The gene product associated with a so-called Genomes UNcoupled (gun) mutant interacts with proteins involved in plastid protein homeostasis.
Abstract
Developmental or metabolic changes in chloroplasts can have profound effects on the rest of the plant cell. Such intracellular responses are associated with signals that originate in chloroplasts and convey information on their physiological status to the nucleus, which leads to large-scale changes in gene expression (retrograde signaling). A screen designed to identify components of retrograde signaling resulted in the discovery of the so-called genomes uncoupled (gun) mutants. Genetic evidence suggests that the chloroplast protein GUN1 integrates signals derived from perturbations in plastid redox state, plastid gene expression, and tetrapyrrole biosynthesis (TPB) in Arabidopsis (Arabidopsis thaliana) seedlings, exerting biogenic control of chloroplast functions. However, the molecular mechanism by which GUN1 integrates retrograde signaling in the chloroplast is unclear. Here we show that GUN1 also operates in adult plants, contributing to operational control of chloroplasts. The gun1 mutation genetically interacts with mutations of genes for the chloroplast ribosomal proteins S1 (PRPS1) and L11. Analysis of gun1 prps1 lines indicates that GUN1 controls PRPS1 accumulation at the protein level. The GUN1 protein physically interacts with proteins involved in chloroplast protein homeostasis based on coimmunoprecipitation experiments. Furthermore, yeast two-hybrid and bimolecular fluorescence complementation experiments suggest that GUN1 might transiently interact with several TPB enzymes, including Mg-chelatase subunit D (CHLD) and two other TPB enzymes known to activate retrograde signaling. Moreover, the association of PRPS1 and CHLD with protein complexes is modulated by GUN1. These findings allow us to speculate that retrograde signaling might involve GUN1-dependent formation of protein complexes.
Developmental or metabolic changes in chloroplasts can have profound effects on the rest of the plant cell. Such intracellular responses are associated with signals that originate in chloroplasts and convey information on their physiological status to the nucleus, which leads to large-scale changes in gene expression (retrograde signaling; Nott et al., 2006; Pogson et al., 2008; Chi et al., 2013). The first mutant screen designed to identify components of retrograde signaling resulted in the discovery of the so-called genomes uncoupled (gun) mutants (Susek et al., 1993). While norflurazon (NF), an inhibitor of carotenoid biosynthesis, efficiently blocks expression of photosynthesis-associated nuclear genes (PhANGs; such as LHCB1.2) in wild-type plants, gun mutants are characterized by their capacity to express PhANGs after exposure to NF. Because the proteins GUN2 to GUN6 are all involved in tetrapyrrole biosynthesis (TPB; Mochizuki et al., 2001; Larkin et al., 2003; Woodson et al., 2011), one of the retrograde signaling pathways is clearly triggered by perturbations in TPB. Besides the TPB pathway, signals derived from plastid gene expression (PGE) and the redox state of the photosynthetic electron chain (Redox; for review, see Nott et al., 2006; Woodson and Chory, 2008; Chi et al., 2013), as well as products of secondary metabolism (Estavillo et al., 2011; Xiao et al., 2012), products of carotenoid oxidation (Ramel et al., 2012), and mobile transcription factors (Sun et al., 2011; Isemer et al., 2012), have been implicated in retrograde signaling. Moreover, retrograde signals contribute both to the developmental regulation of organelle biogenesis (biogenic control; e.g. TPB and PGE signaling) and to rapid adjustments in energy metabolism in response to environmental and developmental constraints (operational control; e.g. Redox signaling; Pogson et al., 2008; Jarvis and López-Juez, 2013).
Genetic evidence suggests that GUN1 signaling activates the nuclear transcription factor ABI4 (Koussevitzky et al., 2007). Because plants that lack GUN1 or ABI4 display a gun phenotype in the presence of both NF (indicative for TPB signaling) and lincomycin (which inhibits PGE; Gray et al., 2003), GUN1 and ABI4 are obviously involved in both pathways. Moreover, differential expression of nuclear marker genes for the Redox signaling pathway requires GUN1 and ABI4 (Koussevitzky et al., 2007). Therefore, GUN1 apparently integrates signals from three different retrograde signaling pathways: TPB, PGE, and Redox. Strikingly, only very young plants show the gun phenotype, so GUN1-ABI4 signaling is thought to operate mainly in the biogenic control circuit (Pogson et al., 2008).
The GUN1 protein contains two domains with putative nucleic acid-binding capacity (Koussevitzky et al., 2007). The first of these belongs to the pentatricopeptide repeat (PPR) family, whose members are thought to bind to RNA and are known to have a range of essential functions in posttranscriptional processes in mitochondria and chloroplasts, including RNA editing, RNA splicing, RNA cleavage, and translation (Barkan and Small, 2014). The second is a small MutS-related (SMR) domain, which is usually found in proteins involved in DNA repair and recombination (Fukui and Kuramitsu, 2011). Indeed, in vitro experiments have suggested that GUN1 binds DNA (Koussevitzky et al., 2007).
We have now pinpointed binding partners of GUN1 and found, surprisingly, that it interacts with proteins. Moreover, we were unable to detect the direct interactions with nucleic acids expected of a PPR-SMR protein. Among the interactors identified by coimmunoprecipitation are several proteins involved in PGE and plastid protein homeostasis. In yeast two-hybrid (Y2H) and bimolecular fluorescence complementation (BiFC) experiments, the GUN1 protein interacts with plastid ribosomal protein S1 (PRPS1) and several enzymes in the TPB pathway. Mutants for three of the latter each display a gun phenotype. Moreover, altered dosage of GUN1 modulates the abundance of PRPS1 and of the D subunit of the Mg chelatase (CHLD), as well as affects their organization into complexes. These findings allow us to speculate that retrograde signaling involving GUN1 might be related to the formation of protein complexes in the chloroplast.
RESULTS
GUN1 Is Coexpressed with PGE Genes and Also Is Relevant for Signaling in Adult Plants
The PPR domain of GUN1 contains 11 canonical amino acid pairs at positions 6 and 1’ that can be used for RNA target-site prediction (Supplemental Fig. S1A). For the inferred RNA target sequence ANAUUCGUCGAA (in the stringent version; Supplemental Fig. S1A), 38 hits comprising six or more nucleotides occur in the reference chloroplast genome, although no perfect match was found. To experimentally identify RNA or DNA targets of GUN1, a combination of RNA and DNA immunoprecipitation was employed (Supplemental Fig. S1, B and C). Surprisingly, no interaction with nucleic acids could be identified with this assay or with one-hybrid experiments (see Supplemental Text S1).
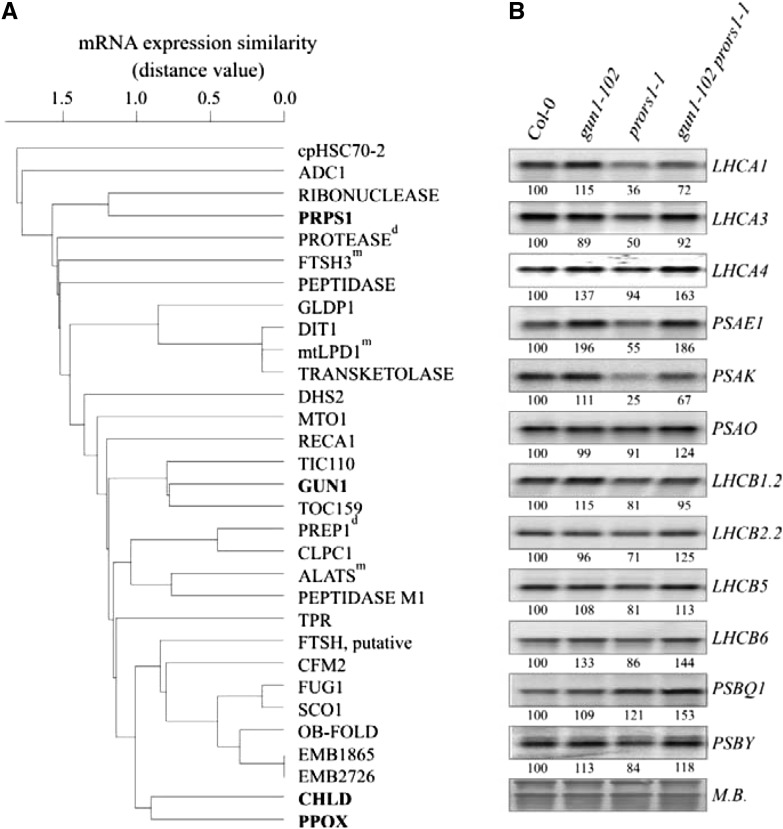
To narrow the search space for possible functions of GUN1 in PGE, we adopted a guilt-by-association approach based on the observation that nuclear PGE genes are highly coregulated at the transcriptional level (Leister et al., 2011). A set of 29 nuclear genes with GUN1-like transcript profiles were identified (Fig. 1A), and found to include genes for enzymes of tetrapyrrole and amino acid biosynthesis, components of the chloroplast import machinery, and factors involved in the synthesis (e.g. PRPS1) or degradation (e.g. FtsH3, ClpC homolog 1, and several peptidases) of chloroplast proteins.
Figure 1.
The GUN1 transcript signature and effects of the gun1-102 mutation on transcription patterns in adult leaves. A, GUN1 is preferentially coexpressed with genes for proteins linked to PGE. Correlations of GUN1 expression with the 29 most highly coexpressed genes were hierarchically clustered (see “Materials and Methods”). The degree of coexpression was measured by the mutual rank test. Low distance values indicate high coexpression. Full names and accession numbers of corresponding proteins encoded are provided in “Materials and Methods.” With the exception of proteins indicated by the superscript “m” (for mitochondrial) or “d” (for dual targeting to mitochondria and chloroplasts), all proteins are predicted or experimentally confirmed chloroplast proteins. Proteins further investigated in this study are indicated in bold. B, Northern analysis of transcripts encoding components of PSI (LHCA1, -2, -3, and -4, and PSAE1, -K, and -O) and PSII (LHCB1.2, -2.2, -5, and -6, and PSBQ1 and -Y), isolated from light-adapted wild-type (Col-0), gun1-102, prors1-1, and gun1-102 prors1-1 plants. To control for RNA loading, blots were stained with methylene blue (M.B.). Quantification of signals (by ImageJ) relative to the wild type (=100%) is provided below each panel. A representative result from three independent experiments is shown.
