Abstract
Background
Interpreting human immunodeficienc virus type 1 (HIV-1) genotypic drug-resistance test results is challenging for clinicians treating HIV-1–infected patients. Multiple drug-resistance interpretation algorithms have been developed, but their predictive value has rarely been evaluated using contemporary clinical data sets.
Methods
We examined the predictive value of 4 algorithms at predicting virologic response (VR) during 734 treatment-change episodes (TCEs). VR was define as attaining plasma HIV-1 RNA levels below the limit of quantification Drug-specifi genotypic susceptibility scores (GSSs) were calculated by applying each algorithm to the baseline genotype. Weighted GSSs were calculated by multiplying drug-specifi GSSs by antiretroviral (ARV) potency factors. Regimen-specifi GSSs (rGSSs) were calculated by adding unweighted or weighted drug-specif c GSSs for each salvage therapy ARV. The predictive value of rGSSs were estimated by use of multivariate logistic regression.
Results
Of 734 TCEs, 475 (65%) were associated with VR. The rGSSs for the 4 algorithms were the variables most strongly predictive of VR. The adjusted rGSS odds ratios ranged from 1.6 to 2.2 (P < .001). Using 10-fold cross-validation, the averaged area under the receiver operating characteristic curve for all algorithms increased from 0.76 with unweighted rGSSs to 0.80 with weighted rGSSs.
Conclusions
Unweighted and weighted rGSSs of 4 genotypic resistance algorithms were the strongest independent predictors of VR. Optimizing ARV weighting may further improve VR predictions.
Retrospective studies have shown that the presence of human immunodeficienc virus type 1 (HIV-1) drug resistance before the start of a new antiretroviral (ARV) regimen is an independent predictor of the virologic response (VR) to that regimen [1]. Prospective controlled studies have shown that patients whose physicians have access to drug-resistance data respond better to therapy than those whose physicians do not [2–5]. The accumulation of such retrospective and prospective data has led to the routine use of genotypic resistance testing in the management of HIV-1–infected patients [6–8].
However, interpreting the results of HIV-1 genotypic drug-resistance tests is one of the most diffi ult challenges facing clinicians who care for HIV-1–infected patients. First, there are many mutations associated with drug resistance [9]. Second, there are synergistic and antagonistic interactions among these drug-resistance mutations [10]. Third, some mutations may not reduce susceptibility by themselves but may compensate for the effect of other mutations [11] or may be sentinel indicators of emerging drug resistance. As a result, several different algorithms have been developed for interpreting HIV-1 genotypic drug-resistance test results.
Several studies have compared the predictive ability of different genotypic drug-resistance algorithms using retrospective clinical data sets [12–17]. In the present study, we evaluate the predictive value of 4 genotypic drug-resistance test interpretation algorithms in a patient population undergoing a wide range of salvage ARV therapy.
METHODS
Study Subjects and Treatment-Change Episodes (TCEs)
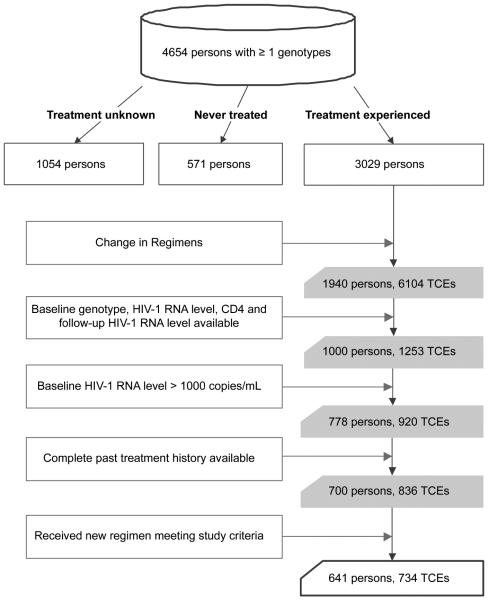
The study subjects were from 16 clinics of the Kaiser-Permanente Medical Care Program–Northern California and had plasma HIV-1 samples sent to Stanford University Hospital for genotypic drug-resistance testing between 1998 and 2007. Eligible subjects included those with a valid TCE, define as a change in ARVs within 24 weeks of a genotypic drug-resistance test and characterized by the administration of a new salvage regimen for ≥4 weeks (figu e 1). Additional requirements included a plasma HIV-1 RNA level of >1000 copies/mL obtained within 8 weeks before the ARV change and ≥2 plasma HIV-1 RNA levels obtained between 4 and 36 weeks after the ARV change. We also required that a complete list of all the ARVs that a patient had ever received be available.
This protocol received expedited approval by institutional review boards at Stanford University and Kaiser-Permanente Medical Care Program–Northern California. A waiver of consent was provided because only anonymized retrospective data were used.
The VR to the new salvage regimen was classifie as sustained for patients with ≥2 subsequent plasma HIV-1 RNA levels below the limit of quantificatio (i.e., <75 copies/mL [hereafter “BLQ”], as determined by use of the Versant bDNA assay [Bayer]), transient for those with a single plasma HIV-1 RNA level BLQ and absent for those without a plasma HIV-1 RNA level BLQ.
Genotypic Susceptibility Scores (GSSs)
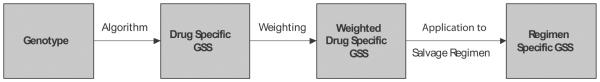
Four genotypic interpretation algorithms were used, including Agence National de Recerche sur le SIDA (hereafter, “ANRS”; version 2007.10) [18], HIV Drug Resistance Database (hereafter, “HIVdb”; version 4.3.1) [19], Rega (version 7.1.1) [20], and ViroSeq (version 2.8) [21]. A 3-step procedure was used to derive a regimen-specifi GSS (rGSS) from the baseline genotype (figu e 2). First, for each algorithm, the drug-specifi GSS for each ARV was calculated. The assigned drug-specif c GSS was 1.0 for ARVs for which HIV-1 was predicted to be fully susceptible and 0 for ARVs for which HIV-1 was predicted to be fully resistant. A drug-specifi GSS between 0 and 1.0 was assigned to ARVs predicted to have intermediate levels of resistance. For each algorithm, the assigned drug-specif c GSS was proportional to the extent of predicted resistance. A drugspecifi GSS of 1.0 was assigned to the fusion inhibitor enfuvirtide if it had not been used previously; otherwise, a GSS of 0 was assigned. Second, because ARVs vary in their potency, define as their antiviral activity and genetic barrier to resistance, we also calculated 2 weighted GSSs. The fi st weighting entailed multiplying the drug-specifi GSSs for boosted protease inhibitors (PIs) by 1.5 (“boosted PI” weighting). This was motivated by the Rega algorithm, the only algorithm of the 4 that provides instructions for creating a weighted GSS. The second “comprehensive” weighting entailed multiplying the drug-specifi GSSs for boosted PIs by 2.0 and those for nucleoside (nucleotide) reverse-transcriptase (RT) inhibitors (NRTIs) by 0.5. The GSSs for nonnucleoside RT inhibitors (NNRTIs), enfuvirtide, and nelfinavi were not weighted. Third, the drugspecifi GSSs (whether weighted or unweighted) for each ARV in a regimen were added to obtain an rGSS. We evaluated each of 12 rGSSs (4 algorithms times 3 weighting schemes) in subsequent analyses.
