A type 1 protein phosphatase, TOPP4, participates in the phyB signaling pathway by dephosphorylating PIF5 and controlling the stability of PIF5 in seedlings.
Abstract
In plants, photoreceptors transfer light signals to phytochrome-interacting factors (PIFs), inducing the rapid phosphorylation and degradation of PIFs to promote photomorphogenesis. However, the phosphatase responsible for PIF dephosphorylation remains unknown. In this study, we identified a type 1 protein phosphatase, TOPP4, that is essential for PIF5 protein stability in Arabidopsis (Arabidopsis thaliana). Compared with the wild type, the dominant-negative mutant, topp4-1, displayed reduced hypocotyl length and larger apical hook and cotyledon opening angle under red light. Overexpression of topp4-1 in the wild type led to defects that were similar to those in the topp4-1 mutant. Red light induced phytochrome B (phyB)-dependent TOPP4 expression in hypocotyls. The topp4-1 mutation weakened the closed cotyledon angle of phyB-9 and phyA-211 phyB-9, while overexpression of TOPP4 significantly repressed the short hypocotyls of phyB-green fluorescent protein seedlings, indicating that TOPP4 and phyB function in an antagonistic way during photomorphogenesis. Protein interaction assays and phosphorylation studies demonstrate that TOPP4 interacts directly with PIF5 and dephosphorylates it. Furthermore, TOPP4 inhibits the red light-induced ubiquitination and degradation of PIF5. These findings demonstrate that dephosphorylation of PIF5 by TOPP4 inhibits its ubiquitin-mediated degradation during photomorphogenesis. These data outline a novel phytochrome signaling mechanism by which TOPP4-mediated dephosphorylation of PIF5 attenuates phytochrome-dependent light responses.
Light is a key environmental cue that controls seed germination, seedling deetiolation, shade avoidance, leaf expansion, circadian rhythms, and flowering and, thus, integrates the plant response throughout its life cycle (Deng and Quail, 1999; Wang and Deng, 2003; Jiao et al., 2007). In the dark, Arabidopsis (Arabidopsis thaliana) seedlings develop a long embryonic stem and a pair of closed embryonic leaves (called cotyledons) that are protected by an apical hook in the primary stem during their emergence from the soil. This developmental program in the absence of light is called skotomorphogenesis. In contrast, a different growth program known as photomorphogenesis is initiated when Arabidopsis seedlings are exposed to light. This process includes the inhibition of hypocotyl elongation, green cotyledons, and leaf expansion (Franklin and Quail, 2010). These morphological differences between dark- and light-grown seedlings are caused by distinctive gene expression (Tepperman et al., 2006; Jiao et al., 2007; Hu et al., 2009; Leivar et al., 2009). Photomorphogenesis is triggered by photoreceptors that perceive different colors of light (Kami et al., 2010). Phytochrome A (phyA) and phyB are the most prominent phytochromes that regulate plant development and growth (Franklin and Quail, 2010; Kami et al., 2010). PhyA plays a dominant role in seedling deetiolation during early red light and continuous far-red light exposure (Sharrock and Clack, 2002; Tepperman et al., 2006), while phyB plays a major role in hypocotyl inhibition that is mediated by continuous red light (Somers et al., 1991; Reed et al., 1993). The ability of the photoreceptor to perceive these signals is achieved by reversible switching between two relatively stable forms, Pr and Pfr. In dark-germinated seedlings, phytochrome predominantly exists in the cytoplasm in the inactive Pr form. Following exposure to red light, Pr converts into the active Pfr form and translocates to the nucleus (Quail, 2002; Tu and Lagarias, 2005). As the subcellular localization of phytochrome changes, early subnuclear speckles, called photobodies, are rapidly formed (Sakamoto and Nagatani, 1996; Kircher et al., 2002; Chen and Chory, 2011). These different patterns of action provide the molecular basis for a variety of responses to different types of light in seedlings that produce different morphologies.
Modification by phosphorylation plays fundamental roles in the regulation of receptor molecules. Phosphorylation of oat (Avena sativa) phyA inhibits its interaction with signal-transducing proteins (Choi et al., 1999; Kim et al., 2004), and phosphorylation of phyB attenuates light signaling via accelerated dark reversion (Medzihradszky et al., 2013). An Arabidopsis Ser/Thr-specific protein phosphatase 2A (FyPP) interacts with phyA, and recombinant FyPP efficiently dephosphorylates oat phyA (Kim et al., 2002). A type 5 protein phosphatase specifically dephosphorylates biologically active Pfr phytochromes and enhances phytochrome-mediated photoresponses (Ryu et al., 2005). Phytochrome-associated protein phosphatase type 2C effectively dephosphorylates phytochromes and indirectly mediates the in vitro dephosphorylation of PHYTOCHROME-INTERACTING FACTOR3 (PIF3; Phee et al., 2008). Therefore, phosphorylation and dephosphorylation are considered to be important mechanisms for modulating phytochrome signaling (Nito et al., 2013).
The regulation of plant development by inducing PIF degradation is a dominant mechanism of action by phytochrome family proteins (Sakamoto and Nagatani, 1996; Yamaguchi et al., 1999; Huq et al., 2003; Nagatani, 2004; Leivar and Quail, 2011). Preferentially, PIFs interact directly with the active forms of phyA and phyB (Leivar and Quail, 2011). PIF proteins interact with phyB via the active phyB-binding motif, a short conserved motif located at their N-terminal domain (Khanna et al., 2004). Within this group of transcription factors, PIF3 is a basic member that strongly interacts with the active Pfr form of phytochrome (Ni et al., 1998, 1999). Following the discovery of PIF3, more members of subfamily 15 of the Arabidopsis basic helix-loop-helix family have been identified (Bailey et al., 2003; Toledo-Ortiz et al., 2003; Khanna et al., 2004). These members include PIF1/PHYTOCHROME-INTERACTING FACTOR3-LIKE5 (PIL5; Huq et al., 2004; Oh et al., 2004, 2006, 2007; Shen et al., 2005), PIF4 (Huq and Quail, 2002), PIF5/PIL6 (Makino et al., 2002; Khanna et al., 2004), PIF6/PIL2 (Salter et al., 2003; Khanna et al., 2004), PIF7 (Leivar et al., 2008a), and PIF8 (Leivar and Quail, 2011).
Recent studies have demonstrated that PIFs directly regulate the expression of thousands of genes and that they act as a signaling hub to initiate photomorphogenesis (Zhang et al., 2013). The phytochrome family plays a central role in inducing the switch from skotomorphogenic to photomorphogenic development, which is achieved primarily through the rapid phytochrome-induced phosphorylation and degradation of most PIFs (Bauer et al., 2004; Shen et al., 2005, 2007; Al-Sady et al., 2006; Oh et al., 2006; Nozue et al., 2007; Lorrain et al., 2008). For instance, red light induces the rapid phosphorylation of PIF5, and a mobility shift in PIF5 is observed prior to its degradation, but this light-induced mobility shift can be restored by alkaline phosphatase treatment (Shen et al., 2007). Furthermore, light also induces the rapid phosphorylation of PIF1, PIF3, and PIF4 prior to their degradation (Al-Sady et al., 2006; Lorrain et al., 2008; Shen et al., 2008). PIF3 interacts with phyA and phyB before their degradation in the subnuclear domains, photobodies (Bauer et al., 2004; Al-Sady et al., 2006). HEMERA is a novel phytochrome signaling element that mediates the formation of photobodies and regulates the degradation of PIFs (Galvão et al., 2012). However, the molecular nature of phytochrome-induced phosphorylation is still unknown. Data from the Quail laboratory show that light-induced phosphorylation of PIF3 triggers its rapid degradation, and PIF3 phosphorylation is required for the rapid ubiquitin modification in light. Phosphorylation is indispensable for the negative feedback modulation of photoactivated phyB levels under prolonged light exposure (Ni et al., 2013). A recent study demonstrated that PIF degradation requires Light-Response Bric-a-Brack/Tramtrack/Broad (LRB) E3 ubiquitin ligases that promote PIF polyubiquitination and its subsequent degradation (Ni et al., 2014). Simultaneously, light-induced phosphorylation of PIF3 promotes the recruitment of LRB E3 ubiquitin ligases to the PIF3-phyB complex (Ni et al., 2014). In the dark, PIF protein levels are positively regulated by DEETIOLATED1, which is a repressor of photomorphogenesis (Dong et al., 2014).
Some kinases are also involved in the phosphorylation of PIFs. Casein kinase II (CK2)-mediated phosphorylation of PIF1 is necessary for its light-induced degradation, although how CK2-mediated phosphorylation affects the light-induced degradation of PIF1 remains unclear (Bu et al., 2011). BRASSINOSTEROID-INSENSITIVE2 (BIN2), a GLYCOGEN SYNTHASE KINASE3 (GSK3)-like kinase reported previously to regulate brassinosteroid signaling, can phosphorylate PIF4. Mutation of the BIN2 phosphorylation consensus sequence leads to strong PIF4 stabilization (Bernardo-García et al., 2014). In conclusion, extensive studies on PIFs have aided in defining the signaling mechanisms involved in mediating PIF degradation. As phosphorylation is a reversible modification, some phosphatases might specifically dephosphorylate PIF. However, until now, the identity of such phosphatases has remained unknown.
PROTEIN PHOSPHATASE1 (PP1), a Ser/Thr protein phosphatase that comprises a catalytic and regulatory subunit, modulates a wide range of cellular processes, such as metabolism, muscle contraction, transcription, cell division, and protein synthesis, in animal systems (Bollen, 2001; Cohen, 2002; Ceulemans and Bollen, 2004). PP1 is expressed mainly in eukaryotic cells (Shi, 2009). Previous studies have demonstrated that PP1 in Vicia faba functions downstream of phototropin1 and phototropin2 in the light signaling pathway (Takemiya et al., 2006). In addition, PP1 also positively regulates blue light signaling for stomatal opening (Takemiya et al., 2006, 2013). Our previous studies have demonstrated that a type 1 protein phosphatase, TOPP4, a member of the PP1 family of proteins found in Arabidopsis, is involved in DELLA-mediated GA signaling pathways and mediates PIN-FORMED1 (PIN1) polarity and trafficking (Qin et al., 2014; Guo et al., 2015). In this study, we demonstrate that TOPP4 is involved in the phyB signaling pathway and in the regulation of hypocotyl elongation and the cotyledon angle of Arabidopsis seedlings. Genetic analyses indicate that TOPP4 acts upstream of PIF5. Further biochemical studies demonstrate that TOPP4 dephosphorylates PIF5 and subsequently regulates its ubiquitination and stability. Our findings reveal that TOPP4-mediated PIF5 dephosphorylation plays a significant role in modulating the flux of PIF-regulated light signaling.
RESULTS
Mutant topp4-1 Shows Shorter Hypocotyl and Larger Apical Hook Angle, While TOPP4 Overexpression Seedlings Display Longer Hypocotyls under Red Light
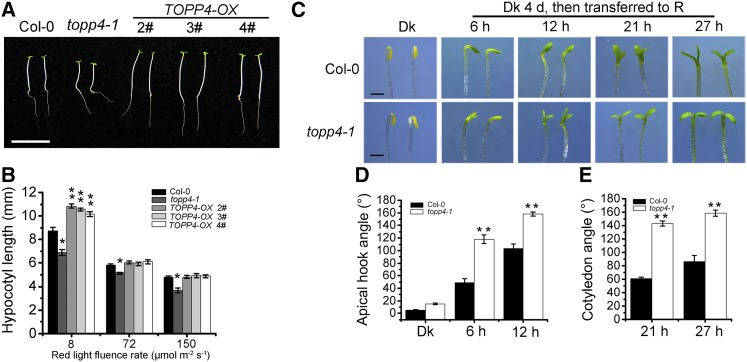
We reported previously that TOPP4 is highly expressed in young seedlings, especially in cotyledons (Qin et al., 2014). Here, we examined the tissue-specific expression of TOPP4 under different light conditions. In dark-grown seedlings, TOPP4 was highly expressed in the apical hook, but expression in the hypocotyl was very weak. However, under white, blue, red, and far-red lights, intense GUS staining was observed in hypocotyls and cotyledons (Supplemental Fig. S1A). Therefore, TOPP4 might play an important role during hypocotyl and cotyledon development under light conditions. To verify this hypothesis, photoresponses of the topp4-1 mutant and TOPP4 -overexpressing lines were assessed under various light conditions (dark, white, red, blue, and far-red light). TOPP4-overexpressing seedlings were generated by overexpressing the FLAG epitope-tagged full-length TOPP4 driven by the constitutive 35S promoter of cauliflower mosaic virus, TOPP4-FLAG #2, which showed the highest expression level. TOPP4 was selected for further analysis and renamed as TOPP4-OX (Supplemental Fig. S2). The topp4-1 mutant displayed a short-hypocotyl phenotype, whereas hypocotyl lengths of TOPP4-OX seedlings were similar to those in the wild type under continuous red light (Supplemental Fig. S1, B and D). The hypocotyls of topp4-1 and TOPP4-OX were almost indistinguishable under continuous white, blue, and far-red light irradiation as well as under dark conditions (Supplemental Fig. S1, B–D). To further confirm the phenotype of topp4-1 and TOPP4-OX under red light, we investigated the hypocotyl lengths of these lines under different red light intensities. Compared with the wild type, hypocotyl elongation in topp4-1 was suppressed under red light of various fluence rates, while TOPP4-OX lines exhibited obviously longer hypocotyls only under 8 μmol m−2 s−1 red light (Fig. 1, A and B). Given that topp4-1 displayed a defect in hypocotyl growth only under red light, we further analyzed the phenotypes of topp4-1 and TOPP4-OX under red light.
