A conserved Mediator complex subunit physically interacts with the transcription factor ETHYLENE INSENSITIVE3-LIKE1 (TaEIL1) to promote ETHYLENE RESPONSE FACTOR1 (TaERF1) expression and repress powdery mildew resistance.
Abstract
Powdery mildew, caused by the biotrophic fungal pathogen Blumeria graminis f. sp. tritici, is a major limitation for the production of bread wheat (Triticum aestivum). However, to date, the transcriptional regulation of bread wheat defense against powdery mildew remains largely unknown. Here, we report the function and molecular mechanism of the bread wheat Mediator subunit 25 (TaMED25) in regulating the bread wheat immune response signaling pathway. Three homoalleles of TaMED25 from bread wheat were identified and mapped to chromosomes 5A, 5B, and 5D, respectively. We show that knockdown of TaMED25 by barley stripe mosaic virus-induced gene silencing reduced bread wheat susceptibility to the powdery mildew fungus during the compatible plant-pathogen interaction. Moreover, our results indicate that MED25 may play a conserved role in regulating bread wheat and barley (Hordeum vulgare) susceptibility to powdery mildew. Similarly, bread wheat ETHYLENE INSENSITIVE3-LIKE1 (TaEIL1), an ortholog of Arabidopsis (Arabidopsis thaliana) ETHYLENE INSENSITIVE3, negatively regulates bread wheat resistance against powdery mildew. Using various approaches, we demonstrate that the conserved activator-interacting domain of TaMED25 interacts physically with the separate amino- and carboxyl-terminal regions of TaEIL1, contributing to the transcriptional activation activity of TaEIL1. Furthermore, we show that TaMED25 and TaEIL1 synergistically activate ETHYLENE RESPONSE FACTOR1 (TaERF1) transcription to modulate bread wheat basal disease resistance to B. graminis f. sp. tritici by repressing the expression of pathogenesis-related genes and deterring the accumulation of reactive oxygen species. Collectively, we identify the TaMED25-TaEIL1-TaERF1 signaling module as a negative regulator of bread wheat resistance to powdery mildew.
Bread wheat (Triticum aestivum; 2n = 42; AABBDD) is a major staple crop worldwide. Global demand for bread wheat is increasing with world population growth. To guarantee global food security, people have been seeking new agronomic traits of bread wheat to improve its yield as well as its capacity to adapt to biotic and abiotic stresses. In bread wheat, powdery mildew is caused by Blumeria graminis f. sp. tritici (Bgt), which is one of the most destructive fungal pathogens worldwide (Bourras et al., 2015; Parlange et al., 2015). To improve bread wheat resistance against powdery mildew, it is vital to uncover the underlying regulatory mechanisms for bread wheat defense responses.
Ethylene (ET), as a gas phytohormone, plays significant roles in plant growth, development, and responses to biotic and abiotic stresses (van Loon et al., 2006; Yang et al., 2015a). Over the past two decades, genetic studies in the dicotyledonous model plant Arabidopsis (Arabidopsis thaliana) have uncovered a linear ET signaling pathway from ET perception in the endoplasmic reticulum to transcriptional regulation in the nucleus (Yang et al., 2015a). In Arabidopsis, ET leads to the activation of ETHYLENE INSENSITIVE3 (EIN3) and ETHYLENE INSENSITIVE3-LIKE1 (EIL1) transcription factors (TFs), which are necessary and sufficient for the induction of ET responses (Chao et al., 1997; Solano et al., 1998; Broekaert et al., 2006). As the primary transcriptional regulation factors in Arabidopsis, EIN3/EIL1 TFs bind to the promoter regions of ETHYLENE RESPONSE ELEMENT-BINDING PROTEIN genes, including ETHYLENE RESPONSE FACTOR1 (ERF1), to promote their expression (Broekaert et al., 2006). However, to our knowledge, the specific transcriptional activation regions of Arabidopsis EIN3/EIL1 and the underlying mechanisms of how EIN3/EIL1 activate target gene expression remain to be clarified. Arabidopsis ERF1, one of the most important downstream targets of EIN3, was reported to play critical roles in ET-related plant development, environmental stress responses, and pathogen resistance (Berrocal-Lobo et al., 2002; Yi et al., 2004; Zhang et al., 2004; Cheng et al., 2013; Schmidt et al., 2013; Zhu et al., 2014). Previous studies in Arabidopsis demonstrated that ET signaling components are involved in plant immune responses to various pathogens (Bent et al., 1992; Knoester et al., 1998; Thomma et al., 1999; Berrocal-Lobo et al., 2002; van Loon et al., 2006; Chen et al., 2009). For instance, the ET-insensitive mutants ein2 and ein3 were found to be resistant to the hemibiotrophic bacterial pathogen Pseudomonas syringae (Bent et al., 1992; Chen et al., 2009) but more susceptible to necrotrophic fungi such as Botrytis cinerea and Plectosphaerella cucumerina (Bent et al., 1992; Thomma et al., 1999; Berrocal-Lobo et al., 2002; Chen et al., 2009). Generally, in Arabidopsis, ET acts synergistically with jasmonate (JA), whereas it acts antagonistically with salicylic acid (SA) to modulate plant-pathogen interactions (Robert-Seilaniantz et al., 2011). However, the ET signaling mechanisms during bread wheat-pathogen interactions remain to be clarified. Two recent studies in bread wheat reported that TaEIL1, a wheat homolog of AtEIN3, acts as a negative regulator in the bread wheat-stripe rust fungus interaction (Duan et al., 2013), and the pathogen-induced TaERF1 mediates host responses to both the necrotrophic pathogen Rhizoctonia cerealis and freezing stresses (Zhu et al., 2014).
Mediator is a conserved multisubunit complex in eukaryotes that promotes transcription by bridging specific TFs with RNA polymerase II (Pol II; Chadick and Asturias, 2005). The biochemical purification of the Arabidopsis Mediator complex identified 21 conserved and six plant-specific subunits (Bäckström et al., 2007). Several Mediator subunits have been functionally characterized in modulating Arabidopsis plant development and abiotic responses. For example, the Arabidopsis Mediator subunit MED25 was shown to play multiple roles in regulating flowering (Cerdán and Chory, 2003), organ size (Xu and Li, 2011), salt stress responses (Elfving et al., 2011), and hormone signaling (Kidd et al., 2009; Chen et al., 2012). Loss-of-function mutants of Arabidopsis MED21 are embryonic lethal (Dhawan et al., 2009), indicating its essential roles in plant growth and development. Arabidopsis MED16, first reported as an essential component in SA- and JA-mediated cold tolerance, flowering, and circadian rhythm, is also a positive regulator of iron homeostasis (Knight et al., 2008, 2009; Yang et al., 2014; Zhang et al., 2014). Nevertheless, the potential function of the Mediator complex in Arabidopsis plant defense signaling was not well characterized until recently. Initially, Arabidopsis MED21, MED25, and MED8 were shown to be essential for JA/ET-mediated defense gene expression and immune responses against the necrotrophic pathogens, such as Alternaria brassicicola and B. cinerea, but confer susceptibility to the hemibiotrophic fungus Fusarium oxysporum (Dhawan et al., 2009; Kidd et al., 2009). Recently, growing evidence reveals complex roles of Mediator in SA- or JA/ET-dependent defense processes in Arabidopsis. For instance, Arabidopsis MED16 controls both SA- and JA-mediated defense gene expression as well as plant immunity to P. syringae (Wathugala et al., 2012; Zhang et al., 2012); meanwhile, it was reported recently that MED16 regulates WRKY33-activated defense gene expression and the basal resistance against the necrotrophic fungal pathogen Sclerotinia sclerotiorum (Wang et al., 2015). Arabidopsis MED19a is involved mainly in SA-triggered immune responses and acts as a positive regulator of plant immunity against the oomycete downy mildew pathogen Hyaloperonospora arabidopsidis (Caillaud et al., 2013). Likewise, Arabidopsis MED14 and MED15 have been shown to impact the SA-mediated plant immunity against biotrophic and hemibiotrophic pathogens (Canet et al., 2012; Zhang et al., 2013). Although our knowledge of the functions of the Mediator complex in Arabidopsis is increasing rapidly, the roles of the Mediator complex in crop plants such as rice (Oryza sativa) and bread wheat need to be studied.
Given that the Mediator complex plays key roles in Arabidopsis plant growth, development, and defense responses, we are interested in elucidating the roles of the Mediator complex in bread wheat defense responses. Among the different Mediator subunits studied in Arabidopsis, the MED25 subunit acts mainly through interacting with specific TFs of plant hormone signaling to regulate plant defense responses (Kidd et al., 2009; Chen et al., 2012). Therefore, we hypothesize that bread wheat MED25 (TaMED25) might regulate the basal resistance to obligate biotrophic fungi such as powdery mildew through integrating the plant hormone signaling pathway. To confirm our hypothesis, in this study, we first cloned TaMED25 and analyzed the biochemical mechanism of TaMED25 in modulating bread wheat resistance against powdery mildew. We show that TaMED25, together with the TF TaEIL1, plays negative regulatory roles in bread wheat resistance against powdery mildew. Our analyses reveal that the conserved activator-interacting domain (ACID) of TaMED25 is sufficient for its interaction with TaEIL1; meanwhile, the separate N- and C-terminal regions of TaEIL1 are able to interact physically with TaMED25. Furthermore, we show that TaMED25 has a positive effect on TaEIL1-mediated activation of the powdery mildew-responsive gene TaERF1. Moreover, we demonstrate that the TaMED25-TaEIL1-TaERF1 signaling module represses the expression of specific pathogenesis-related genes (TaPRs) and deters the accumulation of reactive oxygen species (ROS) in bread wheat leaf cells to fine-tune bread wheat resistance against powdery mildew.
RESULTS
Identification of the Bread Wheat Mediator Subunit TaMED25
In this study, we are interested in characterizing the function and molecular mechanism of the bread wheat Mediator subunit MED25 in modulating bread wheat basal resistance against obligate biotrophic fungi such as powdery mildew. We first identified bread wheat TaMED25 based on the protein sequence of AtMED25 (GenBank accession no. NP_173925) and genome sequences of the bread wheat A and D genome donors (Supplemental Fig. S1; Jia et al., 2013; Ling et al., 2013). Three highly homologous sequences of TaMED25 were isolated from the hexaploid bread wheat ‘Beijing 837’ (Supplemental Fig. S2). Chromosomal locations of TaMED25 genes were further determined using sequences of TaMED25 as query sequences to blast the wheat survey sequences, which include the chromosome-based draft sequence of the hexaploid wheat (https://urgi.versailles.inra.fr/blast; Deng et al., 2007; International Wheat Genome Sequencing Consortium, 2014). The results showed that the three TaMED25 sequences were located on chromosomes 5AL, 5BL, and 5DL, respectively, and were designated as TaMED25-A, TaMED25-B, and TaMED25-D. Further gene structure analysis revealed that TaMED25-A, TaMED25-B, and TaMED25-D all contains 15 exons and 14 introns (Supplemental Figs. S3 and S4).
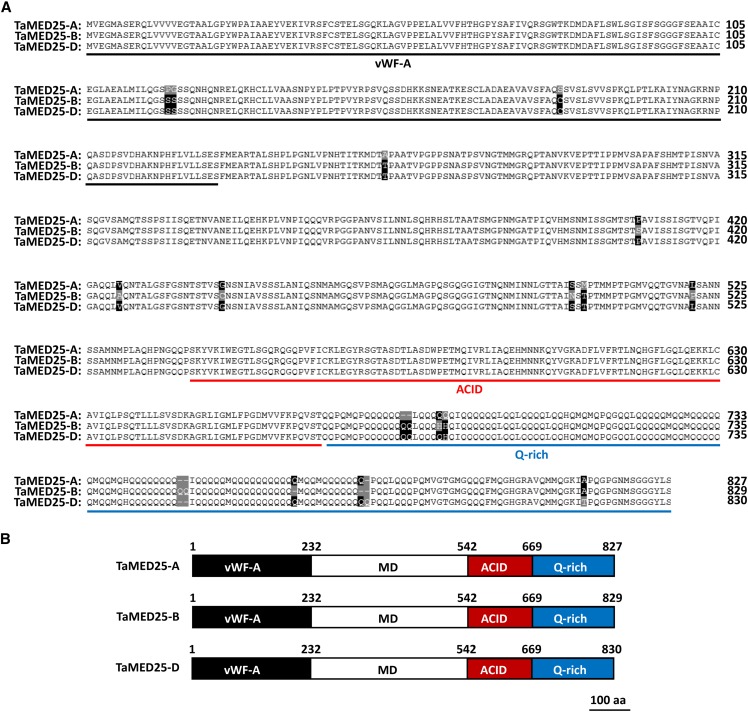
The predicted TaMED25-A, TaMED25-B, and TaMED25-D proteins have 827, 829, and 830 amino acids, respectively (Fig. 1). A sequence alignment with the Arabidopsis MED25 protein revealed that TaMED25 shares the common structural features of AtMED25, containing a von Willebrand factor A domain (vWF-A) for binding to the Mediator complex, a nonconserved middle domain (MD), a conserved ACID for interaction with TFs, and a Gln-rich domain for transcriptional activation (Fig. 1; Supplemental Fig. S1; Chen et al., 2012).
