MiR163 targets PXMT1 transcripts at the early stage of light responses to promote seed germination and root development.
Abstract
Expression of many plant microRNAs is responsive to hormones and environmental stimuli, but none has yet been associated with light. Arabidopsis (Arabidopsis thaliana) miR163 is 24 nucleotides in length and targets mRNAs encoding several S-adenosyl-Met-dependent carboxyl methyltransferase family members. Here, we found that miR163 is highly induced by light during seedling de-etiolation as well as seed germination. Under the same condition, its target PXMT1, encoding a methyltransferase that methylates 1,7-paraxanthine, is down-regulated. Light repression of PXMT1 is abolished in a mir163 null mutant, but the repression can be restored to wild-type levels in complementation lines expressing pri-miR163 gene in the mir163 mutant background. During seed germination, miR163 and its target PXMT1 are predominantly expressed in the radicle, and the expression patterns of the two genes are inversely correlated. Moreover, compared with the wild type, mir163 mutant or PXMT1 overexpression line shows delayed seed germination under continuous light, and seedlings develop shorter primary roots with an increased number of lateral roots under long-day condition. Together, our results indicate that miR163 targets PXMT1 mRNA to promote seed germination and modulate root architecture during early development of Arabidopsis seedlings.
MicroRNAs (miRNAs) are a class of small noncoding RNAs, ranging in size from 21 to 24 nucleotides (nt) and found in plants, animals, and other eukaryotes, that function in transcriptional and posttranscriptional regulation of target genes via mRNA degradation or translational repression (Huntzinger and Izaurralde, 2011; Pasquinelli, 2012). MiRNAs are processed from a primary miRNA (pri-miRNA) that forms a hairpin-like imperfect stem-loop structure. In animal cells, the pri-miRNA transcript is first processed into a stem-loop precursor (premiRNA) by the nuclear RNase III-like Drosha (Lee et al., 2004). Subsequently, premiRNAs are exported to the cytoplasm and further cleaved by the RNase III domain-containing Dicer to yield 21- to 24-nt mature miRNAs (Lee et al., 2003). By contrast, plant miRNA biogenesis differs from animal systems owing to the absence of a Drosha homolog. Instead, the plant Dicer homolog DICER-LIKE1 (DCL1) has both Drosha as well as Dicer functions within the nucleus (Kurihara and Watanabe, 2004). In many cases, these processing events are aided by other RNA-binding proteins, HYPONASTIC LEAVES1 and SERRATE. After DCL1-mediated cleavage, the miRNA/miRNA* duplexes are exported to the cytoplasm by HASTY, a plant homolog of Exportin-5 (Park et al., 2005). Mature miRNAs are then incorporated into an effector complex, RNA-induced silencing complex, to guide target mRNA cleavage (Bartel, 2004).
Plants have evolved functions to sense and respond to environmental signals. Being one of the most important environmental factors, light orchestrates dramatic changes in plant development and growth. Genome-wide studies have shown that the expression of more than 20% of genes in the Arabidopsis (Arabidopsis thaliana) genome is altered by light, and these expression changes likely precede and underpin subsequent light-induced morphological changes (Jiao et al., 2005, 2007; Wang et al., 2014). The reprogramming of gene expression during dark/light transition suggests regulation mainly at the transcriptional level.
In early stages of light response, radicle and cotyledons emerge from the seed coat, leading eventually to the establishment of a seedling. Many miRNAs are expressed in germinating seeds and seedlings of Arabidopsis, but expression of some of them is related to plant hormones such as abscisic acid and auxin (Reyes and Chua, 2007; Martin et al., 2010). Plant miRNAs also display tissue- or developmental stage-specific expression patterns (Breakfield et al., 2012). In addition, the abundance of a particular miRNA varies greatly among different cell types and/or under different conditions, suggesting functional diversification in plants. In Arabidopsis, a large set of plant miRNAs is known to control developmental timing by regulating the abundance of their target mRNAs encoding transcription factor, e.g. the transition from vegetative to reproductive phase (miR156), flowering time and floral organ development (miR172), and lateral root development (miR164; Wu and Poethig, 2006; Jung et al., 2007; Guo et al., 2005; Aukerman and Sakai, 2003). In addition, the expression patterns of many plant miRNAs are responsive to plant hormones and biotic/abiotic stresses. Notable examples are gibberellin-inducible miR159 (Achard et al., 2004), auxin-inducible miR164 (Guo et al., 2005), biotic stress- or abiotic stress-inducible miR393 (Navarro et al., 2006; Sunkar and Zhu, 2004), sulfate starvation-inducible miR395 (Jones-Rhoades and Bartel, 2004), and phosphate starvation-inducible miR399 (Fujii et al., 2005). By contrast, miR167 is down-regulated at high osmotic stress, allowing accumulation of its target mRNA encoding IAA-Ala Resistant3, which releases bioactive auxin to modulate root development (Kinoshita et al., 2012). However, no miRNA has yet been described that responds to light signals.
Whereas most miRNAs are evolutionarily conserved within the plant kingdom, Arabidopsis encodes several nonconserved miRNAs, including miR161 and miR163 (Xie et al., 2005; Jones-Rhoades et al., 2006). MiR163 is atypical in that it is 24 nt in length. However, like other typical 21-nt miRNAs, miR163 can be maturated also by DCL1 (Kurihara and Watanabe, 2004). Target genes of miR163 encode members of the family of plant methyltransferases, paraxanthine methyltransferase 1 (PXMT1) and farnesoic acid methyltransferase (FAMT), which are located on chromosomes 1 and 3 as a cluster (Ng et al., 2011). The cleavage sites of several target transcripts of miR163 have been validated by 5′ RACE (Allen et al., 2004; Ng et al., 2011). MiR163 is highly expressed in all organs of Arabidopsis (Guo et al., 2005; Preuss et al., 2008; Ng et al., 2011) but undetectable in Arabidopsis arenosa (Ng et al., 2011), suggesting it is a recently evolved gene.
Here, we showed that miR163 is light inducible and predominantly expressed in roots during seed germination as well as seedling de-etiolation. PXMT1, a target of miR163, was found not only to be expressed in roots at the same developmental stages but also to be repressed by light. The light inducibility of pri-miR163 was not affected by temperature, but miR163 maturation was attenuated at low temperatures, resulting in stable PXMT1 transcripts. Analysis of mir163 mutant and transgenic plants revealed that PXMT1 mRNA levels were inversely affected by changes in miR163 levels. Mir163 mutant and PXMT1 overexpression line showed delayed radicle emergence during seed germination and shorter primary roots with an increased number of lateral roots in young seedlings. Finally, we found that the function of miR163 at the early stage of light response is to promote seed germination and primary root growth through targeting of PXMT1 transcripts.
