Abstract
Actin filaments are major components of the cytoskeleton in eukaryotic cells and are involved in vital cellular functions such as cell motility and muscle contraction. Tropomyosin is an alpha-helical, coiled coil protein that covers the grooves of actin filaments and stabilizes them. Actin filament length is optimized by tropomodulin, which caps the slow growing (pointed end) of thin filaments to inhibit polymerization or depolymerization. Tropomodulin consists of two structurally distinct regions: the N-terminal and the C-terminal domains. The N-terminal domain contains two tropomyosin-binding sites and one tropomyosin-dependent actin-binding site, whereas the C-terminal domain contains a tropomyosin-independent actin-binding site. Tropomodulin binds to two tropomyosin molecules and at least one actin molecule during capping. The interaction of tropomodulin with tropomyosin is a key regulatory factor for actin filament organization. The binding efficacy of tropomodulin to tropomyosin is isoform-dependent. The affinities of tropomodulin/tropomyosin binding influence the proper localization and capping efficiency of tropomodulin at the pointed end of actin filaments in cells. Tropomodulin and tropomyosin are crucial constituents of the actin filament network, making their presence indispensable in living cells. Here we describe how a small difference in the sequence of the tropomyosin-binding sites of tropomodulin may result in dramatic change in localization of Tmod in muscle cells or morphology of non-muscle cells. We also suggest most promising directions to study and elucidate the role of Tmod-TM interaction in formation and maintenance of sarcomeric and cytoskeletal structure.
Keywords: tropomodulin, tropomyosin, leiomodin, actin filament, pointed end, capping
Introduction
The dynamics of actin filaments is essential for muscle contraction and whole-cell locomotion, and determine the shape of the cell surface (for reviews see (dos Remedios et al. 2003; Winder and Ayscough 2005; Pollard and Cooper 2009)). Actin belongs to a highly conserved family of proteins and is abundant in the cytoplasm of eukaryotic cells. Actin concentration is highest in striated muscles, but non-muscle cells also contain a considerable amount. In vertebrates there are three groups of actin isoforms: α-, β- and γ-actin. Actin monomers, or G-actin, polymerize to form actin filaments (F-actin). The ends of the filament, a fast growing (barbed) and a slow growing (pointed) end have different polymerization rates. Actin filament elongation is a strictly regulated process in eukaryotic cells and it ensues from association and dissociation of actin monomers from either end of the filament. Association is more prevalent at the barbed end, whereas dissociation dominates at the pointed end.
There are a large number of actin-binding proteins. Although they may share the same locus on the surface of the actin filament, actin-binding proteins have distinct functions, including sequestering monomers and severing, depolymerizing, cross-linking, stabilizing, or capping the thin filament.
Dynamic arrangement and organization dictates the precise structure and architecture of actin filaments. This phenomenon is crucial for cellular functions, both in muscle and non-muscle cells. This review investigates the joined function of two actin-binding proteins: tropomodulin (Tmod) and tropomyosin (TM). TM, a rodlike α-helical protein, polymerizes in a head-to-tail fashion to span along the thin filaments and stabilizes them. Tmod binds to and caps the pointed end of actin filaments in a TM-dependent manner (Weber et al. 1994). Tmod regulates the length of thin filaments by inhibiting association or dissociation from this end. Tmod together with TM protect actin filaments from depolymerization and severing by other actin-binding proteins (Yamashiro et al. 2008). We will describe the molecular basis behind the interactions of Tmod, TM and actin, and the fate of cell physiology and functions that depend on their proper action.
Tropomodulin
Tropomodulin Isoforms
Tmod was first detected in the erythrocyte membrane as a TM-binding protein with a molecular mass of 40 kDa (Fowler 1987). It inhibits actin polymerization and depolymerization to fine-tune TM-coated thin filament lengths in muscle and non-muscle cells (Fowler et al. 1993; Gregorio and Fowler 1995). The determination of Tmod’s sequence revealed that the Tmod protein family is different from the other TM-binding protein families (Sung et al. 1992). So far, four Tmod isoforms have been identified (Cox and Zoghbi 2000). Tmod1 (E-Tmod) is mainly found in erythrocytes (Fowler 1987), heart muscle (Sussman et al. 1994a), skeletal muscles (Fowler et al. 1993) and also in other tissues in lesser amounts (Conley et al. 2001). Tmod2 (N-Tmod) is exclusively found in the brain cells (Watakabe et al. 1996). Tmod3 (U-Tmod) is ubiquitously distributed in a variety of cells, and Tmod4 (Sk-Tmod) is the isoform that functions in skeletal muscle cells and replaces Tmod1 during development (Almenar-Queralt et al. 1999; Cox and Zoghbi 2000). Tmod isoforms show 60% identity and 70% similarity in amino acid sequence.
Recently a new Tmod isoform, E-Tmod29, was reported (Yao and Sung 2010). This isoform of 29 kDa is derived from the Tmod1 gene, and is a truncated version of Tmod1 that lacks the N-terminal 102 residues. E-Tmod29 was found in the smooth muscle cell (aorta and uterus in particular) and in the erythrocyte ghost membrane. The authors suggested that expression of E-Tmod29 and Tmod1 may be the result of alternative splicing, multiple transcriptional sites, alternative promoters, promoter switch, protein stability and final cellular destination during cell development and/or differentiation.
Highly ordered organisms such as amphibians, avians and mammals have the genes that code for all Tmod isoforms. However, zebrafish lacks Tmod2, and ascidian, amphioxus and nematode have only one Tmod gene (Bao et al. 2012). Drosophila has one Tmod homolog coded by the so-called sanpodo gene (Dye et al. 1998). The absence of multiple Tmod genes in simpler organisms may indicate that they have appeared in highly ordered organisms as a result of evolution for advanced tissue/organ formation and function. The reason zebrafish lacks Tmod2 may be the adequacy of Tmod1 and Tmod3 for the development of a simpler neural structure and brain. Accordingly, the Tmod2 isoform appeared in vertebrates where a more complex and efficient central nervous system is formed.
Isoforms can replace each other for capping the pointed-ends of actin filaments; it was shown that knocking out Tmod1 resulted in the replacement of Tmod1 with Tmod4 in embryonic mice skeletal muscle (Gokhin et al. 2010). The same study reported that both Tmod3 and Tmod4 were detected at the sarcomeric pointed ends in adult skeletal muscles after Tmod1 knockout. On the other hand, the short Tmod isoform E-Tmod29 (Yao and Sung 2010) does not replace Tmod1 and compensate its action, because Tmod1 knockout was shown to be lethal in embryonic mice (Chu et al. 2003; Fritz-Six et al. 2003; McKeown et al. 2008). Tmod1 is degraded faster than E-Tmod29 due to the unstructured nature of the N-terminal regions. Therefore, E-Tmod29 may be more stable at later stages of red blood cell development and it compensates the action of Tmod1 once the cells are matured (Yao and Sung 2010). In the Tmod1 knock-out mouse Tmod3 was detected in red blood cells, although it is not found there under normal conditions (Moyer et al. 2010). The absence of Tmod1 resulted in transportation of Tmod3 to red blood cells to fulfill Tmod1’s function.
Tmod was shown to nucleate actin monomers (Fowler et al. 2003). Tmod2 and Tmod3 are capable of sequestering and nucleating actin monomers, but this function is performed weakly by Tmod1 and not performed at all by Tmod4 (Fowler et al. 2003; Fischer et al. 2006). Actin nucleation by Tmod is concentration-dependent, and micromolar concentration of Tmod is necessary for this function to take place.
Structural Organization of Tropomodulin
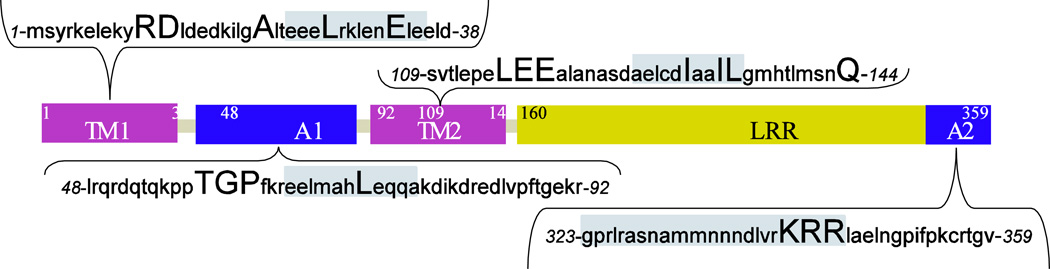
The structure of Tmod1 has been investigated more extensively than the structure of the other isoforms. Tmod1 consists of two distinct structural domains: a compact, globular C-terminal domain and an unstructured, highly disordered N-terminal domain (Kostyukova et al. 2000; Fujisawa et al. 2001; Kostyukova et al. 2001; Krieger et al. 2002). The N-terminal domain of Tmod1 has two TM binding sites and one TM-dependent actin-capping site (Fowler et al. 2003; Greenfield et al. 2005; Kostyukova et al. 2005; Kostyukova et al. 2006) (Figure 1). The elongated and flexible, yet highly disordered N-terminal domain of Tmod1 gains structure upon binding to TM (Kostyukova et al. 2001).
Figure 1.
The schematic view of Tmod1 molecule. LRR: leucine rich repeats. TM1 and TM2 are the tropomyosin-binding sites, A1 and A2 are the actin-binding sites. Amino acids sequences of the sites are presented in the insets with the helical regions indicated by grey color. Residues mutated in the sites are in capital.
The compact C-terminal domain of Tmod bears a TM-independent actin-capping site. This site is crucial for capping actin in the absence of TM in vitro (Fowler et al. 2003; Kostyukova and Hitchcock-DeGregori 2004) and for localizing at the pointed end in sarcomeres of myocytes (Gregorio et al. 1995; Tsukada et al. 2011). The exact location of the C-terminal actin-binding site of Tmod is yet to be mapped, but there is strong evidence that it is located within the C-terminus of Tmod (Kostyukova and Hitchcock-DeGregori 2004; Yamashiro et al. 2010).
The C-terminal domain contains 5 leucine-rich repeats (LRRs) that are 28–30 residues long and it is also called the LRR domain (Krieger et al. 2002). Its structure is alternating α-helices and β-strands. LRR sites are usually involved in protein-protein and ligand interactions (Kobe and Deisenhofer 1994). The members of Tmod protein family are the only known actin-associated proteins with an LRR motif (Krieger et al. 2002).
