Agronomically realistic, long-term drought stress mis-regulates some miRNAs and induces the down-regulation of a set of small RNA loci in the maize leaf.
Abstract
Endogenous small RNAs (sRNAs) contribute to gene regulation and genome homeostasis, but their activities and functions are incompletely known. The maize genome has a high number of transposable elements (TEs; almost 85%), some of which spawn abundant sRNAs. We performed sRNA and total RNA sequencing from control and abiotically stressed B73 wild-type plants and rmr6-1 mutants. RMR6 encodes the largest subunit of the RNA polymerase IV complex and is responsible for accumulation of most 24-nucleotide (nt) small interfering RNAs (siRNAs). We identified novel MIRNA loci and verified miR399 target conservation in maize. RMR6-dependent 23-24 nt siRNA loci were specifically enriched in the upstream region of the most highly expressed genes. Most genes misregulated in rmr6-1 did not show a significant correlation with loss of flanking siRNAs, but we identified one gene supporting existing models of direct gene regulation by TE-derived siRNAs. Long-term drought correlated with changes of miRNA and sRNA accumulation, in particular inducing down-regulation of a set of sRNA loci in the wild-typeleaf.
Plant endogenous small RNAs (sRNAs) range in length from 20 to 24 nucleotides (nts) and contribute to regulate gene expression through RNA-mediated transcriptional gene silencing (TGS) and post-TGS mechanisms. Their activity is essential for the maintenance of genome integrity, the intrinsic normal growth of cells, and proper plant development.
MiRNAs are mostly 21 nt long and are encoded by endogenous MIRNA genes, which are transcribed by polymerase (Pol) II to single-stranded precursors that fold into stem-loop secondary structures. The stems are precisely cleaved by the DICER-LIKE 1 protein, liberating a miRNA/miRNA* duplex (for review, see Rogers and Chen, 2013). After the duplex is loaded into ARGONAUTE 1, the miRNA guide strand is most frequently retained and directs target repression, while the miRNA* is more often rapidly degraded. Most plant miRNAs induce post-TGS of their targets, but some cases of TGS have been described (Bao et al., 2004; Wu et al., 2010; Hu et al., 2014). Plant miRNAs share extensive sequence complementarity with their target RNAs: there are only a few examples with more than five mismatches between the miRNA and the target (Axtell, 2013a). Many known miRNAs and their targets are conserved across different plant species (Montes et al., 2014). In maize, many conserved miRNA targets have been experimentally confirmed (Shen et al., 2013; Zhai et al., 2013; Zhao et al., 2013; Liu et al., 2014), but a few exceptions exist, for example those of miR168 and miR399. MiRNAs are members of multiple regulatory networks controlling plant development, and many miRNA families also play roles in stress responses and tolerance (for review, see Sunkar et al., 2012). For example, miR156, which targets SQUAMOSA PROMOTER BINDING PROTEIN-LIKE genes, coordinates the balance between development and abiotic stress tolerance and is important for heat stress memory in Arabidopsis (Cui et al., 2014; Stief et al., 2014).
A specific class of endogenous small interfering RNA (siRNAs), mostly 24 nts long, participates in the RNA-directed DNA methylation (RdDM) process. Pol IV transcribes target loci, typified by high levels of DNA methylation, into single-stranded RNAs that are copied into short double-stranded RNAs (dsRNAs) by RNA-DEPENDENT RNA POLYMERASE 2 (Blevins et al., 2015; Zhai et al., 2015). These short dsRNAs are then processed by DICER-LIKE 3 into 24-nt siRNAs. Once loaded onto ARGONAUTE 4, these siRNAs are thought to target nascent, chromatin-associated noncoding RNAs transcribed by RNA Pol V. Successful targeting correlates with the deposition of de novo DNA methylation and other repressive epigenetic marks at the target loci, inducing TGS (for review, see Matzke and Mosher, 2014). Pol IV-dependent siRNAs are often produced from transposable elements (TEs), TE-like sequences, and other repeats (Zhang et al., 2007; Mosher et al., 2008). They contribute to the reinforcement of TE silencing (Slotkin et al., 2005; Marí-Ordóñez et al., 2013; Nuthikattu et al., 2013) and in some cases are also essential to control the expression of protein-coding genes in cis or in trans (Liu et al., 2004; Kinoshita et al., 2007; McCue et al., 2013).
In maize the majority of genes are located within 1 kb of an annotated TE (Baucom et al., 2009; Schnable et al., 2009), and loci undergoing RdDM are primarily located in gene flanking regions (Gent et al., 2013, 2014). The enrichment of RdDM-associated siRNAs near maize genes (Wang et al., 2009; Gent et al., 2013; Gent et al., 2014; Xin et al., 2014) has been suggested to ensure the continuous silencing of potentially deleterious TEs and TE-like sequences in an active and accessible chromatin environment required for the Pol II transcription of close genes (Gent et al., 2014). In maize, gene expression can be influenced by gene-proximal TEs and repeats (Erhard et al., 2013), and both genes and TEs can be regulated by the direct competition between Pol IV and Pol II occupancy at their promoters (Hale et al., 2009; Erhard et al., 2015). At the genome-wide level, the expression of maize genes positively correlates with the accumulation levels of upstream 24-nt siRNAs (Gent et al., 2013). In the absence of Pol IV-dependent siRNAs, Pol II transcription globally decreases around the transcription start site and increases at 3′ end of genes (Erhard et al., 2015), and gene-proximal TEs lose DNA methylation (Li et al., 2015). Thus, the available data suggest that Pol IV-dependent 24-nt siRNAs in maize primarily serve to mark and enforce boundaries between areas of transcriptionally active euchromatin and transcriptionally repressed heterochromatin.
RdDM-associated siRNAs may also be important in adaptation to biotic and abiotic stresses. Arabidopsis ago4 mutants are impaired in bacterial disease resistance (Agorio and Vera, 2007), and genome-wide DNA methylation patterns are altered in Arabidopsis plants during bacterial infection (Dowen et al., 2012). RdDM is also required for basal heat tolerance in Arabidopsis (Popova et al., 2013). Environmental stresses trigger the expression of siRNAs that modulate target genes involved in the stress response (Tricker et al., 2012; Wang et al., 2015). RdDM-associated siRNAs can also defend the genome from heat-induced movements of TEs (Ito et al., 2011). Here, we sought to extend our knowledge of maize stress-induced changes in miRNA and siRNA accumulation to agronomically realistic drought and salinity stresses, which are the major environmental stresses worldwide adversely affecting crop productivity, using coupled sRNA and total RNA sequencing.
RESULTS
De Novo Identification of sRNA Loci by High-Throughput Sequencing
A total of 46 sRNA-seq libraries were sequenced from leaves of wild-typeand rmr6-1 plants, grown under control conditions, after 10 d of three different abiotic stress treatments and 7 d poststress recovery (Supplemental Data Set S1). The abiotic stresses were drought, high salinity, and drought and high salinity combined. An additional four libraries were obtained from control and drought-treated wild-type shoot apical meristematic areas. Each sRNA library had two or three biological replicates (Supplemental Data Set S1). The majority of genome-aligned sRNAs from wild-type leaves were 24 nts long (Fig. 1A-C). The trend toward 24-nt RNAs was even more pronounced in the meristematic libraries (Fig. 1A-C). Maize RMR6 encodes the largest subunit of Pol IV (Erhard et al., 2009). As expected, 24-nt RNAs were mostly eliminated in the rmr6-1 mutant, and 22-nt RNAs had a slightly higher accumulation level (Fig. 1A-C). No major alterations in the sRNA size distribution were observed in the stressed samples (Fig. 1A-C).
Figure 1.
Characterization of maize sRNAs. A-C, Size distributions of genome-aligned sRNAs for three biological replicates. Multiple lines within a tissue/genotype represent various control and stress treatments (Supplemental Data set S1). D, Total number of identified non-MIRNA and MIRNA loci, reported separately by predominant RNA size; N, predominant RNA size < 20 nts or > 24 nts.
We performed de novo annotation of maize sRNA loci using the merged set of all aligned sRNA reads. A total of 320,110 clusters were found (Fig. 1D; Supplemental Data Set S2, S3), of which 251,496 were dominated by RNAs 20 to 24 nts in length. The other 68,614 clusters were not examined further, because they were not likely generated by the catalytic activity of DCL proteins. The remaining sRNA clusters were classified based upon their most abundant RNA size (size class; Figure 1D). A total of 48 MIRNA loci and 251,448 non-MIRNA loci were identified, with most frequent size class of 21 nts and 24 nts, respectively (Fig. 1D).
