Abstract
Background
Mutations of WNT3, WNT5A, WNT9B, and WNT11 genes are associated with orofacial birth defects, including non-syndromic cleft lip with cleft palate in humans. However, the source of Wnt ligands and their signaling effects on the orofacial morphogenetic process remain elusive.
Results
Using Foxg1-Cre to impair Wnt secretion through the inactivation of Gpr177/mWls, we investigate the relevant regulation of Wnt production and signaling in nasal–facial development. Ectodermal ablation of Gpr177 leads to severe facial deformities resulting from dramatically reduced cell proliferation and increased cell death due to a combined loss of WNT, FGF and BMP signaling in the developing facial prominence. In the invaginating nasal pit, the Gpr177 disruption also causes a detrimental effect on migration of the olfactory epithelial cells into the mesenchymal region. The blockage of Wnt secretion apparently impairs the olfactory epithelial cells through modulation of JNK signaling.
Conclusions
Our study thus suggests the head ectoderm, including the facial ectoderm and the neuroectoderm, as the source of canonical as well as noncanonical Wnt ligands during early development of the nasal–facial prominence. Both β-catenin–dependent and –independent signaling pathways are required for proper development of these morphogenetic processes.
Keywords: canonical Wnt, Gpr177, Wntless, noncanonical Wnt, nasal pit, JNK
Introduction
The mammalian upper face (lip and nose) arises from several paired and unpaired tissue (buds) during early organogenesis (Jiang et al., 2006; Minoux and Rijli, 2010). On mouse embryonic day 9 (E9.0), cranial neural crest-derived mesenchymal cells under the head ectoderm-derived epithelium populate extensively to form the frontal nasal prominence rostral to the telecephalon. At E10, the nasal placode forms as the focal thickening of the surface ectoderm in the ventrolateral part of the frontonasal prominence. Asymmetric outgrowth and bulge around the periphery of the nasal placode form the horse-shoe shaped lateral (LNP) and medial (MNP) nasal prominences, resulting in formation of the nasal pit (or olfactory pit) at E10.5. The main nasal epithelium extends olfactory nerves which grow toward the main olfactory bulbs. In addition to the outgrowth of nerves, several cells leave the placode region and translocate along similar trajectories as the nerves to constitute neural components of the central and peripheral nervous systems (Brunjes and Frazier, 1986; De Carlos et al., 1996; Wray, 2010). During the formation of the nasal pit, mesenchymal cells rapidly proliferate to conduct the outgrowth of LNP, MNP, and maxillary (MXP) prominences of the first branchial arch, pushing the nasal pits to the medial face. Around E11.0, convergence and fusion in multiple directional junctions with the LNP and MXP, LNP, and MNP result in formation of the nostrils and the lip. The proper thickening and fusion of the nasal epithelium are highly coordinated by various cellular signals including Shh, FGFs, TGF-b, and WNT across the epithelium and the underlying mesenchyme. Interference with activation of these signaling pathways will lead to orofacial clefts (Jiang et al., 2006).
Variation in WNT genes is tightly associated with nonsyndromic cleft lip with or without cleft palate (Chenevix-Trench et al., 1992). The human WNT9B and WNT3 genes are located on chromosome 17q21, one of the first chromosome region implicated in nonsyndromic cleft lip with cleft palate (CLP) using genetic linkage analysis (Chenevix-Trench et al., 1992; Niemann et al., 2004; Menezes et al., 2010). It has also been independently detected in several other linkage and association studies of CLP (Marazita et al., 2004; Juriloff et al., 2006; Lan et al., 2006). In mice, several Wnt ligands, including Wnt3 and Wnt9b (Lan et al., 2006), are expressed in the ectoderm of the early facial prominences. Canonical Wnt signaling reporters such as TopGAL, BATgal, and Axin2-lacZ are also highly active in the facial prominences and their derivatives, implicating the importance of Wnt/b-catenin signaling in facial development (Maretto et al., 2003; Brugmann et al., 2007; Al Alam et al., 2011).
Compound mutation of TCF1/Lef1, the key transcription effectors for Wnt/β-catenin signaling, resulted in regional alterations of facial specification (Brugmann et al., 2007). The mouse deletion of canonical Wnt9b showed significant retardation in the outgrowth of the nasal and maxillary prominences due to reduced proliferation of the mesenchymal cells. This defect led to a failure of physical contact between the facial prominences resulting in cleft lip and cleft palate (Jin et al., 2012). Mice with deficiency of Lrp6 exhibited cleft lip with cleft palate, resembling a common birth defect in humans (Song et al., 2009). The frontonasal and upper jaw prominences cannot be formed upon conditional ablation of β-catenin in the facial ectoderm where the expression of ectodermal Shh and Fgf8 were down-regulated (Reid et al., 2011; Wang et al., 2011). However, the tissue source and effects of Wnt signaling in orofacial regulation remain elusive, most likely due to the dual role of β-catenin in cellular signaling and cell–cell interactions.
In the current study, we investigate the requirement of ectodermal Wnt using Foxg-1-Cre-mediated deletion of Gpr177 (commonly known as Wntless [Wls]), which is essential for Wnt sorting and secretion. The results show that ablation of Wnt secretion in the ectoderm leads to failure of the frontonasal and maxillary prominences to form. These abnormalities phenocopy the orofacial defects observed in Catnb1lox(ex2-6)Foxg1Cre mice (Wang et al., 2011), suggesting the facial ectoderm and the telencephalic neuroepithelium as the source for Wnt in orofacial shaping and development. Our data further uncover that ectodermal Wnt ligands are required for invagination of the nasal pit and migration of olfactory epithelial cells.
Results
Ectodermal Gpr177 is Required for Orofacial Patterning
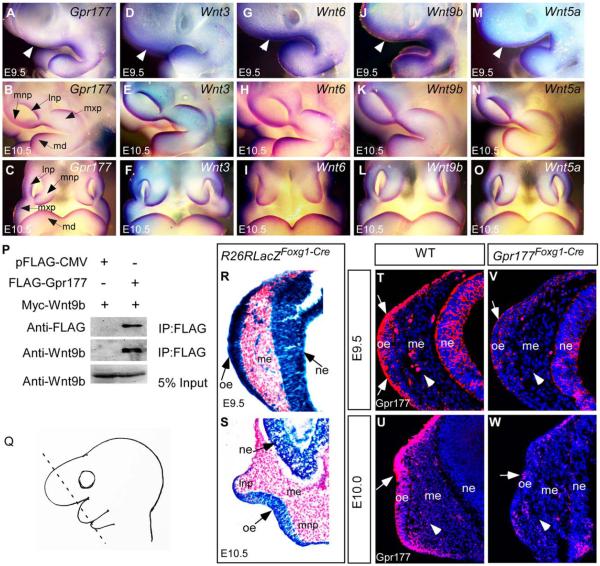
To examine the involvement of Gpr177-mediated Wnt production during formation of LNP, MNP and MXP, whole-mount in situ hybridization was performed on the E9.5 to 10.5 embryos. The Gpr177 transcript was colocalized with those of Wnt3, Wnt6, Wnt9b, and Wnt5a in the nasal prominence and maxillary process of the first branchial arch (Fig. 1A–O). Gpr177 is the mouse orthologue of Drosophila Wntless essential for intracellular Wnt trafficking (Banziger et al., 2006; Fu et al., 2009; Zhu et al., 2013). Previous studies have shown that Gpr177 binds to mouse Wnt3, Wnt6, and Wnt5a (Fu et al., 2009; Zhu et al., 2013). The results of coimmunoprecipitation analysis showed that Gpr177 associated with Wnt9b (Fig. 1P), indicating its roles in regulating Wnt9b sorting and secretion. Here we performed genetic inactivation of Gpr177 to understand the tissue source of Wnt required for its signaling activation in nasal pit development. To delete Gpr177 in the head ectoderm, Foxg1-Cre transgene was crossed into mice homozygous with the Gpr177Fx allele to obtain the Gpr177Foxg1Cre mutants (Hebert and McConnell, 2000; Wang et al., 2011). X-gal staining of embryos carrying a R26R reporter allele revealed that Foxg1-Cre permits site-specifi DNA recombination in the facial ectoderm and the telencephalic neuroepithelium at E9.5 (Fig. 1R). The Cre-mediated recombination became evident in the epithelium of the nasal pits undergoing invagination at E10.5 (Fig. 1S). Highly efficient deletion of Gpr177 occurred at E9.5 in the nasal placode of Gpr177Foxg1Cre mutants (Fig. 1T,V), indicating that Foxg1-Cre is ideal for inactivation of Gpr177 during nasal pit development. Gpr177 was almost eliminated from the facial epithelium at E10.0 (Fig. 1U,W).
Fig. 1.
