Abstract
Tumor necrosis factor receptor-associated factor 6 (TRAF6) has been found to be involved in multiple cancers. However, the effect of small interfering RNA (siRNA)-induced knockdown of TRAF6 on the biological behaviors of cancer cells remains unknown. Thus, the present study aimed to investigate the effect of siRNA-induced knockdown of TRAF6 on the biological behaviors of human lung cancer SPC-A1 cells. The expression of TRAF6 was determined in human lung adenocarcinoma A549, non-small cell lung cancer H1650, human airway epithelial Calu-3 and human lung cancer SPC-A1 cell lines using quantitative RT-PCR (qRT-PCR) and western blotting at the transcriptional and translational levels. TRAF6 expression was knocked down in the SPC-A1 cells using an siRNA technique, and the effects of TRAF6 knockdown on NF-κB activity, cell proliferation, apoptosis, cell cycle, invasion and migration of the SPC-A1 cells were determined using electrophoretic mobility shift assay (EMSA), cell proliferation assay, flow cytometry, Transwell invasion assay and scratch wound assay. In addition, the protein expression of CD24, CXCR4, MMP1, MMP2, MMP9, TWIST, TIMP-2 and Slug was quantified using western blotting assay. Western blotting and qRT-PCR assays showed upregulation of TRAF6 at both the translational and transcriptional levels in the Calu-3 and SPC-A1 cells, and K63-linked ubiquitination of TRAF6 and constitutive NF-κB activation were detected in the SPC-A1 cells. Knockdown of TRAF6 inhibited the migration and invasion and promoted the apoptosis of the SPC-A1 cells, but had little effect on cell proliferation and the cell cycle. In addition, siRNA-induced TRAF6 knockdown caused a marked reduction in the protein expression of CD24 and CXCR4, but had little effect on MMP-1, MMP-2, MMP-9, Twist, TIMP-2 or Slug expression. The present study demonstrated that TRAF6 is upregulated in human lung cancer cells, and siRNA-induced TRAF6 knockdown inhibits the invasion of lung cancer cells and promotes apoptosis. It is suggested that TRAF6 may be a promising target for the therapy of lung cancer.
Keywords: lung cancer, TRAF6, apoptosis, invasion, small interfering RNA, knockdown, nuclear factor-κB
Introduction
Lung cancer is one of the most common cancers worldwide, and it is the leading cause of cancer-related deaths in both men and women (1). The high incidence and mortality rate make this malignancy a major public health issue throughout the world (2). In addition, a recent systematic review showed that lung cancer has become one of the major causes of years of life lost (YLLs) worldwide (3).
The development and progression of lung cancer requires a long period of time, in which a series of molecular events and alterations may occur (4). It has been shown that adhesion molecules (5), angiogenic factors (6), chemokines (7) and growth factors (8,9) appear to be responsible for the invasion and metastasis of lung cancer cells. However, the predominant drivers contributing to lung cancer cell malignancy, migration and invasion, remain to be determined.
Tumor necrosis factor receptor-associated factors (TRAFs) are a family of adaptor proteins that couple tumor necrosis factor receptor family to signaling pathways, and they are also characterized to be signal transducers of Toll/interleukin-1 (IL-1) family members, which play an important role in physiological and pathological processes (10). To date, 7 TRAFs have been identified, which are termed TRAF1 to 7 (11). As a key activator of nuclear factor-κB (NF-κB), TRAF6 functions as a signal transducer in the NF-κB pathway and activates the inhibitor of IκB kinase (IKK) in response to pro-inflammatory cytokines (11). In addition, TRAF6, which acts as an E3 ubiquitin ligase, catalyzes K63 polyubiquitination of TAK1 which is required for IKK activation, and mediates the activation of other downstream signaling molecules, thereby inducing the constitutive activation of NF-κB (12).
Elevated TRAF6 expression has been detected in colon cancer (13), glioma (14), osteosarcoma (15), esophageal squamous cell carcinoma (16), breast cancer (17) and lung cancer tissues (18,19), and TRAF has been found to promote the proliferation of these cancer cells. In addition, inhibition of TRAF6 was reported to result in suppression of lung cancer cell proliferation and tumor formation (20,21), and the TRAF6 status was found to correlate inversely with response to chemotherapy in overall advanced non-small cell lung cancer patients (22). However, the effect of small interfering RNA (siRNA)-induced knockdown of TRAF6 on the biological behaviors of cancer cells remains unknown until now.
Hereby, we knocked down TRAF6 using a specific siRNA targeting TRAF6, and assessed the effect of TRAF6 knockdown on the proliferation, migration, invasion, cell cycle and apoptosis of human lung cancer SPC-A1 cells. In addition, the expression of proteins associated with tumor migration and invasion was determined in the SPC-A1 cells transfected with TRAF6 siRNA, so as to provide new insight into the therapeutic potential of TRAF6 against lung cancer.
Materials and methods
Cell line and culture
The human lung adenocarcinoma A549, non-small cell lung cancer H1650, human airway epithelial Calu-3 and human lung cancer SPC-A1 cell lines were purchased from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China), and were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS) at 37°C containing 5% CO2.
