Abstract
MicroRNAs (miRs) serve a regulatory function in oxidative radical-mediated inflammation and apoptosis during ischemia/reperfusion (IR) injury. Lipocalin 2 (Lcn-2), a target protein of miR-138, is widely involved in the systemic response to IR injury. The aim of the present study was to investigate the association between miR-138 and Lcn-2 in a rat model of cerebral ischemia/reperfusion (CIR) injury and to verify the interaction between miR-138 and Lcn-2 in a PC12 cell model of hypoxia/reoxygenation injury. Reverse transcription-quantitative polymerase chain reaction and western blot analysis were used to detect the mRNA and protein expression levels of miR-138 and Lcn-2. Cell proliferation was determined by MTT assay. The results suggested that the expression of miR-138 was inversely correlated with the expression of Lcn-2 in the CIR rat model and the PC12 cells subjected to hypoxia and reoxygenation. The expression of Lcn-2 was inhibited by miR-138 mimics and enhanced by miR-138 inhibitors, thereby indicating that miR-138 functions as a negative regulator for Lcn-2 expression. This study provides an experimental basis for the further study of miR-138-based therapy for CIR injury.
Keywords: cerebral injury, ischemia/reperfusion, microRNA-138, lipocalin 2
Introduction
Cerebral ischemia/reperfusion injury (CIR) is caused by brain ischemia and further deteriorated by sudden recovery of the blood supply. Oxidative radical-mediated inflammation and apoptosis serve a crucial function in CIR-induced neural injury (1). Therefore, any interventions aimed at inhibiting inflammation and apoptosis may possess therapeutic potential for the treatment of IR-induced neural injury.
MicroRNAs (miRs) are a class of small non-coding and single-stranded RNAs (~22 nucleotides) first identified in Caenorhabditis elegans in 1993 (2). miRs exist in virtually all organisms and are evolutionarily conserved. Their principle function is to regulate target-gene expression at the post-transcriptional level (3). To date, >2,000 miRs have been identified in the human genome, and >60% of protein-coding genes have been found to be regulated by miRs (4,5). miRs are known to have pathophysiological relevance in various diseases, including some that affect the brain (6–8). miR-138 serves a crucial function in various biological processes. The dysregulation of miR-138 has been demonstrated to be a key factor in ischemia/perfusion injury of the heart (9) and lungs (10). In a study using zebrafish, it was found that the disruption of miR-138 function resulted in the ventricular expansion of gene expression generally restricted to the atrioventricular valve region, and ultimately resulted in the disruption of ventricular cardiomyocyte morphology and cardiac function (11). By contrast, the overexpression of miR-138 in zebrafish was found to protect the heart from mycotoxin-induced cardiotoxicity (12). However, the role of miR-138 in the development of CIR has not been fully investigated.
Lipocalin 2 (Lcn-2), which is also known as neutrophil gelatinase-associated lipocalin, has multiple functions including the regulation of cell death/survival, cell migration/invasion, cell differentiation and iron delivery. The expression of Lcn-2 is markedly upregulated in injured spinal cord and brain tissue (13,14). The involvement of Lcn-2 in the systemic response to IR has been documented in previous studies of the heart and kidneys (15–17).
The present study was conducted to investigate the interrelation and interaction of Lcn-2 and miR-138 in a rat model of CIR and PC12 cells, with the aim of providing an experimental basis for the development of an effective therapy for CIR.
Materials and methods
Animals
A total of 60 male Sprague-Dawley (SD) rats were purchased from Weitong Lihua Laboratory Animal Center (SCXK 2006-0009; Beijing, China). All animal procedures were conducted in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals (National Institutes of Health, Bethesda, MA, USA). The Experimental protocols of the present study were approved by the Animal Care Committee at Hubei University of Medicine (Shiyan, China).
Preparation of the CIR rat model
SD rats were randomly allocated to normal, model and sham-operated groups (n=20 per group). Rats in the CIR model group underwent middle cerebral artery occlusion while anesthetized by an intraperitoneal injection with 10% chloral hydrate (350 mg/kg; Shanghai Biomart Technology Co., Ltd., Shanghai, China). An incision was made along the cervical midline followed by the separation of the right common carotid artery (CCA), external carotid artery (ECA) and internal carotid artery (ICA). The CCA and ECA were ligated with suture thread at the proximal and distal ends, respectively. Subsequently, the ICA was clamped with a bulldog clamp at the distal end and an opening was made ~0.5 cm away from the junction of the ECA and ICA and a rounded nylon thread (0.3 mm diameter; Takasago Medical Industry, Co., Ltd., Tokyo, Japan) was immediately inserted into the opening at a depth of ~18 mm, thereby inducing cerebral ischemia. The bulldog clamp was removed and the nylon thread was immobilized along the incision, followed by the closing the skin incision. After 2 h of occlusion, cerebral reperfusion was induced by drawing out the nylon thread by ~10 mm. Rats in the sham-operated group underwent an identical surgical procedure to that conducted in the model group, but without ICA occlusion. Rats in the normal group did not undergo any surgical procedure. All animals were maintained under a 12-h light/dark cycle with ad libitum access to food and water.
