The ALA10 phospholipid flippase reduces phosphatidylcholine desaturation and stimulates galactolipid-tophosphatidylcholine ratio in photosynthetic tissues.
Abstract
The biogenesis of photosynthetic membranes relies on galactoglycerolipids, which are synthesized via pathways that are dispatched over several cell compartments. This membrane biogenesis requires both trafficking of lipid intermediates and a tight homeostatic regulation. In this work, we address the role of ALA10 (for aminophospholipid ATPase), a P4-type ATPase, in a process counteracting the monogalactosyldiacylglycerol (MGDG) shortage in Arabidopsis (Arabidopsis thaliana) leaves. ALA10 can interact with protein partners, ALIS1 (for ALA-interacting subunit1) or ALIS5, leading to differential endomembrane localizations of the interacting proteins, close to the plasma membrane with ALIS1 or to chloroplasts with ALIS5. ALA10 interacts also with FATTY ACID DESATURASE2 (FAD2), and modification of ALA10 expression affects phosphatidylcholine (PC) fatty acyl desaturation by disturbing the balance between FAD2 and FAD3 activities. Modulation of ALA10 expression downstream impacts the fatty acyl composition of chloroplast PC. ALA10 expression also enhances leaf growth and improves the MGDG-PC ratio, possibly through MGDG SYNTHASE1 (MGD1) activation by phosphatidic acid. The positive effect of ALA10 on leaf development is significant in conditions such as upon treatment of plants with Galvestine-1, an inhibitor of MGDG synthases, or when plants are grown at chilling temperature.
Plant cells contain diverse membranes with unique lipid compositions. Most of the membranes have a high proportion of phospholipids, with equivalent prevalence of phosphatidylcholine (PC) and phosphatidylethanolamine (PE), whereas chloroplast membranes contain only a low proportion of phospholipids. In contrast, chloroplast membranes show a specific enrichment in nonphosphorylated galactolipids, with up to 50% monogalactosyldiacylglycerol (MGDG) and 30% digalactosyldiacylglycerol in thylakoids. Interestingly, PE is strictly absent from chloroplasts. In addition, PC, representing less than 5% of total chloroplast lipids is present only in the cytosolic layer of the outer envelope membrane, where it nevertheless represents almost 60% of the lipids (Block et al., 2007; Jouhet et al., 2007).
Galactolipids result from the galactosylation of diacylglycerol (DAG) by UDP-Gal. This step of synthesis occurs in the plastid envelope. In Arabidopsis (Arabidopsis thaliana) chloroplasts, the reaction is catalyzed by MGD1, which is a peripheral protein of the chloroplast envelope inner membrane, essential for the formation of photosynthetic membranes (Boudière et al., 2014). MGD1 is activated by very low levels of phosphatidic acid (PA; Dubots et al., 2010). Its DAG substrate is either de novo synthesized in the chloroplast by the acylation of glycerol-3-phosphate or is formed by a complex conversion of PC from the endoplasmic reticulum (ER). The feeding of galactolipid synthesis by a nonchloroplast source of DAG requires a transfer of lipid between the ER and the chloroplast. The transfer mechanism is unknown, although some components have been identified. The transferred molecules rise from PC after 18:1 to 18:2 desaturation by FATTY ACID DESATURASE2 (FAD2) in the ER (Ohlrogge and Browse, 1995). It may be either PC itself or DAG that is transferred, since, when chloroplasts are incubated with PC and phospholipase C, PC is a source of galactolipid synthesis (Andersson et al., 2004). Since the chloroplast envelope proteins, TRIGALACTOSYLDIACYLGLYCEROL2 (TGD2) and TGD4, bind PA and are required for feeding galactolipid synthesis with a nonchloroplast source of DAG, it has been proposed that PA is transferred from the ER to chloroplasts by the TGD complex (Roston et al., 2011; Wang et al., 2012). The TGD complex is possibly located at previously identified ER-chloroplast membrane contact sites (Andersson et al., 2007). More recently, a report indicated that long-chain acyl-CoA synthetases located in the ER or in the chloroplast outer envelope are important for galactolipid synthesis (Jessen et al., 2015), sustaining a model where acyl-CoA and lysophosphatidylcholine (lysoPC) would traffic between membranes, as relatively water-soluble lipid intermediates, at ER-chloroplast membrane contact sites (Block and Jouhet, 2015). PC would then be restored in the chloroplast envelope from lysoPC and acyl-CoA, as supported by the presence of lysoPC:acyl-CoA acyltransferases in the chloroplast envelope (Bessoule et al., 1995).
The unilateral localization of PC in the cytosolic leaflet of the chloroplast outer envelope membrane might be related to this process. In general, asymmetric distribution of lipids between membrane leaflets has been shown to be important for a variety of cell functions, including responses to abiotic stresses (Devaux et al., 2008; Folmer et al., 2009). Molecularly, phospholipid translocators can contribute to build asymmetry in these membranes. P4-type ATPases, also called flippases, translocate phospholipids between the two leaflets of a membrane in opposition to a gradient concentration. They were first characterized in the yeast Saccharomyces cerevisiae, which holds five P4-type ATPase genes: Defective in ribosome synthesis2, Neomycin resistance1, Drs2 and neomycin family1 (DNF1), DNF2, and DNF3 (Tang et al., 1996; Pomorski et al., 2003; Natarajan et al., 2004; Chen et al., 2006). Deficiency in Drs2p, Dnf1p, or Dnf2p disturbs phospholipid asymmetry at the plasma membrane, and the Δdrs2 and Δdrs2Δdnf1Δdnf2 mutant strains present a low phospholipid translocase activity at the plasma membrane. In addition P4-type ATPases have a role in membrane trafficking (Pomorski et al., 2003; Natarajan et al., 2004; Liu et al., 2008; Paulusma and Elferink, 2010; Coleman et al., 2013; Xu et al., 2013; Lee et al., 2015).
In yeast, P4-type ATPases are present in specific areas of the endomembrane network (Pomorski et al., 2003). Drs2p is mostly present in the post-Golgi network, but the protein is also present in the plasma membrane. Dnf1p and Dnf2p are mostly present in the plasma membrane. They flip specific phospholipids: Drs2p flips more specifically phosphatidylserine (PS), and Dnf1p flips PC and PE (Zhou and Graham, 2009). It should be noticed, however, that most of the translocation assays have been done so far with NBD-modified lipids and not natural ones. The structure of P4-type ATPases is currently unknown, and the way that P4-type ATPases transfer the large polar head group of phospholipids through the hydrophobic membrane barrier is still a matter of study (Vestergaard et al., 2014). Through a combination of genetic screens and directed mutagenesis of Dnf1 and Drs2, Baldridge and Graham (2012, 2013) identified amino acid residues that are critical for the lipid head group recognition. The position of these residues with respect to the membrane allowed the authors to propose a structural model where the entry and exit gates of the phospholipid were located on either side of the membrane, delineating a groove at the protein periphery. The activity of P4-type ATPases was also shown to depend on some protein partners. Cell division cycle50 (Cdc50p), a protein interacting with the P4-type ATPase Drs2p, contributes to the lipid translocation process (Saito et al., 2004; Lenoir et al., 2009; García-Sánchez et al., 2014). Interaction of Cdc50p with a P4-type ATPase would facilitate phospholipid binding and release during the ATPase catalytic cycle even if Cdc50p does not seem absolutely required for flipping activity. Cdc50p also could partake in lipid translocation by sorting Drs2p to a specific membrane location. In Arabidopsis, this was demonstrated for several P4-type ATPases. Upon interaction with a Cdc50p homolog, several Arabidopsis P4-type ATPases initially present in the ER exit the ER and localize to another part of the endomembrane system (López-Marqués et al., 2010, 2012; Poulsen et al., 2015).
In Arabidopsis, the family of P4-type ATPases includes 12 members phylogenetically related to the yeast P4-type ATPases, which are called ALA for aminophospholipid ATPase. Five Cdc50p homolog proteins called ALIS for ALA-interacting subunit are also reported (Gomès et al., 2000). ALAs are large proteins of about 140 kD with 10 transmembrane-spanning segments that have the characteristic motifs of the P4-type ATPase family (Axelsen and Palmgren, 2001). ALA4 to ALA12 group together, whereas ALA1, ALA2, and ALA3 are more divergent. Several members of the ALA family have been studied. Reports indicate for ALA1, ALA2, and ALA3 a regulated localization in the endomembrane system, a phospholipid transport activity, and a physiological role connected with temperature stress tolerance. ALA1 has been shown to rescue the cold tolerance and the disturbed PS transport at the plasma membrane of the yeast Δdrs2 strain (Gomès et al., 2000). It is involved in cold tolerance and targeted to the plasma membrane following association in the ER with ALIS1, ALIS3, or ALIS5 (López-Marqués et al., 2012). ALA2 also can rescue the cold tolerance of the Δdrs2Δdnf1Δdnf2 yeast strain when coexpressed with ALIS1, ALIS3, or ALIS5 (López-Marqués et al., 2010). ALA2 promotes PS internalization in yeast when ALIS1, ALIS3, or ALIS5 is present, and the lipid specificity is independent of the type of ALIS partner. In the absence of an ALIS, ALA2 does not exit the ER, whereas in the presence of ALIS1, ALIS3, or ALIS5, it exits the ER and shows a prevacuolar localization. ALA3 is required for the growth of various tissues: roots, pollen tube, and trichomes (Zhang and Oppenheimer, 2009). Like ALA2, ALA3 requires ALIS1, ALIS3, or ALIS5 for functionality, but its final destination within the cell and lipid specificity appear similar regardless of the ALIS it is associated with (Poulsen et al., 2008). With ALIS1, ALIS3, or ALIS5, ALA3 sorts from the ER to the Golgi and translocates PS, PE, and PC (López-Marqués et al., 2010). ALA3 activity is connected with tolerance of the plant to abiotic stresses and is critical for growth conditions (McDowell et al., 2013). Recent reports also indicate that ALA6 and ALA7 are critical for pollen tube development and seed production under a heat/cold temperature stress and might be associated with a modification of plasma membrane lipid composition of the pollen tube (McDowell et al., 2015). On the contrary, ALA10 cannot rescue the cold tolerance of the Δdrs2Δdnf1Δdnf2 yeast strain even in coexpression with ALIS (Poulsen et al., 2015). ALA10 fused to GFP in the N terminus (Nter; GFP-ALA10) in the presence of ALIS was shown to localize in the plasma membrane of tobacco (Nicotiana tabacum) leaf epidermal cells and to translocate in order of preference PC, PE, PS, and lysoPC (Poulsen et al., 2015).
In another study, transcriptomic analyses revealed that the expression of ALA10 is modified when galactolipid synthesis is disturbed. Chemical inhibition of MGDG synthase by Galvestine-1 was shown to induce a marked increase of ALA10 mRNA expression in leaves (Botté et al., 2011). Considering that ALA10 contributes to the translocation movement of phospholipids and possibly could be involved in a cell process counteracting MGDG shortage, we investigated the role of ALA10 in galactolipid synthesis in Arabidopsis leaves.
