Acylsugar acylhydrolases are carboxylesterases that catalyze the hydrolysis of specific acyl chains from acylsugars found in trichomes.
Abstract
Glandular trichomes of cultivated tomato (Solanum lycopersicum) and many other species throughout the Solanaceae produce and secrete mixtures of sugar esters (acylsugars) on the plant aerial surfaces. In wild and cultivated tomato, these metabolites consist of a sugar backbone, typically glucose or sucrose, and two to five acyl chains esterified to various positions on the sugar core. The aliphatic acyl chains vary in length and branching and are transferred to the sugar by a series of reactions catalyzed by acylsugar acyltransferases. A phenotypic screen of a set of S. lycopersicum M82 × Solanum pennellii LA0716 introgression lines identified a dominant genetic locus on chromosome 5 from the wild relative that affected total acylsugar levels. Genetic mapping revealed that the reduction in acylsugar levels was consistent with the presence and increased expression of two S. pennellii genes (Sopen05g030120 and Sopen05g030130) encoding putative carboxylesterase enzymes of the α/β-hydrolase superfamily. These two enzymes, named ACYLSUGAR ACYLHYDROLASE1 (ASH1) and ASH2, were shown to remove acyl chains from specific positions of certain types of acylsugars in vitro. A survey of related genes in M82 and LA0716 identified another trichome-expressed ASH gene on chromosome 9 (M82, Solyc09g075710; LA0716, Sopen09g030520) encoding a protein with similar activity. Characterization of the in vitro activities of the SpASH enzymes showed reduced activities with acylsugars produced by LA0716, presumably contributing to the high-level production of acylsugars in the presence of highly expressed SpASH genes.
Hundreds of thousands of specialized metabolites are found in plants, and this large number of structurally diverse compounds results from the actions of a variety of enzyme classes, many of which appear to evolve rapidly (Pichersky and Lewinsohn, 2011; Milo and Last, 2012). Acylation of oxygen-containing functional groups to form esters is widespread in metabolism, particularly in plant specialized metabolism. The formation of ester-linked derivatives can alter the chemical and physical properties of a molecule, leading to changes in stability, localization, and solubility, and many studies have focused on the enzymes that catalyze their formation (for review, see D’Auria, 2006; Mugford and Milkowski, 2012). Reverse reactions that hydrolyze ester linkages in metabolism are also prevalent, and the responsible enzymes are referred to as carboxylesterases (CXEs). CXE proteins are classified into three groups based on sequence. Class I and II CXEs contain enzymes belonging to the α/β-hydrolase superfamily, while class III proteins belong to the GDS lipase family of enzymes (Gershater and Edwards, 2007). Plant CXEs play a role in the modification of esters of the signaling molecules jasmonic acid, salicylic acid, and auxin (Stuhlfelder et al., 2004; Forouhar et al., 2005; Yang et al., 2008), regulation of metabolite transport and localization (Kandzia et al., 1998), deprotection of functional groups in alkaloid biosynthesis (Ruppert et al., 2005; Winzer et al., 2012), bioactivation of herbicides (Gershater et al., 2007), and protection against pathogen infection (Pontier et al., 1994).
Plant class I CXE proteins are characterized as containing Interpro domain IPR013094 (Mitchell et al., 2015), have a catalytic triad of Ser, His, and Asp, and are typically inhibited by organophosphates. Twenty class I CXE genes were described from Arabidopsis (Arabidopsis thaliana; Marshall et al., 2003), and a similar number of genes were found in apple (Malus domestica) EST collections (Souleyre et al., 2011). Only a few plant class I CXEs have been biochemically characterized. SlCXE1 from cultivated tomato (Solanum lycopersicum) hydrolyzes volatile acetate esters in fruit, influencing aromatic flavor components (Goulet et al., 2012). Similarly, the apple MdCXE1 protein is expressed during fruit ripening and is thought to hydrolyze acetate esters important for fruit flavor (Souleyre et al., 2011). A poppy (Papivar somniferum) gene, PsCXE1, is part of a cluster of genes encoding enzymes for noscapine biosynthesis (Winzer et al., 2012); this enzyme is suggested to catalyze the hydrolysis of an acetyl group from papaveroxine to form narcotine hemiacetal. Two plant class I CXE enzymes were shown to catalyze in vitro reactions that differ from the typical hydrolytic cleavage expected from CXEs: hydroxyisoflavanone dehydratase from soybean (Glycine max) and Glycyrrhiza echinata catalyzes the 1,2-elimination of water from hydroxyisoflavanones (Akashi et al., 2005), while a tulip (Tulipa gesneriana) CXE catalyzes an intramolecular transesterification reaction resulting in the formation of an antimicrobial lactone product from the Glc ester of 4-hydroxy-2-methylenebutanoate (Nomura et al., 2012). The GA receptor GID1 is a class I CXE that has lost the ability to catalyze hydrolysis reactions and instead functions by binding GA, ultimately leading to the ubiquitination and degradation of transcriptional repressors of GA signaling (Ueguchi-Tanaka et al., 2005). Lastly, there are plant class I CXEs associated with certain plant responses, but the endogenous substrates remain unknown. For example, the Arabidopsis CXE12 protein was demonstrated to convert exogenously applied methyl-2,4-dichlorophenoxyacetate to the active herbicide 2,4-dichlorophenoxyacetic acid (Gershater et al., 2007). Tobacco (Nicotiana tabacum) and pepper (Capsicum annuum) class I CXE genes are induced upon pathogen infection and, while the endogenous substrates are not known, are thought to participate in the hypersensitive response (Baudouin et al., 1997; Ko et al., 2005). Although many class I CXEs remain to be analyzed, all of the enzymatically active class I CXEs from plants with well-characterized activities function in specialized metabolism.
Many plants throughout the nightshade family (Solanaceae) have glandular trichomes that produce and secrete specialized metabolites, including polyacylated sugar esters known as acylsugars (Schilmiller et al., 2008). Due to their presence on the surface of aerial tissues, these acylsugars are positioned to provide resistance against insect herbivores (Simmons and Gurr, 2005; Weinhold and Baldwin, 2011). Acylsugars and related compounds also have commercial and medicinal uses as surfactants (Hill and Rhode, 1999; Dembitsky, 2004), antimicrobial compounds (Chortyk et al., 1993; Ovenden et al., 2005), and antiinflammatory compounds (Pérez-Castorena et al., 2010). These varied properties pique interest in understanding how acylsugars are produced in trichomes and how their levels are regulated.
Within the tomato clade of the genus Solanum, acylsugars consist of a sugar backbone, either Glc or Suc, to which two to five aliphatic acyl chains are esterified. The acyl chains range in length from two to 13 carbons and may be either straight or branched. The types of sugar backbones as well as the lengths and branching of acyl chains on acylsugars vary widely among the species of the Solanaceae and also can display significant variation within accessions of a single species (Kroumova and Wagner, 2003; Kim et al., 2012; Ning et al., 2015; Schilmiller et al., 2015). We recently demonstrated that the biosynthesis of acylsucroses in cultivated and wild (Solanum pennellii) tomato involves BAHD-type acyltransferases that catalyze the transfer of acyl chains from an acyl-CoA donor. In the S. lycopersicum, acylsucrose assembly begins with the addition of a five-carbon acyl chain to the pyranose ring of Suc by the enzyme ACYLSUGAR ACYLTRANSFERASE1 (ASAT1) followed by the addition of a second pyranose ring acyl chain by ASAT2 (Fan et al., 2016). ASAT3 can add an acyl chain to the furanose ring of diacylsucroses or add an acyl chain to the pyranose ring of a monoacylsucrose acceptor, depending on the isoform (Schilmiller et al., 2015). ASAT4 adds an acetyl group to the pyranose ring of a triacylsucrose acceptor (Kim et al., 2012; Schilmiller et al., 2012).
We report the identification of class I CXE family enzymes from cultivated and wild tomato, which we named acylsugar acylhydrolases (ASHs). Introgression of a dominant locus containing two ASH genes from the wild tomato S. pennellii LA0716 into the S. lycopersicum M82 genetic background results in reduced levels of acylsugars (Schilmiller et al., 2010a). The dominant inheritance of the LA0716 locus is due to replacement of an inactive SlASH2 gene with a highly expressed and functional SpASH2 in IL5-3 plants. RNA interference (RNAi)-mediated suppression of the SpASH alleles in the IL5-3 introgression line caused the restoration of acylsugar levels, documenting their in planta function. In vitro assays with recombinant ASH proteins showed their ability to catalyze the hydrolysis of ester-linked acyl chains from specific positions of acylsugars. Assays with LA0716 ASH enzymes demonstrated reduced activity with acylsucroses produced by LA0716 compared with high activity against the structurally different acylsucroses produced by M82 trichomes. These results illustrate a mechanism for a negative-effect locus from S. pennellii LA0716 that should be avoided for breeding increased acylsugar levels and suggest the possibility of a mechanism for the turnover or remodeling of acylsugar molecules in trichomes.