Leiomodins: Homologs of Tropomodulins
Leiomodin (Lmod) is a 64 kDa protein that belongs to the tropomodulin protein family (Conley et al. 2001). Lmod has three isoforms. Lmod1 (SM-Lmod) is found in smooth muscle, Lmod2 (C-Lmod) is restricted to cardiac and skeletal muscle, and lastly, Lmod3 (fetal Lmod) is the fetal isoform (Conley et al. 2001). Lmods share common structural and functional properties with Tmods. Lmod2 is the most investigated isoform and it is thought to be involved in the pathogenesis of hypertrophic cardiomyopathy (Conley et al. 2001). The first two thirds of the Lmod2 domain structure are markedly similar to Tmod1. This region contains one TM-binding site and one actin-binding site at its N-terminus, and an LRR domain. Trials for finding the second TM-binding site of Lmod failed, since Lmod2131–163, the region that is homologous to the second TM-binding site of Tmod, was not able to bind any TM isoforms (Kostyukova 2007). Current opinion is that Lmod2 contains only one TM-binding site (Kostyukova 2007; Chereau et al. 2008; Skwarek-Maruszewska et al. 2010).
Lmods have an extended region at their C-terminus, which accounts for their higher molecular weight than Tmod (Conley et al. 2001). In Lmod2, the C-terminal extension contains a proline-rich region and an actin-binding Wiskott-Aldrich syndrome protein homology 2 (WH2) domain (Chereau et al. 2008). The presence of three-actin binding sites in Lmod2 makes it a very effective actin-nucleating agent and it was shown to nucleate actin polymerization in vitro (Chereau et al. 2008). Lmods bind both muscle and non-muscle TM isoforms with different affinities (Kostyukova 2007). It was proposed that during late stages of development, Lmod2 replaces Tmod1 at the pointed end of thin filaments and allows the actin filament to elongate (Tsukada et al. 2010).
Significance of Tropomodulin/Leiomodin in Cells
Overexpression of Tmod1 results in dilated cardiomyopathy in mouse myocardium due to myofibril degeneration and formation of shorter thin filaments (Sussman et al. 1998a; Sussman et al. 1998b; Littlefield et al. 2001). On the other hand, inadequate concentrations of Tmod1 cause formation of longer actin filaments (Sussman et al. 1998a); the Tmod1 knockout mouse exhibited fragile red blood cells, heart defects, such as the inability to pump, aborted development of the myocardium and eventual embryonic lethality (Chu et al. 2003; Fritz-Six et al. 2003; McKeown et al. 2008). Absence of Tmod1 in embryonic stem cells and cardiac myocytes leads to delayed myofibril assembly (Chu et al. 2003; Ono et al. 2005).
In the brain, Tmod2 is expressed in parallel with TM. Through its interaction with TM and actin, Tmod2 is thought to assist neuronal growth and differentiation by organizing the thin filament architecture of the central nervous system (Sussman et al. 1994b). Supporting this, studies showed altered Tmod2 expression levels during neural disorders such as epilepsy and cerebral ischemia (Sung et al. 1996; Iwazaki et al. 2006; Chen et al. 2007). Deletion of Tmod2 in mice results in behavioral changes such as impaired memory, reduced sensorimotor gating, hyperactivity and impaired learning, but enhances long-term potentiation with no gross morphological or anatomical abnormalities (Cox et al. 2003). Tmod2 expression exceeds normal levels after middle cerebral artery occlusion and this may be related to its influence in increased neuronal repair and synaptic plasticity (Chen et al. 2007).
A study done on another Tmod isoform reports that overexpression of Tmod3 caused decreased endothelial cell motility (Fischer et al. 2003). Deletion of Tmod3 from intestinal epithelial cells resulted in a 30% and 20% decrease in TM and F-actin expression, respectively (Weber et al. 2007). Disassembly of thin filaments was observed due to decreased expression of TM. In this study, it was stated that Tmod3 acts as an agent to consolidate the spectrin-actin membrane skeleton in epithelial cells and maintain epithelial cell morphology.
The knockout of the gene encoding hematopoietic protein-1, which participates in regulation of polymerization of F-actin in erythrocytes, caused decreased expression of Tmod1 in murine erythrocytes (Chan et al. 2013). Simultaneously, the expression of Tmod3 increased. This supports the idea that Tmod3 can compensate the function of Tmod1 and erythrocytes compensate the loss in the amount of Tmod1 by counter-expressing Tmod3.
Tmod, TM and F-actin associate to form protofilaments in erythrocyte membranes (Sung et al. 2000). The participation of Tmod and short TM isoforms in the thin filament structure stabilizes the lens cell cortical cytoskeleton (Fischer et al. 2000). This improved strength in the F-actin framework augments cellular functions, such as adhesion (Watanabe et al. 1992) and intercellular communication (Goodenough 1992; Goodenough et al. 1996) in differentiated lens cells. Developmental regulation of Tmod expression in cells indicates the importance of its interaction with thin filaments in tissue and cell differentiation (Ito et al. 1995).
Recently, Lmod isoforms were detected in non-muscle cells. The presence of Lmod1 was found as a result of steroid induction in human trabecular meshwork cells (Clark et al. 2013). It was demonstrated that phencyclidine (a dissociative drug) induction resulted in up-regulation of Lmod2 in the thalamus of rat brain (Takebayashi et al. 2009). The authors proposed that the expressional change in the level of thalamic Lmod2 is caused by the drug involvement for regulating psychological and motor function, which eventually causes age-dependent onset of drug-induced schizophrenia. This finding is highly intriguing because before there was no evidence of Lmod presence in brain tissue.
These findings taken together strongly suggest that Tmod and Lmod are essential contributors for the proper and healthy functioning of muscle and non-muscle cells. Further experiments are necessary to clarify how certain Tmod and Lmod isoforms appear in cells and tissues, where they are normally not expressed (Takebayashi et al. 2009; Moyer et al. 2010; Clark et al. 2013).
Tropomyosin
Tropomyosin Isoforms
In vertebrates, four distinct genes (α, β, γ, δ) undergo alternative primary RNA splicing to code for more than 40 TM isoforms (for review see (Gunning et al. 2008)). The TM isoforms are classified in two categories: low-molecular weight (LMW) or short, and high-molecular weight (HMW) or long isoforms. Short TMs consist of 248 amino acids (28.5 kDa), whereas long TMs consist of 284 amino acids (33 kDa) (Lin et al. 1997). The gene products from the alternate promoter selection express exons 1a and 2 in long TM isoforms and exon 1b in short TM isoforms for their N-termini, and exons 9(a-d) for their C-termini.
The structural differences among the isoforms, especially at the N-terminus (Greenfield et al. 1998; Greenfield et al. 2001), give the isoforms divergent functional properties such as actin-binding affinities (Mak et al. 1987; Novy et al. 1993; Fanning et al. 1994; Moraczewska et al. 1999), inclination for head-to-tail association (Cote et al. 1978; Dabrowska et al. 1983) and Tmod-binding affinities (Greenfield and Fowler 2002; Uversky et al. 2011). Different TM isoforms are responsible for the regulation of cell motility and the organization of actin filaments in various cell types.
Structural Organization of Tropomyosin
TM is composed of two monomers that form a coiled coil (Perry 2001). TM molecules form one polymer on each side of the actin filament and coat the grooves of the filaments in a head-to-tail fashion: the N-terminal end of one molecule binds the C-terminal end of the following molecule (Leavis and Gergely 1984). Each TM molecule spans six or seven actin monomers, depending on the TM isoform (Cote and Smillie 1981).
The function of TM is governed strongly by its N-terminus (Cho et al. 1990). Deletion of the first 7 or 9 residues caused the loss of TM’s regulatory function and ability to bind to actin (Cho et al. 1990). The N-terminus of TM also contains a Tmod-binding site (Sung and Lin 1994), while the C-terminus has no effect on Tmod binding (Kostyukova and Hitchcock-DeGregori 2004). Also, unlike the N-terminus of TM, the sequence of C-terminus is highly variable between isoforms.
Because of the flexibility of the TM molecule, it was difficult to obtain high resolution crystals; 3.5Å was the best resolution for full-length TM (Meshcheryakov et al. 2008). Therefore, the structure of various TM isoforms was solved using fragments (Greenfield et al. 1998; Greenfield et al. 2001; Brown et al. 2005; Greenfield et al. 2006; Nitanai et al. 2007; Minakata et al. 2008; Greenfield et al. 2009; Meshcheryakov et al. 2011; Rao et al. 2012).
The N-terminus of TM goes through acetylation, which increases the stability of the N-terminal domain (Greenfield et al. 1994). Acetylation also increases the affinity of long TM isoforms for actin (Hitchcock-DeGregori and Heald 1987; Heald and Hitchcock-DeGregori 1988; Cho et al. 1990; Willadsen et al. 1992) and Tmod (Greenfield and Fowler 2002). Replacing the N-terminal acetyl group with Gly reestablishes the formation of the α-helical coiled-coil and restores the ability of TM to bind to Tmod (Greenfield and Fowler 2002).
Significance of Tropomyosin in Cells
TM is a significant regulator of actin filaments in muscle and non-muscle cells. TM regulates muscle contraction by enhancing the binding of troponin and myosin (Patchell et al. 2005). TM also acts as a stiffening and stabilizing agent for thin filaments (Fattoum et al. 1983) and it is responsible for recruiting specific myosins in epithelial cells (Gupton et al. 2005). Actin filaments coated with TM show resistance to bundling (Burgess et al. 1987), severing, branching (Pruliere et al. 1986; Ishikawa et al. 1989; Blanchoin et al. 2001; DesMarais et al. 2002), fragmentation (Wegner 1982; Fattoum et al. 1983) and disassembly (Bernstein and Bamburg 1982; Nishida et al. 1985) by actin-binding proteins.
TM is known to hold significance in vivo, and it regulates actin-filament organization. It was shown that in budding yeast, a single TM gene disruption resulted in reduced cell growth and the disappearance of cytoplasmic actin cables (Liu and Bretscher 1989). Caenorhabditis elegans show disorganized sarcomeric actin filaments and muscle paralysis when TM expression is suppressed (Ono and Ono 2002). Mutagenesis of TM1, a muscle TM isoform causes alterations in force-generating properties in Drosophila (Kreuz et al. 1996). Mutations in exons 2a and 6b of α-TM, in particular D175N, E180G are associated with familial hypertrophic cardiomyopathy (for review see (Gunning et al. 2008)).
Even very small differences in TM isoforms may be responsible for distinct functional properties and the TM isoform selectivity in cells may arise from these minor dissimilarities at their N-termini. The R14Q mutation in the sequence of short non-muscle α-TM drastically decreased its binding to Tmod1 (Kostyukova et al. 2007). In human skeletal muscle, the M8R mutation weakens the binding of TM to actin (Moraczewska et al. 2000) and to Tmod (Greenfield and Fowler 2002), which leads to nemaline myopathy, a skeletal muscle disease associated with muscle weakness (North et al. 1997). Recently, Ochala et al. showed that E181K and R167H mutations in the TPM2 and TPM3 genes, respectively, cause nemaline myopathy (Ochala et al. 2012). The TPM3-R167H mutation decreased cooperative thin filament activation, myosin cross-bridge number and force production. On the other hand, the TPM2-E181K mutation demonstrated the opposite effect and it increased thin filament activation, myosin cross-bridge binding and force production.
