Abstract
Bovine mastitis is a widespread disease in dairy cows, and is often caused by bacterial mammary gland infection. Mastitis causes reduced milk production and leads to excessive use of antibiotics. We present meta-analysis of transcriptional profiles of bovine mastitis from 10 studies and 307 microarrays, allowing identification of much larger sets of affected genes than any individual study. Combining multiple studies provides insight into the molecular effects of Escherichia coli infection in vivo and uncovers differences between the consequences of E. coli vs. Staphylococcus aureus infection of primary mammary epithelial cells (PMECs). In udders, live E. coli elicits inflammatory and immune defenses through numerous cytokines and chemokines. Importantly, E. coli infection causes downregulation of genes encoding lipid biosynthesis enzymes that are involved in milk production. Additionally, host metabolism is generally suppressed. Finally, defensins and bacteria-recognition genes are upregulated, while the expression of the extracellular matrix protein transcripts is silenced. In PMECs, heat-inactivated E. coli elicits expression of ribosomal, cytoskeletal and angiogenic signaling genes, and causes suppression of the cell cycle and energy production genes. We hypothesize that heat-inactivated E. coli may have prophylactic effects against mastitis. Heat-inactivated S. aureus promotes stronger inflammatory and immune defenses than E. coli. Lipopolysaccharide by itself induces MHC antigen presentation components, an effect not seen in response to E. coli bacteria. These results provide the basis for strategies to prevent and treat mastitis and may lead to the reduction in the use of antibiotics.
Introduction
Mastitis is, arguably, the most important disease of dairy cattle [1, 2]. It is often caused by the infection of the mammary gland by various micro-organisms, including E. coli, Streptococcus uberis and Staphylococcus aureus [3–6]. Mastitis causes reduced milk production in affected cows, premature culling, discarding of inferior quality milk, veterinary and labor costs and the pervasive use of antibiotics [7].
Escherichia coli and S. aureus infections result in different symptoms and cellular responses. Escherichia coli infection is typically associated with an acute and severe form of mastitis, while S. aureus causes often a chronic but sub-clinical disease. In bovine primary mammary epithelial cells (PMECs), E. coli infection induces the expression of Toll-like receptor 2 (TLR2) and Toll-like receptor 4 (TLR4), and cytokines Tumor Necrosis Factor-α, Interleukin-1α, Interleukin-6 and Interleukin-8, and activation of the NFκB pathway; on the other hand, while S. aureus infection induces TLR2 expression, other molecular responses are delayed if present at all [8–11].
There have been significant attempts to prevent or ameliorate the consequences of bovine mastitis. For example, lipopolysaccharide (LPS) can be used to stimulate the inflammatory reactions in udders; such treatments may reduce the severity of subsequent infections [12, 13]. Lipopolysaccharide is recognized by TLR4, which may prime the innate immune system to recognize Gram-negative pathogens, such as E. coli. [14]. Mastitis is commonly treated with antibiotics [15], which has disadvantages including development of resistance and the need for increasing dosage [16].
The responses to mastitis infection have been studied using transcriptional profiling, both in infected udders in vivo, as well as by treating PMECs with heat-inactivated bacteria in vitro [17–23]. Drawing conclusions from these studies is hindered by extensive differences in individual responses between cows, even when the cows came from the same herd, with similar genetic backgrounds and similar age [24]. Recently, important gene-wide association studies between DNA polymorphisms and mastitis susceptibility in dairy cows, and these have been correlated with changes in gene expression [25–27]. While, in the same animal, responses are similar between repeated infections [28], different animals will respond inconsistently to E. coli infection [29–31]. Combining data from many studies using meta-analysis can bypass the challenges associated with individual variations, and addresses a much larger set of comparisons than any individual study [32, 33].
Here we assemble and present a meta-analysis comprising 307 microarrays from 10 individual studies of mastitis-related transcriptional profiling of responses to E. coli and S. aureus. Combining multiple studies, we were able to identify large sets of differentially regulated genes, which allowed us insights into the molecular effects of E. coli infection in vivo. Additionally, we found differences between E. coli and S. aureus infections of PMECs. We found that lipid biosynthesis enzymes involved in milk production are repressed under E. coli infection, which provides molecular insight into reduced milk production in infected animals. We defined the specific effects of heat-treated E. coli in vitro, which, we propose, may have prophylactic effects against mastitis. We also identify responses to bacterial LPS that are not elicited by live bacteria. The results provide insight for developing strategies to prevent and treat mastitis and may lead to the reduction in the use of antibiotics in its treatment.
Methods
Downloading the data files
Searching GEO Datasets for the key term “mastitis” and selecting “Bos taurus” as the organism yielded twenty nine data sets as output. From these, we selected studies focused on responses of the epithelial cells to a mastitis-causing bacterium, E. coli or S. aureus, either conducted in vivo (udder tissue) or in vitro (mammary epithelial cells). We did not analyze systemic responses in blood cells. The selected studies used the “Affymetrix Bovine Genome Array” platform containing 24128 genes. Additional studies were found using non-Affymetrix microarrays, but we decided not to include these for the following reasons: 1. such studies mostly used in-house microarrays, which incompletely overlap the Affymetrix arrays, and therefore would significantly reduce the total number of genes studied; 2. Each of the in-house array is used in just a few datasets (at most 3 datasets, e.g., for GPL8776, or GPL6082); 3. They used two-color RNA labeling approach, which yields relative expression values, which are not easily integrated with the Affymetrix studies; 4. The Affymetrix studies can analyze a high number of samples, and employ standardized quality controls and analysis algorithms, which can be used across different studies. The.CEL or.TXT files deposited from these studies were downloaded and unzipped, then log2 transformed. Datasets obtained were combined and analyzed using RMAExpress for quality control [33, 34]. For each study, data obtained from bacteria-treated and untreated, control cells were saved in different columns of Excel spread sheets (Table 1).
Table 1. Studies details.
| No | Acc. No | Total M.A. | M.A. C+T | Bacterial strain | Tissue or Cell type | Treatment time (h) |
|---|---|---|---|---|---|---|
| Live Escerichia coli | ||||||
| 1 | GSE15020 | 10 | 5+5 | E. coli 1303 | Udder biopsy | 24 |
| 2 | GSE15019 | 10 | 5+5 | E. coli 1303 | Udder biopsy | 6 |
| 3 | GSE24217 | 49 | 23+26 | E. coli k2bh2 | Udder biopsy | 24, 192 |
| 4 | GSE50685 | 20 | 5+15 | E. coli ECC-Z | Udder biopsy | 24, 48 |
| Heat-Inactivated Escerichia coli | ||||||
| 5 | GSE24560 | 58 | 27+31 | E. coli 1303 | PMEC | 1, 6, 24 |
| 6 | GSE25413 | 18 | 6+12 | E. coli 1303 | PMEC | 1, 3, 6, 24 |
| 7 | GSE32186 | 12 | 6+6 | E. coli 1303 | PMEC | 6 |
| Heat-Inactivated Staphylococcus aureus | ||||||
| 8 | GSE24560 | 57 | 27+30 | S. aureus M60 | PMEC | 1, 6, 24 |
| 9 | GSE25413 | 18 | 6+12 | S. aureus 1027 | PMEC | 1, 3, 6, 24 |
| Lipopolysaccharide | ||||||
| 10 | GSE32186 | 12 | 6+6 | LPS | PMEC | 6 |
M.A. C+T stands for number of microarrays, control (C) and treated (T); PMEC for primary mammary epithelial cells; Acc. No. for accession number.
Grouping studies for analysis using RankProd software
For global comparison of the expression profiles of E. coli-treated and control samples, we combined microarray data containing the 177 microarrays from the E. coli experiments into a single spreadsheet, using data-loader. We performed four separate analyses: 1) 4 studies comprising 89 microarrays for control and E. coli-infected udder biopsies. Differentially expressed genes in each of the class were recorded [21–23]. 2) Data of heat-inactivated E. coli-treated PMEC containing three data sets with 49 treated samples and 39 controls [18–20]. 3) Microarray data for LPS-treated and untreated samples from one study with 12 microarrays [18]. 4) Two studies with 75 microarrays from treated and control samples for PMEC responses to heat-inactivated S. aureus [19, 20]. Several strains of E. coli and S. aureus were used in these studies, specifically, E. coli 1303, E. coli k2bh2, E. coli ECC-Z, S. aureus M60 and S. aureus 1027 (Table 1). The animals used in these studies are from three different countries, Germany (GSE15020, GSE15019), Denmark (GSE24217) and the USA (GSE50685).