PGE signaling also can occur in adult plants, as shown by Pesaresi et al. (2006) who demonstrated that the prors1-1 mutation, which causes a defect in prolyl-tRNA synthetase activity in chloroplasts and mitochondria, results in down-regulation of PhANG mRNA levels. Indeed, by employing control mutants defective in only PGE (prpl11-1), only mitochondrial gene expression (MGE; mrpl11-1), or in both PGE and MGE (prpl11-1 mrpl11-1), it was found that the prpl11 mrpl11 double mutant, but neither of the single mutants, resulted in strong down-regulation of PhANGs (like that seen in prors1-1; Pesaresi et al., 2006). This implies that, when organellar translation is perturbed, signals derived from both types of organelles cooperate in the regulation of nuclear photosynthetic gene expression (Pesaresi et al., 2006).
Therefore, a loss-of-function allele of GUN1 (gun1-102; see “Materials and Methods”) was combined with prors1-1. In the resulting gun1-102 prors1-1 plants, wild-type-like levels of several PhANG genes (LHCA1, LHCA3, and PSAK) were restored (Fig. 1B), resembling the situation of the mrpl11-1 mutant in which only MGE is perturbed (see above; Pesaresi et al., 2006). Thus, without GUN1 the defect in PGE due to the prors1-1 mutation cannot trigger the corresponding plastid signal that (together with the still-active mitochondrion-derived signal due to the prors1-1-derived defect in MGE) is required to down-regulate PhANGs. This implies that GUN1 also operates in adult leaves and is compatible with the fact that, while GUN1 mRNA is expressed predominantly in young tissues, it also is detectable in older plants (Supplemental Fig. S1D).
Epistasis between gun1 and Mutations Affecting Plastid Ribosomal Proteins
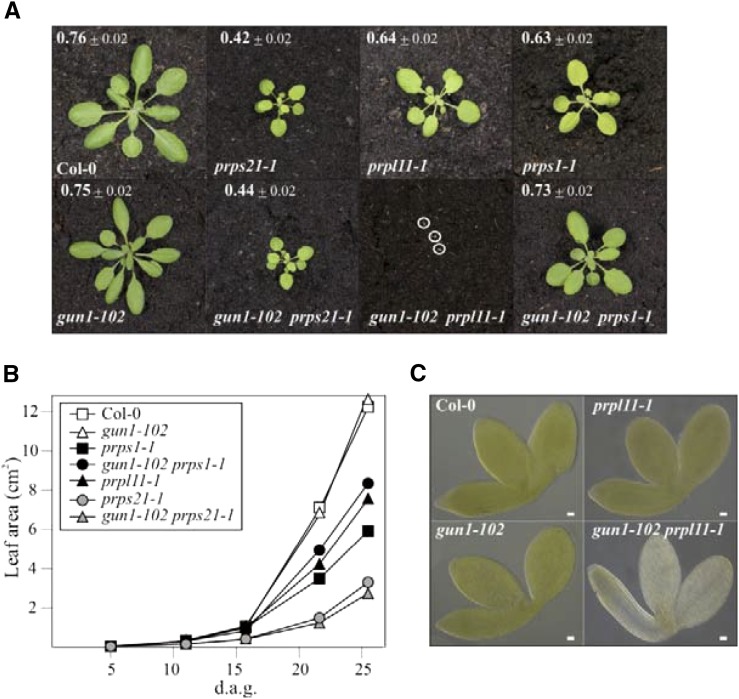
From the genes that are tightly coregulated with GUN1 at the transcript level (Fig. 1A), we selected PRPS1 (coding for plastid ribosomal protein S1) for further analysis. This is because lincomycin, which has been used to define the integrative function of GUN1 (Gray et al., 2003), inhibits plastid ribosome activity at the level of the peptidyltransferase (Sohmen et al., 2009), and PRPS1 is the only plastid ribosomal protein identified by the coexpression analysis (Fig. 1A). To this end, gun1-102 was introduced into genetic backgrounds carrying mutations in genes for PRPS1 and, as controls, for PRPS21 and PRPL11. Notably, while gun1-102 plants displayed wild-type-like growth and photosynthesis, the three ribosomal mutants showed, to varying degrees, perturbations in photosynthetic electron flow and decreased growth rates (Fig. 2A). Furthermore, while the double mutant gun1-102 prps21-1 behaved like the prps21-1 single mutant, gun1-102 prps1-1 displayed an attenuated (suppressor) phenotype and gun1-102 prpl11-1 an exacerbated (enhancer) phenotype (Fig. 2, A and B). Thus, the strongly enhancing effect of gun1-102 resulted in a highly penetrant synthetic seedling-lethal phenotype of gun1-102 prpl11-1 plants (Fig. 2, A and C); only about 3% of the double mutants developed beyond the cotyledon stage. On the contrary, in gun1-102 prps1-1 plants, the negative effect of the prps1-1 mutation on growth and photosynthetic performance was largely suppressed (Fig. 2, A and B), indicating that a functional relationship exists between GUN1 and PRPS1. The epistatic interactions between gun1-102 and mutations affecting plastid ribosomes are specific, because gun2 to gun5 all failed to suppress the prps1-1 growth phenotype and did not induce seedling lethality when combined with the prpl11-1 mutation (Supplemental Fig. S2).
Figure 2.
Interactions between gun1-102 and mutations affecting individual ribosomal proteins. A, Phenotypes of 4-week-old wild-type (Col-0), single (gun1-102, prpl11-1, prps1-1, and prps21-1), and double (gun1-102 prps1-1, gun1-102 prps21-1, and gun1-102 prpl11-1) mutant plants grown in a growth chamber. The effective quantum yield of photosystem II (ΦII) is indicated for each plant (average ± sd; n ≥ 12). Note that the photograph of the albinotic double gun1-102 prpl11-1 mutant plants (highlighted by white circles) was taken at 5 d after germination (d.a.g.). B, Growth kinetics of the different genotypes determined from 5 to 26 d after germination. For each time point, the average leaf area (based on measurements from at least 12 individuals) is provided. sds were <10%. C, Images of fully mature embryos (bent cotyledon stage) from wild-type (Col-0), gun1-102, prpl11-1, and gun1-102 prpl11-1 plants. Albino seedlings also were observed in about 25% of the progeny of gun1-102 PRPL11/prpl11-1 and GUN1/gun1-102 prpl11-1 plants. Scale bars = 20 μm.
GUN1 Controls PRPS1 Accumulation at the Protein Level
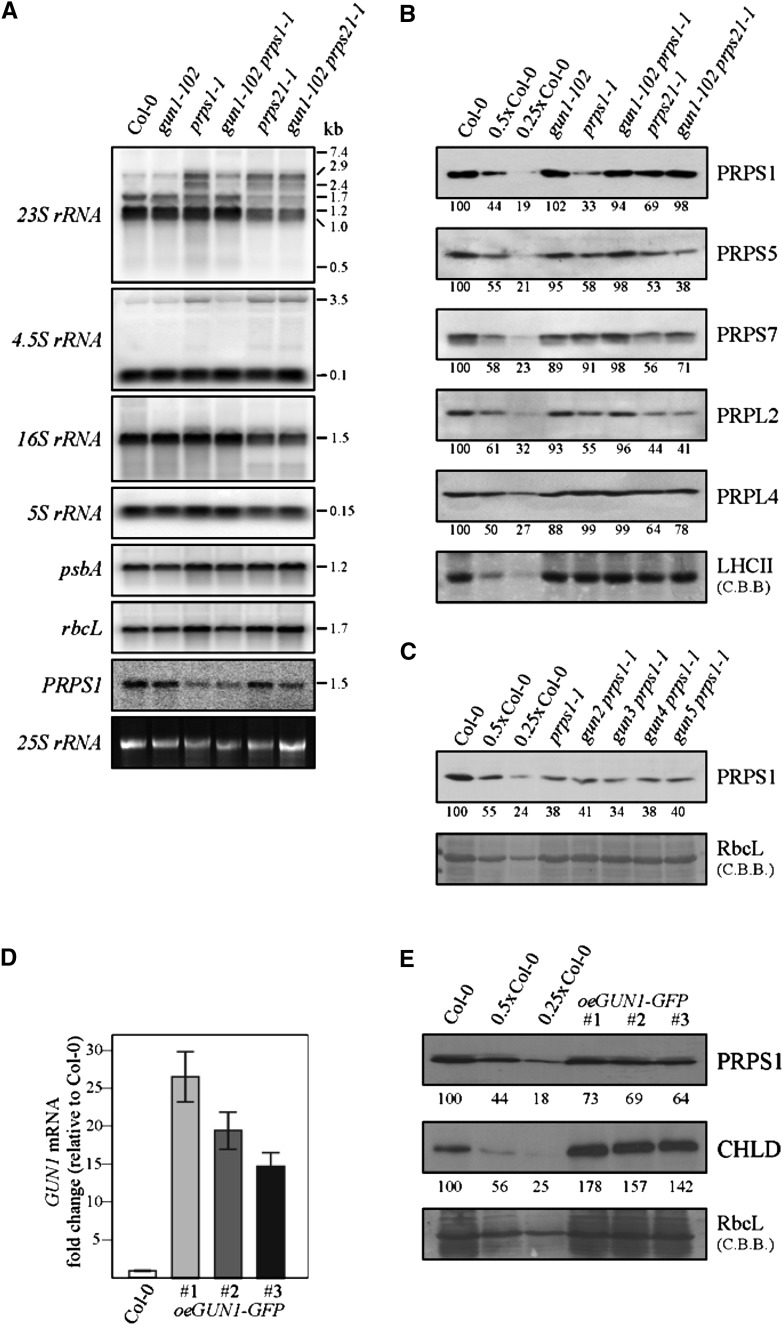
To further study the function of GUN1 in adult plants, we focused on the functional relationship between GUN1 and PRPS1. The prokaryotic ribosomal S1 protein recognizes mRNA leaders and mediates binding of diverse mRNAs to the ribosome at the translation initiation step (for review, see Hajnsdorf and Boni, 2012). Given that this S1 function is conserved in prokaryotes and chloroplasts, complete inactivation of PRPS1 can be expected to result in lethality in Arabidopsis (Arabidopsis thaliana), and such mutants have not been described. Hence, the prps1-1 allele used here is leaky (providing 8% of wild-type PRPS1 transcript levels; Romani et al., 2012). Processing and abundance of plastid rRNAs are not altered in gun1-102, whereas both prps21-1 and prps1-1 strains exhibit aberrant processing of 23S and 4.5S precursor rRNAs (Fig. 3A). These rRNA processing effects are very similar to the ones of other mutants with impaired chloroplast translation (including other plastid ribosomal mutants) and have been interpreted as secondary consequences of impaired chloroplast translation (Tiller et al., 2012). Intriguingly, in gun1-102 prps1-1, but not in gun1-102 prps21-1, the changes in 23S and, to lesser extent, 4.5S processing were largely attenuated (Fig. 3A).