Harvesting of brain specimens
Following 2 h of cerebral ischemia and 22 h of reperfusion, rats were guillotined while anesthetized with chloral hydrate, and brain specimens (0.05–0.11 g) were collected and frozen in liquid nitrogen.
Cell culture
The PC12 cell line was purchased from the Cell Bank at Wuhan University (Wuhan, China) and cultured in RMPI-1640 medium with 10% fetal bovine serum, 2 mM L-glutamine and 1% penicillin-streptomycin solution (Invitrogen; Thermo Fisher Scientific, Shanghai, China). Cells were incubated at 37°C in a humidified, 5% CO2/20% O2 atmosphere. For the induction of hypoxia/reoxygenation (H/R) injury, 6-well plates containing PC12 cells (5×105 cells/well) were placed into a humidified airtight container, with continued flow through of 95% N2/5% CO2 to achieve an oxygen-deficient environment. The sealed chamber was placed into a 37°C incubator for 6 h, then returned to a 20% O2 atmosphere. The cells used in this study were limited to within 3 passages.
PC12 cell transfection and luciferase activity assay
PC12 cells at the logarithmic growth phase were seeded into 6-well plates (5×105 cells/well). On the following day, the PC12 cells were transfected with miR-138 mimics, miR-138 inhibitors (Guangzhou RiboBio, Co., Ltd., Guangzhou, China) and pcDNA 3.1-Lcn-2 (Guangzhou Vipotion Biotechnology Co., Ltd., Guangzhou, China) using Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific) in accordance with the manufacturer's instructions. The final concentrations of miR-138 mimics, miR-138 inhibitors and pcDNA 3.1-Lcn-2 were 100, 100 and 500 nM respectively. A mock transfection was performed in a separate well.
PC12 cells were also seeded into 96-well plates (1×104 cells/well) for the luciferase-labeled Lcn-2 transfection and luciferase activity assay. Briefly, PC12 cells were transfected with luciferase-labeled wild-type Lcn-2 (3′UTR-Lcn2-WT; 500 nM; Guangzhou RiboBio, Co., Ltd.) or luciferase-labeled mutant Lcn-2 (3′UTR-Lcn2-MUT; 500 nM; Guangzhou RiboBio, Co., Ltd.), with or without miR-138 mimics (100 nM; Guangzhou Ribobio, Co., Ltd.), respectively, using Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific, Inc.). On day 2 following transfection, 1 µl D-luciferin (0.15 mg/ml; Caliper Life Sciences, Hopkinton, MA, USA) was added to each well and the cells were incubated for 10 min. Subsequently, bioluminescence was measured using the IVIS Spectrum System (Caliper Life Sciences). The relative bioluminescence intensity was an indicator of the luciferase activity. All experiments in each group were repeated in triplicate.
Target search of miR-138
The complementary binding status between miR-138 and the 3′-untranslated region (3′-UTR) of Lcn-2 was obtained using TargetScan (www.targetscan.org).
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR) detection of miR-138 and Lcn-2
Total RNA was extracted from cerebral tissue and PC12 cells using TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.). The isolated RNA was treated with DNase I (Invitrogen; Thermo Fisher Scientific). First strand cDNA was synthesized using 3 µl RNA and the Super Script III Reverse Transcriptase kit (Invitrogen; Thermo Fisher Scientific) with oligonucleotide dTs (0.5 µg/µl). Then, 1 µg cDNA product was used for the amplification procedure in a 20-µl reaction mixture containing 10 µl SYBR Green PCR Master mix (Invitrogen; Thermo Fisher Scientific, Inc.), 7 µl diethylpyrocarbonate-H2O and 1 µl forward and reverse primers (Table I). The amplification was conducted using the 7900HT Fast Real-Time PCR system (Applied Biosystems; Thermo Fisher Scientific, Inc.). The protocol consisted of an initial denaturation and enzyme activation at 95°C for 10 min, followed by 35 cycles of 30 sec denaturation at 95°C, an attachment of primers for 1 min at 60°C and extension at 72°C for 30 sec, and finally 1 cycle at 72°C for 10 min for final elongation. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) and U6 were used as internal controls for Lcn-2 and miR-138, respectively. Relative expression levels of Lcn-2 and miR-138 were calculated using the 2−∆∆Cq method (18).