RESULTS
Expression of MiR163 and Its Target mRNA PXMT1 during Seedling De-Etiolation
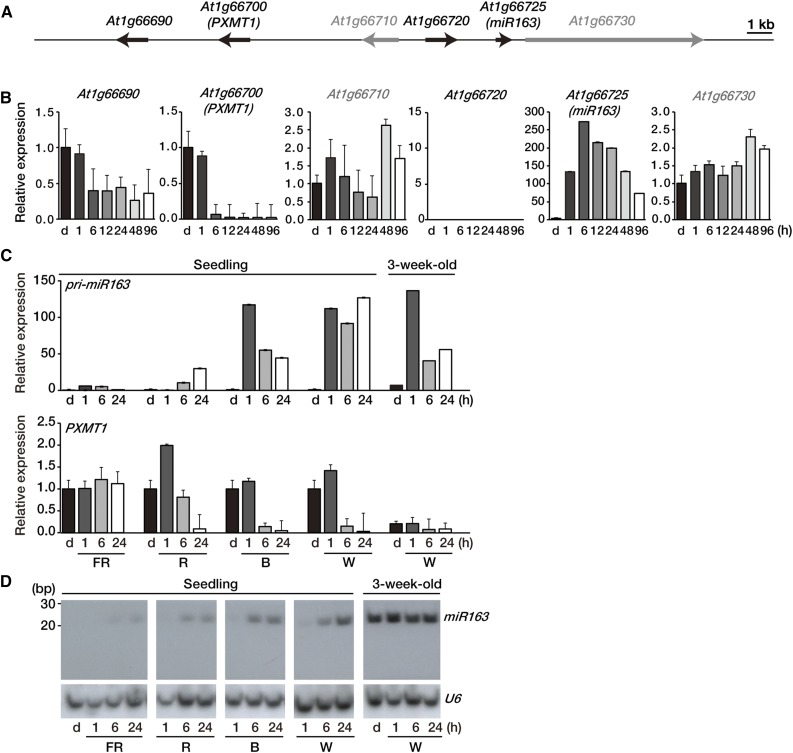
More than 427 miRNAs have so far been reported in Arabidopsis (www.mirbase.org), but none has yet been implicated in light signaling and/or light-dependent growth or development. Using our custom microarrays (Liu et al., 2012) to profile transcriptome changes in etiolated seedlings treated by red (R) light (Wang et al., 2014), we identified several R light-inducible noncoding RNAs, including pri-miRNAs. Among these R light-inducible noncoding RNAs, pri-miR163 displayed the largest induction ratio in expression. Compared with the other canonical miRNAs, miR163 has several characteristics. (1) It is 24 nt in length, which is 3 nt longer than the other canonical miRNAs; and (2) its target transcripts encode S-adenosyl-Met-dependent carboxyl methyltransferase (SAMT)-like proteins and FAMT. Genes for these enzymes are clustered on chromosomes 1 and 3, respectively. (3) Moreover, the pri-miR163 gene is located just adjacent to a cluster of three SAMT-like genes on chromosome 1 (Fig. 1A; Allen et al., 2004; Ng et al., 2011). We examined sequence complementarity between Arabidopsis miR163 and its target mRNAs. We found that miR163 has perfect sequence complementarity to the target sites on At1g66690 and At1g66700 (PXMT1), but it has 1- or 2-bp mismatches to the predicted target sequence of At1g66720, At3g44860 (FAMT), and At3g44870 (Allen et al., 2004; Ng et al., 2011; Supplemental Fig. S1).
Figure 1.
Expression levels of miR163 and PXMT1 during seedling de-etiolation. A, Schematic diagram showing the location of miR163 (At1g66725) and its putative targets on chromosome 1. Putative targets (At1g66690, At1g66700, and At1g66720) of miR163 and neighboring (At1g66710 and At1g66730) genes are shown as black and gray arrows, respectively. The arrows in the diagram indicate transcriptional directions. Bar = 1 kb. B, Expression of pri-miR163, its putative targets, and neighboring genes during seedling de-etiolation. Four-day-old etiolated Arabidopsis seedlings (d) were treated with W light as indicated (1, 6, 12, 24, 48, and 96 h). Expression levels were determined by qRT-PCR with specific gene primer sets and normalized with Actin2 expression level. Values were normalized to that of the etiolated sample (d) of each graph. Error bars indicate sd (n = 3). C, Expression levels of pri-miR163 and PXMT1 during seedling de-etiolation under different light qualities. Four-day-old etiolated Arabidopsis seedlings (d) were treated with FR (5 µmol m−2 s−1), R (20 µmol m−2 s−1), B (10 µmol m−2 s−1), and W (100 µmol m−2 s−1) light as indicated (1, 6, and 24 h). Expression levels were determined by qRT-PCR as described in B. Values were normalized to that of the d sample of each panel. Error bars indicate sd (n = 3). D, Small RNA gel-blot analysis showing miR163 expression during seedling de-etiolation. Total RNA was extracted from the samples described in C. U6 snRNA levels were used as a loading control.
We first investigated the expression pattern of pri-miR163 (At1g66725) and its neighboring genes, including genes encoding putative miR163 targets during seedling de-etiolation. Time-course experiments showed that pri-miR163 transcript was barely detectable in etiolated seedlings, but its expression was rapidly induced by light (miR163; Fig. 1B). After 6 h of light treatment, pri-miR163 expression levels were 250-fold higher than dark levels (miR163; Fig. 1B). Among genes encoding putative target mRNAs surrounding the pri-miR163 locus, only one locus (At1g66700, encoding PXMT1, a methyltransferase that methylates 1,7-paraxanthine) showed an inverse expression pattern compared with that of pri-miR163. Figure 1B shows that PXMT1 transcript accumulated in etiolated seedlings but became down-regulated after 6 h of light (Fig. 1B). The expression pattern of other candidate target genes was not correlated to that of pri-miR163; moreover, some of them were poorly expressed or even undetectable under the same condition (Fig. 1B; Supplemental Fig. S2). Therefore, we considered it is reasonable to select PXMT1 (At1g66700) as a target of miR163 during seedling de-etiolation.
Next, we examined expression of pri-miR163 and PXMT1 upon exposure to different light qualities, far-red (FR), R, blue (B), and white (W) light. Figure 1C shows that pri-miR163 levels increased under all light conditions, especially under B and W after 1 h. By contrast, PXMT1 transcript levels decreased after 6 h under the same light conditions, except under FR where pri-miR163 was not highly induced (Fig. 1C). Mature miR163 was undetectable in etiolated seedlings, but it could be clearly detected after 6 h of exposure to FR, R, B, and W light, correlating with changes in pri-miR163 levels (Seedling; Fig. 1D). The accumulation of miR163 was accompanied by a decrease of PXMT1 transcript abundance under these conditions (Fig. 1, C and D).
It has been previously reported that miR163 is stable in mature Arabidopsis plants with a high expression level in all organs (Guo et al., 2005; Preuss et al., 2008). Although miR163 levels were stable during dark/light transition of 3-week-old mature Arabidopsis plants (3-week-old; Fig. 1D), it was not known whether pri-miR163 levels would change under the same conditions. Figure 1C shows that in 3-week-old Arabidopsis plants kept in the dark for 24 h, pri-miR163 levels were very low and comparable to those of etiolated seedlings but were induced upon light exposure. Our results showed that the expression of pri-miR163, but not of miR163, is light inducible even in mature plants. Furthermore, light does not affect processing and maturation of miR163. Consistent with its regulation by miR163, PXMT1 transcript levels were reduced in mature plants compared with etiolated seedlings (Fig. 1, C and D).
