The structure of human growth differentiation factor 11 (GDF11) determined to a resolution of 1.50 Å reveals subtle conformational differences when compared with the previously determined structure of human myostatin (GDF8) while maintaining a highly similar quaternary architecture.
Keywords: growth differentiation factor 11, GDF11, TGF-β family
Abstract
Members of the TGF-β family of proteins are believed to play critical roles in cellular signaling processes such as those involved in muscle differentiation. The extent to which individual family members have been characterized and linked to biological function varies greatly. The role of myostatin, also known as growth differentiation factor 8 (GDF8), as an inhibitor of muscle differentiation is well understood through genetic linkages. In contrast, the role of growth differentiation factor 11 (GDF11) is much less well understood. In humans, the mature forms of GDF11 and myostatin are over 94% identical. In order to understand the role that the small differences in sequence may play in the differential signaling of these molecules, the crystal structure of GDF11 was determined to a resolution of 1.50 Å. A comparison of the GDF11 structure with those of other family members reveals that the canonical TGF-β domain fold is conserved. A detailed structural comparison of GDF11 and myostatin shows that several of the differences between these proteins are likely to be localized at interfaces that are critical for the interaction with downstream receptors and inhibitors.
1. Introduction
Many cellular proliferation and differentiation processes, notably muscle differentiation, are controlled via signaling pathways involving the TGF-β family of proteins. The TGF-β family may be divided into three subclasses: TGF-β, bone morphogenetic protein (BMP) and activin/inhibin (Innis et al., 2000 ▸). The TGF-β family of proteins share a common protein fold, and significant effort has been expended to understand the structural basis of the differential signaling of individual family members. The TGF-β proteins are expressed with a propeptide region that is subsequently cleaved. The proteins then form disulfide-linked homodimers or heterodimers. All family members exert their influence through interaction with pairs of type I or type II transmembrane receptor serine/threonine kinases.
One of the better characterized members of the TGF-β family is myostatin, also known as growth differentiation factor 8 (GDF8). The role of myostatin as an inhibitor of muscle growth has been established through genetic linkages in multiple species, in which mutations lead to increased muscle growth (McPherron et al., 1997 ▸; McPherron & Lee, 1997 ▸). While the crystal structure of human myostatin alone has not been determined, there are two structures of myostatin bound to extracellular antagonists: follistatin 288 (Fst288; Cash et al., 2009 ▸) and follistatin-like 3 (Fstl3; Cash et al., 2012 ▸). These structures revealed how follistatin mimics the interactions that have previously been described between the TGF-β family and the type I and type II receptors (Greenwald et al., 2004 ▸; Stamler et al., 2008 ▸).
The biological role of growth differentiation factor 11 (GDF11) has recently been the focus of intense debate (Brun & Rudnicki, 2015 ▸; Egerman et al., 2015 ▸; Katsimpardi et al., 2014 ▸; Leinwand & Harrison, 2013 ▸; Loffredo et al., 2013 ▸; Poggioli et al., 2016 ▸; Sinha et al., 2014 ▸; Smith et al., 2015 ▸). It is known that both myostatin and GDF11 bind to activin type II receptors, and that binding and subsequent phosphorylation of these receptors triggers signaling via the Smad2/3 pathway. We determined the structure of human GDF11 in order to obtain structural insights into its distinctive biological role compared with the highly homologous myostatin. A comparison of the structure of GDF11 with other TGF-β family proteins may reveal a structural component of the differential signaling of these highly similar proteins.
2. Materials and methods
2.1. Protein production
Recombinant human GDF11 was obtained from R&D Systems (catalogue No. 1958-GD-10). The protein, consisting of residues Asn299–Ser407 (accession No. O95390), was produced in Escherichia coli as a disulfide-linked homodimer. The protein was shown to be active in a cell-based Smad2/3 assay (data not shown).
