During germination, defective xylose trimming alters xyloglucan anisotropic localization in cell walls during hypocotyl elongation and decreases endosperm resistance to radicle protrusion
Abstract
Cell wall remodeling is an essential mechanism for the regulation of plant growth and architecture, and xyloglucans (XyGs), the major hemicellulose, are often considered as spacers of cellulose microfibrils during growth. In the seed, the activity of cell wall enzymes plays a critical role in germination by enabling embryo cell expansion leading to radicle protrusion, as well as endosperm weakening prior to its rupture. A screen for Arabidopsis (Arabidopsis thaliana) mutants affected in the hormonal control of germination identified a mutant, xyl1, able to germinate on paclobutrazol, an inhibitor of gibberellin biosynthesis. This mutant also exhibited reduced dormancy and increased resistance to high temperature. The XYL1 locus encodes an α-xylosidase required for XyG maturation through the trimming of Xyl. The xyl1 mutant phenotypes were associated with modifications to endosperm cell wall composition that likely impact on its resistance, as further demonstrated by the restoration of normal germination characteristics by endosperm-specific XYL1 expression. The absence of phenotypes in mutants defective for other glycosidases, which trim Gal or Fuc, suggests that XYL1 plays the major role in this process. Finally, the decreased XyG abundance in hypocotyl longitudinal cell walls of germinating embryos indicates a potential role in cell wall loosening and anisotropic growth together with pectin de-methylesterification.
Seed germination is a complex process that begins with the absorption of water and ends when the radicle breaks through the seed coat (or testa). In Arabidopsis (Arabidopsis thaliana), as in most angiosperms, the embryo is surrounded by the triploid endosperm and the seed coat of maternal origin (Nonogaki et al., 2010; North et al., 2010). The completion of germination requires the growth potential of the embryo to overcome the resistance of the endosperm and testa layers, which is controlled by the hormonal balance between abscisic acid (ABA) and gibberellins (GAs). During seed development, ABA induces embryo growth arrest at the transition from embryogenesis to the phase of reserve accumulation and then induces primary dormancy, thus preventing vivipary and allowing seed dispersal in a dormant state. Dormancy delays germination until environmental conditions become favorable for seedling survival and growth (Finkelstein et al., 2008; Nambara et al., 2010; Graeber et al., 2012). Dormancy depth varies among plant species and between Arabidopsis accessions; however, seed dormancy of the most commonly used accession Columbia-0 (Col-0) is relatively low and can be released by a few weeks of after-ripening (dry storage) or stratification (cold imbibition). Shortly after hydration, ABA is rapidly degraded in both dormant and nondormant seeds, but ABA catabolism is more active in nondormant seeds, leading to lower ABA levels and thus allowing GA activation of germination processes (Millar et al., 2006). GA increases the elasticity of the wall, thereby reducing the resistance of the endosperm while triggering the elongation of the hypocotyl (Nonogaki et al., 2010). Radicle protrusion through the micropylar endosperm is also stimulated by ethylene, which has an antagonist action with ABA on endosperm cap weakening (Linkies and Leubner-Metzger, 2012). Microarray analyses highlighted the importance of cell wall remodeling processes during germination in various species (Penfield et al., 2006; Morris et al., 2011; Endo et al., 2012; Martínez-Andújar et al., 2012; Dekkers et al., 2013). These studies provided compelling evidence that the tissue-specific expression of genes encoding cell wall biosynthesis or modification enzymes, and their differential response to hormonal signals in the endosperm and embryo, influences the rate of germination.
Cell walls are constituted of crystalline cellulose microfibrils that are embedded in an amorphous matrix of complex polysaccharides: pectin and hemicelluloses. Xyloglucan (XyG) is the major hemicellulose polymer in the primary cell walls of gymnosperms and most angiosperms, and its binding to cellulose microfibrils by hydrogen bonding contributes to loosening or stiffening of the wall during cell elongation (Cosgrove, 2005). XyG chains can be cleaved and reconnected by endo-transglycosylases/hydrolases (XTH). Other families of proteins also act on XyG chains, such as expansins, which are thought to nonenzymatically modulate XyG interactions with cellulose microfibrils, thereby controlling the distance between the microfibrils. XyG has a backbone of (1→4)-linked β-d-glucopyranosyl residues, which can be substituted with α-d-xylopyranosyl residues at O-6 (Supplemental Fig. S1). The pattern of XyG substitutions is described using a single-letter nomenclature (Fry et al., 1993). The letter G is used for an unsubstituted Glc and X when it is substituted with a Xyl. In Arabidopsis, like in many other dicots, the xylosylation pattern is in general regular, consisting mainly of XXXG-type units. The xylosyl residue can be further substituted at O-2 with a β-galactosyl (L side chain), which in turn can be substituted at O-2 with α-l-fucosyl (F side chain).
Many of the biosynthetic enzymes involved in XyG biosynthesis have been identified, including a glucan synthase, xylosyl, galactosyl, and fucosyltransferases (Scheller and Ulvskov, 2010). Among these, two xylosyltransferases, named XXT1 and XXT2, have been shown to be involved in the synthesis of XyG in Arabidopsis, and the double mutant xxt1 xxt2 lacks detectable XyG (Cavalier et al., 2008). Both belong to the GT34 subfamily of glycosyltransferases, and a third enzyme, XXT5 from a separate clade of GT34, may also be involved in XyG synthesis (Zabotina et al., 2008). These glycosyltransferases are Golgi-localized enzymes, which produce substituted XyG precursors that are secreted into the cell wall. Subsequent trimming of XyG chains is performed by apoplastic glycosidases and determines hemicellulose structure and properties in the wall (Scheller and Ulvskov, 2010). A number of genes involved in the XyG metabolism have been identified, including XYL1, BGAL10, and AXY8 encoding, respectively, α-xylosidase, β-galactosidase, and α-fucosidase (Sampedro et al., 2010; Günl et al., 2011; Günl and Pauly, 2011; Sampedro et al., 2012). Loss of function of these glycosidases results in significant alterations in XyG composition. Although XyG has been proposed to be a major player in cell wall extension and plant growth, mutants with altered XyG composition display only minor growth-related phenotypes. The XyG-deficient double mutant xxt1 xxt2 shows no major growth defect except for deformed root hairs (Cavalier et al., 2008). Nevertheless, it was recently reported that the production of Gal-depleted XyG causes dwarfism in the galactosyltransferase mutant mur3 (Kong et al., 2015) in contrast to xyl1 and bgal10, where increased galactosylation results in shorter but wider siliques (Sampedro et al., 2010; Günl and Pauly, 2011; Sampedro et al., 2012). Phenotypes have not been observed from either reduced or increased fucosylation in the fucosyltransferase mutant mur2 and fucosidase mutant axy8 (Vanzin et al., 2002; Günl et al., 2011). AXY8 overexpression does, however, restore hypocotyl elongation in dwarf AUXIN BINDING PROTEIN1 knockdown seedlings. This demonstrates that in muro removal of Fuc residues can modulate cell elongation (Paque et al., 2014).
In contrast to the numerous studies on the impact of XyG composition on plant growth, little information is available on the role of XyG in seed development or germination. A recent study highlighted the slower germination rate of xxt1 xxt2 mutant seeds compared to wild type, whereas germination rates of the arabinan-deficient arad1 arad2 and putative pectin methyltransferase qua2 mutants were not affected (Lee et al., 2012). As mentioned above, XyG chain hydrolysis and linkage is catalyzed by XTH activities, one of which, AtXTH31/XTR8, is encoded by an endosperm-specific gene. Loss of function leads to faster germination, suggesting that AtXTH31/XTR8 is involved in the reinforcement of the cell wall of the endosperm during germination (Endo et al., 2012). Here, we report the identification of an additional xyl1 allele from a screen designed to isolate mutants impaired in the hormonal control of germination, based on their ability to germinate on the GA biosynthesis inhibitor paclobutrazol. To investigate the role of XyG metabolism in seed dormancy and germination characteristics, xyl1 seed phenotypes were correlated with spatio-temporal XyG accumulation during seed development and germination. Comparative studies using mutants impaired in two other apoplastic glycosidases, BGAL10 and AXY8, indicate a major role for XYL1 in XyG remodelling processes that affect germination.
RESULTS
Identification of the Paclobutrazol-Resistant Mutant xyl1-4
A screen of an ethyl-methanesulfonate (EMS) mutagenized population was carried out for seeds able to germinate on paclobutrazol, and a novel mutant, later named xyl1-4, was isolated. This screen has previously been used to select ABA-deficient mutants (Léon-Kloosterziel et al., 1996; Nambara et al., 1998; North et al., 2007), and seed dormancy was reduced in xyl1-4 as reported for most ABA-deficient mutants (Fig. 1, A and B). ABA contents of dry seeds were, however, slightly higher than wild type, indicating that this mutant was not impaired in ABA biosynthesis (Fig. 1C). Furthermore, the germination rate of xyl1 mutants was slower than that of ABA-deficient mutants (Supplemental Fig. S2; North et al., 2007).
Figure 1.
Seed phenotypes of wild type and xyl1 mutants. A, Paclobutrazol resistance of stratified seeds. B, Germination of freshly harvested seeds. C, ABA content in dry seeds. D, Gene model indicating the position of xyl1 mutations. The number of seedlings with green cotyledons was scored 7 d after sowing (A) or the number of seeds with a protruding radicle after 15 d (B); both were then compared to the total number of seeds sown. Results are means of 3 (A, B) and 4 (C) biological replicates with SE. In D, exons are represented by white rectangles and introns are hatched.