The consequences of altered TM expression and function are not limited to muscle disorders. In the blood cells, an altered ratio of cytoskeletal TM isoforms has been associated with idiopathic hypertension (Dunn et al. 2003). TM is also involved in the formation of neurofibrillary tangles, which are intracellular protein aggregates that cause neurodegenerative disorders such as Alzheimer’s disease (Galloway et al. 1990). TM5NM1 was found to be a major target of oxidative damage in Alzheimer’s disease (Perez-Gracia et al. 2009) and altered levels of TPM3 gene products have been detected in the postmortem brain tissue of subjects diagnosed with schizophrenia (Martins-de-Souza et al. 2009).
In addition to neurological diseases, the growth and spread of cancer is also associated with altered TM expression (for review see (Gunning et al. 2008)). Several studies corroborate that during malignancy the expression of short TM isoforms increases, compensating the decrease in the levels of long TM isoforms. It is believed that short TM isoforms are associated with cancer and are essential for tumor growth (Hughes et al. 2003; Raval et al. 2003; Li et al. 2006; Stehn et al. 2006; Helfman et al. 2008).
The Molecular Basis of Tropomodulin/Tropomyosin Interaction with and without Actin
Tmod-Binding Site of Tropomyosin
An investigation using point mutations made on the N-terminal domain of TM5NM1, a short γ-TM isoform, showed that the Tmod-binding site is located at residues 6–13 (corresponds to residues 7–14 when first Met is present in the sequence) (Vera et al. 2000). To form a coiled coil TMs contain a series of heptad repeats (a, b, c, d, e, f, g), where residues a and d are hydrophobic and link the two alpha helices in TM together. Residues 6, 9 and 13 in the N-terminal region of TM are hydrophobic and they are located at the a and d positions. These residues are responsible for the hydrophobic interaction between Tmod and TM. Mutations that changed the length of the side chains or the hydrophobicity of residues 6 and 13 decreased the binding of γ-TM to Tmod. Additionally, the R11A mutation showed the necessity for a basic residue at the f position of the heptad repeats for Tmod binding (Vera et al. 2000). In this manner, both hydrophobic interactions and ionic interactions play roles in the interaction between TM and Tmod for complex formation.
In long TMs the first 14 N-terminal residues are homologous to residues 6–19 of short TMs and also form a binding site for Tmod (Greenfield and Fowler 2002). Other residues are shown to be responsible for isoform-dependent binding. Mutations S4T and R14Q made in short α-TM were shown to attenuate its affinity to the first TM-binding site in Tmod1 making it similar to that of γ-TM and δ-TM.
TM-Binding Sites of Tropomodulin/Leiomodin
It was originally thought that erythrocyte TMs and skeletal TMs bind to different sites on Tmod, residues 6–94 and 90–184, respectively (Babcock and Fowler 1994). Truncation of Tmod1 showed that residues 1–104 have no ability to bind to short non-muscle γ-TM, but residues 1–127 and longer clones were able to bind (Vera et al. 2005). Later, it was shown that all TM isoforms can bind to both sites though with different affinity (Table 1) (Kostyukova et al. 2007; Uversky et al. 2011). The low affinity of γ-TM for the first TM-binding site of Tmod1 could not be detected earlier due to lower sensitivity of methods used in that study (Babcock and Fowler 1994).
Table 1.
Dissociation constants (Kd) obtained for the complexes of TM and Tmod/Lmod peptides from different isoforms. Tmod peptides contain the TM-binding sites 1 (s1) and 2 (s2). Lmod13–40 and Lmod25–42 were chosen as possible TM-binding sites based on Lmod’s sequence homology with Tmod. TM peptides contain the N-terminal residues of long α-TM (αTM1a), and short α-, γ- and δ-TMs (αTM1b, γTM1b and δTM1b, respectively).
| Peptides | Kd, µM | |||
|---|---|---|---|---|
| αTM1a | αTM1b | γTM1b | δTM1b | |
| Tmod1 s1 | 1.1±0.4a | 0.09±0.02b | Weakc | Weakc |
| Tmod2 s1 | 4.7±0.4a | 0.26±0.2a | 0.6±0.1a | 0.9±0.14a |
| Tmod3 s1 | 1.9±0.3a | 0.24±0.1a | 1.1±0.1a | 1.1±0.1a |
| Tmod4 s1 | 2.4±0.6a | 0.27±0.13a | Weaka | Weaka |
| Lmod13–40 | 1.98d | 0.016±0.009d | 0.61±0.07d | 0.43±0.08d |
| Lmod25–42 | 0.8±0.2d | 0.011±0.008d | 0.6±0.1d | 0.24±0.07d |
| Tmod1 s2 | 1.3±0.3a | 0.003±0.001b | 0.04±0.03c | 0.09±0.02c |
| Tmod2 s2 | 4.4±0.5a | 0.3±0.2a | 0.29±0.11a | 0.14±0.01a |
| Tmod3 s2 | 2.1±0.4a | 0.69±0.07a | 0.86±0.03a | 0.2±0.02a |
| Tmod4 s2 | 3.6±1.2a | 0.43±0.29a | 0.36±0.19a | 2.5±0.5a |
Published by Uversky et al. 2011;
Published by Kostyukova et al. 2006;
Published by Kostyukova et al. 2007;
Published by Kostyukova 2007.
The shortest known fragment of Tmod1 that preserves the function of inhibiting actin filament elongation at the pointed end in the presence of TM is the N-terminal 92 residue fragment, Tmod11–92 (Kostyukova and Hitchcock-DeGregori 2004). Residues 24–35 of this fragment form an amphipathic helix and have a high probability to form a triple coiled coil (Greenfield et al. 2005). Mutations that destabilize the helix (L27G) or destroy the formation of the hydrophobic surface on the helix (L27E) were found to disrupt Tmod11–92’s ability to bind TM (Table 2). A shorter Tmod1 peptide (residues 23–38) that contained only the helical region, did not bind to TM (Kostyukova et al. 2007). Based on these results the first TM-binding site was localized to residues 1–38 of Tmod1.
Table 2.
The effect of mutations on the TM- and actin-binding properties of Tmod.
| Tmod | TM | Mutation in Tmod | Effect of mutation | Method | Reference |
|---|---|---|---|---|---|
| Tmod11–92 | αTM1a/1b zip | L27G | destroyed α-helix formation in TM-binding site 1; caused loss of TM-binding | CD, nondenaturing PAGE | (Greenfield et al. 2005) |
| αTM1a/1b zip | L27E | destroyed the hydrophobic interface in TM-binding site 1; caused loss of TM-binding | |||
| TM5a | L71D | destroyed the hydrophobic interface in actin binding site 1; caused loss of actin binding | pyrene-actin assay | (Kostyukova et al. 2005) | |
| L27G/L71D | caused loss of capping activity | ||||
| T59/G50A/P61A | decreased the flexibility, 10-fold decrease of capping activity | ||||
| Tmod1 | δTM1bzip | A21K/E33V | increased the affinity to αTM1bzip | CD, nondenaturing PAGE | (Uversky et al. 2011; Moroz et al. 2013a) |
| γTM1bzip | |||||
| αTM1bzip | decreased the affinity to αTM1bzip | ||||
| stTM | R11K/D12N/Q144K | decreased the affinity to long α-TM (st TM) | pyrene-actin assay | (Moroz et al. 2013b) | |
| TM5NM1 | L116A | caused loss of binding to γ-TM | IP, solid-phase binding assay | (Vera et al. 2005) | |
| E117R/E118R | |||||
| αTMbzip | I131D | caused loss of binding in TM-binding site 2 | nondenaturing PAGE | (Kostyukova et al. 2006) | |
| TM5NM1 | I134D | decreased TM-binding ability | IP, GST-affinity chromatography | (Kong and Kedes 2006) | |
| L135E | |||||
| L135V | |||||
| Tmod3 | - | K344A | decreased actin nucleating ability and capping ability in the absence of TM | pyrene-actin assay | (Yamashiro et al. 2010) |
| R345A/R346A |
CD, circular dichroism; IP, immunoprecipitation.
A series of point mutations made in the region of residues 90–184 allowed localization of the second TM-binding site. Initially, Vera et al. (2005) stated that residues 105–127 are essential for Tmod’s binding to γ-TM. Later, the L116A and E117R/E118R mutations within this region of Tmod1 were shown to cause a loss of binding to γ-TM (Table 2). The secondary structure prediction for the N-terminal half of Tmod1 stated that residues 126–135 should also form an amphipathic helix (Kostyukova et al. 2006). The I131D mutation that destroyed the hydrophobic surface of this putative helix resulted in a loss of TM-binding ability in the second site (Kostyukova et al. 2006). Other mutations that were done in this helix also affected TM binding (Kong and Kedes 2006). Mutation L135E in Tmod1 caused a loss in its binding ability to short non-muscle γ-TM, TM5NM1, whereas the I134D and L135V mutations caused ~50 % reduction in their binding ability to TM5NM1. Both of these independent studies confirmed that the formation of the amphipathic helix (residues 126–135) is crucial for the second TM-binding site of Tmod (Kong and Kedes 2006; Kostyukova et al. 2006). Finally, the second TM-binding site was localized to residues 109–144 (Kostyukova et al. 2006).
Both TM-binding sites of Tmod show a great conservation among vertebrates. Interestingly, the helix that comprises the first TM-binding site of the only Tmod isoform in amphioxus is significantly less conserved than the second TM-binding site when compared with human Tmod isoforms (Bao et al. 2012). The higher identity of the second TM-binding site of Tmod1 in amphioxus may be explained by the fact that this site has a greater affinity to non-muscle TMs. Since only one Tmod isoform is expressed in these organisms, tight binding of the second TM-binding site to the non-muscle TM isoforms may be crucial for neural system operation. Therefore, the second TM-binding site is evolutionarily more conserved when compared to the first TM-binding site.
To study the affinity of Lmod binding to TM, two Lmod fragments, Lmod13–40 and Lmod25–42, which were chosen based on Lmod’s sequence homology with Tmod, were used (Kostyukova 2007). Lmod-TM interaction is isoform-dependent (Table 1). The binding affinity of Lmod fragments to long TM isoforms was close to that of the first TM-binding site of Tmod1. However, the binding of short α-, γ- and δ-TM isoforms to Lmod fragments was drastically stronger than their binding to Tmod1, with the binding affinity between short α-TM and Lmod fragments being 100-fold higher in particular.