Figure 1.
Homology-based identification of the bread wheat Mediator subunit TaMED25. A, Amino acid sequences of TaMED25. TaMED25-A, TaMED25-B, and TaMED25-D represent TaMED25 proteins from the bread wheat A, B, and D genomes, respectively. The conserved vWF-A, ACID, and Gln-rich (Q-rich) domain are underlined separately in black, red, and blue; amino acid variations among TaMED25-A, TaMED25-B, and TaMED25-D are marked by black- or gray-shaded background. B, Schematic representation of TaMED25-A, TaMED25-B, and TaMED25-D protein structures. Bar = 100 amino acids (aa).
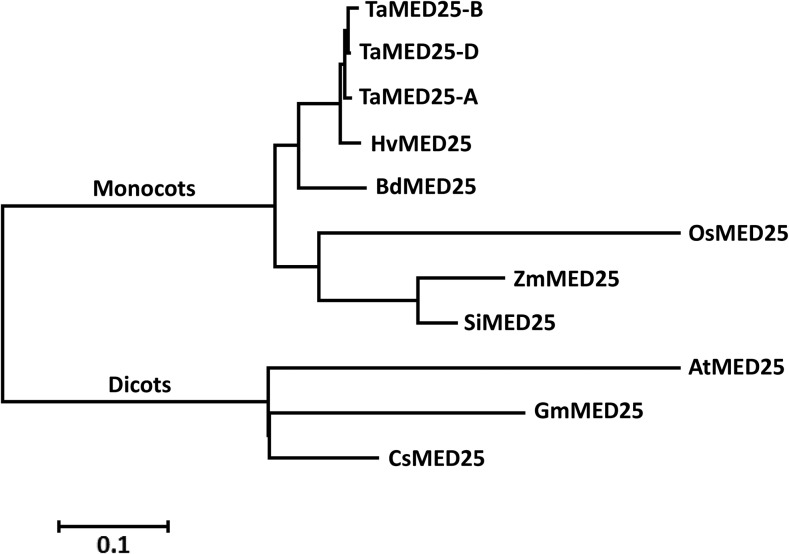
Phylogenetic analysis of the MED25 proteins from various plant species showed that the TaMED25-A, TaMED25-B, and TaMED25-D proteins are closely related to the barley (Hordeum vulgare) MED25 protein HvMED25 (Fig. 2), with a sequence identity of 97%. Based on the above analyses, we conclude that the TaMED25-A, TaMED25-B, and TaMED25-D genes we identified encode TaMED25.
Figure 2.
Phylogenetic tree of MED25 proteins. The phylogenetic tree was constructed based on the neighbor-joining method using MEGA7 software. The evolutionary distances were computed in units of the number of amino acid substitutions per site, as shown by the scale below the tree. Ta, Triticum aestivum; Hv, Hordeum vulgare; Bd, Brachypodium distachyon; Os, Oryza sativa; Zm, Zea mays; Si, Setaria italica; At, Arabidopsis thaliana; Gm, Glycine max; Cs, Citrus sinensis. The GenBank accession numbers of the MED25 proteins or MED25 coding sequences are KU030834 (TaMED25-A), KU030835 (TaMED25-B), KU030836 (TaMED25-D), AK252911 (HvMED25), XP_010238042 (BdMED25), XP_004959349 (SiMED25), XP_008670123 (ZmMED25), EEE69415 (OsMED25), XP_006484420 (CsMED25), XP_006574879 (GmMED25), and NP_173925 (AtMED25).
Knockdown of MED25 Reduces Bread Wheat Susceptibility to Powdery Mildew
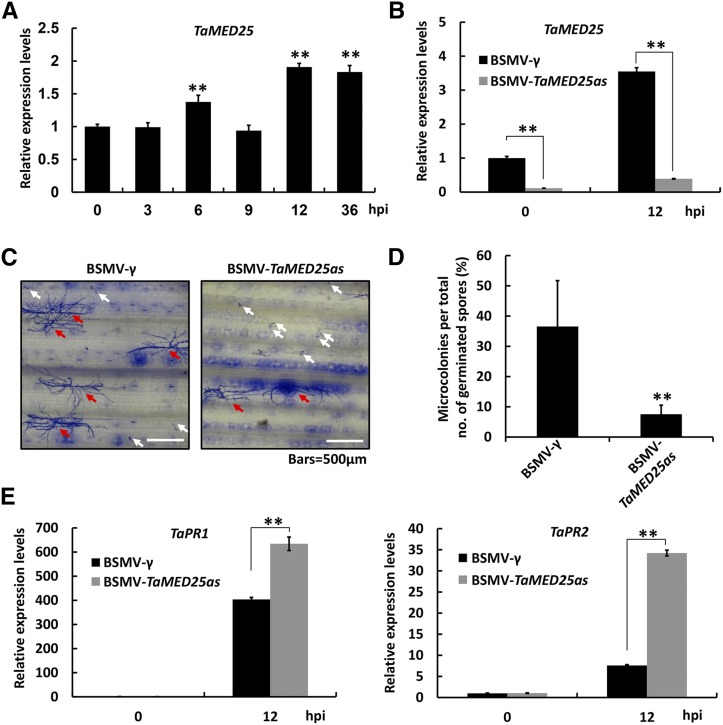
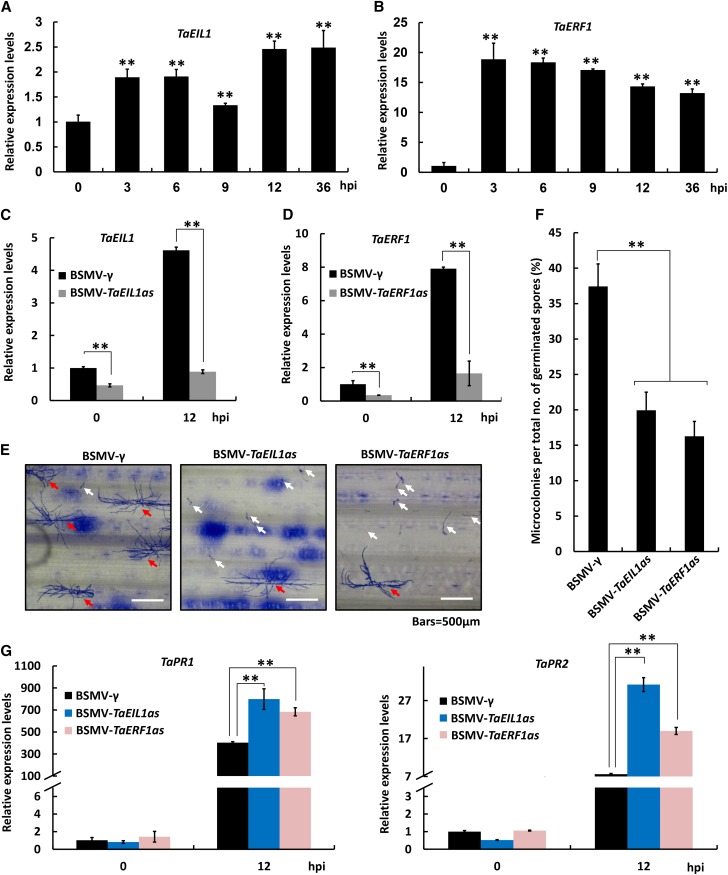
To test whether TaMED25 is involved in regulating the bread wheat defense response against Bgt, we first analyzed the expression pattern of the TaMED25 gene in response to the powdery mildew infection. Quantitative reverse transcription-(qRT)-PCR confirmed that the transcript levels of TaMED25 were marginally elevated in bread wheat seedlings at 6, 12, and 36 h post inoculation (hpi) with powdery mildew (Fig. 3A). Furthermore, we employed a Barley stripe mosaic virus (BSMV) virus-induced gene silencing (VIGS) strategy to knock down all three copies of the endogenous TaMED25 genes (TaMED25-A, TaMED25-B, and TaMED25-D) in the bread wheat ‘Beijing 837’ (Supplemental Figs. S5 and S6; Yuan et al., 2011). qRT-PCR confirmed that endogenous TaMED25 transcription was reduced substantially in the BSMV-VIGS lines (Fig. 3B; BSMV-TaMED25as). Thereafter, we used Bgt strain E09, which is virulent to the bread wheat ‘Beijing 837’ (Zhou et al., 2005), to infect these VIGS lines and employed a microcolony formation index (MI%) to evaluate their susceptibility to Bgt strain E09. In control samples infected with BSMV-γ EV, the MI% was normally around 30% to 40%, while the MI% calculated from the BSMV-TaMED25as-infected leaves was decreased significantly to approximately 8% (Fig. 3, C and D; Supplemental Table S1), illustrating remarkably reduced susceptibility to powdery mildew. PR genes, such as PR1 and PR2, are usually induced by pathogens and SA and play positive roles in plant defense against the biotrophic pathogens (Yalpani et al., 1991; Fu et al., 2014; Yang et al., 2015b). As expected, we observed significant induction of the transcript levels of TaPR1 and TaPR2 in the bread wheat seedlings upon inoculation with Bgt (Supplemental Fig. S7). We further assayed whether the down-regulation of endogenous TaMED25 expression would affect the transcript levels of TaPR1 and TaPR2. Our results showed that, after inoculation with Bgt, the transcripts of TaPR1 and TaPR2 accumulated to extremely higher levels in the BSMV-TaMED25as lines than those in the BSMV-γ negative control (Fig. 3E).
Figure 3.
TaMED25 is a negative regulator of bread wheat resistance to Bgt. A, qRT-PCR analysis of TaMED25 relative expression levels in Bgt-infected bread wheat leaves. RNA samples were isolated from the leaves of Bgt (strain E09)-infected bread wheat ‘Beijing 837’ at 0, 3, 6, 9, 12, and 36 hpi. TaMED25 expression levels were normalized against TaGAPDH. B, Relative transcript levels of TaMED25 in BSMV-VIGS bread wheat leaves. Bread wheat leaves were infected with BSMV-TaMED25as harboring an antisense fragment of TaMED25 or BSMV-γ empty vector (EV) constructs. After typical BSMV symptoms appeared, the leaves were challenged with Bgt spores at a low density. Leaf samples were collected at 0 and 12 hpi for gene expression analyses. C, B. graminis microcolony formation on BSMV-γ or BSMV-TaMED25as bread wheat leaves. Microcolonies were observed microscopically and analyzed 72 hpi. Red arrows indicate successfully colonized spores, and white arrows represent spores that germinated but failed to form a colony. Bars = 500 μm. D, Statistical analysis of B. graminis MI% on the bread wheat BSMV-γ and BSMV-TaMED25as lines. For each treatment, 10 to 15 leaves (3–4 cm in length) were collected independently (the third and fourth leaves from 10–15 BSMV-VIGS bread wheat plants). Then, successfully colonized B. graminis and the spores that did not form a colony were counted separately; the B. graminis MI% represents the percentage of successfully colonized B. graminis out of all analyzed spores. Means and sd were calculated with data from three independent biological replicates. E, Relative transcript levels of the pathogenesis-related genes TaPR1 and TaPR2 in BSMV-γ or BSMV-TaMED25as infected bread wheat leaves. Error bars represent the sd among three independent replicates, and asterisks above the bars represent significant differences between the control and each treatment at P < 0.01 (Student’s t test). All experiments were performed independently three times and gave similar results.
To further explore whether the Mediator subunit MED25 plays a general role in regulating disease resistance to powdery mildew pathogens in Triticeae species, we carried out the well-established BSMV-VIGS assays in barley to evaluate the function of HvMED25 in disease resistance to Blumeria graminis f. sp. hordei (Bgh) during the compatible plant-pathogen interaction (barley ‘Golden Promise’ and Bgh virulent isolate K1; Supplemental Figs. S5 and S8). We first analyzed the expression levels of the HvMED25 gene in response to infection with Bgh. Our results showed that the transcriptional expression of HvMED25 was not changed significantly at our investigated time points upon inoculation with Bgh, with the exception of a reduction at 16 hpi (Supplemental Fig. S9A). Furthermore, we showed that knockdown of HvMED25 significantly reduced the susceptibility of barley ‘Golden Promise’ to Bgh isolate K1, for the MI% in the BSMV-HvMED25as lines (approximately 30%) was markedly lower than that in the BSMV-γ negative control (approximately 50%; Supplemental Fig. S9; Supplemental Table S2). In summary, our results demonstrate that the Mediator subunit MED25 plays a general role in regulating disease resistance to powdery mildew in the Triticeae species bread wheat and barley.