Expression of Gpr177 and multiple Wnts in early facial primordial development and interaction of Gpr177. A–O: Whole-mount in situ hybridization shows that Gpr177, Wnt3, Wnt6, Wnt9b, and Wnt5a are expressed in the nasal prominences of E9.5 as lateral views (A,D,G,J,M, white arrowheads represent the nasal placode region) and E10.5 as lateral views (B,E,H,K,N) and frontal views (C,F,I,LO). P: Gpr177 coimmunoprecipitates with Wnt9b in vivo. HEK293 cells transfected with the indicated vectors were subjected to immunoprecipitation analysis with anti-FLAG antibody-coupled beads. Western blot was performed with anti-FLAG and Wnt9b antibodies. Q: A schematic showing the orientation of the tissue sections. R,S: X-gal staining shows that Foxg1-Cre activity is present in the facial epithelium and neuroepithelium at E9.5 (R) and E10.5 (S). Note the X-Gal positive stains in the mesenchymal cells underlying the thickening nasal placode at E9.5 (R). T–W: Immunofluorescence of Gpr177 expression in the facial epithelium, the neuroepithelium, and the underling mesenchyme (arrowheads) at E9.5 and E10.0. Arrows indicate positive staining of Gpr177 in the olfactory epithelium. Note that Gpr177 is almost deleted in the Gpr177Foxg1-Cre mutant at E10.0 (W). lnp: Lateral nasal process; mnp: medial nasal process; mxp: maxillary process; md: mandibular process; oe, olfactory epithelium; me, mesenchyme; ne, neuroepithelium.
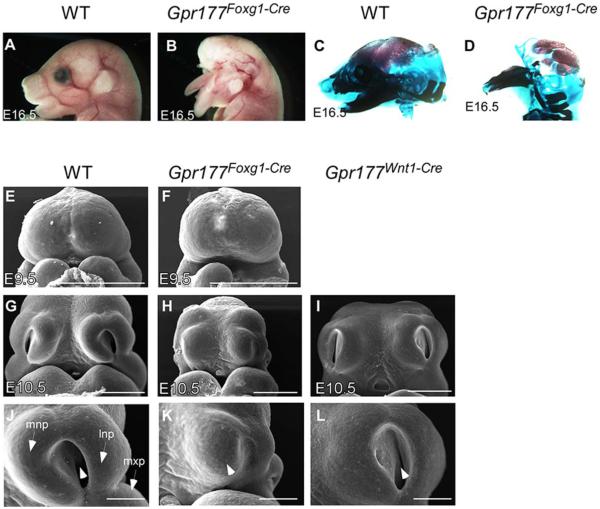
Compared with wild-type embryos (or Gpr177(fx/1)Foxg1-Cre), the Gpr177Foxg1Cre mutants exhibited severe facial deformities, and failed to form several key features within the upper face, including upper nasal, upper jaw, and ocular structures at E16.5 (Fig. 2A–D). Interestingly, these facial defects are highly reminiscent of the phenotype associated with inactivation of β-catenin mediated by Foxg1-Cre (Wang et al., 2011). SEM images revealed that the Gpr177Foxg1Cre mutants successfully develop the frontal nasal prominence at E9.5 (Fig. 2E,F), but are unable to properly form the LNP and MNP at E10.5 (Fig. 2G,H,J,K). These results suggest that Gpr177 might be involved in the invagination processes of the nasal pit. In contrasts, deletion of Gpr177 in the facial mesenchyme by Wnt1-Cre did not cause any noticeable defects during nasal pit formation (Fig. 2I,L), albeit cleft palate were detected (Fu et al., 2011; Liu et al., 2015). Therefore, ectodermal rather than mesenchymal production of Wnt is required for the initiation of LNP and MNP as well as their outgrowth around the nasal pit.
Fig. 2.
Gpr177 is required for facial development. A,B: Foxg1-Cre mediated loss of ectodermal Gpr177 leads to the disruption of craniofacial structures as revealed by gross morphology. C,D: Alizarin Red and Alcian Blue staining of skeletons on embryos at E16.5. E–L: Scanning electron micrographs of the facial prominences at E9.5 and E10.5. Front facial views of wild-type (E,G,J) and Gpr177-deficient (F,H,K). Gpr177Foxg1-Cre embryos display dramatic defects in nasal pit invagination (arrowhead in K vs. J) and outgrowth of mnp, lnp, and mxp (K vs. J) at E10.5. In contrast, mesenchymal deletion of Gpr177 by Wnt1-Cre shows minor defects with formation of the nasal pit (arrowhead in L). lnp, lateral nasal process; mnp, medial nasal process; mx, maxillary process; md, mandibular process. Scale bars ¼ 500 mm in E–I; 200 mm in J–L.
Activation of β-Catenin Signaling in the Nasal Placode Epithelium Requires the Presence of Ectodermal Gpr177
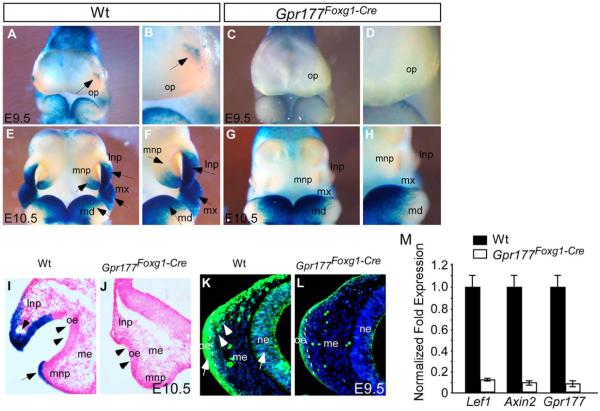
Extensive studies have shown that the loss of Gpr177 impairs canonical Wnt signaling in many organs (Fu et al., 2009, 2011; Chen et al., 2012; Fu and Hsu, 2013; Maruyama et al., 2013; Zhu et al., 2013, 2014). To determine if this is also the case in development of the nasal pit, we analyzed the activity of TOPGAL in Gpr177Foxg1Cre. Whole-mount X-gal staining of the E9.5 wild-type head showed that β-catenin signaling was focally activated in the confined areas surrounding the nasal placode (Fig. 3A,B, arrows) where LNP and MNP will develop later at E10.5 (Fig. 3E,F, arrows). Sections of the TOPGAL stained embryos revealed that β-catenin signaling is activated in the epithelium at E10.5 (Fig. 3I). Immunostaining of the active form of β-catenin further illustrated that the canonical Wnt signaling is activated in the frontal nasal prominence at E9.5 (Fig. 3K). In contrast, both TOPGAL and β-catenin activities were significantly diminished in the Gpr177Foxg1Cre mutants (Fig. 3C,D,G,H,J,L), indicating that canonical Wnt signaling stimulation in the epithelium and mesenchyme of the nasal placode relies on ectodermal secretion of Wnt ligands. Additionally, quantitative real-time polymerase chain reaction (PCR) and expression analyses of Axin2 and Lef1, two target genes of canonical Wnt signaling, showed their reductions in the mutants (Fig. 3M). Consistent with the requirement of ectodermal β-catenin for shaping of the mouse face (Wang et al., 2011), our data support the hypothesis that ectoderm, including the facial ectoderm and the neuroectoderm, is the source of canonical Wnt signaling during nasal pit formation.
Fig. 3.
Ectodermal Gpr177 loss-of-function leads to disruption of canonical Wnt signaling. A–H: Whole-mount X-gal staining shows significant reduction of TopGal activity in E9.5 and E10.5 Gpr177Foxg1-Cre embryos (p E10.5 embryos shows that TopGal signal is present in the epithelium (arrows in I). Note the loss of staining and the inhibited outgrowth of lnp, mnp, and mxp in E10.5 Gpr177Foxg1-Cre embryos (J). The olfactory epithelial invagination of mutant embryos is also significantly inhibited (arrowheads, J compared with wild-type I). K,L: Immunostaining with active β-Catenin shows that the presence of active β-catenin in facial epithelium, neuroepithelium, and facial mesenchyme (arrows and arrowhead in K) is significantly reduced upon the deletion of epithelial Gpr177 (L). The dash line indicates the boundary between olfactory epithelium and mesenchyme. M: Quantitative RT-PCR analysis of gene expression in facial prominence of E10.25 embryos. Expression of Lef1, Axin2, and Gpr177 is significantly down-regulated in Gpr177Foxg1-Cre embryos. lnp, lateral nasal process; mnp, medial nasal process; mx, maxillary process; md, mandibular process; oe, olfactory epithelium; op, olfactory primordium; me, mesenchyme; ne, neuroepithelium.
