Abstract
The vitamin D receptor (VDR) is a member of the nuclear receptor (NR) superfamily. The VDR binds to active vitamin D3 metabolites, which stimulates downstream transduction signaling involved in various physiological activities such as calcium homeostasis, bone mineralization, and cell differentiation. Quercetin is a widely distributed flavonoid in nature that is known to enhance transactivation of VDR target genes. However, the detailed molecular mechanism underlying VDR activation by quercetin is not well understood. We first demonstrated the interaction between quercetin and the VDR at the molecular level by using fluorescence quenching and saturation transfer difference (STD) NMR experiments. The dissociation constant (Kd) of quercetin and the VDR was 21.15 ± 4.31 μM, and the mapping of quercetin subsites for VDR binding was performed using STD-NMR. The binding mode of quercetin was investigated by a docking study combined with molecular dynamics (MD) simulation. Quercetin might serve as a scaffold for the development of VDR modulators with selective biological activities.
Keywords: VDR, Quercetin, Fluorescence, STD-NMR, Molecular dynamics simulation
INTRODUCTION
The vitamin D receptor (VDR) is a ligand-dependent transcription factor that belongs to the nuclear receptor (NR) superfamily. The VDR contains a ligand binding domain (LBD) that recognizes a wide range of compounds, including 1,25-dihydroxyvitamin D3 (1,25(OH)2D3, calcitriol), steroids, secondary bile acids, polyunsaturated fatty acids, and curcuminoids (Belorusova and Rochel, 2014). The ligand-bound VDR associates with the retinoid X receptor (RXR), upon which VDR-RXR heterodimer binds to vitamin D-responsive elements (VDREs) in the promoter regions of VDR-responsive genes in many target organs (Haussler et al., 2011). Binding of VDR-RXR heterodimer to VDREs leads to the recruitment of relevant coactivators and expression of VDR-responsive genes, such as CYP24A1, TRPV6, and CYP3A4 (Yamada and Makishima, 2014). VDR-mediated molecular processes are highly regulated and directly related to various biological functions, such as calcium homeostasis, bone mineralization, cell differentiation, anti-proliferation, and immune modulation. Although more than ten vitamin D analogues have been used clinically to treat bone, skin, and immune disorders (Yamamoto et al., 2014), therapeutic doses of VDR ligands cause serious adverse effects, particularly hypercalcemia, which limit their clinical application. Therefore, new VDR-targeted drugs that minimize adverse effects and allow clinical use should be developed, possibly with a novel scaffold for VDR binding.
Since the first crystal structure of the human VDR-LBD/1,25-(OH)2D3 complex was reported, several dozens of the crystal structure of the VDR-LBD in complex with various synthetic VDR analogues have been determined. Although the reported structural information revealed the active conformation and critical ligand binding residues of the VDR, there were no significant structural distinctions among the VDR structures in their active states. Moreover, no crystal structure of apo-VDR is available, possibly due to the flexibility of the apo state in comparison with the VDR-LBD/agonist complex (Singarapu et al., 2011). Because of these limitations, the conformational change of the VDR from its inactive state into its active state is not fully understood, which hinders the development of biologically selective modulators.
Quercetin is one of the most abundant flavonoids that are found in various fruits, vegetables, leaves, and grains. This natural compound exhibits a wide range of biologically beneficial effects, such as anti-oxidant, anti-inflammatory, and anti-proliferative properties, which are important for the prevention and therapy of obesity, cancer, and cardiovascular diseases (Inoue et al., 2010; Russo et al., 2012). It has been reported that quercetin enhances transactivation of VDR target genes in human embryonic kidney (HEK) 293 and Caco-2 cells, demonstrating that quercetin is a VDR activator (Inoue et al., 2010). One of the most important questions regarding VDR activation by quercetin is whether such activation is exerted by direct VDR-quercetin interaction that triggers downstream signaling events or by altering the expression and molecular organization of VDR-related transcriptional cofactors.
In this study, we first demonstrated direct binding between human VDR and quercetin at the molecular level using fluorescence spectroscopy and saturation transfer difference (STD) NMR. Mapping of quercetin subsites for VDR binding was performed. The conformation of the VDR-quercetin complex was identified by docking and molecular dynamics (MD) simulation, providing new structural information regarding VDR modulation by quercetin.
