TEMPRANILLO repress epidermal trichome initiation by controlling gibberellin accumulation in the mesophyll as well as by acting as link between GA and CK signaling in the epidermis.
Abstract
Plant trichomes are defensive specialized epidermal cells. In all accepted models, the epidermis is the layer involved in trichome formation, a process controlled by gibberellins (GAs) in Arabidopsis rosette leaves. Indeed, GA activates a genetic cascade in the epidermis for trichome initiation. Here we report that TEMPRANILLO (TEM) genes negatively control trichome initiation not only from the epidermis but also from the leaf layer underneath the epidermis, the mesophyll. Plants over-expressing or reducing TEM specifically in the mesophyll, display lower or higher trichome numbers, respectively. We surprisingly found that fluorescently labeled GA3 accumulates exclusively in the mesophyll of leaves, but not in the epidermis, and that TEM reduces its accumulation and the expression of several newly identified GA transporters. This strongly suggests that TEM plays an essential role, not only in GA biosynthesis, but also in regulating GA distribution in the mesophyll, which in turn directs epidermal trichome formation. Moreover, we show that TEM also acts as a link between GA and cytokinin signaling in the epidermis by negatively regulating downstream genes of both trichome formation pathways. Overall, these results call for a re-evaluation of the present theories of trichome formation as they reveal mesophyll essential during epidermal trichome initiation.
Trichomes are epidermal cell protrusions present in most of the vascular plants surfaces that defend the plant against insect herbivores, UV light, and water loss (Traw and Bergelson, 2003; Olsson et al., 2009). Many plant species respond to insect damage by increasing the density and/or number of trichomes on new leaves (Traw and Bergelson, 2003). In Arabidopsis, leaf trichomes are unicellular structures, whose development has been used as a model for addressing crucial questions in plant biology such as control of cell fate specification and differentiation as well as plant defense mechanisms (Traw and Bergelson, 2003; Olsson et al., 2009; Hülskamp, 2004; Gilding and Marks, 2010). Once an epidermal cell precursor is specified to enter the trichome pathway, an elaborated and well-regulated morphogenetic cell transformation occurs in order for it to become a trichome (Gilding and Marks, 2010). First, radial cell expansion occurs centered on the external face of the epidermal trichome precursor that develops into an elongated stalk (Szymanski et al., 1998). Then, cell expansion on the stalk produces branch initiation and growth. Finally, once trichome expansion is complete, the cell wall gets thicker and many papillae form on the outer surface of the trichome, resulting in a mature trichome (Gilding and Marks, 2010).
Trichome proliferation and development process involves diverse genes at different regulatory pathways (Payne et al., 2000; Bernhardt et al., 2005). These include a multimeric complex formed by the R2R3 MYB GLABROUS1 (GL1); two redundant trichome formation bHLH proteins, GLABRA3 (GL3) and ENHANCER OF GLABRA3 (EGL3); and a WD-40 repeat containing protein TRANSPARENT TESTA GLABROUS1 (TTG1) (Bernhardt et al., 2005). Mutations in GL1, TTG1, and both GL3 and EGL3, result in a significant loss of trichomes per leaf (Payne et al., 2000; Bernhardt et al., 2005). This complex has a role not only in trichome initiation but also in later stages of trichome development, because mutations lead to smaller and less-branched trichomes (Payne et al., 2000). Furthermore, hormones play an important role in trichome initiation by controlling essential downstream genes (Schellmann et al., 2002; Gan et al., 2006; Zhao et al., 2008a). In particular, gibberellins (GAs) and cytokinins (CK) overlap in stimulating this process (Nemhauser et al., 2006; Gan et al., 2007; D’Aloia et al., 2011). GAs are required in the epidermis for trichome proliferation in rosette leaves, stem, and inflorescences (Chien and Sussex, 1996; Perazza et al., 1998; An et al., 2012), while CK action is limited to trichome initiation in upper inflorescence organs, including cauline leaves, stems, and sepals (Gan et al., 2007). The GA-dependent pathway acts partially through GLABROUS INFLORESCENCE STEMS (GIS), a C2H2 transcription factor, which positively regulates the trichome activation complex formed by GL1, GL3, EGL3, and TTG1 in the epidermis (Payne et al., 2000; Zhao et al., 2008a). Furthermore, within the CK-dependent pathway, trichome production control requires two C2H2 transcription factors, GLABROUS INFLORESCENCE STEMS2 (GIS2) and ZINC FINGER PROTEIN8 (ZFP8) (Gan et al., 2007). Both genes, ZFP8 and GIS2, are similarly expressed at early stages of inflorescence development but differentially expressed in inflorescence organs (Gan et al., 2007). Mutations in ZFP8 display a reduction in trichome density on the upper cauline leaves and branches, but not in vegetative organs; while mutations in GIS2 give rise to trichome reduction mainly in flowers (Gan et al., 2007). Similarly to GIS, GIS2, and ZFP8 also mediate the regulation of trichome initiation by GA; however, GIS does not play a significant role in CK response (Gan et al., 2007). Therefore, GIS, GIS2, and ZFP8 play partially redundant roles in inflorescence trichome initiation, and integration of CK and GA requires the action of all these genes.
In Arabidopsis, both epidermal GA- and CK-dependent trichome regulatory pathways converge on the activation of GLABROUS2 (GL2), considered as the universal promoter of trichome initiation (Szymanski et al., 1998; Lin and Aoyama, 2012). GL2 is a homeodomain transcription factor required for trichome formation as well as for subsequent trichome morphogenesis phases such as cell expansion, branching, and maturation of trichome cell wall (Szymanski et al., 1998). Indeed, GL2 expression profile shows spatio-temporal variation either in leaves and trichomes. GL2 is strongly expressed in developing trichomes and surrounding cells; however, its expression persists but decreases in mature trichomes (Szymanski et al., 1998).
TEM1 and TEM2 transcription factors, two proteins belonging to the small plant-specific RELATED TO ABI3 AND VP1 (RAV) family, have been previously identified as repressors of floral induction (Castillejo and Pelaz, 2008; Osnato et al., 2012). TEM1 and TEM2 act redundantly to negatively control floral induction by controlling and integrating both the photoperiod and GA signaling pathways during long days and short days (Castillejo and Pelaz, 2008; Osnato et al., 2012). As other transcription factors, it has been suggested that these may control different biological aspects during plant development (Matías-Hernández et al., 2014). Here we report that TEM gene products play pivotal roles during the repression of another developmental process: trichome initiation. TEM1 and TEM2 directly repress trichome initiation by controlling GA accumulation and distribution in the leaf mesophyll as well as by integrating both GA- and CK-dependent regulatory pathways in the epidermis.
RESULTS
TEM1 and TEM2 Repress Trichome Initiation in Leaves and Inflorescences
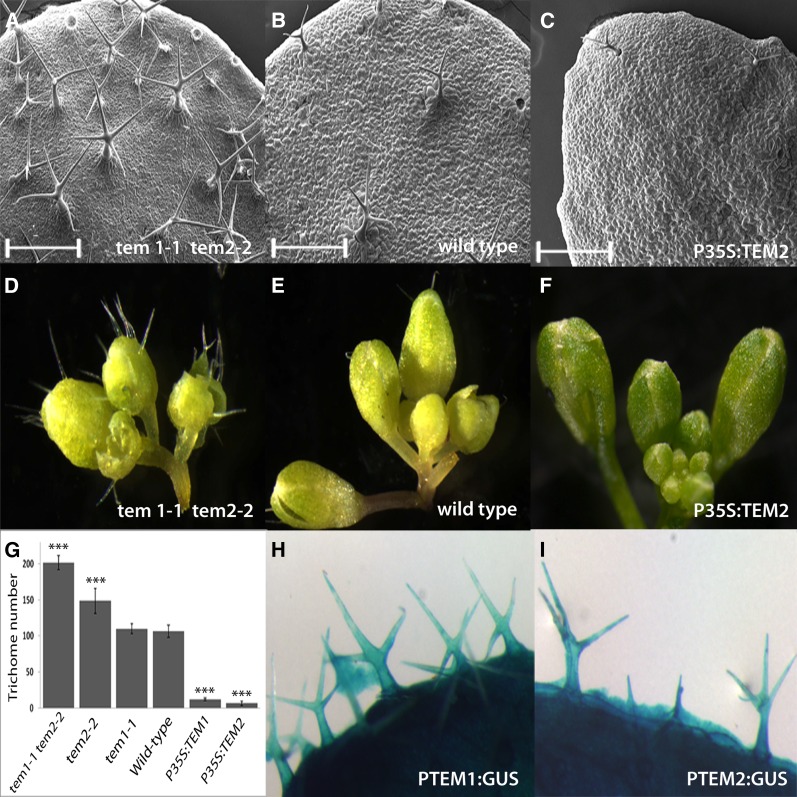
TEM proteins belong to the unique RAV transcription factor family, characterized by the presence of two different DNA-binding domains, an AP2 and a B3 domain (Riechmann et al., 2000; Matías-Hernández et al., 2014). Since TEM1 and TEM2 are consistently expressed in diverse but specific plant tissues and at diverse moments during Arabidopsis development (Castillejo and Pelaz, 2008; Osnato et al., 2012), it has been suggested that TEM may control other plant developmental processes (Matías-Hernández et al., 2014). To investigate possible contributions of TEM toward additional biological processes, phenotypical analyses of diverse TEM loss of function and gain of function were performed. As results show, it was clearly observed that the different mutants and over-expressors analyzed were defected in trichome initiation number and density (Fig. 1, A to G, Supplemental Fig. 1, and Supplemental Table S1). Although tem1-1 mutant did not show a clear trichome phenotype, single tem2-2 mutants produced more trichomes in rosette leaves than wild-type plants (Fig. 1G). This effect was stronger in tem1-1 tem2-2 and in a previously characterized RNAi lines that partially silence both TEM genes (Castillejo and Pelaz, 2008), not only in rosette leaves, but also in inflorescences (Fig. 1, A, D, and G, and Supplemental Fig. S1). In contrast, TEM1 and TEM2 over-expressors (P35S:TEM1, P35S:TEM2) showed almost glabrous leaves, stems, and flowers (Fig. 1, C, F, and G, Supplemental Fig. S1, and Supplemental Table S1). Taken together, these data indicate that, similarly to the effect on floral induction, TEM1 and TEM2 act redundantly to repress trichome initiation. In addition, GUS expression analyses using PTEM1:GUS and PTEM2:GUS reporter lines were conducted. The expression of the β-glucuronidase gene driven by the promoters of TEM1 (Castillejo and Pelaz, 2008) and TEM2 was clearly detected in both the epidermis and the mesophyll layers of rosette leaves (Supplemental Fig. S2). As expected, GUS activity was also detected in trichomes of rosette leaves at different stages of development (Fig. 1, H and I, and Supplemental Fig. S2) consistent with the effect of TEM1 and TEM2 on trichome initiation.
Figure 1.
TEMPRANILLO affects trichome number. A, B, and C, SEM of tem1-1 tem2-2 (A), wild-type (B), and P35S:TEM2 rosette leaves (C). Scale bars represent 500 μm (A–C). D, E, and F, Inflorescences of the same genotypes showing sepal trichomes. tem1-1 tem2-2 shows more trichomes (A and D) than the wild-type (B and E), whereas P35S:TEM2 (C and F) are almost glabrous. G, Trichome number in the 5th-6th rosette leaves of 21 DAG plants of different genetic backgrounds, as indicated. Error bars indicate sd of the mean number of trichomes. Asterisks indicate statistically significant differences (* P ≤ 0.05, ** P ≤ 0.01, *** P ≤ 0.001) obtained using Tukey’s range test. H and I, GUS staining of PTEM1:GUS (H) and PTEM2:GUS (I) reveals a strong expression of both TEM genes in trichomes. Pictures were taken at same magnification (D–F, H, and I).
