Aquaporins located in the vacuolar membrane contribute to the proper development of lateral roots.
Abstract
Aquaporins (AQPs) are water channels allowing fast and passive diffusion of water across cell membranes. It was hypothesized that AQPs contribute to cell elongation processes by allowing water influx across the plasma membrane and the tonoplast to maintain adequate turgor pressure. Here, we report that, in Arabidopsis (Arabidopsis thaliana), the highly abundant tonoplast AQP isoforms AtTIP1;1, AtTIP1;2, and AtTIP2;1 facilitate the emergence of new lateral root primordia (LRPs). The number of lateral roots was strongly reduced in the triple tip mutant, whereas the single, double, and triple tip mutants showed no or minor reduction in growth of the main root. This phenotype was due to the retardation of LRP emergence. Live cell imaging revealed that tight spatiotemporal control of TIP abundance in the tonoplast of the different LRP cells is pivotal to mediating this developmental process. While lateral root emergence is correlated to a reduction of AtTIP1;1 and AtTIP1;2 protein levels in LRPs, expression of AtTIP2;1 is specifically needed in a restricted cell population at the base, then later at the flanks, of developing LRPs. Interestingly, the LRP emergence phenotype of the triple tip mutants could be fully rescued by expressing AtTIP2;1 under its native promoter. We conclude that TIP isoforms allow the spatial and temporal fine-tuning of cellular water transport, which is critically required during the highly regulated process of LRP morphogenesis and emergence.
Plant cell elongation is a developmental process during which cells increase 10- to 100-fold in volume before reaching their final size. It requires a fast and continuous inflow of water, which ends up mainly in the vacuole, in a process that is controlled by gradients in water potential, which itself is generated by the accumulation of solutes (for review, see Fricke and Chaumont, 2007).
The fast and passive movement of water across cell membranes is facilitated by the presence of water channels, called aquaporins (AQPs), which belong to the superfamily of the major intrinsic proteins (Agre et al., 1993; Maurel, 1997; Gomes et al., 2009). AQPs are involved, on the one hand, in the long-distance movement of water during evapo-transpiration and, on the other hand, in the short-distance transport and osmotic adjustment within cells (expansion, opening of stomata, water homeostasis; Maurel et al., 2008; Gomes et al., 2009; Heinen et al., 2009; Chaumont and Tyerman, 2014). They also contribute to the maintenance of a favorable water balance within plants under changing and sometimes hostile environmental conditions. Several AQPs are also important for the facilitated membrane diffusion of other uncharged solutes, such as urea, CO2, NH3, H2O2, and metalloids (Liu et al., 2003; Uehlein et al., 2003; Jahn et al., 2004; Bienert et al., 2007; Bienert et al., 2008; Bienert et al., 2011; Bienert et al., 2014).
The genome of Arabidopsis (Arabidopsis thaliana) encodes 35 major intrinsic proteins that phylogenetically cluster in four subfamilies: plasma membrane intrinsic proteins (PIPs), tonoplast intrinsic proteins (TIPs), small basic intrinsic proteins, and nodulin 26-like intrinsic proteins (Johanson et al., 2001). Arabidopsis AtTIPs (10 isoforms) and AtPIPs (13 isoforms) are mostly found in the vacuolar and plasma membranes, respectively, and act as water channels when heterologously expressed in Xenopus oocytes (Maurel et al., 1993; Daniels et al., 1994; Kammerloher et al., 1994; Maurel et al., 1995; Daniels et al., 1996; Weig et al., 1997; Johanson et al., 2001). The TIP protein sequences are more divergent compared with PIPs and phylogenetically divided into five groups (TIP1–TIP5). TIP1 and TIP2 isoforms are largely expressed in vegetative tissues and are thought to be preferentially associated with the large lytic vacuoles and vacuoles accumulating vegetative storage proteins, respectively (Jauh et al., 1998). TIP3s are expressed in seeds (Vander Willigen et al., 2006; Hunter et al., 2007; Frigerio et al., 2008), and TIP5;1 is exclusively expressed in dry seeds and pollen grains (Vander Willigen et al., 2006; Soto et al., 2008; Wudick et al., 2014).
The importance of TIP AQPs in tissue elongation has been mainly suggested by transcriptional analysis. Arabidopsis plants expressing the GUS marker protein under the transcriptional control of the AtTIP1;1 promoter show high GUS activity in root and stem elongating tissues, but no expression is detected in meristems or older parts of plants (Ludevid et al., 1992). The maize (Zea mays) ZmTIP1;1 homolog is highly expressed in expanding cells in roots, leaves, and reproductive organs (Barrieu et al., 1998; Chaumont et al., 1998). An increased expression of TIPs is also observed in elongating tissues of hypocotyls in soybean (Glycine max), castor bean (Ricinus communis), and radish (Raphanus sativus) seedlings (Higuchi et al., 1998; Suga et al., 2001; Eisenbarth and Weig, 2005) and in protruding seed radicles in canola (Brassica napus; Gao et al., 1999). Furthermore, the abundance of HvTIP1;1 mRNA increases in the barley (Hordeum vulgare) slender mutant, which exhibits a faster leaf elongation rate compared with the wild type (Schünmann and Ougham, 1996). However, even if the expression pattern of these TIP genes suggests an important role in cell elongation, direct proof of this physiological function during plant development is still missing. Some clues come from the overexpression of the cauliflower (Brassica oleracea) or tobacco (Nicotiana tabacum) TIP1;1 gene in tobacco suspension cells, which results in an increase in cell growth and size (Reisen et al., 2003; Okubo-Kurihara et al., 2009).
Roots represent a very useful model to investigate cell elongation and growth processes (De Smet et al., 2012; Petricka et al., 2012). The primary root meristem, located at the organ apex, continuously produces cells that elongate after they leave the apical meristem and, hence, push the meristem forward. The primary root of many dicotyledonous plants repeatedly branches to generate several orders of lateral roots. In Arabidopsis, lateral roots exclusively originate from three files of pericycle founder cells overlying the xylem poles (Dolan et al., 1993). Xylem-pole pericycle cells are first “primed” to become lateral root founder cells close to the apical meristem in a zone termed the basal meristem (De Smet et al., 2007). Lateral roots are initiated when pairs of pericycle founder cells undergo several rounds of anticlinal division to create a new lateral root primordium (LRP) composed of up to 10 cells of equal length designated as stage I (Malamy and Benfey, 1997; Casimiro et al., 2001; Dubrovsky et al., 2001). Next, these cells divide periclinally, giving rise to an inner and an outer layer (termed stage II). Further anticlinal and periclinal divisions create a dome-shaped primordium (stages III–VII) that emerges (stage VIII) from the parental root (Malamy and Benfey, 1997; Péret et al., 2009). The tight correlation of lateral root formation and emergence is controlled by auxin, which acts as a local inductive signal that also triggers cell separation in the overlying tissues (Swarup et al., 2008).
The involvement of PIP AQPs in proper lateral root emergence (LRE) was recently demonstrated in Arabidopsis (Péret et al., 2012). Expression analysis revealed that most of the analyzed AtPIP and AtTIP genes are repressed during LRP formation, a regulatory response that can be mimicked by exogenous treatment with auxin. The latter effect is correlated with a reduction in cell and whole root hydraulic conductivity. AtPIP2;1 overexpressing lines and pip2;1 knockout mutants exhibit delayed LRE, with an accumulation of early LRP developmental stages at 42 h after a gravitropic stimulus. Furthermore, LRPs fail to protrude into the overlying tissues (Péret et al., 2012). This pattern of delayed LRP development and emergence in both AtPIP2;1 overexpressing lines and pip2;1 mutants was predicted by a mathematical model. The interplay of the spatial and temporal control of auxin-dependent cell hydraulic conductivity could be critical during LRP development.
In this work, we investigated the contribution of AtTIP AQPs in this developmental process making use of different tip knockout lines. While the simultaneous lack of AtTIP1;1, AtTIP1;2, and AtTIP2;1 had only a small impact on the primary root growth, it delayed LRE, resulting in a strong reduction in lateral root number. The phenotype could be fully rescued by expressing AtTIP2;1 under its native promoter. These results show that regulation of the tonoplast water permeability plays, in addition to the plasma membrane permeability, an important role in early lateral root growth and development.
RESULTS
Molecular Analysis of the tip Knockout Lines
To gain insight into TIP function, different Arabidopsis lines with T-DNA or transposon insertion in AtTIP1;1, AtTIP1;2, and AtTIP2;1 genes were characterized. For all tip mutant lines, the homozygosity of the insertions was confirmed by PCR using gene-, T-DNA-, or transposon-specific primers (Supplemental Table S1). The T-DNA and defective Suppressor-mutator (dSpm) insertions in single tip1;1, tip1;2, and tip2;1 mutants are illustrated in Supplemental Figure S1 (A, B, and C, respectively). The tip1;2 tip2;1 double mutant was obtained by crossing the two single insertion lines (Supplemental Fig. S1, B and C). The tip1;1 tip1;2 tip2;1 triple mutant was obtained by crossing independent tip single mutants (Supplemental Fig. S1D), which contain insertion at different positions in the AtTIP genes compared with the single tip mutant lines used in this study (as shown in Supplemental Fig. S1, A–C; Schüssler et al., 2008). Quantitative reverse transcription PCR (RT-qPCR) confirmed the absence of the corresponding RNA in the single, double, and triple tip mutant lines (Supplemental Fig. S1).