RESULTS AND DISCUSSION
Identification of an S. pennellii Locus That Causes Reduced Acylsugars
Genetic Mapping of the IL5-3 Low-Acylsugar Trait
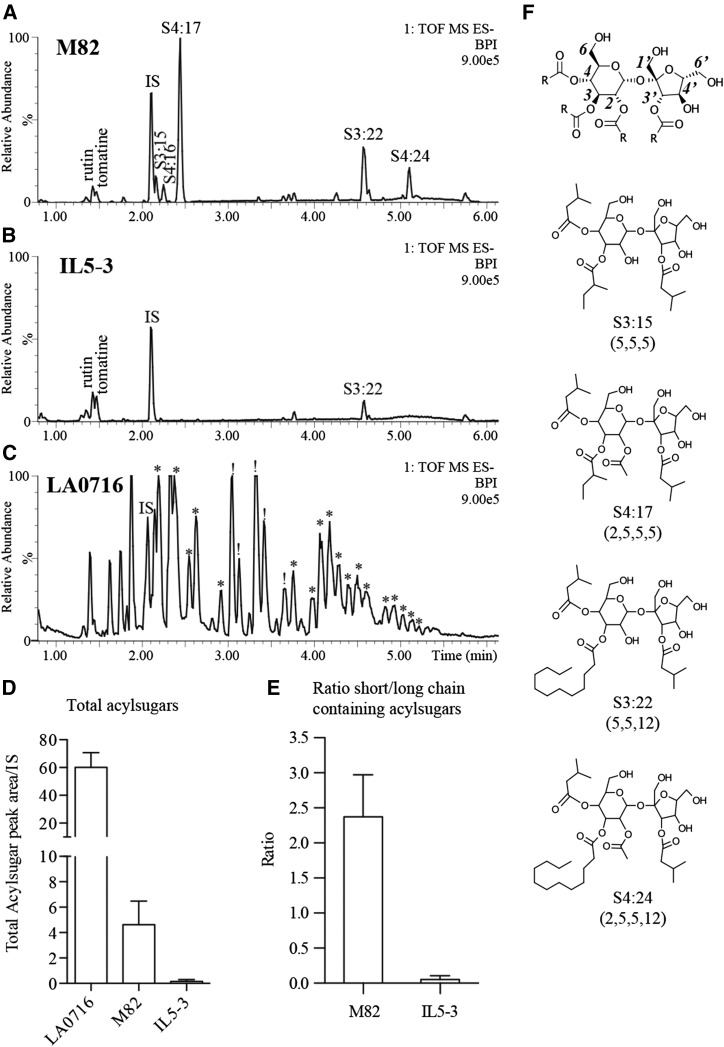
Previous analysis of trichome-specialized metabolites in the S. lycopersicum M82 × S. pennellii LA0716 introgression lines identified a region on chromosome 5 that influences total acylsugar levels (Schilmiller et al., 2010a). The parental S. lycopersicum var M82 produces a variety of triacylsucroses and tetraacylsucroses (Fig. 1; Ghosh et al., 2014). In contrast, IL5-3 leaves accumulated approximately 30-fold lower levels of acylsugars than wild-type M82 plants (Fig. 1, B and D). The effect on acylsugar levels was not equivalent for all acylsucroses: IL5-3 had a larger reduction in short (C5 or less) acyl chain-containing acylsugars compared with acylsugars containing a long (C10 or more) acyl chain (Fig. 1E). Nonacylsugar metabolites that are extracted by the leaf-dip method employed, including rutin and tomatine, were not affected in IL5-3.
Figure 1.
M82 and IL5-3 acylsugar phenotypes. A and B, Base peak intensity (BPI) liquid chromatography-mass spectrometry (LC-MS) chromatograms for M82 (A) and IL5-3 (B) show reduced levels of acylsugars in IL5-3 plants. Levels of rutin (flavonoid glycoside) and tomatine (glycoalkaloid) are similar for M82 and IL5-3. C, Acylsugar profile of LA0716, which produces an abundance of both acylglucoses (*) and acylsucroses (!). Base peak intensity chromatograms are plots of the abundance of the most abundant ion in each scan. The number 9.00e5 at the top right of each chromatogram indicates the ion count represented by 100% relative abundance. IS, Internal standard. D, Total acylsugar peak areas for [M+formate]– ions from extracted ion chromatograms for LA0716, M82, and IL5-3. Error bars indicate sd from three biological replicates. E, Ratio of total peak area for acylsugars containing only short (C5 or less) acyl chains to acylsugars containing one long (C10 or more) acyl chain. The reduced ratio for IL5-3 indicates a stronger reduction in short chain-containing acylsugars compared with acylsugars having a long chain. F, Structures of the most abundant acylsugars of M82 and a general acylsucrose structure showing position numbering for reference. Acylsugar nomenclature, using S3:15 (5,5,5) as an example, is as follows: S indicates an acylsucrose, 3 indicates three acyl chains, 15 indicates 15 total carbons in the three acyl chains, and in parentheses are listed the lengths of each individual acyl chain.
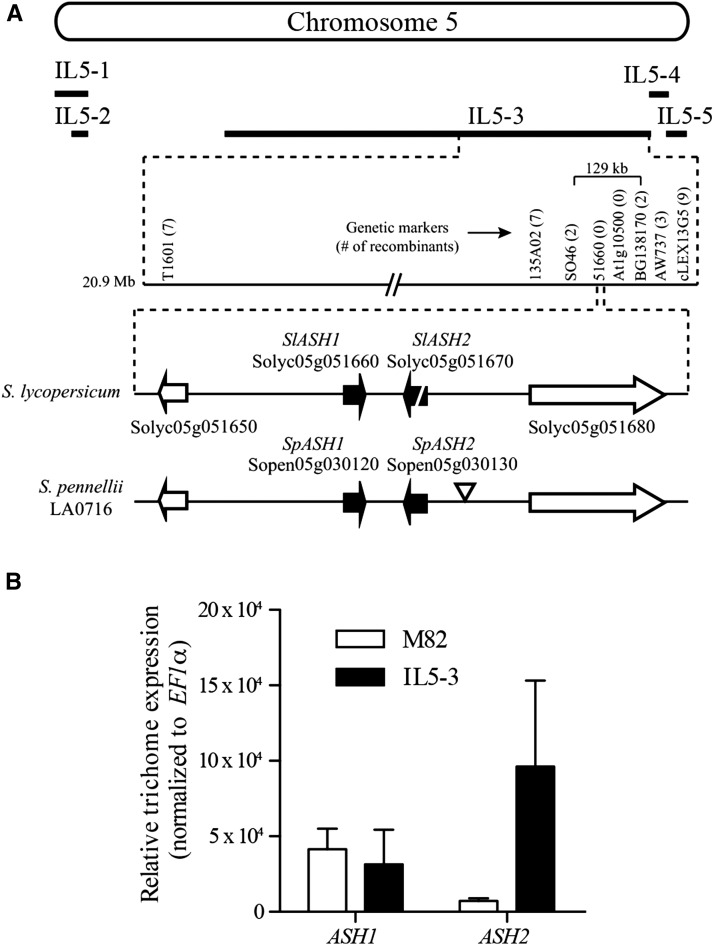
To understand the genetic basis for reduced acylsugars in IL5-3 plants, we crossed IL5-3 with the M82 parent. The F1 plants showed the reduced acylsugar phenotype, consistent with the S. pennellii locus being dominant. F2 progeny were generated, and the low:high progeny showed a ratio of 2.9:1 (χ2 = 0.266, n = 1,055), as expected for a dominant monogenic trait. The F2 progeny were used to perform fine-mapping to narrow the locus from the approximately 48-Mb IL5-3 span, which was predicted to encode 1,099 genes (Bombarely et al., 2011; Chitwood et al., 2013), to a region of approximately 129 kb (Fig. 2A). The annotation of the S. lycopersicum Heinz 1706 genome assembly (Sol Genomics Network [SGN; http://solgenomics.net]; Tomato Genome Consortium, 2012) included 17 predicted open reading frames (ORFs) within the mapping interval (Supplemental Table S1).
Figure 2.
Mapping of the IL5-3 locus and relative expression of ASH transcripts in trichomes of M82 and IL5-3. A, A positional cloning approach using F2 recombinants from an M82 × IL5-3 cross was used to narrow the causative locus to a region spanning approximately 129 kb. Markers are shown, with the number of recombinants in parentheses. A subset of the 129-kb region containing ASH1/2 is shown for the S. lycopersicum Heinz 1706 and S. pennellii LA0716 assemblies. The triangle after SpASH2 indicates a transposon insertion in the LA0716 genome at this position. B, ASH transcript levels were measured by quantitative RT-PCR in trichomes of M82 and IL5-3. The SpASH2 gene is expressed at higher levels in the IL5-3 trichomes compared with the trichome expression level in M82 for SlASH2, which is predicted not to encode a functional protein. Error bars indicate sd among three biological replicates.
Two Genes from This Region Are Expressed in Trichomes
To narrow this list of candidate genes further, we looked for evidence of expression in S. lycopersicum M82 and S. pennellii LA0716 trichomes using available trichome RNA sequencing (RNAseq) data (Schilmiller et al., 2010b; McDowell et al., 2011; Ning et al., 2015). Four genes in the interval showed no evidence of trichome expression, and another nine had only a small number of reads (less than 0.005% of the total reads in the library; reads per kilobase of transcript per million reads mapped < 10) mapped to each gene, indicating a low level of expression in trichomes (Supplemental Table S1). Two genes in the region, Solyc05g051660 (Sopen05g030120) and Solyc05g051670 (Sopen05g030130; renamed ASH1 and ASH2, respectively), had large numbers of reads in the LA0716 trichome complementary DNA (cDNA) library but low read counts in M82 trichome cDNA (Supplemental Table S1). M82 RNAseq data using isolated trichomes from stem tissue and stem tissue after removal of trichomes were analyzed to determine relative levels of expression in trichome versus nontrichome tissue (Ning et al., 2015). The expression of both SlASH1 and SlASH2 was enriched in trichome tissue, with 10-fold (SlASH1) and 127-fold (SlASH2) higher expression in trichomes versus stems and petioles after removal of the trichomes. Quantitative reverse transcription (RT)-PCR using the same cDNA validated the trichome-enriched expression of SlASH1 and SlASH2 (Supplemental Fig. S1). Although SlASH1 expression is highest in trichome tissue, the expression is not trichome specific, in contrast to what is seen for the ASAT3 and ASAT4 genes, which encode proteins that catalyze the transfer of acyl chains to form acylsucroses (Schilmiller et al., 2012, 2015). A SlASH1 unigene (SGN-U566465) is present in the SGN database, and it consists of nine ESTs from various cDNA libraries, including trichomes (one), whole plants (two), developing flower buds (two), suspension cell cultures (two), and seeds (two). Flower buds and whole plants contain trichomes and would be expected to show the expression of trichome-specific transcripts; however, the ESTs from suspension cells and seeds suggest the possibility of additional non-trichome-localized functions for SlASH1. No ESTs for SlASH2 were found in any tomato cDNA library in the SGN database.
