A transcription factor induced by phosphate starvation regulates metabolism genes to enhance starch biosynthesis in microalgae.
Abstract
Many eukaryotic microalgae modify their metabolism in response to nutrient stresses such as phosphorus (P) starvation, which substantially induces storage metabolite biosynthesis, but the genetic mechanisms regulating this response are poorly understood. Here, we show that P starvation-induced lipid and starch accumulation is inhibited in a Chlamydomonas reinhardtii mutant lacking the transcription factor Pi Starvation Response1 (PSR1). Transcriptomic analysis identified specific metabolism transcripts that are induced by P starvation but misregulated in the psr1 mutant. These include transcripts for starch and triacylglycerol synthesis but also transcripts for photosynthesis-, redox-, and stress signaling-related proteins. To further examine the role of PSR1 in regulating lipid and starch metabolism, PSR1 complementation lines in the psr1 strain and PSR1 overexpression lines in a cell wall-deficient strain were generated. PSR1 expression in the psr1 lines was shown to be functional due to rescue of the psr1 phenotype. PSR1 overexpression lines exhibited increased starch content and number of starch granules per cell, which correlated with a higher expression of specific starch metabolism genes but reduced neutral lipid content. Furthermore, this phenotype was consistent in the presence and absence of acetate. Together, these results identify a key transcriptional regulator in global metabolism and demonstrate transcriptional engineering in microalgae to modulate starch biosynthesis.
The need to mitigate the environmental impacts of industry is prompting the development of sustainable industrial methods, such as the applied use of biological processes (Skjånes et al., 2007). Likewise, the contribution of fossil fuels to greenhouse gas emissions has created significant interest in biofuels (Hill et al., 2006; Georgianna and Mayfield, 2012). Algae, including eukaryotic unicellular microalgae, are an attractive feedstock for sustainable industrial biotechnology. These photosynthetic microorganisms can use solar energy to convert CO2 into various metabolites that have applications for many industries, including pharmaceuticals, food, health, materials, and energy (Skjånes et al., 2007; Guedes et al., 2011). To harness the full potential of microalgae for industrial biotechnology, an improved understanding of microalgae metabolism is essential, and tools to engineer algae metabolism for the overproduction of desired products are needed.
Microalgae store carbon principally as lipids in the form of triacylglycerol (TAG) and carbohydrate in the form of starch (Johnson and Alric, 2013). In many species, the biosynthesis of these storage metabolites is highly induced under conditions of environmental stress such as nitrogen (N) or phosphorus (P) starvation, high light, or salinity (Hu et al., 2008; Siaut et al., 2011). Nutrient starvation is a particularly reliable means to induce the accumulation of storage metabolites and has been routinely used as a condition to further our understanding of the transcriptional changes that occur at the onset of carbon storage (Merchant et al., 2012), especially for TAG accumulation in response to N starvation in the model species Chlamydomonas reinhardtii (Miller et al., 2010; Boyle et al., 2012; Blaby et al., 2013). In particular, these studies have demonstrated that genes encoding acyltransferase enzymes, which are responsible for de novo TAG biosynthesis, including type 1 and type 2 diacylglycerol acyltransferases (DGATs) and a phospholipid diacylglycerol acyltransferase, are highly up-regulated following N starvation. In contrast, very little is known about the transcriptional responses to P starvation or the regulators that control these metabolic changes.
P is an essential macronutrient that is needed for various biochemical and cellular processes (Raghothama, 1999). In the biosphere, P is biologically available to most organisms in the form of inorganic phosphate (Pi), but its concentration is often very limited, particularly due to complexation with metal cations and organic particles (Hudson et al., 2000). To cope with this limited availability, plants and microalgae have evolved adaptive mechanisms to facilitate improved acquisition and conservation of P and to allow survival under P starvation conditions (Moseley and Grossman, 2009; Rouached et al., 2010). In higher plants, the P starvation survival mechanism involves metabolic remodeling, including the replacement of phospholipids with sulfolipids, the use of alternative glycolytic pathways that utilize Pi-independent enzymes, and the accumulation of starch, due in part to activation of the Pi-sensitive ADP-Glc pyrophosphorylase (Plaxton and Tran, 2011). The transcriptional and posttranscriptional regulation of these plant P starvation responses is well understood (Rouached et al., 2010; Chiou and Lin, 2011; Sobkowiak et al., 2012), but it is unclear whether the control mechanisms of such responses are conserved in microalgae. In C. reinhardtii, P starvation responses are known to be controlled by the MYB family transcription factor Pi Starvation Response1 (PSR1), which regulates Pi acquisition through the up-regulation of phosphatases, including the alkaline phosphatase encoded by the PHOX gene, and the up-regulation of Pi transporters, in particular the PO43−/Na+ symporters encoded by members of the Pi Transporter B (PTB) gene family (Shimogawara et al., 1999; Wykoff et al., 1999; Moseley et al., 2006). In addition, PSR1 has been demonstrated to regulate processes to maximize P reallocation, such as through the modification of nucleic acid metabolism (Yehudai-Resheff et al., 2007). However, a potential role of PSR1 in regulating P starvation-induced starch and TAG biosynthesis is unclear and has not been studied.
The use of genetic engineering to enhance carbon storage metabolism in microalgae has so far had mixed success (Radakovits et al., 2010; Driver et al., 2014). For example, down-regulation of lipid catabolism or carbohydrate synthesis through targeted knockdown of a lipase gene or a UDP-Glc pyrophosphorylase gene, respectively, provided an increase in TAG yield (Trentacoste et al., 2013; Daboussi et al., 2014). In contrast, overexpression of DGAT genes, encoding the final enzymatic step of TAG synthesis, failed to increase TAG content (La Russa et al., 2012), demonstrating that direct manipulation of individual genes is not always successful. An alternative approach is the manipulation of transcriptional regulators, as this may provide more substantial metabolic modification by controlling multiple steps in a pathway (Grotewold, 2008). However, few transcriptional regulators of lipid and starch metabolism in microalgae have been described. Furthermore, to date, such transcriptional engineering of microalgae to modulate metabolism has not been explored. Transcriptomic analyses of N starvation responses have begun to identify putative transcriptional regulators (Miller et al., 2010; Boyle et al., 2012; Gargouri et al., 2015; Ngan et al., 2015). This includes Nitrogen Response Regulator1 (NRR1), a putative SQUAMOSA promoter-binding protein-type transcription factor from C. reinhardtii, which was demonstrated, through analysis of the nrr1 knockout mutant, to be a regulator of N-induced TAG biosynthesis (Boyle et al., 2012). More recently, PSR1 has also been identified as a regulator of TAG biosynthesis in response to N starvation (Ngan et al., 2015). However, that study did not begin to understand the mechanism of this PSR1 regulation at the level of individual downstream target genes and did not examine the metabolic control role of PSR1 under P starvation. Recently we observed that, under P starvation, mutation of PSR1 not only inhibits lipid accumulation but also significantly abolishes starvation-induced starch accumulation (Bajhaiya et al., 2016); however, the potential regulation of starch metabolism by PSR1 has not yet been examined.
In order to better understand the transcriptional changes underlying starch induction in response to P starvation, we have performed mRNA transcript abundance analysis of selected transcripts from wild-type and psr1 mutant C. reinhardtii under P-replete and P-starvation conditions and identified specific starch and lipid metabolism genes that are PSR1 regulated. To then further validate the role of PSR1 as a global metabolic regulator, we generated psr1:PSR1 complementation lines and PSR1 overexpression lines and demonstrated that PSR1 is able to modulate metabolism through the transcriptional regulation of specific carbon storage genes, leading to the increased accumulation of starch.
RESULTS
Identification of P Starvation-Induced and psr1-Misregulated Transcripts
To examine the transcriptional response to P starvation in wild-type and PSR1 mutant (psr1) plants, a preliminary RNA sequencing (RNA-Seq) data set of psr1 and the parental wild type (CC125) was generated and used to identify target transcripts to be analyzed further (Supplemental Methods S1). Wild-type cells grown in medium containing 10 µm P (low P) completely exhausted the Pi by day 3 of batch-culture growth, while cells in medium containing 1 mm P (high P) were P replete throughout the entire period of growth (Supplemental Fig. S1, A and B). This starvation of Pi was coincident with the start of lipid and starch accumulation that continued over the next 4 d (Supplemental Fig. S1F). Transcripts were quantified in day-3 cells, at the onset of P starvation, and in day-5 cells, 48 h after the onset of P starvation (Supplemental Fig. S2, A and B), as these were the time points where the transcriptional response was elevated (Supplemental Fig. S1, G and H). Of the 17,737 transcripts identified, at day 3, 1,105 transcripts in the wild type were greater than 2-fold up-regulated by low P, and of these, only 139 were greater than 2-fold up-regulated in psr1, while 1,042 transcripts in the wild type were greater than 2-fold down-regulated by low P, and of these, only 112 were greater than 2-fold down-regulated in psr1 (Supplemental Data Set S1). A large proportion of the up-regulated but psr1-misregulated transcripts was classified as encoding Pi homeostasis proteins. As anticipated from a previous microarray analysis (Moseley et al., 2006), many of the known Pi homeostatic transcripts, including PSR1, PTB2, PTB4, and most clearly PHOX, were highly up-regulated by low P at day 3 in the wild type, but this up-regulation was abolished in psr1 (Supplemental Fig. S3A).
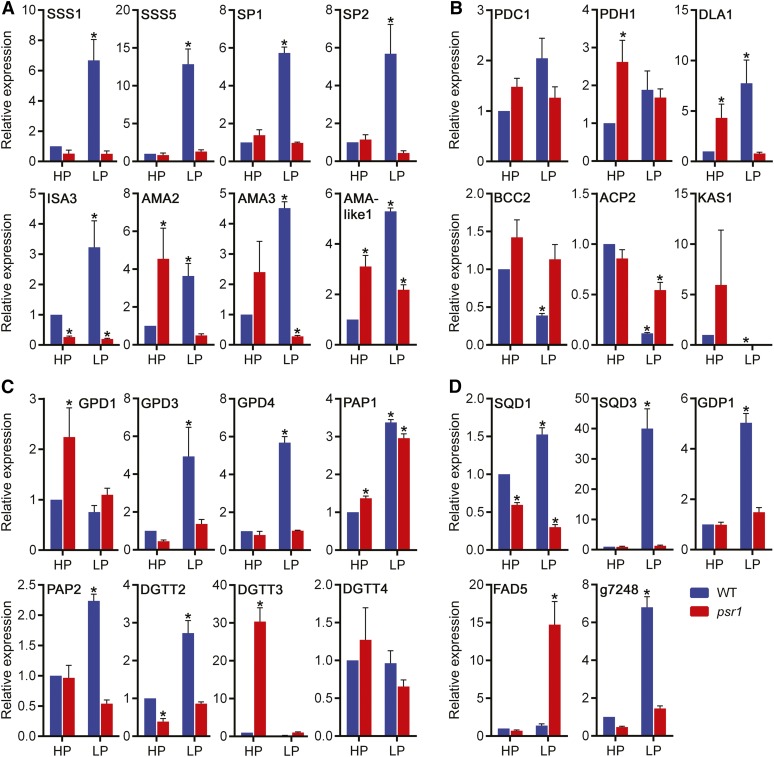
In addition to Pi homeostasis, the preliminary RNA-Seq data indicated that a high proportion of the P starvation-regulated transcripts were predicted to be involved in primary metabolism, especially starch and lipid synthesis, catabolism, and modification. Clusters of lipid and starch metabolism transcripts were indicated by hierarchical clustering to be down-regulated, up-regulated, or had no change in expression in the wild type in response to low P but were misregulated in psr1 (Supplemental Fig. S2C). A number of these transcripts were further examined in multiple replicate samples by quantitative real-time PCR (qPCR). Many of the down-regulated lipid metabolism transcripts were for fatty acid synthesis, including ACCase subunit transcripts such as BCC2 and fatty acid synthase component transcripts such as ACP2 and KAS1 (Fig. 1). Of particular interest were a group of starch and lipid enzyme transcripts, which clustered with the PSR1, PHOX, and PTB transcripts, that were highly up-regulated under low P but showed low or no up-regulation in psr1 under low P. Eight starch metabolism transcripts were confirmed to be highly up-regulated by low P but misregulated in psr1: two soluble starch synthases (SSS1 and SSS5), two starch phosphorylases (SP1 and SP2), an isoamylase (ISA3), and three α-amylases (AMA2, AMA3, and AMA-like1; Fig. 1A). Five lipid metabolism transcripts were also confirmed to be low-P induced and psr1 misregulated: the phosphatidic acid phosphatase PAP2, two glycerol 3-phosphate dehydrogenases (GPD3 and GPD4), one DGAT (DGTT2), and the dihydrolipoamide acetyltransferase DLA1 (Fig. 1C). Other classes of lipid metabolism transcripts were induced by P starvation but psr1 misregulated, including SQD1 and SQD3, required for the synthesis of the sulfolipid sulfoquinovosyldiacylglycerol (SQDG), and the glycerophosphoryl diester phosphodiesterase GDP1 (Fig. 1D). Selected predicted lipase transcripts were also up-regulated by P starvation and appeared to be PSR1 dependent, such as the putative esterase/lipase transcript g7248, although it is not possible to determine whether such lipases are involved in TAG breakdown or the remobilization of membrane lipids into neutral lipids.
Figure 1.
psr1-dependent misregulation of P starvation-induced starch and lipid metabolism genes. Relative expression is shown for selected starch metabolism genes (A), selected fatty acid synthesis-related genes (B), selected TAG synthesis genes (C), and other selected lipid metabolism genes (D) in high-P-treated (HP) and low-P-treated (LP) wild-type (WT; CC125) and psr1 cells at day 3. Expression of the mRNA transcripts by real-time PCR was determined relative to CBLP expression and is shown relative to the high-P-treated wild-type transcript. Data points are means ± se calculated from at least three biological replicates each with three technical replicates. Asterisks denote significant differences (P < 0.05) from high-P-treated wild-type values as determined by one-way ANOVA.