Among patients who received PIs, only those who received nelfinavi or a ritonavir-boosted PI were included because the HIVdb and ViroSeq algorithms provide interpretations for ritonavir-boosted PIs but not unboosted PIs. TCEs from patients who received a combination of ≥2 PIs plus ritonavir (dual-boosted PIs) were excluded. Among patients who received NNRTIs, only those who received efavirenz or nevirapine were included, because ANRS does not provide an interpretation for delavirdine. TCEs for which the new salvage regimen contained raltegravir, maraviroc, and etravirine were excluded because these ARVs were licensed only in the last months of the study.
Statistical Analysis
Potential explanatory variables
For each TCE, the following groups of covariates were analyzed, in addition to the rGSS: (1) age, sex, and race; (2) features of the ARV treatment history before the TCE, including duration of therapy, the number of ARVs received, the number of ARV regimens and classes received, the number of PIs received, the number of NRTIs and NNRTIs received, the number of non–highly active ARV therapy (HAART) regimens received, the year ARV therapy was firs begun, and the patient’s history of ever having attained a VR; (3) plasma HIV-1 RNA level and CD4 cell count at baseline; and (4) features of the salvage regimen, including the year of the TCE and the number of ARVs, new ARVs, and new ARV drug classes received.
Model selection
Univariate analyses were performed to quantify the association between each covariate and VR. Then, for each of the 4 algorithms and 3 GSS weighting schemes (12 models), we performed forward stepwise logistic regression with all covariates that had a statistically signifi ant association with VR in univariate analyses. Among the covariates selected by this approach, those with statistically signifi ant associations (P < .05) in ≥6 of 12 models were considered robust covariates.
Multivariate logistic regression analyses
We performed multivariate logistic regression to evaluate the association between each of 12 rGSSs and VR, adjusted for the robust covariates. The adjusted odds ratios (ORs) for attaining VR for each unit increase in the rGSS and the goodness of fi (1-residual deviance/null deviance) were calculated. The prediction of each multivariate logistic regression model was tested using 10-fold cross-validation. With the R package ROCR (version 1.0-2), the receiver operating characteristic (ROC) curve of each prediction result for the 10 test sets was calculated to analyze the trade-off between the proportion of true-positive (correct VR prediction) and false-positive (incorrect VR prediction) results across the range of possible prediction cutoffs [22].
Classificatio tree
To examine the effects of rGSS and the robust covariates on the numbers of patients who attained VR, we built classificatio trees for each of the 12 models, using the R package Rpart (Recursive partitioning; version 3.1-39) (http://cran.r-project.org/web/packages/rpart/index.html). Tree pruning was performed using the default parameters in Rpart to minimize the complexity of the tree as a function of improvement in classific tion accuracy.
RESULTS
TCE characteristics
Of the 4654 patients who underwent genotypic drug-resistance testing, 3600 had treatment histories available, including 3029 who received ARVs and 571 who received no ARVs. Of the 3029 ARV-treated patients, 641 patients had 734 valid TCEs (figure 3 and the appendix). The median duration between the baseline genotype and the ARV change was 3 weeks (interquartile range, 1–8 weeks). A mean of 3.4 plasma HIV-1 RNA level test results were available during the 36-week follow-up period, including ≥1 plasma HIV-1 RNA level test result available for weeks 1–12, 13–24, and 25–36 for 82%, 80%, and 68% of TCEs, respectively.
Figure 3.
Description of the complete patient population from which the treatment-change episodes (TCEs) were derived. Of 4654 patients who underwent genotypic drug-resistance testing between 1998 and 2007, 3600 patients had a known history of antiretroviral (ARV) treatment. Of these, 3029 had received ≥1 ARV drug treatment regimens, and 641 subsequently met study enrollment criteria. These 641 patients had 734 valid TCEs. HIV, human immunodeficiency virus.
The mean age of the patients was 45 years (range, 18–77 years). Ninety-two percent were male; 58% were white, 17% were African American, 16% were Latino, 5% were Asian, and 4% were of unknown race. All but 9 patients were infected with subtype B viruses. The median year in which the TCE occurred was 2002 (range, 1998–2007). The mean baseline plasma HIV-1 RNA level was 4.2 log copies/mL (range, 3.1 to 15.9 log copies/mL), and the mean CD4 cell count was 280 cells/mL (range, 3–1110 cells/mL). Before the TCE, patients had received ARV therapy for a mean of 5.1 years (range, 1–14 years). Previous ARVs included a mean of 3.5 NRTIs (range, 0–8 NRTIs), 1.7 PIs (range, 0–8 PIs), and 0.6 NNRTIs (range, 0–3 NNRTIs).
Table 1 summarizes the frequency with which different ARVs or ARV combinations were used for salvage therapy. A PI was used in 555 TCEs and an NNRTI in 330 TCEs. One or more NRTIs were used in all but 11 TCEs; the distribution was as follows: 1 NRTI was used in 115 TCEs, 2 in 380 TCEs, 3 in 205 TCEs, and 4 in 22 TCEs. The salvage regimens included NRTIs and a PI for 374 TCEs (51.0%), NRTIs and an NNRTI for 149 TCEs (20.3%), NRTIs and a PI plus an NNRTI for 170 TCEs (23.2%), and NRTIs only for 30 TCEs (4.1%). Twenty patients received enfuvirtide.
Table 1.