The sequences of ASH1 and ASH2 in M82 and LA0716 were examined more closely to understand how they could influence acylsugar levels in IL5-3. A BLAST search showed that both SlASH1 and SlASH2 encode proteins in the carboxylesterase subgroup of the α/β-hydrolase fold superfamily based on the presence of Interpro domain IPR013094 (Hotelier et al., 2004). The SlASH1 ORF is 969 bp with no introns and is predicted to encode a protein with 322 amino acids. The current annotation of the tomato genome (ITAG2.4; http://solgenomics.net) indicates that SlASH2 has two introns and is predicted to result in an 855-bp ORF encoding a protein with 284 amino acids. The S. pennellii LA0716 genomic sequence shows that both SpASH1 and SpASH2 lack introns and are predicted to encode proteins of 323 amino acids in length. Alignment of S. lycopersicum and S. pennellii ASH genomic sequences suggests that the SlASH2 gene is likely to be inactive, because it harbors a 31-bp deletion and a 4-bp insertion that disrupts its predicted ORF (Supplemental Fig. S2). In addition, SlASH2 has 59 single-nucleotide polymorphisms that are unique to SlASH2 (compared with SpASH2 and both ASH1 sequences), whereas SlASH1 has only eight unique single-nucleotide polymorphisms, suggesting that the SlASH2 sequence has been diverging rapidly, as seen commonly for pseudogenes.
In addition to the presence of a functional SpASH2 gene in IL5-3, a published RNAseq analysis of gene expression in the S. pennellii introgression lines suggested that increased expression of the ASHs in IL5-3 may also contribute to the low-acylsugar phenotype. An approximately 3- to 4-fold increase was observed for the expression of SpASH1 and SpASH2 in IL5-3 13-d-old seedlings compared with that of other introgression lines containing the SlASH alleles (Chitwood et al., 2013). To validate these results and test whether this expression difference is also observed in trichomes, quantitative PCR (qPCR) was performed using cDNA from isolated trichomes collected from stems and petioles of 3-week-old M82 and IL5-3 plants. The SpASH2 transcript levels were approximately 13-fold higher in IL5-3 trichome RNA compared with SlASH2 in M82 trichomes (Fig. 2B). In contrast, SpASH1 and SlASH1 transcripts accumulated to similar levels in trichomes (Fig. 2B). Taken together, these results led to the hypothesis that the low-acylsugar phenotype in IL5-3 results from introgression of the more highly expressed SpASH2, replacing the mutated SlASH2 gene in the M82 genome. This proposed mechanism is consistent with the dominance of the S. pennellii locus over the S. lycopersicum M82 locus.
ASH1 and ASH2 Proteins Have Acylsugar Acylhydrolase Activity
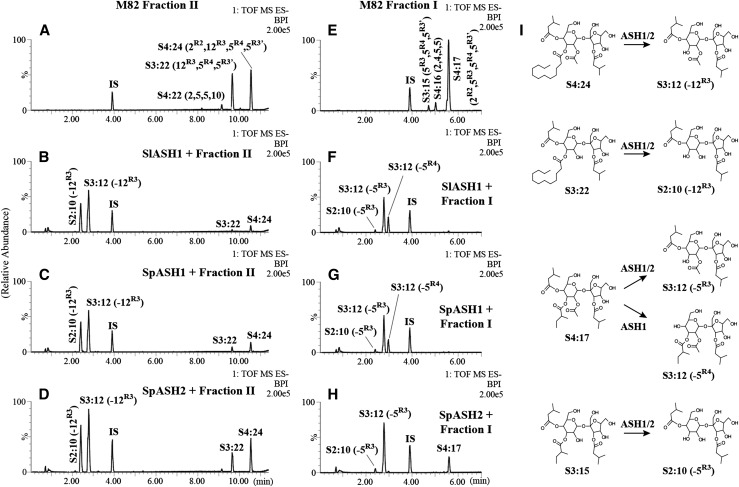
Based upon the homology of ASH1 and ASH2 to carboxylesterases, we tested the hypothesis that the IL5-3 SpASH enzymes hydrolyze S. lycopersicum acylsucrose acyl chains. The SlASH1, SpASH1, and SpASH2 enzymes were expressed in Escherichia coli as 6× His-tagged fusion proteins and assayed using S. lycopersicum acylsugars as substrates. The triacylsucroses of S. lycopersicum have acyl chains at the R3, R4, and R3′ positions, with tetraacylsucroses having an additional acetyl group at the R2 position; the major acylsugars in M82 trichome extracts include S3:15 (5,5,5), S4:17 (2R2,ai5R3,i5R4,i5R3′), S3:22 (ai5R3,i5R4,n12R3′), and S4:24 (2R2,ai5R3,i5R4,n12R3′) (Schilmiller et al., 2010a, 2012; Ghosh et al., 2014). Acylsugar nomenclature is as follows: using S3:15 (5,5,5) as an example, S indicates an acylsucrose, 3 indicates three acyl chains, and 15 indicates 15 total carbons in the three acyl chains. The length of each individual acyl chain is in parentheses, with superscripts indicating the position of the acyl chain, ai = anteiso branched, i = iso branched, and n = normal straight chain (Fig. 1E). Because we observed differences in the impact of IL5-3 on the accumulation of short and long chain-containing acylsucroses, the M82 acylsugar mixture was separated into two fractions prior to their use as substrates. Fraction I contained triacylsucroses and tetraacylsucroses having only short acyl chains (C5 or less), while fraction II consisted of triacylsucroses and tetraacylsucroses having a single long acyl chain (C10–C12).
ASH1 and ASH2 Hydrolyze the R3 Position of the Six-Membered Ring of Fraction II
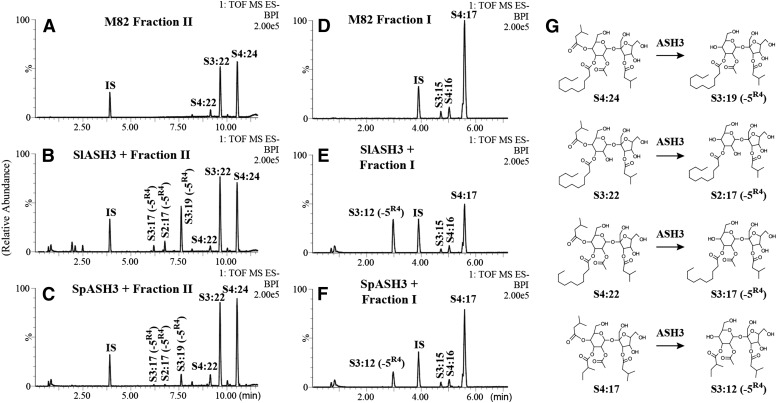
Our results demonstrate that the ASH enzymes hydrolyzed the long chain-containing triacylsucroses and tetraacylsucroses at the R3 position (Fig. 3). Incubation of these two M82 acylsugar fractions with SlASH1, SpASH1, or SpASH2 resulted in the production of new, earlier-eluting peaks, which were identified as S2:10 (5,5) and S3:12 (2,5,5) based on fragmentation in negative-ion mode liquid chromatography-time of flight-mass spectrometry (Fig. 3; Supplemental Fig. S3). These diacylsucrose and triacylsucrose products were not detectable in either of the substrate fractions and, thus, result from hydrolysis of acyl chains from the triacylsucroses and tetraacylsucroses. That the R3 position acyl chain that was removed was most clearly observed in the reactions using the long chain-containing acylsucrose substrate fraction II (Fig. 3, A–D): the S2:10 (5,5) and S3:12 (2,5,5) products that were observed could only result from hydrolysis of the C12 acyl chain of S3:22 (5,5,12) and S4:24 (2,5,5,12), respectively, or from hydrolysis of C10 from S4:22 (2,5,5,10). No hydrolysis products were detected that had a C5 acyl chain removed and the long acyl chain remaining in the ASH1 or ASH2 reaction when fraction II was used as a substrate.
Figure 3.
Evidence that ASH1 and ASH2 hydrolyze acyl chains from the R3 or R4 position of M82 acylsucroses. Base peak intensity (BPI) LC-MS chromatograms are shown for reactions with the ASH1 and ASH2 enzymes using M82 long chain-containing (fraction II) and short chain-containing (fraction I) acylsucrose substrates. Specific positions of acyl chains are given as superscripts when known from NMR-resolved structures (Ghosh et al., 2014); otherwise, only acyl chain lengths are given. IS, Internal standard. Peak labels for ASH products indicate the acyl chain that was hydrolyzed in the reaction. A, Fraction II substrate. B, SlASH1 + fraction II. C, SpASH1 + fraction II. D, SpASH2 + fraction II. E, Fraction I substrate. F, SlASH1 + fraction I. G, SpASH1 + fraction I. H, SpASH2 + fraction I. Structures of acylsucrose substrates and their reaction products are shown in I. The scale of chromatogram signals at 100% (representing the most abundant peak) is indicated by the number at the top right corner of each chromatogram.
ASH2 Hydrolyzes the R3 Position of the Six-Membered Ring of Fraction I
When fraction I, containing short-chain acylsucroses from M82, was tested as a substrate with SpASH2, S2:10 (5,5) and S3:12 (2,5,5) again were the major products observed for each reaction (Fig. 3, E–H). These products result from hydrolysis of a single C5 acyl chain from S3:15 (5,5,5) and S4:17 (2,5,5,5), respectively, or from hydrolysis of a C4 acyl chain from S4:16 (2,4,5,5). Determination of the position of hydrolysis is less straightforward with these substrates than with long-chain acylsugar fraction II, due to the presence of multiple C5 chains, but consideration of additional LC-MS information helped to narrow the likely position. Fragmentation of these products in positive-ion mode, which results in cleavage of the glycosidic bond between the pyranose (six-membered) and furanose (five-membered) rings of the acylsucrose (Ghosh et al., 2014), showed that each product contained a C5 acyl chain on the furanose ring (Supplemental Fig. S4). This indicates that the hydrolyzed acyl chain was on the pyranose ring. The S2:10 and S3:12 products produced by the ASH2-fraction I reaction coeluted with S2:10 and S3:12 from reactions using fraction II, the long chain-containing substrate mixture. Taken together, these data are consistent with the hypothesis that SpASH2 catalyzed a single cleavage, at the R3 position of short chain-containing acylsucroses.
