Chloroplasts modulate programmed cell death via at least two different pathways: photosynthetic activity is required for cell death while a retrograde signal negatively regulates cell death and plant innate immunity.
Abstract
Programmed cell death (PCD) is a crucial process both for plant development and responses to biotic and abiotic stress. There is accumulating evidence that chloroplasts may play a central role during plant PCD as for mitochondria in animal cells, but it is still unclear whether they participate in PCD onset, execution, or both. To tackle this question, we have analyzed the contribution of chloroplast function to the cell death phenotype of the myoinositol phosphate synthase1 (mips1) mutant that forms spontaneous lesions in a light-dependent manner. We show that photosynthetically active chloroplasts are required for PCD to occur in mips1, but this process is independent of the redox state of the chloroplast. Systematic genetic analyses with retrograde signaling mutants reveal that 3′-phosphoadenosine 5′-phosphate, a chloroplast retrograde signal that modulates nuclear gene expression in response to stress, can inhibit cell death and compromises plant innate immunity via inhibition of the RNA-processing 5′-3′ exoribonucleases. Our results provide evidence for the role of chloroplast-derived signal and RNA metabolism in the control of cell death and biotic stress response.
Programmed cell death (PCD) is a universal process in multicellular organisms, contributing to the controlled and active degradation of the cell. In plants, PCD is required for processes as diverse as development, self-incompatibility, and stress response. One well-documented example is the induction of PCD upon pathogen attack, allowing the confinement of the infection, and resistance of the plant. The signaling events leading to the onset of PCD have been extensively studied: pathogen recognition triggers activation of mitogen-activated protein kinase cascades, as well as production of reactive oxygen species (ROS) and salicylic acid (SA), which lead to a hypersensitive response (Coll et al., 2011).
From a cellular point of view, several classes of plant PCD have been described and compared with the ones found in animal cells (van Doorn, 2011). PCD is thought to have evolved independently in plants and animals, and genes underlying these mechanisms are therefore poorly conserved between the two kingdoms. However, most cellular features are conserved between plant and animal PCD that are both characterized by cell shrinkage, chromatin condensation, DNA laddering, mitochondria permeabilization, and depolarization (Dickman and Fluhr, 2013). In animal cells, mitochondria play a central role in the regulation of apoptosis (Czabotar et al., 2014; Mariño et al., 2014), and this role is likely shared between the two kingdoms (Lord and Gunawardena, 2012). That said, additional mitochondria-independent PCD pathways have clearly evolved in plants.
Genetic approaches have greatly contributed to our understanding of cellular pathways governing PCD in plants. For example, the isolation of lesion mimic mutants (LMMs), in which cell death occurs spontaneously, has allowed the identification of several negative regulators of cell death (for review, see Bruggeman et al., 2015b). Interestingly, lesion formation is light dependent in several of these mutants, which include one of the best characterized LMMs—lesions simulating disease1 (lsd1; Dietrich et al., 1994). The LSD1 protein is required for plant acclimation to excess excitation energy (Mateo et al., 2004): when plants are exposed to excessive amounts of light, the redox status of the plastoquinone pool in the chloroplastic electron transfer chain is thought to influence LSD1-dependent signaling to modulate cell death (Mühlenbock et al., 2008). Additionally, we have previously identified the myoinositol phosphate synthase1 (mips1) mutant as a LMM, in which lesion formation is also light dependent (Meng et al., 2009). This mutant is deficient in the myoinositol (MI) phosphate synthase that catalyzes the first committed step of MI biosynthesis and displays pleiotropic defects such as reduced root growth, abnormal vein development, and spontaneous cell death on leaves, together with severe growth reduction after lesions begin to develop (Meng et al., 2009; Donahue et al., 2010). The light-dependent PCD in the mips1 mutant, as observed for lsd1, suggests that chloroplasts may play a role in the MI-dependent cell death regulation. Accumulating evidence suggests that chloroplasts may play a central role in PCD regulation like mitochondria in animal cells (Wang and Bayles, 2013). First, as described in the case of lsd1, excess light energy received by the chloroplast can function as a trigger for PCD. Furthermore, singlet oxygen (1O2), a ROS, can activate the EXECUTER1 (EX1) and EX2 proteins in the chloroplasts to initiate PCD (Lee et al., 2007). Likewise, ROS generated by chloroplasts play a major role for PCD onset during nonhost interaction between tobacco (Nicotiana tabacum) and Xanthomonas campestris (Zurbriggen et al., 2009). Finally, functional chloroplasts have also been shown to be required for PCD in cell suspensions (Gutierrez et al., 2014) and in a number of LMMs (Mateo et al., 2004; Meng et al., 2009; Bruggeman et al., 2015b). Thus, chloroplasts are now recognized as important components of plant defense response against pathogens (Stael et al., 2015) and are proposed to function with mitochondria in the execution of PCD (Van Aken and Van Breusegem, 2015). However, the exact signaling and metabolic contribution of chloroplasts to PCD remain to be elucidated. Furthermore, cross talk between chloroplasts and mitochondria does occur, such as during photorespiration (Sunil et al., 2013), but whether such communication functions sequentially or in parallel in the control of PCD remains to be determined (Van Aken and Van Breusegem, 2015).
To further investigate how chloroplasts contribute to the regulation of cell death, we performed both forward and reverse genetics on the mips1 mutant. An extragenic secondary mutation in divinyl protochlorophyllide 8-vinyl reductase involved in chlorophyll biosynthesis leads to chlorophyll deficiency that abolishes the mips1 cell death phenotype, as do changes in CO2 availability. These findings provide evidence for a link between photosynthetic activity and PCD induction in mips1. Additionally, we investigated the contribution of several retrograde signaling pathways (Chan et al., 2015) to the control of PCD in mips1. This process was independent of GENOMES UNCOUPLED (GUN) and EX signaling pathways, but we found that the SAL1-PAP_XRN retrograde signaling pathway inhibits cell death as well as basal defense reactions in Arabidopsis (Arabidopsis thaliana).
RESULTS
Chlorophyll Accumulation Is Required for the Onset of PCD in mips1
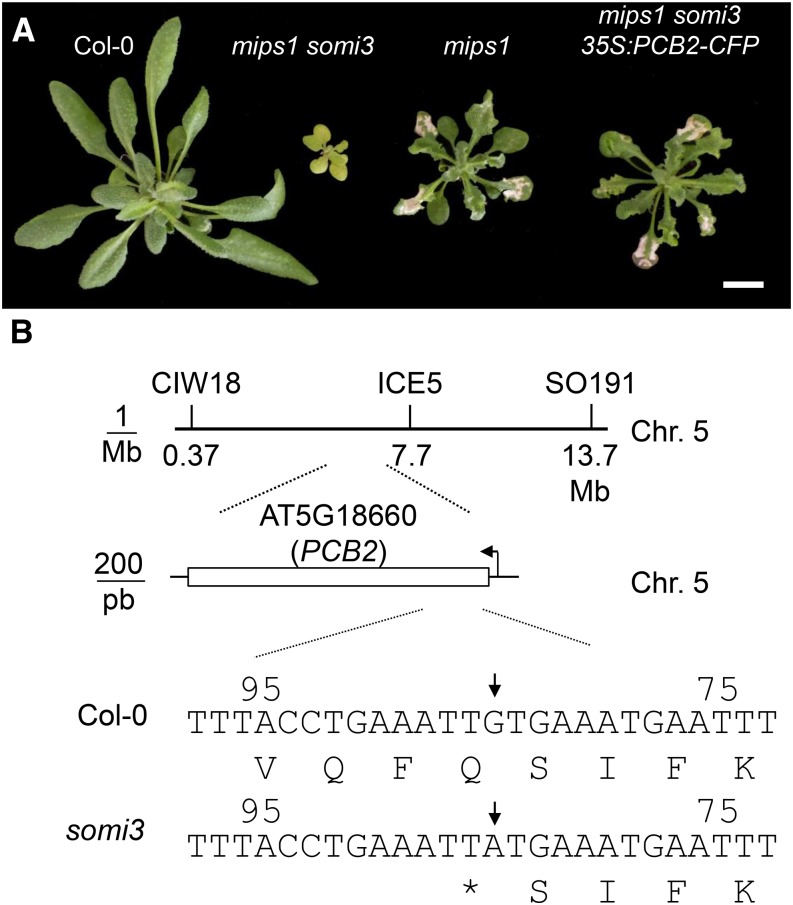
In a genetic screen designed to identify suppressors of the mips1 cell death phenotype, we isolated a mutant severely deficient for pigment accumulation: suppressor of mips1 3 (somi3; Fig. 1A; Supplemental Fig. S1). Rough mapping of the mutation allowed us to position it at the top of chromosome 5 between markers CIW18 and ICE5 (Fig. 1B). SHORE mapping revealed a single mutation in the PALE GREEN AND CHLOROPHYLL B REDUCED2 (PCB2; AT5G18660) gene (Nakanishi et al., 2005). PCB2 encodes a divinyl protochlorophyllide 8-vinyl reductase that catalyzes the conversion of divinyl protochlorophyllide a to monovinyl protochlorophyllide a (Nakanishi et al., 2005). Transformation of mips1 somi3 mutants with a construct encompassing the wild-type PCB2 cDNA restored chlorophyll accumulation and lesion formation (Fig. 1A), confirming that suppression of the mips1 phenotype was due to a mutation in PCB2. Therefore, the somi3 mutant allele is hereafter referred to as pcb2-2.
