The crystal structure of 5-enolpyruvylshikimate-3-phosphate (EPSP) synthase, a component of the shikimate pathway, was determined during its evaluation as a target for new antimicrobials effective against multidrug-resistant, extensively drug-resistant and pan-drug-resistant A. baumannii. This enzyme is essential for the growth and survival of this clinically important pathogen during host infection.
Keywords: shikimate pathway, Acinetobacter baumannii, essential genes, antibiotic targets, multidrug resistance, EPSP synthase
Abstract
The enzyme 5-enolpyruvylshikimate-3-phosphate (EPSP) synthase catalyzes the sixth step of the seven-step shikimate pathway. Chorismate, the product of the pathway, is a precursor for the biosynthesis of aromatic amino acids, siderophores and metabolites such as folate, ubiquinone and vitamin K. The shikimate pathway is present in bacteria, fungi, algae, plants and apicomplexan parasites, but is absent in humans. The EPSP synthase enzyme produces 5-enolpyruvylshikimate 3-phosphate and phosphate from phosphoenolpyruvate and shikimate 3-phosphate via a transferase reaction, and is the target of the herbicide glyphosate. The Acinetobacter baumannii gene encoding EPSP synthase, aroA, has previously been demonstrated to be essential during host infection for the growth and survival of this clinically important drug-resistant ESKAPE pathogen. Prephenate dehydrogenase is also encoded by the bifunctional A. baumannii aroA gene, but its activity is dependent upon EPSP synthase since it operates downstream of the shikimate pathway. As part of an effort to evaluate new antimicrobial targets, recombinant A. baumannii EPSP (AbEPSP) synthase, comprising residues Ala301–Gln756 of the aroA gene product, was overexpressed in Escherichia coli, purified and crystallized. The crystal structure, determined to 2.37 Å resolution, is described in the context of a potential antimicrobial target and in comparison to EPSP synthases that are resistant or sensitive to the herbicide glyphosate.
1. Introduction
Infections owing to drug-resistant Acinetobacter baumannii are becoming increasingly commonplace, with resultant increases in morbidity, mortality and healthcare-associated costs (Spellberg & Bonomo, 2014 ▸; Villar et al., 2014 ▸). The Centers for Disease Control and Prevention (CDC) has designated the threat level for A. baumannii as serious (Centers for Disease Control and Prevention, 2013 ▸), and it has been included as an ESKAPE pathogen to emphasize the threat to public health (Boucher et al., 2013 ▸; Paterson & Harris, 2015 ▸). Recently, up to 50% of A. baumannii isolates were classified as extensively drug-resistant (XDR) in US intensive-care units (Lee et al., 2014 ▸; Spellberg & Bonomo, 2014 ▸). The promise of a post-antibiotic era is on the cusp of being fulfilled by A. baumannii (Garnacho-Montero & Amaya-Villar, 2010 ▸; Kim et al., 2009 ▸; Napier et al., 2013 ▸; Perez et al., 2007 ▸) and true pan-drug-resistant (PDR) strains have been reported (Göttig et al., 2014 ▸; Rolain et al., 2013 ▸). Therefore, our group has focused on the identification and validation of new or underexploited antimicrobial targets within A. baumannii (Russo et al., 2009 ▸, 2010 ▸; Umland et al., 2012 ▸, 2014 ▸). An efficient genetic screen was developed to identify A. baumannii genes that are essential in vivo (i.e. essential for pathogen growth and survival in an infected host; Umland et al., 2012 ▸). The resulting gene set identified by this in vivo essentiality screen included two A. baumannii shikimate-pathway genes: aroA [encoding 5-enolpyruvylshikimate-3-phosphate (EPSP) synthase] and aroC (encoding chorismate synthase). The A. baumannii aroA gene atypically encodes prephenate dehydrogenase (PD) activity in addition to EPSP synthase, producing a PD-EPSP synthase fusion (Adams et al., 2008 ▸). In most prokaryotes, separate enzymes perform these two activities. PD activity is utilized downstream of the shikimate pathway for the biosynthesis of tyrosine. Thus, PD activity is expected to be dependent upon the presence of an intact shikimate pathway. A third A. baumannii gene in the shikimate pathway, aroK (encoding shikimate kinase), has also been demonstrated to be essential in vivo (Sutton et al., 2015 ▸), further demonstrating the importance of this metabolic pathway during infection.
The shikimate pathway is present in bacteria, fungi, apicomplexan parasites and plants, with the product chorismate serving as a precursor of aromatic amino acids and other aromatic metabolites, including folate, ubiquinone and vitamin K (Abell, 1999 ▸; Bentley & Haslam, 1990 ▸; Haslam, 1974 ▸; Herrmann & Weaver, 1999 ▸; McConkey et al., 2004 ▸). Moreover, a number of important bacterial pathogens utilize chorismate-derived siderophores as virulence factors (Miethke & Marahiel, 2007 ▸). Inhibition of EPSP synthase (also referred to as 3-phosphoshikimate-1-carboxyvinyl transferase; PSCVT) is the basis of the widely used herbicide glyphosate. The EPSP synthases have been divided into two classes according to intrinsic glyphosate sensitivity (Franz et al., 1997 ▸; Funke et al., 2006 ▸; Stallings et al., 1991 ▸). Class I EPSP synthases, which are present in plants and some bacteria (e.g. Salmonella typhimurium and Escherichia coli), are inhibited at low concentrations of glyphosate. Class II EPSP synthases, which are present in many bacterial species, are glyphosate-resistant. Examples of species possessing the class II enzyme include Staphylococcus aureus, Streptococcus pneumoniae and, notably, Agrobacterium sp. strain CP4. The ortholog from this latter species was used to create commercial transgenic glyphosate-resistant crops (Padgette et al., 1995 ▸).
EPSP synthase (EC 2.5.1.19) catalyzes the transfer of the enolpyruvyl moiety of phosphoenolpyruvate (PEP) to the 5-hydroxy position of shikimate 3-phosphate (S3P; Bentley & Haslam, 1990 ▸; Levin & Sprinson, 1964 ▸), a requisite step in the biosynthesis of chorismate and ultimately aromatic metabolites. The in vivo essentiality of A. baumannii aroA is at least in part attributed to the EPSP synthase fragment (residues 301–756), as there is no known enzyme substitute or other route to the synthesis of 5-enolpyruvylshikimate 3-phosphate. This pathway is absent from humans, an attractive feature for novel antimicrobial targets (Coggins et al., 2003 ▸). However, the respective A. baumannii pathway enzymes have not been specifically characterized and validated as antimicrobial targets. Here, the expression, purification, crystallization and structure analysis of unliganded recombinant A. baumannii EPSP (AbEPSP) synthase is reported.