ASH1 Cleaves a Single Acyl Chain from the R3 or R4 Position of the Six-Membered Ring of Fraction I
Interpretation of the results from the SlASH1 and SpASH1 enzymatic reactions is more complicated. The S2:10 and S3:12 peaks arising from R3 hydrolysis were also seen with these enzymes; however, a second, later-eluting S3:12 (2,5,5) peak also was observed from the fraction I substrates. Positive-ion mode mass spectrometry analysis revealed that this product also retained the R3′ furanose ring C5 acyl chain (Supplemental Fig. S4). The presence of two chromatographically separable S3:12 (2,5,5) peaks in the ASH1 samples is most simply explained as the two positional isomers, (2R2,5R3,5R3′) and (2R2,5R4,5R3′), resulting from hydrolysis of S4:17 (2R2,5R3,5R4,5R3′) at different positions (Fig. 3; Supplemental Fig. S3). Alternatively, the two S3:12 peaks could correspond to two C5 acyl chain isomers, present as either terminally branched (iso) or subterminally branched (anteiso) chains. However, this scenario is unlikely, because the magnitude of the retention time difference for the two S3:12 peaks suggests a more significant difference in structure than simply branch point isomers of an acyl chain. Clear evidence for R4 hydrolysis by ASH1 comes from results shown below for a related hydrolase (ASH3) that cleaves esters at the R4 position. The later-eluting S3:12 product of ASH1 cochromatographs with the ASH3 S3:12 product. Our results suggest that the length of the acyl chain at the R3 position influences ASH1 hydrolysis of the R4 position. This is because the ASH1 enzymes did not produce S3:19 (2R2,12R3,5R3′) or S2:17 (12R3,5R3′), which are the expected products of R4 hydrolysis of the fraction II substrates. The activity of ASH1 hydrolyzing multiple positions of the short chain-containing acylsugars is consistent with the observed IL5-3 phenotype of substantially lower levels of the short chain-containing acylsugars compared with the long chain-containing acylsugars. Taken together, our data indicate that the ASH1 and ASH2 enzymes are carboxylesterases with hydrolytic activity for the cleavage of acyl chains from specific positions of triacylsucroses and tetraacylsucroses.
Reducing the Expression of SpASH1/2 in IL5-3 Restores Acylsugar Levels
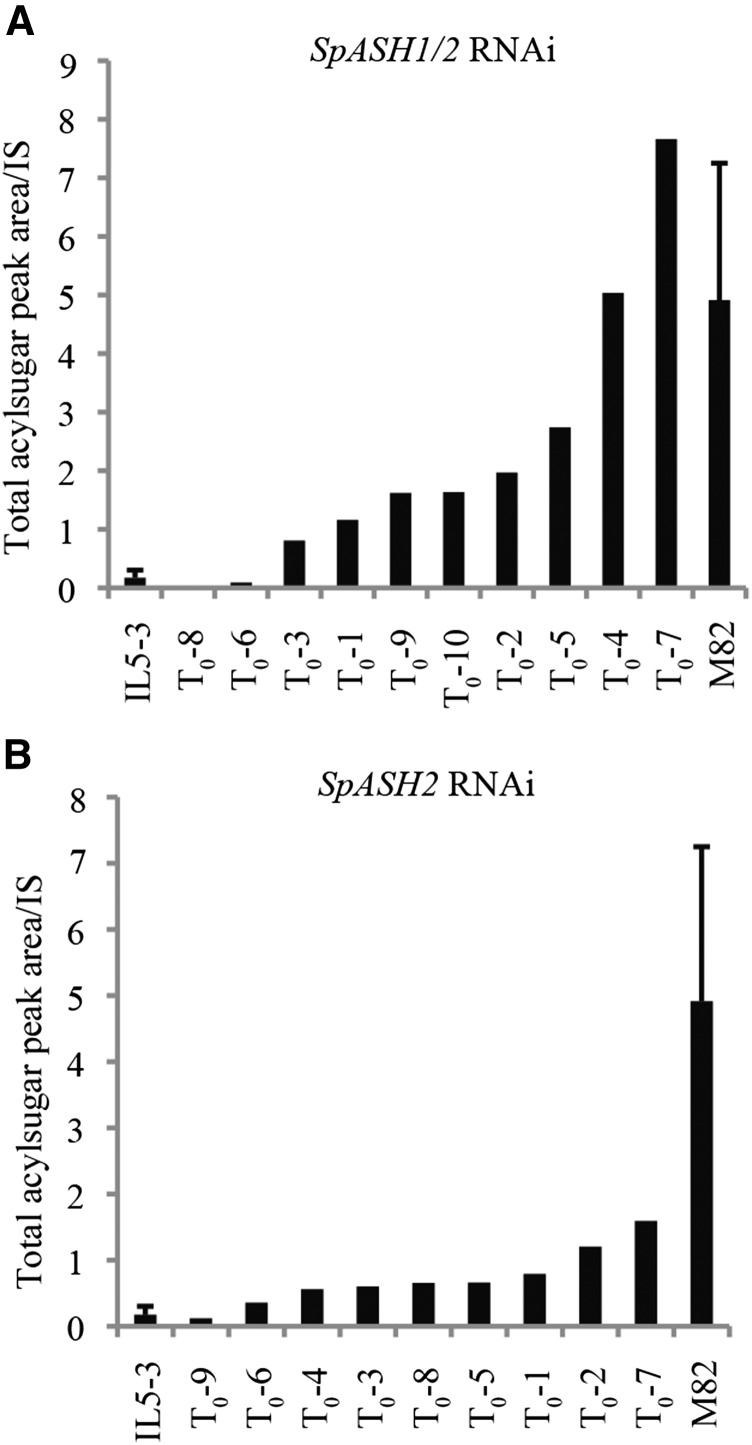
RNAi transgenic line phenotypes were analyzed to test the hypothesis that SpASH introgression caused reduced acylsugar levels. Taking advantage of the observation that SpASH1 and SpASH2 are 95% identical at the nucleotide level, we designed an RNAi construct aimed at silencing both genes (Supplemental Fig. S2). Eight of ten independent transgenic IL5-3 lines transformed with this construct showed increased acylsugar levels, with two having wild-type M82 levels (Fig. 4A). The ratio of acylsugars having all short chains to acylsugars having a long acyl chain also was restored to wild-type M82 levels in seven of 10 transgenics (Supplemental Fig. S5). The restoration of the short-to-long chain-containing acylsugar ratio and the lack of any new acylsugar intermediates in the RNAi lines suggest that ASH1/2 probably act first followed by other hydrolases that further degrade acylsugars. Nine independent transgenic lines in the IL5-3 background were recovered using an RNAi construct targeting SpASH2 specifically. Of these nine lines, seven showed a small increase in acylsugar levels compared with IL5-3, although the levels were still statistically significantly lower than the M82 wild-type levels (Fig. 4B). These results suggest that both S. pennellii ASH genes contribute to reduced acylsugar levels in IL5-3 plants.
Figure 4.
RNAi suppression of SpASH1 and SpASH2 restores acylsugar production in IL5-3. A, When IL5-3 was transformed with an RNAi construct targeting sequence with high identity for both SpASH1 and SpASH2, the primary transformants showed a range of increased total acylsugar levels, with some showing acylsugar levels comparable to the M82 phenotype. B, An RNAi construct targeting specifically SpASH2 did not have a strong impact on acylsugars when transformed into IL5-3 plants. IS, Internal standard.
Taken together, the introgression of SpASH1 and SpASH2 and the increased expression of SpASH2 can explain the IL5-3 low-acylsugar phenotype. However, the in vitro assay results suggest that the SlASH1 enzyme in wild-type S. lycopersicum also should be able to hydrolyze acylsugars in the trichome, yet the wild-type M82 plants produce easily detected levels of acylsucroses in the presence of SlASH1. One hypothesis for explaining the acylsugar phenotype of M82 plants is that the acylsugar levels really are quite low, compared with LA0716, and that this is due in part to the activity of SlASH1 in the trichomes. Based on this logic, silencing of ASH genes could potentially lead to increased acylsugar levels. However, silencing of ASH expression in IL5-3 did not result in acylsugar levels that were significantly greater than the levels in wild-type M82 plants when acylsugars were measured in the primary transformants. This suggests that other modes of acylsugar regulation exist, possibly at the level of biosynthesis or secretion, that result in S. pennellii LA0716 accumulating acylsugars in amounts much higher than S. lycopersicum M82.
Other Tomato CXEs with ASH Activity
The ASH1 and ASH2 enzymes specifically hydrolyze the R3 and R4 positions of acylsugar substrates from S. lycopersicum, yet IL5-3 does not accumulate any detectable diacylsucroses or triacylsucroses lacking acyl chains at these positions. This suggests that other ASHs are expressed in trichomes, and this hypothesis was explored in several ways. First, a phylogeny was constructed using the complete set of S. lycopersicum class I carboxylesterase enzymes containing Interpro domain IPR013094 in the most recent annotation (ITAG2.4; Supplemental Fig. S6). A search was then conducted to explore for evidence of expression of the 33 other predicted tomato CXE genes in the S. lycopersicum and S. pennellii trichome RNAseq data (Schilmiller et al., 2010a; McDowell et al., 2011; Ning et al., 2015). Two other acylsugar acylhydrolase candidates, Solyc09g075710 (Sopen09g030520) and Solyc04g005230 (Sopen04g001210), are clustered with ASH1 and ASH2 and show trichome-enriched expression (Supplemental Fig. S6).