MATERIALS AND METHODS
Cloning and protein purification
The gene encoding the ligand binding domain (LBD) of human VDR (VDR-LBD, residues 118–427) was cloned into the plasmid pET28a with NdeI and XhoI restriction sites. This construct was expressed as a fusion protein with 20 non-native residues containing a hexahistidine tag and thrombin cleavage site at the N-terminus. Overexpression was induced with 0.5 mM isopropyl β-D-thiogalactopyranoside (IPTG) in Escherichia coli BL21-CodonPlus (DE3) cells for 6 h at 20°C. These cells were harvested, resuspended, and sonicated in lysis buffer containing 20 mM Tris (pH 8.0), 300 mM NaCl, 5% glycerol, 5 mM imidazole, 2 mM beta-mercaptoethanol (BME), and EDTA-free Protease Inhibitor Cocktail tablets (Roche, Basel, Switzerland). The supernatant was loaded onto an Ni Sepharose column (GE Healthcare, Cambridge, UK) and proteins were eluted with an imidazole gradient using buffer containing 20 mM Tris (pH 8.0), 5% glycerol, 500 mM NaCl, 500 mM imidazole, and 5 mM BME. The hexahistidine tag was cleaved from the eluted protein by an overnight incubation with thrombin. Further purification was performed using diethylaminoethyl cellulose (DEAE) anion-exchange chromatography (GE Healthcare, Cambridge, UK) by gradient elution with increasing ionic strength (up to 1 M NaCl). The final protein sample was obtained using size exclusion chromatography with Superdex200 (GE Healthcare, Cambridge, UK) in buffer containing 20 mM Tris (pH 7.6), 150 mM NaCl, 0.2 mM Tris (2-carboxyethyl)phosphine (TCEP), and 0.1 mM phenylmethanesulfonylfluoride (PMSF). Site-directed mutagenesis (W286A and R274L/S278G/W286A) of the VDR-LBD was performed using the QuikChange site-directed mutagenesis kit (Agilent Technologies, USA). The VDR-LBD mutants were expressed and purified as described above.
Fluorescence titration experiment
All fluorescence spectra were recorded at 298 K with a SpectraMax Gemini XS Microplate Fluorometer. In titration experiments aimed at determining the tryptophan fluorescence of VDR-LBD, 90 μM VDR-LBD and various concentrations of quercetin (9 μM to 360 μM) were used. The excitation and emission wavelengths of the protein were 290 and 335 nm, respectively. Fluorescence emission spectra for the VDR-LBD bound to quercetin were recorded at wavelengths ranging from 300 to 400 nm. All samples used for the measurements were prepared with 10 mM NaH2PO4/Na2HPO4 buffer (pH 7.5) containing 100 mM NaCl and 5% DMSO. Corrections for background fluorescence changes were made by titrations with buffer. Three measurements were averaged for each titration point. Fluorescence data were plotted as cumulative fluorescence quenching signals with respect to the concentration of added quercetin. MATLAB software was used to perform data fitting. The dissociation constant (Kd) was determined using the following equation (Duurkens et al., 2007):
Where Δobs was the observed fluorescence quenching signal at each titration point, Δmax was the maximal observed fluorescence quenching signal during the titration, Kd was the dissociation constant, L was the concentration of added ligand, and N was the concentration of binding sites.
STD-NMR
All STD-NMR experiments were performed at 298 K on a Bruker AVANCE DRX 600 spectrometer equipped with a cryogenically cooled probe. For NMR sample preparation, a mixture of 200 μM quercetin and 4 μM VDR-LBD was dissolved in 300 μL of buffer containing 10 mM phosphate, 100 mM NaCl, 5% DMSO-d6, and 95% D2O at pH 7.5 (not corrected for D2O). On-resonance irradiation of the protein was performed at a chemical shift of 1 ppm, while off-resonance irradiation was applied at −30 ppm, at which no protein and ligand signals were observed (Meyer and Peters, 2003). STD-NMR experiments for VDR-LBD mutants (W286A and R274L/S278G/W286A) were performed as described above. To quantitatively compare the STD spectra for the wild-type and mutants, the acquisition parameters for receiver gain (RG) and number of scan (NS) were set to be equal. A train of Gaussian-shaped pulses (each 50 ms) was used to selectively saturate the protein. The total length of the saturation train was adjusted for the number of pulses in the train and set to 2 s. A 15-ms spin-lock pulse of 1 W, a so-called “T2 filter,” was applied to eliminate the broad protein signals from the STD spectrum. A spectral window of 12 ppm and 32000 real points were acquired with four dummy scans followed by 6320 transients. A 3-9-19 pulse scheme of gradient tailored excitation (WATERGATE) was employed to suppress the residual water. To process the NMR data, the two serial free-induction decays in the single pseudo-2D dataset were split into two separate 1D datasets (on- and off-resonance). The data were processed with 0.5 Hz line broadening and an exponential window function of free-induction decay prior to Fourier transformation.
