Abstract
Aim:
The current study was the first of its kind taken upon indigenous ecotypes of the Karnataka in order to unravel the diversity details at 20 chicken microsatellite regions.
Materials and Methods:
210 indigenous chicken belonging to six districts of Bangalore and Mysore division formed the target sample for the present study. The genomic deoxyribonucleic acid was isolated by phenol chloroform isoamyl alcohol method. A panel of 20 microsatellite regions, including 14 recommended by FAO and six identified from published scientific literature became the targeted chicken genomic region. 27-33 samples were successfully genotyped in each of the six ecotypes through simplex or multiplex polymerase chain reactions, polyacrylamide gel electrophoresis and silver staining for the selected microsatellite panel.
Results:
The chickens of Ramanagara and Chamrajnagara were most distant with a Nei’s genetic distance value of 0.22. The chickens of Bangalore rural and Mysore were least distant with a value of 0.056. The Ramanagara and Chamrajnagara pair had Nei’s genetic identity value of 0.802, which is least among all pairs of ecotypes. There were five main nodes from which the six ecotypes evolved on the basis 20 microsatellite markers used in this study. This study indicates that the four ecotypes Ramnagara, Bangalore Rural, Chickaballapura and Mysore are genetically identical due to their common ancestral evolution while, Mandya and Chamrajnagara ecotypes formed a relatively different cluster due to a separate common ancestral chicken population and less number of generations since drifting from bifurcation node.
Conclusion:
Twenty microsatellite markers based genetic diversity study on six indigenous ecotypes indicated lower genetic distances as well as lower FST values compared to the distinguished breeds reported. There were two main clusters, which differentiated into six ecotypes. They may differentiate into more distinct varieties if bred in isolation for a longer number of generations.
Keywords: animal genetic resources, ecotypes, microsatellites, polyacrylamide gel
Introduction
Characterization of animal genetic resources (AnGR) involves activities associated with the identification, quantitative and qualitative description, documentation of breed populations, the natural habitats and production systems to which they are not adapted. Since the beginning of the 1990s, molecular data have become more and more relevant for the characterization of genetic diversity [1]. In 1993, FAO working group proposed a global program for characterization of AnGR, including molecular genetic characterization and formulated the secondary guidelines: Measurement of domestic animal diversity.
Indigenous chicken have an inherent scavenging and nesting habit. Years of natural selection under scavenging conditions, has made them tough and resistant to various diseases, especially to those caused by bacteria, and protozoa and other internal and external parasites; they have better survival than the commercial hybrid strains under village production conditions. However, the village chicken is a poor egg producer, laying on average 40-60 eggs per year in three or four clutches, with an average egg weight of around 35-45 g. They generally have small body size; for various chicken breeds, mature body weight varies between 1.3 and 1.9 kg for males and between 1.0 and 1.4 kg for females.
Molecular genetic characterization explores polymorphism in selected protein molecules and deoxyribonucleic acid (DNA) markers in order to measure genetic variation at the population level. To obtain a better knowledge of AnGR, of their present and potential future uses for food and agriculture in defined environments, and their current state as distinct breed populations. In poultry, different genetic marker systems have successfully been used for estimation of genetic variability. They were DNA fingerprints, RAPDs, and microsatellites. Recently, the determination of heterozygosity and genetic distances based on microsatellite markers is regarded as a most convenient tool, and many microsatellite loci are available in chicken. These tandem repeated DNA segments show extensive allelic differences in length based mainly on variation in the number of repeats and, partly, in adjacent regions. These studies are important for understanding the history of species, and for testing hypotheses regarding evolutionary processes. Currently, microsatellites are most commonly used, and successful application of this sort of markers in diversity studies has been reported for all major livestock species [2-6]. Microsatellites have proven to be an extremely valuable tool for genome mapping in many organisms, but their applications span over different areas ranging from forensic DNA studies to population genetics and conservation/management of biological resources.
The Indian chicken ecotypes consisted of different phenotypes of almost centuries of natural selection and reared by smallholder farmers across distinct agro-ecological regions. To date, however, no investigation has yet been directed toward the use of microsatellite markers to study the genetic variability of these ecotypes, despite a large number of microsatellite primers published in scientific literature. Despite the importance of native chicken in tribal/rural areas, information is lacking on their genetic makeup with respect to genetic variability, genetic relationships, performance, adaptability and resistance to diseases. In this context, this study of diversity in indigenous chicken ecotypes based on appropriate microsatellite panel in Karnataka was taken up.