Figure 3.
GUN1 controls PRPS1 accumulation at the protein level. A, RNA gel-blot analyses performed on samples (10 µg) of total RNA from 4-week-old wild-type (Col-0) and mutant (gun1-102, prps1-1, prps21-1, gun1-102 prps1-1, and gun1-102 prps21-1) plants with probes specific for plastid rRNAs (23S, 16S, 5S, and 4.5S) and probes complementary to psbA, rbcL, and PRPS1 mRNAs. The sizes of transcripts (in kb) are shown. Cytosolic 25S rRNA stained with ethidium bromide served as loading control. B, Immunoblot analyses of ribosomal proteins. Nitrocellulose filters carrying fractionated total proteins were probed with antibodies directed against proteins of the 30S (PRPS1, PRPS5, and PRPS7) and 50S (PRPL2 and PRPL5) ribosomal subunits. Decreasing levels of wild-type proteins were loaded in the lanes marked 0.5x Col-0 and 0.25x Col-0. C, Immunoblot analyses as in B were performed on the wild type (Col-0), prps1-1, and double mutants of prps1-1 and gun2-gun5 using a PRPS1-specific antibody. D, Relative expression levels of GUN1 in 4-week-old wild-type (Col-0) and three independent oeGUN1-GFP plants (Col-0 = 100%). The relative level of GUN1 transcript accumulation was determined by real-time PCR of leaf cDNA (three replicates). E, Immunoblot analysis of PRPS1 and CHLD accumulation in the three independent oeGUN1-GFP lines. B, C, and E, To control for loading, gels were stained with Coomassie Brilliant Blue (C.B.B.), and quantification of signals (by ImageJ) relative to the wild type (=100%) is provided below each panel.
At the protein level, reduced PRPS1 accumulation (approximately one-third of wild-type levels) in prps1-1 was associated with decreased levels of PRPS5 and PRPL2 (both ∼ 60% of the wild type; Fig. 3B). In the gun1-102 prps1-1 double mutant, the PRPS1 protein accumulated to wild-type-like levels, and amounts of PRPS5 and PRPL2 were near normal. The T-DNA in prps1-1 disrupts the promoter region of the gene (Romani et al., 2012), and in the gun1-102 prps1-1 mutant PRPS1 mRNA levels were similar to prps1-1 (Fig. 3A), suggesting that the suppressor effects are based on posttranscriptional events. Moreover, in gun1-102 prps21-1, the secondary effect of the prps21-1 mutation on PRPS1 levels was reversed (from ∼ 70% to ∼ 100% of the wild type; Fig. 3B), implying that lack of GUN1 can mitigate decreases in PRPS1 levels irrespective of its original cause: down-regulation of PRPS1 transcription (as in prps1-1) or decreased levels of other ribosomal proteins (as in prps21-1). Again, the suppressor effects observed are specific to gun1-102, because gun2 to gun5 each failed to rescue the accumulation of PRPS1 when combined with the prps1-1 mutation (Fig. 3C). In addition, the decreased formation of polysomes observed in prps1-1 and prps21-1 mutants was attenuated in gun1-102 prps1-1, but not in gun1-102 prps21-1 leaves (Supplemental Fig. S3A). Accordingly, the drop in translation rates observed in prps1-1 was also reversed in gun1-102 prps1-1 plants (Supplemental Fig. S3B).
Because absence of GUN1 can increase PRPS1 levels (at least in the prps1-1 genetic background), it is tempting to assume that higher levels of GUN1 might reduce PRPS1 levels. To test this, lines overexpressing a GUN1:GFP fusion were generated (Fig. 3D; Supplemental Fig. S4, A and B). In fact, the GUN1-GFP protein is functional and can replace GUN1 because the oeGUN1-GFP gun1-102 prpl11-1 mutant is viable and displays prpl11-1-like growth and photosynthesis (Supplemental Fig. S4B). Overexpression of GUN1-GFP in the wild-type background (oeGUN1-GFP plants; Fig. 3D, Supplemental Fig. S4, A and B) reduces accumulation of the PRPS1 protein to about two-thirds of the wild type (Fig. 3E), which supports the idea that GUN1 can negatively regulate PRPS1 levels. Moreover, since amounts of PRPS1 reach already about 175% of wild-type levels in the prpl11-1 mutant (Supplemental Fig. S4C), one plausible explanation for the seedling-lethal phenotype seen in the double mutant gun1-102 prpl11-1 (Fig. 2, A and C) might be that a further increase in PRPS1 levels upon removal of GUN1 in the prpl11-1 background might be lethal. This relates to the observation that overexpression of the ribosomal S1 protein in Escherichia coli leads to the accumulation of “free” S1, which is thought to inhibit translation by sequestering mRNAs, which has a negative impact on the association of mRNAs with ribosomes and the rate of polysome formation (Delvillani et al., 2011). Moreover, this might also explain why previous (Yu et al., 2012) and our own (Supplemental Fig. S4D) attempts to obtain plants overexpressing the PRPS1 protein failed.
Taken together, these results suggest that suppression of the prps1-1 phenotype at the protein level by gun1-102 and exacerbation of the prpl11-1 phenotype by gun1-102 might have the same molecular cause: increased levels of PRPS1 when GUN1 is absent (in the gun1-102 background) and, accordingly, destabilization of PRPS1 when GUN1 is present (in the wild type and oeGUN1-GFP). That the increase in PRPS1 levels in combination with prpl11-1 leads to much more drastic effects than in combination with prps1-1 is a matter of PRPS1 concentrations. In gun1-102 prps1-1, the wild-type-like levels of PRPS1 mitigate the subtle phenotype of the prps1-1 mutation, whereas a further increase in PRPS1 concentrations in gun1-102 prpl11-1 results in lethality, reflecting the effects of overexpression of the S1 protein in E. coli.
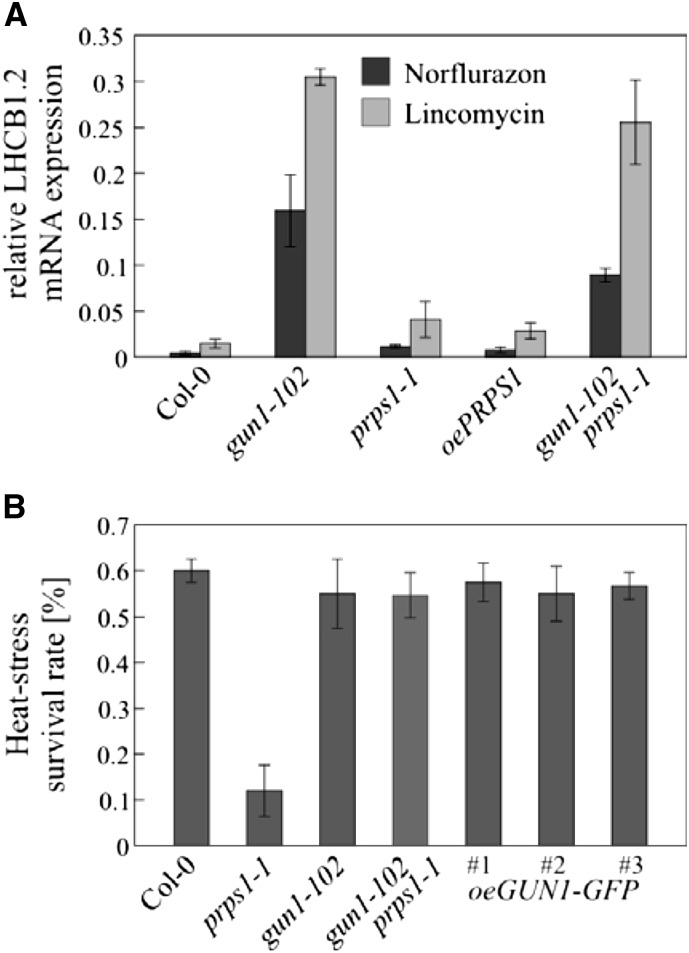
GUN1 and Heat-Responsive Signaling Pathways Are Not Interlinked through PRPS1
Because accumulation of the PRPS1 protein, but not of its RNA template, is induced by heat, and knockdown of PRPS1 results in significant loss of heat tolerance, chloroplast translation capacity has been suggested to be a critical factor in heat-responsive retrograde signaling (Yu et al., 2012). Therefore, both GUN1 function and the cellular response to heat involve PRPS1. To determine whether and how the two pathways are interlinked, we looked at possible functions of (1) PRPS1 in gun signaling and (2) GUN1 in the heat stress response. To this end we generated plants overexpressing the PRPS1 gene (oePRPS1) at the transcript level (Supplemental Fig. S4D). These plants failed to produce increased amounts of the PRPS1 protein—probably because a compensatory mechanism prevents an increase in PRPS1 protein accumulation to avoid harmful interference with translation like the one observed in E. coli where overexpression of RPS1 causes polysome disappearance and translation inhibition (Delvillani et al., 2011). Neither prps1-1 nor oePRPS1 plants show a gun phenotype (Fig. 4A), so we characterized the response to heat stress in plants with different levels of GUN1 expression (gun1-102, Columbia Col-0, and oeGUN1-GFP plants), as well as in prps1-1 and gun1-102 prps1-1 plants as controls. As expected and described earlier (Yu et al., 2012), the survival rate of prps1-1 plants after heat treatment was severely depressed compared with the wild type (Fig. 4B). However, gun1-102 and oeGUN1-GFP lines showed wild-type-like phenotypes after heat challenge (Fig. 4B), indicating that the ∼ 30% drop in PRPS1 levels observed in oeGUN1-GFP plants (Fig. 3F) does not alter heat tolerance sufficiently to be detectable by our assay. As expected from the wild-type-like PRPS1 levels in gun1-102 prps1-1 (Fig. 3B), plants of this genotype displayed a wild-type-like survival rate after heat treatment.
