New methods allow for efficient affinity purification of polymerization-competent tubulin from tobacco and Arabidopsis cell cultures.
Abstract
Microtubules assemble into several distinct arrays that play important roles in cell division and cell morphogenesis. To decipher the mechanisms that regulate the dynamics and organization of this versatile cytoskeletal component, it is essential to establish in vitro assays that use functional tubulin. Although plant tubulin has been purified previously from protoplasts by reversible taxol-induced polymerization, a simple and efficient purification method has yet to be developed. Here, we used a Tumor Overexpressed Gene (TOG) column, in which the tubulin-binding domains of a yeast (Saccharomyces cerevisiae) TOG homolog are immobilized on resin, to isolate functional plant tubulin. We found that several hundred micrograms of pure tubulin can readily be purified from cell suspension cultures of tobacco (Nicotiana tabacum) and Arabidopsis (Arabidopsis thaliana). The tubulin purified by the TOG column showed high assembly competence, partly because of low levels of polymerization-inhibitory phosphorylation of α-tubulin. Compared with porcine brain tubulin, Arabidopsis tubulin is highly dynamic in vitro at both the plus and minus ends, exhibiting faster shrinkage rates and more frequent catastrophe events, and exhibits frequent spontaneous nucleation. Furthermore, our study shows that an internal histidine tag in α-tubulin can be used to prepare particular isotypes and specifically engineered versions of α-tubulin. In contrast to previous studies of plant tubulin, our mass spectrometry and immunoblot analyses failed to detect posttranslational modification of the isolated Arabidopsis tubulin or detected only low levels of posttranslational modification. This novel technology can be used to prepare assembly-competent, highly dynamic pure tubulin from plant cell cultures.
Microtubules (MTs) are important cytoskeletal polymers that are conserved in eukaryotic cells and are assembled from α- and β-tubulin heterodimers (Desai and Mitchison, 1997). In plants, MTs have important functions in essential cellular processes, such as cell division, and in cell morphogenesis. MTs in plant cells adopt several distinct higher order arrays and are remodeled in response to the cell cycle, developmental programs, and environmental cues (Hashimoto, 2015). Genetic, molecular, and cell biological approaches have been used to identify cellular factors that regulate the organization and dynamics of plant MTs. Considerable effort has been devoted to simulating the organization of cortical MT arrays by computational modeling.
Cell-free in vitro studies are essential for the biochemical characterization of various MT regulators and for elucidating the mechanistic principles underlying the versatility of this dynamic polymer in cellular functions. The purification of sufficient amounts of assembly-competent tubulin is a prerequisite for these in vitro studies. Tubulin is traditionally purified from mammalian brains, since these tissues contain sufficiently high concentrations of tubulin to allow MT assembly in crude cell extracts. Polymerized MTs and their associated MT-binding proteins are separated from other cellular proteins by sedimentation. Pelleted MTs are then depolymerized upon drug washout under MT-destabilizing conditions, such as high concentrations of salt and calcium and low temperature. A few rounds of assembly-disassembly cycles highly enrich for tubulin and copurify MT-associated proteins, which can subsequently be removed by column chromatography (Borisy et al., 1975). Tubulin has also been purified from several plant sources (Morejohn and Fosket, 1982; Mizuno, 1985; Jiang et al., 1992; Bokros et al., 1993; Moore et al., 1997). However, since tubulin concentrations are low in plant cells, taxol, which stabilizes MTs, is generally included in the polymerization buffer, and cytoplasm-rich miniprotoplasts, which lack vacuoles, are sometimes used as starting material (Hamada et al., 2013). Since it is technically challenging to isolate assembly-competent pure tubulin from nonneural sources (Sackett et al., 2010), general plant science laboratories may hesitate to prepare plant tubulin themselves.
Although the primary amino acid sequences of eukaryotic tubulins are fairly well conserved and the biophysical mechanisms of MT assembly and disassembly are thought to be similar for all MTs, the kinetics of MT dynamic instability differ for MTs assembled from animal and plant tubulin (Moore et al., 1997). Interactions with MT-interacting proteins may differ for tubulins isolated from different biological sources, as reported for the MT-dependent activation of kinesin (Alonso et al., 2007). Posttranslational modifications of tubulin, which generate distinct tubulin signatures and may modulate the functions of MT-interacting proteins, such as kinesin (Sirajuddin et al., 2014), are extensive in brain tubulin (Janke, 2014) but may be quantitatively and qualitatively different in plant tubulin. Furthermore, MT nucleation by the γ-tubulin ring complex shows a strong preference for tubulin from the same species (Kollman et al., 2015). Thus, it is important to use plant tubulin, and not brain tubulin, for in vitro studies of plant MTs.
Tubulin is folded by a series of molecular chaperones to form an αβ-tubulin heterodimer in which one structural GTP is embedded in the interdimer interface (Lundin et al., 2010). The requirement of these eukaryote-specific chaperones precludes the use of prokaryotic expression systems for synthesizing properly folded and functional tubulin. Bacterially synthesized tubulin can be folded in rabbit reticulocyte lysate to produce functional tubulin, but with moderate yields (Shah et al., 2001). A yeast (Saccharomyces cerevisiae) expression system has been developed to produce modified yeast tubulin (Uchimura et al., 2006; Johnson et al., 2011), but this system is not suitable for the synthesis of animal (Sirajuddin et al., 2014) and plant (our unpublished data) tubulin. A baculovirus-insect cell expression system was recently reported to yield functional human tubulin (Minoura et al., 2013).
Tubulin-binding proteins have been used to develop affinity-purification columns. The TOG domains (named after the human MT regulator, colonic and hepatic Tumor Overexpressed Gene [ch-TOG]) are among the best-characterized tubulin-binding domains. ch-TOG and orthologs from other eukaryotes bind to the growing plus ends of MTs and accelerate MT growth (Al-Bassam and Chang, 2011). TOG domains from the yeast ortholog Stu2 were recently used to affinity purify assembly-competent tubulin from fungal and animal sources (Widlund et al., 2012). In this study, we demonstrate that a TOG-based affinity column can be used to purify functional tubulin from tobacco (Nicotiana tabacum) and Arabidopsis (Arabidopsis thaliana). We examined the posttranslational modifications of the isolated tubulins by mass spectrometry and immunoblot analysis and showed that a His-tagged Arabidopsis tubulin isotype could be purified using this column. These results show that wild-type and recombinant functional tubulin from plant sources can be isolated efficiently.
RESULTS
Preparation of TOG Columns
Widlund et al. (2012) developed an affinity purification column in which the tubulin-binding domains TOG1 and TOG2 of the yeast Stu2 protein were fused in tandem to glutathione S-transferase (GST) and immobilized on resin that was successfully used to purify tubulins from several eukaryotic cells, tissues, and whole organisms. However, they did not evaluate the ability of this column to purify tubulin sources from flowering plants. We essentially followed the procedures of Widlund et al. (2012) to purify the recombinant GST-TOG1/2 fusion from transgenic Escherichia coli and chemically linked the fusion to a Sepharose resin. In this study, the TOG resin was packed in an open column (Fig. 1A). To examine whether the corresponding TOG1 and TOG2 domains of MICROTUBULE ORGANIZATION1 (MOR1; Whittington et al., 2001), an Arabidopsis homolog of yeast Stu2, can be used to isolate tubulin, we also purified a recombinant GST fusion of the TOG1 and TOG2 domains from MOR1 and used this fusion to prepare a TOGMOR1 column.
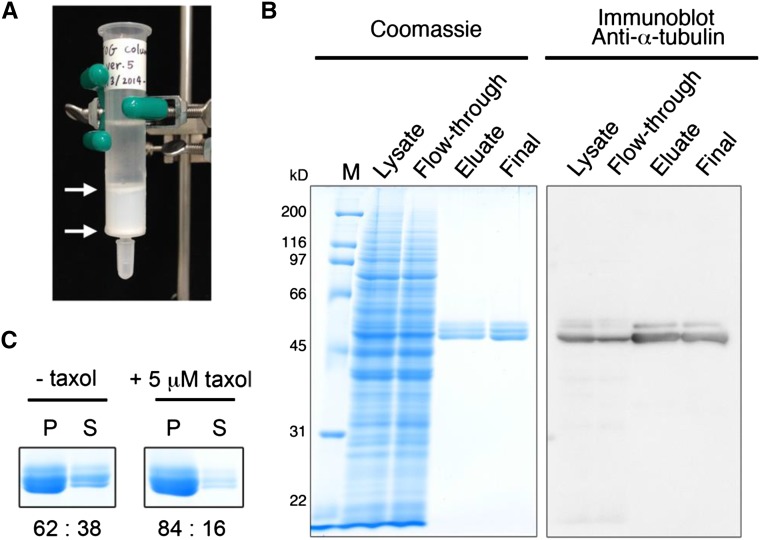
Figure 1.
Affinity purification of Arabidopsis tubulin using the TOG column. A, A gravity-flow TOG column. The GST-TOG1/2-coupled resin (2 mL) is sandwiched between the upper and lower porous plastic filters (arrows) and packed in a 15-mm-diameter column. B, Purification of tubulin from Arabidopsis MM2d cultured cells. Protein samples taken from different purification steps were separated by SDS-PAGE and analyzed by Coomassie Blue staining (left) and immunoblotting with anti-α-tubulin antibody (right). M, Molecular mass marker; Lysate, 5 μL of crude cell extract containing 27 μg of proteins; Flow-through, 5 μL of the flow-through fraction that did not bind to the TOG column; Eluate, 10 μL of the TOG column eluate; Final, 1 μg of the desalted and concentrated tubulin. C, MT sedimentation assay. Purified MM2d tubulin (20 μm) was assembled into MTs in the absence (left) or presence of 5 μm taxol (right). Coomassie Blue-stained tubulin bands in the pellet (P) and supernatant (S) fractions represent polymerized MTs and tubulin, respectively. Proteins were quantified by densitometric analysis, and ratios (%) of the pellet to supernatant fractions are indicated.
We first tested whether 7-d-old Arabidopsis seedlings could be used as a tubulin source for purification, since we were previously able to purify modest amounts of Arabidopsis tubulin from seedlings using the TOG column (Fujita et al., 2013). After crude cell extracts were applied to the TOG or TOGMOR1 column, the resins were washed and the bound proteins were eluted under high-salt conditions (for details, see “Materials and Methods”). When the TOG column was used, we obtained approximately 18 μg of pure tubulin from 15 g fresh weight of Arabidopsis seedlings, with a tubulin recovery rate from total extracted proteins of 0.024% (Table I). In some of our early purification attempts, purified tubulin was partly degraded, probably due to slow handling of the samples. In contrast to the TOG column, the TOGMOR1 column yielded no detectable amount of tubulin (Supplemental Fig. S1). Apparently, the first two TOG domains of MOR1 are not functionally equivalent to those of yeast Stu2 and do not bind to tubulin with high affinity. In subsequent studies, we used the Stu2-derived TOG domains to explore improved conditions for obtaining pure plant tubulin at higher quantities.
Table I. Purification of plant tubulins by the TOG column.
| Material | Culture Duration (Volume of Culture Medium) | Column Sizea | Sample Weight | Total Protein | Purified Tubulin | Percentage in Total Protein |
|---|---|---|---|---|---|---|
| g fresh wt | mg | μg | ||||
| Arabidopsis seedlings | S | 15 | 75 | 18 | 0.024 | |
| Arabidopsis MM2d cultured cells | 4 d (300 mL) | S | 26 | 211 | 590 | 0.28 |
| S | Flow throughb | 253 | 0.12 | |||
| Arabidopsis MM2d cultured cells | 4 d (200 mL) | M | 18 | 150 | 687 | 0.46 |
| Arabidopsis T87 cultured cells | 4 d (200 mL) | S | 19 | 96 | 224 | 0.23 |
| Arabidopsis Alex cultured cells | 3 d (100 mL) | S | 18 | 91 | 26 | 0.029 |
| Tobacco BY-2 cultured cells | 3 to 4 d (300 mL) | S | 10 | 89 | 244 | 0.27 |
| Tobacco BY-2 cultured cells | 5 d (100 mL) | S | 12 | 70 | 58 | 0.083 |
The resin volumes for the small (S) and medium (M) columns are 1 and 2 mL, respectively. bThe flow-through fraction of the above TOG column purification step was recovered and applied to a new TOG column.
Purification of Tubulin from Suspension-Cultured Plant Cells
Next, we evaluated whether suspension-cultured cells could be used as an alternative tubulin source. Three Arabidopsis cell lines (MM2d, T87, and Alex) and the Bright Yellow-2 (BY-2) tobacco cell line were used. When crude extracts of cultured cells in the exponential growth phase (after 3 to 4 d of subculture) were applied to the TOG column, pure tubulin, as judged by Coomassie Blue protein staining, was efficiently isolated and eluted from the TOG column (Fig. 1B; Supplemental Fig. S2). When starting with 10 to 26 g (fresh weight) of Arabidopsis MM2d and T87 cells and tobacco BY-2 cells, we obtained a yield of pure tubulin ranging from 224 to 590 μg, with tubulin recovery rates from total extracted proteins of 0.23% to 0.28% (Table I). These recovery rates are roughly 10-fold higher than that obtained using Arabidopsis seedlings (Fujita et al., 2013). Immunoblot analysis indicated that a considerable portion of tubulin remained unbound in the flow-through fraction (Fig. 1B, flow-through lanes). Reloading the flow-through fraction onto the same TOG column several times did not improve the yield (data not shown). However, considerable amounts of tubulin were recovered from the flow-through fraction when it was applied to another fresh column (Table I), suggesting that the amount of tubulin in the cell extracts surpassed the binding capacity of the TOG column. Indeed, higher amounts of tubulin were recovered when a larger resin volume (2 mL instead of 1 mL) was packed into the column, to generate a medium-sized TOG column (Table I). The growth phase of the cell cultures affected the tubulin yield. When tobacco BY-2 cells were harvested after 5 d of subculture, the tubulin yield was low, representing 0.08% of the total protein amount, compared with the yield of 0.27% obtained from exponentially growing tobacco cells after 3 to 4 d of subculture (Table I). The Arabidopsis suspension-cultured cell line Alex grew rather slowly under our culture conditions compared with the MM2d and T87 cells and had a lower tubulin recovery rate (Table I; Supplemental Fig. S2B).