The mutation affecting germination was mapped to a region on the lower arm of chromosome 1 between 24,699 and 25,665 Mb. Whole-genome sequencing identified a G to A substitution typical of EMS mutagenesis, which introduced a premature stop codon in the At1g68560 gene (Fig. 1D). This locus encodes an α-xylosidase belonging to glycoside hydrolase family 31 (GH31; Cantarel et al., 2009), already known as XYL1 and AXY3 (Sampedro et al., 2010; Günl and Pauly, 2011). In accordance with the nomenclature first used by Sampedro et al. (2001), our mutant allele was named xyl1-4. Although XYL1 is the only α-xylosidase to be described that modifies XyG composition in vitro (Sampedro et al., 2001) and in vivo (Sampedro et al., 2010; Günl and Pauly, 2011), only mild vegetative phenotypes were observed for xyl1-4, as previously described for xyl1 alleles: reduced silique length and increased silique width (Supplemental Fig. S3). Paclobutrazol resistance, reduced dormancy, and ABA levels were analyzed for the T-DNA insertion allele xyl1-2/axy3-2 (Sampedro et al., 2010; Günl and Pauly, 2011) and showed that xyl1-2 and xyl1-4 alleles had similar phenotypes, confirming that these were due to defects in XYL1 function (Fig. 1).
xyl1 Mutations Reduce the Relative Abundance of Fucosylated Residues in Seed Tissues
Mutations in XYL1 have been shown to alter XyG accumulation in vegetative tissues (Sampedro et al., 2010; Günl and Pauly, 2011). To further compare xyl1-4 to xyl1-2 phenotypes and evaluate the effect of these mutations on XyG tissue-specific accumulation in seeds, XyG composition was analyzed by matrix-assisted laser-desorption ionization time of flight mass spectrometry. Measurement of relative contents of major oligosaccharide subunits in each extract was performed after endoglucanase digestion (Fig. 2). Seeds were dissected and separated into embryo and endosperm/testa fractions, either shortly after transfer of stratified seeds to 25°C in continuous light (3 h of imbibition) or at endosperm rupture (ER), when germination sensu stricto is completed and radicle protrusion is observed (after about 20 h of imbibition).
Figure 2.
Matrix-assisted laser-desorption ionization time of flight analysis of XyGs of wild-type (black bars), xyl1-2 (white bars), and xyl1-4 (gray bars) germinating seeds. Analysis was performed on 3-d-stratified seeds after imbibition at 25°C in the light. Embryos were separated from testa and endosperm by dissection after 3 h at 25°C and at ER. Relative abundance of major subunits is shown as means of four biological replicates with SE. Minor subunits (<5% relative abundance) are not shown. Mean values represent acetylated and nonacetylated subunits of XXLG, XXFG, and XLFG combined.
In both embryo and endosperm/testa fractions, a reduction in the relative abundance of fucosylated subunits (XXFG, XLFG) was correlated with an increased abundance of less substituted subunits (XXXG and XXLG) in both xyl1 alleles compared to wild type. Although XyG embryo composition was similar to that reported for seedlings (Günl and Pauly, 2011) or stems (Sampedro et al., 2010), highly substituted XLFG subunits were more abundant in endosperm/testa (Fig. 2B) than in embryo fractions (Fig. 2A).
Both xyl1-2 and xyl1-4 Impair XYL1 Protein Accumulation
The xyl1-2 mutant contains a T-DNA insertion in the second exon of XYL1, and this has been shown to prevent transcript accumulation (Günl and Pauly, 2011). In xyl1-4, the mutation is located at the 3′ end of the third exon, 200 nucleotides upstream of the stop codon (Fig. 1D), introducing a premature stop codon that would lead to the production of a truncated protein. To determine the impact of these two mutations on XYL1 protein accumulation, bidimensional (2D) gel electrophoresis was performed from dry mature seed protein extracts (Fig. 3). XYL1 protein accumulation has been previously observed during wild-type Arabidopsis seed germination but was not detected in the neosynthesised proteome (Galland et al., 2014). Comparative analysis of 2D gels indicated the absence, in dry seeds of both xyl1-2 and xyl1-4 mutants, of 4 protein spots detected in wild-type extracts (Fig. 3B-D). These 4 spots were analyzed by tandem mass spectrometry and confirmed to be the XYL1 protein (Supplemental Table S1). This highlighted the presence of multiple XYL1 isoforms; the shifts in mass and pI suggested that these isoforms might result from posttranslational modifications (Fig. 3B), which could not be determined by our mass spectrometry analysis. Nevertheless, in accordance with observed phenotypic alterations, our data provided evidence that neither xyl1-2 nor xyl1-4 seeds produce a full-length protein.
Figure 3.
XYL1 protein accumulation in dry wild-type (WT) and mutant seeds. Total soluble proteins were separated by 2D gel electrophoresis and silver nitrate stained. For wild type, the selected area of the gel (A) was enlarged in B; for xyl1 alleles, only the enlarged area is shown (C, D). The arrows indicate four spots corresponding to XYL1 isoforms detected in wild-type dry seeds, but absent in xyl mutants. Similar results were obtained in three independent experiments.
Other Modifications of XyG Maturation Pathway Do Not Affect Germination
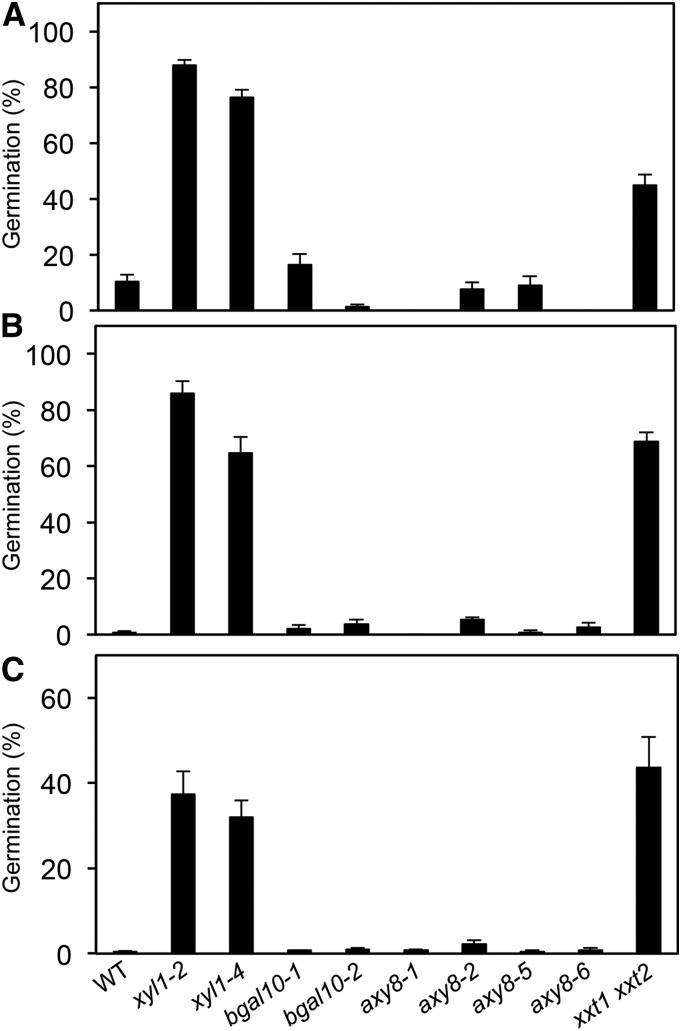
In addition to XYL1, two other glycosidases, BGAL10 and AXY8, have been characterized that trim Gal and Fuc, respectively, off XyG side chains (Günl et al., 2011; Sampedro et al., 2012). To determine whether mutations of BGAL10 and AXY8 affect germination characteristics, we assessed dormancy, sensitivity to paclobutrazol, and thermoinhibition at 34°C of two T-DNA insertion alleles (bgal10-1 and bgal10-2; Sampedro et al., 2012), two EMS (axy8-1 and axy8-2), and two T-DNA insertion alleles (axy8-5 and axy8-6; Günl et al., 2011). Phenotypes of the xylosyltransferase double mutant xxt1 xxt2, which lacks XyG side chains (Cavalier et al., 2008), were also compared with xyl1.
In contrast to the increased tolerance to paclobutrazol and thermoinhibition and reduced dormancy of xyl1 mutant seeds, bgal10 and axy8 alleles displayed similar phenotypes to wild type (Fig. 4). Surprisingly, xxt1 xxt2 mutant seeds, which are defective for the addition of Xyl residues to XyG chains, displayed similar phenotypes to xyl1 mutants, which are affected in their removal. To evaluate whether the absence of bgal10 and axy8 seed phenotypes was correlated with the impact of these mutations on other organs, we measured silique size for two mutant alleles for each gene (Supplemental Fig. S3). As already described (Sampedro et al., 2010, 2012), silique length is reduced in xyl1 and bgal10 alleles. Although no vegetative phenotypes were previously observed in axy8 mutants (Günl et al., 2011), reduced silique length was also detected for both axy8 alleles, axy8-1 being less affected than axy8-2. This was possibly due to differences in allele severity. Thus, in contrast to seeds, altered vegetative phenotypes were observed in all three XyG maturation mutants, together with the biosynthesis mutant xxt1 xxt2. Nevertheless, silique growth was less affected in bgal10 and axy8 than in xyl1 and xxt1 xxt2 mutants, suggesting either gene redundancy within BGAL10 and AXY8 families could compensate their defect in seeds or that XYL1 has a specific function in the control of seed dormancy and germination.