Summary of the Most Commonly Used Antiretrovi-rals (ARVs) for Salvage Therapy
| ARV class and regimen |
|---|
| Protease inhibitora |
| NFV (65) |
| fAPV/r (69) |
| ATV/r (55) |
| IDV/r (69) |
| LPV/r (183) |
| SQV/r (94) |
| TPV/r (12) |
| DRV/r (8) |
| Nucleoside reverse-transcriptase inhibitor |
| ddI + AZT/d4T (93) |
| 3TC/FTC + AZT/d4T (84) |
| TDF + 3TC/FTC (56) |
| AZT/d4T (54) |
| TDF + 3TC/FTC + AZT/d4T (53) |
| ABC + 3TC/FTC + AZT/d4T (47) |
| ABC + AZT/d4T (44) |
| TDF + ddI + 3TC/FTC (40) |
| ABC (27) |
| ddI + 3TC/FTC + AZT/d4T (26) |
| Nonnucleoside reverse-transcriptase inhibitor |
| EFV (235) |
| NVP (95) |
| Fusion inhibitors |
| Enfuvirtide (20) |
NOTE. No. in parentheses indicates the numbers of treatment-change episodes in which the ARV or ARV combination was included in the salvage therapy regimen. 3TC, lamivudine; 3TC/FTC, 3TC or emtricitabine; ABC, abacavir; ATV, atazanavir; AZT, zidovudine; AZT/d4T, AZT or stavudine; ddI, didanosine; DRV, darunavir; EFV, efavirenz; fAPV, fosamprenavir or am-prenavir; IDV, indinavir; LPV, lopinavir; NFV, nelfinavir; NVP, nevirapine; SQV, saquinavir; TDF, tenofovir; TPV, tipranavir.
Ritonavir boosting is denoted by “/r.”
rGSSs by algorithm
The mean unweighted rGSSs for the ANRS, HIVdb, Rega, and ViroSeq algorithms ranged from 1.9 to 2.3; boosted PI–weighted rGSSs ranged from 2.1 to 2.5, and comprehensively weighted rGSSs ranged from 1.8 to 2.2. All 4 algorithms were correlated with R2 values of 0.71–0.88 for the unweighted rGSSs. However, statistically signif cant pairwise differences were observed between the rGSSs of the 4 algorithms (data not shown).
For 278 of 734 TCEs, ≥1 historical genotype was available before the baseline genotype, including 1 genotype for 168 TCEs and ≥2 genotypes for 110 TCEs. For all 4 algorithms, the mean unweighted rGSS calculated using the baseline genotype (range, 1.9–2.4) was higher than the mean unweighted rGSS based on a cumulative genotype containing the union of mutations in the baseline and historical genotypes (range, 1.6–2.0; P < .001).
VR and TCEs
Sixty-fiv percent of the TCEs (475 of 734) were associated with VR, including 51% (n p 378) with a sustained VR, define as ≥2 plasma HIV-1 RNA measurements BLQ within the firs 36 weeks of follow-up, and 13% (n p 97) with a transient VR, define as a single plasma HIV-1 RNA measurement BLQ. Multivariate logistic regression yielded a slightly better goodness of fi when the transient responders were grouped with the sustained responders rather than with the nonresponders (data not shown); therefore, for the analyses that follow, the sustained and transient responders were grouped together. There was no difference between the goodness of fi of models, including all 734 TCEs from 641 patients (77 patients had 11 TCE), and that of models, including only 1 TCE per patient (data not shown). Therefore, for the analyses that follow, we included all 734 TCEs.
Univariate analyses of predictors of VR
Table 2 summarizes the univariate analyses of the association between VR and each of the unweighted rGSSs and the covariates. For each algorithm, a higher rGSS was associated with a greater likelihood of VR. Among the covariates related to past treatment, the number of ARVs and ARV regimens previously received, the number of PIs and ARV classes previously received, and the number of non-HAART regimens received were associated with a decreased likelihood of VR. Among the covariates related to the salvage regimen, the number of ARVs received, the number of new ARVs received, and the number of new ARV classes received were associated with increased VR. A lower baseline plasma HIV-1 RNA level, a higher baseline CD4 cell count, a history of previous VR, and a more recent TCE were also associated with increased VR.
Table 2.
Univariate Analyses of Virologic Response to Treatment-Change Episodes as a Function of Demographic Factors, Baseline Disease Factors, and Salvage Therapy
| Variable | Nonresponders (n = 259) |
Responders (n = 475) |
P a |
|---|---|---|---|
| Characteristic of treatment history | |||
|
| |||
| ARV therapy, years | 5 (3–7) | 5 (3–7) | .3 |
|
| |||
| ARVs, no. | 6 (4–8) | 5 (3–7) | <.001 |
|
| |||
| ARV regimens, no. | 4 (2–6) | 3 (1–4) | <.001 |
|
| |||
| Non-HAART regimens, no. | 2 (1–3) | 1 (0–3) | <.001 |
|
| |||
| Protease inhibitors, no. | 2 (1–3) | 1 (1–2) | <.001 |
|
| |||
| NRTIs, no. | 4 (2–4) | 3 (2–4) | .006 |
|
| |||
| NNRTIs, no. | 1 (0–1) | 0 (0–1) | <.001 |
|
| |||
| ARV classes, no. | 3 (2–3) | 2 (2–3) | <.001 |
|
| |||
| Year ARV therapy begun | 1996 (1994–1997) | 1996 (1994–1998) | <.001 |
|
| |||
| Previous HIV-1 RNA BLQ, % | 20.1 | 41.3 | <.001b |
|
| |||
| Patient characteristic at baseline | |||
|
| |||
| Age, years | 42 (38–49) | 45 (39–51) | .009 |
|
| |||
| Female sex, % | 7.7 | 8.2 | .03b |
|
| |||
| HIV-1 RNA, log copies/mL | 4.4 (4–4.9) | 4.1 (3.6–4.6) | <.001 |
|
| |||
| CD4 cell count, cells/mL | 190 (94.5–327) | 283 (175–407.5) | <.001 |
|
| |||
| Characteristic of salvage regimen | |||
|
| |||
| ARVs, no. | 3 (3–4) | 3 (3–4) | .01 |
|
| |||
| Previously unused ARVs, no. | 1 (1–2) | 2 (1–2) | <.001 |
|
| |||
| Previously unused ARV classes, no. | 0 (0–1) | 1 (0–1) | <.001 |
|
| |||
| Year of treatment-change episode | 2000 (1999–2002) | 2001 (2000–2003) | <.001 |
|
| |||
| Unweighted rGSSc | |||
|
| |||
| ANRS | 2 (1–3) | 3 (2–3) | <.001 |
|
| |||
| HIVdb | 1.5 (0.75–2.25) | 2.25 (1.5–2.75) | <.001 |
|
| |||
| Rega | 2 (1–2.5) | 2.5 (2–3) | <.001 |
|
| |||
| ViroSeq | 1.5 (1–2.5) | 2.5 (2–3) | <.001 |
NOTE. Except for categorical data, given as percentages, data are given as medians, with interquartile ranges in parentheses. ANRS, Agence National de Recerche sur le SIDA; ARV, antiretroviral; BLQ, below the limit of quantification (<75 copies/mL; Versant branched DNA assay); HAART, highly active antiretroviral therapy; HIV-1, human immunodeficiency virus type 1; HIVdb, HIV Drug Resistance Database; NRTIs, nucleoside reverse-transcriptase (RT) inhibitors; NNRTIs, nonnucleoside RT inhibitors; rGSS, regimen-specific genotypic susceptibility score.