TaMED25 Interacts with the Ethylene Signaling TF TaEIL1
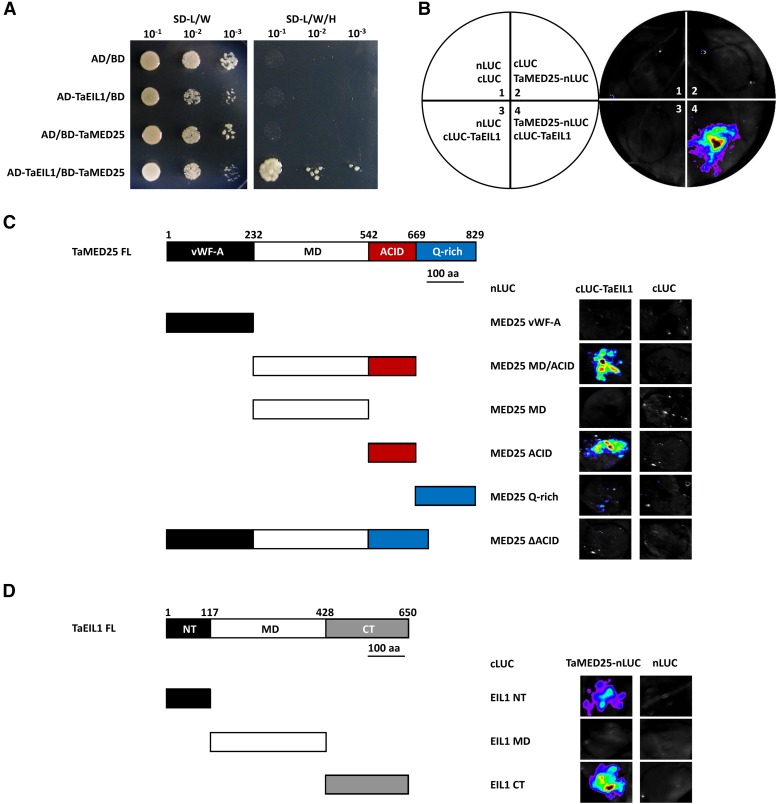
Furthermore, to elucidate the molecular mechanism by which TaMED25 suppresses bread wheat resistance against powdery mildew, we aimed to characterize TaMED25-associating proteins in bread wheat, in particular the plant hormone signaling TFs. A recent investigation showed that TaEIL1 is a bread wheat ortholog of the ethylene signaling TF EIN3 in Arabidopsis, which negatively regulates bread wheat resistance to the obligate biotrophic stripe rust fungus Puccinia striiformis f. sp. tritici (Duan et al., 2013). In this context, we proposed a hypothesis that TaEIL1 also might be involved in the regulation of bread wheat resistance against powdery mildew via physical interaction with TaMED25. Toward this goal, we performed yeast two-hybrid assays between TaMED25 and TaEIL1. Considering that TaMED25-A, TaMED25-B, and TaMED25-D are highly conserved in amino acid sequence (more than 98% identity; Fig. 1A; Supplemental Fig. S2), we used TaMED25-D here as a representative for protein interaction analyses. As shown in Figure 4A, the interaction between TaMED25 and TaEIL1 was obviously detected in the AH109 yeast (Saccharomyces cerevisiae) cells. To further confirm that the interaction can occur in planta, we conducted firefly luciferase (LUC) complementation imaging (LCI) assays in Nicotiana benthamiana (Song et al., 2011). We fused TaMED25 to the N-terminal part of LUC to produce the TaMED25-nLUC construct and TaEIL1 to the C-terminal part of LUC to generate the cLUC-TaEIL1 construct. When TaMED25-nLUC and cLUC-TaEIL1 were coinfiltrated into N. benthamiana leaves, strong LUC activity was observed (Fig. 4B, coinfiltration 4), whereas no LUC signal was detected in the negative controls (Fig. 4B, coinfiltrations 1–3). Together, these results strongly indicate that TaMED25 interacts physically with TaEIL1 in vivo.
Figure 4.
TaMED25 interacts with the TF TaEIL1 through its ACID region. A, Yeast two-hybrid analysis showing the interaction of TaMED25 and TaEIL1. The full-length coding sequences of TaEIL1 and TaMED25 were fused separately to the GAL4-AD and GAL4-BD vectors to generate AD-TaEIL1 and BD-TaMED25 constructs, and these two constructs were cotransformed into yeast AH109 cells. The combinations AD/BD, AD-TaEIL1/BD, and AD/BD-TaMED25 were employed as negative controls. Transformed yeast cells were selected on synthetic dextrose medium lacking Leu and Trp (SD-L/W) and then transferred to synthetic dextrose medium lacking Leu, Trp, and His (SD-L/W/H) with different dilution series (10−1, 10−2, and 10−3). B, LCI assay showing that TaMED25 interacts with TaEIL1 in N. benthamiana. N. benthamiana leaves were infiltrated with Agrobacterium tumefaciens strain GV3101 containing the indicated constructs. The signals were collected 48 h after infiltration. In each experiment, at least 10 independent N. benthamiana leaves were infiltrated and analyzed. C and D, LCI assays of interactions using truncated versions of TaMED25 and TaEIL1 in N. benthamiana. TaMED25 vWF-A (amino acids [aa] 1–232), MD/ACID (233–669), MD (233–542), ACID (543–669), Gln-rich (670–829), and ΔACID (full-length MED25 with the ACID deleted [i.e. 1–542 + 670–829]) were fused with nLUC and coexpressed with cLUC-TaEIL1; TaEIL1 NT (N terminus; amino acids 1–117), MD (118–428), and CT (C terminus; 429–650) were fused with cLUC and coexpressed with TaMED25-nLUC. The signals were collected at 48 h after infiltration. In each experiment, at least 10 N. benthamiana leaves were infiltrated independently and analyzed, and similar signals were observed. All experiments were repeated three times with similar results.
TaMED25 Interacts with TaEIL1 through Its ACID Region
To define the interaction domain of TaMED25 for TaEIL1 binding, we generated the different truncated forms of TaMED25 to fuse with nLUC for the LCI assays (Fig. 4C). As shown in Figure 4C, the MD/ACID and ACID parts of TaMED25 reserved interactions with TaEIL1, whereas the N-terminal part (vWF-A), MD, C-terminal Gln-rich domain, and ACID-deleted TaMED25 (TaMED25ΔACID) could not interact with TaEIL1. Together, these results indicate that the conserved ACID part of TaMED25 is essential and sufficient for its interaction with TaEIL1.
Furthermore, we used the LCI assays to map the domain of TaEIL1 responsible for interaction with TaMED25. Toward this goal, we generated several TaEIL1 derivatives based on previous functional analyses of the EIN3 protein in Arabidopsis (Chao et al., 1997; Fig. 4D). As indicated in Figure 4D, a strong interaction was detected between TaMED25 and the C-terminal part of TaEIL1 containing the putative Gln-rich domain, while a relatively weaker interaction between TaMED25 and the N-terminal part of TaEIL1 was also observed. No obvious interaction signal was detected between TaMED25 and the MD part of TaEIL1 (Fig. 4D).
Physical Interaction with TaMED25 Correlates with the Transcriptional Activity of TaEIL1
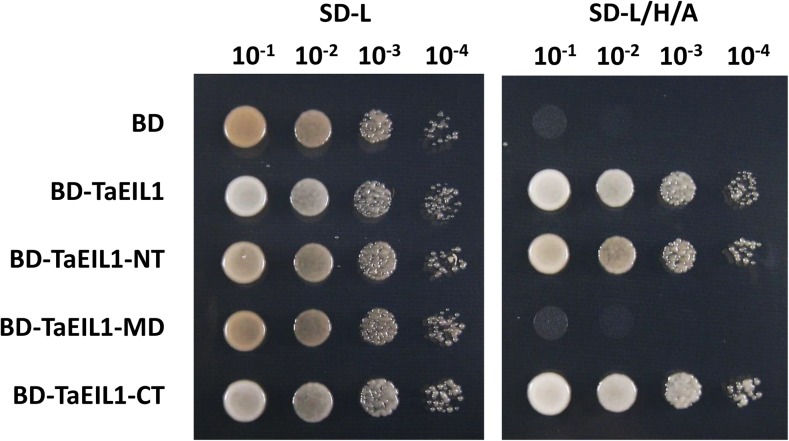
To elucidate the biological significance of the interaction between TaMED25 and TaEIL1, we mapped the transcriptional activation domains of TaEIL1. As shown in Figure 5, we found that the AH109 yeast (Saccharomyces cerevisiae) cells containing BD-TaEIL1, BD-TaEIL1-NT, and BD-TaEIL1-CT, but not BD-TaEIL1-MD, grew well on the selective SD-L/H/A, indicating that both N-terminal and C-terminal parts of TaEIL1 have transcriptional activation activity. The coupling of TaEIL1 transcriptional activation domains and its interaction parts with TaMED25 led us to propose that the interaction with TaMED25 might contribute to the transcriptional activation activity of TaEIL1.
Figure 5.
Transcriptional activity determination of the TaEIL1 protein in yeast. The full-length TaEIL1 as well as TaEIL1 NT (N terminus), MD, and CT (C terminus) were separately fused with GAL4-BD and expressed in yeast strain AH109; the GAL4-BD EV was transformed as a negative control. Yeast cells were selected on synthetic dextrose medium lacking Leu (SD-L) and then transferred to synthetic dextrose medium lacking Leu, His, and adenine (SD-L/H/A) with different dilution series (10−1, 10−2, 10−3, and 10−4).
TaMED25 Colocalizes and Interacts with TaEIL1 in the Bread Wheat Cell Nucleus
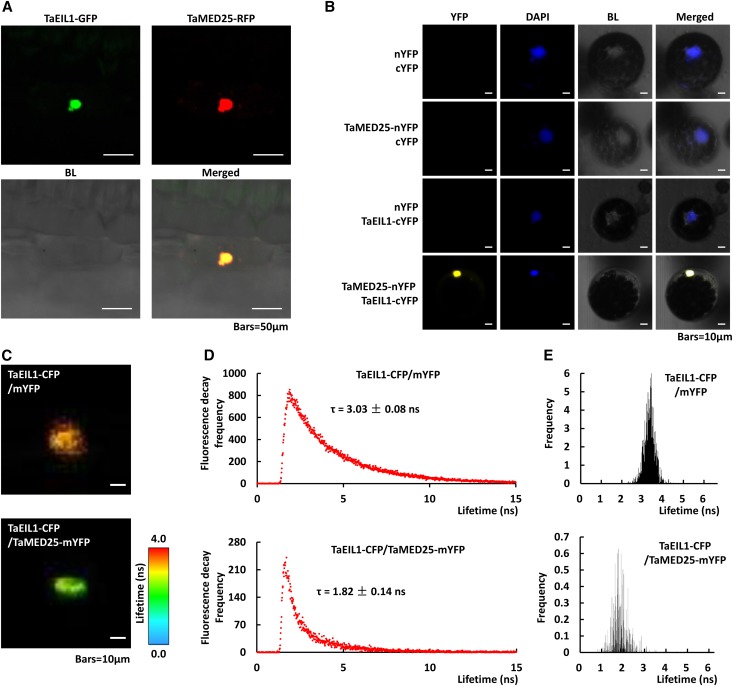
To determine the subcellular localization of TaMED25 and TaEIL1 proteins in bread wheat, we transiently expressed both 35S::TaEIL1-GFP and 35S::TaMED25-RFP (for red fluorescent protein) in the bread wheat epidermal cells by particle delivery and observed the fluorescence with confocal microscopy at 48 h after transformation. The result showed that the fluorescent signals of TaEIL1-GFP and TaMED25-RFP fusion proteins were observed exclusively from the nuclei of bread wheat epidermal cells (Fig. 6A). Importantly, we observed that the green fluorescent signal of TaEIL1-GFP was closely overlapped with the red fluorescent signal of TaMED25-RFP in the cell nuclei (Fig. 6A, bottom right).
Figure 6.