Figure 1.
somi3 harbors a mutation in the PCB2 gene. A, Suppression of the cell death phenotype in the mips1 somi3 mutant obtained by ethyl methanesulfonate mutagenesis of the mips1 mutant. Plants were grown under SD for 2 weeks and transferred to LD for 6 d to induce lesion formation. Complementation of mips1 somi3 with a wild-type PCB2 cDNA restores lesion formation. B, Bulked segregants. Analysis allowed mapping of the somi3 mutation to the region between the CIW18 and ICE5 markers on chromosome 5. Shore map analysis revealed a single point mutation at position 84 of the PCB2 gene.
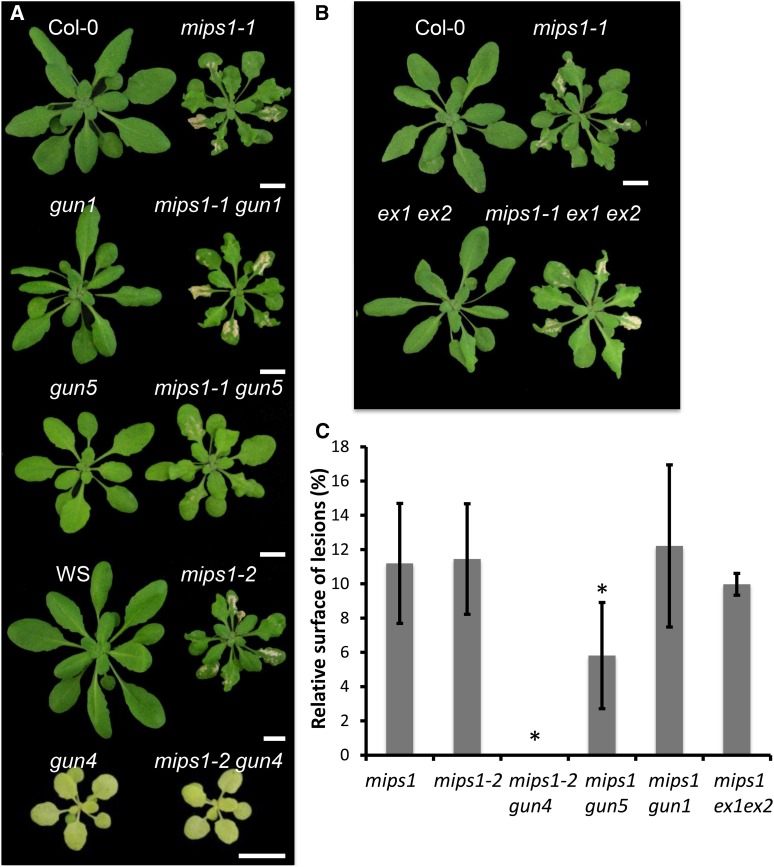
We have previously reported that the mips1 phenotype was suppressed by inactivation of the GUN4 gene via an artificial microRNA (Meng et al., 2009). Because intermediates of tetrapyrrole biosynthesis have been involved in chloroplast to nucleus signaling (Lepistö et al., 2012), the reduction of cell death associated with GUN4 silencing and pcb2-2 mutation may reflect either a role for this retrograde signaling pathway in the control of cell death or the requirement for photosynthetically competent chloroplasts to trigger cell death in the mips1 mutant. To discriminate between these two possibilities, the mips1 mutant was crossed with three gun mutants that all show deregulation in retrograde signaling but are differentially affected in chlorophyll biosynthesis (Susek et al., 1993). As shown in Figure 2, A and C, the gun1 mutation in a pentatricopeptide protein that is hypothesized to integrate GUN-mediated retrograde signaling but not chlorophyll biosynthesis (Koussevitzky et al., 2007) had no effect on the mips1 phenotype. By contrast, mutations in the GUN4 and GUN5 proteins, which are part of an enzyme complex in tetrapyrrole and chlorophyll biosynthesis, rescued the mips1 phenotype to varying degrees. gun5-1, which is a weak allele (Mochizuki et al., 2001), had a mild effect on the mips1 phenotype, whereas the gun4-100 mutation that most severely affects chlorophyll accumulation (Cottage et al., 2008) completely abolished the cell death phenotype in mips1 1 week after transfer to long-day (LD) conditions. Hence, control of PCD in mips1 is independent of GUN signaling, and, instead, a correlation exists between the intensity of lesions and chlorophyll accumulation (Supplemental Fig. S2).
Figure 2.
PCD in mips1 requires chlorophyll accumulation but is not mediated by EX-dependent singlet oxygen production. A, The cell death phenotype of mips1 is suppressed by the gun4 and gun5 mutations, which alter chlorophyll accumulation, but not by the gun1 mutation, which does not (Supplemental Fig. S1). The mips1-1 mutant in the Col-0 background was crossed to the gun1-1 and gun5-1 mutants, whereas the mips1-2 mutant, in the WS background, was crossed to the gun4-100 mutant. Double mutants were identified from the F2 generation. Bars = 1 cm. B, Lesion formation does not depend on EX-mediated singlet oxygen signaling: mips1 ex1 ex2 triple mutant forms lesions under LD like the mips1 mutant. Bars = 1 cm. C, Lesion quantification in mips1, mips1-2, and double mutants. Data are average ± sd from measurements performed on at least 12 plants using Image J software and are representative of two independent experiments. Asterisks indicate significantly different values (Student’s t test, P < 0.001).
Redox Changes Associated with Chloroplast Activity Are Not Responsible for the mips1 Phenotype
The rescue of PCD in mips1 by decreased chlorophyll content could be directly or indirectly caused by a reduction of oxidative stress associated with impaired light harvesting. We therefore tested the contribution of several previously described pathways that can lead to cell death in a light-dependent manner, and have been connected to oxidative stress or changes in the redox balance in chloroplasts.
In the light, chloroplasts of plant cells are a major source of ROS, and EX proteins have been shown to control PCD in response to 1O2 (Lee et al., 2007). We therefore generated mips1 ex1 ex2 triple mutants to determine whether PCD was reduced in this context. Surprisingly, the mips1 ex1 ex2 triple mutant was identical to mips1 (Fig. 2, B and C), indicating that EX proteins do not participate in the PCD signaling in mips1.
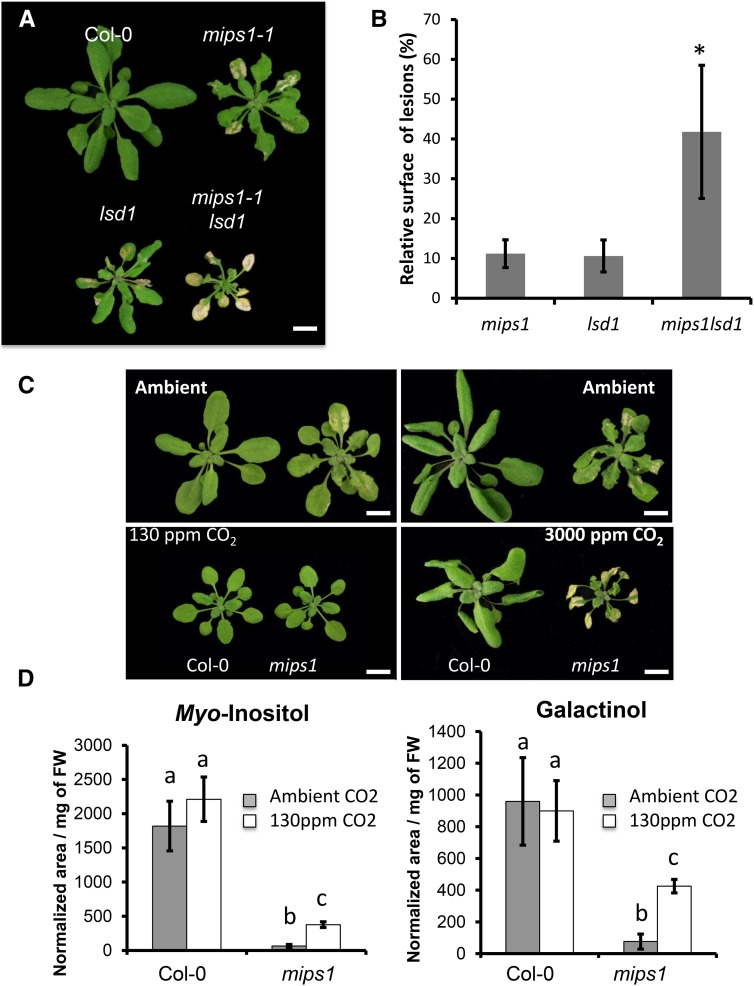
PCD can also be triggered by changes in the redox status of the chloroplast when plants receive excess amounts of light, and the LSD1 protein has been shown to be required for confinement of cell death under these conditions (Mateo et al., 2004): lsd1 mutants show light-dependent lesion formation like the mips1 mutant. To genetically test whether MIPS1 could function in the same pathway, we generated mips1lsd1 double mutants. mips1lsd1 mutants were more severely affected than the two parents: lesions represented a greater proportion of the leaf surface, and plant growth was more severely inhibited (Fig. 3, A and B), pointing to an additive or even a synergistic effect that is compatible with the fact the lsd1 has been described as a propagation mutant that fails to confine cell death once it is initiated (Jabs et al., 1996). Thus, LSD1 and MIPS1 are not epistatic and are likely to mediate PCD via parallel pathways.