Student’s t test was used for all comparisons, except as indicated for categorical data.
Comparisons by χ2 test.
Algorithm versions were as follows: ANRS 2007.10, HIVdb 4.3.1, Rega 7.1.1, and ViroSeq 2.8.
Multivariate logistic regression
Twelve multivariate logistic regression models were developed, 1 for each combination of the 4 algorithms and 3 weighting schemes. In forward selection procedures for all 12 models, there was a statistically significan association between rGSS and VR (P < .001). Four covariates were robustly associated with VR—that is, they were significantl associated with VR in ≥6 of the 12 models. The robust covariates were as follows: (1) baseline plasma HIV-1 RNA level (12 models), (2) history of previous virologic suppression (12 models), (3) number of new ARVs received (12 models), and (4) number of new ARV classes received (6 models).
The adjusted ORs for the rGSSs demonstrated an increased likelihood of VR, with a range of 63%–71% for each 1.0 increase in unweighted rGSS (i.e., the addition of 1 fully active ARV), 81%–89% for each 1.0 increase in the boosted PI–weighted rGSSs (i.e., the addition of a boosted PI predicted to retain two-thirds of its activity or any other fully active ARV), and 108%–124% for each 1.0 increase in the comprehensively weighted rGSS (i.e., the addition of a boosted PI that retained half of its activity, 2 fully active NRTIs, or any other fully active ARV) (table 3).
Table 3.
Multivariate Logistic Regression Analysis Evaluating the Vi-rologic Function of 3 Weighting Schemes
| Algorithma | Mean rGSS ± SD |
Adjusted OR of rGSSb |
Mean AUC ± SDc |
|---|---|---|---|
| Unweighted | |||
|
| |||
| ANRS | 2.26 ± 0.94 | 1.70 | 0.76 ± 0.08 |
|
| |||
| HIVdb | 1.91 ± 0.91 | 1.71 | 0.76 ± 0.08 |
|
| |||
| Rega | 2.22 ± 0.91 | 1.63 | 0.76 ± 0.08 |
|
| |||
| ViroSeq | 2.14 ± 0.96 | 1.66 | 0.76 ± 0.08 |
|
| |||
| Boosted PI weighting | |||
|
| |||
| ANRS | 2.51 ± 1.03 | 1.89 | 0.77 ± 0.05 |
|
| |||
| HIVdb | 2.12 ± 0.99 | 1.88 | 0.77 ± 0.05 |
|
| |||
| Rega | 2.49 ± 0.91 | 1.83 | 0.77 ± 0.06 |
|
| |||
| ViroSeq | 2.37 ± 1.05 | 1.81 | 0.77 ± 0.05 |
|
| |||
| Comprehensive weighting | |||
|
| |||
| ANRS | 2.14 ± 0.97 | 2.14 | 0.79 ± 0.03 |
|
| |||
| HIVdb | 1.82 ± 0.88 | 2.22 | 0.80 ± 0.03 |
|
| |||
| Rega | 2.18 ± 0.91 | 2.15 | 0.80 ± 0.04 |
|
| |||
| ViroSeq | 2.02 ± 0.95 | 2.08 | 0.80 ± 0.03 |
NOTE. For boosted protease inhibitor (PI) weighting, the drug-specific genotypic susceptibility score (GSS) was multiplied by 1.5 before the regimen-specific GSS (rGSS) was calculated. This is the weighting used by the Rega algorithm. For comprehensive weighting, the drug-specific GSS was multiplied by 2.0 for boosted PIs and by 0.5 for nucleoside reverse-transcriptase inhibitors before the rGSS was calculated. ANRS, Agence National de Recerche sur le SIDA; AUC, area under the receiver operating characteristic curve; HIVdb, HIV Drug Resistance Database; OR, odds ratio; SD, standard deviation.
Algorithm versions were as follows: ANRS 2007.10, HIVdb 4.3.1, Rega 7.1.1, and ViroSeq 2.8.
Adjusted ORs for attaining virologic suppression (plasma human immunodeficiency virus type 1 RNA level below the limit of quantification) for each unit increase in rGSS. For all ORs in this column, P values were <.001.
Mean AUCs were calculated from the prediction results of multivariate logistic regression models tested in 10-fold cross-validation.
We assessed the predictive accuracy of each model by measuring the area under the ROC curve (AUC) generated by 10-fold cross-validation testing. For each weighting scheme, the averaged AUCs of 4 multivariate models (1 for each of the 4 algorithms) were nearly identical (table 3). When the unweighted rGSS was used, the averaged AUC was 0.76 for all 4 algorithms. However, when the comprehensively weighted rGSS was used, the averaged AUC was 0.80 for 3 algorithms and 1.79 for the fourth (ANRS) (table 3).
Multivariate models for unweighted GSSs for all 4 algorithms were highly concordant in predicting VR. Of the 475 TCEs with a VR, when a probability of response of ≥0.5 was used 403 of 475 were correctly predicted (85% true-positive), and 47 of 475 were incorrectly predicted (10% false-negative) by all 4 algorithms; only 25 (5%) of 475 yielded discordances among the algorithms. Of the 259 TCEs without a VR, 113 of 259 were correctly predicted (44% true-negative), and 116 of 259 were incorrectly predicted (45% false-positive) by all 4 algorithms; only 30 (11%) of 259 yielded discordances among the algorithms. At a probability-of-response cutoff of 0.8, the false-positive rate decreased from 45% to 10% at the expense of an increased false-negative rate.
Of 734 TCEs, there were 54, 384, and 296 associated with experience with 1, 2, and 3 ARV classes, respectively. Although the VR rate was higher among patients with experience of 2 classes versus those with experience of 3 classes (74% vs. 53%; P < .001), the predictive accuracy was higher among those with experience of 3 classes for all 4 algorithms. For 278 TCEs with ≥1 historical genotype, the predictive accuracy did not differ significantl depending on whether a baseline or a cumulative genotype was used.
Classificatio trees
For all 12 classificatio trees, the rGSS was the optimal firs classifie for VR. Figure 4 shows the tree for the HIVdb algorithm using the comprehensive weighting scheme: VR occurred in 24% of patients (35 of 148) with an rGSS of <1.1, compared with 75% of patients (440 of 586) with an rGSS of ≥1.1. Among those with an rGSS of ≥1.1, a further increase in VR was associated with a baseline HIV-1 RNA level <4.7 log copies/mL, an rGSS ≥1.8, and a history of ever having attained VR. Among those with an rGSS of <1.1, the likelihood of VR was increased in patients receiving ≥1.5 new ARVs who had a history of VR.