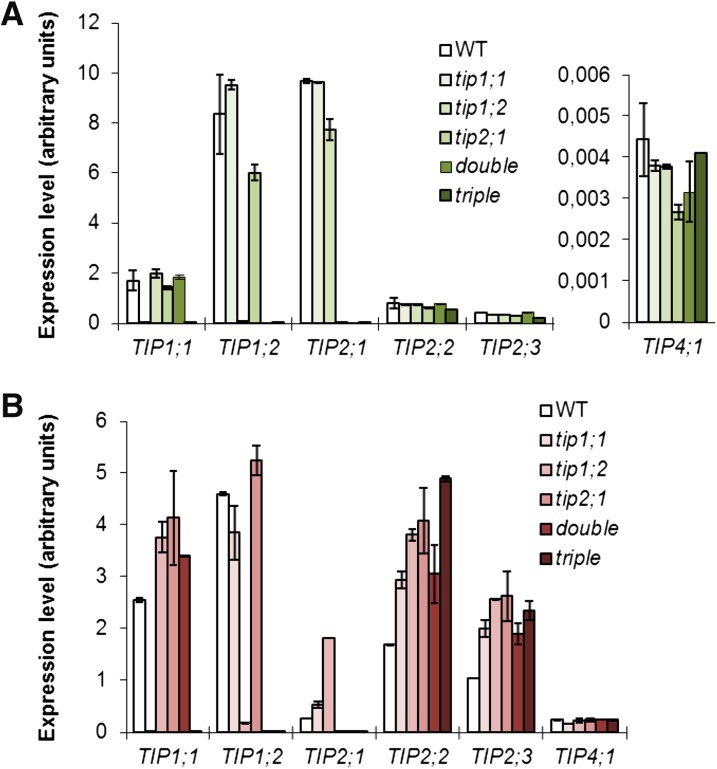
The tip mutant lines did not exhibit any striking phenotypes when grown under standard conditions (data not shown). To investigate whether this observation was due to compensatory up-regulation of other AtTIP genes, we analyzed the mRNA levels of all the AtTIP isoforms in roots and shoots of 12-d-old wild-type and tip mutant seedlings by RT-qPCR using isoform-specific primers (Supplemental Table S2). In both roots and leaves, the transcript levels of the AtTIP genes carrying T-DNA or transposon insertion were down-regulated by at least 64-fold compared with the wild type (Fig. 1). The AtTIP expression profile of the detected isoforms in the wild type corresponded to those shown in the literature (Alexandersson et al., 2005; Schüssler et al., 2008). In roots, no significant up-regulation of the root expressed genes (AtTIP1;1, AtTIP1;2, AtTIP2;1, AtTIP2;2, AtTIP2;3, and AtTIP4;1) was observed in the tip mutant lines, except for the tip1;2 mutant in which AtTIP2;1 exhibited a 3-fold up-regulation (Fig. 1). AtTIP1;3, AtTIP3;1, AtTIP3;2, and AtTIP5;1 transcripts could not be detected in the wild type or any of the tip mutant lines, confirming previous root expression data (Alexandersson et al., 2005).
Figure 1.
Expression levels of AtTIP mRNAs in Arabidopsis wild type (WT) and tip mutant lines. AtTIP mRNA levels in leaves (A) and roots (B) of 12-d-old Arabidopsis grown on vertical plates containing half-strength Murashige and Skoog medium, determined by RT-qPCR. The 10 AtTIP isoforms were tested in the assay. Only the detected isoforms are shown. Data are given as means ± sd. Double, tip1;2 tip2;1 double mutant; triple, tip1;1 tip1;2 tip2;1 triple mutant.
The tip Triple Mutant Exhibits Shorter Roots at 11 d after Germination
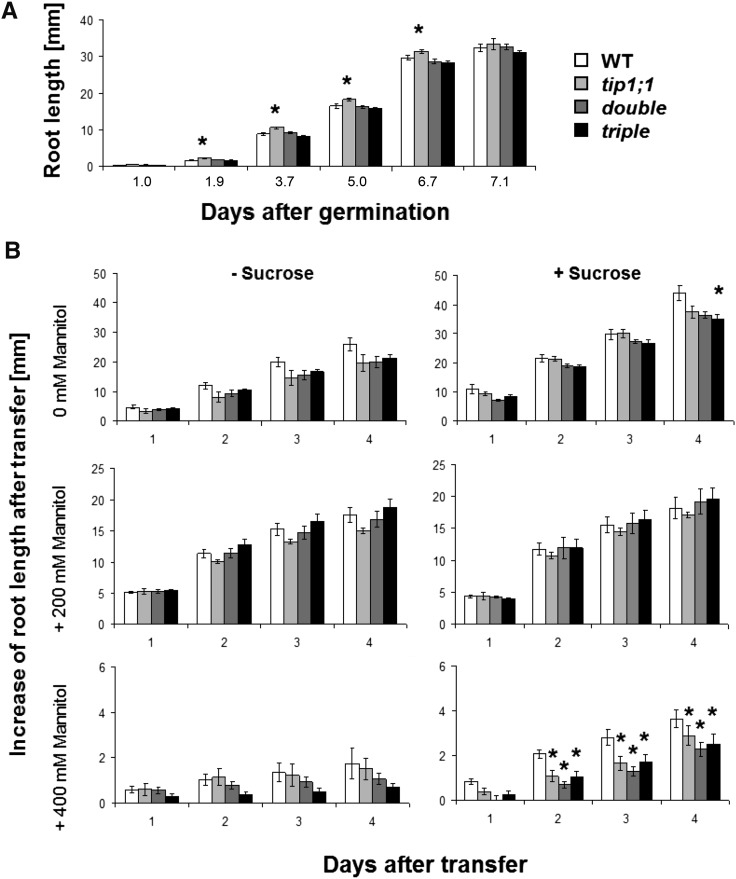
Since AtTIP genes are expressed in elongation zones, the lack of the most highly expressed isoforms could result in a deficiency in cell elongation. As we did not observe any difference in hypocotyl elongation between the different tip mutant lines compared with the wild type (data not shown), we decided to focus on root growth. No root growth phenotype was previously reported for tip1;1 (Schüssler et al., 2008; Beebo et al., 2009) and tip1;2 (Schüssler et al., 2008) single mutants, suggesting a possible functional redundancy of AtTIP isoforms. However, if AtTIPs are important for root growth processes, coinciding mutations in several AtTIP isoforms might cause growth phenotypes. Seedlings of the single tip1;1, double tip1;2 tip2;1, and triple tip1;1 tip1;2 tip2;1 mutants were grown on vertical half-strength Murashige and Skoog plates containing 1.5% (w/v) Suc, and the elongation of the main root was monitored during 7 d (see “Materials and Methods”). No significant difference in root length of double and triple mutants was observed compared with wild-type plants at 7 d after germination (dag; Fig. 2). However, the tip1;1 single mutant exhibited a small but significant increase in root length compared with the wild type at early seedling stage (up to 6 dag).
Figure 2.
Root growth of Arabidopsis tip mutants under normal conditions and osmotic stress. A, Root length of Arabidopsis wild type (WT) and tip mutant lines over time grown on vertical plates containing half-strength Murashige and Skoog medium supplemented with 1.5% (w/v) Suc. Data are given as means of 48 to 72 plants ± se. B, Root length of Arabidopsis tip mutant lines over time after transferring 7-d-old seedlings from A to vertical plates containing half-strength Murashige and Skoog medium in the presence or absence of 1.5% (w/v) Suc supplemented with 0, 200, or 400 mm mannitol. Data are given as means of six to 16 plants ± se. Asterisks indicate significant differences from the wild type (Student’s t test, P < 0.05).
Since cell elongation is driven by high and constant cell turgor, which is obtained by high osmotic concentration of the vacuolar sap, changes in turgor pressure mediated by an osmotic stress alter elongation processes (Fricke and Peters, 2002). Therefore, we investigated root elongation of tip mutants under osmotic stress conditions. At 7 dag, the seedlings from the experiment shown in Figure 2A were transferred to plates containing half-strength Murashige and Skoog medium supplemented with 0, 200, or 400 mm mannitol, in the absence or presence of 1.5% (w/v) Suc. Suc itself could induce an osmotic stress on the seedlings and mask effects by the supplemented mannitol (1.5% [w/v] Suc corresponds to about 44 mm, exhibiting an osmotic pressure of about 0.11 MPa according to van’t Hoff’s law). The root length was measured as described above. To avoid variations resulting from different root lengths of individual seedlings at the time of the transfer, the position of the root tip after the transfer was set to zero, and the increase in length after the transfer was measured. No significant difference in root length increase was observed for all tip mutants compared with the wild type, when they were transferred to plates containing 0, 200, or 400 mm mannitol without Suc (Fig. 2). In the presence of 1.5% (w/v) Suc but absence of mannitol (i.e. under control conditions), only the tip1;1 tip1;2 tip2;1 triple mutant exhibited a significantly lower increase in root length compared with the wild type 4 d after the transfer (corresponding to 11 dag). In the presence of 400 mm mannitol and 1.5% (w/v) Suc, all the tip mutant lines showed significantly shorter roots after the transfer.
The tip Triple Mutant Exhibits Significantly Fewer Lateral Roots
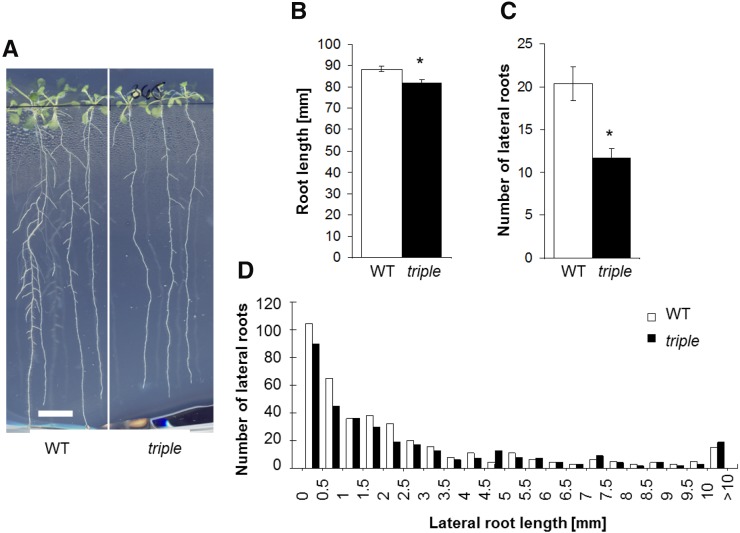
Since the triple tip mutant line had shorter roots at 11 dag under control conditions, we selected this line for a closer investigation of its root phenotype. Arabidopsis wild-type and tip1;1 tip1;2 tip2;1 mutant plants were grown on vertical plates containing half-strength Murashige and Skoog medium supplemented with 1.5% (w/v) Suc. At 12 dag, the length of the main roots was measured, and the lateral roots were counted (Fig. 3). The triple tip mutant exhibited slightly shorter roots (Fig. 3, A and B, about 10% reduction), confirming our previous observations (Fig. 2B). Interestingly, the triple mutant exhibited a 50% reduction in the number of lateral roots compared with the wild type (Fig. 3, A and C). The length of all lateral roots was measured for 50 plants from each genetic background, by drawing a straight line between the tip and the base of individual lateral roots. Whereas the number of lateral roots with a length longer than 3.0 mm was fairly similar between the wild type and the triple tip mutant lines, the latter exhibited a reduced number of newly emerged (short) lateral roots (Fig. 3D), suggesting a defect during early stages of lateral root development. The reduced number of lateral roots in the triple tip mutant was observed on Suc-containing medium, which was previously shown to promote outgrowth of initialized LRPs (Macgregor et al., 2008).
Figure 3.
The Arabidopsis tip1;1 tip1;2 tip2;1 triple mutant shows a reduced number of lateral roots. A, Arabidopsis wild type (WT) and tip1;1 tip1;2 tip2;1 triple mutant (triple) were grown vertically for 12 days on half-strength Murashige and Skoog medium supplemented with 1.5% (w/v) Suc; bar = 10 mm. B, Compared with the wild type, the triple tip mutant exhibits slightly shorter main roots. C, Compared with the wild type, the triple tip mutant exhibits a strong decrease in the number of lateral roots. Means are given ± se, n = 47 plants. Asterisks indicate significant differences from the wild type (Student’s t test, P < 0.05). D, Length distribution of all lateral roots from both, 50 wild-type and tip1;1 tip1;2 tip2;1 triple tip mutant plants (n = 381 and 288 for the wild type and tip1;1 tip1;2 tip2;1, respectively).