Figure 4.
Seed germination phenotypes of wild type (Col-0), XyG hydrolase mutants (xyl1, bgal10, and axy8), and the XyG biosynthesis mutant xxt1 xxt2. A, Paclobutrazol (30 µM) resistance and B, germination at 34°C of stratified seeds. C, Germination of freshly harvested seeds. The number of seeds with protruding radicle (B, C) or seedlings with green cotyledons (A) was scored 5 d after transfer to 25°C and continuous light (A, B) or 14 d after sowing (C) and compared to the total number of seeds. Results are means of three replicates with SE and were obtained for three independent seed batches.
XYL1 Spatio-Temporal Expression in Seeds
XYL1 expression was previously examined in vegetative tissues (Sampedro et al., 2010), but not in seeds. Using a GUS reporter gene transcriptional fusion with a 3-kb XYL1 promoter region, expression was detected in roots, leaves, flowers, and siliques over a range of cell types in three independent transformants. In developing siliques, GUS staining was detected in the valves, style, abscission zone, and pedicels (Sampedro et al., 2010). Here, we used the same three transgenic lines to determine the tissue-specific GUS expression during seed development and germination.
Seeds were isolated from developing siliques and then embryos separated from the surrounding tissues, i.e. testa and endosperm, by dissection. GUS expression was visible at early stages from 6 d after pollination (DAP). Dissection of seeds at 6 DAP was not possible, but GUS staining at 8 and 10 DAP suggested that XYL1 expression at this stage was mainly in the endosperm (Fig. 5A). In contrast to the intense staining detected in this tissue throughout seed development, no GUS expression was observed in the testa. In the embryo, staining was visible ubiquitously at very low levels from 10 DAP and then increased, being strongest at the end of seed development. In imbibed seeds, GUS staining was detected in the embryo and endosperm at similar levels to that observed at the end of seed development up until ER (Fig. 5B). After 48 h of imbibition, strong expression was visible in the tip of the seedling radicle and in the root hair growth region (Fig. 5C). The observed XYL1 expression throughout the embryo and endosperm of developing and imbibed seeds suggests potential roles in both seed development and germination.
Figure 5.
XYL1 expression in seeds. GUS expression from XYL1 promoter in dissected seeds during seed development (A) or imbibition (B) and in germinating seedling (C). In developing seeds from 6 to 20 DAP (A): a, whole seed at 6 DAP; b-d, h- k, embryo; e-g, l-o, endosperm remaining inside the testa. In imbibed seeds 4 and 24 h after sowing (B): a-b, embryo; c-d, testa; and e-f, endosperm remaining around the embryo or inside the testa. C, 48-h germinated seedling. DAP, Days after pollination; HAI, hours after imbibition. Bar = 100 µm for all pictures, as shown in A-o, B-f, and C-b, except for C-a (bar = 50 µm).
XyG Immunolocalization Is Not Altered in xyl1 Developing Seeds
To investigate further the impact of the XYL1 defect on XyG accumulation and its possible link with dormancy and germination phenotypes, the pattern of XyG accumulation was studied in wild-type and xyl1 seeds by immunolocalization using three different antibodies. LM15, LM25, and CCRC-M1 bind specifically to xylosylated (XXXG), galactosylated (XXLG), or fucosylated (XXFG) XyGs, respectively (Puhlmann et al., 1994; Marcus et al., 2008; Pedersen et al., 2012). During seed development, two stages were examined: 10 DAP during the embryo growth phase after the completion of embryogenesis and 14 DAP during reserve accumulation after embryo growth arrest. These stages were chosen as they marked the beginning and end of the observed increase in XYL1 expression in embryos (Fig. 5).
At 10 DAP, all three epitopes were abundant in testa and endosperm cell walls of wild-type seeds (Fig. 6, A and B; Supplemental Fig. S4, A and B). In embryos, XyG were observed in intracellular vesicles and cell walls. Intracellular vesicle labeling (Fig. 6B) suggests a progressive deposition of XyGs in wild-type embryo cell walls during seed development following their synthesis in Golgi vesicles. Furthermore, the stronger labeling in vascular tissues, compared to other hypocotyl cell walls, probably indicates that XyG are deposited earlier in these tissues (Supplemental Fig. S4, B and C). By 14 DAP, XyG appeared intensely labeled and ubiquitously distributed in embryo cell walls (Fig. 6C; Supplemental Fig. S4, C and D). Compared to embryos, endosperm cell walls were relatively more labeled at 10 DAP than at 14 DAP (Fig. 6; Supplemental Fig. S4), also suggesting a differential timing of XyG deposition in these tissues.
Figure 6.
Immunolocalization of XyGs in Arabidopsis wild-type seeds during seed development and early imbibition. Whole seed sections at 10 DAP (A), hypocotyl transverse sections at 10 DAP (B), whole seed sections at 14 DAP (C), and after 3 h of imbibition (D). In magenta: cellulose labeling with Calcofluor White (A1-D1); in green: antibody labeling with LM15 (A2-D2); composite images showing both Calcofluor White and antibody labeling (A3-D3). c, cotyledons; en, endosperm; h, hypocotyl; t, testa; v, vascular bundles. Bar = 50 µm.
As shown in Supplemental Figures S5 and S6, at both stages during seed development, XyG localization in xyl1 mutant cell walls was similar to that of wild type for all seed tissues. Similar relative increases in XyG labeling in embryo cell walls and decreases in endosperm cell walls were observed. These results suggest that during development of both wild-type and xyl1 seeds, embryo growth arrest and transition to reserve accumulation are correlated with an important deposition of XyGs in embryo cell walls and a possible concomitant degradation of XyGs in endosperm cell walls. Nevertheless, there was no clear link between these changes in cellular and tissue-specific XyG distribution and XYL1 spatiotemporal expression (Fig. 5).
In Germinating Embryos, xyl1 Alters Asymmetric Distribution of XyGs But Not Demethylesterified Pectins
To further investigate the evolution of XyG distribution during germination, immunolocalization analyses were performed in mature stratified seeds of wild type and both xyl1 alleles, during imbibition after transfer to 25°C in continuous light, at two time points: 3 h after transfer and at ER. After 3 h, XyG localization in wild-type xyl1 seeds was quite similar in both embryo and endosperm to that observed at 14 DAP, suggesting that no major changes had occurred during late seed development (Fig. 6, C and D; Supplemental Figs. S7 and S8). Furthermore, similar antibody labeling profiles were observed for the three epitopes (XXXG, XXLG, and XXFG) in embryo cell walls. Seed imbibition induced the rupture of the seed coat epidermal cell walls and mucilage release. XyG that were highly accumulated in outer cell walls during seed development were still detected in cell wall fragments (Fig. 6; Supplemental Fig. S7).
Significant variations in epitope abundance were detected in seeds at ER compared to 3-h-imbibed seeds. XyG immunolocalization in germinating wild-type seeds displayed an asymmetric distribution of the three epitopes in hypocotyl cells, with a more intense labeling in transversal than longitudinal cell walls (Fig. 7A). The reduced epitope labeling of longitudinal cell walls was detected only in elongating hypocotyl cells, whereas it remained uniform in cotyledon or root tip cell walls. Furthermore, an accumulation of XyG was also visible in intracellular vesicles (Fig. 7C). In contrast to wild type, the analysis of XyG localization in xyl1 mutants did not show an anisotropic epitope distribution in hypocotyl cell walls (Fig. 7B). Epitope abundance was low in both transversal and longitudinal cell walls, and intense epitope labeling was detected only in intracellular vesicles (Fig. 7D). Although the endosperm layer was not easily visible at this stage due to its rupture, no clear difference in XyG labeling was detected between xyl1 and wild type (Fig. 7). Radicle protrusion was therefore correlated with the disappearance of XyG epitopes in longitudinal cell walls of wild-type hypocotyls and in both transversal and longitudinal cell walls in xyl1.
Figure 7.
Immunolocalization of XyGs and demethylesterified homogalacturonans in wild-type and xyl1-2 germinating seeds at ER. Whole seed sections of wild type (A) and xyl1-2 (B). In magenta: cellulose labeling with Calcofluor White (A1-B1); in green: antibody labeling with LM15 (A2-B2), LM25 (A4-B4), or CCRC-M1 (A5-B5); composite images showing both Calcofluor White and LM15 labeling (A3-B3); enlarged images of CCRC-M1 (C, D) and 2F4 (E, F) labeling in wild type (C, E) and xyl1-2 (D, F) hypocotyls. Similar labeling was observed for both xyl1 alleles, therefore a single representative picture was chosen for each antibody. c, cotyledons; en, endosperm; h, hypocotyl; t, testa; v, vascular bundles. Bar = 50 µm.