2.2. Crystallization and data collection
GDF11 was obtained as a lyophilized powder and was dissolved in 60 mM sodium acetate buffer pH 4.5 at 5 mg ml−1. Initial crystallization conditions were identified from sparse-matrix screens. A series of gradient screens were employed to optimize the final condition, and three-dimensional diffraction-quality crystals were finally grown in sitting drops at 293 K from 0.18 M magnesium acetate, 0.1 M HEPES pH 7.5, 22.5% PEG 3350, 0.01 M strontium chloride. Diamond-shaped crystals appeared after 57 d and continued to grow. The crystals were harvested at 64 d with a nylon loop (Hampton Research) and flash-cooled in liquid nitrogen using 20% glycerol, 25% PEG 3350, 0.2 M magnesium chloride as a cryoprotectant. X-ray diffraction data were collected on beamline X10SA at the Swiss Light Source, Paul Scherrer Institute using a Pilatus 6M-F detector. Crystals were continuously rotated over 180° with collection of images at every 0.5°. The crystal-to-detector distance was 240 mm.
2.3. Structure determination and refinement
Diffraction data were processed using d*TREK (Rigaku) and the structure was determined by molecular replacement (MR). The processed data were carefully analysed using phenix.xtriage. A homology model of GDF11 was generated with MOE (Molecular Operating Environment; Chemical Computing Group, Montreal, Canada) using the closely related myostatin crystal structure (PDB entry 3hh2; Cash et al., 2009 ▸) as a template. The monomeric GDF11 was used as the search model for MR in Phaser (McCoy et al., 2007 ▸) as implemented in the PHENIX software package (Adams et al., 2010 ▸). Iterative refinement and manual model building were performed using REFMAC (Murshudov et al., 2011 ▸; Vagin et al., 2004 ▸; Winn et al., 2011 ▸), phenix.refine (Afonine et al., 2012 ▸) and Coot (Emsley et al., 2010 ▸). Five TLS groups identified by PHENIX, consisting of groupings of amino acids 2–15, 16–44, 45–55, 56–78 and 79–109, were used during the final stages of refinement. The final atomic model consists of amino-acid residues 2–109 (numbered in alignment to GDF8), two polyethylene glycol (PEG) molecules and 142 water molecules. Statistics of data collection, processing and refinement are presented in Table 1 ▸.
Table 1. Summary of data-collection, processing and refinement statistics.
Values in parentheses are for the outer shell.
| Beamline | X10SA, Swiss Light Source |
| Wavelength (Å) | 1.0 |
| Detector | Pilatus 6M-F |
| Space group | P41212 |
| a, b, c (Å) | 32.27, 32.27, 210.60 |
| α, β, γ (°) | 90, 90, 90 |
| Mosaicity (°) | 0.68 |
| Resolution range (Å) | 22.82–1.50 (1.55–1.50) |
| Total No. of reflections | 54719 |
| No. of unique reflections | 17599 |
| Completeness (%) | 92.3 (91.8) |
| Multiplicity | 3.11 (3.01) |
| 〈I/σ(I)〉 | 9.4 (1.9†) |
| R meas | 0.067 (0.560) |
| R merge | 0.056 (0.466) |
| R r.i.m. | 0.068 (0.569) |
| R work/R free | 0.188(0.290)/0.207(0.283) |
| R.m.s. deviations | |
| Bonds (Å) | 0.006 |
| Angles (°) | 1.06 |
| Average B factor (Å2) | 28.40 |
| Ramachandran plot | |
| Most favored (%) | 96 |
| Outliers (%) | 0 |
| MolProbity statistics | |
| Score | 1.16 |
| Clashscore | 1.68 |
| Rotamer outliers (%) | 1.0 |
| No. of non-H atoms | 1069 |
| No. of solvent molecules | 142 |
| PDB code | 5e4g |
I/σ(I) falls below 2.0 at a resolution of 1.54 Å.
2.4. Structural analysis
Structural analysis was performed with the superpose algorithm in PyMOL (v.1.7.4; Schrödinger). Buried surface area calculations were performed using PISA (Krissinel & Henrick, 2007 ▸) in the CCP4 software suite (Krissinel & Henrick, 2007 ▸; Winn et al., 2011 ▸).
2.5. Other software
The figures were prepared with PyMOL.