Figure 1.
The topp4-1 mutant exhibits abnormal hypocotyl elongation, hook opening, and cotyledon unfolding, and TOPP4-OX displays longer hypocotyl length under red light. A, Phenotypes of 5-d-old TOPP4-OX 2#, 3#, and 4# seedlings under 8 μmol m−2 s−1 red light for 5 d. Bar = 1 cm. B, Quantitative analysis of the hypocotyl lengths of approximately 50 seedlings from each plant line under 8, 72, and 150 μmol m−2 s−1 red light for 5 d. C, Phenotypes of the apical hook and cotyledon angle of wild-type Columbia-0 (Col-0) and the topp4-1 mutant. Seedlings were grown in the dark (Dk) for 4 d and subsequently transferred to red light (R) for different times as indicated. Bars = 1 mm. D and E, Quantification of the apical hook angle and cotyledon separation of seedlings under the conditions described in C. In B, D, and E, error bars indicate se (n = 50). Statistical significance was determined by Tukey’s lsd test between the wild type and topp4-1 (*, P < 0.05 and **, P < 0.01).
topp4-1 seedlings grown in the dark had a slightly larger apical hook compared with wild-type seedlings (Fig. 1, C and D). After treatment with red light for 6 and 12 h, an exaggeration in the apical hook curvature was observed in topp4-1 seedlings (Fig. 1, C and D). In seedlings grown under red light for 21 h, the cotyledon opening angle in topp4-1 was 141°, which was more than 2-fold greater than the angle in wild-type seedlings. Similar results were observed when seedlings were treated with red light for 27 h (Fig. 1, C and E). However, the apical hook and cotyledon opening angle of three TOPP4-OX lines were similar to those of the wild-type seedlings (Supplemental Fig. S3, A and B). These results indicate that mutation of TOPP4 also affects the apical hook angle and cotyledon unfolding under red light.
The topp4-1 Phenotype Is Rescued by Overexpression of TOPP4
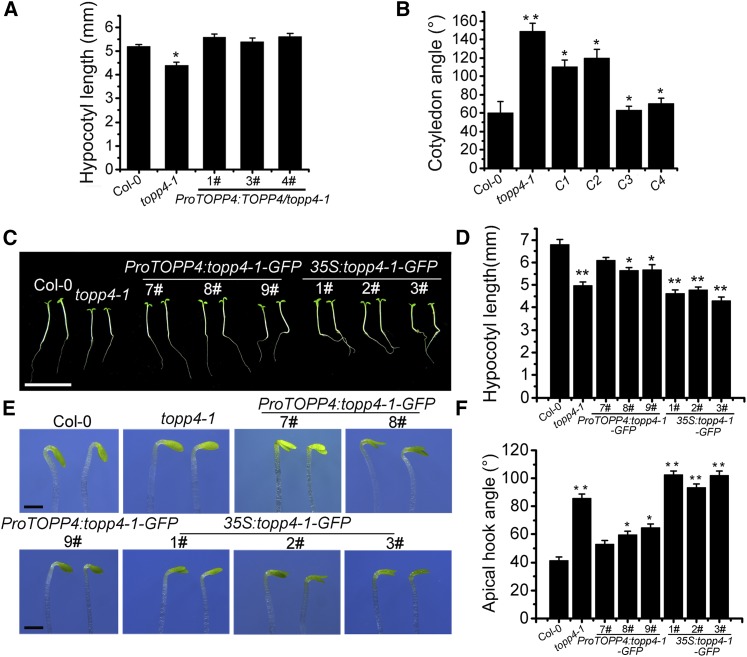
To determine whether TOPP4 was responsible for the defects in topp4-1 seedlings under red light, ProTOPP4:TOPP4/topp4-1 seedlings were analyzed. ProTOPP4:TOPP4 completely rescued the hypocotyl phenotype of topp4-1 (Fig. 2A; Supplemental Fig. S4A), but the cotyledon angle was only partly recovered in ProTOPP4:TOPP4/topp4-1 seedlings (Fig. 2B; Supplemental Fig. S4B). Therefore, the cotyledon angle of 35S:TOPP4/topp4-1 seedlings was analyzed, and the cotyledon angle was completely restored to that seen in wild-type seedlings (Fig. 2B; Supplemental Fig. S4B). The expression of TOPP4 in 35S:TOPP4/topp4-1 6# and 9# was confirmed by quantitative real-time (qRT)-PCR (Supplemental Fig. S5A). These results show that the topp4-1 mutation caused the short-hypocotyl and exaggerated cotyledon angle phenotypes of topp4-1 seedlings. Taken together, these data indicate that TOPP4 is involved in the regulation of Arabidopsis photomorphogenesis under red light.
Figure 2.
Overexpression of TOPP4 in the topp4-1 mutant rescues its cotyledon unfolding defects, and overexpression of topp4-1 in the wild type mimics the defective phenotypes of topp4-1 in Arabidopsis seedlings. A, Quantitative analysis of hypocotyl lengths of ProTOPP4:TOPP4/topp4-1 transgenic seedlings grown under red light for 5 d. B, Quantification of the cotyledon angle of seedlings grown in the dark for 4 d and subsequently transferred to red light for 21 h. C1 and C2 represent ProTOPP4:TOPP4/topp4-1 1# and 3#; C3 and C4 represent 35S:TOPP4/topp4-1 6# and 9#. C, Photographs showing the hypocotyl phenotypes of the indicated genotypes grown under red light for 5 d. Bar = 1 cm. D, Quantitative analysis of hypocotyl lengths of the indicated genotypes. E, Photographs showing the hook angle phenotypes of the indicated genotypes grown in the dark for 3 d and subsequently transferred to red light for 6 h. Bars = 1 mm. F, Quantification of hook opening of the seedlings corresponding to E. In A, B, D, and F, error bars indicate se (n = 50). Statistical significance was determined by Tukey’s lsd test (*, P < 0.05 and **, P < 0.01).
To further investigate the function of TOPP4, we analyzed the hypocotyl length and cotyledon opening angle in two transfer DNA (T-DNA) lines of TOPP4, N466328 and SALK_090980, in which T-DNAs are not inserted in the coding region. The transcript level of TOPP4 in the N466328 line was reduced to 40%, while the insertion did not alter the transcription level in the SALK_090980 line (Qin et al., 2014). Therefore, these T-DNA insertion mutants are not null mutants. The hypocotyl length observed in these two lines were similar to those observed in wild-type seedlings grown under red light (Supplemental Fig. S4, C and D). The seedlings of these two lines exhibited slightly larger cotyledon angles than the wild-type seedlings under red light, but the difference was not statistically significant (Supplemental Fig. S4, B and E). Thus, these lines did not show a major visible phenotype, possibly due to high TOPP4 transcript levels or to functional redundancy among TOPP genes.
Overexpression of topp4-1 in Wild-Type Plants Mimics the Defect Phenotypes of the topp4-1 Mutant
We have shown previously that that topp4-1 protein has a dominant-negative effect on plant development and pavement cell morphogenesis (Qin et al., 2014; Guo et al., 2015). To investigate whether the short-hypocotyl and larger apical hook angle phenotypes of topp4-1 under red light were due to the dominant-negative effect of the topp4-1 protein, the phenotypes of ProTOPP4:topp4-1-GFP and 35S:topp4-1-GFP transgenic lines grown under red light were analyzed. qRT-PCR analysis confirmed that the expression of topp4-1 increased in 35S:topp4-1-GFP transgenic lines 1#, 2#, and 3# (Supplemental Fig. S5B). The three ProTOPP4:topp4-1-GFP lines only partly displayed the defects observed in topp4-1, whereas all three 35S:topp4-1-GFP lines displayed phenotypes that were similar to topp4-1. It is worth noting that 35S:topp4-1-GFP lines showed more notable exaggerated apical hook compared with the topp4-1 mutant (Fig. 2, E and F). These results indicate that topp4-1 affects photomorphogenesis in a dominant-negative manner.
TOPP4 Stability Is Not Affected by Red Light, and the Defects in topp4-1 Seedlings under Red Light Are Not Due to Changes in DELLA Proteins
Light regulates the stability of many proteins via posttranslational modification. We examined the localization of TOPP4 under red light. TOPP4-GFP transgenic plants grown in the dark displayed an intense fluorescent signal mainly in the nucleus and the membrane of hypocotyl cells. The localization and fluorescence intensity of TOPP4-GFP were not influenced by exposure to red light for 1 h (Supplemental Fig. S6A). The levels of TOPP4 protein under continuous dark, red, and far-red light conditions were similar (Supplemental Fig. S6B). Moreover, when dark-grown TOPP4-GFP seedlings were treated with a saturating red light, TOPP4 protein levels did not show any changes (Supplemental Fig. S6C). Similar results were observed for 10-, 30-, and 60-min continuous red light irradiation (Supplemental Fig. S6D). These results suggest that neither TOPP4 protein localization nor its accumulation is sensitive to red light. In addition, to determine if the defects observed in hypocotyls and cotyledon angles of topp4-1 were due to the lack of TOPP4 protein, we analyzed TOPP4 levels in topp4-1 and the wild type under dark and red light conditions and observed that the mutation of TOPP4 did not alter TOPP4 protein level under continuous dark and red light (Supplemental Fig. S7A).
Previous studies have reported that light promotes the accumulation of DELLA proteins and that DELLA proteins restrain plant growth largely through the regulation of the light-responsive transcription factors, PIFs (Alabadí et al., 2004; Achard et al., 2007). We previously reported that the DELLA proteins RGA (for repressor of ga1-3) and GAI (for GA insensitive) accumulate in the topp4-1 mutant, whereas their levels decrease in TOPP4-overexpressing plants (Qin et al., 2014). However, similar levels of these two proteins were observed in 5-d-old topp4-1, TOPP4-OX, and wild-type seedlings under red light (Supplemental Fig. S7, B and C). Similar results were observed for RGA protein levels in RGA-GFP/topp4-1 and RGA-GFP/TOPP4-OX seedlings (Supplemental Fig. S7D), suggesting that the hypocotyl and cotyledon angle defects in topp4-1 and TOPP4-OX seedlings under red light are not due to changes in DELLA protein levels.
TOPP4 Is Involved in the phyB Signaling Pathway
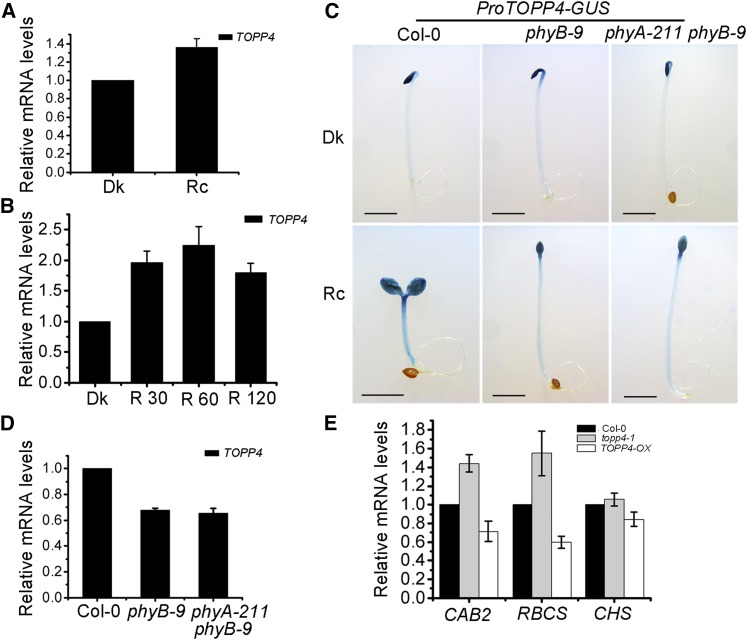
Our results have demonstrated that red light induces TOPP4 expression in hypocotyls. Furthermore, the expression of TOPP4 was 1.3-fold higher in seedlings grown under red light than in dark-grown seedlings (Fig. 3A). When dark-grown seedlings were subjected to 30-, 60-, or 120-min red light irradiation, the expression of TOPP4 was obviously up-regulated (Fig. 3B). These results indicate that red light induces TOPP4 expression in Arabidopsis seedlings. PhyB is the major photoreceptor involved in mediating the response to red light (Franklin and Quail, 2010). To investigate whether red light-induced TOPP4 expression in hypocotyls depends on phytochromes, we transformed ProTOPP4-GUS into phyB-9 and phyA-211 phyB-9 lines. As a result, the hypocotyl-specific TOPP4 expression in phyB-9 and phyA-211 phyB-9 seedlings was lower than that in the wild type under continuous red light (Fig. 3C), suggesting that the red light-induced expression of TOPP4 in hypocotyls is phyB dependent. We also performed qRT-PCR to analyze TOPP4 expression in phyB-9 and phyA-211 phyB-9 mutants. Compared with the wild type, TOPP4 expression was down-regulated in these mutant lines under continuous red light (Fig. 3D), indicating that phyB affects TOPP4 expression in whole seedlings under red light.
Figure 3.