Figure 4.
Relationship between GUN1 signaling and heat-responsive signaling. A, Altered PRPS1 levels are not associated with impairment of NF- or lincomycin-triggered retrograde regulation. Levels of LHCB1.2 mRNA expression were determined by real-time PCR in Col-0, gun1-102, prps1-1, oePRPS1, and gun1-102 prps1-1 seedlings grown for 6 d under continuous light (100 μmol photons m−2 s−1) in the presence of NF or lincomycin. The levels of LHCB1.2 mRNA are expressed relative to those in the Col-0 control (grown in the absence of NF and lincomycin), which were set to 1. The results were normalized to the expression level of At4g36800, which codes for a RUB1-conjugating enzyme. The data are shown as mean values (±sd) from three independent experiments, each done with three technical replicates. B, Altered GUN1 levels do not affect the survival rate of heat-challenged seedlings. Col-0, prps1-1, gun1-102, gun1-102 prps1-1, and oeGUN1-GFP seedlings (2.5 d old) were acclimated to heat at 38°C for 1 h, returned to 22°C for 2 h, and then challenged at 45°C for 3 h. The survival rate of the seedlings was determined after a recovery period of 7 d at 22°C in long-day conditions (16/8 h light-dark cycles). The data are given as mean values (±sd) from two independent experiments, each done with two different plates for every genotype.
The findings presented above demonstrate that a function for GUN1 in regulating heat tolerance appears unlikely. Because prps1-1 and oePRPS1 are not gun mutants, PRPS1 is unlikely to be involved in PGE signaling. However, it cannot be excluded that the alterations in the PRPS1 levels in prps1-1 (which is a leaky mutant allele) and oePRPS1 (with only a very limited, if at all, increase in PRPS1 levels) are insufficient to disrupt PGE signaling to such an extent that a gun phenotype becomes evident. Thus, GUN signaling and heat-responsive signaling appear to be served by different retrograde signaling pathways.
GUN1 Interacts Physically with PRPS1 and Several TPB Enzymes
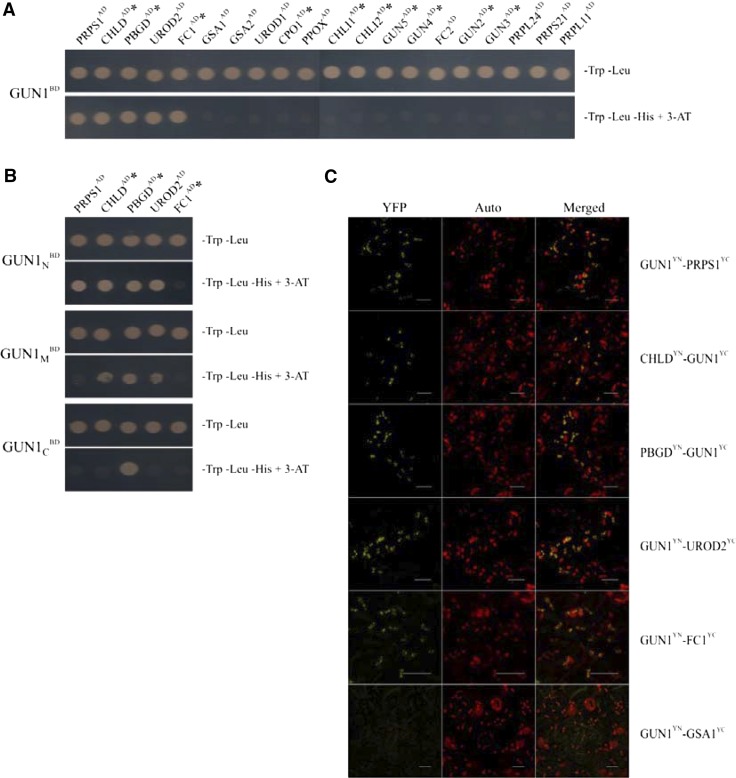
A possible explanation for the negative influence of GUN1 on PRPS1 accumulation is that its signaling function requires physical interactions with components of PGE and TPB. To explore this possibility, we used several approaches to test for physical interactions of GUN1 with PRPS1 and other ribosomal proteins. We also tested for interactions between PRPS1 and several TPB enzymes, including CHLD and protoporphyrinogen oxidase (PPOX)—both of which are tightly coregulated with GUN1 at the transcriptional level (Fig. 1A). In our Y2H analyses, mature GUN1 (GUN143-918) interacted with PRPS1 and CHLD, but not with PPOX or any other ribosomal protein tested (Fig. 5A). In addition, GUN1 interacts with three other TPB enzymes, namely porphobilinogen deaminase (PBGD), uroporphyrinogen III decarboxylase (UROD2), and ferrochelatase I (FC1; Fig. 5A). Interestingly, mutants altered in three of these GUN1 interactors—CHLD, PBGD, and FC1—have been described as gun mutants (Strand et al., 2003; Huang and Li, 2009; Woodson et al., 2011). Two other GUN gene products, coproporphyrinogen III oxidase 1 (CPO1; Strand et al., 2003) and the I subunit of the Mg chelatase (CHLI; Huang and Li, 2009), as well as GUN2 to GUN5, all failed to interact with GUN1 in our Y2H assay (Fig. 5A; Supplemental Fig. S5A). To identify the protein-interacting domain(s) of GUN1, the N-terminal portion of GUN1 (GUN143-251, GUN1N), its PPR-containing domain (GUN1252-687, GUN1M), and the C-terminal segment containing the SMR domain (GUN1688-918, GUN1C) were tested for their capacity to interact with the five proteins that interact with GUN143-918 (Fig. 5B). These experiments showed that all three GUN1 domains can interact with one or more of these proteins, and GUN1N interacts with four of them. GUN1-FC1 interactions might require more than one of the three GUN1 domains tested here because each single one failed to result in interactions with FC1.
Figure 5.
Characterization of protein interactions of GUN1. A and B, Y2H assays. Yeast cells were cotransformed with a plasmid expressing mature GUN1 (GUN143-918; A) or its truncated versions (GUN1N, GUN143-251; GUN1M, GUN1252-687; GUN1C, GUN1688-918; B) as bait proteins and plasmids expressing potential interaction partners as prey proteins (see “Materials and Methods”). Yeast cells were grown on permissive (−Trp −Leu) medium and on selective (−Trp –Leu −His +5 mm 3-AT) medium (which reveals interactions). Reciprocal experiments (exchanging prey and bait) gave identical results. Asterisks indicate GUN gene products. Controls with the empty bait vector are shown in Supplemental Figure S5A. C, BiFC in tobacco leaves detected by fluorescence confocal microscopy. GUN1 and test proteins were either fused to the N-terminal (YN) or the C-terminal (YC) end of the Venus protein, respectively, and cotransformed into tobacco leaves. Reconstitution of YFP fluorescence (signaling positive interaction), chlorophyll autofluorescence (Auto), and their overlay are shown. Reciprocal experiments exchanging YN and YC resulted in identical results, and representative images are shown. Scale bars = 20 μm.
BiFC assays in tobacco-leaf mesophyll cells corroborated the interactions of GUN1 with PRPS1, CHLD, PBGD, UROD2, and FC1, indicating that these interactions also occur in planta (Fig. 5C). The distribution of yellow fluorescence signals resulting from these protein-protein interactions were localized to distinct spots within chloroplasts, resembling the distribution of green fluorescence emitted by the GUN1-GFP construct (Supplemental Fig. S4A). The combination GSA1YN-GUN1YC, used as negative control, failed to produce a YFP signal.
In summary, GUN1 interacts with PRPS1 and several TPB enzymes. Of the latter set, CHLD, PBGD, and FC1 have already been implicated in NF-induced gun signaling.
GUN1 Promotes Formation of Complexes Containing PRPS1 or CHLD
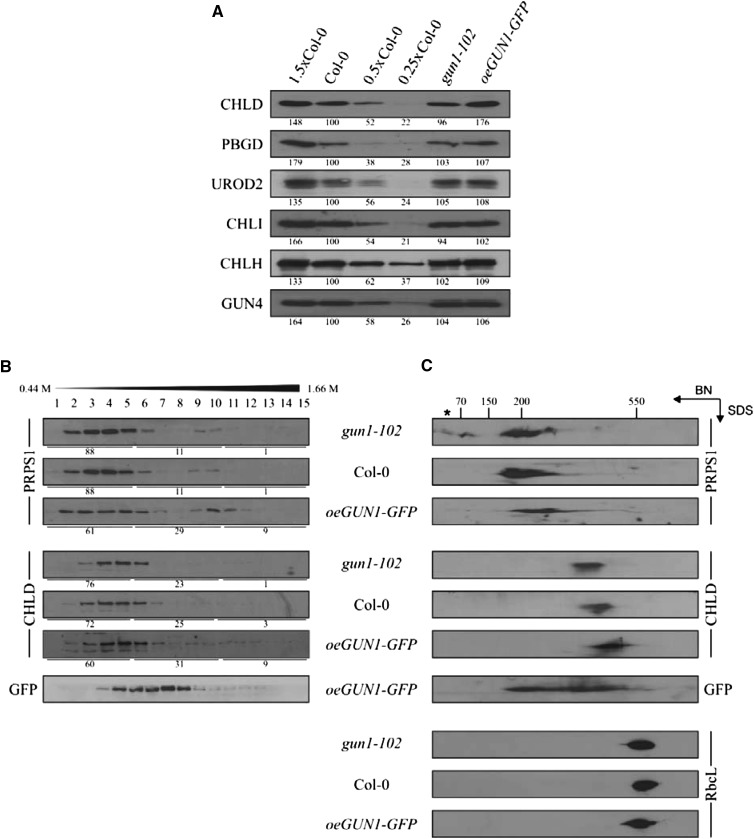
Because (1) GUN1 interacts with PRPS1 and (2) changes in GUN1 levels affect the accumulation of PRPS1 at the protein level (Fig. 3), we tested whether the abundance of the other three GUN1 interactors for which antibodies were available (CHLD, PBGD, and UROD2) also is affected by alterations in GUN1 levels (Fig. 6A). In all three independent oeGUN1-GFP lines, accumulation of CHLD (Fig. 3F)—but not of the corresponding transcript (Supplemental Fig. S5B)—was increased by about 50%, whereas gun1-102 plants have wild-type-like CHLD levels (Fig. 6A). This effect was specific because neither PBGD or UROD2 nor any other protein involved in TPB for which antibodies were available (CHLI, CHLH, and GUN4) varied in its concentration in lines with different levels of GUN1. As expected from the results obtained with gun1-102 prps1-1 plants, also in oeGUN1-GFP plants, the decrease in PRPS1 protein levels is not associated with corresponding variations in PRPS1 mRNA levels (Supplemental Fig. S5B).
