Abstract
Ubiquitination has long been known to regulate fundamental cellular processes through the induction of proteasomal degradation of target proteins. More recently, ‘atypical’ nondegradative types of polyubiquitin chains have been appreciated as important regulatory moieties by modulating the activity or subcellular localization of key signaling proteins. Intriguingly, many of these non-degradative types of ubiquitination regulate the innate sensing pathways initiated by pattern recognition receptors (PRRs), ultimately coordinating an effective antiviral immune response. Here we discuss recent advances in understanding the functional roles of degradative and atypical types of ubiquitination in innate immunity to viral infections, with a specific focus on the signaling pathways triggered by RIG-I-like receptors, Toll-like receptors, and the intracellular viral DNA sensor cGAS.
Keywords: Antiviral immunity, Type-I interferon, RIG-I-like receptors, Toll-like receptors, cGAS, STING, Ubiquitin, E3 ligases, TRIM proteins, TRIM25
Introduction
Infection with viral pathogens triggers an immediate antiviral response in the host cell, commonly termed ‘innate immune response’. This response is characterized by rapid gene expression of a variety of antiviral and inflammation-inducing molecules, including type-I interferons (IFN-α/β), type-III IFNs (IFN-λ or IL-28/29), proinflammatory cytokines and chemokines. Upon secretion and subsequent binding to their respective receptors on the surface of surrounding cells, IFNs lead to the upregulation of more than one hundred different interferon-stimulated genes (ISGs) [1, 2]. ISGs encode for either signaling molecules, including transcription factors, that amplify the innate immune response, or for antiviral effector proteins to block virus replication through multiple mechanisms, such as cleavage of viral RNA or shutdown of host cell translation. Furthermore, secreted proinflammatory cytokines and chemokines produced during the innate immune response are critical for priming and fine-tuning the adaptive immune response [3, 4].
One class of important molecules in the activation of the innate antiviral response are pattern recognition receptors (PRRs), which recognize viral proteins or specific features in the viral nucleic acid, and then trigger immune signaling that results in IFN production [5, 6]. At least three major classes of PRRs recognizing viral nucleic acids have been identified: (1) the cytosolic RIG-I-like receptors (RLRs) sensing viral RNA species produced during both RNA and DNA virus infections; (2) the membrane-bound Toll-like receptors (TLRs) detecting viral RNA or DNA in endolysosomes immediately after virus entry; and (3) a group of structurally-unrelated viral DNA sensors, with cGAS (cyclic GMP-AMP synthase) representing a key sensor of various DNA virus infections. Upon sensing of viral nucleic acid, these sensors activate several kinases belonging to the IKK (inhibitor of nuclear factor kappa-B [IκB] kinase) family, namely the canonical IKKα and IKKβ together with their essential regulatory subunit IKKγ/NEMO, as well as the non-canonical IKKε and TANK-binding kinase-1 (TBK1). IKKα/β/γ and TBK1/IKKε then activate the transcription factors NF-κB and IFN-regulatory factors 3 and 7 (IRF3/7), respectively. In addition, PRRs activate several mitogen-activated protein kinases (MAPK), leading to the activation of AP-1 (activator protein-1). IRF-3/7, NF-κB and AP-1, upon their translocation into the nucleus, transcriptionally induce IFNs and other cytokines, ultimately establishing an antiviral program in the infected host cell or uninfected surrounding cells [7, 8].
Aberrant PRR activation and signaling can lead to chronic inflammation and tissue damage, and potentially cause autoimmune disorders. Indeed, recent findings indicated that some autoimmune diseases, e.g. systemic lupus erythematosus and Aicardi-Goutières Syndrome, are linked to single nucleotide polymorphisms (SNPs) in PRRs that lead to their constitutive activation (reviewed in [9, 10]). To prevent premature or excessive activation of PRR-induced antiviral signaling, an elegant system of regulation is in place. A key host mechanism for modulating the stability and signaling activity of PRRs and their downstream signaling molecules is reversible posttranslational modification (PTM), with phosphorylation and ubiquitination being the most well studied PTMs. Here we focus on the role of ubiquitination and the reversal of this process, deubiquitination, in the regulation of three major innate sensing pathways of viral infections: the RLR, TLR and cGAS-STING pathways.
Ubiquitin conjugation and deubiquitination of proteins
Ubiquitin is a small, 76 amino acid protein that is conserved across eukaryotic organisms and can be covalently attached to lysines or other residues in target proteins to modify their stabilities or activities. Ubiquitin conjugation is completed through step-wise catalysis using three distinct classes of enzymes, termed E1, E2 and E3 [11, 12]. First, E1 activates the ubiquitin molecule in an ATP-dependent manner by forming an intermediate thioester bond between an active cysteine group in the E1 enzyme itself and the ubiquitin C-terminus. The E1 ubiquitin-activating enzyme next binds to the E2 enzyme, also called ubiquitin-conjugating enzyme, which accepts the ubiquitin at a catalytic cysteine residue. Finally, the E3 ubiquitin ligase, in complex with E2, facilitates the transfer of the ubiquitin moiety to the substrate protein by forming an isopeptide bond, usually between the ε-amino group of a lysine in the substrate and the C-terminal glycine residue of the ubiquitin molecule. Given that the E3 ligase determines the substrate specificity and that there are many different substrate proteins for ubiquitination in human cells, it is not surprising that a large number (more than 700) of E3 ligases exists. Furthermore, in humans, there are two E1 enzymes, which usually do not have any specificity for the E2 or E3 enzyme, and ~40 different E2 enzymes, whose primary function is to determine which types of polyubiquitin chains are catalyzed by the E3.
The E3 ubiquitin ligase superfamily can be classified into four major families: RING (Really Interesting New Gene), HECT (homologous to E6-associated protein C-terminus), U-box (UFD2 homology), and RBR (RING-in-between-RING) E3 ligases [13-15]. Members of each E3 ligase family facilitate ubiquitin conjugation to the target protein through different mechanisms. RING E3 ligases, the most prevalent, never directly bind to the ubiquitin moiety. Instead, they serve as mediators for direct transfer of the ubiquitin molecule from the E2 enzyme to the substrate. In contrast, in the case of HECT E3 ligases, an intermediate bond between ubiquitin and a catalytic cysteine on the E3 ligase is formed before transfer of ubiquitin to the target protein. U-box E3 ligases, also dubbed E4 ubiquitin ligases, primarily elongate polyubiquitin chains that have already been begun by another E3 ligase [16]. The recently identified family of RBR E3 ligases (further reviewed in [17]) are structurally characterized by two domains which are bioinformatically similar to RING domains, separated by an intervening sequence called IBR (in-between-RING). RBR E3 ligases catalyze ubiquitin conjugation through a hybrid mechanism in which the first RING domain acts as a canonical RING ligase, interacting with the E2 enzyme, bringing it in proximity to the substrate. The second RING domain, also called Rcat (required for catalysis), then accepts the ubiquitin from the E2 enzyme before transferring it to the substrate, similar to the action of a HECT ligase [17, 18]. Two of the most highly studied RBR ligases are Parkin, an E3 ligase involved in mitochondrial biology and well-known for its implication in Parkinson's disease, and LUBAC, the linear ubiquitination assembly complex, which plays an important role in various antiviral and inflammatory signaling pathways (as discussed in more detail below). The counterplayers of the E2/E3 complex are deubiquitinating enzymes (DUBs), cysteine proteases or metalloproteases that remove monoubiquitin or polyubiquitin chains from substrate proteins. In humans, there are ~100 DUB enzymes which can be further categorized into five main superfamilies dependent on their catalytic domains and mechanisms of action: the ubiquitin-specific proteases (USP), the ovarian tumor (OTU) superfamily, the ubiquitin C-terminal hydrolases (UCH), the Machado-Josephin domain (MJD) superfamily, and the JAMM (JAB1/MPN/Mov34) metalloprotease family [19].
During the past decade, it has become evident that E3 ligases and DUBs play important roles in fine-tuning innate immunity by either modulating the stability of key molecules in the immune system, or by regulating cytokine production through synthesis (or removal) of unconventional types of polyubiquitin that are necessary for innate signal transduction.
Functional roles of different linkage types of polyubiquitination
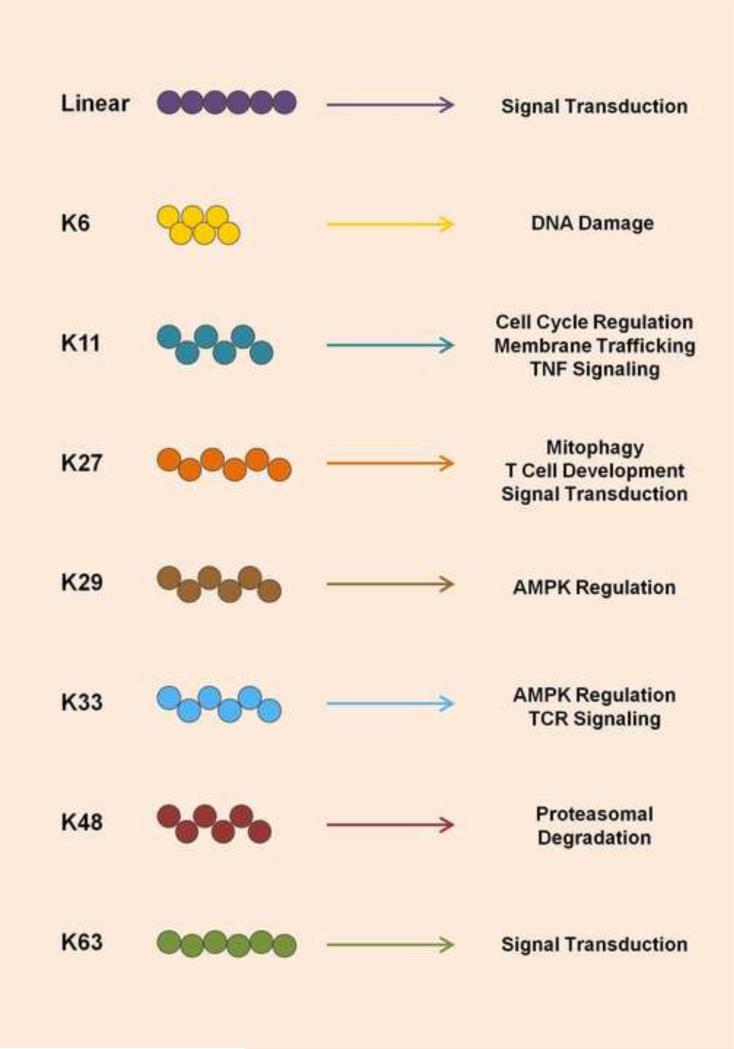
Conjugation of ubiquitin to substrates generally occurs at lysine residues, but may also occur at cysteine, serine, threonine, and tyrosine residues [20]. Residues can be modified with a single ubiquitin moiety (monoubiquitination), two ubiquitin proteins (diubiquitination), or chains of ubiquitin (polyubiquitination). Polyubiquitin chains are usually formed through covalent binding of the C-terminal glycine of one ubiquitin molecule to an internal lysine residue of another ubiquitin molecule. As ubiquitin harbors seven internal lysine residues (K6, K11, K27, K29, K33, K48, and K63), seven different types of polyubiquitin chains can arise (Figure 1) [20, 21]. Among them, K48-linked ubiquitination, the classical ubiquitination involved in proteasomal degradation, and K63-linked polyubiquitination represent the most well studied polyubiquitin types. Furthermore, the C-terminus of one ubiquitin molecule can also bind to the N-terminal methionine of another ubiquitin molecule, giving rise to ‘linear ubiquitination’ (also called ‘head-to-tail’ or Met1 ubiquitination). Polyubiquitin chains are usually covalently linked to the substrate; however, over the past several years, it has become clear that polyubiquitin chains can also be bound non-covalently by substrate proteins.
Figure 1. Functional roles of the different linkage types of polyubiquitination.
The 8 different linkage types of polyubiquitination are illustrated. Known fates of modified substrates as well as key pathways regulated by specific polyubiquitins are shown. The specific details of how different ubiquitin polymers regulate substrate proteins are described in the text.
The cellular fate of ubiquitinated proteins varies greatly based on the linkage type of ubiquitin chains formed on the modified residue (Figure 1) [21, 22]. Classical K48-linked polyubiquitin chains are generally recognized by the proteasome, leading to degradation of the substrate, a mechanism utilized for normal protein turnover in the cell. In contrast, K63-linked ubiquitination does not usually trigger proteasomal degradation, but instead plays an important role in signal transduction pathways. Mechanistically, K63-linked ubiquitination of proteins has been shown to activate signaling pathways by either stabilizing substrates, or by acting as a scaffold for the formation of a signaling multi-complex. Specifically, K63-linked ubiquitination can promote the multimerization of signaling proteins, thereby inducing their active states, allowing for the recruitment of additional interaction partners and ultimately signal transduction. Furthermore, K63-linked ubiquitination can lead to the formation of an active signaling complex through recruitment of binding partners that harbor specific ubiquitin-binding domains (UBDs) that recognize K63-polyubiquitin-modified proteins.
As with K63-linked ubiquitination, linear ubiquitin chains positively regulate signal transduction events rather than leading to protein degradation [23]. Linear ubiquitination is catalyzed by LUBAC, an E3 ligase complex consisting of HOIL-1 (heme-oxidized iron-responsive element binding protein 2 ubiquitin ligase-1) and HOIP (HOIL-1-interacting protein), often complexed with SHARPIN (SHANK-associated RH domain-interacting protein) [24-27].
The functions of the remaining five polyubiquitin linkages are much more enigmatic. Treatment with proteasomal inhibitors leads to cellular enrichment of not only K48-linked polyubiquitin, but also K11-, K29-, and K33-linked polyubiquitin, suggesting that these ubiquitin linkage types may also play roles in protein degradation [28-30]. In contrast, K6-linked ubiquitin chains seem not to be involved in protein degradation, but may be connected to DNA damage [22]; however, exact outcomes of K6-linked ubiquitination of substrates remain largely unclear. K11-linked ubiquitination is thought to specifically regulate the proteasomal degradation of proteins that are involved in cell cycle regulation; consistent with this, the levels of K11-linked ubiquitin chains vary at different time points during the cell cycle [31]. This poses the interesting speculation that perhaps K11-linked ubiquitination specifically targets proteins involved in cell cycle control for degradation, while K48-linked ubiquitination acts as a more general cellular degradation signal. K11-linked polyubiquitination has also been shown to have an activating role in tumor necrosis factor (TNF) signaling [32]. Finally, K11-linked ubiquitination of MHC (major histocompatibility complex) class I mediated by the K5 protein of Kaposi's sarcoma herpes virus (KSHV) leads to receptor internalization and immune evasion [33, 34].