We hypothesize that the decreased abundance of XyG observed in longitudinal cell walls at ER (Fig. 7) might be correlated to asymmetric cell wall loosening, leading to hypocotyl directional growth. Selective pectin demethylesterification in longitudinal cell walls has been recently reported to trigger growth asymmetry in etiolated hypocotyls of dark-grown seedlings (Peaucelle et al., 2015). To determine whether pectin demethylesterification also occurred in elongating hypocotyls at ER, germinating seeds of wild-type and xyl1 were examined after immunolocalization using the 2F4 antibody that recognizes Ca2+-crosslinked homogalacturonan with a low degree of methylesterification (Liners et al., 1989). More intense epitope labeling was observed in longitudinal compared to transversal cell walls of both wild-type and xyl1 hypocotyls (Fig. 7, E and F). However labeling was more heterogeneous than XyG epitope labeling, with stronger labeling at cell corners.
Our results thus suggest an involvement of both XyG removal and pectin demethylesterification in longitudinal cell wall loosening during hypocotyl elongation at ER. Furthermore, although XyG labeling was ubiquitously absent in xyl1 hypocotyls, asymmetric growth was not impaired. Indeed, radicle protrusion was synchronously observed after 20 h of imbibition at 25°C in the light in the stratified seeds of both xyl1 and wild type used here for immunolabeling, and the absence of XyG in transversal cell walls did not notably affect directional hypocotyl growth.
XYL1 Activity in Endosperm Controls Seed Germination
Since XyG alterations in xyl1 embryos were not associated with hypocotyl growth modifications, we investigated whether the permeability or mechanical resistance of the testa or endosperm layers was affected. Increased seed coat permeability could allow increased solute leakage from mutant seeds at imbibition. Despite ABA contents in mutant dry seeds being higher than those of wild type (Fig. 1C), xyl1 seeds exhibited a reduced GA requirement for germination (Fig. 1A); this suggested that ABA might be released upon imbibition. Measurement of ABA contents in dry and imbibed seeds found that similar amounts were leached into the imbibition medium (Supplemental Figure S9). This corresponded to a loss of 15% and 12% of dry seed ABA content on imbibition for wild type and mutant seeds, respectively; further decreases could also occur through ABA catabolism.
Reciprocal crosses enable the maternal determinism of altered germination characteristics to be investigated. Since the testa is derived from the ovule integuments, heterozygous F1 zygotes (embryo and endosperm) are surrounded by a maternal testa. Germination rates for freshly harvested seeds showed that F1 seeds, harvested from a mutant xyl1 mother plant fertilized with wild-type pollen, displayed the same phenotype as wild type and were dormant (Fig. 8). Thus, the altered germination of xyl1 seeds did not result from defective XYL1 in testa layers reducing their mechanical resistance.
Figure 8.
Germination of freshly harvested arvested F1 seeds from crosses between wild type and xyl1 mutant plants. Germination was based on testa rupture, and germinated seeds were scored after 14 d of imbibition. Averages of three independent replicates (at least 40 seeds each) are shown with SE.
To determine whether XYL1 activity in the endosperm may be involved in the control of its rupture during seed germination, we produced xyl1-2 primary transformants that expressed the XYL1 gene under the control of the EPR1 promoter, which was previously shown to be endosperm specific during seed development and germination (Dubreucq et al., 2000). Germinating T2 seeds of most lines showed reduced resistance to paclobutrazol compared to xyl1-2. To facilitate subsequent analyses, we selected transgenic lines, segregating with a 3:1 ratio for hygromycin resistance in T2 seeds as predicted for a single T-DNA insertion. Further analysis of germination characteristics of seeds from four independent homozygous transgenic lines showed that the dormancy of freshly harvested seeds and the resistance of stratified seeds to paclobutrazol and thermoinhibition were similar to wild type (Fig. 9). To determine whether the differential XYL1 expression in embryo and endosperm may affect germination rates, we analyzed the timing of radicle protrusion of after-ripened seeds in which dormancy was released. In the absence of stratification treatment, wild-type and xyl1 seeds exhibited similar germination rates, while in transgenic seeds radicle protrusion was delayed (Fig. 10). This suggested that when endosperm resistance is increased due to XYL1 overexpression, the growth potential of the XYL1-deficient embryo is less effective at overcoming the mechanical restraint imposed by the endosperm.
Figure 9.
Seed phenotypes of wild-type, xyl1-2, and four independent pEPR1:XYL1 homozygous transgenic lines. Paclobutrazol (10 µM) resistance (A) and germination at 34°C (B) of stratified seeds. C, Germination of freshly harvested seeds. The number of seedlings with protruding radicle (B, C) or green cotyledons (A) was scored 5 d (A, B) or 14 d (C) after sowing and compared to the total number of seeds. Results are means of three replicates with SE and were obtained for three independent seed batches.
Figure 10.
Germination rates of after-ripened seeds of wild-type, xyl1-2, and four independent pEPR1:XYL1 homozygous transgenic lines. The number of seeds with protruding radicle was scored over 40 h after sowing and compared to the total number of seeds. Results are means of three replicates with SE.
To confirm that pEPR:XYL1 transformants exhibit differential XYL1 expression in endosperm and embryo, the XyG composition was analyzed. XyG profiles for transgenic embryos were very similar to xyl1-2, at both stages of imbibition, whereas in testa/endosperm fractions they were similar to wild type (Fig. 11). EPR1-driven endosperm-specific XYL1 expression in xyl1-2 mutants was thus able to restore dormancy of freshly harvested seeds and sensitivity of stratified seeds to germination inhibitors and even delay germination of nondormant seeds.
Figure 11.
Matrix-assisted laser-desorption ionization time of flight analysis of XyGs in germinating seeds of wild type (black bars), xyl1-2 (white bars), and four independent pEPR1:XYL1 homozygous transgenic lines (gray bars, from left to right: T1 to T4). Analysis was performed on 3-d-stratified seeds during imbibition at 25°C in the light. Embryos were dissected from testa and endosperm 3 h after sowing and at ER. Relative abundance of major subunits is shown as means of 4 biological replicates with SE. Minor subunits (<5% relative abundance) are not shown. Mean values represent acetylated and nonacetylated subunits of XXLG, XXFG, and XLFG combined.
DISCUSSION
XyG, the most abundant hemicellulose in dicot cell walls, is thought to play a crucial role in cell elongation and also impact cell wall rigidity (Scheller and Ulvskov, 2010). Nevertheless, little is known about its precise contribution to dormancy and germination processes. In this study, we have isolated a xyl1 mutant allele able to germinate on high concentrations of the GA biosynthesis inhibitor paclobutrazol (Fig. 1), thus indicating its lower GA requirement for seed germination. The mutated locus, XYL1, encodes a previously described glycoside hydrolase, which modifies XyGs by removing Xyl residues from XyG side chains (Sampedro et al., 2001). This gene has previously been reported to be expressed in vegetative tissues and involved in cell wall remodeling processes (Sampedro et al., 2010; Günl and Pauly, 2011). Here, we observed that XYL1 is differentially expressed throughout embryo and endosperm tissues during seed development and imbibition (Fig. 5). Accordingly, in addition to plant growth defects, such as shorter and thicker siliques, xyl1 mutants also exhibit altered germination characteristics, such as reduced dormancy and increased tolerance to germination inhibitors and thermoinhibition (Figs. 1 and 4; Supplemental Figs. S2 and S3).
XYL1 Activity in Seeds May Require Posttranslational Modifications
In accordance with mutant germination phenotypes and XYL1 transcript accumulation, the XYL1 protein is accumulated in dry seeds. Proteome analysis by 2D gel electrophoresis of wild-type seed extracts detected four polypeptide spots corresponding to XYL1, which were absent in both the T-DNA allele xyl1-2 and the EMS allele xyl1-4 (Fig. 3; Supplemental Table S1), thus indicating that at least 4 different isoforms of the enzyme are present at this stage. Previous cell wall proteomic studies have also reported the presence of several XYL1 isoforms in vegetative tissues. The XYL1 protein was detected in extracts from stems and etiolated hypocotyls, and the observed mass shifts were predicted to be due to glycosylation at a number of sites (Minic et al., 2007; Albenne et al., 2009; Zhang et al., 2011). Moreover, a pI shift has also been observed due to protein phosphorylation. Such phosphorylation would lead to activation of the protein (Kaida et al., 2010), whereas the glycosylation that takes place in the Golgi apparatus would mediate activity modifications and induce protein secretion to cell walls (Ruiz-May et al., 2012). It is likely, therefore, that the mass and pI shifts detected in dry seed extracts correspond to XYL1 polypeptides targeted to the wall after glycosylation processes in the Golgi as well as differentially phosphorylated isoforms. These protein isoforms most probably represent different states of the translational modification process leading to the mature protein, whose activity has been detected in the apoplastic fluid of Arabidopsis seedlings (Sampedro et al., 2001). A regulatory role for these posttranslational modifications in cell wall remodelling during embryo growth and ER during germination is probable but would require further investigation.
XyG Composition Modulates Endosperm Resistance
Loss of XYL1 function caused alterations in XyG composition (Fig. 2). The defect in Xyl removal caused a relative increase in galactosylated residues at the expense of fucosylated residues in both embryo and testa/endosperm, as previously reported in xyl1 mutant stems and seedlings (Sampedro et al., 2010; Günl and Pauly, 2011; Sampedro et al., 2012). It has been suggested that the sequential removal of substituted sugars is most probably dependent on the previous enzymatic hydrolysis of neighbor residues. In xyl1, the removal of the galactosyl residue may be subject to a previous hydrolysis by XYL1, therefore resulting in an excess of galactosyl residues (Günl et al., 2011).