TEM Controls Trichome Initiation by Affecting GA Biosynthesis
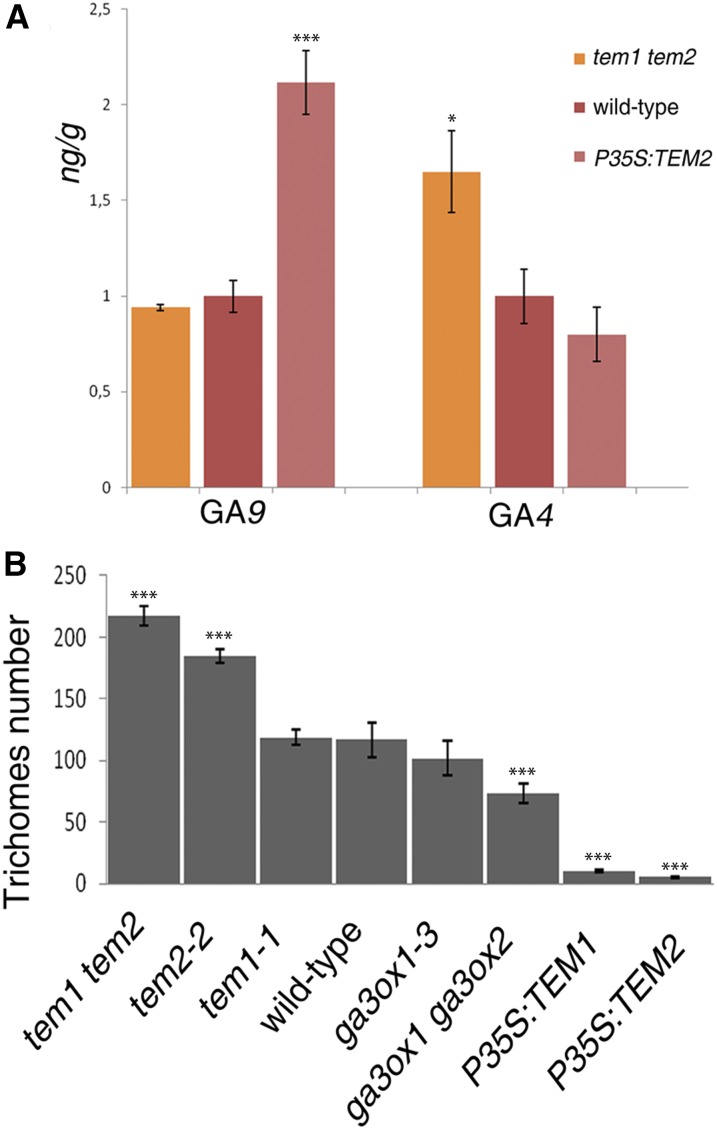
TEM transcription factors are known to repress GA biosynthesis during floral induction (Osnato et al., 2012). TEM1 directly represses the expression of GIBBERELLIN3 OXIDASE1 [GA3ox1) and GA3ox2 (Osnato et al., 2012)], which encode the enzymes that transform GA9 to bioactive GA4 (Mitchum et al., 2006). To further explore whether TEM action affects plant GA biosynthesis, hormone measurements were conducted in Arabidopsis plants collected five days after bolting (DAB). GA4 was significantly increased in tem1-1 tem2-2 plants while it decreased in P35S:TEM2 (Fig. 2A). On the other hand, inactive GA9 accumulated in P35S:TEM2 to higher levels than in the wild type, indicating reduced conversion of GA9 into GA4 in plants over-expressing TEM2 (Fig. 2A). GAs are involved in diverse biological processes and these results may suggest that TEM negative control of GA biosynthesis may affect other important processes within the plant development, including trichome proliferation in the epidermis. Indeed, it is known that ga3ox1-3 and ga3ox2-1 mutants produce almost undetectable levels of bioactive GA4 (Mitchum et al., 2006). Accordingly, these mutants displayed fewer trichomes, in number and density, on the adaxial side of rosette leaves than wild-type plants (Fig. 2B and Supplemental Table S1), and when GA is exogenously added, trichome production is restored (Chien and Sussex, 1996; Traw and Bergelson, 2003). Interestingly, the reduction in trichome number was much stronger in P35S:TEM1 and P35S:TEM2 leaves than in ga3ox1-3 ga3ox2-1 leaves (Fig. 2B); suggesting that TEM affects trichome formation contributing to the control not only of GA biosynthesis but also of other trichome GA-independent pathways.
Figure 2.
TEMPRANILLO regulates GA biosynthesis and trichome initiation. A, GA hormone measurements in mutants and over-expressors of TEM genes 5 DAB, shown with error bars. B, Trichome number in the 5th-6th rosette leaves in different TEM backgrounds and GA biosynthesis mutants at 21 DAG. Error bars indicate sd of the mean number of trichomes. Asterisks indicate statistically significant differences (* P ≤ 0.05, ** P ≤ 0.01, *** P ≤ 0.001) obtained using Tukey’s range test.
Exogenous GA3-Fl Accumulates Exclusively in the Mesophyll and Is Controlled by TEM
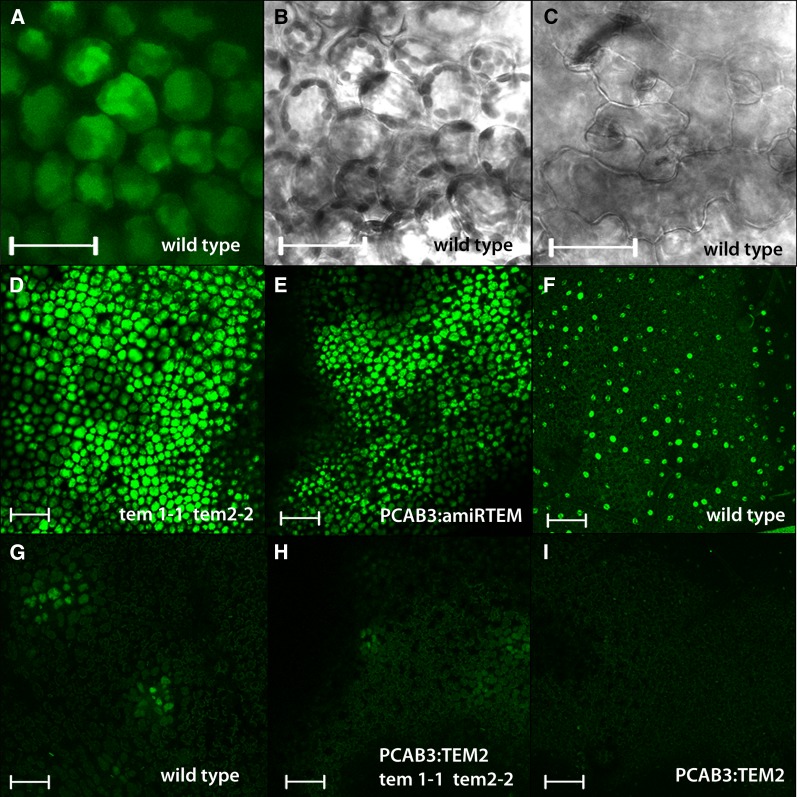
Plants are able to regulate the spatial distribution of their hormones (Wolters and Jürgens, 2009). GA distribution and accumulation are active processes regulated by localized synthesis, transport, and inactivation (Yamaguchi, 2008). GA distribution has been studied in plant roots using a bioactive fluorescent-GA (GA3-Fl), an in vivo stable fluorescent GA-surrogate that retains similar biological activity to GA3 (Shani et al., 2013). To address the issue of GA distribution in rosette leaves, we used confocal microscopy following exogenous overnight application of this fluorescently tagged GA. Unexpectedly, results showed that exogenous GA3-Fl accumulates exclusively in the leaf mesophyll layer (Fig. 3, A and C), distinctly recognizable from epidermal cells by their rounded shape (Deeks and Hussey, 2003; Smith, 2003). In wild-type Arabidopsis rosette leaves, GA3-Fl accumulated in the mesophyll in a highly specific manner at different stages of development (Fig. 3G and Supplemental Fig. S3). Since GA3-Fl was applied on top of leaves (adaxial side) as a water solution, in order for it to reach the mesophyll layer, it had to pass through the epidermis. Nevertheless, we found GA3-Fl mostly in patches that correspond to isolated zones in the mesophyll layer but not in the epidermis (neither adaxial nor abaxial). In only one exceptional case, we observed one stained patch in the adaxial puzzle-shaped epidermal cells, just on top of a mesophyll cell group that showed strong fluorescence (Supplemental Fig. S3). This exception may be due to saturation of the underneath layer. We additionally observed several examples of accumulation in companion cells of the vascular tissue (Supplemental Fig. S3) likely reflecting GA long-distance movement.
Figure 3.
Exogenous GA accumulation in the mesophyll is regulated by TEMPRANILLO. A, B, C, D, E, F, G, H, and I, Arabidopsis rosette leaf confocal sections of the first two rosette leaves of 9 DAG plants. A, B, C, GA3-Fl accumulates exclusively in the mesophyll (A, B) while not in the epidermis (C) of 9 DAG plants. B, C, Images taken in bright field. D, E, GA3-Fl accumulates with a higher intensity compared with wild type (G) and it is distributed in a much bigger area in the mesophyll of tem1-1 tem2-2 (D), and PCAB3:amiRTEM (E) rosette leaf of 9 DAG plants. (F) Exogenous Fl used as a negative control only accumulates in the epidermal stomas but not in the mesophyll. G, H, I, GA3-Fl accumulation in wild type (G), PCAB3:TEM2 tem1-1 tem2-2 (H), and PCAB3:TEM2 (I) mesophyll of 9 DAG rosette leaves. Scale bars from (A to C) represent 50 μm and from (D to I) represent 100 μm.