RESULTS
Expression of ALA10 Is Modified in Response to Chemical Inhibition of MGDG Synthesis
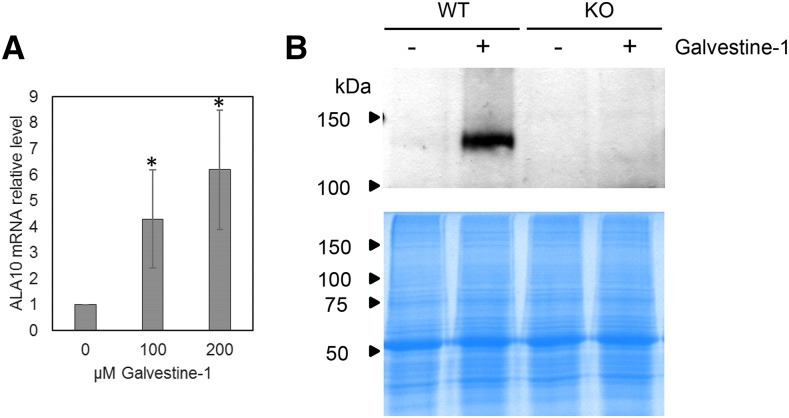
Transcriptomic analyses of Galvestine-1 treatment, used to inhibit MGDG synthases, previously indicated an increase of ALA10 expression (Botté et al., 2011). In a first step, we verified by quantitative real-time reverse transcription PCR (RT-qPCR) that the level of ALA10 mRNA was indeed increased in leaves of Columbia-0 (Col-0) Arabidopsis plants treated with 100 or 200 µm Galvestine-1 (Fig. 1A). In addition, by immunodetection with anti-ALA10 antibodies, we observed that, on a similar loading of leaf proteins, ALA10 signal, which was barely detectable without Galvestine-1, was strongly increased by the 100 µm Galvestine-1 treatment (Fig. 1B).
Figure 1.
Overexpression of ALA10 in plants treated with Galvestine-1. A, Effects of plant treatment with 100 or 200 µm Galvestine-1 on ALA10 mRNA expression in leaves of wild-type plants. The relative level of ALA10 transcripts was analyzed by RT-qPCR in four separate trials and normalized to 1 in the absence of Galvestine-1. Each result is the average ± sd of four biological samples. Asterisks indicate statistically significant differences relative to the control without Galvestine-1 (Student’s t test, P < 0.05). All plants were grown at 20°C on an identical medium containing 2% (w/v) Suc and 0.2% (w/v) Tween 20 except for the Galvestine-1 concentration (see “Materials and Methods”). B, Western-blot immunodetection with antibodies against ALA10 on leaf proteins of wild-type (WT) or knockout (KO) plants treated with 100 µm Galvestine-1. Coomassie Blue staining of samples run on SDS-PAGE is presented below the western blot for a loading control.
ALA10 Localizes to the Endomembrane System with a Pattern Depending on Its ALIS Partner
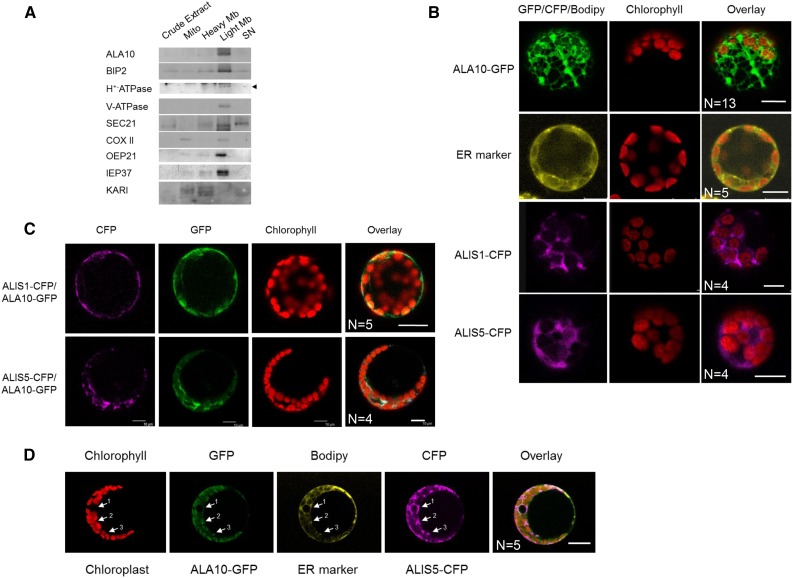
In a second step, we considered the localization of the protein. ALA10 has already been detected in a proteomics analysis of a plasma membrane subfraction (Mitra et al., 2009), but, according to the subcellular proteome database SUBA3 (Tanz et al., 2013), in silico analysis also predicts a potential cleavable N-terminal peptide and a possible sorting to mitochondria or plastid membranes. In subfractions prepared from a culture of Arabidopsis cells, we observed by immunodetection that ALA10 is found predominantly in a light membrane fraction enriched in ER (Fig. 2A). The comparative analysis of subfraction enrichment with markers of different cell membranes suggested that the protein localization was correlated with the localization of BIP2, the reticulum marker, and that ALA10 distribution did not correspond to the pattern of markers for the mitochondrial membranes, the intact plastids, or the Golgi apparatus. However, additional detection of plasma membrane and tonoplast markers in the light membrane fraction did not allow a refined localization of ALA10 in the endomembrane system. Therefore, we combined this subcellular fractionation study with fusion protein imaging. In Col-0 Arabidopsis protoplasts transiently transformed for the expression of ALA10 fused to GFP in the C terminus (Cter; ALA10-GFP), the GFP signal exhibited an ER localization pattern (Fig. 2B). Many reports indicate that P4-type ATPase localization is dependent on a Cdc50-type β-subunit called ALIS in Arabidopsis. Therefore, we analyzed possible interaction of ALA10 with ALIS proteins known to occur in Arabidopsis.
Figure 2.
Analysis of ALA10 and ALIS subcellular localization. A, Western-blot immunodetection of ALA10 in subcellular fractions of Arabidopsis cells. Cell fractionation is described in “Materials and Methods.” The purity of the different fractions was checked by using antibodies against different cell compartment markers: BIP2 (ER), H+-ATPase (plasma membrane), V-ATPase (tonoplast membrane), SEC21 (Golgi apparatus), COXII (mitochondrial inner membrane), OEP21 (chloroplast outer envelope membrane), IEP37 (chloroplast inner envelope membrane), and ketol-acid reductoisomerase (KARI; plastid stroma). Signal at the predicted size for H+-ATPase is indicated by the arrowhead. The mitochondrial inner membrane marker is mostly detected in the mitochondria (Mito) fraction, with a lighter signal in the light membrane (Light Mb) fraction. The stromal marker is mostly detected in the heavy membrane (Heavy Mb) fraction, with additional signals in the mitochondria and supernatant (SN) fractions. B to D, Confocal imaging of protoplasts from Arabidopsis leaves 16 to 28 h after transformation with pUC19:ALA10-GFP, p102:ALIS1-CFP, or p102:ALIS5-CFP (line 1, 3, or 4 respectively) or after 40 min of incubation with the ER marker BODIPY TR Glibenclamide (line 2) in B, with pUC19:ALA10-GFP, p102:ALIS1-CFP, or p102:ALIS5-CFP in C, or with pUC19:ALA10-GFP and p102:ALIS5-CFP and after 40 min of incubation with BODIPY TR Glibenclamide in D. In D, arrows indicate the colocalization of GFP and CFP with the ER marker around the nucleus (arrow 1) and close to chloroplasts (arrows 2 and 3). Green, yellow, and magenta correspond to GFP, BODIPY, and CFP observation, respectively. Red indicates observation of chlorophyll fluorescence, which allows the location of chloroplasts. N, Number of similar images obtained independently. Bars = 10 µm.
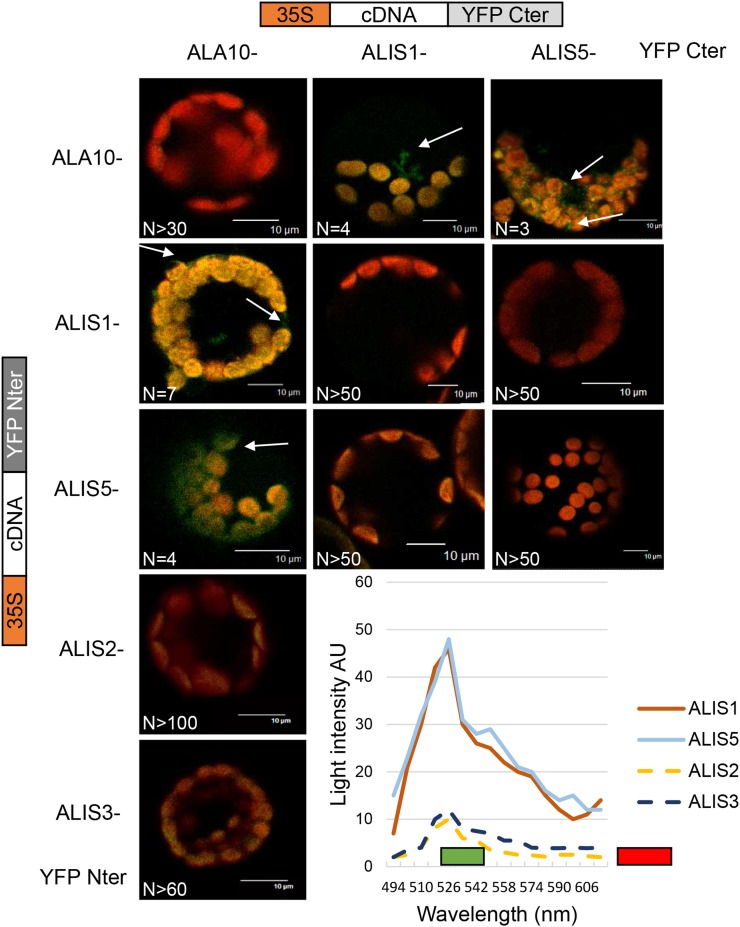
Interactions were first analyzed in vivo by a bimolecular fluorescence complementation (BiFC) assay in Col-0 Arabidopsis protoplasts. N- and C-terminal complementary nonfluorescent fragments of yellow fluorescent protein (YFP) were fused to Cter of ALA10, ALIS1, ALIS2, ALIS3, and ALIS5. The detection of YFP fluorescence reflects the in vivo interaction of the expressed fusion proteins. Cotransformation of protoplasts with a combination of N- and C-terminal YFP constructs led to the detection of YFP fluorescence in a restricted number of cases (Fig. 3). YFP signal was clearly detected when combining ALA10 and either ALIS1 or ALIS5 independently of the YFP fragment each protein was associated with, whereas no clear YFP signal could be detected with ALIS2 or ALIS3. In addition, no YFP was detected when ALA10 was combined with itself, suggesting that no dimerization of ALA10 occurred. These results indicated a preferential interaction of ALA10 with ALIS1 and ALIS5. Furthermore, the YFP interaction signal was observed with a slightly different localization relative to chloroplasts with ALIS1 or ALIS5. With ALIS5, the fluorescence was enriched around chloroplasts, whereas with ALIS1, the signal was detected at the periphery of the protoplast and away from chloroplasts, sometimes close to the plasma membrane. We then compared the localization of fusion proteins by conventional expression of ALA10-GFP, ALIS1-cyan fluorescent protein (CFP), and ALIS5-CFP in protoplasts. When ALIS1-CFP or ALIS5-CFP was expressed alone, we observed an ER pattern of the CFP signal (Fig. 2B). Localization of the CFP signal, however, was different when ALIS-CFP proteins were coexpressed with ALA10-GFP (Fig. 2C). CFP was thus detected preferentially at the periphery of the protoplast for ALIS1-CFP and in the vicinity of chloroplasts for ALIS5-CFP. In addition, GFP showed an overlay with CFP specifically at the periphery of the protoplast for ALIS1-CFP and around chloroplasts for ALIS5-CFP. Altogether, the results indicated that ALA10 is localized in the endomembrane system but with a pattern depending on the coexpressed ALIS. When coexpressed with ALIS5, a concentration around chloroplasts was observed, possibly reflecting the presence of the ER-chloroplast membrane contact sites (Fig. 2D). When coexpressed with ALIS1, ALA10 localized in the endomembrane system, close to and/or in the plasma membrane.