The TOG column can be regenerated by extensive washing with buffer and used repeatedly (see “Materials and Methods”). The tubulin-binding activity of TOG columns did not decrease appreciably after more than 50 uses over 2 years.
Purified tubulin must retain polymerization activity to be suitable for use in subsequent in vitro experiments. To examine the assembly competence of purified plant tubulin, we carried out sedimentation assays with and without the MT stabilizer taxol. After the addition of GTP, 20 μm tubulin was incubated at 30°C to induce MT polymerization, and the assembly mixtures were centrifuged to separate MTs from unpolymerized tubulin. Tubulin in the supernatant (unpolymerized tubulin) and in the pellet (polymerized MT) was analyzed by SDS-PAGE, and the ratio of tubulin in the two fractions was calculated based on densitometry scanning of the Coomassie Blue-stained gels. More than 60% of MM2d tubulin was recovered in the polymer fraction in the absence of taxol, whereas the addition of 5 μm taxol increased the recovery rate to 84% (Fig. 1C). Thus, plant tubulin purified by the TOG column retains its ability to polymerize in vitro.
Arabidopsis MTs Polymerized in Vitro Are Highly Dynamic
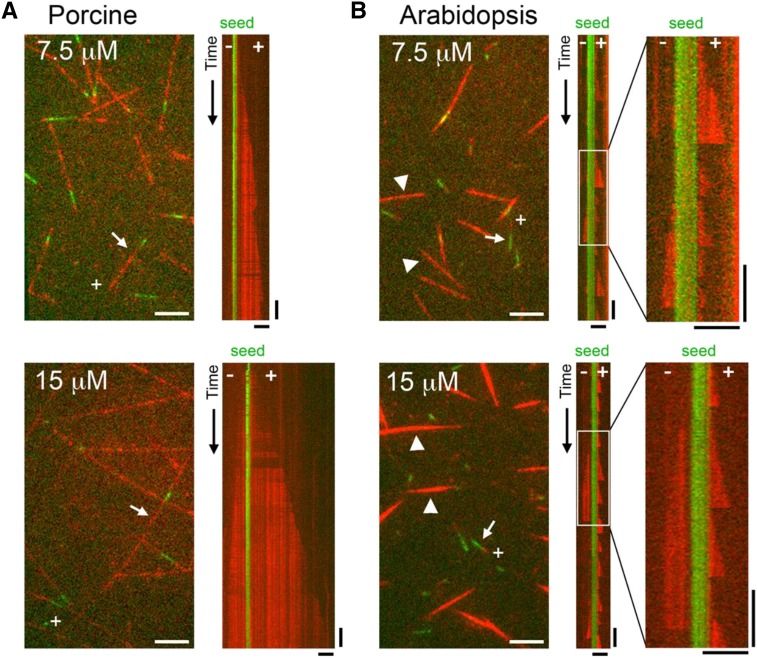
We investigated the behavior of MTs assembled in vitro from the TOG column-purified Arabidopsis tubulin and compared the dynamics of Arabidopsis MTs with those of MTs assembled from porcine brain tubulin purified in the same manner using the TOG column. Tubulins were labeled with rhodamine, applied to a glass slide to which short, stabilized HiLyte 488-labeled MT seeds had been attached, and observed by time-lapse total internal reflection fluorescence (TIRF) microscopy (Fig. 2). In our assay, the MT seeds and dynamic MTs were labeled green and red, respectively. Porcine MTs grew mostly from the seeds and displayed characteristic MT dynamics at both ends (i.e. they had faster growing, more dynamic plus ends and slower growing, less dynamic minus ends; Fig. 2A; Supplemental Movies S1 and S2). When tubulin concentrations were increased from 7.5 to 15 μm, the MT growth rates increased proportionally from 0.39 to 0.84 μm min−1 at the plus end and from 0.07 to 0.19 μm min−1 at the minus end (Table II). The plus end depolymerized at rates of 5.5 to 11.3 μm min−1 and exhibited catastrophe (i.e. transition from growth to shrinkage) and rescue (i.e. transition from shrinkage to regrowth) at rates of 0.15 to 0.49 × 10−3 and 0.038 to 0.059 events s−1, respectively. These dynamics are largely comparable to those reported for MTs assembled from porcine brain tubulin (Grego et al., 2001), except that the rescue frequency calculated in our study was higher, possibly reflecting differences in the purification methods used.
Figure 2.
In vitro MT dynamics assay. MTs polymerized from various concentrations of porcine brain tubulin (A) or Arabidopsis cell culture tubulin (B) were observed using TIRF microscopy. Top images, 7.5 μm tubulin; bottom images, 15 μm tubulin; red, rhodamine-labeled MTs; green, immobilized MT seeds. Kymographs were drawn for the MTs indicated by arrows. MT polarities are shown in the images and kymographs, with + and − indicating the plus and minus ends, respectively. Enlarged kymographs correspond to the boxed areas in the original kymographs in B. Representative MTs nucleated independently of the MT seeds are indicated by arrowheads in B. Horizontal bars = 5 μm and vertical bars = 2 min.
Table II. Dynamic instability parameters of MTs assembled from porcine or Arabidopsis tubulins in vitro.
Growth and shrinkage rates are expressed as means ± sd (n = episodes). Catastrophe and rescue frequencies were calculated from the inverse of the mean time spent in growth and shrinkage, respectively (t = total time spent in growth or shrinkage). N/A, Not applicable (since few shrinkage or rescue events were observed in our assay). *, No shrinking MTs were observed during our assay period.
| Parameters | Plus End | Minus End | ||||
|---|---|---|---|---|---|---|
| Porcine | ||||||
| Tubulin concentration (μm) | 7.5 | 10 | 15 | 7.5 | 10 | 15 |
| Growth rate (μm min−1) | 0.39 ± 0.14 (n = 24) | 0.62 ± 0.16 (n = 15) | 0.84 ± 0.17 (n = 10) | 0.07 ± 0.03 (n = 10) | 0.11 ± 0.02 (n = 10) | 0.19 ± 0.01 (n = 2) |
| Shrinkage rate (μm min−1) | 11.3 ± 7.15 (n = 10) | 5.51 ± 1.01 (n = 3) | N/A* (n = 0) | N/A* (n = 0) | N/A* (n = 0) | N/A* (n = 0) |
| Catastrophe frequency (×10−3 events s−1) | 0.49 (t = 20,495 s) | 0.15 (t = 18,813 s) | N/A (t = 13,450 s) | N/A (t = 17,444 s) | N/A (t = 14,772 s) | N/A (t = 2,819 s) |
| Rescue frequency (events s−1) | 0.059 (t = 169 s) | 0.038 (t = 78 s) | N/A* (t = 0 s) | N/A* (t = 0 s) | N/A* (t = 0 s) | N/A* (t = 0 s) |
| Arabidopsis | ||||||
| Tubulin concentration (μm) | 7.5 | 10 | 15 | 7.5 | 10 | 15 |
| Growth rate (μm min−1) | 0.76 ± 0.19 (n = 43) | 0.79 ± 0.27 (n = 75) | 0.74 ± 0.21 (n = 73) | 0.36 ± 0.08 (n = 24) | 0.36 ± 0.15 (n = 32) | 0.39 ± 0.10 (n = 40) |
| Shrinkage rate (μm min−1) | 49.6 ± 32.6 (n = 40) | 55.0 ± 44.0 (n = 71) | 56.6 ± 48.3 (n = 71) | 30.1 ± 17.7 (n = 23) | 38.3 ± 21.9 (n = 33) | 31.5 ± 16.4 (n = 40) |
| Catastrophe frequency (×10−3 events s−1) | 5.52 (t = 7,247 s) | 7.79 (t = 9,245 s) | 5.72 (t = 12,420 s) | 3.94 (t = 5,839 s) | 5.04 (t = 6,545 s) | 4.31 (t = 9,290 s) |
| Rescue frequency (events s−1) | N/A (t = 115 s) | N/A (t = 170 s) | N/A (t = 192 s) | N/A (t = 76 s) | N/A (t = 74 s) | N/A (t = 116 s) |
In contrast to porcine MTs, significant proportions of Arabidopsis MTs polymerized spontaneously in the absence of MT seeds (Fig. 2B; Supplemental Movies S3 and S4). The proportion of nonseeded MTs increased from 39% of total assembled MTs (n = 80) at a tubulin concentration of 7.5 μm to 56% (n = 117) at 15 μm tubulin. These nonseeded MTs were bundled. The fluorescence intensity of the cross section of the bundled MTs was 2.5 times brighter at the tubulin concentration of 7.5 μm (n = 13) and 6 times brighter at 15 μm tubulin (n = 16) compared with the intensity of single, nonbundled MTs growing from seeds, indicating that these MT structures are composed of several individual MTs. Apparently, single MTs grew and shrank at both ends of the nonseeded MTs. We then analyzed the dynamic behavior of seeded MTs and found that they were highly dynamic and seldom grew longer than 5 μm. The plus ends grew at rates of 0.7 to 0.8 μm min−1, and these rates did not increase markedly at higher tubulin concentrations (Table II). We speculate that spontaneous, nonseeded MT nucleation competes with seeded MT growth for free tubulin and that the actual tubulin concentrations corresponding to the observed dynamic parameters are much lower than 7.5 to 15 μm. The plus ends of Arabidopsis MTs shrank at rates of 50 to 57 μm min−1, much faster than those of porcine MTs. The catastrophe events were strikingly more frequent at the plus ends of Arabidopsis MTs (5.5–7.8 × 10−3 events s−1) than at those of porcine MTs. Since the depolymerizing plus end usually shrank to the stable MT seeds (see the kymographs in Fig. 2B, right), it was not possible to calculate the rescue frequency. The extremely rapid shrinkage rate and the high frequency of catastrophe events explain why MTs that grow from seeds do not reach substantial lengths. The minus ends of Arabidopsis MTs grew (0.3–0.4 μm min−1) and shrank (30–38 μm min−1) more slowly and underwent fewer catastrophe events (3.9–5 × 10−3 events s−1) than the plus ends. Thus, the minus ends of Arabidopsis MTs are more dynamic than the minus ends of porcine MTs.
Posttranslational Modifications of Plant Tubulin
Posttranslational modifications of tubulin are frequent and extensive in animal cells (Song and Brady, 2015), but they have not been critically evaluated in plant cells. Antibodies raised against modified animal tubulin have been used to probe plant tubulin in crude cell extracts and in immunohistochemistry analyses of plant cells (Parrotta et al., 2014). Those previous studies suggest that plant tubulin is posttranslationally modified in a similar manner to animal tubulin. In this study, we used commercial antibodies that have been used frequently for cell biological studies of plant tubulin and examined whether the purified tubulin from Arabidopsis and tobacco cell cultures is posttranslationally modified (Fig. 3).
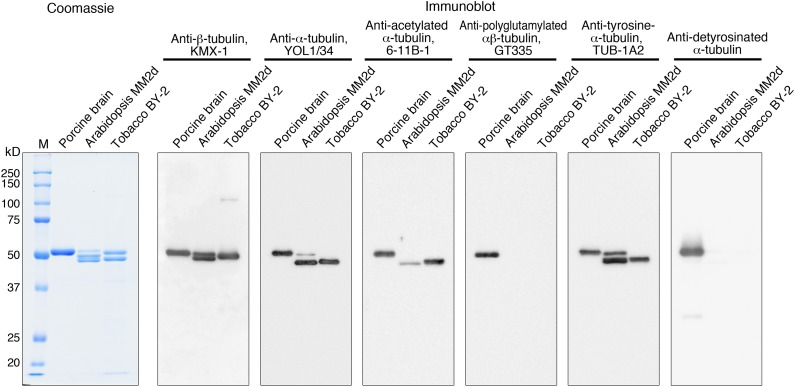
Figure 3.
Posttranslational modifications of purified tubulin evaluated by modification-specific antibodies. Porcine brain tubulin, Arabidopsis MM2d tubulin, and tobacco BY-2 tubulin were separated by SDS-PAGE and analyzed by Coomassie Blue staining (1 μg of tubulin) or immunoblotting (200 ng of tubulin). Primary antibodies were used to detect the indicated forms of tubulin. M, Molecular mass marker.
Immunoblot analysis using antibodies against α-tubulin and β-tubulin showed that purified Arabidopsis tubulin migrated as two α-tubulin bands (a minor band of 50 kD and a major band of 49 kD) and two β-tubulin bands of approximately 50 to 51 kD, whereas tobacco tubulin is composed of 49-kD α-tubulin and 50-kD β-tubulin bands. All the Coomassie Blue-stained proteins were recognized by the tubulin antibodies, confirming that the purified tubulin is pure.