K27-linked ubiquitination has also been implicated in protein degradation; however, its primary role seems to be in the modification of mitochondrial proteins, triggering the induction of mitophagy. This mechanism is particularly well-characterized for Parkin, which leads to the K11-linked ubiquitination of multiple mitochondrial proteins, inducing their degradation [35, 36]. K27-linked ubiquitination has also been shown to regulate the differentiation of regulatory T cells [37]. The least is known about K29- and K33-linked polyubiquitination. Interestingly, members of the AMP-activated protein kinase (AMPK) family can be modified with both K29-and K33-linked ubiquitin chains. These modifications did not affect the protein stability of AMPKs, but inhibited their activities through an unidentified mechanism [38]. One study has also linked K33-linked ubiquitination to non-proteolytic regulation of T-cell receptor signal transduction [39]. Much more work is needed to delve into the fates of these atypical ubiquitin linkages to determine the linkage-specific fates of protein substrates.
In addition to the described homogenously-linked polyubiquitin chains, in the past few years mixed or branched chains of polyubiquitin have been identified [40, 41]. However, to date it remains largely unclear how these mixed ubiquitin linkages are made and what roles they play.
Recently, a role for unanchored or ‘free’ ubiquitin chains has been described. These chains are generated by E2/E3 ligases, but are not covalently conjugated to a substrate protein, and instead are bound non-covalently. To date, three linkage types of unanchored polyubiquitin have been identified, K63-linked, K48-linked and linear ubiquitin chains, all of which play important roles in innate immune signaling. Unanchored K63-linked and linear ubiquitin chains have been described for their roles in promoting NEMO and/or RIG-I signaling (as described in detail below). Furthermore, unanchored K48 ubiquitin chains were recently shown to play a positive regulatory role in type-I IFN receptor (IFNAR) signal transduction. Specifically, upon IFNAR activation, IKKε binds to unanchored K48-linked ubiquitin chains, which activates IKKε to phosphorylate STAT1 (signal transducer and activator of transcription 1), ultimately triggering expression of antiviral ISGs [42].
The role of ubiquitin in RLR signal transduction
The RLRs are a family of DExD/H-box helicases which recognize viral RNA species in the cytosol of infected cells. The two best-studied RLR members are retinoic acid-inducible gene-I (RIG-I) and melanoma differentiation-associated protein 5 (MDA5), critical for the detection of viral 5'triphosphorylated RNA or long dsRNA, respectively (reviewed in detail in [8, 43]). In addition, it has been recently shown that RIG-I can also detect viral RNA containing a diphosphate moiety [44]. Functional studies in RIG-I and MDA5 knockout (KO) cells showed that RIG-I senses influenza viruses, arenaviruses and vesicular stomatitis virus (VSV), while specifically MDA5 detects picornaviruses. Furthermore, many RNA viruses including Flaviviruses, reoviruses and paramyxoviruses are sensed both by RIG-I and MDA5, often in a temporally-distinct manner [7, 45]. RIG-I has also been implicated in the detection of various DNA viruses (e.g. Epstein-Barr virus and adenoviruses) by sensing 5'triphosphorylated small RNAs produced during replication. The third RLR member, LGP2 (laboratory of genetics and physiology 2), is not believed to have sensing capacity but instead, has been shown to have a regulatory role (reviewed in [46]). All three RLR proteins share a helicase and C-terminal domain (CTD), both of which confer RNA binding ability [47, 48]. Additionally, RIG-I and MDA5, but not LGP2, harbor two N-terminal caspase activation and recruitment domains (CARDs), which are critical for downstream signaling and IFN induction. After binding to their RNA ligands, RIG-I and MDA5 oligomerize and interact with MAVS (also known as Cardif, IPS-1 or VISA) on mitochondria via CARD-CARD interactions [49-52]. Activated MAVS then initiates a signaling cascade, which intersects with the signaling pathway induced by several other innate immune receptors (e.g. TLR or cGAS), resulting in the activation of IRF3/7, AP-1 and NF-κB, which function to trigger transcriptional activation of IFNs and proinflammatory cytokines [7, 53].
Activation of RIG-I-MAVS signaling by K63-linked polyubiquitin
Over the past several years, it has become evident that K63-linked polyubiquitination plays a crucial role in promoting RLR signaling (Figure 2). The regulation of RIG-I by K63-linked ubiquitin chains has been particularly well-characterized. Mass spectrometry and biochemical analyses showed that the N-terminal CARDs of RIG-I undergo K63-linked ubiquitination by TRIM25 (also called estrogen-responsive finger protein [EFP]), an IFN-inducible E3 ligase belonging to the tripartite motif (TRIM) protein family [54]. This study showed that upon viral infection, TRIM25 binds using its C-terminal SPRY domain to the first CARD of RIG-I; the RING E3 ligase activity of TRIM25 then conjugates covalent K63-polyubiquitin chains to the residue K172 in the second CARD of RIG-I (as well as to five other lysine residues: K99, K169, K181, K190 and K193). Before TRIM25-RIG-I binding and RIG-I ubiquitination, specific serine and threonine residues in the RIG-I CARDs must be dephosphorylated by PP1α/γ [55-58]. The K63-linked ubiquitin chains on RIG-I promote RIGI's interaction with MAVS, evidenced by mutation of K172 in RIG-I as well as trim25 gene targeting, both of which profoundly reduced the RIG-I CARD ubiquitination and binding of RIG-I to MAVS [54, 59, 60]. Functional studies in mouse embryonic fibroblast cells (MEFs) deficient in the trim25 gene demonstrated that TRIM25 is an important activator of RIG-I and critical for an effective IFN-mediated immune response to various RNA virus infections. Notably, no covalent K63-linked ubiquitination of the MDA5 CARDs, with or without ectopic expression of TRIM25, was detected in human cells [54]. This study showed for the first time that a member of the TRIM family, comprising ~70 proteins in humans, promotes PRR signaling through conjugation of unconventional polyubiquitin, thereby inducing type-I IFNs. Intriguingly, in the past few years, it has been shown that several other TRIM members – all characterized by the RBCC motif comprised of a RING, B-box and Coiled-coil domain (CCD) – regulate PRR signaling pathways by catalyzing K63-linked ubiquitin chains or other non-degradative types of polyubiquitin. Moreover, some TRIM proteins also conjugate degradative K48-linked ubiquitination to key signaling molecules in innate immunity, thereby dampening antiviral and proinflammatory responses. Thus, TRIM proteins represent an important class of immunoregulatory molecules in innate signaling pathways. In addition, several TRIM members act as antiviral restriction factors (e.g. TRIM5α and TRIM79α) by directly interacting with viral proteins to block virus replication [61-63].
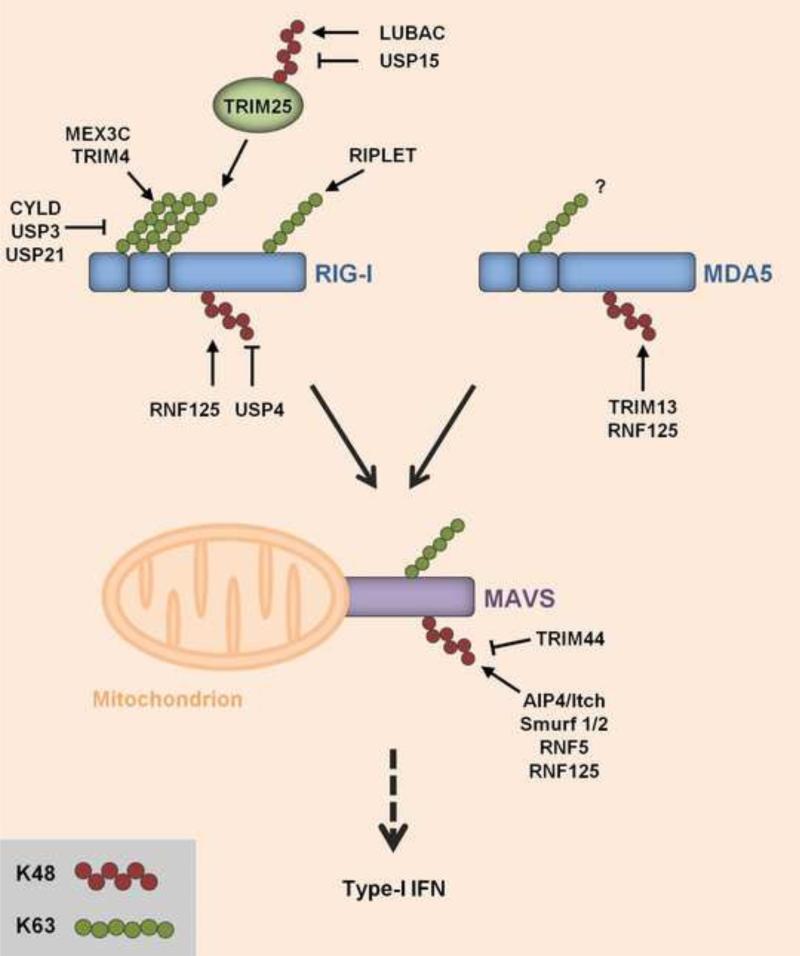
Figure 2. Regulation of RLRs by ubiquitination.
RIG-I and MDA5, members of the RIG-I-like receptor (RLR) family, recognize cytoplasmic viral RNA species and subsequently signal through the adaptor protein MAVS (also called Cardif, IPS-1, or VISA) on mitochondria. Through various steps (not illustrated), MAVS activates downstream signaling, leading to gene expression of type-I IFNs (IFN-α/β). The stability and signaling activities of RIG-I, MDA5 and MAVS are tightly regulated by K48- and K63-linked polyubiquitination, respectively. RIG-I is activated by K63-linked ubiquitination mediated by TRIM25, an IFN-inducible E3 ubiquitin ligase belonging to the large family of TRIM proteins. TRIM25 ubiquitinates several lysines in the N-terminal caspase activation and recruitment domains (CARDs) of RIG-I (not illustrated). TRIM25-mediated ubiquitination specifically at K172 in RIG-I is critical for RIG-I signaling. In addition, TRIM4 and MEX3C were shown to induce K63-linked ubiquitination of the RIG-I CARDs. Furthermore, RIPLET induces K63-linked ubiquitination of K788 (and also other residues) in the C-terminal domain (not illustrated). Both RIG-I and MDA5 have been reported to non-covalently bind unanchored K63-linked ubiquitin chains in vitro; however, the role of MDA5 activation by K63-linked ubiquitin chains remains unclear. TRIM25 itself undergoes K48-linked ubiquitination catalyzed by LUBAC, leading to TRIM25 degradation. Inversely, USP15 antagonizes the LUBAC-induced K48-linked ubiquitination of TRIM25, thereby stabilizing TRIM25 during viral infection, which leads to a sustained IFN response. MAVS is ubiquitinated with K63-linked polyubiquitin chains; however, the E3 ligase for MAVS K63-ubiquitination is unknown. Multiple different E3 ubiquitin ligases have been reported to induce the K48-linked ubiquitination of RLRs and MAVS, triggering their degradation by the proteasome: RNF125 for RIG-I; TRIM13 and RNF125 for MDA5; and AIP4/Itch, Smurf1/2, RNF5 and RNF125 for MAVS. The K48-linked ubiquitination of RIG-I can be actively removed by USP4. Furthermore, TRIM44, an atypical TRIM protein that lacks the RING E3 ligase domain, inhibits the K48-linked ubiquitination of MAVS through an unidentified mechanism.
Besides TRIM25, an additional E3 ligase has been reported to regulate RIG-I through K-63-linked ubiquitination. Riplet (also called RNF135 or REUL) has been shown to ubiquitinate RIG-I, facilitating its activation [64, 65]. In vivo studies confirmed the importance of this E3 ligase in regulating RIG-I signaling: Riplet-deficient mice produced less IFN and were more susceptible to VSV infection than wild-type (WT) mice [66]. While these studies confirmed an important role of Riplet in RIG-I ubiquitination and activation, some of the mechanistic details – specifically which domain and residues in RIG-I are targets of ubiquitination by Riplet – still remain somewhat unclear. While one study suggested that Riplet ubiquitinated residues in the N-terminal CARDs [65], others demonstrated that Riplet ubiquitinated several lysine residues in the CTD of RIG-I, of which K788 seemed to be the functionally important residue for regulating RIG-I signaling [64, 67].
The importance of TRIM25- and Riplet-mediated ubiquitination in RIG-I activation was strengthened by the finding that the non-structural protein 1 (NS1) of influenza A virus (IAV) antagonizes both E3 ligases [60, 68]. Initial studies demonstrated that NS1 interacts with human TRIM25, preventing the K63-linked ubiquitination of the RIG-I CARDs [68]. This was shown to be due to NS1 interacting with the CCD of TRIM25, preventing TRIM25 dimerization, which appears to be critical for TRIM25's enzymatic activity to induce ubiquitination of RIG-I. A recombinant IAV containing an NS1 protein that cannot interact with TRIM25 (E96A/E97A NS1 mutant) did not have an inhibitory effect on the ubiquitin-dependent signaling activity of RIG-I [68]. The lack of conservation of K172 between human and mouse RIG-I led to additional studies looking at the antagonism of RIG-I by the influenza NS1 protein in cells from different host species. This study showed that while NS1 from human, avian, porcine, and murine IAV strains were all able to interact with human TRIM25, none of the tested NS1 proteins interacted with mouse TRIM25. Instead, NS1 efficiently bound to mouse Riplet; however, the ability of NS1 proteins to interact with Riplet was not limited to the murine ortholog. In fact, NS1 proteins from human IAV strains were also able to interact with human Riplet, blocking both TRIM25 and Riplet in human cells, which led to profound inhibition of RIG-I-mediated antiviral signaling [60]. Mechanistically, there is evidence that Riplet induces K63-linked ubiquitination of the CTD of RIG-I first, which likely stabilizes an open conformation of the RIG-I molecule, in which the CARDs are now accessible for TRIM25 binding and TRIM25-mediated ubiquitination, ultimately promoting RIG-I-MAVS interaction [67, 69]. Further studies are needed to fully understand the species-specific roles of TRIM25 and Riplet in RIG-I activation, and how these two E3 ligases act in concert to stimulate RIG-I downstream signaling for a rapid and effective antiviral response.
Recently, there have been reports suggesting a role for unanchored K63-linked polyubiquitin chains in the signal activation of RIG-I. Using a cell free system, Zeng et al. showed that in vitro-generated K63-, but not K48- or linear, polyubiquitin chains were able to bind to RIG-I and facilitate its ability to activate IRF3 [70]. Subsequent studies showed that the MDA5 CARDs are also able to interact with unanchored K63-linked ubiquitin chains in vitro [71], enabling MDA5 to activate IRF3. However, these studies on MDA5 K63-ubiquitin binding were challenged by Wu et al. who did not observe MDA5 activation by K63-polyubiquitin, leaving the role of K63-polyubiquitin in MDA5 activation ambiguous [72].