Docking and molecular dynamics simulation
Docking and scoring calculations for quercetin binding to the VDR-LBD were performed using AutoDock Vina (Trott and Olson, 2010). The crystal structure of the VDR-LBD complexed with 1,25(OH)2D3 (PDB ID: 1DB1) was used as the target structure in the docking study. All crystallographic water molecules were removed for the docking simulation. The file formats of VDR-LBD and quercetin for docking were generated using AutoDockTools (version-1.5.6). All rotatable bonds of the ligand were kept free to allow flexible docking. For blind docking, the grid size was set to accommodate the entire protein inside a 60×60×70 Å grid (x, y, and z). AutoDock Vina with default parameters was used to perform docking and subsequent scoring with an exhaustiveness parameter of 100. The lowest energy-minimized docking model was selected for the best docking conformation of quercetin. The docking model was subjected to molecular dynamics (MD) simulation with GROMACS molecular dynamics package version 4.0.5 (Hess et al., 2008) in conjunction with the GROMOS96 54A7 united atom force field (Schmid et al., 2011). During the MD simulations, all atoms of the protein and ligand were surrounded by explicit water molecules using the simple point charge (SPC) model (Berendsen et al., 1981), while periodic boundary conditions were applied in all directions. A 2-ns MD simulation was performed at constant pressure (1 atm) and temperature (300 K). The electrostatic interactions were calculated by the particle mesh Ewald (PME) algorithm (Darden et al., 1999) and the lengths of all bonds were constrained using the LINCS algorithm (Hess et al., 1997). Topology files in GROMOS and GROMACS format are available for download via the ATB (http://www.compbio.biosci.uq.edu.au/atb/). The modeled structure was inspected visually using the program Pymol (DeLano Scientific LLC) as described in the text.
RESULTS
Quercetin directly binds with the VDR at the molecular level
Because the VDR-LBD has one tryptophan residue that participates in ligand binding (Rochel et al., 2000), intrinsic tryptophan fluorescence was employed to explore whether quercetin binds to the VDR-LBD. The protein was titrated with increasing amounts of quercetin. A general observation is that the tryptophan fluorescence of protein decreases as it directly interacts with ligand, due to the change in the local electronic environments around the tryptophan. As the molar ratio of quercetin to VDR-LBD increased, the fluorescence intensity of the VDR-LBD at the emission maximum (335 nm) decreased (Fig. 1A). Fluorescence was completely quenched by the addition of an approximately 4-fold molar excess of quercetin over the target protein, indicating that the binding was saturated. These results demonstrate that quercetin can bind directly to the VDR-LBD and show that the tryptophan residue is involved in quercetin/VDR-LBD binding.
Fig. 1.
Fluorescence quenching of the VDR-LBD upon quercetin binding. (A) Tryptophan fluorescence spectra of 90 μM VDR-LBD in the absence and presence of quercetin. The concentrations of quercetin were (from top to bottom) 0, 9, 18, 27, 36, 45, 54, 63, 90, 108, 126, 144, 180, 252, 288, and 324 μM. All spectra were recorded at 298 K in 10 mM NaH2PO4/Na2HPO4 buffer (pH 7.5) containing 100 mM NaCl and 5% DMSO. (B) Fluorescence titration curve of the VDR-LBD. The fluorescence quenching values of the VDR-LBD at 335 nm are plotted against the quercetin concentration. The filled circles are experimental data points. The curve was obtained by least-squares fitting to the results, yielding a dissociation constant (Kd) of 21.15 μM.
The reference titration signal (at a wavelength of 335 nm) that was measured only in the presence of VDR-LBD was subtracted from the sample fluorescence signal. The difference in fluorescence represented fluorescence quenching and was plotted against the concentration of quercetin (Fig. 1B). A least-squares fitting search to derive the best value of the dissociation constant (Kd) from the standard binding equation revealed that the data were well fitted to a single-site binding model (Equation in Material and Methods). The estimated Kd value was 21.15 ± 4.31 μM (95% confidence interval), showing approximately three orders of magnitude lower binding affinity than that of active vitamin D3 metabolite 1,25(OH)2D3 (Kd ≈ 10 nM). This result is in good agreement with the previous report that quercetin is a weak VDR activator. We provide the first empirical evidence that quercetin can bind with the VDR with low affinity, substantiating the hypothesis that VDR transactivation is stimulated by direct quercetin binding to the VDR-LBD (Inoue et al., 2010).
STD-NMR detection of quercetin subsites for VDR binding
As quercetin binds to the VDR-LBD with relatively low affinity (Kd in the micromolar range), we employed highly sensitive STD-NMR spectroscopy to analyze the interaction between quercetin and the VDR-LBD. We first performed 1H chemical shift assignments for quercetin by analyzing a reference 1D 1H NMR spectrum of the ligand. The 5 quercetin 1H peaks between 6 and 7.5 ppm corresponded to H6, H8, H5′, H2′, and H6′, respectively, and did not overlap with the residual 1H peaks of water, DMSO, and Tris (Fig. 2A). As a reference, a STD experiment for quercetin in the absence of the VDRLBD was carried out to confirm the absence of STD signals from direct irradiation of quercetin. As predicted, all aromatic resonances of quercetin were not perturbed by the selective on-resonance irradiation (1 ppm), showing no peaks in the reference STD spectrum (Fig. 2B). The STD experiment for a mixture of the VDR-LBD and quercetin with a molar ratio of 1:50 revealed that all assigned proton signals of quercetin were observed in the STD spectrum (Fig. 2C). In contrast, the DMSO peaks from the buffer in the aliphatic region were not affected by the saturation transfer from the VDR-LBD because DMSO does not bind specifically to the protein. The STD peak of DMSO served as an internal control for the STD-NMR experiment and was not observed. These results indicate that quercetin can bind to the VDR-LBD.