The monoclonal antibody (clone 6-11B-1) raised against the acetylated Lys-40 residue of animal α-tubulin detected porcine brain tubulin and α-tubulin from both Arabidopsis and tobacco. Although the signal intensities of porcine and tobacco tubulin were comparable, Arabidopsis tubulin was only weakly recognized by this antibody. Thus, some plant tubulins are acetylated, but the extent of tubulin acetylation may differ among tubulin isotypes and tubulin sources (see “Discussion”).
When the anti-polyglutamylation antibody GT335 was used to detect the polyglutamylation of Glu residues at the C-terminal tails of α- and β-tubulin, positive signals were observed for porcine brain tubulin, as reported previously (Janke and Bulinski, 2011), but not for Arabidopsis and tobacco tubulin, indicating that tubulin from these plant materials is not polyglutamylated.
α-Tubulin from most sources ends with the dipeptide Glu-Tyr at its C terminus, but the terminal Tyr residue can be removed by a carboxypeptidase to expose the Glu residue in animal α-tubulin (Song and Brady, 2015). The C-terminal status of α-tubulin can be assessed using the C-terminal tubulin Tyr antibody, TUB-1A2, and the C-terminal tubulin Glu antibody, AB3201. The Tyr antibody recognized α-tubulin from porcine brain, Arabidopsis, and tobacco in similar patterns as detected by the general α-tubulin antibody, whereas the Glu antibody recognized porcine α-tubulin only. These results indicate that C-terminal detyrosination does not occur in plant α-tubulin. Although the limited but significant divergence in the C-terminal tail sequences of plant and animal α-tubulin warrants caution in interpreting these results (see “Discussion”), mass spectrometry of purified Arabidopsis tubulin (see below) did not detect detyrosinated tubulin either.
α-Tubulin Phosphorylated at Thr-349 Is Inefficiently Polymerized
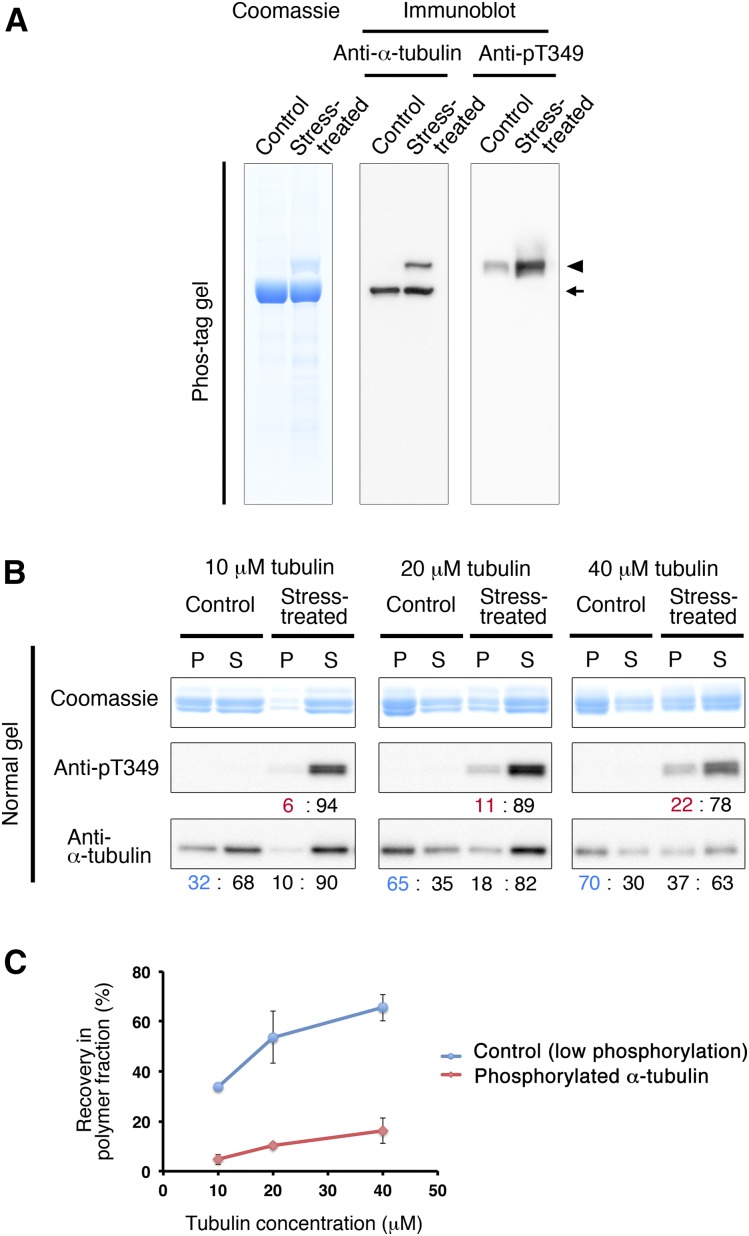
When Arabidopsis seedlings are subjected to hyperosmotic stress, α-tubulin is phosphorylated at Thr-349, which is located at the tubulin interdimer interface, resulting in the inhibition of polymerization (Ban et al., 2013; Fujita et al., 2013). We exposed 4-d-old Arabidopsis cultured MM2d cells to 0.8 m sorbitol and purified tubulin using the TOG column. Phos-tag SDS-PAGE separated phosphorylated and unphosphorylated forms of tubulin (Fig. 4A). Protein staining with Coomassie Brilliant Blue (left) and immunoblotting using anti-α-tubulin antibody (middle) showed that hyperosmotic stress generated a slower migrating band of α-tubulin due to phosphorylation (arrowhead), which represented an amount similar to the nonmodified form (arrow). In the tubulin preparation from control (nonstressed) cells, the slow-migrating phosphorylated α-tubulin form was hardly detectable. We confirmed that the shifted band indeed is phosphorylated α-tubulin, by immunoblotting using a polyclonal antibody raised against a 14-amino acid α-tubulin peptide containing phosphorylated Thr-349 (for details, see “Materials and Methods”). This pT349 antibody specifically recognized the slow-migrating, but not the fast-migrating, bands and was highly sensitive, as shown by the low amount of phosphorylated α-tubulin detected in the control sample (Fig. 4A, right).
Figure 4.
α-Tubulin phosphorylated at Thr-349 does not polymerize efficiently. A, Tubulin was purified from Arabidopsis MM2d cells that had been treated with or without 0.8 m sorbitol using the TOG column and analyzed by Phos-tag SDS-PAGE. Total proteins were detected using Coomassie Blue staining (left), whereas immunostaining (right) with anti-α-tubulin and anti-pT349 antibodies indicates total α-tubulin and Thr-349-phosphorylated α-tubulin, respectively. Phosphorylated tubulin (arrowhead) migrates more slowly than nonphosphorylated tubulin (arrow). B, MT sedimentation assay. Tubulin purified from control and sorbitol-treated MM2d cells was polymerized at three concentrations (10, 20, and 40 μm) and separated by centrifugation into MT (pellet; P) and tubulin (supernatant; S) fractions. Fractionated proteins were analyzed by SDS-PAGE, followed by Coomassie Blue protein staining (top) and immunoblotting using anti-pT349 antibody (middle) and anti-α-tubulin antibody (bottom). Immunoreactive signals were quantified by densitometric scanning and are indicated as the ratios (%) between the pellet and supernatant fractions. Blue numbers indicate total (phosphorylated and nonphosphorylated) tubulin in the nonstressed control sample in the polymer fraction, and red numbers indicate phosphorylated tubulin in the stress-treated sample in the polymer fraction. C, Tubulin preparations containing high levels of phosphorylated α-tubulin do not polymerize efficiently. The percentage of tubulin that was recovered in the pellet fraction was plotted against total tubulin concentration. The blue line represents control tubulin with low phosphorylation, and the red line represents phosphorylated tubulin in the stress-treated sample. Error bars indicate sd. Three independent MT sedimentation experiments were performed.
To assess how phosphorylated tubulin affects the polymerization activity, we performed the MT sedimentation assay in the absence of taxol (Fig. 4B). Tubulin, purified from control and stress-treated cells, was tested at three concentrations (10, 20, and 40 μm), and the tubulin recovered from the supernatant and the pellet was detected by Coomassie Blue staining (total tubulin) and immunoblotting with α-tubulin antibodies and pT349 antibodies (phosphorylated α-tubulin). At the higher tubulin concentrations (20 and 40 μm), more tubulin was recovered in the pellet (polymer) fraction. At all tested tubulin concentrations, the tubulin from stress-treated cells polymerized less efficiently than the control (Fig. 4B, bottom). The α-tubulin phosphorylated at Thr-349 was distributed mainly in the supernatants of all samples rather than in the pellets (Fig. 4B, middle). Quantification of the immunoblot signals (Fig. 4C) confirmed these results and also showed that the phosphorylated tubulin is incorporated into the MT polymer to some extent, especially at higher tubulin concentrations.
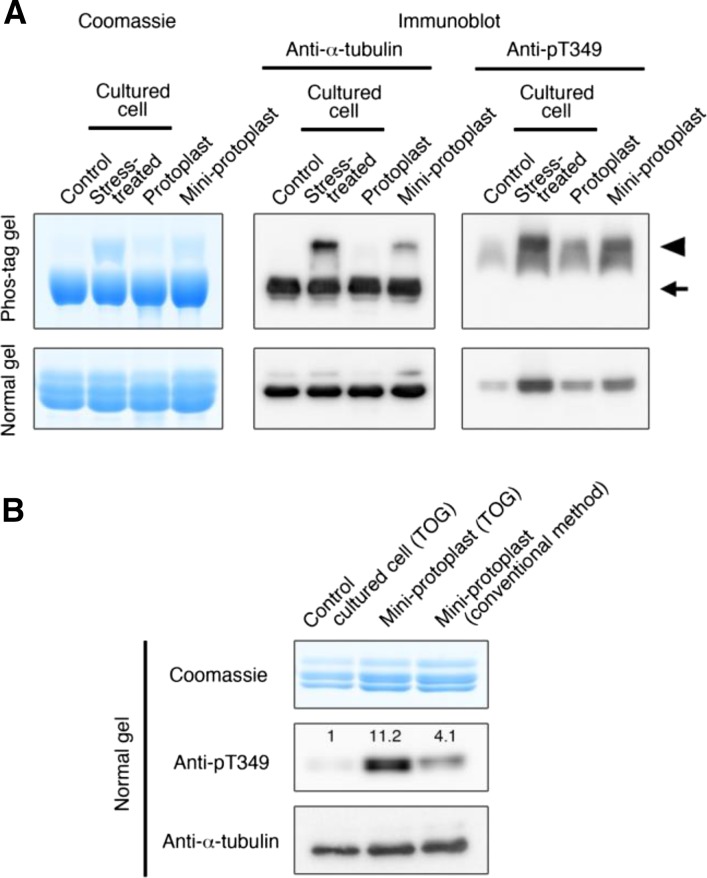
Tubulin Is Phosphorylated during Protoplast Preparation
Standard protocols to purify tubulin and microtubule-associated proteins (MAPs) involve repeated cycles of taxol-assisted tubulin polymerization and MT depolymerization and use protoplasts or cytoplasm-rich miniprotoplasts, from which vacuoles have been removed by Percoll-cushioned centrifugation, as starting material (Hamada et al., 2013). High osmotic pressure is needed to keep protoplasts intact during the procedures. Thus, when we purified tubulin and MAPs from Arabidopsis MM2d cultured cells, we maintained the protoplasts and miniprotoplasts in 0.45 and 0.6 m sorbitol, respectively (Hamada et al., 2013). To evaluate the tubulin phosphorylation status in protoplast and miniprotoplast preparations from Arabidopsis MM2d cells, tubulin was purified using the TOG column (Fig. 5A). Samples, together with tubulin purified from MM2d cells with or without 0.8 m sorbitol treatment, were analyzed by Phos-tag SDS-PAGE or normal SDS-PAGE, followed by immunoblot analysis using anti-α-tubulin antibodies or pT349 antibodies. Phosphorylation at Thr-349 of α-tubulin occurred in both protoplasts and miniprotoplasts. The signal intensities of the phosphorylated tubulin bands indicate that phosphorylation levels increase in the following order: untreated cells, protoplasts, miniprotoplasts, and stress-treated cells.
Figure 5.
Phosphorylation of α-tubulin at Thr-349 occurs during conventional tubulin purification steps from Arabidopsis miniprotoplasts. A, Tubulin phosphorylation in cultured cells, protoplasts, and evacuolated miniprotoplasts of Arabidopsis MM2d cells. Intact protoplasts and miniprotoplasts were maintained in buffer solutions containing 0.45 and 0.6 m sorbitol, respectively. Tubulin, purified by the TOG column, was analyzed with Phos-tag or normal SDS-PAGE followed by Coomassie Blue staining (5.4 μg of tubulin) or immunoblotting (200 ng of tubulin) using anti-α-tubulin or anti-pT349 antibodies. MM2d cultured cells were either not treated (Control) or treated with 0.8 m sorbitol (Stress-treated). The arrow indicates nonphosphorylated tubulin, while the arrowhead shows phosphorylated forms. B, Phosphorylation levels of purified tubulin depend on starting materials and purification methods. Tubulin was purified from nonstressed cultured cells and miniprotoplasts using the TOG column, or from miniprotoplasts by the conventional method (Hamada et al., 2013), and were analyzed by SDS-PAGE (normal gel) followed by Coomassie Blue staining (5.4 μg of tubulin) or by immunoblotting (200 ng of tubulin) using anti-α-tubulin or anti-pT349 antibodies. Relative phosphorylation levels (normalized by total α-tubulin levels on the bottom blot) are indicated on the anti-pT349 blot.
