 1.
Agarose gel electrophoresis stained with ethidium bromide showing the polymerase chain reaction amplification of the 105Val/Ile polymorphism of glutathione S-transferase π 1. M represents the 50 bp ladder. Lanes 1–4: PCR product of 176 bp. Lanes 6, 7, 8 and 12: Homozygous Ile-Ile genotype. Lane 9: Homozygous Val-Val genotype. Lanes 5, 10 and 11: Heterozygous Ile-Val genotype.
Assessment of TLR2 polymorphism
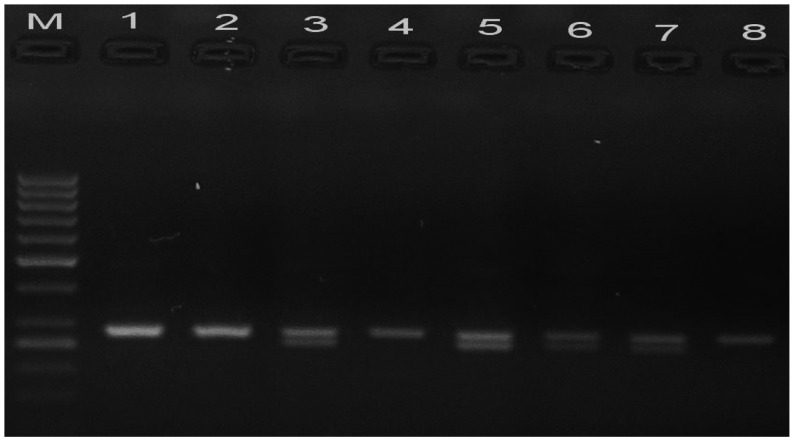
The TLR2 −196 to −174 del/ins polymorphism was assessed using an allele-specific PCR method with the following primers (28): forward, 5′-CACGGAGGCAGCGAGAAA-3′; and reverse, 5′-CTGGGCCGTGCAAAGAAG-3′ (Biosearch Technologies). The procedure was conducted in a total reaction volume of 25 µl, containing 2.5 µl 10X PCR buffer, 2 µl dNTPs (1.25 µmol/l), 0.5 µl MgCl2 (25 mmol/l), 1.25 µl of each primer (25 mmol/l; Sigma-Aldrich, St. Louis, MO, USA), 15.3 µl dH2O, 2 µl DNA (100 ng/µl), and 0.2 µl Taq DNA polymerase (5 U/µl; Invitrogen Life Technologies, Carlsbad, CA, USA). PCR cycling conditions were as follows: Initial denaturation step at 95°C for 5 min; amplification at 95°C for 30 sec, 60°C for 40 sec and 72°C for 40 sec, for 35 cycles; and a final elongation step at 72°C for 7 min. Fig. 2 shows the resulting homozygous (ins/ins; band at 286 bp) and heterozygous (ins/del; bands at 286 and 264 bp) genotypes following 3.5% agarose gel electrophoresis and ethidium bromide staining of the PCR products.
Figure 2.
Agarose gel electrophoresis stained with ethidium bromide showing the amplification of the −196 to −174 del/ins polymorphism of toll-like receptor 2. M represents the 50 bp ladder. Lanes 1, 2, 4 and 8: Homozygous (ins/ins) genotype. Lanes 3, 5, 6 and 7: Heterozygous (ins/del) genotype.
Detection of TLR9 polymorphism
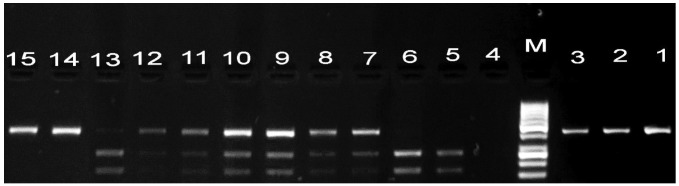
Genotyping of TLR9 was conducted using PCR-RFLP, as previously described (29). PCR reactions consisted of an initial step of 95°C for 5 min, followed by 35 cycles at 95°C 30 sec, 60°C for 30 sec and 72°C for 30 sec, and a final incubation at 72°C for 5 min. Briefly, PCR mixtures (30 µl final volume) contained 20 ng of genomic DNA, 20 pmol of each primer (25 pmol/ml; Intergrated DNA Technologies, Coralville, IA, USA), 3 µl ml of dNTPs (2.5 nM; Thermo Fisher Scientific), 3 µl MgCl2 (25 mM; Thermo Fisher Scientific), 1 ml of NH4 buffer and 0.2 µl of Taq polymerase (Thermo Fisher Scientific). The following primers for TLR9-rs187084 (1237T/C) were used: forward, 5′-ACTATCGAGCCTGCCTGCCATGATACC-3′; and reverse, 5′-ATCCAGCCTTCTTACAAACCTCCCACCC-3′ (Biosearch Technologies). A 15-µl aliquot of each product was digested with 0.5 units of BspTI (Thermo Fisher Scientific). PCR products were subjected to electrophoresis on a 2% agarose gel and visualized by ethidium bromide staining (Fig. 3). The presence of a wild type allele (C allele) for TLR9 resulted in an intact 423-bp band, whereas the RFLP profile of the T variant was characterized by two bands of 172 and 251 bp.
