Root type-specific lateral root branching and pericycle-specific transcriptome reprogramming highlight diverse foraging strategies of maize roots in heterogeneous nitrate environments.
Abstract
The adaptability of root system architecture to unevenly distributed mineral nutrients in soil is a key determinant of plant performance. The molecular mechanisms underlying nitrate dependent plasticity of lateral root branching across the different root types of maize are only poorly understood. In this study, detailed morphological and anatomical analyses together with cell type-specific transcriptome profiling experiments combining laser capture microdissection with RNA-seq were performed to unravel the molecular signatures of lateral root formation in primary, seminal, crown, and brace roots of maize (Zea mays) upon local high nitrate stimulation. The four maize root types displayed divergent branching patterns of lateral roots upon local high nitrate stimulation. In particular, brace roots displayed an exceptional architectural plasticity compared to other root types. Transcriptome profiling revealed root type-specific transcriptomic reprogramming of pericycle cells upon local high nitrate stimulation. The alteration of the transcriptomic landscape of brace root pericycle cells in response to local high nitrate stimulation was most significant. Root type-specific transcriptome diversity in response to local high nitrate highlighted differences in the functional adaptability and systemic shoot nitrogen starvation response during development. Integration of morphological, anatomical, and transcriptomic data resulted in a framework underscoring similarity and diversity among root types grown in heterogeneous nitrate environments.
Sessile plants efficiently exploit nutrients and water by postembryonically formed branched lateral roots. In most plant species, lateral roots are initiated from pericycle cells of the central cylinder that undergo asymmetric divisions (Malamy and Benfey, 1997). Xylem and phloem pole pericycle cells differ in morphological (Beeckman et al., 2001) and ultrastructural (Parizot et al., 2008) characteristics and specific protein and gene expression patterns (Vanneste et al., 2005; Moreno-Risueno et al., 2010). Auxin gradients and their essential downstream molecular components have been identified as driving forces of lateral root development (Vanneste et al., 2005; De Smet et al., 2007, 2008; Dubrovsky et al., 2008; Laskowski et al., 2008). Moreover, a tight control of cell cycle progression in xylem-pole pericycle cells is instrumental for lateral root initiation in Arabidopsis (Arabidopsis thaliana; Himanen et al., 2002, 2004).
In contrast to Arabidopsis, the mechanisms underlying lateral root positioning in the economically important crop species maize (Zea mays) are largely unknown. Lateral root initiation is preceded by auxin response maxima in differentiating xylem cells and in cells surrounding the protophloem vessels in maize (Jansen et al., 2012; Yu et al., 2015a). The maize root system is an ideal model to study organ-specific similarity and diversity because of its spatio-temporal complexity and distinct genetic control of different root types (Hochholdinger and Zimmermann, 2008; Rogers and Benfey, 2015). The maize root system is formed by multiple root types that are of embryonic (primary and seminal roots) and postembryonic (crown and brace roots) origin (Hochholdinger et al., 2004a). These root types are initiated at different stages of development and all form lateral roots (Hochholdinger et al., 2004a). Understanding root system behavior at the scale of specific root types is therefore of great importance in rational breeding approaches aiming on improving crop stress tolerance, efficiency, and productivity (Rogers and Benfey, 2015).
Pericycle cells deeply embedded inside the root tissue can respond to environmental changes to guide lateral root development, thus shaping the highly branched root system in contact with soil (Malamy, 2005; Atkinson et al., 2014). A critical adaptive behavior of lateral roots is foraging of nonuniformly distributed soil resources in both ecological and agronomic contexts (Malamy, 2005; Ruffel et al., 2011; Guan et al., 2014; Yu et al., 2014a). Nitrogen, the most mobile element in soil, determines shoot biomass and yield (Marschner, 1995). Recent results highlighted the superior role of lateral root branching on nitrate and water capturing in maize (Postma et al., 2014; Zhan et al., 2015), suggesting considerable genetic potential to modify this trait. Most studies in Arabidopsis concluded that local nitrate application exerts either no (Zhang and Forde, 1998) or only a relatively small effect (Linkohr et al., 2002) on lateral root number or density. Recently, the auxin receptor AFB3 and the downstream transcription factors NAC4 and OBP4 were identified as key regulators of a nitrate-responsive gene network activating lateral root initiation by controlling cell cycle progression in the pericycle of Arabidopsis (Vidal et al., 2013).
Transcriptome analyses have provided novel insights in lateral root initiation in maize roots by identifying candidate genes associated with early cell divisions (Woll et al., 2005; Dembinsky et al., 2007; Yu et al., 2015a). To obtain an in-depth understanding of nitrate sensing during lateral root branching, this study surveyed the transcriptomic complexity of pericycle cells in the four major root types of maize in response to heterogeneous nitrate application using laser capture microdissection in combination with RNA-sequencing (LCM-RNAseq). We identified substantial differences between the transcriptomic landscapes of pericycle cells of different maize root types, among which functional enrichment was altered as a consequence of changed external nitrate levels. Intriguingly, pericycle cells displayed brace root specific responses to local nitrate stimuli that included unique development-related processes. Finally, by integrating morphological and anatomical observations with cell-specific transcriptome responses, we provided a comprehensive overview of branching mechanisms among maize root types and their response to heterogeneous nitrate during plant development.
RESULTS
Deep Profiling of Lateral Root Branching Response to Local High Nitrate
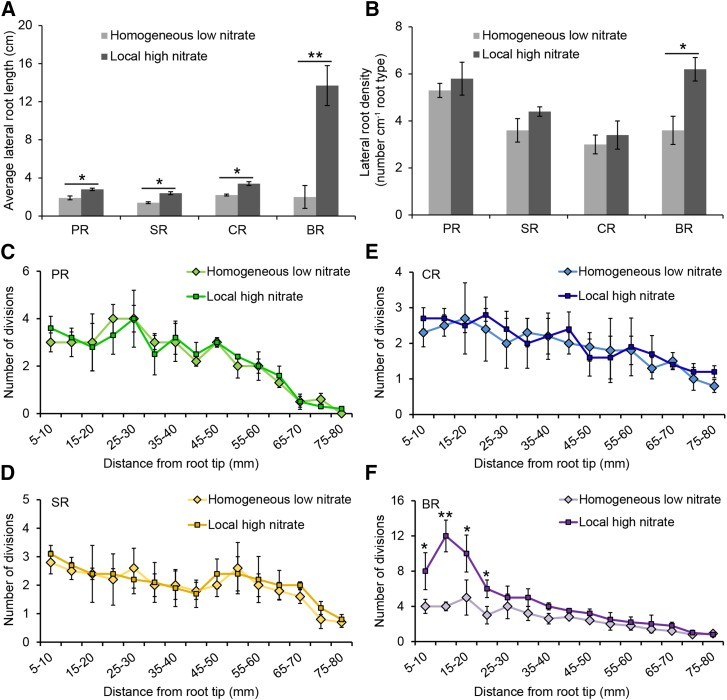
Emerged lateral roots in the branching zone were quantified to determine how heterogeneous nitrate availability contributes to root type-specific lateral root formation in maize. Twenty days after local high nitrate treatment, average lateral root length was significantly increased compared to control experiments in all root types (Fig. 1A). The local effect on the promotion of lateral root elongation in brace roots (600%) was more pronounced than that of the other three root types (50–75%; Fig. 1A). A significant increase in the density of emerged lateral roots was exclusively observed in brace roots (75%) but not in the other three root types (Fig. 1B).
Figure 1.
Effects of homogeneous low nitrate or local high nitrate supplies on lateral root development of four maize root types. Seedlings or adult plants were grown in homogeneous low nitrate nutrient solution before each root type was transferred to local high nitrate nutrient solution (see “Materials and Methods”). Roots were scanned and counted after 20 d of growth in these two nitrate conditions. A and B, Average lateral root length (A) and lateral root density (B) in the lateral root branching zone under homogenous low nitrate and local high nitrate conditions. Error bars indicate means ± se; n = 4 (roots from four different maize plants). C to F, Early pericycle cell divisions in primary (C), seminal (D), crown roots (E), and brace roots (F) 24 h after local high nitrate supply. F is taken from Yu et al. (2015a) as a reference for comparisons with the other root types. Asymmetric pericycle cell divisions were tracked from 5 to 80 mm from root tip after Safranin O and Fast Green staining. Error bars indicate means ± se; n = 4 (roots from four different) maize plants. Asterisks denote significant differences according to paired Student’s t tests (*P < 0.05; **P < 0.01). PR, primary root; SR, seminal root; CR, crown root; BR, brace root. F was adapted from Yu et al. (2015a).
Based on histological analyses, early divisions of pericycle cells were quantified to determine whether local high nitrate influences the stages of lateral root initiation at increasing distances from the root tip. It was previously demonstrated that anticlinal and periclinal divisions of pericycle cells were significantly induced in response to local high nitrate in aboveground shoot-borne brace roots (Yu et al., 2015a). In contrast, no significant induction of cell divisions in phloem pole pericycle cells were detected in this study in any region of the three seedling root types, primary, seminal, and crown roots of 80 mm length, by local high nitrate induction 24 h after treatment (Fig. 1C). Therefore, lateral root branching of brace root was clearly distinct from that of primary, seminal, and crown roots in response to heterogeneous nitrate supplies. This result supports the notion that brace roots are important for water and nutrient acquisition.
Anatomical Characteristics of the Four Maize Root Types
To further understand functional differences between the four root types, microscopic analyses were conducted to investigate anatomical traits in transverse sections of roots in the lateral root initiation zone grown in homogeneous low nitrate. Overall, in brace roots, the total transversal area, the stele transversal area, and the total meta-xylem area were much larger than those in the other three root types (Supplemental Table S1). Remarkably, primary and crown roots displayed similar traits in these transverse sections and differed significantly from those of seminal roots (Supplemental Fig. S1 and Supplemental Table S1). Apparently, brace roots had a higher number of cortical cell layers and meta-xylem elements than the other three root types (Supplemental Fig. S1 and Supplemental Table S1). Subsequently, the length and width of pericycle cells adjacent to phloem and xylem poles were quantified to explore the similarity of pericycle cell size among root types. Pericycle cells adjacent to the xylem or phloem poles in brace roots showed an equal size to those in crown roots but were significantly bigger than those of primary and seminal roots (Supplemental Table S1). Together, these data suggest anatomical diversity of the four maize root types.