Analysis of miRNAs and Their Targets
Of the 48 MIRNA loci that we initially de novo identified (Fig. 1D), 30 were previously known loci annotated in miRBase (version 20; Supplemental Data Set S4). miRBase 20 contains a total of 159 annotated maize MIRNA loci. Our de novo MIRNA discovery method is likely rather insensitive, both because we wish to avoid false positive annotations and because our automated cluster discovery can sometimes incorrectly define the start and stop positions of MIRNA loci. Therefore, we tested the exact coordinates of each of the 159 miRBase (version 20) MIRNA loci against our combined sRNA dataset (Supplemental Data Set S4). A total 67 of the 159 annotated MIRNA loci passed our stringent analyses, but 64 of them failed (Supplemental Data Set S4). The remaining 28 loci had insufficient aligned sRNAs in our data and as such could not be analyzed. The rather low rate of previous MIRNA annotations supported by our data is consistent with observations that many miRBase MIRNA annotations are questionable (Taylor et al., 2014).
The 18 putative MIRNA loci found by our initial analysis were tested for reproducibility. Ten of them independently passed all criteria for MIRNA loci in at least two of our libraries (Supplemental Data Set S5 and S6). As expected for true plant miRNAs, they were either 21 nts or 22 nts long, and their accumulation was not affected in the rmr6-1 background. Three were new members of the miR166 family, and the other seven belong to six new miRNA families without obvious homology with any other previously annotated miRNAs in miRBase.
Maize miR399 was previously predicted to target several mRNAs, including one of unknown function (GRMZM2G165734), mRNAs encoding inorganic phosphate transporters (Zhang et al., 2009), and an mRNA encoding a putative ubiquitin-like 1-activating enzyme E1A (Wang et al., 2014). We found that the putative GRMZM2G165734 gene is actually a MIR399 homolog. We identified more possible target sites in genomic DNA located immediately upstream of GRMZM2G381709, an ortholog of Arabidopsis PHO2 (Calderón-Vázquez et al., 2011). Arabidopsis PHO2 encodes an ubiquitin-conjugating E2 enzyme that plays a role in the maintenance of Pi homeostasis (Bari et al., 2006). The Arabidopsis PHO2 mRNA is targeted by miR399 in multiple sites in its 5′-untranslated region (5′-UTR; Allen et al., 2005). cDNA sequencing demonstrated that the GRMZM2G381709 5′-UTR encompassed the putative miR399 target sites (Fig. 2A). RNA-ligase mediated 5′ RACE found evidence for miR399-directed slicing at two of these sites (Fig. 2A-B). This result indicates that miR399 targeting of PHO2 is conserved across angiosperms (Bari et al., 2006), including maize.
Figure 2.
miR399 targets the 5′-UTR of maize GRMZM2G381709, a PHO2 homolog. A, GRMZM2G381709 gene structure: exons are red blocks, introns are black lines, 5′-UTR and 3′-UTR are brown blocks. Inset shows the 5′-UTR region with predicted miR399 target sites numbered 1-6 and shown in black. Barplot shows the frequencies of 5′-ends recovered from sequenced 5′-RACE products. Four 5′-RACE cleavage products, each sequenced once, are not shown here; they were found approximately 4 kb upstream of the ATG, spanning a region of 343 bp. B, Alignments of miR399 and GRMZM2G381709 at sites 5 and 3. Arrows indicate the terminal positions of the sequenced 5′ RACE cleavage products and the fraction of clones corresponding to these positions.
Long-Term Abiotic Stresses Affect the Accumulation of a Set of miRNAs
The applied abiotic stress treatments mimicked agronomically realistic long-term drought, salinity and combined drought plus salinity stress conditions (Morari et al., 2015). To test their effects on miRNAs, we performed differential expression analysis on mature miRNAs annotated in miRBase and in this work (Supplemental Data Set S5). In total 11 mature miRNAs belonging to ten miRNA families were differentially expressed in at least one pairwise comparison (Fig. 3). Drought stress affected miRNA accumulation mainly in wild-type plants: miR156, miR2275, and miR398 were up-regulated in drought-stressed leaves, while miR166 and miR396 were down-regulated (Fig. 3). Drought stress caused down-regulation of miR399 in wild-type shoot apical meristematic areas (Fig. 3). In contrast, a combined drought and salinity stress effect on miRNA accumulation was mainly detected in the rmr6-1 background, in which four miRNAs were down-regulated (miR164, miR399, miR528, and miR827; Fig. 3). In general, stress-induced effects on miRNA accumulation were not similar between wild-type and rmr6-1.
Figure 3.
Stress-induced changes in miRNA accumulation. Heat map showing expression levels of miRNAs that are differentially expressed in at least one pairwise comparison between control and a stress-treated sample in the leaf or shoot apical meristematic area (M) of wild-type or rmr6-1 mutant plants; C, control; D, drought; S, salinity; D+S, drought and salinity stress, after 10 d of treatment and after 7 d of recovery (+7). The mean reads per million mapped values from the three biological replicates were calculated, after which each row was scaled to have a mean of zero and a sd of 1. Black boxes indicate samples with statistically significant expression changes at a false discovery rate of 0.01. Dendrogram shows hierarchical clustering of rows based on Pearson distance.
Gene-Proximal Regions Are Particularly Enriched for 21-nt, 23-nt ,and 24-nt sRNA Loci
Non-MIRNA loci (Fig. 1D) were examined for their co-occupancy patterns relative to various genomic features (Fig. 4; Supplemental Data Set S7). Repeats account for a very large portion of the maize genome, explaining why they were statistically depleted for sRNA loci although the vast majority of sRNA loci mapped to them. Gene-body regions of mRNAs were highly enriched for loci with predominant RNA sizes of 20 to 22 nts, in contrast their flanking regions were enriched for 21-, 23-, and 24-nt dominated loci. Flanking regions of long noncoding RNAs (lncRNAs) were enriched for 21-, 23-, and 24-nt dominated loci, as for mRNAs, in contrast gene-body regions of lncRNAs were enriched for loci with predominant RNA size of 21 nts. Our results are consistent with previous observations of 24-nt sRNAs concentrated very close to the ends of mRNAs and TEs (Wang et al., 2009; Gent et al., 2013; Xin et al., 2014). Our data demonstrate that this trend extends to lncRNAs as well as mRNAs and TEs in the maize genome.
Figure 4.
Co-occupancy analysis. For each combination of sRNA locus type and genomic feature, the level of enrichment/depletion is reported as the log2 of the observed / expected ratio (observed overlapping nts /expected overlapping nts by random chance). Red values indicate enrichment of overlaps, and blue values represent depletion. -inf: negative infinity (no overlap at all where at least some would be expected by random chance). Dendrograms reflect hierarchical clustering of rows by Pearson distance and of columns by Euclidean distance.
Leaves Have a Lower Ratio of RMR6-Dependent siRNAs to miRNAs Compared to Shoot Apical Meristematic Areas
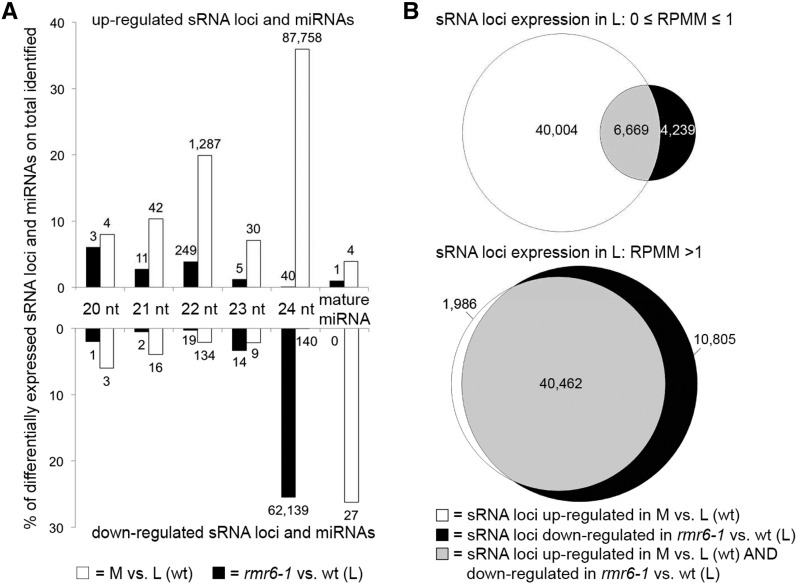
The expression of sRNA loci in the leaf was compared between wild-type and rmr6-1 mutant plants in control conditions at the first time point of sample collection. A total 62,175 sRNA loci (24.7%) were down-regulated in rmr6-1 compared to wt; nearly all of these were dominated by 24-nt RNAs (Fig. 5A). A total 308 loci (0.1%) were up-regulated in rmr6-1 relative to wt, most of which were dominated by 22-nt RNAs (Fig. 5A). Differential expression of sRNAs between wild-type leaves and shoot apical meristematic areas was much different: large numbers of sRNA loci of all size classes were elevated in the shoot apical meristematic areas relative to leaves, while many miRNAs showed the opposite pattern (Fig. 5A).