Some transcripts did not change significantly in response to P status in the wild type but were up-regulated in psr1 either under high-P conditions, such as for the TAG synthesis genes GPD1 and DGTT3, or under low-P conditions, such as for the fatty acid desaturase FAD5 (Fig. 1C). The RNA-Seq data set also indicated that many of the lipid and starch metabolism gene transcripts examined did not show any change in abundance in response to high and low P in the wild type and the psr1 mutant and, thus, were not regulated by P status (91 out of 172 transcripts). These included the pyruvate dehydrogenase complex transcript PDC1 (Fig. 1B) and the DGAT DGTT4 (Fig. 1C). In contrast, a small number of transcripts were up- or down-regulated equally in the wild type and the psr1 mutant, such as PAP1 (Fig. 1C). In contrast to the transcripts up-regulated by low P in the wild type, many transcripts were up-regulated by low P in psr1 but did not show equivalent up-regulation in the wild type, such as FAD5. Many transcripts that were down-regulated in the wild type but unchanged in psr1 were related to fatty acid synthesis, such as the ACCase subunit BCC2 or the acyl carrier ACP2 (Fig. 1B).
Finally, it was suggested that other gene classes, in addition to phosphate homeostasis and lipid and starch metabolism, are regulated by P starvation in a PSR1-dependent manner (Supplemental Data Set S1). It was determined by qPCR that a subset of photosynthesis- and electron transport/redox-related transcripts were misregulated by the absence of PSR1; for example, a light-harvesting complex II transcript (LHCBM9) was highly up-regulated by P starvation in the psr1 mutant, while the ferredoxin isoform FDX2 and a predicted cupredoxin (CSP1) were highly up-regulated by P starvation in the wild type but not in psr1 (Supplemental Fig. S3B). Nutrient starvation, such as N starvation, can also lead to an onset of oxidative stress, which in turn could lead to the induction of autophagic programmed cell death (Jiménez et al., 2009; Pérez-Pérez et al., 2010). In response to P starvation, there was no evidence from the RNA-Seq data set of any changes in transcripts encoding antioxidant enzymes or those associated with autophagy and programmed cell death in either the wild type or psr1 (Supplemental Data Set S1). However, Ca2+ transporters, which have been implicated in regulating abiotic stress responses, potentially as mediators of cellular Ca2+ signals (McAinsh and Pittman, 2009; Pittman et al., 2009), were substantially up-regulated by P starvation and appeared to be misregulated in psr1. In particular, the Ca2+/H+ exchanger isoforms CAX1 and CAX2 had altered transcriptional responses in the psr1 mutant (Supplemental Fig. S3B).
Overall, these data indicate that a subset of primary metabolism genes, which are induced under P starvation conditions, are transcriptionally regulated by PSR1. In higher plants, there are orthologous P starvation response transcription factors, including Phosphate Starvation Response1 (PHR1) from Arabidopsis (Arabidopsis thaliana). The PHR1-binding site (P1BS) cis-element sequence (GNATATNC) was determined previously as the binding motif for PHR1 (Rubio et al., 2001), and this element is present within P starvation-induced genes from various higher plants (Sobkowiak et al., 2012). While PSR1 and PHR1 are closely related but distinct MYB-CC family proteins, as shown by phylogenetic analysis, both the MYB-like DNA-binding and the coiled-coil protein dimerization regions are highly conserved within these proteins (Supplemental Fig. S4), increasing the likelihood that PSR1 may also recognize the P1BS element. To identify putative PSR1-binding elements, the GNATATNC sequence was screened in the promoter and noncoding regions of 500 genes, including 200 known and predicted PSR1-regulated genes and 300 genes predicted from the RNA-Seq data to be non-PSR1 regulated. Copies of the P1BS element were identified within the promoter or intron region of many of the validated genes, including within PTB2, PTB4, and PHOX (Supplemental Table S2). In total, 276 of the 500 genes contained the GNATATNC sequence, with 74% of the PSR1-regulated genes containing at least one copy of the P1BS element while only 42% of the non-PSR1-regulated genes contained the sequence (Supplemental Fig. S4, C and D). The GcATATgC sequence was the most frequently identified. Most of the P1BS-containing genes (58%) have a single copy of the element, 30% have two copies, and 12% have more than two copies, while a copy of the element in present within the promoter region in 66% of the genes and within an intron in 34% of the genes.
Generation of PSR1 Expression Lines
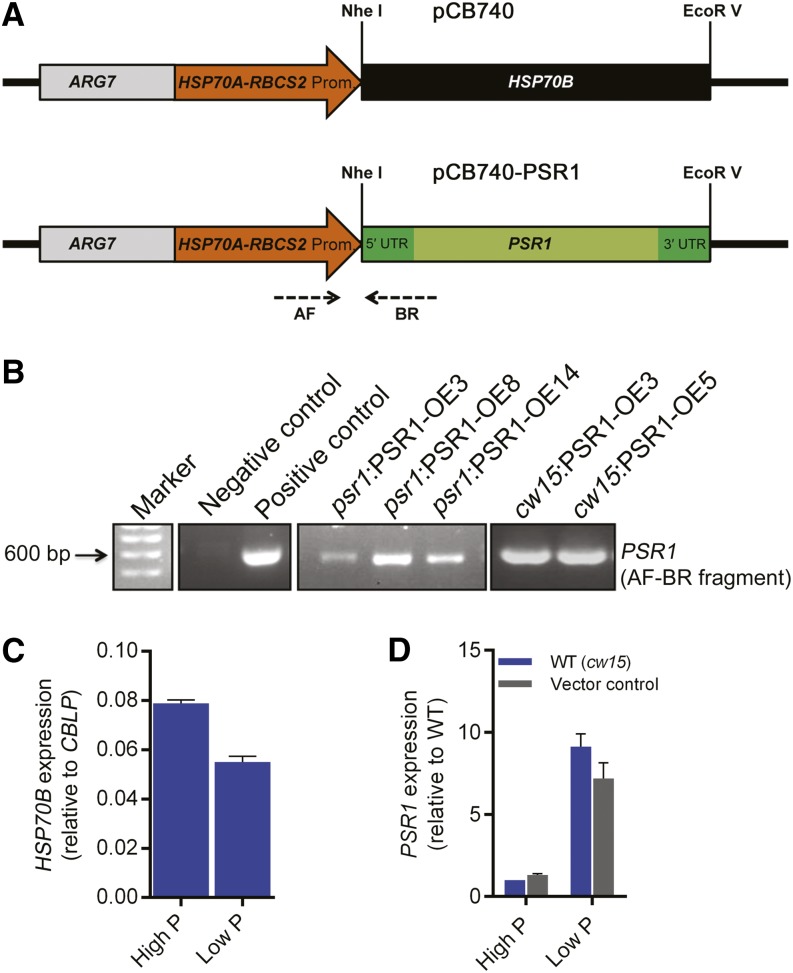
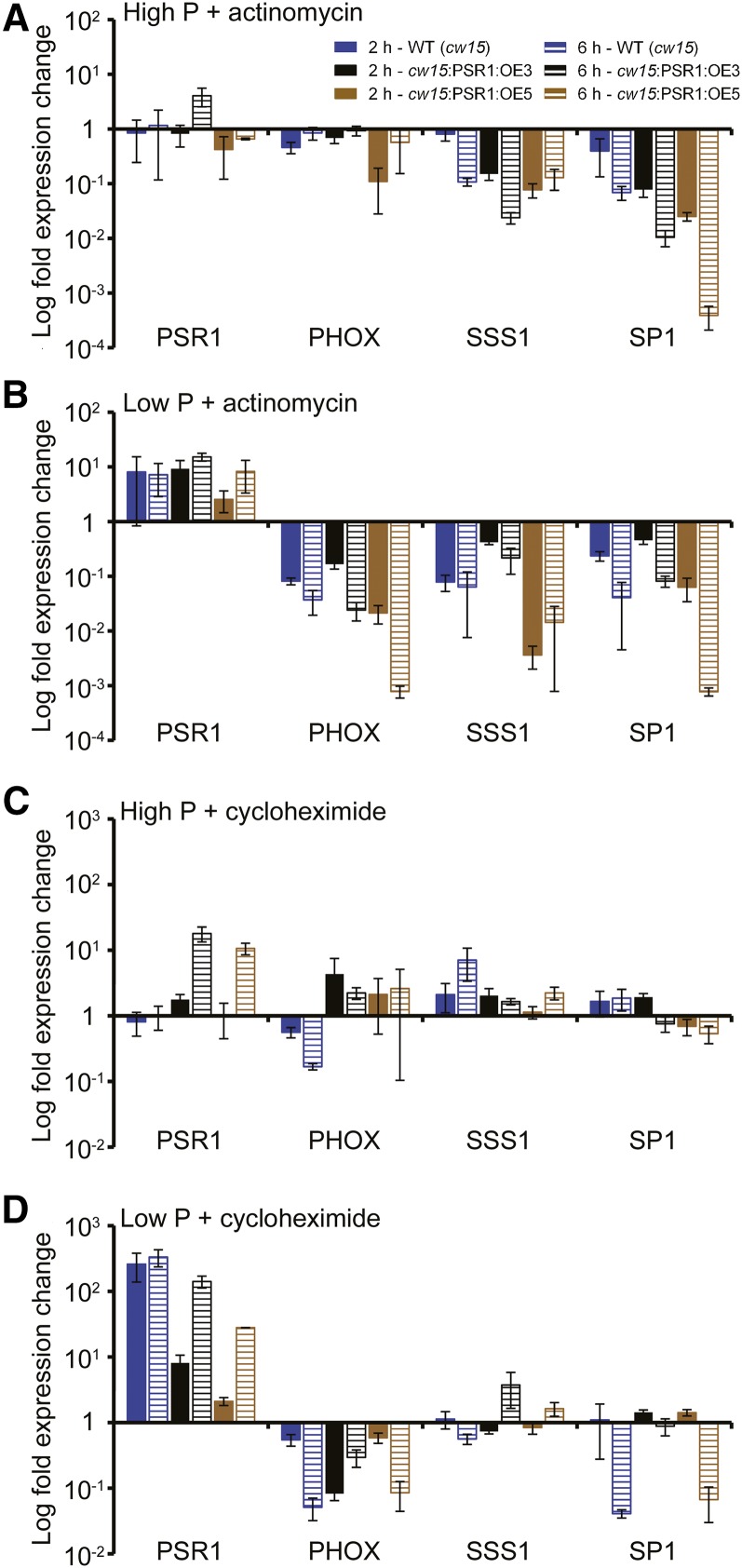
The role of PSR1 in regulating starch and lipid metabolism and the selected metabolism genes was further examined through the generation of PSR1 expression lines. A genomic fragment of PSR1 was fused to the tandem Heat Shock Protein70 (HSP70A)-RBCS2 promoter (Fig. 2A), which was shown previously to elicit the strong and stable expression of nucleus-transformed genes in C. reinhardtii (Schroda et al., 1999, 2000). The pCB740-PSR1 plasmid was transformed in the psr1 mutant background, in order to generate complementation lines and confirm that the expressed PSR1 was functional, and in the wild-type background, in order to examine overexpression phenotypes. For the overexpression lines, the cell wall-deficient cw15 strain was used, as this strain has been shown previously to exhibit high levels of carbon storage, especially starch, possibly due to the lack of cell wall glycoproteins that otherwise form a large carbon sink (Siaut et al., 2011). A number of independent psr1:PSR1-OE complementation and cw15:PSR1-OE overexpression lines were generated, of which three psr1:PSR1-OE lines and two cw15:PSR1-OE lines were examined further (Fig. 2B). All five lines showed significant increases in PSR1 expression relative to psr1 or cw15, while all of the psr1:PSR1-OE lines also showed higher PSR1 expression compared with the CC125 wild type, indicating that these lines not only complemented the psr1 mutation but yielded PSR1 overexpression (Fig. 3, A and B). Altered PSR1 expression was not due to the presence of the empty plasmid (Fig. 2D); however, PSR1 expression in all of the psr1:PSR1-OE and cw15:PSR1-OE lines was enhanced by low-P conditions (Fig. 3, A and B), equivalent to the P starvation-induced expression of native PSR1 (Supplemental Fig. S3A). This was unexpected, as the HSP70A-RBCS2 promoter was expected to provide constitutive expression; indeed, the expression of HSP70B driven by the HSP70A-RBCS2 promoter, tested as a control, did not show an increase under low-P conditions (Fig. 2C). Nevertheless, PSR1 expression was increased significantly relative to the wild type in all lines tested under both high-P and low-P conditions. To examine whether PSR1 mRNA stability was affected by P starvation, high-P- and low-P-grown cw15 and cw15:PSR1-OE cells were incubated with the transcriptional inhibitor actinomycin D and the translational inhibitor cycloheximide for 2 and 6 h before RNA was harvested for analysis. Actinomycin D treatment inhibited the transcription of all genes tested, leading to a reduction in mRNA transcript, with the exception of PSR1. The PSR1 mRNA abundance in all lines, including the wild type, was increased in low-P conditions after both 2 and 6 h of actinomycin D treatment (Fig. 4B) and cycloheximide treatment (Fig. 4D), relative to treatment without inhibitor. Under high-P conditions, there was no change in PSR1 abundance following actinomycin D treatment (Fig. 4A) and a smaller relative increase of transcript abundance following cycloheximide treatment that was only apparent after 6 h and only in the cw15:PSR1-OE lines (Fig. 4C).
Figure 2.
PSR1 expression construct. A, Schematic diagram of the selectable marker-promoter gene region (not to scale) of the PSR1 expression construct. The pCB740 plasmid contains the selectable marker ARG7, the tandem HSP70A-RBCS2 promoter, and the HSP70B gene. HSP70B was replaced with the genomic fragment of PSR1 by NheI and EcoRV restriction enzyme digestion to generate the pCB740-PSR1 plasmid used for transformation into psr1 or wild-type (cw15) cells to generate complementation or overexpression lines, respectively. AF and BR are primer sites used for screening pCB740-PSR1 transformation lines. UTR, Untranslated region. B, PCR genotype analysis of pCB740-PSR1 transformation lines. PSR1 PCR products were amplified using primers AF and BR from genomic DNA isolated from three individual psr1:PSR1-OE lines and two individual cw15:PSR1-OE lines. PCR using a negative control (untransformed cw15 genomic DNA) and a positive control (pCB740+PSR1) is shown. The 600-bp marker is indicated. C and D, Expression in high-P and low-P conditions of HSP70B in wild-type (WT; cw15) cells transformed with pCB740 (C) and of PSR1 in untransformed wild-type (cw15) cells and those transformed with pCB740 (vector control; D). HSP70B and PSR1 expression was determined relative to CBLP expression by real-time PCR. Data points are means ± se calculated from three biological replicates each with three technical replicates.