In regards to RIG-I, the discrepancy between its covalent K63-linked ubiquitination and ability to bind unanchored K63-ubiquitin resulted in the important question of which type of K63-linked polyubiquitin – covalent or non-covalent – is important for RIG-I activation. The recently solved crystal structure of the RIG-I CARDs demonstrated that three K63-ubiquitin chains are bound along the outer rim of the RIG-I 2CARD tetramer, stabilizing the CARDs in a ‘lock-washer’ conformation [73]. Importantly, this study provided several lines of evidence that covalent K63-ubiquitination is important for RIG-I CARD-mediated signaling. First, they showed that residue K172 in RIG-I, previously identified to be covalently attached to K63-ubiquitin [54], is not involved in an interaction with unanchored K63-ubiquitin chains. In fact, the structural analysis showed that K172 is within the covalent linkage distance (< 20 Å) from the C-terminus of ubiquitin, strongly indicating that this residue is covalently modified. Furthermore, this study showed that covalently-attached K63-ubiquitin chains stabilized the signaling-active RIG-I tetramer more efficiently than non-covalent K63-diubiquitin [73].
A recent study has implicated a third ubiquitin E3 ligase, TRIM4, in regulating RIG-I signal transduction. Overexpression of TRIM4 led to increased IFN induction following infection with Sendai virus (SeV), a paramyxovirus known to be detected by RIG-I [74]. More detailed analysis indicated that TRIM4 interacted with RIG-I and led to the K63-linked ubiquitination of K154, K164, and K172 in the CARDs. However, the physiological role and contribution of K63-linked ubiquitination of RIG-I by TRIM4 to innate antiviral immunity has yet to be determined. Another regulatory mechanism of RIG-I activity through K63-linked ubiquitination was recently discovered involving antiviral stress granules (avSG) [75]. The E3 ligase MEX3C was shown to bind to viral RNA, resulting in its association with RIG-I inside avSGs. This study indicated that K48, K99 and K169 of RIG-I were ubiquitinated by MEX3C, and that this ubiquitination increased type-I IFN induction. While this study strengthened the hypothesis that RIG-I's subcellular localization may be important for viral RNA detection, the exact role of avSGs in innate immune signaling remains unclear.
As K63-linked ubiquitination of RIG-I is crucial for its signaling activity in response to virus infection, it is not surprising that several DUBs have been identified that counteract this modification. At least three different DUBs have been implicated in the inhibition of RIG-I signaling through the removal of covalent K63-linked ubiquitin chains (Figure 2). Cylindromatosis (CYLD) was the first DUB identified that led to RIG-I deubiquitination. In uninfected cells, CYLD was shown to keep RIG-I deubiquitinated, preventing any basal activation levels [76]. This study further showed that, upon viral infection, CYLD protein abundance was downregulated, presumably allowing the full ubiquitination and activation of RIG-I. Notably, CYLD's activity was not specific for RIG-I as TBK1 and IKKε were also targets of CYLD-mediated deubiquitination. Similarly, USP21 has been reported to negatively regulate RIG-I by removing K63-linked ubiquitination [77]. USP21 was shown to interact with RIG-I both in uninfected cells and during VSV and SeV infection. This interaction led to RIG-I deubiquitination and a decrease in type-I IFN induction. More recently, USP3 has been shown to deubiquitinate RIG-I, leading to a decrease in IFN-β induction [78]. Upon virus infection, USP3 interacted with RIG-I, likely acting as a negative feedback regulator. Interestingly, this study also showed a negative regulatory effect of USP3 on MDA5 activity; however, the precise mechanism by which USP3 affects MDA5's signaling activity remains unclear, given the elusive role of K63-polyubiquitin in MDA5 activation. Additionally, several studies have shown that viruses encode DUBs to target RIG-I and to evade detection by the innate immune system (reviewed in [79]). This large number of cellular and viral DUBs specifically targeting the K63-linked ubiquitination of RIG-I further confirms the importance of this type of ubiquitination in RIG-I activation.
RIG-I's immediate downstream molecule, the adaptor protein MAVS, has also been shown to be regulated by K63-linked ubiquitination [80]. K63-linked ubiquitination of K500 in MAVS was induced following SeV infection and led to the enhanced recruitment of IKKε, ultimately promoting IRF3 activation and type-I IFN gene expression. The ligase(s) responsible for the K63-linked ubiquitination of MAVS at K500, however, have not yet been identified.
Negative regulation of RLR signaling by K48-linked ubiquitination
In addition to positive regulation through K63-linked ubiquitination, RIG-I has been shown to be degraded in a proteasome-dependent manner based on conjugation of K48-linked polyubiquitin. It was first reported that the RING E3 ligase RNF125 binds to and ubiquitinates RIG-I with K48-linked polyubiquitin chains (Figure 2). This ubiquitin mark led to RIG-I degradation and decreased SeV-induced IFN induction, indicating a negative-feedback loop by regulating RIG-I protein levels [81]. The same study showed that RNF125 does not act specifically on RIG-I, but also interacted with and ubiquitinated MDA5 and MAVS as well - although to a lesser extent [81]. This identified RNF125 as a relatively non-specific negative regulator of the RLR pathway by targeting both RIG-I and MDA5 as well as MAVS for degradation. The K48-linked ubiquitination of RIG-I is counteracted by USP4, stabilizing the protein levels of RIG-I and prolonging IFN induction [82]. Interestingly, USP4 protein levels decrease following virus infection, indicating that it is involved in steady-state regulation of RIG-I.
MDA5 was reported to be negatively regulated by another E3 ubiquitin ligase, TRIM13 [83]. This study showed that overexpression of TRIM13 inhibited MDA5-mediated signaling. Furthermore, infection with encephalomyocarditis virus (EMCV) induced significantly higher type-I IFN levels in trim13 −/− mice than in WT mice [83]. In support of this, trim13 −/− mice had an increased resistance to EMCV infection than their WT littermates. Of note, this study did not look specifically at MDA5 ubiquitination by TRIM13; however, as TRIM13 has a functional RING E3 ligase domain, it is likely that the mechanism involves degradative K48-linked ubiquitination. More detailed studies are needed to define the mechanistic action of TRIM13 in MDA5 signal transduction.
Both HECT and RING E3 ligases have been shown to function as negative regulators of MAVS signaling by inducing its K48-linked ubiquitination. Besides ubiquitination by RNF125, MAVS undergoes K48-linked ubiquitination at residues K371 and K420 by the HECT E3 ligase AIP4 (Atrophin 1 Interacting Protein 4; also called ITCH) [84]. Notably, AIP4/ITCH is not present on the mitochondria during steady state, but is specifically recruited there upon viral infection by the RNA-binding protein PCBP2 (poly(rC) binding protein 2), ultimately triggering the proteasomal degradation of MAVS. Experiments in itch−/− MEFs showed that the absence of AIP4 led to sustained production of several cytokines (e.g. type-I IFNs, TNF, IL6) in response to poly(I:C) transfection or SeV infection [84]. Furthermore, the SMAD ubiquitin regulatory factors (Smurf) 1 and 2 have been shown to induce the K48-linked ubiquitination and degradation of MAVS [85, 86]. In the case of Smurf1, Ndfip1, known for its role in the activation of the Nedd4 family of HECT E3 ligases, was critical for MAVS binding and degradation. Finally, RNF5 has been reported to interact with MAVS upon viral infection, regulating MAVS stability through targeting K362 and K461 in MAVS for K48-linked ubiquitination [87]. Interestingly, TRIM44 has been shown to counteract the K48-polyubiquitin-induced degradation of MAVS [88]. TRIM44 overexpression led to increased MAVS stability and enhanced IFN induction by suppressing the PCBP2/AIP4-induced ubiquitination of MAVS. Of note, TRIM44 is an atypical TRIM protein because it lacks the RING finger domain; instead TRIM44 possesses a ZF-UBP domain, which is typically found in members of the USP family. Further studies will be needed to define the precise mechanism by which TRIM44 stabilizes MAVS, and specifically whether the putative DUB activity of TRIM44 plays a role in MAVS regulation.
Recent studies have identified LUBAC as a negative feedback regulator of the TRIM25-RIG-I signaling complex. LUBAC, composed of the E3 ligases HOIL-1L and HOIP, has been shown to negatively regulate RIG-I signaling utilizing two distinct mechanisms [89]. First, LUBAC induces ubiquitination of the C-terminal SPRY domain of TRIM25, leading to TRIM25 proteasomal degradation. While LUBAC was able to catalyze both linear ubiquitin chains and K48-linked ubiquitin chains on TRIM25 in vitro, cell culture studies indicated that LUBAC regulates TRIM25 stability primarily through classical K48-linked ubiquitination. TRIM25 ubiquitination was depended on the RBR domains of both HOIL-1L and HOIP. Additionally, a second mechanism of how LUBAC inhibits TRIM25 and RIG-I was suggested, which was depended on the NZF (Npl4 zinc finger) domain of HOIL-1L. Specifically, the NZF of HOIL-1L competes with TRIM25 for RIG-I binding, ultimately preventing TRIM25-mediated ubiquitination and activation of RIG-I [89]. USP15, identified as an interaction partner of TRIM25 by mass spectrometry, was recently shown to counteract the inhibitory effect of LUBAC [90]. Mechanistically, USP15 was found to bind to TRIM25 specifically during the later stages of viral infection, removing the LUBAC-induced K48-linked polyubiquitination of TRIM25 at its SPRY domain. This study indicated that USP15 specifically stabilizes the TRIM25 protein levels at later time points during infection, which led to sustained type-I IFN gene expression, facilitating virus clearance.
Regulation of TLR signaling through polyubiquitination
The TLR family consists of multiple members of membrane-bound receptors (TLR 1-10 in humans) that have evolved to recognize a wide array of PAMPs from viruses, bacteria, parasites and fungi. The TLRs responsible for sensing viral infections include TLR2 and TLR4 which sense viral proteins, as well as TLR3, TLR7/8, and TLR9 which recognize virus-derived dsRNA, ssRNA, and unmethylated CpG DNA, respectively [91, 92]. TLR proteins share common structural components including an ectodomain comprising leucine-rich repeats that are responsible for surveillance of ligands, a transmembrane domain, and a cytoplasmic Toll-interleukin-1 receptor (IL-1R) homology (TIR) domain which coordinates downstream signaling. Upon binding to their ligands, the TLRs initiate signaling cascades by binding to one of two key adaptor proteins: MyD88 or TRIF [93, 94]. Classically, MyD88 recruits the IL-1R-associated serine/threonine kinases (IRAKs) 1, 2, and 4. MyD88 and IRAKs then signal to TRAF6 (tumor necrosis factor receptor-associated factor 6), inducing the recruitment and activation of IKKα/β/γ and TAK1 (TGFβ-activated kinase 1), which promote NF-κB activation and proinflammatory cytokine production. On the other hand, TRIF signals downstream through TRAF3, inducing IRF3/7-mediated IFN-α/β induction via TBK1/IKKε. All TLRs, aside from TLR3, activate the MyD88-dependent pathway, while TLR3 signals through TRIF. Furthermore, TLR4 can activate both MyD88- and TRIF-dependent signaling pathways. Over the past few years, ubiquitination has been shown to play an important role in modulating the activities of key molecules in the TLR pathway, both receptor-proximal molecules as well as downstream signaling proteins.
K48-linked ubiquitination of TLRs and their essential adaptor proteins
Multiple E3 ligases have been reported to regulate TLR signaling through K48-linked ubiquitination of the TLRs themselves or their adaptors MyD88 and TRIF (Figure 3). This ubiquitin mark delicately regulates the protein abundance of these signaling proteins, representing a negative feedback loop to prevent extended activation of the innate immune system. Triad3A has been identified as an E3 ligase that binds to and ubiquitinates several members of the TLR family [95]. Triad3A was originally identified as an interactor of TLR9 through yeast two-hybrid screening. More detailed studies showed that Triad3A interacts not only with TLR9 but also with TLR 3, 4, and 5, but not TLR2. Overexpression of Triad3A led to K48-linked polyubiquitination and degradation of TLR 4 and 9, and to a lesser extent TLR 3 and 5. In line with this, overexpression of Triad3A inhibited the signaling abilities of these TLRs. Subsequent studies indicated that Triad3A may also play a role downstream of TLRs (and also RLRs) by targeting two key signaling molecules for degradation: RIP1 (receptor-interacting protein 1) and TRAF3 (TNF receptor-associated factor 3) [96, 97].
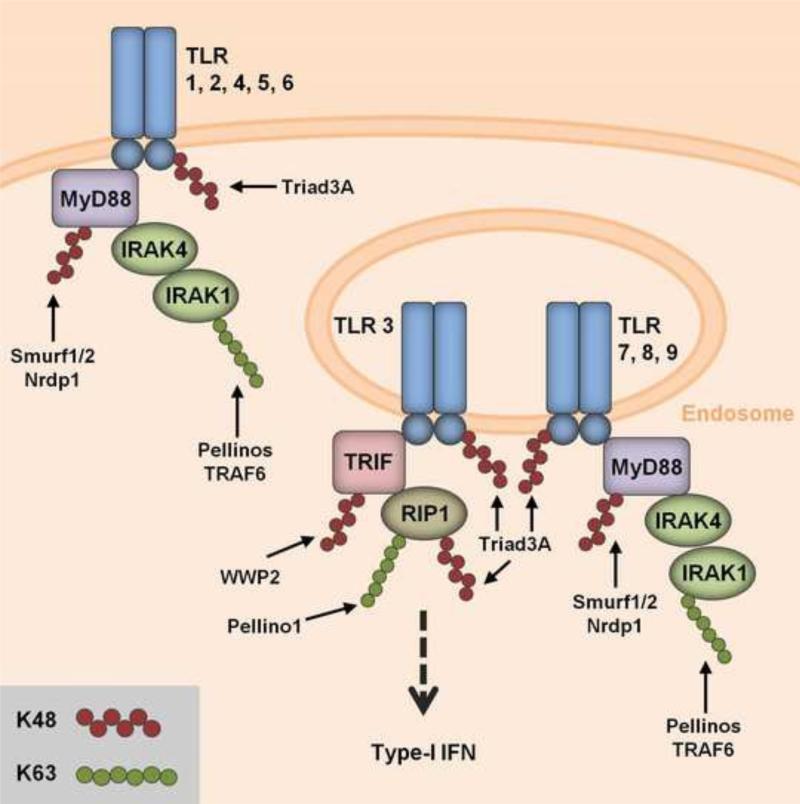
Figure 3. Regulation of TLRs by ubiquitination.
Toll-like receptors (TLRs), found on the cell surface or on endosomal membranes, survey the extracellular milieu for viral nucleic acid or proteins. After binding to their respective viral ligands, TLRs signal through one of two critical adaptor proteins, MyD88/IRAK or TRIF. Signaling by IRAK1 is perpetuated through modification with K63-linked ubiquitination by Pellinos or TRAF6. The activation of TLRs and their adaptor proteins is regulated by degradative K48-linked ubiquitination mediated by the E3 ligases indicated. The specific details of how K48-polyubiquitin regulates TLR signaling are described in the text.