2. Materials and methods
2.1. Cloning, expression and purification of AbEPSP synthase
Primers (sense, 5′-CGCCCGCATATGAATAAGGTGACACA-3′; antisense, 5′-CGAACGGCTCGAGTTATTGGCTAACT-3′) were synthesized (Integrated DNA Technologies, Iowa, USA) and used to amplify via PCR the fragment of the A. baumannii strain 307-0294 aroA gene (ABBFA_001168) encoding EPSP synthase activity (corresponding to amino-acid residues Ala301–Gln756 of the transcribed gene product; Adams et al., 2008 ▸). The PCR product was digested with NdeI and XhoI restriction enzymes and ligated onto the customized expression vector pET-duet-SUMO (Sutton et al., 2015 ▸) to construct pET-SUMO-AbEPSPS. The expression-cassette sequence was verified by DNA sequencing (Roswell Park Cancer Institute Sequencing Facility, New York, USA).
Overexpression of AbEPSP synthase occurred in E. coli Rosetta (DE3) cells grown in LB medium with 100 µg ml−1 ampicillin and 34 µg ml−1 chloramphenicol to an OD600 of 0.8 at 37°C and then induced with 1 mM IPTG and incubated for 4 h. The protein was purified from the crude lysate. The cells were resuspended in lysis buffer consisting of 25 mM HEPES pH 7.5, 250 mM NaCl, 20 mM imidazole, 1 mM β-mercaptoethanol (BME) and were lysed using sonication and a Microfluidizer processor (Microfluidics, Massachusetts, USA). The supernatant was loaded onto an immobilized metal ion-affinity chromatography (IMAC) column (HiTrap, GE Healthcare Life Sciences, Pennsylvania, USA). The His6-SUMO-AbEPSP synthase construct was eluted in 25 mM HEPES pH 7.5, 250 mM NaCl, 1 mM BME with a linear imidazole gradient from 60 mM to 1 M over 100 ml. The His6-SUMO tag was cleaved by overnight incubation with Ulp1 protease using a 500:1 mass ratio. The sample was reapplied onto a HiTrap IMAC column to separate the cleaved affinity tag from the AbEPSP synthase. Gel filtration (Superdex 200 HiLoad 16/60; GE Healthcare Life Sciences) was performed as a final step in 25 mM HEPES pH 7.5, 250 mM NaCl, 1 mM dithiothreitol (DTT). Purified AbEPSP synthase was dialyzed into 25 mM HEPES pH 8.0, 20 mM NaCl, 1 mM DTT and then concentrated to 10 mg ml−1 (Bradford method) by centrifugal ultrafiltration (YM10 Centricon; EMD Millipore, Massachusetts, USA).
2.2. Crystallization
Crystallization screening via microbatch under oil in 1536-multiwell plates was conducted using high-throughput robotics (Luft et al., 2003 ▸). Selected crystallization conditions were optimized via manual hanging-drop vapor diffusion. The crystals used for diffraction data collection were obtained by equilibration at 293 K of a crystallization drop consisting of 3 µl purified AbEPSP synthase at 10.0 mg ml−1 plus 3 µl reservoir solution against 500 µl reservoir solution [100 mM bis-tris propane pH 7.0, 100 mM potassium bromide, 40%(w/v) PEG 8000].
2.3. Diffraction data collection and processing
A crystal was harvested using a nylon loop and then flash-cooled in a cryostream at 100 K. Diffraction data were collected using a Saturn 944+ CCD detector and a MicroMax-007 HF copper rotating-anode X-ray source equipped with Osmic Varimax HF optics with a crystal-to-detector distance of 45 mm and 0.5° oscillation and 30 s exposure per frame. The data were integrated and scaled using HKL-2000 (Otwinowski & Minor, 1997 ▸). Mycobacterium tuberculosis EPSP synthase in complex with S3P (PDB entry 2o0b; Mycobacterium Tuberculosis Structural Proteomics Project, unpublished work) was used as the search model for molecular replacement in MOLREP (Vagin & Teplyakov, 2010 ▸) to generate an initial model. The resulting model was refined using PHENIX (Adams et al., 2010 ▸), including noncrystallographic symmetry (NCS) torsion-based restraints employing a flexible target function that smoothly shuts off to allow local differences between NCS-related chains. Translation–libration–screw rotation model (TLS) parameter refinement (Afonine et al., 2012 ▸) was included at later stages of refinement. Riding H atoms were included during refinement to improve geometry. Iterative manual model building was performed with Coot (Emsley et al., 2010 ▸). Structure analysis and validation made use of PyMOL (Schrödinger), PHENIX (Adams et al., 2010 ▸), jsPISA (Krissinel, 2015 ▸) and the validation tools present in the wwPDB Deposition Tool (http://deposit.wwpdb.org/deposition). WebLogo was used to analyze sequence motifs (Crooks et al., 2004 ▸). The refined coordinates and scaled diffraction data have been deposited in the PDB (PDB entry 5buf).
3. Results and discussion
3.1. Overall structure of AbEPSP synthase
Recombinant AbEPSP synthase, comprising amino-acid residues Ala301–Gln756 of the A. baumannii aroA gene product, was expressed and purified for crystallization by a three-step chromatography strategy. Diffraction-quality rod-shaped crystals were readily obtained, yielding data to 2.37 Å resolution on a rotating-anode X-ray source (Table 1 ▸). Phasing was accomplished by routine molecular replacement using M. tuberculosis EPSP synthase as a search model (PDB entry 2o0b; 22% sequence identity), revealing two protomers to be present within the crystallographic asymmetric unit. The sequence corresponding to AbEPSP synthase was built into the initial electron-density map and the subsequent model was refined. Residues Thr312–Gln756 were defined in electron density for both protomers, with the N-terminal residues Ala301–Val311 presumably disordered. The two protomers were structurally similar, with a root-mean-square deviation (r.m.s.d.) of 1.08 Å for aligned Cα atoms.
Table 1. X-ray data-collection and refinement statistics.