Figure 5.
The ratio of RMR6-dependent 24-nt siRNAs to miRNAs is higher in shoot apical meristematic areas compared to leaves. A, Percentages and tallies of differentially expressed sRNA loci and mature miRNAs for rmr6-1 versus wild type (black bars) and shoot apical meristematic areas (M) versus leaves (L; gray bars). B, Overlaps between sRNA loci down-regulated in rmr6-1 leaves and sRNA loci up-regulated in wild-type shoot apical meristematic areas. Top shows results for lowly expressed sRNA loci (between 0 and 1 reads per million mapped in mean of wild-type control leaves), bottom shows results for loci with higher expression (reads per million mapped) > 1. L, leaf samples; M, shoot apical meristematic area samples; RPMM, reads per million mapped.
sRNA loci were divided into two groups based on their expression levels in wild-type leaves in control conditions at the first time point of sample collection: (1) lowly or non-expressed loci (0 ≤ RPMM ≤ 1; RPMM, Reads Per Million Mapped), and (2) loci with higher expression (RPMM > 1). Relatively few of the lowly expressed loci were significantly down-regulated in rmr6-1, likely because of the low statistical power inherent in calling significant differential expression for lowly expressed loci (Fig. 5B). However, the majority of lowly expressed RMR6-dependent loci in leaves were also up-regulated in the shoot apical meristematic area. Among more highly expressed sRNA loci, the majority were both RMR6-dependent in the leaves and up-regulated in shoot apical meristematic areas (Fig. 5B). Thus, most of the sRNA loci up-regulated in shoot apical meristematic areas are likely RMR6 dependent. Taken together, these observations suggest that the relative activity of the RMR6 sRNA pathway to the miRNA pathway is higher in maize shoot apical meristematic areas compared to leaves.
Highly Expressed mRNAs Are More Likely to Be Flanked by RMR6-Dependent 23- to 24-nt siRNA Loci
RNA-seq was used to measure transcript accumulation levels in wild-type, non-stressed leaves at the first time point of sample collection. mRNAs, TE transcripts, and lncRNAs were binned into 5 groups based on accumulation level. Flanking RMR6-dependent 23- or 24-nt siRNA loci were more numerous at highly expressed transcripts of all types, while non-expressed transcripts had the weakest signal of flanking siRNA loci (Fig. 6). The magnitude of this correlation was greatest for mRNAs and weakest for TE transcripts. We analyzed the strandedness of the RMR6-dependent 23- to 24-nt siRNA loci that flanked mRNAs, TE transcripts, and lncRNAs . Most of the siRNA loci had roughly equal amounts of siRNAs aligned to both genomic strands, as expected for biogenesis from dsRNAs (Supplemental Data Set S8). Among the minority of gene-flanking siRNA loci that did appear to be stranded, there was no correlation between the siRNA strand with the genomic strand of the flanking transcript (Supplemental Data Set S8). These data indicate that, in leaves, siRNA polarity is not correlated with the polarity of the flanking transcript.
Figure 6.
Highly expressed mRNAs are more likely to have flanking RMR6-dependent 23- to 24-nt siRNAs. Metaplots showing distributions of rmr6-1 down-regulated 23- or 24-nt dominated siRNA loci surrounding protein-coding genes (39,392 loci) (A), TE transcripts (33,254 loci) (B), and lncRNA transcripts (13,387 loci) (C). All features are divided into five groups by RNA-seq-based expression levels in wild-type leaves. “RPKM=0” (RPKM, reads per kilobase million). TSS, transcription start site; TTS, transcription termination site.
Loss of Gene-Flanking siRNAs Is Not Generally Predictive of Induction of Gene Expression in rmr6-1
Differential expression analysis of our RNA-seq data (rmr6-1 versus wild-type leaves, control at the first time point) identified 1,013 differentially expressed genes. The majority were up-regulated in rmr6-1, indicating that RMR6 most frequently has a repressive effect on gene expression (Fig. 7A). Nearly one-half of the differentially expressed genes (48.3%; Supplemental Data Set S9) were expressed only in rmr6-1, but not in wt, indicating that the loss of RMR6 function activated many normally repressed genes. Recall that gene-flanking 23- to 24-nt siRNA loci are most strongly enriched near genes actively transcribed in wild type and least enriched near genes that are not transcribed in wild type (Fig. 6). This suggests that loss of gene-flanking siRNAs is most frequently not the cause for de-repressed gene expression in the rmr6-1 background. To test this, we examined the subset of transcripts whose accumulation changed in rmr6-1 leaves. Loss of gene-flanking siRNAs in the rmr6-1 background was equally strong for transcripts that were induced and repressed in rmr6-1 (Fig. 7B). This result suggests that the loss of gene-proximal RMR6-dependent siRNAs is not globally predictive of any expression change of close genes. However, these are genome-wide trends and exceptions are possible in specific cases.
Figure 7.
Fold changes of RMR6-differentially expressed genes and their flanking RMR6-differentially sRNA loci. A, Percentages and tallies of up- and down-regulated genes by transcript type in rmr6-1 leaves. B, Scatterplots showing RMR6-related changes in sRNA accumulation at gene-flanking loci as a function of RMR6-related changes in transcript accumulation. Only genes that were differentially expressed in rmr6-1 leaves relative to wild type and that had a flanking RMR6-altered sRNA cluster are shown. Inf, infinity, genes that lacked any RNA-seq reads in wild type. For potting and regression purposes, these were assigned log2 values of 10. -inf, genes that lacked any RNA-seq reads in rmr6-1. For potting and regression purposes, these were assigned log2 values of -10. R2 values for the indicated linear regressions are shown.
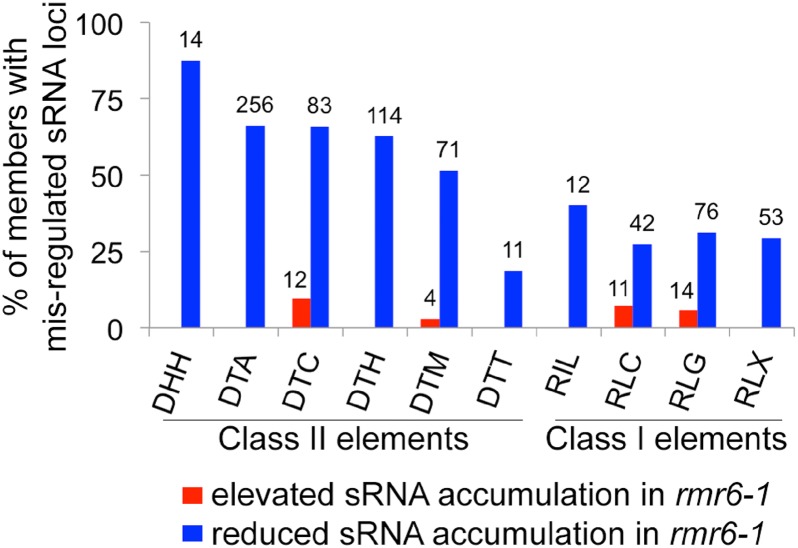
Class II Transposons Are Frequent Sources of RMR6-Dependent siRNAs
Full analysis of TE-derived sRNAs in the maize genome is hampered by the difficulty in aligning sRNAs to very high-copy number elements. As an alternative approach, the original sRNA reads were aligned directly to TE exemplar sequences from the maize transposable element database (http://maizetedb.org/∼maize/). sRNA loci were reannotated with respect to the exemplar TE sequences (Supplemental Data Set S10), and their expression (Supplemental Data Set S11) in the leaf was compared between the wild type and rmr6-1 mutant in control conditions at the first time point of sample collection. Class II (DNA) TEs were frequent sources of RMR6-dependent siRNAs: more than 50% of five of the six class II TE superfamilies produced RMR6-dependent sRNAs (Fig. 8). In contrast, class I TE superfamilies were less-frequent sources of RMR6-dependent sRNAs (Fig. 8). Helitron, hAT, CACTA, PIF/Harbinger, and Mutator DNA TE superfamilies were the most frequent sources of RMR6-dependent siRNAs in leaves.
Figure 8.
