Rice lines with enhanced ADPglc synthesis and import into amyloplasts reveal additional barriers within the stroma that restrict maximum carbon flow into starch.
Abstract
Previous studies showed that efforts to further elevate starch synthesis in rice (Oryza sativa) seeds overproducing ADP-glucose (ADPglc) were prevented by processes downstream of ADPglc synthesis. Here, we identified the major ADPglc transporter by studying the shrunken3 locus of the EM1093 rice line, which harbors a mutation in the BRITTLE1 (BT1) adenylate transporter (OsBt1) gene. Despite containing elevated ADPglc levels (approximately 10-fold) compared with the wild-type, EM1093 grains are small and shriveled due to the reduction in the amounts and size of starch granules. Increases in ADPglc levels in EM1093 were due to their poor uptake of ADP-[14C]glc by amyloplasts. To assess the potential role of BT1 as a rate-determining step in starch biosynthesis, the maize ZmBt1 gene was overexpressed in the wild-type and the GlgC (CS8) transgenic line expressing a bacterial glgC-TM gene. ADPglc transport assays indicated that transgenic lines expressing ZmBT1 alone or combined with GlgC exhibited higher rates of transport (approximately 2-fold), with the GlgC (CS8) and GlgC/ZmBT1 (CS8/AT5) lines showing elevated ADPglc levels in amyloplasts. These increases, however, did not lead to further enhancement in seed weights even when these plant lines were grown under elevated CO2. Overall, our results indicate that rice lines with enhanced ADPglc synthesis and import into amyloplasts reveal additional barriers within the stroma that restrict maximum carbon flow into starch.
Cereal grains contribute a significant portion of worldwide starch production. Unlike other plant tissue, starch biosynthesis in the endosperm storage organ of cereal grains is unique in its dependence on two ADP-Glc pyrophosphorylase (AGPase) isoforms (Denyer et al., 1996; Thorbjørnsen et al., 1996; Sikka et al., 2001), a major cytosolic enzyme and a minor plastidial one, to generate ADP-glucose (ADPglc), the sugar nucleotide utilized by starch synthases in the amyloplast (Cakir et al., 2015). The majority of ADPglc in cereal endosperm is generated in the cytosol from AGPase (Tuncel and Okita, 2013) as well as by Suc synthase (Tuncel and Okita, 2013; Bahaji et al., 2014) and subsequently transported into amyloplasts by the BRITTLE-1 (BT1) protein located at the plastid envelope (Cao et al., 1995; Shannon et al., 1998).
The Bt1 gene, first identified in maize (Zea mays; Mangelsdorf, 1926) and isolated by Sullivan et al. (1991), encodes a major amyloplast membrane protein ranging from 39 to 44 kD (Cao et al., 1995). The BT1 protein and its homologs belong to the mitochondrial carrier family (Sullivan et al., 1991; Haferkamp, 2007), which has a diverse range of substrates (Patron et al., 2004; Leroch et al., 2005; Kirchberger et al., 2008). The assignment of BT1 protein as the ADPglc transporter in cereal endosperms was first proposed by Sullivan et al. (1991), and then it was characterized based on the increased ADPglc levels and reduced ADPglc import rate in endosperms of BT1-deficient maize and barley (Hordeum vulgare) mutants (Tobias et al., 1992; Shannon et al., 1996, 1998; Patron et al., 2004). Biochemical transport studies of the maize BT1 showed that it imported ADPglc by counter exchanging with ADP (Kirchberger et al., 2007). The wheat (Triticum aestivum) BT1 homolog also transports ADPglc but has similar affinities for ADP and AMP as the counter-exchange substrate (Bowsher et al., 2007).
Evidence from previous studies by our laboratory (Sakulsingharoj et al., 2004; Nagai et al., 2009) suggested the potential role of BT1 as well as other downstream processes as a rate-limiting step in starch biosynthesis in the transgenic rice (Oryza sativa) GlgC (CS8) lines overexpressing an up-regulated AGPase (Escherichia coli glgC-TM). In GlgC (CS8) rice lines, grain weights (starch) are elevated up to 15% compared with wild-type plants, indicating that the AGPase-catalyzed reaction is a rate-limiting step in starch biosynthesis under normal conditions. When transgenic GlgC (CS8) plants were grown under elevated CO2 levels, no further increases in grain weight were evident compared with those grown at ambient CO2. As Suc levels are elevated in leaf blades, leaf sheaths, culms (Rowland-Bamford et al., 1990), and peduncle exudates (Chen et al., 1994) in rice plants grown under elevated CO2, developing GlgC (CS8) grains were unable to convert the increased levels of sugars into starch. This lack of increase indicated that the AGPase-catalyzed reaction (ADPglc synthesis) was no longer rate limiting and that one or more downstream processes regulated carbon flux from source tissues in developing GlgC (CS8) endosperm (Sakulsingharoj et al., 2004). This view is also supported by a subsequent metabolite study in which several GlgC (CS8) lines were found to contain up to 46% higher ADPglc levels than wild-type plants (Nagai et al., 2009). As this increase in ADPglc levels was nearly 3-fold higher than the increase in grain weight, starch biosynthesis is saturated with respect to ADPglc levels and carbon flow into starch is restricted by one or more downstream steps. Potential events that may limit the utilization of ADPglc in starch in GlgC (CS8) lines are the import of this sugar nucleotide via the BT1 transporter into amyloplasts and/or the utilization of ADPglc by starch synthases. Mutant analysis of the two major starch synthases indicated no significant impact on grain weight when one of these starch synthases was nonfunctional, suggesting that this enzyme activity, contributed by multiple enzyme isoforms, is present at excessive levels (Fujita et al., 2006, 2007). Therefore, we suspected that BT1 is the likely candidate limiting carbon flow into starch in GlgC (CS8) endosperms.
The aim of this study was to investigate the role of BT1 in mediating the transport of ADPglc into amyloplast and to determine whether this transport activity is rate limiting in rice endosperm. In order to address these questions, we show that BT1 is the major transporter of ADPglc by analysis of the EM1093 rice line, which contains a mutation at the shrunken3 (shr3) locus and, specifically, in the OsBt1-1 gene. Second, we assessed the impact of the expression of the maize ZmBt1 gene in wild-type and GlgC (CS8) seeds to determine the potential limiting role of BT1 transport activity on starch biosynthesis. Our results indicate that BT1 is essential for starch synthesis but is not rate limiting and that one or more stroma-localized processes limit maximum carbon flow into starch.
RESULTS
Genetic and Biochemical Characterization of the shr3 Mutant (EM1093)
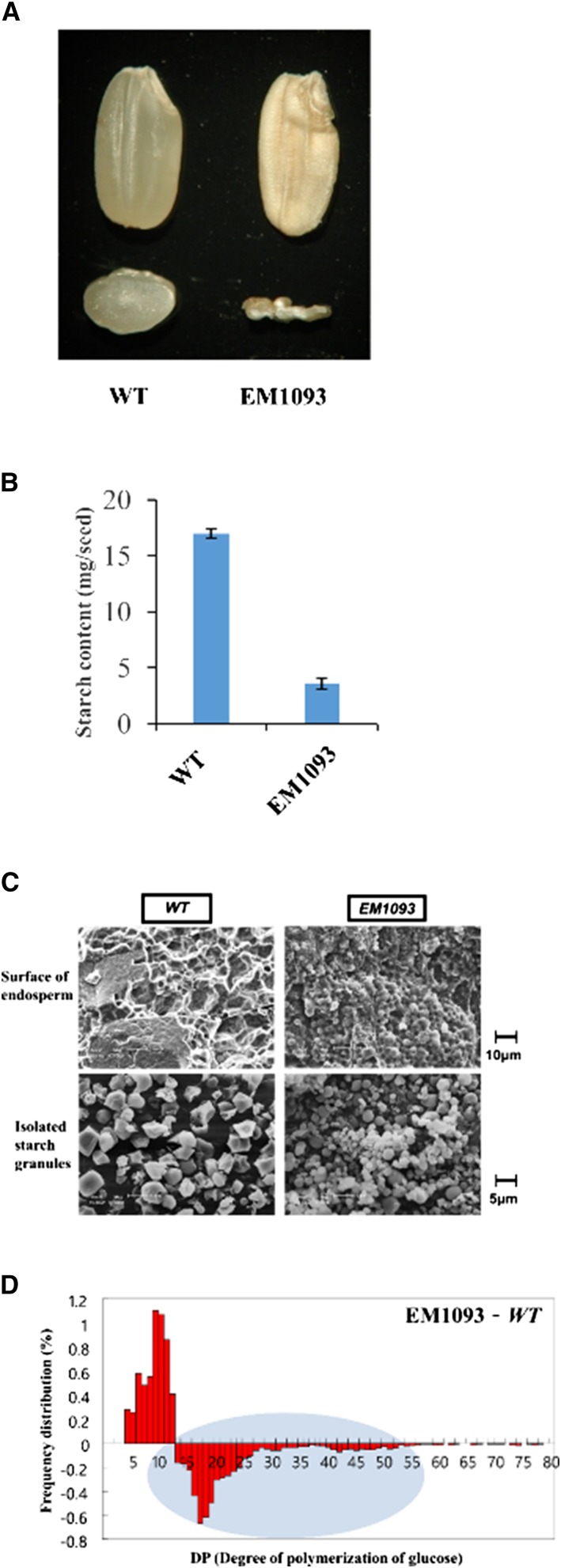
EM1093 plants (shr3) produce seeds that have a shriveled phenotype due to the severe reduction in starch content (Fig. 1A). This phenotype is likely caused by a defect in one of the major enzymes involved in starch biosynthesis (Kawagoe et al., 2005; Tuncel et al., 2014). Starch levels were significantly decreased around 80% in shr3 seeds compared with the wild-type (Fig. 1B). The morphology of starch granules and the chain length distribution of amylopectin in wild-type and shr3 mutant endosperms were investigated by scanning electron microscopy (SEM) and 8-amino-1,3,6-pyrenetrisulfonic capillary electrophoresis, respectively. SEM imaging showed that the starch granules in endosperms of shr3 were smaller and more spherical compared with those of the wild-type (Fig. 1C). Moreover, the shr3 mutation strongly affected the chain length distribution of amylopectin, as the proportion of chains with degree of polymerization less than 13 were elevated, whereas those of chains with degree of polymerization greater than 14 were decreased (Fig. 1D). These results clearly indicate that mutation in the shr3 locus results in depressing starch content as well as altering starch granule morphology and amylopectin structure.