TaMED25 colocalizes and interacts with TaEIL1 in bread wheat cell nucleus. A, Subcellular colocalization of the TaMED25 and TaEIL1 proteins in bread wheat leaf epidermal cells. TaEIL1 and TaMED25 were fused separately with GFP and RFP and cotransformed into bread wheat leaf epidermal cells by particle bombardment. Confocal images were taken at 48 h after transformation. BL, Bright light. In each experiment, more than 50 separate epidermal cells were analyzed, and similar signals were observed. Bars = 50 μm. B, BiFC assay showing the interaction of TaMED25 and TaEIL1 in bread wheat protoplasts. TaMED25 and TaEIL1 were fused with the N-terminal half (nYFP) and C-terminal half (cYFP) of YFP, respectively, and transferred into bread wheat protoplast cells. The fluorescent signals of YFP (yellow) and DAPI (blue; stained by 4,6-diamidino-2-phenylindole to reveal the nuclei) and visible light images (BL) were monitored 24 h after transformation. In each experiment, at least 40 protoplast cells were analyzed. Bars = 10 μm. C to E, FLIM-FRET measurements of the TaEIL1 and TaMED25 interaction. C shows CFP fluorescence lifetime images of the nucleus of a representative cell expressing the indicated proteins, and the fluorescence lifetime is encoded by color as indicated by the scale at right. D represents the CFP fluorescence decay curve, and the average fluorescence lifetimes (τ) are marked. In E, the CFP fluorescence lifetime distributions are shown in column diagrams. For each replication, at least six independent cell nuclei were quantified by confocal microscopy, and three biological replicates were performed. Similar results were observed.
To further characterize the TaMED25-TaEIL1 protein complex in the bread wheat cells, we conducted a bimolecular fluorescence complementation (BiFC) experiment. TaMED25 and TaEIL1 were separately fused with the N- and C-terminal halves of yellow fluorescent protein (YFP) to form TaMED25-nYFP and TaEIL1-cYFP and cotransfected into the bread wheat protoplasts (Fig. 6B). The EVs (nYFP or cYFP) in combination with TaMED25-nYFP or TaEIL1-cYFP were used as negative controls (Fig. 6B). As a result, strong YFP fluorescence was observed exclusively from the nuclei of the bread wheat protoplast cells cotransformed by TaMED25-nYFP and TaEIL1-cYFP, while the negative controls failed to yield any fluorescent signal (Fig. 6B; Supplemental Table S3), indicating that TaMED25 interacts with TaEIL1 in the nuclei of bread wheat cells.
We further conducted Förster resonance energy transfer (FRET) assays to confirm the physical association between TaMED25 and TaEIL1 in bread wheat cells. Here, we adopted quantitative noninvasive fluorescence lifetime imaging (FLIM) to detect FRET. To determine the FRET efficiency between the donor and acceptor proteins, the lifetime of the donor fluorescence is measured in the presence of the acceptor protein and compared with its lifetime in the absence of the acceptor. First, we fused the TaEIL1 and TaMED25 proteins with cyan fluorescent protein (CFP) or mYFP to generate the donor and the acceptor, respectively. Then, coexpression of TaEIL1 and TaMED25 in the bread wheat epidermal cells was carried out with biolistic delivery; meanwhile, TaEIL1-CFP was coexpressed with mYFP as the negative control. We measured the CFP lifetime in the TaEIL1-CFP/mYFP coexpressed negative control sample and found an average lifetime of 3.03 ± 0.08 ns (mean ± sd, n = 6 nuclei; Fig. 6, C–E; Supplemental Table S4). As expected, the average lifetime of CFP in the TaEIL1-CFP/TaMED25-mYFP coexpression cells was 1.82 ± 0.14 ns, which was significantly (P < 0.01) shorter than that of the negative control (Fig. 6, C–E; Supplemental Table S4). These data strongly confirm the physical interaction between TaEIL1 and TaMED25 in the bread wheat cell nuclei.
Silencing of TaEIL1 Reduces Bread Wheat Susceptibility to Powdery Mildew
Based on our finding that TaEIL1 interacts physically with TaMED25, which was shown in this study to be a negative regulator of bread wheat defense against Bgt fungus, we asked whether TaEIL1 is also involved in regulating bread wheat resistance to powdery mildew. To this end, we first analyzed the expression pattern of TaEIL1 in response to infection with Bgt. qRT-PCR results showed that the transcriptional expression of TaEIL1 was induced significantly at an early stage of Bgt infection (3 hpi; Fig. 7A). We further assessed the role of TaEIL1 in bread wheat resistance to Bgt using the BSMV-VIGS approach. qRT-PCR analyses confirmed that the expression of TaEIL1 was indeed reduced in the BSMV-triggered TaEIL1-silencing lines (BSMV-TaEIL1as; Fig. 7C). Remarkably, our results showed that knockdown of TaEIL1 could significantly reduce bread wheat susceptibility to powdery mildew, as the MI% calculated in the BSMV-TaEIL1as-infected leaves was decreased significantly to approximately 13%, compared with approximately 35% for the BSMV-γ EV-infected leaves (Fig. 7, E and F; Supplemental Table S1), reminiscent of the TaMED25 knockdown plants (Fig. 3, C and D; Supplemental Table S1). Furthermore, we showed that, after inoculation with Bgt, the transcript levels of TaPR1 and TaPR2 were enhanced significantly in the BSMV-TaEIL1as lines compared with those in the BSMV-γ negative control lines (Fig. 7G). Together, our results suggest that TaEIL1, resembling TaMED25, negatively regulates bread wheat resistance to powdery mildew.
Figure 7.
TaEIL1 and TaERF1 are negative regulators of bread wheat disease resistance against Bgt. A and B, qRT-PCR analyses of TaEIL1 and TaERF1 relative expression levels after Bgt infection. The relative expression levels of TaEIL1 and TaERF1 were detected at 0, 3, 6, 9, 12, and 36 hpi. The expression levels were normalized against the internal control gene TaGAPDH. Asterisks above the bars represent significant differences in expression levels between the control and each treatment at P < 0.01 (Student’s t test). C and D, Relative transcript levels of TaEIL1 and TaERF1 in BSMV-VIGS bread wheat leaves. Bread wheat leaves were infected separately with BSMV-TaEIL1as and BSMV-TaERF1as derivatives harboring antisense fragments of TaEIL1 and TaERF1 and challenged with Bgt spores at a low density; meanwhile, the BSMV-γ-infected bread wheat leaves were used as negative controls. Leaves were collected and analyzed at 0 and 12 hpi. Asterisks above the bars represent groups with significant differences at P < 0.01 (Student’s t test). E, B. graminis microcolony formation on bread wheat leaves infected with BSMV-γ, BSMV-TaEIL1as, or BSMV-TaERF1as. Red arrows indicate successfully colonized spores, and white arrows represent spores that germinated but failed to form a colony. Bars = 500 μm. F, Statistical analysis of B. graminis MI% on bread wheat leaves after inoculation with BSMV-γ, BSMV-TaEIL1as, or BSMV-TaERF1as. For each treatment, 10 to 15 leaves (3–4 cm in length) were independently collected, and successfully colonized B. graminis as well as spores that did not form a colony were counted microscopically. The B. graminis MI% represents the percentage of successfully colonized B. graminis out of all analyzed spores. Means and sd were calculated with data from three independent biological replicates. G, Relative transcript levels of TaPR1 and TaPR2 in BSMV-VIGS bread wheat leaves. Asterisks in F and G above the bars represent significant differences between the control and each treatment at P < 0.01 (Student’s t test). Error bars represent the sd among at least three independent replicates. All experiments were performed three times with similar results.
TaERF1 Negatively Modulates Bread Wheat Resistance to Powdery Mildew
In Arabidopsis, it has been shown that ERF1 is a direct target of EIN3 (Solano et al., 1998); therefore, we asked whether and how the bread wheat ERF1 ortholog, TaERF1/TaPIE1 (GenBank accession no. ABU62817; Zhu et al., 2014), functions in regulating bread wheat responses to powdery mildew. First, we examined the powdery mildew-triggered transcriptional expression of TaERF1. As shown in Figure 7B, the transcript levels of TaERF1 were elevated rapidly (before 3 hpi) and significantly in the bread wheat seedlings subjected to powdery mildew infection. Next, we generated the VIGS bread wheat plants of TaERF1 (BSMV-TaERF1as) to see whether TaERF1 plays a role in regulating bread wheat resistance against powdery mildew. qRT-PCR analyses showed that the transcriptional expression of TaERF1 was indeed reduced in the BSMV-TaERF1as lines (Fig. 7D). Significantly, our assays showed that knockdown of TaERF1 led to a marked reduction in MI% to approximately 14% compared with the BSMV-γ control plants (Fig. 7, E and F; Supplemental Table S1), indicating that the susceptibility of the BSMV-TaERF1as lines to powdery mildew infection was reduced. To further explore the underlying mechanism, we investigated the effect of TaERF1 on the transcriptional expression of the defense-related genes TaPR1 and TaPR2 following powdery mildew infection. As shown in Figure 7G, the powdery mildew-triggered transcript levels of TaPR1 and TaPR2 were enhanced in the BSMV-TaERF1as-infected lines. In summary, these results confirm that TaERF1 negatively modulates bread wheat resistance to powdery mildew.
TaMED25 and TaEIL1 Synergistically Activate TaERF1 Transcription
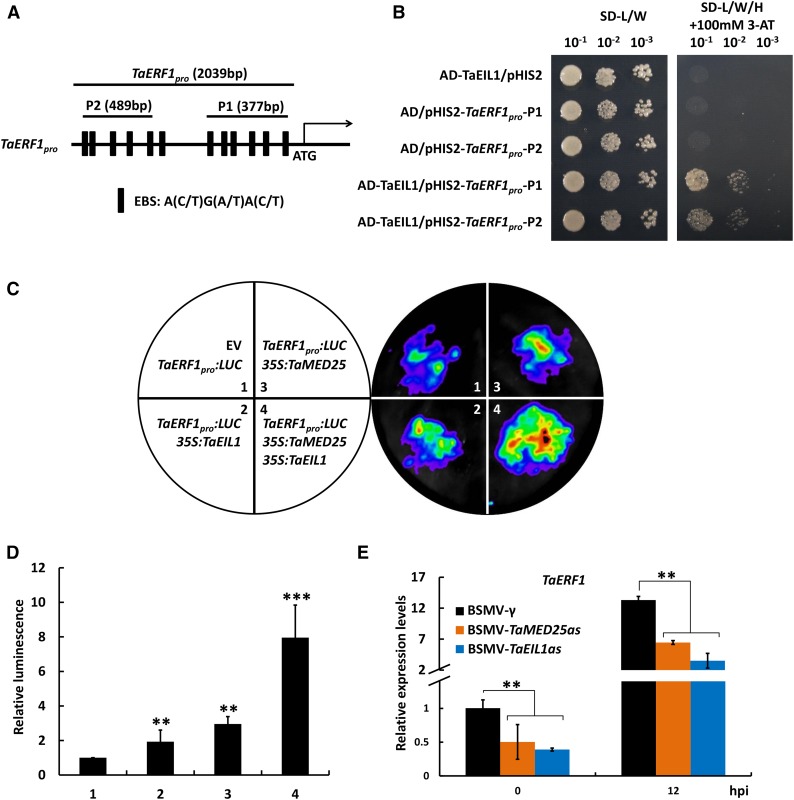
To elucidate the biological implication of the physical interaction between TaMED25 and TaEIL1, we investigated the effect of TaMED25 on TaEIL1-mediated transcriptional regulation of the possible target gene TaERF1. It is well known that the EIN3/EIL TFs in Arabidopsis preferably bind to the EIN3-binding site [A(C/T)G(A/T)A(C/T)CT] in the promoters of their target genes (Broekaert et al., 2006; Chen et al., 2009). Thus, we analyzed the promoter sequence of TaERF1 and found some putative EIN3-binding site sequences [with core sequence A(C/T)G(A/T)A(C/T); Fig. 8A]. To test whether TaEIL1 may actually bind to the TaERF1 promoter region containing EIN3-binding site sequences, we performed yeast one-hybrid assays. Two regions containing EIN3-binding site sequences were selected for analyses (P1 and P2; Fig. 8A; Supplemental Fig. S10). Our results showed that AD-TaEIL1 could indeed bind to the P1 and P2 regions of the TaERF1 promoter to activate downstream reporter gene expression (Fig. 8B).
Figure 8.
TaMED25 and TaEIL1 synergistically activate TaERF1 transcription. A, Illustration of the TaERF1 promoter region showing the presence of the EIN3/EIL-binding site (EBS). The black line represents a 2,039-bp upstream sequence of the TaERF1 gene. The black boxes indicate the positions of EBS DNA motifs. The transcriptional start site (ATG) is indicated. B, Yeast one-hybrid assay showing the direct binding of TaEIL1 to the TaERF1 promoter regions. P1 and P2 regions from the TaERF1 promoter, as shown in A, were fused to pHIS2 to generate pHIS2-TaERF1pro-P1 and pHIS2-TaERF1pro-P2 constructs and cotransformed separately with GAL4-AD-TaEIL1 into the yeast (Saccharomyces cerevisiae) strain AH109. GAL4-AD-TaEIL1/pHIS2, GAL4-AD/pHIS2-TaERF1pro-P1, and GAL4-AD/pHIS2-TaERF1pro-P2 cotransformants were used as negative controls. The strains were first selected on SD-L/W medium and then transferred to SD-L/W/H medium with 100 mm 3-amino-1,2,4-triazole (3-AT) for growth analyses. C, Transient expression assays illustrating that TaMED25 and TaEIL1 synergistically activate TaERF1 transcription. The left circle indicates the combinations of A. tumefaciens strains harboring the indicated constructs. Representative images of N. benthamiana leaves were taken 48 h after infiltration. At least 10 independent N. benthamiana leaves were infiltrated in each experiment. D, Quantification of luminescence intensity in C. At least five independent determinations were analyzed in each experiment. Three biological replicates were performed, and similar results were obtained. E, Relative transcript levels of TaERF1 in TaMED25- or TaEIL1-silenced bread wheat leaves. Error bars in D and E represent sd; asterisks above the bars denote significant differences between the control and each treatment at P < 0.01 (**) and P < 0.001 (***; Student’s t test).