Figure 3.
Lesion formation depends on chloroplast metabolic activity rather than redox status. A, The phenotype of the mips1 mutant is enhanced in the lsd1 background. Bar = 1 cm. B, Lesion quantification in mips1, lsd1, and mips1 lsd1 mutants. We observed 4-fold increase in the relative surface of lesions in the double mutant compared with the parent lines. Data are average ± sd from measurements performed on at least 12 plants using Image J software and are representative of two independent experiments. Asterisks indicate significantly different values (Student’s t test, P < 0.001). C, The severity of the mips1 phenotype is enhanced by CO2 availability. Wild type (Col-0) and mips1 mutant were grown under ambient CO2 and SD conditions for 15 d, and then transferred to LD conditions under ambient, low (130 ppm), or high (3000 ppm) CO2 for 10 and 15 d, respectively. Bars = 1 cm. Under low CO2, lesion formation was completely suppressed, while it was more severe under high CO2. D, Growth under low CO2 partially restores MI and galactinol accumulation in mips1. Values are average ± sd of four biological replicates. Different letters indicate statistically significant differences (Student’s t test, P < 0.001).
Singlet oxygen production and reduction of plastoquinones are not the only redox changes associated with photosynthesis: in the light, chloroplasts also produce superoxide and hydrogen peroxide. Likewise, photorespiration, a metabolic pathway involving the cooperation of chloroplasts, mitochondria, and peroxisomes, also produces hydrogen peroxide in peroxisomes, and deficiency in CATALASE2, the enzyme responsible for the detoxification of photorespiratory H2O2, leads to spontaneous cell death (Queval et al., 2007). We therefore grew the mips1 mutant under high CO2 to lower photorespiration and thereby H2O2 production in peroxisomes (Queval et al., 2007). Surprisingly, this increased the cell death phenotype (Fig. 3C), suggesting that PCD in mips1 correlates with photosynthetic activity rather than with ROS production. To further demonstrate that lesion formation correlates with photosynthetic activity in mips1, we measured the electron transfer rate (ETR) on plants grown in ambient CO2 under short-day (SD) and LD conditions or under low CO2 and LD. In the case of plants grown under LD and ambient CO2, care was taken to measure chlorophyll fluorescence on lesion-free leaves. As shown in Supplemental Figure S3, A and B, in the wild type the ETR was not different between plants grown under SD or LD, but was reduced under low CO2. There was no significant difference between the ETR of wild-type and mutant plants, indicating that photosynthetic activity does not differ from the wild type in mips1, even when plants are grown under LD. Thus, growth under LD does not affect electron transfer in chloroplasts of the mips1 mutant, and lesion formation under LD thus correlates with enhanced carbon fixation per day rather than with increased damage in the electron transfer chain. Finally, we performed gas-exchange measurements on fully expanded leaves of plants grown under SD in ambient air. As shown in Supplemental Figure S3D, photosynthetic activity varied according to CO2 availability in the same way in wild-type and mutant plants: carbon fixation was reduced at a concentration of 100 ppm and reached its maximum around 500 ppm. Taken together, these results confirm that the intensity of lesion formation correlates with carbon fixation in mips1.
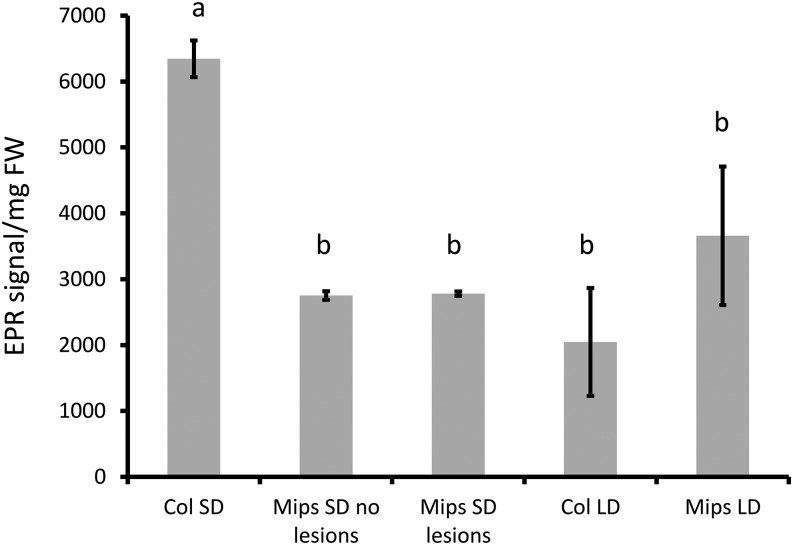
To determine whether growth under low CO2 may affect MI accumulation in mips1 or if accumulation of other metabolites could be modified in these conditions, we performed metabolomic profiling. We found that MI accumulation was 17% of wild-type levels under low CO2, whereas it was below 4% of wild-type levels under ambient CO2 (Fig. 3D). In addition, we found that accumulation of MI derivate galactinol was increased in mips1 under low CO2 (Fig. 3D). We have shown previously that the MIPS2 isoform of MI synthase accounts for residual MI biosynthesis in response to metabolic changes in mips1, whereas the MIPS3 isoform was not involved (Bruggeman et al., 2015a). To determine whether this activity was required for the rescue of cell death under low CO2, mips1 mips2/+ sesquimutants were grown for 1 week under SD and then transferred for 10 d under LD in low or ambient CO2 conditions. As shown in Supplemental Figure S4, the extreme cell death phenotype of the mips1 mips2/+ mutants was rescued under low CO2, indicating that residual MI biosynthesis is likely not responsible for the inhibition of cell death under these conditions. Finally, to clarify whether ROS production was affected in mips1 mutants grown under lesion inducing conditions, we measured reactive oxygen intermediate (ROI) accumulation by electron paramagnetic resonance (EPR) spectroscopy using ethanol/α-(4-pyridyl-1-oxide)-N-tert-butylnitrone (4-POBN) as a specific spin-trapping system for •OH. This method allows the quantification of global production of ROIs in leaves because it detects •OH derived from H2O2 and O2.− in the presence of Fe(II) (Michelet and Krieger-Liszkay, 2012). Because lesion formation is associated with local ROI production, we measured ROI accumulation in the wild type and mips1 mutants grown either in SD with sufficient light intensity to induce mild lesion formation on some leaves, or in LD, where all leaves formed lesions. As shown in Figure 4 and Supplemental Figure S5, in the wild type ROI accumulation was higher in SD than in LD, consistent with previous reports (Michelet and Krieger-Liszkay, 2012). In mips1, we did not detect a significant difference between leaves displaying lesions and healthy leaves. Furthermore, even in LD when lesion formation was severe, we did not observe a sharp increase in ROI accumulation, indicating that ROI production occurs only locally, consistent with previously obtained results with 3,3′-diaminobenzidine staining (Bruggeman et al., 2014).
Figure 4.
ROI production is not enhanced in mips1. Wild-type and mutant plants were grown in SD for 15 d under SD conditions at low light intensity (60 µmol photons m−2 s−1), and then transferred to either SD with high light intensity (about 250 µmol photons m−2 s−1) to induce mild cell death or LD for 10 d. Global ROI production was measured in mature leaves with and without lesions by EPR as described in “Materials and Methods” on whole leaves, and EPR/leaf fresh weight ratios were calculated. Values are average ± sd obtained on three biological replicates. Different letters indicate statistically significant differences (Student t test, P < 0.05).