Figure 1.
A and B, Seed morphology (A) and starch content (B) of the wild-type (WT; cv TC65) and EM1093. C, Scanning electron micrographs of isolated starch granules and the surface of wild-type and EM1093 endosperms. D, Effects of the shr3 mutation on the chain length distribution of amylopectin from EM1093 compared with the wild-type. The chain length distributions were characterized using 8-amino-1,3,6-pyrenetrisulfonic capillary electrophoresis. The data are representative of three different experiments Average starch contents were statistically different based on one-way ANOVA with Tukey’s multiple comparison test (P < 0.05).
The shriveled grain phenotype of shr3 is similar to that seen for the mutant lines EM541 (shr1) and EM22 (shr2), which also produce shrunken seeds due to the lack of large (L2) and small (S2b) subunits, respectively, of the endosperm cytosolic AGPase (Kawagoe et al., 2005; Tuncel et al., 2014). To determine whether EM1093 contained a mutation in one of the structural genes for the major cytoplasmic AGPase, the EM1093 line was crossed with EM541 (Tuncel et al., 2014) or EM22 (Kawagoe et al., 2005), marker lines for the shr1 and shr2 loci, respectively. F1 seeds derived from both the EM1093 × EM541 and EM1093 × EM22 crosses were normal in appearance, whereas F2 seeds demonstrated a ratio of wild-type to shrunken phenotype that fitted well to the expected 9:7 ratio of independent gene inheritance (Table I). This F2 segregation pattern verifies that the shr3 gene locus of EM1093 is independent of both shr1 and shr2 loci. In addition, EM1093 was also crossed with the wild-type parental cv Taichung 65 (TC65). F1 seeds exhibited a wild-type phenotype, while the segregation mode of normal to shrunken seeds fitted well to the expected ratio of a single inheritance, 3:1 (105:31), in the F2 progeny (Table I). These results indicated that the shr3 mutation in EM1093 is controlled by a single gene locus and is unrelated to the shr1 and shr2 loci.
Table I. Segregation analysis of F2 seeds derived from crosses of EM1093 with EM540 and EM22 and of EM1093 with wild-type cv TC65 lines.
χ2 values are based on inheritance ratios of 9:7 for EM crosses and 3:1 for wild-type and EM crosses.
| Cross Combination | F1 Seeds | Segregation in F2 |
Total | χ2 | |
|---|---|---|---|---|---|
| Wild-Type | shr | ||||
| EM540 (shr1) × EM1093 | Normal | 199 | 148 | 347 | 0.17 |
| EM22 (shr2) × EM1093 | Normal | 168 | 142 | 310 | 0.54 |
| Wild-type (cv TC65) × EM1093 | Normal | 105 | 31 | 136 | 0.35 |
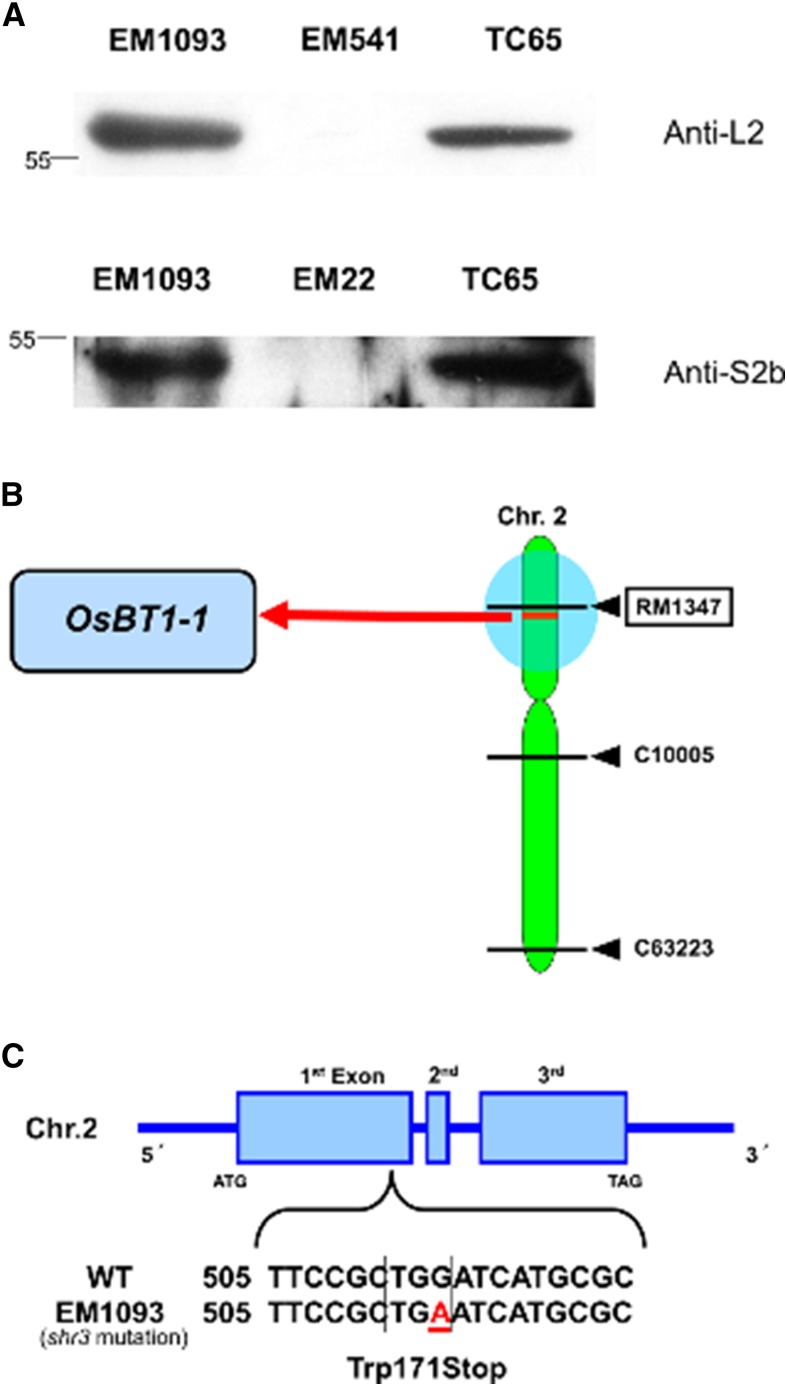
To further verify that the shr3 locus was not an AGPase structural gene, immunoblot studies were conducted. Total proteins, extracted from developing seeds of EM1093, were subjected to immunoblot analysis using anti-S2b (AGPase small subunit) and anti-L2 (AGPase large subunit) antibodies. The results demonstrated that the EM1093 line contains both S2b and L2 proteins (Fig. 2A), suggesting that the shriveled phenotype is likely due to one of the other major starch biosynthetic enzymes.
Figure 2.
Characterization of the shr3 mutant (EM1093). A, Immunoblot analysis of proteins extracted from developing (14-DAF) seeds of the wild-type (cv TC65) and endosperm mutant (EM) lines. EM1093 contains both AGPase subunits, as assessed by immunoblotting using antibodies raised against AGPase L2 and S2b. B, Linkage map of chromosome 2. shr3 maps near the marker RM1347, where OsBt1-1 closely resides. C, Location of the nonsense mutation in the OsBT1-1 gene. A G-to-A mutation at the first exon results in a premature termination codon in EM1093. WT, Wild-type.
In order to identify the proximate chromosome location of shr3, linkage analysis was performed by crossing EM1093 (japonica) with the indica rice cv Kasalath. Mapping studies showed that the shr3 gene cosegregated with three markers, RM1347, C10005, and C63223, on chromosome 2 (Fig. 2B; Supplemental Table S1). The F2 population derived from the cross between rice cv Kasalath and EM1093 indicated that the shr3 mutation cosegregated with the RM1347 marker (Supplemental Table S1). This result clearly demonstrated that the shr3 locus is located on the short arm of chromosome 2 (Fig. 2B).
Mutation in the Bt1 Gene Is Responsible for shr3
Analysis of the rice genomic map for genes located near the RM1347 marker implicated the OsBt1-1 as a potential candidate gene. A mutation in the OsBt1-1 gene, which was initially determined by targeting induced local lesions in genomes (TILLING; Suzuki et al., 2008), was verified by DNA sequencing of OsBt1-1 genomic DNA (GenBank accession no. NM_001052765). In EM1093, a G-to-A replacement at position 513 in the first exon was detected, resulting in a Trp codon being converted to a nonsense codon (Trp-171Stop; Fig. 2C). Such an error generates a truncated polypeptide (170 amino acids) that would be subject to rapid degradation in the cell (Laughlin et al., 1998) or cause nonsense-mediated mRNA decay (Isshiki et al., 2001). In either instance, loss of intact BT1 polypeptide was expected.
To verify the loss of the BT1 polypeptide, amyloplast membrane fractions were prepared from developing wild-type and EM1093 seeds, and extracted proteins were resolved by SDS-PAGE. Polyacrylamide gel slices containing polypeptide in the vicinity of 41 kD, the size of OsBT1, were treated with trypsin, and eluted peptides were analyzed by high-resolution liquid chromatography-tandem mass spectrometry (LC-MS/MS). This analysis showed the presence of unique peptide sequences spanning 45% coverage of the intact BT1 protein in wild-type membranes. By contrast, no BT1 peptides were detected from EM1093 (Table II), indicating that amyloplasts of EM1093 lack intact BT1 protein (Table II). Overall, the results from crossing experiments, proteomic analysis, and DNA sequencing verify that the mutation at the shr3 locus is due to a disruption in the OsBt1-1 gene and a loss of BT1 protein in EM1093.
Table II. Identification of proteins from the wild-type and EM1093.