In a previous study (Fujita et al., 2013), we detected the Thr-349-phosphorylated form of α-tubulin in tubulin preparations that had been purified by conventional polymerization-depolymerization cycles involving taxol-mediated MT stabilization from Arabidopsis MM2d miniprotoplasts (Hamada et al., 2013). Indeed, the tubulin sample prepared by the method described by Hamada et al. (2013) contained 4-fold higher levels of Thr-349-phosphorylated α-tubulin than did tubulin purified directly from cultured cells using the TOG column (Fig. 5B). Thus, phosphorylated tubulin was copolymerized with nonphosphorylated tubulin at high concentrations during taxol-assisted MT sedimentation and remained at considerable levels in the final tubulin preparations.
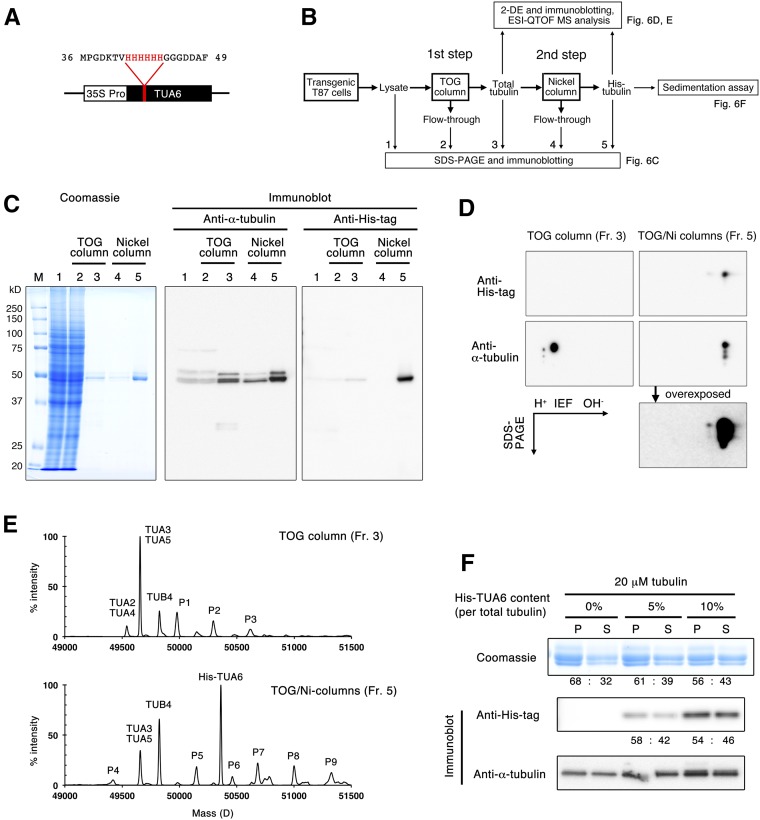
Isolation of a Single α-Tubulin Isotype
Next, we sought to purify a recombinant tubulin protein that can be engineered by introducing targeted mutations or various tags. To this end, we inserted a hexa-His purification tag between Val-42 and Gly-43 of Arabidopsis α-TUBULIN6, generating His-TUA6 (Fig. 6A). This α-tubulin loop region is poorly conserved among various tubulins and faces the lumen of the MT. Insertion of the His tag into this luminal loop was used successfully to purify recombinant tubulin from budding yeast (Sirajuddin et al., 2014). His-TUA6 was expressed constitutively in Arabidopsis T87 cultured cells under the control of the cauliflower mosaic virus 35S promoter and was purified by a two-step procedure (Fig. 6B). Cell extracts were first applied to two TOG columns (small and medium size, containing 1 and 2 mL of resin, respectively) and then further purified by a nickel column (see “Materials and Methods”). Tubulin was efficiently purified by the first two TOG columns, as shown by Coomassie Blue protein staining of the SDS-PAGE gel and the α-tubulin immunoblot (Fig. 6C). Immunoblotting with an anti-poly-His antibody revealed that the hexa-His-tagged tubulin was highly enriched after purification on the nickel column. Starting with 45 to 58 g fresh weight of Arabidopsis T87 cells, 705 to 884 μg of total tubulin was purified, of which 18 to 33 μg was His-TUA6 tubulin (Table III).
Figure 6.
Isolation of a single isotype of α-tubulin using an internal hexa-His tag. A, A hexa-His tag was inserted between Val-42 and Gly-43 of TUA6, and the transgene was expressed under the control of the cauliflower mosaic virus 35S promoter in cultured Arabidopsis T87 cells. B, Experimental flow chart. After total tubulin was purified using the TOG column, His-TUA6 tubulin was recovered using the nickel column. C, Purification of His-TUA6 tubulin by the nickel column. Tubulin samples (fractions 1–5 in B) were separated by SDS-PAGE and detected by Coomassie Blue staining (left) and immunoblotting using anti-α-tubulin and anti-His-tag antibodies (right). M, Molecular mass marker; fraction 1, 4.3 μL of crude cell extract containing 25 μg of proteins; fraction 2, 4.3 μL of the flow-through fraction from the TOG column; fraction 3, 400 ng of total tubulin in the eluate from the TOG column; fraction 4, 8.5 μL of the flow-through fraction from the nickel column; and fraction 5, 700 ng of tubulin in the eluate from the nickel column. D, Two-dimensional gel electrophoresis of tubulin samples (fractions 3 and 5) followed by immunoblotting using anti-α-tubulin and anti-His-tag antibodies. An anti-α-tubulin blot of fraction 5 was overexposed to detect trace amounts of endogenous α-tubulin. E, Mass spectrometry analysis of tubulin in fractions 3 and 5. In addition to unmodified forms of Arabidopsis tubulin and His-TUA6, unknown small peaks (P1–P9) were also detected. For discussion, see text. F, His-TUA6 tubulin polymerizes as efficiently as wild-type tubulin. His-TUA6 tubulin was mixed with Arabidopsis wild-type tubulin at 0%, 5%, or 10% of the total tubulin amount and evaluated using an MT sedimentation assay. Tubulin proteins were separated by SDS-PAGE and detected by Coomassie Blue staining (top) and immunoblotting using anti-His-tag (middle) and anti-α-tubulin (bottom) antibodies. Relative tubulin amounts in the pellet (P) and supernatant (S) are shown.
Table III. Purification of His-TUA6 tubulin from cultured Arabidopsis cells.
Transgenic Arabidopsis cultured T87 cells expressing His-TUA6 were cultured for 4.5 d in 500 mL of culture medium and used in the two-step purification procedure. Two TOG columns (small and medium sizes) were used to purify total tubulin, from which His-TUA6 tubulin was purified by the nickel column. His-TUA6 tubulin was purified three times.
| Experiment | Cell Weight | Total Protein | Purified Tubulin | Percentage in Total Protein | His-TUA6 Tubulin | Percentage in Total Tubulin |
|---|---|---|---|---|---|---|
| g fresh wt | mg | μg | μg | |||
| 1 | 45 | 391 | 705 | 0.18 | 18.1 | 2.6 |
| 2 | 45 | 357 | 884 | 0.25 | 20.8 | 2.4 |
| 3 | 58 | 374 | 848 | 0.23 | 33.3 | 3.9 |
To further evaluate the purity of the sample, proteins were separated by two-dimensional gel electrophoresis consisting of isoelectric focusing (IEF) and SDS-PAGE and analyzed by immunoblotting (Fig. 6D). When tubulin was purified from the His-TUA6-expressing Arabidopsis cells using only the TOG column, two major spots (and a few minor spots of lower molecular mass) were detected by the anti-α-tubulin antibody, whereas no spots were detected by the anti-His-tag antibody. When this tubulin preparation was further purified on the nickel column, one major spot was recognized by both the anti-α-tubulin antibody and the anti-His-tag antibody, with a shift to the right relative to the positions of endogenous wild-type α-tubulin isotypes. This shift of His-TUA6 on the IEF gel is consistent with the expected higher pI value (5.17) of His-TUA6 compared with that of endogenous TUAs (from 4.92 to 4.95). Endogenous α-tubulin proteins were not detected even when the anti-α-tubulin-probed blot was overexposed. These results indicate that the proportion of His-TUA6 in the initial α-tubulin pool was quite low and that the final tubulin sample purified by the two-step TOG/nickel columns contained predominantly His-TUA6.
We further assessed the tubulin composition by electrospray ionization mass spectrometry (Fig. 6E). In the TOG column-purified tubulin preparation, TUA3/5 and TUA2/4 were detected at 49,656 and 49,540 D, respectively (TUA3 and TUA5 have an identical theoretical mass of 49,654 D, while the theoretical mass of TUA2 and TUA4 is 49,541 D), and only TUB4 was observed at 49,825 D (theoretical mass, 49,823 D). The TUA3/5 peak was much larger than the TUA2/4 peak. His-TUA6 was not detectable, as was expected due to its low abundance. By contrast, the final tubulin sample contained a large peak of His-TUA6 at 50,361 D (theoretical mass, 50,361 D) and a smaller peak of TUA3/5 at 49,658 D. Among the β-tubulin isotypes, TUB4 was detected at 49,824 D. We did not expect to detect endogenous TUA3/5 in the final tubulin sample, because the immunoblots after two-dimensional gel electrophoresis (Fig. 6D) failed to detect endogenous α-tubulin in this sample. It is not clear what caused this discrepancy, but the ionization efficiency of TUA3/5 and His-TUA6 in mass spectrometry might differ.
The molecular mass peaks of TUA3/5, TUA2/4, TUB5, and His-TUA6 matched the theoretical sizes of their unmodified forms; no clear peaks were observed at the expected positions of their acetylated, polyglutamylated, or detyrosinated forms. These results are consistent with the results obtained using modification-specific antibodies (Fig. 3) and suggest that posttranslational modifications of Arabidopsis tubulins are limited or absent. In the mass spectra, we did notice several minor peaks that did not correspond to the unmodified, acetylated, polyglutamylated, or detyrosinated forms of Arabidopsis α- and β-tubulin (P1–P9 in Fig. 6E). P1 (49,978 D), P2 (50,295 D), and P3 (50,617 D) are larger than TUA3/5 (49,656 D) by one, two, and three units of approximately 320 D. Similarly, increments of approximately 320 D would generate P7 (50,683 D), P8 (51,000 D), and P9 (51,326 D) from His-TUA6 (50,361 D). We do not know whether these putative α-tubulin adducts actually exist in Arabidopsis cells or represent artifacts from the preparation or analysis of purified tubulin.
To examine whether His-TUA6 tubulin polymerizes as efficiently as wild-type tubulin, we performed an MT sedimentation assay in the absence of taxol. Because the amount of His-TUA6 was rather limited (approximately 30 μg), we did not prepare a tubulin solution that exceeded the critical concentration for spontaneous polymerization. Instead, we mixed His-TUA6 tubulin with wild-type Arabidopsis tubulin to make tubulin solutions in which the tagged tubulin constituted 0%, 5%, and 10% of total tubulin. Total α-tubulin and His-TUA6 were detected by antibodies against α-tubulin and poly-His. In both cases, MTs polymerized at comparable levels to the negative control (0% His-TUA6-containing tubulin; Fig. 6F, Coomassie). His-tagged tubulin was detected by anti-His antibodies in the pellets in both conditions, indicating the incorporation of His-tagged tubulin into MTs (Fig. 6F). The ratios between pellet and supernatant were almost equal on the Coomassie Blue-stained and immunoblot (Anti-His tag) gels for both 5% and 10% tagged tubulin, suggesting that the polymerization competence does not differ between wild-type α-tubulin and His-TUA6. Therefore, we conclude that internally hexa-His-tagged tubulin is functional in vitro.
DISCUSSION
Tubulin Binding by TOG Domains
To purify tubulin from plant cell extracts, TOG domains need to bind free tubulin heterodimers with sufficient affinity and release the captured tubulin in the presence of high salt concentrations or competing ligands. The TOG1 and TOG2 domains of yeast Stu2 protein bind to the slightly curved tubulin heterodimer in the unpolymerized and unbound state but do not bind to the MT lattice (Ayaz et al., 2012, 2014). TOG1 interacts with conserved surfaces that include Glu-415 of α-tubulin and Tyr-106 and Thr-107 of β-tubulin in yeast (Ayaz et al., 2012). These critical residues and their adjacent amino acid residues are highly conserved in plant tubulin, including Arabidopsis α- and β-tubulin (Supplemental Fig. S3), thus providing a mechanistic rationale for why the TOG column can be used to purify tubulin from diverse plant species.
MOR1 is an Arabidopsis homolog of yeast Stu2 (Whittington et al., 2001). Whereas Stu2 has two TOG domains, MOR1 possesses five TOG domains (TOG1–TOG5), as do human ch-TOG and Drosophila melanogaster Msps (Al-Bassam and Chang, 2011). Two critical residues of Stu2 TOG1 (Trp-23 and Arg-200) that are functionally important for TOG1-tubulin interactions (Ayaz et al., 2012) are conserved in TOG1 and TOG2 of MOR1 (Supplemental Fig. S4), indicating that TOG1 and TOG2 of MOR1 might bind unpolymerized tubulin. However, the N-terminal part of MOR1 containing TOG1 and TOG2 bound sheep brain tubulin very weakly when analyzed by gel filtration (Lechner et al., 2012). We also found that our MOR1 TOG column, containing TOG1 and TOG2, does not bind plant tubulin efficiently. The other TOG domains (TOG3–TOG5) of MOR1 might compensate for the apparently weak tubulin interaction of TOG1 and TOG2.