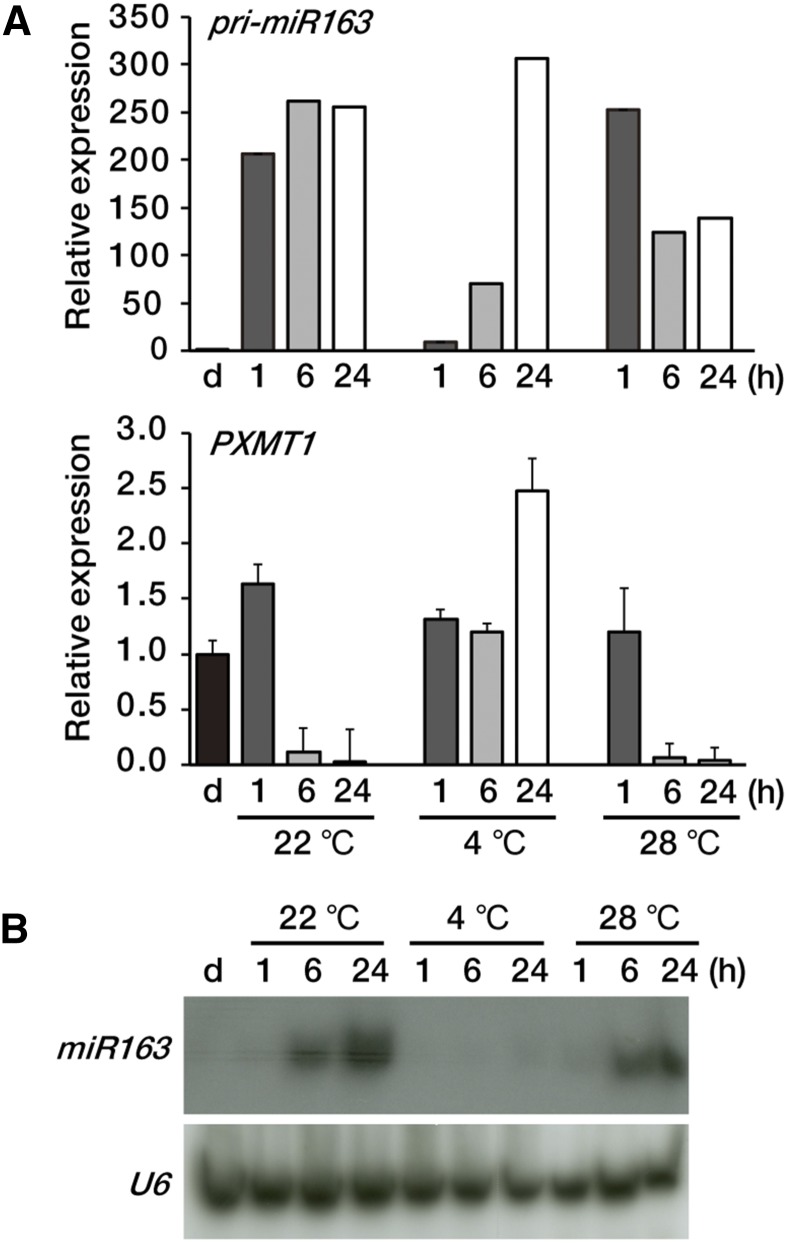
One of the most important environmental stimuli affecting plant growth is temperature (Franklin and Whitelam, 2004; Harmer, 2009). In the natural environment, temperature and light often act together to regulate plant gene expression. We investigated the effects of temperature on pri-miR163 and PXMT1 expression levels. Figure 2A shows that at low temperature (4°C), light induction of pri-miR163 was reduced compared with that at normal growth temperature (22°C) or higher temperature (28°C). Nevertheless, after 24 h of light at 4°C, pri-miR163 levels were high and comparable to those at normal growth temperature. On the other hand, the mature miR163 level remained low at 24 h (Fig. 2B). These results suggest that processing of pri-miR163 may be attenuated at low temperatures. Unlike the situation under 22°C or 28°C, PXMT1 transcripts were unchanged up to 6 h of light under 4°C but increased 2.5-fold after 24 h of light, consistent with the low miR163 levels. Our results showed that pri-miR163 is light inducible regardless of temperature, but miR163 maturation is temperature dependent.
Figure 2.
Expression levels of miR163 and PXMT1 at different temperatures. A, Expression levels of pri-miR163 and PXMT1 at 22°C, 4°C, and 28°C. Etiolated seedlings (d) were incubated at different temperatures (22°C, 4°C, and 28°C) under the same light intensity (100 µmol m−2 s−1) as indicated (1, 6, and 24 h). Values were normalized to that of the d sample of each panel. Error bars indicate sd (n = 3). B, Small RNA gel-blot analysis showing miR163 expression at 22°C, 4°C, and 28°C. Total RNA was extracted from the samples described in A. U6 snRNA levels were used as a loading control.
Spatiotemporal Expression of miR163 during Seedling De-Etiolation
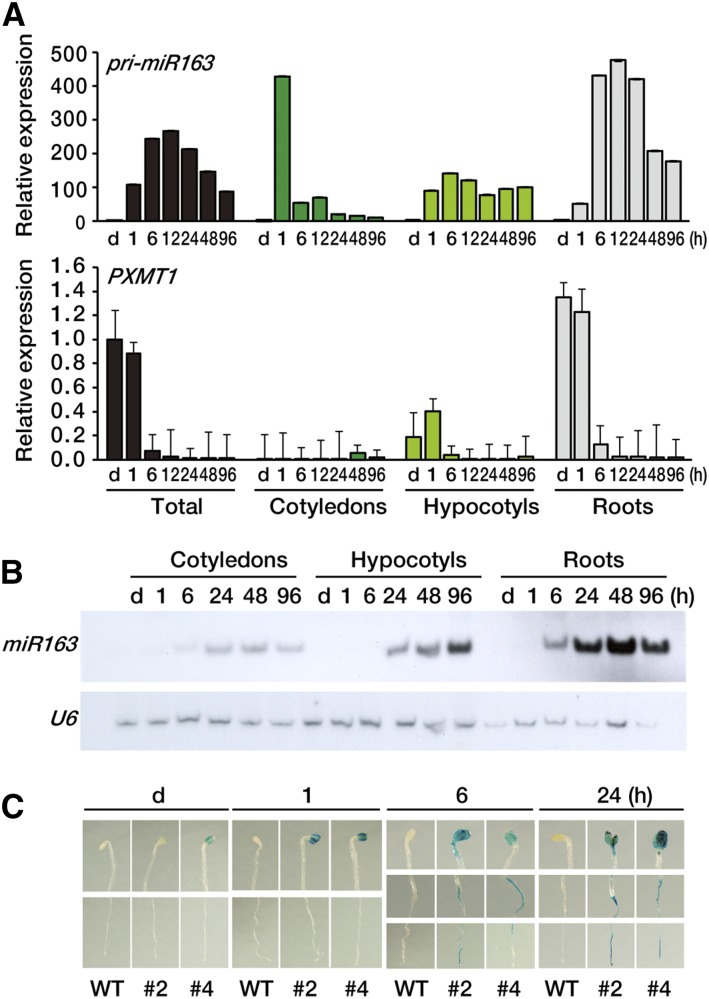
To investigate organ-specific expression pattern of pri-miR163, miR163, and PXMT1 in the early stage of seedling de-etiolation, we dissected the seedlings into three parts, cotyledons, hypocotyls, and roots. Figure 3A shows that pri-miR163 was mainly expressed in roots and highly induced up to more than 400-fold after 6 h of light exposure. Compared with roots, pri-miR163 showed high expression at 1 h of light exposure in cotyledons, but the expression level rapidly decreased thereafter; reduced light induction was detected in hypocotyls. PXMT1 was predominantly expressed in roots of etiolated seedlings rather than in cotyledons or hypocotyls, but its expression decreased after 6 h of light exposure, showing an inverse relationship to that of pri-miR163. These results suggest that miR163 targets PXMT1 in roots in a light-dependent manner. Small RNA blot analysis confirmed that mature miR163 levels were much higher in roots compared with other organs (Fig. 3B).
Figure 3.
Tissue-specific expression pattern of miR163 and PXMT1. A, Expression levels of pri-miR163 and PXMT1 in various tissues during seedling de-etiolation. Seedlings were dissected into three parts (cotyledons, hypocotyls, and roots), and samples were collected indicated. Expression levels were determined by qRT-PCR. The values were normalized to that of the d sample of each panel. Error bars indicate sd (n = 3). B, Small RNA gel-blot analysis showing miR163 expression in cotyledons, hypocotyls, and roots. Total RNA was extracted from the samples described in A. U6 snRNA levels were used as a loading control. C, Histochemical localization of GUS activity in transgenic Arabidopsis seedling carrying the pri-miR163 promoter::GUS-GFP transgene. Four-day-old etiolated seedlings (d) were treated with W light as indicated (1, 6, and 24 h). WT, Wild type.
For a more detailed investigation of the expression profile of pri-miR163, we generated transgenic plants expressing a fusion gene comprising the native pri-miR163 promoter, GUS, and GFP. Approximately 1-kb DNA fragment of the upstream region (−990) from the transcription start site of the pri-miR163 locus (At1g66725) was used to express a GUS-GFP fusion gene. We selected two independent transgenic lines (nos. 2 and 4) showing light inducibility of GUS-GFP fusion proteins by western-blot analysis (Supplemental Fig. S3). Using these selected lines, GUS expression during seedling de-etiolation was analyzed in a time-series experiment. Figure 3C shows that GUS was highly expressed in cotyledons after 1 h of light exposure, and the expression levels remained high until 24 h. However, in the case of roots, GUS activity was detected only after 6 h of light exposure, and the expression level increased at 24 h. This GUS expression pattern in roots was very similar to that of pri-miR163 transcripts. Similar GUS expression pattern was found in both transgenic lines (nos. 2 and 4), and no GUS expression was seen in the nontransgenic wild-type control.