Loss of Ectodermal Gpr177 Interferes With Cell Proliferation and Death in Frontal Nasal Prominences
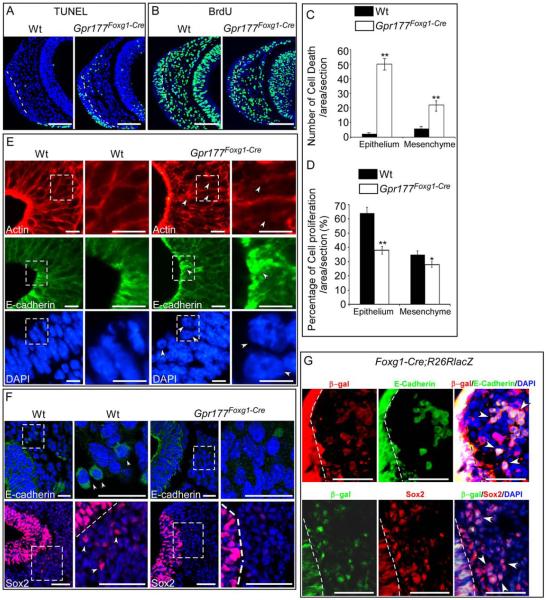
To address the effect of Gpr177 deficiency on cell proliferation and apoptosis in nasal pit development, we examined BrdU labeling and TUNEL staining on sections of the E9.5 rostral nasal prominences. Compared with the control, an increase in apoptosis was evident in the nasal ectoderm as well as the underlying mesenchyme of the Gpr177 mutant (Fig. 4A,C). This alteration was similar to previous finding in the β-catenin mutant (Wang et al., 2011). In addition, BrdU labeling analysis showed a dramatic reduction in the ectoderm and mesenchyme of the mutant nasal prominences (Fig. 4B,D). The defects in cell proliferation and survival likely contributed to the retardation of nasal outgrowth in the Gpr177Foxg1Cre mutant.
Fig. 4.
Deletion of ectodermal Gpr177 leads to increased cell death, reduced cell proliferation, and inhibition of nasal pit invagination. A: TUNEL staining shows that cell death is significantly increased in both the olfactory epithelium and the mesenchyme of Gpr177Foxg1-Cre embryos at E9.5. B: BrdU staining shows reduced cell proliferation in both the olfactory epithelium and the mesenchyme of E9.5 Gpr177Foxg1-Cre embryos. C,D: Comparison of percentage of cell death (C) and cell proliferation (D) in the designated area of the nasal pit in the control and mutant embryos. Standard deviation values are presented as error bars. *P < 0.05. **P < 0.01. E: Immunofluorescence analysis on the nasal pit epithelium of E10.5 embryos for Actin, E-cadherin and DAPI show defective cell shape and cell movement in the nasal pit epithelium of Gpr177Foxg1-Cre embryos. Note the irregular Actin fibers, aggregated E-cadherin, and round-shaped nuclei in the epithelium of Gpr177Foxg1-Cre mutants as indicated by arrowheads. F: We found a dramatic reduction in the number of E-cadherin positive cells in the Gpr177 mutants in the facial mesenchyme as revealed by confocal microscopy analysis at E10.5. Immunostaining of Sox2 for neural stem cells displays a migration defect of E10.5 Gpr177Foxg1-Cre olfactory epithelial cells. The magnified images of the regions within the dotted boxes in E and F are shown to the right of the original photographs. G: Immunofluorescence demonstrates colocalization of Foxg1-Cre positive epithelial cells (β-gal) with E-cadherin or Sox2 positive cells in the mesenchyme of E9.5 embryos. Cells that double-stained with ß-gal and E-cadherin or Sox2 are indicated by arrowheads. The dotted line indicates the boundary between epithelium and mesenchyme. Scale bars ¼ 100 mm in A,B; 10 mm in E; 25 mm in F upper panels,100 mm in lower panels; 50 mm in G.
Epithelial Cell Adhesion and Elongation in the Developing Nasal Pit is Disrupted in the Gpr177Foxg1Cre Mutant
Invagination of cranial placodes, including the optic and nasal placodes, requires dynamic alterations of epithelial cell shape (Sai and Ladher, 2008; Plageman et al., 2011; Metzis et al., 2013). We next investigated the requirement of Gpr177 in the regulation of epithelial cell shape during these morphogenetic processes. Immunostaining analysis revealed that actin filaments are confined to the condensed apical surface of the wild-type nasal pit epithelium; however, severely disorganized apical surface was detected in the nasal pit epithelium of Gpr177Foxg1Cre (Fig. 4E). E-cadherin staining further showed that cell–cell junctions are also disoriented with abnormal aggregation (Fig. 4E), suggesting the importance of Gpr177 in the regulation of epithelial cell shape. DAPI staining revealed the presence of small and round-shaped nuclei in the nasal pit epithelium of Gpr177Foxg1-Cre mutants in contrast to the elongated epithelial nuclei in the wild-type nasal pit (Fig. 4E). Taken together, these data demonstrated an irregular epithelial cell adhesion and elongation within the Gpr177Foxg1Cre nasal pit.
Conversely, Wnt1-Cre mediated deletion of Gpr177 in the mesenchyme did not show any obvious defects in nasal pit cell adhesion and elongation (data not shown), suggesting that the loss of ectodermal, but not mesenchymal, Wnt secretion accounts for the abrogation of the nasal pit invagination. Furthermore, immunostaining of the active form of β-catenin in E9.5 embryos revealed a remarkable decreased active β-catenin in Gpr177 mutant embryos (Fig. 3K,L). The loss of TopGal signaling in Gpr177Foxg1-Cre embryos also indicated a degradation of β-catenin in Gpr177 deficient facial prominence (Fig. 3A–J). Because β-catenin acts as a structural protein at cell–cell adherens junctions as well as the central molecule of canonical Wnt signaling pathway (Sineva and Pospelov, 2014), it is reasonable to deduce that the disrupted cell shape and adhesion in Gpr177Foxg1Cre nasal pit is probably caused by the loss of β-catenin due to inhibition of Wnt trafficking resulting from Gpr177 deficiency.
Cell Motility is Impaired in the Olfactory Epithelium Lacking Wnt
As development proceeds, the nasal or olfactory placodes were subdivided into the sensory and respiratory epithelia within the nasal cavity (Croucher and Tickle, 1989; De Carlos et al., 1996; Toba et al., 2001). The sensory epithelium consists of olfactory sensory neurons among other cell types. We, therefore, tested if motility of the olfactory epithelial cells was altered in the hypoplastic nasal pit epithelium due to an absence of intrinsic Wnt signaling. Examination of Cre recombination activity using R26R/Foxg1-Cre mice showed that X-Gal staining is positive in some mesenchymal cells near the E9.5 nasal placode (Fig. 1R). Immunostaining of E-cadherin identified those olfactory epithelial cells translocating into the nasal pit mesenchyme at E10.5 (Fig. 4F). The results were consistent with previous reports examining olfactory cell migration in the nasal placode in rats (De Carlos et al., 1996; Toba et al., 2001). Sox2, a neural stem cell marker in the nasal epithelium, was also detected in nasal pit mesenchyme of wild-type embryos (Kawauchi et al., 2005) (Fig. 4F).
In contrast, few E-cadherin and Sox2 positive cells were detected in nasal pit mesenchyme of Gpr177Foxg1Cre mice (Fig. 4F). Furthermore, both E-cadherin and Sox2 were found to be colocalized with ß-gal in some cells in mesenchyme of R26R/Foxg1-Cre embryos (Fig. 4G), indicating that those E-cadherin positive cells are also positive for Sox2. Taken together, these results suggest that the migration of olfactory epithelial cells to the mesenchyme is inhibited due to loss of Gpr177. Our findings thus suggest a requirement of intrinsic Wnt for the motility of nasal placode cells. Additionally, significantly increased cell death and reduced cell proliferation present in the olfactory epithelium (Fig. 4A,B) could also lead to a reduction of Gpr177-deleted olfactory epithelial cells that migrate toward the brain.
Loss of Gpr177 in the Head Ectoderm Diminishes FGF Signaling During Invagination of the Nasal Pit Epithelium
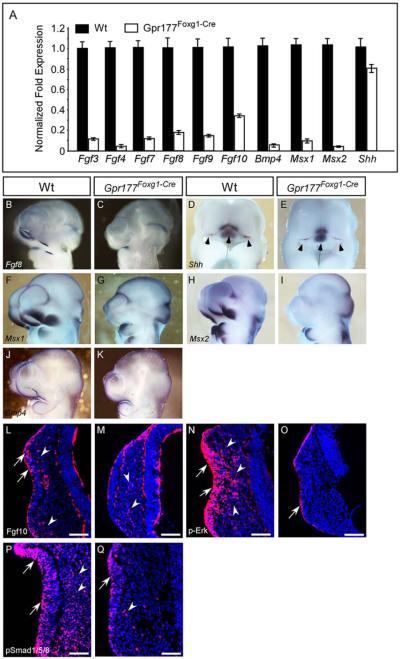
To elucidate molecular mechanisms underlying the nasal pit defects caused by Gpr177 deficiency, we first examined FGF ligands, key downstream signaling regulators of Wnt/β-catenin, critical for mouse facial development. Fgf3, 4, 7, 8, 9, and 10 were expressed in the frontal nasal ectoderm as well as the developing nasal pit (Fig. 5A) (Bachler and Neubuser, 2001; Kawauchi et al., 2005). Fgf8, Fgf10, and Fgf17 were known to be downstream targets of Wnt within the facial ectoderm (Song et al., 2009; Wang et al., 2011; Jin et al., 2012). Using quantitative real-time PCR analysis, we found significant reduction of these genes at E10.25 (Fig. 5A). Closer examination with in situ hybridization and immunostaining revealed that the expression of Fgf8 and Fgf10 is diminished at around E10.25 during nasal pit invagination (Fig. 5A–C,L,M). Moreover, phosphorylated-Erk, a downstream effector of FGF signaling in the developing face (Corson et al., 2003), was detected in both epithelium and mesenchyme in invaginating nasal pit of E10.25 wild-type embryos, but significantly decreased in that of Gpr177 mutants (Fig. 5N,O). FGF signaling (Wang et al., 2011; Jin et al., 2012) may also be required for orchestrating the regulation of epithelial Wnt signal during nasal pit formation.