Figure 4.
Classification tree for the comprehensively weighted version of regimen-specific genotypic susceptibility scores (rGSSs) generated by the HIVdb algorithm. The root node contains all 734 treatment-change episodes (TCEs). The root node and all child nodes show the number of TCEs resulting in virologic failure (left side of node) or virologic success (right side of node). Using the R package Rpart (Recursive partitioning; version 3.1-39), an explanatory variable and a cutoff value were selected at each node to optimally separate virologic responders from nonresponders in the child nodes. Tree pruning was performed using the default parameters in Rpart for minimizing the complexity of the tree as a function of the improvement in classification accuracy. ARV, antiretroviral; HIV, human immunodeficiency virus.
rGSSs and VR by ARV class
To assess the relative observed and predicted activity of boosted PIs compared with NNRTIs, we examined the VR to therapy among 119 patients receiving an initial NNRTI-containing salvage regimen (lacking a PI) and 76 patients receiving an initial boosted PI–containing salvage regimen (but lacking an NNRTI), using the unweighted HIVdb algorithm. Among the 119 patients receiving an initial NNRTI-containing salvage regimen, virologic failure occurred in 35%; in contrast, among 76 patients receiving an initial boosted PI– containing salvage regimen, virologic failure occurred in only 12% (P < .001). This improved VR with the boosted PI–containing regimen occurred even though the mean rGSSs were almost identical for the boosted PI– and NNRTI-containing regimens (rGSS, ~2.3). However, when comprehensive weighting was used, the rGSS of the boosted PI–containing regimen was higher than that of the NNRTI-containing regimen (2.5 vs. 1.7; P < .001).
DISCUSSION
Rationale for genotypic drug-resistance interpretation algorithms
Genotypic drug-resistance testing is part of the standard care of HIV-1–infected patients. However, because the genetic mechanisms of ARV resistance are both numerous and complex, the mutation list generated by resistance testing has meaning only to a small subset of care providers well versed in the HIV resistance literature. The majority of care providers must rely on the results of a genotypic drug-resistance test interpretation that estimates the extent of ARV resistance associated with the list of reported mutations.
A large number of studies have been performed to discover rules that correlate pretherapy drug-resistance mutations with the clinical response to a salvage therapy ARV. The ARV may be a licensed ARV or a new ARV under clinical investigation. In both cases, the ARV is usually given in combination with a background regimen that has been optimized for ARV potency. The RESIST, POWER, and DUET studies are examples of studies that have yielded drug-specifi rules [23–25]. Indeed, there are 150 published studies of this type, including 130 for PIs and 120 for RT inhibitors [26].
However, rules-discovery studies are limited in power because of the large numbers of drug-resistance mutations and covariates that influenc VR. Rules-discovery studies also differ in how VR has been define and in the spectrum of ARVs available for the optimized background regimen [27]. Therefore, several groups have developed integrated genotypic drugresistance test interpretation algorithms that supplement rules discovered from clinical studies with other types of information, including in vitro susceptibility testing and in vitro and in vivo drug selection.
Predictive value of genotypic drug-resistance interpretation algorithms
In this study we found that in multivariate logistic regression analyses, rGSSs as determined by 4 interpretation algorithms were the strongest predictors of VR. Indeed, the adjusted OR for the unweighted rGSS of the 4 algorithms ranged from 1.6 to 1.7. In addition, the rGSS was found to be the optimal firs classifie for VR in all 12 classif cation trees (4 algorithms times 3 weightings).
Other statistically significan covariates included the baseline plasma HIV-1 RNA level, the number of new ARVs and ARV classes in the salvage regimen, and a history of previous virologic suppression. The significanc of baseline plasma HIV-1 RNA levels and of the number of new ARVs and new ARV classes as predictors of VR is consistent with the results of previous studies [12, 15, 17]. A history of previous VR may be a surrogate for patient adherence [12].
Although multivariate logistic regression was useful for estimating the predictive value of the algorithms in this study, the model we developed was not designed to be a clinical tool for predicting VR in individual patients. Moreover, although the AUCs of cross-validated ROC curves are robust indicators of prediction accuracy, maximizing AUCs may not be as clinically meaningful as lowering the false-positive rates.
This study was performed before the widespread impact of 3 second-generation drugs (tipranavir, darunavir, and etravirine) and drugs belonging to 2 new drug classes (the integrase inhibitor raltegravir and the chemokine [C-C motif] receptor 5 inhibitor maraviroc). Therefore, the utility of genotypic drugresistance interpretation algorithms found in this study is particularly relevant to salvage therapy in low- and middle-income countries, where there is less access to the newer, more expensive ARVs that have recently become available in high-income countries.
Weighting of drug-specifi GSSs
The findin that the predictive value of all 4 algorithms improved with weighting suggests that weighting GSSs by ARV potency may improve VR prediction. Two finding in particular supported weighting schemes that attribute increased potency to boosted PIs. First, initial boosted PI–containing salvage regimens lacking an NNRTI were associated with about one-third as many virologic failures (35% vs. 12%) as initial NNRTI-containing salvage regimens lacking a PI. Second, weighting schemes that attributed increased potency to boosted PIs were associated with an increased prediction accuracy for all 4 algorithms. These fi dings are consistent with those of another recent study showing that PI-specifi GSSs were more predictive than NRTI- and NNRTI-specif c GSSs [15].
Although it is possible to incorporate ARV potency into a genotypic drug-resistance interpretation algorithm, this is not consistently done. Instead, most algorithms create ARV-specifi GSSs that primarily indicate the expected activity of an ARV relative to its activity against a wild-type virus (which is how antimicrobial resistance test results are typically reported). Because ARVs differ in their potency, a second layer of logic may be necessary for choosing among ARVs, particularly when multiple ARVs are predicted to be active. However, our study was not optimal for discovering drug potency weights for individual ARVs, because the number of cases per ARV was insuffi ient for many ARVs, and the extent to which each algorithm already incorporated ARV potency was unknown.
Conclusion
In this study, both unweighted and weighted rGSSs were the strongest predictors of VR for 4 genotypic drugresistance interpretation algorithms. Whereas weighting drugspecifi GSSs by ARV potency appears to improve predictive accuracy, the weighting schemes we used in this study represent initial steps toward incorporating ARV potency into interpretation of genotypic drug-resistance test results. Ongoing studies of this type are required as new ARVs are licensed and new knowledge about drug-resistance mutations accrues. Further studies are also needed to explore ARV-specifi weightings that would more faithfully account for the potencies of individual ARVs and ARV combinations.
Figure 1.