Seed extracts from the wild-type and EM1093 were resolved by SDS-PAGE and the polyacrylamide gel area containing 41-kD polypeptides treated with trypsin and resulting peptides analyzed by LC-MS/MS. BT1 peptides were detected only in the wild-type and not in EM1093. Matched peptide values indicate the number of unique peptides matched. Values in parentheses signify the percentage of sequence coverage by peptide mass fingerprinting (PMF) using LC-MS/MS. ND, Not detected.
| Identifier | Protein Name | Matched Peptides |
|
|---|---|---|---|
| Wild-Type | EM1093 | ||
| LOC_Os02g10800 | ADPglc transporter (BT1) | 7 (45) | ND |
| LOC_Os10g26060 | Glutelin | 2 (16) | 3 (21) |
| LOC_Os01g64700 | Tubby-like F-box protein3 | 1 (9) | 1 (12) |
| LOC_Os05g33570 | Pyruvate, phosphate dikinase1 | 1 (11) | 1 (13) |
| LOC_Os07g46460 | Ferredoxin-dependent Glu synthase | 1 (10) | 1 (11) |
Plastids Isolated from shr3 Endosperm Fail to Efficiently Transport ADPglc
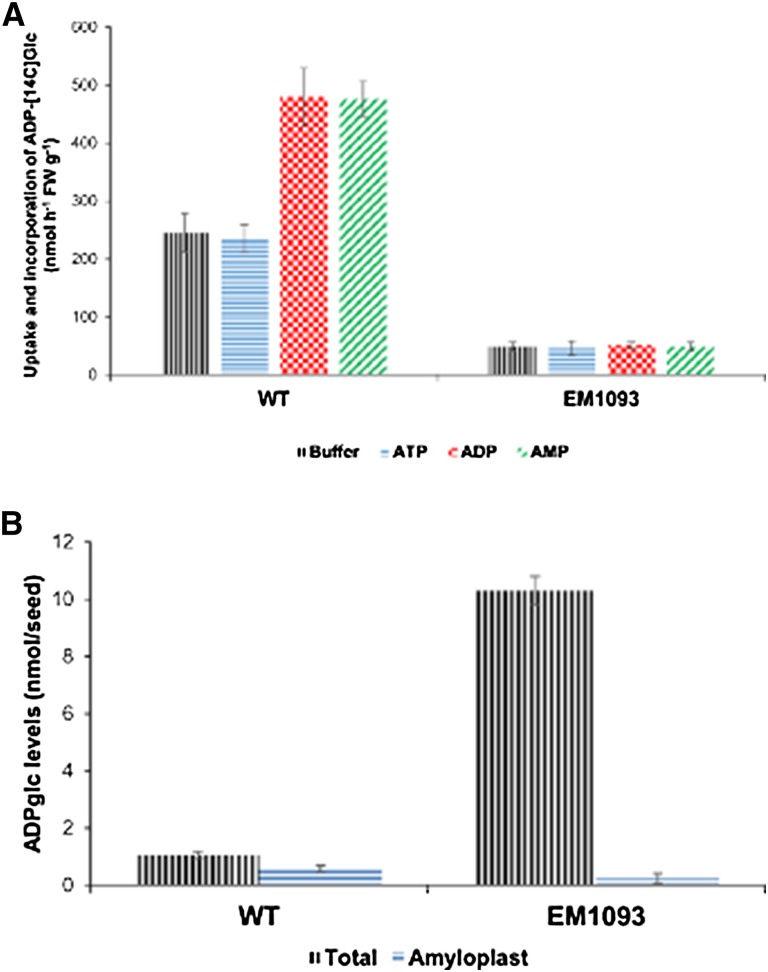
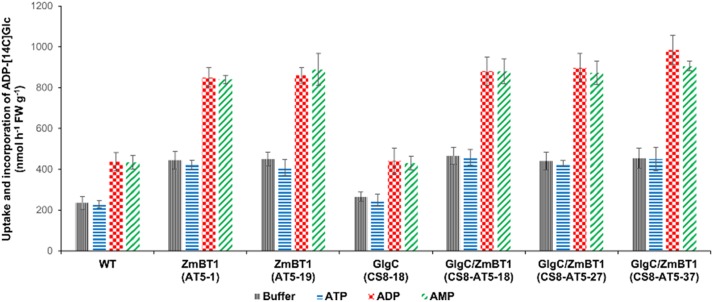
Purified amyloplasts from the wild-type and EM1093 were tested for their transport of ADP-[14C]glc. An approximate ADPglc import rate was determined by measuring the uptake of ADP-[14C]glc and incorporation into starch (Table III; Fig. 3A). Significantly higher levels of ADP-[14C]Glc (approximately 245 nmol g−1 fresh weight) were transported and incorporated into wild-type (cv Kitaake and cv TC65) amyloplasts compared with EM1093 (shr3) endosperms (49 nmol g−1 fresh weight) in the absence of any counter-exchange adenylate nucleotides (Table III; Fig. 3A). ADPglc import rates nearly doubled when wild-type amyloplasts were preincubated with either ADP or AMP, whereas EM1093 amyloplasts failed to exhibit such an increase in ADPglc import (Table III; Fig. 3A). ATP had no influence on ADPglc transport (Fig. 3A), a result similar to those from an earlier study of the wheat BT1 (Bowsher et al., 2007). Overall, these results showed that the loss of BT1 protein in EM1093 results in poor ADPglc import and incorporation into starch by intact amyloplasts and, in turn, lower starch content (Figs. 1 and 3A; Table III).
Table III. Uptake and incorporation of ADP-[14C]glc into starch by purified intact amyloplasts in the presence or absence of the counter-exchange ADP.
Total starch synthase activity was estimated by lysing the amyloplasts and measuring the amount of incorporation of ADP-[14C]glc into starch. Each value represents the mean ± se of three independent experiments.
| Lines | Intact |
Starch Synthase Activity |
||
|---|---|---|---|---|
| Buffer | +ADP | Buffer | +ADP | |
| nmol h−1 g−1 fresh wt | ||||
| Wild-type (cv Kitaake) | 235 ± 33 | 437 ± 45 | 145 ± 13 | 140 ± 11 |
| ZmBT1 (AT5-1) | 445 ± 43 | 850 ± 47 | 153 ± 11 | 152 ± 10 |
| ZmBT1 (AT5-19) | 450 ± 34 | 860 ± 39 | 150 ± 9 | 154 ± 11 |
| GlgC (CS8) | 265 ± 23 | 440 ± 63 | 125 ± 7 | 120 ± 8 |
| GlgC/ZmBT1 (CS8/AT5-18) | 466 ± 41 | 881 ± 70 | 147 ± 12 | 141 ± 16 |
| GlgC/ZmBT1 (CS8/AT5-27) | 440 ± 44 | 897 ± 71 | 144 ± 11 | 154 ± 19 |
| GlgC/ZmBT1 (CS8/AT5-37) | 454 ± 50 | 985 ± 72 | 149 ± 13 | 139 ± 17 |
| Wild-type (cv TC65) | 245 ± 33 | 480 ± 49 | 125 ± 18 | 135 ± 15 |
| EM1093 (cv TC65) | 49 ± 7 | 51 ± 6 | 43 ± 5 | 41 ± 3 |
Figure 3.
ADPglc incorporation and levels in wild-type and EM1093 rice seeds. A, Uptake and incorporation of ADPglc into starch of purified amyloplasts from the wild-type (WT) and EM1093 in the presence or absence of adenylates. Amyloplasts were preloaded with buffer (black vertical lines), 20 mm ATP (blue horizontal lines), 20 mm ADP (red checkered lines), or 20 mm AMP (green crosswise lines) for 10 min. The reaction containing 100 µL of isolated amyloplasts was started by adding 4 mm ADP-[14C]glc. Uptake experiments were performed at 30°C for 30 min. FW, Fresh weight. B, ADPglc levels in wild-type and EM1093 seeds were determined by a reverse direction assay. Data represent means ± se of three independent experiments. Black vertical lines represent total ADPglc levels, and blue horizontal lines represent amyloplast ADPglc levels.
The shr3 Mutant Accumulates Higher Levels of ADPglc
The loss of ADPglc transport activity into amyloplast would likely elevate cytoplasmic ADPglc levels due to their active synthesis by AGPase (Tuncel and Okita, 2013) or by Suc synthase (Bahaji et al., 2014). Estimation of ADPglc levels from developing seed extracts showed that EM1093 contained dramatically increased (approximately 10-fold) ADPglc levels compared with the wild-type (Fig. 3B; Table IV). This result is consistent with an earlier study in maize (Shannon et al., 1996), where bt1 kernels contained 13-fold more ADPglc than normal kernels. Additionally, compartmentalization of ADPglc levels demonstrated that wild-type amyloplasts contained around 52% of the total ADPglc, whereas most of the ADPglc in EM1093 existed in the cytosol (Fig. 3B; Table IV). Thus, the loss of BT1 protein resulted in dramatically decreased ADPglc transport into amyloplasts and, in turn, higher ADPglc levels in cytosol of EM1093 seeds.
Table IV. ADPglc levels in total and amyloplast fractions of wild-type and transgenic lines.
Each value represents the mean ± se of three independent experiments.
| Lines | Total (nmol/seed) | Amyloplast | Percentage of Amyloplast |
|---|---|---|---|
| nmol seed−1 | |||
| Wild-type (cv Kitaake) | 0.87 ± 0.07 | 0.46 ± 0.02 | 52.9 |
| ZmBT1 (AT5-1) | 0.91 ± 0.06 | 0.45 ± 0.04 | 49.5 |
| ZmBT1 (AT5-19) | 0.89 ± 0.04 | 0.44 ± 0.02 | 49.5 |
| GlgC (CS8-18) | 1.17 ± 0.05 | 0.76 ± 0.02 | 65.0 |
| GlgC/ZmBT1 (CS8/AT5-18) | 1.15 ± 0.08 | 0.80 ± 0.02 | 69.6 |
| GlgC/ZmBT1 (CS8/AT5-27) | 1.19 ± 0.09 | 0.83 ± 0.04 | 69.7 |
| GlgC/ZmBT1 (CS8/AT5-37) | 1.16 ± 0.01 | 0.78 ± 0.03 | 67.2 |
| Wild-type (cv TC65) | 0.92 ± 0.09 | 0.47 ± 0.05 | 51.1 |
| EM1093 (cv TC65) | 10.3 ± 0.19 | 0.16 ± 0.03 | 1.6 |
Introduction of ZmBt1 into Wild-Type and GlgC (CS8) Transgenic Rice Lines
Results from previous studies suggested the possibility that the OsBt1 activity limited carbon flux into starch, especially in the starch-overaccumulating GlgC (CS8) rice lines expressing an up-regulated bacterial AGPase (Sakulsingharoj et al., 2004; Nagai et al., 2009). This potential rate-limiting role of the BT1 transporter could be tested directly by overexpressing the native BT1 transporter in wild-type and GlgC (CS8) transgenic rice lines. To verify the expression of native BT1 at the protein level, an antibody was required. Despite repeated attempts, however, we were unable to generate an antibody to the OsBT1 polypeptide or selected peptide fragments. A BT1 antibody was available to the maize ortholog, ZmBT1. Unfortunately, the antibody to the maize transporter did not cross act with the rice OsBT1 protein. In view of these circumstances, the maize ZmBt1 complementary DNA (cDNA) under the control of the Globulin promoter (Wu et al., 1998) was transformed via Agrobacterium tumefaciens into the various rice lines.