Table I.
Primer sequences used for miR and mRNA expression analysis.
| Primer | Primer sequence (5′–3′) |
|---|---|
| miR-138-RT | CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGCGGCCTGAT |
| U6-RT | CGCTTCACGAATTTGCGTGTCAT |
| U6-F | CTCGCTTCGGCAGCACA |
| U6-R | AACGCTTCACGAATTTGCGT |
| miR-138-F | ACACTCCAGCTGGGAGCTGGTGTTG |
| Universal-R | GTGCAGGGTCCGAGGT |
| Lcn-2-F | CTGATCAGTGTGCCCCTGCAG |
| Lcn-2-R | GGAGCTTGGAACGAATGTTCTG |
| GAPDH-F | ACACCCACTCCTCCACCTTT |
| GAPDH-R | TTACTCCTTGGAGGCCATGT |
miR, microRNA; RT, reverse transcription primer; F, forward primer; R, reverse primer; Lcn-2, lipocalin 2; GAPDH, glyceraldehyde 3-phosphate dehydrogenase.
Methyl thiazolyl tetrazolium (MTT) assay
The transfected PC12 cells were cultured in 96-well plates at a density of 1×104 cells per well. After 6 h of incubation under hypoxic conditions, 20 µl MTT (5 mg/ml; Sigma-Aldrich, St. Louis, MO, USA) was added to each well following 0, 24, 48, 72 and 96 h of reoxygenation and incubated for an additional 4 h. Then, the supernatant was removed and dimethyl sulfoxide (150 µl/well) was added to dissolve the blue formazan crystals that had been converted from MTT by live cells. Cell viability was assessed by measuring the optical density at 570 nm using a microplate spectrophotometer (Jupiter G19060; Montréal Biotech, Inc. Dorval, Canada). The absorbance (A) values were calculated as a percentage of the control untreated wells, and used to construct a time-course curve. The survival rate of the PC12 cells following each transfection was calculated as a percentage using the following formula: Growth rate = (Atransfected-Ablank)/(Acontrol-Ablank) × 100. Each group was measured in 5 wells on the same plate in three independent experiments.
Immunoblotting analysis
Immunoblotting analysis was used to detect the Lcn-2 protein expression levels in the cerebral tissues. Protein (50 µg) from the homogenized sample was mixed at a ratio of 1:4 with loading buffer (Invitrogen; Thermo Fisher Scientific, Inc.), separated using 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred to a nitrocellulose membrane (Bio-Rad Laboratories, Inc., Hercules, CA, USA). After blocking the non-specific binding sites with 5% milk powder for 2 h, the membrane was incubated overnight at 4°C with rabbit anti-human Lcn-2 (1:1,000; sc-50351; Santa Cruz Biotechnology, Santa Cruz, CA, USA) and rabbit anti-human GAPDH (1:1,000; sc-25778; Santa Cruz Biotechnology) polyclonal antibodies. After washing three times with Tris-buffered saline supplemented with 0.05% Tween-20 (Abcam, Shanghai, China), the membrane was incubated with horseradish peroxidase-conjugated goat-anti-rabbit IgG (1:5,000; cat no. 1706515; Bio-Rad Laboratories, Inc.) for 1.5 h. The antigen-antibody complex was detected using a standard enhanced chemiluminescence detection system (Wuhan Boster Bio-Engineering Ltd., Co., Wuhan, China). The detected protein was quantified using Gel-pro Analyzer 4.0 software (Media Cybernetics, Inc., Rockville, MD, USA) and was expressed as the ratio to GAPDH protein.
Statistical analysis
Numerical data are expressed as the mean ± standard deviation. Statistical differences between the mean for different groups were assessed using one-way analysis of variance with Prism version 4.0 (GraphPad Software, Inc., La Jolla, CA, USA). P<0.05 was considered to indicate a statistically significant difference.
Results
Expression of miR-138 and Lcn-2 in the cerebral tissue of CIR rats
RT-qPCR was used to detect the expression levels of miR-138 and Lcn-2, while the protein expression levels of Lcn-2 were determined by western blot analysis. As shown in Fig. 1, the expression levels of miR-138 in the CIR model group was significantly reduced compared with those in the control and sham-operated groups (P<0.05). By contrast, the mRNA (Fig. 2A) and protein (Fig. 2B) expression levels of Lcn-2 in the CIR model group were markedly increased compared with those in the control and sham-operated groups (P<0.05). No statistically significant difference was detected in the miR-138 and Lcn-2 expression levels between the control and sham-operated groups.