Figure 3.
Interaction of ALA10 with ALIS proteins analyzed by BiFC. Protoplasts of Arabidopsis leaves were transfected with a combination of ALA10, ALIS1, ALIS2, ALIS3, and ALIS5 fused either with YFP Cter or YFP Nter cloned in pBIFP1 and pBiFP4, respectively. Boxes on top or at left refer to the type of complementary DNA (cDNA) construct used to transfect protoplasts in that column or line, respectively. Confocal imaging was observed with an excitation laser at 488 nm. Overlay of YFP and chlorophyll emission is shown in green for YFP and in red for chlorophyll fluorescence. Typical YFP signal obtained with a combination of ALA10 with ALIS1 or ALIS5 is indicated by the white arrows. No YFP signal was detected for other combinations over more than 50 observations. Representative fluorescence emission spectra of each combination in arbitrary units (AU) over a 10-nm collection window are presented at bottom right: brown line, ALA10-YFP Cter/ALIS1-YFP Nter; blue line, ALA10-YFP Nter/ALIS5-YFP Cter; yellow dotted line, ALA10-YFP Nter/ALIS2-YFP Cter; and dark blue dotted line, ALA10-YFP Nter/ALIS3-YFP Cter. The green bar shows the filter window of YFP observation, and the red bar shows the filter window of chlorophyll observation. N, Number of similar images obtained independently. Bars = 10 µm.
ALA10 Expression Level Has an Impact on Growth
To evaluate the role of ALA10 at the plant level, we analyzed several Arabidopsis plant lines exhibiting different levels of ALA10 expression. To this purpose, a transfer DNA (T-DNA) insertion mutant from the SALK Institute (SALK_024877C; Alonso et al., 2003) was selected for an insertion in the middle of the ALA10 gene (Supplemental Fig. S1). After selection of a homozygous line for this insertion, three backcrosses with wild-type Col-0 were performed before the final selection of a homozygous line for phenotypic studies. RT-qPCR analysis of mRNA levels and protein analyses by immunodectection with anti-ALA10 antibodies indicated the absence of the native ALA10 (Supplemental Fig. S1, C and D). Therefore, we considered this line as a KO line for ALA10. Overexpressing lines were constructed by introduction of a sequence encoding the full-length ALA10 protein fused on its C-terminal end in frame with GFP in Col-0 (L1 and L2) or in the KO line (C1) and selected by a kanamycin resistance cassette. The fusion protein ALA10-GFP was expressed under the control of the cauliflower mosaic virus 35S promoter. Rosette from L1 and L2 lines in the Col-0 background were analyzed for the expression of ALA10. RT-qPCR analysis indicated a slight increase of ALA10 expression in L1 (4-fold) and a high increase in L2 (almost 70-fold) compared with the wild type (Supplemental Fig. S1C). The chimeric ALA10-GFP was expected at a size of 163 kD, and following SDS-PAGE and immunodetection, a signal was observed at this size with anti-GFP antibody in L1 and L2 and with an additional signal just below, possibly resulting from degradation (Supplemental Fig. S1E, arrows). In L2, the protein signal was slightly stronger compared with L1, but not as much as expected from the mRNA level difference (Supplemental Fig. S1). This discrepancy between the mRNA and protein magnitude levels, as well as the reduction of the expression of native ALA10 in the L1 line, supports a posttranslational control of ALA10 in leaves. Furthermore, by confocal imaging, we could not detect GFP fluorescence in L1, but GFP fluorescence was detected in L2 at the leaf cell periphery in epidermal and stomata cells and close to chloroplasts in stomata and mesophyll cells (Supplemental Fig. S1, F and G). In C1, the native ALA10 protein was not detected with the anti-ALA10 antibody as anticipated in a KO genetic background, but a protein was detected at the size expected for ALA10-GFP. In addition, imaging analysis of C1 showed a GFP fluorescence similar to that observed in L2 (Supplemental Fig. S1F).
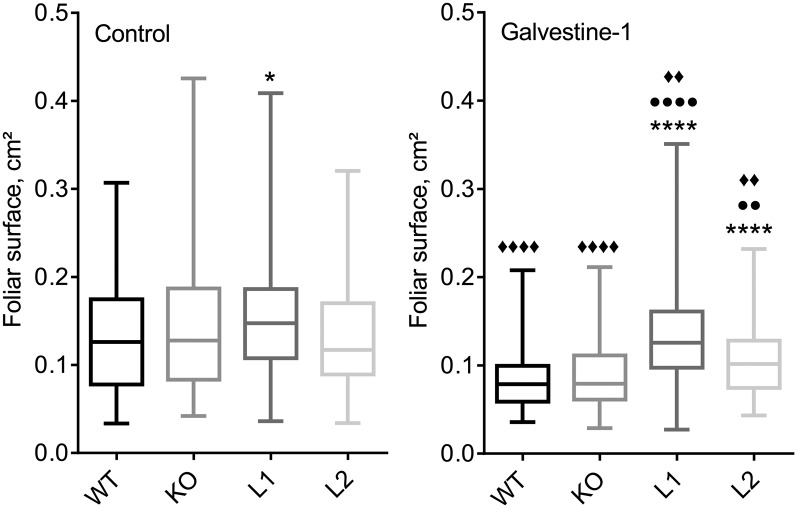
With some variations depending on the experiment, we observed a small but repeated increase of rosette size correlated with the level of ALA10 expression. This difference was significantly visible when plants were grown at 10°C (Supplemental Fig. S2). KO was smaller than the wild type, C1, L1, and L2, whereas L1 and C1 were bigger than the wild type. Considering the effect of MGD inhibition by Galvestine-1 on the expression of ALA10, we analyzed the plant size in relation with Galvestine-1 treatment (Fig. 4). In untreated control plants, the size was relatively similar between the different lines. When plants were treated with 100 µm Galvestine-1, each line showed a reduced size compared with untreated plants (Fig. 4). The size reduction was more significant for the wild type and KO than for L1 and L2. Under Galvestine-1 treatment, L1 and L2 were significantly larger than the wild type and KO. In summary, analysis of the different ALA10 lines indicated a growth phenotype depending on ALA10 expression, probably related to MGDG synthesis, since the growth phenotype was most contrasted under Galvestine-1 treatment. To further address the relation between ALA10 and galactolipid synthesis, we analyzed the lipid profile of each line at the level of leaves of young plantlets.
Figure 4.
Growth phenotypes of ALA10 lines under Galvestine-1 treatment. Plants were treated by 0 (control) or 100 µm Galvestine-1 for 15 d (3 d in the dark followed by 12 d in the light). All plants were grown in parallel at 20°C on medium containing 2% Suc and 0.2% Tween 20 and the indicated Galvestine-1 concentrations. The foliar area was measured using ImageJ software. The distribution of data obtained for each line is presented in a box plot with the median as the solid line inside the box, the first and third quartiles as the lower and upper lines of the box, and the minimum and maximum values as the bottom and top ends, respectively. The number of samples was as follows: for control, 78 wild type (WT), 86 KO, 135 L1, and 103 L2; for Galvestine-1, 121 wild type, 122 KO, 132 L1, and 76 L2. Data were compared with the wild type (asterisks) or the KO line (circles) or with the same line in the control without Galvestine-1 (diamonds). Symbols indicate statistically significant differences (Student’s t test, P < 0.01); four symbols indicate P < 0.0001, two symbols indicate P between 0.01 and 0.001, and one symbol indicates P between 0.1 and 0.01.
ALA10 Expression Enhances the MGDG-PC Ratio
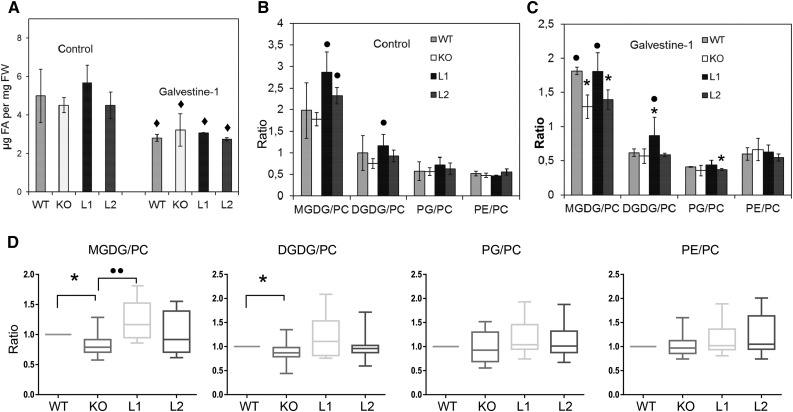
We first analyzed the lipid composition of leaves from the different mutant lines, treated or not with Galvestine-1. In each treatment condition, no significant difference in total fatty acid content per fresh weight was observed between lines (Fig. 5A), indicating that ALA10 does not influence fatty acid synthesis. The lipid profiles of the different lines were not changed severely according to ALA10 expression (Supplemental Fig. S3), but we noticed that the MGDG-PC ratio was increased in the L1 and L2 overexpression lines in the control without Galvestine-1 (Fig. 5). When plants were grown at 10°C, a lipid phenotype concerning MGDG-PC ratio was also observed, with a neat decrease of MGDG-PC ratio in the KO (Supplemental Fig. S2). We evaluated the statistical significance of this result over a larger array of data collected for the same lines compared in several different experiments (Fig. 5D). This large-scale analysis indicated that the MGDG-PC ratio was significantly dependent on ALA10 expression, whereas the proportions of other lipids relative to PC were not impacted significantly. In addition, since MGDG-PC ratio lowering upon Galvestine-1 treatment was higher in the KO line, our conclusion was that ALA10 enhanced MGDG synthesis to the detriment of PC. ALA10 would thus contribute to the protection of chloroplast membrane formation, resulting in growth maintenance.
Figure 5.