RNA Sequencing of Laser Captured Phloem Pole Pericycle Cells from Four Maize Root Types Grown under Different Nitrate Regimes
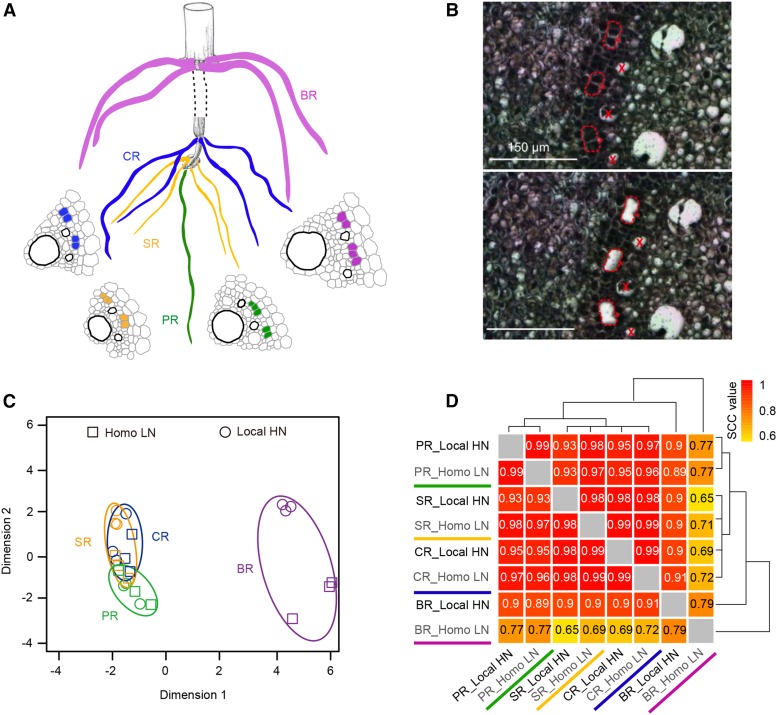
Laser capture microdissection was adopted to isolate phloem pole pericycle cells from four root types of the maize inbred line B73 upon local high nitrate and control treatment (Fig. 2A). Total RNA was extracted and amplified from three independent biological replicates of these captured pericycle cells per root type and treatment resulting in 24 samples (Fig. 2A). Subsequently, RNA populations were reverse-transcribed into cDNA, linearly amplified, and paired-end sequenced using an Illumina HiSeq 2000 platform (see “Materials and Methods”). On average, LCM-RNAseq experiments yielded approximately 23 million 100-bp paired-end reads per sample (Supplemental Table S2). After quality trimming and removal of stacked reads, approximately 68% of all sequences were mapped to the maize reference genome (ZmB73_RefGen_v2; Supplemental Table S2). Finally, approximately 52% of the remaining reads mapped uniquely to the filtered gene set of maize (Supplemental Table S2), which comprises 39,656 high-confidence gene models (FGSv2, release 5b.60).
Figure 2.
Maize root types, cellular organization, and laser captured pericycle cell transcriptomes under homogenous low nitrate or local high nitrate conditions. A, Graphic depiction of pericycle cells (in color) adjacent to phloem poles of the four maize root types primary root (PR), seminal root (SR), crown root (CR), and brace root (BR) subjected to RNA sequencing. B, Representative sections taken before and after laser capture microdissection of phloem pole pericycle cells. Asterisks, phloem poles. C, Multidimensional scaling plot visualizing overall similarity of RNA populations based on gene expression for all 24 root type-specific pericycle cells. Homo LN, homogeneous low nitrate; Local HN, local high nitrate. D, SCC analysis of the eight transcriptomes using log2-transformed FPKM values of all expressed genes. The hierarchical clustering dendrogram was inferred by applying 1-SCC as distance function.
The Transcriptomic Landscape of Pericycle Cells Differs between Root Types under Homogeneous Low or Local High Nitrate Conditions
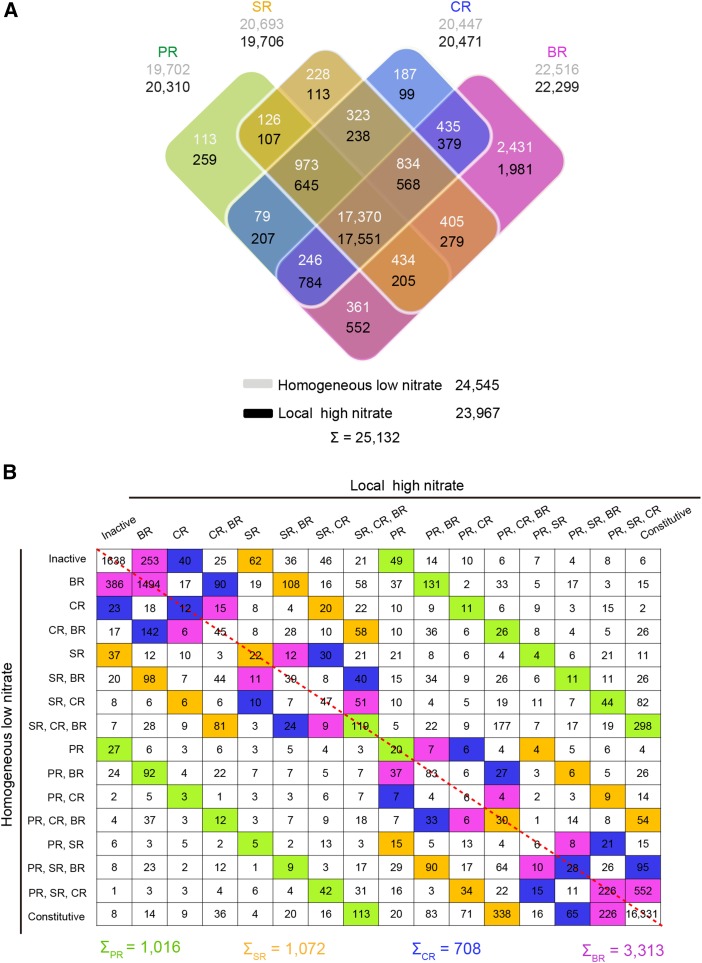
Gene-wise transcriptional activity of pericycle cells for each root type/nitrate treatment combination was computed. To decide if a gene was expressed (“active”) or not (“inactive”) in any root type/nitrate treatment combination, a generalized linear model analysis was conducted (see “Materials and Methods”). In total, 25,132 genes (63% of the filtered gene set; Figure 3A) were declared active in pericycle cells of at least one root type and nitrate condition. The specificity of pericycle transcriptomes in different root types/nitrate combinations was visualized in a Venn diagram. A substantial number of genes were constitutively active in all four tissues under homogeneous low (17,370; 71%) and local high nitrate (17,551; 73%) conditions (Fig. 3A). Notably, a significant number of genes were exclusively active in brace roots under both nitrate conditions (Fig. 3A). In contrast, a smaller number of genes were specifically active in the three seedling root types (Fig. 3A), suggesting root type specificity of genes expressed during early lateral root initiation (Fig. 3A). Both, a multidimensional scaling plot (Fig. 2C) and hierarchical clustering of Spearman correlation coefficient (SCC) analyses (Fig. 2D) revealed high correlation between pericycle transcriptomes of the three seedling root types and a distinct clustering of brace root transcriptomes. Gene-wise transcriptional activity was further computed to explore the functional specificity for each root type/nitrate combination. Remarkably, a substantially higher number of genes were specifically activated by local high nitrate in brace roots (3,313 genes) in comparison to the other root types (primary root, 1,016 genes; seminal root, 1,072 genes; crown root, 708 genes; Fig. 3B). Among the genes only active in brace roots, a majority of them (1,720; 52%) were constitutively active irrespective of the nitrate condition (Fig. 3B). This observation highlights the genetically determined specificity in brace roots, which is different from other root types (Fig. 3B). Hence, our observation might suggest that brace roots are distinct in some specific functions of pericycle cells typically not found in the other root types during lateral root initiation. These results are consistent with the morphological observations on lateral root branching in brace roots.
Figure 3.
Root type-specific gene activity in pericycle cells of maize. A, Venn diagram illustrating the overlap of active genes in pericycle cells of the four root types under homogenous low nitrate or local high nitrate conditions. B, Overview of gene activity patterns and numbers of genes with these patterns in pericycle cells of the four maize root types grown under homogenous low nitrate or local high nitrate conditions. Each colored cell represents a root type-specific gene activity and states the number of active genes. Columns represent patterns under local high nitrate condition, while rows represent expression patterns under homogenous low nitrate condition. Increase of root type-specific gene activity patterns in pericycle cells after local high nitrate stimulus is indicated above the dashed diagonal red line, while increase of such patterns under homogenous low nitrate conditions is summarized below the dashed red line. PR, primary root; SR, seminal root; CR, crown root; BR, brace root.
Dynamic Reprogramming of Pericycle Cell Transcriptomes after Changed External Nitrate
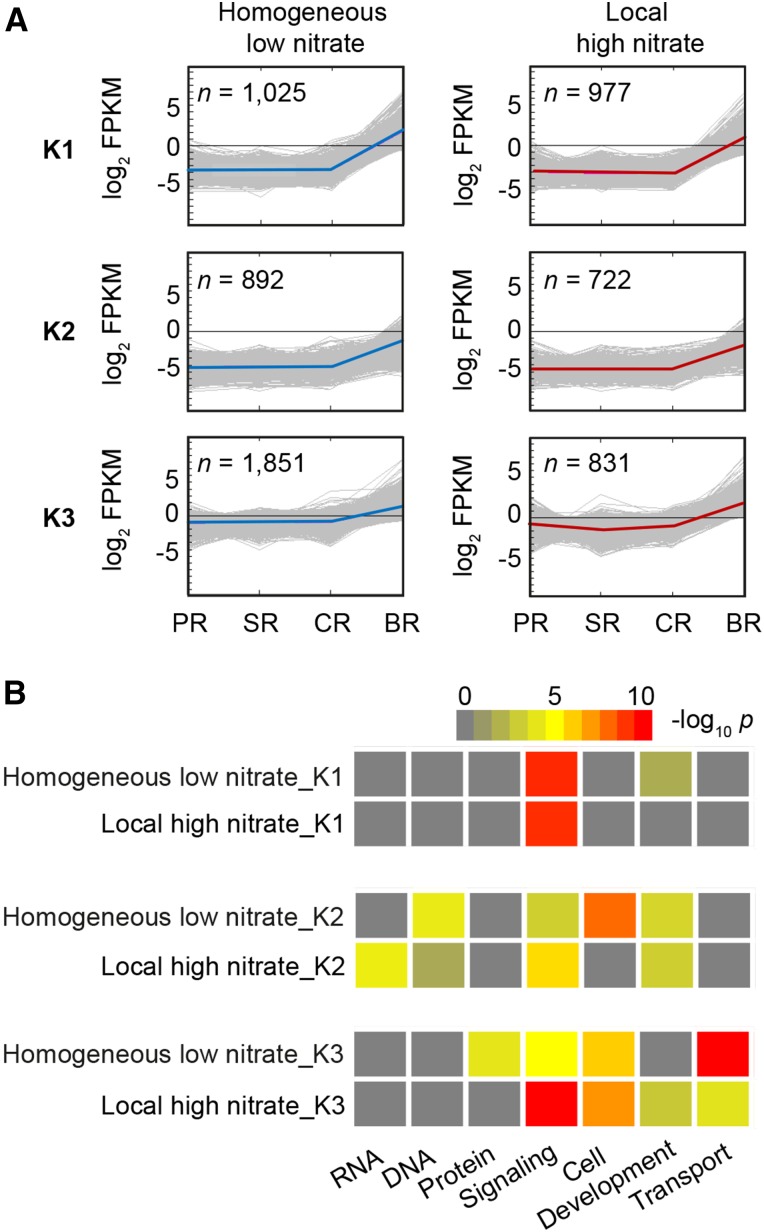
To understand the systemic transcriptome responses of different root types to heterogeneous nitrate distribution, we compared pericycle cell specific transcriptome profiles by K-means clustering. The relationship within clustering representation helped to illustrate the strong similarity and diversity between expression profiles among root types under different nitrate conditions. Thus, we identified 10 clusters among four root types covering all 18,985 expressed genes under homogeneously low nitrate conditions (Supplemental Fig. S2A) and 19,111 expressed genes under local high nitrate conditions (Supplemental Fig. S2B). Among these, three main clusters (K1–K3) representing significant gene expression transitions from low in seedling root types to high in brace roots accounted for 20% of the expressed genes under homogeneous low nitrate and 13% of the expressed genes under local high nitrate (Supplemental Fig. S2A). We then adopted the MapMan functional annotation system to assign the genes in the six clusters (K1 to K3 homogenous low nitrate and K1 to K3 local high nitrate) to functional categories. In total, genes of these six specific clusters were significantly enriched in seven MapMan bins (Fig. 4B). Among those, genes that encode proteins for “signaling” were enriched in all six clusters (Fig. 4B). Moreover, genes that encode proteins related to “DNA” and “transport” were enriched in specific K-means clusters under changing external nitrate levels (Fig. 4B). Finally, genes associated with “RNA” and “protein”-related processes represented enriched functions related to nitrate induction and inhibition, respectively (Fig. 4B). Taken together, this dynamic response established the notion that nitrate availability may reprogram the transcriptome signature of pericycle cells at the root type level.