Assays were performed with recombinant enzymes to test their activities with acylsugar substrates from M82. The Solyc09g075710 enzyme and corresponding S. pennellii LA0716 isoform, renamed SlASH3 and SpASH3, respectively, showed activity cleaving the C5 chain at the R4 position of M82 acylsucroses (Fig. 5). Assays with both ASH3 isoforms using fraction II substrates containing acylsucroses with a long acyl chain resulted in three products, S3:17 (2,5,10), S2:17 (5,12), and S3:19 (2,5,12), from hydrolysis of a C5 acyl chain from S4:22 (2,5,5,10), S3:22 (5,5,12), and S4:24 (2,5,5,12), respectively (Fig. 5, A–C; Supplemental Fig. S7). Each of these products retained the R3′ C5 acyl chain as determined by fragmentation in positive-ion mode LC-MS (Supplemental Fig. S4); therefore, the hydrolyzed chain came from the six-membered pyranose ring. Because the S3:22 (5,5,12) and S4:24 (2,5,5,12) substrates have a single C5 acyl chain on the pyranose ring, we conclude that the cleaved C5 acyl chain is located at the R4 position. Similarly, when the fraction I short chain-containing acylsucroses of M82 were used as substrate, SlASH3 and SpASH3 produced an S3:12 (2,5,5) peak that results from hydrolytic removal of a C5 chain from the six-membered pyranose ring. As mentioned earlier, this product cochromatographs with the later-eluting S3:12 produced in the ASH1 assays that is hypothesized to arise from hydrolysis of the R4 position (Fig. 5, D–F). Taken together, these data show that ASH3 hydrolyzes the C5 chain at the R4 position of triacylsucroses in vitro. In contrast, no activity was detected using the Sopen04g001210 enzyme with either M82 acylsugar fraction I or II.
Figure 5.
Evidence that SlASH3 and SpASH3 hydrolyze the R4 position ester from M82 acylsucroses. Base peak intensity (BPI) LC-MS chromatograms are shown for reactions with the ASH3 enzymes using M82 long chain-containing (fraction II) and short chain-containing (fraction I) acylsucrose substrates. IS, Internal standard. A, Fraction II substrate. B, SlASH3 + fraction II. C, SpASH3 + fraction II. D, Fraction I substrate. E, SlASH3 + fraction I. F, SpASH3 + fraction I. Structures of acylsucrose substrates and their reaction products are shown in G. The scale of chromatogram signals at 100% (representing the most abundant peak) is indicated by the number at the top right corner of each chromatogram.
To test whether ASH3 could use the ASH1 products as substrates, we set up reactions using the M82 fraction I substrate with equal amounts of purified SpASH1 and SpASH3 enzyme. In reactions using only a single ASH enzyme, only one acyl chain was hydrolyzed. When both SpASH1 and SpASH3 were incubated together with M82 fraction I acylsugars, a new and earlier-eluting peak was produced, which corresponds to S2:7 (2R2,5R3′) (Supplemental Fig. S8). This result suggests a pathway where multiple ASH enzymes work sequentially to hydrolyze acylsugars in trichomes.
While introgression of S. pennellii ASH alleles on chromosome 5 resulted in reduced acylsugar levels, no altered acylsugar phenotype was found for IL9-3, the introgression region containing the SpASH3 allele (Schilmiller et al., 2010a). This presumably is the result of several factors. First, the S. lycopersicum and S. pennellii ASH3 isoforms showed the same in vitro activities, which suggests that they have the same in vivo function. Second, the two genes appear to be expressed similarly: no difference in SpASH3 gene expression was found in IL9-3 compared with other introgression lines that have the SlASH3 allele (Chitwood et al., 2013). The low in vitro activity of ASH3 for acylsugar hydrolysis also suggests that ASH3 has a different function in vivo in trichomes.
SpASH Enzymes Display Low or No Activity against LA0716 Acylsugars
RNAseq data indicated that the SpASH genes are highly expressed in S. pennellii LA0716 trichomes, yet plants of this accession produce large amounts of acylsucroses and acylglucoses, estimated to be as high as 20% of leaf dry weight in 17-week-old plants (Fig. 1, C and D; Fobes et al., 1985). To determine how LA0716 plants can accumulate such high levels of acylsugars while expressing genes encoding acylsugar hydrolases in trichomes, we compared the relative activities of S. pennellii ASH enzymes using acylsugars purified from M82 or LA0716 plants.
LA0716 acylsugars consist of both triacylglucoses and triacylsucroses, with the latter having all three acylsucrose acyl chains esterified to the pyranose ring of the Suc backbone (Shapiro et al., 1994; Schilmiller et al., 2015). This is in contrast to the triacylsucroses and tetraacylsucroses produced by M82 trichomes, each of which has a single acyl chain (usually C5) present on the furanose ring of the Suc backbone (Schilmiller et al., 2015). In addition, the M82 tetraacylsucroses have C2 chains at the pyranose ring R2 position, whereas LA0716 acylsugars have longer C5 chains. To determine whether either structural difference inhibits ASH activity, an abundant acylsucrose and an acylglucose were purified from LA0716 for use in SpASH assays. Structures of the purified acylsugars were characterized by NMR spectroscopy: the acylsucrose S3:19 (ai5R2,i10R3,i4R4) and the acylglucose G3:14 (ai5R2,i5R3,i4R4) from LA0716 both have acyl chains in the R2, R3, and R4 positions (Supplemental Tables S2 and S3).
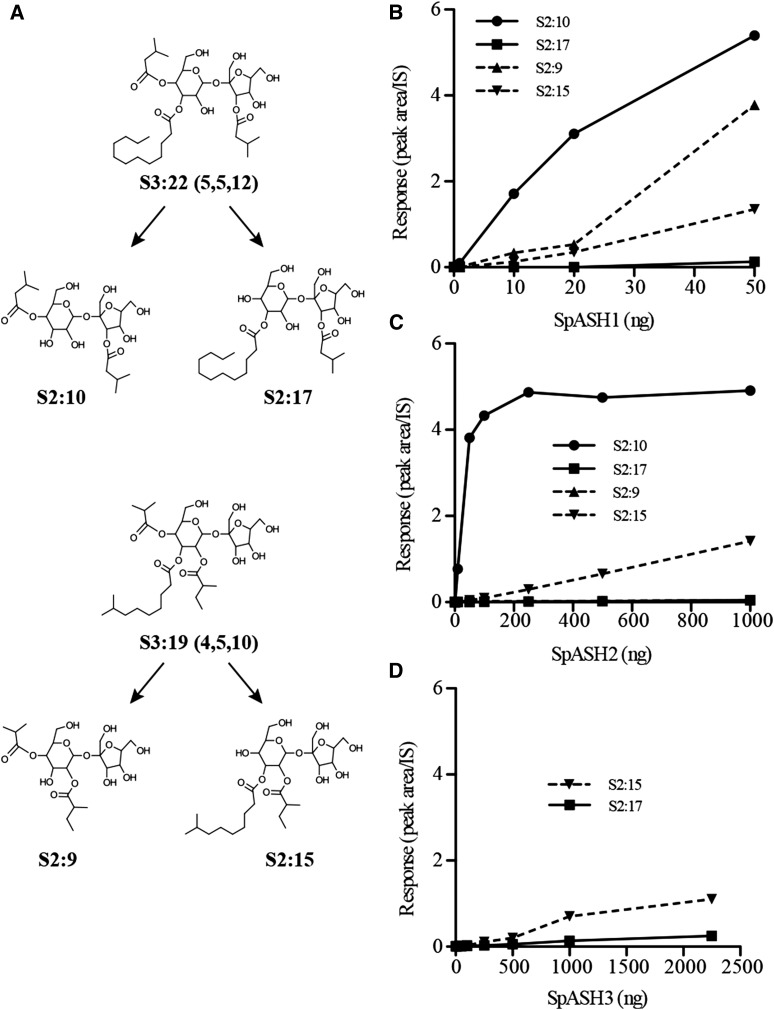
ASH activity assays were first performed using purified acylsucroses with the three recombinant SpASH enzymes. SpASH2 showed the most striking difference in activity: while SpASH2 efficiently hydrolyzed a C5 chain from M82 S3:22 (n12R3,i5R4,i5R3′), only weak activity was observed using LA0716 S3:19 (ai5R2,i10R3,i4R4) as substrate, this despite the use of greater than 20-fold more SpASH2 enzyme than was used in assays with the M82 S3:22 substrate (Fig. 6C). Activity assays with SpASH2 using a variety of purified acylsucroses from Solanum habrochaites with known structures (Ghosh et al., 2014) indicated that the presence of an acyl chain longer than C2 at the R2 position strongly inhibited SpASH2 activity (Supplemental Fig. S9). SpASH1 also showed greater activity with the S. lycopersicum S3:22 (n12R3,i5R4,i5R3′) acylsucrose as substrate compared with S. pennellii S3:19 (ai5R2,i10R3,i4R4), with the difference seen most clearly at lower SpASH1 enzyme concentrations (Fig. 6B). Kinetic analysis of SpASH1 indicated a similar Km for each substrate (Table I; Supplemental Fig. S10; note that SpASH1 can hydrolyze two different positions and that Km values were calculated for each position); however, the maximum velocity was approximately 2.2-fold higher for hydrolysis at the R3 position of S3:22 compared with S3:19 and 6.6-fold higher for hydrolysis at the R4 position. SpASH3 activity with either substrate was detectable only at very high enzyme concentrations, and the specific activity was much lower than that of SpASH1 or SpASH2 (Fig. 6D). These results show that the relatively high in vitro activity of SpASH1 and SpASH2 enzymes with the S. lycopersicum S3:22 is consistent with the observed acylsugar reduction in IL5-3 plants.
Figure 6.
SpASH enzymes show higher activity with an S. lycopersicum acylsucrose compared with an LA0716 acylsucrose. A, Structures of S3:22 from M82 and S3:19 from LA0716 and the respective hydrolysis products from the SpASH reactions. B to D, Product amounts from assays with a fixed concentration (33 μm) of substrate and increasing amounts of SpASH1 (B), SpASH2 (C), or SpASH3 (D) enzyme. IS, Internal standard.
Table I. SpASH1 kinetic parameters.