3. Results and discussion
3.1. Space-group assignment, structure solution and refinement
Initial X-ray data processing suggested that the crystals were likely to belong to a primitive tetragonal Bravais lattice. Data were scaled in multiple space groups from this lattice system, including P41 and P41212, with good merging statistics. However, molecular-replacement attempts with Phaser were unsuccessful in obtaining reliable structure solutions. We decided to lower the symmetry and merge the data in the primitive monoclinic space group P21 (a = 32.23, b = 32.28, c = 210.66 Å, α = γ = 90, β = 89.94°). Molecular replacement generated a clear solution in P21 with two GDF11 dimers in the asymmetric unit. Iterative manual model building and further refinements of the solution led to an R work and R free of 19.5 and 22.0%, respectively. Examination of the metric symmetry indicated it to be close to the primitive tetragonal 422 symmetry. This led to the selection of space group P41212, with a single GDF11 monomer in the asymmetric subunit, and further refinement of the final model. During the refinement, it was observed that the electron density for the side chain of amino-acid residue Cys73 reflected two conformations, only one of which was consistent with the formation of an intermolecular disulfide bond (see §3.3). In addition, pursuing the high-symmetry P41212 space group allowed us to merge only the initial 45° of data with acceptable scaling statistics (Table 1 ▸) to minimize the impact of radiation damage. Refinement of the final atomic model in P41212 yielded an R work and R free of 18.8% and 20.7%, respectively.
3.2. Overall structure
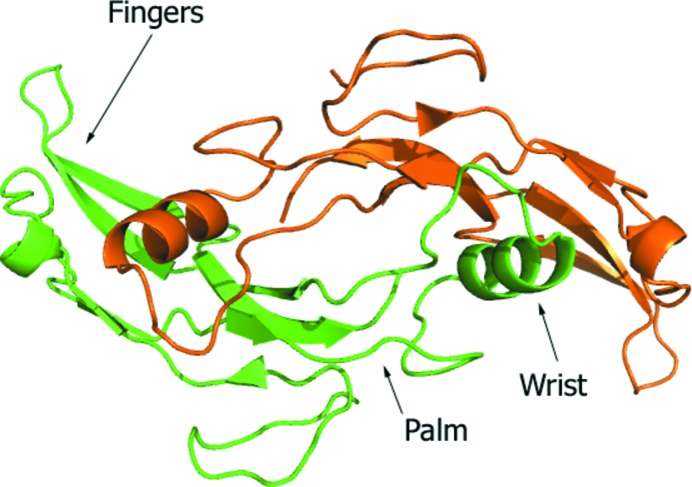
The asymmetric unit consists of a single protein chain of GDF11 and adopts the canonical homodimeric form of the TGF-β family as a result of the crystallographic symmetry. The dimeric human GDF11 exhibits the conserved tertiary structure of the TGF-β family of proteins, which is likened to a ‘hand’ with a four-stranded β-sheet comprising the ‘fingers’, a cystine-knot structure occupying the palm and an α-helix forming the ‘wrist’. In the active, dimeric form, the palms of the monomers are linked through a single disulfide bond in an antiparallel configuration (Fig. 1 ▸). The interleaved packing of the neighboring dimers results in contacts not only between the β-sheet fingers of adjacent molecules, but also between the major helix wrist of the cognate dimer partner and the same fingers of the adjacent molecules. Extensive crystal contacts are observed between the two dimers in the asymmetric unit and with neighboring symmetry-related molecules (Supplementary Fig. S1).
Figure 1.
Overall structure of homodimeric human GDF11 showing the conserved tertiary structure of the TGF-β family of proteins. The biological dimer was created from two copies of the asymmetric unit.
As expected from the high sequence identity between GDF11 and myostatin, the overall structures of the two proteins are very similar. The backbone r.m.s.d. between GDF11 monomers and myostatin monomers from the Fst288 complex (PDB entry 3hh2) ranges between 0.65 and 0.77 Å depending on which monomeric subunits are compared. Aligning the dimers from the same systems, the overall r.m.s.d. is 1.31 Å, largely resulting from the change in the relative orientation of the domains with respect to each other (see below).
