Abstract
Aim:
This study was carried out to determine the prevalence, distribution, and identification of Salmonella serotypes in diarrheagenic infants and young animals, including sewage waste and fresh vegetables.
Materials and Methods:
A total of 550 samples were processed for the isolation of Salmonella spp., using standard microbiological and biochemical tests. Further polymerase chain reaction (PCR) detection of Salmonella genus was carried out using self-designed primers targeting invA gene and thereafter identification of important serotypes namely Salmonella Enterica serovar Typhimurium, Salmonella Enterica serovar Enteritidis, Salmonella Enterica serovar Typhi was performed using published standardized multiplex PCR.
Results:
An overall low prevalence of 2.5% (14/550) was observed. The observed prevalence of Salmonella spp. in diarrheagenic infants was 1.2% (05/400), diarrheagenic young animals 4% (02/50), sewage waste 10% (05/50), and fresh vegetables 4% (02/50), respectively. In diarrheagenic infants, of the five Salmonella isolates identified, two were Salmonella Typhimurium, two Salmonella Enteritidis, and one was unidentified and hence designated as other Salmonella serovar. All the Salmonella isolates identified from diarrheagenic young animals and sewage waste belonged to other Salmonella serovar, whereas, of the two isolates recovered from fresh vegetables, one was identified as other Salmonella serovar, and one as Salmonella Typhimurium, respectively.
Conclusion:
Isolation of Salmonella spp. especially from sewage waste and fresh vegetable is a matter of great concern from public health point of view because these sources can accidentally serve as a potential vehicle for transmission of Salmonella spp. to animals and human beings.
Keywords: inv A, isolation, multiplex polymerase chain reaction, Salmonella
Introduction
Foodborne diseases with its clinical complexities are a potential public health threat worldwide and have a large economic impact across the globe. Foodborne diseases are caused by approximately 250 pathogens including bacteria, viruses, and parasitic organisms [1]. Salmonella species are most frequently reported cause of foodborne illness in both humans and animals. The gastroenteritis caused by non-typhoidal Salmonella contributes to global public health burden with about 93.8 million cases annually [2]. Salmonella infections are largely classified into four clinical types [3]. First, gastroenteritis caused by Salmonella Enterica serovar Typhimurium; second, Bacteremia, osteomyelitis, reactive arthritis due to Salmonella Typhimurium and Salmonella Enteritidis infection; third, enteric fever caused by Salmonella Typhi and Salmonella Paratyphi and lastly, a carrier state in persons with previous infections [4,5]. Non-typhoidal Salmonella is ranked second in its contribution to domestically acquired foodborne illnesses and accounts for 35% of hospitalization and 28% mortality [6]. Of the various Salmonella serotypes, Salmonella Enteritidis and Salmonella Typhimurium are the most common serotypes reported from human clinical cases. In developing countries like India, food-borne illness are mostly under reported; however in the past 29 years (1980-2009) 3485 persons have been affected from 37 Salmonella related outbreaks [7].
Salmonella infections have been recognized in all the countries, but appear to be more prevalent in areas of intensive animal husbandry, especially poultry, cattle, and pig farming [8]. In general, Salmonella contamination is implicated in a wide range of products of animal and plant origin. The primary hosts for non-typhoidal Salmonella include cattle, swine, poultry, wild birds (gulls), and pets which excrete this organism in feces, which in turn, can contaminate various food sources and environment [9]. Many asymptomatic livestock play a role as carriers of human pathogens and their feces may contain high concentrations of the organisms which are rarely detected during routine ante-mortem examination [10].
In India, to the best of our knowledge, studies addressing isolation, identification and distribution of Salmonella serotypes from varied sources are very few and hence the present study was undertaken with an aim to determine the prevalence and distribution pattern of important Salmonella serotypes in diarrheagenic human infants and young animals, sewage waste and fresh vegetables.