PAP Signaling Negatively Regulates Cell Death in mips1 as Well as Plant Basal Immunity
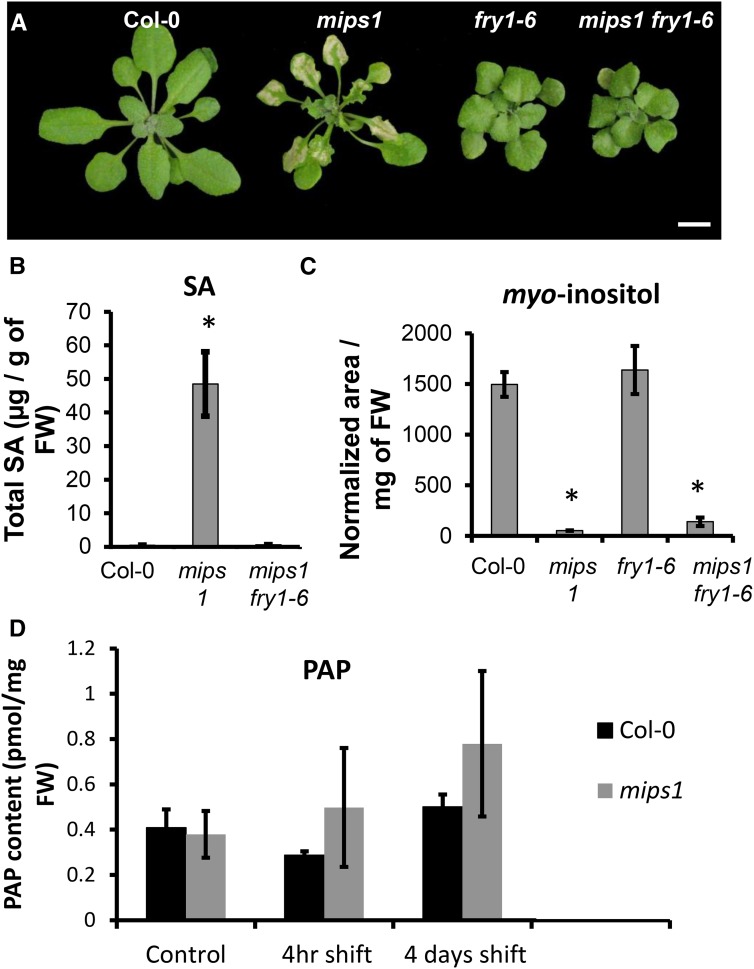
One chloroplast metabolite known to function in retrograde light stress signaling is 3′-phosphoadenosine 5′-phosphate (PAP): this molecule accumulates under high light and drought stress, and is thought to inhibit the activity of exoribonucleases (XRNs), thereby altering RNA metabolism and promoting stress response networks (Estavillo et al., 2011). The sal1/fry1/alx8 mutants lack the enzyme that dephosphorylates PAP into AMP, resulting in constitutively high levels of PAP (Estavillo et al., 2011). We crossed a null allele, fry1-6 (Gy et al., 2007), with mips1 to test the role of PAP signaling in the control of PCD. As shown in Figure 5A, lesion formation was completely abolished in the double mutant as well as the SA accumulation characteristic of the mips1 phenotype (Meng et al., 2009; Fig. 5B), although basal SA levels were unchanged in fry1-6 (Supplemental Fig. S6). Because the mips1 mutant shows pleiotropic defects, we also tested whether the fry1-6 mutation could restore normal root growth and cotyledon development in mips1, but found that these defects were still present or even exacerbated in the double mutants (Supplemental Fig. S7). In agreement with these observations, MI accumulation was not restored in the mips1 fry1-6 mutant (Fig. 5C). One simple hypothesis would be that transfer to LD conditions induces increased PAP production in the wild type and that mips1 mutants are deficient in this regulation. However, PAP quantification did not reveal any changes in PAP accumulation in mips1 compared with the wild type (Fig. 5D), but we were able to detect the expected elevated levels of PAP in fry1-6 and fry1-6 mips1 (Supplemental Fig. S8).
Figure 5.
Lesion formation is suppressed by the fry1-6 mutation. A, The phenotype of the mips1 mutant is rescued by the fry1-6 mutation. Wild type (Col-0), mips1, fry1-6, and mips1 fry1-6 double mutants were grown under SD for 15 d and transferred to LD for 10 d. Bar = 1 cm. B, SA accumulation upon transfer to LD conditions is suppressed in the mips1 fry1-6 mutant. SA accumulation was measured 4 d after transfer to LD conditions. Values are average ± sd obtained on four biological replicates. Asterisks indicate statistically significant differences from the control (Student’s t test, P < 0.001). C, MI accumulation is not restored in the mips1 fry1-6 double mutant. Values are average ± sd from four biological replicates. Asterisks indicate statistically significant differences from the control (Student’s t test, P < 0.001). D, PAP content in the wild type (Col-0) and mips1 4 h and 4 d after transfer to LD conditions. Controls are plants kept under SD conditions. Values are average ± sd obtained from three biological replicates.
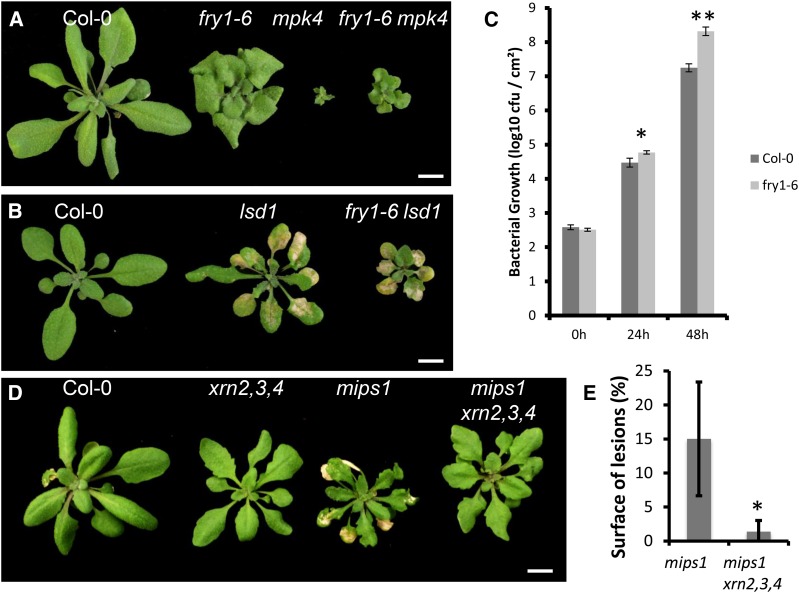
We next asked whether the fry1-6 mutation is a general suppressor of PCD. To this end, we crossed the fry1-6 mutant with lsd1, which promotes PCD independently of mips1, and with map-kinase4 (mpk4), which shows constitutive activation of defense response (Pitzschke et al., 2009). The fry1-6 mutation partially restored the phenotype of the mpk4 mutant (Fig. 6A), but did not prevent lsd1-mediated cell death although PAP levels were identical in fry1-6 and fry1-6 lsd1 mutants (Fig. 6B; Supplemental Fig. S9).
Figure 6.
PAP signaling is not a general suppressor of cell death but a negative regulator of defense response. A, The phenotype of the mpk4 mutant is partially rescued by the fry1-6 mutation. Plants were grown under SD for 2 weeks and transferred to LD for 15 d. Bars = 1 cm. B, The phenotype of the lsd1 mutant is not modified in the fry1-6 background. Plants were grown under SD for 2 weeks and transferred to LD for 7 d. Bars = 1 cm. C, Twenty-eight-day-old Col-0 and fry1-6 plants were grown in SD and infiltrated with a suspension of P. syringae pv tomato DC3000, and bacterial growth was followed for 2 d. The number of colony forming units (cfu) was already significantly higher in fry1-6 than in the wild type (Col-0) 24 h after inoculation, and this difference became more obvious after 48 h. Asterisks indicate significantly different values (Wilcoxon signed-rank test, * P < 0.01, ** P < 0.001). Data are average ± sd from five biological replicates and are representative of two independent experiments. D, Suppression of cell death in mips1 fry1-6 can be reproduced in XRN-deficient mutants. The mips1 mutant was crossed with the xrn2-2 xrn3-3 xrn4-1 triple mutants, and quadruple mutants were selected from the F3 generation. Lesion formation in the quadruple mutant was drastically reduced, while plant growth was restored. Plants were grown under SD for 2 weeks and transferred to LD for 15 d. Bars = 1 cm. E, Lesion quantification in mips1 and mips1 xrn2-2 xrn3-3 xrn4-1 quadruple mutants. Values are average ± sd (n = 12) and are representative of two independent experiments. The relative surface of lesions was significantly reduced in mips1 xrn2 xrn3 xrn4 quadruple mutants (Student’s t test, P < 0.001).
MPK4 is a regulator of plant immunity (Colcombet and Hirt, 2008) and appears to modulate MIPS1 expression in response to the bacterial peptide flagellin (Latrasse et al., 2013). We therefore asked if suppression of PCD in mpk4 by fry1-6 could concurrently impact plant immunity. Wild type and fry1-6 were infiltrated with the virulent strain of Pseudomonas syringae pv tomato DC3000, and bacterial growth was followed for 2 d. As shown in Figure 6C, fry1-6 displayed high susceptibility toward this pathogen, indicating that PAP overaccumulation has a negative impact not only on cell death but also on pathogen defense mechanisms.
Finally, to gain insight into the mechanisms allowing cell death suppression in the mips1 fry1-6 mutant, we tested the contribution of the inhibition of XRN activity by PAP overaccumulation (Estavillo et al., 2011). To this end, the mips1 mutant was crossed with the xrn2-2 xrn3-3 xrn4-1 mutant and quadruple mutants were isolated. As shown in Figure 6, D and E, the loss of XRN2-4 activity drastically reduced lesion formation in mips1, providing evidence for the role of posttranscriptional regulatory mechanisms in the control of cell death. Some lesions could still be observed in quadruple mutants, either because the xrn3-2 mutant is only a knockdown (Gy et al., 2007) or because XRN-independent mechanisms participate in the cell death suppression associated with PAP signaling. To try and identify candidate genes whose deregulation in xrn and fry1 mutants may account for the suppression of the mips1 phenotype, we took advantage of available microarray data. Although we found a significant enrichment for genes associated with chloroplast function and immune response among repressed genes in alx8 (also a null sal1/fry allele; Estavillo et al., 2011) and xrn2 xrn3 mutants (Supplemental Fig. S10A), we did not find significant overlap between deregulated genes in mips1 and alx8 or xrn2 xrn3 mutants (Supplemental Fig. S10B). Hence, the identity of the key actor(s) of the suppression of cell death in mips1 xrn2-4 and mips1 fry1-6 mutants remains a future question.