Values in parentheses are for the highest resolution shell.
| PDB code | 5buf |
| Data collection | |
| Space group | P212121 |
| Unit-cell parameters (Å) | a = 73.9, b = 103.4, c = 113.2 |
| Completeness (%) | 96.9 (99.0) |
| Resolution range (Å) | 34.79–2.37 (2.41–2.37) |
| Total No. of reflections | 261575 |
| No. of unique reflections | 34884 (1759) |
| Multiplicity | 7.5 (6.3) |
| R meas | 0.080 (0.42) |
| 〈I/σ(I)〉 | 17.5 (4.7) |
| Wilson B factor (Å2) | 28.9 |
| Refinement | |
| Resolution range (Å) | 4.79–2.37 (2.44–2.37) |
| Completeness (%) | 97.0 (99.0) |
| No. of reflections, working set | 33045 |
| No. of reflections, test set | 1748 |
| R cryst/R free | 0.1833 (0.2103)/0.2285 (0.2518) |
| No. of non-H atoms | |
| Protein | 6572 |
| Ion | 64 |
| Water | 423 |
| Model geometry (r.m.s. deviations from ideal) | |
| Bonds (Å) | 0.004 |
| Angles (°) | 0.83 |
| Average B factors (Å2) | |
| Protein | 36.2 |
| Ion | 39.0 |
| Water | 33.0 |
| Ramachandran plot† (%) | |
| Favored | 98.0 |
| Allowed | 1.2 |
| Outliers | 0.34 |
| MolProbity clashscore† | 0.53 |
| Rotamer outliers† (%) | 0.72 |
As calculated by MolProbity; MolProbity clashscore corresponds to the 100th percentile (i.e. the best) among structures of comparable resolution.
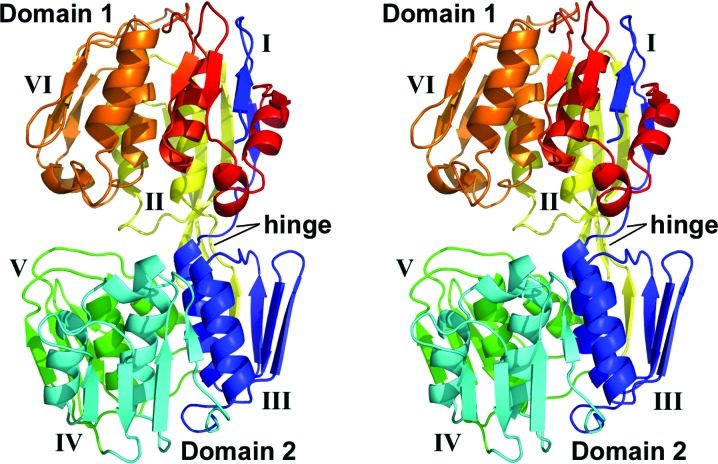
AbEPSP synthase exhibited the twice-repeated domain architecture characteristic of the family. Each domain contains three similar subdomains based upon a βαβαββ core architecture, with the two domains connected by a double hinge comprised of residues Thr329–Asp333 and Val544–Asp547. Domain 1 contained subdomains I, II and VI, and domain 2 contained subdomains III, IV and V (Fig. 1 ▸), as previously observed (Stallings et al., 1991 ▸). Subdomain I contained both the N- and C-termini of the AbEPSP synthase peptide chain owing to the dual crossover between domains. Specifically, the N-terminal residues Gln313–Ile317 form a single β-strand within subdomain I. The remainder of this subdomain is comprised of the C-terminal residues Gly701–Gln756. Likewise, subdomains II and III are formed from a single β-strand (residues Phe324–Phe328 and Thr540–Val544, respectively) interacting with a discontinuous range of residues (residues Asp547–Gly611 and Asp333–Gly388, respectively) that form the remainder of each subdomain. Subdomains I and IV possess an additional 310-helix in the connecting residues between the second β-strand and the second α-helix of the core subdomain fold, resulting in a modified βαβ310αββ topology. Subdomain VI displays an analogous 310-helix, which is likely to participate in the transition between the open (unliganded) and closed (substrate-bound) conformations (Funke et al., 2006 ▸; Park et al., 2004 ▸), plus a second additional helix, to yield a β310αβ310αββ fold. Similar deviations from the core fold have previously been observed in S. pneumoniae EPSP (SpEPSP) synthase and Agrobacterium sp. strain CP4 EPSP (CP4EPSP) synthase (Funke et al., 2006 ▸; Park et al., 2004 ▸). An approximately twofold-symmetric homodimer was formed by the two AbEPSP synthase protomers present within the crystallographic asymmetric unit. However, the dimer interface only involved subdomain IV (domain 2) and buried only ∼550 Å2 per protomer. Thus, this dimer was predicted not to be a stable biological assembly using jsPISA analysis. Moreover, the protein eluted from a Superdex 200 gel-filtration column as expected for a monomer of 48 kDa. Similarly, the crystallized SpEPSP synthase was observed to form oligomers, but analysis of the solution state indicated only monomers to be present for both unliganded and liganded forms (Park et al., 2004 ▸).
Figure 1.
Stereoview of AbEPSP synthase domain organization. The peptide chain is colored sequentially: blue (N-terminus; Thr312–Leu394), cyan (Lys395–Gln468), green (Gln469–Leu537), yellow (Val538–Thr614), orange (Leu615–Gly692) and red (C-terminus; Asp693–Gln756). Individual subdomains are labeled with roman numerals. The N-terminal segment (blue) of the peptide chain participates in the formation of subdomains I, II and III, and a middle segment (yellow) participates in the formation of subdomains II and III. Subdomains IV, V and VI are comprised of contiguous sections of the peptide chain and are colored cyan, green and orange, respectively.