Actin-binding Sites of Tropomodulin
Tmod contains two actin-binding sites, TM-dependent and TM-independent (Fowler et al. 2003; Kostyukova et al. 2005; Kostyukova et al. 2006). The TM-dependent site was localized to residues 48–92 (Greenfield et al. 2005; Kostyukova et al. 2005). It was predicted that residues 65–75 within this region form an amphipathic helix, which was thought to be involved in Tmod-actin binding. The L71D mutation, which destroyed the formation of the hydrophobic surface of this helix, did not affect TM binding but decreased the actin-capping ability (Table 2) (Kostyukova et al. 2005). Tmod138–92, a truncated Tmod1 lacking the first TM-binding site, was shown to be unable to inhibit actin polymerization (Kostyukova et al. 2005). This finding confirmed that binding to TM is crucial for inhibiting actin polymerization and this actin-binding site is TM-dependent. Residues 55–62 in this site were found to have increased flexibility (Greenfield et al. 2005; Kostyukova et al. 2005). It was shown that changing T59, G60 and P61 to alanines caused 10-fold decrease in the ability of Tmod11–92 to inhibit actin polymerization. It was concluded that this flexibility is essential for Tmod’s capping ability. It was suggested that the flexibility is important for the correct positioning of Tmod at the pointed end of thin filaments.
In the recent study of Yamashiro et al. the crosslinking of Tmod3 and G-actin was performed (Yamashiro et al. 2010). It was demonstrated that not only residues within the known N-terminal actin-binding site but also Lys169, a conserved residue located outside of this site, interacted with G-actin. It was concluded that Tmod3 binds actin monomers over an extended interface.
The C-terminal end of Tmod contains another actin-capping site and its function is independent of TM (Fowler et al. 2003; Kostyukova and Hitchcock-DeGregori 2004). Although the exact location of the TM-independent actin-capping site of Tmod is still unknown, it is thought to consist of residues 323–359 in the C-terminal domain (Fowler et al. 2003). Tmod1, without the last 15 C-terminal residues, did not cap actin filaments in the absence of TM; however, it demonstrated the same capping activity as full-length Tmod1 in the presence of TM (Kostyukova and Hitchcock-DeGregori 2004). The current opinion is that the basic residues at the C-terminal end of Tmod are involved in the interaction with actin. Yamashiro et al. showed that mutations K344A and R345A/R346A in Tmod3 had a decreased inhibitory effect for actin polymerization from pointed ends, as well as decreased nucleation ability (Table 2) (Yamashiro et al. 2010). This finding is in concert with the docking model that was proposed earlier for interaction of the C-terminus of Tmod and F-actin (Krieger et al. 2002). On that account, it is plausible that the positively charged cluster (RKRR) composed of residues 340–343 at the C-terminus of Tmod1 is mainly responsible for actin binding. Nonetheless, additional investigations are needed for definitive localization of the TM-independent actin-capping site of Tmod.
Without TM, the affinity of Tmod for the pointed end of actin thin filaments is low, with a Kd of 0.3–0.4 µM (Weber et al. 1994). Tmod is still able to inhibit polymerization and depolymerization of thin filaments without TM, but higher concentrations are necessary. The affinity of Tmod for actin pointed-ends increases dramatically when TM is present and Tmod caps actin filaments with greater efficiency (Kd ~ 50 pM) (Weber et al. 1994; Weber et al. 1999). The affinity of the C-terminal domain (residues 160–359) of Tmod1 for actin is greater than that of the N-terminal domain (residues 1–130) (Fowler et al. 2003). This results in the C-terminal domain having superior actin pointed end-capping activity compared to the N-terminal domain. However, in the presence of TM, the capping activity of the N-terminal domain increases 1000-fold and demonstrates similar affinity levels as the full-length Tmod1, whereas the activity of the C-terminal domain remains unchanged (Fowler et al. 2003).
Although Tmod/TM complex caps the pointed ends of actin filaments tightly in vitro, the capping is transient and dynamic in vivo, and actin monomers are available for exchange from the pointed end (Littlefield et al. 2001).
Sevdali et al. introduced six mutations (G15R, I136M, D154N, V163L, V163M, D292V) to the Drosophila indirect flight muscle-specific actin, which shows 93% homology to human skeletal α-actin (Sevdali et al. 2013). Each of these mutations individually represents a distinct phenotype. All of the heterozygous mutations resulted in changes in muscle structure and function such as muscle weakness, flightlessness, myofiber splitting and disorganization of actin filaments. Although the expression levels and localization of actin and actin-associated proteins (myosin heavy chain, α-actinin, troponin T, TM) was not changed by mutations, surprisingly, Tmod demonstrated disrupted staining and mislocalization in all mutations, except I136M. We believe that actin residues G15, D154, V163 and D292 are important for the interaction of actin with Tmod. Dislocalization of Tmod may be one of the reasons for actin filament disorganization in the myofibrils of Drosophila. In vitro studies on interaction between Tmod and actin with corresponding mutations can elucidate the importance of these residues for Tmod binding.
The Model of Tropomodulin and Tropomyosin Binding at the Pointed End
Kostyukova et al. showed that Tmod1 binds two molecules of the TM peptide containing the N-terminus of short non-muscle α-TM in a cooperative manner (Kostyukova et al. 2006). Later, it was demonstrated that Tmod2 also binds two molecules of TM peptides, which contain the N-terminus of the short non-muscle TM (α, γ or δ), however, there was no cooperativity in the binding (Moroz et al. 2013a).
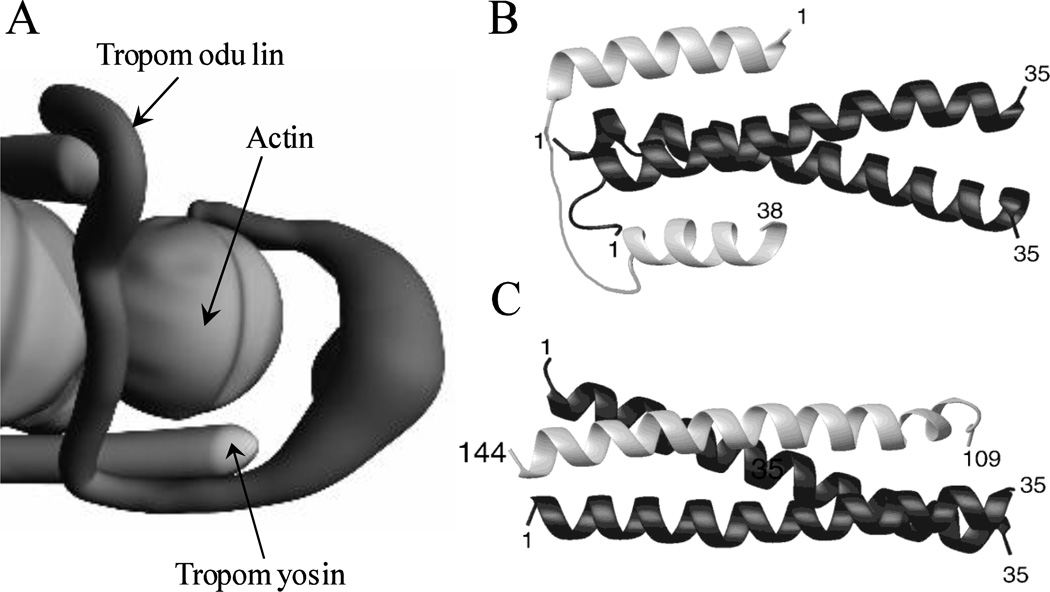
In concordance with the molecular and structural studies, a model for interaction of Tmod1 and short non-muscle α-TM was proposed (Kostyukova et al. 2007). This model suggests that each Tmod molecule binds to the N-termini of two molecules of TM, and interacts with at least one molecule of actin at the pointed end (Figure 2A).
Figure 2.
The model of Tmod’s binding at the pointed end of the actin filament. (A) Cartoon representation of the binding of Tmod to TM and actin molecules at the pointed end. (B) and (C) Ribbon diagrams for the possible binding modes for the first and second TM-binding sites of Tmod, respectively (adapted from (Kostyukova et al. 2007)).
At the first TM-binding site of Tmod1, residues 1–13 lie in an antiparallel fashion on one side of TM and residues 15–26 encircle the N-terminus of TM (Figure 2B). The following residues, 27–38, of Tmod1 close up the loop by binding in parallel with the other side of the TM. The interaction at the second TM-binding site of Tmod1 with TM is executed by formation of a three-helix bundle between residues 121–138 of Tmod1 and residues 6–23 of α-TM (Figure 2C). The orientation of Tmod1 to TM at this site is antiparallel (Kostyukova et al. 2007).
The significance of the localized TM-binding sites were tested in cardiac myocytes (Tsukada et al. 2011). Tmod1, with mutations L27E and I131D, which caused loss of TM-binding in both sites, was found to exhibit either no or very faint pointed end localization. In addition, the expression of single mutations in Tmod1, L27E and I131D revealed that both TM-binding sites are necessary for proper localization of Tmod1 at the pointed ends of thin filaments. However, the second TM-binding site was suggested to be primarily responsible for optimal assembly of Tmod1 in cardiomyocytes (Tsukada et al. 2011). These results support the above-stated binding model.
Lmod was found to localize not only at the pointed ends but also along the thin filaments (Skwarek-Maruszewska et al. 2010; Tsukada et al. 2010). However, the mechanism for the interaction of Lmod with TM and actin at the pointed ends is still not clear. Two hypotheses have been suggested: (1) The final structure and organization of sarcomeres is associated with Lmod through its ability to promote TM-dependent actin filament nucleation (Skwarek-Maruszewska et al. 2010), and (2) Lmod contributes to the elongation of the thin filaments by competing with Tmod1 and antagonizing its capping activity (Tsukada et al. 2010). To create a model for Lmod binding to thin filaments requires more experiments to be done for a better understanding of Lmod’s localization and function on actin filaments. We hypothesize that Lmods utilize different sets of its TM and actin-binding sites: for localization at the pointed end Lmod uses the TM-binding site and first two actin-binding sites, whereas for localization along the actin filament it uses three actin-binding sites only. The different localization patterns of Lmod2 fragments, Lmod44–495 without TM-binding site (Skwarek-Maruszewska et al. 2010) and Lmod21–514 lacking the WH2 domain (Tsukada et al. 2010) support this hypothesis.
Isoform-dependent Interaction of TM and Tmod as a Regulatory Mechanism for Capping Actin Filaments
Various studies suggested that Tmod’s affinity for TM is isoform-specific (Sung et al. 1992; Sung and Lin 1994; Greenfield and Fowler 2002; Kostyukova et al. 2007; Gokhin et al. 2010; Uversky et al. 2011). To investigate the isoform specificity of the Tmod/TM interaction, a thorough analysis of the affinities was done using four TM peptides, which contained the N-terminus of long muscle α-TM and short non-muscle α-, δ- and γ-TMs, and Tmod peptides that corresponded to the first or second TM-binding sites for each Tmod isoform (Uversky et al. 2011) (Table 1). The dissociation constants were obtained for each complex of Tmod/TM peptides. According to these data, short non-muscle δ- and γ-TMs bind to the first TM-binding site of Tmod2 and Tmod3 with a greater affinity than to that of Tmod1 and Tmod4. Whereas, Tmod1’s binding to short non-muscle α-TM is the strongest among all Tmod isoforms (Uversky et al. 2011).