TRAF6 siRNA and transfection
Log-phase SPC-A1 cells were seeded onto 6-well plates (Corning, Inc., Corning, NY, USA) at a density of 1×106 cells/well, and incubated at 37°C for 24 h. Four micrograms of the specific siRNA targeting TRAF6 or control siRNA (both from Santa Cruz Biotechnology, Inc., Santa Cruz, CA, USA) was added to 250 µl of serum-free DMEM and mixed evenly, while 10 µl of Lipofectamine 2000 (Thermo Fisher Scientific, Waltham, MA, USA) was added to another 250 µl of serum-free DMEM, gently mixed evenly and placed at room temperature for 5 min. The siRNA and Lipofectamine 2000 solutions were mixed evenly, and placed at room temperature for 20 min. Then, the mixture was transferred to 6-well plates, mixed evenly and gently and incubated at 37°C. Following a 6-h transfection, the cell culture solution was changed, and to each well h 2 ml medium was added. After 48 h of transfection, the cells were harvested for the subsequent experiments, and non-transfected cells served as blank controls. Transfection efficiency was verified using western blot assay.
Determination of SPC-A1 cell proliferation using a cell proliferation assay
Following a 48-h transfection, the proliferation of SPC-A1 cells was determined using the CellTiter 96 AQueous One Solution Cell Proliferation assay (Promega, Madison, WI, USA) following the manufacturer's instructions. Briefly, TRAF6 and control siRNA-transfected, and non-transfected SPC-A1 cells were seeded onto 96-well plates (Corning, Inc.) at a density of 1×105 cells/well, and then 20 µl of Cell Titer 96 AQueous One Solution was added to each well. Three wells containing DMEM alone were assigned for background subtraction. The cells were then incubated at 37°C for 30 min. The absorbance at 490 nm in each well was then determined using a SpectraMax 340 microplate reader (Molecular Devices Corporation, Sunnyvale, CA, USA). The absorbance of the transfected cells was normalized to that of the non-transfected cells (100% viability). All experiments were replicated in triplicate.
Detection of apoptosis of SPC-A1 cells with flow cytometry
After 48 h of transfection, TRAF6 and control siRNA-transfected and non-transfected SPC-A1 cells were harvested, washed twice in cold PBS and resuspended in 1X Annexin V binding buffer (Calbiochem, San Diego, CA, USA) at a concentration of 1×106 cells/ml. Then, 500 µl of cell suspension (4×105 cells) was transferred to a fluorescence-activated cell sorting (FACS) tube containing 1.25 µl of Annexin V-FITC (Calbiochem). Cells were gently vortexed and incubated at room temperature for 15 min in the dark. Subsequently, 500 µl of cold 1X Annexin V binding buffer and 10 µl of propidium iodide (Calbiochem) were transferred and mixed evenly, and the cells were analyzed on a FACScalibur flow cytometer (BD Biosciences, Burlington, MA, USA). All experiments were repeated in triplicate.
Cell cycle analysis of the SPC-A1 cells using flow cytometry
Following a 48-h transfection, TRAF6 and control siRNA-transfected and non-transfected SPC-A1 cells were harvested and washed twice with PBS. Approximately 6×105 cells were suspended in 150 µl of BD Cytofix/Cytoperm buffer solution (BD Biosciences) at 4°C for 20 min. Subsequently, the cells were washed twice with BD Perm/Wash buffer (BD Biosciences) and incubated in 200 µl of dying buffer containing 0.1 mg/ml of propidium iodide and 2 mg/ml of RNase A at 37°C for 30 min in the dark. Cell cycle was then analyzed on a FACScalibur flow cytometer (BD Biosciences). All procedures were repeated in triplicate.
Transwell migration assay
The invasion of SPC-A1 cells was evaluated using a Transwell migration assay. Briefly, SPC-A1 cells were seeded onto 6-well plates at a density of 1×105/well and incubated at 37°C for 24 h. Then, the cells were transfected with TRAF6 or control siRNA, while non-transfected cells served as blank controls. Following a 24-h incubation, the cells were harvested, digested, centrifuged, and adjusted to a cell density of 4×105 cells/ml. Approximately 0.2 ml of the SPC-A1 cells were transferred to the Transwell chamber pre-coated with 1 µg/ml of fibronectin (Sigma-Aldrich, St. Louis, MO, USA), with DMEM containing 1% inactivated FBS in the upper chamber and DMEM supplemented with 10% inactivated FBS in the lower chamber, and the Transwell chamber was placed in 24-well cell culture inserts at 37°C for 24 h. Following a 48-h incubation, the cells in the chamber were removed, fixed in 90% methanol and stained with 0.1% crystal violet. Five fields of vision were randomly selected, and cells were counted under a microscope at a magnification of ×200 to determine the mean number in five inserts. All experiments were repeated in triplicate.
Scratch wound assay
The migration of the SPC-A1 cells was detected using the scratch wound assay. Briefly, the SPC-A1 cells transfected with TRAF6 and control siRNA, and non-transfected cells at approximately 80% confluency were seeded onto 6-well plates, and incubated at 37°C for 24 h. Then, a vertical scratch wound was made through the center of each well using a 10-µl pipette tip. The cells were then washed three times with PBS to remove the scratched cells, and fresh serum-free medium was transferred. After 12 h, the cells were examined by light microscopy at a magnification of ×200 to determine the resealing of the cell monolayer.