Figure 6.
Formation of complexes containing PRPS1 or CHLD in lines with different GUN1 mRNA levels. A, Immunoblot analysis of TPB proteins (CHLD, PBGD, UROD2, CHLI, CHLH, and GUN4) in total protein extracts from the wild-type (Col-0), gun1-102, and oeGUN1-GFP lines. Decreasing wild-type protein concentrations were loaded into lanes 0.5x Col-0 and 0.25x Col-0. B, Suc-gradient analysis of PRPS1 and CHLD proteins. Total chloroplasts from wild-type (Col-0), gun1-102, and oeGUN1-GFP plants were fractionated on Suc gradients and subjected to immunoblot analysis. Distribution of signal intensities (all fractions in each genotype = 100%) in fractions 1-5, 6-10, and 11-15 obtained by ImageJ analysis are provided below each panel. Representative results of three independent experiments are shown. C, Stromal proteins were fractionated by BN/SDS-PAGE, and PRPS1, CHLD, GUN1-GFP, and, as control, RBCL were detected after immunoblotting. Approximate molecular masses of protein complexes were estimated based on the position of multiprotein complexes in the first dimension (BN) separation. Note that the ∼ 40-kD PRPS1 signal (indicated by an asterisk) might represent PRPS1 monomers. A and B, Representative results of three independent experiments are shown and quantification of signals relative to the wild type (=100%) was performed with ImageJ.
In light of these GUN1-dependent changes in the abundance of PRPS1 and CHLD, we analyzed the distribution of PRPS1 and CHLD in protein complexes by Suc-gradient fractionation and blue native (BN)/SDS-PAGE analysis followed by western analysis in gun1-102, wild-type (Col-0) and oeGUN1-GFP plants (Fig. 6, B and C). The two proteins were detected in molecular species with different masses (PRPS1, ∼200 kD; CHLD, ∼400 kD; Fig. 6C), indicating that they associate with distinct complexes. Interestingly, increased GUN1 dosage enhances the stability, or increases the molecular mass, of protein complexes containing PRPS1 or CHLD, as demonstrated by both Suc-gradient (Fig. 6B) and BN/SDS 2D-PAGE gel (Fig. 6C) analyses. Thus, in gun1-102, but not in the wild type or oeGUN1-GFP, PRPS1 monomers accumulate; in oeGUN1-GFP plants, PRPS1-containing complexes clearly have a higher molecular mass than in the wild type (Fig. 6C). For CHLD, increasing doses of GUN1 resulted in a shift of CHLD-containing complexes toward a higher molecular mass (Fig. 6C).
Taken together, the level of GUN1 has an impact on the formation of protein complexes that contain PRPS1 or CHLD.
Proteins Involved in Plastid Translation and Homeostasis Coimmunoprecipitate with GUN1-GFP
Because GUN1-GFP also accumulates in high-molecular-weight complexes, as demonstrated by analyzing oeGUN1-GFP plants with a GFP-specific antibody (Fig. 6C), we analyzed the associations between GUN1-GFP and chloroplast proteins by coimmunoprecipitation experiments. After testing several protocols, only the addition of the crosslinker 3,3′-dithiobis(sulfosuccinimidylpropionate) (DTSSP) was found to result in reproducible results, indicating that interactions between GUN1-GFP and other chloroplast proteins might be weak or transient. Immunoprecipitations with DTSSP were done in four independent experiments with antibodies against GFP on oeGUN1-GFP and wild-type plants as control. Precipitated proteins were identified and quantified by nano liquid chromatography coupled to tandem mass spectrometry (LC-MS/MS). Mean log2 ratios of abundances of proteins precipitated from oeGUN1-GFP versus wild-type lines and corresponding P values of significance, derived by Student’s t test statistics and subsequent adjustment to control the false discovery rate according to Benjamini and Hochberg (1995), were determined (Supplemental Table S1). Proteins were considered as robustly identified when (1) showing more than 1.5-fold difference in abundance between oeGUN1-GFP and wild-type lines and (2) fulfilling the statistical significance criterion of P ≤ 0.05. Only 22 out of the 271 proteins identified and quantified in the eight coimmunoprecipitations fulfill both criteria (Table I; rank 1 to 22 in Supplemental Table S1; Supplemental Fig. S6). Apparently, the use of the crosslinker during the immunoprecipitation experiment has not massively obscured the results of the experiment (with false positive results) because three of the 22 proteins (FUG1, cpHSC70-2, and ClpC1) match transcripts identified as coexpressed with GUN1 (Fig. 1A). The tentative GUN1-GFP interactors identified were markedly enriched for proteins involved in chloroplast protein synthesis and homeostasis, including four subunits of the plastid ribosome, two subunits of chaperonin 60, two heat shock protein 70 variants, as well as pTAC6, FUG1, and a DEAD-box RNA helicase (Table I). Moreover, PRPS1 also was identified although below the chosen threshold level (rank 56; Supplemental Table S1). Components of the TPB pathway were not included in the 22-protein set and CHLD appeared at rank 89 (Supplemental Table S1). Therefore, GUN1 might actually be at least transiently associated with protein complexes involved in plastid protein synthesis and homeostasis.
Table I. Overview of proteins robustly identified in coimmunoprecipitates of GUN1-GFP.
Of a total of 271 proteins identified and quantified by nanoLC-MS/MS, those 22 proteins are listed that show a more than 1.5-fold difference in abundance in oeGUN1-GFP samples relative to control lines (wild type) and fulfill the statistical significance criterion (P ≤ 0.05). cp, Chloroplast protein.
| Rank | Protein ID | Gene | Annotation |
|---|---|---|---|
| 1 | Q9SHI1 | At1g17220a | cp translation initiation factor IF-2 (cpIF2/FUG1) |
| 2 | Q96291, Q9C5R8 | At3g11630, At5g06290 | cp 2-Cys peroxiredoxin BAS1, BAS1-like |
| 3 | Q9U6Y5 | – | GFP (GUN1-GFP signal) |
| 4 | Q9SIC9 | – | GUN1 (GUN1-GFP signal) |
| 5 | P21238 | At2g28000 | cp chaperonin 60 subunit alpha 1 (CPN60 α1) |
| 6 | Q8L7S8 | At5g26742 | cp DEAD-box ATP-dependent RNA helicase 3 (EMB1138) |
| 7 | Q9SIP7, Q9FJA6, Q9M339 | At2g31610, At5g35530, At3g53870 | 40S ribosomal protein S3-1, S3-3, S3-2 |
| 8 | Q9LTX9 | At5g49910a | cp heat shock protein 70-2 (cpHSC70-2) |
| 9 | P56799 | AtCg00380 | cp ribosomal protein S4 |
| 10 | P21240 | At1g55490 | cp chaperonin 60 subunit beta 1 (CPN60 β1) |
| 11 | Q9XI19 | At1g21600 | Plastid transcriptionally active 6 (pTAC6) |
| 12 | Q94CJ5 | At5g12470 | cp RETICULATA-RELATED4 |
| 13 | Q42112, O04204, P57691 | At3g09200, At2g40010, At3g11250 | 60S acidic ribosomal protein P0-2, P0-1, P0-3 |
| 14 | Q9STW6 | At4g24280 | cp heat shock protein 70-1 (cpHSC70-1) |
| 15 | Q9FY50 | At5g13510 | cp ribosomal protein L10 |
| 16 | Q9M3A8 | At3g49240 | Pentatricopeptide repeat-containing protein (EMB1796) |
| 17 | Q9FI56, F4JF64 | At5g50920a, At3g48870 | cp chaperone protein ClpC1, ClpC2 |
| 18 | P19366 | AtCg00480 | cp ATP synthase β-subunit |
| 19 | P56791 | AtCg00830 | cp ribosomal protein L2 |
| 20 | Q9C5C2 | At5g25980 | Myrosinase 2 |
| 21 | O03042 | AtCg00490 | Rubisco large chain (RbcL) |
| 22 | P56798 | AtCg00800 | cp plastid ribosomal protein S3 |
Note that PRPS1 is listed at rank 56 and CHLD at rank 89 (see Supplemental Table S1). aThese three tentative GUN1 interactors also are coexpressed at the transcript level with the GUN1 gene (Fig. 1A).
DISCUSSION
GUN1 Also Functions Beyond the Seedling Stage
Previous analyses have shown that GUN1 has a function in seedling development (Cottage et al., 2010) and the greening of cotyledons (Ruckle et al., 2007). The seedling-lethal phenotype of gun1-102 prpl11-1 reported here (Fig. 2, A and C) strongly corroborates that GUN1 plays a role in exerting biogenic control. Nevertheless, the negative effects of the prors1-1 mutation on PhANG expression in adult leaves are at least partially reversed in the gun1-102 prors1-1 double mutant (Fig. 1B), GUN1 transcripts are detectable in later developmental stages (Supplemental Fig. S1D), and overexpression of GUN1 affects PRPS1 accumulation in adult leaves (Fig. 3E), implying that GUN1 also functions beyond the seedling stage and might contribute to operational control.
GUN1 Is a PPR Protein That Interacts with Proteins
The GUN1 protein contains PPR and SMR domains that suggest a function related to nucleic acids. Only seven other proteins in Arabidopsis, all located in chloroplasts or mitochondria, are PPR-SMR domain proteins like GUN1 (for review, see Liu et al., 2013). Based on their currently known properties and the endonucleolytic activity of SMR proteins from other organisms, functions in the promotion of transcription or of RNA endonuclease activity have been suggested for them (Liu et al., 2013). However, unlike other PPR-SMR proteins, GUN1 seems to be expressed in very small amounts and has not yet been detected by proteomic approaches (Liu et al., 2013).