Purification of Tagged Tubulin
Ideally, affinity purification tags should not interfere with the polymerization capacity of tubulin and with the binding of MT regulators and motor molecules, such as kinesin, to the polymerized MT. An epitope tag fused to the N terminus of α-tubulin caused severe MT-related defects in transgenic Arabidopsis plants, possibly by interfering with the polymerization-triggered hydrolysis of the exchangeable GTP in β-tubulin (Abe and Hashimoto, 2005). An epitope tag on the β-tubulin N terminus or the α-tubulin C terminus did not result in apparent developmental and morphological abnormalities in Arabidopsis plants (Abe and Hashimoto, 2005), but we cannot exclude the possibility that the functions of some MT-binding proteins and MT motors are affected by modifications of tubulin termini. Indeed, a purification tag at the C terminus of either α- or β-tubulin of yeast origin affected the velocity of kinesin (Sirajuddin et al., 2014). In this study, we adopted a tubulin-tagging strategy that was developed to purify recombinant tubulin from yeast cells (Sirajuddin et al., 2014). In this method, the hexa-His tag is inserted at a poorly conserved loop in α-tubulin, which faces the lumen of the MT. Insertions of up to 17 amino acids into this site were shown not to disrupt MT function in fission yeast (Schatz et al., 1987), and the internal His tag did not appear to affect MT polymerization and motor behavior (Sirajuddin et al., 2014).
The internal His tag in Arabidopsis TUA6 enabled us to purify a tubulin preparation containing a single recombinant α-tubulin isoform from Arabidopsis cell cultures. Since TUB4 is the major β-tubulin isotype in Arabidopsis cell cultures (Fig. 6E), the purified tubulin heterodimer consists mainly of TUA6 and TUB4. While the current method purifies single α-tubulin proteins, a similar strategy may be used to insert a purification tag into a poorly conserved luminal loop region of Arabidopsis β-tubulin. This approach would be useful for characterizing the effect of various tubulin mutations (Ishida et al., 2007; Hashimoto, 2013) and posttranslational modifications (see below) on MT dynamics and for analyzing the interactions of these mutated tubulins with MT regulators in vitro.
In this study, we purified approximately 30 μg of tubulin containing His-TUA6 from 370 mg of crude soluble protein. This modest yield (approximately 0.2%) is due to the rather low percentage of His-TUA6 (approximately 2%–4%) compared with endogenous α-tubulin isoforms (Table III). To improve yield, it may be useful to develop an inducible system for the overexpression of His-TUA6 in cell cultures. This strategy has been used successfully to increase the yield of recombinant tubulin in yeast cells (Johnson et al., 2011). Another method that may be used simultaneously would be to suppress the expression of endogenous tubulin genes. Since TUA6 belongs to a vegetative tubulin subfamily, which is distinct from the vegetative subfamily to which TUA3 and TUA5 belong, and since TUA3 and/or TUA5 constitute a major fraction of α-tubulin expressed in Arabidopsis cell cultures (Fig. 6E), TUA3 and TUA5 may be down-regulated with minimum effect on the expression of the introduced His-TUA6 transgene.
Highly Dynamic Arabidopsis MTs in Vitro
Tubulin purified from Arabidopsis cell suspension cultures using the TOG column is readily recovered in the polymer fraction even in the absence of taxol (Fig. 1C) and spontaneously nucleates bundled MTs in the absence of MT seeds (Fig. 2B). Since porcine brain tubulin purified by the same TOG column method requires MT seeds for efficient nucleation (at least under tubulin concentrations of up to 15 μm; Fig. 2A), the high assembly competence of Arabidopsis tubulin does not result from the use of the TOG column but is an intrinsic property of Arabidopsis tubulin. Both the plus and minus ends of Arabidopsis MTs are extremely dynamic. Compared with porcine MTs, the largest differences are found at the plus end, which exhibits a very rapid shrinkage rate and very high catastrophe frequency (Table II). These characteristic dynamics result in short MTs that frequently alternate cycles of growth and shrinkage at the plus end. When tubulin purified from carrot (Daucus carota) cell suspension culture by a two-step chromatography procedure was assembled onto sea urchin axonemes, carrot MTs underwent numerous catastrophe events and shrank very rapidly (Moore et al., 1997). These dynamic features may be typical of plant tubulins. In interphase Arabidopsis cells, cortical MTs migrate to the cell cortex by polymerization-biased dynamics at the plus end, together with sustained slow shrinkage at the minus end (Shaw et al., 2003; Nakamura et al., 2004). The marked differences in dynamic behaviors of plant MTs in vitro and in vivo suggest that various MAPs regulate the dynamic properties at both ends of MTs in plant cells. Dynamic MTs polymerized from pure plant tubulin will be used in in vitro reconstitution experiments to test whether the combinatorial action of plant MAPs enhances physiological MT dynamics, as first demonstrated in the reconstitution system involving bovine brain tubulin and two major Xenopus laevis MAPs (Kinoshita et al., 2001).
What underlying differences would make porcine and Arabidopsis MTs exhibit highly distinct dynamic properties in vitro? The amino acid sequences of eukaryotic tubulins are fairly well conserved but still display considerable differences. Tubulins of Antarctic fishes accumulate unique amino acid substitutions at positions predicted to be important for MT dynamics and assemble into less dynamic and cold-stable MTs (Detrich et al., 2000). Even distinct tubulin isotypes in a given species show differences in dynamic instability (Hashimoto, 2013). In addition to the variability encoded in the tubulin genes, differences in posttranslational modifications between porcine brain tubulin and Arabidopsis tubulin (see below) might provide another layer of functional divergence in MT dynamics.
Posttranslational Modification of Tubulin
We revisited putative posttranslational modifications of plant tubulin (Fujita et al., 2013; Parrotta et al., 2014) in our purified preparations. The acetylated Lys residue resides in a luminal loop of α-tubulin, very close to the hexa-His insertion site, and both polyglutamylation and detyrosination target the C-terminal tails of tubulin (Janke, 2014). The hyperosmotic stress-induced phosphorylation site is localized to the longitudinal interdimer interface (Fujita et al., 2013). Since these modification sites are not located near the tubulin-interacting regions of the TOG domains (Ayaz et al., 2012), the TOG-mediated affinity purification is unlikely to affect the recovery of these modified tubulin forms.
The monoclonal anti-acetyl-Lys-40 antibody, clone 6-11B-1, reacted strongly with α-tubulin from porcine brain and cultured tobacco cells and weakly with α-tubulin from cultured Arabidopsis cells. This antibody has been used to detect tubulin acetylation, which varies considerably among cell and tissue types (Aström, 1992; Wang et al., 2004; Giannoutsou et al., 2012), at different developmental stages (Nakagawa et al., 2013), and as a result of hormonal treatments (Huang and Lloyd, 1999). Lys-40 is only partially conserved in plant α-tubulin isotypes. For example, Arabidopsis TUA2, TUA4, and TUA6 contain Lys-40, whereas this Lys residue is substituted with Ser or Thr in TUA1, TUA3, and TUA5 (Supplemental Fig. S3). Cellular levels of acetylated MTs may depend on the abundance of Lys-40-type α-tubulin isotypes relative to that of nonacetylatable isotypes. The high abundance of nonacetylatable α-tubulin isotypes (TUA3 and TUA5) in Arabidopsis cell cultures is at least partly responsible for the low acetylation levels of Arabidopsis tubulin (Figs. 3 and 6E). Another source of variability is the activity of tubulin deacetylases, which are still poorly characterized in plants (Tran et al., 2012). Stabilized MTs, but not dynamic MTs, in animal cells are marked by the acetylation of α-tubulin Lys-40 (Szyk et al., 2014). MT acetylation is especially abundant in cilia, flagella, centrioles, and neurons (Song and Brady, 2015). The acetylation of α-tubulin Lys-40 is catalyzed by α-tubulin acetyltransferase (αTAT), which is a homolog of MEC-17 in animals (Akella et al., 2010). Since MEC-17 homologs are absent in fungi and plants (Akella et al., 2010) and plant cells generally lack highly stable MTs (Hashimoto, 2015), it is puzzling how plant cells catalyze this conserved tubulin modification and whether acetylated MTs represent stable and long-lived MT subpopulations in plant cells as well. Overexpression of a nonacetylatable Lys-40-to-Arg α-tubulin mutant does not affect the morphology and development of Arabidopsis plants (Xiong et al., 2013). Functional analyses of acetylated MTs in plant cells require the identification and knockout of plant αTAT genes.
Most eukaryotic α-tubulin genes, such as Arabidopsis TUA genes (Supplemental Fig. S3), encode a C-terminal Tyr residue. In animal cells, this Tyr residue can be enzymatically removed to expose the penultimate Glu residue and religated with tubulin Tyr ligase (Janke, 2014). Detyrosination, like acetylation, is generally found on stable and long-lived MTs (Janke, 2014), and kinetochore motors preferentially track detyrosinated MTs during chromosome segregation in animal cell division (Barisic et al., 2015). Previous studies assessed tubulin detyrosination in plant cells using the anti-Glu antibody, which was raised against the animal α-tubulin tail sequence Gly-Glu-Glu-Glu-Gly-Glu-Glu (Smertenko et al., 1997), or by comparing α-tubulin isoforms detectable by the terminal Tyr-specific antibody and the general α-tubulin antibody (Duckett and Lloyd, 1994; Wiesler et al., 2002; Wang et al., 2004). In this study, the Tyr antibody recognized Arabidopsis and tobacco tubulin from suspension cell cultures, resulting in a similar number and intensity of bands to those detected by the general α-tubulin antibody, whereas the Glu antibody did not (Fig. 3). Moreover, mass spectrometry of purified Arabidopsis tubulin did not detect detyrosinated TUA proteins (Fig. 6E). These results indicate that plant α-tubulin predominantly retains the terminal Tyr residue. The absence of animal-type tubulin Tyr ligase genes (Ersfeld et al., 1993) in plant genomes is also consistent with the presumed lack of the tyrosination-detyrosination cycle in plant cells.
Glu residues are added progressively onto the γ-carboxyl group of one or more Glu residues in the C-terminal tail of both α- and β-tubulin in animal cells (Janke and Bulinski, 2011). While polyglutamylation-specific antibodies detected tubulin-like bands by immunoblot and MT-like structures by immunohistochemistry in tobacco and maize (Zea mays) cells (Smertenko et al., 1997; Wang et al., 2004), we obtained no evidence of the polyglutamylation of purified tubulin from Arabidopsis and tobacco cells using the polyglutamylation antibody (Fig. 3) and mass spectrometry (Fig. 6E). Initial glutamylation and successive polyglutamylation reactions are catalyzed by several related enzymes belonging to the tubulin Tyr ligase-like (TTLL) superfamily (Janke and Bulinski, 2011). The Arabidopsis genome contains only one member (At1g77550), which belongs to the uncharacterized TTLL12 subfamily; no close homologs of the tubulin glutamylation and polyglutamylation enzymes exist in Arabidopsis (Janke et al., 2005). Establishing the molecular identity of (poly)glutamylation enzymes in plants, if they exist, would be a crucial step toward understanding this elusive posttranslational tubulin modification.
The phosphorylation of Thr-349 in α-tubulin is the best-characterized modification of plant tubulin. The Thr residue is located at the longitudinal interface between adjacent tubulin heterodimers in a protofilament (Nogales et al., 1999), and substitution or modification of Thr-349 with a larger residue is expected to impair MT polymerization. Indeed, a GFP-tagged α-tubulin mutant in which Thr-349 was substituted with a phospho-mimic Asp residue was not efficiently incorporated into MTs when expressed in Arabidopsis cells (Fujita et al., 2013). When an active PHS1 tubulin kinase mutant was induced ectopically in Arabidopsis plants, the tubulin purified from this plant source did not polymerize well in vitro (Fujita et al., 2013). In this study, we demonstrated that tubulin phosphorylation significantly decreases the polymerization capacity in vitro but that, at higher tubulin concentrations (e.g. 40 μm), the phosphorylated tubulin copolymerizes with unphosphorylated tubulin to some extent (Fig. 4C). This explains why tubulin preparations purified by taxol-assisted polymerization-depolymerization cycles contain considerable amounts of Thr-349-phosphorylated tubulin (Fujita et al., 2013; this study).
Plants grown under standard conditions without stress contain low levels of phosphorylated tubulin, but, upon exposure to high-salt or osmotic stress conditions, Thr-349 α-tubulin phosphorylation is rapidly and transiently induced (Ban et al., 2013; Fujita et al., 2013). Typical protocols for tubulin purification from plant cell suspension cultures involve preparations of cytoplasm-rich miniprotoplasts, which remain intact in highly osmotic solutions (Hamada et al., 2013). We show here that such miniprotoplasts contain increased levels of phosphorylated tubulin compared with nonstressed plant cells (Fig. 5A). Repeated polymerization-depolymerization cycles did not completely remove the phosphorylated tubulin from the purified tubulin preparations (Fig. 5B). The high polymerization capacity of tubulin purified directly from intact cultured cells by the TOG column may result from low levels of α-tubulin Thr-349 phosphorylation compared with considerable phosphorylation in the tubulin purified from plant protoplasts.