MiR163 Targets PXMT1 in Seedling De-Etiolation
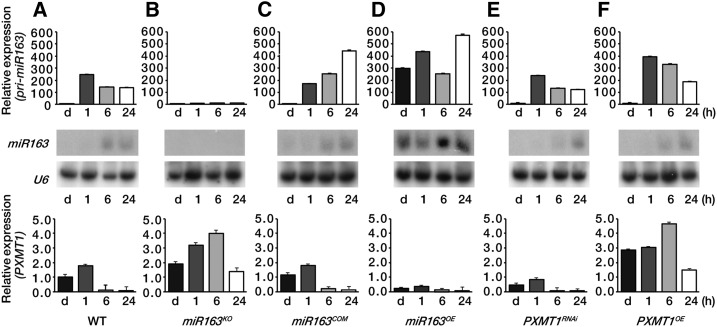
We found an inverse gene expression pattern between miR163 and PXMT1 during seedling de-etiolation, especially in roots (Figs. 1–3 and 4A). To further confirm this observation, we generated transgenic plants constitutively expressing pri-miR163 under the control of a cauliflower mosaic virus 35S promoter and also identified a T-DNA insertion mutant of miR163 (SALK_043556). A complementation line expressing pri-miR163 gene driven by the native pri-miR163 promoter in the mir163 mutant background (miR163COM) was generated as well. Figure 4B shows that in the mir163 mutant (miR163KO), pri-miR163 was not expressed upon light exposure nor was the mature miR163 detected, confirming this mutant is null for miR163 expression. Loss of miR163 in the mir163 mutant resulted in PXMT1 derepression (Fig. 4B). The PXMT1 expression shown in the mir163 mutant could be restored to wild-type levels in the complementation line (miR163COM; Fig. 4C). Moreover, overexpression of miR163 (miR163OE) highly repressed PXMT1 transcript expression, even in the dark, because of miR163 accumulation (Fig. 4D). Taken together, our results confirmed that miR163 targets PXMT1 during seedling de-etiolation. Reduced expression of PXMT1 by RNA interference (RNAi) or PXMT1 overexpression did not alter the light-inducible pri-miR163 or miR163 expression pattern in seedling de-etiolation (Fig. 4, E and F).
Figure 4.
Regulated expression of PXMT1 by miR163 during seedling de-etiolation. A to F, Expression levels of pri-miR163, miR163, and PXMT1 in the wild type (WT; A), mir163 mutant miR163KO (B), miR163 complementation line miR163COM (C), miR163 overexpression line miR163OE (D), PXMT1 RNAi line PXMT1RNAi (E), and PXMT1 overexpression line PXMT1OE (F). Four-day-old etiolated seedlings (d) were treated with W light as indicated (1, 6, and 24 h). All conditions for qRT-PCR and small RNA gel-blot analysis were identical to those described in Figure 1, A and C. Values in A to F were normalized to that of the d sample in the wild type (A). Error bars indicate sd (n = 3).
MiR163 Promotes Seed Germination as Well as Seedling Root Growth
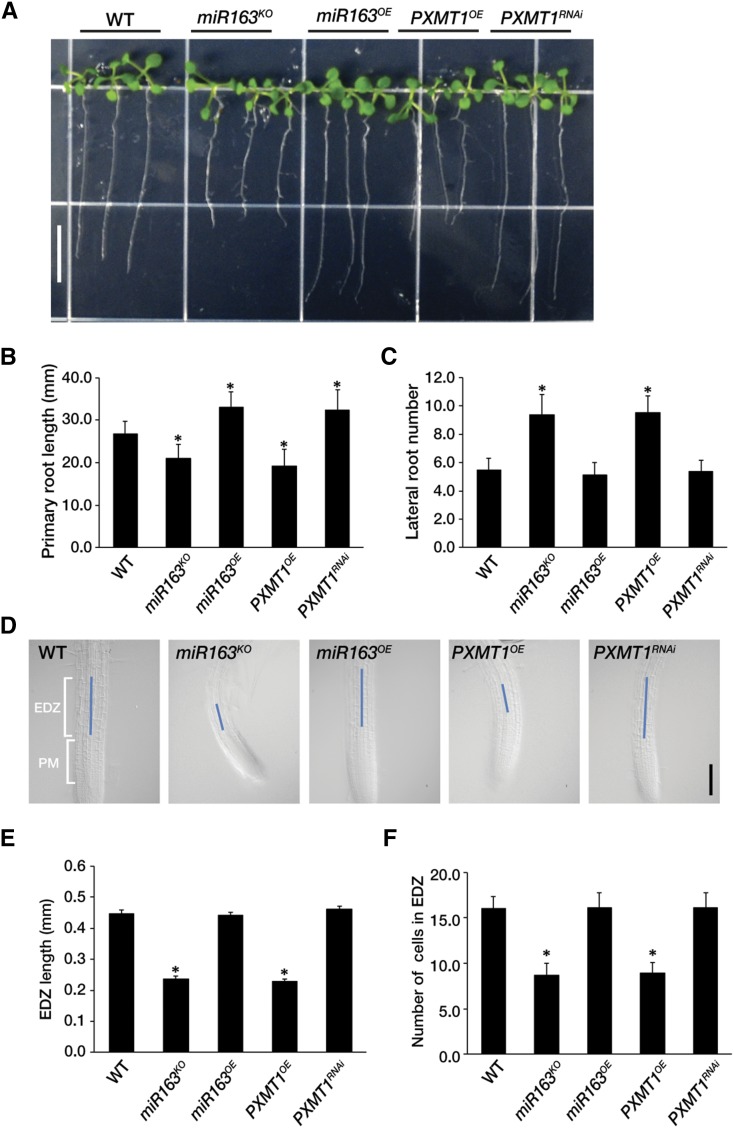
Vaistij et al. (2010) found no obvious phenotypic alterations in the mir163 mutant and its overexpression lines. As pri-miR163, miR163, and its target PXMT1 were predominantly expressed in roots (Fig. 3), we analyzed root phenotypes in all relevant genotypes. On vertically oriented plates under long-day conditions (16 h light/8 h dark), none of these lines displayed any phenotypic differences with respect to the length of primary roots nor the number of lateral roots compared with the wild type (Supplemental Fig. S4). However, under long-day conditions (16 h light/8 h dark), the primary roots of mir163 null mutant (miR163KO) and PXMT1 overexpression line (PXMT1OE) were clearly shorter than those of wild-type control seedlings (Fig. 5, A and B). Conversely, the primary roots of miR163 overexpression seedlings (miR163OE) were significantly longer than those of the wild type and comparable to those of PXMT1 RNAi (PXMT1RNAi) lines. Although the number of lateral roots of miR163OE and PXMT1RNAi lines was not changed, miR163KO and PXMT1OE lines clearly showed an increased number of lateral roots compared with the wild type (Fig. 5C). These results suggest that miR163 mediates seedling root growth through down-regulation of PXMT1.
Figure 5.
MiR163 is involved seedling root growth. A, Root phenotypes of the wild type (WT), mir163 mutant miR163KO, miR163 overexpression line miR163OE, PXMT1 overexpression line PXMT1OE, and PXMT1 RNAi line PXMT1RNAi. The seedlings were grown on Murashige and Skoog medium for 14 d under long-day conditions (16 h light/8 h dark). Scale bar = 1 cm. B, Primary root length of seedlings shown in A. C, Number of lateral roots of seedlings shown in A. D, Elongation/differentiation zone length of the wild type, mir163 mutant miR163KO, miR163 overexpression line miR163OE, PXMT1 overexpression line PXMT1OE, and PXMT1 RNAi line PXMT1RNAi. Blue line indicates the elongation/differentiation zone in root meristem. Scale bar = 0.2 mm. PM, Proximal meristem; EDZ, elongation/differentiation zone. E, Elongation/differentiation zone length shown in D. F, Number of cells in elongation/differentiation zone shown in D. B, C, E, and F, Error bars indicate the mean ± sd (n = 50) with biological replicates, and values (*) are statistically significant from the wild type based on Student’s t test (P < 0.001).