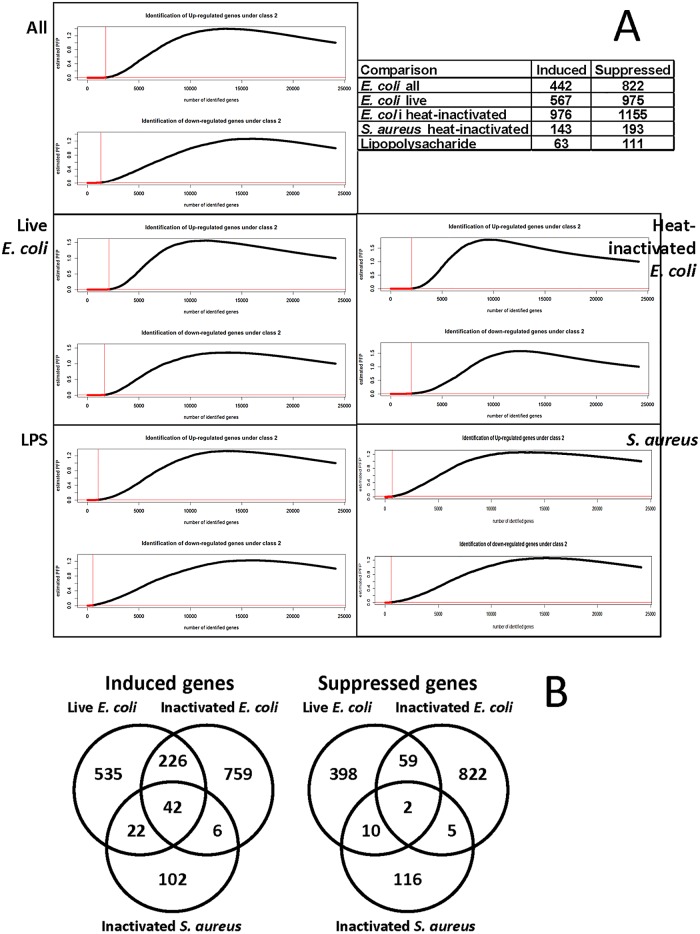
We used the RankProd Software to identify the differentially expressed genes with p-values better than 10−4, when compared with respective controls in the following data sets: global, live E. coli-, heat-inactivated E. coli- and heat-inactivated S. aureus-treated samples. For each analysis, the number of genes induced or suppressed in the respective comparison is recorded in Fig 1.
Fig 1. Selection of regulated genes using nonparametric RankProd evaluation.
A) The genes differentially expressed with a p-value better than 0.01 are marked with dashed line. The table inset shows the numbers of regulated genes used in analysis, selected with a 10−4 cut-off, except for the LPS treatment, where we used 10−3 cut-off because a single study provided statistically less significant values. B) Venn diagrams of overlaps among the selected genes. Note that the more extensive overlaps between the E. coli regulated genes may be due to the larger numbers of such genes, when compared to the list of genes regulated by S. aureus. For studies used in this figure please refer to Table 1.
Ontological Analysis
We chose genes with p-values better than our threshold from RankProd output and used online Database for Annotation, Visualization and Integrated Discovery (DAVID) software for further analysis as described before [33, 35]. For differentially expressed genes in the LPS-treated and control PMEC, we chose those with p-values better than 10−3. We also generated clusters of ontological categories containing extensively overlapping sets of genes, which condensed some redundancies in the regulated ontological categories. We separately identified ontological data for the induced and suppressed ontological clusters and genes in each comparison.
The PRISMA Checklist is included as S1 PRISMA Checklist.
Results
Datasets characterization
We searched GEO DataSets using key terms “mastitis” and “Bos taurus” and selected studies using Affymetrix bovine microarrays platform only. We found that studies describing transcriptional responses to live E. coli strains were conducted in vivo in udder tissues, while the responses to heat-inactivated E. coli, S. aureus or LPS were studied in primary cultures of mammary epithelial cells. We analyzed the gene ontologies upregulated and downregulated in these data sets separately (Table 1). We found ten appropriate studies containing 307 microarrays. In four studies, live E. coli were used in vivo, in three heat-inactivated E. coli was used on PMEC in vitro, in two studies similarly heat-inactivated S. aureus was used and we found a single study using LPS.
The effects of live E. coli
The most prominent cluster of ontological categories induced by live E. coli comprises wound responses, defense and inflammatory responses, Table 2. The defense genes induced are listed in Table 3. Highly prominent in the list are genes encoding CCL and CXCL chemokines, the secreted polypeptides mediating chemotactic signals that attract macrophages, mast cells, eosinophils and neutrophils. Additional genes encoding proinflammatory polypeptides, such as IL-1α, IL-1β and vanin, are also induced. The taxis cluster, the third most prominent cluster induced by E. coli (Table 2), is an element of the wound response. It comprises the set of chemokines listed in Table 3. Similarly, vasculature development/angiogenesis is prominent in the induced categories. We also note the abundant presence of complement components. Importantly, defensins, which can be produced by the epithelia and are directly bactericidal or bacteriostatic, are strongly induced by live E. coli; these include beta-defensins DEFB10, DEFB4A, BNBD-9, as well as defensin genes LAP, LBP, LTF, and LYZ2. Live E. coli infection also upregulates expression of additional constituents of the innate responses, including CD14, TLR2 and PYCARD, proteins that recognize and orchestrate responses to bacterial infection.
Table 2. Clusters of ontological categories suppressed or induced by E. coli infection in cow udders in vivo.
| INDUCED Ontological categories | p Value | SUPRESSED Ontological categories | p Value | ||
|---|---|---|---|---|---|
| 1 | 14.88 | 1 | 3.81 | ||
| response to wounding | 1.59E-16 | polysaccharide binding | 6.77E-05 | ||
| defense response | 2.46E-15 | glycosaminoglycan binding | 9.94E-05 | ||
| inflammatory response | 5.99E-15 | 2 | 3.78 | ||
| 2 | 11.01 | carboxylic acid biosynthetic process | 7.16E-05 | ||
| extracellular region | 2.40E-12 | lipid biosynthetic process | 7.53E-05 | ||
| extracellular space | 1.16E-11 | 3 | 2.99 | ||
| 3 | 5.60 | extracellular region part | 1.01E-04 | ||
| taxis | 6.88E-09 | extracellular matrix | 6.04E-04 | ||
| chemokine receptor binding | 6.12E-06 | 4 | 2.31 | ||
| 4 | 4.82 | glucose transport | 3.03E-03 | ||
| lysosome | 1.73E-06 | hexose transport | 3.96E-03 | ||
| lytic vacuole | 1.73E-06 | 5 | 2.09 | ||
| 5 | 4.61 | skeletal system development | 5.22E-04 | ||
| protein dimerization activity | 1.21E-06 | ossification | 3.80E-03 | ||
| identical protein binding | 6.24E-06 | 6 | 1.91 | ||
| 6 | 4.59 | aromatic compound catabolic process | 3.03E-03 | ||
| vasculature development | 4.64E-06 | L-phenylalanine metabolic process | 7.39E-03 | ||
| blood vessel development | 1.19E-05 | 7 | 1.85 | ||
| 7 | 3.92 | gland development | 4.56E-03 | ||
| carbohydrate binding | 8.70E-06 | mammary gland development | 1.99E-02 | ||
| glycosaminoglycan binding | 1.30E-04 | 8 | 1.53 | ||
| 8 | 3.78 | isoprenoid metabolic process | 8.22E-03 | ||
| melanosome | 4.61E-05 | Cholesterol biosynthesis | 2.85E-02 | ||
| cytoplasmic vesicle | 3.53E-04 | 9 | 1.46 | ||
| 9 | 3.66 | tissue morphogenesis | 2.43E-02 | ||
| endocytosis | 1.11E-05 | epidermis morphogenesis | 2.47E-02 | ||
| phagocytosis | 9.82E-04 | serine/threonine kinase signaling | 2.92E-02 | ||
| 10 | 3.38 | 10 | 1.44 | ||
| negative regulation of apoptosis | 7.46E-06 | Viral myocarditis | 6.94E-03 | ||
| anti-apoptosis | 5.50E-03 | MHC class II protein complex | 1.62E-02 |
The top ten clusters with best enrichment scores are shown. The p-values are noted for individual ontological categories in each cluster.
