Abstract
5-Aza-2′-deoxycytidine (5-Aza-CdR) is currently acknowledged as a demethylation drug, and causes a certain degree of demethylation in a variety of cancer cells, including pancreatic cancer cells. Emodin, a traditional Chinese medicine (TCM), is an effective monomer extracted from rhubarb and has been reported to exhibit antitumor activity in different manners in pancreatic cancer. In the present study, we examined whether emodin caused demethylation and increased the demethylation of three tumor-suppressor genes P16, RASSF1A and ppENK with a high degree of methylation in pancreatic cancer when combined with 5-Aza-CdR. Our research showed that emodin inhibited the growth of pancreatic cancer Panc-1 cells in a dose- and time-dependent manner. Dot-blot results showed that emodin combined with 5-Aza-CdR significantly suppressed the expression of genome 5mC in PANC-1 cells. In order to verify the effect of methylation, methylation-specific PCR (MSP) and bisulfite genomic sequencing PCR (BSP) combined with TA were selected for the cloning and sequencing. Results of MSP and BSP confirmed that emodin caused faint demethylation, and 5-Aza-CdR had a certain degree of demethylation. When emodin was combined with 5-Aza-CdR, the demethylation was more significant. At the same time, fluorescent quantitative PCR and western blot analysis results confirmed that when emodin was combined with 5-Aza-CdR, the expression levels of P16, RASSF1A and ppENK were increased more significantly compared to either treatment alone. In contrast, the expression levels of DNA methyltransferase 1 (DNMT1) and DNMT3a were more significantly reduced with the combination treatment than the control or either agent alone, further proving that emodin in combination with 5-Aza-CdR enhanced the demethylation effect of 5-Aza-CdR by reducing the expression of meth-yltransferases. In conclusion, the present study confirmed that emodin in combination with 5-Aza-CdR enhanced the demethylation by 5-Aza-CdR of tumor-suppressor genes p16, RASSF1A and ppENK by reducing the expression of methyltransferases DNMT1 and DNMT3a.
Keywords: emodin, pancreatic carcinoma, methyltransferase, demethylation
Introduction
Pancreatic cancer is a highly invasive malignant tumor. Its incidence is roughly close to the mortality rates. At present, surgical resection is still the first treatment choice by which to attempt to cure pancreatic cancer. However, only 20% of patients at the time of diagnosis are resectable; 30–40% of patients with pancreatic cancer are unresectable, even though the tumor is confined to the pancreatic area (1). Tumorigenesis is the result of the interaction between multiple genetic and epigenetic factors and abnormal gene methylation is one of the significant factors. High methylation of tumor-suppressor genes results in transcriptional suppression and even loss of expression; while low methylation may result in extremely active transcription and uncontrolled expression that may lead to abnormal differentiation and proliferation, and the development of cancer (2).
Epigenetic modifications have attracted much attention in the research on the development of tumors. Epigenetic modifications mainly include DNA methylation and hydroxylmethylation, histone methylation and acetylation, chromatin remodeling, genomic imprinting and RNA interference. The formation of DNA methylation is catalyzed by DNA methyltransferases (DNMTs), mainly including DNMT1, DNMT3a and DNMT3b, and 5mC is formed under catalysis of these enzymes (3). DNA methylation modification is closely related to transcriptional inactivation, gene imprinting and X chromosome inactivation (4). Research evidence has shown that DNA methylation plays an important role in the development of many malignant tumors.
Similar to other malignancies, the development and progression of pancreatic carcinoma is a multistage process that involves a series of genetic alterations and gene methylation changes. Research has demonstrated that tumor-suppressor genes P16 and RASSF1A are highly methylated in pancreatic cancer, Ueki et al (5) and Fukushima (6) et al reported that ppENK gene methylation levels were increased in more than 90% of pancreatic cancer cases. Schutte et al (7) reported that the P16 gene was inactivated in 95% of cases, and 15% of these cases were correlated with methylation. Moore et al (8) reported that the P16 gene was methylated in 27% of pancreatic cancer cell lines. Dammann et al (9) reported RASSF1A gene methylation levels were increased in 64% of primary pancreatic ductal carcinoma cells, 83% of pancreatic endocrine tumors and 88% of pancreatic cancer cell lines. When pancreatic cancer cells were treated with the demethylation drug 5-Aza-CdR, expression levels of ppENK, P16 and RASSF1A which were downregulated by methylation respectively had different degrees of re-expression to exert an antitumor efficiency. This provided the theoretical basis for antitumor treatment using demethylation drugs in clinical study.
5-Aza-CdR is currently one of the most commonly used demethylation nucleoside analogues (10), and plays a role in methylation mainly by inhibiting the expression and activity of DNMT under a low concentration. 5-Aza-CdR was approved by the food and drug administration (FDA) to be mainly used for the treatment of blood system tumors. Zhang et al (11) reported that, when pancreatic cancer PANC-1 cells treated with 1 µmol/l 5-Aza-CdR for 72 h, the tumor-suppressor genes RASSF1A, P16 and ppENK had different levels of demethylation. In recent years, Chinese herbal medicines have been recognized as having antitumor efficacy. Emodin (1,3,8-trihydroxy-6-methylanthraquinone), one such Chinese herbal medicine, has extensive pharmacological effects such as immune regulation, antibacterial, anti-inflammatory and antitumor activities (12). Liu et al (13) reported that emodin inhibited pancreatic cancer cell growth through different modes of action, yet the detailed mechanism remained unclear. It was reported that emodin caused a certain degree of demethylation in pancreatic cancer PANC-1 cells, but the demethylation intensity was weaker when compared with 5-Aza-CdR.
The etiology of tumors include multiple factors. Comprehensive treatment is the main treatment mode for tumors at present. Drug combinations are currently an important strategy for antitumor treatment. The aim of the present study was to investigate the demethylation efficacy of emodin in combination with 5-Aza-CdR on pancreatic cancer PANC-1 cells. It was demonstrated that emodin combined with 5Aza-CdR enhanced the demethylation by 5-Aza-CdR alone on tumor-suppressor genes RASSF1A, P16 and ppENK in pancreatic cancer cells by reducing the expression of methyltransferases DNMT1 and DNMT3a. This finding provides a new strategy for the clinical treatment of pancreatic cancer.