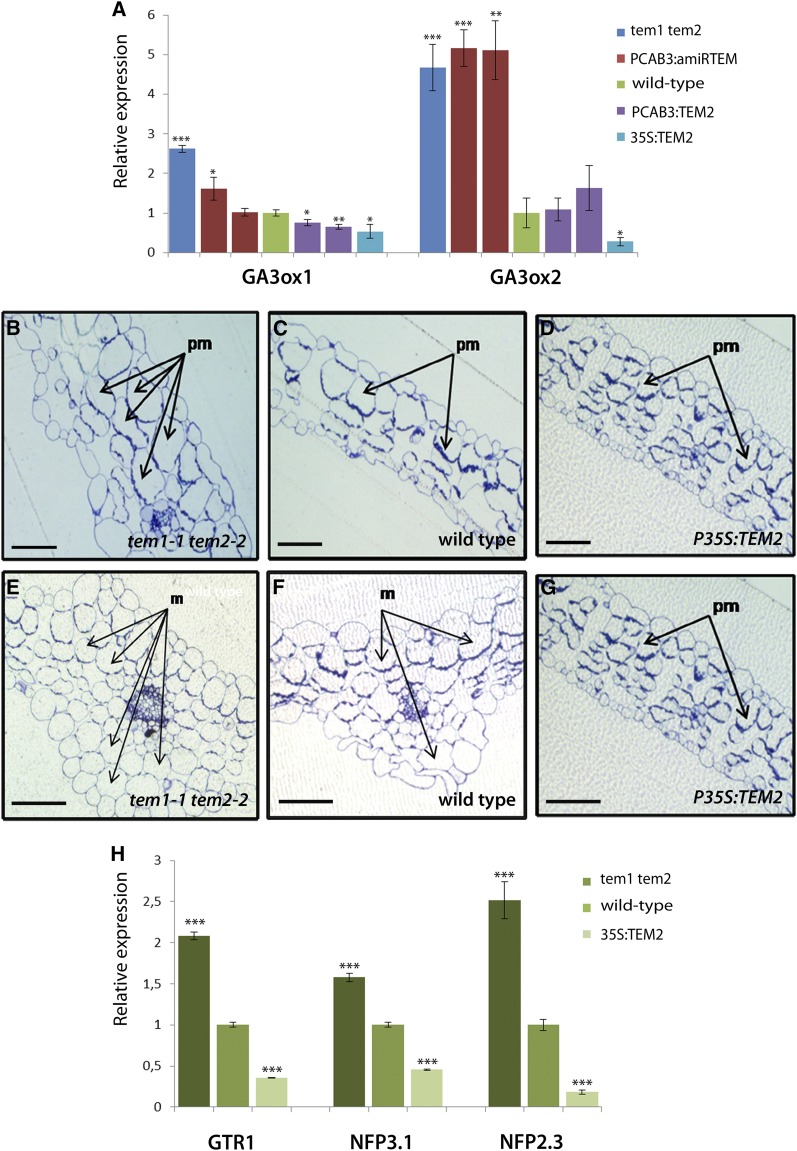
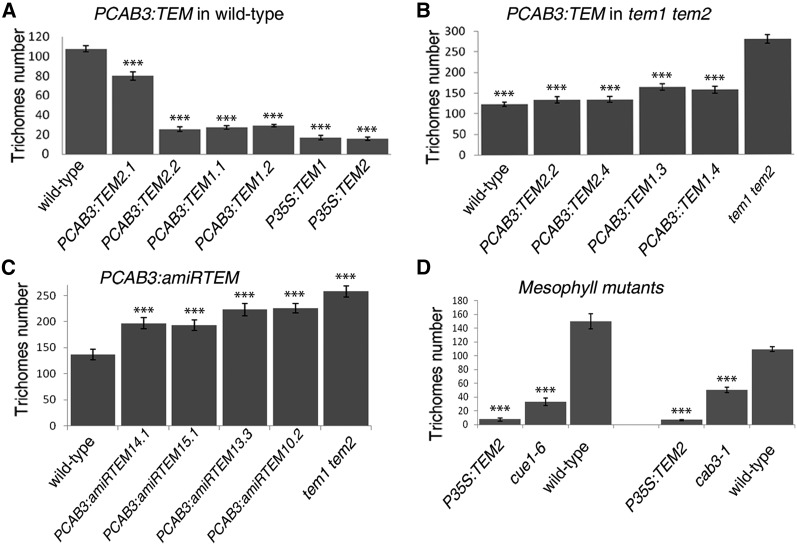
TEM transcription factors are known to repress GA biosynthesis during floral induction (Osnato et al., 2012). Therefore, in order to test whether TEM is also capable of controlling GA’s accumulation in the mesophyll layer, the distribution of GA3-Fl in leaves was studied in different TEM mutants and over-expressors lines. In tem1-1 tem2-2 double mutant plants, GA3-Fl uptake increased in comparison with wild-type plants, as GA3-Fl accumulated and was distributed through a much larger area of the leaf (Fig. 3D and Supplemental Fig. S3). The observed GA3-Fl broader distribution throughout the leaf might be directly related to normal TEM repression of GA biosynthesis in the leaf mesophyll structure and/or due to an effect on GA transport. To further support this, we first cloned an artificial microRNA (amiRNA), previously used for partial silencing of both TEM genes (Osnato et al., 2012), under the mesophyll-specific CHLOROPHYLL A/B BINDING PROTEIN 3 promoter (PCAB3; Endo et al., 2007; Ranjan et al., 2011). Transgenic plants expressing amiRTEM under the mesophyll-specific PCAB3 promoter, showed similar GA3-Fl accumulation patterns to tem1-1 tem2-2. PCAB3:amiRTEM leaves displayed a higher GA3-Fl uptake in the mesophyll in comparison with wild-type leaves (Fig. 3E and Supplemental Fig. S3), while plants over-expressing TEM2 in the mesophyll (PCAB3:TEM2) in tem1-1 tem2-2 background showed a strong reduction in GA3-Fl accumulation compared with tem1-1 tem2-2, reaching a similar pattern to that of wild-type leaves (Fig. 3H and Supplemental Fig. S3). On the other hand, leaves of PCAB3:TEM2 in wild-type background showed an almost complete absence of fluorescence signal compared with wild-type plants (Fig. 3I and Supplemental Fig. S3). As mentioned, TEM transcription factors are known to repress GA biosynthesis by directly repressing the expression of GA3ox1 and GA3ox2 enzymes (Osnato et al., 2012). Therefore, for testing whether the expression or down-regulation of TEM in the mesophyll mimics the effect of P35S:TEM and tem1-1 tem2-2, respectively; expression analyses of GA3ox1 and GA3ox2 in different PCAB3:amiRTEM and PCAB3:TEM2 plant lines were conducted. Indeed, results showed that TEM over-expression or silencing exclusively in the mesophyll is enough to affect GA biosynthesis by affecting these GA biosynthetic enzymes (Fig. 4A).
Figure 4.
TEMPRANILLO controls leaf mesophyll morphology and GA accumulation in the mesophyll. A, Expression analysis of GA biosynthesis enzymes GA3ox1 and GA3ox2 in tem1 tem2, PCAB:amiRTEM, wild-type, PCAB·:TEM2 and P35S:TEM2 seedlings at 11 DAG grown under LD. Two independent lines over-expressing (PCAB3: TEM2.1 and PCAB3: TEM2.2)and partially silencing TEM specifically in the mesophyll (PCAB3:amiRTEM13.3 and PCAB3:amiRTEM10.2) were included in the expression analyses. B, C, D, E, F, and G, Arabidopsis leaf cross sections. B, C, D, Leaf cross sections of tem1-1 tem2-2 (B), wild type (C), and P35S:TEM2 (D) in 11 DAG plants. E, F, G, central leaf cross sections of tem1-1 tem2-2 (E), wild type (F), and P35S:TEM2 (G) in 11 DAG plants. Cross sections show the central area of the rosette leaf including the main vascular bundle. pm, palisade mesophyll cells; m, mesophyll cells. Scale bars represent 100 μm. (H) Relative expression of different NPF transporter genes in TEM mutants and over-expressors 11 DAG. One representative of three biological replicates is shown with error bars of three qPCR replicates. Asterisks indicate statistically significant differences (* P ≤ 0.05, ** P ≤ 0.01, *** P ≤ 0.001) obtained using Student’s t test.
Thus, GA3-Fl uptake in the leaf seems to be a dynamic and well-regulated process that results in specific accumulation in the mesophyll layer, similarly to its accumulation in root endodermal cells (Shani et al., 2013). This GA3-Fl accumulation pattern may reflect the endogenous GA accumulation as it retains similar biological activities in roots (Shani et al., 2013). Thus, this unique accumulation pattern, at least partially controlled by TEM, suggests that GA can move not only over long distances but also locally among neighboring cells. Interestingly, this GA’s accumulation and movement in the mesophyll layer could affect biological processes occurring at other leaf layers, including trichome proliferation in the epidermis.
TEM Affects Palisade Mesophyll Development and GA Transport in the Rosette Leaves
In order to investigate why the GA3-Fl uptake to the mesophyll was higher in tem mutants and reduced in TEM over-expressors plants, we analyzed the mesophyll structure of tem1-1 tem2-2 double mutants and TEM2 over-expressors. Morphological analysis of the mesophyll structure was carried out in the first pair of leaves of 21-day-old Arabidopsis plants. At this stage, the leaf is fully expanded and does not exhibit any evidence of senescence. tem1-1 tem2-2 rosette leaves displayed mesophyll cells with an apparent increased size (Fig. 4, B and E). In contrast, mesophyll cells were smaller in P35S:TEM2 (Fig. 4, D and G) than in wild types (Fig. 4, C and E). Consequently, these palisade mesophyll defects in both tem1-1 tem2-2 and P35S:TEM plants may imply a primary role for TEM genes in rosette leaf development.
Not only GA biosynthesis (Osnato et al., 2012) but also GA mesophyll accumulation and distribution seem to be controlled by TEM. GA3-Fl uptake in the mesophyll of rosette leaves in tem1-1 tem2-2 mutants increased and it was distributed throughout a larger leaf area in comparison with wild-type plants. However, morphological analysis tem1-1 tem2-2 of mesophyll structure showed that these mutant rosette leaves displayed mesophyll cells with an increased size. In order to exclude the possibility that the accumulation of GA3-Fl in tem1-1 tem2-2 mesophyll cells was higher due only to the alteration in the leaf morphology, further analyses were conducted.
Plant hormones are active in plant tissues at very low concentration; therefore the synthesis, transport and signaling of hormones are precisely controlled processes (Chiba et al., 2015). Recent studies suggest that GA movement and distribution might be regulated by a group of GA transporters (Chiba et al., 2015; Saito et al., 2015). The NITRATE TRANSPORTER 1/PEPTIDE TRANSPORTER FAMILY (NPF) is a family of proteins initially identified as nitrate or peptide transporters (Tsay et al., 2007; Léran et al., 2014), and was later found to also transport hormones such as auxin and abcisic acid (Krouk et al., 2010; Kanno et al., 2012). It was recently reported that some members of this NPF family were also efficient transporters of GA and/or Jasmonic acid (Chiba et al., 2015; Saito et al., 2015). Therefore, we investigated the expression of several NPF transporters reported to show yeast-based GA transport function (Chiba et al., 2015; Saito et al., 2015) under TEM genetic manipulation. We first applied in silico analysis to observe that the chosen NPF genes were expressed in the rosette leaf mesophyll (Arabidopsis eFP Browser 2.0). Interestingly, the expression levels of NPF2.3, NPF2.10, and NPF3.1 were clearly down-regulated by TEM in 11-day-old plants (Fig. 4H). This data strongly suggests that TEM seems to affect GA accumulation and distribution in the mesophyll cells by modulating the expression of specific GA transporters, which at the same time would affect the GA transport toward neighboring epidermal cells.
TEM Reveals the Mesophyll as a Layer Controlling Trichome Initiation
For unraveling whether the role of TEM in controlling GA accumulation in the mesophyll layer affects epidermal trichome initiation, transgenic plants expressing TEM1 and TEM2 under the mesophyll-specific CAB3 promoter were analyzed. Surprisingly, we found that plants over-expressing either TEM1 or TEM2 in the mesophyll (PCAB3:TEM), which had reduced GA3-Fl accumulation, showed a strong reduction in trichome number and density, behaving almost as TEM over-expressors (Fig. 5A and Supplemental Table S1).
Figure 5.
TEMPRANILLO controls trichome initiation from the mesophyll. A, B, and C, Trichome average number in the 5th-6th rosette leaves of 21 DAG plants of different independent lines over-expressing TEM specifically in the mesophyll in wild-type (A) and tem1-1 tem2-2 (B) backgrounds, or partially silencing TEM1 or TEM2 (C). D, Trichome average number in cue1-1 and cab3-1 mutants that show defects only in the mesophyll. Error bars indicate sd of the mean number of trichomes. Asterisks indicate statistically significant differences (* P ≤ 0.05, ** P ≤ 0.01, *** P ≤ 0.001) obtained using Tukey’s range test.
Moreover, specific TEM expression in the mesophyll of tem1 tem2 mutants, PCAB3:TEM tem1-1 tem2-2, restored tem1-1 tem2-2 trichome number to the wild-type level (Fig. 5B). These results led us to propose that TEM-specific mesophyll expression might be sufficient for formation of a normal trichome number. To further support this, PCAB3:amiRTEM lines were analyzed. As expected, PCAB3:amiRTEM lines showed an increased production of trichomes, resulting in hairier leaves with normal epidermal cells (Fig. 5C, Supplemental Fig. S4 and Supplemental Table S1). Therefore, these data provide robust and independent evidence that specific mesophyll over-expression or silencing of TEM genes is enough to affect trichome development, indicating that TEM mesophyll expression is necessary and sufficient for normal trichome initiation. This strongly supports that not only the epidermis but also the mesophyll layer is essential to direct epidermal trichome initiation during early stages of development, where a bigger palisade mesophyll cell would promote increased trichome production, while smaller cells would result in reduced number of trichomes.