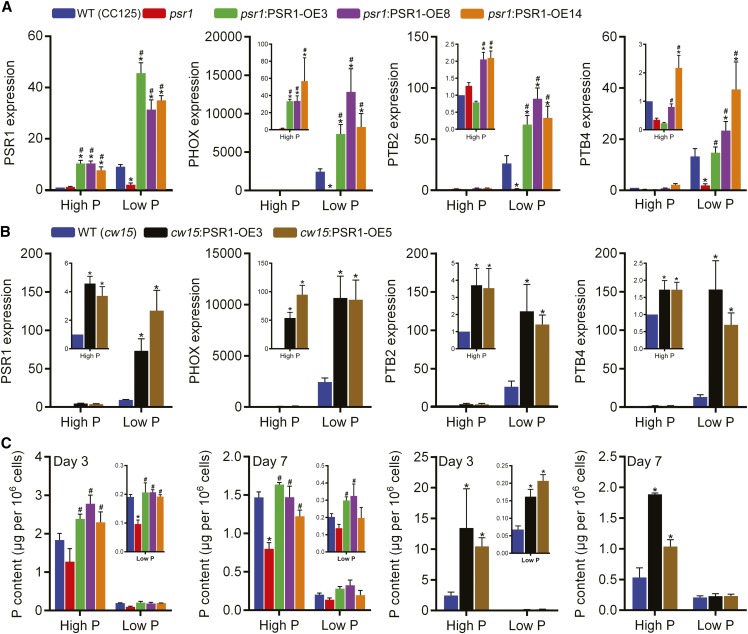
Figure 3.
Altered P homeostasis in PSR1 overexpression lines. A and B, Relative expression of PSR1 was quantified in psr1:PSR1 complementation lines compared with the wild type (WT; CC125) and psr1 (A) and in overexpression lines compared with the wild type (cw15; B) under high-P and low-P conditions at day 3. The relative expression of known PSR1-dependent, low-P-induced P homeostasis genes (PHOX, PTB2, and PTB4) was quantified in psr1:PSR1 complementation lines (A) and overexpression lines (B). Expression of the mRNA transcripts by real-time PCR was determined relative to CBLP expression and is shown relative to high-P-treated wild-type transcript. C, Pi accumulation in psr1:PSR1 complementation lines and overexpression lines compared with the wild type and psr1 in high-P and low-P conditions. Cellular Pi concentrations at days 3 and 7 are shown. Each data point represents the mean ± se of three biological and technical replicate culture flasks and is representative of three independent experiments. Asterisks denote significant differences (P < 0.05) from wild-type values and pound signs denote significant differences (P < 0.05) from psr1 values, both as determined by one-way ANOVA.
Figure 4.
Effect of actinomycin D and cycloheximide treatment on transcript abundance in PSR1 overexpression lines. Change in expression is shown for PSR1, PHOX, SSS1, and SP1 in PSR1 overexpression lines and the wild type (WT; cw15) at day 3 following treatment for 2 h (solid bars) and 6 h (hatched bars) with the transcription inhibitor actinomycin D during high-P (A) and low-P (B) growth and with the translation inhibitor cycloheximide during high-P (C) and low-P (D) growth. Expression of the mRNA transcripts by real-time PCR was determined relative to CBLP expression, and the increase or decrease in transcript abundance is shown relative to expression without inhibitor treatment. Each data point represents the mean ± se of three biological and technical replicate culture flasks.
Increased expression of the known PSR1-regulated PHOX, PTB2, and PTB4 was also observed in all psr1:PSR1-OE and cw15:PSR1-OE lines, compared with psr1 and cw15, respectively, under both high-P and low-P conditions (Fig. 3, A and B). In contrast to the effect on PSR1 abundance, both actinomycin D and cycloheximide treatment caused substantial reductions of PHOX mRNA abundance under low-P conditions in the wild type and cw15:PSR1-OE lines (Fig. 4) compared with PHOX abundance in the absence of inhibitors.
Up-regulation of the PHOX phosphatase and the Pi uptake transporters would be expected to increase Pi content within the cell. At day 3, cellular Pi content was significantly increased in all psr1:PSR1-OE lines compared with psr1 and all cw15:PSR1-OE lines compared with cw15 under both high-P and low-P treatments (Fig. 3C). By day 7 (late exponential growth phase/onset of stationary phase), a significant increase in Pi content was still observable in high-P cells, but in the low-P cells, this was only increased significantly in two of the psr1:PSR1-OE lines, due to the substantial depletion of Pi in the low-P medium by this time (Supplemental Fig. S1B). In all, these findings demonstrate that the expressed PSR1 was functional.
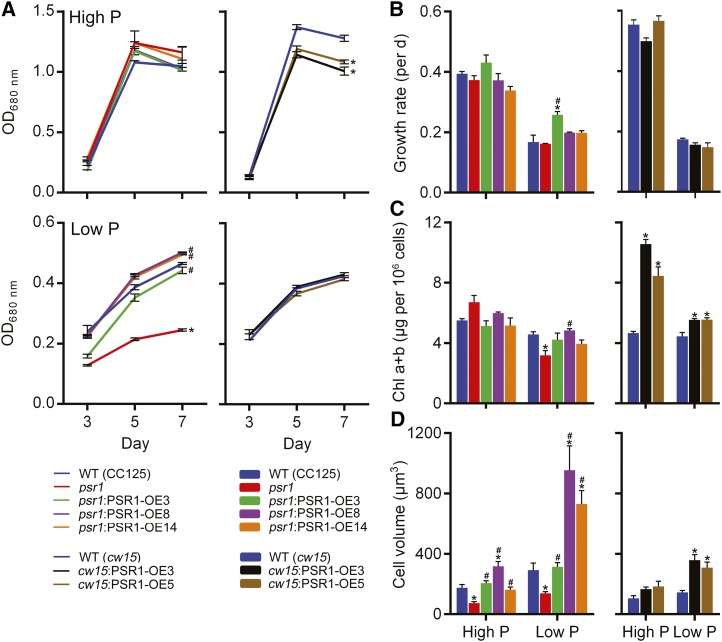
Increased Cell Volume and Starch Accumulation in PSR1 Overexpression Lines
Under high-P conditions, there was no difference in cell density, growth rate, biomass yield, or chlorophyll content between any of the psr1:PSR1-OE lines and the wild type and psr1 (Fig. 5, A–C; Supplemental Fig. S5A). Under low-P conditions, the growth of psr1 cells was inhibited compared with the wild type, as shown previously (Wykoff et al., 1999), and PSR1 expression in psr1 fully rescued the reduction in both cell density (Fig. 5A) and biomass (Supplemental Fig. S5A). In the cw15 background lines, there was no difference in biomass yield or growth rate under either P condition, although cell density was slightly but significantly reduced in the two cw15:PSR1-OE lines in high P (Fig. 5, A and B; Supplemental Fig. S5A). However, there was a large significant increase in total chlorophyll concentration in the cw15:PSR1-OE lines particularly under high-P conditions but also in low P (Fig. 5C). The most marked change in morphology of the PSR1-expressing cells was with regard to cell size, with many lines showing consistent increases in cell volume compared with the wild type and psr1 (Fig. 5D; Supplemental Fig. S6), particularly lines psr1:PSR1-OE8 and psr1:PSR1-OE14 under low-P conditions but also both cw15:PSR1-OE lines under low P.
Figure 5.
Changes in the physiology of PSR1-expressing cells in response to P starvation. Cell density determined by optical density at 680 nm (OD680nm) measurement (A) and specific growth rate determined at exponential phase from optical density at 680 nm values (B), total chlorophyll (Chl a+b; C), and biovolume (D) of PSR1 overexpression lines are shown. Chlorophyll and biovolume were quantified at day 7. Each data point represents the mean ± se of three replicates each with three technical replicates and is representative of three independent experiments. Asterisks denote significant differences (P < 0.05) from wild-type (WT) values and pound signs denote significant differences (P < 0.05) from psr1 values, both as determined by one-way ANOVA.
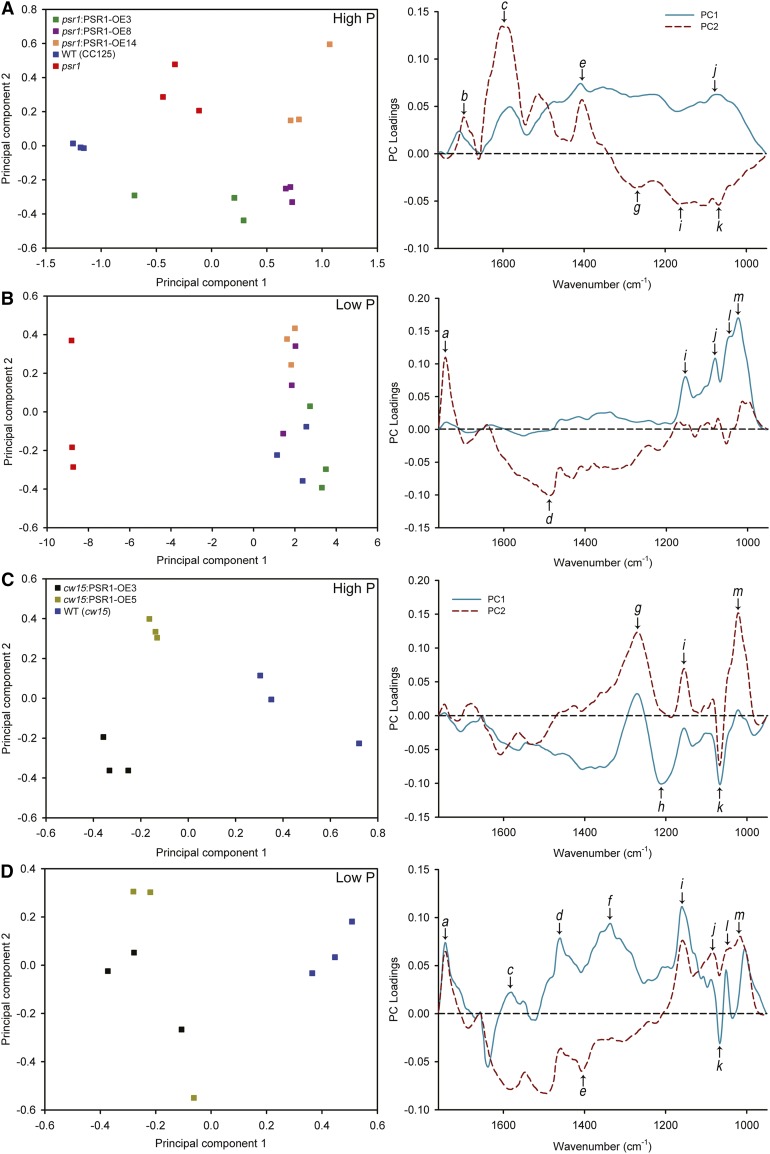
Increase in cell volume has been linked previously to the accumulation of carbon storage products (Dean et al., 2010; Work et al., 2010). Fourier transform infrared (FT-IR) spectroscopy, which is a powerful method to generate metabolic fingerprints (Ellis et al., 2002; Dean et al., 2010; Bajhaiya et al., 2016), was used to determine a global metabolic profile of each cell line at day 7 when end-point storage metabolites begin to accumulate. Three replicates of FT-IR spectra from each line were analyzed by principal component analysis (Fig. 6). All of the psr1:PSR1-OE lines clustered together closely with the wild type but clearly distinct from psr1 under low-P conditions, demonstrating that the successful complementation of psr1 by the PSR1 transgene could be determined at the metabolic level (Fig. 6B). The distinction between psr1 and the other cells was largely indicated by principal component 1, with the loading plot demonstrating that the metabolic difference between psr1 and the wild type or psr1:PSR1-OE cells was due predominantly to an increase in carbohydrates. In contrast, there was a less obvious pattern of clustering in these cells under high-P conditions (Fig. 6A). For the cw15 cells, there was a clear separation between the cw15:PSR1-OE lines and the wild type under high-P and low-P conditions, in both cases primarily determined by principal component 1. This demonstrates that there were significant metabolic differences within the cw15:PSR1-OE lines compared with the wild type, although the loading plots for both P conditions indicated that the metabolic variation was complex and not due to a single metabolite (Fig. 6, C and D). However, a clear change in both P conditions was an increase in a specific undetermined carbohydrate (denoted as band k) in the cw15:PSR1-OE cells. In contrast, under P starvation conditions only, the principal component loading plot indicates that the cw15:PSR1-OE cells had a marked decrease in the abundance of lipid (denoted as band a) compared with the wild-type cells (Fig. 6D).
Figure 6.
FT-IR spectroscopy analysis of PSR1 expression lines. Principal component analysis scores (left) and principal component (PC) loading plots (right) are shown for FT-IR spectra from the wild type (WT; CC125), psr1, and psr1:PSR1 complementation lines (A and B) and wild-type (cw15) and PSR1 overexpression lines (C and D) under high-P (A and C) and low-P (B and D) conditions. Analysis was performed on three replicate spectra for each sample and treatment. Selected bands that show strong changes are highlighted on the loading plots: a, νC=O of ester functional groups from lipids and fatty acids; b, νC=O of amides associated with protein (amide I); c, δ N-H of amides associated with protein (amide II); d, δas CH3 and δas CH2 of lipids and proteins; e, δas CH3 and δas CH2 of proteins, νsC-O of carboxylic groups; f, unknown; g, νasP=O of nucleic acids, phosphoryl group, due to DNA/RNA backbones, phosphorylated proteins, and polyphosphate storage products; h, νC-O-C of polysaccharides; i, νC-O of carbohydrates; j, νC-O of carbohydrates; k, νC-O-C of carbohydrates; l, νC-O of carbohydrates; m, νC-O of carbohydrates.