The protein levels of MyD88 are also tightly regulated via K48-linked ubiquitination, which was first discovered when cells were stimulated with the anti-inflammatory cytokine transforming growth factor-β (TGF-β) [98]. Subsequent studies showed that the E3 ubiquitin ligases Smurf1 and 2 bound to MyD88, an interaction which was dependent on SMAD6 [99]. The interaction of MyD88 with Smurf1/2 led to the ubiquitination and degradation of MyD88, limiting the inflammatory response induced by TLR signaling. Nrdp1 (neuregulin receptor degradation protein 1) is another E3 ligase which leads to MyD88 degradation, but it has a more complex role in TLR signaling. Nrdp1 acts at two steps of the TLR signaling pathway to shift the response from NF-κB- to IRF3-driven gene expression [100]. Mechanistically, Nrdp1 attaches K48-linked polyubiquitin to MyD88, leading to its degradation, and on the other hand, conjugates K63-linked polyubiquitin to TBK1, activating the IRF3-dependent response. Thus, the dual activity of Nrdp1 coordinates distinct signaling of one arm of the TLR response, while dampening the other arm of this pathway.
The adaptor protein TRIF, which specifically mediates signaling by TLR3, has also been shown to be a target of K48-linked ubiquitination. TLR3 senses viral dsRNA and subsequently associates through its TIR domain with TRIF, triggering the activation of TRAF3 and TBK1, ultimately leading to type-I IFN induction. To keep the TLR3-TRIF mediated signal transduction pathway in check, TRIF is targeted for K48-linked ubiquitination and subsequent degradation by the HECT E3 ligase WWP2 (WW domain-containing protein 2) [101]. Wwp2-deficient bone marrow-derived macrophages exhibited increased levels of IFN-β, TNFα, and IL-6 in response to the TLR3 agonist poly(I:C). Consistent with this, Wwp2-knockout mice showed an enhanced susceptibility to poly(I:C)-induced death than WT animals.
Regulation of TLR-proximal signaling molecules by K63-linked ubiquitination
The activities of many signaling molecules downstream of TLRs (and also other PRRs), such as TRAFs, NEMO, and TBK1, are regulated through K63-linked ubiquitination (as discussed in detail below). In addition, several receptor-proximal signaling molecules have been shown to be modified with K63-ubiquitin polymers, among them the well-studied kinase IRAK1 (Figure 3). IRAK1 and also IRAK4 are recruited to MyD88 upon activation of various TLRs, propagating further signal transduction that leads to NF-κB and MAPK activation. Interestingly, several groups have shown that K63-linked ubiquitination of IRAK1 is required for downstream signaling by recruiting TAK1 and NEMO to the TRAF6 complex [102-105]. There has been some discrepancy as to which E3 ubiquitin ligase is responsible for IRAK1 ubiquitination. While several studies indicated that members of the Pellino E3 ligase family induce K63-linked ubiquitination of IRAK1 [102, 106], others have suggested that TRAF6 is involved in IRAK1 ubiquitination [104]. Moreover, Pellino 1 has been shown to mediate the K63-linked ubiquitination of RIP1 upon its recruitment to the TLR3-TRIF complex [107]. Ubiquitinated RIP1 then recruits NEMO and the TAK1 complex, inducing NF-κB activation.
Ubiquitin-mediated regulation of the cGAS-STING pathway
In contrast to the well-characterized cytosolic RNA sensing pathways, the mechanisms of how the cell senses virus-derived DNA, or host DNA from damaged cells, has just begun to be elucidated. A key component of the intracellular DNA sensing pathway is the adaptor protein STING (also called MITA, ERIS, or MPYS) [108-110]. STING is a membrane-resident protein found on the ER or mitochondrion. STING was shown to be activated by viral or immunostimulatory DNA, facilitating its binding to TBK1 and subsequent IRF3-mediated expression of type-I IFNs and other cytokines. Functional studies in sting-deficient cells demonstrated that STING plays a crucial role in dsDNA sensing and antiviral innate immune responses to HSV-1 and Listeria monocytogenes [108]. Infection studies in sting-knockout mice demonstrated that STING is essential for innate immune signaling upon recognition of foreign intracellular dsDNA [110]. Moreover, sting knockout or knockdown also abrogated the IFN-mediated immune response to certain RNA viruses that are sensed by RIG-I.
Because STING does not directly sense DNA, it had been proposed that one or multiple hitherto-unknown DNA sensors mediate STING activation. In recent years, a plethora of putative cytosolic DNA sensors have been identified; however, some of these sensors had great cell-type specificity or little in vivo (in mouse) significance, leaving it somewhat unclear whether some of these molecules are indeed bona fide DNA sensors [111-114]. More recently, a new cytosolic DNA sensor, cGAS, was identified, along with the detailed mechanism of STING activation upon DNA stimulation [115-119]. These studies showed that after cGAS binds to foreign DNA in the cytoplasm, it synthesizes cyclic GMP-AMP (cGAMP) [116, 120, 121], which is then detected by STING. These studies confirmed previous findings, which had shown that STING senses cyclic nucleotides produced during bacterial infection [122]. Furthermore, functional studies in cGas knockout mice as well as structural analyses strengthened that cGAS-STING is a critical sensing pathway of cytoplasmic DNA [120, 123].
STING activation through K63-linked ubiquitination
The signaling activity of STING is tightly controlled by K63-linked ubiquitination mediated by two TRIM family members, TRIM56 and TRIM32 (Figure 4) [124, 125]. A cDNA screen analyzing IFN-β promoter activation after treatment with various stimuli identified TRIM56 as regulatory molecule in the IFN induction pathway [124]. TRIM56 promoted type-I IFN induction in response to poly(dA:dT) and also poly(I:C), consistent with STING's role in both cytoplasmic DNA and RNA detection. However, TRIM56 did not directly bind to poly(dA:dT), ruling out that TRIM56 acts as a dsDNA sensor, thereby inducing STING-dependent signaling. Biochemical analysis showed that TRIM56 binds to the C-terminal domain of STING, and that it induced K63-linked polyubiquitination of STING. Residue K150 in STING was shown to be critical for K63-linked ubiquitination by TRIM56; a K150R mutant of STING was no longer ubiquitinated and was unable to induce IFN-β. Mechanistically, the K63-linked ubiquitination facilitated dimerization of STING and its interaction with TBK1, two critical steps in STING-mediated signaling.
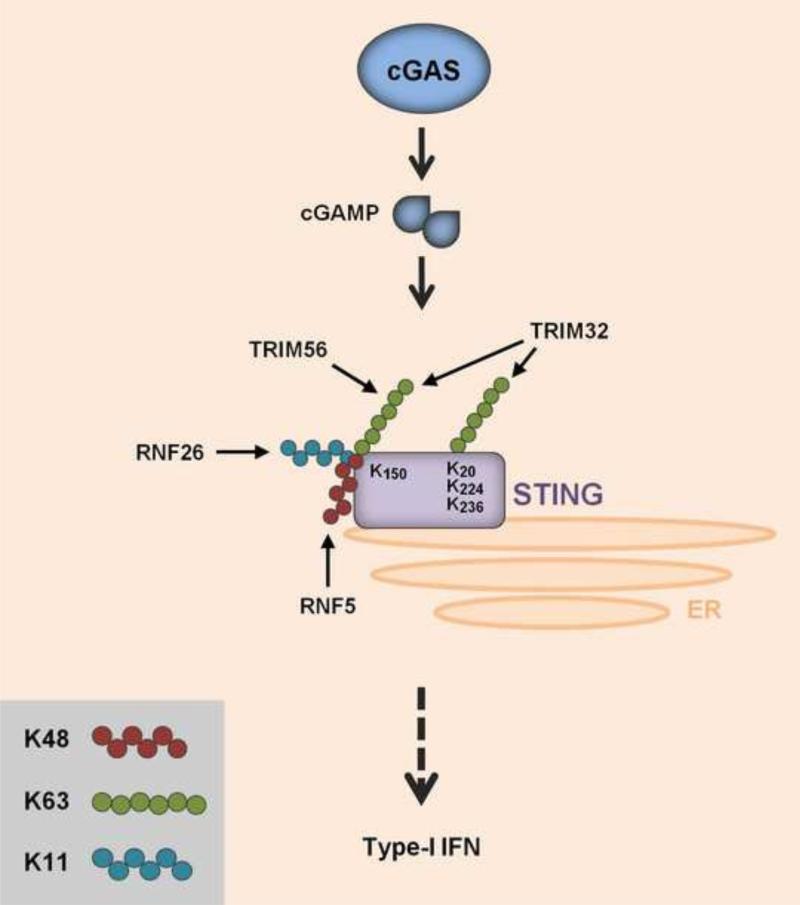
Figure 4. Regulation of the cGAS-STING pathway by ubiquitination.
cGAS recognizes viral DNA in the cytoplasm and synthesizes cyclic GMP-AMP (cGAMP). cGAMP then activates STING on the ER, inducing downstream signaling for type-I IFN induction. STING is regulated by three types of polyubiquitination: K11-linked and K63-linked ubiquitination of K150 by RNF26 and TRIM56 or TRIM32, respectively, facilitating STING activation and type-I IFN gene expression. Besides K500, TRIM32 ubiquitinates three other residues in STING (K20, K224 and K236). Furthermore, K500 in STING is covalently modified by K48-linked ubiquitination mediated by RNF5. RNF5-induced STING ubiquitination leads to STING degradation.
A second E3 ligase mediating STING ubiquitination, TRIM32, was identified through a cDNA screen testing the effects of 352 ubiquitin-related enzymes on STING ubiquitination [125]. TRIM32 induced robust K63-linked ubiquitination of STING, but not of RIG-I, MDA5, and MAVS. Functional studies revealed that TRIM32 is involved both in cytoplasmic poly(I:C)-and poly(dA:dT)-induced IFN responses, but not in TLR3 signal transduction. Biochemical studies showed that the C-terminal NHL (named for NCL-1, HT2A and Lin-41 repeat) domain of TRIM32 interacted with the transmembrane domain of STING, leading to K63-linked ubiquitination of STING. This study identified four lysine residues implicated in TRIM32-mediated STING ubiquitination. Whereas individual mutation of K20R, K150R, K224R, or K236R only partially reduced STING ubiquitination by TRIM32, a mutant of STING in which all four residues were mutated had a total loss of ubiquitination and signaling activity. TRIM32-induced ubiquitination of STING seemed to aid STING binding to TBK1, as depletion of TRIM32 led to a decrease of endogenous TBK1-STING interaction upon infection with SeV or HSV-1 [125].
Regulation of STING stability through K11- and K48-linked ubiquitination
In addition to its positive regulation by TRIM56 and TRIM32, STING undergoes K48-linked ubiquitination, leading to its degradation through the proteasome pathway (Figure 4). Zhong et al. conducted a yeast two-hybrid screen using full-length STING as bait, resulting in the identification of the E3 ubiquitin ligase RNF5 as STING interacting molecule [126]. This interaction was mediated by the respective transmembrane domains of STING and RNF5. Overexpression of WT RNF5, but not its catalytically-inactive RING mutant, promoted K48-linked ubiquitination and degradation of STING. Furthermore, the authors identified K150 in STING as the key residue for K48-linked ubiquitination, and suggested that STING ubiquitination and degradation by RNF5 resulted in a negative-regulatory feedback loop to avoid excessive immune signaling in response to viral infection [126]. Cell fractionation and confocal microscopy experiments showed that RNF5, similar to STING, is localized both at the ER and mitochondria; however, RNF5 targeted specifically mitochondrion-localized STING for ubiquitination and subsequent degradation upon viral infection.
Recently, it has been shown that STING also serves as substrate for K11-linked polyubiquitination catalyzed by RNF26 [127]. Interestingly, this modification was shown to be conjugated to K150 in STING also. To date, the precise fate of K11-linked ubiquitinated proteins has not been fully elucidated; however, RNF26-induced ubiquitination prevented STING degradation by displacing RNF5-mediated K48-linked ubiquitination at K150. In contrast, RNF26 did not affect the K63-linked polyubiquitination of STING at K150. Of note, the authors also observed a negative regulatory role of RNF26 on innate immune signaling specifically at later time points during infection, which was due to autophagic degradation of IRF3 by RNF26. This suggests a model in which RNF26 has a dual role in regulating innate immune signaling. Early during infection RNF26 promotes antiviral signaling by protecting STING from degradation, while at the late phase of viral infection RNF26 may act as a negative feedback regulator by inducing IRF3 degradation [127].
Ubiquitin-dependent regulation of common downstream signaling molecules of PRRs
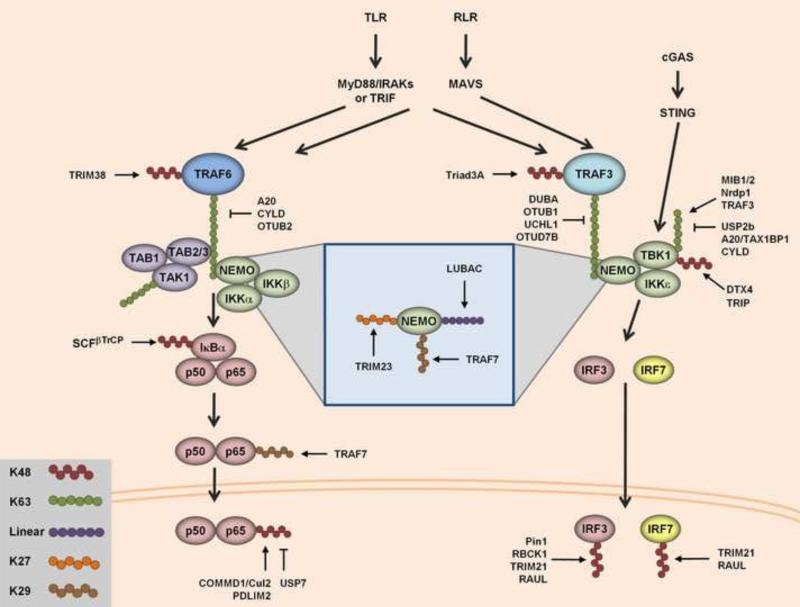
RLRs, TLRs and cGAS signal through distinct adaptor proteins, namely MAVS, MyD88/TRIF and STING, respectively. However, downstream of these adaptors, the PRR signaling pathways converge on common signaling molecules, leading to the activation of the aforementioned transcription factors, NF-κB, IRF3/7 and AP-1 (Figure 5). Among these common downstream signaling molecules are members of the TRAF protein family of ubiquitin E3 ligases – especially TRAF3 and 6 – as well as the kinases TBK1 (or IKKε) and IKKα/β/γ which trigger the activation of the IRF3/7- and NF-κB-induced signaling arm, respectively [53].
Figure 5. Ubiquitin-dependent regulation of common downstream molecules of PRRs.
After recognition of viral nucleic acids, RLRs and TLRs signal through their downstream adaptors MAVS and MyD88/TRIF, respectively. These adaptors then propagate this signal to TRAF6 for NF-κB activation and TRAF3 for IRF3/7 activation. TRAF6 and TRAF3 both induce autoubiquitination, creating a scaffold for downstream signaling partners to interact. K63-linked polyubiquitin on TRAF6 leads to the recruitment of the TAK1/TAB2/3 complex, which in turn recruits NEMO and the IKK complex to phosphorylate IκBα. Phosphorylation of IκBα then leads to its K48-polyubiquitin-dependent degradation. Degradation of IκBα releases the NF-κB subunits p50 and p65, allowing for their translocation into the nucleus to activate transcription of target genes. On the other hand, K63-linked ubiquitination of TRAF3 recruits NEMO specifically complexed TBK1/IKKε. sTING also directly binds to and activates TBK1. TBK1/IKKε then phosphorylate IRF3 and IRF7, leading to their dimerization and translocation to the nucleus to induce transcription of type-I IFN and antiviral genes. Many proteins in these signaling cascades are targets for degradative K48-linked ubiquitination and non-degradative types of polyubiquitination. The E3 ligases involved in these ubiquitination events, as well as the DUBs responsible for removal of polyubiquitin, are indicated. The details of how specific ubiquitin marks regulate the activities of the illustrated signaling molecules are described in the text.