Quantitative RT-PCR (qRT-PCR) assay
The relative mRNA expression of TRAF6 was determined in A549, H1650, Calu-3 and SPC-A1 cells using a qRT-PCR assay. Total RNA was isolated from the four lung cancer cell lines using the RNeasy Mini kit (Qiagen Inc., Westborough, MA, USA) following the manufacturer's protocol, and reverse-transcribed into cDNA using the Promega Reverse Transcription system A3500 (Promega) at 42°C for 15 min, at 95°C for 5 min, and at 4°C for 5 min. qRT-PCR assay was run in 20 µl of the reaction system containing 10 µl of 2X PCR Master Mix, 1 µl of each primer, 1 µl of cDNA, and 7 µl of nuclease-free water, on a LightCycler Roche 480 with DyNAmo Flash SYBR-Green qPCR kit (Thermo Fisher Scientific, Inc.) using the following primers: TRAF6 (1) forward, 5′-CTA TTC ACC AGT TAG AGG-3′ and reverse, 5′-GCT CAC TTA CAT ACA TAC T-3′; TRAF6 (2) forward, 5′-GTT GCT GAA ATC GAA GCA CA-3′ and reverse, 5′-CGG GTT TGC CAG TGT AGA AT-3′; and β-actin forward, 5′-TGG CAC CAC ACC TTC TAC A-3′ and reverse, 5′-AGC ACA GCC TGG ATA GCA-3′ under the following conditions: at 95°C for 15 min; 40 cycles of at 95°C for 15 sec, at 55°C for 30 sec, and at 72°C for 1 sec; finally at 40°C for 1 min, while β-actin served as a reference gene. Relative quantity of mRNA expression was calculated by using the 2−ΔΔCt method. All experiments were repeated in triplicate.
Western blotting assay
The protein expression of TRAF6, MMP-1, MMP-2, MMP-9, Twist, TIMP-2, Slug, CD24 and CXCR4 was determined using western blot analysis. Briefly, total protein was extracted from the human lung cancer cells, and the protein concentration was determined using the bicinchoninic acid (BCA) assay. An amount 25 µg of total protein was separated on a 10% SDS-PAGE gel, and then transferred to the nitrocellulose (NC) membrane for 1 h at 60 V in transfer buffer. The membrane was then blocked with 5% nonfat milk for 4 h at room temperature, and incubated with 1:1,000 primary rabbit ant-human TRAF6 monoclonal antibody (catalogue no: 8028S), rabbit anti-human MMP-1 monoclonal antibody (catalogue no: sc-21731), rabbit anti-human MMP-2 polyclonal antibody (catalogue no: sc-10736), rabbit anti-human MMP-9 polyclonal antibody (catalogue no: sc-10737), rabbit anti-human Twist polyclonal antibody (catalogue no: sc-15393), rabbit anti-human TIMP-2 polyclonal antibody (catalogue no: sc-5539), rabbit anti-human Slug monoclonal antibody (catalogue no: 9585S), rabbit anti-human CD24 polyclonal antibody (catalogue no: sc-11406) (all from Santa Cruz Biotechnology, Inc.) and rabbit anti-human CXCR4 monoclonal antibody (catalogue no: sc-53534; Cell Signaling Technology, Inc., Beverly, MA, USA) overnight at 4°C on a gentle shaker. Following washing with TBST (20 mmol/l Tris-HCl, 150 mmol/l NaCl and 0.05% Tween-20; pH 7.4) three times, for 10 min each time, the membrane was incubated in 1:4,000 anti-rabbit HRP-conjugated IgG antibody (catalogue no: 7074S) or anti-mouse HRP-conjugated IgG antibody (catalogue no: 7076S) (both from Cell Signaling Technology, Inc.) at room temperature for 1 h. The membrane was then washed three times for 10 min with TBST buffer (20 mmol/l Tris-HCl, 150 mmol/l NaCl and 0.05% Tween-20; pH 7.4) and visualized with the SuperSignal West Pico kit (Thermo Fisher Scientific).
Electrophoretic mobility shift assay (EMSA)
Nuclear protein was extracted as described previously, and all nuclear extractions contained 1X Roche Mini Complete protease inhibitor cocktail solution (Roche Applied Science, Burgess Hill, UK). A 22-mer NF-κB consensus oligonucleotide (Promega) was labeled according to the manufacturer's instructions using the DIG gel-shift kit (Roche Applied Science). An amount of 10 mg of nuclear extract was used per binding reaction as detailed in the manufacturer's protocol, with some control reactions using NF-κB unlabeled oligonucleotides or nonspecific negative control OCT1 consensus oligonucleotides (Promega) for competition or the NF-κB p65 antibody (Thermo Fisher Scientific) for supershift reactions. Reactions were run on 6% (v/v) DNA retardation gels (Invitrogen, Paisley, UK), and then blotted and cross-linked onto positively charged nylon membranes (Roche Applied Science) before immunodetection.
Statistical analysis
All data are expressed as mean ± standard deviation (SD), and all statistical analyses were performed using the statistical software SPSS version 16.0 (SPSS Inc., Chicago, IL, USA). Differences in means were tested for statistical significance with the Student's t-test, with a p-value <0.05 indicative of statistical significance.