Fig. 5.
FGF, BMP, and SHH signaling pathways are altered in the facial prominence of Gpr177Foxg1-Cre embryos. A: Quantitative RT-PCR analysis of gene expression in the facial prominences of E10.25 embryos. Expression of Fgf3, Fgf4 Fgf7, Fgf8, Fgf9, Fgf10, Bmp4, Msx1, and Msx2 is significantly down-regulated in Gpr177Foxg1-Cre embryos. By contrast, expression of Shh is mildly decreased. B–K: Whole-mount in situ analysis shows that expression of Fgf8, Shh, Msx1, Msx2, and Bmp4, in the facial prominences of E10.5 Gpr177Foxg1-Cre embryos as presented in sagittal views. Note that the expression of Fgf8, Msx1, and Msx2, is abolished completely in lnp, mnp, and mxp of Gpr177 mutant embryos. D,E: Frontal facial views of the wild-type and mutant embryos show that in Gpr177Foxg1-Cre embryos, the expression of Shh is retained in the FEZ (Arrowheads); however, its expression region in telencephalon and diencephalon becomes smaller (Arrows). Immunofluorescence analysis on sections of E10.25 facial primordia for Fgf10 (L,M), p-Erk (N,O), and pSmad1/5/8 (P,Q). Expression of Fgf10 protein is reduced in both the epithelium (arrows) and the mesenchyme (arrowheads) of embryos lacking epithelial Gpr177. Note that, in the facial primordia of wild-type embryos, distribution of Fgf10 proteins is concentrated at the sub-epithelial mesenchyme where the epithelium recesses (L). However, Fgf10 proteins are scattered in the facial mesenchyme of Gpr177Foxg1-Cre embryos (M). Phosphorylation of Erk of Gpr177Foxg1-Cre embryos is significantly reduced in the epithelium and diminished in the mesenchyme (O vs. wild-type N). Phosphorylation of Smad1/5/8 of Gpr177Foxg1-Cre embryos is reduced in the olfactory epithelium and diminished in the mesenchyme (Q vs. P). Arrows and arrowheads point out positive staining in the epithelium and the mesenchyme, respectively. Scale bars ¼ 100 mm. FEZ, frontonasal ectodermal zone.
Shh Signaling is Disrupted in the Facial Prominence of Gpr177Foxg1Cre
Previous studies have shown that forebrain Shh signaling participates in regulating the onset of facial Shh expression which is required for establishing the signaling properties of the frontonasal ectodermal zone (FEZ), which then patterns outgrowth of the upper jaw (Marcucio et al., 2005, 2011; Hu and Marcucio, 2009; Hu et al., 2015). To determine whether the Shh signaling form forebrain to facial ectoderm is affected in Gpr177Foxg1-cre, we examined the expression of Shh in both Gpr177Foxg1-cre and wild-type embryos. As revealed by quantitative real-time PCR analysis, the expression of Shh was mildly reduced in facial prominence at E10.25 (Fig. 5A). Whole-mount in situ hybridization results showed that Shh expression was retained in FEZ of E10.5 Gpr177Foxg1-cre embryos (Fig. 5D,E, Arrowheads); however, the expression of Shh was reduced in telencephalon and diencephalon of Gpr177 mutant embryos (Fig. 5D,E, Arrows), suggesting that the Shh signaling is impaired in response to deletion of Gpr177 in facial epithelium and neuroepithelium.
Msx1 and Msx2 Expression Was Disrupted in the Invaginating Nasal Pit of Gpr177Foxg1Cre
In formation of the LNP and MNP, we detected focal expression of homeobox transcription factors, Msx1 and Msx2, (Fig. 5A,F,H), which are also targets of the canonical Wnt pathway in development of the orofacial prominences (Song et al., 2009; Jin et al., 2012). The deletion of Gpr177 dramatically reduced the expression of Msx1 and Msx2 in these domains (Fig. 5A,G,I) (Song et al., 2009; Wang et al., 2011), suggesting that failure formation of the nasal pit is associated with the dysregulation of downstream transcription factors modulated by epithelial Wnts.
Bone Morphogenetic Protein Signaling Within the Invaginating Nasal Pit is Inhibited in the Gpr177 Mutants
Bmp4 was normally expressed in the fusion regions of the LNP, MNP, and MXP during nasal lip invagination (Fig. 5J). In situ hybridization showed significant reduction of Bmp4 in LNP of the Gpr177Foxg1Cre mutants (Fig. 5K). In contrast, Bmp4 expression in the upper lip/nasal primordial is not modified in Wnt9b−/− embryos and is only mildly decreased in Lrp6 mutants (Song et al., 2009; Jin et al., 2012). These data suggest a distinct mechanistic regulation of Bmp4 in response to the loss of Wnt production and the loss of Wnt/β-catenin signaling in the nasal placode epithelium. To test the requirement of Gpr177-mediated Wnt production for the maintenance of bonne morphogenetic protein (BMP) signaling, we performed immunostaining of phosphorylated Smad1/5/8 as an indicator for its signaling activity at E10.25. The results showed that activation of BMP4 (Fig. 5J,K) and its signaling (Fig. 5P,Q) in both the epithelium and the mesenchyme of the invaginating nasal pit is significantly reduced in the Gpr177Foxg1Cre mutants.
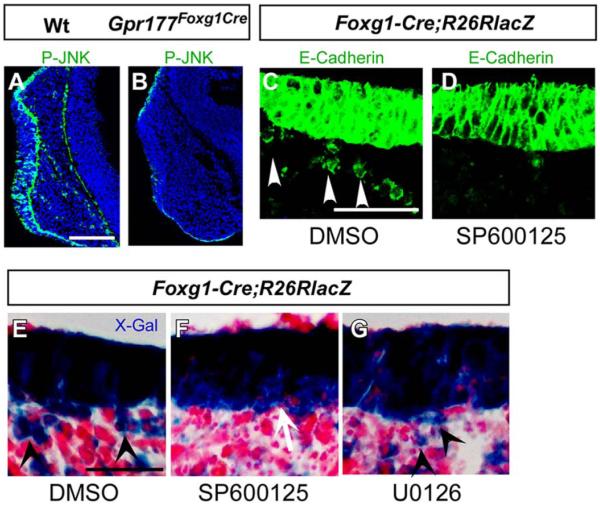
Gpr177 Ablation Results in Inhibition of JNK Essential for Nasal Epithelial Cell Migration
Previous studies suggested that a β-catenin–independent Wnt pathway influences tissue polarity and cell motility through modulation of the JNK (Jun N-terminal kinase) (Davis, 2000; Karner et al., 2009) whose regulation has also been implicated in cell migration and tissue morphogenesis (Kuan et al., 1999; Huang et al., 2003; Weston et al., 2003; Xia and Karin, 2004). To determine if the loss of Gpr177 affects the JNK pathway in olfactory epithelial invagination, we first examined its activation by immunostaining. In the control, cells positive for phosphorylated JNK were found in both the invaginating nasal pit epithelium and the underlying mesenchyme at E10.25 (Fig. 6A). In contrast, we identified a significant reduction of the phosphorylated JNK in the epithelium and mesenchyme of Gpr177Foxg1Cre (Fig. 6B). We were able to further determine the role of JNK in this developmental process because mesenchymal cells double positive for E-cadherin and X-gal staining are actually the migratory epithelial cells (Fig. 4G). Next, using in vitro organ culture with nasal placode explants at E10, we investigated the effects of SP600125, a JNK inhibitor (Huang et al., 2003) on cell migration. Immunostaining of E-cadherin and X-Gal staining for the R26R/Foxg1Cre reporter cells showed that the addition of SP600125 (Fig. 6D,F), but not U0126 (an inhibitor for ERK) (Fig. 6G), efficiently inhibits distribution of the nasal epithelial cells within the underlying mesenchyme (Fig. 6C–G). These data suggest that JNK activation is required for migration of the olfactory epithelial cells during morphogenesis of the nasal pit and that the epithelial noncanonical Wnt ligands are involved in olfactory epithelial cell migration.
Fig. 6.