Red light regulates TOPP4 gene expression dependent on phyB. A and B, Real-time analysis of TOPP4 expression in 5-d-old wild-type seedlings under dark (Dk) and continuous red light (Rc) conditions (A) or in 5-d-old dark-grown wild-type seedlings after being exposed for 30, 60, and 120 min (R 30, R 60, and R 120) of red light (B). C, Histochemical assay of ProTOPP4-GUS expression in Col-0, phyB-9, and phyA-211 phyB-9 grown for 4 d under dark or continuous red light. Bars = 2 mm. D, Real-time analysis of TOPP4 expression in 5-d-old Col-0, phyB-9, and phyA-211 phyB-9 seedlings under continuous red light. E, Real-time analysis of CAB2, RBCS, and CHS expression in Col-0, topp4-1, and TOPP4-OX seedlings. Seedlings were grown under 8 μmol m−2 s−1 red light for 5 d. In A, B, D, and E, PP2A was used as a reference gene.
Light controls plant growth and development through an important mechanism that regulates the expression of specific genes (Tobin and Kehoe, 1994), such as CHLOROPHYLL A/B BINDING PROTEIN (CAB), RUBISCO SMALL SUBUNIT (RBCS), and CHALCONE SYNTHASE (CHS; Sun and Tobin, 1990; Gilmartin et al., 1992). To provide further molecular evidence for TOPP4 regulating photomorphogenesis, the transcript levels of the three light-regulated genes CAB2, RBCS, and CHS were analyzed. The levels of CAB2 and RBCS were clearly up-regulated in the topp4-1 mutant but reduced in TOPP4 overexpression lines under red light (Fig. 3E). However, no obvious change was observed in CHS gene expression in topp4-1 and TOPP4-OX seedlings (Fig. 3E). These data indicate that TOPP4 modulates the expression of light-regulated genes under red light. Thus, TOPP4 is involved in the phyB signaling pathway.
TOPP4 and phyB Regulate Photomorphogenesis in an Antagonistic Way
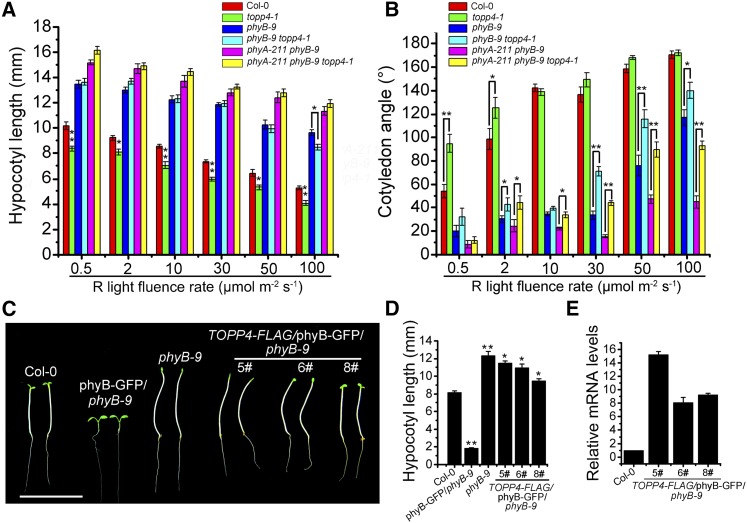
To examine the genetic interaction between phyB and TOPP4, the topp4-1 mutant was introduced into the phyB-9 single mutant and the phyA-211 phyB-9 double mutant. Phenotypic analysis revealed that phyB-9 topp4-1 and the triple mutant phyA-211 phyB-9 topp4-1 exhibited long hypocotyls similar to those observed in phyB-9 and phyA-211 phyB-9 under 0.5, 2, 10, 30, and 50 μmol m−2 s−1 red light. However, when grown under 100 μmol m−2 s−1 red light, the hypocotyl length of phyB-9 topp4-1 was significantly shorter than that of phyB-9 (Fig. 4A; Supplemental Fig. S8A), indicating that the topp4-1 mutation could partly recover the abnormal hypocotyl length of phyB-9. The topp4-1 mutation could also recover the closed cotyledon angle of phyB-9 and phyA-211 phyB-9 to some extent, because phyB-9 topp4-1 and phyA-211 phyB-9 topp4-1 mutants showed greater cotyledon opening angles compared with those of phyB-9 and phyA-211 phyB-9 under continuous 2, 30, 50, and 100 μmol m−2 s−1 red light (Fig. 4B; Supplemental Fig. S8B). In addition, when dark-grown seedlings were exposed to red light for 32 h, the cotyledon angle in phyB-9 topp4-1 was wider than that observed in phyB-9 seedlings (Supplemental Fig. S8, C and D). These results suggest that the suppression of hypocotyl elongation and the promotion of cotyledon unfolding by topp4-1 are dependent on the function of phyA and phyB during photomorphogenesis.
Figure 4.
The topp4-1 mutation partly recovers the closed cotyledon angle of phyB-9 and phyA-211 phyB-9, and overexpression of TOPP4 in the phyB-GFP background significantly weakens the short-hypocotyl phenotype of phyB-GFP. A and B, Quantitative analysis of the hypocotyl length and cotyledon opening angle of 5-d-old seedlings grown under various red (R) light fluence rates. C, Hypocotyl lengths of 5-d-old seedlings grown under 8 μmol m−2 s−1 red light. Bar = 1 cm. D, Quantitative analysis of hypocotyl lengths of the indicated genotypes. E, Real-time analysis of the expression levels of TOPP4 in different TOPP4-FLAG/phyB-GFP phyB-9 lines. In A, B, D, and E, error bars represent se (n = 50). Statistical significance was determined by Tukey’s lsd test (*, P < 0.05 and **, P < 0.01).
To further study the genetic interaction between TOPP4 and phyB, TOPP4-FLAG was transformed into phyB-GFP/phyB-9 seedlings that were constructed by overexpressing PHYB in the phyB-9 background (Medzihradszky et al., 2013). The expression of TOPP4 in TOPP4-FLAG/phyB-GFP/phyB-9 5#, 6#, and 8# was verified by qRT-PCR (Fig. 4E). Compared with phyB-GFP/phyB-9 seedlings that had severely reduced hypocotyls, TOPP4-FLAG/phyB-GFP/phyB-9 lines 5# and 6# exhibited elongated hypocotyls similar to phyB-9 seedlings under red light, while line 8# displayed longer hypocotyls than the wild type but shorter than the phyB-9 mutant (Fig. 4, C and D). These results indicate that overexpression of TOPP4 partially suppresses the severe short-hypocotyl phenotype of phyB-GFP/phyB-9 seedlings. TOPP4 also significantly reduced the high levels of expression of the light-regulated genes CAB2 and RBCS in phyB-GFP/phyB-9 (Fig. 5A). In addition, the rosette leaves in lines 5# and 6# were larger than those in phyB-GFP/phyB-9 seedlings, and line 8# had smaller leaves than lines 5# and 6# (Supplemental Fig. S9A), indicating that the overexpression of TOPP4 weakens the strong response of phyB-GFP/phyB-9 plants to light. Taken together, these results suggest that TOPP4 and phyB act antagonistically to regulate light-dependent growth in Arabidopsis.
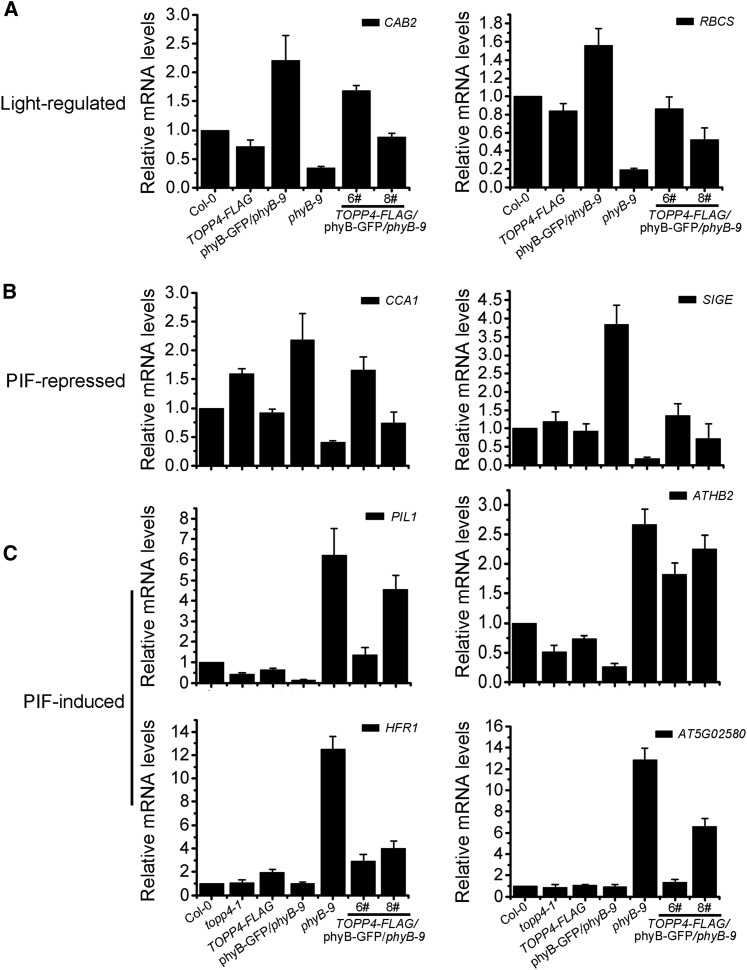
Figure 5.
Expression levels of light-regulated genes and PIF-regulated genes in TOPP4-FLAG/phyB-GFP/phyB-9. A, Real-time analysis of the expression of the light-regulated genes CAB2 and RBCS in different seedlings. B and C, Real-time analysis of the expression of the PIF-repressed genes CCA1 and SIGE (B) and the PIF-induced genes PIL1, ATHB2, HFR1, and AT5G02580 (C) in different seedlings. All seedlings were grown under 8 μmol m−2 s−1 red light for 5 d for expression level analysis. PP2A was used as a reference gene.
Furthermore, immunoblot analysis of phyB levels did not show any visible difference between phyB-GFP/phyB-9 and TOPP4-FLAG/phyB-GFP/phyB-9 lines (Supplemental Fig. S9B), indicating that the promotion of hypocotyl elongation by TOPP4 in phyB-GFP/phyB-9 is not caused by the decrease in phyB protein. We also detected several PIF direct target genes in these lines. Two PIF-repressed genes, CIRCADIAN CLOCK ASSOCIATED1 (CCA1) and SIGMA FACTOR E (SIGE), were down-regulated in TOPP4-FLAG/phyB-GFP/phyB-9 6# and 8# seedlings, while they were highly expressed in phyB-GFP/phyB-9 (Fig. 5B). The expression of four PIF-induced genes, PIL1, ARABIDOPSIS THALIANA HOMEOBOX2 (ATHB2), LONG HYPOCOTYL IN FAR-RED1 (HFR1), and AT5G02580, was repressed in phyB-GFP/phyB-9 seedlings compared with their expression in phyB-9, but it was markedly up-regulated when TOPP4 was overexpressed in phyB-GFP/phyB-9 (Fig. 5C). Moreover, in topp4-1, the PIF-repressed gene CCA1 was strictly up-regulated while the PIF-induced genes PIL1 and ATHB2 were down-regulated. In contrast, only HFR1 was up-regulated 2-fold in TOPP4-overexpressing seedlings, while the expression of other PIF-regulated genes did not change (Fig. 5, B and C). Taken together, these result suggest that TOPP4 overexpression could repress the expression of PIF-repressed genes and up-regulate the expression of PIF-induced genes in TOPP4-FLAG/phyB-GFP/phyB-9.