Figure 4.
Dynamics and functional shift of pericycle cell transcriptomes of the four maize root types upon altered external nitrate levels. A, K-means clusters (K1–K3) showing the dynamics of pericycle transcriptomes among maize root types across homogenous low nitrate and local high nitrate conditions. Expressed genes that follow similar absolute values of Euclidean distance were indicated and clustered as an expression pattern. The K-means support module embedded in the MEV program (http://www.tm4.org/mev) was used to generate clusters. Each gene is plotted as a gray line, and the average expression pattern for each cluster is drawn as a blue or red line. n, the number of genes belonging to each cluster. B, Functional categories (extracted from MapMan bins) analyzed among the six induced K-means clusters depicted in A. Color code from gray to red represents significant enrichment of functional categories calculated by Fisher’s exact test and normalized by -log10 P.
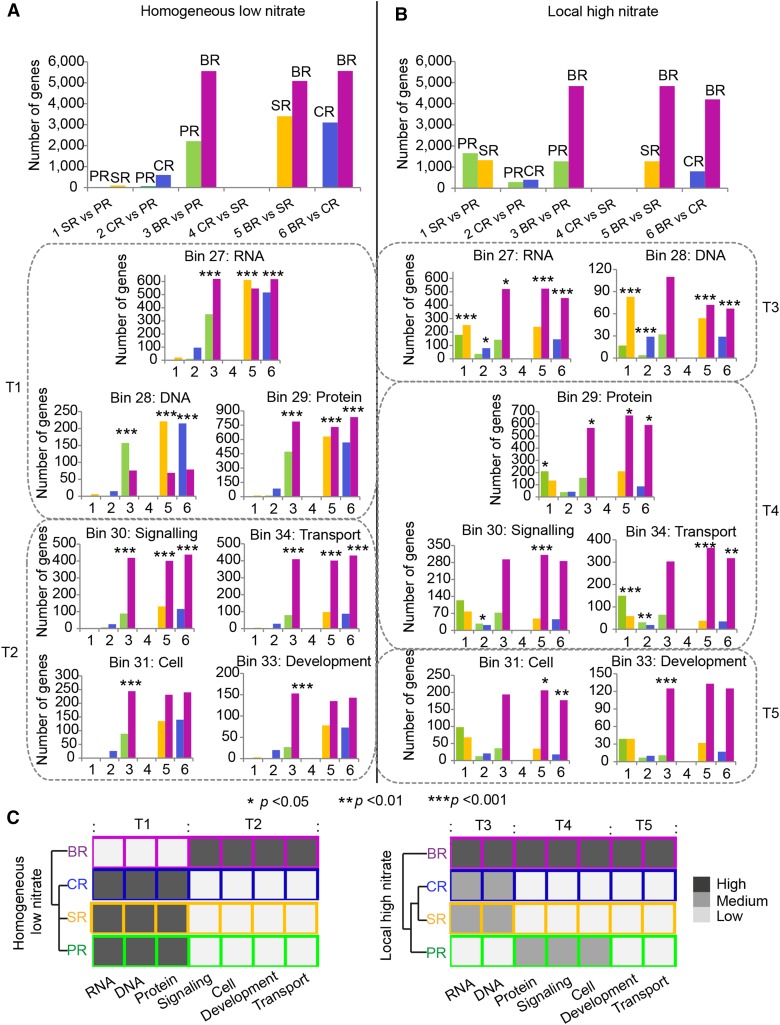
To further analyze the functions of the differentially expressed genes, the MapMan bin classification system was applied to compare the distribution of differentially expressed genes in the six pairwise comparisons of the four root types under homogenous low nitrate (Fig. 5A) and local high nitrate (Fig. 5B) conditions. For each comparison, the differentially expressed genes were split in two columns indicating the root type in which these genes were preferentially expressed (Fig. 5, A and B, upper panels). The ratios of these two columns for all six root type comparisons were defined as the expected values for further analyses. Subsequently, these ratios were determined for each of the six root type comparisons and both nitrate conditions for each of the 35 functional MapMan bins, and these were compared with the expected values by a two-tailed Fisher’s exact test. For both nitrate conditions, these ratios differed significantly from the expected values in the same seven classes (Fig. 5, A and B). These classes were “RNA,” “DNA,” “protein,” “signaling,” “transport,” “cell,” and “development” (Fig. 5, A and B). Under homogeneously low nitrate (Fig. 5A), the functional classes “RNA,” “DNA,” and “protein” displayed a significant enrichment (T1) of genes preferentially expressed in pericycle cells of primary, seminal, and crown roots compared to brace roots (classes 3, 5, and 6) relative to the expected value. By contrast, comparisons of primary, seminal, and crown roots between each other (classes 1, 2, and 4) did not display any significant alterations of differentially expressed genes in these MapMan bins under homogenous low nitrate conditions compared to the reference values (Fig. 5A). These trends have been summarized in Figure 5C. In this panel, primary, seminal, and crown roots are depicted by the same dark color indicating no differences between these roots but an enrichment compared to the light color of brace roots. The opposite transcript accumulation trend (T2) was identified for the functional classes “signaling,” “transport,” “cell,” and “development”: brace root > primary root = seminal root = crown root, with higher accumulation in adult brace roots than the three seedling root types (Fig. 5A). This pattern is also illustrated by the color scheme summary in Figure 5C. The same analysis was repeated for the six comparisons between root types grown under local high nitrate conditions (Fig. 5B) and summarized in Figure 5C. In this analysis another three accumulation trends based on altered transcript ratios were observed. For “RNA” and “DNA,” the enrichment trend was brace root > seminal root = crown root > primary root (T3). For “protein,” “signaling,” and “transport,” the trend was brace root > primary root > seminal root = crown root (T4), while for “cell” and “development,” the trend we brace root > primary root = seminal root = crown root (T5). Taken together, this observation suggests a functional shift in the transcriptome profile of pericycle cells among root types upon change of the external nitrate stimulus.
Figure 5.
Preferentially expressed genes and functional distributions in pericycle cells among six pairwise root type comparisons under homogeneous low nitrate (A) and local high nitrate (B) conditions. All genes preferentially expressed in one root type in the six comparisons were mapped to the 35 MapMan bins. The different color-coded columns in each comparison indicate the number of genes preferentially expressed in the corresponding root type. For each pair of root type comparisons (1–6), functional classes in which the ratio of preferentially expressed genes significantly differed from the expected ratio calculated from all differentially expressed genes were combined based on similar trends in classes T1 to T5. *P < 0.05, **P < 0.01, and ***P < 0.001. C, Root type-specific enrichment trends of MapMan classes according to the results of A and B. The hierarchical clustering dendrogram was inferred by applying PCC as distance function. Same fill colors of the squares indicate no relative enrichment of differentially expressed genes in these MapMan bins between root types while darker colors indicate relative enrichment of differentially expressed genes in root types compared to root types with lighter fill colors. PR, primary root; SR, seminal root; CR, crown root; BR, brace root.
Nitrate-Responsive Genes in Brace Roots of Maize
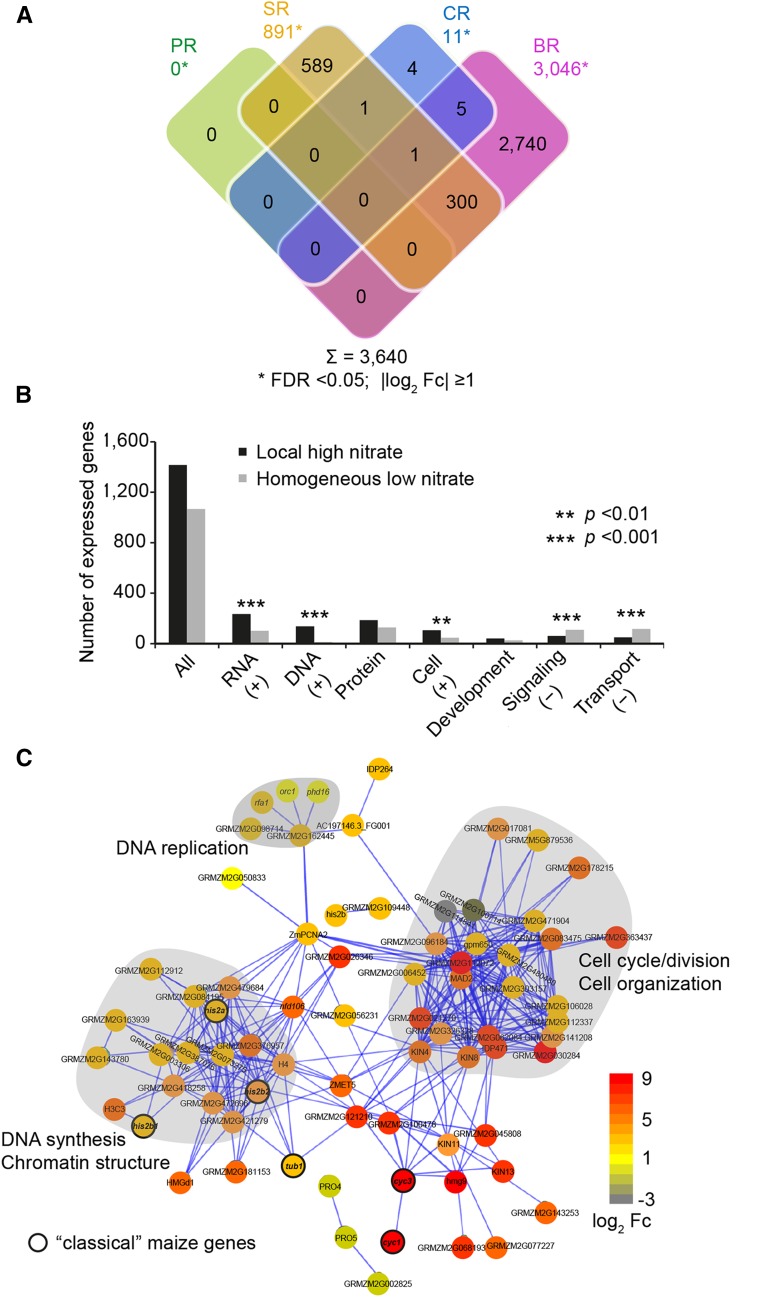
Contrasts were calculated to identify significant changes (false discovery rate [FDR] < 0.05; |log2 Fc| ≥ 1) in response to local high nitrate for each root type using a log-transformed linear model. In total, 3,640 genes expressed in pericycle cells were differentially expressed across the four root types (Fig. 6A). Among those, a majority of 3,046 differentially expressed genes were shown in brace roots (Fig. 6A), while seminal roots contributed 24% of all differentially expressed genes (Fig. 6A). Remarkably, primary and crown roots showed almost no responsive genes when induced by local high nitrate (Fig. 6A). This suggests that upon local high nitrate stimulus, diverse root types showed significantly diverse transcriptome responses to heterogeneous nitrate supplies.