DISCUSSION
As chloroplasts are the sites of light energy capture and a hub for metabolic pathways, they are ideally suited to perceive changes in environmental conditions. Indeed, there is accumulating evidence that these organelles play a critical role in plant response to various types of stress (Xiao et al., 2013) and in the control of PCD (Kim et al., 2012). However, the molecular basis for the chloroplast’s contribution to PCD and the intersection of PCD and abiotic stress signaling by the chloroplast is poorly understood.
Here, we have taken advantage of the conditional mips1 mutant to investigate the role of chloroplasts in the control of PCD, and we show that the photosynthetic capacity of the plant appears to be required for cell death to occur. Similar observations have been reported in a number of LMMs, but the underlying mechanisms likely differ between mutants because our genetic analysis demonstrates that LSD1 and MIPS1 function in different pathways. In the case of the lsd1 mutant, reduced chlorophyll content likely suppresses cell death by avoiding the accumulation of reduced plastoquinone, whereas photosynthetic activity rather than redox changes appears to play a crucial role for PCD onset in mips1. Indeed, the mips1 phenotype is more severe when plants are grown under high CO2, whereas these growth conditions suppress cell death in lsd1 (Mateo et al., 2004) and in catalase2 (Queval et al., 2007), making the mips1 cell death phenotype unique and providing evidence for a yet undescribed PCD regulation pathway in plants. Together, our results suggest that photosynthetic activity itself is the trigger for PCD induction in mips1 and that reducing carbon fixation either by lowering the chlorophyll content in chloroplasts or by growing plants under low CO2 is sufficient to avoid cell death to occur. This was the case even in mips1 mips2/+ sesquimutants, in which MI accumulation is further reduced compared with mips1 single mutants. Data reported here do not support a role for oxidative stress in the onset of PCD in mips1, in line with the finding that the cellular redox status is not altered in mips1 under LD conditions (Bruggeman et al., 2014).
The latter hypothesis is further confirmed by the observation that EX-dependent 1O2 signaling is not involved in triggering cell death in mips1. Recent progress in our understanding of singlet oxygen response in plants has revealed that EX-mediated signaling is not the only pathway that regulates cell death and that most of the singlet oxygen-regulated genes actually respond to loss of chloroplast integrity due to singlet oxygen production. In addition, β-cyclocitral has also been shown to regulate 1O2-responsive genes independently of the EX pathway (Ramel et al., 2012). 1O2 production does not differ between SD- and LD-grown plants (Michelet and Krieger-Liszkay, 2012), or between plants grown under high or low CO2 (Triantaphylidès et al., 2008), suggesting that enhanced 1O2 accumulation is likely not the trigger for PCD onset in mips1. Nevertheless, 1O2 has been shown to play a central role in the onset and execution of excess light energy-induced cell death (Triantaphylidès et al., 2008) and thus likely contributes to later steps of lesion formation in mips1. On the contrary, basal levels of ROS production in mips1 may be required to dampen cell death. Indeed, Laloi and colleagues have shown that improving H2O2 detoxification in the chloroplast by overexpression of the thylakoid-associated ascorbate peroxidase dramatically increases plant cell death and growth inhibition in response to 1O2 production, suggesting that different ROS can have opposing effects on the same cellular response (Laloi et al., 2007). Reduction of H2O2 production under high CO2 may thus contribute to the increased severity of the mips1 phenotype via perturbations in ROS sensing/signaling. In line with this hypothesis, direct measurement of ROI accumulation in mips1 using EPR spectroscopy did not reveal a global increase in ROI production, even after lesions appeared. We have shown that local ROI production does occur at the site of lesion formation (Bruggeman et al., 2014). However, we have also reported that the mips1 mutant is more tolerant than the wild type to oxidative stress (Meng et al., 2009). Together, previous results and data presented here support the view that cell death in mips1 is not triggered by ROI production but rather induces expression of the antioxidant system (Meng et al., 2009), leading to reduced ROI accumulation except in dying cells.
Together, our results show that chloroplast photosynthetic activity rather than changes in redox equilibrium is the trigger for PCD in mips1, but the nature of the signal remains an open question. Several molecules originating from both primary and secondary metabolism have been shown to act as retrograde signaling elements. For example, Vogel et al. have recently shown that transport of triose-phosphates to the cytoplasm is likely instrumental in promoting changes in nuclear gene expression when plants are transferred from low to high light conditions (Vogel et al., 2014), and a wealth of plastid-derived metabolites have been shown to modulate plant response to abiotic stress (Xiao et al., 2013). Among those, PAP plays a role in plant tolerance to high light (Estavillo et al., 2011) while it affects the accumulation of molecules involved in plant defense: sal1 mutants accumulate high levels of jasmonate (Rodríguez et al., 2010) and low levels of glucosinolates (Lee et al., 2012). PAP was thus an excellent candidate to oppose the signaling pathway leading to cell death in mips1. PAP overaccumulation due to the fry1-6 mutation completely abolished lesion formation in mips1, and this effect was specific to the mips1 and mpk4 mutants that have previously been shown to participate in the same pathway (Latrasse et al., 2013), whereas PAP had no effect on cell death induced by lsd1 mutation. This finding indicates that PAP overaccumulation does not antagonize all types of cell death or SA production, but rather opposes cellular responses related to plant defense mechanisms, consistent with the observation that fry1-6 mutants are hypersensitive to P. syringae. Indeed, although the fry1-6 mutation suppresses SA hyperaccumulation in mips1, if PAP signaling directly inhibited PCD by decreasing SA accumulation, the fry1-6 mutation should have also suppressed the lsd1 cell death phenotype that is also SA dependent (Aviv et al., 2002). Hence, we postulate that PAP signaling opposes events occurring upstream of the induction of SA production in mips1 and mpk4 mutants. The question is how is this PAP-dependent complementation accomplished? Accumulation of a number of metabolites and phytohormones is altered by PAP, including MI, putrescine, and jasmonic acid (JA; Rodríguez et al., 2010). However, we did not detect restoration of MI accumulation in mips1 fry1-6, nor did we detect increase in MI accumulation in the fry1-6 allele used in this study. SA and JA signaling are generally regarded as antagonistic pathways for a number of physiological responses (Kunkel and Brooks, 2002), and production of coronatine, a phytotoxin that structurally mimics JA, has been shown to act as a virulence factor for Pseudomonas (Xin and He, 2013). However, if JA overaccumulation were the mechanism, the fry1-6 mutation would be expected to rescue other mutants displaying SA-dependent cell death, such as lsd1. Perturbation of glucosinolate metabolism and/or signaling can cause compensatory enhanced cell death in response to pathogen challenge (Bednarek et al., 2009). Thus, whether the altered glucosinolate homeostasis due to PAP overaccumulation (Lee et al., 2012) also contributes to suppression of mips1 PCD is an open question. However, lesion formation was also suppressed in xrn2 xrn3 xrn4 background in which glucosinolate accumulation is not modified, suggesting that change in glucosinolate levels is not the primary cause for cell death suppression. It is worth noting that cell death was only reduced in mips1 xrn2 xrn3 xrn4 quadruple mutants, whereas it was completely abolished in mips1 fry1 mutants. This could be due either to a contribution of glucosinolates to PCD regulation, or to the fact the xrn3-3 mutant is only a knockdown and not a null mutant (Gy et al., 2007).
Finally, the finding that mutations in XRN2, XRN3, and XRN4 also suppressed lesion formation in mips1 opens the intriguing possibility that posttranscriptional regulations targeting defense response genes are involved in PCD onset in mips1. XRNs have been involved in various posttranscriptional regulatory pathways including microRNA biogenesis and posttranscriptional gene silencing because they are required to degrade biogenesis intermediates (Gy et al., 2007). It is now clear that posttranscriptional regulations play a key role in the control of plant immunity (Staiger et al., 2013) and that XRN4 contributes to plant defenses against viruses (Gy et al., 2007; Jaag and Nagy, 2009), indicating that microRNAs, posttranscriptional gene silencing, or both could thus be involved in the regulation of PCD. We recently reported that the CPSF30 subunit of the polyadenylation complex was required for lesion formation in mips1 (Bruggeman et al., 2014), providing further evidence for the role of posttranscriptional mechanisms in the regulation of plant PCD. However, we showed that CPSF30 is required for cell death in all tested LMMs, suggesting that CPSF30 acts on much more general pathways than PAP.