Class II TEs are more frequent sources of RMR6-dependent siRNAs relative to class I TEs. Percentages and tallies of TE exemplar sequences that have significant up- or down-regulation of sRNA loci in rmr6-1 leaves. DHH, Helitron; DTA, hAT; DTC, CACTA; DTH, PIF/Harbinger; DTM, Mutator; DTT, Tc1/Mariner; RIL, L1; RLC, Copia; RLG, Gypsy; RLX, Unknown LTR. Only TE superfamilies with more than six members are shown.
An RMR6–Up-Regulated Gene Potentially Controlled by an siRNA-Targeted Repeat
Manual inspection of genome-browser visualizations of our data identified a gene induced in rmr6-1 that also showed mis-regulation of intronic 24-nt siRNA-associated TEs. The GRMZM2G168956 gene, encoding a putative 3-ketoacyl-CoA synthase, was up-regulated in rmr6-1 relative to wild type (log2fold change = 1.73, q-value = 0.04; Supplemental Data Set S9). The mutant showed loss of siRNAs and increased antisense transcription within the GRMZM2G168956 intron (Fig. 9). This intron harbors a class I sofi element fragment. We propose that loss of RMR6-dependent siRNAs targeting this intron allowed de-repression of both the intron-located TE and the associated gene. This observation supports the hypothesis that TE-like sequences and repeats targeted by siRNAs can function as controlling elements influencing the expression of close or overlapping genes (Liu et al., 2004; Kinoshita et al., 2007; Erhard et al., 2013).
Figure 9.
RNA and sRNA read distributions at the RMR6-up-regulated GRMZM2G168956 gene. Schematic representation of chromosome 9 region (9:31491700-31495650) comprising the GRMZM2G168956 gene and its putative sofi controlling element. Arrows indicate gene transcription start site and the orientation of the gene and associated repeats. Red boxes are exons, red lines introns, and green boxes repeats. Class I TEs: RLG, Gypsy; RLC, Copia. Class II TEs: DTM, Mutator. For both RNA-seq and sRNA-seq, the coverage in read per million mapped (RPMM) of control leaf wild-type and rmr6-1 mutant samples at the first time point of sample collection is plotted seperately for the positive (+) and negative (-) genomic strands. Both uniquely and repetitively mapping reads are included. The R2+R3 strand-specific replicate for RNA-seq and the R1 replicate for sRNA-seq are plotted. Only sRNA reads with length in the range 20 to 24 nts are plotted.
Long-Term Drought Stress Induces Down-Regulation of Very Small Number of sRNA Loci in Wild-Type Leaves
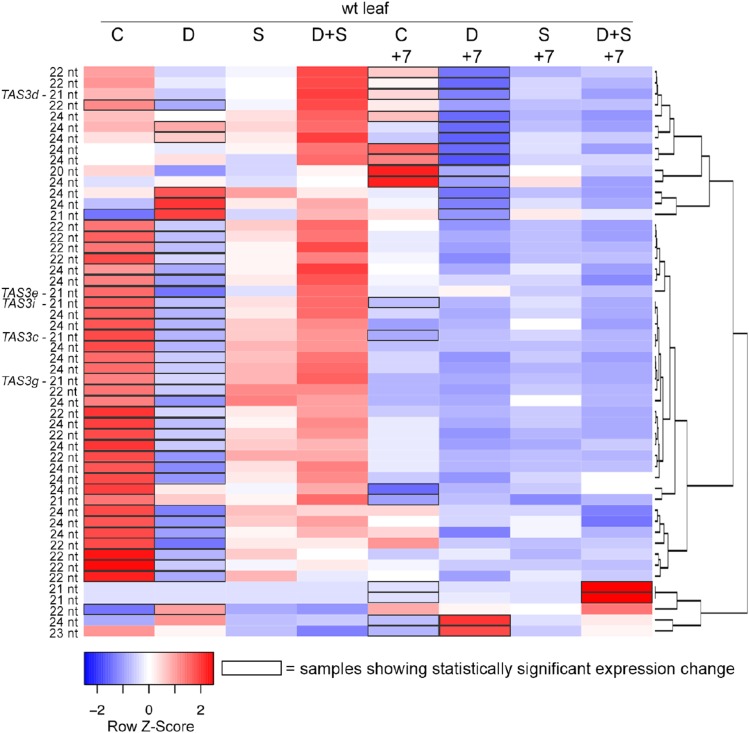
We examined the effects of our long-term stress treatments on leaf-expressed non-MIRNA sRNA loci in wild-type plants. Only 50 of the 251,385 sRNA loci were differentially expressed (0.02%; Supplemental Data Set S12). Most of the differential accumulation was evident in drought-stressed plants (Fig. 10). In leaves of wild-type plants, drought stress mainly caused a decrease of sRNA accumulation both during the stress and after its recovery (Fig. 10). The affected sRNA loci in wild type were a roughly equal mixture of 22-nt and 24-nt–dominated loci. Of the total 23 loci that had a predominant RNA size of 24 nts and that were altered by drought in wild-type leaves, 21 were down-regulated in rmr6-1 (Supplemental Data Set S12). RNA-seq was used to measure transcript accumulation levels in wild-type leaves after 10 d of stress treatments: none of the applied stresses significantly altered the expression of the RMR6 gene (log2fold change < 0.2 and q-value > 0.96 for all the comparisons between one of the stresses and the control). Among the sRNA loci down-regulated in wild type by drought stress (during the treatment or after the recovery), five were TAS3 loci (TAS3c-e, TAS3g, and TAS3i). Plant TAS3 genes are important regulators of leaf polarity and vegetative phase change (Peragine et al., 2004; Nogueira et al., 2007). Our finding that their expression is affected by long-term drought stress implies a connection between control of development and environmental conditions in maize leaves.
Figure 10.
Stress-induced changes in sRNA accumulation. Heat map showing expression levels of sRNA loci that are differentially expressed in at least one pairwise comparison between control and a stress-treated wild-type leaf sample. C, control; D, drought; S, salinity; D+S, drought and salinity stress, after 10 d of treatment and after 7 d of recovery (+7). The mean reads per million mapped values from the three biological replicates were calculated, after which each row was scaled to have a mean of zero and a sd of 1. Black boxes indicate samples with statistically significant expression changes at a false discovery rate of 0.01. Dendrogram shows hierarchical clustering of rows based on Pearson distance. The predominant RNA size of each sRNA locus is indicated on the left.
DISCUSSION
Long-Term Abiotic Stresses Affect Accumulation of Very Few miRNAs and sRNAs
One striking result of our study is that the long-term stresses we applied affected the accumulation of very small numbers of sRNA loci. Considering the stress effects in both the wild-type and rmr6-1 backgrounds, only 11 of 95 mature miRNAs and 101 of 251,385 other sRNA loci had significant changes in accumulation in any of the stress treatments. We conclude that major, global changes in the sRNA profile are not part of the acclimation to the realistic abiotic stresses that were used in our experiments.
In the leaf of wild-type plants, we detected a drought induction of miR156. Similar observations have previously been made in maize (Wei et al., 2009; Kong, 2010; Wang et al., 2014) and in many other plant species (Sunkar et al., 2012). In Arabidopsis, miR156 was proposed to regulate leaf cell number and size (Usami et al., 2009), and its induction under stress conditions was shown to maintain the plant in the juvenile state for a relatively longer period of time, which helps it withstand unfavorable environmental conditions (Cui et al., 2014; Stief et al., 2014). Another two miRNAs involved in the control of normal leaf growth and development were altered by the abiotic stress treatments miR166 and miR396 (Nogueira et al., 2007; Liu et al., 2009). MiR399 controls inorganic phosphate (Pi) homeostasis and its response to Pi is highly specific (Bari et al., 2006). Nonetheless, we detected miR399 down-regulation in the wild-type shoot apical meristematic area during drought stress and in the leaf of rmr6-1 plants during the combined stress. In general, the wild-type and rmr6-1 mutant plants responded differently to the treatments in terms of most effective stress (drought for the wild-type and the combined stress for rmr6-1) and in terms of miRNA families altered, which were mostly different between the two genotypes. These differences were not expected, because miRNAs are transcribed by Pol II (Rogers and Chen, 2013) and in control conditions miRNAs were not altered in rmr6-1. It is possible that these differences might be secondary effects of the mutation.
The small fraction of non-MIRNA sRNA loci that were stress responsive was mostly affected during drought stress, similar to the miRNAs. In leaves of wild-type plants, we detected a bias toward a drought-induced down-regulation of sRNA loci, most of which were dominated by 22-nt or 24-nt RNAs. Only the 24-nt loci were mostly RMR6 dependent (21/23). The expression of the RMR6 gene was not significantly altered by the treatments, and most RMR6-dependent siRNA loci were unaffected by the stresses. These observations suggest that stress-induced changes observed at RMR6-dependent siRNA loci were not caused by a general repression of Pol IV activity during stress. Interestingly, five TAS3 loci were among the sRNA loci down-regulated in drought-stressed plants. This is similar to previous results in Arabidopsis, in which drought and salinity stresses induced the down-regulation of ta-siRNAs produced from TAS1, TAS2, and TAS3 loci. These changes in turn influenced the expression of ARF target genes and downstream genes, finally leading to an altered floral architecture (Matsui et al., 2014).