Materials and Methods
Ethical approval
All the procedures have been carried out in accordance with the guidelines laid down by the Institutional Ethics Committee and in accordance with local laws and regulations. In case of human infants, the diarrheal stool samples were randomly collected, primarily from Medical colleges, pediatric hospitals, and home settings, where the cases of diarrhea were observed. World Health Organization (2009) criteria for acute diarrheal episodes were fulfilled by all the children evaluated at hospitals, health care facilities, and home. Samples were collected only after obtaining informed consent either from the parents of infants or with the help of medical practitioners. The diarrheic fecal samples from young animals were collected from Veterinary dispensaries, organized or unorganized farms after having proper consent from animal owners.
Collection of samples
A total of 550 samples, from Telangana, Chennai, Maharashtra, Goa, Uttar Pradesh, and Rajasthan comprising of 400 stool samples from diarrheagenic infants (<5 years), 50 fecal samples from diarrheic young animals, 50 samples of fresh vegetables viz., mint leaves, tomato, cilantro leaves and 50 samples from sewage, were collected and screened in the present study. All the samples except fresh raw vegetables were collected aseptically using Cary Blair transport swabs (Hi Media Labs, Mumbai, India) and were transported to the laboratory within a week for further isolation and identification studies.
Isolation of Salmonella
Isolation of Salmonella was performed as recommended by FDA [11]. In brief, 1ml of the sample from the transport swab was inoculated in 9 ml of buffered peptone water (Hi Media) and incubated at 37°C for 18 h for pre-enrichment. Further, for selective enrichment 0.1 ml of the pre-enriched inoculum was transferred to 10 ml of Rappaport-Vassiliadis broth (Hi Media) and incubated at 42°C for 24 h. After enrichment, a loopful (10 µl) of inoculums was then streaked on xylose lysine desoxycholate (XLD) agar (Hi Media) and incubated at 37°C for 24 h. The presumptive Salmonella colonies (4-5 colonies/plate) appearing slightly transparent red halo with a black center surrounded by a pink-red zone on XLD agar were screened further for its biochemical characterization.
Identification of Salmonella
Biochemical characterization
The presumptive colonies of Salmonella were further subjected to biochemical tests viz., triple sugar iron (TSI), ortho-nitrophenyl galactosidase (ONPG), urease broth, indole, methyl red, Voges-Proskauer and Citrate test (IMViC) as per the standard test protocol described in Bacteriological Analytical Manual FDA [11].
Genus identification of Salmonella isolates by polymerase chain reaction (PCR)
The biochemically positive Salmonella isolates were reconfirmed for genus Salmonella by employing PCR targeting genus-specific inv A gene, with an amplicon size of 423 bp, using a self-designed primers (Table-1). In brief, the DNA was extracted from Salmonella isolates using QIAamp DNA extraction kit as per the instructions recommended by the manufacturer. The targeted gene amplification by PCR was carried out with following PCR reaction mixture, which comprised of 2.5 ml of 10× PCR buffer (100 mM Tris-HCl buffer, pH 8.3 containing 500 mM KCl, 15 mM MgCl2, and 0.01% gelatin), 1 ml of 2.5 mM dNTP mix (a final concentration of 1 mM), 1 ml of 50 mM MgCl2, and 10 pmol of forward and reverse primer (Eurofins Pvt. Ltd., Bangaluru), 1 U of Taq DNA polymerase (3B Black Bio, Spain), 4 µl of DNA as a template, and nuclease free water to make up the reaction volume of 25 µL. The PCR amplification was performed in Mastercycler Pro Thermocycler (Eppendorf, Germany). The cycling conditions after gradient PCR were optimized with an initial denaturation at 94°C for 5 min, followed by 35 cycles of denaturation at 94°C for 30 s, annealing at 56°C for 1 min, and extension at 72°C for 1 min 30 s, followed by 10 min of final extension at 72°C and hold at 4°C. The amplified PCR products were resolved by agarose gel electrophoresis, using 1.5% agarose gel stained with ethidium bromide (0.5 mg/ml) and visualized and documented using UV gel documentation system (UVP Gel Seq Software, England). The presence of amplicon at 423 bp was confirmed as Salmonella genus (Figure-1).
Table-1.