Surprisingly, E-Tmod29 (Tmod1 lacking the first TM- and actin-binding sites) was shown to bind better to TM5NM1 and to G-actin compared to the full-length Tmod1 (2-fold and 1.3-fold, respectively), while binding to F-actin was lower (Yao and Sung 2010). The authors suggested that E-Tmod29 acts as a reservoir for TM5NM1 and G-actin. E-Tmod29 may sequester and release TM5 and G-actin in erythrocyte membranes, making them accessible for Tmod1. However, the solid-phase binding assay used in these experiments is not a quantitative method. Other binding assays should be used to support these findings on E-Tmod29 interactions with TM and actin.
The hypothesis that isoform-dependent interaction of TM and Tmod is one of the regulatory mechanisms of pointed end capping is very tempting. This hypothesis was tested in several studies where the TM-binding ability of one Tmod isoform was altered by mutations and became similar to that of another isoform. The consequences of such alterations were investigated in two types of cells, skeletal muscle myocytes (Moroz et al. 2013b) and PC12, a model cell line used to study neuritogenesis (Moroz et al. 2013a).
Mutations R11K, D12N and Q144K in both TM-binding sites decreased the affinity of Tmod1 to the skeletal muscle α-TM, stTM, making it similar to that of other isoforms, Tmod3 and Tmod4 (Table 2) (Moroz et al. 2013b). As a result, the actin-capping ability of the mutated Tmod1 in the presence of stTM also decreased 3-fold (Moroz et al. 2013b). Moreover, assembly of this mutant at the pointed end of thin filaments in the sarcomeres of skeletal myocytes was shown to decrease by ~35%. Together, these data indicate that TM-binding affinity influences actin-capping ability and localization of Tmod in myocytes.
In neurons and PC12 cells, a model system for neuronal differentiation, Tmod1 was found to be associated with the F-actin bundles in the lamellipodia and growth cones, whereas diffused Tmod2 was found in the cytoplasmic compartment of the cell body and the central domain of the growth cone (Fath et al. 2011; Moroz et al. 2013a). Overexpression of Tmod1 did not affect neurite formation, while overexpression of Tmod2 caused drastic reduction in the number and the length of neurites. Mutations A21K and E33V were introduced to Tmod1 in the first TM-binding site to alter its TM-binding ability (Moroz et al. 2013a). Earlier these mutations were shown to increase the binding affinity to short δ- and γ-TM isoforms found in brain cells, TM4 and TM5NM1, respectively, making this affinity similar to that of Tmod2 (Table 2) (Uversky et al. 2011). These mutations also were found to decrease binding to short nonmuscle α-TMs (Moroz et al. 2013a). Overexpression of Tmod1 with A21K/E33V mutations in PC12 cells exhibited localization similar to Tmod2 (Moroz et al. 2013a). There was only a slight decrease in the number of neurites per cell, while at the same time, the average length of neurites shortened by 2- to 3-fold.
Conclusions
Tmod and TM are crucial components of the cytoskeleton. Selective binding of Tmod to TM isoforms, which is induced by their specific binding affinities, influences the localization and activity of each Tmod isoform in different cell types. Interaction of Tmod with TM and actin takes place in a precise and strictly regulated manner. Indeed, even minor structural changes, such as a single mutation, in these proteins are adequate for instigating drastic transformations in protein behavior, cytoskeletal architecture and operation of cells. Recent studies have begun to concentrate on the purpose and consequences of isoform specificity in both Tmod and TM isoforms. From our point of view, there are three worthwhile directions to study and elucidate the role of Tmod-TM interplay in actin dynamics. The first direction is further exploration of isoform-dependent interaction between these two proteins by altering Tmod affinity to specific TM isoforms. The second direction is determination of the pointed end structure. It is a very difficult task but it can be done by determining the structures of Tmod/Lmod-actin and Tmod/Lmod-TM complexes (full-length or fragments) separately using x-ray crystallography and/or NMR. These structures then can be combined into one structural model with the help of high-resolution electron microscopy. The third direction is to study the influence of Tmod-TM interactions on the formation of cytoskeleton in non-muscle cells. Future investigations will provide augmented insight on formation of the intracellular network and filament dynamics in living cells.
Acknowledgements
This work was supported by a National Institutes of Health grant GM081688 to ASK.
References
- Almenar-Queralt A, Lee A, Conley CA, Ribas de Pouplana L, Fowler VM. Identification of a novel tropomodulin isoform, skeletal tropomodulin, that caps actin filament pointed ends in fast skeletal muscle. The Journal of biological chemistry. 1999;274(40):28466–28475. doi: 10.1074/jbc.274.40.28466. [DOI] [PubMed] [Google Scholar]
- Babcock GG, Fowler VM. Isoform-specific interaction of tropomodulin with skeletal muscle and erythrocyte tropomyosins. The Journal of biological chemistry. 1994;269(44):27510–27518. [PubMed] [Google Scholar]
- Bao Y, Kake T, Hanashima A, Nomiya Y, Kubokawa K, Kimura S. Actin capping proteins, CapZ (beta-actinin) and tropomodulin in amphioxus striated muscle. Gene. 2012;510(1):78–86. doi: 10.1016/j.gene.2012.07.081. [DOI] [PubMed] [Google Scholar]
- Bernstein BW, Bamburg JR. Tropomyosin binding to F-actin protects the F-actin from disassembly by brain actin-depolymerizing factor (ADF) Cell motility. 1982;2(1):1–8. doi: 10.1002/cm.970020102. [DOI] [PubMed] [Google Scholar]
- Blanchoin L, Pollard TD, Hitchcock-DeGregori SE. Inhibition of the Arp2/3 complex-nucleated actin polymerization and branch formation by tropomyosin. Current biology: CB. 2001;11(16):1300–1304. doi: 10.1016/s0960-9822(01)00395-5. [DOI] [PubMed] [Google Scholar]
- Brown JH, Zhou Z, Reshetnikova L, Robinson H, Yammani RD, Tobacman LS, Cohen C. Structure of the mid-region of tropomyosin: bending and binding sites for actin. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(52):18878–18883. doi: 10.1073/pnas.0509269102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess DR, Broschat KO, Hayden JM. Tropomyosin distinguishes between the two actin-binding sites of villin and affects actin-binding properties of other brush border proteins. The Journal of cell biology. 1987;104(1):29–40. doi: 10.1083/jcb.104.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan MM, Wooden JM, Tsang M, Gilligan DM, Hirenallur SD, Finney GL, Rynes E, Maccoss M, Ramirez JA, Park H, Iritani BM. Hematopoietic protein-1 regulates the actin membrane skeleton and membrane stability in murine erythrocytes. PloS one. 2013;8(2):e54902. doi: 10.1371/journal.pone.0054902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen A, Liao WP, Lu Q, Wong WS, Wong PT. Upregulation of dihydropyrimidinase-related protein 2, spectrin alpha II chain, heat shock cognate protein 70 pseudogene 1 and tropomodulin 2 after focal cerebral ischemia in rats--a proteomics approach. Neurochemistry international. 2007;50(7–8):1078–1086. doi: 10.1016/j.neuint.2006.11.008. [DOI] [PubMed] [Google Scholar]
- Chereau D, Boczkowska M, Skwarek-Maruszewska A, Fujiwara I, Hayes DB, Rebowski G, Lappalainen P, Pollard TD, Dominguez R. Leiomodin is an actin filament nucleator in muscle cells. Science. 2008;320(5873):239–243. doi: 10.1126/science.1155313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho YJ, Liu J, Hitchcock-DeGregori SE. The amino terminus of muscle tropomyosin is a major determinant for function. The Journal of biological chemistry. 1990;265(1):538–545. [PubMed] [Google Scholar]
- Chu X, Chen J, Reedy MC, Vera C, Sung KL, Sung LA. E-Tmod capping of actin filaments at the slow-growing end is required to establish mouse embryonic circulation. American journal of physiology Heart and circulatory physiology. 2003;284(5):H1827–H1838. doi: 10.1152/ajpheart.00947.2002. [DOI] [PubMed] [Google Scholar]
- Clark R, Nosie A, Walker T, Faralli JA, Filla MS, Barrett-Wilt G, Peters DM. Comparative genomic and proteomic analysis of cytoskeletal changes in dexamethasone-treated trabecular meshwork cells. Molecular & cellular proteomics : MCP. 2013;12(1):194–206. doi: 10.1074/mcp.M112.019745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conley CA, Fritz-Six KL, Almenar-Queralt A, Fowler VM. Leiomodins: larger members of the tropomodulin (Tmod) gene family. Genomics. 2001;73(2):127–139. doi: 10.1006/geno.2000.6501. [DOI] [PubMed] [Google Scholar]
- Cote G, Lewis WG, Smillie LB. Non-polymerizability of platelet tropomyosin and its NH2- and COOH-terminal sequences. FEBS letters. 1978;91(2):237–241. doi: 10.1016/0014-5793(78)81181-8. [DOI] [PubMed] [Google Scholar]
- Cote GP, Smillie LB. The interaction of equine platelet tropomyosin with skeletal muscle actin. The Journal of biological chemistry. 1981;256(14):7257–7261. [PubMed] [Google Scholar]
- Cox PR, Fowler V, Xu B, Sweatt JD, Paylor R, Zoghbi HY. Mice lacking Tropomodulin-2 show enhanced long-term potentiation, hyperactivity, and deficits in learning and memory. Molecular and cellular neurosciences. 2003;23(1):1–12. doi: 10.1016/s1044-7431(03)00025-3. [DOI] [PubMed] [Google Scholar]
- Cox PR, Zoghbi HY. Sequencing, expression analysis, and mapping of three unique human tropomodulin genes and their mouse orthologs. Genomics. 2000;63(1):97–107. doi: 10.1006/geno.1999.6061. [DOI] [PubMed] [Google Scholar]
- Dabrowska R, Nowak E, Drabikowski W. Some functional properties of nonpolymerizable and polymerizable tropomyosin. Journal of muscle research and cell motility. 1983;4(2):143–161. doi: 10.1007/BF00712027. [DOI] [PubMed] [Google Scholar]