Materials and Methods
Ethical approval
This research was approved by the Local Ethics Committee of Veterinary College, Bangalore, Karnataka Veterinary, Animal and Fisheries sciences University, as per LPM/IAEC/77/2011 dated 20-01-2011.
Experimental birds
The indigenous chicken belonging to Ramnagara, Bangalore rural and Chikkaballapur districts of Bangalore division and Chamarajnagara, Mysore and Mandya districts of Mysore division of Karnataka state (Figure-1) maintained at AICRP on Poultry for Meat, Veterinary College, Hebbal formed the material for the present study. 35 randomly chosen adult birds from each of the above districts formed the research sample for the present study.
Figure-1.
Study area map
The birds belonging to the six districts were wing banded at the hatch from randomly collected fertile eggs in respective districts. The birds chosen in each district belonged to three hatches and were badged while allocating to breeding pens after growing phase. The wing bands and serial numbers were noted down at the time of blood collection from each bird. The serial numbers for each district were maintained on the 2 ml Eppendorf tubes and subsequent tubes until the successful amplification of desired segment in each marker. The numbers were maintained until genotyping and further statistical analysis.
Primer and DNA collection
20 primer pairs corresponding to microsatellite regions of chicken autosomes 1, 2, 3, 4, 5, 6, 7, 8, 10, 13, 14, 17, and 23 were identified by review of literature (Table-1). The procedure recommended by Khosravinia et al. [2] with little modifications for DNA extraction from whole fresh avian blood was adopted.
Table-1.
The microsatellite primer details.
| Marker name | F/R | Primer sequence (5’-3’) | Primer length | Chromosome number | At (°C) | Product size (bp) |
|---|---|---|---|---|---|---|
| ADL0020 | F R |
GCACTCAAAAGAAAACAAAT TAGATAAAAATCCTTCCCTT |
20 20 |
1 | 55 | 88 |
| ADL0023 | F R |
CTTCTATCCTGGGCTTCTGA CCTGGCTGTGTATGTGTTGC |
20 20 |
5 | 60 | 164 |
| ADL0176 | F R |
TTGTGGATTCTGGTGGTAGC TTCTCCCGTAACACTCGTCA |
20 20 |
2 | 52 | 175-194 |
| MCW007 | F R |
AGCAAAGAAGTGTTCTCTGTTCAT ACCCTGCAAACTGGAAGGGTCTCA |
24 24 |
1 | 60 | 313-349 |
| MCW0 041 | F R |
CCCATGTGCTTGAATAACTTGGG CCAGATTCTCAATAACAATGGCAG |
23 24 |
2 | 55 | 140-162 |
| MCW0014 | F R |
TATTGGCTCTAGGAACTGTC GAAATGAAGGTAAGACTAGC |
20 20 |
6 | 58 | 164-182 |
| MCW0183 | F R |
ATCCCAGTGTCGAGTATCCGA TGAGATTTACTGGAGCCTGCC |
21 21 |
7 | 58 | 296-326 |
| ADL0278 | F R |
CCAGCAGTCTACCTTCCTAT TGTCATCCAAGAACAGTGTG |
20 20 |
8 | 60 | 114-126 |
| MCW0067 | F R |
GCACTACTGTGTGCTGCAGTTT GAGATGTAGTTGCCACATTCCGAC |
22 24 |
10 | 60 | 176-186 |
| MCW0104 | F R |
TAGCACAACTCAAGCTGTGAG AGACTTGCACAGCTGTGTACC |
21 21 |
13 | 60 | 190-234 |
| MCW0123 | F R |
CCACTAGAAAAGAACATCCTC GGCTGATGTAAGAAGGGATGA |
21 21 |
14 | 60 | 76-100 |
| MCW0330 | F R |
TGGACCTCATCAGTCTGACAG AATGTTCTCATAGAGTTCCTGC |
21 22 |
17 | 60 | 256-300 |
| MCW0165 | F R |
CAGACATGCATGCCCAGATGA GATCCAGTCCTGCAGGCTGC |