Disrupted AtTIP Expression Causes Delayed LRP Development
To determine the contribution of the AtTIP genes during early stages of LRP development, we induced lateral root initiation employing a 90° gravitropic stimulus, which is commonly used to synchronize lateral root initiation and investigate its developmental progression (Fig. 4; Ditengou et al., 2008; Lucas et al., 2008; Richter et al., 2009; Péret et al., 2012). Since Suc contained in the culture medium can promote LRP development (Macgregor et al., 2008), we performed this experiment on Suc-free medium to strengthen the LRP phenotype of the tip mutants. In this analysis, in addition to the triple tip mutant, we included the single tip1;1, tip1;2, and tip2;1 and double tip1;2 tip2;1 mutant lines to possibly relate the contribution of single AtTIP isoform to LRP development, even if no phenotype regarding lateral roots was previously reported for tip1;1, tip1;2, and tip1;1 tip1;2 mutants (Schüssler et al., 2008; Beebo et al., 2009). To date no phenotypic data are available for the tip2;1 mutant line.
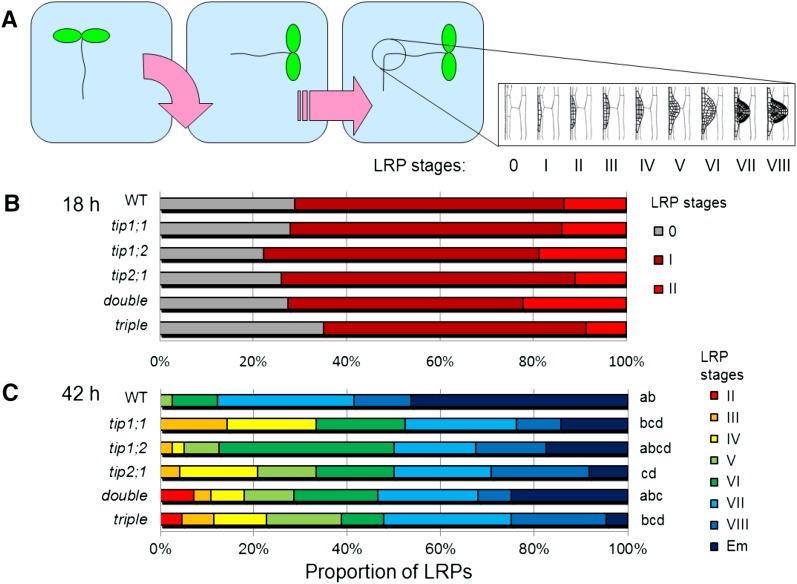
Figure 4.
Disrupted AtTIP expression causes delayed LRP development. A, To synchronize lateral root development, plants were subjected to a 90° gravitropic stimulus. B and C, LRP stages (from 0 to VIII, Em; Malamy and Benfey, 1997) are expressed as percentage of the total number of LRPs, 18 h (B) and 42 h (C) pgi. Double, tip1;2 tip2;1; triple, tip1;1 tip1;2 tip2;1. n = 12 to 24, pooled from two independent experiments. Statistical analysis was performed using standard contingency tables to sequentially compare the lines with each other (see “Materials and Methods”).
In wild-type plants, stage I primordia were first detected 18 h postgravitropic induction (pgi), and the other stages (II–VIII) peaked approximately every 3 h until emergence (stage emerged [Em]) at 42 h pgi (Péret et al., 2012). At 18 h pgi, no significant difference in LRP stage distribution was detected in single, double, or triple tip mutants compared with the wild type (Fig. 4B), indicating that these AtTIP genes were not required for primordia initiation. In contrast, at 42 h pgi, all the tip mutant lines showed a shift in LRP stage distribution toward earlier stages (Fig. 4C). Whereas ∼ 50% of LRPs had emerged in wild-type roots, only 5% to 15% of the lateral roots at the bending site of tip mutants had emerged. In some seedlings of the tip1;2 tip2;1 and tip1;1 tip1;2 tip2;1 mutants, LRPs still remained in stage II, in contrast to the wild-type plants, where the earliest LRP stage found was V. These data strongly indicate an important role of AtTIPs in LRP development. The nonadditive response of the single, double, and triple mutant lines suggests a complex interplay of different AtTIP isoforms during this process.
To verify that the delay in LRP development was due to the T-DNA or transposon insertion in AtTIP genes, we included another independent batch of single tip mutants in our analysis, tip1;1-2, tip1;2-2 (Schüssler et al., 2008), and tip2;1-2, which were used to obtain the triple tip mutant. The T-DNA or transposon insertions in these lines were verified by PCR on genomic DNA (Supplemental Fig. S2). These new single tip mutated lines were subjected to a root bending assay. All of them except the tip1;2-2 line showed a similar delay in LRP development at 42 h pgi (Supplemental Fig. S2) as the previously tested lines (Fig. 4B).
AtTIP Expression Patterns Are Correlated to Lateral Root Formation
A strictly defined spatial and temporal TIP expression pattern might be crucial for the directed growth of complex three-dimensional structures, such as LRPs. During LRE, cells located in the outer layers of the LRP base need to stretch as the LRP increases in volume, suggesting that requirement for a faster water influx across the plasma membrane into the cytosol as well as across the tonoplast into the vacuole, which occupies most of the volume in these cells. It was previously reported that the expression of AtTIP1;1, AtTIP1;2, AtTIP2;2, and AtTIP2;3 was rapidly down-regulated after lateral root gravitropic induction (Péret et al., 2012). To investigate the expression pattern of AtTIP1;1 and AtTIP1;2 in developing LRPs, roots from 5- to 7-d-old seedlings containing proAtTIP1;1:AtTIP1;1-GFP or proAtTIP1;2:AtTIP1;2-GFP constructs were stained for 5 min with propidium iodide (PI), and LRPs were investigated using confocal laser scanning microscopy (Fig. 5). The expression of AtTIP1;1-GFP (Fig. 5, A–H) and AtTIP1;2-GFP (Fig. 5, I–P) under their native promoter appeared to be ubiquitous in the main root. In the zone where lateral root development occurred, the TIP-GFP signals were abundant in the outer cell layers facing the LRPs during early developmental stages (Fig. 5, E, F, M, and N, LRP stages II and IV) but were reduced in these cells at late developmental stages (Fig. 5, G, H, O, and P, LRP stages VI and Em). No expression of AtTIP1;1-GFP or AtTIP1;2-GFP was detected in LRPs perfectly correlating with the decrease in AtTIP1;1 and AtTIP1;2 mRNA levels measured during LRP development (Péret et al., 2012). In addition, while the AtTIP1;2-GFP signal decreased in the cortex cells facing the LRPs during late developmental stages, it increased in the lateral root inner basal tissues and in the stele (Fig. 5, O and P).
Figure 5.
Expression of AtTIP1;1 and AtTIP1;2 during lateral root development. Seven- to eight-day-old roots from proAtTIP1;1:AtTIP1;1-GFP (A–H) and proAtTIP1;2:proAtTIP1;2-GFP (I–P) transgenic seedling were stained with PI for 5 min, and lateral roots at different developmental stages were visualized by confocal laser scanning microscopy. A to D and I to L, Single optical sections of the transmitted light signal showing the developmental state of the lateral root. E to H and M to P, Maximal projections of optical z-stacks of merged signals from GFP (green) and PI fluorescence (red). The dome shape of the LRP is indicated by dotted lines at the developmental stages II, IV, and VI (Malamy and Benfey, 1997). Scale bars = 20 µm.
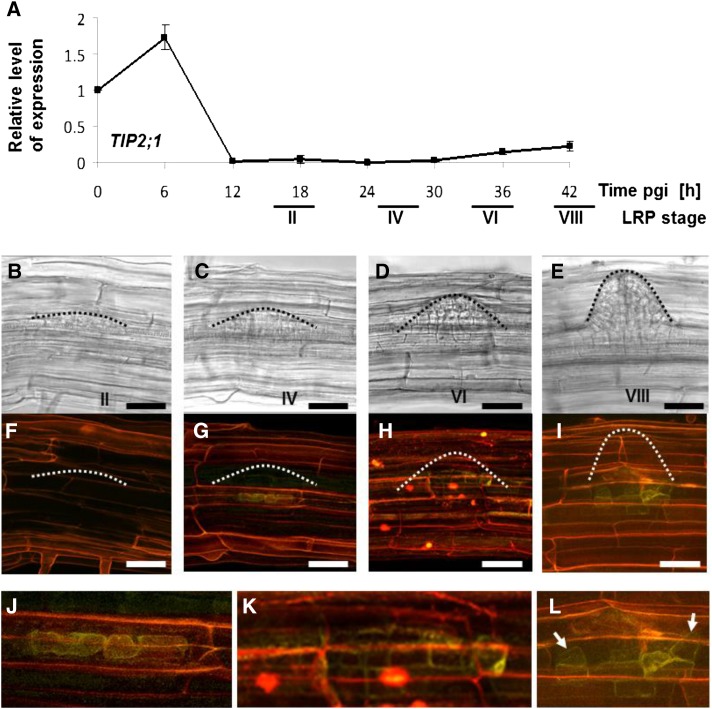
As no data were available for AtTIP2;1 mRNA levels during LRP development, we used RT-qPCR to analyze the expression of AtTIP2;1 in microdissected root bends in response to a gravitropic stimulus (Fig. 6). The level of AtTIP2;1 mRNA increased 1.7-fold at 6 h pgi compared with time 0 and then dramatically decreased at 12 h pgi (Fig. 6A). This expression pattern differs from the one reported for AtTIP1;1, AtTIP1;2, AtTIP2;2, and AtTIP2;3 but is similar to the expression of AtPIP2;4 and AtPIP2;1 (Péret et al., 2012).
Figure 6.
Expression of AtTIP2;1 during LRP development. A, AtTIP2;1 expression level was followed after gravitropic induction of lateral root formation and dissection of the root bend. Expression level is shown as a function of pgi relative to the expression right after the gravitropic induction. Approximate LRP developmental stages (Malamy and Benfey, 1997) at the individual time pgi are indicated. Data are presented as means of two biological replications ± sd. B to L, Roots from proAtTIP2;1:AtTIP2;1-YFP plants (Gattolin et al., 2009) were stained with PI for 5 min, and LRPs at different developmental stages were investigated by confocal laser scanning microscopy. B and F, LRP stage II, approximately corresponding to the developmental status at about 18 h pgi. C and G, LRP stage IV, approximately corresponding to 24 h pgi. D and H, LRP stage VI, approximately corresponding to 36 h pgi. E and I, LRP stage VII, approximately corresponding to 42 h pgi. B to E, Single optical sections of the transmitted light signal showing the developmental state of the LRP. F to I, Maximal projections of optical z-stacks of merged signals from YFP (yellow) and PI fluorescence (red). The dome shape of the LRP is indicated by dotted lines. J to L, Magnification of the AtTIP2:1-YFP expressing cells during LRP stages IV, IV, and VIII. The arrows indicate triangular-shaped cells containing intense AtTIP2;1-YFP signal. The images show representative results for each LRP stage. Scale bars = 50 µm.