Results
TRAF6 expression and K63-linked ubiquitination of TRAF6
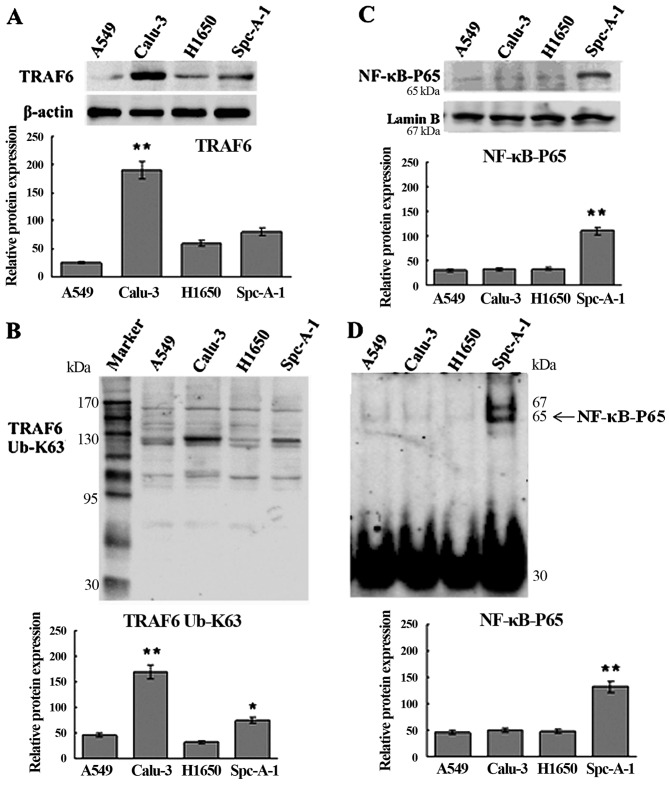
Western blotting and qRT-PCR assays showed higher expression of TRAF6 in the Calu-3 and SPC-A1 cells than the expression level in the A549 and H1650 cells at both the translational and transcriptional levels (Figs. 1A and 2), and K63-linked ubiquitination of TRAF6 was detected in the SPC-A1 cells, as revealed by western blot analysis (Fig. 1B).
Figure 1.
TRAF6 expression, K63-linked ubiquitination and constitutive activation of NF-κB. (A) Western blotting assay shows higher TRAF6 protein expression in the Calu-3 and SPC-A1 cells than that in the A549 and H1650 cells. (B) Western blotting revealed K63-linked ubiquitination of TRAF6 in the SPC-A1 cells. (C and D) EMSA and western blotting revealed constitutive activation of NF-κB in the SPC-A1 cells. *P<0.05 vs. A459 cells; **P<0.01 vs. A549 cells.
Figure 2.
Relative TRAF6 expression in four cell lines. *P<0.05 vs. A459 cells.
High expression of TRAF6 is associated with constitutive activation of NF-κB in the SPC-A1 cells
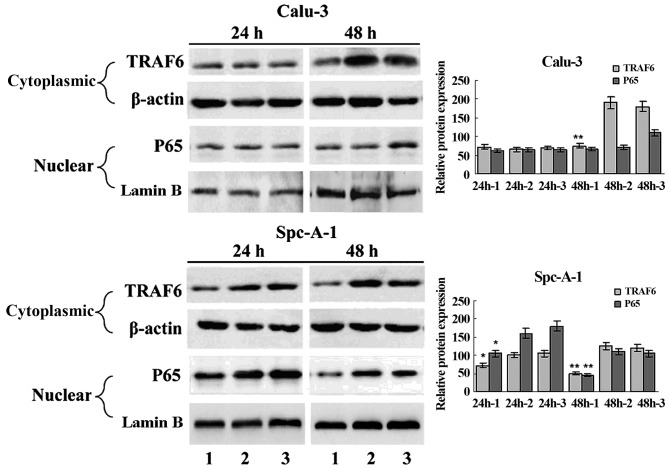
EMSA and western blot analysis showed constitutive NF-κB activation in the SPC-A1 cells (Fig. 1C and D), and a significant inhibition of NF-κB activation in the SPC-A1 cells 48 h post-transfection with TRAF6 siRNA (Fig. 3). These findings indicate that high expression of TRAF6 was associated with constitutive activation of NF-κB in the SPC-A1 cells.
Figure 3.
siRNA-induced TRAF6 knockdown reduces NF-κB activity in the SPC-A1 cells. Decreased NF-κB activation was detected in the TRAF6 siRNA-transfected SPC-A1 cells but not in the Calu-3 cells. SPC-A1 and Calu-3 cells were transfected with TRAF6 siRNA or control siRNA. After 24 or 48 h, cell extracts were immunoblotted to detect the indicated protein. Cytoplasmic lysates were analyzed for TRAF6 expression by western blotting, and β-actin was used as a loading control. Nuclear lysates were analyzed for P65 by western blotting assay and Lamin B was used as a loading control. Lane 1, TRAF6 siRNA; lane 2, non-transfected; lane 3, control siRNA. *P<0.05 vs. the non-transfected group; **P<0.01 vs. the non-transfected group.