21 20 |
23 | 60 | 114-118 |
| MCW0103 | F R |
AACTGCGTTGAGAGTGAATGC TTTCCTAACTGGATGCTTCTG |
21 21 |
3 | 64 | 266-270 |
| MCW0034 | F R |
TGCACGCACTTACATACTTAGAGA TGTCCTTCCAATTACATTCATGGG |
24 24 |
2 | 60 | 212-246 |
| MCW0081 | F R |
GTTGCTGAGAGCCTGGTGCAG CCTGTATGTGGAATTACTTCTC |
21 22 |
5 | 60 | 112-135 |
| MCW0284 | F R |
GCCTTAGGAAAAACTCCTAAGG CAGAGCTGGATTGGTGTCAAG |
22 21 |
4 | 60 | 235-243 |
| MCW0078 | F R |
CCACACGGAGAGGAGAAGGTCT TAGCATATGAGTGTACTGAGCTTC |
22 24 |
5 | 60 | 135-147 |
| ADL0268 | F R |
CTCCACCCCTCTCAGAACTA CAACTTCCCATCTACCTACT |
20 20 |
1 | 60 | 102-116 |
| ADL0112 | F R |
GGCTTAAGCTGACCCATTAT ATCTCAAATGTAATGCGTGC |
20 20 |
10 | 58 | 120-134 |
Polymerase chain reaction (PCR) details
PCR reactions were set up on clean benches with autoclaved lab disposables like micro tips, PCR tubes, 1.5-2.0 ml Eppendorf tubes and autoclaved milliq water ensuring proper thawing, mixing of PCR reaction components and cold chain of around 4°C. The standard PCR protocol was followed for amplifying all the microsatellite regions. The standard 12% non-denaturing polyacrylamide gel (PAGE) was performed for detection of allele size on all PCR products belonging to 20 microsatellite loci.
Genotyping
The silver staining was adopted for staining of PAGEs in this study. The improved staining method developed by Halima et al. [7]. The same technique with little modification was adopted in this study. Each of the lanes with clearer bands was compared with the 50 bp ladder lane to genotype each sample.
Statistical analysis
The data obtained regarding genotypes of 20 microsatellite regions for six ecotypes of Bangalore and Mysore divisions in base pair format was utilized as input file for GenAlex 6.5b5 in order to obtain genetic distances, and Wright’s F-statistics. The dendrogram was obtained from the POPGENE software. The Polymorhic information for each marker was obtained from MS tools programme.
Results and Discussion
Polymorphic information content (PIC) values
The PIC values of 20 loci for all the six ecotypes are presented in the Table-2. The locus MCW0183 was the least polymorphic (0.219-0.5846) while MCW0103 was the most polymorphic (0.8324-0.894) locus across six ecotypes.
Table-2.
Microsatellite loci wise PIC values for six ecotypes.
| Locus | Ecotypes | |||||
|---|---|---|---|---|---|---|
| RNGR | BNGRRL | CBR | MYSORE | MANDYA | CHMNGR | |
| ADL020 | 0.7521 | 0.6220 | 0.7362 | 0.6445 | 0.6513 | 0.7494 |
| ADL023 | 0.7622 | 0.6588 | 0.7216 | 0.7024 | 0.7045 | 0.7846 |
| ADL176 | 0.6487 | 0.4463 | 0.6392 | 0.5812 | 0.5945 | 0.6774 |
| MCW007 | 0.8523 | 0.7938 | 0.8727 | 0.8053 | 0.8272 | 0.8386 |
| MCW041 | 0.7017 | 0.6530 | 0.7440 | 0.7577 | 0.7366 | 0.7923 |
| MCW014 | 0.8373 | 0.7739 | 0.8367 | 0.7829 | 0.7568 | 0.7990 |
| MCW183 | 0.4034 | 0.2190 | 0.4845 | 0.3859 | 0.3907 | 0.5846 |
| ADL278 | 0.7814 | 0.6302 | 0.6933 | 0.7225 | 0.7801 | 0.8115 |
| MCW067 | 0.7252 | 0.6018 | 0.7067 | 0.6998 | 0.6423 | 0.6838 |