Previous studies have found much larger numbers of differentially expressed sRNAs under abiotic stresses in Brachypodium (Wang et al., 2015) and foxtail millet (Qi et al., 2013). Our experiments detected a much smaller number of stress-modulated sRNA loci. This could be due to the different stress protocols: the previous studies applied more severe stress conditions, such as polyethylene glycol-simulated drought conditions (Qi et al., 2013), or measured the stress effects after a much shorter period of treatment (Wei et al., 2009; Wang et al., 2015). There also may be species-specific differences within the grasses. The stress protocols used in this work mimicked real-world, progressive environmental stresses; therefore, they could be useful to study stress tolerance mechanisms.
Gene-Proximal, RMR6-Dependent 24-nt siRNAs Generally Do Not Repress Expression of Their Neighboring Genes
RMR6-dependent 24-nt siRNA loci were enriched both up- and downstream of genes of all types, but especially for protein-coding mRNAs. This enrichment was clearly biased toward genes that were most highly expressed in wild-type and away from non-expressed genes. These observations are consistent with previous studies that demonstrated that maize TEs located near genes are targeted by 24-nt siRNA-dependent RdDM, resulting in local deposition of DNA methylation in the asymmetric CHH context in so-called CHH islands (Gent et al., 2013). Maize mutants that disrupt 24-nt siRNA accumulation show loss of CHH islands as well as instances where the flanking regions also lose CG/CHG methylation (Li et al., 2015). Our RNA-seq data show that that major effect on gene expression in the rmr6-1 mutant is a de-repression of genes that are normally lowly- or non-expressed. It is important to note that this observation is inconsistent with the hypothesis that loss of CHH islands causes de-repression; the siRNAs that cause CHH islands are primarily associated with genes that are already highly expressed in the wild type, not with those that are non-expressed. In addition, we found that the smaller numbers of genes that are down-regulated in the rmr6-1 background also show the same loss of gene-proximal siRNAs. Together, these data support the current model by which the gene-flanking 24-nt siRNAs in maize primarily function to reinforce transcriptional silencing of TEs located near active genes, avoiding the spreading of euchromatin into the close TEs (Gent et al., 2013, 2014; Erhard et al., 2015; Li et al., 2015). The similar occupancy of RMR6-dependent, gene-flanking, 24-nt siRNA loci found for protein-coding genes and lncRNAs suggests that the CHH island system of RdDM is engaged near active genes independently of their coding or noncoding nature.
If loss of CHH islands does not generally explain the differentially expressed transcripts in the rmr6-1 background, what does? We believe that many of these differentially expressed genes might be indirect or secondary effects of the rmr6-1 mutation. For example, cells lacking siRNAs might experience stress-like conditions due to the RdDM impairment, which could induce certain suites of genes.
Putative Direct and Indirect RdDM Targets in Maize
Loss of 24-nt siRNAs in rmr6-1 caused relatively low numbers of TEs to become transcriptionally de-repressed, confirming previous findings in mop1-1 and rmr6 (Jia et al., 2009; Madzima et al., 2014; Erhard et al., 2015) and supporting the hypothesis that RdDM is generally dispensable for stable silencing of the vast majority of TEs (Matzke et al., 2015). It was recently demonstrated that at RdDM loci, mop1-1 decreases DNA methylation in all C contexts but does not completely remove CHH methylation (Gent et al., 2014): lower levels of residual CHH methylation might still be sufficient to ensure DNA silencing at most RdDM loci. The down-regulation of the DNA demethylase ROS1/DML1 might also contribute to buffer the effects of siRNA loss (Huettel et al., 2006; Madzima et al., 2014). We studied the first rmr6-1 homozygous generation; therefore, higher decreases in DNA methylation at RdDM loci and a greater number of influenced genes might be expected in subsequent mutant generations (Woodhouse et al., 2006). Finally, it also seems likely that in the deep heterochromatin that typifies many maize TEs, the persistent de novo DNA methylation caused by siRNAs is not required to maintain a stable heterochromatic state.
We found evidence that an rmr6-1-misregulated gene could be a direct RdDM target: an intron-localized TE that lost siRNAs and became transcriptionally active in the rmr6-1 background, coincident with activated transcription of the enclosing gene. The analysis of additional genomic and chromatin features combined with screenings in other available maize RdDM mutants will be necessary to distinguish between direct and indirect RdDM targets and to better elucidate the role of siRNAs in controlling gene silencing.
MATERIALS AND METHODS
Plant Materials, Stress Protocols, and Tissue Collection
Plants from inbred B73 (Zea mays) and rmr6-1 (loss-of-function allele introgressed in B73; Erhard et al., 2009) stocks were grown in pots in a greenhouse with temperatures between 28°C and 30°C during the day and 20°C to 22°C at night and relative humidity between 60% and 80%. Plants were watered to pot capacity until V5/V6 stage (plants with 5/6 visible leaf collars), when stress treatments were applied as described in detail in Morari et al., 2015, with some changes compared to the original protocol. Briefly, control plants were watered with 75% of disposable water at 0.1 dS/m salt concentration; drought-stressed plants with 25% of disposable water at 0.1 dS/m salt concentration; salinity-stressed plants with 75% of disposable water at 15 dS/m salt concentration; drought plus salinity-stressed plants with 25% of disposable water at 15 dS/m salt concentration. To mimic the composition of highly saline soils, a complex mixture of salts (Cristal Sea Marinemix) was added to water to reach the defined electrical conductivity values. Treatments were applied daily, and on the tenth day of treatment the youngest wrapped leaf and the shoot apical meristematic area (shoot apical meristem and a few surrounding developing leaves) were harvested from a subset of plants. The remaining plants were watered to pot capacity to recover from stress, and on the seventh day of recovery the youngest wrapped leaf was harvested from each plant. Leaf samples of four to five plants for each combination of genotype-treatment-sampling time point were pooled together, flash-frozen in liquid nitrogen, and stored at −80°C. The complete experiment was replicated three times.
sRNA Sequencing and de novo Annotation of sRNA Loci
Total RNA was extracted from frozen tissue with the Spectrum Plant Total RNA Kit (SIGMA), using “Protocol A” with 750µL of Binding Solution, to recover more of the small-sized RNA, and subjected to On-Column DNase Digestion (SIGMA). Then 50 sRNA libraries were produced with the TruSeQ sRNA Sample Preparation kit (Illumina) and sequenced on an Illumina HisEquation 2000 platform by the Istituto di Genomica Applicata (Udine, Italy). 3′ and 5′ adapters were removed with cutadapt (“-m 15–discard-untrimmed;” Martin, 2011). Low complexity sequences, containing only two different nucleobases, were removed through a customized Perl script. FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/) was used to verify that in each library the Q score of the 90th percentile of reads was ≥28 across all bases. ShortStack version 3.3 (“–nostitch–mincov 20–readfile [clean .fastq files];” Axtell, 2013b) was used to de novo annotate the sRNA loci first on the maize B73 genome (ZmB73 AGPv3) and then on the TE exemplar sequences annotated in the maize TE database (http://maizetedb.org/∼maize/).
Total RNA Sequencing and Maize Transcriptome Reassembly
Total RNA previously extracted was subjected to Ribo-Zero rRNA Removal kits, “Plant Leaf” (Epicenter-Illumina). R1 samples libraries were produced with TruSeq RNA Library Prep kit (nondirectional), R2 and R3 samples were pooled together, and libraries prepared with the TruSeq Stranded RNA Library Prep kit (directional): libraries were sequenced on an Illumina HisEquation 2000 platform by the Istituto di Genomica Applicata. Reads were trimmed with cutadapt and aligned to the maize B73 reference genome (ZmB73 AGPv3) with TopHat (Trapnell et al., 2009). The directional RNA-seq libraries were used to obtain a reassembly of the maize transcriptome with Cufflinks (RABT mode; Roberts et al., 2011).