For PRR-mediated NF-κB activation, TRAF6 first recruits the TAK1/TAB1/2 complex. This complex of kinases then phosphorylates NEMO/IKKγ, activating it to serve as a scaffold for the recruitment of IKKα and IKKβ. The IKKα/β/γ complex then recruits a signaling complex consisting of the inhibitor of NF-kappaB-α (IκBα) as well as distinct NF-κB subunits, such as p65 and p50. IKKβ is then able to induce K48-linked ubiquitination and degradation of IκBα, which leads to the release of p65/50, allowing them to translocate into the nucleus to promote transcription of NF-κB target genes.
For the activation of IRF3/7, the E3 ligase TRAF3 is recruited either to MAVS or TRIF [128, 129]. TRAF3 also activates NEMO, which in this case forms a distinct signaling complex with TANK and TBK1 or IKKε. STING also interacts with TBK1, activating the IRF3-mediated response. TBK1/IKKε then phosphorylate IRF3/7, leading to their dimerization and translocation to the nucleus to induce IRF target genes.
The role of K63-linked ubiquitination in the activation of IKKs and TBK1
The role of K63-linked ubiquitin polymers in the regulation of PRR-proximal signaling events and in particular NF-κB activation has been extensively characterized (reviewed in detail in [130-132]). One of the first identified functions of K63-linked ubiquitination was described for the signal-transducing activity of TRAF6 [133]. Following its recruitment to MyD88 or MAVS, TRAF6, which belongs to the RING-finger type E3 ligase family, generates K63-linked polyubiquitin chains (Figure 5). These K63-polyubiquitin chains are essential for the recruitment and activation of the TAK1/TAB and IKK kinase complexes [133, 134]; however, the target protein of K63-polyubiquitin had been unknown for quite some time. It has been demonstrated that TRAF6 catalyzes its own ubiquitination, and also produces unanchored K63-linked polyubiquitin [135]. This ubiquitination serves a dual role in innate signaling by stabilizing TRAF6 protein levels and by serving as a scaffold for TAB2 binding, which then recruits and activates TAK1 [134-136]. The K63-polyubiquitin chains catalyzed by TRAF6 are also responsible for recruiting NEMO, which in turn recruits IKKα and IKKβ to the TAK1 complex [137]. NEMO has also been shown to bind to diubiquitin molecules, both K63-linked and mixed linkages [138, 139]. The ability of NEMO to bind to diubiquitin moieties is important for its ability to activate the IKK complex, likely through stabilization of NEMO upon diubiquitin binding. To inhibit TRAF6-NEMO signal-transducing activities, several molecules harboring DUB activity have been identified that target specifically the K63-linked ubiquitination of TRAF6, including A20, CYLD and the OTU deubiquitinase 2 (OTUB2) [140-146].
For IRF3 activation in response to PRR signaling, TRAF3 is recruited to MAVS and TRIF after RLR and TLR activation, respectively [128, 129]. Similar to TRAF6, TRAF3 also modifies itself with K63-linked polyubiquitin chains, a process which is dependent on the E2 enzyme Ubc5 [147, 148]. This K63-linked ubiquitin chain again serves as a scaffold for the recruitment of NEMO. NEMO then binds a complex consisting of TANK and TBK1/IKKε. This interaction activates TBK1/IKKε to phosphorylate IRF3 and IRF7, inducing the transcription of IGSs. The importance of K63-linked ubiquitination for TRAF3-dependent signaling was strengthened by the identification of several DUBs removing the K63-linked ubiquitination from TRAF3: the deubiquitinating enzyme A (DUBA), OTUB1, as well as the ubiquitin carboxyl-terminal hydrolase L1 (UCHL1) that is specifically subverted by high-risk human papillomaviruses to downregulate IRF3 activation and PRR responses [146, 149-151]. Furthermore, OTUD7B (OTU domain-containing protein 7B) was recently shown to interact with TRAF3, removing its K63-linked ubiquitination, which prevented TRAF3 proteolysis and consequently aberrant non-canonical NF-κB activation [152].
TBK1 is another important target of K63-linked ubiquitination, and at least three E3 ubiquitin ligases have been identified for TBK1 ubiquitination. Mind bomb 1 and 2 (MIB 1/2), identified by a global proteomic analysis of a human innate immunity interactome, induced K63-polyubiquitin conjugation on residues K69, K154 and K372 in TBK1. Detailed analysis demonstrated that TBK1 ubiquitination by MIBs was critical for the recruitment of NEMO and the antiviral response triggered by cytosolic viral RNA [153, 154]. Mechanistically, MIB2 was shown to bind to the adaptor MAVS involving a highly conserved DLAIS motif at amino acid positions 438 to 442 of MAVS; this motif was critical for MIB2-mediated TBK1 ubiquitination and subsequent IRF3/7 phosphorylation by TBK1 [155]. Furthermore, TRAF3 and Nrdp1 have also been shown to mediate TBK1 K63-linked ubiquitination [100, 156]. Recently, structural analysis of the near-full-length TBK1 protein showed that TBK1 forms a dimer and that K63-linked ubiquitination (at K30 and K401) of the dimerized TBK1 is required for TBK1 enzymatic activity [157].
Inversely, cleavage of K63-linked polyubiquitin from TBK1 by USP2b or CYLD inhibited the kinase activity of TBK1 [76, 158]. Furthermore, A20 together with the adaptor protein TAX1BP1 have been shown to disrupt the K63-linked polyubiquitination of TBK1 (and also of IKKε), an activity that was not dependent on the DUB activity of A20 but due to the disruption of the interaction of TBK1/IKKε with TRAF3[156].
K48-linked ubiquitination to modulate NF-κB- and IRF-mediated antiviral gene transcription
Degradative K48-linked ubiquitination has been shown to be essential for NF-κB activation (Figure 5). In unstimulated cells, NF-κB subunits, such as the canonical p50 and p65, are kept in an inactive complex by binding to IκBα. After activation by TAK1/IKKs, IKKβ phosphorylates IκBα on S32 and S36, leading to the recruitment of the SCFβTrCP E3 ligase [159-161]. Upon recruitment, SCFβTrCP conjugates K48-linked ubiquitination on IκBα, thereby targeting it for degradation by the proteasome; this releases the NF-κB subunits and allows them to translocate into the nucleus to activate the transcription of target genes.
In addition, many other downstream signaling proteins in the PRR signaling pathways are regulated through K48-linked ubiquitination and degradation; in these cases, degradation of the proteins dampens the NF-κB- and IRF3-mediated immune response. TRAF6 has been shown to be targeted by TRIM38 for ubiquitination to prevent excessive NF-κB activation in macrophages [162]. Triad3A, known for its role in the degradation of TLRs, has been reported to induce K48-linked ubiquitination of specifically TRAF3 [96], indicating that this E3 ubiquitin ligase targets multiple proteins in innate immunity to downregulate IRF3-mediated antiviral gene expression. Another mechanism to avoid excessive NF-κB-mediated gene transcription is the K48-polyubiquitin-dependent degradation of NF-κB itself by COMMD1 together with an ubiquitin ligase complex comprised of Cullin2 (Cul2), Elongins B and C, and SOCS1 (also known as ECSSOCS1). Activation of p65 involves phosphorylation of S468. Interestingly, this phosphorylation mark also allows the recruitment of COMMD1 and Cul2 to chromatin-bound p65, ultimately inducing p65 degradation to terminate NF-κB transactivation [163-166]. A second nuclear E3 ligase,PDLIM2 (also known as Mystique or SLIM), has been shown to target p65. PDLIM2 binds to p65 and induces its specific relocation to PML-containing intranuclear compartments where p65 undergoes K48-linked ubiquitination and degradation by the 26S proteasome [167]. USP7 has been reported to counteract p65 degradation through removal of K48-linked ubiquitin chains [168].
Furthermore, TBK1 undergoes K48-linked polyubiquitination, which is mediated by the E3 ubiquitin ligase DTX4. Specifically, DTX4 is recruited to TBK1 by NLRP4 (NACHT, LRR and PYD domains-containing protein 4), inducing ubiquitination of TBK1 at K670, ultimately leading to TBK1 destabilization [169]. The TRAF-interacting protein (TRIP) has also been shown to negatively regulate the protein stability of TBK1 by binding to and inducing K48-linked polyubiquitination [170].
IRF3 is also targeted for K48-polyubiquitin-dependent degradation [171]. A negative feedback mechanism for IRF3 activation was first shown for the peptidyl-prolyl isomerase Pin1, which specifically binds to the phosphorylated active form of IRF3 [172]. This interaction was observed specifically upon stimulation of cells with dsRNA, indicating that Pin1 acts as a feedback negative-regulatory molecule to dampen the IRF3 response. Further studies have identified that the E3 ligases RBCK1 (RBCC protein interacting with PKC1; better known as HOIL-1) and TRIM21/Ro52 also modify IRF3 with K48-linked ubiquitin chains, triggering IRF3 degradation and cessation of target gene expression [173, 174]. The gene expression of both E3 ligases is induced upon viral infection, suggesting that these two E3 ligases likely also act as negative feedback regulators of IRF3-dependent transcription. In addition, IRF7 is also ubiquitinated and degraded by TRIM21 after its activation [175]. Finally, RAUL, a HECT-domain E3 ubiquitin ligase, was demonstrated to target both IRF3 and IRF7, comprehensively limiting type-I IFN gene expression [176]. Interestingly, KSHV-encoded RTA (replication and transcription activator), which is the master regulator of KSHV lytic replication, was shown to recruit RAUL to IRFs, decreasing antiviral signaling and ultimately allowing for efficient virus replication [176]. Moreover, several herpesviruses actively induce K48-linked ubiquitination and proteolysis of IRF3/7 using their immediate-early protein ICP0, which exhibits E3 ligase activity [177-179].
Regulation of NEMO by K27-linked, K29-linked and linear polyubiquitination
In addition to its K63-polyubiquitin binding properties (as described above), NEMO has been shown to serve as a substrate for K27-linked polyubiquitination [180]. The RING E3 ligase TRIM23 binds to NEMO and conjugates K27-linked ubiquitin chains to multiple lysine residues (K165, K309, K325, K326 and K344) in NEMO (Figure 5, inset). Ubiquitination of NEMO led to an increase in IFN-β induction, indicating that TRIM23 promotes the signaling activity of NEMO. In support of this, a catalytically-inactive RING mutant of TRIM23 had a dominant-negative effect on ISRE-, IFN-β- and NF-κB-dependent gene transcription. Knockdown studies showed that the signal-transducing activity of NEMO was limited in trim23-depleted cells, resulting in an increase in virus growth [180]. The fate of K27-linked ubiquitinated NEMO and other cellular substrates has not been fully characterized, and in fact, another study has shown that K27-linked ubiquitination of different lysine residues in NEMO by a Shigella effector protein possessing E3 ligase activity (IpaH9.8) leads to the proteasomal degradation of NEMO, a strategy utilized by this bacterium to perturb the host inflammatory response [181]. Further studies will be needed to characterize the effects of K27-linked ubiquitin conjugation in general and on NEMO specifically.
NEMO is also modified by K29-linked ubiquitin chains [182]. Using NEMO as bait in a yeast two-hybrid assay, TRAF7 was identified as a NEMO binding partner. Further characterization showed that expression of TRAF7 decreased NF-κB promoter activation following various stimuli. Using ubiquitin mutants in which all except one of the 7 internal lysines are mutated, the authors further showed that TRAF7 catalyzes specifically K29-linked ubiquitin chains to NEMO as well as to the NF-κB subunit p65. This ubiquitin mark led to reduced protein levels of NEMO and p65, which the authors determined to be due to lysosomal degradation [182]. Of note, this finding is in accordance with other reports that have suggested that K29-linked polyubiquitin represents a signal for protein degradation by the lysosome [22].
NEMO has also been shown to be a target of linear ubiquitin chains [183, 184]. Initial reports showed that NEMO is ubiquitinated by LUBAC at the residues K285 and K309. Conjugated linear polyubiquitin led to the stabilization of the TAK1/TAB and IKK complexes due to recruitment of TAB2 using its NZF domains [183]. While linear ubiquitination activates TAK1 and NF-κB activation, it has also been reported to dampen the type-I IFN response mediated by RIG-I and MAVS [185]. NEMO modified with two or more linear ubiquitin moieties, but not unmodified NEMO, interacted with TRAF3, disrupting the MAVS-TRAF3 complex, which is critical for antiviral IFN induction. This study also showed that in cells deficient in SHARPIN, which is critical for LUBAC function, VSV replicated less efficiently due to a prolonged and increased type-I IFN response. In contrast, NF-κB activation was impaired in SHARPIN-knockout cells [185]. Additional studies have demonstrated that NEMO, using its UBAN (ubiquitin binding in ABIN and NEMO) motif, binds to linear ubiquitin chains, which is independent of its direct ubiquitin conjugation. The crystal structure of the UBAN motif of NEMO bound to linear diubiquitin provided detailed evidence for the specificity of linear ubiquitin binding versus interaction with K63- or K48-linked ubiquitin chains. The specific residues which were involved in linear ubiquitin binding were essential for NF-κB activation, indicating that ubiquitin binding also leads to stabilization of NEMO and its activation [184]. Together, these studies indicate that NEMO-dependent signaling is delicately regulated by at least four different linkage types of polyubiquitin. More detailed studies, however, will be required to fully understand the dynamic interplay of these ubiquitin-dependent regulatory mechanisms for modulating NEMO-mediated antiviral and proinflammatory host responses.
Concluding Remarks
Given the pivotal role of ubiquitination in modulating innate sensing pathways, we eagerly await the identification of disease-relevant mutations in the responsible key enzymes of the ubiquitin conjugation system. Furthermore, while much study has been done looking at the linear, K63- and K48- linked ubiquitination of key proteins in PRR signaling cascades, future studies should be focused on dissecting the role of other atypical polyubiquitin chains as well as branched ubiquitin chains in innate immunity.
Moreover, it remains to be elucidated how various ubiquitin modifications work together (or against each other) to dynamically modulate the signal-transducing activity of individual proteins, and the pathways as a whole. It also remains to be seen how different E3 ligases and DUBs, which often differ in their expression patterns, ubiquitin-linkage specificities, and interaction modes, regulate one particular signaling protein (such as STING or NEMO) to induce an effective antiviral response. Detailed insights into the ubiquitin-dependent regulatory networks in PRR-mediated innate immunity will allow us to exploit this knowledge for the development of new clinical therapies, both for infectious diseases as well as disorders caused by a hyperactive inflammatory response.