| MCW104 | 0.7592 | 0.6937 | 0.7984 | 0.6476 | 0.7307 | 0.8350 |
| MCW123 | 0.6041 | 0.4214 | 0.6678 | 0.5517 | 0.5539 | 0.676 |
| MCW330 | 0.6913 | 0.7043 | 0.7802 | 0.6926 | 0.7718 | 0.8573 |
| MCW165 | 0.7055 | 0.6748 | 0.7821 | 0.7592 | 0.7405 | 0.7947 |
| MCW103 | 0.8813 | 0.8324 | 0.8943 | 0.8550 | 0.8681 | 0.8940 |
| MCW034 | 0.7863 | 0.7742 | 0.8513 | 0.7885 | 0.7398 | 0.8147 |
| MCW081 | 0.6897 | 0.5153 | 0.6868 | 0.6108 | 0.6489 | 0.7335 |
| MCW284 | 0.6650 | 0.6237 | 0.684 | 0.7069 | 0.7371 | 0.7730 |
| MCW078 | 0.7744 | 0.7242 | 0.7235 | 0.7077 | 0.7666 | 0.8002 |
| ADL268 | 0.7744 | 0.7242 | 0.7235 | 0.7077 | 0.7666 | 0.8002 |
| ADL112 | 0.7681 | 0.7339 | 0.7793 | 0.7555 | 0.7547 | 0.8279 |
PIC: Polymorphic information content
Heterozygosity values
The heterozygosity values are depicted in Table-3. The mean H0 over all loci was lowest for Bangalore Rural with a value of 0.855 and highest for Chickaballapura with a value of 0.868. The mean He over all loci was lowest for Bangalore rural with a value of 0.685 and highest for Chamrajnagara with a value of 0.803. The mean uHe over all loci was lowest for Bangalore Rural with a value of 0.698 and highest for Chamrajnagara with a value of 0.815. The mean ‘F’ value over all loci was lowest for Bangalore Rural with a value of −0.256 and highest for Chamrajnagara with a value of −0.066.
Table-3.
Summary of heterozygosity information over all loci for each population.
| Ecotype | N | Na | Ne | I | Ho | He | uHe | F |
|---|---|---|---|---|---|---|---|---|
| RNGR | ||||||||
| Mean | 29.000 | 7.650 | 4.687 | 1.671 | 0.857 | 0.760 | 0.773 | −0.121 |
| SE | 0.000 | 0.568 | 0.368 | 0.074 | 0.042 | 0.022 | 0.022 | 0.049 |
| BNGRL | ||||||||
| Mean | 28.000 | 5.600 | 3.640 | 1.381 | 0.855 | 0.685 | 0.698 | −0.256 |
| SE | 0.000 | 0.336 | 0.286 | 0.081 | 0.041 | 0.031 | 0.031 | 0.046 |
| CBR | ||||||||
| Mean | 30.000 | 7.500 | 4.954 | 1.698 | 0.868 | 0.772 | 0.785 | −0.117 |
| SE | 0.000 | 0.531 | 0.438 | 0.076 | 0.041 | 0.018 | 0.018 | 0.048 |
| MYSORE | ||||||||
| Mean | 30.000 | 6.600 | 4.100 | 1.532 | 0.857 | 0.731 | 0.743 | −0.164 |
| SE | 0.000 | 0.328 | 0.290 | 0.066 | 0.041 | 0.021 | 0.022 | 0.048 |
| MANDYA | ||||||||
| Mean | 27.000 | 6.600 | 4.367 | 1.563 | 0.861 | 0.748 | 0.762 | −0.137 |
| SE | 0.000 | 0.444 | 0.310 | 0.072 | 0.051 | 0.019 | 0.020 | 0.065 |
| CHMNGR | ||||||||
| Mean | 33.000 | 8.600 | 5.518 | 1.835 | 0.861 | 0.803 | 0.815 | −0.066 |
| SE | 0.000 | 0.472 | 0.374 | 0.063 | 0.042 | 0.014 | 0.014 | 0.048 |
Genetic distances
Among the 15 pairs of ecotypes studied, the chickens of Ramanagara and Chamrajnagara were most distant with a Nei’s genetic distance value of 0.22. The chickens of Bangalore rural and Mysore were least distant with a value of 0.056. The Ramanagara and Chamrajnagara pair had Nei’s genetic identity value of 0.802, which is least among all pairs of ecotypes. The Bangalore Rural and Mysore had maximum Nei’s genetic identity with a value of 0.946 among all the pairs of ecotypes (Table-4).
Table-4.