Analyses of mutants defective in sigma factor genes argued already against a prominent role for GUN1 in plastid RNA metabolism (Woodson et al., 2013), and our nucleotide immunoprecipitation on chip NIP-chip and one-hybrid experiments also failed to detect any significant GUN1-nucleic acid interaction. Instead, our study provides multiple lines of evidence for the notion that GUN1 interacts with several chloroplast proteins, in particular ones involved in PGE or chloroplast protein homeostasis. Interestingly, based on Y2H experiments, the part of GUN1 located N-terminal of the PPR domain appears to be involved in most of these interactions.
Does GUN1-Dependent Formation of Protein Complexes Relate to Plastid Signaling?
The most prominent characteristics of GUN1 at the biochemical level discovered in this study are (1) its ability to physically interact with several chloroplast proteins, and (2) its effect on the stability or formation of complexes containing PRPS1 or CHLD. Therefore, the speculation at hand is that perturbations in PGE and TPB might mobilize specific PGE- and TPB-related components, respectively, making them (more) available for interaction with GUN1 to trigger signaling. Indeed, PRPS1 is the only ribosomal protein that can shuttle from a ribosome-bound to an unbound form to associate with mRNAs (Hajnsdorf and Boni, 2012), so it is conceivable that perturbations in PGE and, hence, in ribosome activity might increase the fraction of ribosome-unbound PRPS1, allowing increased interaction with GUN1. Accordingly, PRPS1 accumulates as the free monomer in the gun1-102 mutant (Fig. 6C), as expected if GUN1 normally captures PRPS1 released from ribosomes to facilitate its integration into complexes with other proteins. However, the apparent low abundance of GUN1 raises the questions how the protein can trigger tentative signaling events associated with such complexes, whether GUN1 is part of the PRPS1- or CHLD-containing complexes, and what physiological functions the GUN1-dependent alterations of the molecular mass of the PRPS1- or CHLD-containing complexes might have. We can only speculate on the physiological relevance of the altered molecular masses of these complexes, and one possibility is that these differences are associated with altered translational (PRPS1-containing complexes) or TPB (CHLD-containing complexes) activities. However, such differences in activities among the gun1 mutant, wild-type plants, and oeGUN1-GFP lines must be subtle or conditional given their wild-type-like phenotypes. Concerning the identity of the proteins that are added to or released from the complexes, we can at least exclude GUN1 as a cause for the differences between gun1 and wild-type plants because PRPS1 and CHLD were not prominently detected in our coimmunoprecipitation experiments and the abundance of the GUN1 protein is too low in wild-type plants to account for the different molecular masses. However, the coimmunoprecipitation data from oeGUN1-GFP plants suggest that GUN1 is associated with PGE components, although PRPS1 was not among the top 22 identified interactors (but at rank 56; see Table I). Therefore, further experiments are needed to decide if and how such GUN1-modulated protein complexes are directly involved in retrograde signaling and other chloroplast processes.
Outlook
The role of GUN1 becomes critical for plant development under certain genetic conditions (see gun1-102 prpl11-1), and our data clearly show that GUN1 also plays a prominent role beyond the seedling stage. Its function in adult plants becomes evident under certain genetic conditions (see gun1-102 prps1-1), and future experiments need to identify environmental conditions in which lack of GUN1 becomes phenotypically evident. Here, we shed light on the relationship between GUN1 and three of the products of genes coexpressed with GUN1 (PRPS1, CHLD, and PPOX), and further studies aiming at the molecular functions of GUN1 will focus on the other GUN1-coexpressed genes and on the interactors of the GUN1 protein as a starting point.
Intriguingly, all three GUN1 interactors identified by both coimmunoprecipitation (Table I) and mRNA coexpression analyses (Fig. 1A) have functions related to chloroplast protein homeostasis. The fug1 mutation can suppress leaf variegation in var1 and var2 mutations that are defective in FtsH5 and FtsH2, respectively (Kato et al., 2009); the two near-identical ClpC paralogs ClpC1 and ClpC2 appear to function primarily within the Clp protease, as the principle stromal protease responsible for maintaining homeostasis (Sjögren et al., 2014); and cpHsc70 proteins are important for protein translocation in chloroplasts (Su and Li, 2010). This, together with the function of GUN1 in regulating the abundance of the PRPS1 protein, suggests that GUN1 is embedded in a network of factors that contribute to chloroplast protein homeostasis.
MATERIALS AND METHODS
Plant Material and Cultivation
The Arabidopsis (Arabidopsis thaliana) T-DNA insertion mutant lines gun1-102 (SAIL_290_D09) and prps21-1 (SAIL_1173_CO3) are both from the SAIL mutant collection (Sessions et al., 2002). The regions flanking the T-DNA insertion in the vector pCSA110 were PCR amplified and sequenced (primer sequences in Supplemental Table S2): gun1-102 contains the T-DNA insertion in exon 2 (position 2313 relative to the start codon); the T-DNA in prps21-1 lies in the only intron (position 1154). Both mutations prevent the accumulation of the respective transcripts, as determined by reverse transcription PCR analyses (primer sequences in Supplemental Table S2), and the gun1-102 line shows a gun phenotype (Fig. 4A). The oeGUN1-GFP lines were generated by introducing the GUN1 coding sequence, under the control of the 35S promoter from Cauliflower mosaic virus, into the wild type (Col-0) using the vector pB7FWG2 (Flanders Interuniversity Institute for Biotechnology, Gent, Belgium). The following previously described mutant lines were used in this work: gun2 and gun3 (Susek et al., 1993), gun4 (Larkin et al., 2003), gun5 (Mochizuki et al., 2001), prpl11-1 (Pesaresi et al., 2001), prps1-1, and oePRPS1 (35S:PRPS1 prps1-1; Romani et al., 2012). Arabidopsis plants were grown on soil in a climate chamber as described (Pesaresi et al., 2009).
Coexpression Analyses
To identify genes represented on the ATH1 microarray (22K) chip that show significant coexpression with GUN1, an expression correlation analysis with the CoExSearch tool implemented in ATTED-II (http://atted.jp/; Obayashi et al., 2007; Obayashi et al., 2009) was performed. Hierarchical clustering was carried out with the single linkage method provided by the HCluster tool in ATTED-II. Subcellular localizations for the different proteins were inferred from TAIR (http://arabidopsis.org/) and the “subcellular localization of proteins in Arabidopsis” database (SUBA3; http://suba.plantenergy.uwa.edu.au/).
Chlorophyll a Fluorescence Measurements
In vivo chlorophyll a fluorescence of leaves was measured as described (Pesaresi et al., 2009) employing a Dual-PAM-100 (Walz). Whole-plant chlorophyll a fluorescence was recorded using an imaging chlorophyll fluorometer (Walz) as reported earlier (Armbruster et al., 2013).
Nucleic Acid Analyses
Arabidopsis genomic DNA was isolated (Ihnatowicz et al., 2004), and RNA was purified from total leaf frozen tissue as before (Armbruster et al., 2010). Northern analysis was performed under stringent conditions (Sambrook and Russell, 2001) on 10-µg samples of total RNA. Probes complementary to nuclear and chloroplast genes were used for the hybridizations. Primers used to amplify the probes are listed in Supplemental Table S2. All probes used were cDNA fragments labeled with 32P. Quantitative real-time PCR profiling was done as described previously (Voigt et al., 2010). All reactions were performed in triplicate on three biological replicates, and primers are listed in Supplemental Table S2.
Immunoblot Analyses
Immunoblot analyses were carried out as described (Ihnatowicz et al., 2004) and immunodecorated with specific antibodies. Antibodies directed against plastid ribosomal proteins were obtained from Uniplastomic, the GFP antibody from Life Technologies, and the RbcL antibody from Agrisera. Antibodies specific for TPB enzymes were obtained from R.M. Larkin (Michigan State University; CHLI, CHLD, and CHLH), P.E. Jensen (CHLD), B. Grimm (Humbolt University; GUN4), and A. Smith (University of Cambridge; PBGD and UROD). The level of signals was quantified by the ImageJ software (http://imagej.nih.gov/ij/index.html).
Protein Complex Analyses
For Suc-gradient analysis, intact chloroplasts were isolated and solubilized in extraction buffer (Kunst, 1998) containing 0.6% (v/v) NP-40 detergent (15 min, 4°C). After centrifugation (16,000g for 15 min), the supernatant was layered on a Suc step-gradient (15%–55%, w/v) and centrifuged (5 h, 240,000g). Fifteen fractions were collected and analyzed by SDS-PAGE on a 12% (w/v) PAA gel.
For BN/SDS-PAGE analysis of stromal protein complexes, chloroplasts from 4-week-old leaf material (corresponding to 60 µg of Chl) were isolated as described above; resuspended in 100 µL of 30 mm HEPES-KOH, pH 8.0, 60 mm KOAc, and 10 mm MgOAc; solubilized by adding NP-40 (final concentration 0.5%, v/v); and centrifuged (16,000g, 15 min, 4°C). The supernatant was then analyzed by BN/SDS-PAGE as described previously (Qi et al., 2012).
For two-hybrid assays, the coding sequences for the proteins of interest, devoid of the chloroplast transit peptides (for primer sequences, see Supplemental Table S2), were cloned into pGBKT7 (GUN1) and pGADT7 (PRPS1, PRPS21, PRPL11, and PRPL24; CHLD; FC1 and FC2; PBGD; UROD1 and UROD2; CPO1; GSA1 and GSA2; CHLI1 and CHLI2; PPOX; and GUN2–GUN5) vectors (Clontech) or vice versa. Interactions in yeast were then analyzed as described before (DalCorso et al., 2008).
For coimmunoprecipitation analysis, intact chloroplasts were isolated from 10 g of fresh leaf material in extraction buffer (Kunst, 1998) containing 5 mm DTSSP (Pierce; Thermo Scientific) and incubated for 1 h at 0°C. The reaction was quenched at a final concentration of 30 mm Tris/HCl, pH 7.0, and samples were solubilized by adding NP-40 (final concentration 0.75%, v/v) for 15 min and centrifuged (16,000g, 15 min, 4°C). NaCl was added to the supernatant to a concentration of 200 mm and incubated for 2 h with 30 µL of magnetic GFP-Trap (Chromotek). After three washing steps, samples were resuspended in reducing Laemmli buffer and heated for 5 min at 95°C.