Highlights.
Different linkage types of polyubiquitination regulate PRR signal transduction.
K63-linked ubiquitination plays an important role in promoting RIG-I signaling.
K48- and K63-linked ubiquitination fine-tunes TLR-dependent signal transduction.
Cytosolic DNA sensing is regulated through ubiquitination of STING.
Acknowledgements
We apologize to all colleagues whose relevant work could not be cited due to space constraints. Research on antiviral innate immunity in the Gack laboratory is supported by U.S. National Institutes of Health grants (AI087846, AI097699 and AI104415), The Alexander and Margaret Stewart Trust Foundation, and a John and Virginia Kaneb Fellowship.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Ivashkiv LB, Donlin LT. Regulation of type I interferon responses. Nat Rev Immunol. 2014;14(1):36–49. doi: 10.1038/nri3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liu SY, Sanchez DJ, Cheng G. New developments in the induction and antiviral effectors of type I interferon. Curr Opin Immunol. 2011;23(1):57–64. doi: 10.1016/j.coi.2010.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sadler AJ, Williams BR. Interferon-inducible antiviral effectors. Nat Rev Immunol. 2008;8(7):559–68. doi: 10.1038/nri2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sen GC, Sarkar SN. The interferon-stimulated genes: targets of direct signaling by interferons, double-stranded RNA, and viruses. Curr Top Microbiol Immunol. 2007;316:23350. doi: 10.1007/978-3-540-71329-6_12. [DOI] [PubMed] [Google Scholar]
- 5.Creagh EON. LA TLRs, NLRs and RLRs: A trinity of pathogen sensors that co-operate in innate immunity. Trends Immunol. 2006;27:352–357. doi: 10.1016/j.it.2006.06.003. [DOI] [PubMed] [Google Scholar]
- 6.Takeuchi O, Akira S. Pattern Recognition Receptors and Inflammation. Cell. 2010;140(6):805–820. doi: 10.1016/j.cell.2010.01.022. [DOI] [PubMed] [Google Scholar]
- 7.Loo Y-M, Gale M. Immune signaling by RIG-I-like receptors. Immunity. 2011;34(5):680692. doi: 10.1016/j.immuni.2011.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goubau D, Deddouche S, Reis e Sousa C. Cytosolic sensing of viruses. Immunity. 2013;38(5):855–69. doi: 10.1016/j.immuni.2013.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kato H, Fujita T. Autoimmunity caused by constitutive activation of cytoplasmic viral RNA sensors. Cytokine Growth Factor Rev. 2014;(0) doi: 10.1016/j.cytogfr.2014.08.003. [DOI] [PubMed] [Google Scholar]
- 10.Smith S, Jefferies C. Role of DNA/RNA sensors and contribution to autoimmunity. Cytokine Growth Factor Rev. 2014;(0) doi: 10.1016/j.cytogfr.2014.07.019. [DOI] [PubMed] [Google Scholar]
- 11.Pickart CM, Eddins MJ. Ubiquitin: structures, functions, mechanisms. Biochim Biophys Acta. 2004;1695(1-3):55–72. doi: 10.1016/j.bbamcr.2004.09.019. [DOI] [PubMed] [Google Scholar]
- 12.Chernorudskiy AL, Gainullin MR. Ubiquitin System: Direct Effects Join the Signaling. 2013;6:pe22–pe22. doi: 10.1126/scisignal.2004251. [DOI] [PubMed] [Google Scholar]
- 13.Berndsen CE, Wolberger C. New insights into ubiquitin E3 ligase mechanism. Nat Struct Mol Biol. 2014;21(4):301–7. doi: 10.1038/nsmb.2780. [DOI] [PubMed] [Google Scholar]
- 14.Mattiroli F, Sixma TK. Lysine-targeting specificity in ubiquitin and ubiquitin-like modification pathways. Nat Struct Mol Biol. 2014;21(4):308–16. doi: 10.1038/nsmb.2792. [DOI] [PubMed] [Google Scholar]
- 15.Nagy V, Dikic I. Ubiquitin ligase complexes: from substrate selectivity to conjugational specificity. Biol Chem. 2010;391(2-3):163–9. doi: 10.1515/bc.2010.021. [DOI] [PubMed] [Google Scholar]
- 16.Koegl M, et al. A Novel Ubiquitination Factor, E4, Is Involved in Multiubiquitin Chain Assembly. Cell. 1999;96(5):635–644. doi: 10.1016/s0092-8674(00)80574-7. [DOI] [PubMed] [Google Scholar]
- 17.Spratt DE, Walden H, Shaw GS. RBR E3 ubiquitin ligases: new structures, new insights, new questions. Biochem J. 2014;458(3):421–37. doi: 10.1042/BJ20140006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wenzel DM, et al. UBCH7 reactivity profile reveals parkin and HHARI to be RING/HECThybrids. Nature. 2011;474(7349):105–8. doi: 10.1038/nature09966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reyes-Turcu FE, Ventii KH, Wilkinson KD. Regulation and Cellular Roles of Ubiquitin-Specific Deubiquitinating Enzymes. Annual Review of Biochemistry. 2009;78(1):363–397. doi: 10.1146/annurev.biochem.78.082307.091526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McDowell GS, Philpott A. Non-canonical ubiquitylation: mechanisms and consequences. Int J Biochem Cell Biol. 2013;45(8):1833–42. doi: 10.1016/j.biocel.2013.05.026. [DOI] [PubMed] [Google Scholar]
- 21.Ikeda F, Crosetto N, Dikic I. What determines the specificity and outcomes of ubiquitin signaling? Cell. 2010;143(5):677–81. doi: 10.1016/j.cell.2010.10.026. [DOI] [PubMed] [Google Scholar]
- 22.Kulathu Y, Komander D. Atypical ubiquitylation - the unexplored world of polyubiquitin beyond Lys48 and Lys63 linkages. Nat Rev Mol Cell Biol. 2012;13(8):508–23. doi: 10.1038/nrm3394. [DOI] [PubMed] [Google Scholar]
- 23.Walczak H, Iwai K, Dikic I. Generation and physiological roles of linear ubiquitin chains. BMC Biol. 2012;10:23. doi: 10.1186/1741-7007-10-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kirisako T, et al. A ubiquitin ligase complex assembles linear polyubiquitin chains. Embo J. 2006;25(20):4877–87. doi: 10.1038/sj.emboj.7601360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tokunaga F, et al. SHARPIN is a component of the NF-kappaB-activating linear ubiquitin chain assembly complex. Nature. 2011;471(7340):633–6. doi: 10.1038/nature09815. [DOI] [PubMed] [Google Scholar]
- 26.Gerlach B, et al. Linear ubiquitination prevents inflammation and regulates immune signalling. Nature. 2011;471(7340):591–6. doi: 10.1038/nature09816. [DOI] [PubMed] [Google Scholar]
- 27.Ikeda F, et al. SHARPIN forms a linear ubiquitin ligase complex regulating NF-kappaB activity and apoptosis. Nature. 2011;471(7340):637–41. doi: 10.1038/nature09814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wagner SA, et al. A proteome-wide, quantitative survey of in vivo ubiquitylation sites reveals widespread regulatory roles. Mol Cell Proteomics. 2011;10(10):M111 013284. doi: 10.1074/mcp.M111.013284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim W, et al. Systematic and quantitative assessment of the ubiquitin-modified proteome. Mol Cell. 2011;44(2):325–40. doi: 10.1016/j.molcel.2011.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kravtsova-Ivantsiv Y, Ciechanover A. Non-canonical ubiquitin-based signals for proteasomal degradation. J Cell Sci. 2012;125(Pt 3):539–48. doi: 10.1242/jcs.093567. [DOI] [PubMed] [Google Scholar]
- 31.Matsumoto ML, et al. Kll-linked polyubiquitination in cell cycle control revealed by a Kll linkage-specific antibody. Mol Cell. 2010;39(3):477–84. doi: 10.1016/j.molcel.2010.07.001. [DOI] [PubMed] [Google Scholar]
- 32.Dynek JN, et al. c-IAPl and UbcH5 promote Kll-linked polyubiquitination of RIPl in TNF signalling. 2010;29:4198–4209. doi: 10.1038/emboj.2010.300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Goto E, et al. Contribution of Lysine ll-linked Ubiquitination to MIR2-mediated Major Histocompatibility Complex Class I Internalization. Journal of Biological Chemistry. 2010;285(46):35311–35319. doi: 10.1074/jbc.M110.112763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Boname JM, et al. Efficient Internalization ofMHC I Requires Lysine-11 and Lysine-63 Mixed Linkage Polyubiquitin Chains. Traffic. 2010;11(2):210–220. doi: 10.1111/j.1600-0854.2009.01011.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Glauser L, et al. Parkin promotes the ubiquitination and degradation of the mitochondrial fusion factor mitofusin 1. J Neurochem. 2011;118(4):636–45. doi: 10.1111/j.1471-4159.2011.07318.x. [DOI] [PubMed] [Google Scholar]
- 36.Geisler S, et al. PINKl/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nat Cell Biol. 2010;12(2):119–31. doi: 10.1038/ncb2012. [DOI] [PubMed] [Google Scholar]
- 37.Peng D-J, et al. Noncanonical K27-Linked Polyubiquitination of TIEG1 Regulates Foxp3 Expression and Tumor Growth. The Journal of Immunology. 2011;186(10):5638–5647. doi: 10.4049/jimmunol.1003801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Al-Hakim AK, et al. Control of AMPK-related kinases by USP9X and atypical Lys(29)/Lys(33)-linked polyubiquitin chains. Biochem J. 2008;411(2):249–60. doi: 10.1042/BJ20080067. [DOI] [PubMed] [Google Scholar]
- 39.Huang H, et al. K33-Linked Polyubiquitination of T Cell Receptor-^ Regulates Proteolysis-Independent T Cell Signaling. Immunity. 2010;33(1):60–70. doi: 10.1016/j.immuni.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nakasone MA, et al. Mixed-linkage ubiquitin chains send mixed messages. Structure. 2013;21(5):727–40. doi: 10.1016/j.str.2013.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kirkpatrick DS, et al. Quantitative analysis of in vitro ubiquitinated cyclin B1 reveals complex chain topology. Nat Cell Biol. 2006;8(7):700–710. doi: 10.1038/ncb1436. [DOI] [PubMed] [Google Scholar]
- 42.Rajsbaum R, et al. Unanchored K48-linked polyubiquitin synthesized by the E3-ubiquitin ligase TRIM6 stimulates the interferon-IKKepsilon kinase-mediated antiviral response. Immunity. 2014;40(6):880–95. doi: 10.1016/j.immuni.2014.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schlee M. Master sensors of pathogenic RNA - RIG-I like receptors. Immunobiology. 2013;218(11):1322–1335. doi: 10.1016/j.imbio.2013.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Goubau D, et al. Antiviral immunity via RIG-I-mediated recognition of RNA bearing 5#2370-diphosphates. Nature. 2014 doi: 10.1038/nature13590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kato H, et al. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature. 2006;441(7089):101–105. doi: 10.1038/nature04734. [DOI] [PubMed] [Google Scholar]
- 46.Rodriguez KR, Bruns AM, Horvath CM. MDA5 and LGP2: Accomplices and Antagonists of Antiviral Signal Transduction. J Virol. 2014;88(15):8194–8200. doi: 10.1128/JVI.00640-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yoneyama M, et al. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat Immunol. 2004;5(7):730–7. doi: 10.1038/ni1087. [DOI] [PubMed] [Google Scholar]
- 48.Andrejeva J, et al. The V proteins of paramyxoviruses bind the IFN-inducible RNA helicase, mda-5, and inhibit its activation of the IFN-beta promoter. Proc Natl Acad Sci USA. 2004;101(49):17264–9. doi: 10.1073/pnas.0407639101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kawai T, et al. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat Immunol. 2005;6(10):981–8. doi: 10.1038/ni1243. [DOI] [PubMed] [Google Scholar]
- 50.Meylan E, et al. Cardifis an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature. 2005;437(7062):1167–72. doi: 10.1038/nature04193. [DOI] [PubMed] [Google Scholar]
- 51.Seth RB, et al. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF 3. Cell. 2005;122(5):669–82. doi: 10.1016/j.cell.2005.08.012. [DOI] [PubMed] [Google Scholar]
- 52.Xu LG, et al. VISA is an adapter protein required for virus-triggered IFN-beta signaling. Mol Cell. 2005;19(6):727–40. doi: 10.1016/j.molcel.2005.08.014. [DOI] [PubMed] [Google Scholar]
- 53.Belgnaoui SM, Paz S, Hiscott J. Orchestrating the interferon antiviral response through the mitochondrial antiviral signaling (MAVS) adapter. Curr Opin Immunol. 2011;23(5):56472. doi: 10.1016/j.coi.2011.08.001. [DOI] [PubMed] [Google Scholar]
- 54.Gack M, et al. TRIM25 RING-finger E3 ubiquitin ligase is essential for RIG-I-mediated antiviral activity. Nature. 2007;446(7138):916–920. doi: 10.1038/nature05732. [DOI] [PubMed] [Google Scholar]
- 55.Gack MU, et al. Phosphorylation-mediated negative regulation of RIG-I antiviral activity. J Virol. 2010;84(7):3220–9. doi: 10.1128/JVI.02241-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nistal-Villan E, et al. Negative role of RIG-I serine 8 phosphorylation in the regulation of interferon-beta production. J Biol Chem. 2010;285(26):20252–61. doi: 10.1074/jbc.M109.089912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wies E, et al. Dephosphorylation of the RNA Sensors RIG-I and MDA5 by the Phosphatase PP1 Is Essential for Innate Immune Signaling. Immunity. 2013;38(3):437–49. doi: 10.1016/j.immuni.2012.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Maharaj NP, et al. Conventional protein kinase C-alpha (PKC-alpha) and PKC-beta negatively regulate RIG-I antiviral signal transduction. J Virol. 2012;86(3):1358–71. doi: 10.1128/JVI.06543-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gack M, et al. Roles of RIG-I N-terminal tandem CARD and splice variant in TRIM25-mediated antiviral signal transduction. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(43):16743–16748. doi: 10.1073/pnas.0804947105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rajsbaum R, et al. Species-specific inhibition of RIG-I ubiguitination and IFN induction by the influenza A virus NS1 protein. PLoS Pathogens. 2012;8(11) doi: 10.1371/journal.ppat.1003059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gack M. TRIMming Flavivirus infection. Cell host & microbe. 2011;10(3):175–177. doi: 10.1016/j.chom.2011.08.012. [DOI] [PubMed] [Google Scholar]
- 62.Rajsbaum R, Garcia-Sastre A, Versteeg GA. TRIMmunity: the roles of the TRIM E3-ubiguitin ligase family in innate antiviral immunity. J Mol Biol. 2014;426(6):1265–84. doi: 10.1016/j.jmb.2013.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.McNab F, et al. Tripartite-motif proteins and innate immune regulation. Current opinion in immunology. 2011;23(1):46–56. doi: 10.1016/j.coi.2010.10.021. [DOI] [PubMed] [Google Scholar]
- 64.Oshiumi H, et al. Riplet/RNF135, a RING finger protein, ubiguitinates RIG-I to promote interferon-beta induction during the early phase of viral infection. The Journal of biological chemistry. 2009;284(2):807–817. doi: 10.1074/jbc.M804259200. [DOI] [PubMed] [Google Scholar]