Materials and methods
Chemicals and reagents
Emodin (purity ≥98%), 5-Aza-CdR and dimethylsulfoxide (DMSO) were purchased from Sigma (St. Louis, MO, USA). Emodin was dissolved in DMSO to create a stock solution at concentrations of 10 and 20 mmol/l, which were stored at −70°C. The DMSO concentration was maintained below 0.1% in all of the cell cultures and did not exert any detectable effect on cell growth or cell death. The Cell Counting Kit-8 (CCK-8) was purchased from Gibco. A cell and tissue genomic DNA extraction kit, methylation and FQ-PCR primers were purchased from Fastagen Biotech (Shanghai, China). The EpiTect® Bisulfite and EpiTect® methylation-specific PCR (MSP) kits were purchased from Qiagen. The RNA extraction kit was purchased from Tiangen (Beijing, China). SYBR-Green fluorescent dye was purchased from Invitrogen. The antibodies, anti-RASSF1A and anti-ppENK were purchased from Abcam. The anti-P16/INK4a antibody and anti-β-actin were purchased from Epitomics.
Cell line and culture
Human pancreatic cancer cell line PANC-1 was obtained from the American Type Culture Collection (ATCC; Manassas, VA, USA). The cells were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 100 U/ml penicillin and 100 µg/ml streptomycin. Cells were maintained at 37°C in a humidified atmosphere of 5% CO2. The medium was changed every 2–3 days, and the cells were subcultured when confluency reached 70–80% in 0.25% trypsin at 37°C.
Cell proliferation assay
Cell survival was determined using the CCK-8 kit. Briefly, logarithmic phase PANC-1 cells were plated into 96-well culture plates (~5×103 cells/well). After 24 h of incubation, the cells were treated with the vehicle alone (0.1% DMSO) and various concentrations (10, 20, 40 and 80 µM) of emodin, followed by a 24-, 48- and 72-h cell culture. Each group had 6-wells. A total of 10 µl CCK-8 was added to each well for 1 h before the end of the incubation period. The absorbance at 450 nm was read using the Bio-Tek ELx800 absorbance microplate reader. The experiment was repeated three times. To determine the inhibition rate of the drugs on the cells, the following calculation was performed: Relative % inhibition = 1 − (dosing absorbance - blank absorbance)/(control absorbance − blank absorbance) × 100%.
Dot-blot assay
The PANC-1 cells were treated with the optimal concentration of emodin (40 µM) and 5-Aza-CdR (1 µM), either alone or in combination for 72 h. The control cells were treated with 0.1% DMSO only. The total DNA was isolated from the cultured cells using the cell/tissue genomic DNA extraction kit according to the manufacturer's instructions. The concentrations of DNA were determined by fluorometry using the Qubit® dsDNA HS kit and fluorom-eter (both from Invitrogen). The procedure for the Dot-blot assay was performed with reference to a previous study (14). First, DNA in each group was placed on a nylon membrane (Hybond-N+; GE), and put in ultraviolet light-emitting instruments for 5 min, after being blocked for 1.5 h in 5% skim milk and washed with ice-cold phosphate-buffered saline (PBS) for 5 min. The membrane was then incubated with the desired primary antibody overnight at 4°C. The membrane was then treated with the appropriate peroxidase-conjugated secondary antibody for 1 h at room temperature, and the immune complexes were observed using an enhanced chemiluminescence reagent followed by analysis of the grey value of the dot with ImageJ software.
Methylation-specific PCR (MSP)
The PANC-1 cells were treated with the optimal concentration of emodin (40 µM) and 5Aza-CdR (1 µM), either alone or in combination for 72 h. The control cells were treated with 0.1% DMSO only. Genomic DNA was extracted using the cell/tissue genomic DNA extraction kit according to the manufacturer's instructions. When the DNA was concentrated to 1 µg, bisulfite modification of genomic DNA was performed using the EpiTect® Bisulfite kit. The methylation and unmethylation primer sequences for P16, RASSF1A and ppENK are shown in Table I. Bisulfite-modified DNA (4 µl), the methylation (3 µl) and unmethylation primer (3 µl), 2X Taq PCR Master Mix (25 µl) and RNase-free water (17 µl) were added to achieve a final volume of 50 µl. PCR amplification conditions were as follows: 95°C for 10 min, 94°C for 30 sec, annealing for 30 sec, and extension at 72°C for 45 sec; a total of 35 cycles; followed by a final extension at 72°C for 10 min. A total of 10 µl of the PCR product was separated using 2.5% agarose gel electrophoresis including GoldView I type of nucleic acid stain for 45 min, and the results were photographed and analyzed.
Table I.
Primer sequences for PCR, MSP and bisulfite sequencing.
| Genes | Primer pairs (5′-3′) | Product size (bp) | |
|---|---|---|---|
| P16 | F | GCCGATCCAGGTCATGATGAT | |
| R | GCATCTATGCGGGCATGGTTA | 300 | |
| RASSF1A | F | TGGGGAGGTGAACTGGGAC | |
| R | ACACGGCACGCACTTGG | 217 | |
| ppENK | F | GCGGTTCCTGACACTTTGC | |
| R | GGGTGCTGGTGCCATCTT | 245 | |
| DNMT1 | F | GACCCATCTCTTGAAGGTGGTGTT | |
| R | CCTCGTCATAACTCTCCACCTGCT | 164 | |
| DNMT3a | F | AGGTGGACCGCTACATTGCC | |
| R | GAGATGTCCCTCTTGTCACTAACG | 143 | |
| P16 | MF | TTATTAGAGGGTGGGGCGGATCGC | |
| MR | GACCCCGAACCGCGACCGTAA | 150 | |
| UF | TTATTAGAGGGTGGGGTGGATTGT | ||
| UR | CAACCCCAAACCACAACCATAA | 151 | |
| RASSF1A | MF | GTGTTAACGCGTTGCGTATC | |
| MR | AACCCCGCGAACTAAAAACGA | 93 | |
| UF | TTTGGTTGGAGTGTGTTAATGTG | ||
| UR | CAAACCCCACAAACTAAAAACAA | 105 | |
| ppENK | MF | TGTGGGGAGTTATCGAGC | |
| MR | GCCTTCGCGAAAAAAATCG | 96 | |
| UF | TTGTGTGGGGAGTTATTGAGT | ||
| UR | CACCTTCACAAAAAAAATCAATC | 100 | |
| P16 | BS-F | TTGTTGTTTAGGTTGGAGTGTAGTG | |
| BS-R | TCAAAAACATATATTAATAACAACCATCAA | 256 | |
| ppENK | BS-F | AAAGAGTTTTTGGAAATAGGGGATA | |
| BS-R | CATCAACAATTTCCCACTAAAAAAT | 241 | |
| RASSF1A | BS-F | GTATGTAAGGGTTGGATGTGTAGAGA | |
| BS-R | CCCCAAATAAAATCTCCACAAAAATC | 298 |
MSP, methylation-specific PCR; F, forward; R, reverse; M, methylated; U unmethylated; BS, bisulfite sequence.