We next determined whether TaMED25 facilitates the transcriptional activation activity of TaEIL1. To this end, we carried out transient transcriptional activity assays in N. benthamiana leaves (Sun et al., 2012) using the TaERF1 promoter (2,039 bp; Fig. 8A; Supplemental Fig. S10) fused with the LUC gene as a reporter. A low level of LUC activity could be detected when TaERF1pro:LUC was coinfiltrated with EV into N. benthamiana (Fig. 8, C and D, coinfiltration 1). When 35S:TaEIL1 was coexpressed with TaERF1pro:LUC, an obvious induction in luminescence intensity was observed (Fig. 8, C and D, coinfiltration 2). Importantly, our results showed that coexpression of TaEIL1 and TaMED25 could elevate the LUC reporter activity almost 7 times compared with the EV control (Fig. 8, C and D, coinfiltration 4), indicating that the TaEIL1-TaMED25 interaction can contribute to the efficient enhancement of TaERF1 transcription. Interestingly, the expression of TaMED25 alone also resulted in a 3-fold induction of the LUC reporter activity (Fig. 8, C and D, coinfiltration 3). For this phenomenon, we speculated that the accumulated TaMED25 might interact physically with some EIN3-like TFs in N. benthamiana to activate the TaERF1pro:LUC expression.
Furthermore, we determined the effects of TaMED25 and TaEIL1 on the transcriptional expression of TaERF1 in bread wheat seedlings. qRT-PCR assays showed that the Bgt-triggered transcript levels of TaERF1 were reduced markedly in the TaMED25 and TaEIL1 knockdown lines (BSMV-TaMED25as and BSMV-TaEIL1as) compared with the BSMV-γ control plants (Fig. 8E), suggesting that TaMED25 and TaEIL1 are indeed essential for the transcriptional activation of TaERF1 in bread wheat plants. Taken together, the above results led us to conclude that TaMED25 and TaEIL1 synergistically activate TaERF1 transcription during the bread wheat-powdery mildew interaction.
Silencing of TaMED25, TaEIL1, and TaERF1 Enhances Bgt-Triggered ROS Production in Bread Wheat Leaves
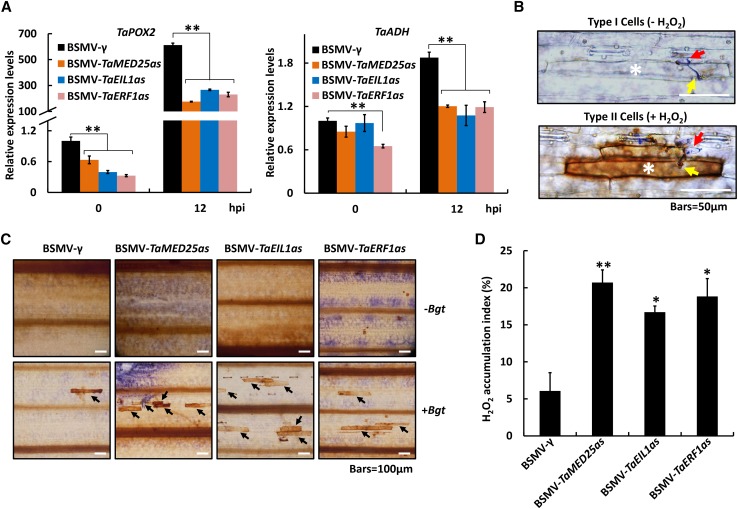
Plant cells generate superoxide or its dismutation product hydrogen peroxide (H2O2) upon successful recognition of pathogens (Torres et al., 2006). ROS is commonly believed to contribute to the establishment of plant defense reactions and the hypersensitive response, which are beneficial for fighting against biotrophic pathogens (Torres et al., 2006; Pastor et al., 2013). A recent study showed that TaERF1 represses stress-induced ROS accumulation by promoting the expression of some ROS-scavenging genes, including TaPOX2 and TaADH (Zhu et al., 2014). Here, we proposed that the negative regulatory role of the TaMED25-TaEIL1-TaERF1 module in bread wheat defense against powdery mildew might be due to the blockage of Bgt-triggered ROS accumulation. To verify this hypothesis, we first detected the expression of TaPOX2 and TaADH in bread wheat after Bgt inoculation and found marked induction of the two ROS-scavenging genes (Supplemental Fig. S11). The transcript levels of TaPOX2 and TaADH were further determined in the BSMV-TaMED25as, BSMV-TaEIL1as, and BSMV-TaERF1as wheat lines. As expected, knockdown of TaMED25, TaEIL1, or TaERF1 largely compromised the Bgt-triggered up-regulation of TaPOX2 and TaADH (Fig. 9A). We next analyzed the ROS accumulation in bread wheat leaves in response to Bgt infection. After the staining of Bgt-infected bread wheat epidermal cells with 3,3′-diaminobenzidine (DAB), two types of Bgt-infected cells could be observed microscopically: type I, cells without H2O2 production; and type II, cells with high-level H2O2 accumulation (Fig. 9B). Our analyses revealed that knockdown of TaMED25, TaEIL1, and TaERF1 all led to obvious increases of type II cells compared with the BSMV-γ control plants (Fig. 9, C and D; Supplemental Table S5), implying that ROS was accumulated more efficiently. These data support the hypothesis that the negative regulatory role of the TaMED25-TaEIL1-TaERF1 signaling module in bread wheat resistance against powdery mildew might be due partially to its repression of the Bgt-triggered ROS accumulation.
Figure 9.
Silencing of the TaMED25-TaEIL1-TaERF1 module leads to the down-regulation of ROS-scavenging gene expression and triggers the accumulation of H2O2 in Bgt-infected bread wheat cells. A, Quantification of TaPOX2 and TaADH transcript levels in TaMED25-, TaEIL1-, or TaERF1-silenced bread wheat leaves by qRT-PCR. B, Histochemical localization of H2O2 in Bgt-infected cells. Bread wheat leaves were stained with DAB to detect the accumulation of H2O2, while Bgt spores were visualized by Coomassie Brilliant Blue R250 staining. Two types of Bgt-infected bread wheat epidermal cells are shown: type I, cells without H2O2 production; and type II, cells with highly accumulated H2O2. White asterisks show the positions of Bgt-infected cells, red arrows point to germinated Bgt spores, and yellow arrows show the infection sites (appressorium) of Bgt. Bars = 50 μm. C, DAB staining showing H2O2 accumulation in BSMV-VIGS bread wheat leaves upon Bgt infection. The top row shows BSMV-VIGS bread wheat leaves without Bgt infection (−Bgt), and the bottom row shows Bgt-infected bread wheat leaves collected at 12 hpi (+Bgt). Black arrows show type II cells as described in B. Bars = 100 μm. D, Statistical analysis of the H2O2 accumulation index in bread wheat epidermal cells. The numbers of type I and type II Bgt-infected bread wheat epidermal cells (as shown in B) were counted microscopically, and the H2O2 accumulation index represents the percentage of type II cells out of all Bgt-infected epidermal cells. In each treatment, at least 10 independent bread wheat leaves (3–4 cm in length) were stained, and all the germinated Bgt spores on these leaves were analyzed microscopically. Three replicates were performed, and similar results were observed. Error bars in A and D represent the sd among at least three independent replicates; asterisks above the bars represent significant differences between the control and each treatment at P < 0.01 (**) and P < 0.05 (*; Student’s t test).
DISCUSSION
Powdery mildew can reduce bread wheat productivity considerably, raising concerns over future food security. Therefore, studies on the molecular mechanism of bread wheat resistance to powdery mildew are agriculturally important. In this study, our investigation of TaMED25 function in regulating bread wheat resistance to powdery mildew is mainly focused on its coaction with the TaEIL1 TF.
TaMED25 Negatively Regulates Bread Wheat Resistance against Powdery Mildew
In previous studies, several negative regulators of powdery mildew resistance were identified from barley and Arabidopsis (Büschges et al., 1997; Vogel et al., 2004; Opalski et al., 2005; Schultheiss et al., 2005, 2008). For instance, the membrane-anchored Mlo protein, first identified by map-based cloning in barley, was shown to repress defenses against Bgh (Büschges et al., 1997). Similarly, simultaneous knockout of all three MLO homoalleles in bread wheat confers heritable resistance to the powdery mildew fungus Bgt (Wang et al., 2014). The barley RAC/ROP (Rho of plants) protein RACB and its interacting protein RIC171, which encodes a ROP-interactive CRIB (CDC42/RAC interactive binding) motif-containing protein, contribute to barley susceptibility to Bgh by supporting entry of the powdery mildew fungus (Opalski et al., 2005; Schultheiss et al., 2005, 2008). In Arabidopsis, loss of function of the POWDERY MILDEW RESISTANT5 gene renders plants resistant to powdery mildew by affecting the pectin composition of host cell wall, a major barrier to pathogen infection (Vogel et al., 2004). Notably, these reported regulators mediate powdery mildew susceptibility mainly by facilitating powdery mildew entry or growth in host plant cells. In this study, we focus on the transcriptional regulation of bread wheat defense responses against powdery mildew pathogens.
At the core of transcriptional regulation is Mediator, a multisubunit complex that serves as a bridge between specific TFs and the RNA polymerase machinery to regulate transcription. Several Mediator subunits were functionally characterized recently in Arabidopsis defense responses (Hemsley et al., 2014). For instance, in Arabidopsis, MED16 regulates both the SA-mediated systemic acquired resistance and the JA/ET-mediated plant defense (Wathugala et al., 2012; Zhang et al., 2012). The Arabidopsis MED25 protein is required for the basal resistance to some necrotrophic fungal pathogens, probably due to its positive regulatory role in JA signaling (Kidd et al., 2009; Chen et al., 2012), and appears to attenuate plant defense against the hemibiotrophic pathogen F. oxysporum (Kidd et al., 2009), indicating that the Arabidopsis MED25 protein may differentially regulate plant immune responses against various pathogens, depending on the lifestyles of the pathogens.
The wheat powdery mildew fungus Bgt is an obligate biotrophic fungus that grows only on living host cells. So far, the potential function of the Mediator complex in crop plant immunity against obligate biotrophic pathogens, such as powdery mildew, has not been reported. In this study, we describe the biological function and molecular mechanism of the bread wheat Mediator subunit TaMED25 in regulating bread wheat responses to powdery mildew. Based on our results, TaMED25 contributes to bread wheat susceptibility to Bgt (Fig. 3; Supplemental Table S1). Moreover, we expanded our assays to barley, a close relative of bread wheat, and demonstrated a general role of the Mediator subunit MED25 in regulating disease resistance to powdery mildew in Triticeae species (Supplemental Fig. S9; Supplemental Table S2). Our results also revealed that knockdown of the endogenous TaMED25 genes leads to an obvious up-regulation of the SA-responsive TaPR1 and TaPR2 genes (Fig. 3E), indicating that TaMED25 might negatively impact the SA signaling-related defense. Consistent with our findings, in med16-1 mutant Arabidopsis plants, S. sclerotiorum infection also triggered significant up-regulation of the PR1 and PR2 transcript levels (Wang et al., 2015), implying a potentially conserved function of Mediator in defense signaling in dicots and monocots. Identification of the transcriptional Mediator subunit TaMED25 in this study should provide new insights for understanding the complex regulatory mechanisms of bread wheat defense against biotrophic pathogens.
Interaction with TaMED25 Facilitates the Transcriptional Activity of TaEIL1
In Arabidopsis, the Mediator subunits MED25 and MED16 modulate defense responses by integrating with JA and SA signaling pathways (Kidd et al., 2009; Chen et al., 2012; Zhang et al., 2012). However, the molecular basis of the Mediator-ET interaction in both model and crop plants remains to be identified. In this study, we provide evidence that both TaMED25 and TaEIL1 play negative regulatory roles in modulating bread wheat resistance against powdery mildew (Figs. 3 and 7). These findings led us to confirm the potential physical interaction between TaMED25 and TaEIL1 in vitro and in planta. Interestingly, our results reveal that the ACID of TaMED25 is sufficient for its interaction with TaEIL1 (Fig. 4C). On the contrary, a recent study in Arabidopsis showed that both the ACID and MD are essential for MED25 interaction with the master TF MYC2 of JA signaling (Chen et al., 2012). These results suggest that MED25 uses distinctive interaction modules for EIN3 and MYC2 binding in modulating the ET and JA signaling pathways in higher plants. Taken together, these results highlight that the Mediator subunit MED25 integrates different hormone signaling pathways by interacting with specific TFs in different plant species.