Vmax values are not given units because actual product amounts could not be quantified in μmol; instead, they are calculated using peak response and are shown relative to Vmax for S3:22 R3 hydrolysis.
| Substrate | Apparent Km | Apparent Vmax |
|---|---|---|
| μm | ||
| S3:22 R3 hydrolysis | 17.4 ± 2.5 | 100 |
| S3:19 R3 hydrolysis | 24.8 ± 10.8 | 45 |
| S3:19 R4 hydrolysis | 44.2 ± 19.7 | 15 |
S. pennellii LA0716 accumulates even higher amounts of acylglucoses than acylsucroses (Shapiro et al., 1994; Ning et al., 2015). This led us to hypothesize that these acylglucoses are also poor substrates for SpASH enzymes. As was the case for the LA0716 S3:19 acylsucrose (Fig. 6), SpASH2 showed very low activity detectable only at high enzyme concentrations when using G3:14 (ai5R2,i5R3,i4R4) as substrate (Supplemental Fig. S11). SpASH1 and SpASH3 both showed higher activity with G3:14 compared with SpASH2; however, the amount of conversion to product was low even at the highest enzyme concentrations (Supplemental Fig. S11). Taken together, the low activity of SpASH enzymes with the S. pennellii acylsucrose and acylglucose is consistent with the high accumulation of acylsugars in LA0716.
CONCLUSION
The in vitro activity of SpASH1 and SpASH2 in hydrolyzing acylsucroses, together with the restoration of acylsugar production following the silencing of SpASH1/2, indicates that these carboxylesterases condition the IL5-3 reduced acylsugar phenotype. While the actual in vivo functions of the ASH enzymes on specific acylsugar substrates in S. lycopersicum and S. pennellii are less clear, several testable hypotheses seem plausible. One possibility is that the ASH enzymes play a role in acylsugar turnover in the trichomes. The commitment of large amounts of Suc or Glc or short-chain fatty acids for acylsugar production could have a negative effect on apical cell function or plant growth under certain environmental conditions, and the ASHs could return acylsugars back into other metabolic pathways. Alternatively, the ASH enzymes could play a role in the remodeling of acylsugar structures; remodeling of lipids via acyl editing has been shown to occur in plant and animal systems (Bates et al., 2007; Robichaud et al., 2013). Similarly, ASHs could remove acyl chains, allowing replacement with an acyl chain of different length or branch structure. Another alternative is that the ASH enzymes could perform the reverse reaction and catalyze the transfer of acyl chains to form an ester-linked acylsugar product rather than hydrolyze ester linkages.
Analysis of the sequences of the ASH enzymes does not indicate a subcellular localization other than cytosolic, which is similar to that of the ASAT BAHD enzymes that produce acylsucroses. Colocalization of biosynthetic enzymes with enzymes capable of hydrolyzing acylsugars would seemingly create the possibility of a futile cycle of synthesis and degradation; however, this does not prevent the accumulation of acylsugars on S. lycopersicum and S. pennellii leaves. More work is needed to understand whether temporal expression or localization differences for the ASAT and ASH enzymes are important for controlling acylsugar production. Labeling experiments similar to that described by Wang and Jones (2014) using IL5-3 or some of the RNAi lines described in this work may help to reveal the dynamics of acylsugar accumulation and the roles that ASH enzymes play.
Due to their role in defense against herbivores, acylsugar breeding programs are under way to create S. lycopersicum varieties with acylsugar levels and compositions that increase resistance to insect pests (Alba et al., 2009; Leckie et al., 2012). Quantitative trait locus (QTL) analyses using populations derived from S. lycopersicum and S. pennellii LA0716 previously identified a negative-effect QTL on chromosome 5 that maps to the same region as ASH1 and ASH2 (Leckie et al., 2012). In fact, one of the markers used to define the QTL is located only approximately 56 kb away from the ASH locus. Our results suggest that SpASH1 and SpASH2 are the causative genes in this negative-effect QTL. The dominance of the S. pennellii alleles is consistent with the observed reduction in acylsugar levels when the chromosome 5 QTL is present in the heterozygous state (Leckie et al., 2012).
Our understanding of plant specialized metabolism is ever increasing as more pathways and enzymes are discovered (Osbourn and Lanzotti, 2009). Many plant specialized metabolites are modified by esterification with a variety of chemical side chains, including acetyl, aliphatic, phenolic, and organic acid groups. These modifications can modulate the chemical and physiological properties of specialized metabolites or serve as blocking or activating groups in metabolic pathway intermediates. Studies focused on the expression (tissue specificity, environmental regulation, etc.) and activities of carboxylesterases will lead to better understanding of how hydrolysis influences the structures, functions, and amounts of these metabolites. Given the important roles in both the synthesis and turnover of metabolites that the small number of characterized CXEs play in plant physiology, a systematic analysis of this gene family will be important for the rational engineering of biosynthetic pathways of valuable products that are not readily chemically synthesized.
MATERIALS AND METHODS
Mapping of the IL5-3 Acylsugar Locus
An F2 mapping population was created by self-pollination of a cultivated tomato (Solanum lycopersicum) M82 × Solanum pennellii IL11-3 F1 line kindly provided by Dani Zamir (Hebrew University Faculty of Agriculture). Genotyping using PCR-based cleaved-amplified polymorphic sequence markers (Supplemental Table S4) and phenotyping by negative-ion mode LC-MS analysis of acylsugars extracted by gentle agitation for 2 min were performed as described previously (Schilmiller et al., 2015).
Quantitative RT-PCR
To compare the expression in M82 versus IL5-3 trichomes, total RNA was extracted from stem and petiole trichomes of 3-week-old M82 and IL5-3 plants using the RNeasy Plant Mini Kit (Qiagen). First-strand cDNA was synthesized from 0.5 μg of total trichome RNA using SuperScript II reverse transcriptase (Invitrogen), and the cDNA was diluted 10-fold for qPCR. To determine the expression levels in M82 trichomes versus the underlying stems and petiole tissue, cDNA was prepared from 1 μg of total RNA that was used for RNAseq as described (Ning et al., 2015). ASH transcripts were quantified using the standard curve method. The qPCR mix consisted of SYBR Select master mix (Life Technologies) with primer concentration of 250 nm and supplemented with 0.25 μL of Taq polymerase (Invitrogen) per 20-μL reaction. Primer sequences are listed in Supplemental Table S4. Reactions were analyzed using an ABI 7500 Fast qPCR machine (Life Technologies). ASH transcript levels were normalized to EF1α (Solyc06g009970) transcript levels for comparison across tissues and genotypes.
Silencing of ASH1/2 Using RNAi
Constructs for RNAi silencing of SpASH1 and SpASH2 in IL5-3 plants were created using the pHELLSGATE-12 binary vector (Eamens and Waterhouse, 2011). A 511-bp fragment of SpASH1 was amplified from IL5-3 genomic DNA and cloned into pENTR/D-TOPO (Invitrogen) followed by recombination into pHELLSGATE-12 (primers are listed in Supplemental Table S4). Similarly, a 351-bp fragment of SpASH2 from the 3′ end of the ORF and untranslated region was amplified from IL5-3 genomic DNA, cloned into pENTR/D-TOPO, and recombined into pHELLSGATE-12. RNAi constructs were transformed into Agrobacterium tumefaciens strain AGL0, and transformation of IL5-3 tomato was performed as described previously (McCormick, 1991).
Phylogenetic Tree Construction
Protein sequences containing Interpro domain IPR013094 from the annotated tomato genome (ITAG2.4) were aligned together with several other plant class I CXEs using the Muscle algorithm in MEGA software (version 6.06; Tamura et al., 2013). The phylogeny was constructed using the maximum-likelihood method with the Le and Gascuel substitution model and bootstrapped with 1,000 replicates.
Preparation of Acylsugar Substrates
Acylsugar mixtures from M82 were collected by extracting leaves and stems from 3-week-old plants using acetonitrile:isopropanol:water (3:3:2, v/v/v) containing 0.1% (v/v) formic acid. The extract was diluted 1:4 with water and then loaded onto a 500-mg C18 solid-phase extraction cartridge (Supelco; catalog no. 57012) equilibrated with 10% (v/v) acetonitrile. Short chain-containing acylsucroses were eluted using 5 mL of 45% (v/v) acetonitrile followed by washing with 5 mL of 90% (v/v) acetonitrile to elute long chain-containing acylsucroses. Fractions were reduced to dryness under vacuum using a Speed-Vac at room temperature, and the resulting residue was dissolved in 50% (v/v) ethanol for use in assays.
The acylsugars S3:22 (5,5,12) from M82 and S3:19 (4,5,10) and G3:14 (4,5,5) from LA0716 were purified as described (Ghosh et al., 2014). NMR spectra were recorded as described (Schilmiller et al., 2015). The concentrations of purified S3:22, S3:19, and S3:14 were determined using a modified version of the method described by Ghosh and Jones (2015). Acyl chains were hydrolyzed by saponification, and the sugar backbone concentration was determined using stable isotope dilution as follows. A 10-μL aliquot of the purified acylsugar stock solution in acetonitrile was transferred to a 250-μL glass vial, and the solvent was removed under vacuum in a Speed-Vac at room temperature. The resulting residue was dissolved in 10 μL of methanol followed by the addition of 10 μL of freshly prepared 1 m sodium hydroxide. After 48 h at room temperature (25°C), 10 μL of water plus 70 μL of methanol were added, giving a final composition of 4:1 (v/v) methanol:water, and samples were further incubated at room temperature for another 24 h. Samples were diluted either 10- or 100-fold with 4:1 (v/v) methanol:water prior to adding either [13C12]Suc or [13C6]Glc internal standard at a final concentration of 0.5 μm. Saponified sugars were analyzed using a Waters Quattro Premier triple quadrupole LC-MS device interfaced to a Waters Acquity ultra-performance liquid chromatography apparatus. Chromatography was performed using a Waters BEH-amide column (2.1 × 100 mm, 1.7-μm particle size) with column oven set to 40°C. Initial conditions were 10% water + 0.1% formic acid (v/v; solvent A)/90% acetonitrile (solvent B) at a flow rate of 0.3 mL min−1 for 1 min, followed by a linear gradient to 70% A:30% B at 5 min, then to 100% A at 6 min, hold at 100% A to 8 min, return to 10% A:90% B at 9 min, and then hold at 10% A:90% B until 10 min. Compounds were ionized by electrospray ionization in negative-ion mode, and mass spectra were acquired using multiple reaction monitoring (MRM). The capillary voltage, extractor voltage, and radiofrequency lens setting were 3 kV, 5 V, and 4.3, respectively. Cone gas and desolvation gas flow rates were 50 and 600 L h−1, and the source and desolvation temperatures were 120°C and 350°C. The source cone potentials and collision energies, respectively, were as follows: for Suc, 35 and 17 V; for Glc, 16 and 10 V. The precursor and product ion masses used for the MRM transitions were 341 > 179 (Suc), 353 > 185 ([13C12]Suc), 179 > 89 (Glc), and 185 > 92 ([13C6]Glc). External standard curves were prepared for Glc and Suc ranging from 0.1 to 125 μm containing 0.5 μm 13C-labeled internal standard, and response factors were calculated based on the MRM peak areas.