Primer details for identification of genus and serotypes of Salmonella isolates.
| Bacteria | Gene targeted | Primers | Primer sequence | Product size (bp) | Reference |
|---|---|---|---|---|---|
| Salmonella | Inv A | Forward | TCG TGA CTC GCG TAA ATG GCG ATA | 423 | This study |
| Reverse | GCA GGC GCA CGC CAT AAT CAA TAA | ||||
| Salmonella Enteritidis | Sdf I | Forward | TGT GTT TTA TCT GAT GCA AGA GG | 304 | De Freitas et al. 2010 |
| Reverse | TGA ACT ACG TTC GTT CTT CTG G | ||||
| Salmonella Typhi | Via B | Forward | CAC GCA CCA TCA TTT CAC CG | 738 | De Freitas et al. 2010 |
| Reverse | AAC AGG CTG TAG CGA TTT AGG | ||||
| Salmonella Typhimurium | Spy | Forward | TTG TTC ACT TTT TAC CCC TGA A | 401 | De Freitas et al. 2010 |
| Reverse | CCC TGA CAG CCG TTA GAT ATT |
Figure-1.
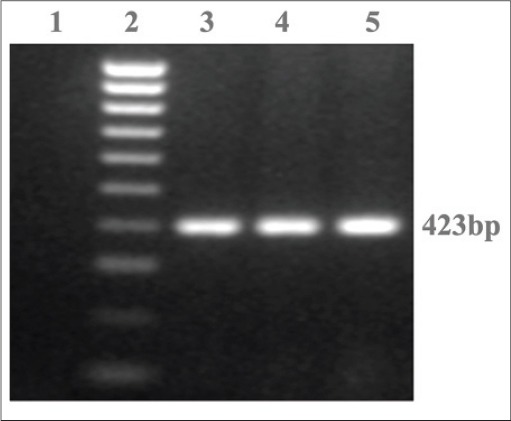
Representative agarose gel analysis of PCR assay targeting invA gene in Salmonella isolates. Lane 1: Negative control, Lane 2: 100 bp DNA ladder, Lane 3: S. Typhimurium, Lane 4-5: Positive samples
Serotype identification of Salmonella isolates by multiplex PCR
The serotype identification namely Salmonella Enteritidis, Salmonella Typhi, and Salmonella Typhimurium for the confirmed Salmonella isolates was carried out using multiplex PCR assay as described earlier [12] with slight modifications. The details of primers, genes targeted and the amplicon size for the serotypes mentioned above are presented in Table-1. In brief, the optimized PCR reaction mixture consisted of 2.5 ml of 10× PCR buffer, 3 µl of 50 mM of MgCl2, 3 µl of 2.5 mM of dNTP mix, 1 U of Taq polymerase, 10 pmol of each set of forward primer, and 10 pmol of each set of reverse primer (Eurofins Pvt. Ltd., Bangaluru), 5 µl of DNA as a template and nuclease free water to make 25 ml of reaction volume. The PCR cycling conditions were programmed with an initial denaturation step of 5 min at 94°C; followed by 35 cycles, each with denaturation at 94°C for 30 s, annealing at 57°C for 1 min., extension at 72°C for 1 min 30 s and finally the final extension was performed at 72°C for 7 min and hold at 4°C. The amplified PCR products were resolved by agarose gel electrophoresis, using 1.5% agarose gel stained with ethidium bromide (0.5 mg/ml) and visualized and documented using UV gel documentation system (UVP Gel Seq Software, England).
Result
The results of isolation of Salmonella spp. and its further serotype identification employing multiplex PCR are presented in Table-2. In brief, on microbiological analysis of 550 samples, 92 samples revealed presumptive Salmonella colonies on XLD agar plate. Further, on biochemical characterization, only 14 samples revealed biochemical profile suggestive of Salmonella genus. All the 14 isolates were TSI positive, urease negative, ONPG negative, indole negative, methyl red positive, Voges Proskauer negative, and Citrate test positive, respectively. All these biochemically confirmed Salmonella isolates were reconfirmed using genus-specific invA gene PCR (Table-2).
Table-2.