To summarize, our results show that chloroplasts can contribute to the regulation of plant PCD via at least two distinct mechanisms: cell death induction in MI-deficient plants requires photosynthetically active chloroplasts, and can be negated by lowering photosynthesis by reducing either chlorophyll accumulation or CO2 availability. In addition, PAP accumulation is a negative regulator of MI-dependent PCD and plant basal immunity via its effect on XRN activity. This result sheds light on the complex question of how plants can deal with multiple stresses, since signaling pathways activated by one stress can have detrimental outcomes to fight other adverse conditions. Because the SAL1 enzyme is located both in chloroplasts and in mitochondria, our results provide an exciting platform to interrogate the specific roles for chloroplasts, and cross talk between chloroplasts and mitochondria, in the control of PCD.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Seeds were surface-sterilized by treatment with Bayrochlore (Bayrol) for 20 min, and washed and imbibed in sterile water for 2 to 4 d at 4°C to obtain homogeneous germination. Seeds were sown on commercially available 0.5× Murashige and Skoog medium (Basal Salt Mixture M0221; Duchefa) supplemented or not with Suc (1%, w/v) solidified with 0.8% (w/v) agar (Phyto-Agar HP696; Kalys) and grown in a LD (16 h light, 8 h night, 21°C) growth chamber. After 2 weeks, the plants were transferred to soil in a glasshouse or in a growth chamber under SD conditions (8 h light 20°C, 16 h night at 18°C) for 2 weeks before being transferred to LD conditions. Seeds of the ex1 (SALK_026475), ex2 (SALK_112295), and fry1-6 (SALK_020882; Alonso et al., 2003) mutants were obtained from the Nottingham Arabidopsis Stock Centre. Seeds of the gun1-1, gun4-100, and gun5-1 mutants were a gift from J. Jouhet. xrn2-2 and xrn3-3 mutants were described by Gy et al. (2007), and xrn4-1 was described by Gazzani et al. (2004). The xrn2 xrn3 xrn4 triple mutant was a gift from A. Mallory. For genotyping, plant genomic DNA was extracted according to Edwards et al. (1991). Primers used for PCR analysis are listed in Supplemental Table S1, and PCR conditions used for genotyping are indicated in Supplemental Table S2.
Bulked Segregant Analysis and SHORE Map
To map the somi3 mutation, mips1-1 somi3 plants in the Columbia (Col)-0 background were crossed with the mips1-2 mutant in the Wassilewskija (WS) background. First, a Bulked Segregant Analysis was performed using F2 plants without (n = 22) or with lesions (n = 22) in LD. Primer sequences that flank polymorphism markers between Col-0 and WS were described by Păcurar et al. (2012). Second, fine mapping of the somi3 mutation was performed by a high-throughput sequencing of nuclear DNA obtained from a bulk of 100 F2 plants without lesions in LD. Whole Arabidopsis (Arabidopsis thaliana) genome resequencing was performed by the company BGI Tech using Illumina HiSeq2000 technology. The software CLC genomic workbench (www.clcbio.com) was used to analyze sequence and identify single-nucleotide polymorphism by comparison with the Col-0 genome as reference (TAIR10).
Cloning Methods
The coding sequence of PCB2 (At5g18660) was amplified from Col-0 genomic DNA using the primers listed in Supplemental Table S1. The PCB2 coding sequence was cloned into the Gateway-compatible pEntr3C vector (Invitrogen) and transferred into the binary vector pB7FWC;2 using LR clonase (Invitrogen) according to manufacturer’s instructions. In this plasmid, the expression of cDNA is under the control of the 35S promoter and encoded proteins are fused with CFP.
SA Quantification, Metabolite Extraction, and GC-MS Analysis
SA was quantified as described by Bruggeman et al. (2014).
For metabolome analysis, analytical procedures were adapted from Roessner-Tunali et al. (2003).
About 100 mg of rosette leaves of the indicated lines were harvested 4 d after transfer in LD conditions. Samples were lyophilized and ground, and metabolites were extracted by adding 500 µL of cold methanol containing 250 µm ribitol as internal standard. Extracts were then warmed at 70°C for 15 min and vortexed regularly. Chloroform (500 µL) was added, and samples were vortexed and warmed at 37°C for 5 min. Then, 450 µL of ultrapure water, containing 250 µm ribitol, was added on extracts that were centrifuged for 5 min at 13,200 rpm, at 4°C, to separate phases. The upper phase was collected (600 µL), and 50 µL of these supernatants was evaporated overnight under vacuum.
Sample Derivatization
Fifty microliters of a pyridine solution containing 20 mg/mL of methoxyamine hydrochloride and 5 µL/mL of a 2 mg/mL n-alkanes mixture (retention index standards: decane, dodecane, pentadecane, octadecane, nonadecane, docosane, octacosane, dotriacontane, and hexatriacontane) was added. The tubes were sealed and heated to 40°C for 90 min. Then 80 µL of N,O-bis(trimethylsilyl)trifluoroacetamide was added for silylation to perform during 30 min at 40°C.
GC-MS Analysis
Analyses were performed by gas chromatography (Agilent 6890) and quadrupole mass spectrometry with electron impact ionization (Agilent 5973 Network). A volume of 1 µL was injected on an Agilent J&W HP-5ms column (diameter: 0.25 mm; film thickness: 0.25 µm; length: 30 m) with a 0.6 mL/min helium flow. The injection parameters were as follows: splitless injection at 230°C with a 1-min purge at 20 mL/min. The temperature gradient was as follows: 1 min at 70°C; 9°C min until 320°C, then 10 min at 320°C; and solvent delay of 5.4 min. The source was set to 250°C and 70 eV, scanning at 20 scans/min, from 70 to 600 m/z. Acquisition was performed with Chemstation software (Agilent).
Data Treatment
Semiautomated integration of chromatograms was performed with Quanlynx software (Waters) after conversion of the raw data into NetCDF format. A model ion was chosen manually to perform a correct integration for 138 peaks, with the following criteria: Gaussian shape, high relative abundance in the mass spectrum, and absence of saturation in any sample. Three very abundant compounds, however, gave saturated signal for any ion: Suc, citrate, and malate; the samples where thus reinjected in split mode (1:30) to integrate these compounds. Metabolites were annotated according to the Chemical Analysis Working Group (Sumner et al., 2007): 41 metabolites identified using pure reference standards, 34 putatively annotated compounds on the basis of both spectral and retention index matches with a public database (The Golm Metabolome Database; Kopka et al., 2005), 41 compounds related to known compounds or chemical classes on the unique basis of spectral match, and 10 unknown analytes. Peak surfaces were then normalized according to internal standard peak surface (ribitol) and sample fresh weight, to obtain semiquantitative data. A fully quantitative analysis was performed for myoinositol and Glc with calibration curves (0, 0.01, 0.05, 0.1, and 1 g/L).
Adenosine Extraction and Quantification
PAP was quantified according to Estavillo et al. (2011). In brief, 100 mg of leaf tissue was flash-frozen in liquid nitrogen, ground to powder with Tissue Lyzer II (Qiagen) at 25 Hz for 1 min, and extracted in 0.1 m HCl for 15 min at 4°C. After two rounds of centrifugation at maximum speed at 4°C, the clarified supernatant was derivatized in a citric-phosphate buffer with chloroacetaldehyde at 80°C for 10 min. Following centrifugation at maximum speed for 45 min, the adenosines were resolved on a reverse-phase C18 column and detected by release of fluorescence at 410 nm following excitation at 280 nm. Authentic PAP standard (Sigma-Aldrich) was concurrently run for quantification.
Spin-Trapping EPR Measurements
Spin-trapping assays with ethanol/4-POBN (Sigma-Aldrich) were carried out using whole leaves. Leaves were vacuum-infiltrated with the buffer containing the spin-trap reagents prior to the illumination and then floating on the same buffer during the illumination. Samples were illuminated for a given time with white light (150 μmol photons m−2 s−1) in the presence of 50 mm 4-POBN, 4% ethanol, 50 μm Fe-EDTA, and buffer (25 mm HEPES, pH 7.5, 5 mm MgCl2, 0.3 m sorbitol). EPR spectra were recorded at room temperature in a standard quartz flat cell using an E-Scan (9.73 GHz) spectrometer (Bruker). The following parameters were used: microwave frequency, 86 GHz; modulation frequency, 86 kHz; modulation amplitude, 1 G; microwave power, 1.4 mW; receiver gain, 5 × 102; time constant, 40.96 ms; and number of scans, 2.
Image Analysis and Statistical Analysis
Quantification of lesion surface was performed using Image J software (rsbweb.nih.gov/ij/). Ratios were calculated for each plant between total lesion surface and total leaf surface of the rosette. All statistical analyses were performed using R software.
Detailed Procedure for Pathogen Resistance Assays
Virulent Pseudomonas syringae pv tomato DC3000 strains obtained from J. Glazebrook (University of Minnesota, St. Paul, MN). Bacteria were grown overnight at 30°C in Luria-Bertani medium with appropriate antibiotics (kanamycin 25 µg mL−1 and rifampicin 50 µg mL−1). Bacteria were washed in 10 mm MgCl2 and prepared for inoculations as described by Langlois-Meurinne et al. (2005). Whole leaves of 6- to 7-week-old plants, grown in SD conditions, were infiltrated with P. syringae pv tomato DC3000 at the concentration of 105 cfu mL−1, using a 1-mL syringe without a needle. Leaf discs (0.28 cm2 each) were harvested from inoculated leaves at 0, 24, and 48 h postinoculation. For each time point, five samples were made by pooling four leaf discs from different treated plants. The leaves of five different plants were harvested at each time point. To monitor bacterial growth, leaf discs were homogenized in 400 µL of sterile water, and six serial dilutions (1:10) were performed for each sample. Eight 10-µL aliquots of these 1:10 serial dilutions were spotted on solid Luria-Bertani medium containing kanamycin (25 µg mL−1) and rifampicin (50 µg mL−1), and colony numbers were quantified after 2 to 3 d of incubation at 30°C. Statistical analyses of the differences between two means of log-transformed data were performed according to Wilcoxon signed-ranked test.