The accumulation of XyG that links cellulose microfibrils has been suggested to increase cell wall stiffness (Hayashi and Kaida, 2011). Variations in the complexity of XyG side chains may modulate endosperm weakening by hydrolases (XTH, glucanases, endo-β-mannanases) or extensibility by expansins, since these activities have been widely reported to facilitate radicle protrusion (Nonogaki et al., 2010; Park and Cosgrove, 2012; Shi et al., 2015). Upon imbibition, the micropylar endosperm has been shown to stretch before its rupture due to embryo growth (Lee et al., 2012). A clear link between germination characteristics and XyG composition in the endosperm has been demonstrated here. A reduction in the relative abundance of highly ramified fucosylated residues in the xyl1 endosperm was correlated with decreased dormancy and GA requirement for germination, whereas XYL1 expression under the control of the endosperm-specific EPR1 promoter in transgenic mutant seeds restored wild-type XyG profiles and germination phenotypes (Figs. 1, 2, 9, and 10). Since the double mutant xxt1 xxt2 exhibited the same phenotypes as xyl1 (Fig. 4), it further suggests that endosperm cell wall resistance is reduced when XyG are either absent or contain less branched side-chains that potentially could influence interactions with cellulose microfibrils (Zhao et al., 2014; Park and Cosgrove, 2015).
The bgal10 and axy8 mutants did not display similar seed phenotypes to those observed for xyl1 (Fig. 4). Nevertheless, transcriptome analyses have revealed that transcript abundance of AXY8, BGAL10, and XYL1 increased during seed imbibition, although XYL1 was more strongly expressed (Nakabayashi et al., 2005; Winter et al., 2007). Moreover, BGAL10 and XYL1 displayed similar expression patterns in response to dormancy cycling, indicating that BGAL10 is also highly regulated in seeds (Cadman et al., 2006). The milder effect of mutating BGAL10 and AXY8 might result from functional redundancy, since residual galactosidase and fucosidase activities, respectively, were detected in these mutants, which contrasts with the complete absence of xyloxidase activity in xyl1 (Sampedro et al., 2010, 2012; Günl et al., 2011). Nonetheless, a reduction in silique length was observed in all three mutants (Supplemental Fig. S3), suggesting that if functional redundancy occurs for galactosidase and fucosidase, it is differentially regulated during plant and seed development. In axy8 vegetative tissues, increased amounts of the fucosylated oligosaccharide XXFG were observed with a concomitant decrease in XXLG (Günl et al., 2011). In bgal10 mutants, the relative abundance of XXFG was lower than in wild type, but higher than in the bgal10 xyl1 double mutant (Sampedro et al., 2012). These data further indicate that XyG side chain complexity is negatively correlated with germination potential in these mutants, as observed in pEPR:XYL1 overexpressors.
Nevertheless, other explanations are possible. An increased accumulation of unbound XyG oligosaccharides was previously measured in xyl1 mutants compared to wild type (Sampedro et al., 2010; Günl et al., 2011), and it is possible that these unbound oligomers interfere with XTH activities, thereby impacting XyG remodeling and germination properties. Alternatively, as described in earlier studies and recently reviewed by Scheller and Ulvskov (2010), XyG oligomers might act as signal molecules. Nevertheless, the accumulation of these oligomers in xyl1 seeds is unlikely to play a major role in the germination phenotypes observed here, as they are not detected in xxt1 xxt2 mutants that have similar phenotypes to xyl1.
Anisotropic XyG Intracellular Distribution May Contribute to Cell Wall Loosening
Immunolocalization of xylosylated, galactosylated, and fucosylated residues indicated that XyG accumulation is developmentally regulated in seeds, but no major alterations were observed prior to ER (Figs. 6 and 7). At ER, a striking difference was observed in growing hypocotyl cells of germinating wild-type embryos with XyG labeling detected in transversal cell walls, but absent from longitudinal walls, suggesting an anisotropic XyG secretion in transversal walls or a specific hydrolysis and/or remobilization from longitudinal walls. The latter seems more likely, since ubiquitous cell wall labeling was observed at earlier stages and decreased XyG abundance correlated with concomitant labeling of intracellular vesicles. In accordance, inhibitors of endocytosis have previously been shown to significantly delay germination, indicating that internalization of extracellular molecules plays an important function (Pagnussat et al., 2012). Interestingly, the differential XyG accumulation was observed in a region just above the radicle tip previously described as the embryo elongation zone during germination (Sliwinska et al., 2009). The XyG localization correlates well, therefore, with cell elongation and may be implicated in cell wall loosening prior to the directional growth of hypocotyls that leads to radicle emergence.
In dark-grown elongating hypocotyls, an asymmetric loosening of longitudinal walls has been observed by atomic force microscopy prior to changes in cortical microtubule orientation (Peaucelle et al., 2015). The selective demethylesterification of cell wall pectin in longitudinal walls has been reported to be responsible for this mechanical asymmetry, which is required for growth symmetry breaking. In accordance, an asymmetrical accumulation of homogalacturonans with a low degree of methylesterification was also observed here in elongating hypocotyls at ER, which was inversely correlated with XyG abundance (Fig. 7). Nevertheless, we cannot exclude that decreased XyG abundance in longitudinal cell walls results from epitope masking, since removal of pectic homogalacturonan has previously been shown to increase LM15 labeling (Marcus et al., 2008). However, almost identical labeling patterns were observed here with three different antibodies (LM15, LM25, and CCRC-M1), implying that if epitope masking occurs, it would affect the recognition of distinct XyG epitopes equally. Furthermore, the intense homogalacturonan labeling of cell corners with 2F4 was not an exact match of the uniform reduction of XyG labeling in longitudinal cell walls. This rules out a direct negative effect of pectin demethylesterification on XyG epitope detection, although remodeling of other cell wall components might still be involved. Nonetheless, the specific spatiotemporal regulation of XyG abundance in hypocotyl cells suggests that decreased XyG abundance contributes to longitudinal cell wall loosening together with pectin demethylesterification.
XYL1 Activity Is Essential for Anisotropic XyG Accumulation in Hypocotyl Elongating Cells
While no differences were detected between xyl1 and wild type from seed maturation to early imbibition, at ER XyG were absent from the longitudinal and transversal walls of xyl1 hypocotyl cells, although they were still detected within intracellular vesicles (Fig. 7). This indicates that alterations to XyG structure in xyl1 mutants may affect polarized XyG secretion or remobilization. Moreover, labeled intracellular vesicles observed in xyl1 hypocotyl cells may contain the excess, unbound XyG detected in previous studies (Sampedro et al., 2010; Günl and Pauly, 2011). In accordance, a recent study on the biosynthesis mutant mur3 that produces less substituted XyG due to defective galactosylation reported that intracellular polysaccharide aggregates were observed in mur3, which were attributed to altered secretion of Gal-depleted XyG (Kong et al., 2015).
Intriguingly, although mutation of XYL1 prevented the establishment of an anisotropic XyG distribution in mutant hypocotyls (Figs. 6 and 7), this did not negatively impact hypocotyl elongation and radicle protrusion (Figs. 1 and 4). Hence, even if decreased XyG contents in hypocotyl longitudinal cell walls are necessary for cell elongation to occur, their anisotropic distribution is not essential for asymmetrical growth. Other mechanisms could generate this asymmetry, such as the degree of pectin demethylesterification, prior to microtubule reorganization (Peaucelle et al., 2015). Yet size reduction in siliques was correlated with increased width in both xyl1 and xxt1 xxt2 and to a lesser extent in bgal10 and axy8, strongly suggesting a more predominant role of XyG metabolism in directional growth in these organs than hypocotyls (Supplemental Fig. S3; Sampedro et al., 2010; Günl and Pauly, 2011).
To conclude, our study provides further evidence that endosperm has a pivotal role as a mechanical barrier to embryo growth in the control of seed dormancy and germination. It also reveals that fine-tuning of XyG hydrolysis in cell walls is essential in the control of micropylar ER in response to exogenous and endogenous signals, such as temperature and GAs. The spatio-temporal regulation of XyG intracellular accumulation was shown to be tightly correlated with hypocotyl cell elongation; however, the mechanisms regulating XyG anisotropic abundance and possible roles in cell wall loosening still remain to be clarified.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Arabidopsis (Arabidopsis thaliana; Col-0 accession) wild-type and mutant seeds were surface sterilized, sown in petri dishes containing Arabidopsis Gamborg B5 medium (Duchefa, http://www.duchefa.com) supplemented with 30 mm Suc, and stratified at 4°C in the dark for 3 d. Petri dishes were then placed for 4 d in a growth chamber (16-h photoperiod, 50-μmol m−2 sec−1 light intensity, 18°C, 60% relative humidity). Germinated seedlings were transferred to soil (Tref Substrates, http://www.trefgroup.com) and, unless otherwise stated, grown in a glasshouse with a minimum photoperiod of 13 h assured by supplementary lighting.
xyl1-2 was obtained from the GABI-KAT collection as GABI_749G08 (Kleinboelting et al., 2012). Seeds of T-DNA mutants bgal10-1 and bgal10-2 (Sampedro et al., 2012) were a gift of Javier Sampedro, and seeds of EMS mutants axy8-1 and axy8-2 and T-DNA mutants axy8-5 and axy8-6 (Günl et al., 2011) were a gift of Markus Pauly.