According to this rationale, mutants that specifically affect the mesophyll layer should exhibit altered trichome numbers. Therefore, we studied the number of trichomes per leaf of two mutants, CHLOROPHYLL A/B-BINDING PROTEIN-UNDEREXPRESSED1-1 mutant (cue1-6) and cab3-1, which display defects only in the leaf mesophyll but not in the epidermis (Li et al., 1995; Lundquist et al., 2014). We found that both mutants showed a significant reduction in the number and density of trichomes resembling pCAB3:TEM plants (Fig. 5D and Supplemental Table S1); strongly supporting the mesophyll as an essential layer that control epidermal trichome initiation.
TEM Integrates GA- and CK-Dependent Trichome Pathways in the Epidermis
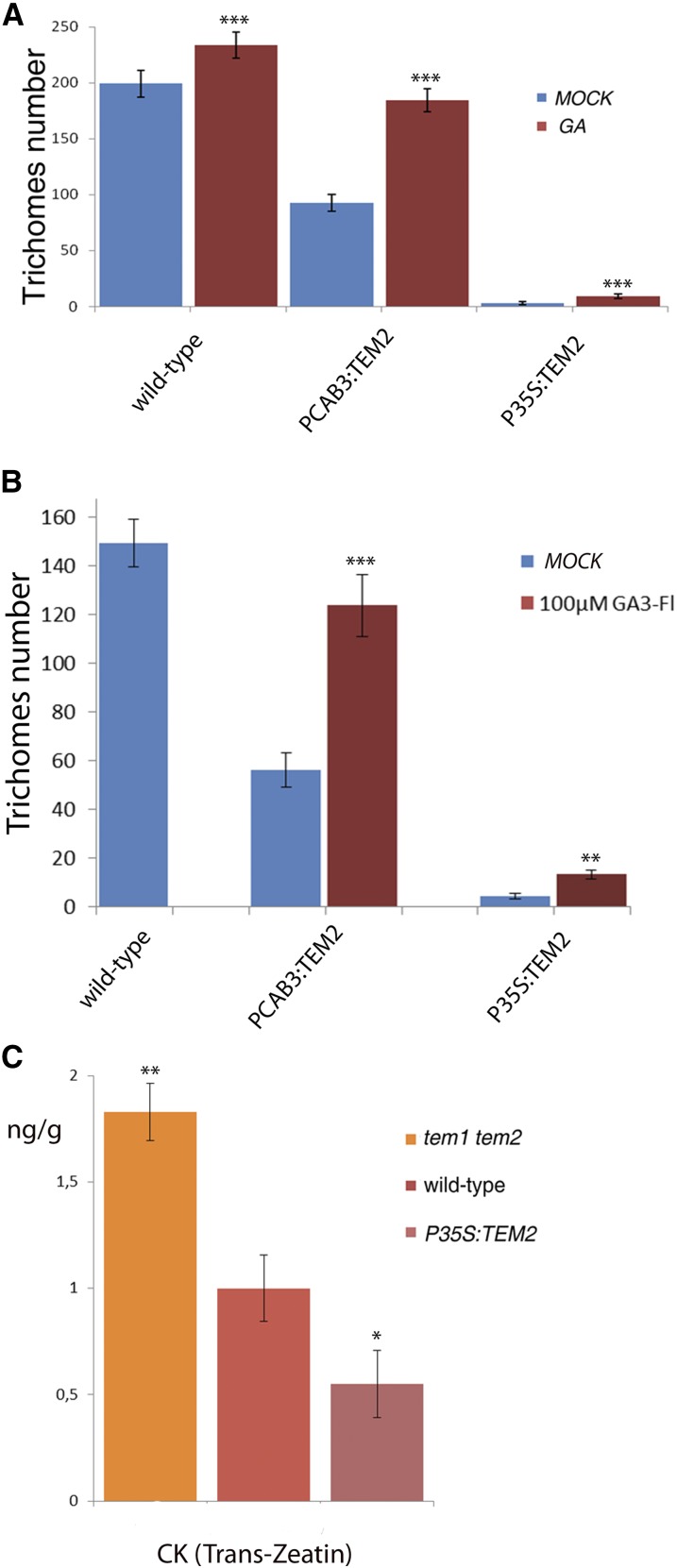
As expected, when exogenous GA is added, trichome production of PCAB3:TEM2 leaves is restored to wild-type level, yet, it was only partially restored in P35S:TEM2 leaves (Fig. 6A and Supplemental Table S1), suggesting that TEM control additional epidermal GA-independent genes involved in trichome initiation. Similar results were obtained when plants were treated exogenously with GA3-Fl at similar concentrations (Fig. 6B and Supplemental Table S1), showing that GA3-Fl retains similar bioactivity than GA3 for trichome induction.
Figure 6.
TEMPRANILLO also affects CK. A, Trichome number in the 7th-8th rosette leaves in different TEM backgrounds after MOCK and GA treatment at 25 DAG. Error bars indicate sd of the mean number of trichomes. B, Trichome number in the 6th-7th rosette leaves in different TEM backgrounds after MOCK and 100 μM GA3-Fl treatment (at 18 DAG). Error bars indicate sd of the mean number of trichomes. C, CK hormone measurements in mutants and over-expressors of TEM genes 5 DAB, shown with error bars. Asterisks indicate statistically significant differences (* P ≤ 0.05, ** P ≤ 0.01, *** P ≤ 0.001) obtained using Student’s t test.
Furthermore, integration of hormone signaling is essential for many processes in plant development (Gan et al., 2007; D’Aloia et al., 2011). GA and CK act antagonistically in leaf formation and shoot meristem maintenance (Weiss and Ori, 2007), where GAs counteract some CK effects on epidermal differentiation as the inflorescence develops (Gan et al., 2006; Nemhauser et al., 2006). However, both phytohormones overlap in stimulating trichome initiation in different plant tissues (Gan et al., 2007). CK trichome production action is limited to upper inflorescence organs (D’Aloia et al., 2011), whereas GA action affects all trichome-producing epidermal tissues (Gan et al., 2006, 2007). Interestingly, the number of trichomes in mutants and over-expressors of TEM genes was clearly affected not only in rosette leaves but also in upper inflorescence organs (Supplemental Fig. S1), where trichome formation is at least partially controlled by CK. Consequently, we hypothesized that TEM may control other hormones involved in trichome proliferation. We therefore analyzed the levels of CK, and detected higher levels of Trans-Zeatin, the bioactive form of CK, in tem1-1 tem2-2 and lower levels in P35S::TEM2 than in wild-type plants 5 DAB (Fig. 6C). These data suggest that TEM may also regulate the CK biosynthetic pathway, which in turn affects trichome formation.
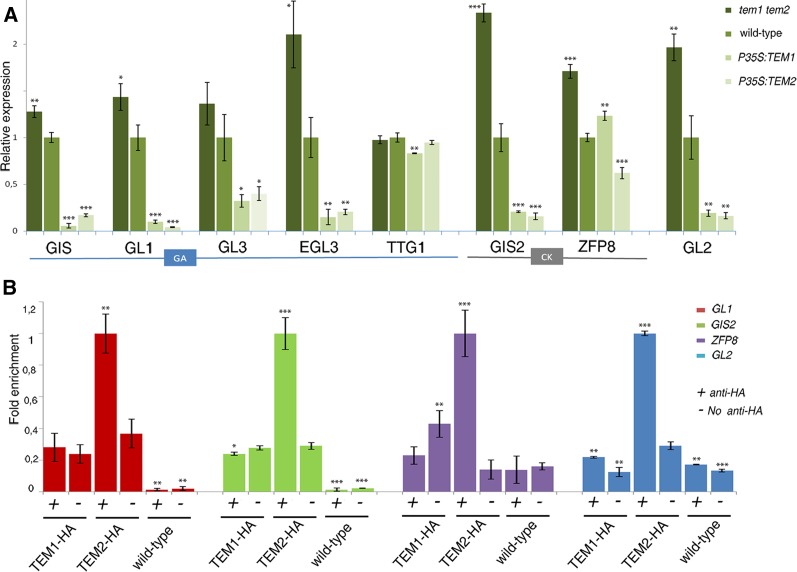
As previously described, the trichome GA-dependent pathway acts in the epidermis partially through GIS, which positively regulates GL1, GL3, EGL3, and TTG1 genes whose proteins form the trichome activation complex (Payne et al., 2000; Zhao et al., 2008a). On the other hand, the trichome CK-dependent pathway acts also in the epidermis and is controlled by GIS2 and ZFP8 (Gan et al., 2007). Mutations in all these main genes result in a significant loss of trichomes, but none of these mutants were as glabrous as TEM over-expressors. TEM is not only widely expressed in the rosette mesophyll but also in the epidermis (Supplemental Fig. S2). To investigate the molecular basis underlying the trichome suppressing effect of TEM in the epidermis, we performed gene expression analysis in TEM mutants and over-expressors 11 days after germination (DAG), when new rosette leaves with new trichomes are proliferating, i.e. before bolting, and 5 DAB. Our results indicated that GIS, GL1, GL3, and EGL3 were clearly repressed by TEM, while TTG1 and some trichome repressor genes, such as CAPRICE (CPC) and SPINDLY (SPY) (Perazza et al., 1999; Kirik et al., 2004), were not affected (Fig. 7A, Supplemental Fig. S5, and Supplemental Fig. S6). As we wondered whether the trichome suppressing effect of TEM through down-regulation of epidermal transcription factors could also take place from the mesophyll, we conducted gene expression analyses on different PCAB3:amiRTEM and PCAB3:TEM2 plant lines at the same stage of development (Supplemental Fig. S5). Results showed that exclusive mesophyll TEM over-expression or silencing clearly affect the expression of trichome GA-dependent genes (Supplemental Fig. S5). In addition, GIS2 and ZFP8 expression was also significantly repressed by TEM at 5 DAB (Fig. 7A) but not so clearly at 11 DAG (Supplemental Fig. S5). At 11 DAG, plants have not yet produced any inflorescence organs, consistent with the fact that CK-dependent genes were less affected at this stage.
Figure 7.
TEMs control expression and binds in vivo to the regulatory regions of several trichome initiation genes. A, Relative expression of GA and CK pathway genes in TEM mutants and over-expressors 5 DAB. One representative of three biological replicates is shown with error bars of three qPCR replicates. B, Relative enrichment of the regulatory regions of GL1, GIS2, ZFP8, and GL2 after immuno-precipitation of TEM1 or TEM2 followed by a qRT PCR in plant collected 5 DAB. One representative of two biological replicates shown with error bars of three qPCR replicates. Asterisks indicate statistically significant differences (* P ≤ 0.05, ** P ≤ 0.01, *** P ≤ 0.001) obtained using Student’s t test.
