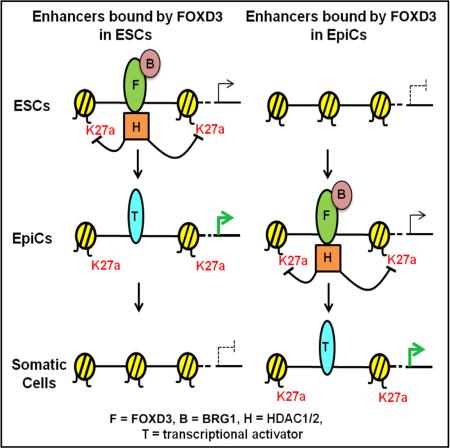
SUMMARY
Early development is governed by the ability of pluripotent cells to retain the full range of developmental potential and respond accurately to developmental cues. This property is achieved in large part by the temporal and contextual regulation of gene expression by enhancers. Here, we evaluated regulation of enhancer activity during differentiation of embryonic stem to epiblast cells and uncovered the forkhead transcription factor FOXD3 as a major regulator of the developmental potential of both pluripo-tent states. FOXD3 bound to distinct sites in the two cell types priming enhancers through a dual-functional mechanism. It recruited the SWI/SNF chromatin remodeling complex ATPase BRG1 to promote nucleosome removal while concurrently inhibiting maximal activation of the same enhancers by recruiting histone deacetylases1/2. Thus, FOXD3 prepares cognate genes for future maximal expression by establishing and simultaneously repressing enhancer activity. Through switching of target sites, FOXD3 modulates the developmental potential of pluripotent cells as they differentiate.
Graphical Abstract
INTRODUCTION
One of the most extraordinary and rapid sequences of cell fate transitions occurs prior to gastrulation. In mammals, these transitions include the establishment of the pluripotent populations of the inner cell mass, the early epiblast, and the late epiblast (Arnold and Robertson, 2009; Rossant and Tam, 2009; Snow, 1977). This period represents a continuum that prepares the pluripotent cells of the epiblast for differentiation into all of the cell types of the adult body. Little is known about the drivers of these transitions and the mechanisms by which these cells retain their developmental potential during this window. One of the primary driving forces of cell fate transitions is the sequence-dependent binding of transcription factors (TFs) and the associated changes in gene expression. TF binding is regulated by DNA accessibility as determined by local chromatin structure. The mechanisms by which TFs regulate the precise developmental timing of gene expression during the pluripotent transition is not well understood.
Enhancers are the major DNA elements that regulate cell-specific gene expression (Bulger and Groudine, 2011; Calo and Wysocka, 2013; Shlyueva et al., 2014). Therefore, to understand the mechanisms that regulate developmental timing of gene expression, it is essential to identify the underlying enhancers driving gene expression and the TFs that bind to and regulate the activity of these enhancers. Enhancers can be identified on a global scale by the coincidence of several histone marks including his-tone 3 lysine 4 monomethylation (H3K4me1), histone 3 lysine 27 acetylation (H3K27ac), and histone 3 lysine 27 trimethylation (H3K27me3) together with the histone acetyltransferase (HAT) P300 (Bulger and Groudine, 2011; Calo and Wysocka, 2013; Shlyueva et al., 2014). Furthermore, the combination of these factors distinguishes multiple enhancer states including active, primed, and poised. Active enhancers have high levels of H3K4me1, H3K27ac, and P300 binding while primed enhancers have H3K4me1 alone. Poised enhancers consist of H3K4me1, H3K27me3, and P300 and are hypothesized to represent a class of enhancers that are not fully activated until H3K27me3 is removed (Creyghton et al., 2010; Rada-Iglesias et al., 2011). Our ability to identify enhancers through these marks has led to an explosion in their discovery across many cell and tissue types. Current estimates predict that there are over a million enhancers in the mammalian genome (ENCODE Project Consortium, 2012). However, our understanding of enhancer dynamics as well as the TFs that drive these changes has been limited to a small number of cell fate transitions (Hawkins et al., 2011; Wamstad et al., 2012; Whyte et al., 2012).