Pairwise genetic identity (above the diagonal) and distance (below the diagonal) values.
| Ecotype | RNGR | BNGRL | CBR | MYSORE | MANDYA | CHMNGR |
|---|---|---|---|---|---|---|
| RNGR | 1.000 0.000 |
0.942 | 0.911 | 0.909 | 0.868 | 0.802 |
| BNGRL | 0.060 | 1.000 0.000 |
0.922 | 0.946 | 0.900 | 0.845 |
| CBR | 0.093 | 0.081 | 1.000 0.000 |
0.858 | 0.825 | 0.829 |
| MYSORE | 0.095 | 0.056 | 0.153 | 1.000 0.000 |
0.936 | 0.863 |
| MANDYA | 0.142 | 0.105 | 0.192 | 0.066 | 1.000 0.000 |
0.930 |
| CHMNGR | 0.220 | 0.169 | 0.187 | 0.148 | 0.073 | 1.000 0.000 |
Among the 15 pairs of ecotypes studied, the chicken of Ramanagara and Chamrajnagara were most distant with an unbiased Nei’s genetic distance value of 0.159. The chicken of Ramanagara and Bangalore Rural were least distant with a value of 0.011. The Ramanagara and Chamrajnagara pair had Unbiased Nei’s genetic identity value of 0.852 which is least among all pairs of ecotypes. The Ramanagara and Bangalore Rural had maximum unbiased Nei’s genetic identity with a value of 0.989 among all the pairs of ecotypes (Table-5). The genetic distance between populations provides a relative estimate of the time elapsed since the subdivisions existed as a single population and helps in characterizing the breeds or lines. Among the 15 pairs of ecotypes, Nei’s genetic distance was in the range of 0.056-0.22. The similar trend was observed for Nei’s unbiased genetic distances as well for the 15 pairs of ecotypes. The Nei’s standard genetic distances reported by Chang et al. [3] were moderate to high (0.59-0.93). The findings of Sangwon et al. [4] were in conformity with our genetic distance values. The genetic distance of such magnitude is predictable for the ecotypes which are not completely isolated from each other for a longer number of generations and they are also not subjected to differential selection pressures. The exchange of genes between populations homogenizes allele frequencies between populations and determines the relative effects of selection and genetic drift. High gene flow precludes local adaptation (i.e. the fixation of alleles, which are favored under local conditions), and will therefore also impede the process of speciation [8].
Table-5.
Pairwise Nei’s unbiased genetic identity (above diagonal) and distance (below diagonal) values.
| Ecotype | RNGR | BNGRL | CBR | MYSORE | MANDYA | CHMNGR |
|---|---|---|---|---|---|---|
| RNGR | 1.000 0.000 |
0.989 | 0.966 | 0.958 | 0.919 | 0.853 |
| BNGRL | 0.011 | 1.000 0.000 |
0.969 | 0.988 | 0.945 | 0.890 |
| CBR | 0.035 | 0.031 | 1.000 0.000 |
0.905 | 0.875 | 0.882 |
| MYSORE | 0.043 | 0.012 | 0.100 | 1.000 0.000 |
0.986 | 0.912 |
| MANDYA | 0.084 | 0.056 | 0.134 | 0.014 | 1.000 0.000 |
0.988 |
| CHMNGR | 0.159 | 0.116 | 0.125 | 0.092 | 0.012 | 1.000 0.000 |
Dendrogram
The dendrogram was constructed on the basis of genetic distance [9] and neighbor-joining methods following un-weighted pair-group method using arithmetic averages. There were five main nodes from which the six ecotypes evolved on the basis 20 microsatellite markers used in this study (Figure-2). Nodes four and two drifted from five and were geographically isolated for a prolonged number of generations represented in time units of 2.24 and 4.03, respectively. The node three and Mysore drifted from four and were geographically separated for a prolonged number of generations represented in time units of 1.56 and 5.45, respectively. The Ramanagara ecotype and node one drifted from node three in the recent past and geographically isolated in time units of 3.88 and 1.10, respectively. Bangalore Rural and Chickaballapura ecotypes drifted from node one, evolved into current type by geographical isolation for a time unit of 2.79. Mandya and Chamrajnagara ecotypes were evolved from node two by drifting and geographical isolation for a time unit of 3.65 (Table-6). The accuracy of a reconstructed tree also depends on a tree making method and distance measures used. Nei [10] suggested that although this result is discouraging it must be accepted due to the stochastic nature of gene substitution and that one cannot be over confident about the evolutionary tree reconstructed from electrophoretic data. There were five main nodes from which the six ecotypes evolved on the basis of 20 microsatellite markers used in this study. Nodes four and two drifted from five and were geographically isolated for a prolonged number of generations represented in time units of 2.24 and 4.03 respectively. The node three and Mysore drifted from four, geographically separated for a prolonged number of generations represented in time units of 1.56 and 5.45 respectively. The Ramanagara ecotype and node one drifted from node three in the recent past and geographically isolated in time units of 3.88 and 1.10 respectively. Bangalore Rural and Chickaballapura ecotypes drifted from Node one, evolved into current type by geographical isolation for a time unit of 2.79. Mandya and Chamrajnagara ecotypes were evolved from node two by drifting and geographical isolation for time unit of 3.65. This study indicates that the four ecotypes Ramnagara, Bangalore Rural, Chickaballapura and Mysore are genetically identical due to their common ancestral evolution while, Mandya and Chamrajnagara ecotypes formed a relatively different cluster due to a separate common ancestral chicken population and less number of generations since drifting from bifurcation node.