Mutant Isolation and Map-Based Cloning
A mutant screen was performed as previously described for the aba4 mutant identified using the same phenotype of germination tolerance to paclobutrazol (North et al., 2007). For mapping, the xyl1-4 mutant (Col-0 accession) was crossed with wild-type plants (Landsberg erecta accession), and 148 homozygous xyl1-4 plants were selected from the F2 progeny based on germination phenotypes. Genomic DNA was extracted from flower buds of selected plants and an approximate genome position determined based on recombination percentages with simple sequence length polymorphisms, as described previously (North et al., 2007). Whole-genome resequencing was carried out on xyl1-4 DNA isolated from 3 g of flower buds with CTAB buffer (2% [w/v] N-cetyl-N,N,N,-trimethyl-ammonium bromide, 1.4 m NaCl, 20 mm EDTA, 100 mm Tris-HCl, pH 8.0, 0.2% [v/v] β- mercaptoethanol) prior to extraction with an equal volume of chloroform:isoamyl alcohol (24:1) and precipitation with 0.7 volumes of isopropanol. After supernatant removal and pellet drying, the pellet was resuspended with 10 mm Tris-HCl, 1 mm EDTA, pH 8.0. DNA was then purified with DNeasy Plant MiniKit (Qiagen) from step 13 including an RNase treatment. Sequencing was performed by Genoscreen (France) using an Illumina HiSeq2000 sequencer.
Germination Experiments
For dormancy assays, freshly harvested seeds were sown in triplicate in Petri dishes containing 0.5% (w/v) agarose and then placed in a growth chamber (continuous light, 25°C, 70% relative humidity). Germination was scored each day based on radicle protrusion. For paclobutrazol resistance tests, surface-sterilized seeds were sown on 0.5% (w/v) agarose supplemented with paclobutrazol (Syngenta, http://www.syngenta-agro.fr), stratified at 4°C for 3 d, then incubated in the same conditions as the dormancy assays for 5 d. Seedlings were scored as resistant if they developed green cotyledons. For thermoinhibition of germination, surface-sterilized seeds were sown on 0.5% (w/v) agarose, stratified at 4°C for 3 d, then incubated for 5 d in a growth chamber at 34°C (continuous light, 34°C, 70% relative humidity). Seeds were scored as resistant if the radicle protruded.
ABA Content Determination
One hundred milligrams of dry seeds was analyzed directly or imbibed in 0.8 mL water, and after 1 or 5 h of imbibition in the light at room temperature with gentle agitation, the medium around imbibed seeds was recovered. Dry seeds, imbibed seeds, and imbibition medium were frozen in liquid nitrogen and then freeze-dried. Dry and imbibed seeds were ground in 2 mL of extraction solvent (acetone/water/acetic acid, 80/19/1, v/v/v) to which 10 ng of deuterated ABA (purchased from Irina Zaharia, Plant Biotechnology Institute, National Research Council Canada, http://www.nrc-cnrc.gc.ca) was added as an internal standard. Samples were centrifuged and the supernatant recovered, the pellet was then resuspended in a further 1 mL of extraction solvent by sonication, recentrifuged, and the supernatants combined. The extraction solvent was then evaporated and the residue resuspended by sonication in 0.2 mL of chromatography mobile phase (acetonitrile/water/acetic acid, 50/50/0.05, v/v/v). Dried imbibition medium was directly resuspended in 0.2 mL chromatography mobile phase with 10 ng deuterated ABA by mixing. Finally, all samples were filtered through a 1.6-mm GFA filter (Whatman, http://www.whatman.com/). ABA was quantified using an LC-ESI-MS-MS system (Quattro LC; Waters, http://www.waters.com).
Reporter Gene Analysis
Three independent transformant lines containing a pXYL1:GUS construct were a gift of Javier Sampedro (Sampedro et al., 2010). Developing seeds were dissected from siliques staged by tagging flowers on the day of pollination and harvested at the specified number of days afterward. Germinating seeds were sown on filters in a growth chamber (continuous light, 25°C, 70% relative humidity) and were harvested after 4 24, and 48 h at two physiological stages: either nongerminated seeds or seeds that had just completed radicle protrusion after ER. Histochemical GUS staining was performed, as previously described (North et al., 2007), after the removal of the embryo from the testa and endosperm and observed under a light microscope (Axioplan 2; Zeiss, http://www.zeiss.com).
Cloning and Plant Transformation
A 0.8-kb promoter fragment for the EPR1 gene (At2g27380) was amplified from wild-type Col-0 genomic DNA by PCR using Pfu Ultra DNA polymerase (Stratagene, Amsterdam, The Netherlands) and the following primers: forward 5′-CCCAAGCTTGGGCTGGAATTTAATTTAAGTTTTGTTTTTGG-3′ and reverse 5′-TTGGCGCGCCAACTTGTAAGTGATTTACTGAGAACT-3′.
Sequencing of the amplified fragment was carried out to confirm that it was identical to the sequence in the databases before introduction into the binary Gateway Destination vector pMDC32 (Life Technologies) after digestion with the restriction enzymes HindIII and AscI.
The XYL1 coding sequence (At1g68560) was amplified from wild-type Col-0 genomic DNA by PCR using the following primers containing attB1 and attB2 recombination sequences: forward primer 5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTTCATGGCTTCCTCTT-3′; reverse primer 5′-GGGGACCACTTTGTACAAGAAAGCTGGGTCTTAATTGATACC-3′.
The resulting PCR product was then recombined into pDONR207 vector with BP clonase (Invitrogen; http://www.invitrogen.com/) according to the manufacturer’s instructions and transformed into the Escherichia coli strain DH10B. The XYL1 gene fragment was sequenced to confirm there were no PCR-induced errors present and then recombined into the modified binary vector pMDC32 containing the EPR1 promoter fragment. The resulting binary vector was transformed into Agrobacterium tumefaciens C58C1pMP90 by electroporation prior to stable transformation of wild-type or xyl1-2 plants.
Proteomic Analysis
Total soluble protein extracts were prepared from 50 mg of dry mature seeds, and the concentrations of total soluble proteins in the lysates were estimated using the Bradford protein assay, as previously described (Rajjou et al., 2011). All samples were stored at −20°C prior to analysis. Isoelectric focusing was performed with 150 μg of protein for each sample. Proteins from the various extracts were separated using immobilized pH gradients gel strips (Immobiline DryStrip, nonlinear pH gradient 3-11, 24 cm; GE Healthcare). Immobilized pH gradients gel strips were incubated for 14 h at 22°C in rehydration buffer (18 mm Tris-HCl, 14 mm Tris base, 7 m urea, 2 m thiourea, 4% [w/v] CHAPS, 20 mm dithiothreitol, 2% [v/v] Triton X-100, and 1% [v/v] pharmalyte pH 3-10 carrier ampholytes) containing 150 μg protein. Bidimensional electrophoresis was carried out as previously described (Rajjou et al., 2011). For each condition analyzed, 2D gels were carried out for three biological replicates. Proteins on the 2D gels were stained by silver nitrate, dried for 2 d at room temperature, and scanned as previously described (Arc et al., 2012).
XyG and Pectin Immunolocalization
Developing siliques were staged by tagging flowers on the day of pollination, in the same way as for reporter gene expression analysis, and harvested at 10 or 14 DAP. Seeds were also sown on filters, placed at 4°C for 3 d in the dark, and then placed in a growth chamber (continuous light, 25°C, 70% relative humidity) prior to harvest after 3 h of imbibition. Seeds that had undergone ER were selected at the start of radicle protrusion, about 20 h after transfer to 25°C. Samples were fixed in a solution of 4% (w/v) paraformaldehyde and 0.1% Triton X-100 under vacuum for 1 h. Samples were then washed three times in phosphate-buffered saline (PBS), dehydrated through a graded ethanol series (30, 50, 70, 90, and 97% [v/v] in PBS), and then incubated in a mixture of 100% wax and 97% ethanol (1:1, v/v) at 40°C overnight and embedded in 100% wax at 40°C. Sections (8 mm) were prepared using a microtome and air-dried onto poly-Lys-coated glass slides. Samples on slides were dewaxed and rehydrated through a degraded ethanol series (97, 90, and 50% [v/v] in PBS or T/Ca/S). At all steps, the PBS buffer was used for LM15, LM25, and CCRC-M1 epitope detection, and the T/Ca/S buffer (20 mm Tris-HCl, pH 8.2, 0.5 mm CaCi2 , and 150 mm NaCl) for 2F4 epitope detection. Samples were blocked with 1% (w/v) bovine serum albumin in PBS or T/Ca/S buffer. Samples were then incubated with antibodies diluted in their respective blocking solution. After three washes of 5 min each in PBS or T/Ca/S buffer without bovine serum albumin, slides were incubated with antirat IgG or antimouse IgG labeled with Alexa Fluor 488 (Molecular Probes, Eugene, OR) in blocking solution. Following antibody labeling, the samples were washed in PBS or T/Ca/S and counterstained for 10 min with Calcofluor White M2R (fluorescent brightener 28; Sigma-Aldrich) at 0.25 mg mL−1 in water. After washing with PBS or T/Ca/S, slides were sealed. As controls, sections were incubated without the primary antibodies and with Alexa Fluor 488-labeled secondary antibodies. Immunofluorescence was observed using a spectral confocal laser-scanning microscope (Zeiss LSM710) at 405-nm excitation and 410- to 490-nm emission for Calcofluor White M2R and at 488-nm excitation and 495- to 550-nm emission for Alexa Fluor 488.