CONCLUSION
The TOG column effectively purifies assembly-competent tubulin from several plant materials in one step. The yield depends on the type of starting material: cell suspension cultures give higher recovery rates than do whole seedlings. As much as 0.7 mg of tubulin can be purified from 18 g (fresh weight) of Arabidopsis cultured cells. An internal hexa-His tag in α-tubulin enables the purification of tubulin that is highly enriched in a single α-tubulin isotype by a two-step affinity purification protocol. In contrast to the heavily modified tubulin isolated from vertebrate brains, plant tubulin exhibits low levels of posttranslational modification. The availability of pure plant tubulin will facilitate in vitro functional studies of tubulin mutants, tubulin posttranslational modifications, and interactions between tubulin and various plant MT regulators and motor molecules.
MATERIALS AND METHODS
Plant Materials
Suspension-cultured cells of Arabidopsis (Arabidopsis thaliana) cell lines (MM2d and T87) and the tobacco (Nicotiana tabacum) cell line BY-2 were grown in modified Linsmaier and Skoog medium on a gyratory shaker at 130 rpm at 27°C in darkness for MM2d and BY-2 cells (Kumagai et al., 2001; Hamada et al., 2013) or at 23°C with a 16-h-light/8-h-dark cycle for T87 cells. The cultures were maintained by transferring 3, 5, and 0.8 mL of 7-d-old MM2d, T87, and BY-2 cells, respectively, to 95 mL of fresh medium in 300-mL flasks. Suspension-cultured cells of an Arabidopsis Alex cell line were maintained as described previously (Oda and Fukuda, 2011). Arabidopsis seedlings (Columbia-0 ecotype) were grown in liquid medium of one-half-strength Murashige and Skoog salts as described previously (Fujita et al., 2013). To harvest cells, cell suspensions were poured onto filter paper on a Buchner funnel and the culture medium was thoroughly removed by aspiration. Seedlings were placed on paper towels to remove the culture medium. After fresh weight was measured, cells and seedlings were frozen in liquid nitrogen and stored at −80°C until used.
Preparation of GST-TOG1/2 Proteins
A bacterial expression plasmid, pGEX-6P-1-Stu2(1-590), that encodes a GST fusion of the TOG1 and TOG2 domains of Stu2 (GST-TOG1/2) was a kind gift from Per Widlund. Recombinant GST-TOG1/2 protein was expressed in Escherichia coli (strain Rosetta) and purified as described (Widlund et al., 2012) with modifications. Bacteria were cultured at 37°C in 500 mL of Terrific Broth (1.2% tryptone, 2.4% yeast extract, 0.4% glycerol, 17 mm KH2PO4, and 72 mm K2HPO4) containing 100 μg mL−1 carbenicillin and 15 μg mL−1 chloramphenicol in a 3-L flask. When the optical density at 600 nm reached 0.6 to 0.8, the flask was placed in ice-cold water for 10 min and shaken at 18°C for another 1 h. Isopropyl β-d-1-thiogalactopyranoside was then added at a final concentration of 0.2 mm, and the culture was incubated at 18°C overnight. Cells were collected by centrifugation (2,500g for 10 min at 18°C) and washed in phosphate-buffered saline (PBS; 140 mm NaCl, 2.7 mm KCl, 10 mm Na2HPO4, and 1.8 mm KH2PO4) in 50-mL tubes. Final bacterial pellets were stored at −80°C until used.
All protein extraction and column chromatography procedures were carried out on ice or at 4°C, except for the elution step. Bacterial pellets from a 1-L culture were lysed with 120 mL of ice-cold bacterial extraction buffer containing 1× PBS, 1 mm MgCl2, 0.5% Triton X-100, 1 mm ATP, 1 mm dithiothreitol (DTT), and 0.2 mm phenylmethylsulfonyl fluoride. After sonication for 5 min, the lysate was centrifuged at 25,000g for 20 min. Cleared lysate was divided in half, and each half (approximately 60 mL) was loaded onto a column containing 5 mL of glutathione Sepharose 4B (GE Healthcare). After washing with 50 mL of wash buffer (1× PBS, 1 mm MgCl2, 1 mm ATP, 1 mm DTT, and 0.2 mm phenylmethylsulfonyl fluoride), the column was moved to room temperature. Elution was carried out with 5 mL of a glutathione solution (1× PBS and 10 mm reduced glutathione). Protein quality was evaluated with SDS-PAGE followed by Coomassie Blue staining, and protein quantity was estimated by the Bradford protein assay using bovine serum albumin as a control. Purified GST-TOG1/2 protein was either frozen with liquid nitrogen and stored at −80°C until used or placed on ice for immediate use.
To prepare a GST fusion of the TOG1 and TOG2 domains of MOR1 (GST-TOG1/2MOR1), a MOR1 complementary DNA fragment encoding the N-terminal 528 amino acid residues was cloned into pGEX-6P-1 (GE Healthcare) at the EcoRI and XhoI sites. Recombinant GST-TOG1/2MOR1 protein was expressed in the E. coli BL21 (DE3) pLysS strain after isopropyl β-d-1-thiogalactopyranoside-mediated induction and purified as described above.
Preparation of TOG Columns
Small (1 mL of resin) and medium (2 mL of resin) TOG columns were prepared from 20 and 40 mg of purified GST-TOG1/2 protein, respectively. Purified GST-TOG1/2 protein was dialyzed in 100 mm NaHCO3 and 100 mm NaCl at 4°C overnight to remove glutathione and then concentrated to 4 mg mL−1 using an ultrafiltration device (Amicon, 10K cutoff; GE Healthcare). After centrifugation at 100,000g for 15 min at 4°C, MgCl2 was added to a final concentration of 80 mm. N-Hydroxysuccinimide (NHS)-activated Sepharose 4 Fast Flow resin (GE Healthcare; 1 mL for a small column) was packed into an empty PD-10 column (GE Healthcare) from which the originally packaged desalting resin had been removed and washed with 3 mL of ice-cold 1 mm HCl. The GST-TOG1/2 solution was then loaded onto the column. The flow-through fraction was loaded back to the same column six to seven times. The upper filter, which was removed with the commercial PD-10 column, was placed on the top surface of the resin bed. Subsequently, the GST-TOG1/2 resin was quenched by loading 3 mL of the quenching solution (0.5 m ethanolamine and 0.5 m NaCl, pH 8.3) onto the column and incubating the column at room temperature for 15 min. The quenching solution was then drained, and this quenching procedure was repeated twice. The GST-TOG1/2 column was washed with 10 mL of 6× PBS and subsequently with 5 mL of 1× PBS and was stored in 1× PBS with 50% glycerol at −20°C. Typically, 76% to 78% of the applied GST-TOG1/2 protein was bound to the resin.
The TOGMOR1 column (a MOR1 version of the TOG column) was prepared in the same way using purified GST-TOG1/2MOR1 protein.
Stress Treatment and Protoplast Preparation
For the hyperosmotic stress treatment, 4-d-old suspension-cultured MM2d cells from three 300-mL flasks were collected on a glass filter (Shibata; 17GP100) in a Buchner funnel and washed gently with 700 mL of culture medium supplemented or not with 0.8 m sorbitol. After the wash step, the cells were resuspended in 100 mL of fresh culture medium with or without 0.8 m sorbitol each in three 300-mL flasks and rotated at 130 rpm for 1 h at 27°C in darkness.
Protoplasts and evacuolated miniprotoplasts were prepared as described (Hamada et al., 2013).
Purification of Endogenous Tubulin
About 30 g fresh weight of cultured cells was ground with liquid nitrogen using a mortar and pestle. Ground cell powder was mixed with 30 mL of 2× extraction buffer containing 160 mm PIPES (pH 6.8), 2 mm EGTA, 2 mm DTT, 200 μm Na3VO4, 10 mm NaF, 100 mm glycerophosphate, and Complete Protease Inhibitor Cocktail EDTA-free (Roche), and proteins were extracted at 4°C for 30 min with gentle agitation. Proteins from protoplasts and miniprotoplasts were extracted by homogenization in 1× extraction buffer on ice. After the crude cell lysate was centrifuged at 25,000g for 10 min at 4°C, the supernatant was passed through a 0.45-μm syringe filter (Iwaki; 2053-025).
All steps of tubulin purification were carried out at 4°C, and all solutions were prechilled on ice. Cleared lysate was loaded onto the column that had been preequilibrated with 20 mL of extraction buffer. The flow-through fraction may be reloaded back on the column once or twice. The column was washed successively with the following wash buffers: Wash #0 (5 mL of 80 mm PIPES, 1 mm EGTA, and 100 μm GTP, pH 6.8); Wash #1 (5 mL of 1× BRB80 [80 mm PIPES, 1 mm EGTA, and 1 mm MgCl2, pH 6.8] and 100 μm GTP); Wash #2 (12 mL of 1× BRB80 with 10 μm GTP); Wash #3 (4 mL of 1× BRB80, 10 mm MgCl2, 100 μm GTP, and 5 mm ATP); and Wash #4 (6 mL of 1× BRB80 with 10 μm GTP). To elute tubulin, 2.5 mL of elution buffer [1× BRB80, 10 μm GTP, and 500 mm (NH4)2SO4] was loaded onto the column. The eluate was applied to a PD-10 desalting column (GE Healthcare) that had been preequilibrated with 25 mL of 1× BRB80. Desalted tubulin was eluted with 3.5 mL of 1× BRB80, concentrated to approximately 50 μL by an ultrafiltration device (Amicon, 10K cutoff), and centrifuged at 2,380g for 75 to 100 min. Tubulin was quantified using the Bradford protein assay, and densitometric analysis of Coomassie Blue-stained SDS-PAGE gels was performed using bovine serum albumin as a standard. When the medium TOG column was used, all the solution volumes were scaled up accordingly. Glycerol was added to the purified tubulin sample to a final concentration of 10%. Purified tubulin was divided into aliquots, snap frozen with liquid nitrogen, and stored at −80°C. The TOG column was regenerated by washing with 10 mL of 1× PBS, followed by 20 mL of 10× PBS, and then by 20 mL of 1× PBS.
Arabidopsis tubulin was also isolated from miniprotoplasts of MM2d culture cells by the conventional taxol-mediated assembly-disassembly method, followed by anion-exchange column chromatography (Hamada et al., 2013).
Porcine brain tubulin was purified by a TOG column or the conventional method consisting of two cycles of assembly-disassembly (Shelanski et al., 1973), followed by ion-exchange chromatography using DEAE Sephacel (GE Healthcare), and used in the in vitro dynamics assay (Fig. 2) or in the immunoblot experiment (Fig. 3), respectively. The TOG column was used essentially in the same manner as described above for plant tubulin. Fresh porcine brains were ground in BRB80 supplemented with 2 mm DTT and Complete Protease Inhibitor Cocktail EDTA-free and centrifuged at 2,380g for 10 min at 4°C. The supernatant was further cleared by ultracentrifugation at 100,000g for 30 min at 4°C and loaded onto a TOG column. The subsequent steps were carried out as described (Shelanski et al., 1973).
Purification of His-Tagged Tubulin
After the coding region of Arabidopsis TUA6 complementary DNA was cloned into the pDONRzeo vector (Invitrogen), a hexa-His-encoding sequence (5′-CATCACCATCATCACCAT-3′) was inserted between Val-42 and Gly-43 of TUA6. This His-TUA6 was cloned into the pGWB2 Gateway vector (Nakagawa et al., 2007) using LR Clonase II (Invitrogen) and then transformed into Agrobacterium tumefaciens strain GV3101 (pMP90). Arabidopsis T87 cells were transformed by exactly the same method as reported for BY-2 cells (Kumagai et al., 2001).
Total tubulin was purified from transgenic T87 cultured cells expressing His-TUA6 using the TOG column as described above, with slight modifications. Since the subsequent nickel column purification is not compatible with EGTA, an additional wash step (Wash #5: 80 mm PIPES, 1 mm MgCl2, and 10 μm GTP, pH 6.8; 6 and 12 mL, respectively, for small and medium columns) was included directly before the elution procedure. EGTA was omitted from the elution buffer. Eluted tubulin solutions were combined, supplemented with 30 mm imidazole, and gently agitated for 30 min at 4°C with 1.2 mL of Ni Sepharose 6 Fast Flow resin (GE Healthcare) that had been preequilibrated with binding buffer [80 mm PIPES, 1 mm MgCl2, 500 mm (NH4)2SO4, and 30 mm imidazole, pH 6.8]. The resin was then collected by centrifugation (1,000g, 2 min, and 4°C), transferred to an empty PD-10 column, and washed with 35 mL of binding buffer supplemented with 10 μm GTP and with 12 mL of wash buffer (80 mm PIPES, 1 mm MgCl2, 30 mm imidazole, and 10 μm GTP, pH 6.8). Tubulin containing His-TUA6 was eluted with 7.5 mL of elution buffer (80 mm PIPES, 1 mm MgCl2, 250 mm imidazole, and 10 μm GTP, pH 6.8), and 1 mm EGTA was added to the eluted tubulin fraction. Desalting and ultrafiltration were performed as described above.