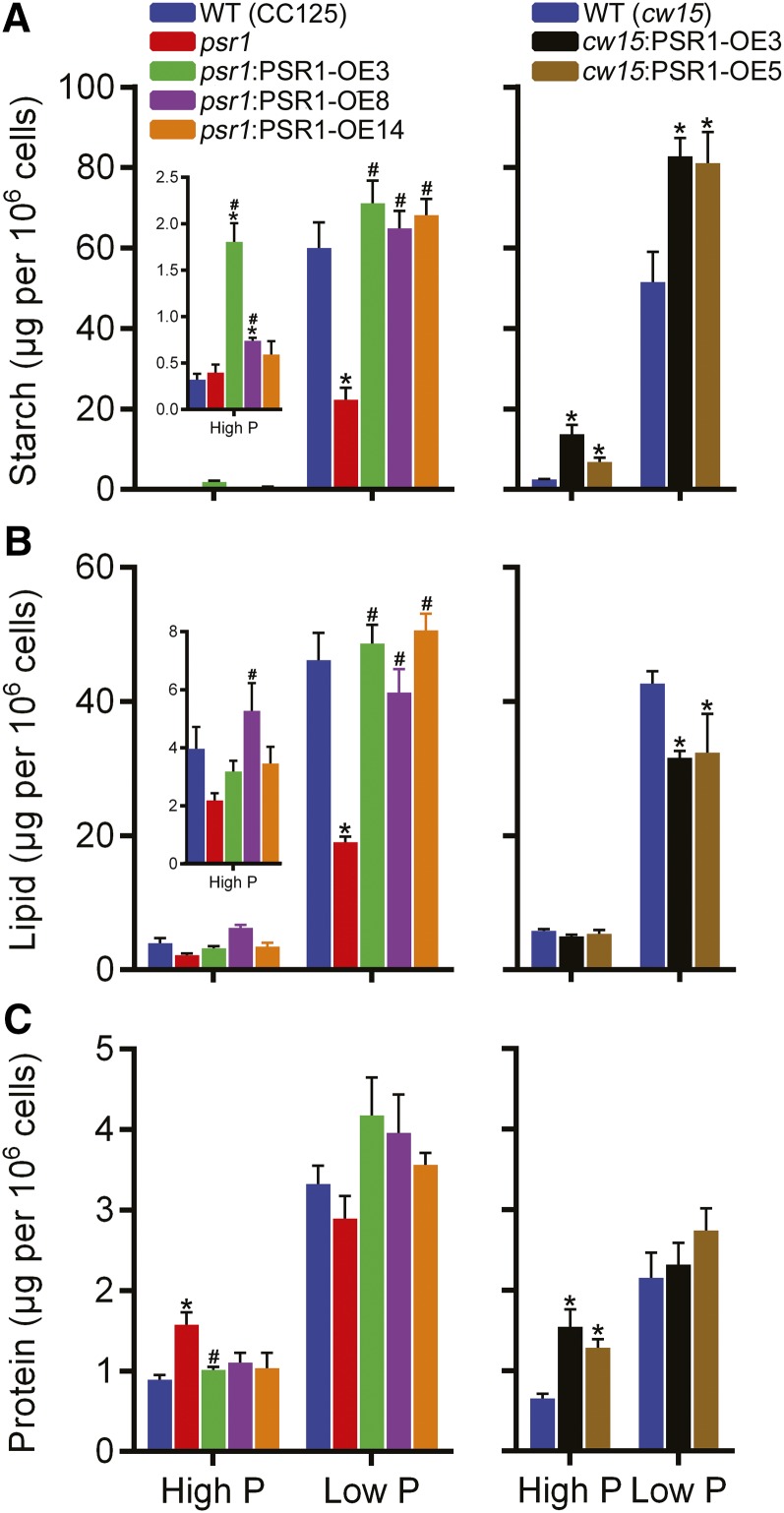
Metabolic changes within the cells were further examined by specific metabolite quantification. All psr1:PSR1-OE lines showed significant increases in starch compared with psr1 in low-P conditions, while two psr1:PSR1-OE lines showed significant starch increases in high-P conditions (Fig. 7A). Likewise, both cw15:PSR1-OE lines showed large significant increases in starch concentration compared with the wild type in both P conditions, with a mean increase (for the two lines) of 59% in low-P conditions and 310% in high-P conditions. For the cw15:PSR1-OE lines in low P, this substantial increase in starch was correlated with a 25% reduction in neutral lipid concentration, while in the psr1:PSR1-OE lines, there was no difference in lipid concentration compared with the wild type, but the lipid concentration was increased significantly compared with psr1 (Fig. 7B). These lipid and starch phenotypes were also observed consistently when quantified on the basis of biomass (Supplemental Fig. S5) and when quantified by FT-IR spectroscopy (Supplemental Fig. S7). Protein concentrations per cell did not change markedly, apart from an increase in both cw15:PSR1-OE lines compared with the wild type in high-P conditions (Fig. 7C), but there was no significant difference in protein concentration between lines and in response to P status when normalized on the basis of biomass (Supplemental Fig. S5B).
Figure 7.
Metabolite contents in PSR1 overexpression lines. Total starch (A), lipid (B), and protein (C) were quantified at day 7 in high-P and low-P growth conditions. Each data point represents the mean ± se of three replicates each with three technical replicates and is representative of three independent experiments. Asterisks denote significant differences (P < 0.05) from wild-type (WT) values and pound signs denote significant differences (P < 0.05) from psr1 values, both as determined by one-way ANOVA.
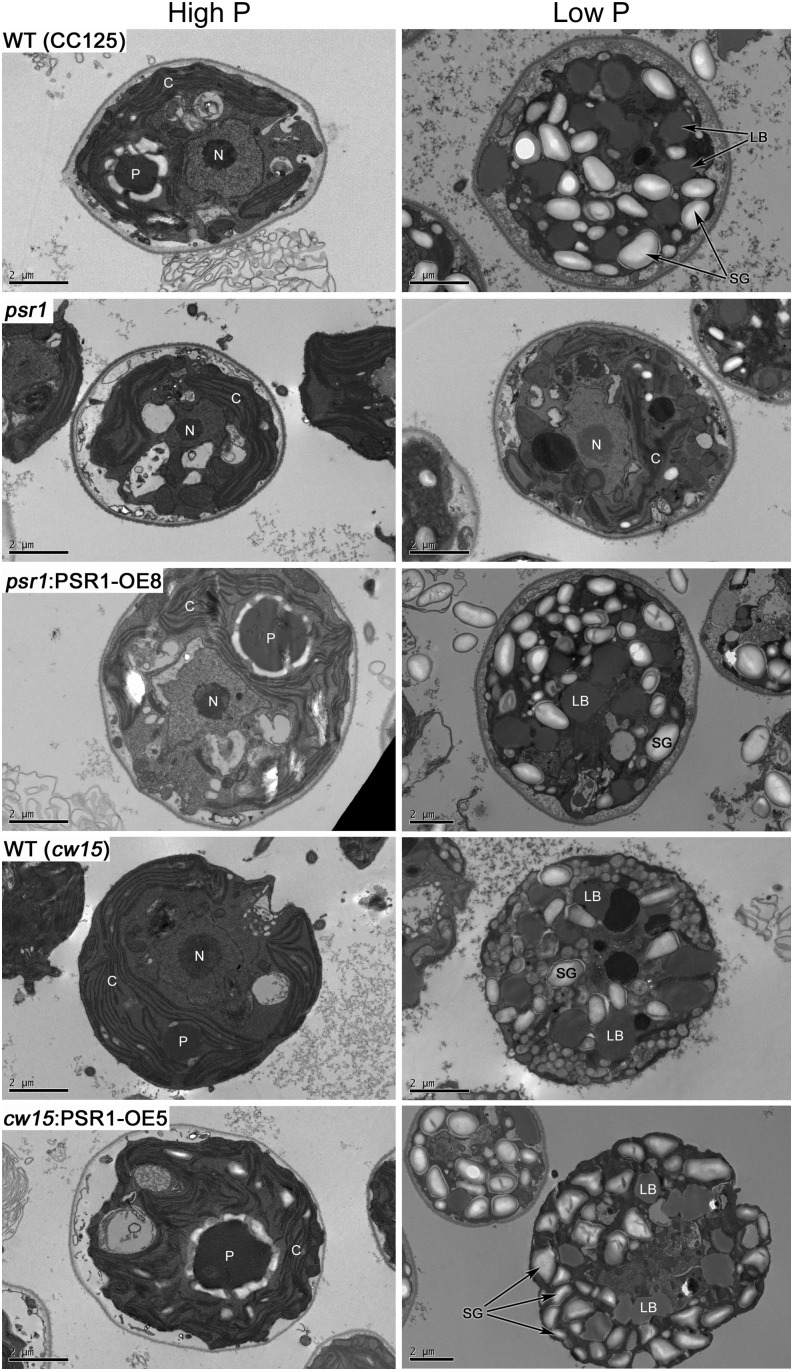
The quantified increases in starch accumulation were confirmed by imaging of starch granules. Detailed imaging of cell morphology by transmission electron microscopy (TEM; Fig. 8) of line psr1:PSR1-OE8 in comparison with the wild type (CC125) and psr1, and line cw15:PSR1-OE5 in comparison with the wild type (cw15), demonstrated the marked intracellular storage metabolite changes that occur in response to P starvation. The accumulation of many starch granules and lipid bodies was observed in all of the low-P-treated cells except for psr1 cells, which looked equivalent to high-P-grown cells and had a significantly reduced number of starch granules per cell compared with the wild type (Supplemental Fig. S8). This psr1 phenotype was rescued in the psr1:PSR1-OE8 cells, which displayed high numbers of starch granules in the low-P-grown cells (Fig. 8) but also a significant increase in starch granule number when grown in high-P conditions. Mean starch granule number was also significantly higher in cw15:PSR1-OE5 cells compared with wild-type cells in both P conditions, although there was no significant difference in mean starch granule size (Supplemental Fig. S8).
Figure 8.
Changes in the morphology of PSR1-expressing cells in response to P starvation. TEM images (representative of 15–20 images) are shown for wild-type (WT; CC125 and cw15), psr1, psr1:PSR1-OE8, and cw15:PSR1-OE5 cells in high-P and low-P conditions at day 7 of growth. Some subcellular structures are labeled: C, chloroplast; LB, lipid body; N, nucleus; P, pyrenoid; SG, starch granule. Bars = 2 µm.
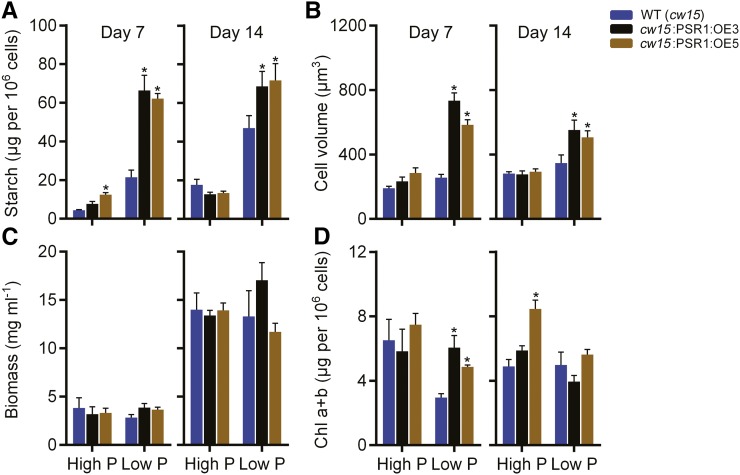
To assess whether the PSR1 overexpression phenotype was dependent on the presence of acetate, cw15 and cw15:PSR1-OE lines were grown without acetate. Cells were harvested and analyzed at days 7 and 14, as growth in the absence of acetate was expected to be slower. Equivalent phenotypes were observed with the acetate-free medium compared with the acetate-containing medium (Fig. 9). At both time points, starch concentration and cell volume were increased significantly in all cw15:PSR1-OE lines compared with the wild type, while there was no significant change in biomass and minor changes in total chlorophyll content.
Figure 9.
Changes in the starch content and physiology of PSR1-expressing cells in response to P starvation in the absence of acetate. Total starch (A), biovolume (B), fresh weight biomass (C), and total chlorophyll (Chl a+b; D) of PSR1 overexpression lines are shown. Parameters were quantified at days 7 and 14 in high-P and low-P growth conditions in the absence of acetate. Each data point represents the mean ± se of three replicates each with three technical replicates and is representative of three independent experiments. Asterisks denote significant differences (P < 0.05) from wild-type (WT) values as determined by one-way ANOVA.
Altered Expression of Starch and Lipid Metabolism Genes in PSR1 Overexpression Lines
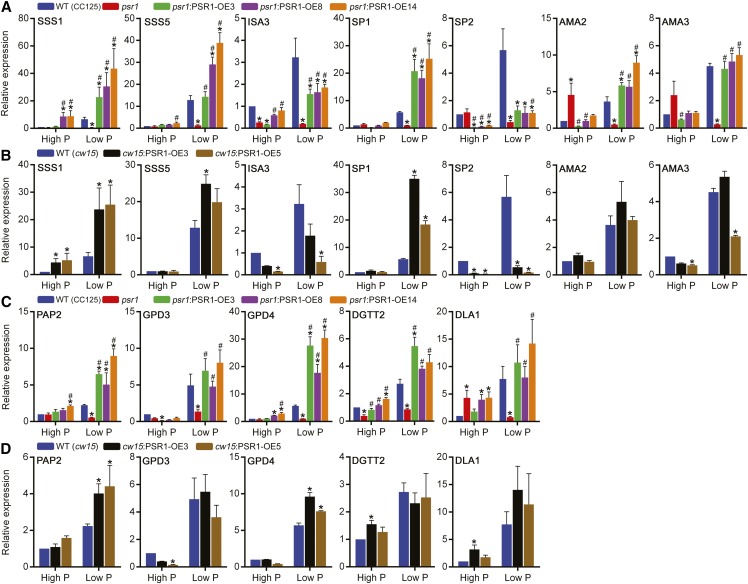
Expression of the starch and lipid metabolism genes shown to be misregulated in psr1 (Fig. 1) was quantified in the psr1:PSR1-OE and cw15:PSR1-OE lines under high-P and low-P conditions. Many of the starch metabolism genes showed altered expression following PSR1 overexpression both in the psr1 background lines (Fig. 10A) and in the cw15 background lines (Fig. 10B). The expression of SSS1, SSS5, and SP1 was increased significantly relative to the wild type in both psr1:PSR1-OE and cw15:PSR1-OE lines. In contrast, the expression of ISA3 was reduced slightly compared with the wild type in both sets of lines, although ISA3 expression was higher than in psr1 in the complemented lines. Expression of SP2, which encodes the PhoB plastidial starch phosphorylase, was repressed significantly in all PSR1 overexpression lines. The expression of AMA2 and AMA3 did not show major changes relative to the wild type, although the expression of both genes was increased relative to psr1 in all psr1:PSR1-OE lines under low-P conditions. Similar to the response of PHOX transcript abundance to transcription inhibition, actinomycin D treatment caused a substantial reduction of SSS1 and SP1 mRNA abundance under both high-P and low-P conditions in the wild type and cw15:PSR1-OE lines compared with transcript abundance in the absence of the inhibitor (Fig. 4, A and B). Translation inhibition by cycloheximide treatment had no significant effect on the abundance of both transcripts under high-P conditions, equivalent to the response of PHOX (Fig. 4C), while under low-P conditions, SSS1 abundance was not changed significantly by cycloheximide treatment, but there was a fold reduction in the abundance of SP1 after 6 h in the wild type and the psr1:PSR1-OE5 line (Fig. 4D).