Bisulfite sequencing PCR (BSP)
The PANC-1 cells were treated with the optimal concentration of emodin (40 µM) and 5Aza-CdR (1 µM), either alone or in combination for 72 h. The control cells were treated with 0.1% DMSO only. Genomic DNA was extracted using the cell/tissue genomic DNA extraction kit according to the manufacturer's instructions, which were respectively modified by sulfite. The modified DNA was used for BSP. The sequences of the primers for P16, RASSF1A and ppENK are shown in Table I. Template DNA (4 µl), primers (each 1.5 µl), 2X Taq PCR Master Mix (12.5 µl) and DEPC-H2O (5.5 µl) were added to achieve a final volume of 25 µl. PCR amplification conditions were as follows: 95°C for 5 min, 94°C for 30 sec, annealing for 45 sec, and extension at 72°C for 45 sec; a total of 40 cycles; followed by a final extension at 72°C for 10 min. A total of 10 µl of the PCR product was separated using 2% agarose gel electrophoresis, and the results were photographed. BSP products were extracted from agarose gel including GoldView I type of nucleic acid stain for 45 min, then purified and sequenced (ShangHai Maipu Biotechnology Co., Ltd., China). The methylation of the sample was analyzed using BiQ Analyzer software.
Fluorescent quantitative PCR (FQ-PCR)
The PANC-1 cells were treated with the optimal concentration of emodin (40 µM) and 5Aza-CdR (1 µM), either alone or in combination for 72 h. The control cells were treated with 0.1% DMSO only. Total RNA was isolated from the cells using TRIzol reagent according to the manufacturer's instructions. Quantitative analysis of RNA was carried out by ELISA and the integrity of the RNA was verified by agarose gel electrophoresis. For reverse transcriptase analysis, 2 µg of total RNA was reversely transcribed using the RevertAid™ First Strand cDNA Synthesis kit. Cell cDNA was diluted three times with DEPC. P16, RASSF1A, ppENK and GAPDH primers were diluted with 1X TBE buffer at 10 pmol/ml. PCR amplification with 10 µl of the reaction system [prepared cDNA 1 µl and diluent primers (upstream and downstream primers each 1 µl), fluorescence PCR water, SYRB-Green 5 µl] was performed with SYBR-Green PCR Master Mix-Plus. Results were analyzed with LightCycler 480 software. All samples were performed in triplicate, and the relative amount of the target gene was normalized to GAPDH.
Western blot analysis
The PANC-1 cells were treated with the optimal concentration of emodin (40 µM) and 5Aza-CdR (1 µM), either alone or in combination for 72 h. The control cells were treated with 0.1% DMSO only. Total proteins were extracted from the cells using precooling lysis buffer (RIPA:PMSF=100:1), and placed on ice for 20 min, and treated with ultrasound (150 W) for 5 sec for an average of three times. After centrifugation at 120,000 x g for 5 min at 4°C, the supernatant was collected and the protein concentration was determined using the BCA protein assay kit according to the manufacturer's instructions. The protein lysates (20 µg/lane) were separated on 10% SDS polyacrylamide gel and transferred onto a nitrocellulose membrane. Each membrane was blocked with 5% skim milk and then incubated with the indicated primary antibodies against P16, RASSF1A, ppENK, DNMT1, DNMT3a, DNMT3b and β-actin overnight at 4°C. Subsequently, the membrane was incubated with the secondary antibodies, goat anti-rabbit and anti-mouse IgG conjugated with HRP, for 1 h at room temperature, and the formed immunocomplex was visualized by enhanced chemiluminescence reagent and exposed to X-ray film. Quantitative data are expressed as a percentage of the mean ± standard deviation (SD) of the relative levels of the objective protein and control β-actin of each group of cells from three independent experiments.
Statistical analysis
All results were repeated in at least three separate experiments. The data are expressed as the means ± SD. Statistical comparisons were carried out using one-way analysis of variance, which revealed significant differences between groups, and a Student's t-test which revealed significant differences between two sample means. Statistical analyses were carried out using SPSS version 17.0 software (SPSS, Inc., Chicago, IL, USA). P<0.05 was considered to indicate a statistically significant difference.
Results
Effect of emodin on PANC-1 cell proliferation
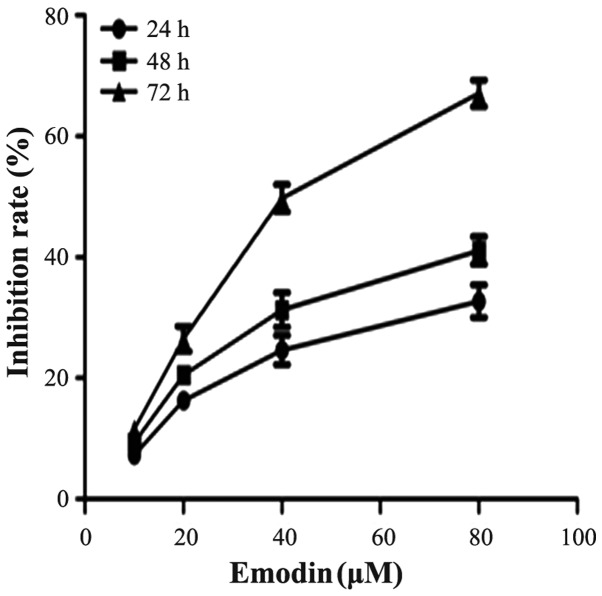
To investigate the effect of emodin on cell growth, PANC-1 cells were cultured with 0, 10, 20, 40 and 80 µM emodin for 24, 48 and 72 h. Cell proliferation was determined by the CCK-8 assay. As demonstrated in Fig. 1, emodin was shown to inhibit the growth of the cells in a dose- and time-dependent manner. The inhibition rate of emodin at a concentration of 40 µM for 72 h was 50.6%, thus 40 µM was close to the half maximal inhibitory concentration (IC50). This result was similar to a previous study (11). Therefore, emodin at a concentration of 40 µM was used for the following experimental research.