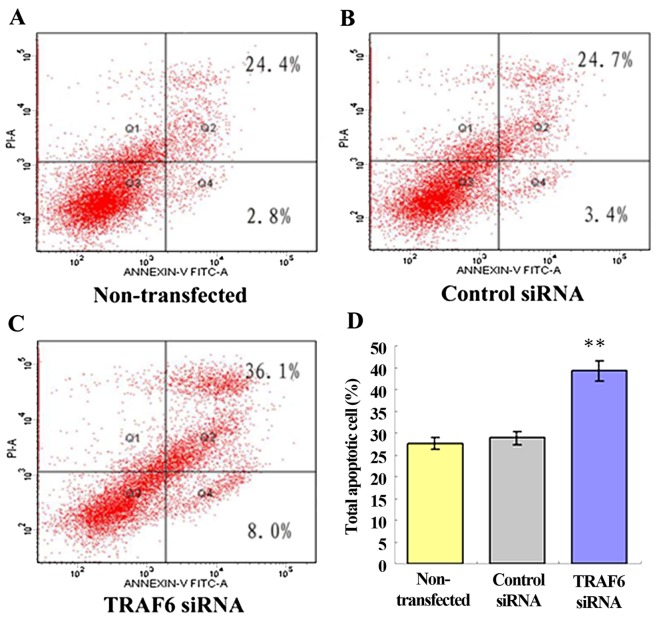
siRNA-induced knockdown of TRAF6 promotes the apoptosis of SPC-A1 cells but does not affect cell proliferation and cell cycle
Following a 48-h transfection, the cell proliferation assay revealed no significant difference in the proliferation of the TRAF6 siRNA-transfected, control siRNA-transfected and non-transfected SPC-A1 cells. Similarly, siRNA-induced knockdown of TRAF6 showed no clear-cut effect on the cell cycle of the SPC-A1 cells (data not shown). After 48 h of transfection, flow cytometry showed a 44.0±0.98% apoptosis rate in the TRAF6 siRNA-transfected SPC-A1 cells, a 27.2±1.86% apoptosis rate in the control siRNA-transfected cells, and a 28.1±1.45% apoptosis rate in the non-transfected cells (Fig. 4). The results demonstrated that siRNA-induced knockdown of TRAF6 resulted in an ~1.5-fold increase in the apoptosis of the SPC-A1 cells (P<0.05).
Figure 4.
Effect of TRAF6 knockdown on the apoptosis of SPC-A1 cells. In the FSC/SSC scattergrams, cancer cells were gated, and the percentage of Annexin V-positive cells in this group of gated cells was determined. PI-negativity indicates early apoptosis, and PI-positivity indicates late apoptosis. (A) Non-transfected group; (B) control siRNA group; (C) TRAF6 siRNA group; (D) percentage of apoptotic cells. siRNA-induced knockdown of TRAF6 resulted in an ~1.5-fold increase in the apoptosis of SPC-A1 cells. These data were analyzed by one-way ANOVA. All experiments were repeated in triplicate. **P<0.05 vs. non-transfected or control siRNA.
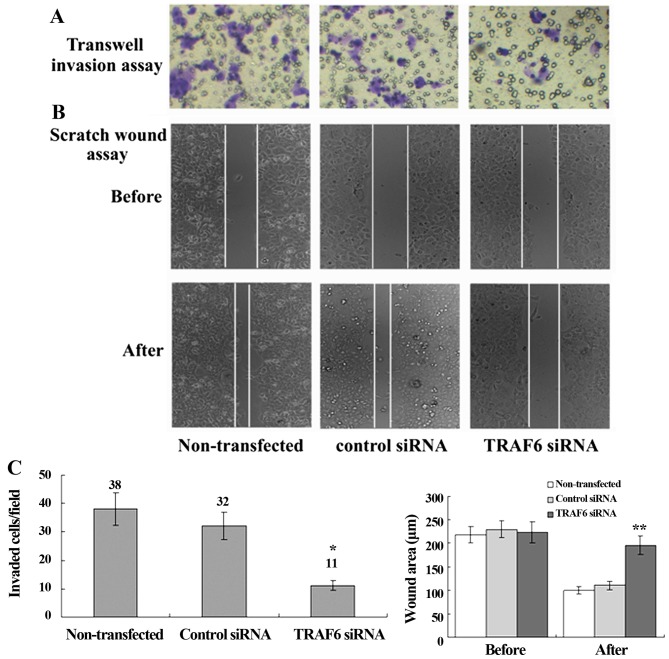
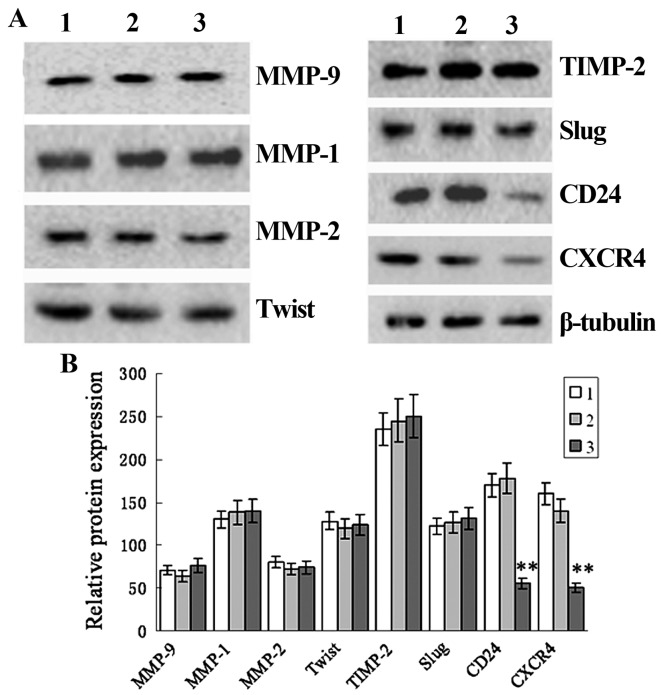
siRNA-induced knockdown of TRAF6 suppresses cell migration and invasion and reduces CD24 and CXCR4 protein expression in SPC-A1 cells
Transwell migration assay revealed a significant reduction in the invasion of the TRAF6 siRNA-transfected SPC-A1 cells as compared to the control siRNA-transfected or non-transfected cells, and scratch wound assay showed a slower migration of the TRAF6 siRNA-transfected cells than the control siRNA-transfected or non-transfected cells (Fig. 5). Western blot analysis revealed a marked reduction in the protein expression of CD24 and CXCR4 in the TRAF6 siRNA-transfected SPC-A1 cells as compared to that in the control siRNA-transfected or the non-transfected cells, while no significant difference was detected in the protein expression of MMP-1, MMP-2, MMP-9, Twist, TIMP-2 or Slug (Fig. 6). These findings demonstrated that siRNA-induced knockdown of TRAF6 reduced CD24 and CXCR4 protein expression in the SPC-A1 cells.