3.3. Quaternary structure
The reported crystal structures of myostatin and other TGF-β family members show the proteins to be intermolecular disulfide-linked dimers. Similarly, GDF11 is believed to form a Cys73 disulfide-linked homodimer in its active state. Examination of the electron density supports two possible conformations for Cys73, with one conformation being consistent with a disulfide bond with the symmetry-equivalent residue. This is suggestive of partial occupancy of a disulfide linkage within the crystal form (Supplementary Fig. S2). However, only the dimeric species was observed in intact mass-spectrometry studies using the same protein preparation, supporting the exclusive presence of a native disulfide linkage (data not shown). Therefore, it is likely that the partial occupancy of this disulfide bond in our structure is the result of radiation damage during data collection. A comparison of the electron density associated with the other disulfide bonds in the molecule also shows weaker than expected density for the thiol side chain, which is suggestive of radiation damage. The lack of electron density in this intermolecular disulfide bond between the monomers may result from the fact that it is the most surface-exposed of all of the disulfide bonds in the assembly.
It has previously been recognized that there is a degree of flexibility in the relative orientations of the subunits of TGF-β family members. Notably, Cash et al. (2012 ▸) reported that the extremities of the myostatin dimer are closer by 4.3 Å and the buried surface area is increased by roughly 10% when bound to Fst288 rather than Fstl3. In the asymmetric unit of the GDF11 crystal, the average buried surface area between the dimer subunits is 1286 Å2. The buried surface area in GDF11 is slightly larger than the buried surfaces areas of 907 and 1005 Å2 observed for myostatin in complex with Fstl3 and Fst288, respectively. It is possible that the more compact nature of the GDF11 dimer reported here results from the protein not being complexed with inhibitors, which may, in the case of myostatin, serve to encircle and ‘open’ the homodimer.
3.4. Comparison with other GDF proteins
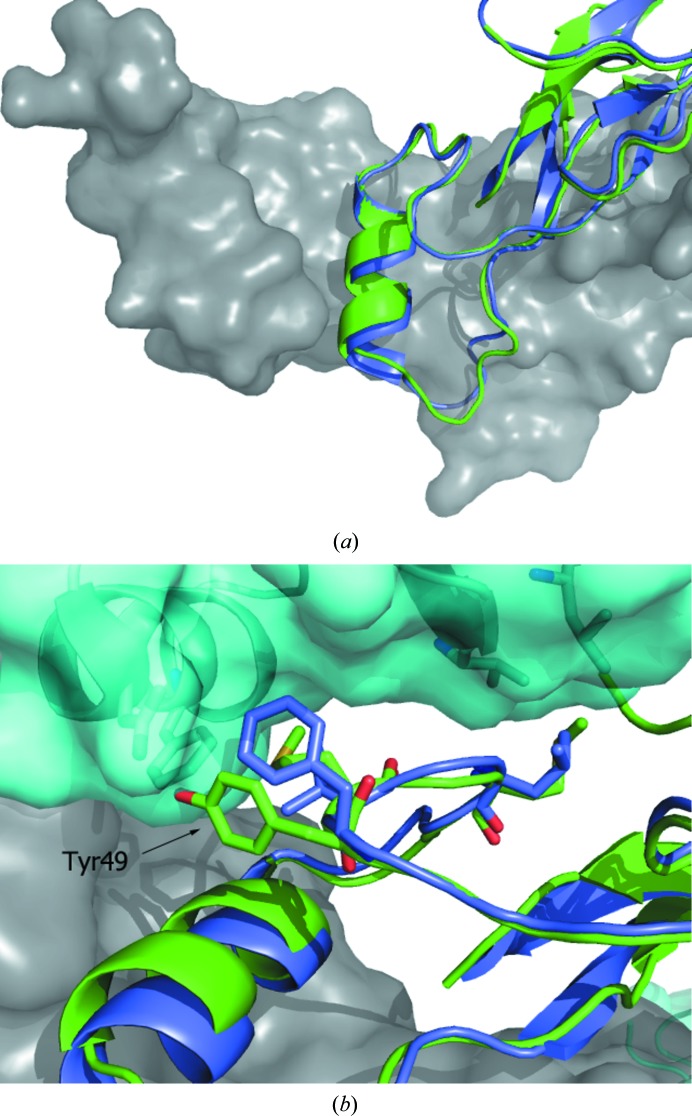
When the crystal structure of myostatin was compared with those of other family members (Cash et al., 2009 ▸), considerable differences were observed in the prehelix loop region (residues 49–55 in myostatin), leading to the suggestion that this region of the protein may be critical in the differential signaling of family members. The prehelix loop of GDF11 adopts a similar backbone conformation (Fig. 2 ▸ a) to that of myostatin (and thus is significantly different from activin A and BMP2 and is more similar to TGF-β; Cash et al., 2012 ▸). However, in spite of the similar backbone trajectories, the residue differences in the prehelix loop could potentially differentiate signaling by the two molecules (Fig. 2 ▸ b). Residue 49 of GDF11 is a tyrosine rather than a phenylalanine. In the Fst288 complex, Phe49 of myostatin is situated in a hydrophobic groove formed by Phe47 and Leu16 of the inhibitor, whereas the more lipophilic tyrosine of GDF11 would not be able to maintain these interactions. At the other end of the prehelix loop, the Leu52 found in myostatin is replaced by a methionine. A three-dimensional alignment of GDF11 and myostatin in the Fst288 complex suggests that the larger methionine residue could be accommodated in the complex because this region of the protein is partly solvent-exposed.