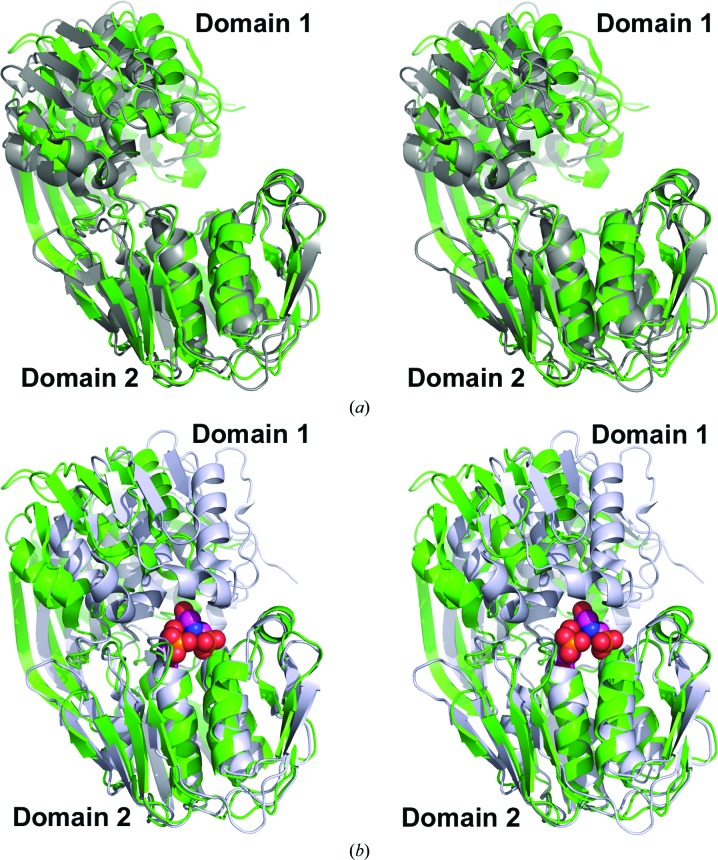
3.2. Substrate-induced conformation change
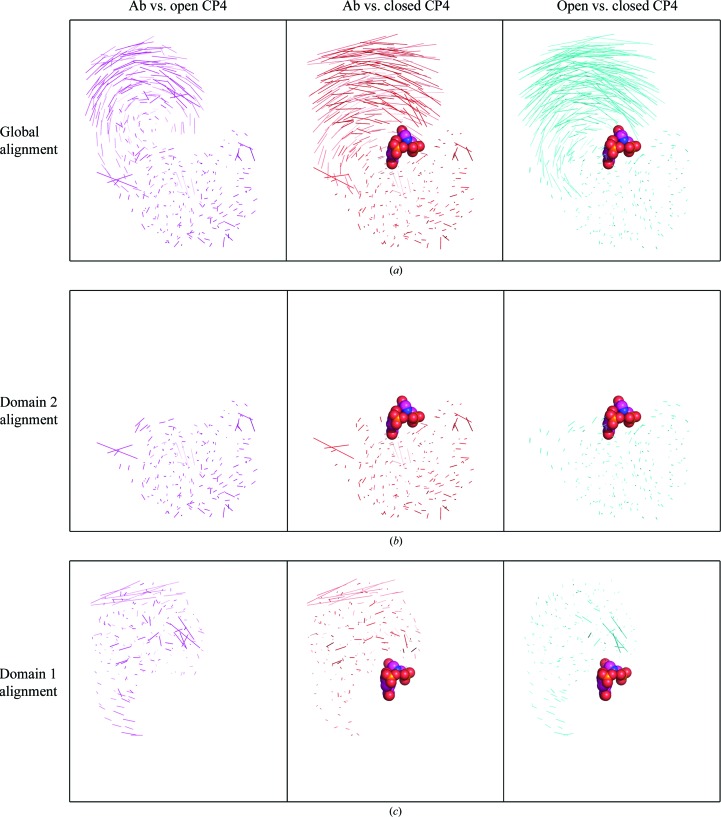
EPSP synthases undergo significant substrate-induced conformational changes upon binding S3P, with complete and productive substrate-binding and active sites formed at the interface between the two domains upon transition to the closed conformation (Funke et al., 2006 ▸; Park et al., 2004 ▸; Schönbrunn et al., 2001 ▸). The AbEPSP synthase structure reported here is in the open conformation, as expected given the absence of bound substrate (Fig. 2 ▸ a). S3P is thought to initially bind to domain 2, triggering the switch to the closed conformation. Superimposing the Cα atoms of domain 2 from AbEPSP synthase (unliganded, open conformation) with those of CP4EPSP synthase (unliganded, open conformation; PDB entry 2gg4; Funke et al., 2006 ▸) and of the CP4EPSP synthase–S3P–glyphosate complex (closed conformation; PDB entry 2gga; Funke et al., 2006 ▸) resulted in r.m.s.d. values of 1.92 and 1.88 Å, respectively, for aligned Cα atoms of domain 2 (Figs. 2 ▸ a and 2 ▸ b). In comparison, the r.m.s.d. was 0.79 Å for the superimposed Cα atoms of domain 2 of the two CP4EPSP synthase structures. Plotting the set of lines connecting paired atoms of two structurally aligned proteins is useful for evaluating both domain-level and localized conformational differences between two members of the same family (i.e. AbEPSP and CP4EPSP synthases) or between the same protein in different states (i.e. open versus closed conformations). The aligned paired-atom lines obtained upon global structural alignments indicated that the largest domain-level conformational differences occur primarily for domain 1 both between EPSP synthase orthologs and between states (Fig. 3 ▸ a). Upon structural alignment of only domain 2, the corresponding paired-atom lines displayed negligible differences within domain 2 both between orthologs and between states (Fig. 3 ▸ b). These analyses suggested that domain 2 is structurally very similar between AbEPSP synthase and CP4EPSP synthase and does not undergo major structural changes upon binding S3P and the subsequent conversion from the open to the substrate-bound closed conformation.