A Gpr177/JNK signaling is required for nasal epithelial cell migration. A,B: Immunofluorescent staining shows that phosphorylated JNK is reduced in the epithelium and completely abolished in the mesenchyme of Gpr177Foxg1-Cre embryos (B) comparing with the wild-type (A). C–G: Immunofluorescence and X-gal staining on the sections of R26RLacZFoxg1-Cre facial explants shows that the epithelial cell migration is inhibited with treatment of SP600125 (D, F vs. C, E). Note that the migrating cells with positive staining are present in dimethyl sulfoxide group (C,E) and U0126 (G), but not in SP600125 (D,F) group. Arrowheads indicate migrating epithelial cells. Scale bars ¼ 100 mm in A,B; 50 mm in C–G.
Discussion
Tissue-specific deletion of Gpr177 in the ectoderm of the rostral head performed in this study provides genetic evidence that facial epithelium and the telencephalic neuroepithelium is the source of Wnt during nasal pit invagination. Our finding is the first to show that defective Wnt production in the ectoderm results in failure of upper face development. Similar to epithelial ablation of β-catenin, these results support the notion that signaling transduced by canonical Wnt ligands within the ectoderm is critical for upper facial development. Additionally, the current study provides evidence for first time that Gpr177-mediated Wnt production within the ectoderm is required for the maintenance of JNK activation tightly linked to nasal epithelial cell motility, which is essential for functional establishment of the olfactory epithelium.
The deletion of Gpr177 in the head ectoderm abolishes secretion of both canonical and noncanonical Wnt ligands within the developing nasal epithelia, including the LNP, MNP and MXP. Genetic analysis of the Gpr177-mediated Wnt production suggests that β-catenin signaling essential for orofacial development depends on Wnt ligands secreted from the head ectoderm. This conclusion is consistent with previous reports on the impaired upper face arising from ablation of β-catenin signaling (Song et al., 2009; Wang et al., 2011; Jin et al., 2012). However, Axin2 expression and β-catenin activation are significantly lower in the Gpr177 mutants, compared with ectoderm-specific deletion of βcatenin, or loss of Wnt9b or Lrp6 (Song et al., 2009; Wang et al., 2011; Jin et al., 2012). In contrast, targeted deletion of Gpr177 in the neural crest-derived mesenchyme (Gpr177Wnt1Cre) did not seem to alter canonical Wnt signaling in the area surrounding the nasal pit (Fu et al., 2011), albeit cleft palate is observed in the mutants (Fu et al., 2011; Liu et al., 2015). The presence of Gpr177 or Gpr177-mediated Wnt sorting and secretion in the facial mesenchyme is thus dispensable for nasal pit formation, but absolutely necessary for palatogenesis. Wnt5a, shown to signal through noncanonical signaling, is the only ligand known to be expressed in the cranial mesenchyme (He et al., 2008; Paiva et al., 2010; Zhu et al., 2013), and plays an essential role in secondary palate development (He et al., 2008).
Our data show that ectodermal deletion of Gpr177 in the nasal prominence and the telencephalon causes even more severe inhibition of FGF gene expression. FGF signaling in the facial ectoderm has been shown to be regulated by Wnt/β-catenin signaling through modulation of mesenchymal cell survival and proliferation (Song et al., 2009; Wang et al., 2011; Jin et al., 2012). Ectodermal FGFs in nasal pit development are modulated by the canonical pathway (Wang et al., 2011; Jin et al., 2012). Fgf3, 8, 17, or 10 was significantly down-regulated in the rostral head tissue in Foxg1-Cre/b-catenin mutant, Wnt9b−/− or Lrp5−/− (Song et al., 2009; Wang et al., 2011; Jin et al., 2012). On the other hand, β-catenin gain of function in the frontal nasal ectoderm up-regulates the FGF and Shh signaling (Wang et al., 2011). Ectodermal deletion of Gpr177 causes down-regulation of FGF genes, including Fgf3, Fgf4, Fgf7, Fgf8, Fgf9, Fgf10, in the rostral head, further supporting this hypothesis.
Reduction of Erk phosphorylation in Gpr177Foxg1Cre is consistent with the requirement of epithelial Wnt in regulating the FGF signaling-dependent outgrowth of nasal pits (Jin et al., 2012). Phosphorylated Erk staining reveals FGF signaling domains including the frontonasal prominences and the branchial arch during embryogenesis (Corson et al., 2003). The activation of Erk1/2 in the facial mesenchyme is affected by Wnt9b mutation (Jin et al., 2012), indicating that the ectodermal Wnt ligands play an important role in the FGF-mediated nasal outgrowth. The mutation of Lrp6, the co-receptor for Wnt9b, showing decreases in cell proliferation also supports our findings (Song et al., 2009).
Enhanced cell apoptosis detected in the invaginating nasal pit due to the loss of Gpr177 is consistent with the ectodermal deletion of β-catenin (Jin et al., 2012), but is not in agreement with the mutation of Wnt9b or Lrp6. It has been suggested that ectodermal FGF regulates cell proliferation and survival in a concentration-dependent manner, in which cell survival requires a lower signaling level (Jin et al., 2012).
Interestingly, Gpr177 in the head ectoderm seems to play role in determining nasal epithelial cell structure critical for invagination. Ectodermal deletion of Gpr177 reveals an essential requirement for Wnt production in maintenance of the epithelial cell shape and organization critical for epithelial motility and morphogenesis (Lienkamp et al., 2012). In mouse organogenesis, the limb outgrowth and lung tube shaping are shown to be associated with the orientation of cellular divisions, regulated by the noncanonical Wnt5a/JNK pathway and/or the FGF/ERK pathway (Oishi et al., 2003; Gros et al., 2010; Tang et al., 2011). Similar defects in epithelial cell shape have recently been shown in the Wnt1Cre;Ptc1c/c mutants, where Ptc1 is deleted in the facial mesenchyme (Metzis et al., 2013), suggesting that a potentially conserved mechanism underlying the interaction of signaling pathways is responsible for the control of placode invagination.
Cell motility is a major contributor to epithelial placode morphogenesis (Streit, 2002; Ahtiainen et al., 2014). The development of cranial placodes involves cell shape changes and morphogenetic movements, allowing the placodes to develop into columnar epithelia, to invaginate, and to give rise to a variety of migratory cells (Schlosser, 2006). It has been shown that olfactory epithelial cells including sensory CNS neurons leave the olfactory epithelium and migrate into the forebrain domain (Wray et al., 1989; Valverde et al., 1993; Wray, 2010). Migration from the olfactory neuroepithelium begins earlier than the superficial ectodermal invagination that gives rise to the olfactory pit (Fornaro et al., 2001). It has been shown that directional cell migration drives hair placode morphogenesis (Ahtiainen et al., 2014); however, whether the epithelial–mesenchymal transition promotes the invagination of the superficial epithelium and consequently contributes to the formation of nasal pit requires further investigation.
The migratory defects of the Gpr177Foxg1Cre olfactory epithelial cells indicate the involvement of Wnt in the regulation of cell motility during development of a functional olfactory organ. However, how the nasal placode responds to the loss of epithelial Wnt production requires further investigation. In normal development of the nasal placode, secreted ligands including canonical and noncanonical Wnts, diffuse within the epithelium or travel across the underlying mesenchyme, to activate the downstream signaling cascades. Mesenchymal deletion of Gpr177, however, results in proper epithelial invagination and formation of normal nasal pits in which production of noncanonical Wnt5a is likely blocked. Although Wnt5a is required for planar cell polarity (PCP) and PCP-dependent cellular movement by means of activation of Erk and JNK (Gros et al., 2010), our data do not support this regulatory mechanism. But, it remains possible for an existence of another noncanonical Wnt pathway which demarcates the motility of nasal epithelial cells in the nasal pit through activation of the JNK pathway to promote cell migration (Gros et al., 2010). This is supported by studies of kidney tubule morphogenesis in which Wnt9b signals through a noncanonical pathway to activate JNK critical for cell migration and morphogenesis (Huang et al., 2003; Zhang et al., 2003; Xia and Karin, 2004; Waetzig et al., 2006; Karner et al., 2009). In the retina, the JNK pathway controls the expression of Bmp4, which in turn regulates Shh and PAX2 critical for optic fissure closure (Weston et al., 2003). Furthermore, the RAS/MAP kinase pathway is the main downstream pathway associated with FGF signaling in which JNK is involved (Raju et al., 2014; Teven et al., 2014), suggesting the possibility that the inhibition of the FGF signaling could also contribute to the loss of JNK activation in Gpr177 deficient embryos.
As BMP signaling is required for upper lip development (Liu et al., 2005), this study supports that activation of BMP4 in nasal pit development is downstream of the epithelial Wnt. The findings are consistent with developmental studies in many other organs including hair follicles, teeth, and the tongue bud (Chen et al., 2012; Fu and Hsu, 2013; Zhu et al., 2013, 2014). However, the loss of β-catenin did not affect Bmp4 expression in the nasal pit epithelium (Reid et al., 2011), suggesting downstream signaling other than β-catenin is required for regulation of BMP signaling in nasal pit development.
In summary, our findings show that Gpr177 mediated Wnt ligands, produced by the head ectoderm, regulate the invagination of the developing nasal pit. We suggest that in addition to the requirement for the well-known Wnt/β-catenin–dependent FGF pathway, Gpr177 mediated noncanonical Wnt production is required for JNK-dependent cellular shaping, cell–cell junction, and cell migration of the invaginating nasal pit epithelium. These are in turn required to complete development of the upper face.