Analysis of the lipid composition of leaves in ALA10 lines. A to C, Lipid composition of plants grown without (control) or with 100 µm Galvestine-1. A, Total fatty acid (FA) content per fresh weight. Diamonds indicate statistically significant differences compared with the same line without Galvestine-1 based on Student’s t test, P < 0.05. FW, Fresh weight. B and C, Lipid-to-PC ratios in control plants (B) and in Galvestine-1-treated plants (C). Each result is the average ± sd of three biological repeats. Asterisks and circles indicate statistically significant differences compared with the wild type (WT) or the KO line (Student’s t test, P < 0.05). All plants were grown in parallel at 20°C as described in Figure 4. Calculation of lipid ratios is based on the lipid composition presented in Supplemental Figure S3. PG, Phosphatidylglycerol. D, Statistical analysis of lipid-to-PC ratios in leaves of ALA10 lines grown in different conditions. The distribution of results is presented in box plots as displayed in Figure 4. Numbers of biological samples are 13 KO, 13 wild type, eight L1, and eight L2 collected in 13 different trials. Data of KO, L1, and L2 were normalized using measures obtained for the wild type grown in the same experiment. They were then compared with the wild type (asterisks) with a statistically significant difference based on a Wilcoxon signed rank test with theoretical median = 1 and P < 0.05 or with the KO line (circles) with a statistically significant difference based on a Mann-Whitney U test with P < 0.05 and a P value between 0.01 and 0.001.
PC and MGDG are possibly linked through different mechanisms. First, PC could provide metabolic intermediates for chloroplast MGDG synthesis. This pathway drives the formation of MGDG with a specific acyl signature, containing an 18-carbon fatty acid instead of a 16-carbon fatty acid at the sn-2 glycerol position, and this ultimately leads to an enrichment of 18:3 containing MGDG relative to 16:3 containing MGDG. Second, PA resulting from PC hydrolysis could enhance MGD1 independently of the DAG feeding source, leading to unaltered MGDG fatty acid composition (Dubots et al., 2010). A third possibility is the enhancement of MGD1 expression in connection with PC metabolism. Because the expression of MGD genes was relatively stable in the different ALA10 lines, we excluded this latter hypothesis (Supplemental Fig. S4). In addition, the fatty acid composition of MGDG was relatively similar in the different lines, with no significant modification of the 18:3-to-16:3 ratio, and PA was increased significantly in the L1 and L2 lines compared with the wild type over Galvestine-1 treatment (Supplemental Fig. S3). Our conclusion, therefore, was that ALA10 might determine an enhancement of MGD1 activity by PC derivatives such as PA.
Chloroplast PC Is Dependent on ALA10
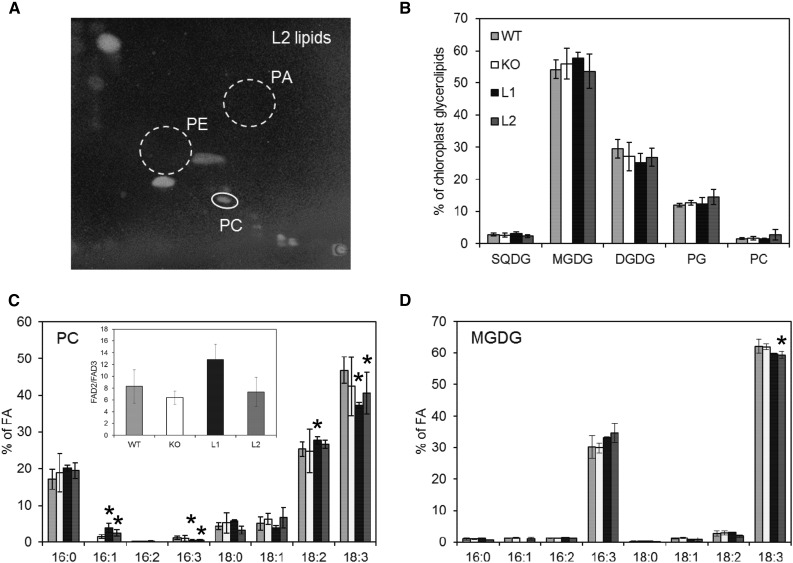
In leaf cells, MGDG synthesis occurs mainly through MGD1 activity in the chloroplast inner envelope membrane. PC is de novo synthesized in the ER and abundant in the extraplastidic membranes. The noteworthy amount of PC that is present in chloroplasts is located in the cytoplasmic leaflet of the outer envelope membrane (Block et al., 2007). To activate MGD1, we hypothesized that lipid transfer possibly operates from the PC-enriched membranes toward the chloroplast inner envelope membrane. In order to evaluate if ALA10 could modify the lipid content of chloroplasts, we extracted chloroplasts from leaves of the ALA10 lines and analyzed their lipid composition. As reported previously, we observed the absence of PE and PA in all chloroplast extracts (Fig. 6, A and B). Since PE is a major component of extraplastidic membranes, we considered that our chloroplast preparations were rather clean of extraplastidic membrane contamination. We observed very little differences in chloroplast lipid composition between lines. However, a nonsignificant increase of PC proportion was visible in L2 chloroplasts compared with other lines. Considering the fatty acid composition of lipids, we observed a change in PC profile of L1 and L2, with an increase of 18:2 and a decrease of 18:3 (Fig. 6C), whereas MGDG profile showed a decrease of 18:3 in L2 (Fig. 6D). Because PC desaturation appeared to be reduced in ALA10 overexpressors, possible hypotheses could be elaborated including an interaction of ALA10 with PC desaturation in the ER and/or a modification of PC transfer from the ER to chloroplasts after PC desaturation. Localization of ALA10 with ALIS5 in the ER close to chloroplasts, possibly at ER-chloroplast membrane contact sites, would support these hypotheses.
Figure 6.
Lipid composition of chloroplasts isolated from ALA10 lines. A, Two-dimensional thin-layer chromatography (2D-TLC) of chloroplast lipids from the L2 line. Lipids were visualized with 8-anilino-1-naphthalenesulfonic acid. Whereas PC is easily detectable, PE and PA are below the detectable level. B, Glycerolipid composition of chloroplasts. C and D, Fatty acid (FA) profiles of PC and MGDG in chloroplasts from the different mutant lines. The inset in C shows the analysis of FAD2/FAD3 desaturation efficiency as explained in Figure 7D. The different lines were grown on soil at 22°C in the same culture room for 5 weeks but over periods shifted in time in order to get in the available space enough leaf material for the batch isolation of chloroplasts. Each result is the mean ± sd of three to four biological repeats. Asterisks indicate statistically significant differences from the wild type (WT; Student’s t test, P < 0.05).
ALA10 Alters the FAD2-FAD3 Activity Ratio
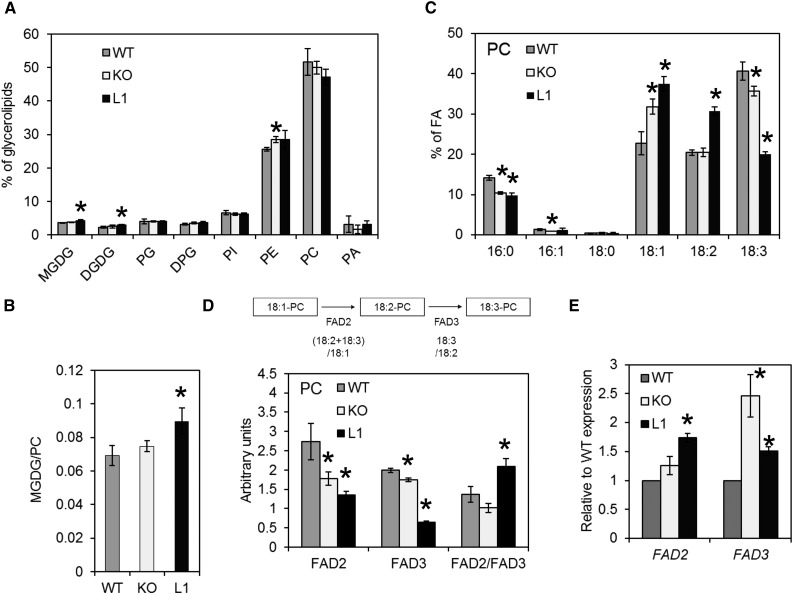
Localization of ALA10 in the endomembrane system finally suggests that the protein first acts on endomembrane lipid composition. To investigate this aspect, we chose to work on cell calli isolated from leaf patches of the wild type and ALA10 mutated lines. When compared with leaf cells, calli cells contain almost no chloroplast and only some amyloplasts, and they are highly enriched in endomembranes. They divide quickly and behave as a relatively fast membrane-synthesizing system. Lipid analysis indicated a very low proportion of galactolipids and a major content in PC and PE in these cells, as expected from their low content in thylakoid (Fig. 7A). No obvious change in glycerolipid composition was detected between ALA10 lines, although a slight but significant increase in the MGDG-PC ratio was observed in L1 (Fig. 7B). The fatty acid composition, however, was very different between lines (Fig. 7C). The desaturation profile of PC was affected, with major changes in proportions of 18:1, 18:2, and 18:3 in KO and L1 relative to the wild type (Fig. 7C), indicating a perturbation of the activity of FAD2 for desaturation of 18:1 and FAD3 for desaturation of 18:2. Both KO and L1 showed decreases in desaturation level compared with the wild type. However, they were differently affected in the FAD2 and FAD3 desaturation steps, suggesting also an alteration in the flux between FAD2 and FAD3.
Figure 7.
Lipid composition of cell cultures from ALA10 lines. A, Glycerolipid composition of wild-type (WT), KO, and L1 cell lines. DPG, Diphosphatidyldiacylglycerol; PG, phosphatidylglycerol; PI, phosphatidylinositol. B, Ratio of MGDG to PC. C, Fatty acid (FA) profile of PC. D, Analysis of the efficiency of PC fatty acid desaturation. Ratios are presented. The FAD2 and FAD3 desaturation efficiencies were evaluated by their respective product-to-substrate ratio in PC for FAD2 ([18:2+18:3]/18:1) and for FAD3 (18:3/18:2). FAD2/FAD3 is the ratio of FAD2 and FAD3 efficiencies. E, Relative expression of FAD2 and FAD3 mRNA. Each result is the average ± sd of three biological repeats cultivated in parallel. Asterisks indicate statistically significant differences compared with the wild type (Student’s t test, P < 0.05).
To analyze FAD2 and FAD3 desaturation, we calculated the product-substrate ratio for the efficiency of each reaction (Fig. 7D); that is, for FAD2, we considered the ratio of the sum of 18:2 and 18:3 to 18:1 in PC, and for FAD3, we considered the ratio 18:3 to 18:2. Both values were reduced in the KO and L1 lines, suggesting that an optimal level of ALA10 was important for both FAD2 and FAD3 to operate correctly. Eventually, the ratio of FAD2 to FAD3 reactions was reduced in KO and enhanced in L1. Since we did not observe a reduced expression but rather an enhanced expression of FAD2 and FAD3 genes in KO and L1 lines (Fig. 7E), indicating that the reduced efficiency of FAD is not due to a down-regulation of their expression, our results supported that ALA10 activity was likely a flipping of PC in the endomembrane, controlling FAD2 and FAD3 activities. In the absence of ALA10, there was a decrease of efficiency of FAD2 relative to FAD3, and when ALA10 was overexpressed, a decrease of efficiency of FAD3 relative to FAD2 was seen.