Embryonic stem cells (ESCs), which are the in vitro counterpart to the early epiblast, provide a starting point from which we can begin to uncover the chromatin changes that occur during early mammalian development (Nichols and Smith, 2011). ESCs can be differentiated into epiblast cells (EpiCs), which have molecular profiles similar to the late epiblast (Hayashi et al., 2011; Kojima et al., 2014). These cells then go on to produce cells of all three germ layers (Bradley et al., 1984; Keller, 2005; Smith, 2001). Recently described reporters that are distinctly expressed in these early cell states make it possible to follow the transition from ESCs to EpiCs at a resolution previously unachievable (Buecker et al., 2014; Parchem et al., 2014).
Here we took advantage of one of these reporter systems to evaluate global enhancer transitions during the tightly controlled ESC to EpiC developmental window. Furthermore, we used the dynamic changes in enhancer states to identify TFs critical to this developmental transition. We uncovered FOXD3 as a major regulator of enhancers during the differentiation of ESCs. FOXD3 bound largely mutually exclusive sites in the ESCs and EpiCs. Its binding promoted nucleosome depletion at enhancers through recruitment of the chromatin remodeler BRG1, yet it prevented full enhancer acetylation and activation by recruitment of histone deacetylases (HDACs). Upon further differentiation, FOXD3 dissociated from the enhancers, allowing increased expression of associated genes. Thus, by acting simultaneously as an initiator and suppressor of bound enhancer sites, FOXD3 fine-tunes gene expression to ensure tight temporal regulation of developmental cell fate transitions.
RESULTS
Chromatin Is Highly Dynamic during the ESC to EpiC Transition, with Enhancers Transitioning between the Active and Primed States
In order to follow enhancer changes during early ESC differentiation, we took advantage of a recently developed reporter system that follows the transition from the ESC to the EpiC state (Parchem et al., 2014). This reporter system uses fluores-cent markers targeted to two miRNA loci, miR-290 (mCherry) and miR-302 (eGFP). In vivo, all cells express these two reporters consecutively—miR-290-mCherry at embryonic day 3.5 (e3.5), both reporters at e5.5, and miR-302-GFP alone at e7.5. These transitions can be recapitulated in vitro starting with ESCs grown in LIF and 2i (GSK and Mek inhibitors), which express mCherry alone (R for red). Removal of LIF and 2i under low-density conditions resulted in retention of mCherry alone for roughly 16 hr (dR for differentiating Red) before a near homogeneous expression of both reporters by 50 hr (dY for differentiating Yellow) followed by a less homogenous transition to GFP alone (dG for differentiating Green) and finally neither reporter (dB for differentiating Black) (Figure 1A, Figure S1). The later two populations were sorted to increase homogeneity. Self-renewing epiblast stem cells (EpiSCs) have been described as an in vitro surrogate for epiblast cells (Chenoweth et al., 2010; Jouneau et al., 2012; Najm et al., 2011). EpiSCs expressing only GFP were derived after multiple passages of differentiating ESCs grown in FGF and Activin (G for Green). Principal component analysis (PCA) on microarray data showed a smooth transition during the successive stages of differentiation (Figure 1B). In contrast, EpiSCs were a distinct population, sharing some, but not all, features of EpiCs, as has previously been described (Hayashi et al., 2011). Going forward, we refer to R cells expressing only mCherry in LIF and 2i growth conditions as ESCs, and dY cells expressing mCherry and eGFP after 50 hr of LIF and 2i removal as EpiCs.