To gain further insight on the expression pattern of AtTIP2;1 and given that AtTIP2;1-YFP expressed under the control of the AtTIP2;1 promoter is detected in a ring-like cell cluster at the base of emerging lateral roots (Gattolin et al., 2009), we decided to closely investigate AtTIP2;1 expression during LRP development using the same translational reporter. Roots from 5- to 7-d-old seedlings expressing the proAtTIP2;1:AtTIP2;1-YFP construct were stained for 5 min with PI, and LRPs were investigated using confocal laser scanning microscopy. The expression of AtTIP2;1-YFP was exclusively restricted to sites of LRP development (Figs. 6 and 7). In LRPs at developmental stage II, no YFP fluorescence was detected (Fig. 6, B and F), whereas in LRPs at stage IV, a fluorescent signal was observed in a single cell file located in the pericycle of the newly established LRP (Fig. 6, C, G, and J). At stage VI, AtTIP2;1-YFP was also detected in about two cell layers surrounding the base of the newly developed LRP (Fig. 6, D, H, and K). At stage VIII, several cell layers at the base of the LRP showed a YFP fluorescent signal that was more intense in the larger, triangular-shaped cells at the borders of the LRP (Fig. 6, E, I, and L, indicated by arrows).
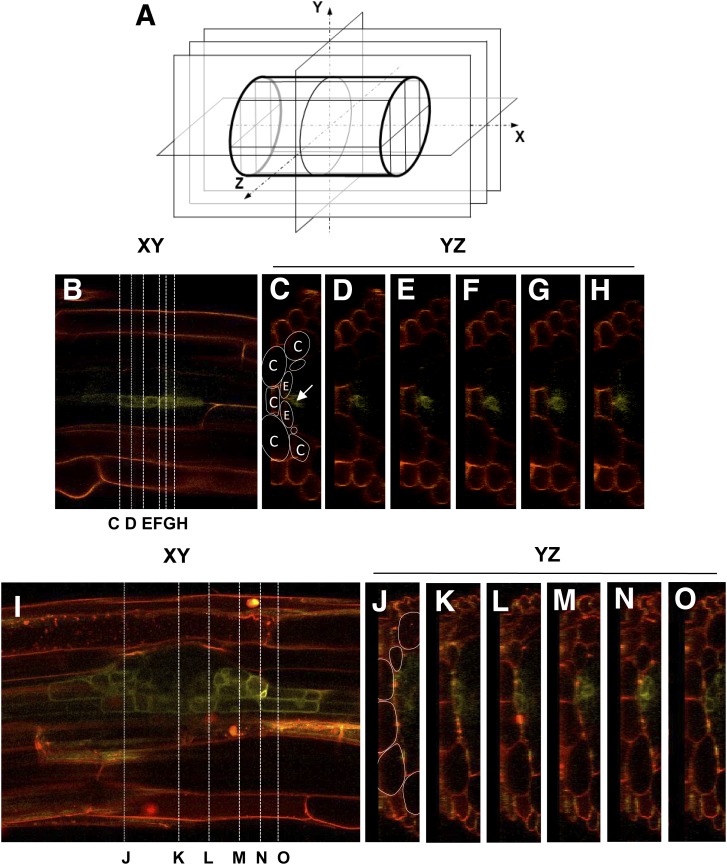
Figure 7.
Orthogonal sections of a LRP in the developmental stages IV and VI. A, Orientation of the space dimensions during confocal imaging. The cylinder represents a root section, which is imaged in several XY planes with different focus depth Z. This so-called z-stack can be cut along XZ or YZ planes, allowing virtual tissue sectioning. B to H, AtTIP2;1-YFP in a stage IV LRP (from Fig 7, C, G, and J). B, XY plane. The lines indicate the positions of the according YZ planes shown in C to H. The scheme in C illustrates the various root cell types in the cross-section: C denotes cortex cells, and E denotes endodermis cells. The arrow points toward the AtTIP2;1-YFP signal, located in newly created LRP cells adjacent to the endodermis. I to O, AtTIP2;1-YFP in a stage VI LRP (from Fig. 7, E, I, and L). I, XY plane. The lines indicate the according positions of the YZ planes shown in J to O. The scheme in J illustrates the positions of cortex cells. The YFP signal is located underneath the cortex cells in the newly developed primordial cells as well as in the neighboring endodermis cells.
As the images shown in Figure 6, F to L, resulted from a three-dimensional reconstruction of single confocal images taken at sequential focus planes, denoted as XY in Figure 7, we virtually cut this image stack orthogonally in YZ planes. This analysis allowed us to determine that, at stage IV of LRP development, AtTIP2;1-YFP was expressed in a row of cells inside of the endodermis, presumably corresponding to pericycle cells (Fig. 7, B–H). In stage VI of LRP development, AtTIP2;1-YFP was also observed in endodermal cells at the ends of the LRP and in newly raised primordial cells located at the base of the LRP, facing cortical cells (Fig. 7, I–O). These results indicate that LRE requires a tight regulation of AtTIP expression. In particular, AtTIP2;1 activity seems to be required for the growth of LRP basal and border cells and the regulation of LRP development and LRE of the main root. LRE appears also to be correlated to a reduction of AtTIP1;1 and AtTIP1;2 protein levels in the cortex cells.
The Delay in LRP Development of the Triple tip Mutant Is Only Rescued When AtTIP2;1 Is Expressed under Its Native Promoter
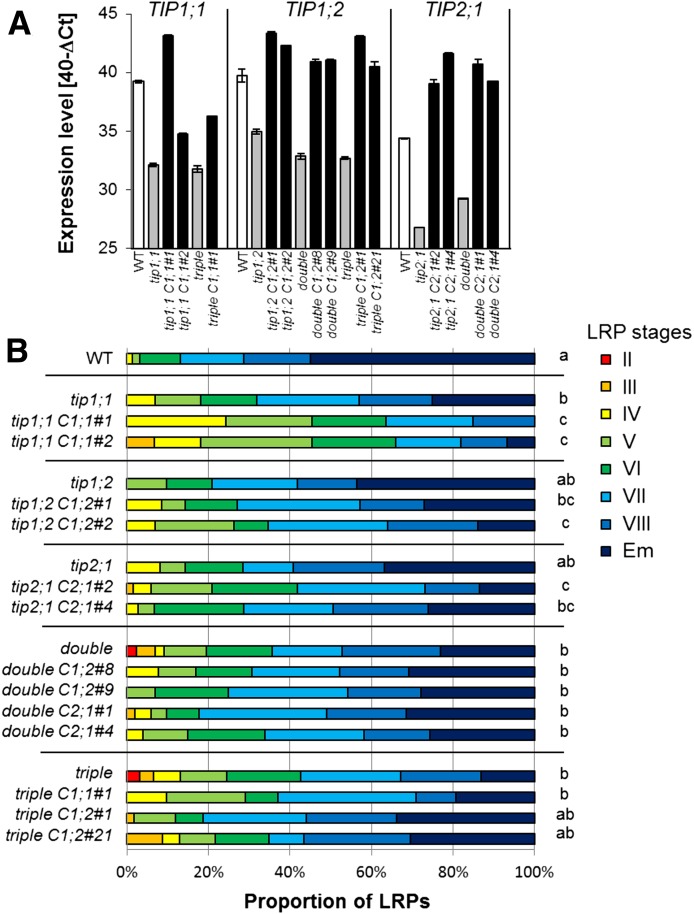
To correlate the LRP phenotype to the lack of AtTIPs, we modulated AtTIP expression in these mutants by expressing the respective AtTIP isoform under the control of the constitutive cauliflower mosaic virus 35S promoter in the tip mutant lines (Fig. 8). For the double and triple mutants, only one of the TIP genes was expressed. The expression of the AtTIP genes in the tip mutated lines was verified by RT-qPCR (Fig. 8A). In all the retransformed lines, we measured a higher level of the respective AtTIP mRNA compared with the mutant. The retransformed tip mutated lines were subjected to a root bending assay, and the LRPs were analyzed 42 h pgi (Fig. 8B). The introduction of a constitutively expressed TIP isoform in the tip1;1, tip1;2, or tip2;1 mutant did not lead to a reversion of the LRP development defect but, instead, to a stronger delay in this process compared with the tip1;1, tip1;2, or tip2;1 single mutant. No change in the pattern of LRP development was detected in the complemented tip1;2 tip2;1 double mutant lines. Finally, the expression of AtTIP1;2 in the tip1;1 tip1;2 tip2;1 background resulted in a shift of the LRP stage distribution toward the one observed in the wild type. However, the distribution pattern of wild-type LRP development could not be reached, probably due to the fact that a fine-tuning of AtTIP expression could not be achieved using the constitutively active 35S promoter (as observed when AtPIP2;1 expression was driven by the 35S promoter in a pip2;1 mutant line; Péret et al., 2012).
Figure 8.
Constitutive expression of AtTIP genes in the tip mutant lines alters LRP development. A, Transgene AtTIP expression in the complemented tip mutant lines determined by RT-qPCR and compared with wild-type (WT) control. Double, tip1;2 tip2;1; triple, tip1;1 tip1;2 tip2;1. Italic C indicates that the line is complemented, followed by the number of the AtTIP isoform and the number of the independently selected line (for example, “triple C1;2#21” represents the tip1;1 tip1;2 tip2;1 mutant complemented with AtTIP1;2, selected line no. 21). B, Developmental stage distribution (from II to VII, Em; Malamy and Benfey, 1997) of LRPs 42 h pgi in Arabidopsis tip mutant and the complemented lines. n = 23 to 102, pooled from two independent experiments. Statistical analysis was performed using standard contingency tables to sequentially compare relevant lines (see “Materials and Methods”). Only plants corresponding to one mutant line (grouped by horizontal lines in the chart) were compared with each other and the wild type.