Accession Numbers
Accession numbers for genes mentioned in this work are as follows: MIPS1 (AT4G39800), FRY1 (AT5G63980), XRN2 (AT5G42540), XRN3 (AT1G75660), XRN4 (AT1G54490), LSD1 (AT4G20380), GUN1 (AT2G31400), GUN4 (AT3G59400), GUN5 (AT5G13630), PCB2 (AT5G18660), MPK4 (AT4G01370), EX1 (AT4G33630), and EX2 (AT1G27510).
Supplemental Data
The following supplemental materials are available.
Supplemental Table S1. Sequence of primers used in this study.
Supplemental Table S2. PCR conditions used for genotyping.
Supplemental Figure S1. Suppression of the cell death phenotype in the mips1 somi3 mutant obtained by ethyl methanesulfonate mutagenesis of the mips1 mutant.
Supplemental Figure S2. Chlorophyll accumulation is differentially affected by the gun1, gun4, and gun5 mutations.
Supplemental Figure S3. Photosynthetic activity in wild-type plants and mips1 mutants grown under various light and CO2 regimes.
Supplemental Figure S4. Low CO2 rescues cell death in the mips1 mips2/+ sesquimutant.
Supplemental Figure S5. ROS detection by spin-trapping EPR using 4-POBN/ethanol as spin trap.
Supplemental Figure S6. Basal levels of SA are not modified in the fry1-6 mutant.
Supplemental Figure S7. mips1 developmental defects are not suppressed by the fry1-6 mutation.
Supplemental Figure S8. PAP content in wild type (Col-0) and mips1 4 h and 4 d after transfer to LD conditions.
Supplemental Figure S9. PAP content is identical in fry1-6 and fry1-6 lsd1 mutants.
Supplemental Figure S10. Comparison of transcriptome data obtained on mips1, alx8, and xrn mutants.
Supplementary Material
Acknowledgments
We are grateful to H. Hirt and his group (URGV, Evry) for constructive discussions on this project, and to Pierre Joliot (IBPC, Paris) for critical reading of the manuscript. We thank A. Mallory (Institut Curie, Paris) for the gift of xrn triple mutants, and J. Jouhet (CEA, Grenoble) for the gift of gun mutants. We thank Y. Dellero (IPS2) for help and advice for gas-exchange measurements. We thank two anonymous reviewers for constructive comments about the article. The IPS2 benefits from the support of the LabEx Saclay Plant Sciences-SPS (ANR-10-LABX-0040-SPS).
Glossary
- PCD
programmed cell death
- ROS
reactive oxygen species
- SA
salicylic acid
- LMM
lesion mimic mutant
- MI
myoinositol
- LD
long day
- ETR
electron transfer rate
- SD
short day
- ROI
reactive oxygen intermediate
- EPR
electron paramagnetic resonance
- 4-POBN
α-(4-pyridyl-1-oxide)-N-tert-butylnitrone
- XRN
exoribonuclease
- JA
jasmonic acid
References
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al. (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Aviv DH, Rustérucci C, Holt BF III, Dietrich RA, Parker JE, Dangl JL (2002) Runaway cell death, but not basal disease resistance, in lsd1 is SA- and NIM1/NPR1-dependent. Plant J 29: 381–391 [DOI] [PubMed] [Google Scholar]
- Bednarek P, Pislewska-Bednarek M, Svatos A, Schneider B, Doubsky J, Mansurova M, Humphry M, Consonni C, Panstruga R, Sanchez-Vallet A, et al. (2009) A glucosinolate metabolism pathway in living plant cells mediates broad-spectrum antifungal defense. Science 323: 101–106 [DOI] [PubMed] [Google Scholar]
- Bruggeman Q, Garmier M, de Bont L, Soubigou-Taconnat L, Mazubert C, Benhamed M, Raynaud C, Bergounioux C, Delarue M (2014) The polyadenylation factor subunit CLEAVAGE AND POLYADENYLATION SPECIFICITY FACTOR30: a key factor of programmed cell death and a regulator of immunity in Arabidopsis. Plant Physiol 165: 732–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruggeman Q, Prunier F, Mazubert C, de Bont L, Garmier M, Lugan R, Benhamed M, Bergounioux C, Raynaud C, Delarue M (2015a) Involvement of Arabidopsis Hexokinase1 in cell death mediated by myo-inositol accumulation. Plant Cell 27: 1801–1814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruggeman Q, Raynaud C, Benhamed M, Delarue M (2015b) To die or not to die? Lessons from lesion mimic mutants. Front Plant Sci 6: 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan KX, Phua SY, Crisp P, McQuinn R, Pogson BJ (2015) Learning the language of the chloroplast: retrograde signaling and beyond. Annu Rev Plant Biol http://dx.doi.org/10.1146/annurev-arplant-043015-111854 [DOI] [PubMed] [Google Scholar]
- Colcombet J, Hirt H (2008) Arabidopsis MAPKs: a complex signalling network involved in multiple biological processes. Biochem J 413: 217–226 [DOI] [PubMed] [Google Scholar]
- Coll NS, Epple P, Dangl JL (2011) Programmed cell death in the plant immune system. Cell Death Differ 18: 1247–1256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cottage AJ, Mott EK, Wang J-H, Sullivan JA, MacLean D, Tran L, Choy M-K, Newell C, Kavanagh TA, Aspinall S, et al. (2008) GUN1 (GENOMES UNCOUPLED1) encodes a pentatricopeptide repeat (PPR) protein involved in plastid protein synthesis-responsive retrograde signaling to the nucleus. In Allen JF, Gantt E, Golbeck JH, Osmond B, Photosynthesis. Energy from Sun. Springer, Dordrecht, The Netherlands, pp 1202–1205 [Google Scholar]
- Czabotar PE, Lessene G, Strasser A, Adams JM (2014) Control of apoptosis by the BCL-2 protein family: implications for physiology and therapy. Nat Rev Mol Cell Biol 15: 49–63 [DOI] [PubMed] [Google Scholar]
- Dickman MB, Fluhr R (2013) Centrality of host cell death in plant-microbe interactions. Annu Rev Phytopathol 51: 543–570 [DOI] [PubMed] [Google Scholar]
- Dietrich RA, Delaney TP, Uknes SJ, Ward ER, Ryals JA, Dangl JL (1994) Arabidopsis mutants simulating disease resistance response. Cell 4: 565–577 [DOI] [PubMed] [Google Scholar]
- Donahue JL, Alford SR, Torabinejad J, Kerwin RE, Nourbakhsh A, Ray WK, Hernick M, Huang X, Lyons BM, Hein PP, et al. (2010) The Arabidopsis thaliana Myo-inositol 1-phosphate synthase1 gene is required for Myo-inositol synthesis and suppression of cell death. Plant Cell 22: 888–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards K, Johnstone C, Thompson C (1991) A simple and rapid method for the preparation of plant genomic DNA for PCR analysis. Nucleic Acids Res 19: 1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estavillo GM, Crisp PA, Pornsiriwong W, Wirtz M, Collinge D, Carrie C, Giraud E, Whelan J, David P, Javot H, et al. (2011) Evidence for a SAL1-PAP chloroplast retrograde pathway that functions in drought and high light signaling in Arabidopsis. Plant Cell 23: 3992–4012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gazzani S, Lawrenson T, Woodward C, Headon D, Sablowski R (2004) A link between mRNA turnover and RNA interference in Arabidopsis. Science 306: 1046–1048 [DOI] [PubMed] [Google Scholar]
- Gutierrez J, Gonzalez-Perez S, Garcia-Garcia F, Daly CT, Lorenzo O, Revuelta JL, McCabe PF, Arellano JB (2014) Programmed cell death activated by Rose Bengal in Arabidopsis thaliana cell suspension cultures requires functional chloroplasts. J Exp Bot 65: 3081–3095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gy I, Gasciolli V, Lauressergues D, Morel JB, Gombert J, Proux F, Proux C, Vaucheret H, Mallory AC (2007) Arabidopsis FIERY1, XRN2, and XRN3 are endogenous RNA silencing suppressors. Plant Cell 19: 3451–3461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaag HM, Nagy PD (2009) Silencing of Nicotiana benthamiana Xrn4p exoribonuclease promotes tombusvirus RNA accumulation and recombination. Virology 386: 344–352 [DOI] [PubMed] [Google Scholar]
- Kim C, Meskauskiene R, Zhang S, Lee KP, Lakshmanan Ashok M, Blajecka K, Herrfurth C, Feussner I, Apel K (2012) Chloroplasts of Arabidopsis are the source and a primary target of a plant-specific programmed cell death signaling pathway. Plant Cell 24: 3026–3039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopka J, Schauer N, Krueger S, Birkemeyer C, Usadel B, Bergmüller E, Dörmann P, Weckwerth W, Gibon Y, Stitt M, et al. (2005) GMD@CSB.DB: the Golm Metabolome Database. Bioinformatics 21: 1635–1638 [DOI] [PubMed] [Google Scholar]
- Koussevitzky S, Nott A, Mockler TC, Hong F, Sachetto-Martins G, Surpin M, Lim J, Mittler R, Chory J (2007) Signals from chloroplasts converge to regulate nuclear gene expression. Science 316: 715–719 [PubMed] [Google Scholar]
- Kunkel BN, Brooks DM (2002) Cross talk between signaling pathways in pathogen defense. Curr Opin Plant Biol 5: 325–331 [DOI] [PubMed] [Google Scholar]