Integration of CK and GA signaling requires the action of all these GA- and CK-dependent genes on GL2 epidermal expression (Szymanski et al., 1998; Gan et al., 2007). GL2 is considered to be the universal activator of trichome proliferation and it is required from the earliest morphogenetic events of trichome growth (Szymanski et al., 1998; Lin and Aoyama, 2012). A highly plant-conserved mechanism that transduces GAs to promote epidermal cell elongation has been recently revealed: GL2 protein interacts with DELLA complex to affect epidermal cell elongation during trichome formation and seed germination (Rombola-Caldentey et al., 2014; Shan et al., 2014). Additionally, it is well known that GL2 acts downstream of the GA-activation complex (GL1/GL3/TTG1/EGL3) as well as the CK-dependent GIS2/ZFP8 genes during early stages of trichome development (Szymanski et al., 1998; Gan et al., 2007). Because our data showed that both GA- and CK-dependent trichome pathways were controlled by TEM, we next investigated whether GL2 expression levels were affected. As expected, GL2 expression was clearly repressed by TEM at both stages of plant development, 11 DAG and 5 DAB (Fig. 7A and Supplemental Fig. S5), suggesting that the strong trichome phenotypes found in tem mutants and P35S:TEM plants are, at least partially, caused by GL2 repression. To substantiate our findings we next investigated whether the spatial pattern of GL2 transcription was affected in tem1-1 tem2-2 and P35S:TEM2 plants. For this purpose, we used PGL2:GUS lines containing the 2,1Kb of the 5′-untranslated promoter region of GL2 (Szymanski et al., 1998) (Supplemental Fig. S7). In wild-type plants we observed PGL2:GUS expression throughout trichome development including surrounding epidermal pavement cells, cells that will not enter to the trichome pathway (Szymanski et al., 1998) (Supplemental Fig. S7). By contrast, PGL2:GUS activity was not detected in the few P35S:TEM2 trichomes that develop (Supplemental Fig. S8). These data support the finding that GL2 is strongly repressed by TEM during trichome initiation and development.
To further identify the molecular mechanism underlying the trichome-suppressing effect of TEM, we next used P35S:TEM1-HA and P35S:TEM2-HA plants for chromatin immuno-precipitation (ChIP) using an anti-HA antibody followed by qPCR. These experiments revealed binding of TEM2 to the RAV binding sites of the regulatory regions of four essential trichome initiation genes: GL1, GIS2, ZFP8, and GL2 at both stages of plant development with enrichments above 2.5 fold (Fig. 7B, Supplemental Fig. S6, and Supplemental Fig. S8). Interestingly, these genes were found to be direct targets of TEM2 but not of TEM1. Altogether, our data suggest that TEM transcriptionally repress trichome initiation in the epidermis by binding in vivo to diverse downstream genes essential to both main trichome initiation pathways, which at the same time are affected by the hormones GA and CK.
DISCUSSION
Trichomes are specialized epidermal protrusions on the surfaces of leaves and other aerial organs of many plants (Olsson et al., 2009), which defend plants against insect herbivores, virus, UV light, and excessive water loss (Traw and Bergelson, 2003). In addition, trichomes are able to synthesize, store, and sometimes secrete large amounts of specialized metabolites with significant commercial value as pharmaceuticals, natural pesticides, or food additives (Schilmiller et al., 2008). Consequently, a better understanding of the molecular mechanisms that control trichome formation will be important for future commercial biopharming applications as trichomes would be able to produce and store such specialized valuable metabolites (Murphy, 2007; Ahmad et al., 2012).
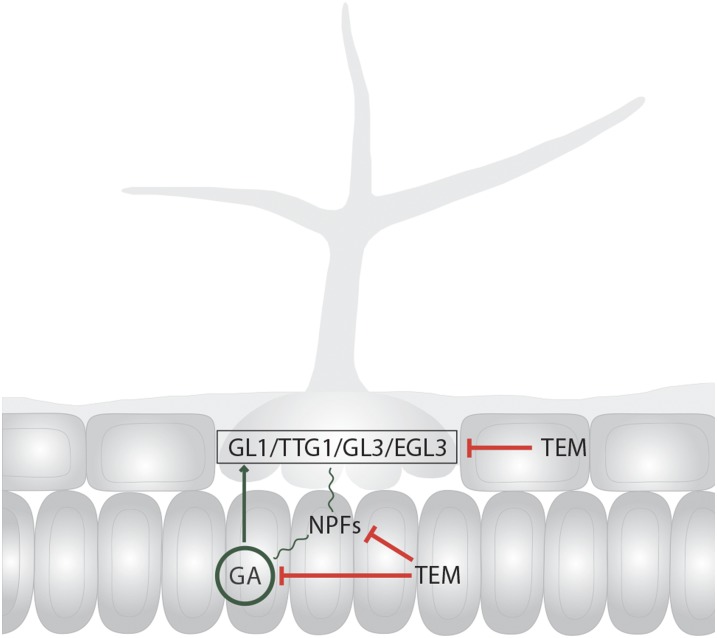
Here we report, to our knowledge, a novel double role for TEM genes in trichome initiation. Our data clearly indicate that, similarly to their effect on floral induction (Castillejo and Pelaz, 2008), TEM1 and TEM2 act redundantly to repress trichome initiation from two leaf tissue layers, epidermis and mesophyll. In the epidermis they directly control trichome gene expression, and in the mesophyll the accumulation and distribution of GA, that ultimately activates the epidermal trichome factors, through the repression of GA biosynthetic genes and GA transporters. The GAs could transiently move to the epidermis, as well as other proteins including TEM, to activate the trichome initiation cascade. In this way, TEMs tightly control trichome formation, acting as secure locks at different steps and locations.
TEM Negatively Affects Trichome Initiation in Arabidopsis
Transcription factors of the RAV family are unique plant-specific proteins, mainly characterized by the presence of two different DNA-binding domains, AP2 and B3 (Riechmann et al., 2000; Matías-Hernández et al., 2014). In Arabidopsis, TEM1 and TEM2, two members of this family, have been previously identified as repressors of floral induction (Castillejo and Pelaz, 2008; Osnato et al., 2012). TEM1 and TEM2 genes act redundantly to repress floral transition by integrating both the photoperiod and GA signaling pathways under long days and short days (Castillejo and Pelaz, 2008; Osnato et al., 2012). In addition, recent results indicated that both TEM1 and TEM2 are regulated by genes acting upstream in other flowering pathways, suggesting a possible role for TEM in integrating information from diverse redundant pathways (Yant et al., 2010; Tao et al., 2012; Marín-González et al., 2015). Similarly to other transcription factor families, it has been suggested that RAV genes may control additional biological aspects during plant development (Matías-Hernández et al., 2014). Indeed, flowering is not the only process controlled by RAV proteins. RAV members have been found to be involved in other plant development aspects such as leaf senescence, pathogen infections, abiotic stresses, and growth regulation (Zhao et al., 2008b; Woo et al., 2010; Matías-Hernández et al., 2014; Fu et al., 2014; Wang et al., 2014).
P35S:TEM1 and P35S:TEM2 showed almost completely glabrous leaves, stems, and flowers, while tem1-1 tem2-2 plants produced an approximately double number of trichomes than wild types. Although TEM1 and TEM2 seemed to play a redundant role in repressing trichome initiation, TEM2 had a stronger effect on this negative regulation. tem2-2 plants produced more trichomes in rosette leaves than tem1-1 and wild-type plants. Additionally, GUS expression analyses showed that both genes are expressed in all plant trichomes: in juvenile rosette leaves, adult leaves, stems, and sepals. However at later stages of leaf development, TEM2 but not TEM1 expression was restricted to trichomes formed at the central part and the periphery of adult leaves.
In Arabidopsis, after initiation, trichomes develop and grow through endoreduplication, a process where cell replicates its genome but without cytokinesis (Szymanski et al., 1998). As a consequence of this endoreduplication, an Arabidopsis plant will form trichomes with three to four branches in the rosette leaves, one to two branches in the stem, and one branch in the sepals (Gilding and Marks, 2010). However, phenotypical analyses conducted in mutants and over-expressors of TEM genes exclude the possibility of TEM being involved in later processes of trichome development as the branching of tem1-1 tem2-2, P35S:TEM1, and P35S:TEM2 trichomes resembled wild-type trichomes.
TEM Reveals the Palisade Mesophyll as Essential for Trichome Initiation
In plants, leaf development meticulously follows a sequence of events that produce a complex organ with diverse kinds of cells types, thereby the morphological differentiation of cells and their relative position patterns are essential for ensuring the correct function of the leaf (Lin and Aoyama, 2012). The outer L1 layer of the SAM will form the leaf epidermis, while L2 and L3 cell layers will become mesophyll and vascular tissue, respectively (Langdale, 1998; Balkunde et al., 2010).
Trichomes are formed by the specification of epidermal cells. Once an epidermal precursor is specified to enter the trichome pathway, an elaborate morphogenetic cell transformation occurs in order for it to become a trichome (Langdale 1998; Kirik et al., 2004). It is widely accepted that patterns of trichome cell fate should be explained by mechanisms of lateral inhibition toward the epidermal surrounding cells that involves cell to cell communication with a spatial and temporal control (Langdale 1998; Kirik et al., 2004). In Arabidopsis, negative regulators of trichome initiation and patterning, such as CPC, and ENHANCER OF TRIPTYCHON AND CAPRICE1 (ETC1) traffic from trichome cell precursor to neighboring epidermal pavement cells in the epidermis to repress trichome formation (Balkunde et al., 2010; Zhao et al., 2008a). This is due to trichome activation factors turning on their inhibitors, which can subsequently move into neighboring epidermal cells to avoid trichome formation (Balkunde et al., 2011). Similar regulation occurs between positive regulators such as GL3 and TTG1 (Zhao et al., 2008a; Savage et al., 2008). It has been shown that TTG1 moves between epidermal cells and when artificially expressed in the subepidermis, freely moves to the epidermis to rescue ttg1 mutant phenotype (Bouyer et al., 2008). In addition, its function as trichome promoter is mediated by a trapping mechanism and translocation to the nucleus in the trichome cells through interaction with GL3 (Balkunde et al., 2011), resulting in TTG1 depletion from nontrichome cells (Bouyer et al., 2008). Although it is widely accepted that in Arabidopsis, the competency to enter the trichome pathway is limited to epidermal cells, our results uncover, to our knowledge, a novel mesophyll-epidermis communication mechanism required to control the transition of epidermal cells into trichomes; and that, consequently, may be at odds with the dogma of trichome epidermal specificity. This mechanism would involve the NPF GA transporters that would allow the GA-dependent epidermis genes be activated from GA specific mesophyll accumulation.
Our data indicate that intercellular signaling between the mesophyll and the epidermis might be affected by TEM, CUE, and CAB3. Although these proteins or other molecules regulated by them may move from mesophyll to epidermal cells, our results suggest that at least GA would use their NPF transports to activate the epidermal trichome initiation genes.
Here we showed that the levels of GA, a mobile hormone (King et al., 2003; Yamaguchi, 2008), are also controlled by TEM (Osnato et al., 2012). GA distribution is an active and highly regulated process (Yamaguchi, 2008; Shani et al., 2013). Our results, using a fluorescently labeled GA, clearly indicate that mesophyll cells have unique features that enable GA accumulation and distribution, suggesting that the mesophyll plays a special role in GA storage. TEM is able to negatively control, not only GA biosynthesis but also NPF-specific GA transporters. These data lead us to propose, to our knowledge, a novel plausible mechanism, where TEM may regulate mesophyll cell development and epidermal trichome initiation, in part, by affecting the GA levels and distribution in the mesophyll as well as the GA transport toward neighbored cells such as epidermal cells (Fig. 8). In the absence of GA accumulation in the mesophyll, as in Arabidopsis lines over-expressing TEM genes, there is a significant reduction in trichome number. This would explain, at least partially, how trichome proliferation is controlled from the mesophyll: the mesophyll plays an essential role in controlling GA storage, distribution, and transport, through the action of TEM, which at the end also controls trichome initiation in the epidermis.
Figure 8.
Mechanisms of epidermal trichome formation controlled by TEM. TEMs repress trichome initiation from two leaf tissue layers, the epidermis and the mesophyll. In the epidermis TEMs directly control trichome gene expression, and in the mesophyll the accumulation and distribution of GA, that ultimately activates the epidermal trichome factors, through the repression of GA biosynthetic genes and GA transporters.
Thus, our data unravel a unique communication channel from one cell layer (mesophyll) to another (epidermis), to inform cell differentiation processes, and that might be mediated by NPF transporters. Potentially, this novel communication channel might also be contributing to additional essential leaf developmental events.