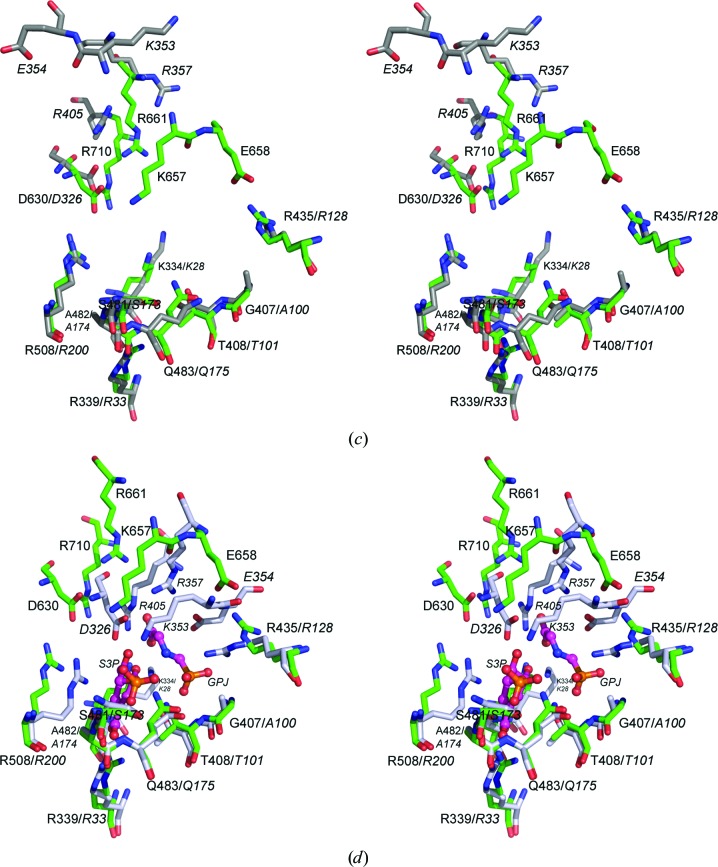
Figure 2.
Stereoviews of AbEPSP synthase and CP4EPSP synthase superimposed. (a, b) Cartoon representations of unliganded AbEPSP synthase (green) superimposed upon (a) the unliganded, open conformation (PDB entry 2gg4; dark gray) and (b) the S3P/glyphosate-bound closed conformation (PDB entry 2gga; gray with ligands displayed as spheres) CP4EPSP synthase structures. (c, d) Predicted ligand-binding residues in AbEPSP synthase (green C atoms) superimposed upon the CP4EPSP synthase residues (italicized residue labels) that participate in S3P and PEP/glyphosate binding in (c) the unliganded open conformation (PDB entry 2gg4; dark gray C atoms) and (d) the S3P/glyphosate-bound closed conformation (PDB entry 2gga; gray C atoms with ligands displayed in ball-and-stick representation with magenta C atoms). In all cases, structures were superimposed using only the Cα atoms of domain 2 to emphasize substrate-induced conformational changes. The orientation of AbEPSP synthase is identical in all panels of the figure.
Figure 3.
Displacements of paired atoms for superimposed EPSP synthase structures. The displayed lines connect atoms paired between superimposed proteins, providing a comparison of global and localized structural differences. Columns are labelled as follows: Ab vs. open CP4, superimposed open (unliganded) AbEPSP and CP4EPSP (PDB entry 2gg4) synthases; Ab vs. closed CP4, superimposed open (unliganded) AbEPSP synthase and closed CP4EPSP synthase–S3P–glyphosate complex (PDB entry 2gga); open vs. closed CP4, superimposed open and closed CP4EPSP synthases (PDB entries 2gg4 and 2gga, respectively). (a) Global alignment: structural alignment over both domains to emphasize gross domain movements. (b) Domain 2: only domain 2 superimposed and displayed to emphasized differences localized within domain 2. (c) Domain 1 alignment: only domain 1 superimposed and displayed to emphasize differences localized within domain 1. SP3 and glyphosate are displayed when present in a structure included in the comparison. The protein orientation is identical to that in Fig. 2 ▸.
Conversely, a similar comparison of superimposed Cα atoms of domain 1 demonstrated that domain 1 exhibits greater localized structural diversity both between orthologs and between the open and closed conformations of the same ortholog (Figs. 2 ▸ b, 3 ▸ a and 3 ▸ c). Specifically, superimposing the domain 1 Cα atoms of AbEPSP synthase (unliganded, open conformation) on those of CP4EPSP synthase (unliganded, open conformation; PDB entry 2gg4) and the CP4EPSP synthase–S3P–glyphosate complex (closed conformation; PDB entry 2gga) resulted in r.m.s.d. values of 3.41 and 3.22 Å, respectively, for aligned domain 1 Cα atoms. By comparison, the r.m.s.d. was 1.65 Å for superimposed Cα atoms of domain 1 of CP4EPSP synthase in open and closed conformations. The paired-atom lines for superimposed Cα atoms of domain 1 of AbEPSP synthase and open CP4EPSP synthase revealed two regions of significant localized differences (Fig. 3 ▸ c). The region distant from the substrate-binding site differed owing to differences in loop structures connecting core secondary-structural elements, and thus is probably functionally unimportant. The second region borders the substrate-binding site and includes AbEPSP synthase residues Thr649–Arg661. Interestingly, these structural differences in domain 1 at the substrate-binding site are largely absent in the alignment of AbEPSP synthase with closed CP4EPSP synthase, but are present for open versus closed CP4EPSP synthase (Fig. 3 ▸ c). This result suggested that the substrate-binding region of domain 1 in unliganded AbEPSP synthase exhibits structural aspects of closed CP4EPSP synthase (Figs. 2 ▸ c and 2 ▸ d).
AbEPSP synthase residues Thr649–Arg661 formed an extended loop that connects the second β-strand to the second α-helix in subdomain VI (domain 1) and has been observed to undergo a conformational change as part of the open-to-closed transition upon S3P binding in CP4EPSP and SpEPSP synthase (Funke et al., 2006 ▸; Park et al., 2004 ▸). This loop contains residues that are conserved in class II EPSP synthases, including several observed to directly participate in S3P and glyphosate binding (Lys353, Glu354 and Arg357 in CP4EPSP synthase; Lys339, Glu340 and Arg343 in SpEPSP synthase; the analogous residues in AbEPSP synthase are Lys657, Glu658 and Arg661; Figs. 2 ▸ c and 2 ▸ d). In both CP4EPSP and SpEPSP synthase this loop converted from an extended structure that is poorly defined by electron density to a compact stabilized conformation, including formation of the previously mentioned 310-helix, upon transition to the closed conformation. Moreover, both ligand-binding interactions plus interdomain interactions (e.g. an Arg128 guanidino–Val352 carbonyl hydrogen bond and an Arg132–Glu349 salt bridge in CP4EPSP synthase) stabilize this region in the closed conformation.