- 65.Gao D, et al. REUL is a novel E3 ubiguitin ligase and stimulator of retinoic-acid-inducible gene-I. PloS one. 2009;4(6):e5760. doi: 10.1371/journal.pone.0005760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Oshiumi H, et al. The ubiguitin ligase Riplet is essential for RIG-I-dependent innate immune responses to RNA virus infection. Cell host & microbe. 2010;8(6):496–509. doi: 10.1016/j.chom.2010.11.008. [DOI] [PubMed] [Google Scholar]
- 67.Oshiumi H, et al. A distinct role of Riplet-mediated K63-Linked polyubiguitination of the RIG-I repressor domain in human antiviral innate immune responses. PLoS Pathogens. 2013;9(8) doi: 10.1371/journal.ppat.1003533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gack M, et al. Influenza A virus NS1 targets the ubiguitin ligase TRIM25 to evade recognition by the host viral RNA sensor RIG-I. Cell host & microbe. 2009;5(5):439–449. doi: 10.1016/j.chom.2009.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gack MU. Mechanisms of RIG-I-Like Receptor Activation and Manipulation by Viral Pathogens. Journal of Virology. 2014;88(10):5213–5216. doi: 10.1128/JVI.03370-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zeng W, et al. Reconstitution of the RIG-I pathway reveals a signaling role of unanchored polyubiguitin chains in innate immunity. Cell. 2010;141(2):315–330. doi: 10.1016/j.cell.2010.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jiang X, et al. Ubiguitin-induced oligomerization of the RNA sensors RIG-I and MDA5 activates antiviral innate immune response. Immunity. 2012;36(6):959–973. doi: 10.1016/j.immuni.2012.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wu B, et al. Structural basis for dsRNA recognition, filament formation, and antiviral signal activation by MDA5. Cell. 2013;152(1-2):276–289. doi: 10.1016/j.cell.2012.11.048. [DOI] [PubMed] [Google Scholar]
- 73.Peisley A, et al. Structural basis for ubiguitin-mediated antiviral signal activation by RIG-I. Nature. 2014;509(7498):110–114. doi: 10.1038/nature13140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yan J, et al. TRIM4 modulates type I interferon induction and cellular antiviral response by targeting RIG-I for K63-linked ubiguitination. Journal of molecular cell biology. 2014;6(2):154–163. doi: 10.1093/jmcb/mju005. [DOI] [PubMed] [Google Scholar]
- 75.Kuniyoshi K, et al. Pivotal role of RNA-binding E3 ubiguitin ligase MEX3C in RIG-I-mediated antiviral innate immunity. Proc Natl Acad Sci USA. 2014;111(15):5646–51. doi: 10.1073/pnas.1401674111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Friedman CS, et al. The tumour suppressor CYLD is a negative regulator of RIG-I-mediated antiviral response. 2008;9:930–936. doi: 10.1038/embor.2008.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Fan Y, et al. USP21 negatively regulates antiviral response by acting as a RIG-I deubiguitinase. The Journal of experimental medicine. 2014;211(2):313–328. doi: 10.1084/jem.20122844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Cui J, et al. USP3 inhibits type I interferon signaling by deubiguitinating RIG-I-like receptors. Cell Res. 2014;24(4):400–416. doi: 10.1038/cr.2013.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chiang JJ, Davis ME, Gack MU. Regulation of RIG-I-like receptor signaling by host and viral proteins. Cytokine & Growth Factor Reviews. 2014;(0) doi: 10.1016/j.cytogfr.2014.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Paz S, et al. Ubiguitin-regulated recruitment of IkappaB kinase epsilon to the MAVS interferon signaling adapter. Mol Cell Biol. 2009;29(12):3401–12. doi: 10.1128/MCB.00880-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Arimoto K.-i., et al. Negative regulation of the RIG-I signaling by the ubiguitin ligase RNF125. Proceedings of the National Academy of Sciences of the United States of America. 2007;104(18):7500–7505. doi: 10.1073/pnas.0611551104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wang L, et al. USP4 Positively Regulates RIG-I-Mediated Antiviral Response through Deubiguitination and Stabilization of RIG-I. Journal of Virology. 2013;87(8):4507–4515. doi: 10.1128/JVI.00031-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Narayan K, et al. TRIM13 is a negative regulator of MDA5-mediated type I IFN production. J Virol. 2014 doi: 10.1128/JVI.02593-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.You F, et al. PCBP2 mediates degradation of the adaptor MAVS via the HECT ubiguitin ligase AIP4. Nat Immunol. 2009;10(12):1300–1308. doi: 10.1038/ni.1815. [DOI] [PubMed] [Google Scholar]
- 85.Wang Y, Tong X, Ye X. Ndfipl Negatively Regulates RIG-I-Dependent Immune Signaling by Enhancing E3 Ligase Smurfl-Mediated MAVS Degradation. The Journal of Immunology. 2012;189(11):5304–5313. doi: 10.4049/jimmunol.1201445. [DOI] [PubMed] [Google Scholar]
- 86.Pan Y, et al. Smurf2 negatively modulates RIG-I-dependent antiviral response by targeting VISA/MAVS for ubiguitination and degradation. J Immunol. 2014;192(10):4758–64. doi: 10.4049/jimmunol.1302632. [DOI] [PubMed] [Google Scholar]
- 87.Zhong B, et al. The E3 Ubiguitin Ligase RNF5 Targets Virus-Induced Signaling Adaptor for Ubiguitination and Degradation. The Journal of Immunology. 2010;184(11):6249–6255. doi: 10.4049/jimmunol.0903748. [DOI] [PubMed] [Google Scholar]
- 88.Yang B, et al. Novel function of Trim44 promotes an antiviral response by stabilizing VISA. J Immunol. 2013;190(7):3613–9. doi: 10.4049/jimmunol.1202507. [DOI] [PubMed] [Google Scholar]
- 89.Inn K-S, et al. Linear ubiguitin assembly complex negatively regulates RIG-I- and TRIM25-mediated type I interferon induction. Molecular cell. 2011;41(3):354–365. doi: 10.1016/j.molcel.2010.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Pauli E-K, et al. The Ubiguitin-Specific Protease USP15 Promotes RIG-I-Mediated Antiviral Signaling by Deubiguitylating TRIM25. 2014;7:ra3–ra3. doi: 10.1126/scisignal.2004577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Medzhitov R. Toll-like receptors and innate immunity. Nat Rev Immunol. 2001;1(2):135–45. doi: 10.1038/35100529. [DOI] [PubMed] [Google Scholar]
- 92.Gay NJ, et al. Assembly and localization of Toll-like receptor signalling complexes. Nat Rev Immunol. 2014;14(8):546–58. doi: 10.1038/nri3713. [DOI] [PubMed] [Google Scholar]
- 93.Akira S, Takeda K. Toll-like receptor signalling. Nat Rev Immunol. 2004;4(7):499–511. doi: 10.1038/nri1391. [DOI] [PubMed] [Google Scholar]
- 94.O'Neill LA, Bowie AG. The family of five: TIR-domain-containing adaptors in Toll-like receptor signalling. Nat Rev Immunol. 2007;7(5):353–64. doi: 10.1038/nri2079. [DOI] [PubMed] [Google Scholar]
- 95.Chuang TH, Ulevitch RJ. Triad3A, an E3 ubiguitin-protein ligase regulating Toll-like receptors. Nat Immunol. 2004;5(5):495–502. doi: 10.1038/ni1066. [DOI] [PubMed] [Google Scholar]
- 96.Nakhaei P, et al. The E3 ubiguitin ligase Triad3A negatively regulates the RIG-I/MAVS signaling pathway by targeting TRAF3 for degradation. PLoS Pathog. 2009;5(11):e1000650. doi: 10.1371/journal.ppat.1000650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Fearns C, et al. Triad3A regulates ubiguitination and proteasomal degradation of RIP1 following disruption of Hsp90 binding. J Biol Chem. 2006;281(45):34592–600. doi: 10.1074/jbc.M604019200. [DOI] [PubMed] [Google Scholar]
- 98.Naiki Y, et al. Transforming growth factor-beta differentially inhibits MyD88-dependent, but not TRAM- and TRIF-dependent, lipopolysaccharide-induced TLR4 signaling. J Biol Chem. 2005;280(7):5491–5. doi: 10.1074/jbc.C400503200. [DOI] [PubMed] [Google Scholar]
- 99.Lee YS, et al. Smad6-specific recruitment of SmurfE3 ligases mediates TGF-betal-induced degradation of MyD88 in TLR4 signalling. Nat Commun. 2011;2:460. doi: 10.1038/ncomms1469. [DOI] [PubMed] [Google Scholar]
- 100.Wang C, et al. The E3 ubiguitin ligase Nrdpl 'preferentially'promotes TLR-mediated production of type I interferon. Nat Immunol. 2009;10(7):744–52. doi: 10.1038/ni.1742. [DOI] [PubMed] [Google Scholar]
- 101.Yang Y, et al. E3 ligase WWP2 negatively regulates TLR3-mediated innate immune response by targeting TRIFfor ubiguitination and degradation. Proc Natl Acad Sci USA. 2013;110(13):5115–20. doi: 10.1073/pnas.1220271110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ordureau A, et al. The IRAK-catalysed activation of the E3 ligase function of Pellino isoforms induces the Lys63-linked polyubiquitination of IRAKl. Biochem J. 2008;409(1):43–52. doi: 10.1042/BJ20071365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Schauvliege R, Janssens S, Beyaert R. Pellino proteins: novel players in TLR and IL-lR signalling. J Cell Mol Med. 2007;11(3):453–61. doi: 10.1111/j.1582-4934.2007.00040.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Conze DB, et al. Lys63-linked polyubiquitination of IRAK-l is required for interleukin-l receptor- and toll-like receptor-mediated NF-kappaB activation. Mol Cell Biol. 2008;28(10):3538–47. doi: 10.1128/MCB.02098-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Jiang Z, et al. Pellino l is required for interleukin-l (IL-l)-mediated signaling through its interaction with the IL-l receptor-associated kinase 4 (IRAK4)-IRAK-tumor necrosis factor receptor-associated factor 6 (TRAF6) complex. J Biol Chem. 2003;278(13):10952–6. doi: 10.1074/jbc.M212112200. [DOI] [PubMed] [Google Scholar]
- 106.Schauvliege R, Janssens S, Beyaert R. Pellino proteins are more than scaffold proteins in TLR/IL-lR signalling: a role as novel RING E3-ubiquitin-ligases. FEBS Lett. 2006;580(19):4697702. doi: 10.1016/j.febslet.2006.07.046. [DOI] [PubMed] [Google Scholar]
- 107.Chang M, Jin W, Sun SC. Pelil facilitates TRIF-dependent Toll-like receptor signaling and proinflammatory cytokine production. Nat Immunol. 2009;10(10):1089–95. doi: 10.1038/ni.1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Ishikawa H, Barber G. STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature. 2008;455(7213):674–678. doi: 10.1038/nature07317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Zhong B, et al. The adaptor protein MITA links virus-sensing receptors to IRF3 transcription factor activation. Immunity. 2008;29(4):538–550. doi: 10.1016/j.immuni.2008.09.003. [DOI] [PubMed] [Google Scholar]
- 110.Ishikawa H, Ma Z, Barber GN. STING regulates intracellular DNA-mediated, type I interferon-dependent innate immunity. Nature. 2009;461(7265):788–92. doi: 10.1038/nature08476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Takaoka A, et al. DAI (DLM-l/ZBPl) is a cytosolic DNA sensor and an activator of innate immune response. Nature. 2007;448(7152):501–5. doi: 10.1038/nature06013. [DOI] [PubMed] [Google Scholar]
- 112.Zhang Z, et al. The helicase DDX4l senses intracellular DNA mediated by the adaptor STING in dendritic cells. Nat Immunol. 2011;12(10):959–65. doi: 10.1038/ni.2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Unterholzner L, et al. IFIl6 is an innate immune sensor for intracellular DNA. Nat Immunol. 2010;11(11):997–1004. doi: 10.1038/ni.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Brunette RL, et al. Extensive evolutionary and functional diversity among mammalian AIM2-like receptors. J Exp Med. 2012;209(11):1969–83. doi: 10.1084/jem.20121960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Zhang X, et al. The cytosolic DNA sensor cGAS forms an oligomeric complex with DNA and undergoes switch-like conformational changes in the activation loop. Cell Rep. 2014;6(3):421–30. doi: 10.1016/j.celrep.2014.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Diner EJ, et al. The innate immune DNA sensor cGAS produces a noncanonical cyclic dinucleotide that activates human STING. Cell Rep. 2013;3(5):1355–61. doi: 10.1016/j.celrep.2013.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Sun L, et al. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science. 2013;339(6121):786–91. doi: 10.1126/science.1232458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Wu J, et al. Cyclic GMP-AMP is an endogenous second messenger in innate immune signaling by cytosolic DNA. Science. 2013;339(6121):826–30. doi: 10.1126/science.1229963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Ablasser A, et al. cGAS produces a 2[prime]-5[prime]-linked cyclic dinucleotide second messenger that activates STING. Nature. 2013;498(7454):380–384. doi: 10.1038/nature12306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Kranzusch PJ, et al. Structure of human cGAS reveals a conserved family of second-messenger enzymes in innate immunity. Cell Rep. 2013;3(5):1362–8. doi: 10.1016/j.celrep.2013.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Sauer JD, et al. The N-ethyl-N-nitrosourea-induced Goldenticket mouse mutant reveals an essential function of Sting in the in vivo interferon response to Listeria monocytogenes and cyclic dinucleotides. Infect Immun. 2011;79(2):688–94. doi: 10.1128/IAI.00999-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Burdette DL, et al. STING is a direct innate immune sensor of cyclic di-GMP. Nature. 2011;478(7370):515–8. doi: 10.1038/nature10429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Li X-D, et al. Pivotal roles of cGAS-cGAMP signaling in antiviral defense and immune adjuvant effects. Science (New York, N.Y.) 2013;341(6152):1390–1394. doi: 10.1126/science.1244040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Tsuchida T, et al. The ubiguitin ligase TRIM56 regulates innate immune responses to intracellular double-stranded DNA. Immunity. 2010;33(5):765–76. doi: 10.1016/j.immuni.2010.10.013. [DOI] [PubMed] [Google Scholar]
- 125.Zhang J, et al. TRIM32 protein modulates type I interferon induction and cellular antiviral response by targeting MITA/STING protein for K63-linked ubiguitination. J Biol Chem. 2012;287(34):28646–55. doi: 10.1074/jbc.M112.362608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Zhong B, et al. The ubiguitin ligase RNF5 regulates antiviral responses by mediating degradation of the adaptor protein MITA. Immunity. 2009;30(3):397–407. doi: 10.1016/j.immuni.2009.01.008. [DOI] [PubMed] [Google Scholar]