Root growth depends on cell division in the proximal meristem and cell elongation in the elongation/differentiation zone of root meristem (Perilli et al., 2012). Therefore, we investigated whether miR163 knockout or PXMT1 overexpression would affect the cell number and/or length of elongation/differentiation zones in Arabidopsis root meristem. Interestingly, we found that in miR163KO and PXMT1OE, there was no change in root cell size, but root cell number was reduced by about 2 times by shortening of the elongation/differentiation zone of root meristem compared with the wild type (Fig. 5, D–F). However, these findings were not clearly observed in the proximal meristem of root. These results suggest that the cell cycle time might be longer in miR163KO and PXMT1OE compared with the wild type because their root meristem produced fewer cells. In miR163OE and PXMT1RNAi, there were no significant changes in both cell number and length of root meristem in comparison to the wild type (Fig. 5, D–F).
There was the remote possibility that even if miR163 was less expressed in cotyledons and hypocotyls than in roots, it may have some function in these tissues. However, there were no significant differences with respect to early light seedling responses such as hypocotyl length reduction, hook opening, and cotyledon greening under several light conditions (Supplemental Fig. S5).
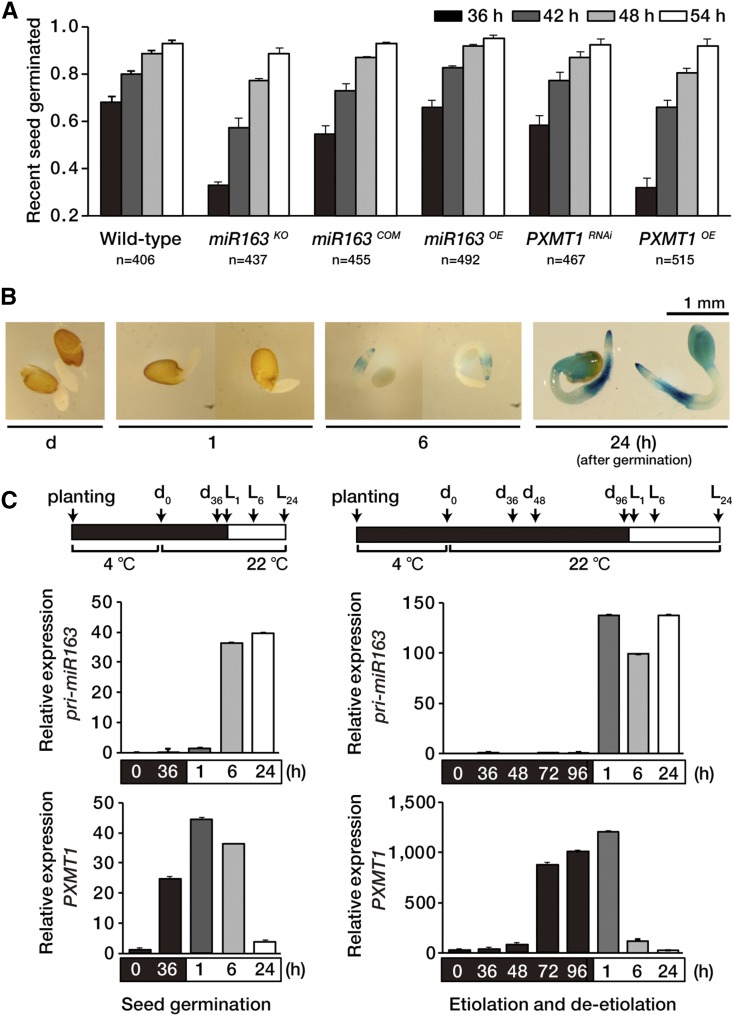
Seed germination is usually defined as the emergence of radicle (embryonic root) after imbibition, and this process is regulated by environmental and hormonal signals. Since a major environmental signal promoting seed germination is light, we hypothesized miR163 may mediate radicle emergence through targeting of PXMT1. For seed germination assays, we used more than 150 seeds per assay with three biological replicates and examined seed germination under continuous light at several early time points (36, 42, 48, and 54 h). Figure 6A shows that approximately 70% of wild-type seeds showed radicle emergence after 36 h, and this number increased to about 80% and 90% after 42 h and 48 h, respectively. By contrast, in the case of mir163 mutant seeds (miR163KO), less than 35% of the seeds showed radicle emergence after 36 h, and radicle emergence was highly delayed until 42 h. Similar results were obtained with PXMT1 overexpression lines (PXMT1OE), indicating that miR163 mediates radicle emergence through targeting of PXMT1 mRNA. The delayed germination of mir163 mutant seeds at early time point was restored to wild-type levels by miR163 expression, confirming miR163 function in radicle emergence (miR163COM). No significant differences were found in radical emergence between miR163 overexpression (miR163OE) seeds or PXMT1 RNAi (PXMT1RNAi) seeds and wild-type seeds because of low PXMT1 expression under this condition.
Figure 6.
MiR163 plays a role in seed germination. A, Seed germination of the wild type, mir163 mutant miR163KO, miR163 complementation line miR163COM, miR163 overexpression line miR163OE, PXMT1 RNAi line PXMT1RNAi, and PXMT1 overexpression line PXMT1OE. Seed germination was scored by radicle emergence at different time points (36, 42, 48, and 54 h) under continuous light (100 µmol m−2 s−1). Seed numbers (n) are represented below the x axis. Error bars indicate sd (n = 3). B, Histochemical localization of GUS activity in germinating transgenic Arabidopsis seeds carrying pri-miR163 promoter::GUS-GFP transgene. GUS staining was performed with de-etiolated seedlings indicated (1, 6, and 24 h). C, Expression levels of pri-miR163 and PXMT1 during seed germination or seedling de-etiolation. Arrows indicate sampling points. Values were normalized to that of the d0 sample of each group with Actin8 expression level. Error bars indicate sd (n = 3).
Next, we investigated GUS activity in transgenic seeds expressing pri-miR163 promoter::GUS-GFP fusion during seed germination. After seed imbibition in the dark for 1 d, seeds were stained for GUS activity under W light and images taken at the times indicated (1, 6, and 24 h). Figure 6B shows that GUS was predominantly expressed in radicle after 6 h of light until 24 h, but the expression was extended to the cotyledons after 24 h. Like seedling de-etiolation, miR163 was induced upon exposure to light during seed germination, whereas PXMT1 expression was decreased under the same condition (Fig. 6C). Taken together, our results suggest miR163 functions in early stages from seed germination to seedling growth through targeting PXMT1 transcripts.
DISCUSSION
Plant miRNAs respond to plant hormones as well as many environmental factors, including high salinity, drought, temperature, nutrients, and UV-B or ozone-induced oxidative stress (Achard et al., 2004; Sunkar and Zhu, 2004; Jones-Rhoades and Bartel, 2004; Guo et al., 2005; Fujii et al., 2005; Navarro et al., 2006; Reyes and Chua, 2007; Zhou et al., 2007; Liu et al., 2008; Iyer et al., 2012); however, miRNAs related to light remain unidentified. This is rather surprising because about 20% of the genes in the Arabidopsis genome are known to be responsive to light (Jiao et al., 2005, 2007; Wang et al., 2014). Four Arabidopsis miRNAs (miR171, miR398, miR168, and miR167) have been shown to oscillate in their expression levels during the diurnal cycle, but there was no inverse correlation with the expression of their target mRNAs under the same conditions (Siré et al., 2009). Here, we show that miR163 levels respond to light during seed germination and seedling de-etiolation. Expression of pri-miR163 can be induced by light of all qualities with the strongest response seen under normal W. The induction seemed to be partially dependent on light intensity since W (100 µmol m−2 s−1) was the highest in light intensity and R (20 µmol m−2 s−1) was second highest. However, the induction of pri-miR163 transcripts under R was less than under B (10 µmol m−2 s−1). Time-course studies showed that miR163 was induced early upon light exposure (1 to 6 h) during seedling de-etiolation or germination, and miR163 accumulated at a higher level in roots compared with cotyledons or hypocotyls. We also confirmed the previous finding that miR163 shows stable, high expression levels in all organs of mature Arabidopsis plants (Guo et al., 2005; Preuss et al., 2008); in addition, we showed that in these plants the pri-miR163 is inducible by light. The light inducibility of pri-miR163 is perhaps not surprising since several light-responsive cis-elements such as GATA/I-box and GT1 consensus (GGTTAA) have been found in the upstream region of the gene (Ng et al., 2011; Supplemental Fig. S6), which may account in part for its light inducibility during seed germination and seedling de-etiolation. Histochemical GUS analyses of pri-miR163 promoter::GUS-GFP fusion in transgenic plants confirmed the early light responsiveness of pri-miR163 observed in quantitative real-time reverse transcription PCR (qRT-PCR) analyses.