- DesMarais V, Ichetovkin I, Condeelis J, Hitchcock-DeGregori SE. Spatial regulation of actin dynamics: a tropomyosin-free, actin-rich compartment at the leading edge. Journal of cell science. 2002;115(Pt 23):4649–4660. doi: 10.1242/jcs.00147. [DOI] [PubMed] [Google Scholar]
- dos Remedios CG, Chhabra D, Kekic M, Dedova IV, Tsubakihara M, Berry DA, Nosworthy NJ. Actin binding proteins: regulation of cytoskeletal microfilaments. Physiological reviews. 2003;83(2):433–473. doi: 10.1152/physrev.00026.2002. [DOI] [PubMed] [Google Scholar]
- Dunn SA, Mohteshamzadeh M, Daly AK, Thomas TH. Altered tropomyosin expression in essential hypertension. Hypertension. 2003;41(2):347–354. doi: 10.1161/01.hyp.0000050646.79785.7c. [DOI] [PubMed] [Google Scholar]
- Dye CA, Lee JK, Atkinson RC, Brewster R, Han PL, Bellen HJ. The Drosophila sanpodo gene controls sibling cell fate and encodes a tropomodulin homolog, an actin/tropomyosin-associated protein. Development. 1998;125(10):1845–1856. doi: 10.1242/dev.125.10.1845. [DOI] [PubMed] [Google Scholar]
- Fanning AS, Wolenski JS, Mooseker MS, Izant JG. Differential regulation of skeletal muscle myosin-II and brush border myosin-I enzymology and mechanochemistry by bacterially produced tropomyosin isoforms. Cell motility and the cytoskeleton. 1994;29(1):29–45. doi: 10.1002/cm.970290104. [DOI] [PubMed] [Google Scholar]
- Fath T, Fischer RS, Dehmelt L, Halpain S, Fowler VM. Tropomodulins are negative regulators of neurite outgrowth. European journal of cell biology. 2011;90(4):291–300. doi: 10.1016/j.ejcb.2010.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fattoum A, Hartwig JH, Stossel TP. Isolation and some structural and functional properties of macrophage tropomyosin. Biochemistry. 1983;22(5):1187–1193. doi: 10.1021/bi00274a031. [DOI] [PubMed] [Google Scholar]
- Fischer RS, Lee A, Fowler VM. Tropomodulin and tropomyosin mediate lens cell actin cytoskeleton reorganization in vitro. Investigative ophthalmology & visual science. 2000;41(1):166–174. [PubMed] [Google Scholar]
- Fischer RS, Quinlan RA, Fowler VM. Tropomodulin binds to filensin intermediate filaments. FEBS letters. 2003;547(1–3):228–232. doi: 10.1016/s0014-5793(03)00711-7. [DOI] [PubMed] [Google Scholar]
- Fischer RS, Yarmola EG, Weber KL, Speicher KD, Speicher DW, Bubb MR, Fowler VM. Tropomodulin 3 binds to actin monomers. The Journal of biological chemistry. 2006;281(47):36454–36465. doi: 10.1074/jbc.M606315200. [DOI] [PubMed] [Google Scholar]
- Fowler VM. Identification and purification of a novel Mr 43,000 tropomyosin-binding protein from human erythrocyte membranes. The Journal of biological chemistry. 1987;262(26):12792–12800. [PubMed] [Google Scholar]
- Fowler VM, Greenfield NJ, Moyer J. Tropomodulin contains two actin filament pointed end-capping domains. The Journal of biological chemistry. 2003;278(41):40000–40009. doi: 10.1074/jbc.M306895200. [DOI] [PubMed] [Google Scholar]
- Fowler VM, Sussmann MA, Miller PG, Flucher BE, Daniels MP. Tropomodulin is associated with the free (pointed) ends of the thin filaments in rat skeletal muscle. The Journal of cell biology. 1993;120(2):411–420. doi: 10.1083/jcb.120.2.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritz-Six KL, Cox PR, Fischer RS, Xu B, Gregorio CC, Zoghbi HY, Fowler VM. Aberrant myofibril assembly in tropomodulin1 null mice leads to aborted heart development and embryonic lethality. The Journal of cell biology. 2003;163(5):1033–1044. doi: 10.1083/jcb.200308164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujisawa T, Kostyukova A, Maeda Y. The shapes and sizes of two domains of tropomodulin, the P-end-capping protein of actin-tropomyosin. FEBS letters. 2001;498(1):67–71. doi: 10.1016/s0014-5793(01)02498-x. [DOI] [PubMed] [Google Scholar]
- Galloway PG, Mulvihill P, Siedlak S, Mijares M, Kawai M, Padget H, Kim R, Perry G. Immunochemical demonstration of tropomyosin in the neurofibrillary pathology of Alzheimer's disease. The American journal of pathology. 1990;137(2):291–300. [PMC free article] [PubMed] [Google Scholar]
- Gokhin DS, Lewis RA, McKeown CR, Nowak RB, Kim NE, Littlefield RS, Lieber RL, Fowler VM. Tropomodulin isoforms regulate thin filament pointed-end capping and skeletal muscle physiology. The Journal of cell biology. 2010;189(1):95–109. doi: 10.1083/jcb.201001125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodenough DA. The crystalline lens. A system networked by gap junctional intercellular communication. Seminars in cell biology. 1992;3(1):49–58. doi: 10.1016/s1043-4682(10)80007-8. [DOI] [PubMed] [Google Scholar]
- Goodenough DA, Goliger JA, Paul DL. Connexins, connexons, and intercellular communication. Annual review of biochemistry. 1996;65:475–502. doi: 10.1146/annurev.bi.65.070196.002355. [DOI] [PubMed] [Google Scholar]
- Greenfield NJ, Fowler VM. Tropomyosin requires an intact N-terminal coiled coil to interact with tropomodulin. Biophysical journal. 2002;82(5):2580–2591. doi: 10.1016/S0006-3495(02)75600-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenfield NJ, Huang YJ, Palm T, Swapna GV, Monleon D, Montelione GT, Hitchcock-DeGregori SE. Solution NMR structure and folding dynamics of the N terminus of a rat non-muscle alpha-tropomyosin in an engineered chimeric protein. Journal of molecular biology. 2001;312(4):833–847. doi: 10.1006/jmbi.2001.4982. [DOI] [PubMed] [Google Scholar]
- Greenfield NJ, Huang YJ, Swapna GV, Bhattacharya A, Rapp B, Singh A, Montelione GT, Hitchcock-DeGregori SE. Solution NMR structure of the junction between tropomyosin molecules: implications for actin binding and regulation. Journal of molecular biology. 2006;364(1):80–96. doi: 10.1016/j.jmb.2006.08.033. [DOI] [PubMed] [Google Scholar]
- Greenfield NJ, Kostyukova AS, Hitchcock-DeGregori SE. Structure and tropomyosin binding properties of the N-terminal capping domain of tropomodulin 1. Biophysical journal. 2005;88(1):372–383. doi: 10.1529/biophysj.104.051128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenfield NJ, Kotlyanskaya L, Hitchcock-DeGregori SE. Structure of the N terminus of a nonmuscle alpha-tropomyosin in complex with the C terminus: implications for actin binding. Biochemistry. 2009;48(6):1272–1283. doi: 10.1021/bi801861k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenfield NJ, Montelione GT, Farid RS, Hitchcock-DeGregori SE. The structure of the N-terminus of striated muscle alpha-tropomyosin in a chimeric peptide: nuclear magnetic resonance structure and circular dichroism studies. Biochemistry. 1998;37(21):7834–7843. doi: 10.1021/bi973167m. [DOI] [PubMed] [Google Scholar]
- Greenfield NJ, Stafford WF, Hitchcock-DeGregori SE. The effect of N-terminal acetylation on the structure of an N-terminal tropomyosin peptide and alpha alpha-tropomyosin. Protein science : a publication of the Protein Society. 1994;3(3):402–410. doi: 10.1002/pro.5560030304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregorio CC, Fowler VM. Mechanisms of thin filament assembly in embryonic chick cardiac myocytes: tropomodulin requires tropomyosin for assembly. The Journal of cell biology. 1995;129(3):683–695. doi: 10.1083/jcb.129.3.683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregorio CC, Weber A, Bondad M, Pennise CR, Fowler VM. Requirement of pointed-end capping by tropomodulin to maintain actin filament length in embryonic chick cardiac myocytes. Nature. 1995;377(6544):83–86. doi: 10.1038/377083a0. [DOI] [PubMed] [Google Scholar]
- Gunning P, O'Neill G, Hardeman E. Tropomyosin-based regulation of the actin cytoskeleton in time and space. Physiological reviews. 2008;88(1):1–35. doi: 10.1152/physrev.00001.2007. [DOI] [PubMed] [Google Scholar]
- Gupton SL, Anderson KL, Kole TP, Fischer RS, Ponti A, Hitchcock-DeGregori SE, Danuser G, Fowler VM, Wirtz D, Hanein D, Waterman-Storer CM. Cell migration without a lamellipodium: translation of actin dynamics into cell movement mediated by tropomyosin. The Journal of cell biology. 2005;168(4):619–631. doi: 10.1083/jcb.200406063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heald RW, Hitchcock-DeGregori SE. The structure of the amino terminus of tropomyosin is critical for binding to actin in the absence and presence of troponin. The Journal of biological chemistry. 1988;263(11):5254–5259. [PubMed] [Google Scholar]
- Helfman DM, Flynn P, Khan P, Saeed A. Tropomyosin as a regulator of cancer cell transformation. Advances in experimental medicine and biology. 2008;644:124–131. doi: 10.1007/978-0-387-85766-4_10. [DOI] [PubMed] [Google Scholar]
- Hitchcock-DeGregori SE, Heald RW. Altered actin and troponin binding of amino-terminal variants of chicken striated muscle alpha-tropomyosin expressed in Escherichia coli. The Journal of biological chemistry. 1987;262(20):9730–9735. [PubMed] [Google Scholar]
- Hughes JA, Cooke-Yarborough CM, Chadwick NC, Schevzov G, Arbuckle SM, Gunning P, Weinberger RP. High-molecular-weight tropomyosins localize to the contractile rings of dividing CNS cells but are absent from malignant pediatric and adult CNS tumors. Glia. 2003;42(1):25–35. doi: 10.1002/glia.10174. [DOI] [PubMed] [Google Scholar]
- Ishikawa R, Yamashiro S, Matsumura F. Differential modulation of actin-severing activity of gelsolin by multiple isoforms of cultured rat cell tropomyosin. Potentiation of protective ability of tropomyosins by 83-kDa nonmuscle caldesmon. The Journal of biological chemistry. 1989;264(13):7490–7497. [PubMed] [Google Scholar]
- Ito M, Swanson B, Sussman MA, Kedes L, Lyons G. Cloning of tropomodulin cDNA and localization of gene transcripts during mouse embryogenesis. Developmental biology. 1995;167(1):317–328. doi: 10.1006/dbio.1995.1026. [DOI] [PubMed] [Google Scholar]