ASH Protein Expression and Enzyme Assays
Recombinant protein for enzyme assays was produced in Escherichia coli as follows. Full-length ORFs for SlASH1, SpASH1, SpASH2, SlASH3, SpASH3, and Sopen04g001210 were amplified using primers (Supplemental Table S4) that added restriction enzyme sites to the ends for cloning into pET28b (EMD Millipore). The resulting constructs, which were designed to add an N-terminal 6× His tag to the protein for affinity purification, were transformed into BL21 Rosetta cells (EMD Millipore). Protein expression was induced in cultures growing in Luria-Bertani medium by adding 0.05 mm isopropyl β-d-1-thiogalactopyranoside followed by shaking at 37°C and 200 rpm overnight. Soluble protein was purified by nickel-affinity chromatography. Cells were lysed by sonication in extraction buffer (50 mm sodium phosphate, pH 6.3, 150 mm sodium chloride, and 10% [v/v] glycerol), and the cleared lysate was added to nickel resin (Qiagen) and incubated for 30 min at 4°C followed by two 15-min washes with extraction buffer + 20 mm imidazole. Protein was eluted from the resin with extraction buffer + 200 mm imidazole. Protein concentration was assayed using the bicinchoninic acid assay (Thermo Scientific).
Acylsugar hydrolase assays using S. lycopersicum M82 acylsucrose fractions were performed in a 30-μL volume by incubating 1 μg of purified protein in 50 mm ammonium acetate buffer (pH 6). Reactions were allowed to proceed for 30 min at 30°C followed by termination by the addition of 60 μL of acetonitrile:isopropanol:formic acid (1:1:0.001, v/v/v) containing 10 μm propyl-4-hydroxybenzoate internal standard and centrifugation at 13,000g for 10 min. Ten microliters of the supernatant was analyzed by LC-MS as described previously (Schilmiller et al., 2015). Assays for determining the activity of SpASH enzymes with purified S3:22 (5,5,12), S3:19 (4,5,10), and G3:14 (4,5,5) were performed by varying the amount of purified enzyme with 33 μm final concentration of acylsugar substrate in a 30-μL volume and incubating at 30°C for 5 min. Assays for determining SpASH1 kinetic constants were performed using 10 ng of purified SpASH1 enzyme with varying concentrations of acylsucrose and incubating at 25°C for 3 min. Five microliters of the reaction mixture was analyzed by LC-MS to determine velocities (in units of peak area of hydrolysis product divided by peak area of internal standard). Apparent Km and Vmax values were determined by nonlinear regression using the Michaelis-Menten kinetics model in GraphPad Prism5 (GraphPad Software).
Sequence data can be found in the GenBank database under the following accession numbers: SlASH1 (KT282356), SlASH2 (KT282357), SlASH3 (KT282358), SpASH1 (KT282359), SpASH2 (KT282360), SpASH3 (KT282361), and Sopen04g001210 (KT282362).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. SlASH transcript levels in isolated trichomes.
Supplemental Figure S2. Alignment of S. lycopersicum M82 and S. pennellii LA0716 ASH1 and ASH2 cDNA sequences.
Supplemental Figure S3. Negative-ion mode mass spectra of ASH1 and ASH2 assay products.
Supplemental Figure S4. Positive-ion mode LC-MS analysis of ASH reaction products using M82 acylsucrose substrates.
Supplemental Figure S5. Ratio of short-to-long chain-containing acylsugars in IL5-3 RNAi lines.
Supplemental Figure S6. Phylogeny of tomato CXE proteins containing Interpro domain IPR013094 with other characterized plant CXE proteins.
Supplemental Figure S7. Mass spectra of ASH3 reaction products using M82 long chain-containing acylsucroses (fraction II) as substrate.
Supplemental Figure S8. Combined SpASH1 and SpASH3 reactions with M82 fraction I substrates.
Supplemental Figure S9. SpASH2 activity with various S. habrochaites acylsucroses.
Supplemental Figure S10. Nonlinear regression analysis of SpASH1 activity to determine kinetic constants.
Supplemental Figure S11. SpASH enzyme activity with the acylglucose G3:14 (5,5,4).
Supplemental Table S1. List of genes in the IL5-3 mapping interval.
Supplemental Table S2. NMR chemical shifts for G3:14.
Supplemental Table S3. NMR chemical shifts for S3:19.
Supplemental Table S4. Primer sequences.
Supplementary Material
Acknowledgments
We thank Amanda Charbonneau for technical assistance, Gaurav Moghe for assistance with RNAseq data analysis, Dani Zamir for IL5-3 × M82 F1 seeds, other trichome project members, and the RTSF Mass Spectrometry and Metabolomics Core Facility at Michigan State University.
Glossary
- RNAi
RNA interference
- SGN
Sol Genomics Network
- ORF
open reading frame
- RNAseq
RNA sequencing
- cDNA
complementary DNA
- RT
reverse transcription
- qPCR
quantitative PCR
- LC-MS
liquid chromatography-mass spectrometry
- QTL
quantitative trait locus
- MRM
multiple reaction monitoring
Footnotes
This work was supported by the National Science Foundation (grant no. IOS–1025636 to A.D.J. and R.L.L. and Major Research Instrumentation grant no. DBI–0619489).
Articles can be viewed without a subscription.
References
- Akashi T, Aoki T, Ayabe S (2005) Molecular and biochemical characterization of 2-hydroxyisoflavanone dehydratase: involvement of carboxylesterase-like proteins in leguminous isoflavone biosynthesis. Plant Physiol 137: 882–891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alba JM, Montserrat M, Fernández-Muñoz R (2009) Resistance to the two-spotted spider mite (Tetranychus urticae) by acylsucroses of wild tomato (Solanum pimpinellifolium) trichomes studied in a recombinant inbred line population. Exp Appl Acarol 47: 35–47 [DOI] [PubMed] [Google Scholar]
- Bates PD, Ohlrogge JB, Pollard M (2007) Incorporation of newly synthesized fatty acids into cytosolic glycerolipids in pea leaves occurs via acyl editing. J Biol Chem 282: 31206–31216 [DOI] [PubMed] [Google Scholar]
- Baudouin E, Charpenteau M, Roby D, Marco Y, Ranjeva R, Ranty B (1997) Functional expression of a tobacco gene related to the serine hydrolase family: esterase activity towards short-chain dinitrophenyl acylesters. Eur J Biochem 248: 700–706 [DOI] [PubMed] [Google Scholar]
- Bombarely A, Menda N, Tecle IY, Buels RM, Strickler S, Fischer-York T, Pujar A, Leto J, Gosselin J, Mueller LA (2011) The Sol Genomics Network (solgenomics.net): growing tomatoes using Perl. Nucleic Acids Res 39: D1149–D1155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chitwood DH, Kumar R, Headland LR, Ranjan A, Covington MF, Ichihashi Y, Fulop D, Jiménez-Gómez JM, Peng J, Maloof JN, et al. (2013) A quantitative genetic basis for leaf morphology in a set of precisely defined tomato introgression lines. Plant Cell 25: 2465–2481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chortyk OT, Severson RF, Cutler HC, Sisson VA (1993) Antibiotic activities of sugar esters isolated from selected Nicotiana species. Biosci Biotechnol Biochem 57: 1355–1356 [DOI] [PubMed] [Google Scholar]
- D’Auria JC. (2006) Acyltransferases in plants: a good time to be BAHD. Curr Opin Plant Biol 9: 331–340 [DOI] [PubMed] [Google Scholar]
- Dembitsky VM. (2004) Astonishing diversity of natural surfactants. 1. Glycosides of fatty acids and alcohols. Lipids 39: 933–953 [DOI] [PubMed] [Google Scholar]
- Eamens AL, Waterhouse PM (2011). Vectors and methods for hairpin RNA and artificial microRNA-mediated gene silencing in plants. In J Birchler, ed, Plant Chromosome Engineering: Methods and Protocols. Humana Press, New York, pp 179–197 [DOI] [PubMed] [Google Scholar]
- Fan P, Miller AM, Schilmiller AL, Liu X, Ofner I, Jones AD, Zamir D, Last RL (2016) In vitro reconstruction and analysis of evolutionary variation of the tomato acylsucrose metabolic network. Proc Natl Acad Sci USA 113: E239–E248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fobes JF, Mudd JB, Marsden MP (1985) Epicuticular lipid accumulation on the leaves of Lycopersicon pennellii (Corr.) D’Arcy and Lycopersicon esculentum Mill. Plant Physiol 77: 567–570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forouhar F, Yang Y, Kumar D, Chen Y, Fridman E, Park SW, Chiang Y, Acton TB, Montelione GT, Pichersky E, et al. (2005) Structural and biochemical studies identify tobacco SABP2 as a methyl salicylate esterase and implicate it in plant innate immunity. Proc Natl Acad Sci USA 102: 1773–1778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gershater MC, Cummins I, Edwards R (2007) Role of a carboxylesterase in herbicide bioactivation in Arabidopsis thaliana. J Biol Chem 282: 21460–21466 [DOI] [PubMed] [Google Scholar]
- Gershater MC, Edwards R (2007) Regulating biological activity in plants with carboxylesterases. Plant Sci 173: 579–588 [Google Scholar]
- Ghosh B, Jones AD (2015) Dependence of negative-mode electrospray ionization response factors on mobile phase composition and molecular structure for newly-authenticated neutral acylsucrose metabolites. Analyst (Lond) 140: 6522–6531 [DOI] [PubMed] [Google Scholar]