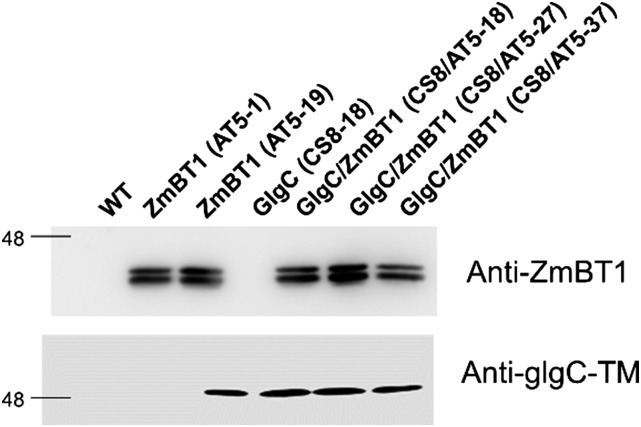
The generated transgenic plants were screened for the expression of both GlgC-TM (bacterial AGPase) and ZmBT1 proteins by immunoblot analysis using anti-GlgC and anti-ZmBT1 antibodies. Transgenic plants expressing the ZmBt1 cDNA (AT5 lines) alone or coexpressed with glgC-TM (CS8) during seed development were identified by immunoblotting (Fig. 4). All ZmBt1-expressing transgenic plants showed two polypeptide bands corresponding to ZmBT1 at approximately 41 kD (Shannon et al., 1998), whereas transgenic plants containing glgC-TM expressed a single band at 48 kD (Fig. 4). These transgenic lines were further propagated until homozygous T3 lines were identified in order to characterize the role of BT1 on starch biosynthesis.
Figure 4.
Immunoblot analysis of proteins extracted from developing (14-DAF) seeds of the wild-type (WT) and GlgC (CS8) and ZmBT1 (AT5) transgenic lines. CS8 lines, harboring the E. coli glgC-TM gene and AT5 lines, expressing the ZmBt1 gene, were analyzed using antibodies raised against GlgC-TM and ZmBT1.
Higher ADPglc Uptake Is Readily Detected in Amyloplasts Isolated from ZmBT1 (AT5) Transgenic Plants
Transgenic Lines Harboring ZmBt1 Exhibit Higher ADPglc Transport Capacity
Intact amyloplasts were isolated from developing rice seeds (12–15 d after pollination [DAP]) to investigate the role of the ZmBT1 transporter on ADPglc uptake capacity. Amyloplasts isolated from the transgenic lines containing ZmBt1 (AT5-1, CS8/AT5-18, CS8/AT5-27, and CS8/AT5-37) exhibited approximately 2-fold higher ADP-[14C]glc transport-incorporation rates than the wild-type and GlgC (CS8; Fig. 5; Table III). Irrespective of the rice line, preincubation of the intact amyloplasts with ADP stimulated ADP-[14C]glc transport-incorporation rates by nearly 2-fold. By contrast, ADP-[14C]glc incorporation into starch by ruptured amyloplasts from the various rice lines was considerably lower than the rates measured for intact amyloplasts. Moreover, the rates of ADP-[14C]glc incorporation by ruptured amyloplasts were not stimulated by ADP, unlike the rates observed for intact amyloplasts (Table III), and did not vary from one rice line to another. Thus, the observed increases in ADP-[14C]glc transport and incorporation into starch by intact amyloplasts from the various ZmBT1-expressing rice lines are due to enhanced transport into the amyloplast stromal compartment and not to differences in starch synthase activities. These results are consistent with the view that overexpression of ZmBT1 leads to an increase in ADPglc uptake across the amyloplast membrane.
Figure 5.
Uptake and incorporation of ADP-[14C]glc into starch by purified amyloplasts from wild-type (WT) and transgenic rice seeds. ADPglc import into amyloplasts is shown for the wild-type, GlgC (CS8), ZmBT1 (AT5), and GlgC/ZmBT1 (CS8/AT5) lines in the presence or absence of adenylates. Amyloplasts were preloaded with buffer (black vertical lines), 20 mm ATP (blue horizontal lines), 20 mm ADP (red checkered lines), or 20 mm AMP (green crosswise lines) for 10 min. Uptake experiments were performed at 30°C for 30 min. Data represent means ± se of three independent experiments. FW, Fresh weight.
ADPglc Levels in Transgenic Seeds
The contribution of elevated ADPglc transport capacity to ADPglc levels in transgenic rice seeds was investigated. Transgenic ZmBT1 (AT5) plants expressing the maize BT1 alone contained ADPglc levels similar to those observed for the wild-type. Consistent with our earlier results (Nagai et al., 2009), total ADPglc levels in GlgC (CS8) plants were elevated around 40% compared with the wild-type (Table IV). ADPglc levels were also increased in the double transgenic lines (GlgC/ZmBT1) to the same extent (approximately 40%) observed in GlgC (CS8) compared with the wild-type (Table IV). Even though ZmBT1 (AT5) lines exhibited higher in vitro transport rates of ADPglc (Fig. 5), these transgenic plants failed to accumulate increased ADPglc levels in amyloplasts compared with the wild-type (Table IV). In both rice lines, about 50% of the total ADPglc was evident in isolated amyloplasts. A much higher proportion (65%–69%) of total ADPglc was associated with the amyloplasts of GlgC (CS8) and GlgC/ZmBT1 (CS8/AT5) endosperms (Table IV).
Analysis of Amylose and Amylopectin Content from Wild-Type and Transgenic Endosperms
To test whether elevated ADPglc levels lead to a modified starch form, the amylose and amylopectin contents were investigated by gel filtration chromatography. GlgC (CS8) and GlgC/ZmBT1 (CS8/AT5) lines contained higher amylose contents (fraction 1) compared with wild-type endosperms (Table V). This is most likely due to elevated ADPglc levels in these transgenic lines (Table IV), since granule-bound starch synthase I (GBSSI), which is responsible for amylose formation, possesses lower affinity toward ADPglc than the other starch synthase isozymes (Clarke et al., 1999). By contrast, the amylose content from ZmBT1 (AT5) lines was not significantly different from that seen for wild-type endosperms (Table V). There were also no significant changes in the fraction III-fraction II ratio between the transgenic lines and the wild-type indicating that the chain length distribution of amylopectin was not affected (Table V).
Table V. Composition of carbohydrate in endosperm fractions of transgenic lines.
Three fractions (I, II, and III) were detected by refractive index detectors. Fraction I is enriched with amylose, whereas fractions II and III are enriched with amylopectin chains. Total carbohydrate content is 100%. Each value represents the mean ± se of three independent experiments.
| Lines | Fraction I | Fraction II | Fraction III | Fraction III:II Ratio |
|---|---|---|---|---|
| weight % | ||||
| Wild-type (cv Kitaake) | 15.1 ± 0.19 | 22.6 ± 0.37 | 62.3 ± 0.42 | 2.76 ± 0.06 |
| GlgC (CS8-3) | 16.9 ± 0.70a | 22.0 ± 0.90 | 61.1 ± 0.30 | 2.79 ± 0.13 |
| ZmBT1 (AT5) | 15.7 ± 0.33 | 22.2 ± 0.27 | 62.1 ± 0.34 | 2.79 ± 0.04 |
| GlgC/ZmBT1(CS8/AT5) | 16.1 ± 0.16a | 21.9 ± 0.08 | 62.0 ± 0.15 | 2.84 ± 0.01 |
Significant differences between transgenic lines and the wild-type by Student’s t test at P < 0.05.
Comparison of Seed Weight and Seed Morphology between Wild-Type and Transgenic Plants
The effect of ZmBt1 introduction on starch biosynthesis was examined by measuring the average weight of mature seeds from transgenic lines grown under ambient and elevated CO2 conditions. Under ambient conditions, ZmBT1 (AT5-1 and AT5-19) transgenic lines did not demonstrate any significant changes in grain weight compared with the wild-type (Table VI). By contrast, as shown earlier (Nagai et al., 2009), the GlgC (CS8) transgenic lines exhibited about an 11% increase in grain weight, an increase that was also reflected in grain size (Table VI). GlgC/ZmBT1 (CS8/AT5) lines produced heavier seeds but were nearly identical in weight to those from the CS8 line (Table VI).
Table VI. Average weights of individual seeds from various rice lines.