Table 3. Defense response genes induced in udder in vivo by E. coli.
| Symbol | Name | Function |
|---|---|---|
| BCL2 | B-cell CLL/lymphoma 2 | Transcription |
| BNBD-9-LIKE | BNBD-9-LIKE | Bactericidal activity |
| C1S | complement component 1, s | Peptidase |
| C3 | complement component 3 | Complement activation |
| C4BPA | complement component 4 bp, alpha | Complement activation |
| C6 | complement component 6 | Lytic complex formation |
| CCL20 | chemokine (C-C motif) ligand 20 | Chemotactic factor |
| CCL3 | chemokine (C-C motif) ligand 3 | inflammation and chemokine |
| CCL4 | chemokine (C-C motif) ligand 4 | inflammation and chemokine |
| CCL5 | chemokine (C-C motif) ligand 5 | Chemotactic factor |
| CCR5 | chemokine (C-C motif) receptor 5 | Chemokine Receptor |
| CD14 | CD14 molecule | Mediates response to LPS |
| CFB | complement factor B | Complement component cleavage |
| COTL1 | coactosin-like 1 (Dictyostelium) | Binding to F-actin |
| CXCL11 | chemokine (C-X-C motif) ligand 11 | Chemotactic factor |
| CXCL16 | chemokine (C-X-C motif) ligand 16 | Chemotactic response |
| CYBA | cytochrome b-245 alpha | Critical in Phagocyte oxidation |
| DEFB10 | beta-defensin 10 | Bactericidal activity |
| DEFB4A | beta-defensin 4 | Bactericidal activity |
| FCER1G | Fc fragment of IgE | Immune response regulation |
| FGR | Gardner-Rasheed feline | Catalysis |
| FN1 | fibronectin 1 | Cell surface and compounds binding |
| HMOX1 | heme oxygenase (decycling) 1 | catalysis |
| IL1A | interleukin 1, alpha | Stimulate thymocyte proliferation |
| IL1B | interleukin 1, beta | Stimulate thymocyte proliferation |
| ITGB6 | integrin, beta 6 | Receptor for fibronectin and cytoactin |
| LAP | lingual antimicrobial peptide | Antibacterial and antifungal activities |
| LBP | lipopolysaccharide binding protein | Bactericidal activity |
| LOC504773 | regakine 1 | Immunoattractant |
| LTF | lactotransferrin | catalytic activity |
| LYZ2 | lysozyme C-2 | catalytic activity |
| NCF1 | neutrophil cytosolic factor 1 | NADPH activation |
| NFKBIZ | NF kappa B-cells inhibitor zeta | NFkB signaling |
| NOS2 | nitric oxide synthase 2 | catalytic activity |
| OLR1 | oxidized LDL receptor 1 | Involved in degradation of oLDL |
| ORM1 | alpha-1 acid glycoprotein | Modulate immune system activity |
| PTAFR | platelet-activating factor receptor | inflammation |
| PYCARD | PYD and CARD domain containing | Promotes caspase-mediated apoptosis |
| RAB27A | member RAS oncogene family | GTPase superfamily |
| S100A12 | S100 calcium binding protein A12 | Belongs to the S-100 family |
| SAA3 | serum amyloid A3 | Major acute phase reactant |
| SELP | selectin P | Receptor for myeloid cells |
| SERPINF2 | serpin peptidase inhibitor | Plasmin, trypsin, chymotrypsin inhibitor |
| THBS1 | thrombospondin 1 | Cell to cell or matrix interaction mediator |
| TLR2 | toll-like receptor 2 | Mediates response to LPS |
| VNN1 | vanin 1 | catalytic activity |
The second most prominent induced cluster comprises genes encoding extracellular proteins (Table 2). The character of the secreted proteins in the induced and suppressed sets is diametrically different: while genes encoding small signaling polypeptides, growth factors, cytokines and chemokines are induced (Table 4A), the much larger basement membrane, extracellular matrix and cell attachment protein genes are suppressed (Table 4B). Essentially, E. coli-infected epithelia express secreted proinflammatory signals and concomitantly relax their attachment to the dermal connective tissue.
Table 4. Genes encoding extracellular proteins.
| Table 4A: Extracellular Region Genes INDUCED by E. coli | |||
| Symbol | Name | Function | |
| ADM | adrenomedullin | Hypotensive peptide controls circulation | Signaling |
| ALB | albumin | allergic reaction in human | |
| ANGPT2 | angiopoietin 2 | counteracts blood vessel maturation | Signaling |
| ANGPTL4 | angiopoietin-like 4 | hypoxia-induced expression in endothelial cells | Signaling |
| APOE | apolipoprotein E | Mediates the binding, internalization, and catabolism of LPS | Signaling |
| C3 | complement 3 | Complement activation | Signaling |
| CALR | calreticulin | interacts with monoglucosylated proteins synthesized in ER | |
| CCL19 | chemokine (C-C) 19 | inflammatory and immunological responses | Signaling |
| CCL2 | chemokine (C-C) 2 | Chemoattractant for monocytes | Signaling |
| CCL20 | chemokine (C-C) 20 | Chemoattractant for lymphocytes and neutrophils | Signaling |
| CCL3 | chemokine (C-C) 3 | inflammatory and chemokinetic properties | Signaling |
| CCL4 | chemokine (C-C) 4 | inflammatory and chemokinetic properties | Signaling |
| CCL5 | chemokine (C-C) 5 | Chemoattractant for monocytes, T-helper cells and eosinophils | Signaling |
| CHI3L1 | chitinase 3-like 1 | defense against pathogens or in tissue remodeling | Signaling |
| COL1A2 | collagen I, alpha 2 | fibrillar forming collagen | ECM |
| CXCL11 | chemokine (C-X-C) 11 | Chemotactic for IL-activated T-cells | Signaling |
| CXCL13 | chemokine (C-X-C) 13 | Chemotactic for B-lymphocytes | Signaling |
| CXCL16 | chemokine (C-X-C)16 | Induces chemotactic response | Signaling |
| ECM1 | extracellular matrix protein 1 | promotes angiogenesis, ossification and endothelial cells prolif. | ECM |
| EDN1 | endothelin 1 | Potent vasoconstrictor | Signaling |
| FGF1 | fibroblast growth factor 1 | angiogenic agents and potent mitogens | Signaling |
| FGL2 | fibrinogen-like 2 | contributes in physiologic lymphocyte functions at mucosal sites | ECM |
| GPX3 | glutathione peroxidase 3 | Protects cells and enzymes from oxidative damage | |
| HP | haptoglobin | protects kidneys from damage by hemoglobin ICAM1 | |
| ICAM1 | intercellular adhesion molecule 1 | ligand for leukocyte adhesion protein LFA-1 | Signaling |
| IFNAR2 | interferon receptor 2 | signal transduction interacting TK-JAK1 | Signaling |
| IGFBP4 | insulin like GF binding protein 4 | inhibit or stimulate growth promoting effects of IGFs | Signaling |
| IL18 | interleukin 18 | Stimulates natural killer cell activity and IFN-ɣ production | Signaling |
| IL1A | interleukin 1, alpha | inflammatory response | Signaling |
| IL1B | interleukin 1, beta | inflammatory response | Signaling |
| IL1RN | interleukin1 receptor antagonist | Inhibits activity of IL-1 | Signaling |
| LBP | LPS binding protein | Binds to LPS | Signaling |
| LGALS1 | lectin galactoside-binding soluble1 | regulates apoptosis, cell proliferation and cell differentiation | Signaling |
| LOC504773 | regakine 1 | Chemotactic for neutrophils and lymphocytes | Signaling |
| MMP9 | matrix metallopeptidase 9 | Functions in bone osteoclastic resorption | ECM |
| ORM1 | alpha-1 acid glycoprotein | modulate immune system during acute-phase reaction | Signaling |
| PDIA3 | disulfide isomerase family A,3 | Catalyzes rearrangement of -S-S- bonds in proteins | |
| PLA2G7 | phospholipase A2, group VII | Modulates action of platelet activating factor | Signaling |
| RBP4 | retinol binding protein 4 | Delivers retinol from liver to peripheral tissues | Signaling |
| SAA3 | serum amyloid A 3 | acute phase reactant, Apolipoprotein of HDL complex | Signaling |
| SERPINA1 | serpin peptidase inhibitor cladeA, 1 | Inhibitor of serine proteases | Signaling |
| SERPINA3-1 | serpin peptidase inhibitor clade A,3 | inhibitor of serine proteases | Signaling |