Analysis of Coimmunoprecipitations by Mass Spectrometry
Proteins immunoprecipitated with antibodies against GFP were separated by SDS-PAGE, and proteins were fixed in the gels by washing in water/ethanol/acetic acid (50:40:10). Protein-containing bands were excised and washed twice in 50 mm NH4HCO3/acetonitrile (50:50). Subsequently, proteins were reduced with dithiothreitol (12 mm) and alkylated using iodoacetamide (50 mm). After dehydration the gels were soaked with trypsin solution (12.5 ng/µL trypsin in 25 mm NH4HCO3) and incubated for 12 h at 37°C. After the addition of formic acid (1% final concentration), tryptic peptides were extracted twice, first using extraction buffer (5% acetonitrile, 1% formic acid) and then using extraction buffer/acetonitrile (50:50). Peptide extracts were dried and dissolved in HPLC buffer A (3% acetonitrile, 0.1% formic acid), and 2 µL or 5 µL were analyzed by LC-MS/MS (Eksigent nanoLC 425 coupled to TripleTOF 6600; ABSciex). Peptides were separated by reversed phase (symmetry C18, 5 µm particles, 180 µm × 20 mm as trapping column and C18-AQ, 1.9 µm particles, 75 µm x 150 mm as analytical column) using a flow rate of 300 nL/min and 30 min or 90 min gradients from 2% to 35% HPLC buffer B (90% acetonitrile, 0.1% formic acid). Peptides were ionized by nanoESI and analyzed in the mass spectrometer, first generating a TOF MS in the m/z range of 350 to 1500, then sequentially fragmenting the 20 most intense multiple charged precursors resulting in a total cycle time of 1.7 s. The selected precursors were excluded from MS/MS analysis for 12 s. Based on the UniProt Knowledgebase for Arabidopsis proteins, peptide identification, protein assembly, and protein quantification were performed with MaxQuant software (Cox and Mann, 2008) using default parameters, but allowing four missed cleavages and variable Lys modification with 3-(carbamidomethylthio)propanoyl caused by crosslinking.
BiFC Analyses
Cloning of genes into pVyNE or pVyCE (Gehl et al., 2009), which carry sequences encoding the N-terminal or the C-terminal portion of the Venus protein (a YFP derivative), respectively, transformation of Agrobacterium tumefaciens, infiltration of Nicotiana benthamiana leaves, and BiFC analyses were performed as described (Richter et al., 2013).
Heat Stress Treatment
The acquired heat tolerance test of seedlings was performed as described (Yu et al., 2012): 2.5-d-old seedlings, grown on half-strength Murashige and Skoog medium, were first acclimated to heat at 38°C for 1 h, returned to 22°C for 2 h, and then challenged at 45°C for 3 h. All heat treatments were performed in the dark. The seedlings were allowed to recover for 7 d in a growth chamber at 22°C under 16/8 h light-dark cycles, before the survival rate was determined.
Accession Numbers
The genes for the following proteins are coexpressed with GUN1: cpHSC70-2, chloroplast heat shock protein 70-2 (At5g49910); ADC1, Arg decarboxylase 1 (At2g16500); RNASE, RNase III family protein (At4g37510); PRPS1, plastid ribosomal protein S1 (At5g30510); PROTEASE, zincin-like metalloproteases family protein (At5g65620); FTSH3, FtsH protease 3 (At2g29080); PEPTIDASE, prolyl oligopeptidase family protein (At2g47390); GLDP1, Gly decarboxylase P-protein 1 (At4g33010); DIT1, dicarboxylate transporter 1 (At5g12860); mtLPD1, mitochondrial lipoamide dehydrogenase 1 (At1g48030); TRANSKETOLASE (At3g60750); DHS2, 3-deoxy-d-arabino-heptulosonate 7-phosphate synthase (At4g33510); MTO1, Met overaccumulation 1 (At3g01120); RECA1, homolog of bacterial RECA (At1g79050); TIC110/TOC159, translocon at the inner/outer envelope membrane of chloroplasts 110/159 (At1g06950/At4g02510); PREP1, presequence protease 1 (At3g19170); CLPC1, ClpC homolog 1 (At5g50920); ALATS, alanyl-tRNA synthetase (At1g50200); PEPTIDASE M1, peptidase M1 family protein (At1g63770); TPR, tetratricopeptide family protein (At1g02150); FTSH, putative FtsH protease (At3g02450); CFM2, CRM family member 2 (At3g01370); FUG1, FU-GAERI1 (At1g17220); SCO1, snowy cotyledon 1 (At1g62750); OB-FOLD, nucleic acid-binding OB-fold-like protein (At1g12800); EMB1865 and EMB2726, embryo defective 1865 and 2726 (At3g18390 and At4g29060); CHLD, magnesium-chelatase subunit D (At1g08520); and PPOX, protoporphyrinogen oxidase (At4g01690).
The genes analyzed by northern-blot and polysome analysis were: 16S rRNA (AtCg00920), 23S rRNA (AtCg01180), 4.5S rRNA (AtCg00960), 5S rRNA (AtCg00970), rbcL (AtCg00490), psbA (AtCg00020), PRPS1 (see above), LHCA1 (At3g54890), LHCA3 (At1g61520), LHCA4 (At3g47470), LHCB1.2 (At1g29910), LHCB2.2 (At2g05070), LHCB5 (At4g10340), LHCB6 (At1g15820), PSAE1 (At4g28750), PSAK (At1g30380), PSAO (At1g08380), PSBQ1 (At4g21280), PSBY (At1g67740), and RBCS (At5g38420).
The following proteins were analyzed by Y2H and in some cases also by BiFC analyses: PRPS1, CHLD, and PPOX (see above); PRPS21 (At3g27160), PRPL11 (At1g32990), and PRPL24 (At5g54600); FC1, ferrochelatase I (At5g26030), and FC2 (At2g30390); PBGD, porphobilinogen deaminase (At5g08280); UROD1, uroporphyrinogen III decarboxylase (At3g14930), and UROD2 (At2g40490); CPO1, coproporphyrinogen III oxidase (At1g03475); GSA1, Glu-1-semialdehyde 2,1-aminomutase (At5g63570), and GSA2 (At3g48730); CHLI1, Mg-chelatase I subunit (At4g18480) and CHLI2 (At5g45930); and GUN2 (At2g26670), GUN3 (At3g09150), GUN4 (At3g59400), and GUN5 (At5g13630).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. The PPR protein GUN1 lacks obvious DNA- or RNA-binding activity and is expressed at several stages in development.
Supplemental Figure S2. Interactions between gun2 to gun5 mutations and two mutations affecting single ribosomal proteins (prpl11-1 and prps1-1).
Supplemental Figure S3. Characterization of plastid translation efficiency in wild-type (Col-0) and mutant plants (gun1-102, prps1-1, gun1-102 prps1-1, prps21-1, and gun1-102 prps21-1).
Supplemental Figure S4. GUN1 and PRPS1 overexpressor plants.
Supplemental Figure S5. Characterization of protein interactions of GUN1.
Supplemental Figure S6. Volcano plot of P values against log2-transformed differences in abundances of coimmunoprecipitated proteins.
Supplemental Table S1. List of all proteins identified in coimmunoprecipitates of GUN1-GFP.
Supplemental Table S2. Primers used in this study.
Supplemental Text S1. Supplemental Materials and Methods; Supplemental Literature Cited.
Supplementary Material
Acknowledgments
We thank Sabrina Finster, Christiane Kupsch, and Christian Schmitz-Linneweber for their support in nucleotide immunoprecipitation on chip NIP-chip and PPR prediction analyses, and Poul Erik Jensen and Pablo Pulido for fruitful discussions.
Glossary
- PGE
plastid gene expression
- TPB
tetrapyrrole biosynthesis
- NF
norflurazon
- PhANG
photosynthesis-associated nuclear gene
- PPR
pentatricopeptide repeat
- SMR
small MutS-related
- BiFC
bimolecular fluorescence complementation
- MGE
mitochondrial gene expression
- Y2H
yeast two-hybrid
- BN
blue native
- DTSSP
3,3′-dithiobis(sulfosuccinimidylpropionate)
- LC-MS/MS
liquid chromatography coupled to tandem mass spectrometry
Footnotes
This work was supported by the Deutsche Forschungsgemeinschaft (FOR 804).