Figure 1.
Effects of emodin on the inhibition of the growth of pancreatic cancer (PANC-1) cells in vitro. PANC-1 cells were treated with vehicle or 10, 20, 40 and 80 µM emodin for 24, 48 and 72 h. The cell proliferation inhibition rates were determined by the Cell Counting Kit-8 (CCK-8) assay, and are presented as the means ± SD from three independent experiments.
Effects of emodin and 5-Aza-CdR on the level of genomic 5mC
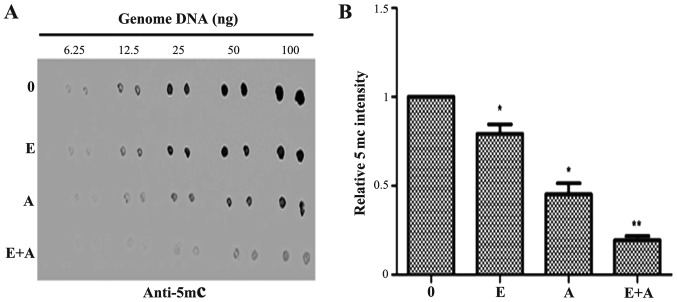
Dot-blot assay was performed to determine the effects of emodin (40 µM) and 5-Aza-CdR (1 µM), alone or in combination on the genomic DNA methylation level in PANC-1 cells. As shown in Fig. 2, emodin (40 µM) and 5-Aza-CdR (1 µM) exhibited an inhibitory effect, respectively and reduced the 5mC level as compared with the level in the control group. The effects observed in the 5-Aza-CdR group were more evident than that in the emodin group. The 5mC level was significantly decreased when the PANC-1 cells were treated with a combination of emodin and 5-Aza-CdR.
Figure 2.
Effect of emodin and 5-Aza-CdR on the levels of genome 5mC in PANC-1 cells. The PANC-1 cells were treated with emodin (40 µM) or 5-Aza-CdR (1 µM) alone or in combination for 72 h, and the levels of 5mC were detected by dot-blot assay. (A) Emodin or 5-Aza-CdR alone or in combination reduced the expression of 5mC. (B) Quantification was performed by calculating the ratio of the experimental value to the control. *P<0.05, **P<0.01 vs. control or cells treated with 5-Aza-CdR alone. 0, the control group; E, 40 µM emodin group; A, 1 µM 5-Aza-CdR group, E+A, combination group.
Effects of emodin and 5-Aza-CdR on the methylation of tumor-suppressor genes P16, RASSF1A and ppENK
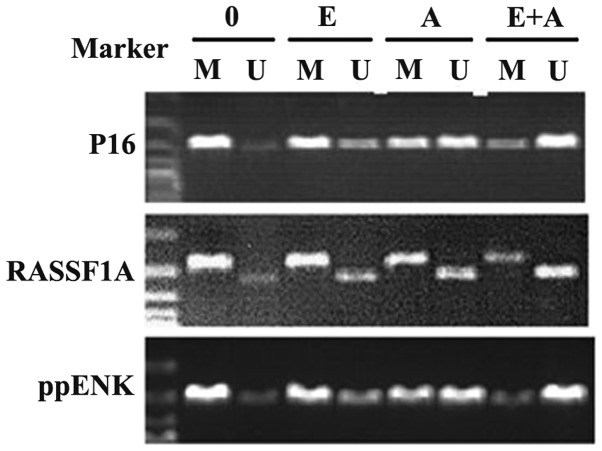
It was reported that methylation of tumor-suppressor genes P16, RASSF1A and ppENK played an important role in the pathogenesis of pancreatic cancer. In order to further clarify the regulatory effect of emodin alone and in combination with 5-Aza-CdR on the methylation of these genes, the MSP assay was used to detect the methylation status of these genes. As shown in Fig. 3, P16, RASSF1A and ppENK presented a high degree of methylation and a faint demethylation state in the pancreatic cancer PANC-1 cells. When the PANC-1 cells were treated with emodin and 5-Aza-CdR alone, the methylation showed different degrees of decline. Simultaneously, demethylation was enhanced at different degrees. In addition, the effect of 5-Aza-CdR was stronger than the effect of emodin. When the PANC-1 cells were treated with emodin in combination with 5-Aza-CdR, methylation of the three tumor-suppressor genes P16, RASSF1A and ppENK was significantly reduced to a greater degree and the demethylation was significantly enhanced when compared with the effect of emodin or 5-Aza-CdR alone. These data showed that emodin or 5-Aza-CdR alone exhibited a certain methylation effect on the P16, RASSF1A and ppENK genes, while the combination of emodin and 5-Aza-CdR had a significantly enhanced methylation effect on these genes.
Figure 3.
Effect of emodin and 5-Aza-CdR on the methylation of tumor-suppressor genes P16, RASSF1A, ppENK in PANC-1 cells. The PANC-1 cells were treated with emodin (40 µM) and 5-Aza-CdR (1 µM) alone or in combination for 72 h, and the methylation of these genes was detected by MSP assay. Emodin and 5-Aza-CdR alone or in combination was observed to weaken the methylation band and gradually strengthened the demethylation band of P16, RASSF1A and ppENK. M, methylation; U, demethylation; 0, control group; E, 40 µM emodin group; A, 1 µM 5-Aza-CdR group; E+A, combination group.