TEM Directly Binds and Represses Genes from both GA- and CK-Dependent Trichome Pathways in the Epidermis
Hormones play an essential role in trichome initiation (D’Aloia et al., 2011; Perazza et al., 1998) and plants use various strategies to orchestrate the competing hormone signals (Nemhauser et al., 2006; Perazza et al., 1998). Among these strategies, a crucial one is the use of specialized regulators that integrate diverse genetic networks in the epidermis (Nemhauser et al., 2006). Indeed, GA and CK hormones have synergistic effects on the constitutive induction of epidermal defensive trichomes and gene regulators may be shared between these two hormonal signaling pathways (Chien and Sussex, 1996; Perazza et al., 1998). Here, we report that TEMs seem to be one of these essential regulators. TEMs affect not only the GA and CK levels within the plants but also genetically control GA- and CK-dependent trichome pathways in the epidermis, which in turn negatively affect trichome formation in all Arabidopsis trichome-producing tissues. TEM1 and TEM2 act redundantly to repress the transcription of most essential positive epidermal regulators in at least two different developmental stages, 11 DAG and 5 DAB, that is before and after bolting, respectively. Among these genes, GL2, considered to be the universal activator of trichome initiation due to the fact that both GA- and CK-dependent trichome pathways converge in its activation, was also repressed. These results suggest that the strong trichome phenotypes found in tem mutants and P35S:TEM plants were, at least partially, caused by alterations in GL2 expression. Interestingly, some of these genes were found to be in vivo direct targets of TEM2 but not of TEM1. Combined with the fact that tem2-2 plants produced more trichomes than tem1-1 and wild-type plants, these suggest that TEM2 may take over TEM1 functions, playing a more important role in trichome initiation. In conclusion, our results strongly suggest that TEM genes control trichome initiation, acting differently in distinct leaf cell layers. In the mesophyll, they seem to regulate trichome formation by controlling the amounts and distribution, through NPF transporters, of GA, known to affect trichome development in the upper cell layer. In that upper layer, the epidermis, they seem to directly repress transcription of well-known transcription factors involved in trichome initiation as GL3, EGL3, or GL2 (Fig. 8). Thus, our data, including, to our knowledge, the novel finding of GA storage, transport, and distribution in the palisade mesophyll and its essentiality for epidermal trichome initiation, reinforces the developmental plasticity found in plant growth and emphasizes the vital role of TEM genes as repressors in several plant developmental programs. As such, TEM may contribute toward additional important processes affected by the GA and CK hormones, such as cell proliferation, leaf formation, or shoot meristem maintenance and formation (Gan et al., 2007; D’Aloia et al., 2011).
MATERIALS AND METHODS
Plant Material and Growth Conditions
Arabidopsis seeds were stratified for 3 d at 4°C, and plants were grown in soil under controlled conditions at 22°C and long days (16 h light/8 h dark). Col-0 was used as wild type in all the experiments. Transgenic, mutant, and control plants used for phenotypic analyses and live-imaging have Col-0 background and were grown together at the growth chamber. For minimizing the effect of environmental fluctuations, each tray was randomly positioned at the growth chambers. tem1-1 and tem2-2 were previously described (Castillejo and Pelaz, 2008; Osnato et al., 2012).
Cloning
For mesophyll-specific TEM expression, we first modified the pENTR-3C vector (Gateway; Invitrogen, Carlsbad, CA) by introducing the PCAB3 and the Nos terminator, resulting in the PCAB3-Nos vector. The Nos terminator was amplified by PCR and cloned in pENTR-3C as a KpnI-EcoRV restriction fragment. The CAB3 promoter was amplified by PCR and cloned in pENTR3C-Nos terminator as a SalI-BamHI fragment.
For mesophyll-specific silencing, we generated PCAB3:amiRTEM. The previously described artificial miRNA (amiRNA) sequence targeted against TEM genes (amiR-TEM) was cloned into the PCAB3-Nos plasmid (Osnato et al., 2012). The resulting plasmid was then linearized with NheI, dephosphorylated with alkaline phosphatase, and recombined by the LR Reaction into pMDC100, a Gateway-compatible binary vector (Curtis and Grossniklaus, 2003).
Similarly, PCAB3:TEM1 and PCAB3:TEM2 plasmids were generated by cloning the TEM1 and TEM2cDNAS as BamHI-NotI restriction fragments into the PCAB3-Nos. These vectors were later recombined by the LR reaction into pMDC99 and pMDC123, respectively.
In addition, for PTEM2:GUS plasmid construction, a 1.9 Kb promoter region of TEM2 was cloned first in pENTRD-TOPO (Gateway; Invitrogen). This plasmid was further recombined by a LR reaction into pBGWFS7, a Gateway plasmid containing GUS (Karimi et al., 2002).
For all PCR reactions, Col-0 genomic DNA was used as template with specific primers as listed in Supplemental Table S1 and in previous publications (Castillejo and Pelaz, 2008; Osnato et al., 2012). All PCR products were verified by sequencing. Agrobacterium tumefaciens (pGV2260 strain) was electroporated with plant expression vectors, and used to transform Col-0 wild-type plants by floral-dip (Clough and Bent, 1998). Between 5 and 10 T1 transgenic lines were selected for each construct on MS1 supplemented with the appropriate antibiotic.
Phenotypic Analyses
For trichome analysis, all the experiments were repeated on soil-grown plants at least twice. Trichome initiation was monitored using a model no. DP71 microscope (Olympus, Melville, NY) by counting all trichomes on the adaxial surface of individual and fully developed rosette leaves (Gan et al., 2007; Yu et al., 2010). 1st-2nd, 3rd-4th, 5th-6th, and 7th-8th rosette leaf trichomes were counted independently given that these leaves showed different trichome production. However, despite the trichome number differences, a similar tendency was observed. Trichome production on the main stem was evaluated by counting trichomes on the first, second, and third inflorescence internodes independently, starting from the bottom to the top. Trichome number was recorded when the main stem reached approximately 17–18 cm in size. Trichome numbers on the first, second, and third cauline leaves of Arabidopsis plants were measured in the whole adaxial area of each fully expanded leaf. In addition, trichome production on sepals was evaluated by counting sepal trichomes in flowers at different developmental stages in plants whose main stem reached approximately 10 cm in size, approximately 5–8 d after flowering. Unless otherwise specified, a minimum of 20 plants was used for trichome analysis for each developmental stage and genotype combination. Data are reported as mean value and sd of the number of trichomes for each genotype.
Hormone Analyses and Treatments
Hormones were measured at the hormone quantification service at the Institute for Plant Molecular and Cell Biology, Valencia, Spain. At least 200 mg of fresh independent Arabidopsis plants for each genotype collected 5 DAB were used. GA was quantified after different steps of extraction and purification including three steps of 80% MeOH-1% acetic acid and a final step using 80% MeOH-1% formic acid. CK was quantified after extraction and purification including 80% MeOH-1% acetic acid and 60% MeOH-5% NH3. GA and CK were finally quantified using an MS-HPLC-Q-Exactive Orbitrap (Thermo Fisher Scientific, Waltham, MA; Glauser et al., 2014). Exogenous GA3 was used for hormone treatment. As described (Gan et al., 2007), different line plants were grown on soil until the first 3–4 leaves had emerged and then plants are sprayed twice a week with MOCK and 100 μM GA4 solutions until plants during 2–3 weeks. For measuring the effect of GA application, 7th-8th rosette leaf trichomes from 20 plants were counted for each genotype combination. Experiments were repeated on soil-grown plants at least twice.
Epidermis and Mesophyll Phenotypic Analyses
SEM was performed as previously described (Sánchez-Chardi et al., 2011). In short, samples were fixed in 2.5% (vol/vol) glutaraldehyde in 0.1 m P-buffer (pH 7.4) for 2 h at 4°C, washed 4 times for 10 min each time in 0.1 m P-buffer, postfixed in 1% (w/t) osmium tetraoxide with 0.7% ferrocyanide in P-buffer, washed in water, dehydrated in an ascending ethanol series (50, 70, 80, 90, and 95% for 10 min each and twice with 100% ethanol), and dried by critical-point drying with CO2 (Julián et al., 2010). On the other side, ultra-thin transverse sections of the central part of rosette leaves 11 DAG were first fixed with osmium, dehydrated with acetone, embedded in Spurr resin and finally stained with toluidine blue (Sánchez-Chardi et al., 2011). For cell morphology analysis, sections of 0.5–1 μm were cut with an ultra-thin microtome using a diamond knife. All measurements and image analyses were done using an AixoPhot DP70 microscope (Olympus).
Exogenous GA3 Bioactivity Assays
For GA3-Fl bioactivity assays and analyses seeds from different genotype were germinated and grown in MS medium under controlled conditions at 22°C and long days (16 h light/8 h dark). Plants were treated with 10 μM GA3-Fl liquid solution for 20 h and then 1st-2nd rosette leaves were imaged (Shani et al., 2013). GA3-Fl bioactivity assays were performed in rosette leaves through development. For that reason, 1st-2nd rosette leaves from at least six plants for each genotype combination lines were imaged at different plant developmental stages 9, 11, 13, 16 DAG. Confocal microscopy (Leica, Wetzlar, Germany) and ImageJ (National Institutes of Health, Bethesda, MD) were used for imaging. Exogenous GA3-Fl was used for hormone treatment. Different line plants were grown in MS medium under controlled conditions at 22°C and long days on soil until the first 2–3 leaves had emerged and then plants are sprayed twice a week with MOCK and 100 μM GA4 solutions during 10 days. For measuring the effect of GA application, 6th-7th rosette leaf trichomes from 20 plants were counted for each genotype combination.
Identification of Putative RAV Binding Site Sequences
The genomic regions located 3 kb upstream of the ATG, 1 kb downstream of the stop codon and in the exons, and introns of the genes involved in GA- and CK-dependent trichome regulatory pathways, were analyzed to identify RAV binding sites sequences (C(A/C/G)ACA(N)2–8(C/A/T)ACCTG). The Fuzznucbioinformatic program available at the Web site (http://www.hpa-bioinfotools.org.uk/pise/fuzznuc.html) allowed us to identify perfect RAV binding sites, and RAV binding sites sequences with one/two mismatches. To restrict the sample further, we selected genes containing at least two putative RAV binding sites sequences within a distance of 300 bp.
Expression Analyses, ChIP, and GUS Assays
Real-time analyses were designed to comply with standards of RT-qPCR (Rieu and Powers, 2009). For RT-qPCR reactions, plants were grown in soil under long days and samples collected at ZT18 at the indicated days. RNA was extracted from a pool of 20 plants 11 DAG or 10 adult plants 5 DAB with PureLink RNA Mini Kit (Ambion; Thermo Fisher Scientific), treated with RNase-free DNaseI (Ambion, Thermo Fisher Scientific) and 1 μg was retrotranscribed with oligo(dT) and SuperScript III (Invitrogen). The expression levels of genes of interest were monitored by qPCR using SYBR Green I Master Mix and Light Cycler 480 (Roche, Basel, Switzerland) with the primers listed in Supplemental Table S2 and below. Data were normalized using the UBQ10 gene as reference. PCR efficiency was calculated and determined as previously described (Talke et al., 2006). For the GA- and CK-dependent pathways, we chose to study only those genes that are involved in the final steps of trichome initiation, which are highly expressed in trichomes, and for which mutants have already been described. Gene-specific primer sequences previously described (Gan et al., 2006, 2007; Balkunde et al., 2010) are listed in Supplemental Table S2 and here:
GIS, 5′-TTCATGAACGTCGAATCCTTCTC-3′ and 5′-ACGAATGGGTTTAGGGTTCTTATCT-3′;
GL1, 5′-CGACTCTCCACCGTCATTGTT-3′ and 5′-TTCTCGTAGATATTTTCTTGTTGATGATG-3′;
GL3, 5′-GGTACCACAGAACATATTACGGAAGA-3′ and 5′-CAAGAACGTTGTCGATGTGATAATC-3′;
EGL3, 5′-ATGGCAACCGGAGAAAACAGAACG-3′ and 5′-TCTCAAGGACTCCTCCAAGAAACG-3′;
TTG1, 5′-ATGGATAATTCAGCTCCAG-3′ and 5′-TCAAACTCTAAGGAGCTGC-3′;
GIS2, 5′-ACCGCCAACAAAACCACATT-3′ and 5′-CGCGTCGTTGATTTGAACAG-3′;
ZFP8, 5′-AAGCCGCCATTATTCGTCTCT-3′ and 5′-CTGCGGATAAGTTGTCGGAGTT-3′;
GL2, 5′-GGACGAGAAGCAAAGACAGC-3′ and 5′-TCTCTAGTTCCGCCTTGAGC-3′;
CPC, 5′-TGGGAAGCTGTGAAGATGTCAG-3′ and 5′-AAGTCTCTTCGTCTGTTGGCA-3′;
SPY, 5′-TGGAAAAGGGATATGCTTGC-3′ and 5′-CTGCCATCAATGCTTTCTG-3′;
UBQ10,5′-AAATCTCGTCTCTGTTATGCTTAAGAAG-3′ and 5′-TTTTACATGAAACGAAACATTGAACTT-3′.