Figure-2.
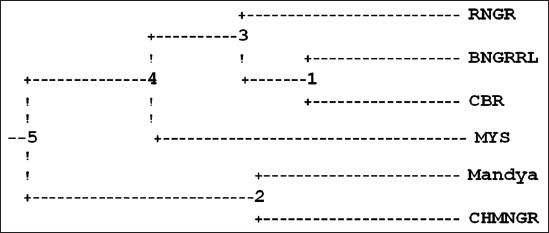
Dendrogram based Nei’s genetic distance for the six ecotypes of indigenous chicken.
Table-6.
The branch lengths among five nodes and ecotypes of the dendrogram.
| Nodes | Nodes/ecotypes | Branch length |
|---|---|---|
| 5 | 4 | 2.24 |
| 4 | 3 | 1.56 |
| 3 | Ramanagara | 3.88 |
| 3 | 1 | 1.10 |
| 1 | Bangalore rural | 2.79 |
| 1 | Chickaballapura | 2.79 |
| 4 | Mysore | 5.45 |
| 5 | 2 | 4.03 |
| 2 | Mandya | 3.65 |
| 2 | Chamrajnagara | 3.65 |
Wright’s F-statistics
The FIS was in the range of −0.475 in MCW123 to 0.551 in MCW183. The FIT was in the range of −0.430 in MCW123 to 0.598 in MCW183. The FST was in the range of 0.012 in MCW123 to 0.104 in MCW183. The FST values calculated by frequency option (Table-7) was highest (0.032) for Bangalore Rural and Chamrajnagara pair while lowest (0.011) for Mandya’s pairing with two other districts of Mysore division. Complimentary to the above trend, the number of migrants (Nm) between the ecotype pairs was lowest (7.587) for Bangalore Rural and Chamrajnagara pair, while highest (around 22) for Mandya’s pairing with two other districts of Mysore division. Fixation indices give an idea about the population structure in terms of the inbreeding coefficient and population differentiation. The F and FIS measures “the Ho expressed as a fraction of He and deviation from 1” for each of the locus/all loci in each ecotype and over all populations for each locus, respectively. The former takes into account individual locus H0 and He values while later considers mean H0 and He values of all ecotypes for each of the locus. They are influenced by the deviation of He from H0. The values were on the positive side for loci with lower H0 than He and are on the negative side for loci with higher H0 than He. The values indicate the extent of fixation of alleles in each of the loci. The F values were positive for only three loci namely MCW183, MCW067 and MCW014 indicating relatively better allele fixation in these loci compared to remaining seventeen loci which had negative F values. The mean F per ecotype over all loci was lowest for Bangalore Rural with a value of −0.256 and highest for Chamrajnagara with a value of −0.066 indicating better allele fixation in Chamrajnagara ecotype among all ecotypes. The values were in conformity with the ranges reported by Pirany et al. [11]. The FIS was in the range of −0.475 in MCW123 to 0.551 in MCW183. The range of FIS in the present study was wider compared to the reports of −0.29 to 0.38 and −0.135 to 0.137 by Pirany et al. [11], respectively. The difference can be ascribed to the difference in the microsatellite markers utilized, mutation rates in genetic groups and varied selection intensities for traits linked to microsatellite markers. The Ht is similar to He except that the former is a measure of expected heterozygosity over all the ecotypes while later is restricted to one population. The behavior of Ht values is similar to that of He. The values are directly proportional to the polymorphism of loci. The similar range of Ht values was reported by Pirany et al. [11]. Fixation indices such as FIS, FIT and FST give an idea about the population structure in terms of inbreeding coefficient and population differentiation [12].
Table-7.