Figure 10.
Altered starch and lipid gene expression in PSR1 overexpression lines. Relative expression of selected starch metabolism genes (A and B) and selected lipid metabolism genes (C and D) in psr1:PSR1 complementation lines is shown compared with the wild type (WT; CC125) and psr1 (A and C) and in overexpression lines compared with the wild type (cw15; B and D) under high-P and low-P conditions at day 3. Expression of the mRNA transcripts by real-time PCR was determined relative to CBLP expression and is shown relative to the high-P-treated wild-type transcript. Each data point represents the mean ± se of three biological replicates each with three technical replicates. Asterisks denote significant differences (P < 0.05) from wild-type values and pound signs denote significant differences (P < 0.05) from psr1 values, both as determined by one-way ANOVA.
Expression changes were also observed for some of the lipid metabolism genes following PSR1 overexpression. In all of the psr1 complementation lines under low-P conditions, the presence of PSR1 induced increased expression of each gene tested relative to psr1, and for some of the genes, particularly PAP2 and GPD4, the expression was significantly higher relative to the wild type (Fig. 10C). Likewise, expression of these two genes was increased significantly under low-P conditions relative to the wild type in both cw15:PSR1-OE lines (Fig. 10D). Under high-P conditions, the expression changes in the psr1:PSR1-OE and cw15:PSR1-OE lines were less clear, although DGTT2 expression was higher relative to psr1 in all lines.
DISCUSSION
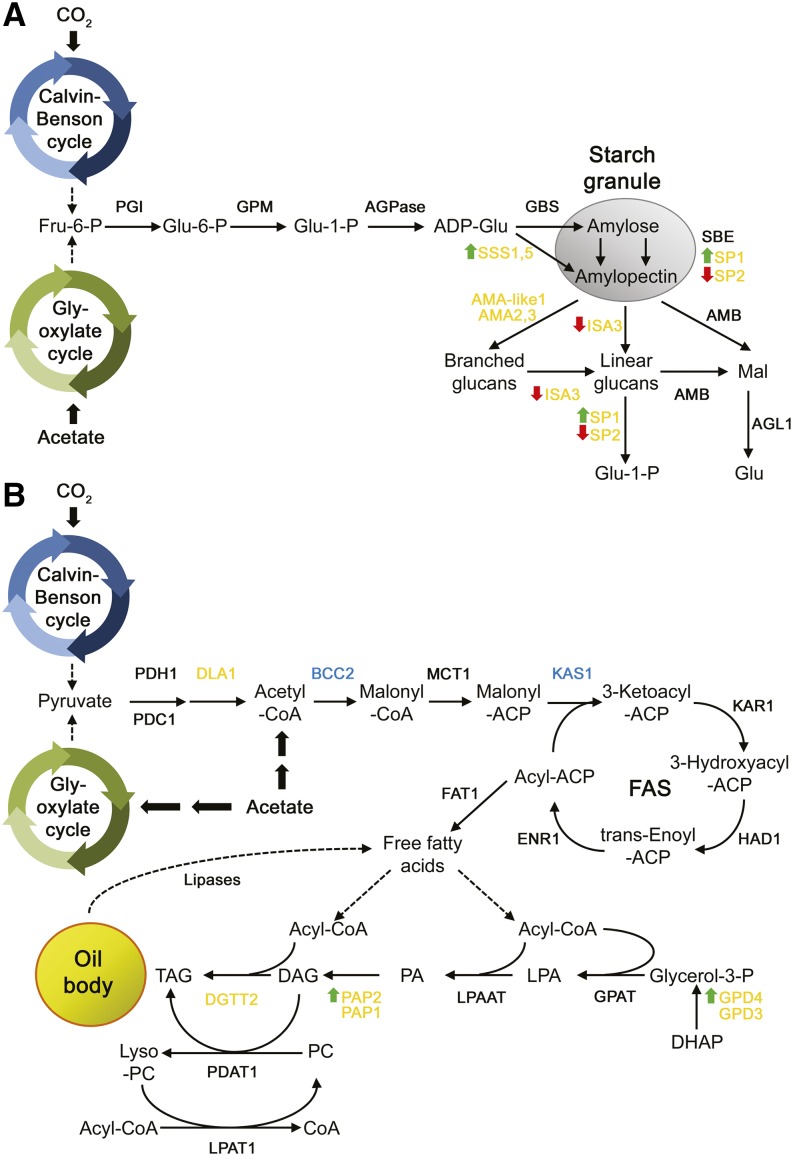
In this study, we provide strong evidence that PSR1 regulates carbon storage metabolism under P starvation conditions through the transcriptional control of specific starch and lipid synthesis genes. In particular, induction or repression of a high number of starch and lipid metabolism genes in response to P starvation was inhibited or abolished in psr1 knockdown lines, while overexpression of PSR1 mediated changes in the expression of some of these genes, resulting in altered partitioning and storage of starch and TAG. The qPCR analysis showed that an equivalent number of PSR1-dependent starch and lipid enzyme genes were modulated by P starvation (Fig. 1), with the psr1 knockdown line showing equivalent inhibition of both starch and lipid accumulation and the psr1:PSR1 complementation lines restoring both metabolites to wild-type levels (Fig. 7). However, the most obvious phenotype of the PSR1 overexpression lines in cw15 cells was a significant increase in starch under both P-replete and P-starvation conditions, while the neutral lipid concentration in these cells was either unchanged or inhibited. This phenotype may be explained partly by the observation that TAG biosynthesis genes, notably PAP2 and GPD4 encoding for the synthesis of DAG and glycerol 3-phosphate, respectively, were only moderately up-regulated in cw15:PSR1-OE cells (Fig. 10). In contrast, two of the SSS isoforms, particularly SSS1, which is most likely required for amylopectin synthesis (Ball and Morell, 2003), was highly up-regulated, as was the PhoA SP encoded by SP1, which has been suggested to play a role in starch synthesis (Dauvillée et al., 2006; Fig. 11). The ISA3 debranching enzyme and AMA3 were slightly down-regulated and thus are likely to inhibit starch catabolism. The other SP isoform (SP2) encoding PhoB was also down-regulated, and although this enzyme has been shown to be involved in starch synthesis (Dauvillée et al., 2006), it is unclear whether PhoB also has a role in starch breakdown.
Figure 11.
Model of the effects of PSR1 overexpression on starch and lipid metabolism. The schematic diagrams represent starch synthesis and breakdown (A) and fatty acid and TAG synthesis (B). Genes in yellow and blue are those up-regulated and down-regulated by P starvation, respectively, as determined by real-time PCR. For gene definitions, see Supplemental Table S1. Green up arrows and red down arrows indicate genes that are positively or negatively regulated by PSR1 overexpression, respectively.
These data also support the model that starch rather than neutral lipids is the major carbon storage molecule in photosynthetic organisms, including C. reinhardtii. Thus, the PSR1-dependent component of the P starvation response of remodeling carbon storage metabolism preferentially increases starch accumulation. This antagonism between starch and lipid synthesis is equivalent to the previous observations that lipid accumulation in C. reinhardtii can be increased through a reduction in starch synthesis, as seen in starchless mutants, including sta6 and sta7 (Work et al., 2010; Blaby et al., 2013; Goodenough et al., 2014), although in each of these cases, under N starvation rather than P starvation conditions. It has been presumed that, in mutant cells that are unable to synthesize starch, during starvation conditions that promote carbon storage, carbon flux instead is directed toward TAG synthesis. However, it is important to remember that these N starvation-induced TAG accumulation phenotypes, both in wild-type strains and starchless mutants, are pronounced in the presence of acetate, and particularly following an acetate boost, while phototrophically grown N-starved cells in the absence of acetate do not exhibit as substantial a TAG yield (Goodson et al., 2011; Goodenough et al., 2014). The PSR1-induced phenotypes observed in this study were seen in cells grown under both photoheterotrophic conditions and photoautotrophic conditions without acetate, with no obvious difference in the degree of starch accumulation (Fig. 9).
This study also allows us to examine how some of the transcriptional responses to P starvation compare with responses to other nutrient starvation conditions quantified previously, including N and sulfur starvation. It has been stated that global responses to macronutrient limitation are very similar in terms of growth inhibition, protease up-regulation, and lipid and starch accumulation when comparing N, P, and sulfur limitation (Schmollinger et al., 2014). However, gas chromatography-mass spectrometry-based metabolomic analysis has indicated that the metabolic differences between P-starved and nutrient-replete cells are distinct from the metabolic differences observed following N and sulfur starvation (Bölling and Fiehn, 2005). Observation of the sulfur starvation transcriptome indicates that this stress does not elicit substantial changes in carbon storage metabolism compared with the marked changes seen here for P starvation (González-Ballester et al., 2010). In particular, although sulfur starvation enhances starch accumulation greatly in microalgae (Brányiková et al., 2011), there appeared to be no significant increase in the abundance of starch metabolism transcripts in response to this stress (González-Ballester et al., 2010). In contrast, N starvation, like P starvation, modifies the expression of many primary metabolism and carbon storage metabolism genes (Miller et al., 2010; Boyle et al., 2012; Blaby et al., 2013; Goodenough et al., 2014; Ngan et al., 2015). Our transcriptomic data generated by qPCR indicate that there are some identical responses between N and P starvation; for example, specific isoforms of starch and TAG synthesis, including SSS5, GPD4, and PAP1, are up-regulated under P starvation (Fig. 1) and N starvation conditions (Miller et al., 2010; Goodenough et al., 2014). However, there are a number of differences between the stresses with regard to fatty acid and TAG synthesis, which is substantially modified under N starvation. Various components of fatty acid synthesis are induced by N starvation, while these are largely down-regulated by P starvation. Variation is seen at the end point of TAG synthesis from diacylglycerol, which is controlled by two acyltransferase pathways: the DGAT and the acyl-CoA-independent phospholipid diacylglycerol acyltransferase pathways. The type 1 DGAT DGAT1 and the type 2 DGAT DGTT1 are both highly up-regulated by N starvation (Boyle et al., 2012), while RNA-Seq analysis indicated that neither of these transcripts appears to be induced either at all or substantially by P starvation, although this needs to be confirmed by qPCR. In contrast, DGTT2 was induced and DGTT3 and DGTT4 were maintained at basal levels under P starvation conditions (Fig. 1). With regard to starch metabolism, both the N and P starvation transcriptomes showed clear starch synthesis gene changes, but these appear to be generally distinct; for example, N starvation mediates significant up-regulation of granule-bound starch synthase genes and starch-branching enzyme genes (Goodenough et al., 2014) that was not seen under P starvation. Recently, Schmollinger et al. (2014) reported a direct comparison of RNA-Seq data derived from N, P, and sulfur limitation treatments and indicated that N and sulfur limitation induces more common transcriptional responses (approximately 23% similarity) compared with N and P limitation (approximately 5% similarity in response). No transcripts related to photosynthesis and tetrapyrrole biosynthesis were reported to be common between P and N limitation, whereas such transcripts showed similar responses between N- and sulfur-limited cells (Schmollinger et al., 2014). Both N and P starvation elicits a reduction in photosynthetic activity, including decreased oxygen evolution and photosynthetic electron transport (Peltier and Schmidt, 1991; Wykoff et al., 1998), but the impact of P starvation on photosynthesis may be less severe. These transcriptomic observations, along with the metabolomic responses to N, P, and sulfur (Bölling and Fiehn, 2005), indicate clear distinctions between different macronutrient limitation conditions.
These PSR1 experiments have also reaffirmed the importance of PSR1 in controlling P homeostasis in C. reinhardtii. We were able to confirm many of the results found in the previous microarray-based transcriptomic analysis that allowed coverage of approximately 3,000 C. reinhardtii transcripts (Moseley et al., 2006). In that study, the microarray data and qPCR validation demonstrated the PSR1-dependent P starvation-induced increase of PHOX, PSR1, and PTB2 to PTB5 and the decrease in transcript abundance of the PTA-type PO43−/H+ symporter isoforms PTA1 and PTA3, many of which are confirmed here. Other P homeostatic and P conservation changes were also apparent at the transcriptional level. Under P limitation, the abundance of the phospholipid phosphatidylglycerol decreases, while the sulfolipid SQDG increases (Riekhof et al., 2003). The SQD1 enzyme that performs the first step of SQDG synthesis is essential for sulfolipid metabolism (Riekhof et al., 2003), and the SQD1 transcript, as well as transcripts for other sulfolipid genes, including SQD3, were found to be highly up-regulated by P starvation in a PSR1-dependent manner (Fig. 1). Another PSR1-dependent transcript up-regulated by P starvation was GDP1, encoding a putative GDP. One assumption could be that this enzyme mediates the scavenging and recycling of P through the hydrolysis of lipid phosphodiester from phospholipids such as phosphatidylcholine. A transcriptomic and proteomic analysis of the marine diatom Thalassiosira pseudonana in response to P limitation also observed an induction of transcripts or proteins for SQDG synthesis and phosphodiesterase activity, in addition to increased alkaline phosphatase and Pi transport (Dyhrman et al., 2012). Our study has gone further and shown that overexpression of PSR1 increases the abundance of the P homeostatic components and, consequently, increases Pi uptake into the cell, including under P-replete conditions (Fig. 3C). This indicates that the P starvation response can be deregulated, allowing excessive P accumulation even when not needed by the cell. Microalgae are able to accumulate and store excess Pi as polyphosphate; thus, increased P accumulation is less likely to be toxic to the cell. Indeed, total biomass yield was unchanged in the cw15:PSR1-OE cells under high-P conditions, although there was a slight but significant reduction in cell density and total chlorophyll content was increased significantly. Thus, there may be applications for high-Pi-accumulating microalgae, such as their use in the bioaccumulation and recovery of waste Pi from wastewater (Pittman et al., 2011).