Isolation and identification of Salmonella isolates from various sources.
| Sampling subjects | Source of sample | Number of sample screened | Positive samples on microbiological analysis | Positive samples on biochemical analysis | Genus specific PCR positive | Serotype identified by multiplex PCR |
|---|---|---|---|---|---|---|
| Human infants (<5 years) | Male | 200 | 23 | 2 | 2 | Salmonella Enteritidis (n=1), other Salmonella serotypes (n=1) |
| Female | 200 | 30 | 3 | 3 | Salmonella Enteritidis (n=1), Salmonella Typhimurium (n=2) | |
| Young animals (<6 months) | Canine | 25 | 8 | 2 | 2 | Other Salmonella serotypes (n=2) |
| Bovine | 15 | 3 | 0 | 0 | ND | |
| Equine | 10 | 0 | 0 | 0 | ND | |
| Fresh vegetables | Cilantro leaves | 10 | 5 | 1 | 1 | Salmonella Typhimurium (n=1) |
| Tomato | 20 | 3 | 1 | 1 | Other Salmonella serotypes (n=1) | |
| Mint leaves | 20 | 0 | 0 | 0 | ND | |
| Sewage waste | 50 | 20 | 5 | 5 | Other Salmonella serotypes (n=5) |
ND=Not detected, PCR=Polymerase chain reaction
Overall, in the present study a low prevalence of 2.5% (14/550) was observed for Salmonella spp. The observed source wise isolation rate of Salmonella spp. in diarrheagenic infants was 1.2% (05/400), diarrheagenic young animals 4% (02/50), sewage waste 10% (05/50), and fresh vegetables 4% (02/50), respectively. The details of serotype identified from the respective source are presented in Table-2 and Figure 2.
Figure-2.
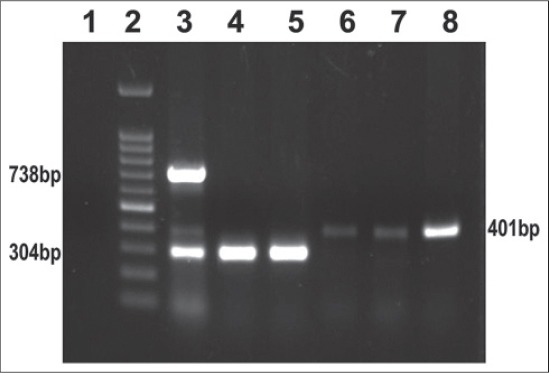
Representative gel analysis of multiplex PCR for identification of Salmonella serotypes. Lane 1: Negative control, Lane 2: 100 bp DNA ladder, Lane 3: Standards (S. Typhi 738 bp, S. Typhimurium 401 bp and S. Enteritidis 304 bp), Lane 4 – 5: S. Enteritidis isolates, Lane 6 – 8: S. Typhimurium isolates
Discussion
In developing countries, Salmonella is considered to be the prime etiological agent in causing foodborne diseases and childhood morbidity and mortality [2,13]. In the present study, all the screened samples, including infant stools were found negative for Salmonella Typhi. This absence of Salmonella Typhi in the present study could be due to effective breastfeeding [14,15]. Moreover, it has been reported that the lower incidence of Salmonellosis in Asian countries has been observed in human infants due to their association with higher rates of breastfeeding among their mothers [16,17].
In India, Salmonellosis is endemic in humans, animals, and also associated with foods of animal origin [18-20]. In our study, the overall prevalence of Salmonella in diarrheagenic human infants was 1.2% and 4% in diarrheic young animals, which obviously was on the lower side as compared to earlier studies [18-20]. Beside this, in the present study, the serotypes namely Salmonella Enteritidis and Salmonella Typhimurium were identified from human infants and fresh vegetables. Similar serotype isolation/identification of Salmonella Enteritidis and Salmonella Typhimurium has also been reported by several authors from human infants and vegetables [21,22]. Almost all the isolates recovered from animals and sewage wastes were considered as other Salmonella serotypes because only three important serotypes namely Salmonella Enteritidis, Salmonella Typhimurium and Salmonella Typhi were detected by multiplex PCR employed in the present study. These results are in partial agreement with other authors wherein besides Salmonella Enteritidis and Salmonella Typhimurium, other Salmonella serotypes have also been identified in animals and sewage waste [22,23]. Further, in our study bovine and equine samples failed to yield Salmonella isolates, although earlier studies [24,25] have revealed a higher incidence of Salmonella spp. in the said species. There can be several factors behind their absence, but the most important would be the limited number of samples screened in the present study and further it has been also reported that diseased animals shed Salmonella intermittently, and therefore a minimal 5 consecutive negative fecal cultures is recommended before declaring the animal negative for Salmonellosis [26-28].