Schematic definition of a treatment-change episode (TCE). Brackets indicate 3 TCE requirements: (1) a baseline genotypic drug-resistance test result obtained within 24 weeks before the treatment change, (2) a baseline plasma human immunodeficiency virus type 1 (HIV-1) RNA level obtained within 8 weeks before the treatment change, and (3) ≥2 plasma HIV-1 RNA levels obtained between 4 and 36 weeks after the treatment change. Additional requirements were a CD4 cell count obtained within 24 weeks before the treatment change and ≥4 weeks of salvage therapy.
Figure 2.
Three-step method for calculating regimen-specific genotypic susceptibility scores (GSSs): (1) calculate drug-specific GSSs, applying a genotypic drug-resistance interpretation algorithm to the mutations present in the baseline genotype to estimate the activity of an antiretroviral (ARV) relative to its activity against a wild-type virus; (2) calculate the weighted drug-specific GSSs, multiplying drug-specific GSSs by a factor that accounts for differences in ARV activity between ARVs belonging to different ARV classes; and (3) calculate regimen-specific GSSs by adding the weighted drug-specific GSSs of the drugs used for salvage therapy.
APPENDIX.
The complete set of data comprising each TCE and the R code used for analysis will be made available at http://hivdb.stanford.edu at the time of publication. The baseline genotypes are in GenBank. The accession numbers are listed below.
FJ983142–FJ983573, AF096886, AF514062, AF514081, FJ983142-FJ983573, AF096886, AF514062, AF514081, AF514082, AF514084, AF514103, AF514109, AF514122, AF514123, AF514126, AF514131, AF514134, AF514155, AF514159, AF514162, AF514163, AF514164, AF514168, AF514171, AF514176, AF514183, AF514184, AF514202, AF514203, AF514205, AF514231, AF514258, AF514272, AF514274, AF544488, AF544489, AF544492, AF544504, AF544506, AF544519, AF544523, AF544533, AF544535, AF544539, AF544546, AF544552, AF544562, AF544563, AF544565, AF544575, AF544588, AF544606, AF544620, AY030508, AY030534, AY030564, AY030570, AY030573, AY030583, AY030585, AY030591, AY030592, AY030597, AY030604, AY030608, AY030617, AY030619, AY030620, AY030621, AY030622, AY030624, AY030625, AY030636, AY030643, AY030652, AY030668, AY030671, AY030677, AY030678, AY030681, AY030685, AY030686, AY030690, AY030706, AY030707, AY030710, AY030712, AY030717, AY030723, AY030728, AY030735, AY030739, AY030751, AY030752, AY030757, AY030758, AY030763, AY030767, AY030770, AY030773, AY030776, AY030782, AY030790, AY030798, AY030804, AY030828, AY030834, AY030837, AY030842, AY030844, AY030848, AY030858, AY030859, AY030860, AY030866, AY030884, AY030891, AY030898, AY030911, AY030917, AY030961, AY030966, AY030967, AY030968, AY030982, AY030983, AY031055, AY031057, AY031068, AY031083, AY031091, AY031118, AY031123, AY031136, AY031141, AY031143, AY031151, AY031168, AY031171, AY031179, AY031180, AY031194, AY031197, AY031205, AY031215, AY031226, AY031233, AY031236, AY031240, AY031241, AY031249, AY031250, AY031255, AY031271, AY031272, AY031288, AY031305, AY031308, AY031329, AY031337, AY031343, AY031357, AY031363, AY031369, AY031375, AY031381, AY031387, AY031413, AY031416, AY031417, AY031431, AY031433, AY031441, AY031442, AY031446, AY031451, AY031464, AY031475, AY031476, AY031481, AY031510, AY031516, AY031521, AY031528, AY031531, AY031536, AY031548, AY031565, AY031575, AY031577, AY031588, AY031591, AY031594, AY031620, AY031645, AY031654, AY031666, AY031677, AY031679, AY031694, AY031702, AY031715, AY031720, AY031740, AY031742, AY031767, AY031768, AY031777, AY031781, AY031796, AY031797, AY031801, AY031806, AY031814, AY031832, AY031837, AY031848, AY031872, AY031874, AY031896, AY031907, AY031929, AY031935, AY031946, AY031961, AY031971, AY031975, AY031990, AY032007, AY032022, AY032023, AY032024, AY032041, AY032053, AY032099, AY032126, AY032142, AY032148, AY032197, AY032202, AY032225, AY032234, AY032236, AY032242, AY032248, AY032266, AY032271, AY032274, AY032312, AY032315, AY032331, AY032332, AY032359, AY032363, AY032372, AY032377, AY032380, AY032390, AY032393, AY032397, AY032402, AY032406, AY032412, AY032413, AY032418, AY032423, AY032439, AY032445, AY032469, AY032470, AY032474, AY032476, AY032480, AY032489, AY032491, AY032495, AY032501, AY032510, AY032525, AY032531, AY032537, AY032541, AY032547, AY032567, AY032571, AY032579, AY047456, AY047457, AY047458, AY305919, AY305924, AY305925, AY305928, AY305929, AY305933, AY305953, AY305957, AY305966, AY305976, AY305981, AY305983, AY305993, AY305994, AY305995, AY351775, AY796606, AY796607, AY796658, AY796681, AY796702, AY796712, AY796717, AY796721, AY796756, AY796760, AY796761, AY796774, AY796775, AY796789, AY796791, AY796793, AY796795, AY796828, AY796831, AY796838, AY796840, AY796860, AY796861, AY796865, AY796894, AY796903, AY796905, AY796909, AY796922, AY796935, AY796943, AY796947, AY796953, AY796954, AY796956, AY796968, AY796972, AY796985, AY796987, AY796992, AY797004, AY797027, AY797028, AY797043, AY797050, AY797059, AY797060, AY797068, AY797071, AY797072, AY797082, AY797094, AY797104, AY797106, AY797114, AY797129, AY797134, AY797143, AY797148, AY797152, AY797163, AY797164, AY797171, AY797172, AY797173, AY797176, AY797177, AY797178, AY797179, AY797180, AY797193, AY797195, AY797196, AY797211, AY797212, AY797218, AY797220, AY797229, AY797231, AY797239, AY797245, AY797250, AY797255, AY797259, AY797261, AY797262, AY797264, AY797269, AY797286, AY797290, AY797292, AY797300, AY797305, AY797317, AY797328, AY797332, AY797338, AY797355, AY797363, AY797375, AY797376, AY797390, AY797398, AY797411, AY797414, AY797420, AY797429, AY797443, AY797448, AY797456, AY797457, AY797459, AY797460, AY797462, AY797465, AY797473, AY797480, AY797482, AY797487, AY797491, AY797496, AY797497, AY797501, AY797503, AY797507, AY797510, AY797515, AY797533, AY797542, AY797544, AY797559, AY797579, AY797585, AY797586, AY797597, AY797603, AY797604, AY797608, AY797611, AY797621, AY797626, AY797630, AY797646, AY797650, AY797662, AY797666, AY797679, AY797685, AY797689, AY797701, AY797704, AY797708, AY797709, AY797727, AY797728, AY797735, AY797741, AY797750, AY797754, AY797758, AY797763, AY797770, AY797774, AY797777, AY797790, AY797796, AY797798, AY797801, AY797803, AY797813, AY797830, AY797831, AY797832, AY797846, AY797847, AY797848, AY797854, AY797856, AY797860, AY797863, AY797877, AY797879, AY797880, AY797883, AY797884, AY797891, AY797893, AY797902, AY797903, AY797913, AY797929, AY797936, AY797941, AY797949, AY797973, AY797977, AY797982, AY797983, AY797984, AY797986, AY797988, AY797998, AY798003, AY798010, AY798012, AY798023, AY798025, AY798029, AY798030, AY798034, AY798035, AY798041, AY798051, AY798060, AY798062, AY798078, AY798081, AY798083, AY798087, AY798099, AY798102, AY798116, AY798126, AY798133, AY798152, AY798157, AY798160, AY798163, AY798175, AY798178, AY798202, AY798206, AY798207, AY798209, AY798229, AY798231, AY798235, AY798256, AY798257, AY798277, AY798287, AY798311, AY798421, AY798433, AY798437, AY798446, AY798493, AY800855, AY800856, AY800934, AY800960, AY800975, AY800979, AY800982, AY801017, AY801022, AY801023, AY801036, AY801037, AY801051, AY801052, AY801054, AY801056, AY801080, AY801085, AY801096, AY801099, AY801117, AY801120, AY801146, AY801155, AY801157, AY801160, AY801172, AY801182, AY801198, AY801203, AY801210, AY801211, AY801213, AY801227, AY801240, AY801244, AY801253, AY801278, AY801279, AY801299, AY801308, AY801309, AY801315, AY801318, AY801319, AY801342, AY801350, AY801352, AY801360, AY801377, AY801382, AY801393, AY801398, AY801402, AY801412, AY801413, AY801420, AY801421, AY801422, AY801424, AY801426, AY801427, AY801428, AY801429, AY801430, AY801442, AY801444, AY801445, AY801460, AY801461, AY801467, AY801469, AY801479, AY801482, AY801490, AY801496, AY801500, AY801505, AY801508, AY801509, AY801512, AY801515, AY801519, AY801536, AY801540, AY801542, AY801552, AY801557, AY801568, AY801579, AY801583, AY801588, AY801597, AY801605, AY801625, AY801626, AY801639, AY801648, AY801660, AY801663, AY801669, AY801679, AY801696, AY801701, AY801710, AY801711, AY801713, AY801714, AY801716, AY801719, AY801727, AY801734, AY801736, AY801741, AY801745, AY801746, AY801749, AY801750, AY801754, AY801756, AY801760, AY801763, AY801768, AY801785, AY801793, AY801795, AY801812, AY801833, AY801839, AY801840, AY801850, AY801856, AY801857, AY801861, AY801864, AY801876, AY801881, AY801885, AY801901, AY801905, AY801918, AY801922, AY801935, AY801941, AY801945, AY801957, AY801960, AY801964, AY801965, AY801983, AY801984, AY801991, AY801997, AY802007, AY802011, AY802016, AY802021, AY802028, AY802032, AY802035, AY802049, AY802055, AY802060, AY802062, AY802073, AY802090, AY802091, AY802092, AY802105, AY802106, AY802107, AY802113, AY802115, AY802119, AY802122, AY802137, AY802139, AY802140, AY802143, AY802144, AY802151, AY802153, AY802162, AY802163, AY802173, AY802189, AY802196, AY802200, AY802208, AY802232, AY802236, AY802241, AY802242, AY802243, AY802245, AY802247, AY802259, AY802264, AY802271, AY802273, AY802284, AY802286, AY802290, AY802291, AY802295, AY802296, AY802302, AY802312, AY802321, AY802323, AY802338, AY802341, AY802343, AY802347, AY802359, AY802362, AY802376, AY802386, AY802393, AY802412, AY802417, AY802420, AY802423, AY802435, AY802438, AY802461, AY802465, AY802466, AY802468, AY802488, AY802490, AY802494, AY802516, AY802517, AY802539, AY802549, AY802574, AY802575, AY802693, AY802708, AY802714, AY802717, AY802724.
Footnotes
Potential conflicts of interest: R.W.S. is a consultant for and received funding from Celera. N.M.M. and C.M.R. are shareholders in Celera. R.A.R. is a shareholder in Abbott. All other authors report no potential conflicts.
Financial support: National Institute of Allergy and Infectious Diseases (grant AI06858 to S.Y.R., T.F.L., J.T., and R.W.S.); Abbott Laboratories (funding to S.Y.R. and R.W.S. to assist with data collection and curation); the Belgian AIDS Reference Laboratory fund (to A.V. and K.L.); the Belgian Fonds voor Wetenschappelijk Onderzoek (F.W.O. G.0611.09 to A.V. and K.L.); the Programme for Interuniversitaire Attractiepolen (UAP P6/41 to A.V. and K.L.).