- Iwazaki T, McGregor IS, Matsumoto I. Protein expression profile in the striatum of acute methamphetamine-treated rats. Brain research. 2006;1097(1):19–25. doi: 10.1016/j.brainres.2006.04.052. [DOI] [PubMed] [Google Scholar]
- Kobe B, Deisenhofer J. The leucine-rich repeat: a versatile binding motif. Trends in biochemical sciences. 1994;19(10):415–421. doi: 10.1016/0968-0004(94)90090-6. [DOI] [PubMed] [Google Scholar]
- Kong KY, Kedes L. Leucine 135 of tropomodulin-1 regulates its association with tropomyosin, its cellular localization, and the integrity of sarcomeres. The Journal of biological chemistry. 2006;281(14):9589–9599. doi: 10.1074/jbc.M512064200. [DOI] [PubMed] [Google Scholar]
- Kostyukova A, Maeda K, Yamauchi E, Krieger I, Maeda Y. Domain structure of tropomodulin: distinct properties of the N-terminal and C-terminal halves. European journal of biochemistry / FEBS. 2000;267(21):6470–6475. doi: 10.1046/j.1432-1327.2000.01738.x. [DOI] [PubMed] [Google Scholar]
- Kostyukova AS. Leiomodin/tropomyosin interactions are isoform specific. Archives of biochemistry and biophysics. 2007;465(1):227–230. doi: 10.1016/j.abb.2007.05.012. [DOI] [PubMed] [Google Scholar]
- Kostyukova AS, Choy A, Rapp BA. Tropomodulin binds two tropomyosins: a novel model for actin filament capping. Biochemistry. 2006;45(39):12068–12075. doi: 10.1021/bi060899i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostyukova AS, Hitchcock-DeGregori SE. Effect of the structure of the N terminus of tropomyosin on tropomodulin function. The Journal of biological chemistry. 2004;279(7):5066–5071. doi: 10.1074/jbc.M311186200. [DOI] [PubMed] [Google Scholar]
- Kostyukova AS, Hitchcock-Degregori SE, Greenfield NJ. Molecular basis of tropomyosin binding to tropomodulin, an actin-capping protein. Journal of molecular biology. 2007;372(3):608–618. doi: 10.1016/j.jmb.2007.05.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostyukova AS, Rapp BA, Choy A, Greenfield NJ, Hitchcock-DeGregori SE. Structural requirements of tropomodulin for tropomyosin binding and actin filament capping. Biochemistry. 2005;44(12):4905–4910. doi: 10.1021/bi047468p. [DOI] [PubMed] [Google Scholar]
- Kostyukova AS, Tiktopulo EI, Maeda Y. Folding properties of functional domains of tropomodulin. Biophysical journal. 2001;81(1):345–351. doi: 10.1016/S0006-3495(01)75704-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreuz AJ, Simcox A, Maughan D. Alterations in flight muscle ultrastructure and function in Drosophila tropomyosin mutants. The Journal of cell biology. 1996;135(3):673–687. doi: 10.1083/jcb.135.3.673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krieger I, Kostyukova A, Yamashita A, Nitanai Y, Maeda Y. Crystal structure of the C-terminal half of tropomodulin and structural basis of actin filament pointed-end capping. Biophysical journal. 2002;83(5):2716–2725. doi: 10.1016/S0006-3495(02)75281-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leavis PC, Gergely J. Thin filament proteins and thin filament-linked regulation of vertebrate muscle contraction. CRC critical reviews in biochemistry. 1984;16(3):235–305. doi: 10.3109/10409238409108717. [DOI] [PubMed] [Google Scholar]
- Li DQ, Wang L, Fei F, Hou YF, Luo JM, Wei C, Zeng R, Wu J, Lu JS, Di GH, Ou ZL, Xia QC, Shen ZZ, Shao ZM. Identification of breast cancer metastasis-associated proteins in an isogenic tumor metastasis model using two-dimensional gel electrophoresis and liquid chromatography-ion trap-mass spectrometry. Proteomics. 2006;6(11):3352–3368. doi: 10.1002/pmic.200500617. [DOI] [PubMed] [Google Scholar]
- Lin JJ, Warren KS, Wamboldt DD, Wang T, Lin JL. Tropomyosin isoforms in nonmuscle cells. International review of cytology. 1997;170:1–38. doi: 10.1016/s0074-7696(08)61619-8. [DOI] [PubMed] [Google Scholar]
- Littlefield R, Almenar-Queralt A, Fowler VM. Actin dynamics at pointed ends regulates thin filament length in striated muscle. Nature cell biology. 2001;3(6):544–551. doi: 10.1038/35078517. [DOI] [PubMed] [Google Scholar]
- Liu HP, Bretscher A. Disruption of the single tropomyosin gene in yeast results in the disappearance of actin cables from the cytoskeleton. Cell. 1989;57(2):233–242. doi: 10.1016/0092-8674(89)90961-6. [DOI] [PubMed] [Google Scholar]
- Mak AS, Roseborough G, Baker H. Tropomyosin from human erythrocyte membrane polymerizes poorly but binds F-actin effectively in the presence and absence of spectrin. Biochimica et biophysica acta. 1987;912(2):157–166. doi: 10.1016/0167-4838(87)90084-7. [DOI] [PubMed] [Google Scholar]
- Martins-de-Souza D, Gattaz WF, Schmitt A, Novello JC, Marangoni S, Turck CW, Dias-Neto E. Proteome analysis of schizophrenia patients Wernicke's area reveals an energy metabolism dysregulation. BMC psychiatry. 2009;9:17. doi: 10.1186/1471-244X-9-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKeown CR, Nowak RB, Moyer J, Sussman MA, Fowler VM. Tropomodulin1 is required in the heart but not the yolk sac for mouse embryonic development. Circulation research. 2008;103(11):1241–1248. doi: 10.1161/CIRCRESAHA.108.178749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meshcheryakov V, Nitanai Y, Maytum R, Geeves MA, Maeda Y. Crystallization and preliminary X-ray crystallographic analysis of full-length yeast tropomyosin 2 from Saccharomyces cerevisiae. Acta crystallographica Section F, Structural biology and crystallization communications. 2008;64(Pt 6):528–530. doi: 10.1107/S1744309108013110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meshcheryakov VA, Krieger I, Kostyukova AS, Samatey FA. Structure of a tropomyosin N-terminal fragment at 0.98 A resolution. Acta crystallographica Section D, Biological crystallography. 2011;67(Pt 9):822–825. doi: 10.1107/S090744491102645X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minakata S, Maeda K, Oda N, Wakabayashi K, Nitanai Y, Maeda Y. Two-crystal structures of tropomyosin C-terminal fragment 176–273: exposure of the hydrophobic core to the solvent destabilizes the tropomyosin molecule. Biophysical journal. 2008;95(2):710–719. doi: 10.1529/biophysj.107.126144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moraczewska J, Greenfield NJ, Liu Y, Hitchcock-DeGregori SE. Alteration of tropomyosin function and folding by a nemaline myopathy-causing mutation. Biophysical journal. 2000;79(6):3217–3225. doi: 10.1016/S0006-3495(00)76554-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moraczewska J, Nicholson-Flynn K, Hitchcock-DeGregori SE. The ends of tropomyosin are major determinants of actin affinity and myosin subfragment 1-induced binding to F-actin in the open state. Biochemistry. 1999;38(48):15885–15892. doi: 10.1021/bi991816j. [DOI] [PubMed] [Google Scholar]
- Moroz NA, Guillaud L, Desai B, Kostyukova AS. Mutations changing tropomodulin affinity for tropomyosin alter neurite formation and extension. PeerJ. 2013a;1(7) doi: 10.7717/peerj.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moroz NA, Novak SM, Azevedo R, Colpan M, Uversky VN, Gregorio CC, Kostyukova AS. Alteration of Tropomyosin-binding Properties of Tropomodulin-1 Affects Its Capping Ability and Localization in Skeletal Myocytes. The Journal of biological chemistry. 2013b;288(7):4899–4907. doi: 10.1074/jbc.M112.434522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyer JD, Nowak RB, Kim NE, Larkin SK, Peters LL, Hartwig J, Kuypers FA, Fowler VM. Tropomodulin 1-null mice have a mild spherocytic elliptocytosis with appearance of tropomodulin 3 in red blood cells and disruption of the membrane skeleton. Blood. 2010;116(14):2590–2599. doi: 10.1182/blood-2010-02-268458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishida E, Muneyuki E, Maekawa S, Ohta Y, Sakai H. An actin-depolymerizing protein (destrin) from porcine kidney. Its action on F-actin containing or lacking tropomyosin. Biochemistry. 1985;24(23):6624–6630. doi: 10.1021/bi00344a049. [DOI] [PubMed] [Google Scholar]
- Nitanai Y, Minakata S, Maeda K, Oda N, Maeda Y. Crystal structures of tropomyosin: flexible coiled-coil. Advances in experimental medicine and biology. 2007;592:137–151. doi: 10.1007/978-4-431-38453-3_13. [DOI] [PubMed] [Google Scholar]
- North KN, Laing NG, Wallgren-Pettersson C. Nemaline myopathy: current concepts. The ENMC International Consortium and Nemaline Myopathy. J Med Genet. 1997;34(9):705–713. doi: 10.1136/jmg.34.9.705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novy RE, Sellers JR, Liu LF, Lin JJ. In vitro functional characterization of bacterially expressed human fibroblast tropomyosin isoforms and their chimeric mutants. Cell motility and the cytoskeleton. 1993;26(3):248–261. doi: 10.1002/cm.970260308. [DOI] [PubMed] [Google Scholar]
- Ochala J, Ravenscroft G, Laing NG, Nowak KJ. Nemaline myopathy-related skeletal muscle alpha-actin (ACTA1) mutation, Asp286Gly, prevents proper strong myosin binding and triggers muscle weakness. PloS one. 2012;7(9):e45923. doi: 10.1371/journal.pone.0045923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono S, Ono K. Tropomyosin inhibits ADF/cofilin-dependent actin filament dynamics. The Journal of cell biology. 2002;156(6):1065–1076. doi: 10.1083/jcb.200110013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono Y, Schwach C, Antin PB, Gregorio CC. Disruption in the tropomodulin1 (Tmod1) gene compromises cardiomyocyte development in murine embryonic stem cells by arresting myofibril maturation. Developmental biology. 2005;282(2):336–348. doi: 10.1016/j.ydbio.2005.03.015. [DOI] [PubMed] [Google Scholar]
- Patchell VB, Gallon CE, Evans JS, Gao Y, Perry SV, Levine BA. The regulatory effects of tropomyosin and troponin-I on the interaction of myosin loop regions with F-actin. The Journal of biological chemistry. 2005;280(15):14469–14475. doi: 10.1074/jbc.M414202200. [DOI] [PubMed] [Google Scholar]