The analogous unliganded AbEPSP synthase loop exhibited higher than average temperature factors, but was present in a conformation similar to that expected for the substrate-induced closed conformation, including the formation of the 310-helix. However, because the overall protein was in the open conformation this loop lacked stabilizing contacts with domain 1 residues or bound ligand. This behavior has previously been reported for unliganded CP4EPSP synthase crystallized in the presence of the monovalent cations K+ or Rb+. The pre-formation of the substrate-binding sites may be related to the catalytic activity enhancement observed for class II EPSP synthases in the presence of monovalent cations (Du et al., 2000 ▸; Funke et al., 2006 ▸). Crystallization of AbEPSP synthase occurred in the presence of NaCl and KBr, and this structural observation suggests that AbEPSP synthase activity will be also enhanced by monovalent cations. However, as in CP4EPSP synthase, no specific cation-binding sites were observed in the crystal structure.
3.3. AbEPSP synthase is predicted to belong to the class II subfamily
The sequence and tertiary structure of AbEPSP synthase were analyzed to predict whether it belonged to the class I (glyphosate-sensitive) or the class II (glyphosate-resistant) subfamily, as it has not previously been classified. CP4EPSP synthase is a prototypical class II EPSP synthase owing to its ability to withstand high concentrations of the inhibitor while maintaining high catalytic efficiency. Residue Ala100 of CP4EPSP synthase, which is located in a short loop within subdomain IV (domain 2), has been identified as a key determinant of glyphosate resistance (Funke et al., 2006 ▸). The Cβ atom of Ala100 protrudes into the glyphosate-binding site, resulting in reduced glyphosate affinity while minimally affecting the binding of the smaller PEP molecule (Funke et al., 2006 ▸; Eschenburg et al., 2002 ▸). In contrast, AbEPSP synthase has a glycine (Gly407) at this position. Glycine, lacking a side chain, would not be expected to cause a steric clash with bound glyphosate. However, CP4EPSP synthase is an outlier in the class II subfamily, as most members possess a glycine rather than an alanine at this position. Thus, the presence of this glycine is not a strong predictor of glyphosate sensitivity. Rather, the presence of Ala100 in CP4EPSP synthase or other EPSP synthases serves as a marker of enhanced glyphosate resistance (Eschenburg et al., 2002 ▸; Funke et al., 2006 ▸; Sost & Amrhein, 1990 ▸).
Structure–function studies strongly support that glyphosate sensitivity is largely dictated by second-sphere residues near the overlapping PEP- and glyphosate-binding sites causing subtle changes in binding-site architecture rather than primarily owing to notable differences in residues directly contacting PEP or glyphosate (Eschenburg et al., 2002 ▸; Funke et al., 2009 ▸; Healy-Fried et al., 2007 ▸; Priestman et al., 2005 ▸; Sammons & Gaines, 2014 ▸). Several class-differentiating motifs have been reported (Funke et al., 2009 ▸; Li et al., 2009 ▸). Class I enzymes have a highly conserved motif corresponding to E. coli EPSP (EcEPSP) synthase residues 90LFLGN AGTAM RPLAA104 (Fig. 4 ▸ a; Funke et al., 2009 ▸). Mutations within this conserved motif can confer glyphosate resistance (e.g. EcEPSP synthase G96A or the double mutant T97I/P101S; Eschenburg et al., 2002 ▸; Funke et al., 2009 ▸; Healy-Fried et al., 2007 ▸). The EcEPSP synthase G96A mutant mimics the glyphosate-resistance determinant Ala100 in CP4EPSP synthase. However, this EcEPSP synthase mutant significantly reduced the affinity for both glyphosate and PEP. Importantly, neither Thr97 nor Pro101 directly contact glyphosate, but the dual mutation shifted the position of Gly96 to interfere with glyphosate binding while minimally altering PEP utilization. AbEPSP synthase has a substantially different sequence (401LYMGN SGTSM RLLSG415) to that of the conserved class I motif. Moreover, this motif is not conserved in CP4EPSP synthase and other class II EPSP synthases (Figs. 4 ▸ b and 4 ▸ c), suggesting that AbEPSP synthase does not belong to the glyphosate-sensitive class I.
Figure 4.
AbEPSP synthase lacks a motif conserved within class I (glyphosate-sensitive) EPSP synthases. (a) A logo representation of a motif adjacent to the PEP/glyphosate-binding site that is highly conserved within class I EPSP synthases (Funke et al., 2009 ▸). The residue numbering is based upon the E. coli ortholog. (b) Logo representation of the corresponding region in class II EPSP synthases, with residue numbering based on the Agrobacterium sp. strain CP4 ortholog crystal structures. [The class II logo was constructed using EPSP synthase sequences from Agrobacterium sp. CP4 (Q0R4E4), Pseudomonas sp. PG2982 (P0A2Y4), Achromobacter sp. LBAA (P0A2Y5), Pseudomonas stutzeri A1501 (ABP79994), Streptococcus pneumoniae (Q9S400) and Staphylococcus aureus (Q05615).] (c) The sequence of the corresponding region in AbEPSP synthase, with asterisks indicating residues identical to the class I motif.
Class II EPSP synthases possess a conserved RPMXR motif that is required for both catalytic activity and glyphosate insensitivity. The leading arginine residue forms a salt bridge to the phosphate group of PEP (e.g. Arg128 of CP4EPSP synthase; PDB entry 2gga; Funke et al., 2006 ▸; Li et al., 2009 ▸). This motif is present in AbEPSP synthase (435RPMER439), further supporting the classification of AbEPSP synthase as a class II (glyphosate-resistant) subfamily member. The nonconserved fourth position in this motif accommodates a variety of amino-acid types, including Gly, Asn, Asp, Arg and Lys (Li et al., 2009 ▸). The glutamate residue present in AbEPSP synthase further expands the allowed repertoire of this variable position. The equivalent residues in class I enzymes display similarity at the N-terminus of the motif and become dissimilar towards the C-terminus (RPhXX, where h is a hydrophobic residue).
4. Conclusions
Antimicrobials effective against the ESKAPE pathogens, including A. baumannii, are urgently needed, especially those that employ new mechanisms of action. Several properties of AbEPSP synthase are appealing as an antimicrobial target, including the in vivo essentiality of both itself and other enzymes in the shikimate metabolic pathway and the lack of a human homolog. Furthermore, the effectiveness of glyphosate as an herbicide targeting EPSP synthases suggests that the family is druggable. The unliganded crystal structure of AbEPSP synthase was determined as part of its evaluation as an antimicrobial target. The AbEPSP synthase structure and sequence suggested that it belongs to the glyphosate-resistant class II subfamily. However, it is likely not to be as tolerant to high glyphosate concentrations as CP4EPSP synthase owing to the presence of a glycine (Gly407) rather than an alanine at the PEP- and glyphosate-binding site. It was also predicted that monovalent cations enhance the catalytic activity of AbEPSP synthase, based upon the presence of a 310-helix in a key ligand-binding loop within subdomain VI in the unliganded state.