An unexpected result from these experiments was the observation that PSR1 expression in the pCB740-PSR1-transformed cells was significantly higher under low-P conditions compared with high-P conditions (Fig. 3). This response does not appear to be due to the HSP70A-RBCS2 promoter, and it is unclear whether this phenotype is due to the induction of PSR1 by the P-limited status or the repression of PSR1 under P-replete conditions, such as through instability of the PSR1 mRNA transcript. The inhibition of transcription by actinomycin D reduced the abundance of all transcripts tested with the exception of PSR1, for which mRNA transcript was still highly abundant, particularly during P starvation (Fig. 4). Furthermore, treatment with the translational inhibitor cycloheximide did not affect the increase of PSR1 transcript abundance under low-P conditions, and transcript abundance remained high, as mRNA stability is well known to be maintained by the inhibition of protein synthesis (Baker and Liggit, 1993). In fact, cycloheximide treatment caused an elevation in the accumulation of the PSR1 transcript (Fig. 4D), which has been seen previously for other C. reinhardtii transcripts (Aksoy et al., 2013), probably due to the inhibition of turnover or the ribosomal protection of transcript. Interestingly, this cycloheximide-induced elevation of PSR1 accumulation was greater under low-P conditions, suggesting increased stability and/or increased transcription in response to P starvation. Furthermore, this result suggests that protein synthesis was not required for the induction of PSR1. In contrast, a substantial reduction of PHOX and SP1 transcript abundance under P starvation following cycloheximide treatment was observed (Fig. 4D), indicating that protein synthesis was required for PHOX and SP1 induction and that the inhibition of PSR1 protein synthesis in turn inhibited the transcriptional activation of these genes. Distinct tiers of regulation, either requiring or not requiring protein synthesis, were observed previously with respect to sulfur limitation (Aksoy et al., 2013). One possible explanation for the P-responsive regulation of PSR1 in the absence of the native promoter could be that PSR1 can still be regulated via an intronic cis-regulatory element present within the gene sequence or that other as yet unknown factors are involved in PSR1 regulation. The higher plant homologs of PSR1, including Arabidopsis PHR1, may be regulated in part posttranslationally, through sumoylation (Miura et al., 2005). When PHR1 was overexpressed in Arabidopsis under the control of a strong constitutive promoter, PHR1 transcript abundance was equivalent between P-limiting and P-replete conditions, but even in the wild-type plant, PHR1 expression was not highly induced by P starvation (Nilsson et al., 2007), indicating differences between the regulation of PSR1/PHR1 in plants versus microalgae.
The potential of oil accumulation in microalgae for bioenergy use has been widely discussed (Georgianna and Mayfield, 2012; Merchant et al., 2012; Driver et al., 2014), and likewise, some authors have suggested that starch accumulation in microalgae might also have potential as a feedstock for biofuel production (Brányiková et al., 2011). Furthermore, there are additional potential nutraceutical and industrial applications of metabolites from microalgae (Guedes et al., 2011). However, for such applications to be viable in the future, tools to enhance and modify specific metabolites will be needed. Transcriptional engineering has been proposed as a potentially more efficient approach to engineer metabolic pathways instead of targeting individual enzymes (Grotewold, 2008). Previously, the putative transcription factor NRR1 was identified, which is induced specifically under N starvation and when knocked out causes substantial reduction of N starvation-induced TAG synthesis (Boyle et al., 2012). A recent study also identified PSR1 as a highly induced transcription factor under N and sulfur starvation, while disruption of PSR1 inhibited nutrient starvation-induced lipid accumulation (Ngan et al., 2015). We have further observed that PSR1 mutation inhibits lipid accumulation in response to P starvation (Bajhaiya et al., 2016; Fig. 7B). Ngan et al. (2015) showed that overexpression of a PSR1 complementary DNA (cDNA) construct in CC125 enhanced lipid accumulation, a function that we have confirmed here, as complementation of the psr1 mutant strain with PSR1 genomic DNA increased lipid content relative to psr1 lipid level (Fig. 7B) and is coincident with an increase in the abundance of various lipid metabolism genes (Fig. 10C). Interestingly, we did not observe a significant increase in lipid concentration when PSR1 was overexpressed in cw15 cells, suggesting that there may be some variation in lipid induction mediated by PSR1 depending on strain background. However, we have clearly shown that overexpression of PSR1 can also markedly enhance starch content, a phenotype not evaluated by Ngan et al. (2015).
Transcriptional regulators for starch metabolism have been identified in higher plants, and manipulation of these transcription factors through overexpression was shown to modify the composition or accumulation of these metabolites (Cernac and Benning, 2004; Fu and Xue, 2010). For example, RSR1 was found to be a negative regulator of starch biosynthesis in rice (Oryza sativa), modulating starch yield and starch granule morphology (Fu and Xue, 2010), while OsbZIP58 was identified as another transcriptional regulator of rice starch metabolism (Wang et al., 2013). However, overexpression of RSR1 did not significantly alter total starch content, while overexpression of OsbZIP58 was not tested. Here, we demonstrated that manipulation of a transcriptional regulator in C. reinhardtii has the potential to increase the content of metabolites such as starch under both nutrient-replete and starvation conditions. However, we also demonstrated that the PSR1 transcription factor is not specific in its modulation of lipid or starch but rather is a global regulator and, therefore, that manipulation with PSR1 would likely alter many processes and thus impact the long-term fitness of the strain. PSR1 is not the only transcriptional regulator of lipid metabolism (Boyle et al., 2012) and is unlikely to be the only starch regulator. Other potential transcription factors induced by nutrient starvation have been identified (Miller et al., 2010; Gargouri et al., 2015; Ngan et al., 2015), some of which are likely to function as metabolic regulators. Therefore, future efforts are needed to identify more metabolism-specific transcriptional regulators.
MATERIALS AND METHODS
Chlamydomonas reinhardtii Strains and Growth Conditions
The C. reinhardtii wild-type strain CC125 (obtained from CCAP; stock no. CCAP11/32C) and the cell wall-deficient strain cw15 (cw15 arg7-8, referred to throughout as cw15; obtained from the Chlamydomonas Resource Center; stock no. CC4351) were used as wild-type control strains. The cw15 arg7 Arg auxotrophic strain has an identical metabolic profile under P-starvation and P-replete conditions compared with the cw15 ARG7 strain (CCAP11/32CW15+) lacking the Arg mutation, as determined by FT-IR spectroscopy (Supplemental Fig. S9). The psr1 knockdown strain was kindly provided by Arthur Grossman. CC125 is the isogenic parental strain to psr1. psr1 was originally isolated from a UV light mutagenesis screen (Shimogawara et al., 1999), although the specific mutation site is unknown. PSR1 expression is severely inhibited in the mutant but not completely abolished (Supplemental Fig. S3A); thus, psr1 is a knockdown mutant rather than a complete null. Furthermore, coverage of RNA-Seq reads indicates that the full-length PSR1 transcript is made and is not truncated in psr1. The PSR1 complementation lines and overexpression lines were created in this study. The strains were grown up to 7 d photoheterotrophically in batch culture in standard Tris-acetate-phosphate (TAP) medium at pH 7 (Harris, 1989; high-P medium) containing 1 mm P in 200-mL glass flasks on an orbital shaker rotating at 2 Hz or in 50-mL Nunc flasks at 25°C under cool-white fluorescent lights (150 µmol m−2 s−1) with a 16-h/8-h light/dark regime. In some experiments, strains were grown photoautotrophically in the absence of acetate in Tris-phosphate medium for up to 14 d, with medium pH adjusted to 7 by the addition of HCl. P deficiency was imposed by growing cells in low-P TAP or Tris-phosphate medium containing 10 µm P by reducing the volume of potassium Pi (pH 7) solution (K2HPO4/KH2PO4) while maintaining a uniform concentration of potassium by the addition of KCl. Before starting cultivation in low-P medium, late exponential phase P-replete cells were washed twice with low-P medium by centrifugation at 3,000g for 1 min to remove externally bound P. Washed cells were inoculated into fresh low-P medium to give an initial cell count of approximately 65 × 103 cells mL−1. In some experiments, strains were grown in high-P or low-P TAP medium until day 3 and then exposed to the transcription inhibitor actinomycin D in 2% (v/v) dimethyl sulfoxide (DMSO) at a final concentration of 100 µg mL−1 or the cytoplasmic 80S ribosome translation inhibitor cycloheximide in DMSO at a final concentration of 10 µg mL−1, or a DMSO control, for 2 and 6 h, before cells were harvested and RNA was isolated, as described below.
Generation of PSR1 Complementation and Overexpression Lines
Genomic DNA was isolated from wild-type CC125 using cetyl trimethyl ammonium bromide (CTAB). A dense culture of cells (2 mL) was centrifuged at 3,000g, and the cell pellet was resuspended in 0.5 mL of CTAB buffer containing 2% (w/v) CTAB, 100 mm Tris-HCl (pH 8), 1.4 m NaCl, 20 mm EDTA, and 2% (v/v) β-mercaptoethanol, then incubated at 65°C for 1 h. The solution was mixed with an equal volume of phenol:chloroform:isoamyl alcohol (25:24:1), and the aqueous phase, obtained following centrifugation (10,000g for 10 min), was incubated with an equal volume of ice-cold isopropanol to precipitate DNA. A 3,764-bp PSR1 genomic DNA fragment, including all exon and intron regions and some 5′ and 3′ untranslated region sequence, was amplified from genomic DNA using primers PSR1F (5′-AAAACTAGTTCGCTATGCAACGATCTACG-3′; SpeI restriction enzyme site underlined) and PSR1R (5′-AAAGATATCGCTGCCGTGAACAGTACAAA-3′; EcoRV restriction enzyme site underlined). The PCR fragment was digested with SpeI and EcoRV and then ligated into the NheI and EcoRV sites of pCB740 (Schroda et al., 1999), so that PSR1 was under the control of the HSP70A-RBCS2 tandem promoter and regulated with the native PSR1 terminator (Fig. 2A). The junction site of the HSP70A-RBCS2 tandem promoter fragment with the PSR1 gene at the 5′ end (NheI site) was 110 bp before the ATG start codon, and the junction site of the PSR1 gene at the 3′ end with the pCB740 plasmid (EcoRV site) was 765 bp after the TAG stop codon. The PSR1 gene within the resulting pCB740-PSR1 plasmid was confirmed by sequencing using the PSR1 primers (Supplemental Table S4). The pCB740 and pCB740-PSR1 plasmids were introduced into cw15 to generate PSR1 overexpression lines and vector control lines and into psr1 to generate psr1:PSR1 complementation lines, using biolistic bombardment as described (Boynton and Gillham, 1993), except using 0.6-µm gold microcarriers and 900-p.s.i. rupture disks (Bio-Rad). cw15:PSR1-OE lines were selected on TAP medium without Arg. The selection of psr1 transformants (psr1:PSR1-OE lines) was based on tolerance to low-P medium by efficient growth on low-P TAP agar plates. All lines were genotyped using primers AF and BR (Fig. 2, A and B; Supplemental Table S4). Of the 25 cw15 and 25 psr1 transformants screened, eight to 10 positive transformants were identified for each genotype, of which lines psr1:PSR1-OE3, psr1:PSR1-OE8, and psr1:PSR1-OE14 as well as lines cw15:PSR1-OE3 and cw15:PSR1-OE5 were studied further.
Cell Growth Analysis
Cell density by optical density measurement and exponential phase growth rate was determined as described (Osundeko et al., 2013). Cell counts were determined using a Nexcelom Cellometer T4 (Nexcelom Biosciences). Fresh weight algal biomass was determined following centrifugation of a 50-mL sample at 1,500g for 20 min in a preweighed tube. Cell dimension measurements of 10 to 39 individual cells taken from bright-field light microscopy images were determined using ImageJ (http://rsbweb.nih.gov/ij). Cell biovolume was calculated using these measurements by approximating the cells to a sphere using the formula given by Hillebrand et al. (1999).
RNA Isolation, cDNA Synthesis, and qPCR
Total RNA was isolated from replicate samples of liquid N2-frozen day-3, -5, or -7 cultures of high-P- and low-P-treated cells using Trizol reagent (Life Technologies) and further purified by phenol:chloroform:isoamyl alcohol (25:24:1) extraction and precipitation with ice-cold isopropanol. RNA quality was checked by NanoDrop UV-Vis spectrophotometer (Thermo Scientific) analysis. First-strand cDNA was synthesized from DNase-treated RNA using BioScript reverse transcriptase (Bioline) and an oligo(dT) primer. The gene expression of selected transcripts was determined by qPCR using a SYBR Green master mix (Roche) and an ABI Prism 7000 machine (Applied Biosystems) with the SYBR Green detection program and normalized to CBLP gene expression and then further normalized against high-P-treated wild-type expression. Primer pairs used for qPCR analyses are listed in Supplemental Table S5. The sizes of the amplification products were 100 to 200 bp, and the primer pairs were confirmed to be specific to the target transcript. Between three and nine independent biological replicate reactions were run, each with three technical replicates, and PCR efficiencies were checked using LinRegPCR (Ruijter et al., 2009). Melting curves were produced for each experiment to ensure that single products were amplified. Relative gene expression was determined using the 2−∆∆CT method (Pfaffl, 2001). Relative amplification efficiency obtained by qPCR varied between 98% and 99%, and the obtained r2 values of all the qPCRs were greater than 0.98. A standard curve using a dilution series of genomic DNA was made for all primer sets, and the efficiency of the qPCR assay was determined along with the r2 value.