TOPP4 Acts Upstream of PIFs in Red Light Signaling
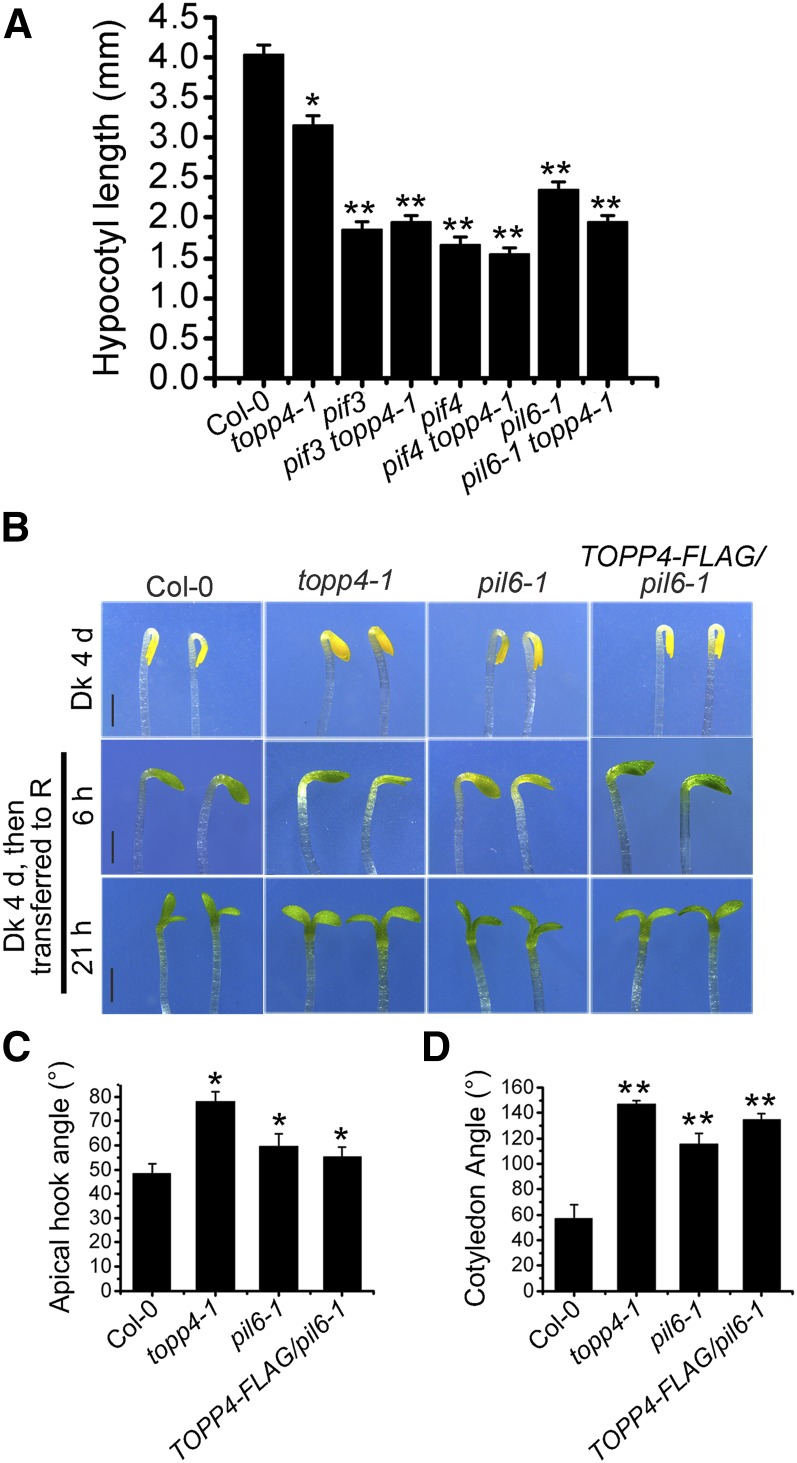
To investigate the epistatic interaction between TOPP4 and PIFs, the hypocotyl lengths of various combinations of pifs and topp4-1 double mutants were examined. The double mutants pif3 topp4-1, pif4 topp4-1, and pil6-1 topp4-1 had similar hypocotyl lengths to the pif3, pif4, and pil6-1 single mutants, respectively (Fig. 6A). Moreover, when TOPP4-FLAG was introduced into pil6-1 and pif3 mutants, the hypocotyl lengths of all transgenic lines were significantly shorter than that of the wild type but similar to those of pil6-1 and pif3 mutants (Supplemental Fig. S10, A–D). PIF5-hemagglutinin (HA) was also introduced into topp4-1 by genetic crossing. PIF5-HA/topp4-1 had long hypocotyls that were similar to those of PIF5-HA seedlings (Supplemental Fig. S10, E and F). These results suggest that PIF is genetically epistatic to TOPP4.
Figure 6.
TOPP4 acts upstream of PIF5 in regulating the growth of Arabidopsis seedlings. A, Quantitative analysis of the hypocotyl lengths of 5-d-old Col-0, topp4-1, pif3, pif4, pil6-1, pif3 topp4-1, pif4 topp4-1, and pil6-1 topp4-1 grown under red light. B, Images of the apical hooks and cotyledon angles of Col-0, topp4-1, pil6-1, and TOPP4-FLAG/pil6-1. Samples were grown 4 d in the dark (Dk) and then exposed for 6 and 21 h of red light (R). Bars = 1 mm. C and D, Quantitative analysis of the apical hook angle (C) and cotyledon angle (D) of different lines as in B. In A, C, and D, error bars represent se (n = 50). Statistical significance was determined by Tukey’s lsd test (*, P < 0.05 and **, P < 0.01).
The apical hook is important for protecting the meristematic primordia in seedlings that emerge through the soil and is an essential feature in photomorphogenesis. The maintenance of the apical hook and the inhibition of cotyledon opening require PIF5 function (Khanna et al., 2007; Gallego-Bartolomé et al., 2011). The apical hook angle and cotyledon opening percentages of TOPP4-FLAG/pil6-1 transgenic lines were consistent with those observed in pil6-1 after treatment with red light for 6 and 21 h (Fig. 6, B–D). Interestingly, topp4-1 exhibited similar patterns to pil6-1 under the same treatment (Fig. 6, B–D). Taken together, these results indicate that TOPP4 acts upstream of PIF during the regulation of photomorphogenesis.
TOPP4 Interacts Physically with PIF3 and PIF5
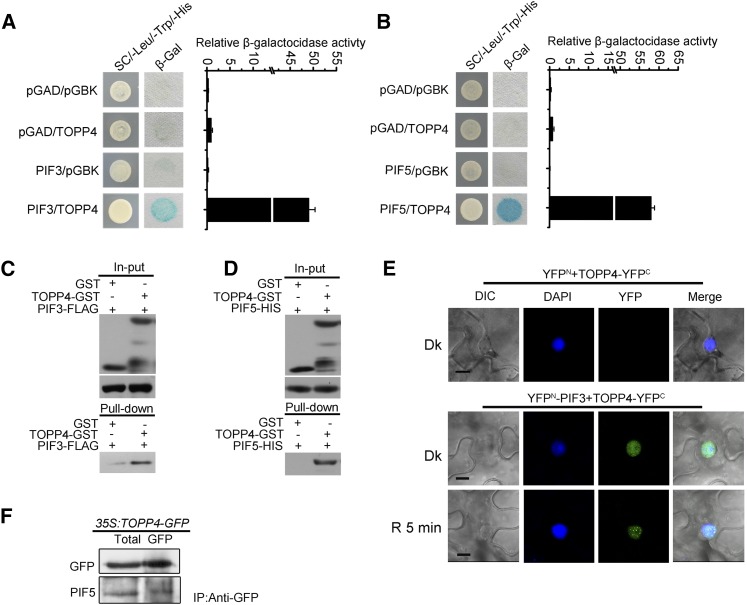
We further analyzed whether TOPP4 interacts directly with PIF. Yeast (Saccharomyces cerevisiae) two-hybrid assays indicated that TOPP4 interacts physically with PIF3 and PIF5 (Fig. 7, A and B). A pull-down assay also demonstrated the interaction between TOPP4, PIF3, and PIF5 in vitro (Fig. 7, C and D). In a bimolecular fluorescence complementation (BiFC) assay, strong nuclear fluorescence was observed following the expression of the TOPP4-YFPC and PIF3-YFPN proteins in tobacco (Nicotiana tabacum) leaves under dark conditions; however, this was not observed in the control expressing TOPP4-YFPC and YFPN. Following irradiation with red light for 5 min, large nuclear bodies were detected in PIF3-YFPN and TOPP4-YFPC coexpressing cells (Fig. 7E). A coimmunoprecipitation assay confirmed that TOPP4 associates with PIF5 in vivo (Fig. 7F). Thus, TOPP4 interacts with PIF3 and PIF5 in vitro and in vivo. Additionally, the pull-down and yeast two-hybrid analyses showed that topp4-1 could interact with PIF5 (Supplemental Fig. S11).
Figure 7.
Interaction of TOPP4 with PIF3 and PIF5. A and B, Yeast two-hybrid assays showing that TOPP4 interacts with PIF3 and PIF5. SC, Synthetic complete medium. Empty vectors were used as negative controls. C, Pull-down assays using glutathione S-transferase (GST)-TOPP4 and GST pull-down PIF3-FLAG. D, Pull-down assays using GST-TOPP4 and GST pull-down PIF5-HIS. E, BiFC assays showing that TOPP4 interacts with PIF3 in tobacco leaf cells. DIC, Differential interference contrast; Dk, samples were incubated in the dark for 48 h; R 5 min, samples were transferred from dark to red light (228 μmol m−2 s−1) for 5 min. Similar results were obtained in three independent experiments. Bars = 10 μm. F, Coimmunoprecipitation of PIF5 with TOPP4 in 35S:TOPP4-GFP seedlings. GFP indicates immunoprecipitation (IP) by anti-GFP antibody, and Total indicates total protein.
TOPP4 Dephosphorylates PIF5 In Vitro and In Vivo
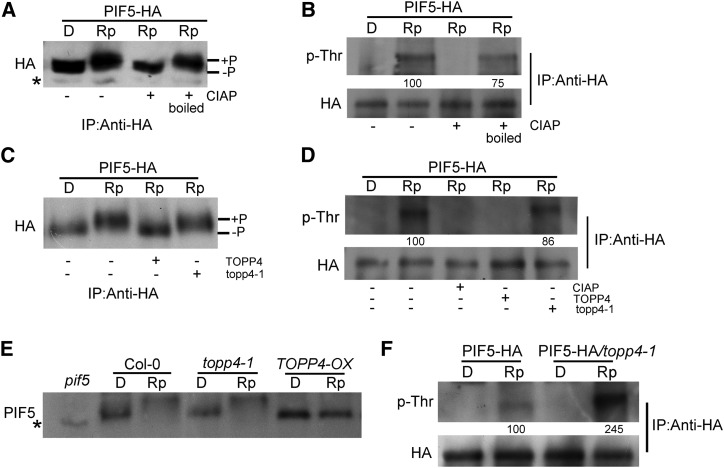
Since TOPP4 interacts physically with PIF5, we examined whether phosphorylated PIF5 is a substrate for TOPP4. When PIF5-HA transgenic lines were exposed to a saturating red light, the PIF5 protein showed a clear light-induced mobility shift (Fig. 8A), and the shifted band was identified as phosphorylated PIF5 by phospho-Thr antibodies (Fig. 8B). After the light-treated sample was incubated with CIAP, the phosphorylated band could no longer be detected, but it could be detected if CIAP was boiled (Fig. 8, A and B). These results indicate that PIF5 is clearly phosphorylated after red light irradiation. Subsequently, phosphorylated PIF5 was treated with TOPP4 or topp4-1. The phosphorylation band was abolished by TOPP4 but not by topp4-1 treatment (Fig. 8, C and D), indicating that TOPP4 could effectively dephosphorylate PIF5 in vitro.
Figure 8.
TOPP4 dephosphorylates PIF5 in vitro and in vivo. A and B, PIF5-HA seedlings were pretreated with MG132 for 4 h and then either kept in darkness (D) or treated with a red light pulse (7,500 μmol m−2 in 33 s) and returned to darkness for 5 min (Rp). Then, samples were extracted with MOPS buffer, immunoprecipitated (IP) using HA-agarose beads prior to calf intestinal alkaline phosphatase (CIAP) treatment, and subjected to western-blot analysis of the mobility shift of PIF5 using anti-PIF5 (A). Samples were also analyzed using anti-phospho-Thr antibodies (p-Thr; B). +, Active CIAP was added; + boiled, heat-denatured CIAP was added. C and D, PIF5-HA seedlings were treated as in A before being treated with TOPP4 and topp4-1. Rp samples were treated with TOPP4 or topp4-1 and subjected to western-blot analysis of the mobility shift using anti-PIF5 (C). Rp samples were then treated with CIAP, TOPP4, or topp4-1 and subjected to western-blot analysis using p-Thr (D). E, Dark-grown Col-0, topp4-1, and TOPP4-OX seedlings were treated with MG132 for 4 h in the dark and exposed to a red light pulse (7,500 μmol m−2 in 33 s), followed by return to darkness for 5 min before extraction by hot denaturing buffer. Protein samples were subjected to western-blot analysis using anti-PIF5 antibody. A pif5 null mutant was used as a negative control. F, Dark-grown PIF5-HA and PIF5-HA/topp4-1 seedlings were treated as in A. D and Rp samples were extracted with MOPS buffer, immunoprecipitated using HA-agarose beads, and subjected to western-blot analysis using p-Thr. In B, D, and F, total proteins were quantified using ImageJ software. PIF5-HA protein levels were normalized with anti-HA after immunoprecipitation for the p-Thr blot, and the value of the starting point was set to 100. Numbers under lanes indicate relative band intensities that were quantified and normalized for each panel. Asterisks indicate nonspecific bands.
Next, we examined the effect of TOPP4 on PIF5 dephosphorylation in vivo. The red light-induced mobility shift of PIF5 was observed in topp4-1 and wild-type samples but not in TOPP4-OX seedlings (Fig. 8E), which was similar to that in the phyA phyB double mutant (Shen et al., 2007), suggesting that endogenous PIF5 is abundantly dephosphorylated by TOPP4 in the TOPP4 overexpression line. A distinct difference in the mobility shift between Col-0 and topp4-1 was not observed, perhaps due to the difficulty of detecting the change in total protein samples. Therefore, PIF5-HA was immunoprecipitated from red light-irradiated PIF5-HA/topp4-1 and PIF5-HA seedlings, and phosphorylated PIF5 was detected using phospho-Thr antibodies. The level of phosphorylated PIF5 in PIF5-HA/topp4-1 seedlings was 2-fold higher than that in the wild type (Fig. 8F). These results indicate that PIF5 is dephosphorylated by TOPP4 in vivo.