References
- Armbruster U, Rühle T, Kreller R, Strotbek C, Zühlke J, Tadini L, Blunder T, Hertle AP, Qi Y, Rengstl B, et al. (2013) The photosynthesis affected mutant68-like protein evolved from a PSII assembly factor to mediate assembly of the chloroplast NAD(P)H dehydrogenase complex in Arabidopsis. Plant Cell 25: 3926–3943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armbruster U, Zühlke J, Rengstl B, Kreller R, Makarenko E, Rühle T, Schünemann D, Jahns P, Weisshaar B, Nickelsen J, et al. (2010) The Arabidopsis thylakoid protein PAM68 is required for efficient D1 biogenesis and photosystem II assembly. Plant Cell 22: 3439–3460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkan A, Small I (2014) Pentatricopeptide repeat proteins in plants. Annu Rev Plant Biol 65: 415–442 [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Roy Stat Soc B 57: 289–300 [Google Scholar]
- Chi W, Sun X, Zhang L (2013) Intracellular signaling from plastid to nucleus. Annu Rev Plant Biol 64: 559–582 [DOI] [PubMed] [Google Scholar]
- Cottage A, Mott EK, Kempster JA, Gray JC (2010) The Arabidopsis plastid-signalling mutant gun1 (genomes uncoupled1) shows altered sensitivity to sucrose and abscisic acid and alterations in early seedling development. J Exp Bot 61: 3773–3786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox J, Mann M (2008) MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol 26: 1367–1372 [DOI] [PubMed] [Google Scholar]
- DalCorso G, Pesaresi P, Masiero S, Aseeva E, Schünemann D, Finazzi G, Joliot P, Barbato R, Leister D (2008) A complex containing PGRL1 and PGR5 is involved in the switch between linear and cyclic electron flow in Arabidopsis. Cell 132: 273–285 [DOI] [PubMed] [Google Scholar]
- Delvillani F, Papiani G, Dehò G, Briani F (2011) S1 ribosomal protein and the interplay between translation and mRNA decay. Nucleic Acids Res 39: 7702–7715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estavillo GM, Crisp PA, Pornsiriwong W, Wirtz M, Collinge D, Carrie C, Giraud E, Whelan J, David P, Javot H, et al. (2011) Evidence for a SAL1-PAP chloroplast retrograde pathway that functions in drought and high light signaling in Arabidopsis. Plant Cell 23: 3992–4012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukui K, Kuramitsu S (2011) Structure and function of the small MutS-related domain. Mol Biol Int 2011: 691735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehl C, Waadt R, Kudla J, Mendel RR, Hänsch R (2009) New GATEWAY vectors for high throughput analyses of protein-protein interactions by bimolecular fluorescence complementation. Mol Plant 2: 1051–1058 [DOI] [PubMed] [Google Scholar]
- Gray JC, Sullivan JA, Wang JH, Jerome CA, MacLean D (2003) Coordination of plastid and nuclear gene expression. Philos Trans R Soc Lond B Biol Sci 358: 135–144; discussion 144–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajnsdorf E, Boni IV (2012) Multiple activities of RNA-binding proteins S1 and Hfq. Biochimie 94: 1544–1553 [DOI] [PubMed] [Google Scholar]
- Huang YS, Li HM (2009) Arabidopsis CHLI2 can substitute for CHLI1. Plant Physiol 150: 636–645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ihnatowicz A, Pesaresi P, Varotto C, Richly E, Schneider A, Jahns P, Salamini F, Leister D (2004) Mutants for photosystem I subunit D of Arabidopsis thaliana: effects on photosynthesis, photosystem I stability and expression of nuclear genes for chloroplast functions. Plant J 37: 839–852 [DOI] [PubMed] [Google Scholar]
- Isemer R, Mulisch M, Schäfer A, Kirchner S, Koop HU, Krupinska K (2012) Recombinant Whirly1 translocates from transplastomic chloroplasts to the nucleus. FEBS Lett 586: 85–88 [DOI] [PubMed] [Google Scholar]
- Jarvis P, López-Juez E (2013) Biogenesis and homeostasis of chloroplasts and other plastids. Nat Rev Mol Cell Biol 14: 787–802 [DOI] [PubMed] [Google Scholar]
- Kato Y, Miura E, Ido K, Ifuku K, Sakamoto W (2009) The variegated mutants lacking chloroplastic FtsHs are defective in D1 degradation and accumulate reactive oxygen species. Plant Physiol 151: 1790–1801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koussevitzky S, Nott A, Mockler TC, Hong F, Sachetto-Martins G, Surpin M, Lim J, Mittler R, Chory J (2007) Signals from chloroplasts converge to regulate nuclear gene expression. Science 316: 715–719 [PubMed] [Google Scholar]
- Kunst L. (1998) Preparation of physiologically active chloroplasts from Arabidopsis. Methods Mol Biol 82: 43–48 [DOI] [PubMed] [Google Scholar]
- Larkin RM, Alonso JM, Ecker JR, Chory J (2003) GUN4, a regulator of chlorophyll synthesis and intracellular signaling. Science 299: 902–906 [DOI] [PubMed] [Google Scholar]
- Leister D, Wang X, Haberer G, Mayer KF, Kleine T (2011) Intracompartmental and intercompartmental transcriptional networks coordinate the expression of genes for organellar functions. Plant Physiol 157: 386–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Melonek J, Boykin LM, Small I, Howell KA (2013) PPR-SMRs: ancient proteins with enigmatic functions. RNA Biol 10: 1501–1510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochizuki N, Brusslan JA, Larkin R, Nagatani A, Chory J (2001) Arabidopsis genomes uncoupled 5 (GUN5) mutant reveals the involvement of Mg-chelatase H subunit in plastid-to-nucleus signal transduction. Proc Natl Acad Sci USA 98: 2053–2058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nott A, Jung HS, Koussevitzky S, Chory J (2006) Plastid-to-nucleus retrograde signaling. Annu Rev Plant Biol 57: 739–759 [DOI] [PubMed] [Google Scholar]
- Obayashi T, Hayashi S, Saeki M, Ohta H, Kinoshita K (2009) ATTED-II provides coexpressed gene networks for Arabidopsis. Nucleic Acids Res 37: D987–D991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obayashi T, Kinoshita K, Nakai K, Shibaoka M, Hayashi S, Saeki M, Shibata D, Saito K, Ohta H (2007) ATTED-II: a database of co-expressed genes and cis elements for identifying co-regulated gene groups in Arabidopsis. Nucleic Acids Res 35: D863–D869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pesaresi P, Hertle A, Pribil M, Kleine T, Wagner R, Strissel H, Ihnatowicz A, Bonardi V, Scharfenberg M, Schneider A, et al. (2009) Arabidopsis STN7 kinase provides a link between short- and long-term photosynthetic acclimation. Plant Cell 21: 2402–2423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pesaresi P, Masiero S, Eubel H, Braun HP, Bhushan S, Glaser E, Salamini F, Leister D (2006) Nuclear photosynthetic gene expression is synergistically modulated by rates of protein synthesis in chloroplasts and mitochondria. Plant Cell 18: 970–991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pesaresi P, Varotto C, Meurer J, Jahns P, Salamini F, Leister D (2001) Knock-out of the plastid ribosomal protein L11 in Arabidopsis: effects on mRNA translation and photosynthesis. Plant J 27: 179–189 [DOI] [PubMed] [Google Scholar]
- Pogson BJ, Woo NS, Förster B, Small ID (2008) Plastid signalling to the nucleus and beyond. Trends Plant Sci 13: 602–609 [DOI] [PubMed] [Google Scholar]
- Qi Y, Armbruster U, Schmitz-Linneweber C, Delannoy E, de Longevialle AF, Rühle T, Small I, Jahns P, Leister D (2012) Arabidopsis CSP41 proteins form multimeric complexes that bind and stabilize distinct plastid transcripts. J Exp Bot 63: 1251–1270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramel F, Birtic S, Ginies C, Soubigou-Taconnat L, Triantaphylidès C, Havaux M (2012) Carotenoid oxidation products are stress signals that mediate gene responses to singlet oxygen in plants. Proc Natl Acad Sci USA 109: 5535–5540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter AS, Peter E, Rothbart M, Schlicke H, Toivola J, Rintamäki E, Grimm B (2013) Posttranslational influence of NADPH-dependent thioredoxin reductase C on enzymes in tetrapyrrole synthesis. Plant Physiol 162: 63–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romani I, Tadini L, Rossi F, Masiero S, Pribil M, Jahns P, Kater M, Leister D, Pesaresi P (2012) Versatile roles of Arabidopsis plastid ribosomal proteins in plant growth and development. Plant J 72: 922–934 [DOI] [PubMed] [Google Scholar]
- Ruckle ME, DeMarco SM, Larkin RM (2007) Plastid signals remodel light signaling networks and are essential for efficient chloroplast biogenesis in Arabidopsis. Plant Cell 19: 3944–3960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Russell DW (2001) Molecular Cloning: A Laboratory Manual, Ed 3. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Sessions A, Burke E, Presting G, Aux G, McElver J, Patton D, Dietrich B, Ho P, Bacwaden J, Ko C, et al. (2002) A high-throughput Arabidopsis reverse genetics system. Plant Cell 14: 2985–2994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjögren LL, Tanabe N, Lymperopoulos P, Khan NZ, Rodermel SR, Aronsson H, Clarke AK (2014) Quantitative analysis of the chloroplast molecular chaperone ClpC/Hsp93 in Arabidopsis reveals new insights into its localization, interaction with the Clp proteolytic core, and functional importance. J Biol Chem 289: 11318–11330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohmen D, Harms JM, Schlunzen F, Wilson DN (2009) SnapShot: antibiotic inhibition of protein synthesis I. Cell 138: 1248–1248.e1 [DOI] [PubMed] [Google Scholar]
- Strand A, Asami T, Alonso J, Ecker JR, Chory J (2003) Chloroplast to nucleus communication triggered by accumulation of Mg-protoporphyrinIX. Nature 421: 79–83 [DOI] [PubMed] [Google Scholar]
- Su PH, Li HM (2010) Stromal Hsp70 is important for protein translocation into pea and Arabidopsis chloroplasts. Plant Cell 22: 1516–1531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X, Feng P, Xu X, Guo H, Ma J, Chi W, Lin R, Lu C, Zhang L (2011) A chloroplast envelope-bound PHD transcription factor mediates chloroplast signals to the nucleus. Nat Commun 2: 477. [DOI] [PubMed] [Google Scholar]
- Susek RE, Ausubel FM, Chory J (1993) Signal transduction mutants of Arabidopsis uncouple nuclear CAB and RBCS gene expression from chloroplast development. Cell 74: 787–799 [DOI] [PubMed] [Google Scholar]
- Tiller N, Weingartner M, Thiele W, Maximova E, Schöttler MA, Bock R (2012) The plastid-specific ribosomal proteins of Arabidopsis thaliana can be divided into non-essential proteins and genuine ribosomal proteins. Plant J 69: 302–316 [DOI] [PubMed] [Google Scholar]
- Voigt C, Oster U, Börnke F, Jahns P, Dietz KJ, Leister D, Kleine T (2010) In-depth analysis of the distinctive effects of norflurazon implies that tetrapyrrole biosynthesis, organellar gene expression and ABA cooperate in the GUN-type of plastid signalling. Physiol Plant 138: 503–519 [DOI] [PubMed] [Google Scholar]
- Woodson JD, Chory J (2008) Coordination of gene expression between organellar and nuclear genomes. Nat Rev Genet 9: 383–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodson JD, Perez-Ruiz JM, Chory J (2011) Heme synthesis by plastid ferrochelatase I regulates nuclear gene expression in plants. Curr Biol 21: 897–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodson JD, Perez-Ruiz JM, Schmitz RJ, Ecker JR, Chory J (2013) Sigma factor-mediated plastid retrograde signals control nuclear gene expression. Plant J 73: 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Y, Savchenko T, Baidoo EE, Chehab WE, Hayden DM, Tolstikov V, Corwin JA, Kliebenstein DJ, Keasling JD, Dehesh K (2012) Retrograde signaling by the plastidial metabolite MEcPP regulates expression of nuclear stress-response genes. Cell 149: 1525–1535 [DOI] [PubMed] [Google Scholar]
- Yu HD, Yang XF, Chen ST, Wang YT, Li JK, Shen Q, Liu XL, Guo FQ (2012) Downregulation of chloroplast RPS1 negatively modulates nuclear heat-responsive expression of HsfA2 and its target genes in Arabidopsis. PLoS Genet 8: e1002669. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.