Figure 6.
Functional specificity of the brace root specific transcriptome. A, Overlap of differentially expressed genes (FDR < 0.05, |log2 Fc| ≥ 1) between pericycle transcriptomes of the four root types (PR, primary root; SR, seminal root; CR, crown root; BR, brace root) in response to local high nitrate supply versus homogenous low nitrate as control. B, Functional categories in which the ratio of preferentially expressed genes differed significantly from the ratio of all expressed genes. The expected number of genes for each functional category was calculated based on the distribution of functional categories among all expressed genes. To determine if significantly more overrepresented (+) or underrepresented (−) genes than expected were detected for each individual category, a χ2 test for independence with Yates’ continuity correction was performed. **P < 0.01 and ***P < 0.001. C, Network view of genes with high coexpression scores of ≥0.7 generated using STRING v10 prediction algorithm. The visible genes associated with the categories “DNA” and “cell” were connected based on 640 genes extracted from overrepresented MapMan bins (“RNA,” “DNA,” and “cell” according to B). Three clusters of direct interaction partners are marked by gray background and correspond to DNA synthesis and chromatin structure, DNA replication, cell cycle/division, and cell organization, respectively. Thickened node outlines represent “classical” maize genes. Color codes from gray to red indicate expression level of nitrate-responsive transcripts normalized as log2 algorithm of fold changes (log2 Fc).
Subsequently, the 2,740 genes that displayed differential expression upon nitrate stimulation exclusively in brace roots were functionally classified according to Gene Ontology (GO) terms using agriGO. To identify overrepresented functional categories, a singular enrichment analysis (SEA) was performed. SEA compares each annotated gene to all annotated expressed genes. In total, 45 GO terms displayed overrepresentation (FDR < 0.001) (Supplemental Fig. S3). In parallel, differentially expressed genes were further assigned to MapMan functional categories to compare the distribution of overrepresented and underrepresented functional classes between nitrate treatments. Overall, 640 distinct genes were significantly overrepresented in MapMan bins “RNA,” “DNA,” and “cell,” and 311 distinct genes were significantly underrepresented in MapMan bins “signaling” and “transport” (Fig. 6B). These functional analyses are consistent with those of the transcriptome expression patterns and suggest that brace root pericycle cells likely acts uniquely in comparison to the other root types. To gain insights into the biological role of the enriched functional classes, we investigated the complete set of 640 identified overrepresented genes for whether they were functionally predictive and associated them with the aid of the STRING algorithm. Coexpression analyses with high confidence of ≥0.7 were performed to construct a functionally connected network. Direct interactions of transcripts were clustered and corresponded to DNA synthesis and chromatin structure, DNA replication, cell cycle/division, and cell organization (Fig. 6C). This network supports a role of pericycle cells adjacent to phloem poles in cell cycle and cell division during lateral root initiation in brace roots.
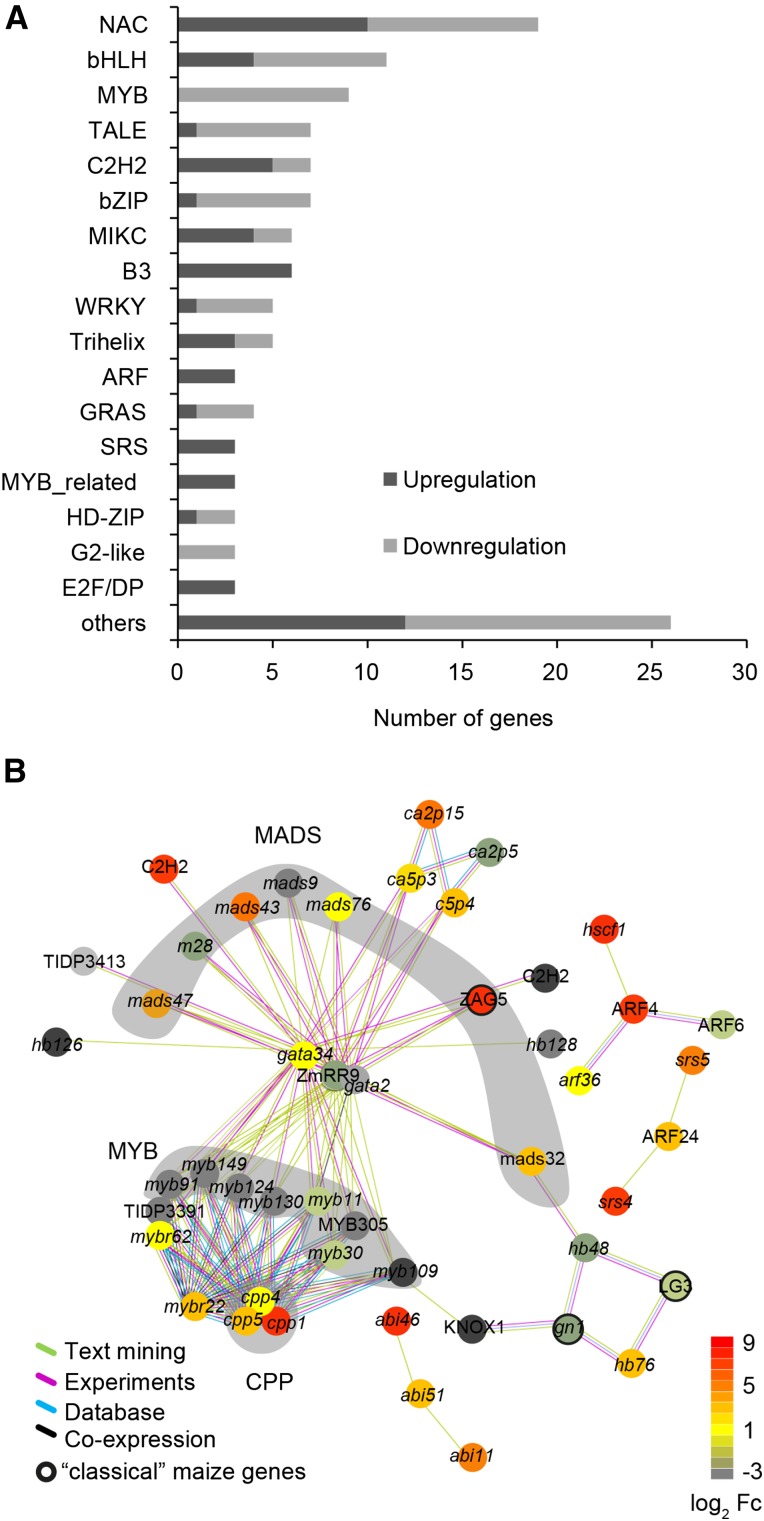
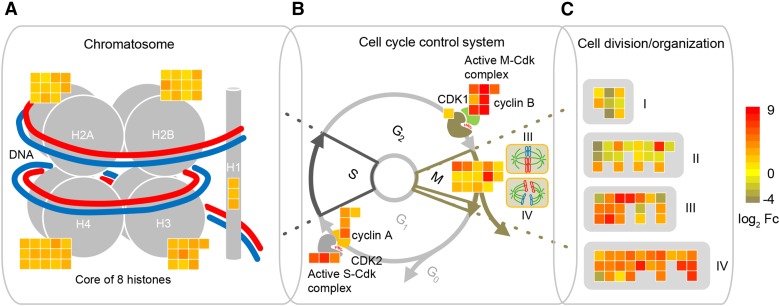
Among the 2,740 nitrate-regulated brace root specific genes, 60 “classical” maize genes with experimentally confirmed functions (Schnable and Freeling, 2011) were determined (Supplemental Table S3). Twelve of them were related to DNA synthesis and cell cycle (Supplemental Table S3). Specifically, six “classical” maize genes were associated with histones (his2a1, his2b1, and his2b2) and cell cycle control (cyc1, cyc3, and tub1; Fig. 6C). Finally, we investigated transcription factors (TFs) that are key regulators of growth, development, and cell fate among the 2,740 nitrate-regulated brace root specific genes. Most differentially regulated TFs belonged to the NAC, bHLH, MYB, TALE, C2H2, and bZIP TF families (Fig. 7A; Supplemental Table S4). All these TFs were further assigned to STRING v10 to predict their functional connectivity. Only hits with high confidence of ≥0.7 were used to construct the network (Fig. 7B). Interactions comprised the MYB, MADS box, and CPP families, which were interconnected by response regulator (RR) ZmRR9 and maize GDB, curated “classical” genes ZmGATA34 and ZmGATA2 (Fig. 7B). The genes enriched in MapMan bins “DNA” and “cell” were used to develop a model to illustrate cell cycle control during lateral root initiation in brace roots (Fig. 8). The four histone families (H2A, H2B, H3, and H4) and linker histone H1, which constitute the chromatosome core are associated with DNA synthesis (MapMan bin classification) and were moderately induced by local high nitrate application (Fig. 8A). A heat map associated with the transition from metaphase to anaphase of mitosis showed upregulated genes encoding several spindle checkpoint proteins (MITOTIC ARREST DEFICIENT2 [MAD2]) and proteins involved in the ANAPHASE-PROMOTING COMPLEX (APC), SMC, and CELL DIVISION CYCLE (CDC) (Fig. 8B). At last, genes enriched in cell division and organization processes were classified into four groups: I, subunits of microtubules; II, structural constituents of the cytoskeleton; III, microtubule binding and associated protein; IV, genes coding kinesin/like motor proteins (Fig. 8C). The complete list of genes associated with chromatin structure and core histones extracted from bin28 and genes associated with cell cycle, cell division, and cell organization extracted from bin31 of the MapMan analysis are shown in Supplemental Tables S5 and S6, respectively.