| SERPINF1 | serpin peptidase inhibitor clade F, 1 | induces neuronal differentiation and inhibitor of angiogenesis | Signaling |
| SRGN | serglycin | lytic vacuole | Signaling |
| THBS1 | thrombospondin 1 | mediates cell-to-cell and cell-to-matrix interactions | ECM |
| VEGFC | vascular endothelial growth factor C | Belongs to the PDGF/VEGF growth factor family | Signaling |
| Table 4B: Extracellular Region Genes SUPRESSED by E. coli | |||
| CCDC80 | coiled-coil domain containing 80 | regulation of cell-substrate adhesion | ECM |
| CMTM8 | CKLF-like MARVEL domain 8 | cytokine activity | Signaling |
| COL17A1 | collagen type 17 alpha 1 | hemidesmosome integrity and basal keratinocytes attachment | ECM |
| COL1A2 | collagen type I alpha 2 | Focal adhesion | ECM |
| CRISPLD2 | cysteine-rich protein LCCL domain2 | Promotes matrix assembly | ECM |
| FMOD | fibromodulin | Affects fibrils formation rate | ECM |
| EGFLAM | EGF-like fibronectin typeIII & laminin G domains | Carbohydrate binding | ECM |
| FGL1 | fibrinogen like 1 | hepatocyte mitogenic activity | ECM |
| HAPLN1 | hyaluronan and proteoglycan link protein1 | Stabilizes aggregates of proteoglycan with hyaluronic acid | ECM |
| KERA | keratocan | functions in corneal transparency and stromal matrix structure | ECM |
| KIT | v-kit Hardy-Zuckerman 4 | catalytic activity in oocyte growth | |
| LOXL1 | lysyl oxidase like 1 | Active on elastin and collagen substrates | ECM |
| LOXL4 | lysyl oxidase like 4 | modulate formation of collagenous extracellular matrix | ECM |
| LPL | lipoprotein lipase | catalytic activity | |
| LPO | lactoperoxidase | catalytic activity | |
| LUM | lumican | important in development of tissue engineered cartilage | ECM |
| MFAP4 | microfibrillar associated protein 4 | involved in Ca-dependent cell adhesion or intercell. interactions | ECM |
| MFGE8 | milk fat globule-EGF factor 8 | Binds to phosphatidylserine cell surfaces | |
| MSR1 | macrophage scavenger receptor 1 | mediate endocytosis of diverse group of macromolecules | |
| MSTN | myostatin | Cytokin and growth factor activity | Signaling |
| MYOC | myocilin | trabecular meshwork inducible glucocorticoid response | ECM |
| NTN4 | netrin 4 | neuron remodeling | Signaling |
| OGN | osteoglycin | Induces bone formation | Signaling |
| POSTN | periostin osteoblast specific factor | important in extracellular matrix mineralization | ECM |
| PRELP | proline/arginine-rich end leucine-rich repeat | anchor basement membranes to underlying connective tissue | ECM |
| PRSS2 | protease serine, 2 | catalytic activity | |
| TFF3 | trefoil factor 3 | Functions as motogen and maintenance and repair of intestinal muc. | ECM |
| TGFB2 | transforming growth factor beta 2 | suppressive effects on IL-2 dependent T-cell growth | Signaling |
| THBS1 | thrombospondin 1 | mediates cell-to-cell and cell-to-matrix interactions | ECM |
| VLDLR | very low density lipoprotein receptor | receptor-mediated endocytosis of specific ligands | Signaling |
A) INDUCED by E. coli. B) SUPRESSED by E. coli. Most of the induced genes encode cytokines and related small signaling polypeptides, whereas most of the suppressed genes encode large extracellular matrix proteins. Data derive from the in vivo experiments.
Escherichia coli induces in vivo the expression of several types of genes encoding intracellular vesicle proteins, lysosomal, melanocytic and endo-phagocytotic (Table 2). We also note that the anti-apoptotic genes are induced in the infected tissue.
Prominent clusters comprise extracellular matrix proteins, as already described. However, particularly remarkable is the second cluster, comprising the carboxylic acid/lipid biosynthesis enzymes: of the 20 genes in this cluster, 11 are directly related to milk production (Table 5). This result clearly identifies the molecular mechanism responsible for the reduced milk production in cows affected by mastitis.
Table 5. Metabolic enzymes suppressed by E. coli.
| Symbol | Function | |
|---|---|---|
| ACACA | sheep milk | Milk-related |
| ACSM1 | Gland development | |
| AGPAT1 | Milk production | Milk-related |
| AGPAT6 | Milk production | Milk-related |
| ALOX15 | Inflammatory responses | |
| BCAT2 | Cellular a.a. catabolism | |
| CBS | Sulphur a.a. metabolism | |
| COQ2 | ubiquinone biosynthesis | |
| FASN | effects milk fat content | Milk-related |
| FDFT1 | Imp for Milk yield and quality | Milk-related |
| GPAM | Milk production | Milk-related |
| HMGCR | Cholestrol synthesis | |
| LPL | Present in milk | Milk-related |
| LTA4H | FA Biosynthesis | Milk-related |
| MVK | FA Biosynthesis | Milk-related |
| PEMT | required for lactation and pregnancy | Milk-related |
| PSAT1 | VitB6 (comp of milk) metabolism | Milk-related |
| PYCR1 | Arginine and proline metabolism | |
| SCD | biosynthesis of unsaturated FA | |
| TM7SF2 | Steroid biosynthesis |
Many genes necessary for milk production are downregulated under E. coli infection. Data derive from the in vivo experiments.
Furthermore, E. coli infection in vivo suppresses several metabolic processes: glucose transport, amino acid and cholesterol metabolism, etc. In addition, E. coli infection suppresses the differentiation of epithelial cells, specifically keratinocyte differentiation. Collectively, in the epithelial cells E. coli infection compromises milk-production and homeostasis at the transcriptional level.
The effects of heat-inactivated E. coli
We analyzed a set of experiments performed with heat-inactivated E. coli to define their effects on PMECs in vitro [18–20]. It is important to note that the heat-inactivated E. coli was used in vitro, with monocultures of PMEC, while the live E. coli was used in vivo in cow udders, which are complex multi-tissue organs. Therefore, we cannot, at this point, distinguish the differences due to the heat-inactivation of the bacteria from those due to the in vivo/in vitro dichotomy. Table 6 lists the regulated ontological categories. The most prominently induced category comprises genes encoding ribosomal proteins. Detailed study of the category shows enhanced ribosomal structural gene expression. The second most prominent category comprises genes encoding cytoskeletal proteins. In contrast to the in vivo results with live E. coli, a prominent upregulated ontological category is programmed cell death, which contains genes involved in positive regulation of apoptosis, namely caspases, hydrolases, peptidases and apoptotic mitochondrial genes. We found some bacterial toxin-response genes in this category as well. Similarly to the in vivo results, PMECs react to E. coli treatment by upregulating secreted signaling polypeptides, in particular angiogenic ones. This category includes genes contributing to cell attachment, morphogenesis and wound healing. We also found that ontological categories of “pigment granules” or “melanocytes” are significantly overrepresented; however, it is important to note that the genes present in these categories are principally heat shock proteins and chaperones, which bind to LPS of bacterial origin and initiate inflammatory response, including TNFα secretion; on the other hand, the encoded proteins may not be directly involved in melanogenesis. Transcription of the proteasome complex, containing threonine-type endopeptidases involved in protein degradation, is also increased.