- Perez-Gracia E, Blanco R, Carmona M, Carro E, Ferrer I. Oxidative stress damage and oxidative stress responses in the choroid plexus in Alzheimer's disease. Acta neuropathologica. 2009;118(4):497–504. doi: 10.1007/s00401-009-0574-4. [DOI] [PubMed] [Google Scholar]
- Perry SV. Vertebrate tropomyosin: distribution, properties and function. Journal of muscle research and cell motility. 2001;22(1):5–49. doi: 10.1023/a:1010303732441. [DOI] [PubMed] [Google Scholar]
- Pollard TD, Cooper JA. Actin, a central player in cell shape and movement. Science. 2009;326(5957):1208–1212. doi: 10.1126/science.1175862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruliere G, d'Albis A, der Terrossian E. Effect of tropomyosin on the interactions of actin with actin-binding proteins isolated from pig platelets. European journal of biochemistry / FEBS. 1986;159(3):535–547. doi: 10.1111/j.1432-1033.1986.tb09920.x. [DOI] [PubMed] [Google Scholar]
- Rao JN, Rivera-Santiago R, Li XE, Lehman W, Dominguez R. Structural analysis of smooth muscle tropomyosin alpha and beta isoforms. The Journal of biological chemistry. 2012;287(5):3165–3174. doi: 10.1074/jbc.M111.307330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raval GN, Bharadwaj S, Levine EA, Willingham MC, Geary RL, Kute T, Prasad GL. Loss of expression of tropomyosin-1, a novel class II tumor suppressor that induces anoikis, in primary breast tumors. Oncogene. 2003;22(40):6194–6203. doi: 10.1038/sj.onc.1206719. [DOI] [PubMed] [Google Scholar]
- Sevdali M, Kumar V, Peckham M, Sparrow J. Human congenital myopathy actin mutants cause myopathy and alter Z-disc structure in Drosophila flight muscle. Neuromuscular disorders : NMD. 2013;23(3):243–255. doi: 10.1016/j.nmd.2012.11.013. [DOI] [PubMed] [Google Scholar]
- Skwarek-Maruszewska A, Boczkowska M, Zajac AL, Kremneva E, Svitkina T, Dominguez R, Lappalainen P. Different localizations and cellular behaviors of leiomodin and tropomodulin in mature cardiomyocyte sarcomeres. Molecular biology of the cell. 2010;21(19):3352–3361. doi: 10.1091/mbc.E10-02-0109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stehn JR, Schevzov G, O'Neill GM, Gunning PW. Specialisation of the tropomyosin composition of actin filaments provides new potential targets for chemotherapy. Current cancer drug targets. 2006;6(3):245–256. doi: 10.2174/156800906776842948. [DOI] [PubMed] [Google Scholar]
- Sung KL, Yang L, Whittemore DE, Shi Y, Jin G, Hsieh AH, Akeson WH, Sung LA. The differential adhesion forces of anterior cruciate and medial collateral ligament fibroblasts: effects of tropomodulin, talin, vinculin, and alpha-actinin. Proceedings of the National Academy of Sciences of the United States of America. 1996;93(17):9182–9187. doi: 10.1073/pnas.93.17.9182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung LA, Fowler VM, Lambert K, Sussman MA, Karr D, Chien S. Molecular cloning and characterization of human fetal liver tropomodulin A tropomyosin-binding protein. The Journal of biological chemistry. 1992;267(4):2616–2621. [PubMed] [Google Scholar]
- Sung LA, Gao KM, Yee LJ, Temm-Grove CJ, Helfman DM, Lin JJ, Mehrpouryan M. Tropomyosin isoform 5b is expressed in human erythrocytes: implications of tropomodulin-TM5 or tropomodulin-TM5b complexes in the protofilament and hexagonal organization of membrane skeletons. Blood. 2000;95(4):1473–1480. [PubMed] [Google Scholar]
- Sung LA, Lin JJ. Erythrocyte tropomodulin binds to the N-terminus of hTM5, a tropomyosin isoform encoded by the gamma-tropomyosin gene. Biochemical and biophysical research communications. 1994;201(2):627–634. doi: 10.1006/bbrc.1994.1747. [DOI] [PubMed] [Google Scholar]
- Sussman MA, Baque S, Uhm CS, Daniels MP, Price RL, Simpson D, Terracio L, Kedes L. Altered expression of tropomodulin in cardiomyocytes disrupts the sarcomeric structure of myofibrils. Circulation research. 1998a;82(1):94–105. doi: 10.1161/01.res.82.1.94. [DOI] [PubMed] [Google Scholar]
- Sussman MA, Sakhi S, Barrientos P, Ito M, Kedes L. Tropomodulin in rat cardiac muscle. Localization of protein is independent of messenger RNA distribution during myofibrillar development. Circulation research. 1994a;75(2):221–232. doi: 10.1161/01.res.75.2.221. [DOI] [PubMed] [Google Scholar]
- Sussman MA, Sakhi S, Tocco G, Najm I, Baudry M, Kedes L, Schreiber SS. Neural tropomodulin: developmental expression and effect of seizure activity. Brain research Developmental brain research. 1994b;80(1–2):45–53. doi: 10.1016/0165-3806(94)90088-4. [DOI] [PubMed] [Google Scholar]
- Sussman MA, Welch S, Cambon N, Klevitsky R, Hewett TE, Price R, Witt SA, Kimball TR. Myofibril degeneration caused by tropomodulin overexpression leads to dilated cardiomyopathy in juvenile mice. The Journal of clinical investigation. 1998b;101(1):51–61. doi: 10.1172/JCI1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takebayashi H, Yamamoto N, Umino A, Nishikawa T. Developmentally regulated and thalamus-selective induction of leiomodin2 gene by a schizophrenomimetic, phencyclidine, in the rat. The international journal of neuropsychopharmacology / official scientific journal of the Collegium Internationale Neuropsychopharmacologicum. 2009;12(8):1111–1126. doi: 10.1017/S1461145709009997. [DOI] [PubMed] [Google Scholar]
- Tsukada T, Kotlyanskaya L, Huynh R, Desai B, Novak SM, Kajava AV, Gregorio CC, Kostyukova AS. Identification of residues within tropomodulin-1 responsible for its localization at the pointed ends of the actin filaments in cardiac myocytes. The Journal of biological chemistry. 2011;286(3):2194–2204. doi: 10.1074/jbc.M110.186924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukada T, Pappas CT, Moroz N, Antin PB, Kostyukova AS, Gregorio CC. Leiomodin-2 is an antagonist of tropomodulin-1 at the pointed end of the thin filaments in cardiac muscle. Journal of cell science. 2010;123(Pt 18):3136–3145. doi: 10.1242/jcs.071837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uversky VN, Shah SP, Gritsyna Y, Hitchcock-DeGregori SE, Kostyukova AS. Systematic analysis of tropomodulin/tropomyosin interactions uncovers fine-tuned binding specificity of intrinsically disordered proteins. Journal of molecular recognition : JMR. 2011;24(4):647–655. doi: 10.1002/jmr.1093. [DOI] [PubMed] [Google Scholar]
- Vera C, Lao J, Hamelberg D, Sung LA. Mapping the tropomyosin isoform 5 binding site on human erythrocyte tropomodulin: further insights into E-Tmod/TM5 interaction. Archives of biochemistry and biophysics. 2005;444(2):130–138. doi: 10.1016/j.abb.2005.10.002. [DOI] [PubMed] [Google Scholar]
- Vera C, Sood A, Gao KM, Yee LJ, Lin JJ, Sung LA. Tropomodulin-binding site mapped to residues 7–14 at the N-terminal heptad repeats of tropomyosin isoform 5. Archives of biochemistry and biophysics. 2000;378(1):16–24. doi: 10.1006/abbi.2000.1802. [DOI] [PubMed] [Google Scholar]
- Watakabe A, Kobayashi R, Helfman DM. N-tropomodulin: a novel isoform of tropomodulin identified as the major binding protein to brain tropomyosin. Journal of cell science. 1996;109(Pt 9):2299–2310. doi: 10.1242/jcs.109.9.2299. [DOI] [PubMed] [Google Scholar]
- Watanabe M, Kobayashi H, Yao R, Maisel H. Adhesion and junction molecules in embryonic and adult lens cell differentiation. Acta ophthalmologica Supplement. 1992;(205):46–52. doi: 10.1111/j.1755-3768.1992.tb02180.x. [DOI] [PubMed] [Google Scholar]
- Weber A, Pennise CR, Babcock GG, Fowler VM. Tropomodulin caps the pointed ends of actin filaments. The Journal of cell biology. 1994;127(6 Pt 1):1627–1635. doi: 10.1083/jcb.127.6.1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber A, Pennise CR, Fowler VM. Tropomodulin increases the critical concentration of barbed end-capped actin filaments by converting ADP.P(i)-actin to ADP-actin at all pointed filament ends. The Journal of biological chemistry. 1999;274(49):34637–34645. doi: 10.1074/jbc.274.49.34637. [DOI] [PubMed] [Google Scholar]
- Weber KL, Fischer RS, Fowler VM. Tmod3 regulates polarized epithelial cell morphology. Journal of cell science. 2007;120(Pt 20):3625–3632. doi: 10.1242/jcs.011445. [DOI] [PubMed] [Google Scholar]
- Wegner A. Kinetic analysis of actin assembly suggests that tropomyosin inhibits spontaneous fragmentation of actin filaments. Journal of molecular biology. 1982;161(2):217–227. doi: 10.1016/0022-2836(82)90149-8. [DOI] [PubMed] [Google Scholar]
- Willadsen KA, Butters CA, Hill LE, Tobacman LS. Effects of the amino-terminal regions of tropomyosin and troponin T on thin filament assembly. The Journal of biological chemistry. 1992;267(33):23746–23752. [PubMed] [Google Scholar]
- Winder SJ, Ayscough KR. Actin-binding proteins. Journal of cell science. 2005;118(Pt 4):651–654. doi: 10.1242/jcs.01670. [DOI] [PubMed] [Google Scholar]
- Yamashiro S, Cox EA, Baillie DL, Hardin JD, Ono S. Sarcomeric actin organization is synergistically promoted by tropomodulin, ADF/cofilin, AIP1 and profilin in C. elegans. Journal of cell science. 2008;121(Pt 23):3867–3877. doi: 10.1242/jcs.040477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashiro S, Speicher KD, Speicher DW, Fowler VM. Mammalian tropomodulins nucleate actin polymerization via their actin monomer binding and filament pointed end-capping activities. The Journal of biological chemistry. 2010;285(43):33265–33280. doi: 10.1074/jbc.M110.144873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao W, Sung LA. Erythrocyte tropomodulin isoforms with and without the N-terminal actin-binding domain. The Journal of biological chemistry. 2010;285(41):31408–31417. doi: 10.1074/jbc.M110.130278. [DOI] [PMC free article] [PubMed] [Google Scholar]