Figure 2.
Comparison of the prehelix loop regions of human GDF11 and myostatin (Cash et al., 2009 ▸). The A chains of the GDF11 (green) and myostatin (blue) homodimers were superimposed. The B chain of the GDF11 biological dimer is shown as a surface (gray). (a) The observed conformations of the prehelix loops of GDF11 and myostatin are highly similar. (b) View of the prehelix loop proximal to the follistatin binding site. The surface of Fst288 is shown in cyan. The side chains of residues that differ between GDF11 (Met50 and Met52) and myostatin (Val50 and Leu52) are represented as sticks. The figure illustrates a possible mismatch between Tyr49 of GDF11 and proximal residues of Fst288.
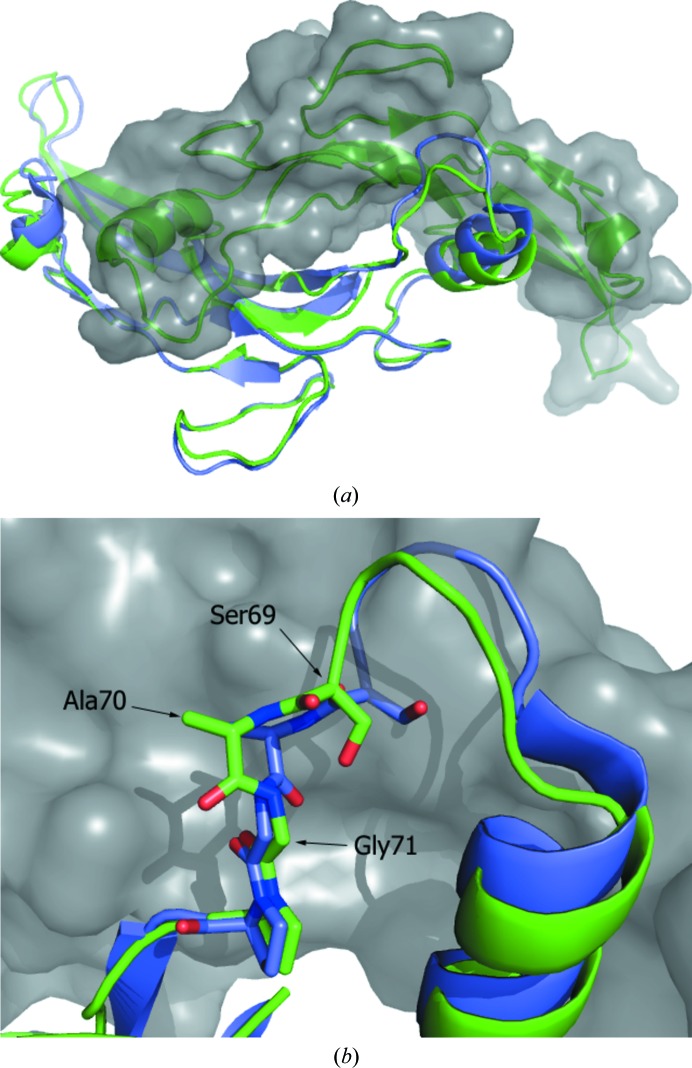
The highly compact and interlocking nature of the TGF-β family dimers dictates that structural changes in one monomer are likely to result in compensating changes in the other subunit. Thus, structural changes in the wrist region of one monomer are likely to be reflected by changes in the finger region of the other monomer. One of the largest differences observed between the GDF11 and myostatin structures occurs in the loop comprised of residues 57–72 immediately following the palm helix (Fig. 3 ▸ a). In GDF11, the backbone in this region is at a distance of up to 4 Å from the position observed in myostatin when bound to Fst288, with the loop being displaced away from the other monomeric unit. Comparing the loops of GDF11 and myostatin, in both the Fst288 and Fstl3 complexes, the major conformational difference appears to be the peptide bond between Ala70 and Gly71, the dihedral angle of which is rotated ∼120° between the two structures (Fig. 3 ▸ b). Since the residues of this loop in myostatin are proximal to follistatin domains, it is not clear whether a lack of these interactions results in the altered conformation of the loop.