Table 6. Top 10 Clusters of ontological categories suppressed or induced by heat-inactivated E. coli.
| Table 6: Ontological Categories in PMECs Treated with Heat-Inactivated E. coli | |||||
|---|---|---|---|---|---|
| INDUCED | SUPRESSED | ||||
| Ontological categories | p Value | Ontological categories | p Value | ||
| 1 | 20.38 | 1 | 14.42 | ||
| Ribosome | 1.84E-30 | organelle inner membrane | 2.51E-20 | ||
| translation | 1.66E-22 | Oxidative phosphorylation | 1.62E-15 | ||
| 2 | 11.81 | 2 | 4.23 | ||
| structural molecule activity | 2.62E-20 | vesicle | 4.18E-05 | ||
| cytoskeleton | 2.28E-04 | melanosome | 7.65E-05 | ||
| 3 | 7.11 | 3 | 3.77 | ||
| apoptosis | 2.07E-08 | cell cycle | 4.29E-07 | ||
| programmed cell death | 3.66E-08 | mitosis | 6.18E-04 | ||
| 4 | 5.11 | 4 | 3.72 | ||
| pigment granule | 8.05E-08 | NADH dehydrogenase activity | 4.34E-05 | ||
| melanosome | 8.05E-08 | oxidoreductase activity | 1.57E-04 | ||
| 5 | 4.76 | 5 | 3.60 | ||
| vasculature development | 7.03E-07 | membrane-enclosed lumen | 3.00E-07 | ||
| angiogenesis | 5.44E-04 | nuclear lumen | 2.07E-03 | ||
| 6 | 4.57 | 6 | 3.34 | ||
| proteasome complex | 3.62E-08 | extracellular structure organization | 2.79E-04 | ||
| proteasome core complex, alpha-subunit complex | 1.23E-02 | collagen fibril organization | 8.73E-04 | ||
| 7 | 3.80 | 7 | 3.14 | ||
| extracellular region part | 8.07E-06 | translation factor activity, nucleic acid binding | 2.70E-04 | ||
| extracellular region | 2.09E-02 | translation initiation factor activity | 6.43E-04 | ||
| 8 | 3.68 | 8 | 2.85 | ||
| regulation of protein kinase cascade | 3.35E-05 | cell-matrix adhesion | 3.97E-06 | ||
| regulation of I-kappaB kinase/NF-kappaB cascade | 6.59E-05 | integrin binding | 1.74E-04 | ||
| 9 | 3.57 | 9 | 2.72 | ||
| positive regulation of cell motion | 7.29E-05 | vacuole | 9.78E-04 | ||
| regulation of cell motion | 7.64E-05 | lytic vacuole | 1.05E-03 | ||
| 10 | 3.28 | 10 | 2.67 | ||
| regulation of apoptosis | 2.78E-06 | extracellular matrix part | 6.25E-05 | ||
| positive regulation of programmed cell death | 2.49E-04 | proteinaceous extracellular matrix | 1.05E-04 | ||
| 14 | 2.68 | ||||
| defense response | 8.29E-04 | ||||
| inflammatory response | 2.20E-03 | ||||
| response to wounding | 5.11E-03 | ||||
| 24 | 2.14 | ||||
| epithelial cell differentiation | 7.30E-04 | ||||
| keratinocyte differentiation | 6.94E-02 | ||||
| 25 | 2.12 | ||||
| Toll-like receptor signaling pathway | 9.90E-05 | ||||
| RIG-I-like receptor signaling pathway | 7.95E-02 | ||||
Additional three clusters, ranked 14, 24 and 25th are shown in the induced category for comparison with the data in Table 2. All these have enrichment scores better than 2. The p-values are noted for individual ontological categories in each cluster.
Inflammatory, defense, wound healing and bacterial recognition mechanisms, both the Toll-like and the RIG-like (retinoic-acid-inducible protein 1-like) receptor signaling pathways, are upregulated but less prominent in heat-inactivated E. coli-treated PMECs (Table 6B), where production of membrane-enclosed organelles and vesicles, in particular mitochondria, is suppressed. Notably, genes encoding nuclear and cell cycle proteins are also suppressed. This is distinct from the processes suppressed by live E. coli in vivo. As in vivo, the genes encoding extracellular matrix and basement membrane proteins are suppressed by the heat-inactivated E. coli.
Overall, the heat-inactivated E. coli regulates a different set of genes from the one regulated by live E. coli: specifically 1) the metabolic enzymes of lipid biosynthesis and sugar transport are not suppressed and 2) inflammation- and defense-related genes are much attenuated in response to heat-inactivated E. coli.
The effects of S. aureus
Infections with S. aureus tend to be milder and cause less significant mastitis morbidity than those with E. coli [3, 7]. Several studies reported the transcriptional profiles of heat-inactivated S. aureus treatment of PMECs [9, 10, 19, 20]. These are directly comparable with the profiles of E. coli-treated PMECs shown above. In the S. aureus treated PMECs, the most prominently induced cluster comprises inflammatory, immune and defense responses (Table 7). Heat-inactivated S. aureus is much more proficient in eliciting these responses than is E. coli. The defense responses include extracellular signaling peptides, cell adhesion molecules, inducers of acute inflammation, regulators of lymphocyte-mediated immunity, etc. We also note quite prominent induction of receptors responsible for recognition of microbes by innate immunity, namely NOD- and Toll-like receptors.
Table 7. Clusters of ontological categories suppressed or induced by S. aureus.
| Table 7: Ontological Categories in PMECs Treated with Heat-Inactivated S. aureus | |||||
|---|---|---|---|---|---|
| INDUCED | SUPRESSED | ||||
| Ontological categories | P-Value | Ontological categories | P-Value | ||
| 1 | 10.70 | 1 | 4.33 | ||
| inflammatory response | 9.84E-14 | cell migration | 2.36E-05 | ||
| defense response | 6.59E-13 | localization of cell | 4.06E-05 | ||
| immune response | 1.01E-12 | Cell Motility | 4.06E-05 | ||
| 2 | 6.54 | 2 | 2.27 | ||
| extracellular space | 5.94E-08 | extracellular space | 2.12E-03 | ||
| extracellular region | 1.24E-07 | extracellular region | 1.98E-02 | ||
| 3 | 4.46 | 3 | 2.24 | ||
| acute inflammatory response | 2.23E-07 | plasma membrane | 2.72E-05 | ||
| positive regulation of cell component organization | 3.54E-04 | plasma membrane part | 8.98E-05 | ||
| 4 | 2.87 | 4 | 2.09 | ||
| Graft-versus-host disease | 6.01E-06 | striated muscle tissue development | 1.48E-03 | ||
| Cell adhesion molecules (CAMs) | 5.94E-03 | striated muscle cell differentiation | 5.55E-02 | ||
| 5 | 2.61 | 5 | 1.71 | ||
| positive regulation of immune system process | 8.80E-08 | receptor tyrosine kinase signaling | 6.68E-05 | ||
| positive regulation of cell proliferation | 4.73E-03 | response to peptide hormone stimulus | 1.91E-02 | ||
| 6 | 2.34 | 6 | 1.64 | ||
| acute inflammatory response | 2.23E-07 | receptor complex | 1.27E-02 | ||
| positive regulation of response to stimulus | 1.58E-04 | integral to plasma membrane | 2.85E-02 | ||
| 7 | 2.12 | 7 | 1.45 | ||
| Graft-versus-host disease | 6.01E-06 | Focal adhesion | 1.30E-03 | ||
| positive regulation of developmental process | 1.03E-03 | cell junction assembly | 3.01E-03 | ||
| 8 | 2.11 | 8 | 1.44 | ||
| NOD-like receptor signaling pathway | 1.27E-03 | tissue homeostasis | 2.17E-02 | ||
| response to bacterium | 2.03E-03 | multicellular organismal homeostasis | 3.71E-02 | ||
| 9 | 1.87 | 9 | 1.39 | ||
| skeletal system development | 7.64E-03 | enzyme linked receptor signaling | 8.86E-07 | ||
| ossification | 1.77E-02 | growth factor binding | 6.41E-03 | ||
| 10 | 1.51 | 10 | 1.37 | ||
| regulation of immune effector process | 1.36E-02 | MHC protein complex | 1.68E-02 | ||
| regulation of lymphocyte mediated immunity | 4.22E-02 | antigen processing and presentation | 2.45E-02 | ||
| 11 | 1.48 | ||||
| positive regulation of response to stimulus | 1.58E-04 | ||||
| Toll-like receptor signaling pathway | 1.81E-04 | ||||
The top 10 and top 11 clusters are given for the suppressed and induced genes, respectively.