Figure 3.
Agarose gel electrophoresis stained with ethidium bromide showing the amplification of the rs187084 polymorphism of toll-like receptor 9 (1237T/C). M represents the 50 bp ladder. Lanes 1–3: polymerase chain reaction product of 423 bp. Lanes 5 and 6: Homozygous (TT) genotype. Lanes 14 and 15: Homozygous (CC) genotype. Lanes 7–13: Heterozygous (CT) genotype.
Statistical analysis
The χ2 test was employed to examine differences in genotypic and allelic distribution between breast cancer patients and controls. The odds ratio (OR) and 95% confidence intervals (CI) were calculated. P<0.05 was considered to indicate statistically significant differences. Statistical tests were performed with GraphPad Prism software version 5.0 (GraphPad Software, Inc, La Jolla, CA, USA).
Results
Patient characteristics
The breast cancer group (n=72) had a median age of 46.2 years (range, 27–76 years), with postmenopausal status in 61.1%, positive lactation history in 95.8%, positive family history of breast cancer in 8.3% and metastasis in 20.8%. The staging and other clinical and pathological characteristics are shown in Table I.
GSTP1 genotype distribution
The distribution of the different genotypes of GSTP1 is presented in Table II. A significantly lower percentage of the GSTP1 Ile/Val (heterozygous) genotype was observed in women with breast cancer compared with healthy individuals (26.4 vs. 41%, respectively; P=0.0297; OR=2.112). There was also a significant decrease in the combined number of Val/Val and Ile/Val genotypes frequencies in breast cancer patients compared with controls, at rates of 36.1 and 51%, respectively (P=0.0283; OR=1.995). Furthermore, the GSTP1 Val allele frequency was decreased in breast cancer patients relative to controls, at rates of 22.9 and 32.5%, respectively.
Table II.
GSTP1 genotypes and allele distribution in breast cancer patients and control subjects.
| Breast cancer patients (n=72) | Controls (n=100) | ||||||
|---|---|---|---|---|---|---|---|
| GSTP1 genotype | n | % | n | % | OR | 95% CI | P-value |
| Ile/Ile | 46 | 63.9 | 47 | 47.0 | Reference | ||
| Ile/Val | 19 | 26.4 | 41 | 41.0 | 2.112 | 1.071–4.165 | 0.0297 |
| Val/Val | 7 | 9.7 | 12 | 12.0 | 1.678 | 0.607–4.639 | 0.315 |
| Ile/Val + Val/Val | 26 | 36.1 | 53 | 53.0 | 1.995 | 1.073–3.712 | 0.0283 |
| Ile frequency | 111 | 77.1 | 135 | 67.5 | |||
| Val frequency | 33 | 22.9 | 65 | 32.5 | 1.619 | 0.994–2.64 | 0.052 |
GSTP1, glutathione S transferase π; OR, odds ratio; CI, confidence interval; Ile, isoleucine; Val, valine.
TLR genotype distribution
The genotype and allele frequencies of TLR2 did not differ significantly between breast cancer patients and controls (Table III). However, the frequency of the TLR9 CC and CT genotypes were significantly lower in women with breast cancer compared with healthy individuals [CC, 12.5% vs. 20%, respectively (P=0.0016; OR=0.316); CT, 47.2% vs. 63%, respectively (P=0.0069; OR=0.264)]. A significant decrease was also observed in the overall TLR9 C allele frequency in breast cancer patients compared with controls, at rates of 36.1 and 51.5%, respectively (P=0.0047; OR=0.532).