Figure 3.
Structural differences reflected across the GDF11 biological dimer relative to the myostatin structure. (a) Illustration of the related structural differences in the two subunits that make up the native dimer. The A chains of GDF11 (green) and myostatin (Cash et al., 2009 ▸; blue) were superimposed. The B chain of the GDF11 biological dimer is shown as a surface (gray). The conformational differences of the loop within the wrist are illustrated, as are the related differences in the ‘finger’ regions of the other monomer. (b) Difference in backbone peptide conformations of GDF11 and myostatin in the ‘palm’ region of the proteins.
The difference in the conformation of the helix and the subsequent loop in the palm of GDF11 relative to myostatin results in a corresponding displacement of the finger regions of the protein. In the GDF11 structure reported here the fingers appear to adopt a more open conformation. The displacement of the palm helix away from the centre of the dimer is reflected by an ∼2.8 Å perturbation of the loop (residues 24–32) away from the core of the protein relative to its position in the myostatin–Fst288 structure. Analysis of the differences in the conformations of the finger regions of the proteins between GDF11 and myostatin is complicated by the location of thefingers of myostatin, which are between the second follistatin domain (FSD2) of one follistatin monomer and the N-terminal domain of the other. Therefore, when we observe that the loop comprised of residues 87–92 is displaced outwards from the centre of the molecule relative to Fst288-bound myostatin, it must be considered that Fst288 is influencing the orientation of this region. There are, however, sequence differences between myostatin and GDF11 in this region, and a possible explanation for the relative outward displacement of the loop in GDF11 is the accommodation of an aspartic acid at residue 89 in GDF11 as opposed to a glycine at the same position in myostatin.
4. Conclusions
We have determined the atomic resolution structure of GDF11 and shown that it adopts the canonical homodimeric structure exhibited by other TGF-β family members. A detailed comparison of the GDF11 structure with the closest published homologous protein, myostatin, is complicated by the fact that the structure of myostatin has not been determined in an uncomplexed state. In contrast, the published myostatin structures are reported as complexes with Fst288 and Fstl3. The structures of these two complexes show that there is conformational flexibility within myostatin that is necessary for it to form these distinct complexes. Such adaptations in structure have been observed in other ligand complexes of TGF-β family members.
The most significant differences between the GDF11 and myostatin structures are associated with the amino-acid sequence difference and reflect compensatory changes in loop regions. One exception, which is not closely related to a sequence difference, is the observed change in peptide-bond conformation between Ala70 and Gly71, which leads to a displacement of the adjacent loop structure.
The minor conformational differences between GDF11 and myostatin presented in this study suggest that in order to fully understand the differential signaling of these highly similar proteins, it will be necessary to determine co-structures with additional signaling molecules to further elucidate the role that minor sequence differences play in defining the function of these molecules.
Supplementary Material
PDB reference: human GDF11, 5e4g
Supporting Information.. DOI: 10.1107/S2053230X16001588/rr5115sup1.pdf
Acknowledgments
We thank Julius Kahn and Ashraf Khalil for determining the activity of the protein in the Smad2/3 assay and Dr Eric Larson for his input into the manuscript.