Along with light, temperature is one of most important environmental stimuli in plant growth and development. miR163 was one of six ambient temperature-responsive miRNAs that express differentially at two different temperatures, 16°C and 23°C, being up-regulated at 23°C (Lee et al., 2010). We found that the expression of pri-miR163 is light dependent even if its expression is differently responsive at low or high temperature (Fig. 2). However, miR163 maturation is temperature dependent as its processing is attenuated at low temperature regardless of the high accumulation levels of pri-miR163 (Fig. 2). We note that the overexpression of pri-miR163 transcripts could be processed into mature miR163 even in the dark (d of miR163OE; Fig. 4D), confirming that this process operates independently of light. In general, siRNA biogenesis is temperature dependent in plants (Szittya et al., 2003); on the other hand, temperature does not affect the accumulation of miRNAs (Szittya et al., 2003). These results suggest that the four plant DCLs may have distinct temperature dependency. Although pri-miR163 is processed by DCL1, which mediates maturation of most plant miRNAs, it should be noted that unlike other typical 21-nt miRNAs, miR163 is an atypical miRNA with 24 nt (Kurihara and Watanabe, 2004).
Our results on the expression pattern of miR163 suggest a potential role of miR163 in response to early developmental stages triggered by light, which regulates the expression of target genes encoding SAMT family members. We found that among the six gene members of the SAMT family, only one, PXMT1, is responsive to light. The expression of PXMT1 is down-regulated by light in an inverse manner to that of pri-miR163. PXMT1 expression was altered by loss or overexpression of miR163 upon light exposure during seed germination and seedling de-etiolation. Although miR163 was found to be responsive to a fungal elicitor, alamethicin, or to nonstress ambient temperature (Lee et al., 2010; Ng et al., 2011), there was no inverse gene expression pattern between miR163 and PXMT1 in these conditions, implying that miR163 may not target PXMT1 in these responses. Here, we have provided four lines of evidence to support the view that light-inducible miR163 targets PXMT1 transcripts to promote seed germination and primary root elongation. (1) Anticorrelated gene expression patterns between miR163 and PXMT1 are found during seed germination under light and seedling de-etiolation. (2) Mutant plants lacking miR163 accumulate higher PXMT1 mRNA levels and show delayed seed germination rate and shorter primary root length with an increased number of lateral roots. (3) These phenotypes are also found in plants overexpressing PXMT1. (4) Delayed seed germination of mir163 mutant can be rescued by the expression of 35S::pri-miR163. More detailed analysis showed that a change in cell number but not cell length is responsible for the shorter root length in miR163 knockout or PXMT1 overexpression plants.
mRNA encoding FAMT (At3g44860), which converts farnesoic acid to methyl farnesoate, has been identified as a target of miR163 in mature Arabidopsis plant (Ng et al., 2011). Nevertheless, we did not detect any inverse gene expression pattern between this mRNA and pri-miR163 in early light-mediated development (Supplemental Fig. S2), suggesting FAMT mRNA plays little or no role in early seedling development. This observation is not surprising since FAMT mediates the synthesis of methyl farnesoate, an insect juvenile hormone that functions in plant defense against insects (Toong et al., 1988).
Although we have evidence that PXMT1 mRNA is negatively regulated by miR163, the precise role of this enzyme in early seedling development is not clear at present. PXMT1 belongs to the plant SABATH (salicylic acid/benzoic acid/theobromine) protein family of methyltransferases that catalyze the AdoMet-dependent methylation of a variety of natural chemicals found in plants, such as plant hormones and signaling molecules (Chen et al., 2003; Zhao et al., 2008). PXMT1 may function before seed germination and/or in darkness during seedling growth since its mRNA is degraded upon light exposure in these stages. Based on Arabidopsis genome annotation, PXMT1 is a putative 1,7-paraxanthine methyltransferase using paraxanthine (1,7-dimethylxanthine) as a substrate. Paraxanthine, along with theophylline (1,3-dimethylxanthine) and theobromine (3,7-dimethylxanthine), can potentially serve as precursors for caffeine (1,3,7-trimethylxanthine) biosynthesis (Ashihara et al., 2008; Ashihara et al., 2011); however, the putative 1,7-paraxanthine methyltransferase activity of PXMT1 has not been verified in vitro or in vivo. Coffee (Coffea arabica) cells contain only limited amounts of paraxanthine (Ashihara et al., 2008). In this plant, caffeine seems to be synthesized from theobromine rather than paraxanthine as caffeine synthase DXMT1 encoding 3,7-dimethylxanthine methyltransferase has been identified (Uefuji et al., 2003; Ashihara et al., 2008; Ashihara et al., 2011). Although Arabidopsis encodes PXMT1, it is unclear if this enzyme can synthesize paraxanthine; besides, the Arabidopsis PXMT1 does not share significant amino acid sequence similarity with the coffee DXMT1. One possible interpretation is that perhaps PXMT1 may not use paraxanthine as a substrate or use paraxanthine for synthesis of other alkaloids rather than caffeine in Arabidopsis. We found that increase in the putative PXMT1 either by miR163 knockout or by overexpression of the gene encoding this enzyme leads to shortening of the elongation/differentiation zone of root meristem with reduced root cell number. Irrespective of the catalytic activity of PXMT1, we can propose two hypotheses to explain the mechanism of root growth inhibition. The substrate of PXMT1 may be needed for normal root elongation, and its depletion would lead to shorter roots. On the other hand, it is possible that accumulation of the product of this enzyme may inhibit root elongation as well. Future experiments should be aimed at the biochemical characterization of the PXMT1 activity as well as metabolomics of plants of different genotypes described here.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
We used Arabidopsis (Arabidopsis thaliana) wild type (Columbia-0) and mir163 T-DNA insertion mutant (Salk_043556) from the Arabidopsis Biological Resource Center.
For etiolation or de-etiolation experiments, surface-sterilized seeds were incubated on Murashige and Skoog medium supplemented with 1% (w/v) Suc at 4°C for 3 d. Vernalized seeds were exposed to W fluorescent light (100 µmol m−2 s−1) for 1 h and germinated in the dark for 4 d at 22°C. After sampling of etiolated seedlings, plates were transferred to continuous W (100 µmol m−2 s−1), FR (5 µmol m−2 s−1), R (20 µmol m−2 s−1), or B (10 µmol m−2 s−1) light for the time period described (1, 6, or 24 h) in the figure legends.
For germination tests, approximately 150 sterilized seeds were plated on Murashige and Skoog medium, incubated in the dark at 4°C for 3 d, and then transferred to continuous W light (100 µmol m−2 s−1). Germination rate was scored by radicle emergence after light exposure for the time period indicated (36, 42, 48, or 54 h). Each germination test was performed with three biological replicates.
To measure the primary root length, the number of lateral roots, the number of cells in the elongation/differentiation zone, and the length of the elongation/differentiation zone (Perilli and Sabatini, 2010; Perilli et al., 2012), 7-d-old seedlings were allowed to grow for another 7 d or transferred onto fresh Murashige and Skoog medium for further vertical growth for 7 d. All root phenotypes were photographed by Zeiss WF Upright (Carl Zeiss). The length of the primary root and the elongation/differentiation zone of root meristem were measured from the root images using ImageJ (National Institutes of Health; http://rsb.info.nih.gov/ij). Statistical significance was evaluated by Student’s t test analysis.
cDNA Synthesis and qRT-PCR
Total RNA was isolated from Arabidopsis seedlings using RNeasy plant mini kit (Qiagen). Total RNA (2.5 µg) was used for cDNA synthesis using SuperScript III reverse transcription kit (Invitrogen) and oligo(dT). For qRT-PCR analysis, cDNA was amplified using SYBR Premix ExTaq (TaKaRa) with gene-specific primers in a Bio-Rad CFX96 real-time system. Actin2 and Actin8 were used as internal controls in each qRT-PCR experiment for seedlings and germinated seeds, respectively.