Figure 1.
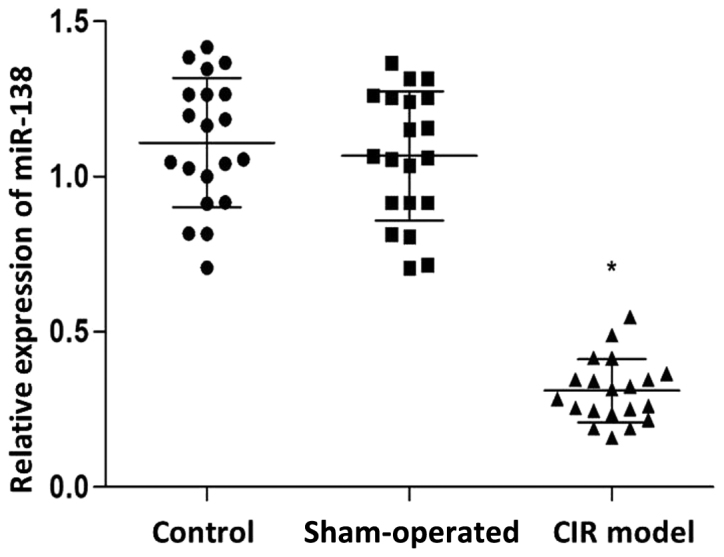
Reverse transcription-quantitative polymerase chain reaction analysis of relative miR-138 mRNA expression in the brain tissues in the normal control, sham-operated control and CIR model groups. Individual data are presented with the indication of mean ± standard deviation. *P<0.05 vs. the control. CIR, cerebral ischemia/reperfusion; miR, microRNA.
Figure 2.
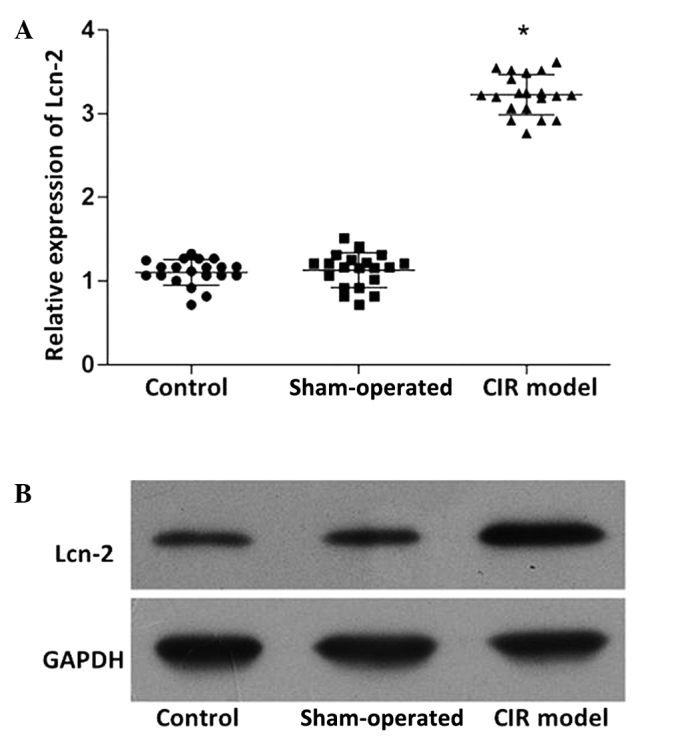
Lcn-2 expression in the brain tissues of rats in the normal control, sham-operated and CIR model groups. (A) Reverse transcription-quantitative polymerase chain reaction analysis of relative Lcn-2 mRNA expression levels. Individual data are presented with the indication of the mean ± standard deviation. *P<0.05 vs. the control. (B) Western blot analysis of Lcn-2 protein expression. A representative image is presented using GAPDH as a loading reference. Lcn-2, lipocalin 2; CIR, cerebral ischemia/reperfusion; GAPDH, glyceraldehyde 3-phosphate dehydrogenase.