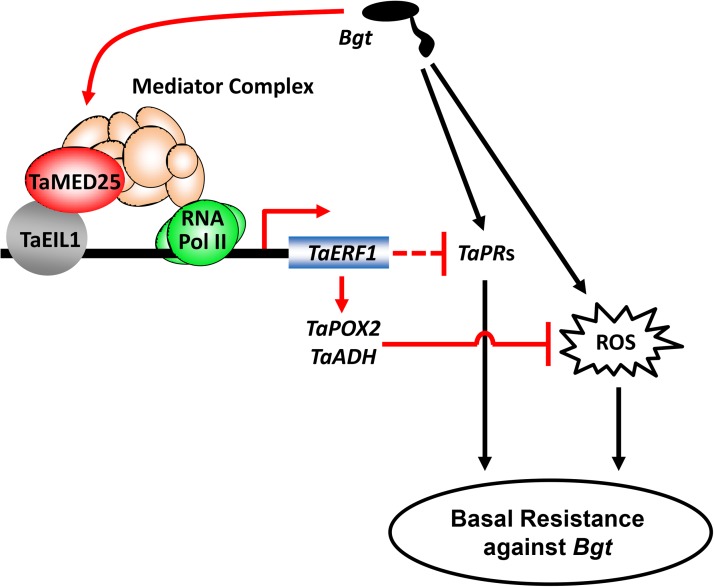
Significantly, we discovered a coaction mechanism of TaMED25 and TaEIL1 to achieve the transcriptional activation activity of TaEIL1. First, we show that the N-terminal and C-terminal domains of TaEIL1 have transcriptional activation activity in yeast cells (Fig. 5). Second, corresponding to the transcriptional activation activities, the two domains of TaEIL1 bind differentially to TaMED25 (Fig. 4D). Third, we demonstrate that TaMED25 and TaEIL1 synergistically activate TaERF1 transcription in N. benthamiana leaves (Fig. 8, C and D). These results are expected because Mediator is believed to contribute to the recruitment of Pol II for transcriptional activation; therefore, the interaction strength of the N- or C-terminal domain of TaEIL1 with TaMED25 might correspond to their transcriptional activation activity, respectively. Although a recent investigation in Arabidopsis provided evidence that MED25 interacts physically with EIN3, their interaction mechanism and the underlying biological significance in plant disease resistance remain unknown (Yang et al., 2014). Here, we propose a working model to elucidate the coaction mechanism of TaMED25 and TaEIL1 in regulating bread wheat immune responses: physical interaction between TaEIL1 and TaMED25 may contribute to the recruitment of Pol II to the promoter regions of TaEIL1 target genes such as TaERF1, consequently fine-tuning bread wheat immune responses to powdery mildew (Fig. 10).
Figure 10.
Proposed working model of the TaMED25-TaEIL1-TaERF1 module in the bread wheat immune response against powdery mildew fungus. When bread wheat leaves are infected by the powdery mildew fungus Bgt, the endogenous PR genes, together with ROS, are highly induced to enhance the bread wheat immune response against Bgt (represented by black lines). Meanwhile, physical interaction between TaEIL1 and TaMED25 may lead to the recruitment of Pol II to promote the transcription of TaERF1. TaERF1 may further repress the expression of PR genes and the accumulation of ROS to partly attenuate bread wheat basal resistance against the Bgt fungus (represented by red lines).
The TaEIL1-TaERF1 Module Antagonizes Bread Wheat Resistance against Powdery Mildew
Previous elegant studies have revealed that phytohormones such as ET, JA, and SA are important regulators of plant immunity (Robert-Seilaniantz et al., 2011). It is generally believed that ET/JA signaling, which synergistically regulates plant tolerance to the necrotrophic pathogens, counteracts SA signaling, which mediates plant defense against biotrophic pathogens (Knoester et al., 1998; Li et al., 2004; Broekaert et al., 2006; Chen et al., 2009; Robert-Seilaniantz et al., 2011; Yi et al., 2014). Consistent with studies in Arabidopsis, a recent work revealed that TaERF1 positively regulates bread wheat resistance to the necrotrophic pathogen R. cerealis (Zhu et al., 2014), manifesting a conserved role of ET-mediated resistance against necrotrophic pathogens in different plant species. However, the molecular basis of ET-mediated bread wheat immune responses to the obligate biotrophic fungus powdery mildew is poorly understood. Here, we show that TaEIL1 activates TaERF1 transcription to negatively modulate bread wheat immunity to powdery mildew, highlighting that TaERF1 differentially regulates bread wheat defense to diverse types of pathogens.
The production of ROS is one of the earliest events in pathogen-infected plant cells. The accumulation of ROS in plant cells may trigger callose deposition, cell wall strengthening, defense gene activation, and, more importantly, hypersensitive cell death (also termed the hypersensitive response; Torres et al., 2006; Luna et al., 2011; Kim et al., 2012; Lehmann et al., 2015). ROS seems to play complicated, even opposite, roles in response to biotrophic and necrotrophic pathogens. For instance, the ROS burst is beneficial for potato (Solanum tuberosum) to fight against the near-obligate hemibiotrophic pathogen Phytophthora infestans but increases susceptibility to the necrotrophic pathogen Alternaria solani (Kobayashi et al., 2012). In bread wheat, TaERF1 was recently reported to down-regulate pathogen-triggered ROS accumulation by promoting the expression of ROS-scavenging genes such as TaPOX2 and TaADH and, consequently, enhance bread wheat resistance to the necrotrophic pathogen R. cerealis (Zhu et al., 2014). However, in our study, the TaERF1-mediated repression of ROS actually compromises bread wheat immunity against powdery mildew fungus to a certain extent (Figs. 3, 7, and 9).
Our results also raise an important question: what is the biological significance of TaERF1 action in the bread wheat-powdery mildew interaction? Considering that ROS overaccumulation as well as the hypersensitive immune responses may be potential threats to plant fitness (Torres et al., 2006; Lehmann et al., 2015), it is reasonable to propose that the TaERF1-mediated attenuation of ROS may play key roles in dampening bread wheat defense signaling, thus minimizing the costs of plant growth.
Together, our findings support the notion that the TaEIL1-TaERF1 module antagonizes bread wheat resistance to the obligate biotrophic fungus powdery mildew. This understanding could drive strategies to enhance the disease resistance of bread wheat in the future.
MATERIALS AND METHODS
Plant and Fungal Materials
Bread wheat (Triticum aestivum ‘Beijing 837’) and barley (Hordeum vulgare ‘Golden Promise’), which were used for gene cloning and powdery mildew infection, were grown in a growth chamber under a 16-h/8-h, 20°C/18°C day/night cycle; Nicotiana benthamiana was grown in a greenhouse at 22°C ± 1°C with a 16-h/8-h light period.
Blumeria graminis f. sp. tritici isolate E09 and Blumeria graminis f. sp. hordei isolate K1 were maintained separately on wheat ‘Beijing 837’ and barley ‘I10’ and kept at 70% relative humidity and a 16-h/8-h, 20°C/18°C day/night cycle.
DNA Constructs and Primers
DNA constructs used in this study were generated following standard molecular biology protocols or Gateway technology (Invitrogen). More details of the DNA constructs are listed in Supplemental Table S6. All primers used for vector construction in the study are listed in Supplemental Table S7.
Cloning of TaMED25 and Sequence Analyses
TaMED25 was cloned by the homology cloning strategy. First, the Arabidopsis (Arabidopsis thaliana) MED25 protein sequence was used as a query to find its ortholog protein in bread wheat according to the predicted protein database of Triticum urartu and Aegilops tauschii (Jia et al., 2013; Ling et al., 2013). Specific primers (Supplemental Table S7) were designed according to the genome sequences of T. urartu and A. tauschii (scaffold61769 and scaffold61247; Supplemental Fig. S3), and a nested PCR strategy was employed to obtain TaMED25 coding sequences from bread wheat ‘Beijing 837’ complementary DNA. The PCR product was ligated to cloning vector pJET1.2 (Thermo; K1232) for sequencing. The sequencing results were then aligned with the bread wheat ‘Chinese Spring’ genome and coding sequences using the service provided by the International Wheat Genome Sequencing Consortium (http://wheat-urgi.versailles.inra.fr/Seq-Repository/BLAST), and the TaMED25 gene structure and predicted chromosomal locations were analyzed. The obtained sequences were deposited in GenBank.
RNA Extraction and Gene Expression Analyses
For gene expression assays in bread wheat plants, seeds of bread wheat ‘Beijing 837’ were planted in soil. The 7-d-old seedlings were treated with powdery mildew for different times and then harvested. For qRT-PCR analyses, total RNA was extracted using Trizol (Invitrogen) reagent. About 2 μg of total RNA and Moloney murine leukemia virus reverse transcriptase (Promega) were further used for reverse transcription. SYBR Premix Ex Taq (Perfect Real Time; TaKaRa) was used for qRT-PCR assay, and the expression levels of target genes were normalized to TaGAPDH. For the expression assay in barley, cv Golden Promise was collected after certain treatments, and 2 μg of total RNA was further used for reverse transcription. The expression levels of genes were normalized to the internal control gene HvACTIN1. Primers for qRT-PCR assays are listed in Supplemental Table S7. The statistical significance was evaluated by Student’s t test.
Yeast Experiments
For yeast one-hybrid assay, the pHIS2 derivatives were cotransformed into the yeast (Saccharomyces cerevisiae) strain AH109 with the construct GAL4-AD-TaEIL1 or GAL4-AD EV. The transformants first grew on SD-L/W and then were transferred to SD-L/W/H supplemented with 100 mm 3-amino-1,2,4-triazole.
For yeast two-hybrid analysis, GAL4-AD and GAL4-BD derivatives were cotransformed into the yeast (Saccharomyces cerevisiae) strain AH109 and grown on SD-L/W. Subsequently, the cotransformed yeast strains were transferred to SD-L/W/H for interaction analysis.
For transcriptional activation activity assays, GAL4-BD derivatives were separately transformed into yeast and grown on synthetic dextrose medium lacking Leu. The transformants were then dropped on SD-L/H/A for transcriptional activation activity evaluation according to their growth status.
BSMV-Mediated Gene Silencing
Constructs of pCaBS-γ-LIC derivatives (pCaBS-γ-TaMED25as, pCaBS-γ-HvMED25as, pCaBS-γ-TaEIL1as, and pCaBS-γ-TaERF1as; Supplemental Fig. S5) harboring approximately 300-bp antisense fragments of TaMED25, HvMED25, TaEIL1, and TaERF1 (Supplemental Figs. S6 and S8) were used to transiently silence endogenous TaMED25, HvMED25, TaEIL1, and TaERF1 in bread wheat or barley plants, as described previously (Yuan et al., 2011). About 15 d later, the newly grown upper wheat leaves with virus symptoms were collected for further analyses.
Powdery Mildew Infection and Microcolony Formation Analyses
Powdery mildew infection and microscopic analyses were performed as reported previously (Shen et al., 2007) with some modifications. Briefly, Bgt strain E09 was used for the inoculation of wheat leaves and Bgh strain K1 was used for the inoculation of barley leaves. Three days post inoculation (72 hpi), leaves were collected and fixed with ethanol:acetic acid solution (1:1, v/v), followed by a destaining process with lactoglycerol solution (lactic acid:glycerol:water, 1:1:1, v/v/v). Finally, leaves were stained with 0.1% (w/v) Coomassie Brilliant Blue R250, and the MI% was microscopically analyzed as reported previously with modifications (Shen et al., 2007). Briefly, the successfully colonized B. graminis as well as spores that did not form colonies were counted separately, and the B. graminis MI% represents the percentage of successfully colonized B. graminis out of all analyzed spores. For each treatment, the spores from 10 to 15 independent leaves (the third and fourth leaves from 10–15 BSMV-VIGS bread wheat plants, 3–4 cm in length) were analyzed separately, and the mean MI% values were calculated. Three independent replications were conducted, and the significant difference level of MI% values between BSMV-γ control and each treatment was evaluated by Student’s t test.
LCI Assays
The LCI assays for the interaction between TaMED25 and TaEIL1 were performed in N. benthamiana leaves as described previously (Sun et al., 2013) with modifications. The full-length TaMED25 coding region or truncated derivatives were fused with the N-terminal part of the luciferase reporter gene LUC. Similarly, the full-length TaEIL1 coding region or truncated derivatives were fused with the C-terminal part of LUC. Agrobacteria harboring nLUC and cLUC derivative constructs were coinfiltrated into N. benthamiana, and the infiltrated leaves were analyzed for LUC activity 48 h after infiltration.