FT-IR Spectroscopy
A 0.5-mL sample from each triplicate flask for each line and treatment at day 7 of cultivation was centrifuged at 1,500g for 20 min, the supernatant was removed, and the cells were weighed and normalized to 60 mg mL−1 by resuspension in Milli-Q (Millipore) water. A 30-μL sample was deposited on a 96-well silicon microplate and oven dried at 40°C overnight. The plate was placed in an HTS-XT high-throughput microplate extension, and FT-IR spectra were collected using a Bruker Equinox 55 FT-IR spectrometer equipped with a deuterated triglycerine sulfate detector. Spectra were collected over the wave number range 4,000 to 600 cm−1. Band assignments were determined as described previously (Giordano et al., 2001; Dean and Sigee, 2006; Murdock and Wetzel, 2009). Generated data were imported into MATLAB version 2010a (MathWorks) for processing and multivariate statistical analysis. Spectra were preprocessed using extended multiplicative signal correction (Martens and Stark, 1991). The spectral data were analyzed by principal component analysis essentially as described previously (Bajhaiya et al., 2016).
Starch, Lipid, Protein, Pi, and Chlorophyll Measurement
Starch measurements were performed, using methods by Ball et al. (1991) and Grant et al. (2006), by sampling cells (5-mL volume) at day 3, 5, or 7, pelleting by centrifugation at 1,500g for 20 min, and then extracting chlorophyll by washing in 80% (v/v) ethanol, incubating at 85°C for 5 min, and centrifuging at 13,000g for 10 min, as described (Ball et al., 1991). The chlorophyll-free cell pellets were resuspended in 200 µL of 80% (v/v) ethanol and 500 µL of DMSO and incubated by shaking at 90°C for 1 h (shaker from Thermo Scientific) to break the cells and solubilize the starch (Grant et al., 2006). The extract was then submitted to complete amyloglucosidase digestion (Ball et al., 1991), and total starch was quantified using a Total Starch Assay Kit (Megazyme) according to the manufacturer’s instructions. Starch concentration was determined using a d-Glc standard curve, and values were multiplied by 162/180 (adjustment for free d-Glc to 1,6-anhydro-β-D-Glc) to calculate total starch. Neutral lipid was quantified in day-3, -5, or -7 cells using the fluorescent dye Nile Red essentially as described (Dean et al., 2010; Osundeko et al., 2013) using a triolein (Sigma-Aldrich) standard and by quantification of lipid peaks from FT-IR spectra by calculating lipid-amide I peak height ratio values. Total protein was determined by resuspending harvested day-7 cells in extraction buffer containing 30 mm Tris-HCl, pH 7.5, and 1 µL of protease inhibitor cocktail (Sigma-Aldrich) followed by two rounds of freezing/thawing in liquid N2. The extract was centrifuged at 12,000g for 15 min, and the supernatant was used for protein estimation using the Bradford dye assay kit (Bio-Rad). For total chlorophyll (chlorophyll a and b) measurement, day-7 cells (5-mL volume) were harvested and centrifuged as described above, resuspended and incubated in 80% (v/v) acetone, and vortexed to extract the pigments. Cellular debris were pelleted by centrifugation (13,000g for 10 min), then chlorophyll a and b concentrations were determined spectrophotometrically using the method and formula described previously (Porra et al., 1989). For cellular Pi measurement, cells (5-mL volume) at different growth points, as described in “Results,” were harvested and centrifuged as described above, washed with 1 mm EDTA to remove any externally bound Pi and further washed with Milli-Q (Millipore) deionized water, and pelleted by centrifugation at 1,500g for 20 min. Pellets were dried for 24 h at 60°C and then digested with 0.5 mL of 67% (v/v) ultrapure nitric acid at 100°C for 3 h. Digested samples (to determine cellular Pi) and 0.45-µm filtered medium samples (to determine medium Pi) were diluted in Milli-Q water and analyzed using an Autoanalyser3 (Seal Analytical) fitted with an XY2 autosampler and AA3 digital colorimeter module HR4. The generated peaks were calibrated against drift and baseline peaks. All starch, lipid, protein, chlorophyll, and Pi measurements were determined on a per cell basis from cell counts or were normalized to fresh weight biomass.
Bright-Field Light Microscopy and TEM
Bright-field differential interference contrast microscope images of day-7 high-P- and low-P-treated cells were observed using a Leica DMR microscope and a 100× oil-immersion objective, and images were taken with a SPOT Xplorer CCD camera (model 17.4; Diagnostic Instruments). The same cells were used for TEM. The cells were fixed with 4% (v/v) formaldehyde and 2.5% (v/v) glutaraldehyde in 0.1 m HEPES buffer (pH 7.2) and then postfixed with 1% (w/v) osmium tetroxide and 1.5% (w/v) potassium ferrocyanide in 0.1 m cacodylate buffer (pH 7.2) for 1 h, then in 1% (v/v) tannic acid in 0.1 m cacodylate buffer (pH 7.2) for 1 h, and finally in 1% (v/v) uranyl acetate in water for 1 h. The samples were dehydrated in an ethanol series infiltrated with TAAB 812 resin and polymerized for 24 h at 60°C. Sections were cut with a Reichert Ultracut ultramicrotome and observed using a Technai-12 BioTwin microscope (FEI Instruments) at 100-kV accelerating voltage. Images were taken with a Gatan Orius SC1000 CCD camera. From the TEM images, starch granule characteristics were quantified using CellProfiler version 2.1 (http://www.cellprofiler.org).
Bioinformatic Sequence Analysis
C. reinhardtii genomic sequence and gene model information was obtained from Phytozome version 9.1 (http://phytozome.jgi.doe.gov/pz/portal.html) using version 5.3 of the C. reinhardtii genome annotations. Multiple sequence alignments were performed using ClustalW2 (http://www.ebi.ac.uk/Tools/msa/clustalw2). Phylogenetic analysis was performed essentially as described previously (Emery et al., 2012) using a ClustalW2 alignment and the maximum likelihood method under the WAG + F model of amino acid substitution and using the fast bootstrap approach to determine tree confidence, using the RAxML version 7.1 program. For bootstrapping, 100 iterations were used. The tree was viewed using the FigTree program (http://tree.bio.ed.ac.uk/software/figtree). For the identification of putative PSR1-binding sites, the P1BS cis-element sequence (GNATATNC) was screened in the noncoding regions, including intronic regions of 500 selected PSR1-regulated genes and non-PSR1-regulated genes (Supplemental Table S3), as determined from RNA-Seq data, using the NewPLACE database (http://sogo.dna.affrc.go.jp/cgi-bin/sogo.cgi?page=analysis&lang=eu; Higo et al., 1999).
Statistical Analysis
Differences between treatments and cell lines were assessed using one-way ANOVA performed using IBM SPSS Statistics version 20. When significant differences were detected at the 95% confidence level, the Tukey posthoc test was applied.
Sequence data from this article can be found in the EMBL-EBI ArrayExpress data library under accession number E-MTAB-2556.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Time-course profile of Pi availability, growth rate, lipid and starch accumulation, and P starvation gene expression in response to P starvation in wild-type cells.
Supplemental Figure S2. Transcriptional responses to P-replete and P-starvation conditions in wild-type and psr1 cells.
Supplemental Figure S3. Expression of selected genes in response to P starvation.
Supplemental Figure S4. Conservation of PSR1 DNA-binding domains and abundance of predicted PSR1-binding cis-elements.
Supplemental Figure S5. Metabolite content in PSR1 overexpression lines normalized by biomass.
Supplemental Figure S6. Changes in morphology of PSR1-expressing cells in response to P starvation.
Supplemental Figure S7. Lipid and carbohydrate contents of PSR1-expressing cells in response to P starvation determined by FT-IR spectroscopy.
Supplemental Figure S8. Starch granule characteristics.
Supplemental Figure S9. Metabolite profile comparison of cw15 ARG7 and cw15 arg7 strains by FT-IR spectroscopy.
Supplemental Table S1. Gene definitions for selected transcripts and gene families described in this study.
Supplemental Table S2. Predicted PSR1-binding cis-elements in PSR1-regulated genes.
Supplemental Table S3. Genes used for P1BS cis-element analysis.
Supplemental Table S4. Primer sequences used for genotyping and sequencing of the pCB740-PSR1 plasmid.
Supplemental Table S5. Primer sequences used for real-time PCR.
Supplemental Data Set S1. Summary of the expression levels and the DESeq normalization for high-P- and low-P-treated wild-type and psr1 mutant cells.
Supplemental Methods S1. RNA-Seq and data analysis.
Acknowledgments
We thank Andy Hayes and Claire Haslam in the Faculty of Life Sciences Genomic Technologies Facility for RNA-Seq processing, Dr. Aleksandr Mironov in the Faculty of Life Sciences Electron Microscopy Facility for TEM assistance, Debbie Ashworth for assistance with Autoanalyser Pi measurements, Thomas Driver and Javiera Ziehe Moreira for critical comments to the article, Thomas Driver for providing some FT-IR spectroscopy data analysis, and Arthur Grossman (Carnegie Institution) for providing the psr1 strain.
Glossary
- TAG
triacylglycerol
- N
nitrogen
- P
phosphorus
- DGAT
diacylglycerol acyltransferase
- Pi
inorganic phosphate
- RNA-Seq
RNA sequencing
- qPCR
quantitative real-time PCR
- SQDG
sulfoquinovosyldiacylglycerol
- P1BS
PHR1-binding site
- FT-IR
Fourier transform infrared
- TEM
transmission electron microscopy
- cDNA
complementary DNA
- TAP
Tris-acetate-phosphate
- DMSO
dimethyl sulfoxide
- CTAB
cetyl trimethyl ammonium bromide
Footnotes
This work was supported by the Carbon Trust (grant no. 082 to J.K.P.), the Leverhulme Trust (grant no. F/00 120/BG to J.K.P.), the Government of India (Ph.D. studentship to A.K.B.), and the Wellcome Trust (equipment grant support to the electron microscopy facility).
Articles can be viewed without a subscription.
References
- Aksoy M, Pootakham W, Pollock SV, Moseley JL, González-Ballester D, Grossman AR (2013) Tiered regulation of sulfur deprivation responses in Chlamydomonas reinhardtii and identification of an associated regulatory factor. Plant Physiol 162: 195–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajhaiya AK, Dean AP, Driver T, Trivedi DK, Rattray NJW, Allwood JW, Goodacre R, Pittman JK (2016) High-throughput metabolic screening of microalgae genetic variation in response to nutrient limitation. Metabolomics 12: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker EJ, Liggit P (1993) Accelerated poly(A) loss and mRNA stabilization are independent effects of protein synthesis inhibition on α-tubulin mRNA in Chlamydomonas. Nucleic Acids Res 21: 2237–2246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ball S, Marianne T, Dirick L, Fresnoy M, Delrue B, Decq A (1991) A Chlamydomonas reinhardtii low-starch mutant is defective for 3-phosphoglycerate activation and orthophosphate inhibition of ADP-glucose pyrophosphorylase. Planta 185: 17–26 [DOI] [PubMed] [Google Scholar]
- Ball SG, Morell MK (2003) From bacterial glycogen to starch: understanding the biogenesis of the plant starch granule. Annu Rev Plant Biol 54: 207–233 [DOI] [PubMed] [Google Scholar]
- Blaby IK, Glaesener AG, Mettler T, Fitz-Gibbon ST, Gallaher SD, Liu B, Boyle NR, Kropat J, Stitt M, Johnson S, et al. (2013) Systems-level analysis of nitrogen starvation-induced modifications of carbon metabolism in a Chlamydomonas reinhardtii starchless mutant. Plant Cell 25: 4305–4323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bölling C, Fiehn O (2005) Metabolite profiling of Chlamydomonas reinhardtii under nutrient deprivation. Plant Physiol 139: 1995–2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle NR, Page MD, Liu B, Blaby IK, Casero D, Kropat J, Cokus SJ, Hong-Hermesdorf A, Shaw J, Karpowicz SJ, et al. (2012) Three acyltransferases and nitrogen-responsive regulator are implicated in nitrogen starvation-induced triacylglycerol accumulation in Chlamydomonas. J Biol Chem 287: 15811–15825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boynton JE, Gillham NW (1993) Chloroplast transformation in Chlamydomonas. Methods Enzymol 217: 510–536 [DOI] [PubMed] [Google Scholar]
- Brányiková I, Maršálková B, Doucha J, Brányik T, Bišová K, Zachleder V, Vítová M (2011) Microalgae: novel highly efficient starch producers. Biotechnol Bioeng 108: 766–776 [DOI] [PubMed] [Google Scholar]
- Cernac A, Benning C (2004) WRINKLED1 encodes an AP2/EREB domain protein involved in the control of storage compound biosynthesis in Arabidopsis. Plant J 40: 575–585 [DOI] [PubMed] [Google Scholar]
- Chiou TJ, Lin SI (2011) Signaling network in sensing phosphate availability in plants. Annu Rev Plant Biol 62: 185–206 [DOI] [PubMed] [Google Scholar]
- Daboussi F, Leduc S, Maréchal A, Dubois G, Guyot V, Perez-Michaut C, Amato A, Falciatore A, Juillerat A, Beurdeley M, et al. (2014) Genome engineering empowers the diatom Phaeodactylum tricornutum for biotechnology. Nat Commun 5: 3831. [DOI] [PubMed] [Google Scholar]
- Dauvillée D, Chochois V, Steup M, Haebel S, Eckermann N, Ritte G, Ral JP, Colleoni C, Hicks G, Wattebled F, et al. (2006) Plastidial phosphorylase is required for normal starch synthesis in Chlamydomonas reinhardtii. Plant J 48: 274–285 [DOI] [PubMed] [Google Scholar]
- Dean AP, Sigee DC (2006) Molecular heterogeneity in Aphanizomenon flos-aquae and Anabaena flos-aquae (Cyanophyta): a synchrotron-based Fourier-transform infrared study of lake micropopulations. Eur J Phycol 41: 201–212 [Google Scholar]
- Dean AP, Sigee DC, Estrada B, Pittman JK (2010) Using FTIR spectroscopy for rapid determination of lipid accumulation in response to nitrogen limitation in freshwater microalgae. Bioresour Technol 101: 4499–4507 [DOI] [PubMed] [Google Scholar]
- Driver T, Bajhaiya A, Pittman JK (2014) Potential of bioenergy production from microalgae. Current Sustainable/Renewable Energy Reports 1: 94–103 [Google Scholar]