Mean weight values (mg) were measured from five plants per line. Asterisks indicate that average seed weights were statistically different at the 95% confidence level compared with the wild-type. Percentage values indicate the percentage of seed weight compared with wild-type cv Kitaake, with the exception of EM1093, which was compared with wild-type cv TC65. ND, Not determined.
| Lines | Ambient CO2 |
High CO2 |
||
|---|---|---|---|---|
| Seed Weight | Percentage | Seed Weight | Percentage | |
| Wild-type (cv Kitaake) | 23.3 ± 0.6 | 100 | 24.4 ± 0.8 | 100 |
| ZmBT1 (AT5-1) | 23.5 ± 0.7 | 101 | 25.3 ± 0.9 | 103 |
| ZmBT1 (AT5-19) | 23.6 ± 0.6 | 101 | 24.9 ± 0.7 | 102 |
| GlgC (CS8-18) | 25.8 ± 0.4* | 111 | 27.1 ± 0.9* | 111 |
| GlgC/ZmBT1 (CS8/AT5-18) | 26.1 ± 0.7* | 112 | 27.4 ± 1.0* | 112 |
| GlgC/ZmBT1 (CS8/AT5-27) | 26.3 ± 0.5* | 113 | 27.6 ± 1.4* | 113 |
| GlgC/ZmBT1 (CS8/AT5-37) | 25.7 ± 0.5* | 110 | 27.1 ± 0.8* | 111 |
| Wild-type (cv TC65) | 22.9 ± 0.7 | 100 | ND | ND |
| EM1093 (cv TC65) | 5.7 ± 0.3 | 25 | ND | ND |
As sugars provided by photosynthetic source leaves may be limiting under ambient conditions, transgenic plants were grown under elevated CO2 levels (1000 µl L−1) during the reproductive stage to investigate whether these plants could utilize the extra Suc and incorporate it into starch. Similar to the results obtained under ambient CO2 levels, ZmBT1 (AT5) transgenic lines exhibited the same grain weight as those from the wild-type (Table VI). Although GlgC (CS8) lines produced heavier seeds, the increases in seed weights were similar to those produced under ambient CO2 conditions (Table VI). Likewise, GlgC/ZmBT1 (CS8/AT5) lines also showed a similar increase in grain weight, varying 11% to 13% from the wild-type, but this increase was nearly identical to those obtained under ambient conditions. Overall, ZmBT1 (AT5) and GlgC/ZmBT1 (CS8/AT5) lines failed to incorporate the excess assimilate generated by higher photosynthesis under elevated CO2 into starch compared with the wild-type and CS8 lines, respectively.
Starch Turnover Does Not Account for the Lack of Increased Starch Synthesis When ADPglc Is in Excess
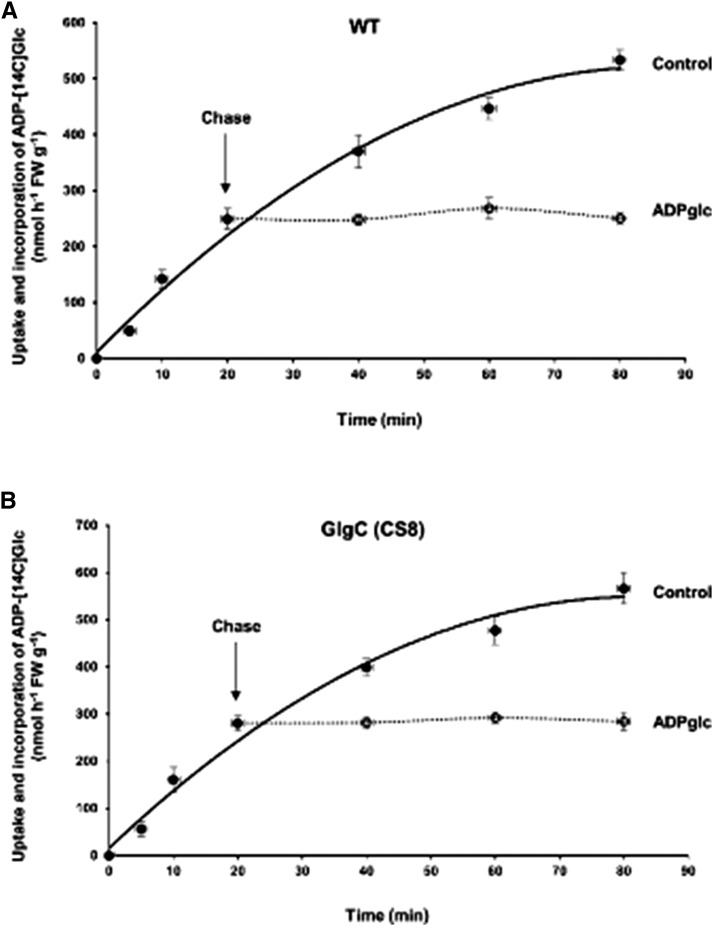
The expression of a plastidial targeted bacterial AGPase gene in Russet Burbank potato (Solanum tuberosum) tubers resulted in a dramatic elevation of starch content (Stark et al., 1992). A comparable study (Sweetlove et al., 1996) in the potato cv Desiree, however, failed to observe enhanced starch content. Metabolic labeling analysis indicated that the failure of cv Desiree tubers to accumulate more starch was likely due to increased starch turnover (Sweetlove et al., 1996). To directly assess whether starch turnover is a major factor limiting carbon flux into starch in rice, ADP-[14C]glc pulse-chase labeling studies were conducted (Fig. 6). Purified amyloplasts were incubated with ADP-[14C]glc for 20 min and then chased with excess nonradioactive ADPglc. No obvious loss of radioactivity from starch was detected in both wild-type and CS8 amyloplasts up to 60 min of incubation. Hence, starch turnover rates are very low and not responsible for the inability of CS8 rice lines to increase starch production when ADPglc is in excess.
Figure 6.
Pulse-chase studies using ADP-[14C]glc to assess starch turnover in intact amyloplast. Wild-type (WT; A) and GlgC (CS8; B) amyloplasts, preloaded with ADP, were incubated in the presence of 0.1 mm ADP-[14C]glc. After 20 min of incubation, a 100-fold excess (10 mm) of unlabeled ADPglc was added to the amyloplast mixture, and the extent of radioactivity in starch was assessed. Data represent means ± se of three independent experiments. FW, Fresh weight.
DISCUSSION
In cereals, the majority of the endosperm ADPglc is synthesized in the cytosol. Therefore, cereals are dependent on the BT1 transporter to import cytosolic ADPglc into amyloplast, which is subsequently incorporated into starch (Denyer et al., 1996; Thorbjørnsen et al., 1996; Sikka et al., 2001). This view is supported by BT1-deficient mutants of maize kernels (Shannon et al., 1998) and barley seeds (Patron et al., 2004), which had drastically reduced starch content. Until now, however, a detailed analysis of BT1 function in starch biosynthesis in rice endosperms was lacking.
In this study, EM1093 seeds contained dramatically decreased starch content due to mutation in the shr3 locus. OsBt1-1 was initially identified as a potential candidate gene underlying the shr3 locus by TILLING. Subsequent DNA sequencing studies showed the presence of a nonsense mutation in the OsBt1-1 gene from EM1093 plants, resulting in a premature stop codon and, in turn, a loss of protein (Fig. 2C). Proteomic analysis confirmed the absence of tryptic fragments corresponding to intact OsBT1 from amyloplast membranes from EM1093 (Table II). Moreover, amyloplasts from EM1093 (shr3) exhibited very low rates of ADPglc transport into amyloplast (Fig. 3A; Table III), which is consistent with the decreased seed weight. The severe reduction in grain weight also indicates that the hexose sugar (e.g. Glc-1-P and Glc-6-P) transporters, located in the plastidial membrane, as well as the amyloplast-localized AGPase are unable to compensate for the loss of a functional BT1 transporter in shr3 endosperm. Although EM1093 plants accumulated dramatically increased ADPglc levels in the endosperm compared with wild-type plants (Fig. 3B), their seeds only contained around 20% of normal starch content due to the loss of BT1 protein and its transport activity. Overall, BT1 is required for a normal rate of starch synthesis during rice seed development.
ZmBT1 is targeted to the amyloplast via the presence of a plastid-targeting leader sequence. Expression of ZmBT1 produced a polypeptide doublet at approximately 41 kD in developing rice seeds as viewed by immunoblotting (Fig. 4). An identical polypeptide doublet was also observed in developing maize kernels by Shannon et al. (1998), who suggested that the ZmBT1 precursor contained two peptide leader processing sites.
In vitro transport and starch incorporation studies demonstrated that ADPglc transport was stimulated by the presence of ADP or AMP (Figs. 3A and 5). This result is consistent with an earlier study from wheat (Bowsher et al., 2007), in which ADPglc transport was enhanced by ADP and AMP as counter-exchange substrates. Interestingly, ATP had no impact on the ADPglc uptake rate. As shown earlier (Bowsher et al., 2007), ATP is the primary substrate for ADP/ATP carriers on the plastid membranes but not for BT1. Under physiological conditions, ADPglc is most likely exchanged with ADP, since this adenylate nucleotide is generated by the action of starch synthases during the polymerization of Glc residues.
The transgenic lines, ZmBT1 (AT5) and GlgC/ZmBT1 (CS8/AT5), had significantly higher ADP-[14C]glc transport and starch incorporation rates by intact amyloplasts than those isolated from the wild-type and GlgC rice lines lacking the maize BT1 transporter (Fig. 5; Table III). Despite the significantly elevated ADP-[14C]glc transport and starch incorporation rates seen for amyloplasts from the ZmBT1 (AT5) transgenic lines, these did not translate into higher amyloplast content of ADPglc and higher seed weights, which were no different from the wild-type values (Table VI). Hence, ADP-[14C]glc transport and starch incorporation rates by purified amyloplasts are poor indicators of the potential for seed weight increases. Better indicators of increased carbon flux into starch and, in turn, higher seed weights are total ADPglc levels per seed and amounts in amyloplasts. While ADPglc levels in the ZmBT1 rice line were no different from the quantities measured in the wild-type, significantly elevated levels of total ADPglc and amounts in amyloplasts were readily evident in GlgC and GlgC/ZmBT1, rice lines that bore seeds of 11% to 13% higher weight. Overall, these results indicate that OsBT1 is not a limiting factor in controlling carbon flux into starch.
The measured levels of ADPglc in developing rice seeds were 0.87 nmol seed−1 (38.8 nmol g−1 fresh weight) for the wild-type and up to 1.19 nmol seed−1 for the GlgC transgenic lines (Table IV). These estimated amounts of ADPglc in developing rice seed (approximately 40 nmol g−1 fresh weight) are about 3- to 4-fold lower than the amounts (146 nmol g−1 fresh weight) enzymatically estimated in developing barley grains (Tiessen et al., 2012). Moreover, the majority of ADPglc (approximately 80%) was located in the cytosol of barley endosperms, whereas approximately 50% of total ADPglc was found in the nonamyloplast fraction in developing rice seeds (Table IV). It should be pointed out that measured ADPglc levels in this study were similar to the amounts estimated by LC-MS/MS analysis in an earlier study from this laboratory (Nagai et al., 2009). For example, the wild-type was estimated to contain about 0.72 nmol seed−1 by LC-MS/MS, an amount similar to the 0.87 nmol seed−1 measured in this study. Hence, the large 3- to 4-fold differences in ADPglc levels between developing rice and barley grains likely represent the innate properties of each plant species and are not due to differences in the way the metabolite samples were prepared or measured between the two studies.