Previous research on EPSP synthases primarily focused on the overlapping PEP- and glyphosate-binding sites owing to the role of the enzyme as a commercially valuable herbicide target. Importantly, these efforts identified a number of natural and engineered glyphosate-resistant EPSP synthases. However, many of these engineered mutants were simultaneously catalytically less efficient. This observation suggests that a small-molecule antimicrobial designed to bind competitively with PEP may be subject to the development of resistant mutants, some of which may also exhibit decreased biofitness. The S3P binding site has been less explored as an inhibitor- or drug-binding site. It offers potential for the development of a multi-target therapeutic, as the S3P substrate shares chemical similarity with the substrates of other members, shikimate dehydrogenase and shikimate kinase, of the essential shikimate pathway (Hsu et al., 2013 ▸), which could hold the promise of increased durability. Therefore, next-stage studies in the assessment of AbEPSP are warranted.
Supplementary Material
PDB reference: EPSP synthase from A. baumannii, 5buf
Acknowledgments
This work was supported in part by a Telemedicine and Advance Technical Research Center (TATRC) Cooperative Agreement (W23RYX1055N607; TAR, LWS and TCU), an Interdisciplinary Grant from the University at Buffalo (TAR and TCU) and a VA Merit Review grant from the Department of Veterans Affairs (TAR). We wish to thank Ms Jessica Graham, Ms Changyi Ji, Mr Ritwik Nandagiri and Mr Hong Guo for assistance in AbEPSP synthase expression-vector preparation. The Ulp1 protease expression vector was a kind gift from Dr Christopher Lima (Sloan-Kettering Institute).
References
- Abell, C. (1999). Comprehensive Natural Products Chemistry, edited by D. Barton, K. Nakanishi & O. Meth-Cohn, pp. 573–607. Oxford: Pergamon.
- Adams, P. D. et al. (2010). Acta Cryst. D66, 213–221.
- Adams, M. D., Goglin, K., Molyneaux, N., Hujer, K. M., Lavender, H., Jamison, J. J., MacDonald, I. J., Martin, K. M., Russo, T., Campagnari, A. A., Hujer, A. M., Bonomo, R. A. & Gill, S. R. (2008). J. Bacteriol. 190, 8053–8064. [DOI] [PMC free article] [PubMed]
- Afonine, P. V., Grosse-Kunstleve, R. W., Echols, N., Headd, J. J., Moriarty, N. W., Mustyakimov, M., Terwilliger, T. C., Urzhumtsev, A., Zwart, P. H. & Adams, P. D. (2012). Acta Cryst. D68, 352–367. [DOI] [PMC free article] [PubMed]
- Bentley, R. & Haslam, E. (1990). Crit. Rev. Biochem. Mol. Biol. 25, 307–384. [DOI] [PubMed]
- Boucher, H. W., Talbot, G. H., Benjamin, D. K. Jr, Bradley, J., Guidos, R. J., Jones, R. N., Murray, B. E., Bonomo, R. A. & Gilbert, D. (2013). Clin. Infect. Dis. 56, 1685–1694. [DOI] [PMC free article] [PubMed]
- Centers for Disease Control and Prevention (2013). Antibiotic Resistance Threats in the United States, 2013, pp. 59–60. http://www.cdc.gov/drugresistance/threat-report-2013. Atlanta: Centers for Disease Control and Prevention.
- Coggins, J. R., Abell, C., Evans, L. B., Frederickson, M., Robinson, D. A., Roszak, A. W. & Lapthorn, A. P. (2003). Biochem. Soc. Trans. 31, 548–552. [DOI] [PubMed]
- Crooks, G. E., Hon, G., Chandonia, J.-M. & Brenner, S. E. (2004). Genome Res. 14, 1188–1190. [DOI] [PMC free article] [PubMed]
- Du, W., Wallis, N. G., Mazzulla, M. J., Chalker, A. F., Zhang, L., Liu, W.-S., Kallender, H. & Payne, D. J. (2000). Eur. J. Biochem. 267, 222–227. [DOI] [PubMed]
- Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. (2010). Acta Cryst. D66, 486–501. [DOI] [PMC free article] [PubMed]
- Eschenburg, S., Healy, M. L., Priestman, M. A., Lushington, G. H. & Schönbrunn, E. (2002). Planta, 216, 129–135. [DOI] [PubMed]
- Franz, J. E., Mao, M. K. & Sikorski, J. A. (1997). Glyphosate: A Unique Global Herbicide. Washington: American Chemical Society.
- Funke, T., Han, H., Healy-Fried, M. L., Fischer, M. & Schönbrunn, E. (2006). Proc. Natl Acad. Sci. USA, 103, 13010–13015. [DOI] [PMC free article] [PubMed]
- Funke, T., Yang, Y., Han, H., Healy-Fried, M., Olesen, S., Becker, A. & Schönbrunn, E. (2009). J. Biol. Chem. 284, 9854–9860. [DOI] [PMC free article] [PubMed]
- Garnacho-Montero, J. & Amaya-Villar, R. (2010). Curr. Opin. Infect. Dis. 23, 332–339. [DOI] [PubMed]
- Göttig, S., Gruber, T. M., Higgins, P. G., Wachsmuth, M., Seifert, H. & Kempf, V. A. (2014). J. Antimicrob. Chemother. 69, 2578–2579. [DOI] [PubMed]
- Haslam, E. (1974). The Shikimate Pathway, edited by E. Haslam, pp. 128–185. London: Butterworth-Heinemann.