- Laloi C, Stachowiak M, Pers-Kamczyc E, Warzych E, Murgia I, Apel K (2007) Cross-talk between singlet oxygen- and hydrogen peroxide-dependent signaling of stress responses in Arabidopsis thaliana. Proc Natl Acad Sci USA 104: 672–677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langlois-Meurinne M, Gachon CMM, Saindrenan P (2005) Pathogen-responsive expression of glycosyltransferase genes UGT73B3 and UGT73B5 is necessary for resistance to Pseudomonas syringae pv tomato in Arabidopsis. Plant Physiol 139: 1890–1901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latrasse D, Jégu T, Meng PH, Mazubert C, Hudik E, Delarue M, Charon C, Crespi M, Hirt H, Raynaud C, et al. (2013) Dual function of MIPS1 as a metabolic enzyme and transcriptional regulator. Nucleic Acids Res 41: 2907–2917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee B-R, Huseby S, Koprivova A, Chételat A, Wirtz M, Mugford ST, Navid E, Brearley C, Saha S, Mithen R, et al. (2012) Effects of fou8/fry1 mutation on sulfur metabolism: is decreased internal sulfate the trigger of sulfate starvation response? PLoS One 7: e39425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KP, Kim C, Landgraf F, Apel K (2007) EXECUTER1- and EXECUTER2-dependent transfer of stress-related signals from the plastid to the nucleus of Arabidopsis thaliana. Proc Natl Acad Sci USA 104: 10270–10275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepistö A, Toivola J, Nikkanen L, Rintamäki E (2012) Retrograde signaling from functionally heterogeneous plastids. Front Plant Sci 3: 286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lord CEN, Gunawardena AH (2012) Programmed cell death in C. elegans, mammals and plants. Eur J Cell Biol 91: 603–613 [DOI] [PubMed] [Google Scholar]
- Mariño G, Niso-Santano M, Baehrecke EH, Kroemer G (2014) Self-consumption: the interplay of autophagy and apoptosis. Nat Rev Mol Cell Biol 15: 81–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateo A, Mühlenbock P, Rustérucci C, Chang CC, Miszalski Z, Karpinska B, Parker JE, Mullineaux PM, Karpinski S (2004) LESION SIMULATING DISEASE 1 is required for acclimation to conditions that promote excess excitation energy. Plant Physiol 136: 2818–2830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng PH, Raynaud C, Tcherkez G, Blanchet S, Massoud K, Domenichini S, Henry Y, Soubigou-Taconnat L, Lelarge-Trouverie C, Saindrenan P, et al. (2009) Crosstalks between myo-inositol metabolism, programmed cell death and basal immunity in Arabidopsis. PLoS One 4: e7364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelet L, Krieger-Liszkay A (2012) Reactive oxygen intermediates produced by photosynthetic electron transport are enhanced in short-day grown plants. Biochim Biophys Acta 1817: 1306–1313 [DOI] [PubMed] [Google Scholar]
- Mochizuki N, Brusslan JA, Larkin R, Nagatani A, Chory J (2001) Arabidopsis genomes uncoupled 5 (GUN5) mutant reveals the involvement of Mg-chelatase H subunit in plastid-to-nucleus signal transduction. Proc Natl Acad Sci USA 98: 2053–2058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mühlenbock P, Szechynska-Hebda M, Plaszczyca M, Baudo M, Mateo A, Mullineaux PM, Parker JE, Karpinska B, Karpinski S (2008) Chloroplast signaling and LESION SIMULATING DISEASE1 regulate crosstalk between light acclimation and immunity in Arabidopsis. Plant Cell 20: 2339–2356; erratum Mühlenbock P, Szechynska-Hebda M, Plaszczyca M, Baudo M, Mateo A, Mullineaux PM, Parker JE, Karpinska B, Karpinski S (2008) Plant Cell 20: 3480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakanishi H, Nozue H, Suzuki K, Kaneko Y, Taguchi G, Hayashida N (2005) Characterization of the Arabidopsis thaliana mutant pcb2 which accumulates divinyl chlorophylls. Plant Cell Physiol 46: 467–473 [DOI] [PubMed] [Google Scholar]
- Păcurar DI, Păcurar ML, Street N, Bussell JD, Pop TI, Gutierrez L, Bellini C (2012) A collection of INDEL markers for map-based cloning in seven Arabidopsis accessions. J Exp Bot 63: 2491–2501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitzschke A, Schikora A, Hirt H (2009) MAPK cascade signalling networks in plant defence. Curr Opin Plant Biol 12: 421–426 [DOI] [PubMed] [Google Scholar]
- Queval G, Issakidis-Bourguet E, Hoeberichts FA, Vandorpe M, Gakière B, Vanacker H, Miginiac-Maslow M, Van Breusegem F, Noctor G (2007) Conditional oxidative stress responses in the Arabidopsis photorespiratory mutant cat2 demonstrate that redox state is a key modulator of daylength-dependent gene expression, and define photoperiod as a crucial factor in the regulation of H2O2-induced cell death. Plant J 52: 640–657 [DOI] [PubMed] [Google Scholar]
- Ramel F, Birtic S, Ginies C, Soubigou-Taconnat L, Triantaphylidès C, Havaux M (2012) Carotenoid oxidation products are stress signals that mediate gene responses to singlet oxygen in plants. Proc Natl Acad Sci USA 109: 5535–5540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez VM, Chételat A, Majcherczyk P, Farmer EE (2010) Chloroplastic phosphoadenosine phosphosulfate metabolism regulates basal levels of the prohormone jasmonic acid in Arabidopsis leaves. Plant Physiol 152: 1335–1345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roessner-Tunali U, Hegemann B, Lytovchenko A, Carrari F, Bruedigam C, Granot D, Fernie AR (2003) Metabolic profiling of transgenic tomato plants overexpressing hexokinase reveals that the influence of hexose phosphorylation diminishes during fruit development. Plant Physiol 133: 84–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stael S, Kmiecik P, Willems P, Van Der Kelen K, Coll NS, Teige M, Van Breusegem F (2015) Plant innate immunity--sunny side up? Trends Plant Sci 20: 3–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staiger D, Korneli C, Lummer M, Navarro L (2013) Emerging role for RNA-based regulation in plant immunity. New Phytol 197: 394–404 [DOI] [PubMed] [Google Scholar]
- Sumner LW, Amberg A, Barrett D, Beale MH, Beger R, Daykin CA, Fan TW, Fiehn O, Goodacre R, Griffin JL, et al. (2007) Proposed minimum reporting standards for chemical analysis Chemical Analysis Working Group (CAWG) Metabolomics Standards Initiative (MSI). Metabolomics 3: 211–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunil B, Talla SK, Aswani V, Raghavendra AS (2013) Optimization of photosynthesis by multiple metabolic pathways involving interorganelle interactions: resource sharing and ROS maintenance as the bases. Photosynth Res 117: 61–71 [DOI] [PubMed] [Google Scholar]
- Susek RE, Ausubel FM, Chory J (1993) Signal transduction mutants of Arabidopsis uncouple nuclear CAB and RBCS gene expression from chloroplast development. Cell 74: 787–799 [DOI] [PubMed] [Google Scholar]
- Triantaphylidès C, Krischke M, Hoeberichts FA, Ksas B, Gresser G, Havaux M, Van Breusegem F, Mueller MJ (2008) Singlet oxygen is the major reactive oxygen species involved in photooxidative damage to plants. Plant Physiol 148: 960–968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Aken O, Van Breusegem F (2015) Licensed to kill: mitochondria, chloroplasts, and cell death. Trends Plant Sci 20: 754–766 [DOI] [PubMed] [Google Scholar]
- van Doorn WG. (2011) Classes of programmed cell death in plants, compared to those in animals. J Exp Bot 62: 4749–4761 [DOI] [PubMed] [Google Scholar]
- Vogel MO, Moore M, König K, Pecher P, Alsharafa K, Lee J, Dietz K-J (2014) Fast retrograde signaling in response to high light involves metabolite export, MITOGEN-ACTIVATED PROTEIN KINASE6, and AP2/ERF transcription factors in Arabidopsis. Plant Cell 26: 1151–1165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Bayles KW (2013) Programmed cell death in plants: lessons from bacteria? Trends Plant Sci 18: 133–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Y, Wang J, Dehesh K (2013) Review of stress specific organelles-to-nucleus metabolic signal molecules in plants. Plant Sci 212: 102–107 [DOI] [PubMed] [Google Scholar]
- Xin X-F, He SY (2013) Pseudomonas syringae pv. tomato DC3000: a model pathogen for probing disease susceptibility and hormone signaling in plants. Annu Rev Phytopathol 51: 473–498 [DOI] [PubMed] [Google Scholar]
- Zurbriggen MD, Carrillo N, Tognetti VB, Melzer M, Peisker M, Hause B, Hajirezaei MR (2009) Chloroplast-generated reactive oxygen species play a major role in localized cell death during the non-host interaction between tobacco and Xanthomonas campestris pv. vesicatoria. Plant J 60: 962–973 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.