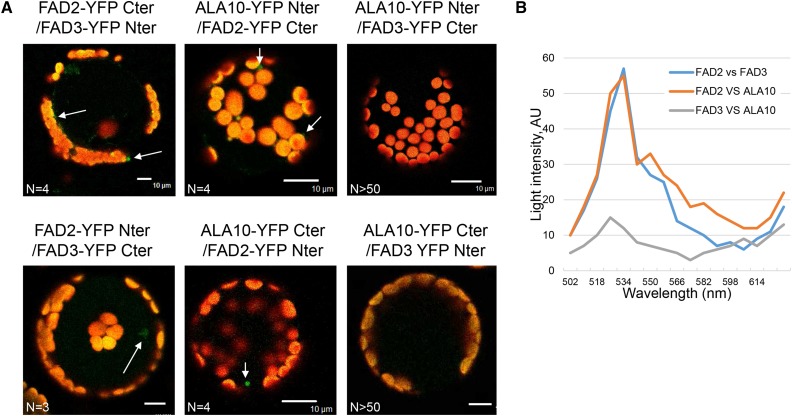
Therefore, ALA10 seemed to act downstream FAD2 but upstream of FAD3, its activity leading to a diversion of 18:2-rich PC from FAD3. Considering that it was recently shown that FAD2 and FAD3 work as homodimers and heterodimers and that their heteromeric dimerization modulates PC desaturation (Lou et al., 2014), we addressed the question of whether ALA10 might also interact with FAD2 or FAD3. We analyzed the interaction by BiFC as conducted previously for the ALA10-ALIS interaction. As shown by Lou et al. (2014), we observed an interaction of FAD2 with FAD3. In addition, we detected an interaction signal between ALA10 and FAD2 but not between ALA10 and FAD3 (Fig. 8). Our conclusion, therefore, was that ALA10 interacted specifically with FAD2 and not with FAD3. A consequence of the ALA10-FAD2 interaction could be the destabilization of the desaturation channeling process going through FAD2 and FAD3.
Figure 8.
Interaction of ALA10 with FAD2 and FAD3 proteins analyzed by BiFC. A, Protoplasts of Arabidopsis leaves were cotransfected with ALA10, FAD2, and FAD3 fused with either YFP Cter or YFP Nter cloned in pBIFP1 and pBiFP4, respectively. Overlay of the YFP signal in green and the autofluorescence of chlorophyll in red are shown. YFP signal appeared to be detectable only for FAD2 versus FAD3 and for ALA10 versus FAD2 in both combinations (white arrows). No YFP signal was found for ALA10 versus FAD3 (n > 50). Bar = 10 µm. B, Representative emission spectra of the protoplasts shown in arbitrary units (AU), here FAD2-YFP Cter/FAD3-YFP Nter, ALA10-YFP Nter/FAD2-YFP Cter, and ALA10-YFP Nter/FAD3-YFP Cter. N, Number of similar images obtained independently.
DISCUSSION
Arabidopsis ALA10 belongs to the P4-type ATPase family, which groups several phospholipid flippases. We show here that ALA10 is present in the endomembrane network, where it can interact with ALIS1 or ALIS5. Like other ALAs, ALA10 fused to GFP in Cter or in Nter is localized in the ER (Fig. 3; Poulsen et al., 2015), whereas when it is coexpressed with ALIS, its localization is altered. We show here that, in cells containing chloroplast and when ALA10 is fused to the GFP in Cter, ALA10 can sort to specific domains of the endomembrane system dependent on the ALIS it interacts with, either close to the plasma membrane with ALIS1 or close to the chloroplasts with ALIS5. These different locations are reflected in vivo in our L2 and C1 lines (Supplemental Fig. S1), where, in nongreen tissues such as the epidermis, ALA10-GFP is localized at the vicinity of the plasma membrane, as observed by Poulsen et al. (2015), whereas in green tissues such as the stomata or the mesophyll, ALA10-GFP is localized in the ER in proximity to the chloroplasts. In green tissues, its localization close to chloroplasts may be related to its role in MGDG formation. We indeed observed that ALA10 enhances the MGDG-PC ratio of plant leaves (Figs. 5 and 7B; Supplemental Fig. S2C). This effect likely belongs to a feedback mechanism related to an alteration of the level of MGDG synthesis, since, when MGDG synthesis is inhibited by Galvestine-1, ALA10 expression is strongly increased (Fig. 1) and since, under Galvestine-1 treatment, the MGDG-PC ratio is clearly deficient in the ALA10 KO line (Fig. 5C). ALA10 could enhance MGDG synthesis in critical physiological conditions such as plant culture at chilling temperature. We observed that, at 10°C, plant growth and MGDG-PC ratio are sustained by ALA10. In Arabidopsis, several ALA proteins are important for growth at cold temperature (Gomès et al., 2000; López-Marqués et al., 2012; McDowell et al., 2013, 2015). However, they were never reported to enhance galactolipid formation in any culture conditions. On the contrary, an increase of digalactosyldiacylglycerol was reported in the pollen tube of the double KO mutant ala6 ala7 (McDowell et al., 2015). Considering that the different ALAs interact with several common ALIS partners, as demonstrated here for ALA10 with ALIS1 and ALIS5 (Fig. 3), and that the subcellular localization of the ALAs is dependent of the expression of an ALIS (López-Marqués et al., 2010, 2012; Poulsen et al., 2015; this work), ALAs likely compete for ALISs. Depending on ALA and ALIS expression, this competition may ultimately affect the protein localization and activity of each flippase in a specific domain of the endomembrane system. In other words, by modulating ALA and ALIS protein amounts, an ALA can be localized in different cell membrane domains, depending on the tissue, the developmental stage, or environmental stresses. This can make more complex the analysis of protein localization and plant mutant lipid composition.
Our results indicate that the effect of ALA10 on the stimulation of MGDG synthesis goes through several different steps. First, ALA10 interacts with FAD2 in the ER. Indeed, FAD2 is located in the ER, and we observed FAD2-ALA10 in vivo interaction by BiFC (Fig. 8). Inversely, no interaction was observed between FAD3 and ALA10, although FAD3 interacted with FAD2 as reported previously (Lou et al., 2014). A possible consequence of the ALA10-FAD2 interaction could be the destabilization of the FAD2-FAD3 protein interaction. This would then lead to the release of 18:2-containing PC from the desaturation channeling process. In addition, the ALA10-FAD2 interaction suggests that both proteins handle the same phospholipid. Indeed, FAD2 desaturates 18:1 when bound on phospholipid, most likely on PC (Somerville and Browse, 1991). Moreover, Poulsen et al. (2015) have shown that ALA10 preferentially translocates PC. From the ALA10-FAD2 interaction, we may further speculate that ALA10 can translocate from one leaflet of the ER membrane to the other, either 18:1-rich PC or 18:2-rich PC. The lipid analysis of calli of the different lines suggested that ALA10 could affect the desaturation of PC at both FAD2 and FAD3 desaturation steps, with no reduced expression of FAD2 and FAD3. However, this inhibition does not correlate with ALA10 expression. On the contrary, the relative efficiency of FAD2 over FAD3, in other words the FAD2-FAD3 balance, shows more correlation with ALA10 expression (Figs. 6C and 7D). This suggests that ALA10 acts between FAD2 and FAD3 desaturation and translocates 18:2-rich PC. Remarkably, this indicates that ALA10 flips a phospholipid from the lumen side toward the cytosolic side of the ER membrane. Since either a decrease or an increase of ALA10 expression leads to a reduction of both FAD2 and FAD3 efficiency, it is possible that changes in the concentration of PC desaturation-intermediate species in the cytosolic leaflet destabilize FAD activities. Since FAD2 and FAD3 are transmembrane proteins, the elaboration of a model requires the accurate orientation of these enzymes (i.e. on which side of the ER membrane are the polar head and the acyl moieties of the phospholipid that FAD2 and FAD3 desaturate). A putative diiron center and three His boxes on the cytosolic side of the membrane suggest that some part of the electron transfer occurs on the cytosolic side (Dyer and Mullen, 2001; Schwartzbeck et al., 2001; Gagné et al., 2009); however, the lipid-binding site is not yet well defined. A mutagenesis study of FAD2-like enzymes also suggests that a portion of the third transmembrane helix positioned relatively closer to the lumen side is important for the acyl selectivity (Gagné et al., 2009). Here, we observed that ALA10 interaction with FAD2 modifies the balance between FAD2 and FAD3 efficiencies to the detriment of FAD3, but the functional mechanism remains uncertain. We propose that ALA10 prevents FAD3 desaturation either by destabilization of the FAD2-FAD3 interaction or by driving 18:2-containing PC away from FAD3. Flipping 18:2-containing PC away from the FAD3-binding site should finally lead to an enrichment of the cytosolic side of the ER in 18:2-containing PC. The resulting asymmetric distribution of PC in the ER is probably confined to specific domains, which could be related to the formation of the ER-chloroplast membrane contact sites.
Indeed, in a second step, ALA10 modifies the lipid transfer from the ER to the chloroplast. Chloroplast lipid analysis of the ALA10 lines shows an increase in 18:2 and a decrease in 18:3 in chloroplast PC when ALA10 is overexpressed (Fig. 6C). These data support an enhanced transfer of 18:2-containing PC from ER to chloroplast that likely occurs at the ER-chloroplast membrane contact sites (Block and Jouhet, 2015). Several P4-type ATPases have been shown to play a role in membrane trafficking (for review, see Coleman et al., 2013). By building an asymmetric distribution of phospholipids in a membrane, they likely favor membrane curvature and membrane fission, which can be amplified by the transfer of polyunsaturated phospholipids (Pinot et al., 2014). In a similar way, by generating an enrichment of the ER cytoplasmic leaflet in FAD2-desaturated PC, ALA10 could create favorable conditions for the transfer of 18:2-containing PC to chloroplasts. This could support the localization at the site of transfer of enzymes such as phospholipase A and long-chain acyl-CoA synthetases to channel specific pools of lysoPC and acyl-CoA between the membranes, before the reconstitution of PC by lysoPC:acyl-CoA acyltransferases in the chloroplast envelope outer membrane. Localization of the LACS and lysoPC:acyl-CoA acyltransferase enzymes in the outer envelope membrane and of PC in the outer leaflet of this membrane are in support of this hypothesis (Block and Jouhet, 2015).
Finally, PC transferred to the chloroplast envelope should lead to enhancement of the MGDG level. The more obvious way would be to increase the feeding of galactolipid synthesis by a nonchloroplast source of DAG issued from PC. However, considering 16:3 in MGDG as a signature of DAG formed in chloroplasts, we observed in the overexpressing lines no decrease in the proportion of 16:3, suggesting no increased feeding by a DAG source coming from the ER (Fig. 6D; Supplemental Fig. S3C). In addition, the expression of MGD1 was stable. Therefore, our data are more consistent with an activation of the MGD1 enzyme. By activation of the cytosolic phospholipase D, PC transferred to the chloroplast envelope may give rise to PA that would further activate MGD1 (Dubots et al., 2010, 2012; Maréchal and Bastien, 2014). In support of ALA10 driving an activation of MGD1 by PA, one of the best ALA10-coexpressed genes encodes the PLDγ1 phospholipase D (http://atted.jp/; Obayashi et al., 2014). PLDγ1 is predicted to be excluded from the ER, being rather cytosolic and possibly associated with either plasma membrane or chloroplast (SUBA3; Tanz et al., 2013).