The most conspicuous ontological categories suppressed by S. aureus involve cell migration (Table 7). Relatedly, genes encoding extracellular matrix proteins and focal adhesion components are suppressed. Proteins embedded in the plasma membrane, including growth factor-binding receptor tyrosine kinases, are also prominent.
On the whole, the transcriptional responses to S. aureus differ from those to E. coli by a significantly stronger induction of proinflammatory and immunomodulatory genes, and stronger suppression of cell attachment and motility genes. At the same time, S. aureus does not suppress the metabolic and milk lipid producing enzymes that E. coli does.
The effects of LPS
While S. aureus is Gram-positive, E. coli is Gram-negative and thus E. coli produces copious amounts of lipopolysaccharide, LPS. In epithelial and other cells, LPS is recognized by TLR4, which initiates a series of responses to infections with Gram-negative bacteria [14]. We hypothesized that treating PMECs with LPS would cause a subset of transcriptional responses caused by E. coli. We found a single study that treats PMECs with LPS [18] and consequently the statistical significance of the regulated genes is markedly reduced (Table 8). Nevertheless, we find that LPS treatment induces immune, inflammatory and defense response in PMECs, including the antigen processing machinery (Table 8). Proteolysis is also induced by LPS. Interestingly, apoptosis related genes seem to be induced. Very few ontological categories suppressed by LPS reached statistical significance, but we note that the genes encoding extracellular matrix proteins seem suppressed.
Table 8. Clusters of ontological categories suppressed or induced by LPS.
| Table 8: Ontological Categories in PMECs Challenged with Lipopolysaccharide | |||||
|---|---|---|---|---|---|
| INDUCED | SUPRESSED | ||||
| Ontological categories | p Value | Ontological categories | p Value | ||
| 1 | 2.59 | 1 | 1.52 | ||
| # | immune response | 2.78E-08 | # | extracellular region | 1.31E-02 |
| positive regulation of immune system process | 5.27E-03 | extracellular region part | 1.91E-02 | ||
| 2 | 2.55 | 2 | 1.48 | ||
| # | Antigen processing and presentation | 3.05E-05 | calcium ion binding | 8.55E-03 | |
| peptide or polysaccharide antigen via MHC class II | 3.62E-03 | metal ion binding | 4.75E-02 | ||
| 3 | 2.35 | ||||
| # | defense response | 2.19E-06 | |||
| immune effector process | 3.29E-05 | ||||
| 4 | 2.02 | ||||
| # | extracellular region | 1.63E-03 | |||
| inflammatory response | 2.24E-03 | ||||
| 5 | 1.83 | ||||
| # | positive regulation of endocytosis | 2.10E-03 | |||
| regulation of vesicle-mediated transport | 1.83E-02 | ||||
| 6 | 1.56 | ||||
| ISG15-protein conjugation | 5.72E-07 | ||||
| proteolysis | 1.52E-02 | ||||
| 7 | 1.25 | ||||
| serine-type peptidase activity | 2.61E-02 | ||||
| peptidase activity, acting on L-amino acid peptides | 4.39E-02 | ||||
| 8 | 1.03 | ||||
| apoptosis | 7.89E-02 | ||||
| programmed cell death | 8.28E-02 | ||||
Only clusters with enrichment scores better than 1.0 are given. Note the significantly higher p-values due to a smaller set of microarrays analyzed. The subset of clusters regulated similarly by E. coli is marked with # signs.
We looked specifically at the set of LPS-induced genes involved in defense and immunity (Table 9). We find that many of these (6 out of 11) are components of the complement system and anti-bacterial defense genes also induced by live E. coli (cf. Table 4A). Of the LPS-induced genes not induced by live E. coli, the majority are involved in MHC antigen presentation process (Table 9). It is of interest that LPS has been proposed as a potential preventive treatment for E. coli-caused mastitis [36]. One potential mechanism may include boosting the antigen presentation machinery, which does not occur after infection with live E. coli.
Table 9. Defense and immunity Genes induced in LPS-challenged PMECs.
| Symbol | Name | Function | |
|---|---|---|---|
| CCL5 | chemokine (C-C motif) ligand 5 | Chemotactic factor | # |
| C2 | complement component 2 | Catalytic activity | MHC |
| C3 | complement component 3 | Complement activation | # |
| CFB | complement factor B | Complement component cleavage | # |
| LTF | lactotransferrin | Catalytic activity | # |
| LAP | lingual antimicrobial peptide | Antibacterial and antifungal activities | # |
| BOLA-RDA | MHC II, DR alpha | Antigen prsentation via MHC II | MHC |
| PTX3 | pentraxin related gene | Regulates innate resistance to pathogens | |
| SAA3 | serum amyloid A3 | Major acute phase reactant | # |
| RSAD2 | radical S-adenosyl methionine domain 2 | Involved in antiviral defense | |
| TAP1 | transporter 1 | Peptide transmembrane transport | MHC |
The genes also induced by live E. coli (Table 3) are marked with #. Note the abundance of MHC-related genes among those NOT induced by E. coli.
Overall, these results support our hypothesis that the effects of LPS generally represent a subset of the effects of E. coli. This subset is marked with a number sign in Table 8.
Discussion
The results presented in this work attest to the power of meta-analysis: the highly variable individual responses to mastitis bacteria could be overcome by assembling multiple analyses and thus increasing the studied population. Importantly, meta-analysis confirmed the most important findings in individual studies, namely response to wounding, inflammatory and defense responses [17–23]. Moreover, this meta-analysis provided many additional details, for example by identifying the cytokines and additional secreted signaling polypeptides produced.
Perhaps the most important novel finding from this meta-analysis concerns the specific suppression of milk-producing metabolic enzymes (Table 5). The infection would be expected to slow down anabolic processes in most cases, as the tissue has to divert energy to fighting infection. However, the unique aspect of this slow-down in bovine mastitis is reduction of milk fat production. The seven marked enzymes in Table 5 are those that are directly and specifically devoted to milk production. It is quite likely that additional enzymes, e.g., those for amino acid biosynthesis, also play important role in milk production.
Additional novel ontological categories shown to be induced in mastitis include cellular taxis, cytoplasmic vesicles and anti-apoptosis agents. Cellular taxis is predominantly related to the leucocyte infiltrates caused by copious production of chemokines and cytokines; at present we cannot exclude enhanced taxis of epithelial cells as well, which will have to be examined with laboratory-based, as well as in-the-field experiments. The vesicle-associated proteins include those related to lysosomes, endocytosis and even melanosomes. The affected cell types are probably diverse, although it should be noted that genes encoding melanosomal proteins are also induced in the primary mammary epithelial cells.
Conversely, mastitis suppresses several aspects of basic epithelial biology, including extracellular matrix biosynthesis, mammary gland development markers and epidermis morphogenesis, including cholesterol biosynthesis, an integral component of epidermal differentiation [37]. Importantly, however, the seven milk production-related enzymes mentioned above are not integral to epidermal differentiation and thus represent a specific metabolic category suppressed in mastitis.
The effects of heat-inactivated E. coli on mammary epithelial cells in vitro are quite different from the in vivo effects. For example, the inflammatory response, and cytotaxis are much attenuated; these are, presumably, induced in vivo in the leucocyte compartment, and so are missing from pure cultures of mammary epithelial cells. We do see induction of melanosomal genes, vesicles specific for the epidermal tissue. In these cells, apoptosis is induced as a defensive mechanism. Interestingly, the innate immunity response, an important function of keratinocytes, is induced; this includes the NFκB pathway as well as the Toll-like and RIG-like receptor signaling pathways. Importantly, heat-inactivated E. coli seem not to suppress the transcription of metabolic enzymes, including those involved in production of milk lipids.