Table III.
Genotypes and alleles distribution of TLR2 and TLR9 in breast cancer patients and control subjects.
| Breast cancer patients (n=72) | Controls (n=100) | ||||||
|---|---|---|---|---|---|---|---|
| Genotypes | n | % | n | % | OR | 95% CI | P-value |
| TLR2 | |||||||
| Ins/Ins | 44 | 61.1 | 61 | 61.0 | Reference | ||
| Del/Ins | 22 | 30.6 | 33 | 33.0 | 1.082 | 0.557–2.103 | 0.8667 |
| Del/Del | 6 | 8.3 | 6 | 6.0 | 0.721 | 0.218–2.386 | 0.7956 |
| Ins frequency | 110 | 76.4 | 155 | 77.5 | |||
| Del frequency | 34 | 23.6 | 45 | 22.5 | 0.939 | 0.565–1.561 | 0.809 |
| TLR9 | |||||||
| TT | 29 | 40.3 | 17 | 17.0 | Reference | ||
| CT | 34 | 47.2 | 63 | 63.0 | 0.316 | 0.153–0.656 | 0.0016 |
| CC | 9 | 12.5 | 20 | 20.0 | 0.264 | 0.098–0.709 | 0.0069 |
| T frequency | 92 | 63.9 | 97 | 48.5 | |||
| C frequency | 52 | 36.1 | 103 | 51.5 | 0.532 | 0.343–0.825 | 0.0047 |
OR, odds ratio; CI, confidence interval; TLR2, toll-like receptor 2; Ins, insertion; Del, deletion; TLR9, toll-like receptor 9; T, thymine; C, cytosine.
Discussion
The present study investigated whether polymorphisms of GSTP1, TLR2 and TLR9 are associated with the development of breast cancer in the Egyptian population. The results of the GSTP1 105 Ile/Val genetic polymorphism assessment revealed a decreased frequency of the Val allele among breast cancer patients relative to that of control subjects. In disagreement with the current results, Shea et al (30) observed no association between GSTP1 genotypes and breast cancer susceptibility in the American population. This discordance of data may be partly attributed to unknown genetic or environmental exposures that may modify the effects of GSTP1 genes in a certain population (31). This result may be suggestive of the differential effect of GSTP1 genotypes on cellular proliferation and apoptosis, which may be translated into cell viability (32). Adler and Pincus (33) reported that peptides covering amino acid residues 99–121 influence the binding of GSTP1 to c-Jun N-terminal kinase (JNK) 1, suggesting that the Ile105Val substitution has no effect on cell proliferation, but protects cells from apoptosis through a JNK-dependent mechanism.
The present investigation into the TLR2 −174 to −196 del polymorphism indicated that the insertion/deletion (ins/del) and del/del genotypes (d allele) are not associated with breast cancer. In agreement with these findings, Etokebe et al (34) failed to demonstrate any correlation between the TLR2 d allele and breast cancer development. However, a study by Theodoropoulos et al (35) reported a positive association between this polymorphism and an increased risk of breast cancer in the Greek population. The reason for this discordance may be attributed to different genetic backgrounds between the studied populations, varying sample sizes and heterogeneity of the tumors examined (36).
The results of the current study indicated that the TLR9 rs187084 (1237T/C) polymorphism was associated with an increased risk of breast cancer in women, based on an Egyptian patient population. To date, no functional data is available regarding the rs187084 polymorphism, however, due to its location in the promoter region of the gene, it may alter the function of the promoter (37). The variant alleles of this TLR9 polymorphism may alter the response to DNA from both microbial and endogenous sources, and thereby influence the production of more pro-inflammatory mediators. A recent study revealed that the high activity T allele of rs187084 appears to be a risk factor for the development of endometrial cancer (38). By contrast, the C allele of the rs187084 and T allele of the rs352140 TLR9 polymorphisms were recently reported to be associated with an increased risk of cervical cancer in Poland (19).
In conclusion, the current findings indicate significant roles for TLR9 rs187084 (1237T/C) and GSTP1 (Ile/Val) polymorphic variants in the susceptibility to breast cancer of an Egyptian female population, whilst no association was observed for the TLR2 −196 to −174 del/ins polymorphism. Thus, polymorphisms in GSTP1 and TLR9, but not TLR2, may be important in the development of breast cancer.