In conclusion, our work shows that ALA10, a member of the Arabidopsis P4-type ATPase protein family, contributes to the regulation of galactolipid synthesis in leaves. Modification of ALA10 expression primarily affects PC desaturation by disturbing the balance between FAD2 and FAD3 in the ER. Then, ALA10 modifies the PC content of chloroplasts. Finally, in some conditions where leaf growth is artificially limited with Galvestine-1, an inhibitor of plant MGDG synthases, or when growth is slowed down by cold, ALA10 can boost the MGDG-PC ratio. This stimulation of MGDG synthesis likely occurs through MGD1 activation by PA. In green tissue, ALA10 can interact with ALIS1 or ALIS5 with different consequences (Figs. 2 and 3). With ALIS1, ALA10 localizes close to the plasma membrane, whereas with ALIS5, it localizes close to chloroplasts, possibly at the ER-chloroplast membrane contact sites. These alternative locations suggest different functions of ALA10 that may be complementary or concurrent. Our work suggests a function in chloroplast biogenesis associated with ALIS5. With ALIS1, we can suppose that ALA10 function may be connected with the role of the plasma membrane, as suggested by Poulsen et al. (2015). The relative availability of ALA10, ALIS1, and ALIS5 might be critical points to control these functions. Altogether, ALA10 appears as a key and multifunctional player in the complex regulation, which connects chloroplast membrane development with nonplastid ones and contributes to the maintenance of membrane lipid homeostasis in leaf cells. Characterization of ALA10 activity and its partners should provide information to help us understand the mechanisms involved.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
The Arabidopsis (Arabidopsis thaliana) insertion mutant SALK_024877 was issued from the SALK Institute collection supplied by the Nottingham Arabidopsis Stock Centre (Alonso et al., 2003). A mutant line with homozygous insertion in At3g25610 was backcrossed three times with the Arabidopsis Col-0 wild-type line before selection of the working homozygous mutant line. To identify homozygous plants, PCR analyses were carried out on genomic DNA using a primer for the T-DNA (LB, 5′-GGCAATCAGCTGTTGCCCGTCTCACTGGTG-3′) and a gene-specific primer (ALA10R1, 5′-GCTTACTTATCAGCTCCCTTAGAC-3′) for detection of an interrupted copy of the gene and two gene-specific primers (ALA10R1, 5′-GCTTACTTATCAGCTCCCTTAGAC-3′; and ALA10F1, 5′-GAACGAAGAACAGAGAGATGG-3′) for detection of a wild-type copy of the gene. The amplified bands were sequenced to confirm T-DNA location and orientation in ALA10.
In standard conditions, plants were grown on solid agar medium containing Murashige and Skoog (Caisson Labs; MSP09) growth medium supplemented with 0.5% (w/v) Suc, 0.8% (w/v) agar, and 0.5 g L−1 MES, pH 5.7. After 2 d of stratification at 4°C, plates were placed in a growth chamber for 2 weeks at 20°C with white light (100 μmol m−2 s−1) and a 16/8-h photoperiod. For Galvestine-1 treatment, plants were grown on a similar medium but with 2% (w/v) Suc and 0.2% (w/v) Tween 20 and, as specified, 0, 100, or 200 µm Galvestine-1. After stratification, they were first placed at 20°C for 3 d in the dark before 12 d of exposure to white light (100 μmol m−2 s−1) and a 16/8-h photoperiod. For cold treatment, plants were grown for 7 weeks on the standard medium containing 0.5% (w/v) Suc and no Tween 20 at 10°C with white light (80 μmol m−2 s−1) and a 16/8-h photoperiod. The determination of foliar surface was achieved using ImageJ. Basically, each image is binarized and then processed using the Analyze Particle tool, which determines the number and size of a pixel in each leaf. Then, the conversion from pixel to centimeter is achieved using the diameter of the plate as a reference.
Calli were produced by cutting fragments of leaves of 10-d-old seedlings grown in standard conditions and setting them in sterile conditions on solid agar medium containing Murashige and Skoog growth medium supplemented with 3% (w/v) Suc, 0.8% (w/v) agar, and 1.2 mg L−1 sodium 2,4-dichlorophenoxyacetate. After 1 month, calli were transferred into liquid medium and grown in liquid medium (Murashige and Skoog growth medium supplemented with 3 mm KH2PO4, 1.5% [w/v] Suc, and 1.2 mg L−1 sodium 2,4-dichlorophenoxyacetate) renewed every week. Cells were maintained as 200-mL cultures on a rotational shaker (125 rpm) with permanent light (100 μmol m−2 s−1) with a callus starting density of 2.5% (v/v).
Construction and Analysis of Transgenic Plants
To construct the vector for ALA10-GFP expression in Arabidopsis, the coding region of ALA10 was PCR amplified from an Arabidopsis cDNA library using the two flanking primers (ALA10F2, 5′-ATGGCTGGTCCAAGTCGGAGAAGAAG-3′; and ALA10R2, 5′-TTAGACACCGACAAGATCCTTATAGATCTGATCGTG-3′). The cDNA fragment was cloned in pUC18 upstream of and in frame with the GFP S65T gene under the control of the cauliflower mosaic virus 35S promoter and of the nos terminator before transfer of the whole construct into the pEL103 binary vector containing a kanamycin resistance cassette (Mittler and Lam, 1995). The plasmid used for Agrobacterium tumefaciens transformation was prepared using the QIAfilter Plasmid Midi Kit (Qiagen Laboratories). Wild-type Col-0 ecotype Arabidopsis plants were transformed by dipping the floral buds of 4-week-old plants into an A. tumefaciens (C58 strain) solution containing a surfactant (Silwet L-77) according to Clough and Bent (1998). Primary transformants were selected on Murashige and Skoog medium containing 50 µm kanamycin. Lines segregating 3:1 for the resistance to kanamycin were selected for further analysis. Primary transformants were then self-pollinated to obtain plants homozygous for the insertion.
Chloroplast Preparation and Cell Fractionation
For chloroplast preparation, plants were grown on soil for 5 weeks in a growth chamber at 22°C with white light (120 μmol m−2 s−1) and a 10/14-h photoperiod. Before leaf harvest, plants were transferred to the dark for 16 h. Chloroplasts were prepared at 4°C from about 100 g of leaves, which was ground in 300 mL of 0.4 m sorbitol, 10 mm EDTA, 10 mm NaHCO3, 0.15% (w/v) bovine serum albumin (BSA), and 20 mm Tricine-KOH, pH 8.4, according to Seigneurin-Berny et al. (2008). Intact chloroplasts were isolated on a linear density gradient preformed by centrifugation of 50% Percoll (Amersham Biosciences) in resuspension buffer (0.4 m sorbitol, 2.5 mm EDTA, 10 mm NaHCO3, 5 mm MgCl2, 0.15% [w/v] BSA, and 20 mm HEPES-KOH, pH 7.6) for 1 h at 40,000g. After 10 min of centrifugation at 10,000g, chloroplasts were collected in the low part of the Percoll gradient and washed out of Percoll twice by gentle dilution and centrifugation in resuspension buffer.
For cell fractionation, about 200 g of Arabidopsis cells (Axelos et al., 1992) was filtered and resuspended in 156 mL of ice-cold grinding buffer (0.3 m mannitol, 15 mm MOPS, 2 mm EGTA, 0.6% [w/v] polyvinylpyrrolidone 25, 0.5% BSA, 10 mm dithiothreitol, 1 mm phenylmethylsulfonyl fluoride [PMSF], 5 mm α-aminocaproic acid, and 1 mm benzamidine, pH 8). A total of 31 mL of sand was added, and cells were crushed with a mortar. Ground cells were centrifuged at 150g for 10 min, and the supernatant corresponding to the fraction called crude extract was collected. The fraction called Mito was enriched in mitochondria (Jouhet et al., 2004) by a three-step centrifugation from the crude extract: two centrifugations at 3,000g for 5 min and one centrifugation at 18,000g for 15 min. Mitochondrial enriched pellet was resuspended in washing buffer (0.3 m mannitol, 10 mm MOPS, 1 mm PMSF, 5 mm α-aminocaproic acid, and 1 mm benzamidine, pH 7.4) and loaded on top of a three-layer Percoll gradient (18% [v/v], 23% [v/v], and 40% [v/v] Percoll in 0.3 m mannitol, 10 mm MOPS, 1 mm EGTA, and 0.1% BSA, pH 7.2). Crude mitochondria were isolated from the 23%/40% interphase after centrifugation for 45 min at 70,000g before being twice washed in washing buffer for 15 min at 17,000g. After one washing, this crude mitochondrial fraction was purified on a continuous 28% Percoll gradient (0.3 m mannitol, 10 mm MOPS, 1 mm EGTA, and 0.1% BSA, pH 7.2) by centrifugation for 1 h at 40,000g, collected in the middle of the gradient, and washed out of Percoll. The heavy membrane fraction was purified by pelleting cell extract twice at 3,000g for 5 min. The heavy membrane pellet was resuspended in washing buffer and centrifuged twice at 3,000g for 5 min. The presence of intact plastids in this fraction was ascertained by codetection of protein markers for the three plastid compartments: the plastid stroma (KARI), the outer envelope, and the inner envelope. The light membrane fraction was enriched by a four-step centrifugation from crude extract: two centrifugations at 3,000g for 5 min, one centrifugation at 18,000g for 15 min, and one centrifugation at 100,000g for 45 min. The supernatant of the last centrifugation corresponds to the SN fraction. The pellet was resuspended in washing buffer and loaded onto a four-layer Suc gradient (45% [w/v], 38% [w/v], 30% [w/v], and 2% [w/v]) and centrifuged at 80,000g for 4 h. The light membrane-enriched fraction was collected from the top of the gradient. After dilution four times in washing buffer, the fraction was centrifuged to a pellet for 1 h at 175,000g.
RT-qPCR
Total RNA was extracted from shoot samples using the RNeasy Plant Mini Kit (Qiagen) according to the manufacturer’s instructions. RNA integrity was gel tested and quantified using a Nanodrop 2000 (Thermo Scientific). Total RNA was treated with DNA-free Turbo DNase (Ambion). Total RNA (1 µg) was used to synthesize cDNA with the Verso cDNA Reverse Transcriptase Kit (Thermo Scientific) and oligo(dT) primers in a total reaction volume of 20 µL. At the end of the reaction, 180 µL of water was added. RT-qPCR was performed with 5 µL of diluted cDNA mixture, in triplicate, on a Rotor-Gene 3000 instrument (Corbett Research) using SYBR Green JumpStart Taq ReadyMix (Sigma-Aldrich). Primers were designed using Primer BLAST (http://www.ncbi.nlm.nih.gov; Ye et al., 2012). Primers for each gene are listed in Supplemental Table S1. A series of five dilutions of a mixture of the different cDNA samples was used to establish the standard curve of each measurement. All primer pairs gave close to 1-efficiency. Negative controls without cDNA or without enzyme were performed in each case. The relative level of transcripts was calculated with the 2−ΔΔCt method using the mean value of technical triplicates and, as a reference, the cycle threshold value of ACT8 (At1g49240) for measurements on plants and the mean cycle threshold value of ACT8 (At1g49240), UBQ10 (At4g05320), and TIP4 (At2g25810) for measurements on cell cultures.