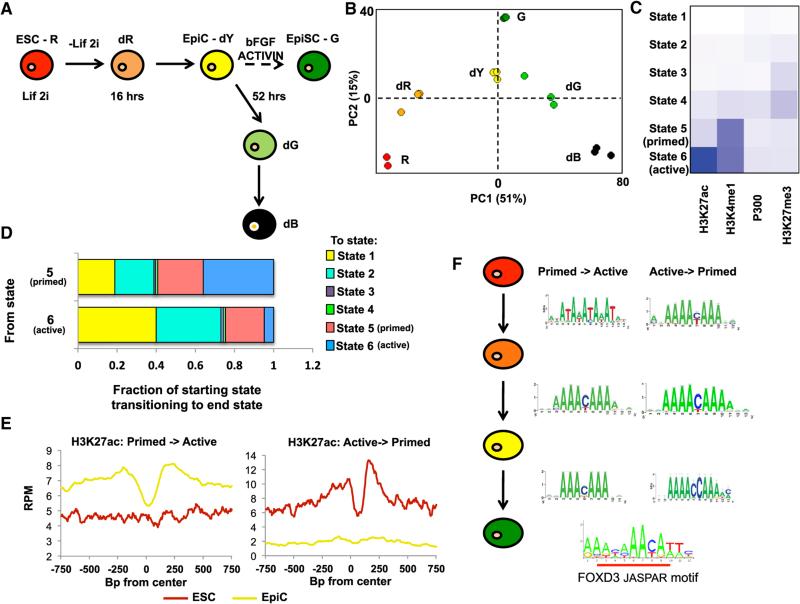
Figure 1. Profiling of Changing Enhancers Identifies FOXD3 as a Key Player during the Transition from the ESC to EpiC State.
(A) Schematic of transitions followed during early differentiation from ESCs (R) to EpiCs (dY). Colors represent expression from miR-290 locus (Red or R, mCherry), miR-302 locus (Green or G, eGFP), both (Yellow or Y), or neither (Black or B). The letter “d” prior to color represents cells undergoing differentiation from ESCs (R) to dR (16 hr off LIF and 2i) to EpiCs (dY) (52 hr off LIF and 2i). Subsequent differentiation yields non-homogenous transition of cells toward dG (green) and dB (black, no reporters), which are FACS sorted for purity. G is a derived self-renewing EpiSC state grown in FGF and Activin. It requires multiple passages in FGF and Activin prior to establishment, represented as a broken line.
(B) Principal component analysis of microarray data of the populations in (D). Array data can be found at GEO: GSE54341 (Parchem et al., 2014).
(C) Heatmap of six states of chromatin determined by ChromHMM using H3K4me1, H3K27ac, H3K27me3, and P300 ChIP-seq data in R, dR, dY, and G cells. States 1–3 represent the majority of the genome and show a general lack of these markers (percentages of genome in states 1 and 2 are shown in pie chart in Figure S4). State 3 shows increased H3K27me3 consistent with heterochromatin. State 4 similarly shows increased H3K27me3, but also detectable levels of H3K4me1. States 5 and 6 show low H3K27me3 and increased H3K4me1. State 5 shows low levels of H3K27ac, while state 6 shows elevated H3K27ac. Intensity of blue indicates average ChIP-seq signal for each modification in each state.
(D) Genome-wide transitions from states 5 and 6 to all states 1–6. x axis shows the two starting states 5 and 6, and the bar graphs represent the distribution of the end states (1–6) for the matching regions following transition between ESC and EpiC.
(E) Metagene analysis of H3K27ac levels centered around enhancers that transition from state 5 (“primed”) to state 6 (“active”) (left), or vice versa (right). Enhancers were defined by the identification of nucleosome-depleted regions in state 6 chromatin as determined by an adapted algorithm (Wamstad et al., 2012) (see Experimental Procedures). Metagene analysis for a 750 bp region surrounding the defined enhancer is shown, with a moving window average of 500 bp windows and 100 bp steps.
(F) RSAT (regulatory sequence analysis tool) was used to identify enriched motifs within nucleosome-depleted regions of identified enhancers that transition between primed and active during ESC to EpiC differentiation (Thomas-Chollier et al., 2011). Shown is the most commonly identified motif found in all transitions between poised and active enhancers, which significantly overlaps with the JASPAR motif for FOXD3 (overlap underlined in red). Significance and enrichment of overlap with FOXD3 and other uncovered TF binding are shown in Table S2.