- Healy-Fried, M. L., Funke, T., Priestman, M. A., Han, H. & Schonbrunn, E. (2007). J. Biol. Chem. 282, 32949–32955. [DOI] [PubMed]
- Herrmann, K. M. & Weaver, L. M. (1999). Annu. Rev. Plant Physiol. Plant Mol. Biol. 50, 473–503. [DOI] [PubMed]
- Hsu, K.-C., Cheng, W.-C., Chen, Y.-F., Wang, W.-C. & Yang, J.-M. (2013). PLoS Comput. Biol. 9, e1003127. [DOI] [PMC free article] [PubMed]
- Kim, B.-N., Peleg, A. Y., Lodise, T. P., Lipman, J., Li, J., Nation, R. & Paterson, D. L. (2009). Lancet Infect. Dis. 9, 245–255. [DOI] [PMC free article] [PubMed]
- Krissinel, E. (2015). Nucleic Acids Res. 43, W314–W319. [DOI] [PMC free article] [PubMed]
- Lee, H.-Y., Chen, C.-L., Wu, S.-R., Huang, C.-W. & Chiu, C.-H. (2014). Crit. Care Med. 42, 1081–1088. [DOI] [PubMed]
- Levin, J. G. & Sprinson, D. B. (1964). J. Biol. Chem. 239, 1142–1150. [PubMed]
- Li, L., Lu, W., Han, Y., Ping, S., Zhang, W., Chen, M., Zhao, Z., Yan, Y., Jiang, Y. & Lin, M. (2009). J. Biotechnol. 144, 330–336. [DOI] [PubMed]
- Luft, J. R., Collins, R. J., Fehrman, N. A., Lauricella, A. M., Veatch, C. K. & DeTitta, G. T. (2003). J. Struct. Biol. 142, 170–179. [DOI] [PubMed]
- McConkey, G. A., Pinney, J. W., Westhead, D. R., Plueckhahn, K., Fitzpatrick, T. B., Macheroux, P. & Kappes, B. (2004). Trends Parasitol. 20, 60–65. [DOI] [PubMed]
- Miethke, M. & Marahiel, M. A. (2007). Microbiol. Mol. Biol. Rev. 71, 413–451. [DOI] [PMC free article] [PubMed]
- Napier, B. A., Burd, E. M., Satola, S. W., Cagle, S. M., Ray, S. M., McGann, P., Pohl, J., Lesho, E. P. & Weiss, D. S. (2013). MBio, 4, e00021-13. [DOI] [PMC free article] [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol. 276, 307–326. [DOI] [PubMed]
- Padgette, S. R., Kolacz, K. H., Delannay, X., Re, D. B., LaVallee, B. J., Tinius, C. N., Rhodes, W. K., Otero, Y. I., Barry, G. F., Eichholtz, D. A., Peschke, V. M., Nida, D. L., Taylor, N. B. & Kishore, G. M. (1995). Crop Sci. 35, 1451–1461.
- Park, H., Hilsenbeck, J. L., Kim, H. J., Shuttleworth, W. A., Park, Y. H., Evans, J. N., Kang, C. (2004). Mol. Microbiol. 51, 963–971. [DOI] [PubMed]
- Paterson, D. L. & Harris, P. N. (2015). Clin. Infect. Dis. 61, 155–156.
- Perez, F., Hujer, A. M., Hujer, K. M., Decker, B. K., Rather, P. N. & Bonomo, R. A. (2007). Antimicrob. Agents Chemother. 51, 3471–3484. [DOI] [PMC free article] [PubMed]
- Priestman, M. A., Healy, M. L., Funke, T., Becker, A. & Schönbrunn, E. (2005). FEBS Lett. 579, 5773–5780. [DOI] [PubMed]
- Rolain, J.-M., Diene, S. M., Kempf, M., Gimenez, G., Robert, C. & Raoult, D. (2013). Antimicrob. Agents Chemother. 57, 592–596. [DOI] [PMC free article] [PubMed]
- Russo, T. A., Luke, N. R., Beanan, J. M., Olson, R., Sauberan, S. L., MacDonald, U., Schultz, L. W., Umland, T. C. & Campagnari, A. A. (2010). Infect. Immun. 78, 3993–4000. [DOI] [PMC free article] [PubMed]
- Russo, T. A., MacDonald, U., Beanan, J. M., Olson, R., MacDonald, I. J., Sauberan, S. L., Luke, N. R., Schultz, L. W. & Umland, T. C. (2009). J. Infect. Dis. 199, 513–521. [DOI] [PubMed]
- Sammons, R. D. & Gaines, T. A. (2014). Pest Manag. Sci. 70, 1367–1377. [DOI] [PMC free article] [PubMed]
- Schönbrunn, E., Eschenburg, S., Shuttleworth, W. A., Schloss, J. V., Amrhein, N., Evans, J. N. & Kabsch, W. (2001). Proc. Natl Acad. Sci. USA, 98, 1376–1380. [DOI] [PMC free article] [PubMed]
- Sost, D. & Amrhein, N. (1990). Arch. Biochem. Biophys. 282, 433–436. [DOI] [PubMed]
- Spellberg, B. & Bonomo, R. A. (2014). Crit. Care Med. 42, 1289–1291. [DOI] [PMC free article] [PubMed]
- Stallings, W. C., Abdel-Meguid, S. S., Lim, L. W., Shieh, H.-S., Dayringer, H. E., Leimgruber, N. K., Stegeman, R. A., Anderson, K. S., Sikorski, J. A., Padgette, S. R. & Kishore, G. M. (1991). Proc. Natl Acad. Sci. USA, 88, 5046–5050. [DOI] [PMC free article] [PubMed]
- Sutton, K. A., Breen, J., MacDonald, U., Beanan, J. M., Olson, R., Russo, T. A., Schultz, L. W. & Umland, T. C. (2015). Acta Cryst. D71, 1736–1744. [DOI] [PubMed]
- Umland, T. C., Schultz, L. W., MacDonald, U., Beanan, J. M., Olson, R. & Russo, T. A. (2012). MBio, 3, e00113-12. [DOI] [PMC free article] [PubMed]
- Umland, T. C., Schultz, L. W. & Russo, T. A. (2014). Future Microbiol. 9, 1113–1116. [DOI] [PubMed]
- Vagin, A. & Teplyakov, A. (2010). Acta Cryst. D66, 22–25. [DOI] [PubMed]
- Villar, M., Cano, M. E., Gato, E., Garnacho-Montero, J., Cisneros, J. M., Ruíz de Alegría, C., Fernández-Cuenca, F., Martínez-Martínez, L., Vila, J., Pascual, A., Tomás, M., Bou, G. & Rodríguez-Baño, J. (2014). Medicine, 93, 202–210. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: EPSP synthase from A. baumannii, 5buf