In our study, Salmonella isolates were also recovered from fresh vegetables (Tomato, Cilantro leaves), which in most of the countries including India are often consumed raw. Thus, the presence of this pathogen in such vegetables is a matter of concern from food safety point of view. Similar reports on detection of Salmonella from fresh vegetable have also been reported by several authors [29-32]. In general, these pathogenic bacteria are brought into aquatic environments mainly through treated or untreated wastewater release, surface runoffs, and soil leaching which in turn poses a substantial risk of widespread occurrence of diseases [33]. Besides this, Salmonella also have the ability to attach to plant tissue and can survive under adverse temperature conditions due to their effective biofilm formation capability [31,34].
Conclusion
Isolation of Salmonella spp. especially from sewage waste and fresh vegetables is a matter of great concern from public health point of view because these sources can accidentally serve as a vehicle for transmission of Salmonella spp. to animals and human beings. However, in the light of results from the present study, epidemiological studies addressing the source of transmission in human and animals needs to devised and executed. Furthermore, more focused intervention studies are required to control this pathogen in sewage waste and fresh vegetables.
Authors’ Contributions
DBR and TB have designed and supervised the study. AN has carried out bacterial isolation and molecular characterization. TB, VM, MK, and JV have collected the samples and also helped in characterizing the isolates. AN and VM drafted and reviewed the manuscript. DBR and SVSM have edited the manuscript. All authors read and approved the final manuscript.
Acknowledgments
The authors are thankful to the Director, Indian Veterinary Research Institute, Izatnagar for providing the necessary facilities to carry out this work. The research work supported by grants from the Department of Biotechnology, Government of India (BT/PR15148/GBD/27/339/2011) to DBR is also duly acknowledged.
Competing Interests
The authors declare that they have no competing interests.
References
- 1.Linscott A.J. Food-borne illnesses. Clin. Microbiol. Newsl. 2011;33(6):41–5. [Google Scholar]
- 2.Majowicz S.E, Musto J, Scallan E, Angulo F.J, Kirk M, O'Brien S.J, Hoekstra R.M. The global burden of non-typhoidal Salmonella gastroenteritis. Clin. Infect. Dis. 2010;50(6):882–889. doi: 10.1086/650733. [DOI] [PubMed] [Google Scholar]
- 3.Bisi-Johnson M.A, Obi C.L. Escherichia coli and Salmonella species: Molecular landscape and therapeutic considerations: A review. Adv. Med. Sci. 2012;1(1):1–16. [Google Scholar]
- 4.Owens M.D, Warren D.A. Salmonella Infection. Emedicine: Emergency Medicine. 2009. [Accessed on 15-04-2015]. Available from: http://www.emedicine.medscapecom/article/7ↁ4-overview.html .
- 5.Klotchko A, Wallace M.R. Salmonellosis: Treatment and Medication. 2009. [Accessed on 15-04-2015]. Available from: http://www.emedicine.medscape.com/article/228174-overview .
- 6.CDC. Estimates of Foodborne Illness in the United States. 2011. [Accessed on 15-04-2015]. Available from: http://www.cdc.gov/foodborneburden/2011-foodborne-estimates.html .
- 7.Sudershan R.V, Kumar R.N, Polasa K. Foodborne diseases in India – A review. Br. Food J. 2012;114(5):661–680. [Google Scholar]
- 8.OIE. Salmonellosis – Chapter 2.9.9. In: OIE Terrestrial Manual. 2008. [Accessed on 15-04-2015]. Available from: http://www.oie.int/fileadmin/Home/eng/Health_standards/tahm/2008/pdf/2.09.09_SALMONELLOSIS.pdf .