Figure 7.
TF analyses among the brace root specific nitrate-responsive genes. A, Distribution of the 132 nitrate-responsive TFs according to families. TFs were classified according to PlantTFDB3.0 database annotation. B, Prediction of connectivity of 132 TFs identified from brace root specific nitrate-responsive genes using STRING v10. Color codes indicate expression values of transcripts normalized as log2 algorithm of fold changes. A green line indicates text mining evidence, a purple line indicates experimental evidence, a blue line indicates database evidence, and a black line indicates coexpression evidence. Clustered groups associated with “MADS box,” “MYB,” and “CPP” families were highlighted accordingly. Outlines of nodes representing “classical” maize transcription factors were thickened.
Figure 8.
Nitrate-responsive regulatory genes and key regulators involved in the cell cycle control system regulating G1-S and G2-M transitions during pericycle cell division. A, Schematic illustration of a chromatosome consisting of a histone octamer. Histones H2A, H2B, H3, and H4, core histones; Histone H1, linker histone. B, S-phase and mitosis-specific cell division (III, metaphase; IV, anaphase) regulation by cell cycle control system. Two activated cyclin and cyclin-dependent protein kinase (CDK) complexes progressing into S-phase and M-phase, respectively. A heat map associated with the transition from metaphase to anaphase showing upregulated genes on spindle checkpoint protein (MAD2), APC, SMC, and CDC protein. C, Transcripts enriched in cell division and organization processes were divided into four groups (I, subunits of microtubule; II, structural constituents of cytoskeleton; III, microtubule binding and associated protein; IV, genes encoding kinesin/like motor proteins). Color code from blue to yellow indicates expression level of nitrate-responsive cell cycle genes normalized as log2 algorithm of fold changes (log2 Fc).
DISCUSSION
Lateral root branching is a key adaptive strategy in establishing a root system architecture that can optimally exploit heterogeneously distributed nutrients from soil (Ruffel et al., 2011; Guan et al., 2014; Postma et al., 2014; Zhan et al., 2015). Highly branched crop root systems are composed of multiple root types formed at different stages of development under the genetic control of distinct genes (Hochholdinger et al., 2004a). The formation of embryonic primary and seminal and postembryonic shoot-borne and lateral roots in monocot cereals is more divergent than root development of dicot root systems (Hochholdinger et al., 2004b). In this study, integration of anatomical, morphological, and a LCM-RNAseq-based specific transcriptome analyses in pericycle cells demonstrated that maize root types showed similar but also divergent responses to heterogeneous nitrate. In particular, brace roots displayed unique cell cycle control mechanisms controlling lateral root formation in response to local high nitrate compared to the other root types.
Local High Nitrate Supply Modulates the Transcriptome Response of Pericycle Cells of Different Root Types in Maize
Determination of specific gene expression in pericycle cells profiles prior to lateral root initiation (Figs. 2 and 3) in combination with morphological and histological analyses (Fig. 1; Supplemental Table S1) allow extrapolation on the plasticity of crop responses under heterogeneous nitrate environments. Cell type-specific RNA-seq experiments revealed that the transcriptomes of pericycle cells isolated from diverse root types represented distinct functions irrespective of external nitrate levels (Figs. 4 and 5). This is consistent with the observation that several maize mutants defective in lateral root formation showed root type-specific phenotypes but did not affect all major root types (Hochholdinger et al., 2004a). The mutants rum1 (Woll et al., 2005) and lrt1 (Hochholdinger and Feix, 1998) are both defective in the formation of lateral roots in primary roots but not affected in the shoot-borne root system. Consistent with this observation, profiling of pericycle transcriptomes revealed exceptional behavior of these cells in shoot-borne brace roots, which are very different from the seedling root types (Figs. 2, 3, and 4A). In rice (Oryza sativa), functionally distinct transcriptome profiles were identified between different root types in response to arbuscular mycorrhizal fungi (Gutjahr et al., 2015). Functional differences of pericycle transcriptomes between the four studied root types were reprogrammed in response to local high nitrate stimulation in maize (Fig. 5C). These results have important implications for understanding root adaptability in sensing environmental nitrate in plants. Remarkably, brace roots exhibited a high transcriptome activity in all seven enriched functional classes in response to local nitrate compared to seedling root types (Fig. 5C). In agricultural systems, adult brace roots contribute more to the branching response and participate in acquisition of nitrate than the seedling primary and seminal roots during the whole life cycle (Yu et al., 2014a, 2014b, 2015b). In particular, the shoot-borne root system, including brace roots, plays a crucial role in resource capturing, yield increase and lodging resistance via their branched roots throughout plant life compared to the seedling root system, which is eminent only during the early stages of plant development (Hochholdinger, 2009; Rogers and Benfey, 2015). Therefore, our findings suggest functional adaptability of the transcriptome signature of diverse root types. This highlights the notion of unique characteristics of the complex molecular networks in monocot cereals compared to the dicot model species Arabidopsis.
Heterogeneous Nitrate Supply Promotes Lateral Root Initiation by Triggering the Pericycle Cell Cycle Control System
Cell cycle control is a fundamental mechanism to regulate lateral root initiation by allowing specific cells to divide or not (Himanen et al., 2002, 2004; Vidal et al., 2013). Our study revealed that cell cycle control is integrated in brace roots by highly connective nitrate-responsive genes unique to this root type (Figs. 6 and 8). Functional analyses indicated enriched coregulated genes involved in nucleosome/chromatin assembly and microtubule motor activity/movement during mitosis (Supplemental Fig. S3; McIntosh et al., 2002). This is consistent with the enrichment of genes encoding proteins involved in DNA binding and microtubule movement upon auxin-induced lateral root initiation in maize (Jansen et al., 2013). The increased transcription of these genes involved at the G1-S phase boundary might be required to provide enough histone mRNA to keep pace with the need for new chromatin synthesis during S phase (Osley, 1991). Moreover, enrichment of genes required for the transition from metaphase to anaphase during mitosis was coordinated with enrichment of genes encoding microtubules and its associated motor proteins (Fig. 8C). This result is in line with the recent finding of active pericycle cell division in the stele of brace roots in the lateral root initiation zone in response to local high nitrate supply (Yu et al., 2015b). TFs play a key role in regulatory networks shaping growth and development. Therefore, identification of nitrate-responsive TFs enabled a better understanding of nitrate during lateral root initiation (Berckmans and De Veylder, 2009; Canales et al., 2014). We identified three TFs (ZmGATA34, ZmGATA2, and ZmRR9) that connected MYB and MADS box families as inferred from network predictions (Fig. 7B). In Arabidopsis, GATA23 plays a key role in specifying pericycle cells to become lateral root founder cells prior to lateral root initiation (De Rybel et al., 2010). Moreover, MADS box transcription factors are implicated in nitrate-induced changes of root architecture (Zhang and Forde, 1998). The observation of MYB transcription factors supports their role in cell cycle regulation and lateral root formation both in Arabidopsis (Ito et al., 2001; Shin et al., 2007; Mu et al., 2009) and maize (Jansen et al., 2013). Moreover, CPP-like TFs play an important role in development of reproductive tissues and control of cell division in plants (Yang et al., 2008), which is correlated to the distinct transcriptomic response in adult brace root. These results revealed strong transcriptional control in nitrate-responsive gene networks during lateral root initiation in brace roots.
By integrating MapMan outputs and network interactions, a model was devised to highlight how nitrate triggers pericycle cell cycle progression during lateral root initiation in brace roots of maize (Fig. 8). Accordingly, G1/S phase-specific genes, such as histone H4, were induced together with the CYCD3;1 cyclin and DNA replication genes by auxin during lateral root initiation in Arabidopsis (Himanen et al., 2002, 2004). In addition, active S-CDK (cyclin-dependent protein kinases) and M-CDK complexes were progressing into the S-phase and M-phase, respectively (Fig. 8B; Polyn et al., 2015). Moreover, a link between MAD2 and the cell cycle regulatory proteins APC and CDC proteins has provided support for the notion that anaphase is initiated (Elledge, 1998; Yu et al., 1999). These results are consistent with genes identified in this study associated with the transition from metaphase to anaphase of mitosis (Fig. 8B; Supplemental Table S6). Dynamic instability of microtubule subunits (TUBLIN A and TUBLIN B) and associated actin-binding proteins (ADF, VILLIN, and PROFILIN4) suggest a driving force for movements of microtubules during cell division (Smith, 2001). Furthermore, microtubule-based molecular motors are functionally important in organelle transport in the cytoplasm and in cell division (Asada and Collings, 1997). Taken together, root type-specific responses induced by local high nitrate supply were demonstrated to be involved in cell cycle control during lateral root initiation, supporting a unique lateral root branching strategy for maize brace roots.
Lateral Root Foraging Strategies Depend on the Developmental Phases and Internal Nitrogen Status
The present study underscores the plasticity of different root types during lateral root initiation at different stages of maize development. The model in Figure 9 supports the notion of highly divergent developmental complexity of maize root system development compared to Arabidopsis. Shoot-borne roots initiate from the elongation stage onward and contribute to the vast majority of total nitrogen uptake in maize plants (Shane and McCully, 1999; Peng et al., 2010). Notably, primary and crown roots showed no changes of their pericycle transcriptomes in response to local nitrate stimulus (Fig. 6A). Their root type-specific reaction to nitrate may suggest that pericycle cells of maize respond differently to environmental nitrate stimulation. A microarray analysis of pericycle cells suggested common mechanisms for lateral root initiation in maize primary and crown roots (Jansen et al., 2013). Remarkably, divergent responses of seminal versus primary and crown roots might be related to their evolutionary origin because cereal species such as rice are lacking seminal roots entirely (Rogers and Benfey, 2015). Thus, we hypothesize that nitrate-induced functional shifts between root types reflect diverse responses on lateral root foraging (Fig. 5C), as illustrated by brace root-specific distribution of functional classes and constitutively active genes in response to local high nitrate stimulus (Fig. 3B).