Acknowledgements
The authors wish to acknowledge the laboratory support of the Clinical Immunology Unit, Clinical Pathology Department, Faculty of Medicine of Mansoura University for facilitating the molecular genetic studies.
References
- 1.Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D, Bray F. Cancer incidence and mortality worldwide: Sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E359–E386. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 2.Howard JH, Bland KI. Current management and treatment strategies for breast cancer. Curr Opin Obstet Gynecol. 2012;24:44–48. doi: 10.1097/GCO.0b013e32834da4b1. [DOI] [PubMed] [Google Scholar]
- 3.Itano JK, Brant J, Conde F, Saria M. Core Curriculum for Oncology Nursing. 5th. Philadelphia, PA: Elsevier Health Sciences; 2015. Breast Cancer; p. 75. [Google Scholar]
- 4.Weigelt B, Peterse JL, van't Veer LJ. Breast cancer metastasis: Markers and models. Nat Rev Cancer. 2005;5:591–602. doi: 10.1038/nrc1670. [DOI] [PubMed] [Google Scholar]
- 5.Ulrich CM, Robien K, McLeod HL. Cancer pharmacogenetics: Polymorphisms, pathways and beyond. Nat Rev Cancer. 2003;3:912–920. doi: 10.1038/nrc1233. [DOI] [PubMed] [Google Scholar]
- 6.Klaunig JE, Kamendulis LM, Hocevar BA. Oxidative stress and oxidative damage in carcinogenesis. Toxicol Pathol. 2010;38:96–109. doi: 10.1177/0192623309356453. [DOI] [PubMed] [Google Scholar]
- 7.Hayes JD, Pulford DJ. The glutathione S-transferase supergene family: Regulation of GST and the contribution of the isoenzymes to cancer chemoprotection and drug resistance. Crit Rev Biochem Mol Biol. 1995;30:445–600. doi: 10.3109/10409239509083492. [DOI] [PubMed] [Google Scholar]
- 8.Yang G, Shu XO, Ruan ZX, Cai QY, Jin F, Gao YT, Zheng W. Genetic polymorphisms in glutathione-S-transferase genes (GSTM1, GSTT1, GSTP1) and survival after chemotherapy for invasive breast carcinoma. Cancer. 2005;103:52–58. doi: 10.1002/cncr.20729. [DOI] [PubMed] [Google Scholar]
- 9.Johansson AS, Stenberg G, Widersten M, Mannervik B. Structure-activity relationships and thermal stability of human glutathione transferase P1-1 governed by the H-site residue 105. J Mol Biol. 1998;278:687–698. doi: 10.1006/jmbi.1998.1708. [DOI] [PubMed] [Google Scholar]
- 10.Zhang BL, Sun T, Zhang BN, Zheng S, Lü N, Xu BH, Wang X, Chen GJ, Yu DK, Lin DX. Polymorphisms of GSTP1 is associated with differences of chemotherapy response and toxicity in breast cancer. Chin Med J (Engl) 2011;124:199–204. [PubMed] [Google Scholar]
- 11.Medvedev AE. Toll-like receptor polymorphisms, inflammatory and infectious diseases, allergies, and cancer. J Interferon Cytokine Res. 2013;33:467–484. doi: 10.1089/jir.2012.0140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xie W, Wang Y, Huang Y, Yang H, Wang J, Hu Z. Toll-like receptor 2 mediates invasion via activating NF-kappaB in MDA-MB-231 breast cancer cells. Biochem Biophys Res Commun. 2009;379:1027–1032. doi: 10.1016/j.bbrc.2009.01.009. [DOI] [PubMed] [Google Scholar]
- 13.Huang B, Zhao J, Unkeless JC, Feng ZH, Xiong H. TLR signaling by tumor and immune cells: A double-edged sword. Oncogene. 2008;27:218–224. doi: 10.1038/sj.onc.1210904. [DOI] [PubMed] [Google Scholar]
- 14.Noguchi E, Nishimura F, Fukai H, Kim J, Ichikawa K, Shibasaki M, Arinami T. An association study of asthma and total serum immunoglobin E levels for Toll-like receptor polymorphisms in a Japanese population. Clin Exp Allergy. 2004;34:177–183. doi: 10.1111/j.1365-2222.2004.01839.x. [DOI] [PubMed] [Google Scholar]