These results lead us to suggest that the treatment of cow udders with heat-inactivated E. coli may have a prophylactic effect against mastitis. While development of vaccines to achieve acquired immunity to mastitis in cattle, though challenging, is progressing [7, 38, 39], the approaches that target the innate immunity may also prove promising. The heat-inactivated E. coli could activate the innate immunity responses with attenuated inflammatory responses, thus priming the tissue to fight subsequent infection, without the concomitant damage due to inflammation. Treatment with heat-inactivated E. coli, if effective, would have major benefits in avoiding widespread use of antibiotics, reducing the costs of treatment and, notably, fighting mastitis in the third world. In underdeveloped areas, where the use of antibiotics is unavailable or prohibitively expensive, heat-inactivation treatments could be properly and easily performed locally.
A related approach using endotoxin to elicit a mild form of mastitis in hope of avoiding subsequent infections had a limited success [13]. The lipopolysaccharide treatment of mammary epithelial cells induced immune response genes, particularly those related to the acquired immunity, including antigen processing by keratinocytes. This is very different from the responses to heat-inactivated E. coli bacteria.
As noted before, we see significant differences in responses to E. coli vs. S. aureus [9, 10, 19, 20]. While both cause robust proinflammatory and immune responses, S. aureus also induces Toll-like and NOD-like innate immunity in mammary epithelia, while suppressing cell motility, antigen presentation and receptor signaling in general, hallmarks of acquired immunity responses. These differences may account for comparatively much milder and sub-acute sequelae of S. aureus-triggered mastitis.
Escherichia coli and S. aureus are not the only bacterial species important in causing mastitis; our study did not include significant microarray studies with Streptococcus uberis [5, 6] because of limited compatibility of GPL8776 microarrays with the Affymetrix platform. However, we want to emphasize that these studies identified important differences between cows fed ad libitum and those with negative energy balance, showing increased expression of lipid metabolism genes in underfed cows [5, 6].
We must emphasize several caveats of our meta-analysis. Given the very individual responses in cows [24, 40–42], our ‘forest’ view may be inapplicable to ‘trees’. Second, there are two important distinctions between our largest data sets: one uses live E. coli in vivo, the other heat-inactivated E. coli on cultured cells. We cannot, from this perspective, distinguish the in vivo/in vitro from the live/heat-inactivated dichotomies, especially as the in vivo studies include mixed populations of cells in their microarrays, while the in vitro studies use pure populations. Third, the LPS-responsive study is compromised by its relatively small size. Fourth, all original data are obtained in western academic settings; this may inadequately represent the conditions in the field, especially in less developed agricultural areas. And fifth, in this meta-analysis we have grouped expression data from short-term, 1–3 hrs., to long-term, 8 day treatments (Table 1); we realize that mastitis-causing infections are dynamic processes and that much additional data needs to be generated before any claims regarding the course of mastitis infection can be described in detail.
Nevertheless, the meta-analysis based on large amount of original data represents an important contribution to our understanding of bovine mastitis in various aspects and provides a solid foundation for the development of new treatments for mastitis.
Supporting Information
(DOC)
Acknowledgments
We thank Lili M. Blumenberg for careful editing of the manuscript.
Data Availability
All relevant data are within the paper.
Funding Statement
The authors have no support or funding to report.
References
- 1.Thompson-Crispi K, Atalla H, Miglior F, Mallard BA. Bovine mastitis: frontiers in immunogenetics. Front Immunol. 2014;5:493 10.3389/fimmu.2014.00493 eCollection 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Seegers H, Fourichon C, Beaudeau F. Production effects related to mastitis and mastitis economics in dairy cattle herds. Vet Res. 2003;34(5):475–91. [DOI] [PubMed] [Google Scholar]
- 3.Wellnitz O, Bruckmaier RM. The innate immune response of the bovine mammary gland to bacterial infection. Vet J. 2012;192(2):148–52. 10.1016/j.tvjl.2011.09.013 Epub 2 Apr 10. [DOI] [PubMed] [Google Scholar]
- 4.Bradley A. Bovine mastitis: an evolving disease. Vet J. 2002;164(2):116–28. [DOI] [PubMed] [Google Scholar]
- 5.Moyes KM, Drackley JK, Morin DE, Bionaz M, Rodriguez-Zas SL, Everts RE, et al. Gene network and pathway analysis of bovine mammary tissue challenged with Streptococcus uberis reveals induction of cell proliferation and inhibition of PPARgamma signaling as potential mechanism for the negative relationships between immune response and lipid metabolism. BMC Genomics. 2009;10:542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moyes KM, Drackley JK, Morin DE, Rodriguez-Zas SL, Everts RE, Lewin HA, et al. Mammary gene expression profiles during an intramammary challenge reveal potential mechanisms linking negative energy balance with impaired immune response. Physiol Genomics. 2010;41(2):161–70. 10.1152/physiolgenomics.00197.2009 Epub 2010 Jan 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deb R, Kumar A, Chakraborty S, Verma AK, Tiwari R, Dhama K, et al. Trends in diagnosis and control of bovine mastitis: a review. Pak J Biol Sci. 2013;16(23):1653–61. [DOI] [PubMed] [Google Scholar]
- 8.Blum S, Sela N, Heller ED, Sela S, Leitner G. Genome analysis of bovine-mastitis-associated Escherichia coli O32:H37 strain P4. J Bacteriol. 2012;194(14):3732 10.1128/JB.00535-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fu Y, Zhou E, Liu Z, Li F, Liang D, Liu B, et al. Staphylococcus aureus and Escherichia coli elicit different innate immune responses from bovine mammary epithelial cells. Vet Immunol Immunopathol. 2013;155(4):245–52. 10.1016/j.vetimm.2013.08.003 Epub Aug 24. [DOI] [PubMed] [Google Scholar]
- 10.Yang W, Zerbe H, Petzl W, Brunner RM, Gunther J, Draing C, et al. Bovine TLR2 and TLR4 properly transduce signals from Staphylococcus aureus and E. coli, but S. aureus fails to both activate NF-kappaB in mammary epithelial cells and to quickly induce TNFalpha and interleukin-8 (CXCL8) expression in the udder. Mol Immunol. 2008;45(5):1385–97. Epub 2007 Oct 22. [DOI] [PubMed] [Google Scholar]
- 11.Bouchard D, Peton V, Almeida S, Le Marechal C, Miyoshi A, Azevedo V, et al. Genome sequence of Staphylococcus aureus Newbould 305, a strain associated with mild bovine mastitis. J Bacteriol. 2012;194(22):6292–3. 10.1128/JB.01188-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shuster DE, Harmon RJ. Lactating cows become partially refractory to frequent intramammary endotoxin infusions: recovery of milk yield despite a persistently high somatic cell count. Res Vet Sci. 1991;51(3):272–7. [DOI] [PubMed] [Google Scholar]
- 13.Lohuis JA, Kremer W, Schukken YH, Smit JA, Verheijden JH, Brand A, et al. Growth of Escherichia coli in milk from endotoxin-induced mastitic quarters and the course of subsequent experimental Escherichia coli mastitis in the cow. J Dairy Sci. 1990;73(6):1508–14. [DOI] [PubMed] [Google Scholar]
- 14.Miyake K. Innate recognition of lipopolysaccharide by Toll-like receptor 4-MD-2. Trends Microbiol. 2004;12(4):186–92. [DOI] [PubMed] [Google Scholar]