Fig. 2.
STD analysis of quercetin. (A) Reference 1H NMR spectrum of quercetin. (B) STD-NMR spectrum of 200 μM quercetin alone. (C) STD-NMR spectrum of a mixture of quercetin (200 μM) and the VDR-LBD (4 μM) at a ratio of 50:1. (D, E) STD-NMR spectrum of 200 μM quercetin in the presence of 4 μM VDR-LBD mutants (W286A and R274L/S278G/W286A).
Interaction between quercetin and VDR mutants
To corroborate the fluorescence quenching data demonstrating that the Trp286 residue of the VDR-LBD is involved in quercetin binding, site-directed mutagenesis of this residue to alanine was performed and the single mutant (W286A) was used in a STD-NMR experiment. A similar experiment was carried out on the triple mutant VDR-LBD (R274L/S278G/W286A) to confirm that quercetin binds to the ligand binding site of the VDR. The STD peaks of quercetin in the presence of each mutant VDR-LBD were significantly decreased in comparison with those of the wild-type VDR-LBD (Fig. 2D, 2E). This difference is due to reduced binding affinity between quercetin and the mutant VDR-LBDs. The reductions in peak intensity for the triple mutant were more striking than those for the single mutant. The mutagenesis data indicated that VDR residues Arg274, Ser278, and Trp286 are important for quercetin binding.
Relative involvement of quercetin protons in VDR binding
Based on the STD value derived from each proton peak of quercetin, we performed group epitope mapping to predict the conformation of quercetin in the binding site of the VDR. Because the STD effects on individual quercetin protons could be quantitatively derived from the differences in the integral peak volumes of the on- and off-resonance spectra, the STD values accounted for the individual atomic contributions to the quercetin-VDR interaction in solution under thermodynamic equilibrium (Mayer and Meyer, 2001). For group epitope mapping, the STD value of the H6 proton (the largest STD value of the quercetin protons) was set to 100%, from which the relative STD values for the H8, H5′, H2′, and H6′ protons were calculated as 93%, 70%, 74%, and 81%, respectively (Fig. 3). Interestingly, the H6 and H8 protons, both located in the A ring segment of quercetin, showed the largest STD values of the quercetin protons, suggesting that this subsite binds in close proximity to the surface of the ligand binding pocket of the VDR. A medium STD value for H6′ on the B-ring could indicate that H6’ interacts with the VDR in a dynamic mode, compared to H6 and H8.
Fig. 3.

Group epitope mapping (GEM) of quercetin. The relative STD values for H6, H8, H2′, H5′, and H6′ are labeled on the structure of quercetin and indicated with dotted circles. In all NMR experiments, a 15-ms T2 filter and WATERGATE pulse were applied to remove protein and water signals, respectively.
Docking model of quercetin with the VDR-LBD
A docking study of quercetin with the VDR-LBD was performed to validate the results of our previous fluorescence and STD-NMR experiments, as well as to establish a binding model of quercetin with the VDR-LBD. The Gibbs free energy of the final docking of quercetin with the VDR-LBD was calculated to be −10.3 kcal/mol. The docking model of quercetin with the VDR-LBD was subjected to MD simulation to validate the model. The MD simulation showed that the A/C ring fragment of quercetin was well buried in the ligand binding pocket of the VDR (Fig. 4A). The VDR-LBD residues that are spatially close to quercetin, including Trp286, Tyr143, Ser278, Leu233, Ser275, and Cys288, are shown in two-dimensional interaction mode in Fig. 4B. In addition, the 3′-OH group of the B ring fragment of quercetin formed two hydrogen bonds with the hydroxyl groups of Tyr143 and Ser278.
Fig. 4.
(A) Predicted binding mode of quercetin in the ligand binding pocket of VDR as optimized by a 2-ns molecular dynamics simulation. (B) Two-dimensional representation of the interaction between quercetin and VDR-LBD using the LigPlot+ program. (C) Crystal structure of the VDR-LBD in complex with 1,25(OH)2D3 (PDB ID: 1DB1). (D) Two-dimensional representation of the interaction between 1,25(OH)2D3 and VDR-LBD using the LigPlot+ program. The VDR-LBD structures are represented as cartoon models, while the key residues for ligand binding are shown as green sticks. Quercetin and 1,25(OH)2D3 are depicted as purple sticks (A, C). The hydrogen bonds between the VDR-LBD and the ligands are shown as pink dotted lines (B, D). Nitrogen, oxygen, hydrogen, and sulfur atoms are colored blue, red, white, and orange, respectively.