- 15.Nischalke HD, Coenen M, Berger C, Aldenhoff K, Müller T, Berg T, Krämer B, Körner C, Odenthal M, Schulze F, et al. The toll-like receptor 2 (TLR2) −196 to −174 del ins polymorphism affects viral loads and susceptibility to hepatocellular carcinoma in chronic hepatitis C. Int J Cancer. 2012;130:1470–1475. doi: 10.1002/ijc.26143. [DOI] [PubMed] [Google Scholar]
- 16.Tahara T, Arisawa T, Wang F, Shibata T, Nakamura M, Sakata M, Hirata I, Nakano H. Toll-like receptor 2–196 to 174del polymorphism influences the susceptibility of Japanese people to gastric cancer. Cancer Sci. 2007;98:1790–1794. doi: 10.1111/j.1349-7006.2007.00590.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Latz E, Schoenemeyer A, Visintin A, Fitzgerald KA, Monks BG, Knetter CF, Lien E, Nilsen NJ, Espevik T, Golenbock DT. TLR9 signals after translocating from the ER to CpG DNA in the lysosome. Nat Immunol. 2004;5:190–198. doi: 10.1038/ni1028. [DOI] [PubMed] [Google Scholar]
- 18.Tian J, Avalos AM, Mao SY, Chen B, Senthil K, Wu H, Parroche P, Drabic S, Golenbock D, Sirois C, et al. Toll-like receptor 9-dependent activation by DNA-containing immune complexes is mediated by HMGB1 and RAGE. Nat Immunol. 2007;8:487–496. doi: 10.1038/ni1457. [DOI] [PubMed] [Google Scholar]
- 19.Ilvesaro JM, Merrell MA, Li L, Wakchoure S, Graves D, Brooks S, Rahko E, Jukkola-Vuorinen A, Vuopala KS, Harris KW, Selander KS. Toll-like receptor 9 mediates CpG oligonucleotide-induced cellular invasion. Mol Cancer Res. 2008;6:1534–1543. doi: 10.1158/1541-7786.MCR-07-2005. [DOI] [PubMed] [Google Scholar]
- 20.Roszak A, Lianeri M, Sowińska A, Jagodziński PP. Involvement of Toll-like receptor 9 polymorphism in cervical cancer development. Mol Biol Rep. 2012;39:8425–8430. doi: 10.1007/s11033-012-1695-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kutikhin AG. Association of polymorphisms in TLR genes and in genes of the Toll-like receptor signaling pathway with cancer risk. Hum Immunol. 2011;72:1095–1116. doi: 10.1016/j.humimm.2011.07.307. [DOI] [PubMed] [Google Scholar]
- 22.Zhang L, Qin H, Guan X, Zhang K, Liu Z. The TLR9 gene polymorphisms and the risk of cancer: Evidence from a meta-analysis. PLoS One. 2013;8:e71785. doi: 10.1371/journal.pone.0071785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Robinson IA, McKee G, Nicholson A, D'Arcy J, Jackson PA, Cook MG, Kissin MW. Prognostic value of cytological grading of fine-needle aspirates from breast carcinomas. Lancet. 1994;343:947–949. doi: 10.1016/S0140-6736(94)90066-3. [DOI] [PubMed] [Google Scholar]
- 24.Singletary SE, Connolly JL. Breast cancer staging: Working with the sixth edition of the AJCC Cancer Staging Manual. CA Cancer J. Clin. 2006;56:37–47. doi: 10.3322/canjclin.56.1.37. [DOI] [PubMed] [Google Scholar]
- 25.Høgdall EV, Christensen L, Høgdall CK, Blaakaer J, Gayther S, Jacobs IJ, Christensen IJ, Kjaer SK. Prognostic value of estrogen receptor and progesterone receptor tumor expression in Danish ovarian cancer patients: From the 'MALOVA' ovarian cancer study. Oncol Rep. 2007;18:1051–1059. [PubMed] [Google Scholar]
- 26.Wolff AC, Hammond ME, Hicks DG, Dowsett M, McShane LM, Allison KH, Allred DC, Bartlett JM, Bilous M, Fitzgibbons P, et al. American Society of Clinical Oncology; College of American Pathologists: Recommendations for human epidermal growth factor receptor 2 testing in breast cancer: American Society of Clinical Oncology/College of American Pathologists clinical practice guideline update. J. Clin Oncol. 2013;31:3997–4013. doi: 10.1200/JCO.2013.50.9984. [DOI] [PubMed] [Google Scholar]