Protein Immunodetection
Leaf proteins were phenol extracted from approximately 50 mg of tissues according to Hurkman and Tanaka (1986) with the addition of 50 mm EDTA, 2% β-mercaptoethanol, 2 mm PMSF, and complete protease inhibitor cocktail as specified by the supplier (Roche). A total of 25 µg of protein was then resolved by SDS-PAGE on a 10% acrylamide gel before transfer to a nitrocellulose membrane. Immunodetection was done by peroxidase-coupled detection. Antibodies against ALA10 were obtained by rabbit immunization with the ALA10 C-terminal peptide RSARFHDQIYKDLVGV fused to the Nter of ovalbumin. ALA10 antibodies were affinity purified on the peptide. Antibodies against ALA10 or against GFP (Santa Cruz) were used at a dilution of 1:100. In cases where ALA10 and ALA10-GFP proteins were present only in low amounts on the blot, in order to improve signal over background, anti-ALA10 and anti-GFP antibody solutions were preincubated before final use with a blot of proteins extracted from the ala10 KO mutant and the wild type, respectively. The purity of cell fractions was checked using different Agrisera antibodies against membrane markers: H+-ATPase (plasma membrane) at 1:1,000, V-ATPase (tonoplast membrane) 1:2,000, BIP2 (ER) at 1:5,000, COXII (mitochondrial inner membrane) at 1:2,000, and SEC21 (Golgi) at 1:1,000. Serum of anti-KARI was used at 1:10,000 as described (Yamaryo et al., 2008).
Glycerolipid Analysis
Glycerolipids were extracted from approximately 60 mg fresh weight of material according to Folch et al. (1957). Tissue or cells were frozen in liquid nitrogen immediately after harvest, freeze dried, and ground in 4 mL of boiling ethanol for 5 min followed by the addition of 2 mL of methanol and 8 mL of chloroform. After saturation with argon and filtration through glass wool, the remains were rinsed with 3 mL of chloroform:methanol (2:1, v/v) and lipids were further extracted by the addition of 5 mL of 1% NaCl. For chloroplast analyses, lipids were recovered by the method of Bligh and Dyer (1959). The chloroform phase was dried under argon before resolubilization of the lipid extract in pure chloroform. Lipids were analyzed on silica gel plates (Merck) by 2D-TLC. The first solvent was chloroform:methanol:water (65:25:4, v/v), while the second one, after a 90° rotation, was chloroform:acetone:methanol:acetic acid:water (50:20:10:10:5, v/v). Lipids were then visualized under UV light after pulverization of 8-anilino-1-naphthalenesulfonic acid at 0.2% in methanol. They were then scraped off from the 2D-TLC plates for by gas chromatography analyses. For lipid quantification, fatty acids were methylated using 3 mL of 2.5% H2SO4 in methanol during 1 h at 100°C (including standard amounts of 21:0). The reaction was stopped by the addition of 3 mL of water and 3 mL of hexane. The hexane phase was analyzed by gas liquid chromatography (Perkin-Elmer) on a BPX70 (SGE) column. Methylated fatty acids were identified by comparison of their retention times with those of standards and quantified by the surface peak method using 21:0 for calibration. Extraction and quantification were done on three biological replicates or as specified. Statistical relevance was based on Student’s t test or as indicated in the figure legends.
Confocal Microscopy
Arabidopsis protoplasts were prepared according to Yoo et al. (2007). Briefly, protoplasts were isolated from leaves of wild-type plants growing on soil for 4 weeks. Cell walls were removed using 1.5% (w/v) Cellulase R10 and 0.4% (w/v) Macerozyme (Yakult Pharmaceutical) in 0.4 m mannitol and 20 mm KCl for 3 h. Polyethylene glycol-calcium-mediated transfection of 100 µL of protoplast suspension was done with 10 µg of DNA followed by 16 to 24 h of incubation before confocal imaging.
Imaging slides were observed with an immersion 40× objective at room temperature by confocal laser scanning microscopy using a TCS-SP2 operating system (Leica). CFP, GFP, BODIPY TR Glibenclamide, MitoTracker, and chlorophyll were excited and signal was collected sequentially (400 Hz, line by line, two to eight lines average) using the 458- and 488-nm line of an argon laser for CFP and GFP, respectively, the 543-nm line of a helium-neon laser for BODIPY and MitoTracker, and the 633-nm line of a helium-neon laser for chlorophyll. Fluorescence was collected between 460 and 480 nm, 505 and 514 nm, 580 and 615 nm, 553 and 607 nm, and 658 and 700 nm for CFP, GFP, BODIPY, MitoTracker, and chlorophyll, respectively.
For localization of ALIS-CFP proteins, sequence encoding the ALIS protein was cloned into p102 vector (Earley et al., 2006) in front of and in frame with the CFP-HA tag using the gateway technic.
BiFC Analysis
Sequences encoding ALA10, ALIS1, ALIS2, ALIS3, and ALIS5 were amplified and cloned in pBiFP1 and pBiFP4 for fusion with YFP Nter and YFP Cter, respectively. Primers for various constructs are listed in Supplemental Table S1. Different constructs were coexpressed in Arabidopsis protoplasts, and fluorescence signals were checked by confocal microscopy with an excitation line at 488 nm and a 10-nm collection window (average line by line of two) for XY λ scan or a 522- to 532-nm collection window (average line by line of eight) for YFP and a 658- to 700-nm collection window for chlorophyll.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers ALA10 : NM_113459.1; ALIS1 : NM_112110.3; ALIS2 : NM_123984.3; ALIS3 : NM_104310.3; ALIS5 : NM_106593.3.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Description of the ALA10 lines analyzed in this study.
Supplemental Figure S2. Growth phenotype and lipid composition of ALA10 plant lines cultured at 10°C.
Supplemental Figure S3. Lipid composition of leaves in ALA10 lines under Galvestine-1 treatment.
Supplemental Figure S4. Comparison of MGD expression in the ALA10 lines.
Supplemental Table S1. List of primers used for BiFC constructs and RT-qPCR analysis.
Supplementary Material
Acknowledgments
We thank Drs. Fabrice Rébeillé and Denis Falconet for supportive discussion of the work and in-depth reading of the article and Melissa Conte for English reading.
Glossary
- PC
phosphatidylcholine
- PE
phosphatidylethanolamine
- MGDG
monogalactosyldiacylglycerol
- DAG
diacylglycerol
- PA
phosphatidic acid
- ER
endoplasmic reticulum
- lysoPC
lysophosphatidylcholine
- PS
phosphatidylserine
- Nter
N terminus
- RT-qPCR
quantitative real-time reverse transcription PCR
- Cter
C terminus
- Col-0
Columbia-0
- BiFC
bimolecular fluorescence complementation
- T-DNA
transfer DNA
- KO
knockout
- cDNA
complementary DNA
- BSA
bovine serum albumin
- PMSF
phenylmethylsulfonyl fluoride
- 2D-TLC
two-dimensional thin-layer chromatography
Footnotes
This work was supported by the Agence Nationale de la Recherche (Young Scientist grant no. ANR–12–JSV2–0001) and the Labex Grenoble Alliance for Integrated Structural Cell Biology.
References
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al. (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Andersson MX, Goksör M, Sandelius AS (2007) Optical manipulation reveals strong attracting forces at membrane contact sites between endoplasmic reticulum and chloroplasts. J Biol Chem 282: 1170–1174 [DOI] [PubMed] [Google Scholar]
- Andersson MX, Kjellberg JM, Sandelius AS (2004) The involvement of cytosolic lipases in converting phosphatidyl choline to substrate for galactolipid synthesis in the chloroplast envelope. Biochim Biophys Acta 1684: 46–53 [DOI] [PubMed] [Google Scholar]
- Axelos M, Curie C, Mazzolini L, Bardet C, Lescure B (1992) A protocol for transient gene expression in Arabidopsis thaliana protoplasts isolated from cell suspension cultures. Plant Physiol Biochem 30: 123–128 [Google Scholar]
- Axelsen KB, Palmgren MG (2001) Inventory of the superfamily of P-type ion pumps in Arabidopsis. Plant Physiol 126: 696–706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldridge RD, Graham TR (2012) Identification of residues defining phospholipid flippase substrate specificity of type IV P-type ATPases. Proc Natl Acad Sci USA 109: E290–E298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldridge RD, Graham TR (2013) Two-gate mechanism for phospholipid selection and transport by type IV P-type ATPases. Proc Natl Acad Sci USA 110: E358–E367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bessoule JJ, Testet E, Cassagne C (1995) Synthesis of phosphatidylcholine in the chloroplast envelope after import of lysophosphatidylcholine from endoplasmic reticulum membranes. Eur J Biochem 228: 490–497 [PubMed] [Google Scholar]
- Bligh EG, Dyer WJ (1959) A rapid method of total lipid extraction and purification. Can J Biochem Physiol 37: 911–917 [DOI] [PubMed] [Google Scholar]
- Block MA, Douce R, Joyard J, Rolland N (2007) Chloroplast envelope membranes: a dynamic interface between plastids and the cytosol. Photosynth Res 92: 225–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Block MA, Jouhet J (2015) Lipid trafficking at endoplasmic reticulum-chloroplast membrane contact sites. Curr Opin Cell Biol 35: 21–29 [DOI] [PubMed] [Google Scholar]
- Botté CY, Deligny M, Roccia A, Bonneau AL, Saïdani N, Hardré H, Aci S, Yamaryo-Botté Y, Jouhet J, Dubots E, et al. (2011) Chemical inhibitors of monogalactosyldiacylglycerol synthases in Arabidopsis thaliana. Nat Chem Biol 7: 834–842 [DOI] [PubMed] [Google Scholar]
- Boudière L, Michaud M, Petroutsos D, Rébeillé F, Falconet D, Bastien O, Roy S, Finazzi G, Rolland N, Jouhet J, et al. (2014) Glycerolipids in photosynthesis: composition, synthesis and trafficking. Biochim Biophys Acta 1837: 470–480 [DOI] [PubMed] [Google Scholar]
- Chen S, Wang J, Muthusamy BP, Liu K, Zare S, Andersen RJ, Graham TR (2006) Roles for the Drs2p-Cdc50p complex in protein transport and phosphatidylserine asymmetry of the yeast plasma membrane. Traffic 7: 1503–1517 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Coleman JA, Quazi F, Molday RS (2013) Mammalian P4-ATPases and ABC transporters and their role in phospholipid transport. Biochim Biophys Acta 1831: 555–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devaux PF, Herrmann A, Ohlwein N, Kozlov MM (2008) How lipid flippases can modulate membrane structure. Biochim Biophys Acta 1778: 1591–1600 [DOI] [PubMed] [Google Scholar]
- Dubots E, Audry M, Yamaryo Y, Bastien O, Ohta H, Breton C, Maréchal E, Block MA (2010) Activation of the chloroplast monogalactosyldiacylglycerol synthase MGD1 by phosphatidic acid and phosphatidylglycerol. J Biol Chem 285: 6003–6011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubots E, Botté C, Boudière L, Yamaryo-Botté Y, Jouhet J, Maréchal E, Block MA (2012) Role of phosphatidic acid in plant galactolipid synthesis. Biochimie 94: 86–93 [DOI] [PubMed] [Google Scholar]