ChIP experiments were performed as a modified version of a previously reported protocol (Matías-Hernández et al., 2010). The direct binding of TEM1 and TEM2 to the regulatory regions of putative targets was assayed using the P35S:TEM1-HA lines previously described (Osnato et al., 2012) and P35S:TEM2-HA. Wild-type plants were used as negative controls. The cross-linked DNA was immuno-precipitated with an anti-HA antibody (Sigma-Aldrich, St. Louis, MO), purified using Protein A-Agarose resin (Millipore, Billerica, MA), and tested by qPCR using different primer sets specific for putative direct targets, as listed in Supplemental Table S2.
Enrichment of the target region was determined using a Sybr Green Assay (SYBR Green Supermix; Roche). The quantitative real-time PCR assay was conducted in triplicate and was performed in a LightCycler480 System (Roche). Relative enrichment was calculated normalizing the amount of immunoprecipitated DNA against a UBIQUITIN (UBQ10) fragment and against total INPUT DNA. In particular, for the binding of TEM1 and TEM2 to the selected genomic regions, the affinity of the purified sample obtained in the P35S:TEM1-HA and P35S:TEM2-HA lines background was compared with the affinity-purified sample obtained in the wild-type background, which was used as negative control. Fold enrichment was calculated using the following formulas, where Ct.tg is target gene mean value, Ct.i is input DNA mean value, and Ct.nc is ubiquitin (negative control) mean value: dCT.tg = CT.i-CT.tg and dCT.nc = CT. i-CT.nc. The propagated error values of these CTs are calculated: dSD.tg = sqrt((SD.i)^2+ (SD.tg^2)/sqrt(n) and dSD.nc = sqrt((SD.i)^2+ (SD.nc^2)/sqrt(n), where n = number of replicate per sample. Fold-change over negative control (ubiquitin and wild-type plants) was calculated finding the “delta delta CT” of the target region as follows: ddCT = dCT.tg-dCT.nc and ddSD = sqrt((dSD.tg)^2+ (dSD.nc)^2. The transformation to linear fold-change values is obtained as follows: FC = 2^(ddCT) and FC.error = ln(2)*ddSD*FC.
On the other side, PGL2:GUS seedlings were used for GUS expression analyses. PGL2:GUS plants were crossed with tem1-1 tem2-2 and P35S:TEM2, and three plant generations (F3) were grown in order to obtain homozygous PGL2:GUStem1 tem2 and PGL2:GUSP35S:TEM2. All GUS staining assays were performed overnight as described previously (Blázquez et al., 1997; Liljegren et al., 2000). Samples were incubated in clearing solution, dissected, and observed using a model no. DP71 microscope equipped with DIC optics (Olympus).
In silico analyses of NPF genes for checking if they were expressed in the rosette mesophyll was done using Arabidopsis eFP Browser 2.0:
http://bar.utoronto.ca/efp2/Arabidopsis/Arabidopsis_eFPBrowser2.html.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Trichome number on different upper inflorescence organs.
Supplemental Figure S2. PTEM1:GUS and PTEM2:GUS are highly expressed through trichome development.
Supplemental Figure S6. Regulation of other trichome genes by TEMPRANILLO.
Supplemental Figure S7. TEM affects trichome pGL2:GUS activity.
Supplemental Table S1. Trichome number and density on different TEM genetic backgrounds.
Supplemental Table S2. List of primers used for cloning and ChIP.
Supplemental Table S3. Statistical analyses of data from Figures 1, 2, 4, 5, 6, and 7.
Supplementary Material
Acknowledgments
We thank Rossana Henriques and Josep Casacuberta for helpful discussions and critical reading of the manuscript; Monste Amenós and the High Resolution Imaging Facility at the Universitat Autònoma de Barcelona for help with confocal microscopy and the ultra-thin transverse sections and imaging, respectively; and Jose Luis Micol for providing cue1-6 seeds and NASC for seeds. A.E.A.-J. performed this work within the framework of a Ph.D. Program of the Universitat Autònoma de Barcelona.
Glossary
- amiRNA
artificial microRNA
- cab3-1
CHLOROPHYLL A/B BINDING PROTEIN
- ChIP
chromatin immuno-precipitation
- CK
creatine kinase
- CPC
CAPRICE
- cue1-6
CHLOROPHYLL A/B-BINDING PROTEIN-UNDEREXPRESSED1-1 mutant
- DAB
days after bolting
- DAG
days after germination
- EGL3
ENHANCER OF GLABRA3
- ETC1
ENHANCER OF TRIPTYCHON AND CAPRICE1
- GA
gibberellin
- GIS
GLABROUS INFLORESCENCE STEMS
- GIS2
GLABROUS INFLORESCENCE STEMS2
- GL1
R2R3 MYB GLABROUS1
- GL2
GLABROUS2
- GL3
GLABRA3
- NPF
NITRIDE PEPTIDE TRANSPORTER FAMILY
- PCAB3
CHLOROPHYLL A/B BINDING PROTEIN 3
- RAV
R̲ELATED TO A̲BI3 AND V
 P1
P1- SPY
SPINDLY
- TEM
TEMPRANILLO
- TTG1
TRANSPARENT TESTA GLABROUS1
- UBQ10
UBITIQUIN
- ZFP8
ZINC FINGER PROTEIN8
Footnotes
This work was supported by a MINECO/FEDER grant (no. BFU2012-33746). The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Articles can be viewed without a subscription.
References
- Ahmad P, Ashraf M, Younis M, Hu X, Kumar A, Akram NA, Al-Qurainy F (2012) Role of transgenic plants in agriculture and biopharming. Biotechnol Adv 30: 524–540 [DOI] [PubMed] [Google Scholar]
- An L, Zhou Z, Su S, Yan A, Gan Y (2012) GLABROUS INFLORESCENCE STEMS (GIS) is required for trichome branching through gibberellic acid signaling in Arabidopsis. Plant Cell Physiol 53: 457–469 [DOI] [PubMed] [Google Scholar]
- Balkunde R, Bouyer D, Hülskamp M (2011) Nuclear trapping by GL3 controls intercellular transport and redistribution of TTG1 protein in Arabidopsis. Development 138: 5039–5048 [DOI] [PubMed] [Google Scholar]
- Balkunde R, Pesch M, Hülskamp M (2010) Trichome patterning in Arabidopsis thaliana from genetic to molecular models. Curr Top Dev Biol 91: 299–321 [DOI] [PubMed] [Google Scholar]
- Bernhardt C, Zhao M, Gonzalez A, Lloyd A, Schiefelbein J (2005) The bHLH genes GL3 and EGL3 participate in an intercellular regulatory circuit that controls cell patterning in the Arabidopsis root epidermis. Development 132: 291–298 [DOI] [PubMed] [Google Scholar]
- Blázquez MA, Soowal LN, Lee I, Weigel D (1997) LEAFY expression and flower initiation in Arabidopsis. Development 124: 3835–3844 [DOI] [PubMed] [Google Scholar]
- Bouyer D, Geier F, Kragler F, Schnittger A, Pesch M, Wester K, Balkunde R, Timmer J, Fleck C, Hülskamp M (2008) Two-dimensional patterning by a trapping/depletion mechanism: the role of TTG1 and GL3 in Arabidopsis trichome formation. PLoS Biol 6: e141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillejo C, Pelaz S (2008) The balance between CONSTANS and TEMPRANILLO activities determines FT expression to trigger flowering. Curr Biol 18: 1338–1343 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Curtis MD, Grossniklaus U (2003) A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol 133: 462–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiba Y, Shimizu T, Miyakawa S, Kanno Y, Koshiba T, Kamiya Y, Seo M (2015) Identification of Arabidopsis thaliana NRT1/PTR FAMILY (NPF) proteins capable of transporting plant hormones. J Plant Res 128: 679–686 [DOI] [PubMed] [Google Scholar]
- Chien JC, Sussex IM (1996) Differential regulation of trichome formation on the adaxial and abaxial leaf surfaces by gibberellins and photoperiod in Arabidopsis thaliana (L.) Heynh. Plant Physiol 111: 1321–1328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Aloia M, Bonhomme D, Bouché F, Tamseddak K, Ormenese S, Torti S, Coupland G, Périlleux C (2011) Cytokinin promotes flowering of Arabidopsis via transcriptional activation of the FT paralogue TSF. Plant J 65: 972–979 [DOI] [PubMed] [Google Scholar]
- Deeks MJ, Hussey PJ (2003) Arp2/3 and ‘the shape of things to come’. Curr Opin Plant Biol J 6: 561–567 [DOI] [PubMed] [Google Scholar]
- Endo M, Mochizuki N, Suzuki T, Nagatani A (2007) CRYPTOCHROME2 in vascular bundles regulates flowering in Arabidopsis. Plant Cell 19: 84–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu M, Kang HK, Son SH, Kim SK, Nam KH (2014) A subset of Arabidopsis RAV transcription factors modulates drought and salt stress responses independent of ABA. Plant Cell Physiol 55: 1892–1904 [DOI] [PubMed] [Google Scholar]
- Gan Y, Kumimoto R, Liu C, Ratcliffe O, Yu H, Broun P (2006) GLABROUS INFLORESCENCE STEMS modulates the regulation by gibberellins of epidermal differentiation and shoot maturation in Arabidopsis. Plant Cell 18: 1383–1395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan Y, Liu C, Yu H, Broun P (2007) Integration of cytokinin and gibberellin signalling by Arabidopsis transcription factors GIS, ZFP8 and GIS2 in the regulation of epidermal cell fate. Development 134: 2073–2081 [DOI] [PubMed] [Google Scholar]
- Gilding EK, Marks MD (2010) Analysis of purified glabra3-shapeshifter trichomes reveals a role for NOECK in regulating early trichome morphogenic events. Plant J 64: 304–317 [DOI] [PubMed] [Google Scholar]
- Glauser G, Vallat A, Balmer D (2014) Hormone profiling. Methods Mol Biol 1062: 597–608 [DOI] [PubMed] [Google Scholar]
- Hülskamp M. (2004) Plant trichomes: a model for cell differentiation. Nat Rev Mol Cell Biol 5: 471–480 [DOI] [PubMed] [Google Scholar]
- Julián E, Roldán M, Sánchez-Chardi A, Astola O, Agustí G, Luquin M (2010) Microscopic cords, a virulence-related characteristic of Mycobacterium tuberculosis, are also present in nonpathogenic mycobacteria. J Bacteriol 192: 1751–1760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanno Y, Hanada A, Chiba Y, Ichikawa T, Nakazawa M, Matsui M, Koshiba T, Kamiya Y, Seo M (2012) Identification of an abscisic acid transporter by functional screening using the receptor complex as a sensor. Proc Natl Acad Sci USA 109: 9653–9658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi M, Inzé D, Depicker A (2002) GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci 7: 193–195 [DOI] [PubMed] [Google Scholar]
- King RW, Evans LT, Mander LN, Moritz T, Pharis RP, Twitchin B (2003) Synthesis of gibberellin GA6 and its role in flowering of Lolium temulentum. Phytochemistry 62: 77–82 [DOI] [PubMed] [Google Scholar]
- Kirik V, Simon M, Huelskamp M, Schiefelbein J (2004) The ENHANCER OF TRY AND CPC1 gene acts redundantly with TRIPTYCHON and CAPRICE in trichome and root hair cell patterning in Arabidopsis. Dev Biol 268: 506–513 [DOI] [PubMed] [Google Scholar]