- Ghosh B, Westbrook TC, Jones AD (2014) Comparative structural profiling of trichome specialized metabolites in tomato (Solanum lycopersicum) and S. habrochaites: acylsugar profiles revealed by UHPLC/MS and NMR. Metabolomics 10: 496–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goulet C, Mageroy MH, Lam NB, Floystad A, Tieman DM, Klee HJ (2012) Role of an esterase in flavor volatile variation within the tomato clade. Proc Natl Acad Sci USA 109: 19009–19014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill K, Rhode O (1999) Sugar-based surfactants for consumer products and technical applications. Fett Lipid 101: 25–33 [Google Scholar]
- Hotelier T, Renault L, Cousin X, Negre V, Marchot P, Chatonnet A (2004) ESTHER, the database of the α/β-hydrolase fold superfamily of proteins. Nucleic Acids Res 32: D145–D147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandzia R, Grimm R, Eckerskorn C, Lindemann P, Luckner M (1998) Purification and characterization of lanatoside 15′-O-acetylesterase from Digitalis lanata Ehrh. Planta 204: 383–389 [DOI] [PubMed] [Google Scholar]
- Kim J, Kang K, Gonzales-Vigil E, Shi F, Jones AD, Barry CS, Last RL (2012) Striking natural diversity in glandular trichome acylsugar composition is shaped by variation at the Acyltransferase2 locus in the wild tomato Solanum habrochaites. Plant Physiol 160: 1854–1870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko MK, Jeon WB, Kim KS, Lee HH, Seo HH, Kim YS, Oh BJ (2005) A Colletotrichum gloeosporioides-induced esterase gene of nonclimacteric pepper (Capsicum annuum) fruit during ripening plays a role in resistance against fungal infection. Plant Mol Biol 58: 529–541 [DOI] [PubMed] [Google Scholar]
- Kroumova AB, Wagner GJ (2003) Different elongation pathways in the biosynthesis of acyl groups of trichome exudate sugar esters from various solanaceous plants. Planta 216: 1013–1021 [DOI] [PubMed] [Google Scholar]
- Leckie BM, De Jong DM, Mutschler MA (2012) Quantitative trait loci increasing acylsugars in tomato breeding lines and their impacts on silverleaf whiteflies. Mol Breed 30: 1621–1634 [Google Scholar]
- Marshall SD, Putterill JJ, Plummer KM, Newcomb RD (2003) The carboxylesterase gene family from Arabidopsis thaliana. J Mol Evol 57: 487–500 [DOI] [PubMed] [Google Scholar]
- McCormick S. (1991) Transformation of tomato with Agrobacterium tumefaciens. In Lindsey K, ed, Plant Tissue Culture Manual. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp 1–9 [Google Scholar]
- McDowell ET, Kapteyn J, Schmidt A, Li C, Kang JH, Descour A, Shi F, Larson M, Schilmiller A, An L, et al. (2011) Comparative functional genomic analysis of Solanum glandular trichome types. Plant Physiol 155: 524–539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milo R, Last RL (2012) Achieving diversity in the face of constraints: lessons from metabolism. Science 336: 1663–1667 [DOI] [PubMed] [Google Scholar]
- Mitchell A, Chang HY, Daugherty L, Fraser M, Hunter S, Lopez R, McAnulla C, McMenamin C, Nuka G, Pesseat S, et al. (2015) The InterPro protein families database: the classification resource after 15 years. Nucleic Acids Res 43: D213–D221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mugford ST, Milkowski C (2012) Serine carboxypeptidase-like acyltransferases from plants. Methods Enzymol 516: 279–297 [DOI] [PubMed] [Google Scholar]
- Ning J, Moghe G, Leong B, Kim J, Ofner I, Wang Z, Adams C, Jones AD, Zamir D, Last RL (2015) A feedback insensitive isopropylmalate synthase affects acylsugar composition in cultivated and wild tomato. Plant Physiol 169: 1821–1835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura T, Ogita S, Kato Y (2012) A novel lactone-forming carboxylesterase: molecular identification of a tuliposide A-converting enzyme in tulip. Plant Physiol 159: 565–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osbourn A, Lanzotti V, editors (2009) Plant-Derived Natural Products: Synthesis, Function, and Application. Springer-Verlag, New York [Google Scholar]
- Ovenden SPB, Yu J, Bernays J, Wan SS, Christophidis LJ, Sberna G, Tait RM, Wildman HG, Lebeller D, Lowther J, et al. (2005) Physaloside A, an acylated sucrose ester from Physalis viscosa. J Nat Prod 68: 282–284 [DOI] [PubMed] [Google Scholar]
- Pérez-Castorena AL, Martínez M, Maldonado E (2010) Labdanes and sucrose esters from Physalis sordida. J Nat Prod 73: 1271–1276 [DOI] [PubMed] [Google Scholar]
- Pichersky E, Lewinsohn E (2011) Convergent evolution in plant specialized metabolism. Annu Rev Plant Biol 62: 549–566 [DOI] [PubMed] [Google Scholar]
- Pontier D, Godiard L, Marco Y, Roby D (1994) hsr203J, a tobacco gene whose activation is rapid, highly localized and specific for incompatible plant/pathogen interactions. Plant J 5: 507–521 [DOI] [PubMed] [Google Scholar]
- Robichaud PP, Boulay K, Munganyiki JÉ, Surette ME (2013) Fatty acid remodeling in cellular glycerophospholipids following the activation of human T cells. J Lipid Res 54: 2665–2677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruppert M, Woll J, Giritch A, Genady E, Ma X, Stöckigt J (2005) Functional expression of an ajmaline pathway-specific esterase from Rauvolfia in a novel plant-virus expression system. Planta 222: 888–898 [DOI] [PubMed] [Google Scholar]
- Schilmiller A, Shi F, Kim J, Charbonneau AL, Holmes D, Jones AD, Last RL (2010a) Mass spectrometry screening reveals widespread diversity in trichome specialized metabolites of tomato chromosomal substitution lines. Plant J 62: 391–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schilmiller AL, Charbonneau AL, Last RL (2012) Identification of a BAHD acetyltransferase that produces protective acyl sugars in tomato trichomes. Proc Natl Acad Sci USA 109: 16377–16382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schilmiller AL, Last RL, Pichersky E (2008) Harnessing plant trichome biochemistry for the production of useful compounds. Plant J 54: 702–711 [DOI] [PubMed] [Google Scholar]
- Schilmiller AL, Miner DP, Larson M, McDowell E, Gang DR, Wilkerson C, Last RL (2010b) Studies of a biochemical factory: tomato trichome deep expressed sequence tag sequencing and proteomics. Plant Physiol 153: 1212–1223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schilmiller AL, Moghe GD, Fan P, Ghosh B, Ning J, Jones AD, Last RL (2015) Functionally divergent alleles and duplicated loci encoding an acyltransferase contribute to acylsugar metabolite diversity in Solanum trichomes. Plant Cell 27: 1002–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro JA, Steffens JC, Mutschler MA (1994) Acylsugars of the wild tomato Lycopersicon pennellii in relation to geographic distribution of the species. Biochem Syst Ecol 22: 545–561 [Google Scholar]
- Simmons AT, Gurr GM (2005) Trichomes of Lycopersicon species and their hybrids: effects on pests and natural enemies. Agric For Entomol 7: 265–276 [Google Scholar]
- Souleyre EJF, Marshall SDG, Oakeshott JG, Russell RJ, Plummer KM, Newcomb RD (2011) Biochemical characterisation of MdCXE1, a carboxylesterase from apple that is expressed during fruit ripening. Phytochemistry 72: 564–571 [DOI] [PubMed] [Google Scholar]
- Stuhlfelder C, Mueller MJ, Warzecha H (2004) Cloning and expression of a tomato cDNA encoding a methyl jasmonate cleaving esterase. Eur J Biochem 271: 2976–2983 [DOI] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 30: 2725–2729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomato Genome Consortium (2012) The tomato genome sequence provides insights into fleshy fruit evolution. Nature 485: 635–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueguchi-Tanaka M, Ashikari M, Nakajima M, Itoh H, Katoh E, Kobayashi M, Chow TY, Hsing YI, Kitano H, Yamaguchi I, et al. (2005) GIBBERELLIN INSENSITIVE DWARF1 encodes a soluble receptor for gibberellin. Nature 437: 693–698 [DOI] [PubMed] [Google Scholar]
- Wang Z, Jones AD (2014) Profiling of stable isotope enrichment in specialized metabolites using liquid chromatography and multiplexed nonselective collision-induced dissociation. Anal Chem 86: 10600–10607 [DOI] [PubMed] [Google Scholar]
- Weinhold A, Baldwin IT (2011) Trichome-derived O-acyl sugars are a first meal for caterpillars that tags them for predation. Proc Natl Acad Sci USA 108: 7855–7859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winzer T, Gazda V, He Z, Kaminski F, Kern M, Larson TR, Li Y, Meade F, Teodor R, Vaistij FE, et al. (2012) A Papaver somniferum 10-gene cluster for synthesis of the anticancer alkaloid noscapine. Science 336: 1704–1708 [DOI] [PubMed] [Google Scholar]
- Yang Y, Xu R, Ma CJ, Vlot AC, Klessig DF, Pichersky E (2008) Inactive methyl indole-3-acetic acid ester can be hydrolyzed and activated by several esterases belonging to the AtMES esterase family of Arabidopsis. Plant Physiol 147: 1034–1045 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.