The lack of further enhancement in seed weights by CS8 or ZmBT1/CS8 transgenic rice lines grown under elevated CO2 conditions compared with those grown under ambient conditions (Table VI) clearly indicate that BT1 transport or AGPase activities do not limit carbon flux into starch. The expression of maize sh2 AGPase genes in maize (Hannah et al., 2012), rice (Smidansky et al., 2003), and wheat (Smidansky et al., 2002) increased crop yields by increasing the number of grains produced per plant. Analyses of the various GlgC and/or ZmBT1 transgenic plants cultured in growth chambers indicate no significant increases in total seed number or total number of panicles per plant (data not shown). We should emphasize, however, that such yield studies require field-grown plants to eliminate limitations in photosynthesis and root growth from attaining maximum seed yields per plant.
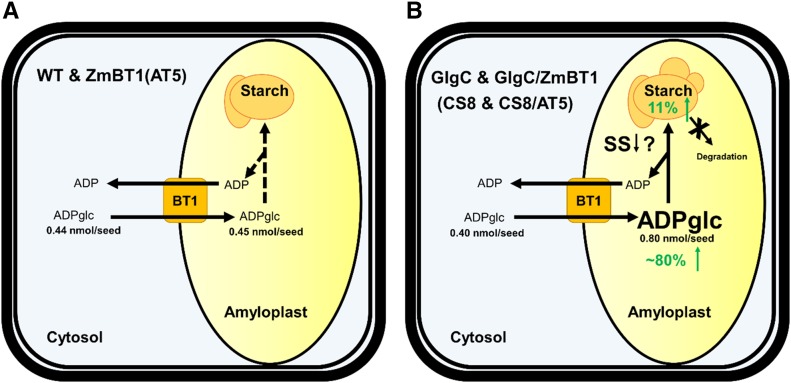
Since the increases in ADPglc amounts in CS8 transgenic lines (Table IV) did not result in corresponding increases in grain weight (Table VI), these results point out that one or more processes located in the stroma control starch synthesis when ADPglc is in excess (Fig. 7). If ADPglc transport activity and levels are not barriers, what is responsible for limiting carbon flow into starch in the GlgC (CS8) rice lines? One possible factor limiting the carbon flux into starch could be simultaneously occurring degradation of starch coupled with synthesis, as observed in GlgC-overexpressing cv Desiree tubers (Sweetlove et al., 1996). Similarly, down-regulation of glucan, water-dikinase, an enzyme that facilitates starch degradation, resulted in enhanced wheat grain yields and increases in biomass (Ral et al., 2012). Hence, increases in starch turnover, which may counterbalance increases in starch synthesis, may be responsible for the absence of any further increases in starch synthesis and accumulation, especially when rice plants are grown under elevated CO2 conditions. Results from the ADP-[14C]glc pulse-chase study using purified amyloplasts indicate, however, that very little, if any, starch turnover occurs in the CS8 line (Fig. 6). Hence, starch turnover is unlikely to be the causal basis for the inability of CS8 lines to increase starch production.
Figure 7.
Summary of ADPglc compartmentalization in the wild-type and transgenic lines. A, ADPglc levels are present at nearly equal amounts in the cytosol and amyloplasts in wild-type (WT) and ZmBT1 (AT5) lines. B, ADPglc levels in GlgC (CS8) and GlgC/ZmBT1 (CS8/AT5) lines are increased significantly (approximately 80%) in amyloplasts while decreased slightly (approximately 9%) in the cytosol compared with the wild-type and ZmBT1 (AT5) lines. This nearly doubling of ADPglc levels, however, only translated into an approximately 11% increase (on average based on three independent transgenic lines) in starch content, indicating that the maximum flux of carbon into starch is tightly regulated. This slowing in net starch synthesis, mediated by elevated ADPglc levels, is probably not caused by starch turnover but more likely by a reduction in one or more starch synthase (SS) isoform enzyme activities.
A second process that may limit carbon flow into starch centers on the starch synthases. We had earlier discounted this possibility and focused on the ADPglc transporter, as genetic mutant studies showed that loss of action of the major starch synthase isoforms, SSI and SSIII, did not have a significant impact on starch synthesis and accumulation. Mutant lines that lack one of these major starch synthase isoforms had starch levels similar to that seen for the wild-type (Fujita et al., 2006, 2007). The loss or reduction in catalytic activity of one of these enzymes is apparently compensated by one or more of the other starch synthase isoforms.
Although genetic studies (Fujita et al., 2006, 2007) imply that the major starch synthases (SSI and SSIIIa) do not limit starch synthesis in endosperm under normal conditions, their catalytic activities may be rate determining in the transgenic rice GlgC (CS8) lines, where ADPglc is in excess (Fig. 7). Indeed, this view is supported by the intracellular distribution of ADPglc in the GlgC (CS8) transgenic lines (Table IV). Irrespective of the presence or absence of maize BT1, the transgenic GlgC (CS8) lines showed elevated amounts of ADPglc in the amyloplast compared with the wild-type or ZmBT1 (AT5) lines (Table IV). This result supports the view that the catalytic activity of one or more of the starch synthase isoforms is insufficient to accommodate the excess ADPglc present in the amyloplast.
A significant reduction in starch synthase activity in the GlgC (CS8) lines may be the result of the plant’s responses to excess ADPglc. Earlier metabolite analysis showed that CS8 lines not only contain elevated levels of ADPglc but also proportional increases in upstream metabolites, UDPglc, Fru, Glc-6-P, and Glc-1-P, and possibly Glc and Suc (Nagai et al., 2009). The hexose sugars and Suc are well-documented signaling molecules that control gene and protein expression (Tognetti et al., 2013; Sheen, 2014), and the elevation of these sugars will likely lead to the reprogramming of gene expression. Indeed, preliminary RNA sequencing studies of the wild-type and CS8 lines show significant alterations in the steady-state RNA levels for several starch biosynthetic enzymes, including significant depression of SSIII expression and an increase in invertase RNA sequences. Moreover, elevated expression of a starch-binding domain protein (SBDP) was observed. This SBDP gene has only been identified by computational analysis of the rice genome; hence, nothing is known about whether it has a role in starch synthesis. Other studies have demonstrated that nonenzymatic carbohydrate-binding proteins play major roles in starch biosynthesis. A novel gene, FLOURY ENDOSPERM6 (FLO6), encodes for a CBM48-containing protein, which interacts with ISOAMYLASE1 and presumably facilitates its debranching activity in rice (Peng et al., 2014). Transgenic plants lacking the FLO6 protein have lower starch content and modified starch granules. Additionally, a novel protein containing a CBM48 domain localizes the GBSS to starch granules (Seung et al., 2015). The absence of this starch-targeting protein resulted in amylose-free starch in Arabidopsis (Arabidopsis thaliana) leaves. Therefore, other carbohydrate-binding module-containing proteins may take part in starch biosynthesis, either as accessory proteins to known starch biosynthetic enzymes, which normally lack carbohydrate-binding domains, or potentially as regulators controlling the flux of carbon into starch.
Overall, our results demonstrated that ADPglc transport by BT1 is essential for the normal rate of starch synthesis in rice endosperm. On the other hand, enhanced ADPglc import did not further increase the sink strength of developing rice seeds, indicating that one or more biochemical processes in the amyloplast stroma control carbon flux into starch. Ongoing efforts by this laboratory are directed at identifying these limiting factors and elucidating their roles in regulating starch biosynthesis as a means to increase the yields of this cereal grain that feeds a large proportion of the world’s population.
MATERIALS AND METHODS
Generation and Screening of the Endosperm Mutant
The sh3 mutation in the rice (Oryza sativa) EM1093 line was generated by N-methyl-N-nitrosourea treatment of independent fertilized egg cells of the japonica cv TC65, as described previously (Satoh and Omura, 1979). Segregation analysis was performed by crossing shr3 with cv TC65 (wild type), EM541 (shr1), and EM22 (shr2). RFLP analysis was performed by crossing the indica cv Kasalath with EM1093. The rice plants were grown under natural conditions at the Kyushu University experimental field plots. A mutation in the OsBt1-1 gene of EM1093 was initially identified by TILLING (Suzuki et al., 2008) and subsequently confirmed by DNA sequencing of the OsBt1-1 gene.
Plasmid Construction and Rice Transformation
The plant expression vector (pAT5) was generated by cloning the ZmBt1 cDNA under the control of the rice endosperm-specific Globulin promoter (Zheng et al., 1993) into pCAMBIA 1303. pAT5 was then transformed into wild-type cv Kitaake and the transgenic CS8 line via Agrobacterium tumefaciens AGL1 (Toki, 1997). Identification of the transgenic plants expressing both the glgC-TM (Sakulsingharoj et al., 2004) and ZmBT1 genes was carried out by histochemical GUS staining. These plants were further screened for the expression of both GlgC-TM and ZmBT1 proteins by immunoblot analysis using anti-GlgC and anti-ZmBT1 antibodies as described previously (Cao et al., 1995; Sakulsingharoj et al., 2004). T3 lines homozygous for both transgenes were used in this study.
Plant Growth under the Elevated CO2 Condition
Wild-type, single transgenic (ZmBT1 overexpressing), and double transgenic (ZmBT1 and GlgC-TM overexpressing) rice plants were grown in a controlled chamber under a 12-h photoperiod at 26°C during the day and 22°C at night. The relative humidity and photosynthetic flux density was set to 70% and 2,000 µmol m−2 s−1, respectively. The plants were grown at ambient CO2 levels (400 µl L−1) until the reproductive stage and then were subjected to elevated CO2 levels (1000 µl L−1) to maturity.
Seed Weight Measurements
At maturity, all seeds from individual plants were harvested, dried, and measured for total yield and average grain weight as described previously (Nagai et al., 2009). Briefly, panicles from transgenic plants were grown in a growth chamber for two consecutive periods and then harvested. The average grain weight of each transgenic plant was measured by weighing 100 seeds from five panicles.