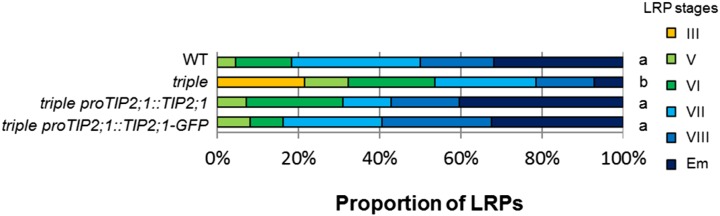
As AtTIP2;1 exhibits an exclusively LRP-related expression pattern, we decided to investigate the ability of this particular isoform to rescue the LRP phenotype in the triple tip mutant. We created transgenic triple tip mutant lines containing proAtTIP2;1:AtTIP2;1 or proAtTIP2;1:AtTIP2;1:GFP construct, and subjected them to a root bending assay as described above (Fig. 9). Both the nonfluorescent and the GFP-tagged AtTIP2;1 expressed under the native promoter completely restored the wild type-like LRP development of the triple tip mutant. Altogether, these data suggest that a fine-tuning of the spatiotemporal distribution (and activity) of AtTIP2;1 is needed to promote LRP development and that ectopic expression of any AtTIP does not rescue or even negatively impacts this developmental process.
Figure 9.
Expression of AtTIP2;1 under the control of its own promoter rescues the LRE phenotype of the triple tip mutant. Developmental stage distribution (from III to VIII, Em; Malamy and Benfey, 1997) of LRPs 42 h pgi from seedlings of the wild type (WT), the tip1;1 tip1;2 tip2;1 triple mutant (triple), transformed triple tip mutant with proTIP2;1:TIP2;1 or proTIP2;1:TIP2;1-GFP fusions lines. n = 22 to 38. Statistical analysis was performed using standard contingency tables to sequentially compare relevant lines (see “Materials and Methods”).
DISCUSSION
TIPs Play a Role in Lateral Root Formation
In this study we show that TIP AQPs are essential for the proper development of LRPs. As TIPs are known to be efficient water channels (Maurel et al., 1993; Maurel et al., 1995; Daniels et al., 1996; Gerbeau et al., 1999), their presence in the tonoplast is expected to allow rapid water exchange between the vacuole and the cytoplasm, an important process during cell expansion, which is driven by the turgor pressure. Since the development of LRPs is a rapid process with a time scale of hours and requires a complex set of cell divisions and expansions, tight control of TIP abundance and activity in the tonoplast of the different LRP cells is probably important for this developmental process.
In a recent study, Péret et al. (2012) showed a similar action of plasma membrane AQPs during lateral root development. The authors developed a computational model that predicts the effect of changes in cell plasma membrane water permeability in different LRP tissues. This model did not include the possibility to distinguish between water fluxes across the plasma membrane and the tonoplast; therefore, it likely also holds for water fluxes across the tonoplast. Indeed, regulation of turgor pressure involved water flows across the plasma membrane as well as the tonoplast since the vacuole occupies the major volume of the cell. Comparing this process with an electrical circuit, PIPs in the plasma membrane and TIPs in the tonoplast act as serial conductors, allowing the fast water exchange between the apoplast, the cytosol, and the vacuole. If the conductance of one of these membranes is reduced by a decreased amount of functional AQPs located in the respective membrane, the turgor regulation is expected to be affected.
The plant hormone auxin is a key regulator of lateral root development (Péret et al., 2009). Xylem pole pericycle cells form LRPs after local auxin levels are elevated, triggering a series of asymmetric cell divisions (De Smet et al., 2007; Dubrovsky et al., 2008). Auxin also promotes the LRP emergence by triggering the expression of cell wall remodeling genes in the overlying tissues (Laskowski et al., 2006; Swarup et al., 2008; Kumpf et al., 2013; Lucas et al., 2013). The local distribution pattern of auxin is created by specialized efflux and influx transport proteins, which cause the accumulation of auxin at the apex and in the overlying tissues of the LRP (Swarup et al., 2008). LRP emergence and the concomitant physical modification of the overlying tissues must be tightly coregulated. Péret et al. (2012) showed that auxin regulates the expression of plasma membrane AQPs, mostly by repressing their expression, and therefore decreasing tissue hydraulic properties, which facilitates LRP emergence. These data suggest that auxin fine-tunes the spatial and temporal control of root tissue hydraulics through the regulation of AQP distribution. In their study, the authors also observed a decrease of expression of AtTIP1;1, AtTIP1;2, AtTIP2;2, and AtTIP2;3 genes upon LRP development. In this work, we observed that the expression of AtTIP2;1 is first up-regulated during early stages of LRP formation before subsequently being down-regulated, suggesting that AtTIP2;1 has a specific role during this process. However, the RT-qPCR experiments reported in this study and by Péret et al. (2012) were performed from total root sections, which prevented conclusion about the exact location of AQP expression. Therefore, we analyzed the expression of fluorescently tagged TIP proteins expressed under the control of their own promoter (Figs. 5–7). The absence of AtTIP1;1-GFP and AtTIP1;2-GFP proteins in the emerging LRP was in accordance with the decrease in mRNA level detected in RT-qPCR. Interestingly, an increase in AtTIP2;1-YFP signal observed in the pericycle, endodermis, and new primordial cells located at the base of the developing LRP was correlated with the transient increase in AtTIP2;1 mRNA measured by RT-qPCR.
LRP developmental process depends on the proper temporal and spatial expression of TIPs. Indeed, attempts to rescue the LRP emergence phenotype in the different mutated tip lines, by ectopic expression (using the 35S promoter) of AtTIP genes, were not successful, except for a partial recovery in the triple tip mutant by AtTIP1;2. The full recovery of the LRP emergence phenotype was only obtained when AtTIP2;1 was expressed under its endogenous promoter. As AtTIP2;1 is exclusively expressed in cells located at the base of developing LRPs, these data demonstrate an important role of AtTIP2;1 in these specific locations, possibly facilitating water movement from the stele to the LRP and the fast growth of these cells and/or the full LRP through the underlying tissue. The exact role of AtTIP1;1 during LRP development remains vague, as single mutants were only slightly affected in LRP formation. AtTIP1;1 expression pattern remained rather constant throughout LRP development, except with a decrease in expression in the cortex cells facing the LRPs at late stages. In contrast, AtTIP1;2 abundance appears to be more tightly linked to LRP development. Its expression was indeed gradually modified with a higher expression in the stele tissues and a lower or no expression in the epidermis and cortex cells facing the developing LRP. Altogether, the AtTIP expression patterns suggest the presence of a gradient in AtTIP abundance and, therefore, in tonoplast water permeability between the LRP and its surrounding tissues (Fig. 10). This could contribute to the maintenance of a high turgor pressure in the cells at the base of the LRP by an influx of water from the stele and probably from the surrounding tissues, giving rise to cell growth, and a decrease in turgor pressure in the overlying cortex cells to allow LRP emergence. AtTIPs could also contribute to the transcellular water flow from the vascular tissues to expanding cell territories of the LRPs. These data highlight the importance of the cell type specificity and timing of AtTIP expression and are in accordance with the results showing that disrupting cell water permeability by ectopic expression of AtPIP2;1 delayed LRP emergence in a wild-type background (Péret et al., 2012).
Figure 10.
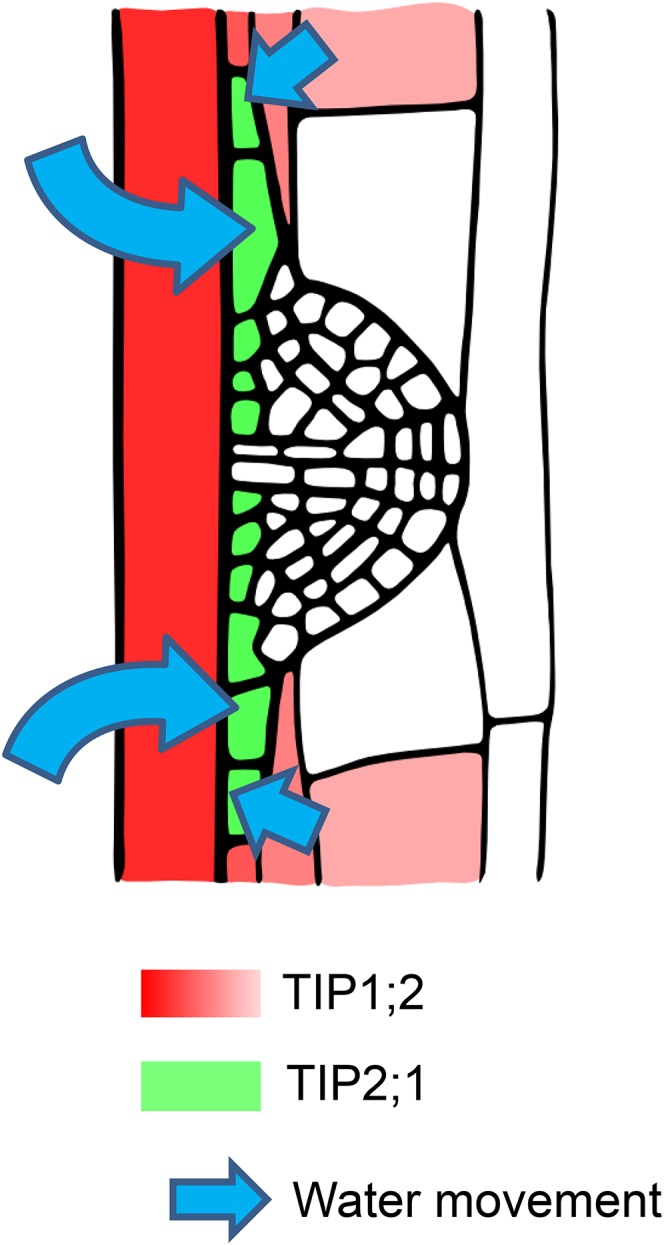
A tightly regulated TIP expression pattern is required for fast LRP development. After the LRP development is initialized, a gradient of TIP1;2 expression (red; dark, high expression; light, low expression) is observed from inner to outer root tissues and down-regulated in cortex cells facing the initialized LRP. At the same time, TIP2;1 expression (green) is up-regulated in cells located at the base of the LRP. This could result in a gradient in vacuolar water permeability between the base of the LRP on one side and the LRP surrounding tissues on the other side. The cells at the base of the LRP therefore could expand rapidly and allow the fast growth and emergence of the LRP throughout the overlying tissues.
While Péret et al. (2012) reported flattened LRP domes in the pip2;1 mutant, we observed only a small fraction of flattened LRPs in all of the analyzed tip mutant lines, the majority of them appearing normal even during early developmental stages. Furthermore, Péret et al. (2012) did not report any change in root architecture in the pip2;1 mutant, as was found for the triple tip mutant. It could be speculated that PIPs play a role in mediating the water influx into the LRP structure, whereas TIPs could be important during cell expansion processes, in particular during late developmental stages after the LRP breaks the casparian strip and rapidly emerges out of the root. This slightly differentiated functionality of PIPs and TIPs during LRP emergence could account for the less pronounced occurrence of flattened LRPs in the tip mutants compared with the pip2;1 mutant (Péret et al., 2012), as well as for the reduced number of LRs in the triple tip mutant.