XyG Analysis
XyG analysis was based on the rapid phenotyping method using enzymatic oligosaccharide fingerprinting previously described (Lerouxel et al., 2002). After 3 d of stratification (T0), seeds were transferred to 25°C in continuous light. Embryos were separated from testa and endosperm by dissection after 3 h at 25°C and at ER. Samples were conserved in ethanol. After ethanol removal and rehydration, XyG oligosaccharides were generated by treating samples with endoglucanase in 50 mm sodium acetate buffer, pH 5, overnight at 37°C. Matrix-assisted laser-desorption ionization time of flight mass spectrometry of the XyG oligosaccharides was recorded with a MALDI/TOF Bruker Reflex III using super-DHB (9:1 mixture of 2,5-dihydroxy-benzoic acid and 2-hydroxy-5-methoxy-benzoic acid; Sigma-Aldrich, sigmaaldrich.com) as matrix.
Accession Numbers
The AGI locus identifier for XYL1 is At1g68560.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Nomenclature and schematic representation of XyG.
Supplemental Figure S2. Germination rates of freshly harvested seeds of wild type, xyl1-2, and xyl1-4.
Supplemental Figure S3. Silique length of xyl1, bgal10, axy8, and xxt1 xxt2 mutants compared to wild type (WT).
Supplemental Figure S4. Immunolocalization of XyGs in Arabidopsis wild-type developing seed sections at 10 and 14 DAP.
Supplemental Figure S5. Immunolocalization of XyGs in Arabidopsis xyl1 developing seed sections at 10 DAP.
Supplemental Figure S6. Immunolocalization of XyGs in Arabidopsis xyl1 developing seed sections at 14 DAP.
Supplemental Figure S7. Immunolocalization of XyGs in Arabidopsis wild-type imbibed seed sections at 3 HAI.
Supplemental Figure S8. Immunolocalization of XyGs in Arabidopsis xyl1 imbibed seed sections at 3 HAI.
Supplemental Figure S9. ABA levels in seeds and imbibition medium.
Supplemental Table S1. XYL1 protein identification after 2D gel electrophoresis and peptide analysis by tandem mass spectrometry.
Supplementary Material
Acknowledgments
We are grateful to Amélie Degueuse, Bruno Letarnec, and Joël Talbotec for plant culture, to Thierry Balliau (Génétique quantitative and Evolution, Le Moulon, France) for tandem mass spectrometry analysis, to Bertrand Daudon for technical aid with xyl1 mapping, and to Rodnay Sormani for help with XYL1 cloning. We thank Javier Sampedro (Universidad de Santiago, Santiago de Compostela, Spain) for providing bgal10 mutants and pXYL1:GUS lines, and Markus Pauly (University of California, Berkeley, USA) for providing axy8 mutants. We also thank Herman Höfte and Jonathan Griffiths for critical reading of the manuscript.
Glossary
- ABA
abscisic acid
- Col-0
Columbia-0
- 2D
bidimensional
- DAP
days after pollination
- EMS
ethyl-methanesulfonate
- ER
endosperm rupture
- GA
gibberellin
- PBS
phosphate-buffered saline
- XyG
xyloglucan
- XTH
endo-transglycosylases/hydrolase
References
- Albenne C, Canut H, Boudart G, Zhang Y, San Clemente H, Pont-Lezica R, Jamet E (2009) Plant cell wall proteomics: mass spectrometry data, a trove for research on protein structure/function relationships. Mol Plant 2: 977–989 [DOI] [PubMed] [Google Scholar]
- Arc E, Chibani K, Grappin P, Jullien M, Godin B, Cueff G, Valot B, Balliau T, Job D, Rajjou L (2012) Cold stratification and exogenous nitrates entail similar functional proteome adjustments during Arabidopsis seed dormancy release. J Proteome Res 11: 5418–5432 [DOI] [PubMed] [Google Scholar]
- Cadman CSC, Toorop PE, Hilhorst HWM, Finch-Savage WE (2006) Gene expression profiles of Arabidopsis Cvi seeds during dormancy cycling indicate a common underlying dormancy control mechanism. Plant J 46: 805–822 [DOI] [PubMed] [Google Scholar]
- Cantarel BL, Coutinho PM, Rancurel C, Bernard T, Lombard V, Henrissat B (2009) The Carbohydrate-Active EnZymes database (CAZy): an expert resource for glycogenomics. Nucleic Acids Res 37: D233–D238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalier DM, Lerouxel O, Neumetzler L, Yamauchi K, Reinecke A, Freshour G, Zabotina OA, Hahn MG, Burgert I, Pauly M, Raikhel NV, Keegstra K (2008) Disrupting two Arabidopsis thaliana xylosyltransferase genes results in plants deficient in xyloglucan, a major primary cell wall component. Plant Cell 20: 1519–1537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosgrove DJ. (2005) Growth of the plant cell wall. Nat Rev Mol Cell Biol 6: 850–861 [DOI] [PubMed] [Google Scholar]
- Dekkers BJ, Pearce S, van Bolderen-Veldkamp RP, Marshall A, Widera P, Gilbert J, Drost HG, Bassel GW, Müller K, King JR, Wood AT, Grosse I, et al. (2013) Transcriptional dynamics of two seed compartments with opposing roles in Arabidopsis seed germination. Plant Physiol 163: 205–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubreucq B, Berger N, Vincent E, Boisson M, Pelletier G, Caboche M, Lepiniec L (2000) The Arabidopsis AtEPR1 extensin-like gene is specifically expressed in endosperm during seed germination. Plant J 23: 643–652 [DOI] [PubMed] [Google Scholar]
- Endo A, Tatematsu K, Hanada K, Duermeyer L, Okamoto M, Yonekura-Sakakibara K, Saito K, Toyoda T, Kawakami N, Kamiya Y, Seki M, Nambara E (2012) Tissue-specific transcriptome analysis reveals cell wall metabolism, flavonol biosynthesis and defense responses are activated in the endosperm of germinating Arabidopsis thaliana seeds. Plant Cell Physiol 53: 16–27 [DOI] [PubMed] [Google Scholar]
- Finkelstein R, Reeves W, Ariizumi T, Steber C (2008) Molecular aspects of seed dormancy. Annu Rev Plant Biol 59: 387–415 [DOI] [PubMed] [Google Scholar]
- Fry SC, York WS, Albersheim P, Darvill A, Hayashi T, Joseleau JP, Kato Y, Lorences EP, Maclachlan GA, McNeil M, Mort AJ, Reid JSG, et al. (1993) An unambiguous nomenclature for xyloglucan‐ derived oligosaccharides. Physiol Plant 89: 1–3 [Google Scholar]
- Galland M, Huguet R, Arc E, Cueff G, Job D, Rajjou L (2014) Dynamic proteomics emphasizes the importance of selective mRNA translation and protein turnover during Arabidopsis seed germination. Mol Cell Proteomics 13: 252–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graeber K, Nakabayashi K, Miatton E, Leubner-Metzger G, Soppe WJJ (2012) Molecular mechanisms of seed dormancy. Plant Cell Environ 35: 1769–1786 [DOI] [PubMed] [Google Scholar]
- Günl M, Neumetzler L, Kraemer F, de Souza A, Schultink A, Pena M, York WS, Pauly M (2011) AXY8 encodes an α-fucosidase, underscoring the importance of apoplastic metabolism on the fine structure of Arabidopsis cell wall polysaccharides. Plant Cell 23: 4025–4040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Günl M, Pauly M (2011) AXY3 encodes a α-xylosidase that impacts the structure and accessibility of the hemicellulose xyloglucan in Arabidopsis plant cell walls. Planta 233: 707–719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi T, Kaida R (2011) Functions of xyloglucan in plant cells. Mol Plant 4: 17–24 [DOI] [PubMed] [Google Scholar]
- Kaida R, Serada S, Norioka N, Norioka S, Neumetzler L, Pauly M, Sampedro J, Zarra I, Hayashi T, Kaneko TS (2010) Potential role for purple acid phosphatase in the dephosphorylation of wall proteins in tobacco cells. Plant Physiol 153: 603–610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinboelting N, Huep G, Kloetgen A, Viehoever P, Weisshaar B (2012) GABI-Kat SimpleSearch: new features of the Arabidopsis thaliana T-DNA mutant database. Nucleic Acids Res 40: D1211–D1215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong Y, Peña MJ, Renna L, Avci U, Pattathil S, Tuomivaara ST, Li X, Reiter WD, Brandizzi F, Hahn MG, Darvill AG, York WS, et al. (2015) Galactose-depleted xyloglucan is dysfunctional and leads to dwarfism in Arabidopsis. Plant Physiol 167: 1296–1306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KJD, Dekkers BJ, Steinbrecher T, Walsh CT, Bacic A, Bentsink L, Leubner-Metzger G, Knox JP (2012) Distinct cell wall architectures in seed endosperms in representatives of the Brassicaceae and Solanaceae. Plant Physiol 160: 1551–1566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Léon-Kloosterziel KM, Gil MA, Ruijs GJ, Jacobsen SE, Olszewski NE, Schwartz SH, Zeevaart JAD, Koornneef M (1996) Isolation and characterization of abscisic acid-deficient Arabidopsis mutants at two new loci. Plant J 10: 655–661 [DOI] [PubMed] [Google Scholar]