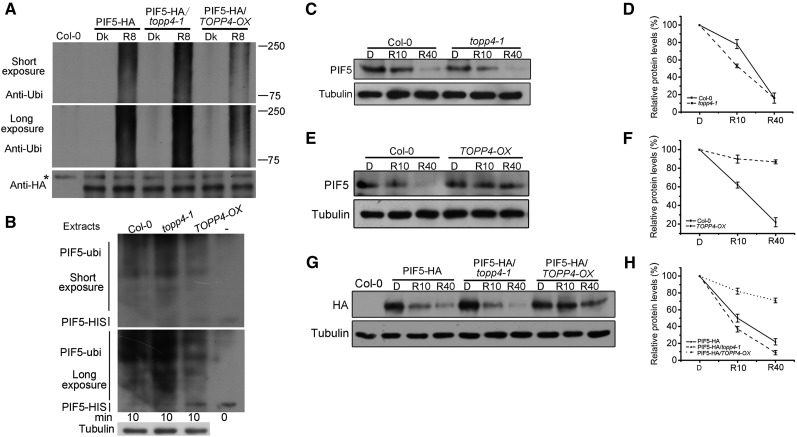
TOPP4 Plays a Dominant Role in Red Light-Induced PIF5 Ubiquitination and Degradation
Phosphorylated PIF3 is the substrate for the Cullin E3 ligase substrate-recognition subunit LRB2 (Ni et al., 2014). In order to determine if TOPP4 is involved in regulating PIF5 ubiquitination, the red light-induced PIF5 ubiquitination in PIF5-HA, PIF5-HA/topp4-1, and PIF5-HA/TOPP4-OX seedlings was analyzed. PIF5-HA/TOPP4-OX was constructed by genetic crossing. As a result, the amount of ubiquitinated PIF5-HA protein was increased in topp4-1 but was markedly reduced in TOPP4-OX compared with PIF5-HA seedlings (Fig. 9A). We further investigated PIF5 ubiquitination in vitro and observed that, compared with the wild type, incubation with protein extracts from topp4-1 seedlings led to a higher degree of ubiquitination of PIF5-HIS, while incubation with TOPP4-OX extracts caused only slight ubiquitination (Fig. 9B). These data support the conclusion that TOPP4-mediated PIF5 dephosphorylation represses its ubiquitination.
Figure 9.
TOPP4 regulates the red light-dependent ubiquitination and degradation of PIF5. A, PIF5 protein in the indicated transgenic seedlings was purified with HA-agarose beads and then blotted with either anti-HA or anti-ubiquitin (anti-Ubi) antibody. Dk, Dark; R8, Dark-grown seedlings were irradiated with a 2-min saturating red light pulse followed by 8 min in the dark. B, Purified PIF5-HIS was treated with Col-0, topp4-1, and TOPP4-OX seedling extracts for 10 min in vitro. The results were analyzed by western blot with anti-ubiquitin antibody. Endogenous tubulin is shown as an equal loading control. C, Four-day-old dark-grown Col-0 and topp4-1 seedlings were exposed to a saturating red light pulse (7,500 μmol m−2 in 33 s), followed by return to darkness (D) for the indicated times before extraction (R10 and R40 indicate 10 and 40 min from the initial red light pulse), and subjected to western-blot analysis using anti-PIF5 or anti-tubulin antibody. D, Total proteins were quantified using ImageJ software. PIF5 protein levels were normalized to tubulin, and the value of the starting point was set to 100. E and F, Dark-grown Col-0 and TOPP4-OX seedlings were treated as in C, analyzed by western blot using anti-PIF5 or anti-tubulin antibody (E), and quantified (F) as in D. G and H, Dark-grown PIF5-HA, PIF5-HA/topp4-1, and PIF5-HA/TOPP4-OX seedlings were treated as in C, analyzed by immunoblot (G), and quantified (H) as in D. Quantitative data are shown as means ± se (n = 3).
Light-induced phosphorylated PIF3 can presumably be identified by the ubiquitin proteasome system, thereby resulting in its degradation (Ni et al., 2013). To determine whether TOPP4 plays a role in PIF5 degradation, PIF5 degradation in topp4-1 and TOPP4-OX lines was assessed. It was found that PIF5 degradation was accelerated in topp4-1 but inhibited in TOPP4-OX seedlings following incubation in the dark for 10 min (Fig. 9, C–F). Moreover, the stability of PIF5 in PIF5-HA/topp4-1 and PIF5-HA/TOPP4-OX was compared, and a similar result was obtained (Fig. 9, G and H). These data suggest that TOPP4 inhibits PIF5 degradation.
To determine whether TOPP4 regulates PIF5 protein stability during or after translation, de novo protein synthesis was blocked by cycloheximide treatment. Compared with wild-type seedlings, PIF5 degradation was markedly delayed in TOPP4-OX seedlings but accelerated in topp4-1 under cycloheximide treatment (Supplemental Fig. S12, A and B). This result suggests that TOPP4 regulates PIF5 stability after translation in response to red light. Phosphomimic mutations in light-induced phosphorylation sites in PIF3 promote its phosphorylation and degradation in vivo in the dark (Ni et al., 2013). We have demonstrated that the amount of phosphorylated PIF5 did not increase in topp4-1 in the dark (Fig. 8, E and F). We treated dark-grown topp4-1 seedlings with the proteasome inhibitor MG132 to examine whether topp4-1 displays preexisting accelerated PIF5 degradation in the dark. PIF5 protein abundance in topp4-1 did not differ from that in the wild type (Supplemental Fig. S12C), indicating that PIF5 is not degraded in topp4-1 in the dark. In addition, we also detected PIF3 degradation in topp4-1 and TOPP4-OX seedlings. Both red light-stimulated migration and degradation of PIF3 in these seedlings were similar to those in the wild-type seedlings (Supplemental Fig. S13), indicating that TOPP4 may not regulate the phosphorylation and degradation of PIF3.
DISCUSSION
The current model suggests that PIF phosphorylation is triggered by photoactivated phytochromes (Bauer et al., 2004; Shen et al., 2005, 2007; Al-Sady et al., 2006; Oh et al., 2006). Recent research has shown that the GSK3-like kinase BIN2 phosphorylates the light-regulated PIF4 transcription factor to mark it for proteasome degradation (Bernardo-García et al., 2014). However, the specific phosphatase involved in PIF dephosphorylation remains to be identified. Our results demonstrate that the phosphatase TOPP4 is involved in phytochrome-mediated photoresponses, where it dephosphorylates PIF5.
TOPP4 and phyB Act Antagonistically to Regulate Arabidopsis Photomorphogenesis
PhyB plays a unique role in the initiation of photomorphogenic responses (Deng and Quail, 1999). According to our findings, TOPP4 regulates red light-mediated photomorphogenic development, and the topp4-1 mutation rescues the closed cotyledon angle phenotype of phyB-9 and phyA-211 phyB-9 mutants to some extent (Fig. 4B; Supplemental Fig. S8). Furthermore, overexpression of TOPP4 weakens the enhanced photomorphogenetic phenotype of phyB-GFP/phyB-9 seedlings (Fig. 4, C and D). Based on these results, we propose that TOPP4 acts as a negative regulator of photomorphogenesis in Arabidopsis, indicating that TOPP4 and phyB act as antagonistic signals in the regulation of Arabidopsis photomorphogenesis. Comparison of the transcript levels of PIF-regulated genes also suggested that overexpression of TOPP4 significantly reduces the effect of phyB on the control of the expression of PIF-regulated genes in phyB-GFP/phyB-9. However, the expression of PIF-regulated genes was barely altered in TOPP4-FLAG seedlings (Fig. 5, B and C). A previous study showed that, at fluence rates of red light higher than 8 μmol m−2 s−1, more than 43% of phyB is estimated to be Pfr (Chen et al., 2003). Large nuclear bodies are formed only when the percentage of the Pfr form of phyB is high. Many large phyB nuclear bodies were observed in phyB-GFP seedlings under 8 μmol m−2 s−1 red light (Chen et al., 2003), indicating that the activity of phyB is very strong. Therefore, under this condition, TOPP4-OX seedlings exhibited a defective response to a lower degree than expected (Supplemental Fig. S3) and only a small change in the expression of PIF-regulated genes.
TOPP4 Modulates PIF5 Ubiquitination and Stability through Direct Dephosphorylation
Previous research has shown that PIF5 acts downstream of phyB (Khanna et al., 2004). In our study, epistasis analysis indicated that PIF5 acts downstream of TOPP4 (Fig. 6; Supplemental Fig. S10). Upon light activation, phytochromes accumulate in the nucleus and trigger the phosphorylation and rapid degradation of PIF5 by the ubiquitin-proteasome system (Shen et al., 2007). Our results provide vital evidence that TOPP4 specifically dephosphorylates the red light-induced phosphorylated form of PIF5 and enhances PIF5 stability (Figs. 8 and 9). The degree of ubiquitination of PIF5 in topp4-1 or TOPP4-OX corresponds to their degradation rate (Fig. 9). The function of TOPP4 in dephosphorylating PIF5 and inhibiting phyB-dependent growth suppression in seedlings was demonstrated by data obtained from the topp4-1 mutant and TOPP4-OX overexpression lines (Fig. 1). The short-hypocotyl and large cotyledon opening angle phenotypes of topp4-1 seedlings are consistent with increased levels of phospho-PIF5. The degradation of PIF5 in TOPP4-OX plants was significantly slower than that in the wild type, mainly because of the excessive dephosphorylation of PIF5 (Figs. 8 and 9). However, light-induced PIF5 degradation in TOPP4-OX was not completely abolished but occurred at a slower rate, while in topp4-1, this process accelerated moderately, suggesting the existence of partial functional redundancy between TOPP4 and other unknown phosphatases in the control of PIF5 stability.
Previous studies have demonstrated that multisite phosphorylation of substrate proteins is essential for E3 ligase recognition (Deshaies and Ferrell, 2001; Deshaies and Joazeiro, 2009; Bao et al., 2010; Varedi K et al., 2010). The nonphosphorylated protein PIF3-A20 almost failed to bind to LRB2, while PIF3 with phospho-mimic mutations showed high binding affinity toward LRB2, indicating that light-induced phosphosites in PIF3 are required for high-affinity binding to LRB2 (Ni et al., 2014). In our study, the highly abundant phosphorylated PIF5 was ubiquitinated in the topp4-1 mutant, leading to faster PIF5 degradation. In contrast, TOPP4-OX lines exhibited reduced PIF5 ubiquitination (Fig. 9, A and B), suggesting that the dephosphorylated state of the PIF5 protein may not be easily recognized by E3 ubiquitin ligase, thereby leading to its subsequent accumulation in seedlings. Taken together, these results serve as evidence that the phosphorylated PIFs are the likely substrates for the related E3 ubiquitin ligase, while the dephosphorylated ones are not.
Notably, we demonstrate that the mutant protein topp4-1 fails to dephosphorylate PIF5 (Fig. 8, C and D) but still interacts with PIF5 (Supplemental Fig. S11). It is possible that the topp4-1 protein competitively binds to PIF5 with a wild-type TOPP4 protein or other proteins and acts as a competitive inhibitor, thus leading to the wild-type protein TOPP4 and other proteins being unable to play their normal biological function. This action subsequently inhibits or blocks the signal transduction. This could explain why the overexpression of topp4-1 in wild-type plants has a dominant-negative effect on seedling growth and leads to defects that are similar to those in topp4-1 mutants.
Action of TOPP4 in Regulating Photomorphogenesis
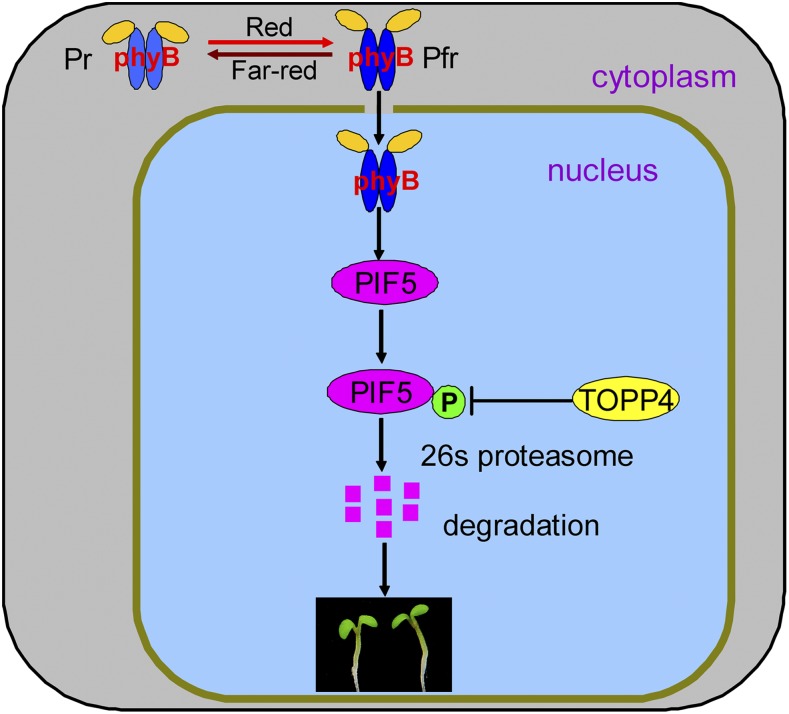
Based on the above results, we propose a model that illustrates the involvement of TOPP4 in phyB signaling (Fig. 10). PIF5 is a photomorphogenesis-repressing factor that binds directly to the Pfr form of phyB strongly and specifically (Khanna et al., 2004; Shen et al., 2007). Upon red light illumination, the phyB-PIF5 interaction triggers PIF5 phosphorylation and destabilization (Shen et al., 2007), while TOPP4 interacts with PIF5 and dephosphorylates the phosphorylated PIF5. Therefore, TOPP4 and phyB modulate the phosphorylation state of PIF5 in an antagonistic way to control photomorphogenesis. Our data reinforce the notion that early steps in phyB signaling primarily involve changes in protein phosphorylation and support a novel phytochrome signaling mechanism whereby TOPP4 negatively regulates PIF5 proteolysis to modulate photomorphogenesis in Arabidopsis.
Figure 10.
Model depicting the proposed role of TOPP4 in red light-induced PIF5 degradation. In darkness, phyB is inactive and localized to the cytoplasm. When exposed to red light, light-activated phyB (Pfr) translocates to the nucleus and induces PIF5 phosphorylation, ubiquitination, and subsequent degradation by the 26S proteasome, while TOPP4 dephosphorylates the phosphorylation state of PIF5 and leads to PIF5 accumulation. These modifications result in PIF5 levels in a dynamic balance. Therefore, TOPP4 and phyB regulate photomorphogenesis in an antagonistic way (adapted from Castillon et al., 2007; Shen et al., 2007).