- Dyer JM, Mullen RT (2001) Immunocytological localization of two plant fatty acid desaturases in the endoplasmic reticulum. FEBS Lett 494: 44–47 [DOI] [PubMed] [Google Scholar]
- Earley K, Haag JR, Pontes O, Opper K, Juehne T, Song K, Pikaard CS (2006) Gateway-compatible vectors for plant functional genomics and proteomics. Plant J 45: 616–629 [DOI] [PubMed] [Google Scholar]
- Folch J, Lees M, Sloane Stanley GH (1957) A simple method for the isolation and purification of total lipides from animal tissues. J Biol Chem 226: 497–509 [PubMed] [Google Scholar]
- Folmer DE, Elferink RP, Paulusma CC (2009) P4 ATPases: lipid flippases and their role in disease. Biochim Biophys Acta 1791: 628–635 [DOI] [PubMed] [Google Scholar]
- Gagné SJ, Reed DW, Gray GR, Covello PS (2009) Structural control of chemoselectivity, stereoselectivity, and substrate specificity in membrane-bound fatty acid acetylenases and desaturases. Biochemistry 48: 12298–12304 [DOI] [PubMed] [Google Scholar]
- García-Sánchez S, Sánchez-Cañete MP, Gamarro F, Castanys S (2014) Functional role of evolutionarily highly conserved residues, N-glycosylation level and domains of the Leishmania miltefosine transporter-Cdc50 subunit. Biochem J 459: 83–94 [DOI] [PubMed] [Google Scholar]
- Gomès E, Jakobsen MK, Axelsen KB, Geisler M, Palmgren MG (2000) Chilling tolerance in Arabidopsis involves ALA1, a member of a new family of putative aminophospholipid translocases. Plant Cell 12: 2441–2454 [PMC free article] [PubMed] [Google Scholar]
- Hurkman WJ, Tanaka CK (1986) Solubilization of plant membrane proteins for analysis by two-dimensional gel electrophoresis. Plant Physiol 81: 802–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jessen D, Roth C, Wiermer M, Fulda M (2015) Two activities of long-chain acyl-coenzyme A synthetase are involved in lipid trafficking between the endoplasmic reticulum and the plastid in Arabidopsis. Plant Physiol 167: 351–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jouhet J, Maréchal E, Baldan B, Bligny R, Joyard J, Block MA (2004) Phosphate deprivation induces transfer of DGDG galactolipid from chloroplast to mitochondria. J Cell Biol 167: 863–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jouhet J, Maréchal E, Block MA (2007) Glycerolipid transfer for the building of membranes in plant cells. Prog Lipid Res 46: 37–55 [DOI] [PubMed] [Google Scholar]
- Lee S, Uchida Y, Wang J, Matsudaira T, Nakagawa T, Kishimoto T, Mukai K, Inaba T, Kobayashi T, Molday RS, et al. (2015) Transport through recycling endosomes requires EHD1 recruitment by a phosphatidylserine translocase. EMBO J 34: 669–688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenoir G, Williamson P, Puts CF, Holthuis JC (2009) Cdc50p plays a vital role in the ATPase reaction cycle of the putative aminophospholipid transporter Drs2p. J Biol Chem 284: 17956–17967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu K, Surendhran K, Nothwehr SF, Graham TR (2008) P4-ATPase requirement for AP-1/clathrin function in protein transport from the trans-Golgi network and early endosomes. Mol Biol Cell 19: 3526–3535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Marqués RL, Poulsen LR, Hanisch S, Meffert K, Buch-Pedersen MJ, Jakobsen MK, Pomorski TG, Palmgren MG (2010) Intracellular targeting signals and lipid specificity determinants of the ALA/ALIS P4-ATPase complex reside in the catalytic ALA alpha-subunit. Mol Biol Cell 21: 791–801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Marqués RL, Poulsen LR, Palmgren MG (2012) A putative plant aminophospholipid flippase, the Arabidopsis P4 ATPase ALA1, localizes to the plasma membrane following association with a β-subunit. PLoS ONE 7: e33042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lou Y, Schwender J, Shanklin J (2014) FAD2 and FAD3 desaturases form heterodimers that facilitate metabolic channeling in vivo. J Biol Chem 289: 17996–18007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maréchal E, Bastien O (2014) Modeling of regulatory loops controlling galactolipid biosynthesis in the inner envelope membrane of chloroplasts. J Theor Biol 361: 1–13 [DOI] [PubMed] [Google Scholar]
- McDowell SC, López-Marqués RL, Cohen T, Brown E, Rosenberg A, Palmgren MG, Harper JF (2015) Loss of the Arabidopsis thaliana P4-ATPases ALA6 and ALA7 impairs pollen fitness and alters the pollen tube plasma membrane. Front Plant Sci 6: 197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDowell SC, López-Marqués RL, Poulsen LR, Palmgren MG, Harper JF (2013) Loss of the Arabidopsis thaliana P₄-ATPase ALA3 reduces adaptability to temperature stresses and impairs vegetative, pollen, and ovule development. PLoS ONE 8: e62577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra SK, Clouse SD, Goshe MB (2009) Enrichment and preparation of plasma membrane proteins from Arabidopsis thaliana for global proteomic analysis using liquid chromatography-tandem mass spectrometry. Methods Mol Biol 564: 341–355 [DOI] [PubMed] [Google Scholar]
- Mittler R, Lam E (1995) Identification, characterization, and purification of a tobacco endonuclease activity induced upon hypersensitive response cell death. Plant Cell 7: 1951–1962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natarajan P, Wang J, Hua Z, Graham TR (2004) Drs2p-coupled aminophospholipid translocase activity in yeast Golgi membranes and relationship to in vivo function. Proc Natl Acad Sci USA 101: 10614–10619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obayashi T, Okamura Y, Ito S, Tadaka S, Aoki Y, Shirota M, Kinoshita K (2014) ATTED-II in 2014: evaluation of gene coexpression in agriculturally important plants. Plant Cell Physiol 55: e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlrogge J, Browse J (1995) Lipid biosynthesis. Plant Cell 7: 957–970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulusma CC, Elferink RP (2010) P4 ATPases: the physiological relevance of lipid flipping transporters. FEBS Lett 584: 2708–2716 [DOI] [PubMed] [Google Scholar]
- Pinot M, Vanni S, Pagnotta S, Lacas-Gervais S, Payet LA, Ferreira T, Gautier R, Goud B, Antonny B, Barelli H (2014) Lipid cell biology: polyunsaturated phospholipids facilitate membrane deformation and fission by endocytic proteins. Science 345: 693–697 [DOI] [PubMed] [Google Scholar]
- Pomorski T, Lombardi R, Riezman H, Devaux PF, van Meer G, Holthuis JC (2003) Drs2p-related P-type ATPases Dnf1p and Dnf2p are required for phospholipid translocation across the yeast plasma membrane and serve a role in endocytosis. Mol Biol Cell 14: 1240–1254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulsen LR, López-Marqués RL, McDowell SC, Okkeri J, Licht D, Schulz A, Pomorski T, Harper JF, Palmgren MG (2008) The Arabidopsis P4-ATPase ALA3 localizes to the Golgi and requires a β-subunit to function in lipid translocation and secretory vesicle formation. Plant Cell 20: 658–676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulsen LR, López-Marqués RL, Pedas PR, McDowell SC, Brown E, Kunze R, Harper JF, Pomorski TG, Palmgren M (2015) A phospholipid uptake system in the model plant Arabidopsis thaliana. Nat Commun 6: 7649. [DOI] [PubMed] [Google Scholar]
- Roston R, Gao J, Xu C, Benning C (2011) Arabidopsis chloroplast lipid transport protein TGD2 disrupts membranes and is part of a large complex. Plant J 66: 759–769 [DOI] [PubMed] [Google Scholar]
- Saito K, Fujimura-Kamada K, Furuta N, Kato U, Umeda M, Tanaka K (2004) Cdc50p, a protein required for polarized growth, associates with the Drs2p P-type ATPase implicated in phospholipid translocation in Saccharomyces cerevisiae. Mol Biol Cell 15: 3418–3432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartzbeck JL, Jung S, Abbott AG, Mosley E, Lewis S, Pries GL, Powell GL (2001) Endoplasmic oleoyl-PC desaturase references the second double bond. Phytochemistry 57: 643–652 [DOI] [PubMed] [Google Scholar]
- Seigneurin-Berny D, Salvi D, Dorne AJ, Joyard J, Rolland N (2008) Percoll-purified and photosynthetically active chloroplasts from Arabidopsis thaliana leaves. Plant Physiol Biochem 46: 951–955 [DOI] [PubMed] [Google Scholar]
- Somerville C, Browse J (1991) Plant lipids: metabolism, mutants, and membranes. Science 252: 80–87 [DOI] [PubMed] [Google Scholar]
- Tang X, Halleck MS, Schlegel RA, Williamson P (1996) A subfamily of P-type ATPases with aminophospholipid transporting activity. Science 272: 1495–1497 [DOI] [PubMed] [Google Scholar]
- Tanz SK, Castleden I, Hooper CM, Vacher M, Small I, Millar HA (2013) SUBA3: a database for integrating experimentation and prediction to define the subcellular location of proteins in Arabidopsis. Nucleic Acids Res 41: D1185–D1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vestergaard AL, Coleman JA, Lemmin T, Mikkelsen SA, Molday LL, Vilsen B, Molday RS, Dal Peraro M, Andersen JP (2014) Critical roles of isoleucine-364 and adjacent residues in a hydrophobic gate control of phospholipid transport by the mammalian P4-ATPase ATP8A2. Proc Natl Acad Sci USA 111: E1334–E1343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Xu C, Benning C (2012) TGD4 involved in endoplasmic reticulum-to-chloroplast lipid trafficking is a phosphatidic acid binding protein. Plant J 70: 614–623 [DOI] [PubMed] [Google Scholar]
- Xu P, Baldridge RD, Chi RJ, Burd CG, Graham TR (2013) Phosphatidylserine flipping enhances membrane curvature and negative charge required for vesicular transport. J Cell Biol 202: 875–886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaryo Y, Dubots E, Albrieux C, Baldan B, Block MA (2008) Phosphate availability affects the tonoplast localization of PLDzeta2, an Arabidopsis thaliana phospholipase D. FEBS Lett 582: 685–690 [DOI] [PubMed] [Google Scholar]
- Ye J, Coulouris G, Zaretskaya I, Cutcutache I, Rozen S, Madden TL (2012) Primer-BLAST: a tool to design target-specific primers for polymerase chain reaction. BMC Bioinformatics 13: 134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo SD, Cho YH, Sheen J (2007) Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat Protoc 2: 1565–1572 [DOI] [PubMed] [Google Scholar]
- Zhang X, Oppenheimer DG (2009) IRREGULAR TRICHOME BRANCH 2 (ITB2) encodes a putative aminophospholipid translocase that regulates trichome branch elongation in Arabidopsis. Plant J 60: 195–206 [DOI] [PubMed] [Google Scholar]
- Zhou X, Graham TR (2009) Reconstitution of phospholipid translocase activity with purified Drs2p, a type-IV P-type ATPase from budding yeast. Proc Natl Acad Sci USA 106: 16586–16591 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.