Expression of miR-138 in PC12 cells and miR-138-induced cell proliferation
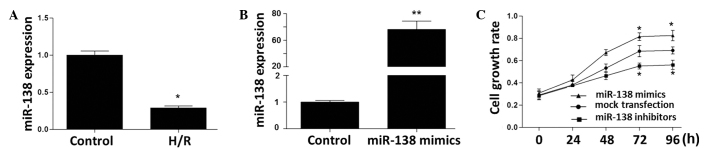
At 24 h of reoxygenation following 6 h of hypoxia, PC12 cells showed markedly reduced expression levels of miR-138 (Fig. 3A). The expression level of miR-138 in the miR-138 mimic-transfected PC12 cells was increased by 68-fold compared with that in the untransfected cells (Fig. 3B). miR-138 expression in the miR-138 inhibitor-transfected PC12 cells was reduced to a non-detectable level (data not shown). As assessed by MTT assay, the PC12 cell growth rate was stimulated by transfection with miR-138 mimics and inhibited by miR-138 inhibitors (Fig. 3C).
Figure 3.
Effects of miR-138 mimics and inhibitors on miR-138 expression and cell proliferation in PC12 cells. The expression levels of miR-138 in PC12 cells with (A) H/R treatment and (B) transfection with mimics were quantified by reverse transcription-quantitative polymerase chain reaction and normalized against U6. Data are expressed as the mean ± standard deviation and analyzed using one-way analysis of variance (*P<0.05, **P<0.01 vs. the control). (C) Cell growth rate was assessed using an MTT assay. miR-138 mimics and inhibitors induced time-dependent changes of cell growth rate (*P<0.05 vs. the mock transfection). miR, microRNA; H/R, hypoxia/reoxygenation; MTT, methyl thiazolyl tetrazolium.
Action of miR-138 on Lcn-2
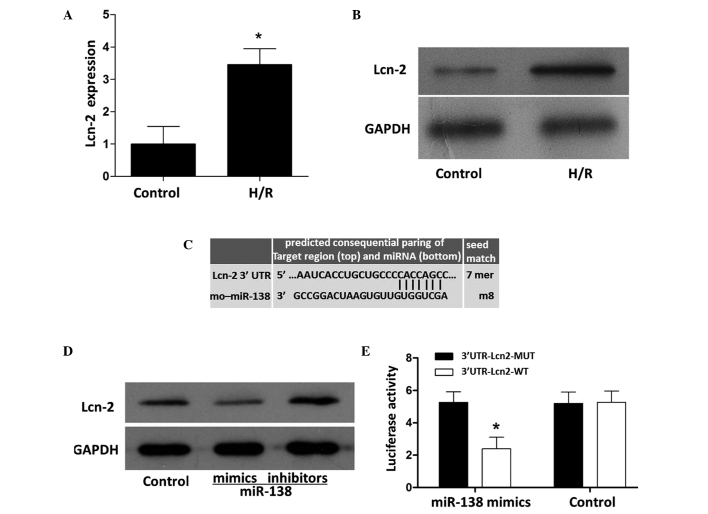
As verified by RT-qPCR and western blot analysis, the expression levels of Lcn-2 in the PC12 cells was significantly increased by H/R (Fig. 4A and B). A TargetScan search indicated that the seed sequence of miR-138 was base-paired with the 3′-UTR region of Lcn-2 via the nucleotides at positions 25–31 (Fig. 4C). The expression of Lcn-2 was inhibited by miR-138 mimics and stimulated by miR-138 inhibitors (Fig. 4D). As shown in Fig. 4E, miR-138 inhibited wild-type Lcn-2 (Lcn-2 WT)-induced luciferase activity without acting on mutant Lcn-2 (Lcn-2 MUT), functionally demonstrating the specificity of miR-138.
Figure 4.
Effects of miR-138 mimics and inhibitors on Lcn-2 expression in PC12 cells. The (A) mRNA (*P<0.05 vs. the control) and (B) protein expression levels of Lcn-2 were detected by reverse transcription-quantitative polymerase chain reaction and western blot analysis, respectively. (C) Seed match status. (D) Compared with the control, the expression of Lcn-2 was inhibited by miR-138 mimics and enhanced by miR-138 inhibitors. (E) miR-138 mimics attenuated 3′UTR-Lcn2-WT-induced luciferase activity without affecting 3′UTR-Lcn2-MUT-induced luciferase activity. Numerical data are expressed as the mean ± standard deviation and analyzed by one-way analysis of variance (*P<0.05 vs. the 3′UTR-Lcn2-MUT). Lcn-2, lipcalin-2; H/R, hypoxia/reperfusion; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; UTR, untranslated region; miR, microRNA; MUT, mutant, WT, wild-type.