Effects of emodin and 5-Aza-CdR on the methylation of tumor-suppressor genes P16, RASSF1A and ppENK
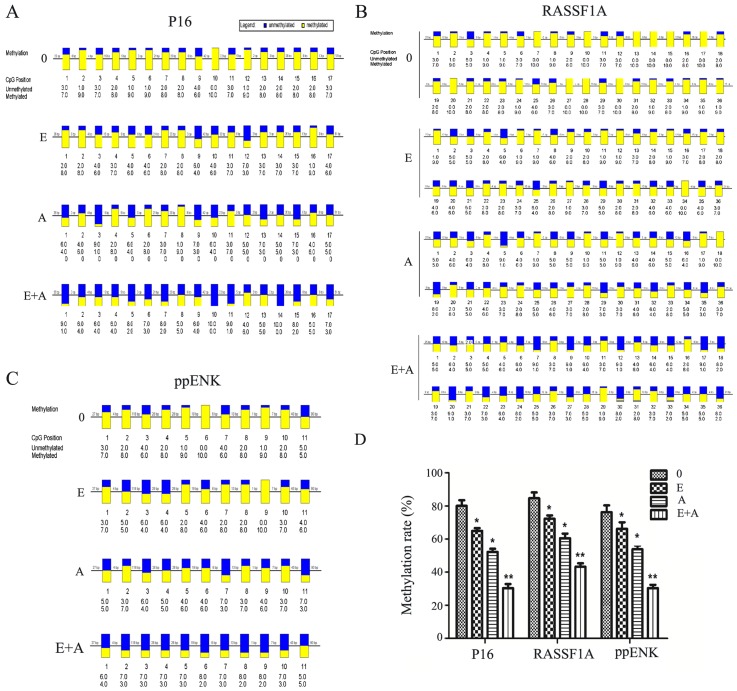
In order to verify the reliability of the above results, the BSP method was used to verify the methylation levels of the three tumor-suppressor genes P16, RASSF1A and ppENK. As shown in Fig. 4, when the PANC-1 cells were treated with emodin or 5-Aza-CdR, alone or in combination for 72 h, the methylation levels of the three tumor-suppressor genes P16, RASSF1A and ppENK were reduced and the demethylation levels were increased compared with the controls. Scopes of P16, ppENK and RASSF1A gene sequencing were respectively composed of 17, 11 and 36 CpG islands, 10 of which were randomly selected to clone and sequence. The methylation rates of the P16 gene were 80, 66.5, 51.8 and 30%, respectively; the meth-ylation rates of the ppENK gene were 76.4, 66.4, 53.6 and 30%, respectively; the methylation rates of the RASSF1A gene were 85, 72.7, 60.3 and 43.6%, respectively. These results further confirmed the above results that emodin and 5-Aza-CdR decreased the methylation levels and increased the demethylation levels of the promoter region of key tumor-suppressor genes RASSF1A, P16 and ppENK in pancreatic cancer. The effect was more apparent when a combination of both were used, indicating that emodin enhanced the demethylation efficiency of 5-Aza-CdR on tumor-suppressor genes RASSF1A, P16 and ppENK.
Figure 4.
Effect of emodin and 5-Aza-CdR on the methylation levels of the promoter region of P16, RASSF1A and ppENK in PANC-1 cells. (A-C) PANC-1 cells were treated with emodin (40 µM) and 5-Aza-CdR (1 µM) alone or in combination for 72 h, and methylation of these genes in the promoter region was detected by BSP. (D) The methylation rates (%) are presented as the mean ± SD of three independent experiments. *P<0.05, **P<0.01 vs. control or cells treated with 5-Aza-CdR alone. Yellow indicates methylation, blue indicates unmethylation; 0, control group; E, 40 µM emodin group; A, 1 µM 5-Aza-CdR group; E+A, combination group.
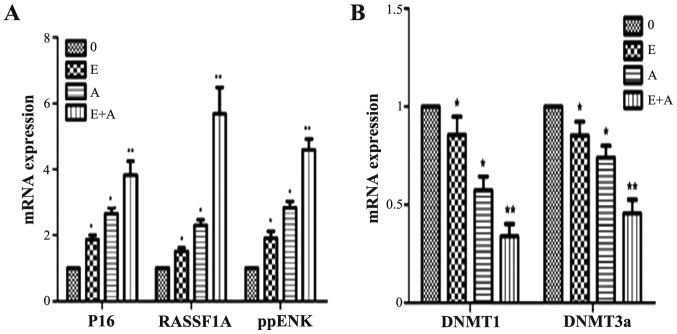
Effects of emodin and 5-Aza-CdR on the mRNA expression levels of tumor-suppressor genes P16, RASSF1A, ppENK and DNMTs
In order to demonstrate whether emodin and 5Aza-CdR have an effect at the transcription level, PCR was used to detect gene transcription levels of tumor-suppressor genes P16, RASSF1A, ppENK and DNMTs. As shown in Fig. 5, when the PANC-1 cells were treated with emodin or 5Aza-CdR alone or in combination for 72 h, compared with the control group, the expression levels of P16, RASSF1A and ppENK were increased to different degrees. In addition, the effect of 5-Aza-CdR was stronger than the effect of emodin. When the PANC-1 cells were treated with emodin in combination with 5-Aza-CdR, the mRNA expression levels of P16, RASSF1A and ppENK were more significantly enhanced when compared with the levels following treatment with emodin or 5-Aza-CdR alone. In contrast, mRNA expression levels of DNMT1 and DNMT3a were reduced at different degree. The effect was the most obvious when a combination was used. However, the mRNA expression levels of DNMT3b had no change (data not shown).
Figure 5.
Effects of emodin and 5-Aza-CdR on the mRNA expression of P16, RASSF1A, ppENK, DNMT1 and DNMT3a in the PANC-1 cells. (A) FQ-PCR analysis showed that emodin or 5-Aza-CdR alone or in combination increased the mRNA expression levels of P16, RASSF1A and ppENK. (B) FQ-PCR analysis showed that emodin or 5-Aza-CdR alone or in combination decreased the mRNA expression levels of DNMT1 and DNMT3a. *P<0.05, **P<0.01 vs. control or cells treated with 5-Aza-CdR alone. 0, the control group; E, 40 µM emodin group; A, 1 µM 5-Aza-CdR group; E+A, combination group.