References
- Adams, P. D. et al. (2010). Acta Cryst. D66, 213–221.
- Afonine, P. V., Grosse-Kunstleve, R. W., Echols, N., Headd, J. J., Moriarty, N. W., Mustyakimov, M., Terwilliger, T. C., Urzhumtsev, A., Zwart, P. H. & Adams, P. D. (2012). Acta Cryst. D68, 352–367. [DOI] [PMC free article] [PubMed]
- Brun, C. E. & Rudnicki, M. A. (2015). Cell Metab. 22, 54–56. [DOI] [PubMed]
- Cash, J. N., Angerman, E. B., Kattamuri, C., Nolan, K., Zhao, H., Sidis, Y., Keutmann, H. T. & Thompson, T. B. (2012). J. Biol. Chem. 287, 1043–1053. [DOI] [PMC free article] [PubMed]
- Cash, J. N., Rejon, C. A., McPherron, A. C., Bernard, D. J. & Thompson, T. B. (2009). EMBO J. 28, 2662–2676. [DOI] [PMC free article] [PubMed]
- Egerman, M. A. et al. (2015). Cell Metab. 22, 164–174. [DOI] [PMC free article] [PubMed]
- Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. (2010). Acta Cryst. D66, 486–501. [DOI] [PMC free article] [PubMed]
- Greenwald, J., Vega, M. E., Allendorph, G. P., Fischer, W. H., Vale, W. & Choe, S. (2004). Mol. Cell, 15, 485–489. [DOI] [PubMed]
- Innis, C. A., Shi, J. & Blundell, T. L. (2000). Protein Eng. 13, 839–847. [DOI] [PubMed]
- Katsimpardi, L., Litterman, N. K., Schein, P. A., Miller, C. M., Loffredo, F. S., Wojtkiewicz, G. R., Chen, J. W., Lee, R. T., Wagers, A. J. & Rubin, L. L. (2014). Science, 344, 630–634. [DOI] [PMC free article] [PubMed]
- Krissinel, E. & Henrick, K. (2007). J. Mol. Biol. 372, 774–797. [DOI] [PubMed]
- Leinwand, L. A. & Harrison, B. C. (2013). Cell, 153, 743–745. [DOI] [PubMed]
- Loffredo, F. S. et al. (2013). Cell, 153, 828–839. [DOI] [PMC free article] [PubMed]
- McCoy, A. J., Grosse-Kunstleve, R. W., Adams, P. D., Winn, M. D., Storoni, L. C. & Read, R. J. (2007). J. Appl. Cryst. 40, 658–674. [DOI] [PMC free article] [PubMed]
- McPherron, A. C., Lawler, A. M. & Lee, S. J. (1997). Nature (London), 387, 83–90. [DOI] [PubMed]
- McPherron, A. C. & Lee, S. J. (1997). Proc. Natl Acad. Sci. USA, 94, 12457–12461.
- Murshudov, G. N., Skubák, P., Lebedev, A. A., Pannu, N. S., Steiner, R. A., Nicholls, R. A., Winn, M. D., Long, F. & Vagin, A. A. (2011). Acta Cryst. D67, 355–367. [DOI] [PMC free article] [PubMed]
- Poggioli, T. et al. (2016). Circ. Res. 118, 29–37. [DOI] [PMC free article] [PubMed]
- Sinha, M. et al. (2014). Science, 344, 649–652.
- Smith, S. C. et al. (2015). Circ. Res. 117, 926–932. [DOI] [PMC free article] [PubMed]
- Stamler, R., Keutmann, H. T., Sidis, Y., Kattamuri, C., Schneyer, A. & Thompson, T. B. (2008). J. Biol. Chem. 283, 32831–32838. [DOI] [PMC free article] [PubMed]
- Vagin, A. A., Steiner, R. A., Lebedev, A. A., Potterton, L., McNicholas, S., Long, F. & Murshudov, G. N. (2004). Acta Cryst. D60, 2184–2195. [DOI] [PubMed]
- Winn, M. D. et al. (2011). Acta Cryst. D67, 235–242.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: human GDF11, 5e4g
Supporting Information.. DOI: 10.1107/S2053230X16001588/rr5115sup1.pdf