As shown in Fig. 5A, the miR-138-induced inhibition of Lcn-2 was attenuated when the cells were co-transfected with miR-138 mimics and Lcn-2 expression vector. In addition, the miR-138-induced PC12 cell proliferation was attenuated when the cells were co-transfected with miR-138 mimics and Lcn-2 expression vectors (Fig. 5B). These results further demonstrated the inhibitory effect of Lcn-2 on PC12 cell growth.
Figure 5.
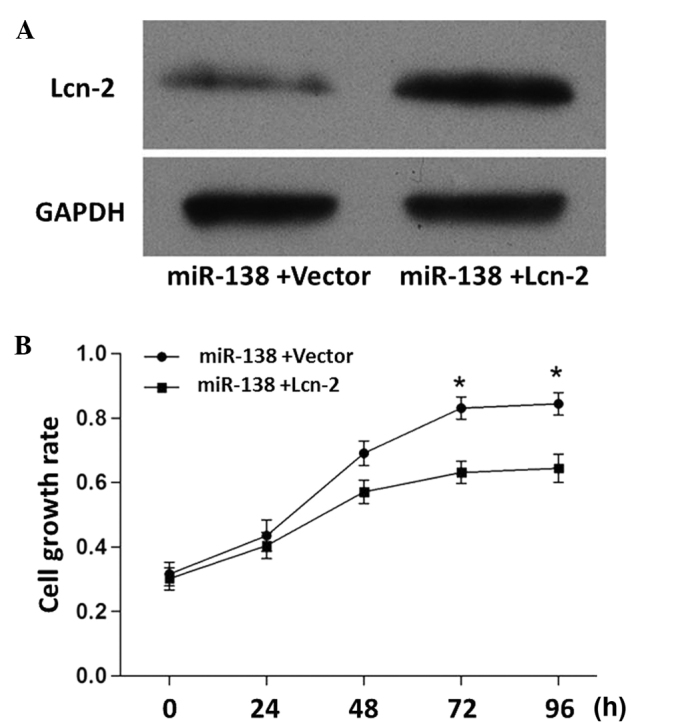
Effects of Lcn-2 overexpression on miR-138-induced cell proliferation. (A) Protein expression levels of Lcn-2 in PC12 cells, as assessed by western blot analysis. PC12 cells were harvested for western blot analysis at 72 h after transfection with miR-138 mimics (100 nM) + pcDNA3.1 (500 nM) or miR-138 mimics (100 nM) + pcDNA3.1-Lcn-2 (500 nM). (B) Cell growth rate was determined by MTT assay, and time-dependent changes are presented as the mean ± standard deviation and analyzed by one-way analysis of variance (*P<0.05 vs. miR-138 + Lcn-2). Lcn-2, lipocalin 2; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; miR, microRNA; MTT, methyl thiazolyl thiazolium.
Discussion
Cerebral injury induced by ischemia/reperfusion (CIR) is notably more severe than damage induced by ischemia alone. It is speculated that oxidative radical-induced inflammation is crucially involved during the progression of CIR injury (17). Since miRs were identified as common regulators of DNA transcription and mRNA translation, numerous miRs have been associated with oxidative radical-mediated pathophysiological alterations (19). As a crucial protein in IR-induced injury, Lcn-2 is included in the list of miR-138 targets (20). In the present CIR rat model, the expression of miR-138 was significantly reduced, while the Lcn-2 was markedly upregulated. The inverse correlation between miR-138 and Lcn-2 suggests that miR-138 functions as a negative regulator of Lcn-2 expression. Diverse mechanisms underlie miR activity (21); for example, miRNAs may increase the translation of target mRNAs by recruiting protein complexes to the AU rich elements of the mRNA, or they may switch the regulation from repression to activation of target gene translation in conditions of cell cycle arrest (22). As miRs frequently target hundreds of mRNAs, miR regulatory pathways are complex. In the present study, in vitro synthesized miR-138-specific mimics and inhibitors were used in PC12 cells to verify the association between miR-138 and Lcn-2.