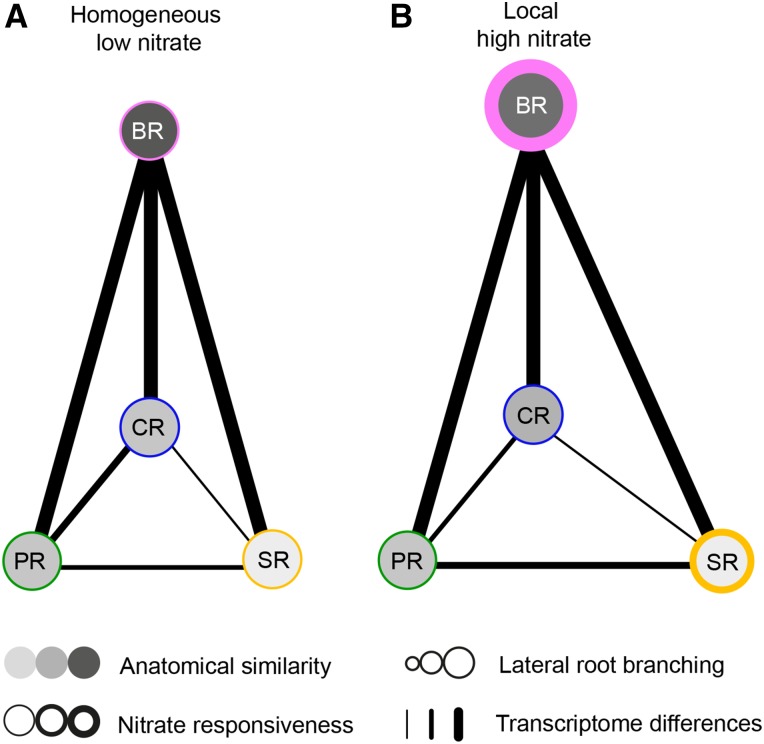
Figure 9.
Nitrate-modulated similarities and differences of lateral root branching in maize root types. Network view of morphology, anatomy, and transcriptome profiling in relation to the four maize root types under homogenous low nitrate (A) and local high nitrate (B) conditions. Grey shades of nodes correspond to the similarity of anatomical traits among the four root types (according to Supplemental Fig. S1 and Supplemental Table S1). Node size corresponds to the degree of lateral root branching response to local nitrate stimulus (according to Fig. 1). Thickened node outlines indicate more nitrate-responsive genes induced by local high nitrate stimulation (according to Figs. 3B and 6A). Line thickness between two nodes is proportional to the number of significantly changing genes (according to Fig. 5).
Systemic morphological and histological analyses revealed that all seedling root types and early shoot-borne roots specifically exploit heterogeneous nitrate by promoting lateral root elongation, whereas shoot-borne roots initiated at late vegetative growth stage (60 d after germination) display more plastic responses showing both increased length and density of lateral roots (Yu et al., 2014a, 2014b, 2015a, 2015b). Such directed lateral root development depends on regulatory networks that integrate both local and systemic signals to coordinate them with the overall plant internal nitrogen status (Ruffel et al., 2011; Guan et al., 2014). Economizing the costs for root development is pivotal for a resource-efficient strategy in nutrient acquisition, as illustrated by the impact of the internal nitrogen status-dependent regulatory module CLAVATA3/EMBRYO-SURROUNDING REGION-related peptides-CLAVATA1 leucine-rich repeat receptor-like kinase (Araya et al., 2014; Tabata et al., 2014). In particular, hormones such as cytokinin, which serves as a long-distance systematic messenger mediated by nitrate, can signal the nitrogen demand of the whole plant (Ruffel et al., 2011). The nitrogen concentration of the shoot shows decreasing progression from seedling to flowering stage, which indicates systemic nitrogen starvation responses intensify gradually (Yu et al., 2014a, 2015b). It was suggested that root growth and plasticity depend on the systemic nutrient status and shoot growth potential (Forde, 2002; Gojon et al., 2009). This notion can explain the more substantial morphological and transcriptome responses to local high nitrate application in brace roots compared to the other root types.
Therefore, this work extends the current knowledge of root type-specific functions of pericycle cells in maize under changing nitrate availability. Mechanistic interactions between plant development and root plasticity highlight specific root foraging responses that depend on the root developmental age and endogenous nitrogen status of the plant (Araya et al., 2014; Tabata et al., 2014). Understanding the reprogramming of these transcriptomic networks under limited nitrate conditions in brace roots in the context of yield acquisition and adult fitness can be relevant for rational breeding approaches. To this end, while cell cycle control mechanisms are likely evolutionary conserved during lateral root initiation across angiosperms, root type-specific transcriptomic networks of pericycle cells are functionally diverse during lateral root initiation in maize.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Seeds of the maize (Zea mays) inbred line B73 were surface sterilized and germinated in paper rolls in distilled water as previously described (Hetz et al., 1996). Seven days after germination, the endosperm of each seedling was excised and uniform seedlings with two visible leaves and four embryonic roots were transferred into low nitrate (0.5 mm NO3−) nutrient solution and grown until a root length of 7 cm per root type. One primary, seminal, or crown root was separately treated for 24 h with homogeneous low nitrate (0.5 mm NO3−) or local high nitrate (4 mm NO3−) nutrient solution in a two-compartment split-root system (Yu et al., 2014a). For brace root treatment, maize plants were cultured in a 20-liter container until brace roots were initiated. Two neighboring brace roots were kept in a small plastic bag with low (0.5 mm NO3−) or high (4 mm NO3−) nitrate nutrient solution for 24 h. These split-root systems were aerated continuously to maintain the oxygen content. The experiment was conducted in a growth chamber in a 16-h-light, 25°C, and 8-h-dark, 21°C cycle. Setup of split nitrate supply to four root types was described in detail by Yu et al. (2015b).
Morphological, Histological, and Anatomical Analyses
To comprehensively determine the morphological and histological responses of lateral root branching, local nitrate-treated primary, seminal, crown, and brace roots were harvested after 20 d of treatment. The average length and density of lateral roots and early pericycle cell divisions for seedling root types were examined according to Yu et al. (2015a).
To determine the anatomical structure of the four maize root types, transverse sections collected at 3 mm from the root tip of 7-d-old primary roots, 9-d-old seminal roots, 15-d-old crown roots, and 60-d-old brace roots were prepared with a VT1200 Vibratome (Leica). Areas of total transverse, stele, meta-xylem vessels, and size of pericycle cells adjacent to phloem and xylem poles were measured under an AxioCam MRc microscope (Carl Zeiss Microimaging) and then documented with Axio-Imager software (Carl Zeiss Microimaging).
Laser Capture Microdissection of Pericycle Cells from Four Root Types
Root segments of 3 to 5 mm from the root tip were dissected and fixed immediately (Yu et al., 2015a). The strength of infiltration was adjusted to 400 MPa for seedling root types and 300 MPa for brace roots to maintain the morphology of the transversal sections and maximize the effects of the fixative and the cryoprotective during sample preparation. The fixative was replaced and vacuum infiltration/swirl steps were repeated twice. The fixative was then replaced by fresh Suc (Sigma-Aldrich) solution as cryoprotective and infiltrated as described previously to minimize the formation of ice crystals (Yu et al., 2015a). In addition, concentrations of Suc were adjusted from 10% and 15% steps for primary, seminal, and crown root to 15% and 40% steps for brace root. The root segments of the three individual seedling root types and brace roots from the adult root system were embedded in 7 × 7 × 5-mm disposable molds (Polysciences Europe) with PolyFreeze-clear tissue freezing medium (Polysciences Europe). About 2,000 phloem pole pericycle cells were collected per root type and nitrate condition in three biological replicates by laser microdissection according to Yu et al. (2015a). The fine-scale capturing of phloem pole pericycle cells is shown in Figure 2B.
RNA Isolation and Amplification
RNA extraction, purification, and quality determination were performed according to Yu et al. (2015a). Only samples with a RIN value ≥6.5 were used for downstream amplification (Supplemental Fig. S4). For each of the 24 samples, 20 ng of high-quality RNA was used for cDNA synthesis and amplification. cDNA synthesis with oligo(dT) and random primers and cDNA amplifications were performed using the SMARTer PCR cDNA synthesis kit (Clontech Laboratories) following the manufacturer’s protocol. The quality and profile of the RNA and amplified cDNA samples were checked on an Agilent 2100 Bioanalyzer (Agilent Technologies) using an Agilent RNA 6000 Pico Kit (Agilent Technologies) and an Agilent High Sensitivity DNA Kit (Agilent Technologies), respectively.
cDNA Library Construction and Illumina Sequencing
The cDNA libraries for Illumina sequencing were constructed according to the manufacturer (TruSeq DNA Sample Prep Kit V3; Illumina). An Agilent 2100 Bioanalyzer (Agilent DNA 1000 LabChip) and ABI StepOnePlus real-time PCR system (Applied Biosystems) were used for quantification and qualification of the sample libraries. For sequencing, 24 libraries were arranged randomly on four flow cell lanes. Cluster preparation and paired-end read sequencing were performed according to the manufacturer’s guidelines (HiSeq 2000; Illumina). After the raw sequencing reads were generated, adapter sequences were trimmed using the Trimmomatic program (Bolger et al., 2014). Quality of the trimmed reads was checked using the FastQC program (Andrews, 2010).
Processing and Mapping of Illumina Sequencing Reads
Raw sequencing reads were processed and subsequently mapped with CLC Genomics Workbench software (version 8.0.1). Reads with more than one mismatch in the adapter sequence were excluded. By quality trimming, low-quality and ambiguous nucleotides of sequence ends and adapter contaminations were removed. Only ≥40-bp reads were retained for further analyses. These were initially mapped to the maize B73 reference genome sequence (RefGen_v2; ftp://ftp.gramene.org/pub/gramene/maizesequence.org/release-5b/assembly/), allowing large gaps of up to 50 kb to span introns. To be mapped, at least 90% of each read had to fit with 90% similarity to the reference. Stacked reads, i.e. redundant reads sharing the same start and end coordinate, sequencing direction, and sequence, were merged into one. The remaining reads were further projected to the “filtered gene set” (FGS v2; release 5b) of the B73 reference genome. Thereby, reads had to fit at least with 80% of their length comprising 90% similarity to the reference. Only reads uniquely mapping to the reference genome were further analyzed.
Statistical Procedures for Determining Active and Inactive Genes
We used a generalized linear model with a negative binomial response to model gene-wise transcriptional activity for each root type and nitrate treatment combination (Opitz et al., 2015). The log of the mean read count was assumed to be the sum of a normalization factor and a linear function of the fixed effects for each combination of root type and nitrate treatment. Normalization factors were calculated separately for each experimental unit by fitting a smoothing spline with response log(count+1) and with GC content and log gene length as explanatory variables, then using the fitted values of each estimated function at each gene’s GC content and length.
The vector of fixed effects for each gene was assumed to be a draw from a multivariate normal distribution with an unknown and unrestricted mean and an unknown diagonal variance-covariance matrix. The log of the negative binomial dispersion parameter was assumed to be constant within a gene and a draw from a normal distribution with unknown mean and variance. The unknown parameters in the distributions of the fixed effects and negative binomial dispersion were estimated using an empirical Bayes procedure via the R package ShrinkBayes (Van De Wiel et al., 2013), which uses integrated nested Laplace approximation (Rue et al., 2009) to approximate the posterior distribution for the fixed effect associated with gene g, root type r, and nitrate condition c.