Transactivation Assays in N. benthamiana Leaves
The transactivation assays were performed in N. benthamiana leaves as described previously (Sun et al., 2012). The 2-kb TaERF1 promoter was amplified by PCR and fused with the luciferase reporter gene LUC through Gateway reactions (Invitrogen) into the plant binary vector pGWB35 (Nakagawa et al., 2007) to generate the reporter construct TaERF1pro:LUC. For the effector constructs 35S:TaMED25-Myc and 35S:TaEIL1-Myc, the full-length TaMED25 and TaEIL1 coding sequences were amplified by PCR and cloned into the plant binary vector pGWB17 (Nakagawa et al., 2007). Agrobacteria harboring reporter and effector constructs were coinfiltrated into N. benthamiana, and the LUC signals were analyzed 48 h after infiltration.
Colocalization of TaMED25 and TaEIL1 in Bread Wheat Epidermal Cells
The HBT-TaEIL1-GFP and HBT-TaMED25-RFP constructs were transiently coexpressed in bread wheat epidermal leaf cells by biolistic delivery as described previously (Liu et al., 2014). Fluorescence signals were observed with a confocal microscope (Leica TCS-SP4) 48 h after biolistic delivery.
BiFC Assay in Bread Wheat Protoplasts
The HBT-nYFP/cYFP derivative constructs carrying TaMED25 and TaEIL1 coding sequences were cotransfected into wheat protoplast cells as described previously (Yoo et al., 2007). About 24 h after transfection, 4,6-diamidino-2-phenylindole solution was added into the protoplast solution for nuclei staining, and YFP fluorescence was observed with a confocal microscope (Leica TCS-SP4).
FLIM-FRET Assay in Bread Wheat Epidermal Cells
TaEIL1 and TaMED25 were fused to CFP and mYFP, respectively, to generate donor and receptor proteins. The experiments were performed as described previously (Shen et al., 2007) with modifications. CFP fluorescence was recorded with the confocal channel using a 480- to 520-nm bandpass filter, and FLIM images were recorded using the PicoHarp 300 TCSPC module. Acceptor photobleaching was performed within a region of interest (ROI). CFP intensities in a ROI were averaged and plotted as a function of time.
DAB Staining for H2O2 Detection
The accumulation of H2O2 was detected by DAB staining. Briefly, wheat leaves were collected 12 hpi and stained in 1 mg mL−1 DAB solution for 10 min of vacuum infiltration and 8 h of incubation at room temperature. The leaves were destained by boiling in acetic acid:glycerol:ethanol solution (1:1:3, v/v/v) for 5 min. H2O2 accumulation in Bgt-infected epidermal cells was analyzed microscopically. Briefly, two types of Bgt-infected bread wheat epidermal cells were calculated microscopically: type I, cells without H2O2 production; and type II, cells with highly accumulated H2O2 (Fig. 9B). The H2O2 accumulation index represents the percentage of type II cells out of all Bgt-infected epidermal cells. In each treatment, at least 10 independent bread wheat leaves (3–4 cm in length) were stained, and all the germinated Bgt spores on these leaves were analyzed. The statistical significance was evaluated by Student’s t test based on three independent replicates.
Sequence data from this study can be found in the GenBank database (http://www.ncbi.nlm.nih.gov/) under the following accession numbers: TaMED25-A, KU030834; TaMED25-B, KU030835; TaMED25-D, KU030836; TaEIL1, KU030837; TaERF1, EF583940; HvMED25, AK252911; BdMED25, XP_010238042; SiMED25, XP_004959349; ZmMED25, XP_008670123; OsMED25, EEE69415; CsMED25, XP_006484420; GmMED25, XP_006574879; and AtMED25, NP_173925.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Sequence comparison of AtMED25 and TaMED25 proteins.
Supplemental Figure S2. Allelic variation at the TaMED25-A, TaMED25-B, and TaMED25-D coding regions.
Supplemental Figure S3. Nucleotide sequences of scaffold61769 from T. urartu and scaffold61247 from A. tauschii that contain the TaMED25 gene.
Supplemental Figure S4. Schematic diagram of TaMED25 gene structures.
Supplemental Figure S5. Construction of BSMV-VIGS vectors.
Supplemental Figure S6. Nucleotide sequences from TaMED25, TaEIL1, and TaERF1 that were used for BSMV-mediated silencing.
Supplemental Figure S7. TaPR genes are induced by Bgt infection.
Supplemental Figure S8. Coding sequence of HvMED25.
Supplemental Figure S9. Knockdown of barley MED25 reduces barley susceptibility to the Bgh fungus.
Supplemental Figure S10. Promoter sequence of TaERF1 used for yeast one-hybrid analysis and transactivation assay.
Supplemental Figure S11. TaERF1 target genes TaPOX2 and TaADH are induced by Bgt infection.
Supplemental Table S1. Statistical analysis of B. graminis MI% on the BSMV-VIGS bread wheat lines.
Supplemental Table S2. Statistical analysis of B. graminis MI% on the BSMV-VIGS barley lines.
Supplemental Table S3. BiFC assay showing the interaction of TaMED25 and TaEIL1 in bread wheat protoplasts.
Supplemental Table S4. CFP fluorescence decay frequency within ROI in the FLIM-FRET experiment to detect the interaction of TaEIL1 and TaMED25.
Supplemental Table S5. Statistical analysis of the H2O2 accumulation index in Bgt-infected bread wheat epidermal cells.
Supplemental Table S6. DNA constructs in this study.
Supplemental Table S7. Primers used in this study.
Supplementary Material
Acknowledgments
We thank Dr. Long Mao for providing the powdery mildew E09 strain and bread wheat ‘Beijing837’, Dr. Dawei Li for sharing the BSMV silencing system, Dr. Qian-hua Shen and Dr. Guangyao Zhao for technical help, and Dr. Yan Liang for advice in writing the article.
Glossary
- Bgt
Blumeria graminis f. sp. tritici
- ET
ethylene
- TF
transcription factor
- JA
jasmonate
- SA
salicylic acid
- ACID
activator-interacting domain
- ROS
reactive oxygen species
- vWF-A
von Willebrand factor A domain
- MD
middle domain
- qRT
quantitative reverse transcription
- hpi
hours post inoculation
- VIGS
virus-induced gene silencing
- BSMV
Barley stripe mosaic virus
- MI%
microcolony formation index
- EV
empty vector
- Bgh
Blumeria graminis f. sp. hordei
- LCI
luciferase complementation imaging
- BiFC
bimolecular fluorescence complementation
- FRET
Förster resonance energy transfer
- FLIM
fluorescence lifetime imaging
- H2O2
hydrogen peroxide
- DAB
3,3′-diaminobenzidine
- ROI
region of interest
- SD-L/W
synthetic dextrose medium lacking Leu and Trp
- SD-L/W/H
synthetic dextrose medium lacking Leu, Trp, and His
- SD-L/H/A
synthetic dextrose medium lacking Leu, His, and adenine
Footnotes
This work was supported by the Institute of Crop Science and the Agricultural Science and Technology Innovation Program of the Chinese Academy of Agricultural Sciences, the National Natural Science Foundation of China (grant no. 31501297), and the Ministry of Agriculture of China (grant no. 2016ZX08009–003–003).
References
- Bäckström S, Elfving N, Nilsson R, Wingsle G, Björklund S (2007) Purification of a plant mediator from Arabidopsis thaliana identifies PFT1 as the Med25 subunit. Mol Cell 26: 717–729 [DOI] [PubMed] [Google Scholar]
- Bent AF, Innes RW, Ecker JR, Staskawicz BJ (1992) Disease development in ethylene-insensitive Arabidopsis thaliana infected with virulent and avirulent Pseudomonas and Xanthomonas pathogens. Mol Plant Microbe Interact 5: 372–378 [DOI] [PubMed] [Google Scholar]
- Berrocal-Lobo M, Molina A, Solano R (2002) Constitutive expression of ETHYLENE-RESPONSE-FACTOR1 in Arabidopsis confers resistance to several necrotrophic fungi. Plant J 29: 23–32 [DOI] [PubMed] [Google Scholar]
- Bourras S, McNally KE, Ben-David R, Parlange F, Roffler S, Praz CR, Oberhaensli S, Menardo F, Stirnweis D, Frenkel Z, et al. (2015) Multiple avirulence loci and allele-specific effector recognition control the Pm3 race-specific resistance of wheat to powdery mildew. Plant Cell 27: 2991–3012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broekaert WF, Delauré SL, De Bolle MF, Cammue BP (2006) The role of ethylene in host-pathogen interactions. Annu Rev Phytopathol 44: 393–416 [DOI] [PubMed] [Google Scholar]
- Büschges R, Hollricher K, Panstruga R, Simons G, Wolter M, Frijters A, van Daelen R, van der Lee T, Diergaarde P, Groenendijk J, et al. (1997) The barley Mlo gene: a novel control element of plant pathogen resistance. Cell 88: 695–705 [DOI] [PubMed] [Google Scholar]
- Caillaud MC, Asai S, Rallapalli G, Piquerez S, Fabro G, Jones JD (2013) A downy mildew effector attenuates salicylic acid-triggered immunity in Arabidopsis by interacting with the host mediator complex. PLoS Biol 11: e1001732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canet JV, Dobón A, Tornero P (2012) Non-recognition-of-BTH4, an Arabidopsis mediator subunit homolog, is necessary for development and response to salicylic acid. Plant Cell 24: 4220–4235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerdán PD, Chory J (2003) Regulation of flowering time by light quality. Nature 423: 881–885 [DOI] [PubMed] [Google Scholar]
- Chadick JZ, Asturias FJ (2005) Structure of eukaryotic Mediator complexes. Trends Biochem Sci 30: 264–271 [DOI] [PubMed] [Google Scholar]
- Chao Q, Rothenberg M, Solano R, Roman G, Terzaghi W, Ecker JR (1997) Activation of the ethylene gas response pathway in Arabidopsis by the nuclear protein ETHYLENE-INSENSITIVE3 and related proteins. Cell 89: 1133–1144 [DOI] [PubMed] [Google Scholar]
- Chen H, Xue L, Chintamanani S, Germain H, Lin H, Cui H, Cai R, Zuo J, Tang X, Li X, et al. (2009) ETHYLENE INSENSITIVE3 and ETHYLENE INSENSITIVE3-LIKE1 repress SALICYLIC ACID INDUCTION DEFICIENT2 expression to negatively regulate plant innate immunity in Arabidopsis. Plant Cell 21: 2527–2540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R, Jiang H, Li L, Zhai Q, Qi L, Zhou W, Liu X, Li H, Zheng W, Sun J, et al. (2012) The Arabidopsis mediator subunit MED25 differentially regulates jasmonate and abscisic acid signaling through interacting with the MYC2 and ABI5 transcription factors. Plant Cell 24: 2898–2916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng MC, Liao PM, Kuo WW, Lin TP (2013) The Arabidopsis ETHYLENE RESPONSE FACTOR1 regulates abiotic stress-responsive gene expression by binding to different cis-acting elements in response to different stress signals. Plant Physiol 162: 1566–1582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng W, Nickle DC, Learn GH, Maust B, Mullins JI (2007) ViroBLAST: a stand-alone BLAST web server for flexible queries of multiple databases and user’s datasets. Bioinformatics 23: 2334–2336 [DOI] [PubMed] [Google Scholar]
- Dhawan R, Luo H, Foerster AM, Abuqamar S, Du HN, Briggs SD, Mittelsten Scheid O, Mengiste T (2009) HISTONE MONOUBIQUITINATION1 interacts with a subunit of the mediator complex and regulates defense against necrotrophic fungal pathogens in Arabidopsis. Plant Cell 21: 1000–1019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan X, Wang X, Fu Y, Tang C, Li X, Cheng Y, Feng H, Huang L, Kang Z (2013) TaEIL1, a wheat homologue of AtEIN3, acts as a negative regulator in the wheat-stripe rust fungus interaction. Mol Plant Pathol 14: 728–739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elfving N, Davoine C, Benlloch R, Blomberg J, Brännström K, Müller D, Nilsson A, Ulfstedt M, Ronne H, Wingsle G, et al. (2011) The Arabidopsis thaliana Med25 mediator subunit integrates environmental cues to control plant development. Proc Natl Acad Sci USA 108: 8245–8250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, Duan X, Tang C, Li X, Voegele RT, Wang X, Wei G, Kang Z (2014) TaADF7, an actin-depolymerizing factor, contributes to wheat resistance against Puccinia striiformis f. sp. tritici. Plant J 78: 16–30 [DOI] [PubMed] [Google Scholar]