Vector Constructions for MiR163 and PXMT1
Primer pairs listed in Supplemental Table S1 for cloning of full-length cDNA were designed based on cDNA sequences in a public database (http://www.arabidopsis.org). Genomic DNA extracted from Arabidopsis seedlings with the DNeasy plant mini kit (Qiagen) was used to amplify the pri-miR163 promoter region.
For vector constructions, we used a Gateway cloning system (Invitrogen). Full-length or partial cDNA fragment of each gene or genomic DNA fragment of promoter region was PCR amplified by KOD hot start DNA polymerase (Novagen). The amplified fragments were cloned into pENTR/D-TOPO vector (Invitrogen). After sequence verification, the pENTR/D-TOPO clone harboring each gene or DNA fragment was integrated into the destination vector by LR clonase (Invitrogen).
For 35S::pri-miR163 and 35S::PXMT1, full-length sequences of pri-miR163 (At1g66725, 568 bp) and of PXMT1 (At1g66700, 1059 bp) were cloned into the destination vector, pBA002 expression vector, which contained a cauliflower mosaic virus 35S promoter (Kost et al., 1998).
For PXMT1 RNAi construction, 236-bp fragment (+200 to +435) of PXMT1 cDNA was cloned into the RNAi destination vector pH7GWIWG2(II) (Karimi et al., 2002).
For pri-miR163 promoter::GUS-GFP expression vector, the promoter region of pri-miR163 (982 bp, −989 to −8) was cloned into the pKGWFS7 destination vector, which contained GUS-GFP reporter genes (Karimi et al., 2002).
For pri-miR163 promoter::pri-miR163 construction, the promoter plus transcribed region (1558 bp, −989 to +569) of pri-miR163 was cloned into the pH2GW7.0 destination vector (Karimi et al., 2002).
Arabidopsis plants were transformed by the floral dip method using Agrobacterium strain GV3101 (Clough and Bent, 1998; Zhang et al., 2006).
Small RNA Gel-Blot Analysis
Total RNAs were extracted using Trizol (Invitrogen) for small RNA gel-blot analysis. Total RNA (7.5 µg) was resolved in 10% (w/v) PAGE with 8 m urea, transferred to Hybond-N+ (GE Healthcare), and followed by cross-linking using a UV Stratalinker (Stratagene). An antisense probe corresponding to the mature miR163 or an antisense U6 oligonucleotide probe was end-labeled with [γ-32P]ATP using T4 polynucleotide kinase (New England Biolabs), and probes were hybridized with membranes at 42°C in ULTRAhyb-oligo hybridization buffer (Ambion) for 16 h. Membranes were washed twice with 2× SSC (0.3 m NaCl, 0.03 m sodium citrate, pH 7.0) containing 0.1% (w/v) SDS at 42°C for 20 min. The sequences of the small RNA probes are listed in Supplemental Table S1.
GUS Assay
Transgenic plants expressing pri-miR163 promoter::GUS-GFP transgene were stained in a solution containing 5-bromo-4-chloro-3-indolyl β-d-glucuronide cyclohexamine salt (X-Gluc; Rose Scientific, ES-1007-001) at room temperature. Seedlings were prefixed in ice-cold acetone (90% [v/v]) under vacuum for 10 min, rinsed twice with 100 mm sodium phosphate buffer (pH 7.0), infiltrated with a 1 mm X-Gluc staining solution containing 0.1% (v/v) Triton X-100, 0.5 mm K3(Fe(CN)6), 0.5 mm K4(Fe(CN)6), and 10 mm EDTA under vacuum for 10 min, and incubated at 37°C for 12 h. Stained seedlings were cleared by several washings of 70% (v/v) ethanol. Cleaned seedlings were mounted on slides and examined under a light microscope.
To visualize GUS expression, seeds were imbibed in the dark at 22°C for 1.5 d. Seed coat was removed after fixing and washing, and the naked seed was incubated with the staining solution.
Accession Numbers
Sequence data can be found in the Arabidopsis Genome Initiative or GenBank database under the following accession numbers: miR163 gene, At1g66725; and SAMT-like genes, At1g66690, At1g66700, At1g66720, At3g44860, and At3g44870.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Core sequence alignment of miR163 with its target mRNAs.
Supplemental Figure S2. Expression levels of candidate target genes of miR163 during seedling de-etiolation.
Supplemental Figure S3. Immunoblot analysis of transgenic Arabidopsis plants expressing pri-miR163 promoter::GUS-GFP.
Supplemental Figure S4. Root growth of the wild type, mir163 mutant miR163KO, miR163 overexpression line miR163OE, PXMT1 RNAi line PXMT1RNAi, and PXMT1 overexpression line PXMT1OE grown on vertically oriented plate.
Supplemental Figure S5. Phenotypes of mir163 mutant, overexpression line of miR163 or PXMT1, and PXMT1 RNAi line under different light qualities.
Supplemental Figure S6. Schematic map of miR163 promoter showing the distribution of light-responsive elements.
Supplemental Table S1. Primer sequences used in this study.
Supplementary Material
Acknowledgments
P.J.C. was supported by the National Research Foundation of Korea Grant funded by the Korean Government (NRF-2011-357-F00015 and NRF-2013R1A6A3A04060627).
Glossary
- miRNA
microRNA
- pri-miRNA
primary miRNA
- FAMT
farnesoic acid methyltransferase
- SAMT
S-adenosyl-Met-dependent carboxyl methyltransferase
- RNAi
RNA interference
- qRT-PCR
quantitative reverse transcription PCR
Footnotes
Articles can be viewed without a subscription.
References
- Achard P, Herr A, Baulcombe DC, Harberd NP (2004) Modulation of floral development by a gibberellin-regulated microRNA. Development 131: 3357–3365 [DOI] [PubMed] [Google Scholar]
- Allen E, Xie Z, Gustafson AM, Sung GH, Spatafora JW, Carrington JC (2004) Evolution of microRNA genes by inverted duplication of target gene sequences in Arabidopsis thaliana. Nat Genet 36: 1282–1290 [DOI] [PubMed] [Google Scholar]
- Ashihara H, Kato M, Crozier A (2011) Distribution, biosynthesis and catabolism of methylxanthines in plants. Handbook Exp Pharmacol 200: 11–31 [DOI] [PubMed] [Google Scholar]
- Ashihara H, Sano H, Crozier A (2008) Caffeine and related purine alkaloids: biosynthesis, catabolism, function and genetic engineering. Phytochemistry 69: 841–856 [DOI] [PubMed] [Google Scholar]
- Aukerman MJ, Sakai H (2003) Regulation of flowering time and floral organ identity by a microRNA and its APETALA2-like target genes. Plant Cell 15: 2730–2741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP. (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116: 281–297 [DOI] [PubMed] [Google Scholar]
- Breakfield NW, Corcoran DL, Petricka JJ, Shen J, Sae-Seaw J, Rubio-Somoza I, Weigel D, Ohler U, Benfey PN (2012) High-resolution experimental and computational profiling of tissue-specific known and novel miRNAs in Arabidopsis. Genome Res 22: 163–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F, D’Auria JC, Tholl D, Ross JR, Gershenzon J, Noel JP, Pichersky E (2003) An Arabidopsis thaliana gene for methylsalicylate biosynthesis, identified by a biochemical genomics approach, has a role in defense. Plant J 36: 577–588 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Franklin KA, Whitelam GC (2004) Light signals, phytochromes and cross-talk with other environmental cues. J Exp Bot 55: 271–276 [DOI] [PubMed] [Google Scholar]
- Fujii H, Chiou TJ, Lin SI, Aung K, Zhu JK (2005) A miRNA involved in phosphate-starvation response in Arabidopsis. Curr Biol 15: 2038–2043 [DOI] [PubMed] [Google Scholar]
- Guo HS, Xie Q, Fei JF, Chua NH (2005) MicroRNA directs mRNA cleavage of the transcription factor NAC1 to downregulate auxin signals for arabidopsis lateral root development. Plant Cell 17: 1376–1386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmer SL. (2009) The circadian system in higher plants. Annu Rev Plant Biol 60: 357–377 [DOI] [PubMed] [Google Scholar]
- Huntzinger E, Izaurralde E (2011) Gene silencing by microRNAs: contributions of translational repression and mRNA decay. Nat Rev Genet 12: 99–110 [DOI] [PubMed] [Google Scholar]