Experimental Procedures
Animal Models
The construction of conditional targeted Gpr177 mouse line, the Gpr177-Fx (Gpr177fx/fx), has been described previously (Fu et al., 2009). The conditional targeted Gpr177 line was maintained in B6/C57 background as homozygous. Foxg1-Cre mouse was used to induce the tissue specific deletion of Gpr177 in the facial ectoderm during embryonic facial development. Briefly, Gpr177fx/fx mice were crossed with Foxg1-Cre to obtain Gpr177(fx/+)Foxg1-Cre mice which were back-crossed with Gpr177fx/fx for Gpr177 conditional knockout mice. Wild-type or Gpr177(fx/+)Foxg1-Cre littermates were used as control embryos. Mouse lines of R26R reporter, TOPGAL, Foxg1-Cre, and Wnt1-Cre were purchased from the Jackson Laboratory, Maine. All animal experimental protocols were approved by the Animal Users Committee of Hangzhou Normal University, China.
Samples of Scanning Electronic Microscope
Embryonic heads were collected and fixed in 0.25% glutaraldehyde. The standard procedures were followed for SME observation as described previously (Zhu et al., 2013).
Immunofluorescence and Co-immunoprecipitation
Embryonic heads were fixed in 4% paraformaldehyde for 30 min, washed three times in phosphate buffered saline (PBS), and processed for either paraffin sections or cryostat sections. For cryostat sections, samples were treated in 5% sucrose and 15% sucrose, 2 hr each, and in 30% sucrose. Samples were embedded in OCT and sectioned at 20 mm. To conduct immunohistochemical staining, sections were washed 3 times in PBST (0.1%Triton X-100/PBS), then blocked in 5% bovine serum albumin (BSA) for 30 min, and incubated with primary antibodies diluted with 5% BSA at 4 degC overnight in a humid chamber. Sections were subsequently washed in PBST, 3 times for 10 min each. Secondary antibodies (1:1000) and DAPI (1:500) diluted in 5% BSA were applied for 30 min in the dark. Following application of secondary antibodies, the sections were washed several times with PBST, for 10 min for each, mounted with Mowiol (Sigma) and stored at 4 degC.
Co-immunoprecipitation was performed as previously described (Zhu et al., 2013).
Primary antibodies used in this study were commercially purchased: E-cadherin (3199, Cell Signaling Technology), p-JNK (9255, Cell Signaling Technology), p-Erk (4695, Cell Signaling Technology), Sox2 (ab97959, Abcam), pSmad1/5/8 (9511, Cell Signaling Technology), Fgf10 (sc-7375, Santa Cruz), Actin (3700, cell signaling technology), active β-catenin (ABC, 05-665, Millipore), β-gal (Z3781, Promega).
Assays of Cell Apoptosis, BrdU Incorporation
Cell apoptosis was detected with TUNEL assay kit (Roche). Cell proliferation was analyzed by incorporation of BrdU using in situ cell proliferation detection kits (Roche). BrdU was injected intraperitoneally to pregnant female mice for 30 min. Embryonic heads were fixed and processed for sections as described above. For statistical analysis, ≥ 20 consecutive fields from three samples for both wild-type and mutant were calculated. Two-tailed Student’s t-tests were used for statistical analysis. A P value < 0.05 was considered statistically significant. Data are represented as mean 6 standard deviation (SD).
Quantitative Real-Time PCR and In Situ Hybridization
To obtain total RNA for RNA expression analysis by quantitative real-time reverse transcriptase PCR (RT-PCR), facial primordial tissue at E9.5 and E10.5 were dissected, and total RNA was isolated using RNAqueous-4PCR kit (Ambion). cDNA was synthesized from 2 mg of total RNA from each sample using PrimeScript 1st strand cDNA Synthesis Kit (Takara). The synthesized cDNA was diluted to 100 ml and an aliquot of 2 ml was used as template in a 20 ml quantitative real-time PCR reaction system. Real-time RT-PCR was performed using following primer sequences: Axin2: 5′-ACGCACTGACCGACGATT-3′ and 5′-AAGGCAGCAGGTTCCA CA-3′; Lef1: 5′-AACGAGTCCGAAATCATCCCA-3′ and 5′-GCCA GAGTAACTGGAGTAGGA-3′; Gpr177: 5′-GAGAAAATGCA GAAATTTCCATGG-3′ and 5′-GGTCTTGGGGGATGTAAAAGTA CAT-3′; Bmp4: 5′-GACTTCGAGGCGACACTTCTA-3′ and 5′-GAA TGACGGCGCTCTTGCTA-3′; Fgf3: 5′-TACAACGCAGAGTGTGA GTTTG-3′ and 5′-CACCGACACGTACCAAGGTC-3′; Fgf4: 5′-T GGGCCTCAAAAGGCTTCG-3′ and 5′-CGTCGGTAAAGAAAGGCA CAC-3′; Fgf7: 5′-CAGAACAAAAGTCAAGGA-GCAACCG-3′ and 5′-GTCGCTCGGGGCTGGAACAG-3′; Fgf8: 5′-CCGAGGA-GGGA TCTAAGGAAC-3′ and 5′-CTTCCAAAAGTATCGGTCTCCAC-3′; Fgf9: 5′-CCCAACGGTACTATCCAGGGA-3′ and 5′-AGGCCCA CTGCTATA-CTGATAAA-3′; Fgf10: 5′-TCAGCGGGACCAAGAA TGAAG-3′ and 5′-CGGC-AACAACTCCGATTTCC-3′; Msx1: 5′-TC ATGGCCGATCACAGGAAG-3′ and 5′-GGAGTCCTCCGACTGA GAAATG-3′; Msx2: 5′-TTCACCACATCCCAGCTTCTA-3′ and 5′-TTGCAGTCTTTTCGCCTTAGC-3′; 18S 5′-TAGAGGGACAA GTGGCGTTC-3′ and 5′-CGCTGAGCCAGTCAGTGT-3′. 18S rRNA was used as a reference gene. Real-time PCR was performed in triplicate using SsoFast EvaGreen Supermix with CFX96 real-time PCR Detection System (Bio-Rad Laboratories). Data were analyzed with the CFXManagerTM software and were represented as mean 6 SEM. Data from the wild-type sample were set as control and were normalized to one.
For in situ hybridizations, embryonic heads were collected at E9.5, E10.5, and E11.5. Samples were fixed in 4% PFA overnight. Nonradioactive ribopobes for Bmp4, Msx1, Msx2, Fgf8, Gpr177, Wnt3, Wnt5a, Wnt6, and Wnt9b were labeled and whole-mount in situ hybridization was performed as described (Zhu et al., 2013, 2014).
In Vitro Organ Culture
Facial prominences from E9.5 embryos were microdissected and placed on a Nucleopore Track-Etch Membrane in a Trowell-type organ culture dish as previously described (Zhu et al., 2014). Facial explants were cultured for 1day in the presence of 25 μM SP600126 or U0126, harvested and processed for immunostaining or X-gal staining. Three or four embryos were used for each group in experiments.
Acknowledgments
The authors thank all members of the Zhang laboratory at the Institute of Developmental and Regenerative Biology, Hangzhou Normal University, for their suggestions during the generation of these data.
Grant sponsor: Natural Scientific Foundation of China; Grant numbers: 31071287, 31371471, 31201090; Grant sponsor: Natural Scientific Foundation of Zhejiang; Grant numbers: LZ12C12002, LY12C1200, NIH DE015654.