Amyloplast Isolation from Developing Rice Endosperm
Amyloplasts were isolated from developing rice seeds (12–15 DAP) as described previously (Sikka et al., 2001). After removal of the seed hull, the ends of developing seeds (approximately 2 g) were carefully removed with forceps and transferred into 5 mL of extraction buffer (0.8 m sorbitol, 50 mm HEPES-KOH, pH 7.5, 1 mm EDTA, 2 mm MgCl2, 10 mm KCl, 5 mm dithiothreitol, and 0.1% (w/v) bovine serum albumin) for plasmolysis. After incubation in the extraction buffer on ice for 30 min, the seeds were gently chopped with a razor blade. The homogenate was filtered through four layers of Miracloth (Calbiochem), and the filtrate was kept on ice for 3 h to sediment amyloplasts via gravity. The pellet, which consists of enriched amyloplasts, was resuspended in extraction buffer. To check the intactness of the plastids, one-half of the isolated amyloplasts were disrupted by the addition of 0.5% Triton X-100 (v/v), agitated for 1 min, and centrifuged at 20,000g for 10 min. The yield of intact amyloplasts was estimated at about 25%, based on the relative amounts of the plastid enzyme marker, inorganic pyrophosphatase, recovered in the amyloplast fraction, as described previously (Sikka et al., 2001).
Analysis of Starch Granules of cv TC65 and EM1093 Endosperms
Starch granule morphology was investigated by SEM as described previously (Wong et al., 2003). The chain length distribution of amylopectin isolated from wild-type and EM1093 endosperms was performed by a modified method (O’Shea et al., 1998) as described previously (Nakamura et al., 2002).
Size Fractionation of Debranched Starch and Amylose Content Analysis from Wild-Type and Transgenic Lines
Powdered mature rice endosperm samples were gelatinized and debranched by Pseudomonas spp. isoamylase (Hayashibar) as described previously (Crofts et al., 2012). The samples were then analyzed using a Toyopearl HW55S gel filtration column (300 × 20 mm) connected to three Toyopearl HW50S columns (300 × 20 mm; Fujita et al., 2007). The eluted glucans were detected with a refractive index detector (Tosoh RI-8020).
Synthesis of ADP-[14C]glc
ADP-[14C]glc was prepared from [14C]Glc-1-P as described previously (Preiss and Greenberg, 1972), except that the product was separated by thin-layer chromatography (TLC) instead of paper chromatography. Briefly, 25 µCi of [14C]Glc-1-P was converted to ADP-[14C]glc using purified potato (Solanum tuberosum) AGPase as described (Hwang et al., 2004). The final reaction mixture was resolved by TLC for 7 h in solvent containing 95% ethanol:1 m ammonium acetate, pH 7.5 (5:2). Hexose sugars were detected by spraying the TLC plate with 10% (v/v) H2SO4 in methanol and then drying at 120°C for 10 min to visualize the sugars. The top of the chromatogram containing ADPglc was collected and then washed with absolute ethanol for 3 h to remove residual ammonium acetate. After drying, ADP-[14C]glc was eluted with water. Based on the initial starting amount of [14C]Glc-1-P, the yield of ADP-[14C]glc was about 70%, as determined by absorption at 260 nm and liquid scintillation spectrometry.
ADP-[14C]glc Transport Assay
The transport of ADPglc by intact amyloplasts was performed as described previously (Shannon et al., 1998). Isolated amyloplasts (100 µL) were transferred into a reaction mixture containing 100 mm Bicine, 0.5 m sorbitol, 12.5 mm EDTA, 50 mm potassium acetate, 10 mm glutathione, and 4 mm ADP-[14C]glc (specific activity of 150 cpm nmol−1). When evaluating the counter-exchange substrates, amyloplasts were preloaded with 20 mm AMP, ADP, or ATP before carrying out uptake-starch incorporation assays. The reaction was carried out at 30°C for 30 min and terminated by the addition of 2 mL of 75% methanol containing 1% (w/v) KCl. The insoluble starch was collected by centrifugation at 2,000g for 10 min and washed twice with methanol/KCl to remove excess substrate. The pellet was then resuspended in water, and the amount of ADP-[14C]glc converted into starch was quantified by liquid scintillation spectrometry.
Starch turnover was investigated by performing pulse-chase studies. Briefly, amyloplasts (100 µL), preincubated with 20 mm ADP, were used for the incorporation assay in the presence of 0.1 mm ADP-[14C]glc (specific activity of 900 cpm nmol−1). After 20 min of uptake and incorporation, a 100-fold excess of unlabeled ADPglc was added, and the reaction was incubated for up to an additional 60 min.
Measurement of ADPglc Levels
Purified amyloplasts were isolated from 50 developing rice seeds (10–13 DAP), lysed by the addition of 0.5% [v/v] Triton X-100, and then boiled for 2 min to inactivate native AGPase activity. ADPglc content was then estimated for the purified amyloplast fraction and total seed extract by performing the reverse AGPase reaction coupled with phosphoglucomutase and Glc-6-P dehydrogenase reactions as described previously (Seferoglu et al., 2014). The reaction was initiated by the addition of excess amounts of near homogenously pure potato AGPase and performed at 37°C for 20 min in reaction buffer containing 100 mm HEPES, pH 7, 5 mm dithiothreitol, 5 mm 3-phosphoglyceric acid, 10 mm MgCl2, 0.6 mm NAD+, 0.4 mg mL−1 bovine serum albumin, 0.5 units of Glc-6-P dehydrogenase, and 0.5 units of phosphoglucomutase. The reaction was terminated at 100°C and clarified by centrifugation, and the amount of NADH was determined by measuring the A340. A standard curve for determining the amounts of ADPglc was generated by incubating known amounts of ADPglc in the coupled enzyme reaction mixtures. The total amount of ADPglc in amyloplasts was estimated by factoring in the percentage recovery of intact amyloplasts using inorganic pyrophosphatase as a plastid marker enzyme as described earlier.
Supplemental Data
The following supplemental materials are available.
Supplementary Material
Acknowledgments
We thank Thomas D. Sullivan (University of Wisconsin School of Medicine) for providing the maize ZmBt1 cDNA and BT1 antibody.
Glossary
- SEM
scanning electron microscopy
- TC65
Taichung 65
- TILLING
targeting induced local lesions in genomes
- cDNA
complementary DNA
- DAP
days after planting
- LC-MS/MS
liquid chromatography-tandem mass spectrometry
- TLC
thin-layer chromatography
Footnotes
This work was supported by grants from the Japan Society for the Promotion of Science (to H.S.), the Division of Chemical Sciences, Geosciences, and Biosciences, Office of Basic Energy Sciences, U.S. Department of Energy (grant no. DE–FG02–12ER20216 to S.-K.H. and T.W.O.), the U.S. Department of Agriculture National Institute of Food and Agriculture (project no. WNP00590), and the Agricultural Research Center, College of Agricultural, Human, and Natural Resource Sciences, Washington State University.
Articles can be viewed without a subscription.
References
- Bahaji A, Li J, Sánchez-López ÁM, Baroja-Fernández E, Muñoz FJ, Ovecka M, Almagro G, Montero M, Ezquer I, Etxeberria E, et al. (2014) Starch biosynthesis, its regulation and biotechnological approaches to improve crop yields. Biotechnol Adv 32: 87–106 [DOI] [PubMed] [Google Scholar]
- Bowsher CG, Scrase-Field EF, Esposito S, Emes MJ, Tetlow IJ (2007) Characterization of ADP-glucose transport across the cereal endosperm amyloplast envelope. J Exp Bot 58: 1321–1332 [DOI] [PubMed] [Google Scholar]
- Cakir B, Tuncel A, Hwang SK, Okita T (2015) Increase of grain yields by manipulating starch biosynthesis. In Nakamura Y, ed, Starch. Springer, Tokyo Japan, pp 371–395 [Google Scholar]
- Cao H, Sullivan TD, Boyer CD, Shannon JC (1995) Btl, a structural gene for the major 39-44 kDa amyloplast membrane polypeptides. Physiol Plant 95: 176–186 [Google Scholar]
- Chen C, Li C, Sung J (1994) Carbohydrate metabolism enzymes in CO2‐enriched developing rice grains of cultivars varying in grain size. Physiol Plant 90: 79–85 [Google Scholar]
- Clarke BR, Denyer K, Jenner CF, Smith AM (1999) The relationship between the rate of starch synthesis, the adenosine 5′-diphosphoglucose concentration and the amylose content of starch in developing pea embryos. Planta 209: 324–329 [DOI] [PubMed] [Google Scholar]
- Crofts N, Abe K, Aihara S, Itoh R, Nakamura Y, Itoh K, Fujita N (2012) Lack of starch synthase IIIa and high expression of granule-bound starch synthase I synergistically increase the apparent amylose content in rice endosperm. Plant Sci 193-194: 62–69 [DOI] [PubMed] [Google Scholar]
- Denyer K, Dunlap F, Thorbjørnsen T, Keeling P, Smith AM (1996) The major form of ADP-glucose pyrophosphorylase in maize endosperm is extra-plastidial. Plant Physiol 112: 779–785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita N, Yoshida M, Asakura N, Ohdan T, Miyao A, Hirochika H, Nakamura Y (2006) Function and characterization of starch synthase I using mutants in rice. Plant Physiol 140: 1070–1084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita N, Yoshida M, Kondo T, Saito K, Utsumi Y, Tokunaga T, Nishi A, Satoh H, Park JH, Jane JL, et al. (2007) Characterization of SSIIIa-deficient mutants of rice: the function of SSIIIa and pleiotropic effects by SSIIIa deficiency in the rice endosperm. Plant Physiol 144: 2009–2023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haferkamp I. (2007) The diverse members of the mitochondrial carrier family in plants. FEBS Lett 581: 2375–2379 [DOI] [PubMed] [Google Scholar]