For a given threshold T, the posterior distributions for gene g were used to find Pgrc (T), the posterior probability that the fixed effect for gene g, root type r, and condition c was larger than T. Gene g was called “active” for root type r and condition c if Pgrc (T) > 0.5 and “inactive” otherwise. Because fixed effects were estimated while accounting for sequencing differences from sample to sample, gene length, and GC content differences, classifying genes as active or inactive based on the posterior distribution of fixed effects is more meaningful than attempting to do so based on a single raw read count threshold applied to all genes. In terms of counts, the selected threshold T resulted in calling a gene of average length and average GC content active if the expected number of reads per million mapped reads was approximately one or more. The read count threshold was, in effect, adjusted up or down for other genes depending on gene length, GC content, and the observed empirical distribution between these variables and read count.
Statistical Procedures for Analyzing Differential Gene Expression
For the following analyses, only genes were considered that were represented by a minimum of five mapped reads in all three replicates of at least one sample. These genes were declared as “expressed.” The raw sequencing reads were normalized by sequencing depth and were log2-transformed to meet the assumptions of linear models. Furthermore, the mean variance relationship within the count data were estimated and precision weights for each observation were computed (Law et al., 2014). Prior to further data analyses, sample relations were analyzed based on multidimensional scaling using the plotMDS function of the Bioconductor package limma (Smyth, 2005) in R (R version 3.1.1 2014-07-10, limma_3.20.9). The distance between each pair of samples was estimated as the root mean square deviation for the top genes with the largest standard deviations between samples. The data were further analyzed according to a completely randomized design including a fixed effect for treatment and a random error term. After model fit, an empirical Bayes approach was applied to shrink the sample variances toward a common value (Smyth, 2004). Hypotheses tests were performed using the contrasts.fit function of the Bioconductor package limma (Smyth, 2005). The resulting P-values of the performed pairwise t tests were used to determine the total number of differentially expressed genes for each comparison by controlling the FDR < 0.05 to adjust for multiple testing (Benjamini and Hochberg, 1995).
GO and Metabolic Pathway Analyses
The Web-based agriGO software was used to assign expressed or differentially expressed genes to GO functional categories (Du et al., 2010). With SEA, overrepresented categories were computed by comparing the lists of differentially expressed and all expressed genes. Multiple testing was corrected by FDR (Benjamini and Yekutieli, 2001), and a cutoff was introduced at 0.05, 0.01, and 0.001. The MapMan software (Thimm et al., 2004) assigned differentially expressed genes to metabolic pathways and subsequently visualized them based on the functional annotation file ZmB73_5b_FGS_cds_2012. Fisher’s exact test was used to determine if the observed number of genes in each of the 35 major MapMan bins significantly deviated from the expected distribution of all expressed genes.
Correlation Analysis on Global Transcriptomes
SCC analysis was used to quantify the relationship among the transcriptomes under a given root type. SCCs were calculated from log2-transformed FPKM (fragments per kilobase of exon per million fragments mapped) values of expressed genes using R. The hierarchical clustering dendrogram was inferred by applying 1-SCC as distance function.
Expression Profile Correlation and Clustering among Root Types
K-means clustering was performed with the K-means support module embedded in the MEV program (http://www.tm4.org/mev). A scalar quantity, called the figure of merit (Yeung et al., 2001), was used to assess the quality of the clustering algorithm and to determine the optimal number of clusters. The K-means analysis was performed six times with each run generating 10 clusters using Euclidean distance by controlling FDR < 0.05 among the expressed genes of four root types under homogeneous low and local high nitrate conditions.
Known and Predicted Protein-Protein Interactions and Network Analyses
Association networks of the identified genes significantly enriched in MapMan bins (“RNA,” “DNA,” “signaling,” “cell,” and “transport”) were constructed with the aid of the online analysis tool STRING v10 (Szklarczyk et al., 2015). Coexpression or similar functional connections were determined at high confidence of ≥0.7. All predicted connections available in STRING v10 were also computed to predict the connectivity among all the 132 transcription factors (according to the Plant Transcription Factor Database v3.0; http://planttfdb.cbi.pku.edu.cn/) detected from the 2,740 genes that were specifically nitrate regulated in brace roots.
Accession Numbers
Raw sequencing data are stored at the Sequence Read Archive (http://www.ncbi.nlm.nih.gov/sra) under accession number SRP062897.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Transverse sections from the tip of maize root types.
Supplemental Figure S2. Expression patterns of the pericycle transcriptomes of maize root types.
Supplemental Figure S3. Simplified hierarchical tree graphs of overrepresented GO terms.
Supplemental Figure S4. Quality assessment of the 24 LCM-dissected cell samples.
Supplemental Table S1. Anatomical characteristics of the four maize root types.
Supplemental Table S3. “Classical” maize genes detected among nitrate-responsive genes.
Supplemental Table S4. List of 132 transcription factor genes.
Supplemental Table S5. List of genes associated with DNA synthesis/chromatin structure.
Supplemental Data Set S1. Complete list of 20,179 expressed genes.
Supplemental Data Set S2. Complete list of 26,770 active/inactive genes.
Supplemental Data Set S3. Complete list of clustered (K1-K3) genes by K-means.
Supplementary Material
Acknowledgments
We thank Mikio Nakazono (Nagoya University) for stimulating discussions on the optimization of laser capture microdissection and Nina Opitz (University of Bonn), Stefan Hey (University of Bonn), and Huanhuan Tai (University of Bonn) for helpful discussions on the data analyses.
Glossary
- LCM-RNAseq
laser capture microdissection in combination with RNA-sequencing
- SCC
Spearman correlation coefficient
- FDR
false discovery rate
- GO
Gene Ontology
- SEA
singular enrichment analysis
- TF
transcription factor
- FPKM
fragments per kilobase of exon per million fragments mapped
Footnotes
Articles can be viewed without a subscription.
References
- Andrews S (2010) FastQC: A quality control tool for high throughput sequence data. Available at www.bioinformatics.babraham.ac.uk/projects/fastqc/
- Araya T, Miyamoto M, Wibowo J, Suzuki A, Kojima S, Tsuchiya YN, Sawa S, Fukuda H, von Wirén N, Takahashi H (2014) CLE-CLAVATA1 peptide-receptor signaling module regulates the expansion of plant root systems in a nitrogen-dependent manner. Proc Natl Acad Sci USA 111: 2029–2034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asada T, Collings D (1997) Molecular motors in higher plants. Trends Plant Sci 2: 29–37 [Google Scholar]
- Atkinson JA, Rasmussen A, Traini R, Voß U, Sturrock C, Mooney SJ, Wells DM, Bennett MJ (2014) Branching out in roots: uncovering form, function, and regulation. Plant Physiol 166: 538–550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beeckman T, Burssens S, Inzé D (2001) The peri-cell-cycle in Arabidopsis. J Exp Bot 52: 403–411 [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: A practical and powerful approach to multiple testing. J R Stat Soc B 57: 289–300 [Google Scholar]
- Benjamini Y, Yekutieli D (2001) The control of the false discovery rate in multiple testing under dependency. Ann Stat 29: 1165–1188 [Google Scholar]
- Berckmans B, De Veylder L (2009) Transcriptional control of the cell cycle. Curr Opin Plant Biol 12: 599–605 [DOI] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B (2014) Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30: 2114–2120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canales J, Moyano TC, Villarroel E, Gutiérrez RA (2014) Systems analysis of transcriptome data provides new hypotheses about Arabidopsis root response to nitrate treatments. Front Plant Sci 5: 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dembinsky D, Woll K, Saleem M, Liu Y, Fu Y, Borsuk LA, Lamkemeyer T, Fladerer C, Madlung J, Barbazuk B, et al. (2007) Transcriptomic and proteomic analyses of pericycle cells of the maize primary root. Plant Physiol 145: 575–588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Rybel B, Vassileva V, Parizot B, Demeulenaere M, Grunewald W, Audenaert D, Van Campenhout J, Overvoorde P, Jansen L, Vanneste S, et al. (2010) A novel aux/IAA28 signaling cascade activates GATA23-dependent specification of lateral root founder cell identity. Curr Biol 20: 1697–1706 [DOI] [PubMed] [Google Scholar]
- De Smet I, Tetsumura T, De Rybel B, Frei dit Frey N, Laplaze L, Casimiro I, Swarup R, Naudts M, Vanneste S, Audenaert D, et al. (2007) Auxin-dependent regulation of lateral root positioning in the basal meristem of Arabidopsis. Development 134: 681–690 [DOI] [PubMed] [Google Scholar]
- De Smet I, Vassileva V, De Rybel B, Levesque MP, Grunewald W, Van Damme D, Van Noorden G, Naudts M, Van Isterdael G, De Clercq R, et al. (2008) Receptor-like kinase ACR4 restricts formative cell divisions in the Arabidopsis root. Science 322: 594–597 [DOI] [PubMed] [Google Scholar]
- Du Z, Zhou X, Ling Y, Zhang Z, Su Z (2010) agriGO: a GO analysis toolkit for the agricultural community. Nucleic Acids Res 38: W64–W70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubrovsky JG, Sauer M, Napsucialy-Mendivil S, Ivanchenko MG, Friml J, Shishkova S, Celenza J, Benková E (2008) Auxin acts as a local morphogenetic trigger to specify lateral root founder cells. Proc Natl Acad Sci USA 105: 8790–8794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elledge SJ. (1998) Mitotic arrest: Mad2 prevents sleepy from waking up the APC. Science 279: 999–1000 [DOI] [PubMed] [Google Scholar]
- Forde BG. (2002) Local and long-range signaling pathways regulating plant responses to nitrate. Annu Rev Plant Biol 53: 203–224 [DOI] [PubMed] [Google Scholar]
- Gojon A, Nacry P, Davidian JC (2009) Root uptake regulation: a central process for NPS homeostasis in plants. Curr Opin Plant Biol 12: 328–338 [DOI] [PubMed] [Google Scholar]
- Guan P, Wang R, Nacry P, Breton G, Kay SA, Pruneda-Paz JL, Davani A, Crawford NM (2014) Nitrate foraging by Arabidopsis roots is mediated by the transcription factor TCP20 through the systemic signaling pathway. Proc Natl Acad Sci USA 111: 15267–15272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutjahr C, Sawers RJH, Marti G, Andrés-Hernández L, Yang SY, Casieri L, Angliker H, Oakeley EJ, Wolfender JL, Abreu-Goodger C, Paszkowski U (2015) Transcriptome diversity among rice root types during asymbiosis and interaction with arbuscular mycorrhizal fungi. Proc Natl Acad Sci USA 112: 6754–6759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetz W, Hochholdinger F, Schwall M, Feix G (1996) Isolation and characterization of rtcs, a maize mutant deficient in the formation of nodal roots. Plant J 10: 845–857 [Google Scholar]