References
- Ahtiainen L, Lefebvre S, Lindfors PH, Renvoise E, Shirokova V, Vartiainen MK, Thesleff I, Mikkola ML. Directional cell migration, but not proliferation, drives hair placode morphogenesis. Dev Cell. 2014;28:588–602. doi: 10.1016/j.devcel.2014.02.003. [DOI] [PubMed] [Google Scholar]
- Al Alam D, Green M, Tabatabai Irani R, Parsa S, Danopoulos S, Sala FG, Branch J, El Agha E, Tiozzo C, Voswinckel R, Jesudason EC, Warburton D, Bellusci S. Contrasting expression of canonical Wnt signaling reporters TOPGAL, BATGAL and Axin2(LacZ) during murine lung development and repair. PLoS One. 2011;6:e23139. doi: 10.1371/journal.pone.0023139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachler M, Neubuser A. Expression of members of the Fgf family and their receptors during midfacial development. Mech Dev. 2001;100:313–316. doi: 10.1016/s0925-4773(00)00518-9. [DOI] [PubMed] [Google Scholar]
- Banziger C, Soldini D, Schutt C, Zipperlen P, Hausmann G, Basler K. Wntless, a conserved membrane protein dedicated to the secretion of Wnt proteins from signaling cells. Cell. 2006;125:509–522. doi: 10.1016/j.cell.2006.02.049. [DOI] [PubMed] [Google Scholar]
- Brugmann SA, Goodnough LH, Gregorieff A, Leucht P, ten Berge D, Fuerer C, Clevers H, Nusse R, Helms JA. Wnt signaling mediates regional specification in the vertebrate face. Development. 2007;134:3283–3295. doi: 10.1242/dev.005132. [DOI] [PubMed] [Google Scholar]
- Brunjes PC, Frazier LL. Maturation and plasticity in the olfactory system of vertebrates. Brain Res. 1986;396:1–45. doi: 10.1016/s0006-8993(86)80188-3. [DOI] [PubMed] [Google Scholar]
- Chen D, Jarrell A, Guo C, Lang R, Atit R. Dermal beta-catenin activity in response to epidermal Wnt ligands is required for fibroblast proliferation and hair follicle initiation. Development. 2012;139:1522–1533. doi: 10.1242/dev.076463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chenevix-Trench G, Jones K, Green AC, Duffy DL, Martin NG. Cleft lip with or without cleft palate: associations with transforming growth factor alpha and retinoic acid receptor loci. Am J Hum Genet. 1992;51:1377–1385. [PMC free article] [PubMed] [Google Scholar]
- Corson LB, Yamanaka Y, Lai KM, Rossant J. Spatial and temporal patterns of ERK signaling during mouse embryogenesis. Development. 2003;130:4527–4537. doi: 10.1242/dev.00669. [DOI] [PubMed] [Google Scholar]
- Croucher SJ, Tickle C. Characterization of epithelial domains in the nasal passages of chick embryos: spatial and temporal mapping of a range of extracellular matrix and cell surface molecules during development of the nasal placode. Development. 1989;106:493–509. doi: 10.1242/dev.106.3.493. [DOI] [PubMed] [Google Scholar]
- Davis RJ. Signal transduction by the JNK group of MAP kinases. Cell. 2000;103:239–252. doi: 10.1016/s0092-8674(00)00116-1. [DOI] [PubMed] [Google Scholar]
- De Carlos JA, Lopez-Mascaraque L, Valverde F. Early olfactory fiber projections and cell migration into the rat telencephalon. Int J Dev Neurosci. 1996;14:853–866. doi: 10.1016/s0736-5748(96)00055-x. [DOI] [PubMed] [Google Scholar]
- Fornaro M, Geuna S, Fasolo A, Giacobini-Robecchi MG. Evidence of very early neuronal migration from the olfactory placode of the chick embryo. Neuroscience. 2001;107:191–197. doi: 10.1016/s0306-4522(01)00334-7. [DOI] [PubMed] [Google Scholar]
- Fu J, Hsu W. Epidermal Wnt controls hair follicle induction by orchestrating dynamic signaling crosstalk between the epidermis and dermis. J Invest Dermatol. 2013;133:890–898. doi: 10.1038/jid.2012.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu J, Ivy Yu HM, Maruyama T, Mirando AJ, Hsu W. Gpr177/ mouse Wntless is essential for Wnt-mediated craniofacial and brain development. Dev Dyn. 2011;240:365–371. doi: 10.1002/dvdy.22541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu J, Jiang M, Mirando AJ, Yu HM, Hsu W. Reciprocal regulation of Wnt and Gpr177/mouse Wntless is required for embryonic axis formation. Proc Natl Acad Sci U S A. 2009;106:18598–18603. doi: 10.1073/pnas.0904894106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gros J, Hu JK, Vinegoni C, Feruglio PF, Weissleder R, Tabin CJ. WNT5A/JNK and FGF/MAPK pathways regulate the cellular events shaping the vertebrate limb bud. Curr Biol. 2010;20:1993–2002. doi: 10.1016/j.cub.2010.09.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He F, Xiong W, Yu X, Espinoza-Lewis R, Liu C, Gu S, Nishita M, Suzuki K, Yamada G, Minami Y, Chen Y. Wnt5a regulates directional cell migration and cell proliferation via Ror2-mediated noncanonical pathway in mammalian palate development. Development. 2008;135:3871–3879. doi: 10.1242/dev.025767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebert JM, McConnell SK. Targeting of cre to the Foxg1 (BF-1) locus mediates loxP recombination in the telencephalon and other developing head structures. Dev Biol. 2000;222:296–306. doi: 10.1006/dbio.2000.9732. [DOI] [PubMed] [Google Scholar]
- Hu D, Marcucio RS. Unique organization of the frontonasal ectodermal zone in birds and mammals. Dev Biol. 2009;325:200–210. doi: 10.1016/j.ydbio.2008.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu D, Young NM, Xu Q, Jamniczky H, Green RM, Mio W, Marcucio RS, Hallgrimsson B. Signals from the brain induce variation in avian facial shape. Dev Dyn. 2015 doi: 10.1002/dvdy.24284. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C, Rajfur Z, Borchers C, Schaller MD, Jacobson K. JNK phosphorylates paxillin and regulates cell migration. Nature. 2003;424:219–223. doi: 10.1038/nature01745. [DOI] [PubMed] [Google Scholar]
- Jiang R, Bush JO, Lidral AC. Development of the upper lip: morphogenetic and molecular mechanisms. Dev Dyn. 2006;235:1152–1166. doi: 10.1002/dvdy.20646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin YR, Han XH, Taketo MM, Yoon JK. Wnt9b-dependent FGF signaling is crucial for outgrowth of the nasal and maxillary processes during upper jaw and lip development. Development. 2012;139:1821–1830. doi: 10.1242/dev.075796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juriloff DM, Harris MJ, McMahon AP, Carroll TJ, Lidral AC. Wnt9b is the mutated gene involved in multifactorial nonsyndromic cleft lip with or without cleft palate in A/WySn mice, as confirmed by a genetic complementation test. Birth Defects Res A Clin Mol Teratol. 2006;76:574–579. doi: 10.1002/bdra.20302. [DOI] [PubMed] [Google Scholar]
- Karner CM, Chirumamilla R, Aoki S, Igarashi P, Wallingford JB, Carroll TJ. Wnt9b signaling regulates planar cell polarity and kidney tubule morphogenesis. Nat Genet. 2009;41:793–799. doi: 10.1038/ng.400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawauchi S, Shou J, Santos R, Hebert JM, McConnell SK, Mason I, Calof AL. Fgf8 expression defines a morphogenetic center required for olfactory neurogenesis and nasal cavity development in the mouse. Development. 2005;132:5211–5223. doi: 10.1242/dev.02143. [DOI] [PubMed] [Google Scholar]
- Kuan CY, Yang DD, Samanta Roy DR, Davis RJ, Rakic P, Flavell RA. The Jnk1 and Jnk2 protein kinases are required for regional specific apoptosis during early brain development. Neuron. 1999;22:667–676. doi: 10.1016/s0896-6273(00)80727-8. [DOI] [PubMed] [Google Scholar]
- Lan Y, Ryan RC, Zhang Z, Bullard SA, Bush JO, Maltby KM, Lidral AC, Jiang R. Expression of Wnt9b and activation of canonical Wnt signaling during midfacial morphogenesis in mice. Dev Dyn. 2006;235:1448–1454. doi: 10.1002/dvdy.20723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lienkamp SS, Liu K, Karner CM, Carroll TJ, Ronneberger O, Wallingford JB, Walz G. Vertebrate kidney tubules elongate using a planar cell polarity-dependent, rosette-based mechanism of convergent extension. Nat Genet. 2012;44:1382–1387. doi: 10.1038/ng.2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Sun X, Braut A, Mishina Y, Behringer RR, Mina M, Martin JF. Distinct functions for Bmp signaling in lip and palate fusion in mice. Development. 2005;132:1453–1461. doi: 10.1242/dev.01676. [DOI] [PubMed] [Google Scholar]
- Liu Y, Wang M, Zhao W, Yuan X, Yang X, Li Y, Qiu M, Zhu XJ, Zhang Z. Gpr177-mediated Wnt signaling is required for secondary palate development. J Dent Res. 2015;94:961–967. doi: 10.1177/0022034515583532. [DOI] [PubMed] [Google Scholar]