References
- 1.DeGruttola V, Dix L, D’Aquila R, et al. The relation between baseline HIV drug resistance and response to antiretroviral therapy: re-analysis of retrospective and prospective studies using a standardized data analysis plan. Antivir Ther. 2000;5:41–48. doi: 10.1177/135965350000500112. [DOI] [PubMed] [Google Scholar]
- 2.Durant J, Clevenbergh P, Halfon P, et al. Drug-resistance genotyping in HIV-1 therapy: the VIRADAPT randomised controlled trial. Lancet. 1999;353:2195–9. doi: 10.1016/s0140-6736(98)12291-2. [DOI] [PubMed] [Google Scholar]
- 3.Baxter JD, Mayers DL, Wentworth DN, et al. A randomized study of antiretroviral management based on plasma genotypic antiretroviral resistance testing in patients failing therapy. CPCRA 046 Study Team for the Terry Beirn Community Programs for Clinical Research on AIDS. AIDS. 2000;14:F83–93. doi: 10.1097/00002030-200006160-00001. [DOI] [PubMed] [Google Scholar]
- 4.Tural C, Ruiz L, Holtzer C, et al. Clinical utility of HIV-1 genotyping and expert advice: the Havana trial. AIDS. 2002;16:209–18. doi: 10.1097/00002030-200201250-00010. [DOI] [PubMed] [Google Scholar]
- 5.Cingolani A, Antinori A, Rizzo MG, et al. Usefulness of monitoring HIV drug resistance and adherence in individuals failing highly active antiretroviral therapy: a randomized study (ARGENTA) AIDS. 2002;16:369–79. doi: 10.1097/00002030-200202150-00008. [DOI] [PubMed] [Google Scholar]
- 6.Hirsch MS, Brun-Vezinet F, Clotet B, et al. Antiretroviral drug resistance testing in adults infected with human immunodeficienc virus type 1: 2003 recommendations of an International AIDS Society–USA panel. Clin Infect Dis. 2003;37:113–28. doi: 10.1086/375597. [DOI] [PubMed] [Google Scholar]
- 7.Vandamme AM, Sonnerborg A, Ait-Khaled M, et al. Updated European recommendations for the clinical use of HIV drug resistance testing. Antivir Ther. 2004;9:829–48. [PubMed] [Google Scholar]
- 8.United States Department of Health and Human Services Panel on Clinical Practices for Treatment of HIV Infection Guidelines for the use of antiretroviral agents in HIV-1-infected adults and adolescents (the living document, January 2008) doi: 10.1310/hct.2000.1.1.008. Available at: http://aidsinfo.nih.gov/. Accessed 3 June 2009. [DOI] [PubMed]
- 9.Shafer RW, Schapiro JM. HIV-1 drug resistance mutations: an updated framework for the second decade of HAART. AIDS Rev. 2008;10:67–84. [PMC free article] [PubMed] [Google Scholar]
- 10.Larder BA. Interactions between drug resistance mutations in human immunodeficienc virus type 1 reverse transcriptase. J Gen Virol. 1994;75:951–7. doi: 10.1099/0022-1317-75-5-951. [DOI] [PubMed] [Google Scholar]
- 11.Quinones-Mateu ME, Moore-Dudley DM, Jegede O, Weber J, Arts EJ. Viral drug resistance and f tness. Adv Pharmacol. 2008;56:257–96. doi: 10.1016/S1054-3589(07)56009-6. [DOI] [PubMed] [Google Scholar]
- 12.De Luca A, Cingolani A, Di Giambenedetto S, et al. Variable prediction of antiretroviral treatment outcome by different systems for interpreting genotypic human immunodeficienc virus type 1 drug resistance. J Infect Dis. 2003;187:1934–43. doi: 10.1086/375355. [DOI] [PubMed] [Google Scholar]
- 13.Ormaasen V, Sandvik L, Asjo B, Holberg-Petersen M, Gaarder PI, Bruun JN. An algorithm-based genotypic resistance score is associated with clinical outcome in HIV-1-infected adults on antiretroviral therapy. HIV Med. 2004;5:400–6. doi: 10.1111/j.1468-1293.2004.00244.x. [DOI] [PubMed] [Google Scholar]
- 14.Cabrera C, Cozzi-Lepri A, Phillips AN, et al. Baseline resistance and virological outcome in patients with virological failure who start a regimen containing abacavir: EuroSIDA study. Antivir Ther. 2004;9:787–800. [PubMed] [Google Scholar]
- 15.Fox ZV, Geretti AM, Kjaer J, et al. The ability of four genotypic interpretation systems to predict virological response to ritonavir-boosted protease inhibitors. AIDS. 2007;21:2033–42. doi: 10.1097/QAD.0b013e32825a69e4. [DOI] [PubMed] [Google Scholar]
- 16.Helm M, Walter H, Ehret R, et al. Differences of nine drug resistance interpretation systems in predicting short-term therapy outcomes of treatment-experienced HIV-1 infected patients: a retrospective observational cohort study. Eur J Med Res. 2007;12:231–42. [PubMed] [Google Scholar]
- 17.Assoumou L, Brun-Vezinet F, Cozzi-Lepri A, et al. Initiatives for developing and comparing genotype interpretation systems: external validation of existing systems for didanosine against virological response. J Infect Dis. 2008;198:470–80. doi: 10.1086/590156. [DOI] [PubMed] [Google Scholar]
- 18.Agence National de Recerche sur le SIDA (ANRS) ANRS genotypic resistance guidelines (version 13) Available at: http://www.hivfrench resistance.org/;. Accessed 3 June 2009.
- 19.Liu TF, Shafer RW. Web resources for HIV type 1 genotypic-resistance test interpretation. Clin Infect Dis. 2006;42:1608–18. doi: 10.1086/503914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Van Laethem K, De Luca A, Antinori A, Cingolani A, Perna CF, Vandamme AM. A genotypic drug resistance interpretation algorithm that significantl predicts therapy response in HIV-1-infected patients. Antivir Ther. 2002;7:123–9. [PubMed] [Google Scholar]
- 21.Eshleman SH, Hackett J, Jr, Swanson P, et al. Performance of the Celera Diagnostics ViroSeq HIV-1 Genotyping System for sequence-based analysis of diverse human immunodeficienc virus type 1 strains. J Clin Microbiol. 2004;42:2711–7. doi: 10.1128/JCM.42.6.2711-2717.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sing T, Sander O, Beerenwinkel N, Lengauer T. ROCR: visualizing classifi r performance in R. Bioinformatics. 2005;21:3940–1. doi: 10.1093/bioinformatics/bti623. [DOI] [PubMed] [Google Scholar]
- 23.Baxter JD, Schapiro JM, Boucher CA, et al. Genotypic changes in human immunodeficienc virus type 1 protease associated with reduced susceptibility and virologic response to the protease inhibitor tipranavir. J Virol. 2006;80:10794–801. doi: 10.1128/JVI.00712-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.De Meyer S, Dierynck I, Lathouwers E, et al. Identificatio of mutations predictive of a diminished response to darunavir/ritonavir: analysis of data from treatment-experienced patients in POWER 1, 2, 3, and DUET-1 and DUET-2 [abstract 54] Program and abstracts of the 6th European HIV Drug Resistance Workshop; Budapest, Hungary: 2008. [Google Scholar]
- 25.Vingerhoets J, Buelens M, Peeters M, et al. Impact of baseline mutations on the virological response to TMC125 in the phase III clinical trials DUET-1 and DUET-2 [abstract 32] Antivir Ther. 2007;12:S34. [Google Scholar]
- 26.Stanford University HIV Drug Resistance Database Genotype–clinical outcome correlations. Available at: http://hivdb.stanford.edu/pages/ genotype-clinical.html. Accessed 3 June 2009.
- 27.Brun-Vezinet F, Costagliola D, Khaled MA, et al. Clinically validated genotype analysis: guiding principles and statistical concerns. Antivir Ther. 2004;9:465–78. [PubMed] [Google Scholar]