- 9.CDC. Section VII-agents summary statements, section VIIA bacterial agents. In: Richmond J.Y, McKinney R.W, editors. Biosafety in Microbiological and Biomedical Laboratories. 4th ed. Washington, U.S: Government Printing Office; 1999. p. 109. [Google Scholar]
- 10.EFSA Panel on Biological Hazards. Scientific opinion on the public health hazards to be covered by inspection of meat (Bovine animals) EFSA, J. 2013;11(6):3266. doi: 10.2903/j.efsa.2013.3265. 1-261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Andrews W.H, Jacobson A, Hammack T.S. Salmonella In: Bacteriological Analytical Manual. 8th ed. 1998. [Accessed on16-04-2015]. Revision A. Available from: http://www.fda.gov/Food/FoodScienceResearch/LaboratoryMethods/ucm2006949.html .
- 12.De Freitas C.G, Santana ÂP, Da Silva P.H.C, Gonçalves V.S.P, Barros M.D. A.F, Torres F.A.G, Perecmanis S. PCR multiplex for detection of Salmonella enteritidis, Typhi and Typhimurium and occurrence in poultry meat. Int. J. Food Microbiol. 2010;139(1-2):15–22. doi: 10.1016/j.ijfoodmicro.2010.02.007. [DOI] [PubMed] [Google Scholar]
- 13.De Freitas Neto O.C, Penha Filho R.A.C, Barrow P, Berchieri A., Jr Sources of human non-typhoid salmonellosis – A review. Rev. Bras. Ciênc. Avíc. 2010;12(1):1–11. [Google Scholar]
- 14.Rowe S.Y, Rocourt J.R, Shiferaw B, Kassenborg H.D, Segler S.D, Marcus R, Slutsker L. Breast-feeding decreases the risk of sporadic salmonellosis among infants in food net sites. Clin. Infect. Dis. 2004;38(3):S262–S270. doi: 10.1086/381595. [DOI] [PubMed] [Google Scholar]
- 15.Ryan A.S, Zhou W, Gaston M.H. Regional and socio-demographic variation of breastfeeding in the United States, 2002. Clin. Pediatr. 2004;43(9):815–824. doi: 10.1177/000992280404300905. [DOI] [PubMed] [Google Scholar]
- 16.CDC. Breastfeeding Among U.S. Children Born 2001-2011. In: National Immunization Survey NIS Data. Provisional breastfeeding rates by socio-demographic factors, among children born in 2007. 2010. [Accessed 16-04-2015]. Available from: http://www.cdc.gov/breastfeeding/data/nis_data .
- 17.Cheng L.H, Crim S.M, Cole C.R, Shane A.L, Henao O.L, Mahon B.E. Epidemiology of infant salmonellosis in the United States, 1996-2008: A foodborne diseases active surveillance network study. J. Pediatr. Infect. Dis. Soc. 2013;4(1):1–8. doi: 10.1093/jpids/pit020. [DOI] [PubMed] [Google Scholar]
- 18.Verma J.C, Singh V.P, Singh B.R, Gupta B.R. Occurrence of Salmonella serotypes in animals in India VII. Indian, J. Comp. Microbiol Immunol. Infect. Dis. 2001;22(1):51–55. [Google Scholar]
- 19.Thapliyal D.C. Diseases caused by viruses. In: Diseases of animals transmissible to man. 1st ed. Lucknow: International Book Distributing Company; 1999. pp. 57–71. [Google Scholar]
- 20.WHO. Laboratory Protocol Isolation of Salmonella spp. From Food and Animal Faeces. In: Global Foodborne Infections Network. 5th ed. Atlanta, GA; USA: WHO; 2010. pp. 4–8. [Google Scholar]
- 21.Rahman H, Streckel W, Prager R, Tschäpe H. Presence of sopE gene and its phenotypic expression among different serovars of Salmonella isolated from man and animals. Indian, J. Med. Res. 2004;120(1):35–38. [PubMed] [Google Scholar]