- 15.Swinkels JM, Hilkens A, Zoche-Golob V, Kromker V, Buddiger M, Jansen J, et al. Social influences on the duration of antibiotic treatment of clinical mastitis in dairy cows. J Dairy Sci. 2015;11(15):00087–9. [DOI] [PubMed] [Google Scholar]
- 16.Bengtsson B, Unnerstad HE, Ekman T, Artursson K, Nilsson-Ost M, Waller KP. Antimicrobial susceptibility of udder pathogens from cases of acute clinical mastitis in dairy cows. Vet Microbiol. 2009;136(1–2):142–9. 10.1016/j.vetmic.2008.10.024 Epub Oct 31. [DOI] [PubMed] [Google Scholar]
- 17.Rinaldi M, Li RW, Bannerman DD, Daniels KM, Evock-Clover C, Silva MV, et al. A sentinel function for teat tissues in dairy cows: dominant innate immune response elements define early response to E. coli mastitis. Funct Integr Genomics. 2010;10(1):21–38. 10.1007/s10142-009-0133-z Epub 2009 Aug 29. [DOI] [PubMed] [Google Scholar]
- 18.Gunther J, Petzl W, Zerbe H, Schuberth HJ, Koczan D, Goetze L, et al. Lipopolysaccharide priming enhances expression of effectors of immune defence while decreasing expression of pro-inflammatory cytokines in mammary epithelia cells from cows. BMC Genomics. 2012;13:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gunther J, Esch K, Poschadel N, Petzl W, Zerbe H, Mitterhuemer S, et al. Comparative kinetics of Escherichia coli- and Staphylococcus aureus-specific activation of key immune pathways in mammary epithelial cells demonstrates that S. aureus elicits a delayed response dominated by interleukin-6 (IL-6) but not by IL-1A or tumor necrosis factor alpha. Infect Immun. 2011;79(2):695–707. 10.1128/IAI.01071-10 Epub 2010 Nov 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brand B, Hartmann A, Repsilber D, Griesbeck-Zilch B, Wellnitz O, Kuhn C, et al. Comparative expression profiling of E. coli and S. aureus inoculated primary mammary gland cells sampled from cows with different genetic predispositions for somatic cell score. Genet Sel Evol. 2011;43:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sipka A, Klaessig S, Duhamel GE, Swinkels J, Rainard P, Schukken Y. Impact of intramammary treatment on gene expression profiles in bovine Escherichia coli mastitis. PLoS One. 2014;9(1):e85579 10.1371/journal.pone.0085579 eCollection 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Buitenhuis B, Rontved CM, Edwards SM, Ingvartsen KL, Sorensen P. In depth analysis of genes and pathways of the mammary gland involved in the pathogenesis of bovine Escherichia coli-mastitis. BMC Genomics. 2011;12:130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mitterhuemer S, Petzl W, Krebs S, Mehne D, Klanner A, Wolf E, et al. Escherichia coli infection induces distinct local and systemic transcriptome responses in the mammary gland. BMC Genomics. 2010;11:138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Akerstedt M, Forsback L, Larsen T, Svennersten-Sjaunja K. Natural variation in biomarkers indicating mastitis in healthy cows. J Dairy Res. 2011;78(1):88–96. 10.1017/S0022029910000786 Epub 2010 Dec 7. [DOI] [PubMed] [Google Scholar]
- 25.Sahana G, Guldbrandtsen B, Thomsen B, Holm LE, Panitz F, Brondum RF, et al. Genome-wide association study using high-density single nucleotide polymorphism arrays and whole-genome sequences for clinical mastitis traits in dairy cattle. J Dairy Sci. 2014;97(11):7258–75. 10.3168/jds.2014-8141 Epub 2014 Aug 22. [DOI] [PubMed] [Google Scholar]
- 26.Wang X, Ma P, Liu J, Zhang Q, Zhang Y, Ding X, et al. Genome-wide association study in Chinese Holstein cows reveal two candidate genes for somatic cell score as an indicator for mastitis susceptibility. BMC Genet. 2015;16:111 10.1186/s12863-015-0263-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen L, Han Y, Chen Y, Li Z, Wang H, Liu Y, et al. Eight SNVs in NF-kappaB pathway genes and their different performances between subclinical mastitis and mixed Chinese Holstein cows. Gene. 2015;555(2):242–9. 10.1016/j.gene.2014.11.011 Epub Nov 15. [DOI] [PubMed] [Google Scholar]
- 28.Hirvonen J, Eklund K, Teppo AM, Huszenicza G, Kulcsar M, Saloniemi H, et al. Acute phase response in dairy cows with experimentally induced Escherichia coli mastitis. Acta Vet Scand. 1999;40(1):35–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vallimont JE, Dechow CD, Sattler CG, Clay JS. Heritability estimates associated with alternative definitions of mastitis and correlations with somatic cell score and yield. J Dairy Sci. 2009;92(7):3402–10. [DOI] [PubMed] [Google Scholar]
- 30.De Schepper S, De Ketelaere A, Bannerman DD, Paape MJ, Peelman L, Burvenich C. The toll-like receptor-4 (TLR-4) pathway and its possible role in the pathogenesis of Escherichia coli mastitis in dairy cattle. Vet Res. 2008;39(1):5 Epub 2007 Nov 20. [DOI] [PubMed] [Google Scholar]
- 31.Burvenich C, Bannerman DD, Lippolis JD, Peelman L, Nonnecke BJ, Kehrli ME Jr., et al. Cumulative physiological events influence the inflammatory response of the bovine udder to Escherichia coli infections during the transition period. J Dairy Sci. 2007;90(Suppl 1):E39–54. [DOI] [PubMed] [Google Scholar]
- 32.Hong F, Breitling R, McEntee CW, Wittner BS, Nemhauser JL, Chory J. RankProd: a bioconductor package for detecting differentially expressed genes in meta-analysis. Bioinformatics. 2006;22(22):2825–7. Epub 006 Sep 18. [DOI] [PubMed] [Google Scholar]
- 33.Mimoso C, Lee DD, Zavadil J, Tomic-Canic M, Blumenberg M. Analysis and meta-analysis of transcriptional profiling in human epidermis. Methods Mol Biol. 2014;1195:61–97. 10.1007/7651_2013_60 [DOI] [PubMed] [Google Scholar]
- 34.Gautier L, Cope L, Bolstad BM, Irizarry RA. affy—analysis of Affymetrix GeneChip data at the probe level. Bioinformatics. 2004;20(3):307–15. 10.1093/bioinformatics/btg405 PubMed PMID: . [DOI] [PubMed] [Google Scholar]
- 35.Dennis G Jr., Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, et al. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4(5):P3 Epub 2003 Apr 3. [PubMed] [Google Scholar]
- 36.Petzl W, Gunther J, Pfister T, Sauter-Louis C, Goetze L, von Aulock S, et al. Lipopolysaccharide pretreatment of the udder protects against experimental Escherichia coli mastitis. Innate Immun. 2012;18(3):467–77. 10.1177/1753425911422407 Epub 2011 Oct 11. [DOI] [PubMed] [Google Scholar]
- 37.Jozic I, Stojadinovic O, Kirsner RS, Tomic-Canic M. Stressing the steroids in skin: paradox or fine-tuning? J Invest Dermatol. 2014;134(12):2869–72. 10.1038/jid.2014.363 [DOI] [PubMed] [Google Scholar]
- 38.Erskine RJ. Vaccination strategies for mastitis. Vet Clin North Am Food Anim Pract. 2012;28(2):257–70. 10.1016/j.cvfa.2012.03.002 Epub Apr 13. [DOI] [PubMed] [Google Scholar]
- 39.Middleton JR. Staphylococcus aureus antigens and challenges in vaccine development. Expert Rev Vaccines. 2008;7(6):805–15. 10.1586/14760584.7.6.805 [DOI] [PubMed] [Google Scholar]
- 40.Burvenich C, Van Merris V, Mehrzad J, Diez-Fraile A, Duchateau L. Severity of E. coli mastitis is mainly determined by cow factors. Vet Res. 2003;34(5):521–64. [DOI] [PubMed] [Google Scholar]
- 41.Green BB, McKay SD, Kerr DE. Age dependent changes in the LPS induced transcriptome of bovine dermal fibroblasts occurs without major changes in the methylome. BMC Genomics. 2015;16(1):30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Benjamin AL, Green BB, Hayden LR, Barlow JW, Kerr DE. Cow-to-cow variation in fibroblast response to a toll-like receptor 2/6 agonist and its relation to mastitis caused by intramammary challenge with Staphylococcus aureus. J Dairy Sci. 2015;15(15):00029–6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOC)
Data Availability Statement
All relevant data are within the paper.