- 127.Qin Y, et al. RNF26 Temporally Regulates Virus-Triggered Type I Interferon Induction by Two Distinct Mechanisms. PLoS Pathog. 2014;10(9):e1004358. doi: 10.1371/journal.ppat.1004358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Oganesyan G, et al. Critical role of TRAF3 in the Toll-like receptor-dependent and -independent antiviral response. Nature. 2006;439(7073):208–211. doi: 10.1038/nature04374. [DOI] [PubMed] [Google Scholar]
- 129.Saha S, et al. Regulation of antiviral responses by a direct and specific interaction between TRAF3 and Cardif. The EMBO journal. 2006;25(14):3257–3263. doi: 10.1038/sj.emboj.7601220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Hayden MS, Ghosh S. Shared principles in NF-kappaB signaling. Cell. 2008;132(3):34462. doi: 10.1016/j.cell.2008.01.020. [DOI] [PubMed] [Google Scholar]
- 131.Chen J, Chen ZJ. Regulation of NF-kappaB by ubiguitination. Curr Opin Immunol. 2013;25(1):4–12. doi: 10.1016/j.coi.2012.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Bhoj VG, Chen ZJ. Ubiguitylation in innate and adaptive immunity. Nature. 2009;458(7237):430–7. doi: 10.1038/nature07959. [DOI] [PubMed] [Google Scholar]
- 133.Deng L, et al. Activation of the IkappaB kinase complex by TRAF6 reguires a dimeric ubiguitin-conjugating enzyme complex and a unigue polyubiguitin chain. Cell. 2000;103(2):351–61. doi: 10.1016/s0092-8674(00)00126-4. [DOI] [PubMed] [Google Scholar]
- 134.Wang C, et al. TAK1 is a ubiguitin-dependent kinase of MKK and IKK. Nature. 2001;412(6844):346–51. doi: 10.1038/35085597. [DOI] [PubMed] [Google Scholar]
- 135.Xia ZP, et al. Direct activation of protein kinases by unanchored polyubiguitin chains. Nature. 2009;461(7260):114–9. doi: 10.1038/nature08247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Jensen LE, Whitehead AS. Ubiguitin activated tumor necrosis factor receptor associated factor-6 (TRAF6) is recycled via deubiguitination. FEBS letters. 2003;553(1-2):190–194. doi: 10.1016/s0014-5793(03)00998-0. [DOI] [PubMed] [Google Scholar]
- 137.Ea CK, et al. Activation of IKK by TNFalpha reguires site-specific ubiguitination of RIP1 and polyubiguitin binding by NEMO. Mol Cell. 2006;22(2):245–57. doi: 10.1016/j.molcel.2006.03.026. [DOI] [PubMed] [Google Scholar]
- 138.Ivins FJ, et al. NEMO oligomerization and its ubiguitin-binding properties. Biochem J. 2009;421(2):243–51. doi: 10.1042/BJ20090427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Lo YC, et al. Structural basis for recognition of diubiguitins by NEMO. Mol Cell. 2009;33(5):602–15. doi: 10.1016/j.molcel.2009.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Boone DL, et al. The ubiguitin-modifying enzyme A20 is reguiredfor termination of Toll-like receptor responses. Nat Immunol. 2004;5(10):1052–60. doi: 10.1038/ni1110. [DOI] [PubMed] [Google Scholar]
- 141.Wertz IE, et al. De-ubiguitination and ubiguitin ligase domains of A20 downregulate NF-kappaB signalling. Nature. 2004;430(7000):694–9. doi: 10.1038/nature02794. [DOI] [PubMed] [Google Scholar]
- 142.Wang YY, et al. A20 is a potent inhibitor of TLR3- and Sendai virus-induced activation of NF-kappaB and ISRE and IFN-beta promoter. FEBS Lett. 2004;576(1-2):86–90. doi: 10.1016/j.febslet.2004.08.071. [DOI] [PubMed] [Google Scholar]
- 143.Shembade N, Ma A, Harhaj EW. Inhibition of NF-kappaB signaling by A20 through disruption of ubiguitin enzyme complexes. Science. 2010;327(5969):1135–9. doi: 10.1126/science.1182364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Zhang M, et al. Regulation of IkappaB kinase-related kinases and antiviral responses by tumor suppressor CYLD. J Biol Chem. 2008;283(27):18621–6. doi: 10.1074/jbc.M801451200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Zhang J, et al. Impaired regulation of NF-kappaB and increased susceptibility to colitis-associated tumorigenesis in CYLD-deficient mice. J Clin Invest. 2006;116(11):3042–9. doi: 10.1172/JCI28746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Li S, et al. Regulation of virus-triggered signaling by 0TUB1- and 0TUB2-mediated deubiguitination of TRAF3 and TRAF6. J Biol Chem. 2010;285(7):4291–7. doi: 10.1074/jbc.M109.074971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Paz S, et al. A functional C-terminal TRAF3-binding site in MAVS participates in positive and negative regulation of the IFN antiviral response. Cell Res. 2011;21(6):895–910. doi: 10.1038/cr.2011.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Zeng W, et al. Key role of Ubc5 and lysine-63 polyubiguitination in viral activation of IRF3. Mol Cell. 2009;36(2):315–25. doi: 10.1016/j.molcel.2009.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Peng Y, Xu R, Zheng X. HSCARG negatively regulates the cellular antiviral RIG-I like receptor signaling pathway by inhibiting TRAF3 ubiguitination via recruiting 0TUB1. PLoS Pathog. 2014;10(4):e1004041. doi: 10.1371/journal.ppat.1004041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Kayagaki N, et al. DUBA: a deubiguitinase that regulates type I interferon production. Science. 2007;318(5856):1628–32. doi: 10.1126/science.1145918. [DOI] [PubMed] [Google Scholar]
- 151.Karim R, et al. Human papillomavirus (HPV) upregulates the cellular deubiguitinase UCHL1 to suppress the keratinocyte's innate immune response. PLoS Pathog. 2013;9(5):e1003384. doi: 10.1371/journal.ppat.1003384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Hu H, et al. 0TUD7B controls non-canonical NF-kappaB activation through deubiguitination of TRAF3. Nature. 2013;494(7437):371–4. doi: 10.1038/nature11831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Li S, et al. Mapping a dynamic innate immunity protein interaction network regulating type I interferon production. Immunity. 2011;35(3):426–40. doi: 10.1016/j.immuni.2011.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Wang L, Li S, Dorf ME. NEMO binds ubiguitinated TANK-binding kinase 1 (TBK1) to regulate innate immune responses to RNA viruses. PLoS One. 2012;7(9):e43756. doi: 10.1371/journal.pone.0043756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Ye JS, et al. Lysine 63-Linked TANK-Binding Kinase 1 Ubiguitination by Mindbomb E3 Ubiguitin Protein Ligase 2 Is Mediated by the Mitochondrial Antiviral Signaling Protein. J Virol. 2014;88(21):12765–76. doi: 10.1128/JVI.02037-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Parvatiyar K, Barber GN, Harhaj EW. TAX1BP1 and A20 inhibit antiviral signaling by targeting TBK1-IKKikinases. J Biol Chem. 2010;285(20):14999–5009. doi: 10.1074/jbc.M110.109819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Tu D, et al. Structure and ubiguitination-dependent activation of TANK-binding kinase 1. Cell Rep. 2013;3(3):747–58. doi: 10.1016/j.celrep.2013.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Zhang L, et al. Ubiguitin-specific protease 2b negatively regulates IFN-beta production and antiviral activity by targeting TANK-binding kinase 1. J Immunol. 2014;193(5):2230–7. doi: 10.4049/jimmunol.1302634. [DOI] [PubMed] [Google Scholar]
- 159.Yaron A, et al. Identification of the receptor component of the I[kappa]B[alpha]-ubiguitin ligase. Nature. 1998;396(6711):590–594. doi: 10.1038/25159. [DOI] [PubMed] [Google Scholar]
- 160.Heissmeyer V, et al. Shared pathways ofIkappaB kinase-induced SCF(betaTrCP)-mediated ubiguitination and degradation for the NF-kappaB precursor p105 and IkappaBalpha. Mol Cell Biol. 2001;21(4):1024–35. doi: 10.1128/MCB.21.4.1024-1035.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Maniatis T. A ubiguitin ligase complex essential for the NF-kappaB, Wnt/Wingless, and Hedgehog signaling pathways. Genes Dev. 1999;13(5):505–10. doi: 10.1101/gad.13.5.505. [DOI] [PubMed] [Google Scholar]
- 162.Zhao W, et al. E3 ubiguitin ligase tripartite motif 38 negatively regulates TLR-mediated immune responses by proteasomal degradation of TNF receptor-associated factor 6 in macrophages. J Immunol. 2012;188(6):2567–74. doi: 10.4049/jimmunol.1103255. [DOI] [PubMed] [Google Scholar]
- 163.Geng H, et al. Phosphorylation of NF-kappaB p65 at Ser468 controls its C0MMD1-dependent ubiguitination and target gene-specific proteasomal elimination. EMBO Rep. 2009;10(4):3816. doi: 10.1038/embor.2009.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Saccani S, et al. Degradation of promoter-bound p65/RelA is essential for the prompt termination of the nuclear factor kappaB response. J Exp Med. 2004;200(1):107–13. doi: 10.1084/jem.20040196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Maine GN, et al. C0MMD1 promotes the ubiguitination of NF-kappaB subunits through a cullin-containing ubiguitin ligase. Embo J. 2007;26(2):436–47. doi: 10.1038/sj.emboj.7601489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Thoms HC, et al. Nucleolar targeting of RelA(p65) is regulated by C0MMD1-dependent ubiguitination. Cancer Res. 2010;70(1):139–49. doi: 10.1158/0008-5472.CAN-09-1397. [DOI] [PubMed] [Google Scholar]
- 167.Tanaka T, Grusby MJ, Kaisho T. PDLIM2-mediated termination of transcription factor NF-kappaB activation by intranuclear sequestration and degradation of the p65 subunit. Nat Immunol. 2007;8(6):584–91. doi: 10.1038/ni1464. [DOI] [PubMed] [Google Scholar]
- 168.Colleran A, et al. Deubiquitination of NF-kappaB by Ubiquitin-Specific Protease-7 promotes transcription. Proc Natl Acad Sci USA. 2013;110(2):618–23. doi: 10.1073/pnas.1208446110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Cui J, et al. NLRP4 negatively regulates type I interferon signaling by targeting the kinase TBK1 for degradation via the ubiquitin ligase DTX4. Nat Immunol. 2012;13(4):387–95. doi: 10.1038/ni.2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Zhang M, et al. TRAF-interacting protein (TRIP) negatively regulates IFN-beta production and antiviral response by promoting proteasomal degradation of TANK-binding kinase 1. J Exp Med. 2012;209(10):1703–11. doi: 10.1084/jem.20120024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Lin R, et al. Virus-dependent phosphorylation of the IRF-3 transcription factor regulates nuclear translocation, transactivation potential, andproteasome-mediated degradation. Mol Cell Biol. 1998;18(5):2986–96. doi: 10.1128/mcb.18.5.2986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Saitoh T, et al. Negative regulation of interferon-regulatory factor 3-dependent innate antiviral response by the prolyl isomerase Pinl. Nat Immunol. 2006;7(6):598–605. doi: 10.1038/ni1347. [DOI] [PubMed] [Google Scholar]
- 173.Higgs R, et al. The E3 ubiquitin ligase Ro52 negatively regulates IFN-beta production post-pathogen recognition by polyubiquitin-mediated degradation of IRF3. J Immunol. 2008;181(3):1780–6. doi: 10.4049/jimmunol.181.3.1780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Zhang M, et al. Negative feedback regulation of cellular antiviral signaling by RBCKl-mediated degradation of IRF3. Cell Res. 2008;18(11):1096–104. doi: 10.1038/cr.2008.277. [DOI] [PubMed] [Google Scholar]
- 175.Young JA, et al. Fas-associated death domain (FADD) and the E3 ubiquitin-protein ligase TRIM21 interact to negatively regulate virus-induced interferon production. J Biol Chem. 2011;286(8):6521–31. doi: 10.1074/jbc.M110.172288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Yu Y, Hayward GS. The ubiquitin E3 ligase RAUL negatively regulates type i interferon through ubiquitination of the transcription factors IRF7 and IRF3. Immunity. 2010;33(6):86377. doi: 10.1016/j.immuni.2010.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Lin R, et al. The herpes simplex virus ICPO RING finger domain inhibits IRF3- and IRF7-mediated activation of interferon-stimulated genes. J Virol. 2004;78(4):1675–84. doi: 10.1128/JVI.78.4.1675-1684.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Henderson G, Zhang Y, Jones C. The Bovine herpesvirus 1 gene encoding infected cell protein O (bICPO) can inhibit interferon-dependent transcription in the absence of other viral genes. J Gen Virol. 2005;86(Pt 10):2697–702. doi: 10.1099/vir.0.81109-0. [DOI] [PubMed] [Google Scholar]
- 179.Saira K, Zhou Y, Jones C. The infected cell protein 0 encoded by bovine herpesvirus 1 (bICPO) induces degradation of interferon response factor 3 and, consequently, inhibits beta interferon promoter activity. J Virol. 2007;81(7):3077–86. doi: 10.1128/JVI.02064-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Arimoto K.-i., et al. Polyubiquitin conjugation to NEMO by triparite motif protein 23 (TRIM23) is critical in antiviral defense. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(36):15856–15861. doi: 10.1073/pnas.1004621107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Ashida H, et al. A bacterial E3 ubiquitin ligase IpaH9.8 targets NEMO/IKKgamma to dampen the host NF-kappaB-mediated inflammatory response. Nat Cell Biol. 2010;12(1):66–73. doi: 10.1038/ncb2006. sup pp 1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Zotti T, et al. TRAF7protein promotes Lys-29-linked polyubiquitination ofIkappaB kinase (IKKgamma)/NF-kappaB essential modulator (NEMO) and p65/RelA protein and represses NF-kappaB activation. J Biol Chem. 2011;286(26):22924–33. doi: 10.1074/jbc.M110.215426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Tokunaga F, et al. Involvement of linear polyubiquitylation of NEMO in NF-kappaB activation. Nat Cell Biol. 2009;11(2):123–32. doi: 10.1038/ncb1821. [DOI] [PubMed] [Google Scholar]
- 184.Rahighi S, et al. Specific recognition of linear ubiquitin chains by NEMO is important for NF-kappaB activation. Cell. 2009;136(6):1098–109. doi: 10.1016/j.cell.2009.03.007. [DOI] [PubMed] [Google Scholar]
- 185.Belgnaoui S, et al. Linear ubiquitination of NEMO negatively regulates the interferon antiviral response through disruption of the MAVS-TRAF3 complex. Cell host & microbe. 2012;12(2):211–222. doi: 10.1016/j.chom.2012.06.009. [DOI] [PubMed] [Google Scholar]