CIR was simulated in PC12 cells by H/R treatment in the current study. The PC12 cell line originates from rat pheochromocytoma, representing the most widely used system for the study of regulatory mechanisms of neuronal survival and apoptosis (23). The expression patterns of miR-138 and Lcn-2 in PC12 cells were comparable with those observed in the CIR rat model. The application of miR-138 mimics downregulated the expression of Lcn-2, while miR-138 inhibitors upregulated Lcn-2 expression. Accordingly, the cell growth rate was enhanced by miR-138 mimics and depressed by inhibitors. The interaction of miRs with the 3′-UTR of protein-coding genes via RNA-RNA base-pairing is considered to be the primary mechanism underlying the actions of miRs, which usually leads to a reduction in protein output, either by mRNA degradation or by translational repression (21). Furthermore, previous studies have suggested that miRs may interact with the 5′UTR of protein-coding genes via complementarity and cause the translational repression or activation of the targeted proteins. Similarly, miRs could target the coding sequence and thereby repress gene translation (24–26). The specificity of miR targeting is determined by the complementary extent of the ‘seed’ sequence (positions 2–8 from the 5′ end of the miR) and the ‘seed-match’ sequence (generally in the 3′UTR of the target mRNA) (27). The base-pairing between miR-138 and Lcn-2 mRNA is in accordance with that described above. To verify the direct interaction between miR-138 and Lcn-2, miR-138 mimics and inhibitors were applied to PC12 cells transfected with luciferase-labeled wild-type Lcn-2 (3′UTR-Lcn-2-WT) or luciferase-labeled mutant Lcn-2 (3′UTR-Lcn-2-MUT). miR-138 specifically attenuated the luciferase activity in PC12 cells transfected with 3′UTR-Lcn-2-WT. In addition, miR-138-induced alterations of Lcn-2 expression and cell growth rate were neutralized by Lcn-2 overexpression.
In conclusion, the results of the present study demonstrated that the expression levels of miR-138 were inversely correlated with the expression levels of Lcn-2 in the CIR rat model and H/R-treated PC12 cells. The expression of Lcn-2 was inhibited by miR-138 mimics and enhanced by miR-138 inhibitors, thereby indicating that miR-138 functions as a negative regulator for Lcn-2 expression. Therefore, the present study provides an experimental basis for the use of miR-138-based therapy for the treatment of CIR injury.
Acknowledgements
The present study was supported by the Taihe Hospital Foundation. The authors would like to thank Mr. James Dai (LJ Resources Co., Ltd., Vancouver, Canada) for his assistance in preparing the manuscript and English proofreading.
References
- 1.Linkermann A, Hackl MJ, Kunzendorf U, Walczak H, Krautwald S, Jevnikar AM. Necroptosis in immunity and ischemia-reperfusion injury. Am J Transplant. 2013;13:2797–2804. doi: 10.1111/ajt.12448. [DOI] [PubMed] [Google Scholar]
- 2.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-Y. [DOI] [PubMed] [Google Scholar]
- 3.Yates LA, Norbury CJ, Gilbert RJ. The long and short of microRNA. Cell. 2013;153:516–519. doi: 10.1016/j.cell.2013.04.003. [DOI] [PubMed] [Google Scholar]
- 4.Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Callegari E, Elamin BK, Sabbioni S, Gramantieri L, Negrini M. Role of microRNAs in hepatocellular carcinoma: A clinical perspective. Onco Targets Ther. 2013;6:1167–1178. doi: 10.2147/OTT.S36161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Smits M, Nilsson J, Mir SE, van der Stoop PM, Hulleman E, Niers JM, de Witt Hamer PC, Marquez VE, Cloos J, Krichevsky AM, et al. miR-101 is down-regulated in glioblastoma resulting in EZH2-induced proliferation, migration and angiogenesis. Oncotarget. 2010;1:710–720. doi: 10.18632/oncotarget.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Malizia AP, Wang DZ. MicroRNAs in cardiomyocyte development. Wiley Interdiscip Rev Syst Biol Med. 2011;3:183–190. doi: 10.1002/wsbm.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mosakhani N, Guled M, Lahti L, Borze I, Forsman M, Pääkkönen V, Ryhänen J, Knuutila S. Unique microRNA profile in Dupuytren's contracture supports deregulation of β-catenin pathway. Mod Pathol. 2010;23:1544–1552. doi: 10.1038/modpathol.2010.146. [DOI] [PubMed] [Google Scholar]