MT Sedimentation Assay
MT assembly was induced by incubating 10, 20, or 40 μm tubulin in 1× BRB80 supplemented with 10% glycerol and 1 mm GTP at 27°C or 30°C for 45 min. Taxol was not included in the polymerization buffer, except for the experiments shown in Figure 1C, in which paclitaxel (Wako) was added to the solution to a final concentration of 5 μm. Polymerized MTs were separated from tubulin by centrifugation at 100,000g for 30 min at 27°C or 30°C. Pellet and supernatant fractions were mixed with the SDS-PAGE sample buffer and analyzed by SDS-PAGE, followed by Coomassie Blue staining. Proteins on the gel were quantified by densitometric scanning.
Fluorescent Labeling of Tubulins
MM2d tubulin polymerization was induced by the addition of 1/100th volume of 100 mm GTP and subsequent incubation at 27°C for 1 h. The MT solution (approximately 650 μL) was overlaid on top of an equal volume of cushion buffer (1× BRB80, 0.1 mm GTP, and 60% glycerol) and centrifuged at 100,000g for 30 min at 27°C. After removing the supernatant and the cushion buffer, the MT pellet was suspended at 27°C in 300 μL of labeling solution (50 mm PIPES, 1 mm EGTA, 1 mm MgCl2, 0.5 mm GTP, and 40% glycerol, pH 8). Then, 4.4 μL of 10 mg mL−1 5-(and-6)-carboxytetramethylrhodamine succinimidyl ester (Invitrogen; referred to as rhodamine in this study) in dimethyl sulfoxide (approximately 10-fold molar excess to tubulin dimer) was added to the MT suspension and mixed by vortexing. The labeling mixture was incubated at 27°C for 30 min in darkness, overlaid on top of a cushion buffer, and centrifuged at 100,000g for 30 min at 27°C. The labeled MT pellet was resuspended in 900 μL of ice-cold 1.1× BRB80 and placed on ice for more than 1 h in darkness to disassemble the MTs. After the remaining MTs were removed by centrifugation at 100,000g for 30 min at 4°C, the tubulin supernatant was concentrated to approximately 120 μL by ultrafiltration (Amicon, 10K cutoff). Glycerol was added to a final concentration of 10%, and the tubulin solution (containing approximately 350 μg of rhodamine-labeled tubulin) was divided into aliquots, snap frozen in liquid nitrogen, and stored at −80°C.
Porcine brain tubulin purified by the conventional assembly-disassembly cycle method (Shelanski et al., 1973) was labeled with rhodamine with the same ratio of tubulin to dye as described above. After labeling, another round of assembly-disassembly was performed to select polymerization-competent tubulin. HiLyte 488-labeled porcine brain tubulin was purchased from Cytoskeleton.
In Vitro MT Dynamics Assay with TIRF Microscopy
The in vitro MT dynamics assay was performed as described previously (Gell et al., 2010; Li et al., 2012), with some modifications. MT seeds were prepared by two cycles of the assembly-disassembly procedure (Gell et al., 2010), using 20 μm porcine brain tubulin mix consisting of 80% nonlabeled tubulin (prepared with the assembly-disassembly method; Shelanski et al., 1973), 10% HiLyte 488-labeled tubulin, and 10% biotinylated tubulin (Cytoskeleton) in BRB80 supplemented with 1 mm guanosine-5′-[(α,β)-methylene] triphosphate (Jena Bioscience), and stored at −80°C until use. A coverslip was cleaned, silanized, and assembled into a reaction chamber consisting of a slide glass and two strips of double-sided tape. The surface of the coverslip was coated with 8 μg mL−1 neutravidin (Thermo Fisher Scientific) and then blocked with 1% pluronic F127 (Invitrogen). Biotinylated MT seeds were removed from the freezer, immediately heated at 37°C, diluted in prewarmed MRB80 (80 mm PIPES, 1 mm EGTA, and 4 mm MgCl2, pH 6.8), and loaded into the chamber. Meanwhile, reaction solutions (7.5, 10, or 15 μm porcine or Arabidopsis tubulin containing 6.7% rhodamine-labeled porcine or Arabidopsis tubulin in MRB80 supplemented with 1 mm GTP, 0.1% methylcellulose [Sigma-Aldrich], 0.5 mg mL−1 k-casein [Sigma-Aldrich], 50 mm Glc [Sigma-Aldrich], 400 μg mL−1 Glc-oxidase [Sigma-Aldrich], 800 μg mL−1 catalase [Sigma-Aldrich], 4 mm DTT [Sigma-Aldrich], and 1.42% glycerol [carried from tubulin stock]) was prepared and preincubated on ice for 5 min and subsequently at room temperature for 3 min before loading into the chamber. Chamber openings were sealed with Vaseline. Time-lapse observations with a 200-ms exposure were carried out at 3-s intervals for 30 min (total, 601 frames) using a TIRF microscope (Olympus) equipped with an UApo N, 100×, numerical aperture 1.49 objective lens, a DV2 beam splitter (Photometrics) with a 488/561-nm laser, and an iXon3 897 EMCCD camera (Andor Technology). The sample temperature was maintained at 27.5°C throughout the experiments using an objective lens heater and a thermoplate (Tokai Hit). Image processing and kymograph analyses were performed with ImageJ 1.46r (National Institutes of Health). Dynamic instability parameters were measured as described (Grego et al., 2001). To evaluate the bundling of Arabidopsis MTs, the mean fluorescence intensity of the rhodamine channel was quantified for each MT bundle and divided by the average intensity of multiple nonbundled MTs extending from the MT seeds after background subtraction. The fluorescence intensities of the original images presented in Figure 2 were differently processed in Adobe Photoshop CS4 and, thus, are not comparable in brightness.
Electrophoresis and Immunoblot Analyses
Conventional 10% Laemmli gels were used for normal SDS-PAGE. Phos-tag SDS-PAGE was performed as described (Fujita et al., 2013), except that the acrylamide concentration was 10% in this study. Gels were stained with colloidal Coomassie Blue solution [0.16% {w/v} Coomassie Brilliant Blue G250, 1.6% {v/v} H3PO4, 8% {w/v} (NH4)2SO4, and 20% {v/v} methanol] and scanned with a film scanner (GT-X900; Epson).
Two-dimensional gel electrophoresis was performed as described (Fukao et al., 2009) with minor modifications. Proteins were separated by IEF using immobilized pH gradient gels (Immobiline Drystrip, pH 4–7, 18 cm; GE Healthcare) in the first dimension and by 10% SDS-PAGE in the second dimension. DTT was not added to the filter paper at the positive side of the IEF gel, and iodoacetamide treatment was omitted after IEF.
For immunoblot analyses, separated proteins were blotted onto polyvinylidene difluoride membranes (Immobilon-P; Merck Millipore) and detected with antibodies using standard protocols. The following commercially available primary antibodies were used: rat monoclonal anti-α-tubulin antibody YOL1/34 (Merck Millipore; CBL270; 1:10,000 dilution), mouse monocolonal anti-α-tubulin antibody DM1A (Abcam; ab7291; 1:4,000 dilution [used only for the results presented in Fig. 6D]), mouse monoclonal anti-β-tubulin antibody KMX-1 (Merck Millipore; MAB3408; 1:8,000 dilution), mouse monoclonal anti-acetylated Lys-40 of α-tubulin antibody 6-11B-1 (Sigma-Aldrich; T7451; 1:10,000 dilution), mouse monoclonal anti-polyglutamylated αβ-tubulin antibody GT335 (Adipogen; AG-20B-0020; 1:8,000 dilution), mouse monoclonal anti-α-tubulin Tyr antibody TUB-1A2 (Sigma-Aldrich; T9028; 1:10,000 dilution), rabbit polyclonal anti-detyrosinated tubulin antibody (Merck Millipore; AB3201; 1:1,200 dilution), and rabbit polyclonal anti-His antibody (MBL; PM032; 1:1,000 dilution). To generate rabbit polyclonal antibody TUApT349, the C terminus of the 14-amino acid polypeptide (N-FVDWCPTGFKCGIN-C) containing a phosphorylated Thr (underlined) was conjugated to keyhole limpet hemocyanin by 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide and used to immunize rabbits. The TUApT349 antibody was purified by binding to and eluting from the phosphorylated peptide-conjugated column, followed by adsorbing to the nonphosphorylated peptide-conjugated column, and was used at 1:10,000 dilution. Horseradish peroxidase-conjugated secondary antibodies were used at 1:10,000 dilution for primary antibodies raised in rabbit (GE Healthcare; NA934), mouse (GE Healthcare; NA931), or rat (Santa Cruz Biotechnology; sc-2006). Signal was visualized with Immobilon Western Chemiluminescent HRP Substrate (Merck Millipore; WBKLS0100) and detected with LAS4000 (Fujifilm). Images were processed with Adobe Photoshop CS4. Densitometric analyses for Coomassie Blue-stained gels and immunoblots were performed using ImageJ 1.46r.
Mass Spectrometry
Full-length polypeptides of purified Arabidopsis tubulin (200–250 pmol per sample) were analyzed with a quadrupole time-of-flight mass spectrometer (QSTAR; AB/MDS SCIEX) as described (Minoura et al., 2013). Raw mass spectra were deconvolved.
Sequence data from this article can be found in the GenBank/EMBL data libraries under the accession numbers At2g35630 (MOR1) and At4g14960 (TUA6).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. A TOGMOR1 column does not bind tubulin.
Supplemental Figure S2. Purification of tubulin from various plant cell cultures.
Supplemental Figure S3. Sequence alignments of α-tubulin and β-tubulin proteins from pig and Arabidopsis.
Supplemental Figure S4. Comparison of the TOG1 and TOG2 domains of Stu2 and Arabidopsis MOR1.
Supplemental Movie S1. Time-lapse movie of MT dynamics for porcine tubulin at 7.5 μm, corresponding to Figure 2A, top.
Supplemental Movie S2. Time-lapse movie of MT dynamics for porcine tubulin at 15 μm, corresponding to Figure 2A, bottom.
Supplemental Movie S3. Time-lapse movie of MT dynamics for Arabidopsis tubulin at 7.5 μm, corresponding to Figure 2B, top.
Supplemental Movie S4. Time-lapse movie of MT dynamics for Arabidopsis tubulin at 15 μm, corresponding to Figure 2B, bottom.
Supplementary Material
Acknowledgments
We thank Per O. Widlund (Max Planck Institute for Molecular Cell Biology and Genetics) for pGEX-6P-1-Stu2(1-590), Tsuyoshi Nakagawa (Shimane University) for the pGWB vectors, Takanori Maeno and Naoyuki Inagaki (Nara Institute of Science and Technology) for the 6-11B-1 and TUB-1A2 antibodies, Takashi Moriwaki and Gohta Goshima (Nagoya University) for technical advice on the TIRF assay, Arata Yoneda (Nara Institute of Science and Technology) for the T87 cell line and advice on genetic transformation, Yoshihisa Oda (National Institute of Genetics) for the Alex cell line, Takahiro Hamada (University of Tokyo) for MM2d tubulin purified by the conventional method and advice on miniprotoplast preparation, Misato Ohtani (Nara Institute of Science and Technology) for advice on two-dimensional gel electrophoresis, and the Support Unit for Biomaterial Analysis at the RIKEN Brain Science Institute Research Resources Center for mass analysis.
Glossary
- MT
microtubule
- BY-2
Bright Yellow-2
- TIRF
total internal reflection fluorescence
- MAP
microtubule-associated protein
- IEF
isoelectric focusing
- PBS
phosphate-buffered saline
- DTT
dithiothreitol
Footnotes
This work was supported by a Grant-in-Aid for Young Scientists (B) from the Japan Society for the Promotion of Science (grant no. 26840095 to T.Ho.) and a Grant-in-Aid for Scientific Research on Innovative Areas from the Ministry of Education, Culture, Sports, Science, and Technology (grant no. 24114004 to T.Ha.).