TIPs Are Not Essential for Elongation of the Main Root and the Hypocotyl
Single mutants for tip1;1 and tip1;2 as well as the tip1;1 tip1;2 double mutant were previously carefully analyzed, and no phenotype was observed with respect to macroscopic appearance and water balance (Schüssler et al., 2008; Beebo et al., 2009). In addition, tip1;1 mutant did not show any compensatory effect by an up-regulation of the expression of other AtTIP genes (Schüssler et al., 2008; Beebo et al., 2009), which is in agreement with the current study. Under our growth conditions, we only observed a significant up-regulation of AtTIP2;1 in the tip1;2 mutant compared with the wild type, which might partially compensate for the loss of AtTIP1;2. However, in the tip1;2 tip2;1 double mutant and the tip1;1 tip1;2 tip2;1 triple mutant, no such up-regulation of any AtTIP gene was detected. The absence of a clear elongation defect of the main root of the triple tip mutant indicates a minor role of the AtTIP1;1, AtTIP1;2, and AtTIP2;1 isoforms in this process. Since the tonoplast was shown to be about 10 times more permeable to water than the plasma membrane (Maurel et al., 1997; Niemietz and Tyerman, 1997), probably due to the high amount of TIPs in the tonoplast, it remains to be determined why no more drastic phenotype was observed in the triple tip mutant, where basically most of the TIP isoforms are absent. It could mean that the residual water permeability of the tonoplast in these lines was sufficient to promote root elongation in the tested conditions. It would be interesting to compare the water permeability of intact vacuoles of the triple tip mutant and wild-type plants by determining the swelling/shrinking rates upon osmotic changes as described (Morillon and Lassalles, 1999).
Similarly to what was observed for the main roots, AtTIP1;1, AtTIP1;2, and AtTIP2;1 did not seem to play a major role during hypocotyl growth (Gendreau et al., 1997; Raz and Koornneef, 2001). On the tissue level, the process of hypocotyl growth is very similar to root elongation: various layers of cells elongate homogeneously in a single-dimensional manner. The hypocotyls gain about 10 mm in length during 4 d in the dark, which is comparable to that of the root. After 4 d under standard growth conditions, no difference of various tip mutant lines compared with the wild type was detected, except for the tip1;1 mutant, which exhibited slightly longer roots. These data were quite unexpected as the expression pattern of AtTIP genes in Arabidopsis and other plant species suggested an important functional role of these proteins in the main root and hypocotyl growths. The promoter of Arabidopsis AtTIP1;1 was shown to be active in the hypocotyl and root elongation zone (Ludevid et al., 1992). Maize ZmTIP1;1 is highly expressed in expanding cells in roots and leaves (Barrieu et al., 1998; Chaumont et al., 1998). An increased expression of TIPs is also observed in elongating tissues of hypocotyls in soybean, castor bean, and radish seedlings (Higuchi et al., 1998; Suga et al., 2001; Eisenbarth and Weig, 2005). Once again, the basal water permeability of the tonoplast in the Arabidopsis tip mutated lines could be sufficient to allow the generation and maintenance of the turgescence essential for the cell elongation process under standard growth conditions. Interestingly, we observed that increasing the medium osmolarity by adding 400 mm mannitol resulted in a decrease of the main root length of the tip mutants compared with wild-type plants (Fig. 3). In these conditions, higher tonoplast water permeability would be required for the equilibrium of the cell water status during the cell growth process.
MATERIALS AND METHODS
Plant Material and Growth Conditions
The Arabidopsis (Arabidopsis thaliana) tip1;1 mutant is a T-DNA insertion mutant from the Salk Institute collection (Alonso et al., 2003) and was obtained from the Nottingham Arabidopsis Stock Centre (NASC) (seed stock N519003). The Arabidopsis tip1;2 and tip2;1 mutants were previously identified in the SLAT collection (Tissier et al., 1999) using the Sequenced Insertion Site database, and seed stocks (N116827, N118785) were obtained from the NASC. The tip1;2 and tip2;1 mutants display a dSpm insertion within the first and second exons, respectively (Supplemental Fig. S1). RT-qPCR experiments showed that AtTIP1;2 and AtTIP2;1 mRNA levels were reduced to less than 3% in tip1;2 and tip2;1 mutants, respectively, compared with the level in wild-type plants (data not shown). To obtain a tip1;2 tip2;1 double mutant line, the F2 progeny of a cross between the two mutant lines was PCR screened to identify plants with a recombinant chromosome bearing the two mutations using the AtTIP-specific primers TIP12F-652 and TIP12UTR3R371 or TIP21F-683 and TIP21UTR3R371 for AtTIP1;2 or AtTIP2;1, respectively, along with the transposon-specific primer dSpm1 to amplify wild-type and mutant alleles (Supplemental Table S1). A plant homozygous for the tip2;1 mutation and heterozygous for the tip1;2 mutation was identified and then allowed to self-pollinate. Plants homozygous for both mutations were identified in the offspring by PCR. Reduction in AtTIP mRNA levels was checked by RT-qPCR in the next generation (data not shown).
The tip1;1 tip1;2 tip2;1 triple mutant was generated by crossing homozygote tip1;1 tip1;2 and tip1;1 tip2;1 double mutants, which were generated from the insertion line SM_3_32402 (tip1;1) and GABI_880H12 (tip1;2; Schüssler et al., 2008), and SM_3_32402 (tip1;1) and SM_3_38811 (tip2;1) by floral crossing. The tip2;1 insertion mutant was kindly supplied by N. von Wirén (University of Hohenheim, Stuttgart, Germany) and originated from the NASC (http://arabidopsis.info). Homozygote insertion was verified by PCR using the primers TIP21F132 and TIP21R395 to amplify the genomic sequence of the wild-type AtTIP2;1 and the primers dSpm1 and TIP21R729 to verify the transposon insertion (Supplemental Table S1).
The Arabidopsis line expressing AtTIP2;1 fused to YFP under the control of the endogenous AtTIP2;1 promoter (∼ 1.2 kb) was described by Gattolin et al. (2009). Arabidopsis wild-type Columbia-0 and tip mutant seeds were surface sterilized in 3% (v/v) commercial bleach and rinsed five times with sterile distilled water. All seeds were placed on 60 mL of half-strength Murashige and Skoog medium at pH 5.8, solidified with 0.85% (w/v) agar in squared plates (approximately 120 × 120 mm). The seeds were stratified for 2 to 4 d at 4°C to obtain homogenous germination. For phenotyping and expression analysis, the medium was supplemented by 1.5% (w/v) Suc, and plants were grown at 60 µmol photons m−2 s−1 under long-day conditions (16 h light/8 h dark). For LRE experiments, plants were grown without the addition of Suc at 150 µmol photons m−2 s−1 under long-day conditions.
Plasmid Constructions and Plant Transformation
The cDNAs of AtTIP1;1, AtTIP1;2, and AtTIP2;1 were amplified by PCR (for primers, see Supplemental Table S3) from single-strand cDNA. PCR fragments were cloned using an uracil excision-based improved high-throughput USER cloning technique into the USER-compatible vector pCambia2300 35S u (Nour-Eldin et al., 2006). The constructs were verified by sequencing. To obtain proAtTIP1;2:AtTIP1;2-GFP and proAtTIP2;1:AtTIP2;1-GFP constructs, the genomic sequences of AtTIP1;2 and AtTIP2;1 starting ∼ 3 kb upstream of the translational start codon and ending before the stop codon were amplified by PCR (for primers, see Supplemental Table S3), cloned in the Gateway entry vector pDONR221 (Invitrogen), and transferred using LR recombination in front of the GFP in the modified pMDC83 binary destination vector (Curtis and Grossniklaus, 2003) in which the double 35S promoter was deleted. The genomic sequence of AtTIP2;1 (including the ∼ 3 kb promoter region) inserted in pDONR221 was recombined into the pGWB1 vector (Nakagawa et al., 2007) to create an untagged construct used to complement the triple tip mutant line. The binary vectors were introduced into Agrobacterium tumefaciens strain GV3101 by electroporation, and recombinant Agrobacterium lines were used to transform Arabidopsis plants through the floral dip method (Clough and Bent, 1998). Transgenic plants were selected on half-strength Murashige and Skoog agar plates containing kanamycin (50 mg/L).
RT-qPCR
Total RNA was extracted from roots and shoots of 12-d-old seedlings using RNeasy RNA plant extraction mini kit (Qiagen), and chromosomal DNA was digested on the column during the RNA isolation process using DNase I (Qiagen) according to manufacturer’s recommendation. RNA was quantified by spectrophotometry at 260 nm. The RNA quality was checked on agarose gel. One microgram of total RNA was transcribed into cDNA with oligo(dT) primers for 1 h at 42°C with M-MuLV reverse transcriptase (Promega), followed by 5 min at 85°C in a thermo cycler as described previously (Hachez et al., 2006). Primer pairs were as described by Alexandersson et al. (2005) or designed using Quantprime (www.quantprime.de; Arvidsson et al., 2008; Supplemental Table S2). RT-qPCR was performed using 1 µL of 1:10 diluted cDNA samples in a 25 µL reaction volume containing 300 nm gene-specific primers and 12.5 µL of SYBR Green 1 PCR mix (Eurogentec) in a StepOnePlus thermocycler (Applied Biosystems) with the following program: (1) 50°C for 2 min, (2) 95°C for 10 min, (3) 95°C for 15 s, and (4) 60°C for 30 s (3 and 4, 40 cycles). No template controls were included in the assay, and the absence of genomic DNA was verified by PCR using primers targeting an intronic region of the control gene MADS AFFECTING FLOWERING5 (MAF5, At5g65080; Supplemental Table S2). Melting curve analyses were performed for all samples by elevating the temperature from 55°C to 95°C. Normalization was performed using geometric averaging of three control genes: actin, α-tubulin, and ubiquitin (Bustin et al., 2009).
Growth Measurements
Six or four seedlings of two or three genotypes, respectively, were grown per plate, resulting in 12 seedlings per plate. Genotypes were spatially randomized, and each genotype appeared on at least three plates. For root growth experiment, the position of the root tips was marked once per day using a razor blade. At the end of the experiments, the plates were scanned, and root lengths were measured on the images assuming straight lines between the root base and marks of the petri dish using ImageJ (http://imagej.nih.gov/ij/index.html).
Root Bending Assay
Five-day-old seedlings grown on vertical half-strength Murashige and Skoog plates were subjected to a gravitropic stimulus by a 90° turn. As it was previously reported that lateral root formation is affected by the uptake of Suc from the medium (Macgregor et al., 2008), plants were grown without Suc. Seedlings were harvested 18 and 42 h pgi of lateral root formation, the tissue was cleared, and LRP stages at the bending site were recorded (Malamy and Benfey, 1997).