Genomic Features Classification and Annotation
Repeats were recovered from the ZmB73 RepeatMasked AGPv3 (excluding “dust” and “trf”) and from the ZmB73 Annotation AGPv3.20 (“transposable_element”). Protein-coding genes annotated in the ZmB73 Annotation AGPv3.20 (“protein_coding”) and confirmed in the transcriptome reassembly used were selected. The annotation of lncRNA transcripts was retrieved from the transcriptome reassembly used, available at the Gene Expression Omnibus (GEO) database at the series GSE71046. Transcripts of the transcriptome reassembly used that overlapped repeats for their entire length on the same strand were annotated as TEs. We used the TE superfamilies classification reported in the ZmB73 RepeatMasked AGPv3, except for unknown TEs (here named “TXX”) and unknown repetitive regions (here named “XXX”). Redundant classifications were found: 236 protein-coding genes had at least one spliced transcript classified as TE, 2,985 protein-coding genes had at least one spliced transcript classified as lncRNA, and 2,007 TE transcripts were also classified as lncRNAs. The 484 chromatin-associated transcripts reported in the Chromatin Database (Gendler et al., 2008) were mapped to the loci of the transcriptome reassembly (85% identity and 95% coverage) to identify their matching transcripts. Best Arabidopsis and rice BLASTP hits (Altschul et al., 1990) of translated genes were obtained from the Phytozome v10.0 Annotation v6a. Gene Ontology functional annotation of genes and transcripts was obtained from Phytozome v10.0 Gene Ontology Annotation v6a. An sRNA locus overlapping repeats for at least 50% of its length, without respect to strand, was annotated as matching repeats.
MicroRNA Analysis and Target Validation for GRMZM2G381709
To analyze the MIRNA loci annotated in miRBase version 20, ShortStack was run on the merged .bam file obtained aligning sRNA to the genome (“--locifile [miRBase MIRNA loci coordinates file]”--mincov 20). To evaluate the consistency of the putative novel MIRNA loci, ShortStack was run separately on the .bam file of each library (“--locifile [putative novel MIRNA loci coordinates file]”). Novel mature miRNAs were aligned against themselves and against miRNAs reported in miRBase with BLASTN (“-strand plus;” Altschul et al., 1990) to find miRNAs belonging to the same family (at most three mismatches). MiRNA targets were predicted with TargetFinder (“-c 2.5” or “-c 3.5;” Fahlgren et al., 2007). The GRMZM2G381709 gene was cloned and sequenced, and its predicted targeting by miR399 was validated through 5′ RACE. Total RNA was extracted from a pool of three first leaves of 7-d-old B73 wild-type seedlings with the Spectrum Plant Total RNA kit (SIGMA), using “Protocol A,” and subjected to On-Column DNase Digestion (SIGMA). cDNA synthesis was performed with the SuperScript III reverse transcriptase kit (Invitrogen) according to the manufacturer’s instructions. One microgram of total RNA was used as a template together with 2 pmol of the gene-specific primer 5′-TCACACTGACCGCAGAAGAC-3′. The GRMZM2G381709 gene was amplified from the cDNA pool using Platinum Taq DNA Polymerase High Fidelity (Invitrogen) with the following specific primers: forward 5′-GCAATTGGACCCGACAGACC-3′ and reverse 5′-TCACACTGACCGCAGAAGAC-3′. The purified product was cloned into pCRII-TOPO vector (Invitrogen) and used as a template for Sanger sequencing. On the total RNA extracted from the pool of first leaves, the 5' RACE assay was performed with the GeneRacer kit (Invitrogen) according to the manufacturer’s instructions but without treating RNA with the calf intestinal phosphatase. The 5′ ends of GRMZM2G381709 were amplified from the cDNA pool with Platinum Taq DNA Polymerase High Fidelity (Invitrogen): in the first PCR the gene-specific forward primer 5′-CACGTCCCAGCTGACCTGAGGGATCAG-3′ and the GeneRacer 5′ Primer were used; in the nested PCR the gene-specific forward primer 5′-GCCAATGCTCTCATCCAGACTGGAC-3′ and the GeneRacer 5′ Nested Primer were used. PCR products were gel purified, incubated with Taq DNA Polymerase recombinant (Invitrogen) at 72°C for 15 min to add 3′ A-overhangs, and cloned into pCRT4-TOPO vector (Invitrogen) and used as a template for Sanger sequencing.
Co-Occupancy Analysis and Distribution of sRNA Loci in Gene Flanking Regions
In the co-occupancy analysis, the count of observed non-redundant overlapping nts, without respect to strand, between an sRNA locus category and a genomic feature was obtained with a customized Perl script. The expected number of non-redundant overlapping nts, without respect to strand, was calculated as follows: ([total non-redundant nt of genomic feature/genome size]*[total non-redundant nt of sRNA loci category/genome size])*genome size. Enrichment/depletion was calculated as follows: log2 (observed overlapping nt/expected overlapping nt). To plot the distribution of sRNA loci in gene flanking regions, protein-coding genes, TE, and lncRNA transcripts were divided into five groups based on their RNA-seq determined expression. The first group included non-expressed features (“RPKM = 0”), corresponding to 21.6%, 72.9%, and 51.3% of the total protein-coding genes, TE, and lncRNA transcripts, respectively. The other four groups included expressed features, divided into four equivalent quartiles, from lowest to highest RPKM values: (1) “low” expression (0 < RPKM ≤ 0.41 for protein-coding genes, 0 < RPKM ≤ 0.04 for TEs and 0 < RPKM ≤ 0.24 for lncRNAs); (2) “mid-low” expression (0.41 < RPKM ≤ 3.04 for protein-coding genes, 0.04 < RPKM ≤ 0.21 for TEs and 0.24 < RPKM ≤ 0.96 for lncRNAs; (3) “mid-high” expression (3.04 < RPKM ≤ 9.07 for protein-coding genes, 0.21 < RPKM ≤ 1.21 for TEs and 0.96 < RPKM ≤ 3.39 for lncRNAs); and (4) “high” expression (9.07 < RPKM ≤ 13530.58 for protein-coding genes, 1.21 < RPKM ≤ 90685.5 for TEs and 3.39 < RPKM ≤ 1108.09 for lncRNAs). The BEDTools (Quinlan and Hall, 2010) function “coverageBed” (“-d”) was used to calculate the presence/absence of 23- to 24-nt siRNA loci in each individual position of gene flanking regions, and a customized Perl script was used to calculate the fraction of genes for each group having close siRNA loci in each 50-bp interval of their flanking regions.
Differential Expression Analysis
Mature miRNA counts and sRNA loci counts were subjected to pairwise differential expression analysis with edgeR: counts were normalized based on total miRNAs or sRNA loci counts, the FDR was set <1% with the Benjamini and Hochberg’s algorithm, and only miRNAs/sRNA loci with log2 fold-change ≥ |1| were selected. Gene counts were subjected to pairwise differential expression analysis using Cuffdiff with the transcriptome reassembly used in this work (“--multi-read-correct--library-type fr-unstranded--compatible-hits-norm--dispersion-method per-condition--library-norm-method quartile”, FDR < 5% as default, http://cole-trapnell-lab.github.io/cufflinks/), and only loci with log2 fold-change ≥ |1| were selected. The distribution of RNA-seq reads (R2+R3 directional sequencing) within each individual RMR6-altered gene was visualized with IGV (Robinson et al., 2011) to verify that the vast majority of them (89.2%) had homogeneous read distribution across their exons according to their annotated strand. The RMR6-altered genes were divided in: protein-coding, TE, lncRNA (for which at least one transcript isoform was a TE or a lncRNA respectively), and the remaining unclassified genes. For 46 RMR6-altered genes, this classification was redundant. Overlaps between RMR6-altered sRNA loci and flanking regions of genes (total or RMR6-altered) were calculated with the BEDTools function “intersectBed” (“-wo”).
Accession Numbers
Small RNA sequencing data have been deposited at NCBI GEO under accession GSE70487. Total RNA sequencing data, including the transcriptome reassembly used in this work, have also been deposited at NCBI GEO under accession number GSE71046. The cDNA sequence of the GRMZM2G381709 gene, determined by cloning and Sanger-sequencing, is deposited at GenBank under accession number KT345709.
Supplemental Data
The following supplemental materials are available.
Supplemental Data Set S1. Summary of sRNA sequencing.
Supplemental Data Set S2. Details of identified sRNA loci in the maize genome.
Supplemental Data Set S3. Raw read counts of identified sRNA loci in the maize genome.
Supplemental Data Set S4. miRBase MIRNA loci evaluation with sRNA-seq data.
Supplemental Data Set S5. Maize MIRNA loci identified in this study.
Supplemental Data Set S6. Details of maize MIRNA loci identified in this study.
Supplemental Data Set S7. Co-occupancy analysis.
Supplemental Data Set S8. Strandedness of RMR6-dependent siRNA loci (predominant RNA size of 23 nts and 24 nts) overlapping with flanking regions of genomic features.