References
- Abe T, Hashimoto T (2005) Altered microtubule dynamics by expression of modified alpha-tubulin protein causes right-handed helical growth in transgenic Arabidopsis plants. Plant J 43: 191–204 [DOI] [PubMed] [Google Scholar]
- Akella JS, Wloga D, Kim J, Starostina NG, Lyons-Abbott S, Morrissette NS, Dougan ST, Kipreos ET, Gaertig J (2010) MEC-17 is an α-tubulin acetyltransferase. Nature 467: 218–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Bassam J, Chang F (2011) Regulation of microtubule dynamics by TOG-domain proteins XMAP215/Dis1 and CLASP. Trends in Cell Biology 21: 604–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso MC, Drummond DR, Kain S, Hoeng J, Amos L, Cross RA (2007) An ATP gate controls tubulin binding by the tethered head of kinesin-1. Science 316: 120–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aström H. (1992) Acetylated α-tubulin in the pollen tube microtubules. Cell Biol Int Rep 16: 871–881 [PubMed] [Google Scholar]
- Ayaz P, Munyoki S, Geyer EA, Piedra F-A, Vu ES, Bromberg R, Otwinowski Z, Grishin NV, Brautigam CA, Rice LM (2014) A tethered delivery mechanism explains the catalytic action of a microtubule polymerase. Elife 3: e03069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayaz P, Ye X, Huddleston P, Brautigam CA, Rice LM (2012) A TOG:αβ-tubulin complex structure reveals conformation-based mechanisms for a microtubule polymerase. Science 337: 857–860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ban Y, Kobayashi Y, Hara T, Hamada T, Hashimoto T, Takeda S, Hattori T (2013) α-Tubulin is rapidly phosphorylated in response to hyperosmotic stress in rice and Arabidopsis. Plant Cell Physiol 54: 848–858 [DOI] [PubMed] [Google Scholar]
- Barisic M, Silva e Sousa R, Tripathy SK, Magiera MM, Zaytsev AV, Pereira AL, Janke C, Grishchuk EL, Maiato H (2015) Microtubule detyrosination guides chromosomes during mitosis. Science 348: 799–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bokros CL, Hugdahl JD, Hanesworth VR, Murthy JV, Morejohn LC (1993) Characterization of the reversible taxol-induced polymerization of plant tubulin into microtubules. Biochemistry 32: 3437–3447 [DOI] [PubMed] [Google Scholar]
- Borisy GG, Marcum JM, Olmsted JB, Murphy DB, Johnson KA (1975) Purification of tubulin and associated high molecular weight proteins from porcine brain and characterization of microtubule assembly in vitro. Ann N Y Acad Sci 253: 107–132 [DOI] [PubMed] [Google Scholar]
- Desai A, Mitchison TJ (1997) Microtubule polymerization dynamics. Annual Rev Cell Dev Biol 13: 83–117 [DOI] [PubMed] [Google Scholar]
- Detrich HW III, Parker SK, Williams RC Jr, Nogales E, Downing KH (2000) Cold adaptation of microtubule assembly and dynamics: structural interpretation of primary sequence changes present in the alpha- and beta-tubulins of Antarctic fishes. J Biol Chem 275: 37038–37047 [DOI] [PubMed] [Google Scholar]
- Duckett CM, Lloyd CW (1994) Gibberellic acid-induced microtubule reorientation in dwarf peas is accompanied by rapid modification of an α-tubulin isotype. Plant J 5: 363–372 [Google Scholar]
- Ersfeld K, Wehland J, Plessmann U, Dodemont H, Gerke V, Weber K (1993) Characterization of the tubulin-tyrosine ligase. The Journal of Cell Biology 120: 725–732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita S, Pytela J, Hotta T, Kato T, Hamada T, Akamatsu R, Ishida Y, Kutsuna N, Hasezawa S, Nomura Y, et al. (2013) An atypical tubulin kinase mediates stress-induced microtubule depolymerization in Arabidopsis. Curr Biol 23: 1969–1978 [DOI] [PubMed] [Google Scholar]
- Fukao Y, Ferjani A, Fujiwara M, Nishimori Y, Ohtsu I (2009) Identification of zinc-responsive proteins in the roots of Arabidopsis thaliana using a highly improved method of two-dimensional electrophoresis. Plant Cell Physiol 50: 2234–2239 [DOI] [PubMed] [Google Scholar]
- Gell C, Bormuth V, Brouhard GJ, Cohen DN, Diez S, Friel CT, Helenius J, Nitzsche B, Petzold H, Ribbe J, et al. (2010) Microtubule dynamics reconstituted in vitro and imaged by single-molecule fluorescence microscopy. Methods Cell Biol 95: 221–245 [DOI] [PubMed] [Google Scholar]
- Giannoutsou E, Galatis B, Zachariadis M, Apostolakos P (2012) Formation of an endoplasmic reticulum ring associated with acetylated microtubules in the angiosperm preprophase band. Cytoskeleton (Hoboken) 69: 252–265 [DOI] [PubMed] [Google Scholar]
- Grego S, Cantillana V, Salmon ED (2001) Microtubule treadmilling in vitro investigated by fluorescence speckle and confocal microscopy. Biophys J 81: 66–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamada T, Nagasaki-Takeuchi N, Kato T, Fujiwara M, Sonobe S, Fukao Y, Hashimoto T (2013) Purification and characterization of novel microtubule-associated proteins from Arabidopsis cell suspension cultures. Plant Physiol 163: 1804–1816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto T. (2013) Dissecting the cellular functions of plant microtubules using mutant tubulins. Cytoskeleton (Hoboken) 70: 191–200 [DOI] [PubMed] [Google Scholar]
- Hashimoto T. (2015) Microtubules in plants. The Arabidopsis Book 13: e0179 doi: 10.1199/tab.0179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang RF, Lloyd CW (1999) Gibberellic acid stabilises microtubules in maize suspension cells to cold and stimulates acetylation of α-tubulin. FEBS Lett 443: 317–320 [DOI] [PubMed] [Google Scholar]
- Ishida T, Kaneko Y, Iwano M, Hashimoto T (2007) Helical microtubule arrays in a collection of twisting tubulin mutants of Arabidopsis thaliana. Proc Natl Acad Sci USA 104: 8544–8549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janke C. (2014) The tubulin code: molecular components, readout mechanisms, and functions. J Cell Biol 206: 461–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janke C, Bulinski JC (2011) Post-translational regulation of the microtubule cytoskeleton: mechanisms and functions. Nat Rev Mol Cell Biol 12: 773–786 [DOI] [PubMed] [Google Scholar]
- Janke C, Rogowski K, Wloga D, Regnard C, Kajava AV, Strub JM, Temurak N, van Dijk J, Boucher D, van Dorsselaer A, et al. (2005) Tubulin polyglutamylase enzymes are members of the TTL domain protein family. Science 308: 1758–1762 [DOI] [PubMed] [Google Scholar]
- Jiang CJ, Sonobe S, Shibaoka H (1992) Assembly of microtubules in a cytoplasmic extract of tobacco BY-2 miniprotoplasts in the absence of microtubule-stabilizing agents. Plant Cell Physiol 33: 497–501 [Google Scholar]
- Johnson V, Ayaz P, Huddleston P, Rice LM (2011) Design, overexpression, and purification of polymerization-blocked yeast αβ-tubulin mutants. Biochemistry 50: 8636–8644 [DOI] [PubMed] [Google Scholar]
- Kinoshita K, Arnal I, Desai A, Drechsel DN, Hyman AA (2001) Reconstitution of physiological microtubule dynamics using purified components. Science 294: 1340–1343 [DOI] [PubMed] [Google Scholar]
- Kollman JM, Greenberg CH, Li S, Moritz M, Zelter A, Fong KK, Fernandez JJ, Sali A, Kilmartin J, Davis TN, et al. (2015) Ring closure activates yeast γTuRC for species-specific microtubule nucleation. Nat Struct Mol Biol 22: 132–137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumagai F, Yoneda A, Tomida T, Sano T, Nagata T, Hasezawa S (2001) Fate of nascent microtubules organized at the M/G1 interface, as visualized by synchronized tobacco BY-2 cells stably expressing GFP-tubulin: time-sequence observations of the reorganization of cortical microtubules in living plant cells. Plant Cell Physiol 42: 723–732 [DOI] [PubMed] [Google Scholar]
- Lechner B, Rashbrooke MC, Collings DA, Eng RC, Kawamura E, Whittington AT, Wasteneys GO (2012) The N-terminal TOG domain of Arabidopsis MOR1 modulates affinity for microtubule polymers. J Cell Sci 125: 4812–4821 [DOI] [PubMed] [Google Scholar]
- Li W, Moriwaki T, Tani T, Watanabe T, Kaibuchi K, Goshima G (2012) Reconstitution of dynamic microtubules with Drosophila XMAP215, EB1, and Sentin. J Cell Biol 199: 849–862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundin VF, Leroux MR, Stirling PC (2010) Quality control of cytoskeletal proteins and human disease. Trends Biochem Sci 35: 288–297 [DOI] [PubMed] [Google Scholar]
- Minoura I, Hachikubo Y, Yamakita Y, Takazaki H, Ayukawa R, Uchimura S, Muto E (2013) Overexpression, purification, and functional analysis of recombinant human tubulin dimer. FEBS Lett 587: 3450–3455 [DOI] [PubMed] [Google Scholar]
- Mizuno K. (1985) In vitro assembly of microtubules from tubulins of several higher plants. Cell Biol Int Rep 9: 13–21 [DOI] [PubMed] [Google Scholar]
- Moore RC, Zhang M, Cassimeris L, Cyr RJ (1997) In vitro assembled plant microtubules exhibit a high state of dynamic instability. Cell Motil Cytoskeleton 38: 278–286 [DOI] [PubMed] [Google Scholar]
- Morejohn LC, Fosket DE (1982) Higher plant tubulin identified by self-assembly into microtubules in vitro. Nature 297: 426–428 [DOI] [PubMed] [Google Scholar]
- Nakagawa T, Kurose T, Hino T, Tanaka K, Kawamukai M, Niwa Y, Toyooka K, Matsuoka K, Jinbo T, Kimura T (2007) Development of series of Gateway Binary Vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J Biosci Bioeng 104: 34–41 [DOI] [PubMed] [Google Scholar]
- Nakagawa U, Kamemura K, Imamura A (2013) Regulated changes in the acetylation of α-tubulin on Lys40 during growth and organ development in fast plants, Brassica rapa L. Biosci Biotechnol Biochem 77: 2228–2233 [DOI] [PubMed] [Google Scholar]
- Nakamura M, Naoi K, Shoji T, Hashimoto T (2004) Low concentrations of propyzamide and oryzalin alter microtubule dynamics in Arabidopsis epidermal cells. Plant Cell Physiol 45: 1330–1334 [DOI] [PubMed] [Google Scholar]
- Nogales E, Whittaker M, Milligan RA, Downing KH (1999) High-resolution model of the microtubule. Cell 96: 79–88 [DOI] [PubMed] [Google Scholar]
- Oda Y, Fukuda H (2011) Dynamics of Arabidopsis SUN proteins during mitosis and their involvement in nuclear shaping. Plant J 66: 629–641 [DOI] [PubMed] [Google Scholar]
- Parrotta L, Cresti M, Cai G (2014) Accumulation and post-translational modifications of plant tubulins. Plant Biol (Stuttg) 16: 521–527 [DOI] [PubMed] [Google Scholar]
- Sackett DL, Werbovetz KA, Morrissette NS (2010) Isolating tubulin from nonneural sources. Methods Cell Biol 95: 17–32 [DOI] [PubMed] [Google Scholar]
- Schatz PJ, Georges GE, Solomon F, Botstein D (1987) Insertions of up to 17 amino acids into a region of α-tubulin do not disrupt function in vivo. Mol Cell Biol 7: 3799–3805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah C, Xu CZQ, Vickers J, Williams R (2001) Properties of microtubules assembled from mammalian tubulin synthesized in Escherichia coli. Biochemistry 40: 4844–4852 [DOI] [PubMed] [Google Scholar]
- Shaw SL, Kamyar R, Ehrhardt DW (2003) Sustained microtubule treadmilling in Arabidopsis cortical arrays. Science 300: 1715–1718 [DOI] [PubMed] [Google Scholar]
- Shelanski ML, Gaskin F, Cantor CR (1973) Microtubule assembly in the absence of added nucleotides. Proc Natl Acad Sci USA 70: 765–768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirajuddin M, Rice LM, Vale RD (2014) Regulation of microtubule motors by tubulin isotypes and post-translational modifications. Nat Cell Biol 16: 335–344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smertenko A, Blume Y, Viklický V, Opatrný Z, Dráber P (1997) Post-translational modifications and multiple tubulin isoforms in Nicotiana tabacum L. cells. Planta 201: 349–358 [DOI] [PubMed] [Google Scholar]
- Song Y, Brady ST (2015) Post-translational modifications of tubulin: pathways to functional diversity of microtubules. Trends Cell Biol 25: 125–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szyk A, Deaconescu AM, Spector J, Goodman B, Valenstein ML, Ziolkowska NE, Kormendi V, Grigorieff N, Roll-Mecak A (2014) Molecular basis for age-dependent microtubule acetylation by tubulin acetyltransferase. Cell 157: 1405–1415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran HT, Nimick M, Uhrig RG, Templeton G, Morrice N, Gourlay R, DeLong A, Moorhead GBG (2012) Arabidopsis thaliana histone deacetylase 14 (HDA14) is an α-tubulin deacetylase that associates with PP2A and enriches in the microtubule fraction with the putative histone acetyltransferase ELP3. Plant J 71: 263–272 [DOI] [PubMed] [Google Scholar]
- Uchimura S, Oguchi Y, Katsuki M, Usui T, Osada H, Nikawa J, Ishiwata S, Muto E (2006) Identification of a strong binding site for kinesin on the microtubule using mutant analysis of tubulin. EMBO J 25: 5932–5941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Vignani R, Scali M, Sensi E, Cresti M (2004) Post-translational modifications of alpha-tubulin in Zea mays L are highly tissue specific. Planta 218: 460–465 [DOI] [PubMed] [Google Scholar]
- Whittington AT, Vugrek O, Wei KJ, Hasenbein NG, Sugimoto K, Rashbrooke MC, Wasteneys GO (2001) MOR1 is essential for organizing cortical microtubules in plants. Nature 411: 610–613 [DOI] [PubMed] [Google Scholar]
- Widlund PO, Podolski M, Reber S, Alper J, Storch M, Hyman AA, Howard J, Drechsel DN (2012) One-step purification of assembly-competent tubulin from diverse eukaryotic sources. Mol Biol Cell 23: 4393–4401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiesler B, Wang QY, Nick P (2002) The stability of cortical microtubules depends on their orientation. Plant J 32: 1023–1032 [DOI] [PubMed] [Google Scholar]
- Xiong X, Xu D, Yang Z, Huang H, Cui X (2013) A single amino-acid substitution at lysine 40 of an Arabidopsis thaliana α-tubulin causes extensive cell proliferation and expansion defects. J Integr Plant Biol 55: 209–220 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.