The docking model was compared to the crystal structure of the VDR-LBD in complex with its natural ligand 1,25(OH)2D3 (Rochel et al., 2000). The A ring 1-OH group of 1,25(OH)2D3 forms two hydrogen bonds with the side-chain hydroxyl groups of Ser237 and Arg274, while the adjacent 3-OH group forms two hydrogen bonds with the side-chain hydroxyl groups of Tyr143 and Ser278 (Fig. 4C, 4D). The 25-OH group of the aliphatic chain is hydrogen-bonded to His305 and His397. The comparison of the docking model and the structure of the VDR-LBD in complex with 1,25(OH)2D3 revealed that quercetin and 1,25(OH)2D3 utilize similar hydrogen bonding networks between the hydroxyl group of the six-membered ring of the ligand and the Tyr143 and Ser278 residues of the VDR-LBD. The side-chain hydroxyl groups of Arg274 and Ser237 are located more distantly from quercetin than from 1,25(OH)2D3, making hydrogen-bonding interactions with quercetin more difficult. In addition, the α face of the C ring of 1,25(OH)2D3 participates in a hydrophobic interaction with Trp286, similar to the A/C ring of quercetin. Other parts of 1,25(OH)2D3 interact predominantly with the hydrophobic residues lining the ligand binding pocket. However, quercetin forms far fewer inter-molecular contacts with the VDR-LBD than 1,25(OH)2D3, and consequently has a relatively high degree of freedom, with relaxed binding specificity inside the ligand binding pocket (Fig. 4A, 4B). This flexible binding mode could explain the weak binding affinity between quercetin and the VDR-LBD.
MD simulation analysis
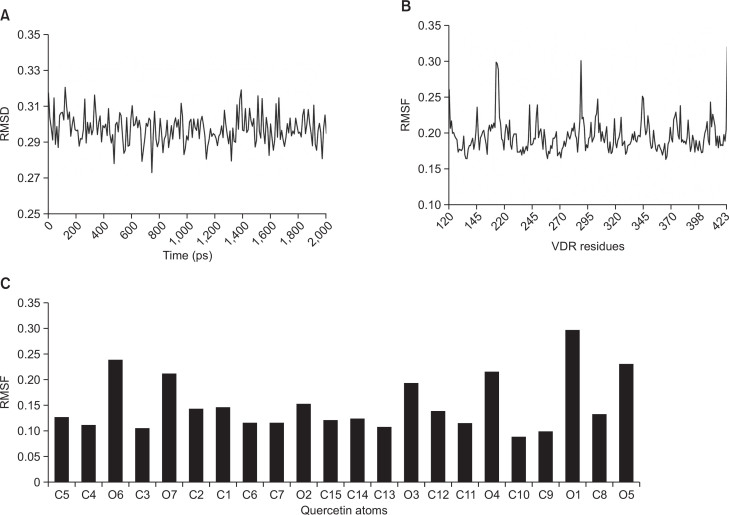
The number of hydrogen bonds formed between VDR-ligand binding pocket (LBP) and quercetin during 2-ns MD simulation was calculated with respect to 200 simulation trajectories. One and two hydrogen bonds between the hydroxyl groups of quercetin and the VDR-LBP were predominantly formed during the first 1.1-ns, and then three hydrogen bonds were formed and retained till the end of the simulation (Fig. 5A). The total number of interatomic contacts within 3.5 Å between the VDR-LBP and quecetin was also calculated (Fig. 5B). From four to seven interatomic contacts are predominantly formed throughout the simulation. Root-mean square deviation (RMSD) values from corresponding starting conformations for the VDR-LBD in complex with quercetin were calculated with regard to simulation time steps, showing a range from 0.27 to 0.33 nm (Fig. 6A). As shown in Fig. 6B, root-mean square fluctuation (RMSF) values of VDR-LBD α-carbons were calculated with regard to three fragments of VDR-LBD (residues 120–164, 216–374, and 378–423). In general, residues with RMSF values above 0.2 nm are considered mobile. The most fluctuation values occur in glycine residues in the loop region (Gly162, Gly163, Gly164, and Gly289). Similarly, RMSF values for quercetin were calculated with regard to carbon and oxygen atoms (Fig. 6C). Overall, oxygen atoms are more fluctuating than carbon atoms, possibly because the carbons comprise the backbone structure of quercetin. The maximum fluctuation value was observed in oxygen atom O1 on the C ring of quercetin while other oxygen atoms, except for O2 of the backbone, show similar fluctuations. The minimum fluctuation value belongs to carbon atom C10 and C9 on the A/C ring.
Fig. 5.
Number of interactions between VDR-LBP and quercetin as a function of simulation time steps. (A) Number of hydrogen bonds formed between the VDR-LBP and quercetin. (B) Number of interatomic contacts within 3.5 Å between the VDR-LBP and quercetin.
Fig. 6.