- Dyhrman ST, Jenkins BD, Rynearson TA, Saito MA, Mercier ML, Alexander H, Whitney LP, Drzewianowski A, Bulygin VV, Bertrand EM, et al. (2012) The transcriptome and proteome of the diatom Thalassiosira pseudonana reveal a diverse phosphorus stress response. PLoS ONE 7: e33768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis DI, Broadhurst D, Kell DB, Rowland JJ, Goodacre R (2002) Rapid and quantitative detection of the microbial spoilage of meat by Fourier transform infrared spectroscopy and machine learning. Appl Environ Microbiol 68: 2822–2828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emery L, Whelan S, Hirschi KD, Pittman JK (2012) Protein phylogenetic analysis of Ca2+/cation antiporters and insights into their evolution in plants. Front Plant Sci 3: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu FF, Xue HW (2010) Coexpression analysis identifies Rice Starch Regulator1, a rice AP2/EREBP family transcription factor, as a novel rice starch biosynthesis regulator. Plant Physiol 154: 927–938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gargouri M, Park JJ, Holguin FO, Kim MJ, Wang H, Deshpande RR, Shachar-Hill Y, Hicks LM, Gang DR (2015) Identification of regulatory network hubs that control lipid metabolism in Chlamydomonas reinhardtii. J Exp Bot 66: 4551–4566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgianna DR, Mayfield SP (2012) Exploiting diversity and synthetic biology for the production of algal biofuels. Nature 488: 329–335 [DOI] [PubMed] [Google Scholar]
- Giordano M, Kansiz M, Heraud P, Beardall J, Wood B, McNaughton D (2001) Fourier transform infrared spectroscopy as a novel tool to investigate changes in intracellular macromolecular pools in the marine microalga Chaetoceros muellerii (Bacillariophyceae). J Phycol 37: 271–279 [Google Scholar]
- González-Ballester D, Casero D, Cokus S, Pellegrini M, Merchant SS, Grossman AR (2010) RNA-seq analysis of sulfur-deprived Chlamydomonas cells reveals aspects of acclimation critical for cell survival. Plant Cell 22: 2058–2084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodenough U, Blaby I, Casero D, Gallaher SD, Goodson C, Johnson S, Lee JH, Merchant SS, Pellegrini M, Roth R, et al. (2014) The path to triacylglyceride obesity in the sta6 strain of Chlamydomonas reinhardtii. Eukaryot Cell 13: 591–613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodson C, Roth R, Wang ZT, Goodenough U (2011) Structural correlates of cytoplasmic and chloroplast lipid body synthesis in Chlamydomonas reinhardtii and stimulation of lipid body production with acetate boost. Eukaryot Cell 10: 1592–1606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant AJ, Rémond M, Starke-Peterkovic T, Hinde R (2006) A cell signal from the coral Plesiastrea versipora reduces starch synthesis in its symbiotic alga, Symbiodinium sp. Comp Biochem Physiol A Mol Integr Physiol 144: 458–463 [DOI] [PubMed] [Google Scholar]
- Grotewold E. (2008) Transcription factors for predictive plant metabolic engineering: are we there yet? Curr Opin Biotechnol 19: 138–144 [DOI] [PubMed] [Google Scholar]
- Guedes AC, Amaro HM, Malcata FX (2011) Microalgae as sources of high added-value compounds: a brief review of recent work. Biotechnol Prog 27: 597–613 [DOI] [PubMed] [Google Scholar]
- Harris EH. (1989) The Chlamydomonas Sourcebook. Academic Press, San Diego [Google Scholar]
- Higo K, Ugawa Y, Iwamoto M, Korenaga T (1999) Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res 27: 297–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill J, Nelson E, Tilman D, Polasky S, Tiffany D (2006) Environmental, economic, and energetic costs and benefits of biodiesel and ethanol biofuels. Proc Natl Acad Sci USA 103: 11206–11210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillebrand H, Durselen CD, Kirschtel D, Pollingher U, Zohary T (1999) Biovolume calculation for pelagic and benthic microalgae. J Phycol 35: 403–424 [Google Scholar]
- Hu Q, Sommerfeld M, Jarvis E, Ghirardi M, Posewitz M, Seibert M, Darzins A (2008) Microalgal triacylglycerols as feedstocks for biofuel production: perspectives and advances. Plant J 54: 621–639 [DOI] [PubMed] [Google Scholar]
- Hudson JJ, Taylor WD, Schindler DW (2000) Phosphate concentrations in lakes. Nature 406: 54–56 [DOI] [PubMed] [Google Scholar]
- Jiménez C, Capasso JM, Edelstein CL, Rivard CJ, Lucia S, Breusegem S, Berl T, Segovia M (2009) Different ways to die: cell death modes of the unicellular chlorophyte Dunaliella viridis exposed to various environmental stresses are mediated by the caspase-like activity DEVDase. J Exp Bot 60: 815–828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson X, Alric J (2013) Central carbon metabolism and electron transport in Chlamydomonas reinhardtii: metabolic constraints for carbon partitioning between oil and starch. Eukaryot Cell 12: 776–793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Russa M, Bogen C, Uhmeyer A, Doebbe A, Filippone E, Kruse O, Mussgnug JH (2012) Functional analysis of three type-2 DGAT homologue genes for triacylglycerol production in the green microalga Chlamydomonas reinhardtii. J Biotechnol 162: 13–20 [DOI] [PubMed] [Google Scholar]
- Martens H, Stark E (1991) Extended multiplicative signal correction and spectral interference subtraction: new preprocessing methods for near infrared spectroscopy. J Pharm Biomed Anal 9: 625–635 [DOI] [PubMed] [Google Scholar]
- McAinsh MR, Pittman JK (2009) Shaping the calcium signature. New Phytol 181: 275–294 [DOI] [PubMed] [Google Scholar]
- Merchant SS, Kropat J, Liu B, Shaw J, Warakanont J (2012) TAG, you’re it! Chlamydomonas as a reference organism for understanding algal triacylglycerol accumulation. Curr Opin Biotechnol 23: 352–363 [DOI] [PubMed] [Google Scholar]
- Miller R, Wu G, Deshpande RR, Vieler A, Gärtner K, Li X, Moellering ER, Zäuner S, Cornish AJ, Liu B, et al. (2010) Changes in transcript abundance in Chlamydomonas reinhardtii following nitrogen deprivation predict diversion of metabolism. Plant Physiol 154: 1737–1752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura K, Rus A, Sharkhuu A, Yokoi S, Karthikeyan AS, Raghothama KG, Baek D, Koo YD, Jin JB, Bressan RA, et al. (2005) The Arabidopsis SUMO E3 ligase SIZ1 controls phosphate deficiency responses. Proc Natl Acad Sci USA 102: 7760–7765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moseley JL, Chang CW, Grossman AR (2006) Genome-based approaches to understanding phosphorus deprivation responses and PSR1 control in Chlamydomonas reinhardtii. Eukaryot Cell 5: 26–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moseley JL, Grossman AR (2009) Phosphorus limitation from the physiological to the genomic. In Stern DB, ed, The Chlamydomonas Sourcebook: Organellar and Metabolic Processes, Vol 2 Academic Press, San Diego, pp 189–215 [Google Scholar]
- Murdock JN, Wetzel DL (2009) FT-IR microspectroscopy enhances biological and ecological analysis of algae. Appl Spectrosc Rev 44: 335–361 [Google Scholar]
- Ngan CY, Wong CH, Choi C, Yoshinaga Y, Louie K, Jia J, Chen C, Bowen B, Cheng H, Leonelli L, et al. (2015) Lineage-specific chromatin signatures reveal a regulator of lipid metabolism in microalgae. Nature Plants 1: 15107. [DOI] [PubMed] [Google Scholar]
- Nilsson L, Müller R, Nielsen TH (2007) Increased expression of the MYB-related transcription factor, PHR1, leads to enhanced phosphate uptake in Arabidopsis thaliana. Plant Cell Environ 30: 1499–1512 [DOI] [PubMed] [Google Scholar]
- Osundeko O, Davies H, Pittman JK (2013) Oxidative stress-tolerant microalgae strains are highly efficient for biofuel feedstock production on wastewater. Biomass Bioenergy 56: 284–294 [Google Scholar]
- Peltier G, Schmidt GW (1991) Chlororespiration: an adaptation to nitrogen deficiency in Chlamydomonas reinhardtii. Proc Natl Acad Sci USA 88: 4791–4795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Pérez ME, Florencio FJ, Crespo JL (2010) Inhibition of target of rapamycin signaling and stress activate autophagy in Chlamydomonas reinhardtii. Plant Physiol 152: 1874–1888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl MW. (2001) A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29: e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pittman JK, Dean AP, Osundeko O (2011) The potential of sustainable algal biofuel production using wastewater resources. Bioresour Technol 102: 17–25 [DOI] [PubMed] [Google Scholar]
- Pittman JK, Edmond C, Sunderland PA, Bray CM (2009) A cation-regulated and proton gradient-dependent cation transporter from Chlamydomonas reinhardtii has a role in calcium and sodium homeostasis. J Biol Chem 284: 525–533 [DOI] [PubMed] [Google Scholar]
- Plaxton WC, Tran HT (2011) Metabolic adaptations of phosphate-starved plants. Plant Physiol 156: 1006–1015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porra RJ, Thompson WA, Kriedemann PE (1989) Determination of accurate extinction coefficients and simultaneous-equations for assaying chlorophyll-a and chlorophyll-b extracted with 4 different solvents: verification of the concentration of chlorophyll standards by atomic-absorption spectroscopy. Biochim Biophys Acta 975: 384–394 [Google Scholar]
- Radakovits R, Jinkerson RE, Darzins A, Posewitz MC (2010) Genetic engineering of algae for enhanced biofuel production. Eukaryot Cell 9: 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghothama KG. (1999) Phosphate acquisition. Annu Rev Plant Physiol Plant Mol Biol 50: 665–693 [DOI] [PubMed] [Google Scholar]
- Riekhof WR, Ruckle ME, Lydic TA, Sears BB, Benning C (2003) The sulfolipids 2′-O-acyl-sulfoquinovosyldiacylglycerol and sulfoquinovosyldiacylglycerol are absent from a Chlamydomonas reinhardtii mutant deleted in SQD1. Plant Physiol 133: 864–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouached H, Arpat AB, Poirier Y (2010) Regulation of phosphate starvation responses in plants: signaling players and cross-talks. Mol Plant 3: 288–299 [DOI] [PubMed] [Google Scholar]
- Rubio V, Linhares F, Solano R, Martín AC, Iglesias J, Leyva A, Paz-Ares J (2001) A conserved MYB transcription factor involved in phosphate starvation signaling both in vascular plants and in unicellular algae. Genes Dev 15: 2122–2133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruijter JM, Ramakers C, Hoogaars WMH, Karlen Y, Bakker O, van den Hoff MJB, Moorman AFM (2009) Amplification efficiency: linking baseline and bias in the analysis of quantitative PCR data. Nucleic Acids Res 37: e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmollinger S, Mühlhaus T, Boyle NR, Blaby IK, Casero D, Mettler T, Moseley JL, Kropat J, Sommer F, Strenkert D, et al. (2014) Nitrogen-sparing mechanisms in Chlamydomonas affect the transcriptome, the proteome, and photosynthetic metabolism. Plant Cell 26: 1410–1435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroda M, Blöcker D, Beck CF (2000) The HSP70A promoter as a tool for the improved expression of transgenes in Chlamydomonas. Plant J 21: 121–131 [DOI] [PubMed] [Google Scholar]
- Schroda M, Vallon O, Wollman FA, Beck CF (1999) A chloroplast-targeted heat shock protein 70 (HSP70) contributes to the photoprotection and repair of photosystem II during and after photoinhibition. Plant Cell 11: 1165–1178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimogawara K, Wykoff DD, Usuda H, Grossman AR (1999) Chlamydomonas reinhardtii mutants abnormal in their responses to phosphorus deprivation. Plant Physiol 120: 685–694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siaut M, Cuiné S, Cagnon C, Fessler B, Nguyen M, Carrier P, Beyly A, Beisson F, Triantaphylidès C, Li-Beisson Y, et al. (2011) Oil accumulation in the model green alga Chlamydomonas reinhardtii: characterization, variability between common laboratory strains and relationship with starch reserves. BMC Biotechnol 11: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skjånes K, Lindblad P, Muller J (2007) BioCO2: a multidisciplinary, biological approach using solar energy to capture CO2 while producing H2 and high value products. Biomol Eng 24: 405–413 [DOI] [PubMed] [Google Scholar]
- Sobkowiak L, Bielewicz D, Malecka EM, Jakobsen I, Albrechtsen M, Szweykowska-Kulinska Z, Pacak A (2012) The role of the P1BS element containing promoter-driven genes in Pi transport and homeostasis in plants. Front Plant Sci 3: 58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trentacoste EM, Shrestha RP, Smith SR, Glé C, Hartmann AC, Hildebrand M, Gerwick WH (2013) Metabolic engineering of lipid catabolism increases microalgal lipid accumulation without compromising growth. Proc Natl Acad Sci USA 110: 19748–19753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JC, Xu H, Zhu Y, Liu QQ, Cai XL (2013) OsbZIP58, a basic leucine zipper transcription factor, regulates starch biosynthesis in rice endosperm. J Exp Bot 64: 3453–3466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Work VH, Radakovits R, Jinkerson RE, Meuser JE, Elliott LG, Vinyard DJ, Laurens LML, Dismukes GC, Posewitz MC (2010) Increased lipid accumulation in the Chlamydomonas reinhardtii sta7-10 starchless isoamylase mutant and increased carbohydrate synthesis in complemented strains. Eukaryot Cell 9: 1251–1261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wykoff DD, Davies JP, Melis A, Grossman AR (1998) The regulation of photosynthetic electron transport during nutrient deprivation in Chlamydomonas reinhardtii. Plant Physiol 117: 129–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wykoff DD, Grossman AR, Weeks DP, Usuda H, Shimogawara K (1999) Psr1, a nuclear localized protein that regulates phosphorus metabolism in Chlamydomonas. Proc Natl Acad Sci USA 96: 15336–15341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yehudai-Resheff S, Zimmer SL, Komine Y, Stern DB (2007) Integration of chloroplast nucleic acid metabolism into the phosphate deprivation response in Chlamydomonas reinhardtii. Plant Cell 19: 1023–1038 [DOI] [PMC free article] [PubMed] [Google Scholar]