- Lerouxel O, Choo TS, Séveno M, Usadel B, Faye L, Lerouge P, Pauly M (2002) Rapid structural phenotyping of plant cell wall mutants by enzymatic oligosaccharide fingerprinting. Plant Physiol 130: 1754–1763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liners F, Letesson JJ, Didembourg C, Van Cutsem P (1989) Monoclonal antibodies against pectin: recognition of a conformation induced by calcium. Plant Physiol 91: 1419–1424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linkies A, Leubner-Metzger G (2012) Beyond gibberellins and abscisic acid: how ethylene and jasmonates control seed germination. Plant Cell Rep 31: 253–270 [DOI] [PubMed] [Google Scholar]
- Marcus SE, Verhertbruggen Y, Hervé C, Ordaz-Ortiz JJ, Farkas V, Pedersen HL, Willats WGT, Knox JP (2008) Pectic homogalacturonan masks abundant sets of xyloglucan epitopes in plant cell walls. BMC Plant Biol 8: 60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Andújar C, Pluskota WE, Bassel GW, Asahina M, Pupel P, Nguyen TT, Takeda-Kamiya N, Toubiana D, Bai B, Górecki RJ, Fait A, Yamaguchi S, et al. (2012) Mechanisms of hormonal regulation of endosperm cap-specific gene expression in tomato seeds. Plant J 71: 575–586 [DOI] [PubMed] [Google Scholar]
- Millar AA, Jacobsen JV, Ross JJ, Helliwell CA, Poole AT, Scofield G, Reid JB, Gubler F (2006) Seed dormancy and ABA metabolism in Arabidopsis and barley: the role of ABA 8′-hydroxylase. Plant J 45: 942–954 [DOI] [PubMed] [Google Scholar]
- Minic Z, Jamet E, Négroni L, Arsene der Garabedian P, Zivy M, Jouanin L (2007) A sub-proteome of Arabidopsis thaliana mature stems trapped on Concanavalin A is enriched in cell wall glycoside hydrolases. J Exp Bot 58: 2503–2512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris K, Linkies A, Müller K, Oracz K, Wang X, Lynn JR, Leubner-Metzger G, Finch-Savage WE (2011) Regulation of seed germination in the close Arabidopsis relative Lepidium sativum: a global tissue-specific transcript analysis. Plant Physiol 155: 1851–1870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakabayashi K, Okamoto M, Koshiba T, Kamiya Y, Nambara E (2005) Genome-wide profiling of stored mRNA in Arabidopsis thaliana seed germination: epigenetic and genetic regulation of transcription in seed. Plant J 41: 697–709 [DOI] [PubMed] [Google Scholar]
- Nambara E, Kawaide H, Kamiya Y, Naito S (1998) Characterization of an Arabidopsis thaliana mutant that has a defect in ABA accumulation: ABA-dependent and ABA-independent accumulation of free amino acids during dehydration. Plant Cell Physiol 39: 853–858 [DOI] [PubMed] [Google Scholar]
- Nambara E, Okamoto M, Tatematsu K, Yano R, Seo M, Kamiya Y (2010) Abscisic acid and the control of seed dormancy and germination. Seed Sci Res 20: 55 [Google Scholar]
- Nonogaki H, Bassel GW, Bewley JD (2010) Germination - Still a mystery. Plant Sci 179: 574–581 [Google Scholar]
- North H, Baud S, Debeaujon I, Dubos C, Dubreucq B, Grappin P, Jullien M, Lepiniec L, Marion-Poll A, Miquel M, Rajjou L, Routaboul JM, et al. (2010) Arabidopsis seed secrets unravelled after a decade of genetic and omics-driven research. Plant J 61: 971–981 [DOI] [PubMed] [Google Scholar]
- North HM, De Almeida A, Boutin JP, Frey A, To A, Botran L, Sotta B, Marion-Poll A (2007) The Arabidopsis ABA-deficient mutant aba4 demonstrates that the major route for stress-induced ABA accumulation is via neoxanthin isomers. Plant J 50: 810–824 [DOI] [PubMed] [Google Scholar]
- Pagnussat L, Burbach C, Baluška F, de la Canal L (2012) Rapid endocytosis is triggered upon imbibition in Arabidopsis seeds. Plant Signal Behav 7: 416–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paque S, Mouille G, Grandont L, Alabadí D, Gaertner C, Goyallon A, Muller P, Primard-Brisset C, Sormani R, Blázquez MA, Perrot-Rechenmann C (2014) AUXIN BINDING PROTEIN1 links cell wall remodeling, auxin signaling, and cell expansion in arabidopsis. Plant Cell 26: 280–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park YB, Cosgrove DJ (2012) Changes in cell wall biomechanical properties in the xyloglucan-deficient xxt1/xxt2 mutant of Arabidopsis. Plant Physiol 158: 465–475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park YB, Cosgrove DJ (2015) Xyloglucan and its interactions with other components of the growing cell wall. Plant Cell Physiol 56: 180–194 [DOI] [PubMed] [Google Scholar]
- Peaucelle A, Wightman R, Höfte H (2015) The control of growth symmetry breaking in the Arabidopsis hypocotyl. Curr Biol 25: 1746–1752 [DOI] [PubMed] [Google Scholar]
- Pedersen HL, Fangel JU, McCleary B, Ruzanski C, Rydahl MG, Ralet MC, Farkas V, von Schantz L, Marcus SE, Andersen MCF, Field R, Ohlin M, et al. (2012) Versatile high resolution oligosaccharide microarrays for plant glycobiology and cell wall research. J Biol Chem 287: 39429–39438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penfield S, Li Y, Gilday AD, Graham S, Graham IA (2006) Arabidopsis ABA INSENSITIVE4 regulates lipid mobilization in the embryo and reveals repression of seed germination by the endosperm. Plant Cell 18: 1887–1899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puhlmann J, Bucheli E, Swain MJ, Dunning N, Albersheim P, Darvill AG, Hahn MG (1994) Generation of monoclonal antibodies against plant cell-wall polysaccharides. I. Characterization of a monoclonal antibody to a terminal α-(1-->2)-linked fucosyl-containing epitope. Plant Physiol 104: 699–710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajjou L, Belghazi M, Catusse J, Ogé L, Arc E, Godin B, Chibani K, Ali-Rachidi S, Collet B, Grappin P, Jullien M, Gallardo K, et al. (2011) Proteomics and posttranslational proteomics of seed dormancy and germination. Methods Mol Biol 773: 215–236 [DOI] [PubMed] [Google Scholar]
- Ruiz-May E, Kim SJ, Brandizzi F, Rose JK (2012) The secreted plant N-glycoproteome and associated secretory pathways. Front Plant Sci 3: 117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampedro J, Gianzo C, Iglesias N, Guitián E, Revilla G, Zarra I (2012) AtBGAL10 is the main xyloglucan β-galactosidase in Arabidopsis, and its absence results in unusual xyloglucan subunits and growth defects. Plant Physiol 158: 1146–1157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampedro J, Pardo B, Gianzo C, Guitián E, Revilla G, Zarra I (2010) Lack of α-xylosidase activity in Arabidopsis alters xyloglucan composition and results in growth defects. Plant Physiol 154: 1105–1115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampedro J, Sieiro C, Revilla G, González-Villa T, Zarra I (2001) Cloning and expression pattern of a gene encoding an α-xylosidase active against xyloglucan oligosaccharides from Arabidopsis. Plant Physiol 126: 910–920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheller HV, Ulvskov P (2010) Hemicelluloses. Annu Rev Plant Biol 61: 263–289 [DOI] [PubMed] [Google Scholar]
- Shi YZ, Zhu XF, Miller JG, Gregson T, Zheng SJ, Fry SC (2015) Distinct catalytic capacities of two aluminium-repressed Arabidopsis thaliana xyloglucan endotransglucosylase/hydrolases, XTH15 and XTH31, heterologously produced in Pichia. Phytochemistry 112: 160–169 [DOI] [PubMed] [Google Scholar]
- Sliwinska E, Bassel GW, Bewley JD (2009) Germination of Arabidopsis thaliana seeds is not completed as a result of elongation of the radicle but of the adjacent transition zone and lower hypocotyl. J Exp Bot 60: 3587–3594 [DOI] [PubMed] [Google Scholar]
- Vanzin GF, Madson M, Carpita NC, Raikhel NV, Keegstra K, Reiter WD (2002) The mur2 mutant of Arabidopsis thaliana lacks fucosylated xyloglucan because of a lesion in fucosyltransferase AtFUT1. Proc Natl Acad Sci USA 99: 3340–3345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter D, Vinegar B, Nahal H, Ammar R, Wilson GV, Provart NJ (2007) An “Electronic Fluorescent Pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS One 2: e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zabotina OA, van de Ven WT, Freshour G, Drakakaki G, Cavalier D, Mouille G, Hahn MG, Keegstra K, Raikhel NV (2008) Arabidopsis XXT5 gene encodes a putative α-1,6-xylosyltransferase that is involved in xyloglucan biosynthesis. Plant J 56: 101–115 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Giboulot A, Zivy M, Valot B, Jamet E, Albenne C (2011) Combining various strategies to increase the coverage of the plant cell wall glycoproteome. Phytochemistry 72: 1109–1123 [DOI] [PubMed] [Google Scholar]
- Zhao Z, Crespi VH, Kubicki JD, Cosgrove DJ, Zhong L (2014) Molecular dynamics simulation study of xyloglucan adsorption on cellulose surfaces: effects of surface hydrophobicity and side-chain variation. Cellulose 21: 1025–1039 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.