(A) RMSD of VDR-LBD in complex with quercetin as a function of simulation time steps. (B) RMSF of VDR-LBD α-carbons. VDR-LBD residues correspond to 120–164, 216–374, and 378–423. (C) RMSF of carbon and oxygen atoms for quercetin. Nomenclature of quercetin is the same as in Fig. 4B. RMSD and RMSF are in unit of nm.
DISCUSSION
In this study, the interaction between the VDR and quercetin was first identified at the molecular level, revealing that VDR activation by quercetin is exerted by direct interaction, triggering subsequent downstream signaling events. This finding suggests that the diverse biological activities of quercetin, including its anti-inflammatory and anti-proliferative effects, could be related to VDR binding.
Our biophysical and computational results explain the different binding affinities of 1,25(OH)2D3 and quercetin with the VDR. In the crystal structure of the VDR/1,25(OH)2D3 complex, three pairs of hydrogen bonds between residues of the VDR and the hydroxyl groups of 1,25(OH)2D3 strengthen their interaction. Ser237/Arg274, Tyr143/Ser278, and His305/His397 of the VDR bind to the 1-OH, 3-OH, and 25-OH groups of 1,25(OH)2D3, respectively (Fig. 4C, 4D) (Rochel et al., 2000; Yamagishi et al., 2006; Motoyoshi et al., 2010). These residues could be ranked according to different hydrogen-bonding strengths calculated from fragment molecular orbital (FMO)-interfragment interaction energies analysis, indicating that Arg274, Tyr143, and His397 are the most important anchoring residues for 1,25(OH)2D3 binding (Yamagishi et al., 2006). This result has been supported by alanine scanning mutation analysis (ASMA) that shows the remarkable reduction of transcriptional activities for these mutants (Yamamoto et al., 2000; Choi et al., 2001; Choi et al., 2003; Shimizu et al., 2004; Yamamoto et al., 2007). In particular, Arg274 is thought to form the strongest hydrogen bond with 1,25(OH)2D3 and its genetic mutation to leucine is known to bring about type II hereditary rickets (Feldman and Malloy, 2014). However, the docking model of the VDR/quercetin complex shows hydrogen bonds only between the hydroxyl group of quercetin and the Tyr143/Ser278 residues of the VDR. The interactions between these residues and ligands are known to stimulate VDR-mediated overexpression of TRVP6 that is a calcium channel for renal and intestinal calcium absorption, inducing hypercalcemia (Inoue et al., 2010).
Among the hydrophobic residues that are located within 4 Å of 1,25(OH)2D3, the tryptophan residue (Trp286) is thought to play an important role in the VDR activation, despite its weak binding compared to those of the six hydrophilic residues in the ligand binding pocket (Yamamoto et al., 2000; Choi et al., 2001; Choi et al., 2003; Shimizu et al., 2004; Yamagishi et al., 2006; Yamamoto et al., 2007; Motoyoshi et al., 2010). The other hydrophobic residues showed a pattern of unfavorable interaction energies with 1,25(OH)2D3 (Yamagishi et al., 2006), facilitating a high degree of conformational freedom of both the receptor and the ligand inside the ligand binding pocket. This dynamic property might be a major contributing factor to the functional conformational change upon ligand binding (Yamamoto et al., 2007; Yamamoto et al., 2014). However, fewer hydrophobic interactions contribute to the formation of the VDR-quercetin than contribute to the formation of the VDR-1,25(OH)2D3 complex because quercetin is a class of polyphenols that are deficient of hydrophobic moieties (Fig. 4). The different VDR binding modes between quercetin and 1,25(OH)2D3 is well consistent with the previous report that 100 μM quercetin stimulates VDR activity with efficacy similar to that of 10 pM 1,25(OH)2D3.
The specific ligand binding induces a folding of helix 12 (H12) of the VDR onto the core domain, thereby forming the activation function 2 (AF2) surface that provides a docking platform for the recruitment of cognate coactivators. The structure-function relationships of the VDR have proposed that the aliphatic side-chain of 1,25(OH)2D3 analogues is important for the repositioning of the H12 that controls the VDR activity (Mizwicki et al., 2010; Yamamoto et al., 2014). However, quercetin does not adopt the elongated conformation that includes the aliphatic side-chain, which suggests that lower VDR activity of quercetin is caused by an equilibrium shift toward the closed H12 form of the VDR.
Recently, nonsteroidal VDR ligands have received significant attention because they are derived from nutritional sources and have VDR activities different from those of steroidal analogues (Stayrook et al., 2011; Yamada and Makishima, 2014). For instance, curcumin is a weak VDR ligand that induces expression of VDR target genes, including CYP3A4, CYP24, p21, and TRPV6 (Bartik et al., 2010; Belorusova and Rochel, 2014). Certain retinoids and carotenoids transactivate CYP3A4 through a VDR/RXR-mediated signaling pathway in Caco-2 and HepG2 cells (Wang et al., 2008). Taken together, nonsteroidal natural quercetin has a high pharmaceutical potential in that its simple structure serves as a starting fragment with a high combinatorial possibility for the development of VDR modulators with selective biological activities and fewer adverse effects. Quercetin-based VDR modulators could represent novel therapies for various VDR-related disorders.