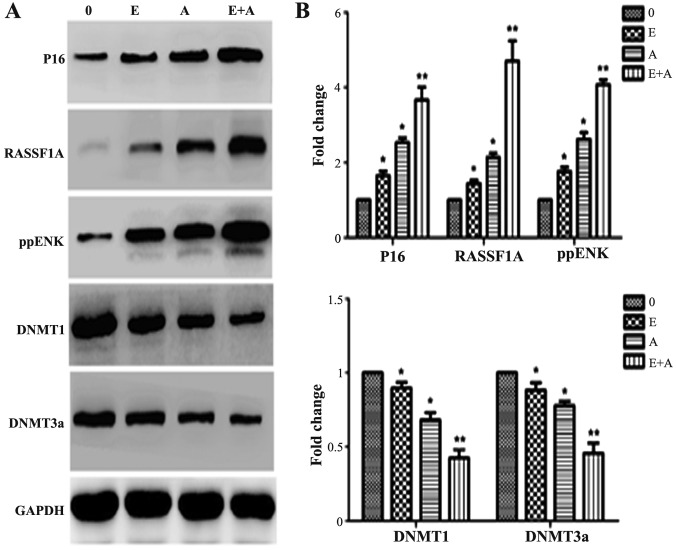
Effects of emodin and 5-Aza-CdR on the protein expression levels of tumor-suppressor genes P16, RASSF1A, ppENK and DNMTs
In order to further demonstrate the effects of emodin and 5-Aza-CdR at the protein level. western blotting was used to detect protein levels of P16, RASSF1A, ppENK and DNMTs. As shown in Fig. 6, when the PANC-1 cells were treated with emodin or 5-Aza-CdR alone or in combination for 72 h, compared with control group, the protein expression of P16, RASSF1A and ppENK was significantly increased. When the PANC-1 cells were treated with emodin in combination with 5-Aza-CdR, the protein expression levels of P16, RASSF1A and ppENK were more significantly enhanced than these levels following treatment with emodin or 5-Aza-CdR alone. At the same time, the expression levels of DNMT1 and DNMT3a were reduced; the expression levels of DNMT1 and DNMT3a were more significantly decreased when a combination of emodin and 5-Aza-CdR was used when compared to treatment with each agent alone. These findings were consistent with the FQ-PCR results.
Figure 6.
Effects of emodin and 5-Aza-CdR on the protein expression levels of P16, RASSF1A, ppENK, DNMT1 and DNMT3a in the PANC-1 cells. (A) Protein expression levels were detected by western blot analysis. Protein expression levels of P16, RASSF1A and ppENK were increased, while protein expression levels of DNMT1 and DNMT3a were decreased. The effect of emodin in combination with 5-Aza-CdR was stronger than the effect of either agent alone in the above experiments. (B) The quantification was performed assigning a value of 1 to the control group. The results obtained from three separate experiments are expressed as mean ± SD. *P<0.05, **P<0.01 vs. control or cells treated with 5-Aza-CdR alone. 0, the control group; E, 40 µM emodin group; A, 1 µM 5-Aza-CdR group; E+A, combination group.
Discussion
In recent years, the incidence of pancreatic cancer has increased yearly. Most patients have advanced disease at the time of diagnosis and hence a dismal prognosis. Gemcitabine has been the standard systemic therapy for the palliative treatment of pancreatic cancer over the last decade, although the 1-year survival rate of ~20% remains unsatisfactory. Evidence suggests that many types of cancer are associated with epigenetic changes and gene mutations, which initiate the process of tumor development. Epigenetic changes refer to heritable changes in gene expression (active versus inactive genes) that do not involve changes to the underlying DNA sequence; a change in phenotype without a change in genotype. DNA methylation is one of the important gene epigenetic modifications. DNA methylation is catalyzed by DNA methyltransferase (DNMT) and S-adenosine methionine (SAM) as a methyl donor which is transferred to a specific base, that does not change the DNA sequences; the genetic code is reversible. DNMTs mainly consist of DNMT1, DNMT3a and DNMT3b. New synthetic single-strand DNA is methylated by DNMT1, and methylation information is transmitted to daughter cells; while DNMT3a and DNMT3b are new methyltransferases for which DNA methylation patterns are established in the process of embryonic development, and are involved in the generation of methylation (15). High expression of DNMTs in tumors results in the high methylation of tumor-suppressor genes (TSGs) and inactivation, and eventually promotes tumorigenesis (16). In the oncogenome, the demethylation of gene promoter CpG islands promote the expression of TSGs, thus they are likely to become new targets for the gene therapy of pancreatic cancer.
The P16 gene can encode the inhibitional proteins of cell cyclin-dependent kinase 4 (CDK4) and plays an important role in cell cycle regulation. P16 is a type of negative regulatory factor, that inhibits the expression of transcription factors, DNA replication and tumorigenesis. In nude mouse transfection experiments, high CpG island of P16 gene was heavily methylated, with loss of expression (17). Peng et al (18) reported that the P16 and APC genes were highly methylated in patients with pancreatic ductal adenocarcinoma. Yang et al (19) analyzed and evaluated the expression and methylation of the P16 gene in 46 cases of human pancreatic cancer and adjacent tissues. The results showed that the rate of P16 protein expression in pancreatic cancer was 41.3% (19/46), and that in the adjacent tissues was 95.7% (44/46); the difference was statistically significant. Moreover, cytosine methylation was not detected in 19 cases of P16 protein-positive pancreatic cancer specimens, while cytosine methylation was detected in 18 cases of 27 cases of p16 protein-negative specimens; the methylation rate was 39.1%. This was in accordance with a previous study (20). When 25 cases of pancreatic cancer were compared with normal tissues, P16 expression was decreased in 80%, 65% of which was caused by the high methylation of the promoter. RASSF1A, whose encoded protein mainly acts on the cell signaling transduction pathways related to Ras proteins, induces cell apoptosis and blocks the accumulation of endogenous cyclin D1 and D3 and inhibits cell malignant transformation via combined with the Ras protein. Shimizu et al (21) reported that in the tumor tissue of hamster pancreatic ductal adenocarcinoma, the expression of RASSF1A was significantly less than that in the normal tissue. When 4 groups of significantly decreased tissues were used to study the methylation status, it was found that the 5′-terminal CpG island of the RASSF1A gene was almost highly methylated, and not mutated. Dammann et al (9) found that in 64% of primary pancreatic cancers and in 83% of pancreatic endocrine tumor cell lines, the RASSF1A promoter was highly methylated. Ueki et al (5) reported that the ppENK gene contained 42 CpG islands, and the methylation rate of seven CpG islands in a pancreatic cancer cell line was 93%, while no methylation was noted in normal pancreatic tissues, showing that abnormal methylation and transcription inhibition of ppENK are ordinary incidents in the development of pancreatic cancer. Fukushima et al (6) found 14 cases of ppENK methylation in 15 cases of invasive ductal adenocarcinoma, while no methylation was noted in the nontumor pancreas epithelium. In Panc-l cells, the ppENK methylation ratio increased with higher classification, suggesting that ppENK methylation was a middle or late event in the development of pancreatic cancer. However, compared with other diseases, ppENK methylation was more easily detected in pancreatic cancer. Thus it can be used as a potential malignant biomarker of the pancreatic epithelial cells.