Supplemental Data Set S9. Differentially expressed genes in rmr6-1 mutant compared to wild type (control conditions at the first time point of sample collection).
Supplemental Data Set S10. Details of identified sRNA loci in TE exemplar sequences.
Supplemental Data Set S11. Raw counts of identified sRNA loci in TE exemplar sequences.
Supplemental Data Set S12. Differentially expressed sRNA loci (nonMIRNA loci) altered by stress treatments and/or time point +7.
Supplementary Material
Acknowledgments
We thank E. De Paoli and M. Stam for helpful discussions on this report.
Glossary
- dsRNA
double-stranded RNA
- GEO
Gene Expression Omnibus
- lncRNA
long noncoding RNA
- nt
nucleotide
- Pol
polymerase
- RdDM
RNA-directed DNA methylation
- siRNA
small interfering RNA
- sRNA
small RNA
- TE
transposable element
- TGS
transcriptional gene silencing
- 5′-UTR
5′-untranslated region
Footnotes
This work was supported by EU FP7 Project AENEAS and Italian EPIGEN Flagship CNR Project. A.L. was granted by a CARIPARO PhD fellowship.
Articles can be viewed without a subscription.
References
- Agorio A, Vera P (2007) ARGONAUTE4 is required for resistance to Pseudomonas syringae in Arabidopsis. Plant Cell 19: 3778–3790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen E, Xie Z, Gustafson AM, Carrington JC (2005) microRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell 121: 207–221 [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215: 403–410 [DOI] [PubMed] [Google Scholar]
- Axtell MJ. (2013a) Classification and comparison of small RNAs from plants. Annu Rev Plant Biol 64: 137–159 [DOI] [PubMed] [Google Scholar]
- Axtell MJ. (2013b) ShortStack: comprehensive annotation and quantification of small RNA genes. RNA 19: 740–751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao N, Lye KW, Barton MK (2004) MicroRNA binding sites in Arabidopsis class III HD-ZIP mRNAs are required for methylation of the template chromosome. Dev Cell 7: 653–662 [DOI] [PubMed] [Google Scholar]
- Bari R, Datt Pant B, Stitt M, Scheible W-R (2006) PHO2, microRNA399, and PHR1 define a phosphate-signaling pathway in plants. Plant Physiol 141: 988–999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baucom RS, Estill JC, Chaparro C, Upshaw N, Jogi A, Deragon JM, Westerman RP, Sanmiguel PJ, Bennetzen JL (2009) Exceptional diversity, non-random distribution, and rapid evolution of retroelements in the B73 maize genome. PLoS Genet 5: e1000732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blevins T, Podicheti R, Mishra V, Marasco M, Tang H, Pikaard CS (2015) Identification of Pol IV and RDR2-dependent precursors of 24 nt siRNAs guiding de novo DNA methylation in Arabidopsis. eLife 4: e09591 10.7554/eLife.09591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calderón-Vázquez C, Sawers RJH, Herrera-Estrella L (2011) Phosphate deprivation in maize: genetics and genomics. Plant Physiol 156: 1067–1077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui L-G, Shan J-X, Shi M, Gao J-P, Lin H-X (2014) The miR156-SPL9-DFR pathway coordinates the relationship between development and abiotic stress tolerance in plants. Plant J 80: 1108–1117 [DOI] [PubMed] [Google Scholar]
- Dowen RH, Pelizzola M, Schmitz RJ, Lister R, Dowen JM, Nery JR, Dixon JE, Ecker JR (2012) Widespread dynamic DNA methylation in response to biotic stress. Proc Natl Acad Sci USA 109: E2183–E2191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erhard KF Jr, Parkinson SE, Gross SM, Barbour J-ER, Lim JP, Hollick JB (2013) Maize RNA polymerase IV defines trans-generational epigenetic variation. Plant Cell 25: 808–819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erhard KF Jr, Stonaker JL, Parkinson SE, Lim JP, Hale CJ, Hollick JB (2009) RNA polymerase IV functions in paramutation in Zea mays. Science 323: 1201–1205 [DOI] [PubMed] [Google Scholar]
- Erhard KF Jr, Talbot J-ERB, Deans NC, McClish AE, Hollick JB (2015) Nascent transcription affected by RNA polymerase IV in Zea mays. Genetics 199: 1107–1125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahlgren N, Howell MD, Kasschau KD, Chapman EJ, Sullivan CM, Cumbie JS, Givan SA, Law TF, Grant SR, Dangl JL, Carrington JC (2007) High-throughput sequencing of Arabidopsis microRNAs: evidence for frequent birth and death of MIRNA genes. PLoS One 2: e219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendler K, Paulsen T, Napoli C (2008) ChromDB: the chromatin database. Nucleic Acids Res 36: D298–D302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gent JI, Ellis NA, Guo L, Harkess AE, Yao Y, Zhang X, Dawe RK (2013) CHH islands: de novo DNA methylation in near-gene chromatin regulation in maize. Genome Res 23: 628–637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gent JI, Madzima TF, Bader R, Kent MR, Zhang X, Stam M, McGinnis KM, Dawe RK (2014) Accessible DNA and relative depletion of H3K9me2 at maize loci undergoing RNA-directed DNA methylation. Plant Cell 26: 4903–4917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale CJ, Erhard KF Jr, Lisch D, Hollick JB (2009) Production and processing of siRNA precursor transcripts from the highly repetitive maize genome. PLoS Genet 5: e1000598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W, Wang T, Xu J, Li H (2014) MicroRNA mediates DNA methylation of target genes. Biochem Biophys Res Commun 444: 676–681 [DOI] [PubMed] [Google Scholar]
- Huettel B, Kanno T, Daxinger L, Aufsatz W, Matzke AJM, Matzke M (2006) Endogenous targets of RNA-directed DNA methylation and Pol IV in Arabidopsis. EMBO J 25: 2828–2836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito H, Gaubert H, Bucher E, Mirouze M, Vaillant I, Paszkowski J (2011) An siRNA pathway prevents transgenerational retrotransposition in plants subjected to stress. Nature 472: 115–119 [DOI] [PubMed] [Google Scholar]
- Jia Y, Lisch DR, Ohtsu K, Scanlon MJ, Nettleton D, Schnable PS (2009) Loss of RNA-dependent RNA polymerase 2 (RDR2) function causes widespread and unexpected changes in the expression of transposons, genes, and 24-nt small RNAs. PLoS Genet 5: e1000737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita Y, Saze H, Kinoshita T, Miura A, Soppe WJJ, Koornneef M, Kakutani T (2007) Control of FWA gene silencing in Arabidopsis thaliana by SINE-related direct repeats. Plant J 49: 38–45 [DOI] [PubMed] [Google Scholar]
- Kong Y. (2010) Differential expression of microRNAs in maize inbred and hybrid lines during salt and drought stress. Am J Plant Sci 01: 69–76 [Google Scholar]
- Li Q, Gent JI, Zynda G, Song J, Makarevitch I, Hirsch CD, Hirsch CN, Dawe RK, Madzima TF, McGinnis KM, Lisch D, Schmitz RJ, et al. (2015) RNA-directed DNA methylation enforces boundaries between heterochromatin and euchromatin in the maize genome. Proc Natl Acad Sci USA 112: 14728–14733 10.1073/pnas.1514680112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D, Song Y, Chen Z, Yu D (2009) Ectopic expression of miR396 suppresses GRF target gene expression and alters leaf growth in Arabidopsis. Physiol Plant 136: 223–236 [DOI] [PubMed] [Google Scholar]
- Liu H, Qin C, Chen Z, Zuo T, Yang X, Zhou H, Xu M, Cao S, Shen Y, Lin H, He X, Zhang Y, et al. (2014) Identification of miRNAs and their target genes in developing maize ears by combined small RNA and degradome sequencing. BMC Genomics 15: 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, He Y, Amasino R, Chen X (2004) siRNAs targeting an intronic transposon in the regulation of natural flowering behavior in Arabidopsis. Genes Dev 18: 2873–2878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madzima TF, Huang J, McGinnis KM (2014) Chromatin structure and gene expression changes associated with loss of MOP1 activity in Zea mays. Epigenetics 9: 1047–1059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marí-Ordóñez A, Marchais A, Etcheverry M, Martin A, Colot V, Voinnet O (2013) Reconstructing de novo silencing of an active plant retrotransposon. Nat Genet 45: 1029–1039 [DOI] [PubMed] [Google Scholar]
- Martin M. (2011) Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal 17: 10 [Google Scholar]
- Martín-Trillo M, Cubas P (2010) TCP genes: a family snapshot ten years later. Trends Plant Sci 15: 31–39 [DOI] [PubMed] [Google Scholar]