Acknowledgments
This work was supported by grants provided by the National Research Foundation of Korea (NRF) funded by the Ministry of Education, Science and Technology (2012R1A1A1039738, 2014R1A1A2054691, 2014R1A1A1002828, and 2015R1A2A-1A 05001894).
REFERENCES
- Bartik L, Whitfield GK, Kaczmarska M, Lowmiller CL, Moffet EW, Furmick JK, Hernandez Z, Haussler CA, Haussler MR, Jurutka PW. Curcumin: a novel nutritionally derived ligand of the vitamin D receptor with implications for colon cancer chemoprevention. J Nutr Biochem. 2010;21:1153–1161. doi: 10.1016/j.jnutbio.2009.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belorusova AY, Rochel N. Modulators of vitamin D nuclear receptor: recent advances from structural studies. Curr Top Med Chem. 2014;14:2368–2377. doi: 10.2174/1568026615666141208095937. [DOI] [PubMed] [Google Scholar]
- Berendsen HJC, Postma JPM, van Gunsteren WF, Hermans J. Interaction Models for Water in Relation to Protein Hydration. In: Pullman B, editor. Intermolecular Forces. Vol. 14. Springer; Netherlands: 1981. pp. 331–342. [DOI] [Google Scholar]
- Choi M, Yamamoto K, Itoh T, Makishima M, Mangelsdorf DJ, Moras D, DeLuca HF, Yamada S. Interaction between vitamin D receptor and vitamin D ligands: two-dimensional alanine scanning mutational analysis. Chem Biol. 2003;10:261–270. doi: 10.1016/S1074-5521(03)00050-4. [DOI] [PubMed] [Google Scholar]
- Choi M, Yamamoto K, Masuno H, Nakashima K, Taga T, Yamada S. Ligand recognition by the vitamin D receptor. Bioorg Med Chem. 2001;9:1721–1730. doi: 10.1016/S0968-0896(01)00060-8. [DOI] [PubMed] [Google Scholar]
- Darden T, Perera L, Li L, Pedersen L. New tricks for modelers from the crystallography toolkit: the particle mesh Ewald algorithm and its use in nucleic acid simulations. Structure. 1999;7:R55–R60. doi: 10.1016/S0969-2126(99)80033-1. [DOI] [PubMed] [Google Scholar]
- Duurkens RH, Tol MB, Geertsma ER, Permentier HP, Slotboom DJ. Flavin binding to the high affinity riboflavin transporter RibU. J Biol Chem. 2007;282:10380–10386. doi: 10.1074/jbc.M608583200. [DOI] [PubMed] [Google Scholar]
- Feldman D, Malloy PJ. Mutations in the vitamin D receptor and hereditary vitamin D-resistant rickets. BoneKEy Rep. 2014;3:510. doi: 10.1038/bonekey.2014.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haussler MR, Jurutka PW, Mizwicki M, Norman AW. Vitamin D receptor (VDR)-mediated actions of 1alpha,25(OH)(2) vitamin D(3): genomic and non-genomic mechanisms. Best Pract Res Clin Endocrinol Metab. 2011;25:543–559. doi: 10.1016/j.beem.2011.05.010. [DOI] [PubMed] [Google Scholar]
- Hess B, Bekker H, Berendsen HJC, Fraaije JGEM. LINCS: A linear constraint solver for molecular simulations. J Comput Chem. 1997;18:1463–1472. doi: 10.1002/(SICI)1096-987X(199709)18:12<1463::AID-JCC4>3.0.CO;2-H. [DOI] [Google Scholar]
- Hess B, Kutzner C, van der Spoel D, Lindahl E. GROMACS 4: algorithms for highly efficient, load-balanced, and scalable molecular simulation. J Chem Theory Comput. 2008;4:435–447. doi: 10.1021/ct700301q. [DOI] [PubMed] [Google Scholar]
- Inoue J, Choi JM, Yoshidomi T, Yashiro T, Sato R. Quercetin enhances VDR activity, leading to stimulation of its target gene expression in Caco-2 cells. J. Nutr. Sci. Vitaminol. (Tokyo) 2010;56:326–330. doi: 10.3177/jnsv.56.326. [DOI] [PubMed] [Google Scholar]
- Mayer M, Meyer B. Group epitope mapping by saturation transfer difference NMR to identify segments of a ligand in direct contact with a protein receptor. J Am Chem Soc. 2001;123:6108–6117. doi: 10.1021/ja0100120. [DOI] [PubMed] [Google Scholar]
- Meyer B, Peters T. NMR spectroscopy techniques for screening and identifying ligand binding to protein receptors. Angew Chem Int Ed Engl. 2003;42:864–890. doi: 10.1002/anie.200390233. [DOI] [PubMed] [Google Scholar]