- Krouk G, Lacombe B, Bielach A, Perrine-Walker F, Malinska K, Mounier E, Hoyerova K, Tillard P, Leon S, Ljung K, Zazimalova E, Benkova E, et al. (2010) Nitrate-regulated auxin transport by NRT1.1 defines a mechanism for nutrient sensing in plants. Dev Cell 18: 927–937 [DOI] [PubMed] [Google Scholar]
- Langdale JA. (1998) Cellular differentiation in the leaf. Curr Opin Cell Biol 10: 734–738 [DOI] [PubMed] [Google Scholar]
- Léran S, Varala K, Boyer JC, Chiurazzi M, Crawford N, Daniel-Vedele F, David L, Dickstein R, Fernandez E, Forde B, Gassmann W, Geiger D, et al. (2014) A unified nomenclature of NITRATE TRANSPORTER 1/PEPTIDE TRANSPORTER family members in plants. Trends Plant Sci 19: 5–9 [DOI] [PubMed] [Google Scholar]
- Li H, Culligan K, Dixon RA, Chory J (1995) CUE1: a mesophyll cell-specific positive regulator of light-controlled gene expression in Arabidopsis. Plant Cell 7: 1599–1610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liljegren SJ, Ditta GS, Eshed Y, Savidge B, Bowman JL, Yanofsky MF (2000) SHATTERPROOF MADS-box genes control seed dispersal in Arabidopsis. Nature 404: 766–770 [DOI] [PubMed] [Google Scholar]
- Lin Q, Aoyama T (2012) Pathways for epidermal cell differentiation via the homeobox gene GLABRA2: update on the roles of the classic regulator. J Integr Plant Biol 54: 729–737 [DOI] [PubMed] [Google Scholar]
- Lundquist PK, Rosar C, Bräutigam A, Weber AP (2014) Plastid signals and the bundle sheath: mesophyll development in reticulate mutants. Mol Plant 7: 14–29 [DOI] [PubMed] [Google Scholar]
- Marín-González E, Matías-Hernández L, Aguilar-Jaramillo A-E, Lee J-H, Ahn J-H, Suárez-López P, Pelaz S (2015) SHORT VEGETATIVE PHASE up-regulates TEMPRANILLO2 floral repressor at low ambient temperatures. Plant Physiol 169: 1214–1224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matías-Hernández L, Aguilar-Jaramillo AE, Marín-González E, Suárez-López P, Pelaz S (2014) RAV genes: regulation of floral induction and beyond. Ann Bot (Lond) 114: 1459–1470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matías-Hernandez L, Battaglia R, Galbiati F, Rubes M, Eichenberger C, Grossniklaus U, Kater MM, Colombo L (2010) VERDANDI is a direct target of the MADS domain ovule identity complex and affects embryo sac differentiation in Arabidopsis. Plant Cell 22: 1702–1715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchum MG, Yamaguchi S, Hanada A, Kuwahara A, Yoshioka Y, Kato T, Tabata S, Kamiya Y, Sun TP (2006) Distinct and overlapping roles of two gibberellin 3-oxidases in Arabidopsis development. Plant J 45: 804–818 [DOI] [PubMed] [Google Scholar]
- Murphy DJ. (2007) Improving containment strategies in biopharming. Plant Biotechnol J 5: 555–569 [DOI] [PubMed] [Google Scholar]
- Nemhauser JL, Hong F, Chory J (2006) Different plant hormones regulate similar processes through largely nonoverlapping transcriptional responses. Cell 126: 467–475 [DOI] [PubMed] [Google Scholar]
- Olsson ME, Olofsson LM, Lindahl AL, Lundgren A, Brodelius M, Brodelius PE (2009) Localization of enzymes of artemisinin biosynthesis to the apical cells of glandular secretory trichomes of Artemisia annua L. Phytochemistry 70: 1123–1128 [DOI] [PubMed] [Google Scholar]
- Osnato M, Castillejo C, Matías-Hernández L, Pelaz S (2012) TEMPRANILLO genes link photoperiod and gibberellin pathways to control flowering in Arabidopsis. Nat Commun 3: 808. [DOI] [PubMed] [Google Scholar]
- Payne CT, Zhang F, Lloyd AM (2000) GL3 encodes a bHLH protein that regulates trichome development in arabidopsis through interaction with GL1 and TTG1. Genetics 156: 1349–1362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perazza D, Herzog M, Hülskamp M, Brown S, Dorne AM, Bonneville JM (1999) Trichome cell growth in Arabidopsis thaliana can be derepressed by mutations in at least five genes. Genetics 152: 461–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perazza D, Vachon G, Herzog M (1998) Gibberellins promote trichome formation by Up-regulating GLABROUS1 in arabidopsis. Plant Physiol 117: 375–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranjan A, Fiene G, Fackendahl P, Hoecker U (2011) The Arabidopsis repressor of light signaling SPA1 acts in the phloem to regulate seedling de-etiolation, leaf expansion and flowering time. Development 138: 1851–1862 [DOI] [PubMed] [Google Scholar]
- Riechmann JL, Heard J, Martin G, Reuber L, Jiang C, Keddie J, Adam L, Pineda O, Ratcliffe OJ, Samaha RR, Creelman R, Pilgrim M, et al. (2000) Arabidopsis transcription factors: genome-wide comparative analysis among eukaryotes. Science 290: 2105–2110 [DOI] [PubMed] [Google Scholar]
- Rieu I, Powers SJ (2009) Real-time quantitative RT-PCR: design, calculations, and statistics. Plant Cell 21: 1031–1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rombolá-Caldentey B, Rueda-Romero P, Iglesias-Fernández R, Carbonero P, Oñate-Sánchez L (2014) Arabidopsis DELLA and two HD-ZIP transcription factors regulate GA signaling in the epidermis through the L1 box cis-element. Plant Cell 26: 2905–2919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito H, Oikawa T, Hamamoto S, Ishimaru Y, Kanamori-Sato M, Sasaki-Sekimoto Y, Utsumi T, Chen J, Kanno Y, Masuda S, Kamiya Y, Seo M, et al. (2015) The jasmonate-responsive GTR1 transporter is required for gibberellin-mediated stamen development in Arabidopsis. Nat Commun 6: 6095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez-Chardi A, Olivares F, Byrd TF, Julián E, Brambilla C, Luquin M (2011) Demonstration of cord formation by rough Mycobacterium abscessus variants: implications for the clinical microbiology laboratory. J Clin Microbiol 49: 2293–2295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savage NS, Walker T, Wieckowski Y, Schiefelbein J, Dolan L, Monk NA (2008) A mutual support mechanism through intercellular movement of CAPRICE and GLABRA3 can pattern the Arabidopsis root epidermis. PLoS Biol 6: e235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schellmann S, Schnittger A, Kirik V, Wada T, Okada K, Beermann A, Thumfahrt J, Jürgens G, Hülskamp M (2002) TRIPTYCHON and CAPRICE mediate lateral inhibition during trichome and root hair patterning in Arabidopsis. EMBO J 21: 5036–5046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schilmiller AL, Last RL, Pichersky E (2008) Harnessing plant trichome biochemistry for the production of useful compounds. Plant J 54: 702–711 [DOI] [PubMed] [Google Scholar]
- Shan CM, Shangguan XX, Zhao B, Zhang XF, Chao LM, Yang CQ, Wang LJ, Zhu HY, Zeng YD, Guo WZ, Zhou BL, Hu GJ, et al. (2014) Control of cotton fibre elongation by a homeodomain transcription factor GhHOX3. Nat Commun 5: 5519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shani E, Weinstain R, Zhang Y, Castillejo C, Kaiserli E, Chory J, Tsien RY, Estelle M (2013) Gibberellins accumulate in the elongating endodermal cells of Arabidopsis root. Proc Natl Acad Sci USA 110: 4834–4839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith LG. (2003) Cytoskeletal control of plant cell shape: getting the fine points. Curr Opin Plant Biol 6: 63–73 [DOI] [PubMed] [Google Scholar]
- Szymanski DB, Jilk RA, Pollock SM, Marks MD (1998) Control of GL2 expression in Arabidopsis leaves and trichomes. Development 125: 1161–1171 [DOI] [PubMed] [Google Scholar]
- Talke IN, Hanikenne M, Krämer U (2006) Zinc-dependent global transcriptional control, transcriptional deregulation, and higher gene copy number for genes in metal homeostasis of the hyperaccumulator Arabidopsis halleri. Plant Physiol 142: 148–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao Z, Shen L, Liu C, Liu L, Yan Y, Yu H (2012) Genome-wide identification of SOC1 and SVP targets during the floral transition in Arabidopsis. Plant J 70: 549–561 [DOI] [PubMed] [Google Scholar]
- Traw MB, Bergelson J (2003) Interactive effects of jasmonic acid, salicylic acid, and gibberellin on induction of trichomes in Arabidopsis. Plant Physiol 133: 1367–1375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsay YF, Chiu CC, Tsai CB, Ho CH, Hsu PK (2007) Nitrate transporters and peptide transporters. FEBS Lett 581: 2290–2300 [DOI] [PubMed] [Google Scholar]
- Wang XHLQ, Li QT, Chen HW, Zhang WK, Ma B, Chen SY, Zhang JS (2014) Trihelix transcription factor GT-4 mediates salt tolerance via interaction with TEM2 in Arabidopsis. BMC Plant Biol 14: 339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss D, Ori N (2007) Mechanisms of cross talk between gibberellin and other hormones. Plant Physiol 144: 1240–1246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolters H, Jürgens G (2009) Survival of the flexible: hormonal growth control and adaptation in plant development. Nat Rev Genet 10: 305–317 [DOI] [PubMed] [Google Scholar]
- Woo HR, Kim JH, Kim J, Kim J, Lee U, Song IJ, Kim JH, Lee HY, Nam HG, Lim PO (2010) The RAV1 transcription factor positively regulates leaf senescence in Arabidopsis. J Exp Bot 61: 3947–3957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi S. (2008) Gibberellin metabolism and its regulation. Annu Rev Plant Biol 59: 225–251 [DOI] [PubMed] [Google Scholar]
- Yant L, Mathieu J, Dinh TT, Ott F, Lanz C, Wollmann H, Chen X, Schmid M (2010) Orchestration of the floral transition and floral development in Arabidopsis by the bifunctional transcription factor APETALA2. Plant Cell 22: 2156–2170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu N, Cai WJ, Wang S, Shan CM, Wang LJ, Chen XY (2010) Temporal control of trichome distribution by microRNA156-targeted SPL genes in Arabidopsis thaliana. Plant Cell 22: 2322–2335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L, Luo Q, Yang C, Han Y, Li W (2008b) A RAV-like transcription factor controls photosynthesis and senescence in soybean. Planta 227: 1389–1399 [DOI] [PubMed] [Google Scholar]
- Zhao M, Morohashi K, Hatlestad G, Grotewold E, Lloyd A (2008a) The TTG1-bHLH-MYB complex controls trichome cell fate and patterning through direct targeting of regulatory loci. Development 135: 1991–1999 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.