- Matsui A, Mizunashi K, Tanaka M, Kaminuma E, Nguyen AH, Nakajima M, Kim J-M, Nguyen DV, Toyoda T, Seki M (2014) tasiRNA-ARF pathway moderates floral architecture in arabidopsis plants subjected to drought stress. BioMed Res Int 2014: 303451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matzke MA, Kanno T, Matzke AJ (2015) RNA-directed DNA methylation: the evolution of a complex epigenetic pathway in flowering plants. Annu Rev Plant Biol 66: 243–267 [DOI] [PubMed] [Google Scholar]
- Matzke MA, Mosher RA (2014) RNA-directed DNA methylation: an epigenetic pathway of increasing complexity. Nat Rev Genet 15: 394–408 [DOI] [PubMed] [Google Scholar]
- McCue AD, Nuthikattu S, Slotkin RK (2013) Genome-wide identification of genes regulated in trans by transposable element small interfering RNAs. RNA Biol 10: 1379–1395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montes RAC, de Fátima Rosas-Cárdenas F, De Paoli E, Accerbi M, Rymarquis L a, Mahalingam G, Marsch-Martínez N, Meyers BC, Green PJ, de Folter S (2014) Sample sequencing of vascular plants demonstrates widespread conservation and divergence of microRNAs. Nat Commun 5: 1–15 [DOI] [PubMed] [Google Scholar]
- Morari F, Meggio F, Lunardon A, Scudiero E, Forestan C, Farinati S, Varotto S (2015) Time course of biochemical, physiological, and molecular responses to field-mimicked conditions of drought, salinity, and recovery in two maize lines. Front Plant Sci 6: 314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosher RA, Schwach F, Studholme D, Baulcombe DC (2008) PolIVb influences RNA-directed DNA methylation independently of its role in siRNA biogenesis. Proc Natl Acad Sci USA 105: 3145–3150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogueira FTS, Madi S, Chitwood DH, Juarez MT, Timmermans MCP (2007) Two small regulatory RNAs establish opposing fates of a developmental axis. Genes Dev 21: 750–755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuthikattu S, McCue AD, Panda K, Fultz D, DeFraia C, Thomas EN, Slotkin RK (2013) The initiation of epigenetic silencing of active transposable elements is triggered by RDR6 and 21-22 nucleotide small interfering RNAs. Plant Physiol 162: 116–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peragine A, Yoshikawa M, Wu G, Albrecht HL, Poethig RS (2004) SGS3 and SGS2/SDE1/RDR6 are required for juvenile development and the production of trans-acting siRNAs in Arabidopsis. Genes Dev 18: 2368–2379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popova OV, Dinh HQ, Aufsatz W, Jonak C (2013) The RdDM pathway is required for basal heat tolerance in Arabidopsis. Mol Plant 6: 396–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi X, Xie S, Liu Y, Yi F, Yu J (2013) Genome-wide annotation of genes and noncoding RNAs of foxtail millet in response to simulated drought stress by deep sequencing. Plant Mol Biol 83: 459–473 [DOI] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM (2010) BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26: 841–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts A, Pimentel H, Trapnell C, Pachter L (2011) Identification of novel transcripts in annotated genomes using RNA-Seq. Bioinformatics 27: 2325–2329 [DOI] [PubMed] [Google Scholar]
- Robinson J, Thorvaldsdóttir H, Winckler W, Guttman M, Lander E, Getz G, Mesirov J (2011) Integrative Genomics Viewer. Nat Biotechnol 29: 24–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers K, Chen X (2013) Biogenesis, turnover, and mode of action of plant microRNAs. Plant Cell 25: 2383–2399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnable PS, Ware D, Fulton RS, Stein JC, Wei F, Pasternak S, Liang C, Zhang J, Fulton L, Graves TA, Minx P, Reily AD, et al. (2009) The B73 maize genome: complexity, diversity, and dynamics. Science 326: 1112–1115 [DOI] [PubMed] [Google Scholar]
- Shen Y, Jiang Z, Lu S, Lin H, Gao S, Peng H, Yuan G, Liu L, Zhang Z, Zhao M, Rong T, Pan G (2013) Combined small RNA and degradome sequencing reveals microRNA regulation during immature maize embryo dedifferentiation. Biochem Biophys Res Commun 441: 425–430 [DOI] [PubMed] [Google Scholar]
- Slotkin RK, Freeling M, Lisch D (2005) Heritable transposon silencing initiated by a naturally occurring transposon inverted duplication. Nat Genet 37: 641–644 [DOI] [PubMed] [Google Scholar]
- Stief A, Altmann S, Hoffmann K, Pant BD, Scheible W-R, Bäurle I (2014) Arabidopsis miR156 regulates tolerance to recurring environmental stress through SPL transcription factors. Plant Cell 26: 1792–1807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunkar R, Li YF, Jagadeeswaran G (2012) Functions of microRNAs in plant stress responses. Trends Plant Sci 17: 196–203 [DOI] [PubMed] [Google Scholar]
- Taylor RS, Tarver JE, Hiscock SJ, Donoghue PCJ (2014) Evolutionary history of plant microRNAs. Trends Plant Sci 19: 175–182 [DOI] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL (2009) TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25: 1105–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tricker PJ, Gibbings JG, Rodríguez López CM, Hadley P, Wilkinson MJ (2012) Low relative humidity triggers RNA-directed de novo DNA methylation and suppression of genes controlling stomatal development. J Exp Bot 63: 3799–3813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usami T, Horiguchi G, Yano S, Tsukaya H (2009) The more and smaller cells mutants of Arabidopsis thaliana identify novel roles for SQUAMOSA PROMOTER BINDING PROTEIN-LIKE genes in the control of heteroblasty. Development 136: 955–964 [DOI] [PubMed] [Google Scholar]
- Wang HL, Dinwiddie BL, Lee H, Chekanova JA (2015) Stress-induced endogenous siRNAs targeting regulatory intron sequences in Brachypodium. RNA 21: 145–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Elling AA, Li X, Li N, Peng Z, He G, Sun H, Qi Y, Liu XS, Deng XW (2009) Genome-wide and organ-specific landscapes of epigenetic modifications and their relationships to mRNA and small RNA transcriptomes in maize. Plant Cell 21: 1053–1069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y-G, An M, Zhou S-F, She Y-H, Li W-C, Fu F-L (2014) Expression profile of maize microRNAs corresponding to their target genes under drought stress. Biochem Genet 52: 474–493 [DOI] [PubMed] [Google Scholar]
- Wei L, Zhang D, Xiang F, Zhang Z (2009) Differentially expressed miRNAs potentially involved in the regulation of defense mechanism to drought stress in maize seedlings. Int J Plant Sci 170: 979–989 [Google Scholar]
- Woodhouse MR, Freeling M, Lisch D (2006) The mop1 (mediator of paramutation1) mutant progressively reactivates one of the two genes encoded by the MuDR transposon in maize. Genetics 172: 579–592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, Zhou H, Zhang Q, Zhang J, Ni F, Liu C, Qi Y (2010) DNA methylation mediated by a microRNA pathway. Mol Cell 38: 465–475 [DOI] [PubMed] [Google Scholar]
- Xin M, Yang R, Yao Y, Ma C, Peng H, Sun Q, Wang X, Ni Z (2014) Dynamic parent-of-origin effects on small interfering RNA expression in the developing maize endosperm. BMC Plant Biol 14: 192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai J, Bischof S, Wang H, Feng S, Lee TF, Teng C, Chen X, Park SY, Liu L, Gallego-Bartolome J, Liu W, Henderson IR, et al. (2015) A one precursor one siRNA model for Pol IV-dependent siRNA biogenesis. Cell 163: 445–455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai L, Liu Z, Zou X, Jiang Y, Qiu F, Zheng Y, Zhang Z (2013) Genome-wide identification and analysis of microRNA responding to long-term waterlogging in crown roots of maize seedlings. Physiol Plant 147: 181–193 [DOI] [PubMed] [Google Scholar]
- Zhang L, Chia JM, Kumari S, Stein JC, Liu Z, Narechania A, Maher CA, Guill K, McMullen MD, Ware D (2009) A genome-wide characterization of microRNA genes in maize. PLoS Genet 5: e1000716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Henderson IR, Lu C, Green PJ, Jacobsen SE (2007) Role of RNA polymerase IV in plant small RNA metabolism. Proc Natl Acad Sci USA 104: 4536–4541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Xu Z, Mo Q, Zou C, Li W, Xu Y, Xie C (2013) Combined small RNA and degradome sequencing reveals novel miRNAs and their targets in response to low nitrate availability in maize. Ann Bot (Lond) 112: 633–642 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.