- Mizwicki MT, Menegaz D, Yaghmaei S, Henry HL, Norman AW. A molecular description of ligand binding to the two overlapping binding pockets of the nuclear vitamin D receptor (VDR): structure-function implications. J Steroid Biochem Mol Biol. 2010;121:98–105. doi: 10.1016/j.jsbmb.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motoyoshi S, Yamagishi K, Yamada S, Tokiwa H. Ligand-dependent conformation change reflects steric structure and interactions of a vitamin D receptor/ligand complex: a fragment molecular orbital study. J Steroid Biochem Mol Biol. 2010;121:56–59. doi: 10.1016/j.jsbmb.2010.03.024. [DOI] [PubMed] [Google Scholar]
- Rochel N, Wurtz JM, Mitschler A, Klaholz B, Moras D. The crystal structure of the nuclear receptor for vitamin D bound to its natural ligand. Mol. Cell. 2000;5:173–179. doi: 10.1016/S1097-2765(00)80413-X. [DOI] [PubMed] [Google Scholar]
- Russo M, Spagnuolo C, Tedesco I, Bilotto S, Russo GL. The flavonoid quercetin in disease prevention and therapy: facts and fancies. Biochem Pharmacol. 2012;83:6–15. doi: 10.1016/j.bcp.2011.08.010. [DOI] [PubMed] [Google Scholar]
- Schmid N, Eichenberger AP, Choutko A, Riniker S, Winger M, Mark AE, van Gunsteren WF. Definition and testing of the GROMOS force-field versions 54A7 and 54B7. Eur Biophys J. 2011;40:843–856. doi: 10.1007/s00249-011-0700-9. [DOI] [PubMed] [Google Scholar]
- Shimizu M, Yamamoto K, Mihori M, Iwasaki Y, Morizono D, Yamada S. Two-dimensional alanine scanning mutational analysis of the interaction between the vitamin D receptor and its ligands: studies of A-ring modified 19-norvitamin D analogs. J. Steroid Biochem. Mol. Biol. 2004;89–90:75–81. doi: 10.1016/j.jsbmb.2004.03.088. [DOI] [PubMed] [Google Scholar]
- Singarapu KK, Zhu J, Tonelli M, Rao H, Assadi-Porter FM, Westler WM, DeLuca HF, Markley JL. Ligand-specific structural changes in the vitamin D receptor in solution. Biochemistry. 2011;50:11025–11033. doi: 10.1021/bi201637p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stayrook KR, Carson MW, Ma YL, Dodge JA. Chapter 78 - Non-secosteroidal Ligands and Modulators. In: Adams DFWPS, editor. Vitamin D. Third Edition. Academic Press; San Diego: 2011. pp. 1497–1508. [DOI] [Google Scholar]
- Trott O, Olson AJ. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Chen S, Xie W, Wan YJ. Retinoids induce cytochrome P450 3A4 through RXR/VDR-mediated pathway. Biochem Pharmacol. 2008;75:2204–2213. doi: 10.1016/j.bcp.2008.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada S, Makishima M. Structure-activity relationship of nonsecosteroidal vitamin D receptor modulators. Trends Pharmacol Sci. 2014;35:324–337. doi: 10.1016/j.tips.2014.04.008. [DOI] [PubMed] [Google Scholar]
- Yamagishi K, Yamamoto K, Yamada S, Tokiwa H. Functions of key residues in the ligand-binding pocket of vitamin D receptor: Fragment molecular orbital-interfragment interaction energy analysis. Chem Phys Lett. 2006;420:465–468. doi: 10.1016/j.cplett.2005.12.078. [DOI] [Google Scholar]
- Yamamoto K, Anami Y, Itoh T. Development of vitamin D analogs modulating the pocket structure of vitamin D receptor. Curr Top Med Chem. 2014;14:2378–2387. doi: 10.2174/156802661421141223091909. [DOI] [PubMed] [Google Scholar]
- Yamamoto K, Choi M, Abe D, Shimizu M, Yamada S. Alanine scanning mutational analysis of the ligand binding pocket of the human Vitamin D receptor. J Steroid Biochem Mol Biol. 2007;103:282–285. doi: 10.1016/j.jsbmb.2006.12.018. [DOI] [PubMed] [Google Scholar]
- Yamamoto K, Masuno H, Choi M, Nakashima K, Taga T, Ooizumi H, Umesono K, Sicinska W, VanHooke J, DeLuca HF, Yamada S. Three-dimensional modeling of and ligand docking to vitamin D receptor ligand binding domain. Proc Natl Acad Sci USA. 2000;97:1467–1472. doi: 10.1073/pnas.020522697. [DOI] [PMC free article] [PubMed] [Google Scholar]