- Marazita ML, Murray JC, Lidral AC, Arcos-Burgos M, Cooper ME, Goldstein T, Maher BS, Daack-Hirsch S, Schultz R, Mansilla MA, Field LL, Liu YE, Prescott N, Malcolm S, Winter R, Ray A, Moreno L, Valencia C, Neiswanger K, Wyszynski DF, Bailey-Wilson JE, Albacha-Hejazi H, Beaty TH, McIntosh I, Hetmanski JB, Tuncbilek G, Edwards M, Harkin L, Scott R, Roddick LG. Meta-analysis of 13 genome scans reveals multiple cleft lip/palate genes with novel loci on 9q21 and 2q32-35. Am J Hum Genet. 2004;75:161–173. doi: 10.1086/422475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcucio RS, Cordero DR, Hu D, Helms JA. Molecular interactions coordinating the development of the forebrain and face. Dev Biol. 2005;284:48–61. doi: 10.1016/j.ydbio.2005.04.030. [DOI] [PubMed] [Google Scholar]
- Marcucio RS, Young NM, Hu D, Hallgrimsson B. Mechanisms that underlie co-variation of the brain and face. Genesis. 2011;49:177–189. doi: 10.1002/dvg.20710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maretto S, Cordenonsi M, Dupont S, Braghetta P, Broccoli V, Hassan AB, Volpin D, Bressan GM, Piccolo S. Mapping Wnt/beta-catenin signaling during mouse development and in colorectal tumors. Proc Natl Acad Sci U S A. 2003;100:3299–3304. doi: 10.1073/pnas.0434590100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama EO, Yu HM, Jiang M, Fu J, Hsu W. Gpr177 deficiency impairs mammary development and prohibits Wnt-induced tumorigenesis. PLoS One. 2013;8:e56644. doi: 10.1371/journal.pone.0056644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menezes R, Letra A, Kim AH, Kuchler EC, Day A, Tannure PN, Gomes da Motta L, Paiva KB, Granjeiro JM, Vieira AR. Studies with Wnt genes and nonsyndromic cleft lip and palate. Birth Defects Res A Clin Mol Teratol. 2010;88:995–1000. doi: 10.1002/bdra.20720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzis V, Courtney AD, Kerr MC, Ferguson C, Rondon Galeano MC, Parton RG, Wainwright BJ, Wicking C. Patched1 is required in neural crest cells for the prevention of orofacial clefts. Hum Mol Genet. 2013;22:5026–5035. doi: 10.1093/hmg/ddt353. [DOI] [PubMed] [Google Scholar]
- Minoux M, Rijli FM. Molecular mechanisms of cranial neural crest cell migration and patterning in craniofacial development. Development. 2010;137:2605–2621. doi: 10.1242/dev.040048. [DOI] [PubMed] [Google Scholar]
- Niemann S, Zhao C, Pascu F, Stahl U, Aulepp U, Niswander L, Weber JL, Muller U. Homozygous WNT3 mutation causes tetra-amelia in a large consanguineous family. Am J Hum Genet. 2004;74:558–563. doi: 10.1086/382196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oishi I, Suzuki H, Onishi N, Takada R, Kani S, Ohkawara B, Koshida I, Suzuki K, Yamada G, Schwabe GC, Mundlos S, Shibuya H, Takada S, Minami Y. The receptor tyrosine kinase Ror2 is involved in non-canonical Wnt5a/JNK signalling pathway. Genes Cells. 2003;8:645–654. doi: 10.1046/j.1365-2443.2003.00662.x. [DOI] [PubMed] [Google Scholar]
- Paiva KB, Silva-Valenzuela M, Massironi SM, Ko GM, Siqueira FM, Nunes FD. Differential Shh, Bmp and Wnt gene expressions during craniofacial development in mice. Acta Histochem. 2010;112:508–517. doi: 10.1016/j.acthis.2009.05.007. [DOI] [PubMed] [Google Scholar]
- Plageman TF, Jr, Chauhan BK, Yang C, Jaudon F, Shang X, Zheng Y, Lou M, Debant A, Hildebrand JD, Lang RA. A Trio-RhoA-Shroom3 pathway is required for apical constriction and epithelial invagination. Development. 2011;138:5177–5188. doi: 10.1242/dev.067868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raju R, Palapetta SM, Sandhya VK, Sahu A, Alipoor A, Balakrishnan L, Advani J, George B, Kini KR, Geetha NP, Prakash HS, Prasad TS, Chang YJ, Chen L, Pandey A, Gowda H. A network map of FGF-1/FGFR signaling system. J Signal Transduct. 2014;2014(2014):962962. doi: 10.1155/2014/962962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid BS, Yang H, Melvin VS, Taketo MM, Williams T. Ectodermal Wnt/beta-catenin signaling shapes the mouse face. Dev Biol. 2011;349:261–269. doi: 10.1016/j.ydbio.2010.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sai X, Ladher RK. FGF signaling regulates cytoskeletal remodeling during epithelial morphogenesis. Curr Biol. 2008;18:976–981. doi: 10.1016/j.cub.2008.05.049. [DOI] [PubMed] [Google Scholar]
- Schlosser G. Induction and specification of cranial placodes. Dev Biol. 2006;294:303–351. doi: 10.1016/j.ydbio.2006.03.009. [DOI] [PubMed] [Google Scholar]
- Sineva GS, Pospelov VA. beta-Catenin in pluripotency: adhering to self-renewal or Wnting to differentiate? Int Rev Cell Mol Biol. 2014;312:53–78. doi: 10.1016/B978-0-12-800178-3.00002-6. [DOI] [PubMed] [Google Scholar]
- Song L, Li Y, Wang K, Wang YZ, Molotkov A, Gao L, Zhao T, Yamagami T, Wang Y, Gan Q, Pleasure DE, Zhou CJ. Lrp6-mediated canonical Wnt signaling is required for lip formation and fusion. Development. 2009;136:3161–3171. doi: 10.1242/dev.037440. [DOI] [PubMed] [Google Scholar]
- Streit A. Extensive cell movements accompany formation of the otic placode. Dev Biol. 2002;249:237–254. doi: 10.1006/dbio.2002.0739. [DOI] [PubMed] [Google Scholar]
- Tang N, Marshall WF, McMahon M, Metzger RJ, Martin GR. Control of mitotic spindle angle by the RAS-regulated ERK1/2 pathway determines lung tube shape. Science. 2011;333:342–345. doi: 10.1126/science.1204831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teven CM, Farina EM, Rivas J, Reid RR. Fibroblast growth factor (FGF) signaling in development and skeletal diseases. Genes Dis. 2014;1:199–213. doi: 10.1016/j.gendis.2014.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toba Y, Ajiki K, Horie M, Sango K, Kawano H. Immunohistochemical localization of calbindin D-28k in the migratory pathway from the rat olfactory placode. J Neuroendocrinol. 2001;13:683–694. doi: 10.1046/j.1365-2826.2001.00685.x. [DOI] [PubMed] [Google Scholar]
- Valverde F, Heredia M, Santacana M. Characterization of neuronal cell varieties migrating from the olfactory epithelium during prenatal development in the rat. Immunocytochemical study using antibodies against olfactory marker protein (OMP) and luteinizing hormone-releasing hormone (LH-RH) Brain Res Dev Brain Res. 1993;71:209–220. doi: 10.1016/0165-3806(93)90173-8. [DOI] [PubMed] [Google Scholar]
- Waetzig V, Zhao Y, Herdegen T. The bright side of JNKs-Multitalented mediators in neuronal sprouting, brain development and nerve fiber regeneration. Prog Neurobiol. 2006;80:84–97. doi: 10.1016/j.pneurobio.2006.08.002. [DOI] [PubMed] [Google Scholar]
- Wang Y, Song L, Zhou CJ. The canonical Wnt/beta-catenin signaling pathway regulates Fgf signaling for early facial development. Dev Biol. 2011;349:250–260. doi: 10.1016/j.ydbio.2010.11.004. [DOI] [PubMed] [Google Scholar]
- Weston CR, Wong A, Hall JP, Goad ME, Flavell RA, Davis RJ. JNK initiates a cytokine cascade that causes Pax2 expression and closure of the optic fissure. Genes Dev. 2003;17:1271–1280. doi: 10.1101/gad.1087303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wray S. From nose to brain: development of gonadotrophin-releasing hormone-1 neurones. J Neuroendocrinol. 2010;22:743–753. doi: 10.1111/j.1365-2826.2010.02034.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wray S, Grant P, Gainer H. Evidence that cells expressing luteinizing hormone-releasing hormone mRNA in the mouse are derived from progenitor cells in the olfactory placode. Proc Natl Acad Sci U S A. 1989;86:8132–8136. doi: 10.1073/pnas.86.20.8132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia Y, Karin M. The control of cell motility and epithelial morphogenesis by Jun kinases. Trends Cell Biol. 2004;14:94–101. doi: 10.1016/j.tcb.2003.12.005. [DOI] [PubMed] [Google Scholar]
- Zhang L, Wang W, Hayashi Y, Jester JV, Birk DE, Gao M, Liu CY, Kao WW, Karin M, Xia Y. A role for MEK kinase 1 in TGF-beta/activin-induced epithelium movement and embryonic eyelid closure. EMBO J. 2003;22:4443–4454. doi: 10.1093/emboj/cdg440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X, Liu Y, Zhao P, Dai Z, Yang X, Li Y, Qiu M, Zhang Z. Gpr177-mediated Wnt Signaling is Required for Fungiform Placode Initiation. J Dent Res. 2014;93:582–588. doi: 10.1177/0022034514531985. [DOI] [PubMed] [Google Scholar]
- Zhu X, Zhao P, Liu Y, Zhang X, Fu J, Ivy Yu HM, Qiu M, Chen Y, Hsu W, Zhang Z. Intra-epithelial requirement of canonical Wnt signaling for tooth morphogenesis. J Biol Chem. 2013;288:12080–12089. doi: 10.1074/jbc.M113.462473. [DOI] [PMC free article] [PubMed] [Google Scholar]