F-statistics and estimates of Nm over all ecotypes for each locus.
| Locus | Ht | Mean He | Mean H0 | FIS | FIT | FST | Nm |
|---|---|---|---|---|---|---|---|
| ADL020 | 0.745 | 0.728 | 0.737 | −0.012 | 0.011 | 0.023 | 10.703 |
| ADL023 | 0.777 | 0.752 | 0.771 | −0.024 | 0.008 | 0.031 | 7.738 |
| ADL176 | 0.682 | 0.661 | 0.937 | −0.419 | −0.375 | 0.031 | 7.849 |
| MCW007 | 0.862 | 0.849 | 1.000 | −0.178 | −0.160 | 0.015 | 16.400 |
| MCW041 | 0.785 | 0.764 | 0.972 | −0.273 | −0.238 | 0.028 | 8.782 |
| MCW014 | 0.839 | 0.822 | 0.790 | 0.039 | 0.058 | 0.020 | 11.955 |
| MCW183 | 0.516 | 0.462 | 0.207 | 0.551 | 0.598 | 0.104 | 2.159 |
| ADL278 | 0.798 | 0.768 | 0.797 | −0.038 | 0.001 | 0.037 | 6.446 |
| MCW067 | 0.746 | 0.723 | 0.631 | 0.126 | 0.154 | 0.031 | 7.761 |
| MCW104 | 0.805 | 0.773 | 0.932 | −0.206 | −0.157 | 0.040 | 5.955 |
| MCW123 | 0.664 | 0.644 | 0.949 | −0.475 | −0.430 | 0.030 | 7.947 |
| MCW330 | 0.816 | 0.781 | 0.972 | −0.245 | −0.192 | 0.043 | 5.590 |
| MCW165 | 0.799 | 0.776 | 0.960 | −0.238 | −0.202 | 0.029 | 8.357 |
| MCW103 | 0.893 | 0.882 | 1.000 | −0.133 | −0.120 | 0.012 | 21.264 |
| MCW034 | 0.850 | 0.817 | 1.000 | −0.224 | −0.177 | 0.039 | 6.138 |
| MCW081 | 0.713 | 0.691 | 0.728 | −0.054 | −0.021 | 0.032 | 7.681 |
| MCW284 | 0.757 | 0.743 | 1.000 | −0.346 | −0.322 | 0.018 | 13.755 |
| MCW078 | 0.811 | 0.780 | 0.906 | −0.162 | −0.118 | 0.038 | 6.328 |
| ADL268 | 0.811 | 0.780 | 0.906 | −0.162 | −0.118 | 0.038 | 6.328 |
| ADL112 | 0.814 | 0.798 | 1.000 | −0.253 | −0.229 | 0.019 | 12.807 |
The FIT was in the range of −0.430 in MCW123 to 0.598 in MCW183 with a mean of −0.101 over all loci and ecotypes in the present study. For the interpretation of FST, it has been suggested that a value lying in the range 0-0.05 indicates little genetic differentiation; a value between 0.05 and 0.15, moderate differentiation; a value between 0.15 and 0.25, great differentiation; and values above 0.25, very great genetic differentiation. Balloux et al. [12] elaborated that a FST of 0.05 will generally be considered as reasonably low, and investigators may interpret that structuring between sub populations is weak. While such an interpretation may turn out to be correct, it may also not be representative at all of the real population differentiation because of the fact that expectation of FST, under complete differentiation will not always be one. In fact, in the great majority of cases, it will not be one because the effect of polymorphism (due to mutations) drastically deflates FST expectations. Hence, a seemingly low FST of 0.05 may, in fact, indicate very important genetic differentiation. This point was already stressed by Barton and Hewitt [8], who wrote that differentiation is by no means negligible if FST is as small as 0.05 or even less. The FST was in the range of 0.012 in MCW123 to 0.104 in MCW183 with a mean of 0.033 over all loci and ecotypes. The mean FIS, FIT and FST for all the microsatellites together in the overall population was −0.18, −0.085 and 0.083, respectively for five chicken populations including two indigenous breeds and three pure lines of Indian Rural synthetics in the reports of Chatterjee et al. [13]. The FST values in the present study were lower than those reported by FAO and Nei [9,14].
Conclusions
Twenty microsatellite markers based genetic diversity study on six indigenous ecotypes indicated lower genetic distances as well as lower FST values compared to those values between distinguished breeds reported. There were two main clusters, which differentiated into six ecotypes. They may differentiate into more distinct varieties if bred in isolation for a longer number of generations.