Tissue Clearing and Microscopy Analysis
For LRP observation, root tissues were cleared using acidified methanol (7:2:1H2O:methanol:acetic acid) for 20 min at 55°C, followed by a basic ethanol (7% [w/v] NaOH, 60% [v/v] ethanol) treatment for 15 min at room temperature. The tissue was rehydrated by an ethanol series of 40%, 20%, and 10% (v/v) for 15 min for each concentration at room temperature. For storage and observation, roots were kept in 50% glycerol. Seedlings, immersed in 50% (v/v) glycerol, were placed on microscope slides, and the developmental stage of the LRP was rated using standard light microscope with 20× or 40× objective (Leica DMR). The expression of fluorescent-tagged AtTIPs under the control of their endogenous promoter was observed in nontreated roots using a confocal microscope (Zeiss LSM710). YFP was excited at 514 nm, and the fluorescence signal was recorded in the range from 524 to 580 nm; GFP was excited at 488 nm, and the signal was detected from 499 to 553 nm. PI was excited using the laser according to the fluorescent protein observed (488 or 514 nm for GFP or YFP, respectively), and the signal was detected in the range from 596 to 710 nm.
Statistical Analysis
Student’s t test was applied to determine the significance of differences of average values between the wild type and mutant lines. For statistical analysis of the developmental stage distribution of LRPs, we applied Pearson’s independency test on standard contingency tables for sequential pairwise comparisons (Zar, 2010) of the developmental stage distribution of LRPs between relevant genetic backgrounds.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative of GenBank/EMBL databases under the following accession numbers: AtTIP1;1, At2g36830; AtTIP1;2, At3g26520; AtTIP1;3, At4g01470; AtTIP2;1, At3g16240; AtTIP2;2, At4g17340; AtTIP2;3, At5g47450; AtTIP3;1, At1g73190; AtTIP3;2, At1g17810; AtTIP4;1, At2g25810; AtTIP5;1, At3g47440; ACT1, At2g37620; TUA4, At1g04820; UBQ10, At4g05320; and MAF5, At5g65080.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Verification of the homozygosity of the insertion in the different tip mutant lines.
Supplemental Figure S2. Independent tip mutants exhibit delay in LRP development.
Supplemental Table S1. Primers used to create multiple tip mutant lines and verify the obtained lines.
Supplemental Table S2. Primers used for RT-qPCR to determine the expression level of all TIP isoforms in the tip mutant lines and the wild type.
Supplemental Table S3. Primers used for cloning.
Supplementary Material
Acknowledgments
We thank the imaging platform IMABIOL.
Glossary
- AQP
aquaporin
- LRPs
lateral root primordia
- LRP
lateral root primordium
- PIP
plasma membrane intrinsic protein
- TIP
tonoplast intrinsic protein
- LRE
lateral root emergence
- RT-qPCR
quantitative real-time PCR
- dag
d after germination
- pgi
postgravitropic induction
- Em
Emerged
- PI
propidium iodide
Footnotes
This work was supported by grants from the Belgian National Fund for Scientific Research (to F.C. and C.H.), the Interuniversity Attraction Poles Programme-Belgian Science Policy (IAP7/29 to F.C. and M.J.B.), the Belgian French community ARC11/16-036 project (to F.C.), the Biotechnology and Biological Sciences Research Council (BBSRC) and Engineering and Physical Sciences Research Council (EPSRC) (BB/D019613/1 and BB/H020314/1 to U.V, K.S., and M.J.B). H.R. was a Research Fellow at the “Fonds pour la Formation à la Recherche dans l’Industrie et dans l’Agriculture.” U.V. was supported by the BBSRC Professorial Research Fellowship funding to M.J.B. (BB/G023972/1).
Articles can be viewed without a subscription.
References
- Agre P, Preston GM, Smith BL, Jung JS, Raina S, Moon C, Guggino WB, Nielsen S (1993) Aquaporin CHIP: the archetypal molecular water channel. Am J Physiol 265: F463–F476 [DOI] [PubMed] [Google Scholar]
- Alexandersson E, Fraysse L, Sjövall-Larsen S, Gustavsson S, Fellert M, Karlsson M, Johanson U, Kjellbom P (2005) Whole gene family expression and drought stress regulation of aquaporins. Plant Mol Biol 59: 469–484 [DOI] [PubMed] [Google Scholar]
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al. (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Arvidsson S, Kwasniewski M, Riaño-Pachón DM, Mueller-Roeber B (2008) QuantPrime--a flexible tool for reliable high-throughput primer design for quantitative PCR. BMC Bioinformatics 9: 465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrieu F, Chaumont F, Chrispeels MJ (1998) High expression of the tonoplast aquaporin ZmTIP1 in epidermal and conducting tissues of maize. Plant Physiol 117: 1153–1163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beebo A, Thomas D, Der C, Sanchez L, Leborgne-Castel N, Marty F, Schoefs B, Bouhidel K (2009) Life with and without AtTIP1;1, an Arabidopsis aquaporin preferentially localized in the apposing tonoplasts of adjacent vacuoles. Plant Mol Biol 70: 193–209 [DOI] [PubMed] [Google Scholar]
- Bienert GP, Bienert MD, Jahn TP, Boutry M, Chaumont F (2011) Solanaceae XIPs are plasma membrane aquaporins that facilitate the transport of many uncharged substrates. Plant J 66: 306–317 [DOI] [PubMed] [Google Scholar]
- Bienert GP, Heinen RB, Berny MC, Chaumont F (2014) Maize plasma membrane aquaporin ZmPIP2;5, but not ZmPIP1;2, facilitates transmembrane diffusion of hydrogen peroxide. Biochim Biophys Acta (1 Pt B) 1838: 216–222 [DOI] [PubMed] [Google Scholar]
- Bienert GP, Møller ALB, Kristiansen KA, Schulz A, Møller IM, Schjoerring JK, Jahn TP (2007) Specific aquaporins facilitate the diffusion of hydrogen peroxide across membranes. J Biol Chem 282: 1183–1192 [DOI] [PubMed] [Google Scholar]
- Bienert GP, Thorsen M, Schüssler MD, Nilsson HR, Wagner A, Tamás MJ, Jahn TP (2008) A subgroup of plant aquaporins facilitate the bi-directional diffusion of As(OH)3 and Sb(OH)3 across membranes. BMC Biol 6: 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, Mueller R, Nolan T, Pfaffl MW, Shipley GL, et al. (2009) The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem 55: 611–622 [DOI] [PubMed] [Google Scholar]
- Casimiro I, Marchant A, Bhalerao RP, Beeckman T, Dhooge S, Swarup R, Graham N, Inzé D, Sandberg G, Casero PJ, et al. (2001) Auxin transport promotes Arabidopsis lateral root initiation. Plant Cell 13: 843–852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaumont F, Barrieu F, Herman EM, Chrispeels MJ (1998) Characterization of a maize tonoplast aquaporin expressed in zones of cell division and elongation. Plant Physiol 117: 1143–1152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaumont F, Tyerman SD (2014) Aquaporins: highly regulated channels controlling plant water relations. Plant Physiol 164: 1600–1618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Curtis MD, Grossniklaus U (2003) A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol 133: 462–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels MJ, Chaumont F, Mirkov TE, Chrispeels MJ (1996) Characterization of a new vacuolar membrane aquaporin sensitive to mercury at a unique site. Plant Cell 8: 587–599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels MJ, Mirkov TE, Chrispeels MJ (1994) The plasma membrane of Arabidopsis thaliana contains a mercury-insensitive aquaporin that is a homolog of the tonoplast water channel protein TIP. Plant Physiol 106: 1325–1333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Smet I, Tetsumura T, De Rybel B, Frei dit Frey N, Laplaze L, Casimiro I, Swarup R, Naudts M, Vanneste S, Audenaert D, et al. (2007) Auxin-dependent regulation of lateral root positioning in the basal meristem of Arabidopsis. Development 134: 681–690 [DOI] [PubMed] [Google Scholar]
- De Smet I, White PJ, Bengough AG, Dupuy L, Parizot B, Casimiro I, Heidstra R, Laskowski M, Lepetit M, Hochholdinger F, et al. (2012) Analyzing lateral root development: how to move forward. Plant Cell 24: 15–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ditengou FA, Teale WD, Kochersperger P, Flittner KA, Kneuper I, van der Graaff E, Nziengui H, Pinosa F, Li X, Nitschke R, et al. (2008) Mechanical induction of lateral root initiation in Arabidopsis thaliana. Proc Natl Acad Sci USA 105: 18818–18823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolan L, Janmaat K, Willemsen V, Linstead P, Poethig S, Roberts K, Scheres B (1993) Cellular organisation of the Arabidopsis thaliana root. Development 119: 71–84 [DOI] [PubMed] [Google Scholar]
- Dubrovsky JG, Rost TL, Colón-Carmona A, Doerner P (2001) Early primordium morphogenesis during lateral root initiation in Arabidopsis thaliana. Planta 214: 30–36 [DOI] [PubMed] [Google Scholar]
- Dubrovsky JG, Sauer M, Napsucialy-Mendivil S, Ivanchenko MG, Friml J, Shishkova S, Celenza J, Benková E (2008) Auxin acts as a local morphogenetic trigger to specify lateral root founder cells. Proc Natl Acad Sci USA 105: 8790–8794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenbarth DA, Weig AR (2005) Dynamics of aquaporins and water relations during hypocotyl elongation in Ricinus communis L. seedlings. J Exp Bot 56: 1831–1842 [DOI] [PubMed] [Google Scholar]
- Fricke W, Chaumont F (2007) Solute and water relations of growing plant cells. In Verbelen JP, Vissenberg K, eds, The Expanding Cell, Vol 5 Springer, Berlin, pp 7–31 [Google Scholar]
- Fricke W, Peters WS (2002) The biophysics of leaf growth in salt-stressed barley. A study at the cell level. Plant Physiol 129: 374–388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frigerio L, Hinz G, Robinson DG (2008) Multiple vacuoles in plant cells: rule or exception? Traffic 9: 1564–1570 [DOI] [PubMed] [Google Scholar]
- Gao YP, Young L, Bonham-Smith P, Gusta LV (1999) Characterization and expression of plasma and tonoplast membrane aquaporins in primed seed of Brassica napus during germination under stress conditions. Plant Mol Biol 40: 635–644 [DOI] [PubMed] [Google Scholar]
- Gattolin S, Sorieul M, Hunter PR, Khonsari RH, Frigerio L (2009) In vivo imaging of the tonoplast intrinsic protein family in Arabidopsis roots. BMC Plant Biol 9: 133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendreau E, Traas J, Desnos T, Grandjean O, Caboche M, Höfte H (1997) Cellular basis of hypocotyl growth in Arabidopsis thaliana. Plant Physiol 114: 295–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerbeau P, Güçlü J, Ripoche P, Maurel C (1999) Aquaporin Nt-TIPa can account for the high permeability of tobacco cell vacuolar membrane to small neutral solutes. Plant J 18: 577–587 [DOI] [PubMed] [Google Scholar]