- 27.Harries LW, Stubbins MJ, Forman D, Howard G, Wolf CR. Identification of genetic polymorphisms at the glutathione S-transferase Pi locus and association with susceptibility to bladder, testicular and prostate cancer. Carcinogenesis. 1997;18:641–644. doi: 10.1093/carcin/18.4.641. [DOI] [PubMed] [Google Scholar]
- 28.de Oliveira JG, Silva AE. Polymorphisms of the TLR2 and TLR4 genes are associated with risk of gastric cancer in a Brazilian population. World J Gastroenterol. 2012;18:1235–1242. doi: 10.3748/wjg.v18.i11.1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Etem EO, Elyas H, Ozgocmen S, Yildirim A, Godekmerdan A. The investigation of toll-like receptor 3, 9 and 10 gene polymorphisms in Turkish rheumatoid arthritis patients. Rheumatol Int. 2011;31:1369–1374. doi: 10.1007/s00296-010-1472-8. [DOI] [PubMed] [Google Scholar]
- 30.Shea TC, Claflin G, Comstock KE, Sanderson BJ, Burstein NA, Keenan EJ, Mannervik B, Henner WD. Glutathione transferase activity and isoenzyme composition in primary human breast cancers. Cancer Res. 1990;50:6848–6853. [PubMed] [Google Scholar]
- 31.Sharma A, Pandey A, Sharma S, Chatterjee I, Mehrotra R, Sehgal A, Sharma JK. Genetic polymorphism of glutathione S-transferase P1 (GSTP1) in Delhi population and comparison with other global populations. Meta Gene. 2014;2:134–142. doi: 10.1016/j.mgene.2013.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Holley SL, Fryer AA, Haycock JW, Grubb SE, Strange RC, Hoban PR. Differential effects of glutathione S-transferase pi (GSTP1) haplotypes on cell proliferation and apoptosis. Carcinogenesis. 2007;28:2268–2273. doi: 10.1093/carcin/bgm135. [DOI] [PubMed] [Google Scholar]
- 33.Adler V, Pincus MR. Effector peptides from glutathione-S-transferase-pi affect the activation of jun by jun-N-terminal kinase. Ann Clin Lab Sci. 2004;34:35–46. [PubMed] [Google Scholar]
- 34.Etokebe GE, Knezević J, Petricević B, Pavelić J, Vrbanec D, Dembić Z. Single-nucleotide polymorphisms in genes encoding toll-like receptor −2,-3,-4 and −9 in case-control study with breast cancer. Genet Test Mol Biomarkers. 2009;13:729–734. doi: 10.1089/gtmb.2009.0045. [DOI] [PubMed] [Google Scholar]
- 35.Theodoropoulos GE, Saridakis V, Karantanos T, Michalopoulos NV, Zagouri F, Kontogianni P, Lymperi M, Gazouli M, Zografos GC. Toll-like receptors gene polymorphisms may confer increased susceptibility to breast cancer development. Breast. 2012;21:534–538. doi: 10.1016/j.breast.2012.04.001. [DOI] [PubMed] [Google Scholar]
- 36.Ng CK, Pemberton HN, Reis-Filho JS. Breast cancer intratumor genetic heterogeneity: Causes and implications. Expert Rev Anticancer Ther. 2012;12:1021–1032. doi: 10.1586/era.12.85. [DOI] [PubMed] [Google Scholar]
- 37.Novak N, Yu CF, Bussmann C, Maintz L, Peng WM, Hart J, Hagemann T, Diaz-Lacava A, Baurecht HJ, Klopp N, et al. Putative association of a TLR9 promoter polymorphism with atopic eczema. Allergy. 2007;62:766–772. doi: 10.1111/j.1398-9995.2007.01358.x. [DOI] [PubMed] [Google Scholar]
- 38.Ashton KA, Proietto A, Otton G, Symonds I, McEvoy M, Attia J, Scott RJ. Toll-like receptor (TLR) and nucleosome-binding oligomerization domain (NOD) gene polymorphisms and endometrial cancer risk. BMC Cancer. 2010;10:382. doi: 10.1186/1471-2407-10-382. [DOI] [PMC free article] [PubMed] [Google Scholar]