TOPP4 Regulation of Photomorphogenesis Does Not Involve the Control of DELLA Protein Levels
The DELLA proteins RGA and GAI are the two main repressors of GA responses. They control hypocotyl growth and stem elongation in Arabidopsis (Peng et al., 1997; Dill et al., 2001). In the absence of GA, DELLA proteins bind to the DNA-recognition domain of PIF3 and PIF4 to block PIF transcriptional activity and subsequently abolish the PIF-mediated light control of hypocotyl elongation. In the presence of GA, DELLA proteins are degraded and their negative effect on PIF is abolished. Subsequently, PIF promotes hypocotyl elongation (de Lucas et al., 2008; Feng et al., 2008). Our previous studies showed that TOPP4 controls the degradation of RGA and GAI, thus indicating its involvement in the GA signaling pathway. DELLA accumulation is altered in 20-d-old topp4-1 and TOPP4-OX plants (Qin et al., 2014). However, RGA and GAI levels are not altered by mutating or overexpressing TOPP4 in 5-d-old seedlings under red light (Supplemental Fig. S7, B–D). This implies that TOPP4 regulates photomorphogenesis by regulating PIF5 stability rather than by indirectly affecting PIF levels through DELLA proteins. The regulatory mechanism by which TOPP4 controls protein accumulation (e.g. RGA, GAI, and PIF5) at different developmental stages in plants might be important for the effective and precise performance of dephosphorylation by TOPP4 (Qin et al., 2014).
Novel Function of PP1 in Regulating Plant Growth and Development
One of the most abundant phosphatases, PP1, regulates many cellular events in animals (Shi, 2009). Previous studies have revealed similarities between the biochemical properties and subcellular localization of PP1 in animals and plants, suggesting that the activity and function of PP1 in animals and plants might also be similar (Cohen, 2002). Nine PP1 genes (TOPP1–TOPP9) have been identified in Arabidopsis (Lin et al., 1998; Qin et al., 2014). However, the functions of these genes remain unclear. In our previous study, we reported that one of the nine TOPP proteins, TOPP4, is involved in GA signal transduction in Arabidopsis (Qin et al., 2014). Subsequently, we demonstrated that TOPP4-regulated PIN1 polar targeting is crucial for the morphogenesis of pavement cells in Arabidopsis (Guo et al., 2015). In this study, we verified that TOPP4 participates in phyB signaling by dephosphorylating and controlling the stability of PIF5 in Arabidopsis seedlings. Based on these results, we conclude that TOPP4 performs the function of dephosphorylation of proteins in different organs and tissues and at various developmental stages. Moreover, the ubiquitous expression of TOPP4 in various organs throughout plant developmental processes implies that TOPP4 participates in many other signaling pathways (Qin et al., 2014). However, there is a high degree of functional redundancy among TOPP genes (Lin et al., 1998), which may be one reason why two T-DNA insertion mutant lines of TOPP4 fail to show an obvious phenotype. topp loss-of-function mutants are required to elucidate the function of TOPP in plants.
The number of kinases in Arabidopsis and other eukaryotic genomes is higher than that of phosphatases (Manning et al., 2002; Tchieu et al., 2003; Kerk et al., 2008). Therefore, a phosphatase usually dephosphorylates many physiological substrates (Luan, 2003). Our findings have demonstrated that TOPP4 regulates several cellular responses by dephosphorylating different substrates in Arabidopsis. This type of regulation is consistent with the mechanism of PP1 in mammalian systems (Johnson et al., 1996, 1997). As a catalytic subunit of protein phosphatase, PP1 forms a functional enzyme to participate in multiple signaling pathways by binding to different regulatory subunits (Tian and Wang, 2002). The catalytic activity of PP1 is determined by many regulatory subunits that target the catalytic subunit to specific subcellular compartments and modulate the phosphorylation state of some proteins (Cohen, 2002). Recently, three regulatory subunits of PP1 were identified in Arabidopsis. One of them, PP1 REGULATORY SUBUNIT 2-LIKE PROTEIN1, regulates blue light signaling in stomata (Takemiya et al., 2013). Our data have demonstrated that TOPP4 functions upstream of PIF in the red light signaling pathway. Therefore, a regulatory subunit of PP1 is expected to mediate this signaling transduction. Further study is needed to identify of the corresponding PP1 regulatory subunit to elucidate PP1’s mode of action in response to red light.
MATERIALS AND METHODS
Plant Material, Seedling Growth, and Measurements
The wild-type Arabidopsis (Arabidopsis thaliana) plants used were ecotype Col-0. The phyB-9 and phyA-211 phyB-9 mutants in the Col-0 ecotype were described previously (Kang et al., 2009). pif3-3 was described previously (Monte et al., 2004). pif3 (SALK_030753) was also described previously (Kim et al., 2003). Seeds of the mutants pif4 (SALK_140393), pif5 (SALK_072306), and pil6-1 (cs66044) in the Col-0 ecotype were ordered from the Arabidopsis Biological Resource Center. The mutant topp4-1 and two T-DNA insertion mutant lines, N466328 and SALK_090980, were described previously (Qin et al., 2014). PIF5-HA and phyB-GFP were described previously (Shen et al., 2007; Medzihradszky et al., 2013). Transgenic lines 35S:TOPP4-GFP, ProTOPP4:topp4-1-GFP, 35S:topp4-1-GFP, ProTOPP4:TOPP4/topp4-1, 35S:TOPP4/topp4-1, RGA-GFP, RGA-GFP/topp4-1, RGA-GFP/TOPP4-OX, and ProTOPP4-GUS were described previously (Qin et al., 2014). Arabidopsis seeds were sterilized and vernalized for 3 d at 4°C in darkness and induced to germinate with 3 h of white light and a 5-min far-red light pulse (60 μmol m−2 s−1) before exposure to different light conditions on growth medium as reported (Leivar et al., 2008b). The seedlings were grown under different light conditions as indicated, the fluence rate of red light (56 μmol m−2 s−1) was used, and some specific light fluence rates were described as indicated in the figure legend. Light treatments were performed in growth chambers equipped with light-emitting diodes (model E30LED; Percival Scientific). Light fluence rates were measured using a spectrometer (model PS-200; Apogee). The hypocotyl length, apical hook angle, and cotyledon angle were measured with ImageJ software.
Plasmid Construction and Generation of Transgenic Lines
35S:TOPP4-FLAG was generated by amplifying the complementary DNA of Col-0 with the primer set 5′-AAAAAAGCAGGCTTCATGGCGACGACGACGA-3′ and 5′-CAAGAAAGCTGGGTTTCAAATCTTTGTGGACATC-3′. The amplified fragment was constructed into pBIB-BASTA-35S-GWR-FLAG. The plasmids were introduced into Agrobacterium tumefaciens strain GV3101 and then transformed into Col-0 and phyB-GFP seedlings; the T1 transgenic lines were selected by BASTA. Phenotypic analyses were conducted using homozygous T3 transgenic plants.
GUS Activity Assay and GFP Localization
The transgenic line assays for GUS activity were performed as described previously (Qin et al., 2014). Briefly, transgenic plants were grown in the dark or under red light for 4 d, and intact whole seedlings were immersed in GUS staining solution at 37°C in the dark condition for 20 h as required. After incubation, the samples were transferred to 70% ethanol to remove the chlorophylls. GUS expression patterns were photographed with a stereomicroscope (Nikon). TOPP4-GFP subcellular localization was observed by confocal microscopy (Olympus FluoView FV1000 MPE).
Immunoprecipitation, CIAP Treatment, and Dephosphorylation Assay in Vitro
Four-day-old dark-grown PIF5-HA seedlings were treated with 30 μm MG132 for 4 h and extracted with MOPS buffer (Ni et al., 2014), and PIF5-HA was immunoprecipitated from supernatants with HA-agarose beads (Abmart); the immunoprecipitation and CIAP (Roche) treatments were performed essentially as described previously (Shen et al., 2007). For the in vitro dephosphorylation assay, the pellets were washed two times with 1× phosphate-buffered saline wash buffer, followed by two times with another buffer (50 mm HEPES, 10 mm NaCl, 2 mm dithiothreitol, 0.01% Brij 35 [pH 7.5], and 1 mm protease inhibitors), then the pellets were added with TOPP4 and topp4-1, which were immunoprecipitated from transgenic Arabidopsis seedlings 35S:TOPP4-GFP or 35S:topp4-1-GFP using anti-GFP antibodies and incubated at 30°C for 1 h. Pellets were boiled in 5× SDS sample buffer and subjected to western-blot analysis.
For immunoblot analysis to detect PIF5 phosphorylation, immunoprecipitated proteins were separated on an 8% SDS-PAGE gel, the membranes were blocked with 1× Tris-buffered saline plus Tween 20 (TBST) plus 1% bovine serum albumin (BSA) buffer, followed by incubation with anti-phospho-Thr antibodies (Cell Signaling) in 1× TBST plus 1% BSA. For secondary antibody, peroxidase-labeled goat anti-rabbit antibody (KPL) in a 1:2,000 dilution with 1× TBST plus 1% BSA was used.
In Vivo and in Vitro Ubiquitination Assay
For the in vivo ubiquitination assay, 4-d-old dark-grown PIF5-HA, PIF5-HA/topp4-1, and PIF5-HA/TOPP4-OX seedlings were treated with 30 μm MG132 for 4 h and irradiated with a 2-min saturating red light pulse followed by 8 min in the dark before extract. Fusion proteins were extracted into MOPS buffer plus 0.1% SDS (Ni et al., 2014) and immunoprecipitated from supernatants with HA-agarose beads. Immunoprecipitation products were separated by SDS-PAGE, blotted, and probed with anti-ubiquitin antibody (Agrisera UBQ11).
For in vitro ubiquitination assay, 5-d-old Arabidopsis Col-0, topp4-1, and TOPP4-OX seedlings grown under continuous red light were pretreated with 30 mm MG132 for 4 h and extracted in degradation buffer containing 25 mm Tris-HCl, pH 7.5, 10 mm NaCl, 10 mm MgCl2, 4 mm phenylmethylsulfonyl fluoride, 5 mm dithiothreitol, 10 mm ATP, and 50 mm MG132 (Naidoo et al., 1999; Osterlund et al., 2000; Yanagisawa et al., 2003). The supernatant was collected by two 10-min centrifugations at 14,000 rpm and 4°C. The recombinant PIF5-HIS protein, which was bounded to nickel-Sepharose 6 Fast Flow (GE), was incubated at 22°C with equal amounts of extracts of Col-0, topp4-1, and TOPP4-OX seedlings for 10 min. The supernatants containing PIF5-HIS and PIF5-HIS-ubiquitin were separated on an SDS-PAGE gel and detected by western blotting using anti-ubiquitin. The total protein extracts were adjusted to equal concentration in the degradation buffer for each assay.
Protein Extraction and Western-Blot Analysis
For protein analysis, all the seedlings were grown under different light conditions as indicated in the figure legend, and total protein was extracted with hot denaturing buffer according to the protocol described previously (Shen et al., 2007). For the purpose of investigating the PIF5 protein mobility shift, the seedlings were treated with 30 μm MG132 for 4 h before extraction, and PhosStop cocktail (Roche) was added correctly to the hot denaturing buffer. Thirty micrograms of protein was run on a 6.5% SDS-PAGE gel, blotted onto a nitrocellulose membrane, and probed with the following primary antibodies as described previously (Qin et al., 2014), anti-PIF5 and anti-PIF3 from Agrisera, anti-HA from Tiangen, anti-GFP from Roche, anti-RGA from Agrisera, anti-GAI from our laboratory (Qin et al., 2014), with anti-tubulin from Sigma as a loading control. The secondary antibody was anti-rabbit or anti-mouse antibody in a 1:10,000 dilution. For western-blot band detection, a KPL Protein Detector kit (54-13-50) was used. ImageJ software was used for protein quantification, and targeted proteins were normalized to the loading control, tubulin. For in vivo degradation assays, the relative protein levels at the start were set to 100.
Isolation of RNA and qRT-PCR
Total RNA was isolated using an extraction kit (E.Z.N.A. Plant RNA Kit; Omega) from Arabidopsis seedlings under different conditions as indicated in the figure legend. One microgram of total RNA was reverse transcribed using the PrimeScript RT Reagent Kit from Takara Bio, and the first-strand complementary DNA was used as a template for PCR amplification. qRT-PCR was performed with SYBR Premix Ex Taq (Takara Bio). We used the PP2A gene as a normalization control. The primers used for qRT-PCR are listed in Supplemental Table S1.
Yeast Two-Hybrid Assay
For the yeast two-hybrid assay, pGBKT7:TOPP4 and pGBKT7:topp4-1 were as described (Qin et al., 2014). PIF3 and PIF5 were constructed into pGAD (Clontech). The yeast strain was Y190, and the assay was performed according to the Matchmaker Two-Hybrid System 3 (Clontech). β-Galactosidase activity was determined according to the manufacturer’s protocol (Clontech).
GST Pull-Down Assays
GST pull-down assays were performed according to the procedure described previously (Qin et al., 2014). Briefly, TOPP4-GST and topp4-1-GST were generated in the pGEX-4T-3 vector. PIF5-HIS was generated in the pET28a vector. PIF3 was amplified by PCR and constructed into pFLAG-MAC vector. All fusion proteins were harvested and purified using glutathione-Sepharose 4B beads. The beads with the immobilized GST fusion proteins were washed six times with phosphate-buffered saline supplemented with 0.1% Nonidet P-40. Bound proteins were eluted by boiling in 1× SDS loading buffer.