Figure 5.
Effect of TRAF6 knockdown on SPC-A1 cell invasion. Light microscopy images are shown immediately after scratching of the monolayer and 48 h later. Cells moved more slowly over the wound after TRAF6 knockdown. The crystal violet staining indicates the SPC-A-1 cells that passed through the polycarbonate membrane. (A) Transwell invasion assay; (B) scratch wound assay; (C) number of invaded/migrated cells without transfecion, transfected with control siRNA and TRAF6 siRNA. *P<0.05 vs. control siRNA; **P<0.01 vs. control siRNA.
Figure 6.
siRNA-induced TRAF6 knockdown decreases CD24 and CXCR4 expression in the SPC-A1 cells. SPC-A1 cells were transfected by TRAF6 siRNA or control siRNA. After 48 h, cell extracts were immunoblotted to detect the indicated protein. β-actin was used as a loading control. Lane 1, TRAF6 siRNA; lane 2, non-transfected; lane 3, control siRNA. (A) Western blot analysis shows the protein expression of MMP-9, MMP-1, MMP-2, Twist, TIMP-2, Slug, CD24, CXCR4 and β-tubulin. (B) Relative protein expression of CD24 and CXCR4 in non-transfected cells and cells transfected with control siRNA and TRAF6 siRNA. 1, TRAF6 siRNA; 2, non-transfected; 3, control siRNA. **P<0.01 vs. the non-transfected group.
Discussion
As a member of the TRAF family, TRAF6 expression has been shown to be upregulated in cancer cells (14–16). Tumor tissue assay analysis demonstrated that TRAF6 was upregulated in colon cancers when compared with that in adjacent non-cancerous tissues (13); western blotting revealed upregulated TRAF6 protein expression in glioma U251, U-87MG, LN-18, and U373 cell lines as compared to that in the noncancerous human glial SVG p12 cell line (14). Using RT-PCR and western blot analysis, TRAF6 expression was found to be markedly upregulated in osteosarcoma tissues than that in normal bone tissues at both the transcriptional and translational levels (P<0.05) (15), and ELISA assay showed significantly higher serum TRAF6 expression in triple-negative breast cancer patients when compared with that in an obese control group (0.90 vs. 0.73 ng/ml; P=0.033) (17). In the present study, we determined the mRNA and protein expression of TRAF6 in human lung adenocarcinoma A549, non-small cell lung cancer H1650, human airway epithelial Calu-3 and human lung cancer SPC-A1 cell lines, and western blotting and qRT-PCR assays showed clear-cut upregulation of TRAF6 mRNA and protein expression in the Calu-3 and SPC-A1 cells when compared with that in the A549 and H1650 cells. The results are consistent with a previous study reporting significantly higher TRAF6 expression in lung cancer tissues than that in normal lung tissues, as revealed by immunohistochemical detection (18). Our findings further validated the important role of TRAF6 in the development of lung cancer.
TRAF6 contains an amino terminal RING domain that comprises the core of the ubiquitin ligase catalytic domain (10). As signal transducers of Toll/IL-1 family members, TRAF6 is recruited to the receptor complexes and forms oligomers upon ligand binding to IL-1R or Toll-like receptors (TLRs), and TRAF6 oligomerization activates its ligase activity, leading to K63-linked polyubiquitination of targets including TRAF6 itself (23). TRAF6 appears to participate in the activation of NF-κB (24), which plays an important role in the regulation of many genes involved in inflammation, the immune response, cellular proliferation, apoptosis, tumorigenesis and invasion (25–27). It has been shown that TRAF6 is overexpressed and bridges RAS and NF-κB signaling in lung cancer tumors, which is important for RAS-mediated oncogenesis (21). In the present study, K63-linked ubiquitination of TRAF6 was detected in human lung cancer SPC-A1 cells, and EMSA and western blot analysis revealed constitutive NF-κB activation in the SPC-A1 cells, and a marked inhibition of NF-κB activation in the SPC-A1 cells 48 h post-transfection with TRAF6 siRNA, indicating that high expression of TRAF6 is associated with constitutive activation of NF-κB in SPC-A1 cells. Our findings further confirm the oncogenic role of TRAF6 in human lung cancer.