- Hemsley PA, Hurst CH, Kaliyadasa E, Lamb R, Knight MR, De Cothi EA, Steele JF, Knight H (2014) The Arabidopsis mediator complex subunits MED16, MED14, and MED2 regulate mediator and RNA polymerase II recruitment to CBF-responsive cold-regulated genes. Plant Cell 26: 465–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Wheat Genome Sequencing Consortium (2014) A chromosome-based draft sequence of the hexaploid bread wheat (Triticum aestivum) genome. Science 345: 1251788. [DOI] [PubMed] [Google Scholar]
- Jia J, Zhao S, Kong X, Li Y, Zhao G, He W, Appels R, Pfeifer M, Tao Y, Zhang X, et al. (2013) Aegilops tauschii draft genome sequence reveals a gene repertoire for wheat adaptation. Nature 496: 91–95 [DOI] [PubMed] [Google Scholar]
- Kidd BN, Edgar CI, Kumar KK, Aitken EA, Schenk PM, Manners JM, Kazan K (2009) The mediator complex subunit PFT1 is a key regulator of jasmonate-dependent defense in Arabidopsis. Plant Cell 21: 2237–2252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim C, Meskauskiene R, Zhang S, Lee KP, Lakshmanan Ashok M, Blajecka K, Herrfurth C, Feussner I, Apel K (2012) Chloroplasts of Arabidopsis are the source and a primary target of a plant-specific programmed cell death signaling pathway. Plant Cell 24: 3026–3039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight H, Mugford SG, Ulker B, Gao D, Thorlby G, Knight MR (2009) Identification of SFR6, a key component in cold acclimation acting post-translationally on CBF function. Plant J 58: 97–108 [DOI] [PubMed] [Google Scholar]
- Knight H, Thomson AJ, McWatters HG (2008) Sensitive to freezing6 integrates cellular and environmental inputs to the plant circadian clock. Plant Physiol 148: 293–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoester M, van Loon LC, van den Heuvel J, Hennig J, Bol JF, Linthorst HJ (1998) Ethylene-insensitive tobacco lacks nonhost resistance against soil-borne fungi. Proc Natl Acad Sci USA 95: 1933–1937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi M, Yoshioka M, Asai S, Nomura H, Kuchimura K, Mori H, Doke N, Yoshioka H (2012) StCDPK5 confers resistance to late blight pathogen but increases susceptibility to early blight pathogen in potato via reactive oxygen species burst. New Phytol 196: 223–237 [DOI] [PubMed] [Google Scholar]
- Lehmann S, Serrano M, L’Haridon F, Tjamos SE, Metraux JP (2015) Reactive oxygen species and plant resistance to fungal pathogens. Phytochemistry 112: 54–62 [DOI] [PubMed] [Google Scholar]
- Li J, Brader G, Palva ET (2004) The WRKY70 transcription factor: a node of convergence for jasmonate-mediated and salicylate-mediated signals in plant defense. Plant Cell 16: 319–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling HQ, Zhao S, Liu D, Wang J, Sun H, Zhang C, Fan H, Li D, Dong L, Tao Y, et al. (2013) Draft genome of the wheat A-genome progenitor Triticum urartu. Nature 496: 87–90 [DOI] [PubMed] [Google Scholar]
- Liu J, Cheng X, Liu D, Xu W, Wise R, Shen QH (2014) The miR9863 family regulates distinct Mla alleles in barley to attenuate NLR receptor-triggered disease resistance and cell-death signaling. PLoS Genet 10: e1004755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luna E, Pastor V, Robert J, Flors V, Mauch-Mani B, Ton J (2011) Callose deposition: a multifaceted plant defense response. Mol Plant Microbe Interact 24: 183–193 [DOI] [PubMed] [Google Scholar]
- Nakagawa T, Kurose T, Hino T, Tanaka K, Kawamukai M, Niwa Y, Toyooka K, Matsuoka K, Jinbo T, Kimura T (2007) Development of series of Gateway Binary Vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J Biosci Bioeng 104: 34–41 [DOI] [PubMed] [Google Scholar]
- Opalski KS, Schultheiss H, Kogel KH, Hückelhoven R (2005) The receptor-like MLO protein and the RAC/ROP family G-protein RACB modulate actin reorganization in barley attacked by the biotrophic powdery mildew fungus Blumeria graminis f.sp. hordei. Plant J 41: 291–303 [DOI] [PubMed] [Google Scholar]
- Parlange F, Roffler S, Menardo F, Ben-David R, Bourras S, McNally KE, Oberhaensli S, Stirnweis D, Buchmann G, Wicker T, et al. (2015) Genetic and molecular characterization of a locus involved in avirulence of Blumeria graminis f. sp. tritici on wheat Pm3 resistance alleles. Fungal Genet Biol 82: 181–192 [DOI] [PubMed] [Google Scholar]
- Pastor V, Luna E, Ton J, Cerezo M, García-Agustín P, Flors V (2013) Fine tuning of reactive oxygen species homeostasis regulates primed immune responses in Arabidopsis. Mol Plant Microbe Interact 26: 1334–1344 [DOI] [PubMed] [Google Scholar]
- Robert-Seilaniantz A, Grant M, Jones JDG (2011) Hormone crosstalk in plant disease and defense: more than just jasmonate-salicylate antagonism. Annu Rev Phytopathol 49: 317–343 [DOI] [PubMed] [Google Scholar]
- Schmidt R, Mieulet D, Hubberten HM, Obata T, Hoefgen R, Fernie AR, Fisahn J, San Segundo B, Guiderdoni E, Schippers JH, et al. (2013) Salt-responsive ERF1 regulates reactive oxygen species-dependent signaling during the initial response to salt stress in rice. Plant Cell 25: 2115–2131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultheiss H, Hensel G, Imani J, Broeders S, Sonnewald U, Kogel KH, Kumlehn J, Hückelhoven R (2005) Ectopic expression of constitutively activated RACB in barley enhances susceptibility to powdery mildew and abiotic stress. Plant Physiol 139: 353–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultheiss H, Preuss J, Pircher T, Eichmann R, Hückelhoven R (2008) Barley RIC171 interacts with RACB in planta and supports entry of the powdery mildew fungus. Cell Microbiol 10: 1815–1826 [DOI] [PubMed] [Google Scholar]
- Shen QH, Saijo Y, Mauch S, Biskup C, Bieri S, Keller B, Seki H, Ulker B, Somssich IE, Schulze-Lefert P (2007) Nuclear activity of MLA immune receptors links isolate-specific and basal disease-resistance responses. Science 315: 1098–1103 [DOI] [PubMed] [Google Scholar]
- Solano R, Stepanova A, Chao Q, Ecker JR (1998) Nuclear events in ethylene signaling: a transcriptional cascade mediated by ETHYLENE-INSENSITIVE3 and ETHYLENE-RESPONSE-FACTOR1. Genes Dev 12: 3703–3714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song S, Qi T, Huang H, Ren Q, Wu D, Chang C, Peng W, Liu Y, Peng J, Xie D (2011) The jasmonate-ZIM domain proteins interact with the R2R3-MYB transcription factors MYB21 and MYB24 to affect jasmonate-regulated stamen development in Arabidopsis. Plant Cell 23: 1000–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Qi L, Li Y, Chu J, Li C (2012) PIF4-mediated activation of YUCCA8 expression integrates temperature into the auxin pathway in regulating Arabidopsis hypocotyl growth. PLoS Genet 8: e1002594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Qi L, Li Y, Zhai Q, Li C (2013) PIF4 and PIF5 transcription factors link blue light and auxin to regulate the phototropic response in Arabidopsis. Plant Cell 25: 2102–2114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomma BP, Eggermont K, Tierens KF, Broekaert WF (1999) Requirement of functional Ethylene-Insensitive 2 gene for efficient resistance of Arabidopsis to infection by Botrytis cinerea. Plant Physiol 121: 1093–1102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres MA, Jones JD, Dangl JL (2006) Reactive oxygen species signaling in response to pathogens. Plant Physiol 141: 373–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Loon LC, Geraats BP, Linthorst HJ (2006) Ethylene as a modulator of disease resistance in plants. Trends Plant Sci 11: 184–191 [DOI] [PubMed] [Google Scholar]
- Vogel JP, Raab TK, Somerville CR, Somerville SC (2004) Mutations in PMR5 result in powdery mildew resistance and altered cell wall composition. Plant J 40: 968–978 [DOI] [PubMed] [Google Scholar]
- Wang C, Yao J, Du X, Zhang Y, Sun Y, Rollins J, Mou Z (2015) The Arabidopsis Mediator complex subunit16 is a key component of basal resistance against the necrotrophic fungal pathogen Sclerotinia sclerotiorum. Plant Physiol 169: 856–872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Cheng X, Shan Q, Zhang Y, Liu J, Gao C, Qiu JL (2014) Simultaneous editing of three homoeoalleles in hexaploid bread wheat confers heritable resistance to powdery mildew. Nat Biotechnol 32: 947–951 [DOI] [PubMed] [Google Scholar]
- Wathugala DL, Hemsley PA, Moffat CS, Cremelie P, Knight MR, Knight H (2012) The Mediator subunit SFR6/MED16 controls defence gene expression mediated by salicylic acid and jasmonate responsive pathways. New Phytol 195: 217–230 [DOI] [PubMed] [Google Scholar]
- Xu R, Li Y (2011) Control of final organ size by Mediator complex subunit 25 in Arabidopsis thaliana. Development 138: 4545–4554 [DOI] [PubMed] [Google Scholar]
- Yalpani N, Silverman P, Wilson TM, Kleier DA, Raskin I (1991) Salicylic acid is a systemic signal and an inducer of pathogenesis-related proteins in virus-infected tobacco. Plant Cell 3: 809–818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C, Lu X, Ma B, Chen SY, Zhang JS (2015a) Ethylene signaling in rice and Arabidopsis: conserved and diverged aspects. Mol Plant 8: 495–505 [DOI] [PubMed] [Google Scholar]
- Yang L, Li B, Zheng XY, Li J, Yang M, Dong X, He G, An C, Deng XW (2015b) Salicylic acid biosynthesis is enhanced and contributes to increased biotrophic pathogen resistance in Arabidopsis hybrids. Nat Commun 6: 7309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Ou B, Zhang J, Si W, Gu H, Qin G, Qu LJ (2014) The Arabidopsis Mediator subunit MED16 regulates iron homeostasis by associating with EIN3/EIL1 through subunit MED25. Plant J 77: 838–851 [DOI] [PubMed] [Google Scholar]
- Yi SY, Kim JH, Joung YH, Lee S, Kim WT, Yu SH, Choi D (2004) The pepper transcription factor CaPF1 confers pathogen and freezing tolerance in Arabidopsis. Plant Physiol 136: 2862–2874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi SY, Shirasu K, Moon JS, Lee SG, Kwon SY (2014) The activated SA and JA signaling pathways have an influence on flg22-triggered oxidative burst and callose deposition. PLoS ONE 9: e88951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo SD, Cho YH, Sheen J (2007) Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat Protoc 2: 1565–1572 [DOI] [PubMed] [Google Scholar]
- Yuan C, Li C, Yan L, Jackson AO, Liu Z, Han C, Yu J, Li D (2011) A high throughput barley stripe mosaic virus vector for virus induced gene silencing in monocots and dicots. PLoS ONE 6: e26468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Zhang D, Chen J, Yang Y, Huang Z, Huang D, Wang XC, Huang R (2004) Tomato stress-responsive factor TSRF1 interacts with ethylene responsive element GCC box and regulates pathogen resistance to Ralstonia solanacearum. Plant Mol Biol 55: 825–834 [DOI] [PubMed] [Google Scholar]
- Zhang X, Wang C, Zhang Y, Sun Y, Mou Z (2012) The Arabidopsis mediator complex subunit16 positively regulates salicylate-mediated systemic acquired resistance and jasmonate/ethylene-induced defense pathways. Plant Cell 24: 4294–4309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Yao J, Zhang Y, Sun Y, Mou Z (2013) The Arabidopsis Mediator complex subunits MED14/SWP and MED16/SFR6/IEN1 differentially regulate defense gene expression in plant immune responses. Plant J 75: 484–497 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Wu H, Wang N, Fan H, Chen C, Cui Y, Liu H, Ling HQ (2014) Mediator subunit 16 functions in the regulation of iron uptake gene expression in Arabidopsis. New Phytol 203: 770–783 [DOI] [PubMed] [Google Scholar]
- Zhou R, Zhu Z, Kong X, Huo N, Tian Q, Li P, Jin C, Dong Y, Jia J (2005) Development of wheat near-isogenic lines for powdery mildew resistance. Theor Appl Genet 110: 640–648 [DOI] [PubMed] [Google Scholar]
- Zhu X, Qi L, Liu X, Cai S, Xu H, Huang R, Li J, Wei X, Zhang Z (2014) The wheat ethylene response factor transcription factor pathogen-induced ERF1 mediates host responses to both the necrotrophic pathogen Rhizoctonia cerealis and freezing stresses. Plant Physiol 164: 1499–1514 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.