BiFC Assay
The vectors for BiFC, pEearleygate201-YN and pEearleygate202-YC, were described previously (Lu et al., 2010; Song et al., 2010). TOPP4-YC and PIF5-YC were obtained by PCR amplification of the related full-length fragments and cloning into the pEearleygate202-YC vector, the full-length TOPP4 and PIF3 were amplified and cloned into pEearleygate201-YN, and the BiFC assay was used as described (Qin et al., 2014).
Sequence data from this article can be found in the Arabidopsis Genome Initiative database under the following accession numbers: TOPP4 (AT2G39840), PIF3 (AT1G09530), PIF4 (AT2G43010), PIF5 (AT3G59060), CHS (AT5G13930), CAB2 (AT1G29920), RBCS (AT1G67090), CCA1 (AT2G46830), SIGE (AT5G24120), ATHB2 (AT4G16780), HFR1 (AT1G02340), and PIL1 (AT2G46970).
Supplemental Data
The supplemental following materials are available.
Supplemental Figure S1. Analysis of TOPP4 expression and photoresponses of topp4-1 mutant and TOPP4 overexpression seedlings under different light conditions.
Supplemental Figure S2. Expression levels of TOPP4 in TOPP4-FLAG overexpression seedlings.
Supplemental Figure S3. Quantitative analysis of the apical hook and cotyledon angle of TOPP4-OX seedlings under red light.
Supplemental Figure S4. Overexpression of TOPP4 in the topp4-1 mutant rescues the cotyledon opening defects; the T-DNA lines of TOPP4 did not show the seedling growth defect.
Supplemental Figure S5. Quantification of TOPP4/topp4-1 expression levels in 35S:TOPP4 and 35S:topp4-1-GFP seedlings.
Supplemental Figure S6. The localization and accumulation of TOPP4 are not affected by red light.
Supplemental Figure S7. The accumulation of TOPP4 is not altered in the topp4-1 mutant, and DELLA proteins are not affected by mutation or overexpression of TOPP4 in Arabidopsis seedlings under red light.
Supplemental Figure S8. The topp4-1 mutation could partly recover the closed cotyledon angle of phyB-9 and phyA-211 phyB-9.
Supplemental Figure S9. Overexpression of TOPP4 in phyB-GFP seedlings has no notable effect on phyB protein levels.
Supplemental Figure S10. PIF is genetically epistatic to TOPP4.
Supplemental Figure S11. topp4-1 interacts with PIF5.
Supplemental Figure S12. TOPP4 regulates PIF5 stability after translation in response to red light, and the PIF5 proteins are not degraded in topp4-1 in the absence of light.
Supplemental Figure S13. The dephosphorylation and stability of PIF3 could not be affected by TOPP4.
Supplemental Table S1. List of primers used for real-time PCR analyses.
Supplementary Material
Acknowledgments
We thank Peter H. Quail for PIF5-HA seeds, HongQuan Yang for phyA-211, phyB-9, and phyA-211 phyB-9 mutants, Ferenc Nagy for phyB-GFP seeds, XingWang Deng for pif3 (SALK_030753), Jia Li for pBIB-BASTA-35S-GWR-FLAG and pFLAG-MAC vectors, Ralan Nelson for measuring light fluence rates, WeiMing Ni and HaoDong Cheng for assistance with the PIF5 degradation assay, HongJu Yin for the detection of protein phosphorylation, and Kai He and ZhongLiang Wu for article revision.
Glossary
- qRT
quantitative real-time
- T-DNA
transfer DNA
- BiFC
bimolecular fluorescence complementation
- CIAP
calf intestinal alkaline phosphatase
- Col-0
Columbia-0
- TBST
Tris-buffered saline plus Tween 20
- BSA
bovine serum albumin
Footnotes
This work was supported by the Ministry of Agriculture of the People’s Republic of China (grant no. 2016ZX08009–003–002) and the National Science Foundation (grant nos. 31470372, 31271460, and 91017002).
References
- Achard P, Liao L, Jiang C, Desnos T, Bartlett J, Fu X, Harberd NP (2007) DELLAs contribute to plant photomorphogenesis. Plant Physiol 143: 1163–1172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alabadí D, Gil J, Blázquez MA, García-Martínez JL (2004) Gibberellins repress photomorphogenesis in darkness. Plant Physiol 134: 1050–1057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Sady B, Ni W, Kircher S, Schäfer E, Quail PH (2006) Photoactivated phytochrome induces rapid PIF3 phosphorylation prior to proteasome-mediated degradation. Mol Cell 23: 439–446 [DOI] [PubMed] [Google Scholar]
- Bailey PC, Martin C, Toledo-Ortiz G, Quail PH, Huq E, Heim MA, Jakoby M, Werber M, Weisshaar B (2003) Update on the basic helix-loop-helix transcription factor gene family in Arabidopsis thaliana. Plant Cell 15: 2497–2502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao MZ, Shock TR, Madhani HD (2010) Multisite phosphorylation of the Saccharomyces cerevisiae filamentous growth regulator Tec1 is required for its recognition by the E3 ubiquitin ligase adaptor Cdc4 and its subsequent destruction in vivo. Eukaryot Cell 9: 31–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer D, Viczián A, Kircher S, Nobis T, Nitschke R, Kunkel T, Panigrahi KC, Adám E, Fejes E, Schäfer E, et al. (2004) Constitutive photomorphogenesis 1 and multiple photoreceptors control degradation of phytochrome interacting factor 3, a transcription factor required for light signaling in Arabidopsis. Plant Cell 16: 1433–1445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernardo-García S, de Lucas M, Martínez C, Espinosa-Ruiz A, Davière JM, Prat S (2014) BR-dependent phosphorylation modulates PIF4 transcriptional activity and shapes diurnal hypocotyl growth. Genes Dev 28: 1681–1694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollen M. (2001) Combinatorial control of protein phosphatase-1. Trends Biochem Sci 26: 426–431 [DOI] [PubMed] [Google Scholar]
- Bu Q, Zhu L, Dennis MD, Yu L, Lu SX, Person MD, Tobin EM, Browning KS, Huq E (2011) Phosphorylation by CK2 enhances the rapid light-induced degradation of phytochrome interacting factor 1 in Arabidopsis. J Biol Chem 286: 12066–12074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillon A, Shen H, Huq E (2007) Phytochrome interacting factors: central players in phytochrome-mediated light signaling networks. Trends Plant Sci 12: 514–521 [DOI] [PubMed] [Google Scholar]
- Ceulemans H, Bollen M (2004) Functional diversity of protein phosphatase-1, a cellular economizer and reset button. Physiol Rev 84: 1–39 [DOI] [PubMed] [Google Scholar]
- Chen M, Chory J (2011) Phytochrome signaling mechanisms and the control of plant development. Trends Cell Biol 21: 664–671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Schwab R, Chory J (2003) Characterization of the requirements for localization of phytochrome B to nuclear bodies. Proc Natl Acad Sci USA 100: 14493–14498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi G, Yi H, Lee J, Kwon YK, Soh MS, Shin B, Luka Z, Hahn TR, Song PS (1999) Phytochrome signalling is mediated through nucleoside diphosphate kinase 2. Nature 401: 610–613 [DOI] [PubMed] [Google Scholar]
- Cohen PT. (2002) Protein phosphatase 1: targeted in many directions. J Cell Sci 115: 241–256 [DOI] [PubMed] [Google Scholar]
- de Lucas M, Davière JM, Rodríguez-Falcón M, Pontin M, Iglesias-Pedraz JM, Lorrain S, Fankhauser C, Blázquez MA, Titarenko E, Prat S (2008) A molecular framework for light and gibberellin control of cell elongation. Nature 451: 480–484 [DOI] [PubMed] [Google Scholar]
- Deng XW, Quail PH (1999) Signalling in light-controlled development. Semin Cell Dev Biol 10: 121–129 [DOI] [PubMed] [Google Scholar]
- Deshaies RJ, Ferrell JE Jr (2001) Multisite phosphorylation and the countdown to S phase. Cell 107: 819–822 [DOI] [PubMed] [Google Scholar]
- Deshaies RJ, Joazeiro CA (2009) RING domain E3 ubiquitin ligases. Annu Rev Biochem 78: 399–434 [DOI] [PubMed] [Google Scholar]
- Dill A, Jung HS, Sun TP (2001) The DELLA motif is essential for gibberellin-induced degradation of RGA. Proc Natl Acad Sci USA 98: 14162–14167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong J, Tang D, Gao Z, Yu R, Li K, He H, Terzaghi W, Deng XW, Chen H (2014) Arabidopsis DE-ETIOLATED1 represses photomorphogenesis by positively regulating phytochrome-interacting factors in the dark. Plant Cell 26: 3630–3645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng S, Martinez C, Gusmaroli G, Wang Y, Zhou J, Wang F, Chen L, Yu L, Iglesias-Pedraz JM, Kircher S, et al. (2008) Coordinated regulation of Arabidopsis thaliana development by light and gibberellins. Nature 451: 475–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin KA, Quail PH (2010) Phytochrome functions in Arabidopsis development. J Exp Bot 61: 11–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallego-Bartolomé J, Arana MV, Vandenbussche F, Zádníková P, Minguet EG, Guardiola V, Van Der Straeten D, Benkova E, Alabadí D, Blázquez MA (2011) Hierarchy of hormone action controlling apical hook development in Arabidopsis. Plant J 67: 622–634 [DOI] [PubMed] [Google Scholar]
- Galvão RM, Li M, Kothadia SM, Haskel JD, Decker PV, Van Buskirk EK, Chen M (2012) Photoactivated phytochromes interact with HEMERA and promote its accumulation to establish photomorphogenesis in Arabidopsis. Genes Dev 26: 1851–1863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmartin PM, Memelink J, Hiratsuka K, Kay SA, Chua NH (1992) Characterization of a gene encoding a DNA binding protein with specificity for a light-responsive element. Plant Cell 4: 839–849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X, Qin Q, Yan J, Niu Y, Huang B, Guan L, Li Y, Ren D, Li J, Hou S (2015) TYPE-ONE PROTEIN PHOSPHATASE4 regulates pavement cell interdigitation by modulating PIN-FORMED1 polarity and trafficking in Arabidopsis. Plant Physiol 167: 1058–1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W, Su YS, Lagarias JC (2009) A light-independent allele of phytochrome B faithfully recapitulates photomorphogenic transcriptional networks. Mol Plant 2: 166–182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huq E, Al-Sady B, Hudson M, Kim C, Apel K, Quail PH (2004) Phytochrome-interacting factor 1 is a critical bHLH regulator of chlorophyll biosynthesis. Science 305: 1937–1941 [DOI] [PubMed] [Google Scholar]
- Huq E, Al-Sady B, Quail PH (2003) Nuclear translocation of the photoreceptor phytochrome B is necessary for its biological function in seedling photomorphogenesis. Plant J 35: 660–664 [DOI] [PubMed] [Google Scholar]
- Huq E, Quail PH (2002) PIF4, a phytochrome-interacting bHLH factor, functions as a negative regulator of phytochrome B signaling in Arabidopsis. EMBO J 21: 2441–2450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Y, Lau OS, Deng XW (2007) Light-regulated transcriptional networks in higher plants. Nat Rev Genet 8: 217–230 [DOI] [PubMed] [Google Scholar]
- Johnson D, Cohen P, Chen MX, Chen YH, Cohen PT (1997) Identification of the regions on the M110 subunit of protein phosphatase 1M that interact with the M21 subunit and with myosin. Eur J Biochem 244: 931–939 [DOI] [PubMed] [Google Scholar]
- Johnson DF, Moorhead G, Caudwell FB, Cohen P, Chen YH, Chen MX, Cohen PT (1996) Identification of protein-phosphatase-1-binding domains on the glycogen and myofibrillar targetting subunits. Eur J Biochem 239: 317–325 [DOI] [PubMed] [Google Scholar]
- Kami C, Lorrain S, Hornitschek P, Fankhauser C (2010) Light-regulated plant growth and development. Curr Top Dev Biol 91: 29–66 [DOI] [PubMed] [Google Scholar]