- Himanen K, Boucheron E, Vanneste S, de Almeida Engler J, Inzé D, Beeckman T (2002) Auxin-mediated cell cycle activation during early lateral root initiation. Plant Cell 14: 2339–2351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himanen K, Vuylsteke M, Vanneste S, Vercruysse S, Boucheron E, Alard P, Chriqui D, Van Montagu M, Inzé D, Beeckman T (2004) Transcript profiling of early lateral root initiation. Proc Natl Acad Sci USA 101: 5146–5151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochholdinger F. (2009) The maize root system: morphology, anatomy, and genetics. In Bennetzen JL, Hake SC, eds, Handbook of Maize: Its Biology. Springer, New York: pp 145–160 [Google Scholar]
- Hochholdinger F, Feix G (1998) Early post-embryonic root formation is specifically affected in the maize mutant lrt1. Plant J 16: 247–255 [DOI] [PubMed] [Google Scholar]
- Hochholdinger F, Park WJ, Sauer M, Woll K (2004b) From weeds to crops: genetic analysis of root development in cereals. Trends Plant Sci 9: 42–48 [DOI] [PubMed] [Google Scholar]
- Hochholdinger F, Woll K, Sauer M, Dembinsky D (2004a) Genetic dissection of root formation in maize (Zea mays) reveals root-type specific developmental programmes. Ann Bot (Lond) 93: 359–368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochholdinger F, Zimmermann R (2008) Conserved and diverse mechanisms in root development. Curr Opin Plant Biol 11: 70–74 [DOI] [PubMed] [Google Scholar]
- Ito M, Araki S, Matsunaga S, Itoh T, Nishihama R, Machida Y, Doonan JH, Watanabe A (2001) G2/M-phase-specific transcription during the plant cell cycle is mediated by c-Myb-like transcription factors. Plant Cell 13: 1891–1905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen L, Hollunder J, Roberts I, Forestan C, Fonteyne P, Van Quickenborne C, Zhen RG, McKersie B, Parizot B, Beeckman T (2013) Comparative transcriptomics as a tool for the identification of root branching genes in maize. Plant Biotechnol J 11: 1092–1102 [DOI] [PubMed] [Google Scholar]
- Jansen L, Roberts I, De Rycke R, Beeckman T (2012) Phloem-associated auxin response maxima determine radial positioning of lateral roots in maize. Philos Trans R Soc Lond B Biol Sci 367: 1525–1533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskowski M, Grieneisen VA, Hofhuis H, Hove CA, Hogeweg P, Marée AF, Scheres B (2008) Root system architecture from coupling cell shape to auxin transport. PLoS Biol 6: e307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law CW, Chen Y, Shi W, Smyth GK (2014) voom: Precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol 15: R29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linkohr BI, Williamson LC, Fitter AH, Leyser HM (2002) Nitrate and phosphate availability and distribution have different effects on root system architecture of Arabidopsis. Plant J 29: 751–760 [DOI] [PubMed] [Google Scholar]
- Malamy JE. (2005) Intrinsic and environmental response pathways that regulate root system architecture. Plant Cell Environ 28: 67–77 [DOI] [PubMed] [Google Scholar]
- Malamy JE, Benfey PN (1997) Organization and cell differentiation in lateral roots of Arabidopsis thaliana. Development 124: 33–44 [DOI] [PubMed] [Google Scholar]
- Marschner H. (1995) Mineral Nutrition of Higher Plants. Academic Press, London [Google Scholar]
- McIntosh JR, Grishchuk EL, West RR (2002) Chromosome-microtubule interactions during mitosis. Annu Rev Cell Dev Biol 18: 193–219 [DOI] [PubMed] [Google Scholar]
- Moreno-Risueno MA, Van Norman JM, Moreno A, Zhang J, Ahnert SE, Benfey PN (2010) Oscillating gene expression determines competence for periodic Arabidopsis root branching. Science 329: 1306–1311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mu RL, Cao YR, Liu YF, Lei G, Zou HF, Liao Y, Wang HW, Zhang WK, Ma B, Du JZ, et al. (2009) An R2R3-type transcription factor gene AtMYB59 regulates root growth and cell cycle progression in Arabidopsis. Cell Res 19: 1291–1304 [DOI] [PubMed] [Google Scholar]
- Opitz N, Marcon C, Paschold A, Malik WA, Lithio A, Brandt R, Piepho H-P, Nettleton D, Hochholdinger F (2015) Extensive tissue-specific transcriptomic plasticity in maize primary roots upon water deficit. J Exp Bot pii: erv453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osley MA. (1991) The regulation of histone synthesis in the cell cycle. Annu Rev Biochem 60: 827–861 [DOI] [PubMed] [Google Scholar]
- Parizot B, Laplaze L, Ricaud L, Boucheron-Dubuisson E, Bayle V, Bonke M, De Smet I, Poethig SR, Helariutta Y, Haseloff J, et al. (2008) Diarch symmetry of the vascular bundle in Arabidopsis root encompasses the pericycle and is reflected in distich lateral root initiation. Plant Physiol 146: 140–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Y, Niu J, Peng Z, Zhang F, Li C (2010) Shoot growth potential drives N uptake in maize plants and correlates with root growth in the soil. Field Crops Res 115: 85–93 [Google Scholar]
- Polyn S, Willems A, De Veylder L (2015) Cell cycle entry, maintenance, and exit during plant development. Curr Opin Plant Biol 23: 1–7 [DOI] [PubMed] [Google Scholar]
- Postma JA, Dathe A, Lynch JP (2014) The optimal lateral root branching density for maize depends on nitrogen and phosphorus availability. Plant Physiol 166: 590–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers ED, Benfey PN (2015) Regulation of plant root system architecture: implications for crop advancement. Curr Opin Biotechnol 32: 93–98 [DOI] [PubMed] [Google Scholar]
- Rue H, Martino S, Chopin N (2009) Approximate Bayesian inference for latent Gaussian models by using integrated nested Laplace approximations. J R Stat Soc B 71: 319–392 [Google Scholar]
- Ruffel S, Krouk G, Ristova D, Shasha D, Birnbaum KD, Coruzzi GM (2011) Nitrogen economics of root foraging: transitive closure of the nitrate-cytokinin relay and distinct systemic signaling for N supply vs. demand. Proc Natl Acad Sci USA 108: 18524–18529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnable JC, Freeling M (2011) Genes identified by visible mutant phenotypes show increased bias toward one of two subgenomes of maize. PLoS One 6: e17855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shane MW, McCully ME (1999) Root xylem embolisms: implications for water flow to the shoot in single-rooted maize plants. Aust J Plant Physiol 26: 107–114 [Google Scholar]
- Shin R, Burch AY, Huppert KA, Tiwari SB, Murphy AS, Guilfoyle TJ, Schachtman DP (2007) The Arabidopsis transcription factor MYB77 modulates auxin signal transduction. Plant Cell 19: 2440–2453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith LG. (2001) Plant cell division: building walls in the right places. Nat Rev Mol Cell Biol 2: 33–39 [DOI] [PubMed] [Google Scholar]
- Smyth GK. (2004) Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol 3: 1–25 [DOI] [PubMed] [Google Scholar]
- Smyth GK. (2005) Limma: linear models for microarray data. In RC Gentleman, VJ Carey, W Huber, RA Irizarry, S Dudoit, eds, Bioinformatics and Computational Biology Solutions Using R and Bioconductor. Springer, New York, pp 397–420 [Google Scholar]
- Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos A, Tsafou KP, et al. (2015) STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res 43: D447–D452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabata R, Sumida K, Yoshii T, Ohyama K, Shinohara H, Matsubayashi Y (2014) Perception of root-derived peptides by shoot LRR-RKs mediates systemic N-demand signaling. Science 346: 343–346 [DOI] [PubMed] [Google Scholar]
- Thimm O, Bläsing O, Gibon Y, Nagel A, Meyer S, Krüger P, Selbig J, Müller LA, Rhee SY, Stitt M (2004) MAPMAN: a user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J 37: 914–939 [DOI] [PubMed] [Google Scholar]
- Van De Wiel MA, Leday GGR, Pardo L, Rue H, Van Der Vaart AW, Van Wieringen WN (2013) Bayesian analysis of RNA sequencing data by estimating multiple shrinkage priors. Biostatistics 14: 113–128 [DOI] [PubMed] [Google Scholar]
- Vanneste S, De Rybel B, Beemster GTS, Ljung K, De Smet I, Van Isterdael G, Naudts M, Iida R, Gruissem W, Tasaka M, et al. (2005) Cell cycle progression in the pericycle is not sufficient for SOLITARY ROOT/IAA14-mediated lateral root initiation in Arabidopsis thaliana. Plant Cell 17: 3035–3050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal EA, Moyano TC, Riveras E, Contreras-López O, Gutiérrez RA (2013) Systems approaches map regulatory networks downstream of the auxin receptor AFB3 in the nitrate response of Arabidopsis thaliana roots. Proc Natl Acad Sci USA 110: 12840–12845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woll K, Borsuk LA, Stransky H, Nettleton D, Schnable PS, Hochholdinger F (2005) Isolation, characterization, and pericycle-specific transcriptome analyses of the novel maize lateral and seminal root initiation mutant rum1. Plant Physiol 139: 1255–1267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Gu S, Wang X, Li W, Tang Z, Xu C (2008) Molecular evolution of the CPP-like gene family in plants: insights from comparative genomics of Arabidopsis and rice. J Mol Evol 67: 266–277 [DOI] [PubMed] [Google Scholar]
- Yeung KY, Fraley C, Murua A, Raftery AE, Ruzzo WL (2001) Model-based clustering and data transformations for gene expression data. Bioinformatics 17: 977–987 [DOI] [PubMed] [Google Scholar]
- Yu HG, Muszynski MG, Kelly Dawe R (1999) The maize homologue of the cell cycle checkpoint protein MAD2 reveals kinetochore substructure and contrasting mitotic and meiotic localization patterns. J Cell Biol 145: 425–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu P, Eggert K, von Wirén N, Li C, Hochholdinger F (2015a) Cell-type specific gene expression analyses by RNA-Seq reveal local high nitrate triggered lateral root initiation in shoot-borne roots of maize by modulating auxin-related cell cycle-regulation. Plant Physiol 169: 690–704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu P, Hochholdinger F, Li C (2015b) Root-type-specific plasticity in response to localized high nitrate supply in maize (Zea mays). Ann Bot (Lond) 116: 751–762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu P, Li X, Yuan L, Li C (2014a) A novel morphological response of maize (Zea mays) adult roots to heterogeneous nitrate supply revealed by a split-root experiment. Physiol Plant 150: 133–144 [DOI] [PubMed] [Google Scholar]
- Yu P, White PJ, Hochholdinger F, Li C (2014b) Phenotypic plasticity of the maize root system in response to heterogeneous nitrogen availability. Planta 240: 667–678 [DOI] [PubMed] [Google Scholar]
- Zhan A, Schneider H, Lynch JP (2015) Reduced lateral root branching density improves drought tolerance in maize. Plant Physiol 168: 1603–1615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Forde BG (1998) An Arabidopsis MADS box gene that controls nutrient-induced changes in root architecture. Science 279: 407–409 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.