It has been demonstrated that TRAF6 promotes cancer cell proliferation, while knockdown of TRAF6 may decrease cell viability, suppress cell proliferation, invasion and migration, and promote cell apoptosis (14,15). To assess the role of TRAF6 in proliferation, apoptosis, cell cycle, invasion and migration of lung cancer cells, we generated a human lung cancer SPC-A1 cell line in which TRAF6 was depleted by using the technique of RNAi. Subsequently, the effects of TRAF6 knockdown on cell viability, apoptosis, cell cycle, invasion and migration of the SPC-A1 cells were determined using a cell proliferation assay, flow cytometry, Transwell invasion assay and scratch wound assay. Our findings showed that siRNA-induced knockdown of TRAF6 promoted the apoptosis of SPC-A1 cells, but had little effect on cell proliferation or cell cycle. Our results are different from a previous study reporting that depletion of TRAF6 expression with short-hairpin RNA (shRNA) decreased viability, suppressed proliferation and invasion, and promoted apoptosis of human lung adenocarcinoma A549 cells (20). Such a difference may be attributed to the following factors: i) the different cell lines used; and ii) experimental errors. Thus, further studies are required to validate TRAF6 knockdown on the biological behaviors of lung cancers. However, Sun and colleagues (13) also observed little effect of TRAF6 knockdown on the survival of colon cancer cells. In the present study, we found that knockdown of TRAF6 inhibited the migration and invasion of SPC-A1 cells, which was in agreement with the findings reported in human lung adenocarcinoma A549 cells (20). Similar results were observed in colon cancer (13), glioma (14), osteosarcoma (15) and esophageal squamous cells (16).
To address the mechanisms underlying the promotion of SPC-A1 cell apoptosis and inhibition of cell migration and invasion by TRAF6 knockdown, we determined the expression of proteins associated with tumor migration and invasion in TRAF6 siRNA-transfected SPC-A1 cells. Western blot analysis revealed that knockdown of TRAF6 caused a marked reduction in the protein expression of CD24 and CXCR4, but had little effect on the protein expression of MMP-1, MMP-2, MMP-9, Twist, TIMP-2 or Slug. CD24 and CXCR4 are known targets of the NF-κB signaling pathway (12), and NF-κB signaling has been demonstrated to promote cancer cell apoptosis by mediating CD24 and CXCR4 expression (28–30). It is therefore postulated that siRNA-induced knockdown of TRAF6 may affect the apoptosis of SPC-A1 cells by mediating CD24 and CXCR4 expression; however, further studies are required to investigate the exact mechanisms responsible for the promotion of SPC-A1 cell apoptosis induced by TRAF6 knockdown.
In conclusion, the results of the present study demonstrated that TRAF6 is upregulated in human lung cancer cells, and TRAF6 may be involved in the apoptosis, migration and invasion of SPC-A1 cells. In addition, siRNA-induced TRAF6 knockdown inhibits the invasion of lung cancer cells and promotes apoptosis through the NF-κB-CD24/CXCR4 signaling pathway. These findings suggest that TRAF6 may be a promising target for the therapy of lung cancer. However, the present study was designed using an in vitro cell assay. Further studies are required in lung cancer animal models to validate the findings and hypotheses from the present study.
Acknowledgments
The present study was funded by the National Clinical Key Specialty Construction Program, the Medical Innovation Foundation of Fujian Province (grant no. 2011-cxb-16) and the Natural Science Foundation of Fujian Province (grant no. 2013J01284).
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015;65:5–29. doi: 10.3322/caac.21254. [DOI] [PubMed] [Google Scholar]
- 2.Herbst RS, Heymach JV, Lippman SM. Lung cancer. N Engl J Med. 2008;359:1367–1380. doi: 10.1056/NEJMra0802714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.GBD 2013 Mortality and Causes of Death Collaborators Global, regional, and national age-sex specific all-cause and cause-specific mortality for 240 causes of death, 1990–2013: A systematic analysis for the Global Burden of Disease Study 2013. Lancet. 2015;385:117–171. doi: 10.1016/S0140-6736(14)61682-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Miller YE. Pathogenesis of lung cancer: 100 year report. Am J Respir Cell Mol Biol. 2005;33:216–223. doi: 10.1165/rcmb.2005-0158OE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Syrigos KN, Katirtzoglou N, Kotteas E, Harrington K. Adhesion molecules in lung cancer: Implications in the pathogenesis and management. Curr Pharm Des. 2008;14:2173–2183. doi: 10.2174/138161208785740162. [DOI] [PubMed] [Google Scholar]
- 6.Gazdar AF, Minna JD. Angiogenesis and the multistage development of lung cancers. Clin Cancer Res. 2000;6:1611–1612. [PubMed] [Google Scholar]
- 7.Balkwill F. Cancer and the chemokine network. Nat Rev Cancer. 2004;4:540–550. doi: 10.1038/nrc1388. [DOI] [PubMed] [Google Scholar]
- 8.Hodkinson PS, Mackinnon A, Sethi T. Targeting growth factors in lung cancer. Chest. 2008;133:1209–1216. doi: 10.1378/chest.07-2680. [DOI] [PubMed] [Google Scholar]