- Hannah LC, Futch B, Bing J, Shaw JR, Boehlein S, Stewart JD, Beiriger R, Georgelis N, Greene T (2012) A shrunken-2 transgene increases maize yield by acting in maternal tissues to increase the frequency of seed development. Plant Cell 24: 2352–2363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang SK, Salamone PR, Kavakli H, Slattery CJ, Okita TW (2004) Rapid purification of the potato ADP-glucose pyrophosphorylase by polyhistidine-mediated chromatography. Protein Expr Purif 38: 99–107 [DOI] [PubMed] [Google Scholar]
- Isshiki M, Yamamoto Y, Satoh H, Shimamoto K (2001) Nonsense-mediated decay of mutant waxy mRNA in rice. Plant Physiol 125: 1388–1395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawagoe Y, Kubo A, Satoh H, Takaiwa F, Nakamura Y (2005) Roles of isoamylase and ADP-glucose pyrophosphorylase in starch granule synthesis in rice endosperm. Plant J 42: 164–174 [DOI] [PubMed] [Google Scholar]
- Kirchberger S, Leroch M, Huynen MA, Wahl M, Neuhaus HE, Tjaden J (2007) Molecular and biochemical analysis of the plastidic ADP-glucose transporter (ZmBT1) from Zea mays. J Biol Chem 282: 22481–22491 [DOI] [PubMed] [Google Scholar]
- Kirchberger S, Tjaden J, Neuhaus HE (2008) Characterization of the Arabidopsis Brittle1 transport protein and impact of reduced activity on plant metabolism. Plant J 56: 51–63 [DOI] [PubMed] [Google Scholar]
- Laughlin MJ, Chantler SE, Okita TW (1998) N- and C-terminal peptide sequences are essential for enzyme assembly, allosteric, and/or catalytic properties of ADP-glucose pyrophosphorylase. Plant J 14: 159–168 [DOI] [PubMed] [Google Scholar]
- Leroch M, Kirchberger S, Haferkamp I, Wahl M, Neuhaus HE, Tjaden J (2005) Identification and characterization of a novel plastidic adenine nucleotide uniporter from Solanum tuberosum. J Biol Chem 280: 17992–18000 [DOI] [PubMed] [Google Scholar]
- Mangelsdorf PC. (1926) Genetics and Morphology of Some Endosperm Characters in Maize. Connecticut Agricultural Experiment Station, New Haven [Google Scholar]
- Nagai YS, Sakulsingharoj C, Edwards GE, Satoh H, Greene TW, Blakeslee B, Okita TW (2009) Control of starch synthesis in cereals: metabolite analysis of transgenic rice expressing an up-regulated cytoplasmic ADP-glucose pyrophosphorylase in developing seeds. Plant Cell Physiol 50: 635–643 [DOI] [PubMed] [Google Scholar]
- Nakamura Y, Sakurai A, Inaba Y, Kimura K, Iwasawa N, Nagamine T (2002) The fine structure of amylopectin in endosperm from Asian cultivated rice can be largely classified into two classes. Starke 54: 117–131 [Google Scholar]
- O’Shea MG, Samuel MS, Konik CM, Morell MK (1998) Fluorophore-assisted carbohydrate electrophoresis (FACE) of oligosaccharides: efficiency of labelling and high-resolution separation. Carbohydr Res 307: 1–12 [Google Scholar]
- Patron NJ, Greber B, Fahy BF, Laurie DA, Parker ML, Denyer K (2004) The lys5 mutations of barley reveal the nature and importance of plastidial ADP-Glc transporters for starch synthesis in cereal endosperm. Plant Physiol 135: 2088–2097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng C, Wang Y, Liu F, Ren Y, Zhou K, Lv J, Zheng M, Zhao S, Zhang L, Wang C, et al. (2014) FLOURY ENDOSPERM6 encodes a CBM48 domain-containing protein involved in compound granule formation and starch synthesis in rice endosperm. Plant J 77: 917–930 [DOI] [PubMed] [Google Scholar]
- Preiss J, Greenberg E (1972) ADP-[14C]glucose. Methods Enzymol 28: 279–281 [Google Scholar]
- Ral JP, Bowerman AF, Li Z, Sirault X, Furbank R, Pritchard JR, Bloemsma M, Cavanagh CR, Howitt CA, Morell MK (2012) Down-regulation of glucan, water-dikinase activity in wheat endosperm increases vegetative biomass and yield. Plant Biotechnol J 10: 871–882 [DOI] [PubMed] [Google Scholar]
- Rowland-Bamford AJ, Allen LH Jr, Baker JT, Boote KJ (1990) Carbon dioxide effects on carbohydrate status and partitioning in rice. J Exp Bot 41: 1601–1608 [Google Scholar]
- Sakulsingharoj C, Choi SB, Hwang SK, Edwards GE, Bork J, Meyer CR, Preiss J, Okita TW (2004) Engineering starch biosynthesis for increasing rice seed weight: the role of the cytoplasmic ADP-glucose pyrophosphorylase. Plant Sci 167: 1323–1333 [Google Scholar]
- Satoh H, Omura T (1979) Induction of mutation by the treatment of fertilized egg cell with N-methyl-N-nitrosourea in rice. Journal of the Faculty of Agriculture-Kyushu University (Japan) 24: 165–174 [Google Scholar]
- Seferoglu AB, Koper K, Can FB, Cevahir G, Kavakli IH (2014) Enhanced heterotetrameric assembly of potato ADP-glucose pyrophosphorylase using reverse genetics. Plant Cell Physiol 55: 1473–1483 [DOI] [PubMed] [Google Scholar]
- Seung D, Soyk S, Coiro M, Maier BA, Eicke S, Zeeman SC (2015) PROTEIN TARGETING TO STARCH is required for localising GRANULE-BOUND STARCH SYNTHASE to starch granules and for normal amylose synthesis in Arabidopsis. PLoS Biol 13: e1002080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon JC, Pien FM, Cao H, Liu KC (1998) Brittle-1, an adenylate translocator, facilitates transfer of extraplastidial synthesized ADP-glucose into amyloplasts of maize endosperms. Plant Physiol 117: 1235–1252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon JC, Pien FM, Liu KC (1996) Nucleotides and nucleotide sugars in developing maize endosperms (synthesis of ADP-glucose in brittle-1). Plant Physiol 110: 835–843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheen J. (2014) Master regulators in plant glucose signaling networks. J Plant Biol 57: 67–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikka VK, Choi SB, Kavakli IH, Sakulsingharoj C, Gupta S, Ito H, Okita TW (2001) Subcellular compartmentation and allosteric regulation of the rice endosperm ADPglucose pyrophosphorylase. Plant Sci 161: 461–468 [Google Scholar]
- Smidansky ED, Clancy M, Meyer FD, Lanning SP, Blake NK, Talbert LE, Giroux MJ (2002) Enhanced ADP-glucose pyrophosphorylase activity in wheat endosperm increases seed yield. Proc Natl Acad Sci USA 99: 1724–1729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smidansky ED, Martin JM, Hannah LC, Fischer AM, Giroux MJ (2003) Seed yield and plant biomass increases in rice are conferred by deregulation of endosperm ADP-glucose pyrophosphorylase. Planta 216: 656–664 [DOI] [PubMed] [Google Scholar]
- Stark DM, Timmerman KP, Barry GF, Preiss J, Kishore GM (1992) Regulation of the amount of starch in plant tissues by ADP glucose pyrophosphorylase. Science 258: 287–292 [DOI] [PubMed] [Google Scholar]
- Sullivan TD, Strelow LI, Illingworth CA, Phillips RL, Nelson OE Jr (1991) Analysis of maize brittle-1 alleles and a defective Suppressor-mutator-induced mutable allele. Plant Cell 3: 1337–1348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T, Eiguchi M, Kumamaru T, Satoh H, Matsusaka H, Moriguchi K, Nagato Y, Kurata N (2008) MNU-induced mutant pools and high performance TILLING enable finding of any gene mutation in rice. Mol Genet Genomics 279: 213–223 [DOI] [PubMed] [Google Scholar]
- Sweetlove LJ, Burrell MM, ap Rees T (1996) Starch metabolism in tubers of transgenic potato (Solanum tuberosum) with increased ADPglucose pyrophosphorylase. Biochem J 320: 493–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorbjørnsen T, Villand P, Denyer K, Olsen OA, Smith AM (1996) Distinct isoforms of ADPglucose pyrophosphorylase occur inside and outside the amyloplasts in barley endosperm. Plant J 10: 243–250 [Google Scholar]
- Tiessen A, Nerlich A, Faix B, Hümmer C, Fox S, Trafford K, Weber H, Weschke W, Geigenberger P (2012) Subcellular analysis of starch metabolism in developing barley seeds using a non-aqueous fractionation method. J Exp Bot 63: 2071–2087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobias RB, Boyer CD, Shannon JC (1992) Alterations in carbohydrate intermediates in the endosperm of starch-deficient maize (Zea mays L.) genotypes. Plant Physiol 99: 146–152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tognetti JA, Pontis HG, Martínez-Noël GM (2013) Sucrose signaling in plants: a world yet to be explored. Plant Signal Behav 8: e23316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toki S. (1997) Rapid and efficient Agrobacterium-mediated transformation in rice. Plant Mol Biol Rep 15: 16–21 [Google Scholar]
- Tuncel A, Kawaguchi J, Ihara Y, Matsusaka H, Nishi A, Nakamura T, Kuhara S, Hirakawa H, Nakamura Y, Cakir B, et al. (2014) The rice endosperm ADP-glucose pyrophosphorylase large subunit is essential for optimal catalysis and allosteric regulation of the heterotetrameric enzyme. Plant Cell Physiol 55: 1169–1183 [DOI] [PubMed] [Google Scholar]
- Tuncel A, Okita TW (2013) Improving starch yield in cereals by over-expression of ADPglucose pyrophosphorylase: expectations and unanticipated outcomes. Plant Sci 211: 52–60 [DOI] [PubMed] [Google Scholar]
- Wong KS, Kubo A, Jane JL, Harada K, Satoh H, Nakamura Y (2003) Structures and properties of amylopectin and phytoglycogen in the endosperm of sugary-1 mutants of rice. J Cereal Sci 37: 139–149 [Google Scholar]
- Wu CY, Adach T, Hatano T, Washida H, Suzuki A, Takaiwa F (1998) Promoters of rice seed storage protein genes direct endosperm-specific gene expression in transgenic rice. Plant Cell Physiol 39: 885–889 [Google Scholar]
- Zheng Z, Kawagoe Y, Xiao S, Li Z, Okita T, Hau TL, Lin A, Murai N (1993) 5′ distal and proximal cis-acting regulator elements are required for developmental control of a rice seed storage protein glutelin gene. Plant J 4: 357–366 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.