- Gomes D, Agasse A, Thiébaud P, Delrot S, Gerós H, Chaumont F (2009) Aquaporins are multifunctional water and solute transporters highly divergent in living organisms. Biochim Biophys Acta 1788: 1213–1228 [DOI] [PubMed] [Google Scholar]
- Hachez C, Moshelion M, Zelazny E, Cavez D, Chaumont F (2006) Localization and quantification of plasma membrane aquaporin expression in maize primary root: a clue to understanding their role as cellular plumbers. Plant Mol Biol 62: 305–323 [DOI] [PubMed] [Google Scholar]
- Heinen RB, Ye Q, Chaumont F (2009) Role of aquaporins in leaf physiology. J Exp Bot 60: 2971–2985 [DOI] [PubMed] [Google Scholar]
- Higuchi T, Suga S, Tsuchiya T, Hisada H, Morishima S, Okada Y, Maeshima M (1998) Molecular cloning, water channel activity and tissue specific expression of two isoforms of radish vacuolar aquaporin. Plant Cell Physiol 39: 905–913 [DOI] [PubMed] [Google Scholar]
- Hunter PR, Craddock CP, Di Benedetto S, Roberts LM, Frigerio L (2007) Fluorescent reporter proteins for the tonoplast and the vacuolar lumen identify a single vacuolar compartment in Arabidopsis cells. Plant Physiol 145: 1371–1382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahn TP, Møller AL, Zeuthen T, Holm LM, Klaerke DA, Mohsin B, Kühlbrandt W, Schjoerring JK (2004) Aquaporin homologues in plants and mammals transport ammonia. FEBS Lett 574: 31–36 [DOI] [PubMed] [Google Scholar]
- Jauh GY, Fischer AM, Grimes HD, Ryan CA Jr, Rogers JC (1998) delta-Tonoplast intrinsic protein defines unique plant vacuole functions. Proc Natl Acad Sci USA 95: 12995–12999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johanson U, Karlsson M, Johansson I, Gustavsson S, Sjövall S, Fraysse L, Weig AR, Kjellbom P (2001) The complete set of genes encoding major intrinsic proteins in Arabidopsis provides a framework for a new nomenclature for major intrinsic proteins in plants. Plant Physiol 126: 1358–1369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kammerloher W, Fischer U, Piechottka GP, Schäffner AR (1994) Water channels in the plant plasma membrane cloned by immunoselection from a mammalian expression system. Plant J 6: 187–199 [DOI] [PubMed] [Google Scholar]
- Kumpf RP, Shi CL, Larrieu A, Stø IM, Butenko MA, Péret B, Riiser ES, Bennett MJ, Aalen RB (2013) Floral organ abscission peptide IDA and its HAE/HSL2 receptors control cell separation during lateral root emergence. Proc Natl Acad Sci USA 110: 5235–5240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskowski M, Biller S, Stanley K, Kajstura T, Prusty R (2006) Expression profiling of auxin-treated Arabidopsis roots: toward a molecular analysis of lateral root emergence. Plant Cell Physiol 47: 788–792 [DOI] [PubMed] [Google Scholar]
- Liu LH, Ludewig U, Gassert B, Frommer WB, von Wirén N (2003) Urea transport by nitrogen-regulated tonoplast intrinsic proteins in Arabidopsis. Plant Physiol 133: 1220–1228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas M, Godin C, Jay-Allemand C, Laplaze L (2008) Auxin fluxes in the root apex co-regulate gravitropism and lateral root initiation. J Exp Bot 59: 55–66 [DOI] [PubMed] [Google Scholar]
- Lucas M, Kenobi K, von Wangenheim D, Vobeta U, Swarup K, De Smet I, Van Damme D, Lawrence T, Peret B, Moscardi E, et al. (2013) Lateral root morphogenesis is dependent on the mechanical properties of the overlaying tissues. Proc Natl Acad Sci USA 110: 5229–5234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludevid D, Höfte H, Himelblau E, Chrispeels MJ (1992) The expression pattern of the tonoplast intrinsic protein gamma-TIP in Arabidopsis thaliana is correlated with cell enlargement. Plant Physiol 100: 1633–1639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macgregor DR, Deak KI, Ingram PA, Malamy JE (2008) Root system architecture in Arabidopsis grown in culture is regulated by sucrose uptake in the aerial tissues. Plant Cell 20: 2643–2660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malamy JE, Benfey PN (1997) Organization and cell differentiation in lateral roots of Arabidopsis thaliana. Development 124: 33–44 [DOI] [PubMed] [Google Scholar]
- Maurel C. (1997) Aquaporins and water permeability of plant membranes. Annu Rev Plant Physiol Plant Mol Biol 48: 399–429 [DOI] [PubMed] [Google Scholar]
- Maurel C, Kado RT, Guern J, Chrispeels MJ (1995) Phosphorylation regulates the water channel activity of the seed-specific aquaporin alpha-TIP. EMBO J 14: 3028–3035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurel C, Reizer J, Schroeder JI, Chrispeels MJ (1993) The vacuolar membrane protein gamma-TIP creates water specific channels in Xenopus oocytes. EMBO J 12: 2241–2247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurel C, Tacnet F, Güclü J, Guern J, Ripoche P (1997) Purified vesicles of tobacco cell vacuolar and plasma membranes exhibit dramatically different water permeability and water channel activity. Proc Natl Acad Sci USA 94: 7103–7108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurel C, Verdoucq L, Luu DT, Santoni V (2008) Plant aquaporins: membrane channels with multiple integrated functions. Annu Rev Plant Biol 59: 595–624 [DOI] [PubMed] [Google Scholar]
- Morillon R, Lassalles JP (1999) Osmotic water permeability of isolated vacuoles. Planta 210: 80–84 [DOI] [PubMed] [Google Scholar]
- Nakagawa T, Kurose T, Hino T, Tanaka K, Kawamukai M, Niwa Y, Toyooka K, Matsuoka K, Jinbo T, Kimura T (2007) Development of series of gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J Biosci Bioeng 104: 34–41 [DOI] [PubMed] [Google Scholar]
- Niemietz CM, Tyerman SD (1997) Characterization of water channels in wheat root membrane vesicles. Plant Physiol 115: 561–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nour-Eldin HH, Hansen BG, Nørholm MH, Jensen JK, Halkier BA (2006) Advancing uracil-excision based cloning towards an ideal technique for cloning PCR fragments. Nucleic Acids Res 34: e122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okubo-Kurihara E, Sano T, Higaki T, Kutsuna N, Hasezawa S (2009) Acceleration of vacuolar regeneration and cell growth by overexpression of an aquaporin NtTIP1;1 in tobacco BY-2 cells. Plant Cell Physiol 50: 151–160 [DOI] [PubMed] [Google Scholar]
- Péret B, De Rybel B, Casimiro I, Benková E, Swarup R, Laplaze L, Beeckman T, Bennett MJ (2009) Arabidopsis lateral root development: an emerging story. Trends Plant Sci 14: 399–408 [DOI] [PubMed] [Google Scholar]
- Péret B, Li G, Zhao J, Band LR, Voß U, Postaire O, Luu DT, Da Ines O, Casimiro I, Lucas M, et al. (2012) Auxin regulates aquaporin function to facilitate lateral root emergence. Nat Cell Biol 14: 991–998 [DOI] [PubMed] [Google Scholar]
- Petricka JJ, Winter CM, Benfey PN (2012) Control of Arabidopsis root development. Annu Rev Plant Biol 63: 563–590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raz V, Koornneef M (2001) Cell division activity during apical hook development. Plant Physiol 125: 219–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reisen D, Leborgne-Castel N, Ozalp C, Chaumont F, Marty F (2003) Expression of a cauliflower tonoplast aquaporin tagged with GFP in tobacco suspension cells correlates with an increase in cell size. Plant Mol Biol 52: 387–400 [DOI] [PubMed] [Google Scholar]
- Richter GL, Monshausen GB, Krol A, Gilroy S (2009) Mechanical stimuli modulate lateral root organogenesis. Plant Physiol 151: 1855–1866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schünmann PHD, Ougham HJ (1996) Identification of three cDNA clones expressed in the leaf extension zone and with altered patterns of expression in the slender mutant of barley: a tonoplast intrinsic protein, a putative structural protein and protochlorophyllide oxidoreductase. Plant Mol Biol 31: 529–537 [DOI] [PubMed] [Google Scholar]
- Schüssler MD, Alexandersson E, Bienert GP, Kichey T, Laursen KH, Johanson U, Kjellbom P, Schjoerring JK, Jahn TP (2008) The effects of the loss of TIP1;1 and TIP1;2 aquaporins in Arabidopsis thaliana. Plant J 56: 756–767 [DOI] [PubMed] [Google Scholar]
- Soto G, Alleva K, Mazzella MA, Amodeo G, Muschietti JP (2008) AtTIP1;3 and AtTIP5;1, the only highly expressed Arabidopsis pollen-specific aquaporins, transport water and urea. FEBS Lett 582: 4077–4082 [DOI] [PubMed] [Google Scholar]
- Suga S, Imagawa S, Maeshima M (2001) Specificity of the accumulation of mRNAs and proteins of the plasma membrane and tonoplast aquaporins in radish organs. Planta 212: 294–304 [DOI] [PubMed] [Google Scholar]
- Swarup K, Benková E, Swarup R, Casimiro I, Péret B, Yang Y, Parry G, Nielsen E, De Smet I, Vanneste S, et al. (2008) The auxin influx carrier LAX3 promotes lateral root emergence. Nat Cell Biol 10: 946–954 [DOI] [PubMed] [Google Scholar]
- Tissier AF, Marillonnet S, Klimyuk V, Patel K, Torres MA, Murphy G, Jones JD (1999) Multiple independent defective suppressor-mutator transposon insertions in Arabidopsis: a tool for functional genomics. Plant Cell 11: 1841–1852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uehlein N, Lovisolo C, Siefritz F, Kaldenhoff R (2003) The tobacco aquaporin NtAQP1 is a membrane CO2 pore with physiological functions. Nature 425: 734–737 [DOI] [PubMed] [Google Scholar]
- Vander Willigen C, Postaire O, Tournaire-Roux C, Boursiac Y, Maurel C (2006) Expression and inhibition of aquaporins in germinating Arabidopsis seeds. Plant Cell Physiol 47: 1241–1250 [DOI] [PubMed] [Google Scholar]
- Weig A, Deswarte C, Chrispeels MJ (1997) The major intrinsic protein family of Arabidopsis has 23 members that form three distinct groups with functional aquaporins in each group. Plant Physiol 114: 1347–1357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wudick MM, Luu DT, Tournaire-Roux C, Sakamoto W, Maurel C (2014) Vegetative and sperm cell-specific aquaporins of Arabidopsis highlight the vacuolar equipment of pollen and contribute to plant reproduction. Plant Physiol 164: 1697–1706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zar JH. (2010) Biostatistical Analysis, Ed 5. Pearson Prentice-Hall, Upper Saddle River, NJ [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.