- 9.He S, Liu P, Jian Z, Li J, Zhu Y, Feng Z, Xiao Y. miR-138 protects cardiomyocytes from hypoxia-induced apoptosis via MLK3/JNK/c-jun pathway. Biochem Biophys Res Commun. 2013;441:763–769. doi: 10.1016/j.bbrc.2013.10.151. [DOI] [PubMed] [Google Scholar]
- 10.Li S, Ran Y, Zhang D, Chen J, Li S, Zhu D. MicroRNA-138 plays a role in hypoxic pulmonary vascular remodelling by targeting Mst1. Biochem J. 2013;452:281–291. doi: 10.1042/BJ20120680. [DOI] [PubMed] [Google Scholar]
- 11.Morton SU, Scherz PJ, Cordes KR, Ivey KN, Stainier DY, Srivastava D. microRNA-138 modulates cardiac patterning during embryonic development. Proc Natl Acad Sci USA. 2008;105:17830–17835. doi: 10.1073/pnas.0804673105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu TS, Yang JJ, Yu FY, Liu BH. Cardiotoxicity of mycotoxin citrinin and involvement of microRNA-138 in zebrafish embryos. Toxicol Sci. 2013;136:402–412. doi: 10.1093/toxsci/kft206. [DOI] [PubMed] [Google Scholar]
- 13.Rathore KI, Berard JL, Redensek A, Chierzi S, Lopez-Vales R, Santos M, Akira S, David S. Lipocalin 2 plays an immunomodulatory role and has detrimental effects after spinal cord injury. J Neurosci. 2011;31:13412–13419. doi: 10.1523/JNEUROSCI.0116-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chia WJ, Dawe GS, Ong WY. Expression and localization of the iron-siderophore binding protein lipocalin 2 in the normal rat brain and after kainate-induced excitotoxicity. Neurochem Int. 2011;59:591–599. doi: 10.1016/j.neuint.2011.04.007. [DOI] [PubMed] [Google Scholar]
- 15.Aigner F, Maier HT, Schwelberger HG, Wallnöfer EA, Amberger A, Obrist P, Berger T, Mak TW, Maglione M, Margreiter R, et al. Lipocalin-2 regulates the inflammatory response during ischemia and reperfusion of the transplanted heart. Am J Transplant. 2007;7:779–788. doi: 10.1111/j.1600-6143.2006.01723.x. [DOI] [PubMed] [Google Scholar]
- 16.Parikh CR, Jani A, Mishra J, Ma Q, Kelly C, Barasch J, Edelstein CL, Devarajan P. Urine NGAL and IL-18 are predictive biomarkers for delayed graft function following kidney transplantation. Am J Transplant. 2006;6:1639–1645. doi: 10.1111/j.1600-6143.2006.01352.x. [DOI] [PubMed] [Google Scholar]
- 17.Cheng L, Xing H, Mao X, Li L, Li X, Li Q. Lipocalin-2 promotes m1 macrophages polarization in a mouse cardiac ischaemia-reperfusion injury model. Scand J Immunol. 2015;81:31–38. doi: 10.1111/sji.12245. [DOI] [PubMed] [Google Scholar]
- 18.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCt method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 19.Chen Z, Wen L, Martin M, Hsu CY, Fang L, Lin FM, Lin TY, Geary MJ, Geary GG, Zhao Y, et al. Oxidative stress activates endothelial innate immunity via sterol regulatory element binding protein 2 (SREBP2) transactivation of microRNA-92a. Circulation. 2015;131:805–814. doi: 10.1161/CIRCULATIONAHA.114.013675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pontrelli P, Accetturo M, Gesualdo L. miRNome analysis using real-time PCR. Methods Mol Biol. 2014;1186:201–232. doi: 10.1007/978-1-4939-1158-5_11. [DOI] [PubMed] [Google Scholar]
- 21.Ling H, Fabbri M, Calin GA. MicroRNAs and other non-coding RNAs as targets for anticancer drug development. Nat Rev Drug Discov. 2013;12:847–865. doi: 10.1038/nrd4140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vasudevan S, Tong Y, Steitz JA. Switching from repression to activation: microRNAs can up-regulate translation. Science. 2007;318:1931–1934. doi: 10.1126/science.1149460. [DOI] [PubMed] [Google Scholar]
- 23.de Posse Chaves EI. Sphingolipids in apoptosis, survival and regeneration in the nervous system. Biochim Biophys Acta. 2006;1758:1995–2015. doi: 10.1016/j.bbamem.2006.09.018. [DOI] [PubMed] [Google Scholar]
- 24.Tay Y, Zhang J, Thomson AM, Lim B, Rigoutsos I. MicroRNAs to Nanog, Oct4 and Sox2 coding regions modulate embryonic stem cell differentiation. Nature. 2008;455:1124–1128. doi: 10.1038/nature07299. [DOI] [PubMed] [Google Scholar]
- 25.Lytle JR, Yario TA, Steitz JA. Target mRNAs are repressed as efficiently by microRNA-binding sites in the 5′ UTR as in the 3′ UTR. Proc Natl Acad Sci USA. 2007;104:9667–9672. doi: 10.1073/pnas.0703820104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Knoll M, Lodish HF, Sun L. Long non-coding RNAs as regulators of the endocrine system. Nat Rev Endocrinol. 2015;11:151–160. doi: 10.1038/nrendo.2014.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bartel DP. MicroRNAs: Target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]