- 9.Sethi T, Woll PJ. Growth factors and lung cancer. Cancer Treat Res. 1995;72:111–130. doi: 10.1007/978-1-4615-2630-8_5. [DOI] [PubMed] [Google Scholar]
- 10.Bradley JR, Pober JS. Tumor necrosis factor receptor-associated factors (TRAFs) Oncogene. 2001;20:6482–6491. doi: 10.1038/sj.onc.1204788. [DOI] [PubMed] [Google Scholar]
- 11.Chung JY, Park YC, Ye H, Wu H. All TRAFs are not created equal: Common and distinct molecular mechanisms of TRAF-mediated signal transduction. J Cell Sci. 2002;115:679–688. doi: 10.1242/jcs.115.4.679. [DOI] [PubMed] [Google Scholar]
- 12.Hayden MS, Ghosh S. Shared principles in NF-kappaB signaling. Cell. 2008;132:344–362. doi: 10.1016/j.cell.2008.01.020. [DOI] [PubMed] [Google Scholar]
- 13.Sun H, Li X, Fan L, Wu G, Li M, Fang J. TRAF6 is upregulated in colon cancer and promotes proliferation of colon cancer cells. Int J Biochem Cell Biol. 2014;53:195–201. doi: 10.1016/j.biocel.2014.04.010. [DOI] [PubMed] [Google Scholar]
- 14.Peng Z, Shuangzhu Y, Yongjie J, Xinjun Z, Ying L. TNF receptor-associated factor 6 regulates proliferation, apoptosis, and invasion of glioma cells. Mol Cell Biochem. 2013;377:87–96. doi: 10.1007/s11010-013-1573-2. [DOI] [PubMed] [Google Scholar]
- 15.Meng Q, Zheng M, Liu H, Song C, Zhang W, Yan J, Qin L, Liu X. TRAF6 regulates proliferation, apoptosis, and invasion of osteosarcoma cell. Mol Cell Biochem. 2012;371:177–186. doi: 10.1007/s11010-012-1434-4. [DOI] [PubMed] [Google Scholar]
- 16.Han Q, Yao F, Zhong C, Zhao H. TRAF6 promoted the metastasis of esophageal squamous cell carcinoma. Tumour Biol. 2014;35:715–721. doi: 10.1007/s13277-013-1098-z. [DOI] [PubMed] [Google Scholar]
- 17.Bilir C, Engin H, Can M, Likhan S, Demirtas D, Kuzu F, Bayraktaroglu T. Increased serum tumor necrosis factor receptor-associated factor-6 expression in patients with non-metastatic triple-negative breast cancer. Oncol Lett. 2015;9:2819–2824. doi: 10.3892/ol.2015.3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang XL, Dang YW, Li P, Rong MH, Hou XX, Luo DZ, Chen G. Expression of tumor necrosis factor receptor-associated factor 6 in lung cancer tissues. Asian Pac J Cancer Prev. 2014;15:10591–10596. doi: 10.7314/APJCP.2014.15.24.10591. [DOI] [PubMed] [Google Scholar]
- 19.Lin G, Huang C, Su G, Hu H, Xu H, Huang C. Effect of TRAF6 downregulation on malignant biological behavior of lung cancer cell lines. Zhongguo Fei Ai Za Zhi. 2015;18:661–667. doi: 10.3779/j.issn.1009-3419.2015.11.01. In Chinese. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhong L, Cao F, You Q. Effect of TRAF6 on the biological behavior of human lung adenocarcinoma cell. Tumour Biol. 2013;34:231–239. doi: 10.1007/s13277-012-0543-8. [DOI] [PubMed] [Google Scholar]
- 21.Starczynowski DT, Lockwood WW, Deléhouzée S, Chari R, Wegrzyn J, Fuller M, Tsao MS, Lam S, Gazdar AF, Lam WL, et al. TRAF6 is an amplified oncogene bridging the RAS and NF-κB pathways in human lung cancer. J Clin Invest. 2011;121:4095–4105. doi: 10.1172/JCI58818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu H, Zhang T, Ye J, Li H, Huang J, Li X, Wu B, Huang X, Hou J. TNF receptor-associated factor 6 in advanced non-small cell lung cancer: Clinical and prognostic implications. J Cancer Res Clin Oncol. 2012;138:1853–1863. doi: 10.1007/s00432-012-1255-6. [DOI] [PubMed] [Google Scholar]
- 23.Chen ZJ. Ubiquitin signalling in the NF-kappaB pathway. Nat Cell Biol. 2005;7:758–765. doi: 10.1038/ncb0805-758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hoesel B, Schmid JA. The complexity of NF-κB signaling in inflammation and cancer. Mol Cancer. 2013;12:86. doi: 10.1186/1476-4598-12-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.DiDonato JA, Mercurio F, Karin M. NF-κB and the link between inflammation and cancer. Immunol Rev. 2012;246:379–400. doi: 10.1111/j.1600-065X.2012.01099.x. [DOI] [PubMed] [Google Scholar]
- 26.Karin M. Nuclear factor-kappaB in cancer development and progression. Nature. 2006;441:431–436. doi: 10.1038/nature04870. [DOI] [PubMed] [Google Scholar]
- 27.Karin M. NF-kappaB as a critical link between inflammation and cancer. Cold Spring Harb Perspect Biol. 2009;1:a000141. doi: 10.1101/cshperspect.a000141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Esencay M, Newcomb EW, Zagzag D. HGF upregulates CXCR4 expression in gliomas via NF-kappaB: Implications for glioma cell migration. J Neurooncol. 2010;99:33–40. doi: 10.1007/s11060-010-0111-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ju JH, Jang K, Lee KM, Kim M, Kim J, Yi JY, Noh DY, Shin I. CD24 enhances DNA damage-induced apoptosis by modulating NF-κB signaling in CD44-expressing breast cancer cells. Carcinogenesis. 2011;32:1474–1483. doi: 10.1093/carcin/bgr173. [DOI] [PubMed] [Google Scholar]
- 30.LaRocca TJ, Fabris F, Chen J, Benhayon D, Zhang S, McCollum L, Schecter AD, Cheung JY, Sobie EA, Hajjar RJ, et al. Na+/Ca2+ exchanger-1 protects against systolic failure in the Akitains2 model of diabetic cardiomyopathy via a CXCR4/NF-κB pathway. Am J Physiol Heart Circ Physiol. 2012;303:H353–H367. doi: 10.1152/ajpheart.01198.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]