5-Aza-CdR is currently one of the most commonly used demethylation nucleoside analogues, and plays a role in demethylation mainly through inhibiting the expression and activation of DNMTs at a low concentration, yet resulted in many side-effects. Drug toxicity and bone marrow suppression are the most obvious side-effects, which limit its application in the clinic. Thus, the identification of drugs with good specificity, high safety, few side-effects and demethylation effects has become an urgent task. According to Zhang et al (11), emodin caused a certain degree of methylation in pancreatic cancer PANC-1 cells, while the degree of demethylation was weaker than that of 5-Aza-CdR. At present, drug combinations are the main mode for the treatment of tumors. Thus, we aimed to ascertain whether the effect of emodin combined with 5-Aza-CdR on pancreatic cancer cells would be stronger than each agent alone. In the present study, we found that emodin inhibited the growth of pancreatic cancer cells in a time- and dose-dependent manner. When the PANC-1 cells were treated with 80 µM emodin for 72 h, the growth inhibition rate was 65.2%, and there was a significant change in cell morphology. These findings were consistent with a previous study by Lin et al (22). When the PANC-1 cells were treated with 40 µM emodin for 72 h, the growth inhibition rate was 50.6%, and there was no significant change in cell morphology. Thus, emodin at a concentration of 40 µM was used in the following experiment. Dot-blot assay showed that emodin at a concentration of 40 µM or 5Aza-CdR at 1 µM reduced the 5mC level when compared with the 5mC level in the control group. Yet, emodin (40 µM) in combination with 5Aza-CdR (1 µM) significantly reduced the 5mC level, showing that emodin enhanced the demethylation effect by 5-Aza-CdR on pancreatic cancer cells, further causing the downregulation of 5mC expression. BSP assay was used to further verify the result of our experiment and showed that emodin combined with 5-Aza-CdR caused demethylation of the P16, RASSF1A and ppENK genes; the drug combination was more effective than either drug alone, consistent with the results of the MSP assay. The loss of expression of tumor-suppressor genes is often caused by methylation. The expression is inversely proportional to the methylation density of the CpG island, and the loss of expression of 67–90% of tumor-suppressor genes are caused by low levels of methylation. Moreover, complete loss of expression of tumor-suppressor genes are caused by high density methylation of CpG islands (23). The FQ-PCR results also confirmed that the re-expression of P16, RASSF1A and ppENK genes, inactivated by methylation, was stronger when emodin was used in combination with 5-Aza-CdR. Moreover, the western blotting results were consistent with the FQ-PCR results, further demonstrating that emodin in combination with 5-Aza-CdR enhanced the demethylation effect of 5-Aza-CdR on pancreatic cancer cell tumor-suppressor genes RASSF1A, P16 and ppENK.
In vivo, methylation is mainly catalyzed by methyltransferases (DNMT1, DNMT3a and DNMT3b), and inhibition of methyltransferase activity and reduction in methyltransferase expression are the two main strategies for the demethylation of tumor suppressor genes. According to the experimental results of FQ-PCR and WB, emodin combined with 5-Aza-CdR significantly reduced the expression of DNMT1 and DNMT3a, and the effect was stronger than either agent alone. Therefore, we speculated that emodin combined with 5-Aza-CdR could enhance the demethylation by 5-Aza-CdR of pancreatic cancer cell tumor-suppressor genes P16 and RASSF1A by reducing the expression of methyltransferase DNMT1 and DNMT3a. In addition, whether emodin and 5-Aza-CdR have effects on methyltransferase activity needs further investigation. Our research is mainly confined to the cellular level, and whether emodin and 5-Aza-CdR also play a role in demethylation in vivo also requires further research.
In summary, Dot-blot assay confirmed that emodin combined with 5-Aza-CdR reduced the genomic 5mC level and the effect was stronger than either agent used alone. Emodin combined with 5-Aza-CdR had a demethylation effect on tumor-suppressor genes RASSF1A, P16 and ppENK in pancreatic cancer PANC-1 cells as confirmed by the results of MSP and BSP assays. FQ-PCR and western blotting results further confirmed that emodin combined with 5-Aza-CdR increased the expression of p16, RASSF1A and ppENK, showing that emodin enhanced the demethylation by 5Aza-CdR on pancreatic cancer Panc-1 cells by inhibiting the expression of DNMT1 and DNMT3a. These findings provide new insight into the clinical treatment of pancreatic cancer with the understanding that the epigenetic demethylation pathway is an important mechanism of action.
Acknowledgments
We are grateful for the financial support from the Zhejiang Medical Science and Technology Project (grant no. 2016KYB286), the key subject of Jiaxing Medicine (General Surgery, grant no. 04-F-15), the Jiaxing Science and Technology Projects (grant no. 2013AY21042-5), the Jiaxing Science and Technology Innovation Team Project (grant no. 2013-03), the Administration of Traditional Chinese Medicine of Zhejiang, China (grant no. 2013ZQ026), the Science and Technology Development Program of Hangzhou (grant no. 20140733Q34), and the Jiaxing Science and Technology Projects (grant no. 2015C23012).