- Kang CY, Lian HL, Wang FF, Huang JR, Yang HQ (2009) Cryptochromes, phytochromes, and COP1 regulate light-controlled stomatal development in Arabidopsis. Plant Cell 21: 2624–2641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerk D, Templeton G, Moorhead GB (2008) Evolutionary radiation pattern of novel protein phosphatases revealed by analysis of protein data from the completely sequenced genomes of humans, green algae, and higher plants. Plant Physiol 146: 351–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khanna R, Huq E, Kikis EA, Al-Sady B, Lanzatella C, Quail PH (2004) A novel molecular recognition motif necessary for targeting photoactivated phytochrome signaling to specific basic helix-loop-helix transcription factors. Plant Cell 16: 3033–3044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khanna R, Shen Y, Marion CM, Tsuchisaka A, Theologis A, Schäfer E, Quail PH (2007) The basic helix-loop-helix transcription factor PIF5 acts on ethylene biosynthesis and phytochrome signaling by distinct mechanisms. Plant Cell 19: 3915–3929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DH, Kang JG, Yang SS, Chung KS, Song PS, Park CM (2002) A phytochrome-associated protein phosphatase 2A modulates light signals in flowering time control in Arabidopsis. Plant Cell 14: 3043–3056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Yi H, Choi G, Shin B, Song PS, Choi G (2003) Functional characterization of phytochrome interacting factor 3 in phytochrome-mediated light signal transduction. Plant Cell 15: 2399–2407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JI, Shen Y, Han YJ, Park JE, Kirchenbauer D, Soh MS, Nagy F, Schäfer E, Song PS (2004) Phytochrome phosphorylation modulates light signaling by influencing the protein-protein interaction. Plant Cell 16: 2629–2640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kircher S, Gil P, Kozma-Bognár L, Fejes E, Speth V, Husselstein-Muller T, Bauer D, Adám E, Schäfer E, Nagy F (2002) Nucleocytoplasmic partitioning of the plant photoreceptors phytochrome A, B, C, D, and E is regulated differentially by light and exhibits a diurnal rhythm. Plant Cell 14: 1541–1555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P, Monte E, Al-Sady B, Carle C, Storer A, Alonso JM, Ecker JR, Quail PH (2008a) The Arabidopsis phytochrome-interacting factor PIF7, together with PIF3 and PIF4, regulates responses to prolonged red light by modulating phyB levels. Plant Cell 20: 337–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P, Monte E, Oka Y, Liu T, Carle C, Castillon A, Huq E, Quail PH (2008b) Multiple phytochrome-interacting bHLH transcription factors repress premature seedling photomorphogenesis in darkness. Curr Biol 18: 1815–1823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P, Quail PH (2011) PIFs: pivotal components in a cellular signaling hub. Trends Plant Sci 16: 19–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P, Tepperman JM, Monte E, Calderon RH, Liu TL, Quail PH (2009) Definition of early transcriptional circuitry involved in light-induced reversal of PIF-imposed repression of photomorphogenesis in young Arabidopsis seedlings. Plant Cell 21: 3535–3553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Q, Li J, Smith RD, Walker JC (1998) Molecular cloning and chromosomal mapping of type one serine/threonine protein phosphatases in Arabidopsis thaliana. Plant Mol Biol 37: 471–481 [DOI] [PubMed] [Google Scholar]
- Lorrain S, Allen T, Duek PD, Whitelam GC, Fankhauser C (2008) Phytochrome-mediated inhibition of shade avoidance involves degradation of growth-promoting bHLH transcription factors. Plant J 53: 312–323 [DOI] [PubMed] [Google Scholar]
- Lu Q, Tang X, Tian G, Wang F, Liu K, Nguyen V, Kohalmi SE, Keller WA, Tsang EW, Harada JJ, et al. (2010) Arabidopsis homolog of the yeast TREX-2 mRNA export complex: components and anchoring nucleoporin. Plant J 61: 259–270 [DOI] [PubMed] [Google Scholar]
- Luan S. (2003) Protein phosphatases in plants. Annu Rev Plant Biol 54: 63–92 [DOI] [PubMed] [Google Scholar]
- Makino S, Matsushika A, Kojima M, Yamashino T, Mizuno T (2002) The APRR1/TOC1 quintet implicated in circadian rhythms of Arabidopsis thaliana. I. Characterization with APRR1-overexpressing plants. Plant Cell Physiol 43: 58–69 [DOI] [PubMed] [Google Scholar]
- Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S (2002) The protein kinase complement of the human genome. Science 298: 1912–1934 [DOI] [PubMed] [Google Scholar]
- Medzihradszky M, Bindics J, Ádám É, Viczián A, Klement É, Lorrain S, Gyula P, Mérai Z, Fankhauser C, Medzihradszky KF, et al. (2013) Phosphorylation of phytochrome B inhibits light-induced signaling via accelerated dark reversion in Arabidopsis. Plant Cell 25: 535–544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monte E, Tepperman JM, Al-Sady B, Kaczorowski KA, Alonso JM, Ecker JR, Li X, Zhang Y, Quail PH (2004) The phytochrome-interacting transcription factor, PIF3, acts early, selectively, and positively in light-induced chloroplast development. Proc Natl Acad Sci USA 101: 16091–16098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagatani A. (2004) Light-regulated nuclear localization of phytochromes. Curr Opin Plant Biol 7: 708–711 [DOI] [PubMed] [Google Scholar]
- Naidoo N, Song W, Hunter-Ensor M, Sehgal A (1999) A role for the proteasome in the light response of the timeless clock protein. Science 285: 1737–1741 [DOI] [PubMed] [Google Scholar]
- Ni M, Tepperman JM, Quail PH (1998) PIF3, a phytochrome-interacting factor necessary for normal photoinduced signal transduction, is a novel basic helix-loop-helix protein. Cell 95: 657–667 [DOI] [PubMed] [Google Scholar]
- Ni M, Tepperman JM, Quail PH (1999) Binding of phytochrome B to its nuclear signalling partner PIF3 is reversibly induced by light. Nature 400: 781–784 [DOI] [PubMed] [Google Scholar]
- Ni W, Xu SL, Chalkley RJ, Pham TND, Guan S, Maltby DA, Burlingame AL, Wang ZY, Quail PH (2013) Multisite light-induced phosphorylation of the transcription factor PIF3 is necessary for both its rapid degradation and concomitant negative feedback modulation of photoreceptor phyB levels in Arabidopsis. Plant Cell 25: 2679–2698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni W, Xu SL, Tepperman JM, Stanley DJ, Maltby DA, Gross JD, Burlingame AL, Wang ZY, Quail PH (2014) A mutually assured destruction mechanism attenuates light signaling in Arabidopsis. Science 344: 1160–1164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nito K, Wong CC, Yates JR III, Chory J (2013) Tyrosine phosphorylation regulates the activity of phytochrome photoreceptors. Cell Reports 3: 1970–1979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nozue K, Covington MF, Duek PD, Lorrain S, Fankhauser C, Harmer SL, Maloof JN (2007) Rhythmic growth explained by coincidence between internal and external cues. Nature 448: 358–361 [DOI] [PubMed] [Google Scholar]
- Oh E, Kim J, Park E, Kim JI, Kang C, Choi G (2004) PIL5, a phytochrome-interacting basic helix-loop-helix protein, is a key negative regulator of seed germination in Arabidopsis thaliana. Plant Cell 16: 3045–3058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh E, Yamaguchi S, Hu J, Yusuke J, Jung B, Paik I, Lee HS, Sun TP, Kamiya Y, Choi G (2007) PIL5, a phytochrome-interacting bHLH protein, regulates gibberellin responsiveness by binding directly to the GAI and RGA promoters in Arabidopsis seeds. Plant Cell 19: 1192–1208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh E, Yamaguchi S, Kamiya Y, Bae G, Chung WI, Choi G (2006) Light activates the degradation of PIL5 protein to promote seed germination through gibberellin in Arabidopsis. Plant J 47: 124–139 [DOI] [PubMed] [Google Scholar]
- Osterlund MT, Hardtke CS, Wei N, Deng XW (2000) Targeted destabilization of HY5 during light-regulated development of Arabidopsis. Nature 405: 462–466 [DOI] [PubMed] [Google Scholar]
- Peng J, Carol P, Richards DE, King KE, Cowling RJ, Murphy GP, Harberd NP (1997) The Arabidopsis GAI gene defines a signaling pathway that negatively regulates gibberellin responses. Genes Dev 11: 3194–3205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phee BK, Kim JI, Shin DH, Yoo J, Park KJ, Han YJ, Kwon YK, Cho MH, Jeon JS, Bhoo SH, et al. (2008) A novel protein phosphatase indirectly regulates phytochrome-interacting factor 3 via phytochrome. Biochem J 415: 247–255 [DOI] [PubMed] [Google Scholar]
- Qin Q, Wang W, Guo X, Yue J, Huang Y, Xu X, Li J, Hou S (2014) Arabidopsis DELLA protein degradation is controlled by a type-one protein phosphatase, TOPP4. PLoS Genet 10: e1004464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quail PH. (2002) Phytochrome photosensory signalling networks. Nat Rev Mol Cell Biol 3: 85–93 [DOI] [PubMed] [Google Scholar]
- Reed JW, Nagpal P, Poole DS, Furuya M, Chory J (1993) Mutations in the gene for the red/far-red light receptor phytochrome B alter cell elongation and physiological responses throughout Arabidopsis development. Plant Cell 5: 147–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu JS, Kim JI, Kunkel T, Kim BC, Cho DS, Hong SH, Kim SH, Fernández AP, Kim Y, Alonso JM, et al. (2005) Phytochrome-specific type 5 phosphatase controls light signal flux by enhancing phytochrome stability and affinity for a signal transducer. Cell 120: 395–406 [DOI] [PubMed] [Google Scholar]
- Sakamoto K, Nagatani A (1996) Nuclear localization activity of phytochrome B. Plant J 10: 859–868 [DOI] [PubMed] [Google Scholar]
- Salter MG, Franklin KA, Whitelam GC (2003) Gating of the rapid shade-avoidance response by the circadian clock in plants. Nature 426: 680–683 [DOI] [PubMed] [Google Scholar]
- Sharrock RA, Clack T (2002) Patterns of expression and normalized levels of the five Arabidopsis phytochromes. Plant Physiol 130: 442–456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen H, Moon J, Huq E (2005) PIF1 is regulated by light-mediated degradation through the ubiquitin-26S proteasome pathway to optimize photomorphogenesis of seedlings in Arabidopsis. Plant J 44: 1023–1035 [DOI] [PubMed] [Google Scholar]
- Shen H, Zhu L, Castillon A, Majee M, Downie B, Huq E (2008) Light-induced phosphorylation and degradation of the negative regulator PHYTOCHROME-INTERACTING FACTOR1 from Arabidopsis depend upon its direct physical interactions with photoactivated phytochromes. Plant Cell 20: 1586–1602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Khanna R, Carle CM, Quail PH (2007) Phytochrome induces rapid PIF5 phosphorylation and degradation in response to red-light activation. Plant Physiol 145: 1043–1051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y. (2009) Serine/threonine phosphatases: mechanism through structure. Cell 139: 468–484 [DOI] [PubMed] [Google Scholar]
- Somers DE, Sharrock RA, Tepperman JM, Quail PH (1991) The hy3 long hypocotyl mutant of Arabidopsis is deficient in phytochrome B. Plant Cell 3: 1263–1274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y, Wu K, Dhaubhadel S, An L, Tian L (2010) Arabidopsis DNA methyltransferase AtDNMT2 associates with histone deacetylase AtHD2s activity. Biochem Biophys Res Commun 396: 187–192 [DOI] [PubMed] [Google Scholar]
- Sun L, Tobin EM (1990) Phytochrome-regulated expression of genes encoding light-harvesting chlorophyll a/b-protein in two long hypocotyl mutants and wild type plants of Arabidopsis thaliana. Photochem Photobiol 52: 51–56 [DOI] [PubMed] [Google Scholar]
- Takemiya A, Kinoshita T, Asanuma M, Shimazaki K (2006) Protein phosphatase 1 positively regulates stomatal opening in response to blue light in Vicia faba. Proc Natl Acad Sci USA 103: 13549–13554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takemiya A, Yamauchi S, Yano T, Ariyoshi C, Shimazaki K (2013) Identification of a regulatory subunit of protein phosphatase 1 which mediates blue light signaling for stomatal opening. Plant Cell Physiol 54: 24–35 [DOI] [PubMed] [Google Scholar]
- Tchieu JH, Fana F, Fink JL, Harper J, Nair TM, Niedner RH, Smith DW, Steube K, Tam TM, Veretnik S, et al. (2003) The PlantsP and PlantsT functional genomics databases. Nucleic Acids Res 31: 342–344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tepperman JM, Hwang YS, Quail PH (2006) phyA dominates in transduction of red-light signals to rapidly responding genes at the initiation of Arabidopsis seedling de-etiolation. Plant J 48: 728–742 [DOI] [PubMed] [Google Scholar]
- Tian Q, Wang J (2002) Role of serine/threonine protein phosphatase in Alzheimer’s disease. Neurosignals 11: 262–269 [DOI] [PubMed] [Google Scholar]
- Tobin EM, Kehoe DM (1994) Phytochrome regulated gene expression. Semin Cell Biol 5: 335–346 [DOI] [PubMed] [Google Scholar]
- Toledo-Ortiz G, Huq E, Quail PH (2003) The Arabidopsis basic/helix-loop-helix transcription factor family. Plant Cell 15: 1749–1770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu SL, Lagarias JC (2005) The phytochromes. In Handbook of Photosensory Receptors, W.R. Briggs and J.L. Spudich, eds., Weinheim, Germany: Wiley-VCH, pp. 121–145. [Google Scholar]
- Varedi K SM, Ventura AC, Merajver SD, Lin XN (2010) Multisite phosphorylation provides an effective and flexible mechanism for switch-like protein degradation. PLoS ONE 5: e14029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Deng XW (2003) Dissecting the phytochrome A-dependent signaling network in higher plants. Trends Plant Sci 8: 172–178 [DOI] [PubMed] [Google Scholar]
- Yamaguchi R, Nakamura M, Mochizuki N, Kay SA, Nagatani A (1999) Light-dependent translocation of a phytochrome B-GFP fusion protein to the nucleus in transgenic Arabidopsis. J Cell Biol 145: 437–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagisawa S, Yoo SD, Sheen J (2003) Differential regulation of EIN3 stability by glucose and ethylene signalling in plants. Nature 425: 521–525 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Mayba O, Pfeiffer A, Shi H, Tepperman JM, Speed TP, Quail PH (2013) A quartet of PIF bHLH factors provides a transcriptionally centered signaling hub that regulates seedling morphogenesis through differential expression-patterning of shared target genes in Arabidopsis. PLoS Genet 9: e1003244. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.