Authors’ Contributions
This is part of the Ph.D. thesis submitted by the first author, consequently research, analysis and manuscript preparation was done by the same. HNNM suggested material collection plan, MRJ instructed format for recording of laboratory data, CSN suggested PCR trouble shooting methods, AMK helped in caliberating Thermal cycler for all primer pairs and SMB provided the key information for interpretation of laboratory results. All authors read and approved the final manuscript.
Acknowledgments
First author is indebted to the administration of Karnataka Veterinary, Animal and Fisheries Sciences University, Bidar for granting deputation to pursue Ph.D. and funding through research contingency for Department of Animal Gentics and Breeding, Veterinary College, Bangalore, Karnataka, India.
Competing Interests
The authors declare that they have no competing interests.
References
- 1.Groeneveld L.F, Lenstra J.A, Eding H, Toro M.A, Scherf B, Pilling D, Negrini R, Finlay E.K, Jianlin H, Groeneveld E, Weigend S, The Globaldiv Consortium Genetic diversity in livestock breeds – A review. Anim. Genet. 2010;41(Suppl 1):6–31. doi: 10.1111/j.1365-2052.2010.02038.x. [DOI] [PubMed] [Google Scholar]
- 2.Khosravinia H, Narasimha Murthy H.N, Therthaparasad D, Pirany N. Optimizing factors influencing DNA extraction from fresh whole avian blood. Afr. J. Biotech. 2007;6(4):481–486. [Google Scholar]
- 3.Chang C.S, Chen C.F, Berthouly Salazar C, Chazara O, Lee Y.P, Chang C.M, Chang K.H, Bedhom B, Tixier-Boichard M. A global analysis of molecular marker and phenotypic traits in local chicken breeds in Taiwan. Anim. Genet. 2011;43:172–182. doi: 10.1111/j.1365-2052.2011.02226.x. [DOI] [PubMed] [Google Scholar]
- 4.Sangwon S, Sharma A, Lee S, Cho C.Y, Kim J.H, Choi S.B, Kim H, Seong H.H, Yeon S.H, Kim D.H, Ko Y.G. Genetic diversity and relationships of Korean chicken breeds based on 30 microsatellite markers. Asian Aust. J. Anim. Sci. 2014;27(10):1399–1405. doi: 10.5713/ajas.2014.14016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Choi N.R, Hoque R, Seo D.W, Sultana H, Park H.B, Lim H.T, Heo K.N, Kang B.S, Jo C, Lee J.H. ISAG recommennded microsatellite marker analysis among five korean native chicken lines. J. Anim. Sci. Technol. 2012;54(6):401–409. [Google Scholar]
- 6.Eltanany M, Philipp U, Weigend S, Distl O. Genetic diversity of ten Egyptian chicken strains using 29 microsatellite markers. Anim. Genet. 2011;42:666–669. doi: 10.1111/j.1365-2052.2011.02185.x. [DOI] [PubMed] [Google Scholar]
- 7.Halima B, Jean-Marie J, Jean-Pierre B, Guy M. Optimization of a reliable, fast, cheap and sensitive silver staining method to detect SSR markers in polyacrylamide gels. Biotechnol. Agron. Soc. Environ. 2006;10(2):77–81. [Google Scholar]
- 8.Barton N.H, Hewitt G.M. Analysis of hybrid zones. Ann. Rev. Ecol. Syst. 1985;16:113–148. [Google Scholar]
- 9.Nei M. Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics. 1978;89:583–590. doi: 10.1093/genetics/89.3.583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nei M. Molecular Evolutionary Genetics. New York: Columbia Press; 1987. [Google Scholar]
- 11.Pirany N, Romanov M.N, Ganpule S.P, Devegowda G, Prasad D.T. Microsatellite analysis of genetic diversity in Indian chicken populations. J. Poult. Sci. 2007;44:19–28. [Google Scholar]
- 12.Balloux F, Lugon-Moulin N. The estimation of population differentiation with microsatellite markers. Mol. Ecol. 2002;11:155–165. doi: 10.1046/j.0962-1083.2001.01436.x. [DOI] [PubMed] [Google Scholar]
- 13.Chatterjee R.N, Niranjan M, Sharma R.P, Dange M, Bhattacharya T.K. Estimation of genetic heterogeneity of chicken germplasm being used for development of rural varieties utilizing DNA markers. J. Genet. 2010;89:e33–e37. [PubMed] [Google Scholar]
- 14.FAO. Molecular genetic characterization of animal genetic resources. Animal Production and Health Guidelines. No. 9. 14. Rome: FAO; 2011. [Google Scholar]