References
- 1.Saif MW. Pancreatic neoplasm in 2011: An update. JOP. 2011;12:316–321. [PubMed] [Google Scholar]
- 2.Miranda TB, Jones PA. DNA methylation: The nuts and bolts of repression. J Cell Physiol. 2007;213:384–390. doi: 10.1002/jcp.21224. [DOI] [PubMed] [Google Scholar]
- 3.Cheng X, Blumenthal RM. Mammalian DNA methyltransferases: A structural perspective. Structure. 2008;16:341–350. doi: 10.1016/j.str.2008.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 5.Ueki T, Toyota M, Skinner H, Walter KM, Yeo CJ, Issa JP, Hruban RH, Goggins M. Identification and characterization of differentially methylated CpG islands in pancreatic carcinoma. Cancer Res. 2001;61:8540–8546. [PubMed] [Google Scholar]
- 6.Fukushima N, Sato N, Ueki T, Rosty C, Walter KM, Wilentz RE, Yeo CJ, Hruban RH, Goggins M. Aberrant methylation of preproenkephalin and p16 genes in pancreatic intraepithelial neoplasia and pancreatic ductal adenocarcinoma. Am J Pathol. 2002;160:1573–1581. doi: 10.1016/S0002-9440(10)61104-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schutte M, Hruban RH, Geradts J, Maynard R, Hilgers W, Rabindran SK, Moskaluk CA, Hahn SA, Schwarte-Waldhoff I, Schmiegel W, et al. Abrogation of the Rb/p16 tumor-suppressive pathway in virtually all pancreatic carcinomas. Cancer Res. 1997;57:3126–3130. [PubMed] [Google Scholar]
- 8.Moore PS, Sipos B, Orlandini S, Sorio C, Real FX, Lemoine NR, Gress T, Bassi C, Klöppel G, Kalthoff H, et al. Genetic profile of 22 pancreatic carcinoma cell lines. Analysis of K-ras, p53, p16 and DPC4/Smad4. Virchows Arch. 2001;439:798–802. doi: 10.1007/s004280100474. [DOI] [PubMed] [Google Scholar]
- 9.Dammann R, Schagdarsurengin U, Liu L, Otto N, Gimm O, Dralle H, Boehm BO, Pfeifer GP, Hoang-Vu C. Frequent RASSF1A promoter hypermethylation and K-ras mutations in pancreatic carcinoma. Oncogene. 2003;22:3806–3812. doi: 10.1038/sj.onc.1206582. [DOI] [PubMed] [Google Scholar]
- 10.Brueckner B, Kuck D, Lyko F. DNA methyltransferase inhibitors for cancer therapy. Cancer J. 2007;13:17–22. doi: 10.1097/PPO.0b013e31803c7245. [DOI] [PubMed] [Google Scholar]
- 11.Zhang H, Chen L, Bu HQ, Yu QJ, Jiang DD, Pan FP, Wang Y, Liu DL, Lin SZ. Effects of emodin on the demethylation of tumor-suppressor genes in pancreatic cancer PANC-1 cells. Oncol Rep. 2015;33:3015–3023. doi: 10.3892/or.2015.3914. [DOI] [PubMed] [Google Scholar]
- 12.Li-Weber M. Targeting apoptosis pathways in cancer by Chinese medicine. Cancer Lett. 2013;332:304–312. doi: 10.1016/j.canlet.2010.07.015. [DOI] [PubMed] [Google Scholar]
- 13.Liu A, Chen H, Tong H, Ye S, Qiu M, Wang Z, Tan W, Liu J, Lin S. Emodin potentiates the antitumor effects of gemcitabine in pancreatic cancer cells via inhibition of nuclear factor-κB. Mol Med Rep. 2011;4:221–227. doi: 10.3892/mmr.2011.414. [DOI] [PubMed] [Google Scholar]
- 14.Xu W, Yang H, Liu Y, Yang Y, Wang P, Kim SH, Ito S, Yang C, Wang P, Xiao MT, et al. Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of α-ketoglutarate-dependent dioxygenases. Cancer Cell. 2011;19:17–30. doi: 10.1016/j.ccr.2010.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bestor TH. The DNA methyltransferases of mammals. Hum Mol Genet. 2000;9:2395–2402. doi: 10.1093/hmg/9.16.2395. [DOI] [PubMed] [Google Scholar]
- 16.Baylin SB, Herman JG. DNA hypermethylation in tumorigenesis: Epigenetics joins genetics. Trends Genet. 2000;16:168–174. doi: 10.1016/S0168-9525(99)01971-X. [DOI] [PubMed] [Google Scholar]
- 17.Hanaoka M, Shimizu K, Shigemura M, Kato A, Fujii H, Honoki K, Tsujiuchi T. Cloning of the hamster p16 gene 5′ upstream region and its aberrant methylation patterns in pancreatic cancer. Biochem Biophys Res Commun. 2005;333:1249–1253. doi: 10.1016/j.bbrc.2005.06.034. [DOI] [PubMed] [Google Scholar]
- 18.Peng DF, Kanai Y, Sawada M, Ushijima S, Hiraoka N, Kitazawa S, Hirohashi S. DNA methylation of multiple tumor-related genes in association with overexpression of DNA methyltransferase 1 (DNMT1) during multistage carcinogenesis of the pancreas. Carcinogenesis. 2006;27:1160–1168. doi: 10.1093/carcin/bgi361. [DOI] [PubMed] [Google Scholar]
- 19.Yang WH, Wang CY, Zhu QS, et al. Gene methylation changes and protein expression analysis of Pl6 in pancreatic cancer. Chinese J of General Surgery. 2007;16:46–450. In Chinese. [Google Scholar]
- 20.Attri J, Srinivasan R, Majumdar S, Radotra BD, Wig J. Alterations of tumor suppressor gene pl6INK4a in pancreatic duetal carcinoma. BMC Gastroenterol. 2005;5:22. doi: 10.1186/1471-230X-5-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shimizu K, Itsuzaki Y, Fujii H, Honoki K, Tsujiuchi T. Reduced expression of the Rassf1a gene and its aberrant DNA methylation in pancreatic duct adenocarcinomas induced by N-nitrosobis(2-oxopropyl)amine in hamsters. Mol Carcinog. 2008;47:80–87. doi: 10.1002/mc.20250. [DOI] [PubMed] [Google Scholar]
- 22.Lin SZ, Wei WT, Chen H, Chen KJ, Tong HF, Wang ZH, Ni ZL, Liu HB, Guo HC, Liu DL. Antitumor activity of emodin against pancreatic cancer depends on its dual role: Promotion of apoptosis and suppression of angiogenesis. PLoS One. 2012;7:e42146. doi: 10.1371/journal.pone.0042146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]