- Iyer NJ, Jia X, Sunkar R, Tang G, Mahalingam R (2012) microRNAs responsive to ozone-induced oxidative stress in Arabidopsis thaliana. Plant Signal Behav 7: 484–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Y, Lau OS, Deng XW (2007) Light-regulated transcriptional networks in higher plants. Nat Rev Genet 8: 217–230 [DOI] [PubMed] [Google Scholar]
- Jiao Y, Ma L, Strickland E, Deng XW (2005) Conservation and divergence of light-regulated genome expression patterns during seedling development in rice and Arabidopsis. Plant Cell 17: 3239–3256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones-Rhoades MW, Bartel DP (2004) Computational identification of plant microRNAs and their targets, including a stress-induced miRNA. Mol Cell 14: 787–799 [DOI] [PubMed] [Google Scholar]
- Jones-Rhoades MW, Bartel DP, Bartel B (2006) MicroRNAS and their regulatory roles in plants. Annu Rev Plant Biol 57: 19–53 [DOI] [PubMed] [Google Scholar]
- Jung JH, Seo YH, Seo PJ, Reyes JL, Yun J, Chua NH, Park CM (2007) The GIGANTEA-regulated microRNA172 mediates photoperiodic flowering independent of CONSTANS in Arabidopsis. Plant Cell 19: 2736–2748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi M, Inzé D, Depicker A (2002) GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci 7: 193–195 [DOI] [PubMed] [Google Scholar]
- Kinoshita N, Wang H, Kasahara H, Liu J, Macpherson C, Machida Y, Kamiya Y, Hannah MA, Chua NH (2012) IAA-Ala Resistant3, an evolutionarily conserved target of miR167, mediates Arabidopsis root architecture changes during high osmotic stress. Plant Cell 24: 3590–3602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kost B, Spielhofer P, Chua NH (1998) A GFP-mouse talin fusion protein labels plant actin filaments in vivo and visualizes the actin cytoskeleton in growing pollen tubes. Plant J 16: 393–401 [DOI] [PubMed] [Google Scholar]
- Kurihara Y, Watanabe Y (2004) Arabidopsis micro-RNA biogenesis through Dicer-like 1 protein function. Proc Natl Acad Sci USA 24: 12573–12578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H, Yoo SJ, Lee JH, Kim W, Yoo SK, Fitzgerald H, Carrington JC, Ahn JH (2010) Genetic framework for flowering-time regulation by ambient temperature-responsive miRNAs in Arabidopsis. Nucleic Acids Res 38: 3081–3093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Lee J, Provost P, Rådmark O, Kim S, et al. (2003) The nuclear RNase III Drosha initiates microRNA processing. Nature 425: 415–419 [DOI] [PubMed] [Google Scholar]
- Lee Y, Kim M, Han J, Yeom KH, Lee S, Baek SH, Kim VN (2004) MicroRNA genes are transcribed by RNA polymerase II. EMBO J 23: 4051–4060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu HH, Tian X, Li YJ, Wu CA, Zheng CC (2008) Microarray-based analysis of stress-regulated microRNAs in Arabidopsis thaliana. RNA 14: 836–843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Jung C, Xu J, Wang H, Deng S, Bernad L, Arenas-Huertero C, Chua NH (2012) Genome-wide analysis uncovers regulation of long intergenic noncoding RNAs in Arabidopsis. Plant Cell 24: 4333–4345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin RC, Liu PP, Goloviznina NA, Nonogaki H (2010) microRNA, seeds, and Darwin? Diverse function of miRNA in seed biology and plant responses to stress. J Exp Bot 61: 2229–2234 [DOI] [PubMed] [Google Scholar]
- Navarro L, Dunoyer P, Jay F, Arnold B, Dharmasiri N, Estelle M, Voinnet O, Jones JDG (2006) A plant miRNA contributes to antibacterial resistance by repressing auxin signaling. Science 312: 436–439 [DOI] [PubMed] [Google Scholar]
- Ng DW, Zhang C, Miller M, Palmer G, Whiteley M, Tholl D, Chen ZJ (2011) cis- and trans-Regulation of miR163 and target genes confers natural variation of secondary metabolites in two Arabidopsis species and their allopolyploids. Plant Cell 23: 1729–1740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park MY, Wu G, Gonzalez-Sulser A, Vaucheret H, Poethig RS (2005) Nuclear processing and export of microRNAs in Arabidopsis. Proc Natl Acad Sci USA 102: 3691–3696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasquinelli AE. (2012) MicroRNAs and their targets: recognition, regulation and an emerging reciprocal relationship. Nat Rev Genet 13: 271–282 [DOI] [PubMed] [Google Scholar]
- Perilli S, Di Mambro R, Sabatini S (2012) Growth and development of the root apical meristem. Curr Opin Plant Biol 15: 17–23 [DOI] [PubMed] [Google Scholar]
- Perilli S, Sabatini S (2010) Analysis of root meristem size development. Methods Mol Biol 655: 177–187 [DOI] [PubMed] [Google Scholar]
- Preuss SB, Costa-Nunes P, Tucker S, Pontes O, Lawrence RJ, Mosher R, Kasschau KD, Carrington JC, Baulcombe DC, Viegas W, et al. (2008) Multimegabase silencing in nucleolar dominance involves siRNA-directed DNA methylation and specific methylcytosine-binding proteins. Mol Cell 32: 673–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes JL, Chua NH (2007) ABA induction of miR159 controls transcript levels of two MYB factors during Arabidopsis seed germination. Plant J 49: 592–606 [DOI] [PubMed] [Google Scholar]
- Siré C, Moreno AB, Garcia-Chapa M, López-Moya JJ, San Segundo B (2009) Diurnal oscillation in the accumulation of Arabidopsis microRNAs, miR167, miR168, miR171 and miR398. FEBS Lett 583: 1039–1044 [DOI] [PubMed] [Google Scholar]
- Sunkar R, Zhu JK (2004) Novel and stress-regulated microRNAs and other small RNAs from Arabidopsis. Plant Cell 16: 2001–2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szittya G, Silhavy D, Molnár A, Havelda Z, Lovas A, Lakatos L, Bánfalvi Z, Burgyán J (2003) Low temperature inhibits RNA silencing-mediated defence by the control of siRNA generation. EMBO J 22: 633–640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toong YC, Schooley DA, Baker FC (1988) Isolation of insect juvenile hormone III from a plant. Nature 333: 170–171 [Google Scholar]
- Uefuji H, Ogita S, Yamaguchi Y, Koizumi N, Sano H (2003) Molecular cloning and functional characterization of three distinct N-methyltransferases involved in the caffeine biosynthetic pathway in coffee plants. Plant Physiol 132: 372–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaistij FE, Elias L, George GL, Jones L (2010) Suppression of microRNA accumulation via RNA interference in Arabidopsis thaliana. Plant Mol Biol 73: 391–397 [DOI] [PubMed] [Google Scholar]
- Wang H, Chung PJ, Liu J, Jang IC, Kean MJ, Xu J, Chua NH (2014) Genome-wide identification of long noncoding natural antisense transcripts and their responses to light in Arabidopsis. Genome Res 24: 444–453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G, Poethig RS (2006) Temporal regulation of shoot development in Arabidopsis thaliana by miR156 and its target SPL3. Development 133: 3539–3547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z, Allen E, Fahlgren N, Calamar A, Givan SA, Carrington JC (2005) Expression of Arabidopsis MIRNA genes. Plant Physiol 138: 2145–2154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Henriques R, Lin SS, Niu QW, Chua NH (2006) Agrobacterium-mediated transformation of Arabidopsis thaliana using the floral dip method. Nat Protoc 1: 641–646 [DOI] [PubMed] [Google Scholar]
- Zhao N, Ferrer JL, Ross J, Guan J, Yang Y, Pichersky E, Noel JP, Chen F (2008) Structural, biochemical, and phylogenetic analyses suggest that indole-3-acetic acid methyltransferase is an evolutionarily ancient member of the SABATH family. Plant Physiol 146: 455–467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Wang G, Zhang W (2007) UV-B responsive microRNA genes in Arabidopsis thaliana. Mol Syst Biol 3: 103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.