- 22.Cellucci T, Seabrook J.A, Chagla Y, Bannister S.L, Salvadori M.I. A 10-year retrospective review of Salmonella infections at the children's hospital in London, Ontario. Can. J. Infect. Dis. Med. Microbiol. 2010;21(2):78. doi: 10.1155/2010/968960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Smith-Palmer A, Stewart W.C, Mather H, Greig A, Cowden J.M, Reilly W.J. Epidemiology of Salmonella enterica serovars enteritidis and typhimurium in animals and people in Scotland between 1990 and 2001. J. Vet. Rec. 2003;153(17):517–520. doi: 10.1136/vr.153.17.517. [DOI] [PubMed] [Google Scholar]
- 24.Bagudo A.I, Tambuwal F.M, Faleke O.O, Egwu O.O, Aliero A.A. Prevalence of Salmonella serotypes in Sokoto abattoir effluents and vegetables cultivated around the abattoir. Microbiol. Res. Int. 2014;2:13–17. [Google Scholar]
- 25.Henriksen S.W.M, Orsel K, Wagenaar J.A, Miko A, Duijkeren E. Animal-to-human transmission of Salmonella typhimurium DT104A variant. Emerg. Infect. Dis. 2004;10(12):2225–2257. doi: 10.3201/eid1012.040286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Alao F, Kester C, Gbagba B, Fakilede F. Comparison of prevalence and antimicrobial sensitivity of Salmonella typhimurium in apparently healthy cattle and goat in Sango-Ota, Nigeria. Internet J. Microbiol. 2012;10(2) [Google Scholar]
- 27.Zahran R, El-Behiry A. Prevalence, molecular identification and virulence attributes of Salmonella serovars isolated from feces of diarrheic cow and buffalo-calves. Int. J. Curr. Microbiol App. Sci. 2014;3(11):9–27. [Google Scholar]
- 28.Smith B.P, Reina-Guerra M, Hardy A.J. Prevalence and epizootiology of equine salmonellosis. J. Am. Vet. Med. Assoc. 1978;172(3):353–356. [PubMed] [Google Scholar]
- 29.Quiroz-Santiago C, Rodas-Suárez O.R, Vázquez Q, Carlos R, Fernández F.J, Quinones-Ramirez E.I, Vazquez-Salinas C. Prevalence of Salmonella in vegetables from Mexico. J. Food Prot. 2009;72(6):1279–1282. doi: 10.4315/0362-028x-72.6.1279. [DOI] [PubMed] [Google Scholar]
- 30.Singh B.R, Singh P, Agrawal S, Teotia U, Verma A, Sharma S, Kant Agarwal R. Prevalence of multidrug resistant Salmonella in Coriander mint, carrot, and radish in Bareilly Kanpur Northern India. Foodborne Pathog. Dis. 2007;4(2):233–240. doi: 10.1089/fpd.2006.0082. [DOI] [PubMed] [Google Scholar]
- 31.Sant'Ana A.S, Barbosa M.S, Destro M.T, Landgraf M, Franco B.D. Growth potential of Salmonella spp. and Listeria monocytogenes in nine types of ready-to-eat vegetables stored at variable temperature conditions during shelf-life. Int. J. Food Microbiol. 2012;157(1):52–58. doi: 10.1016/j.ijfoodmicro.2012.04.011. [DOI] [PubMed] [Google Scholar]
- 32.Nillian E, Ching C.L, Fung P.C, Robin T, Anyi U, Chilek T.Z.T, Nishibuchi M. Simultaneous detection of Salmonella spp Salmonella enteritidis and Salmonella typhimurium in raw salad vegetables and vegetarian burger patties. Food Nutr. Sci. 2011;2(10):1077–1081. [Google Scholar]
- 33.James A.E, Ian P, Helen S, Sojka R.E. Polyacrylamide +Al2 (SO4)3 and polyacrylamide +CaO remove coliform bacteria and nutrients from swine wastewater. Environ. Res. 2003;121(3):453–462. doi: 10.1016/s0269-7491(02)00225-7. [DOI] [PubMed] [Google Scholar]
- 34.Sheffield C.L, Crippen T.L. Invasion and survival of Salmonella in the environment: The role of biofilms. In: Kumar Y, editor. Publisher In Tech; 2012. [Accessed on 16-04-2015]. Available from: http://www.Intechopen.com/articles/show/title/invasion-and-survival-of-Salmonella-in-the-environment-the-role-of-biofilms . ISBN d978-953-307-781-9, 576. DOI:10.5772/2471. [Google Scholar]