To characterize enhancers during early ESC differentiation, we performed Chromatin IP (ChIP)-seq for H3K4me1, H3K27ac, H3K27me3, H3K4me3, and P300 in the homogenous cell populations (ESCs, dR, EpiCs, and EpiSCs) (Table S1). We also performed ChIP-seq for RNA Polymerase II (Pol2) to follow transcriptional activity. Klf4 and Fgf5 represent two genes whose expression is well characterized during this transition, with Klf4 being highly expressed in ESCs and Fgf5 being highly expressed in EpiCs (Lanner and Rossant, 2010; Marks et al., 2012). The patterns of H3K4me3 and Pol2 deposition at the transcription start sites were as expected with an increase at Fgf5 and a decrease at Klf4 during the ESC to EpiC transition (Data S1). Fgf5 showed a gain of H3K27ac and redistribution of H3K4me1 at its promoter regions as well as a gain of both H3K4me1 and H3K27ac at three latent enhancers in ESCs that become active enhancers in EpiCs (Data S1). Klf4 showed an opposing pattern, with a mild reduction of the broad region of H3K4me1 spanning the gene and the loss of a single peak of H3K27ac in the promoter region (Data S1). H3K27me3 changed little in the regions surrounding both genes during the ESC to EpiC transition (Data S1). The major known HAT found at enhancer regions is the protein P300 (Calo and Wysocka, 2013; Shlyueva et al., 2014). Even though H3K27ac was highly dynamic at both Fgf5 and Klf4, there was little change in the levels of bound P300 (Data S1). These findings confirm the validity of our in vitro differentiation system and show highly dynamic changes in H3K27ac, surprisingly in the absence of altered P300 binding.
FOXD3 Binding Motif Is Enriched at Sites of H3K27ac Changes
We used ChromHMM to evaluate the global distribution of the four enhancer marks, H3K4me1, H3K27ac, H3K27me3, and P300 binding (Ernst and Kellis, 2012) during the transition between ESCs and EpiCs. This uncovered six states consisting of different combinations of these marks (Figure 1C, Table S1). States 1 and 2 were relatively devoid of the four marks tested. As expected, these two states made up the vast majority of the genome (Figures S2A and S2B). State 3 showed an enrichment of H3K27me3 in a relative absence of the other marks, thus representing heterochromatin. State 4 contained a combination of enriched H3K27me3 and low levels of H2K27ac, P300, and H3K4me1, potentially reflecting a form of poised enhancers (Calo and Wysocka, 2013; Shlyueva et al., 2014). This state was relatively rare except in EpiSCs (Figures S2A and S2B). States 5 and 6 were of specific interest: both have P300 and H3K4me1, but state 6 also has high levels of H3K27ac. Therefore, states 5 and 6 are consistent with primed and active enhancers, respectively. Of note, H3K27ac changes occurred only at sites that maintained H3K4me1, in keeping with ordered events in histone changes at enhancers with H3K4me1 preceding and lingering after H3K27ac (Figure 1D). A majority of state 5 and 6 chromatin was in intronic and intergenic regions as expected for enhancers (Figure S2C). Interestingly, while H3K27ac binding and P300 levels correlated at these sites, H3K27ac changes between states were far more dynamic than P300 (Figure S2D).
The dynamic and interchangeable nature of states 5 and 6 suggested an important functional role for these enhancers in regulating cell-specific gene expression between the ESC and EpiC fates. To identify potential regulating TFs, we performed a de novo motif search within the nucleosome-depleted regions of these enhancer sites (Wamstad et al., 2012). The nucleo-some-depleted regions were identified as a “dip” in the ChIP-seq signal at the center of the H3K27ac peaks (Figure 1E, Table S1). Focusing on the transitions between ESCs, dR, EpiCs, and EpiSCs, the RSAT and HOMER peak finding algorithms uncovered a number of recognizable TF binding motifs (Table S2) (Nagy et al., 2013; Thomas-Chollier et al., 2011). Of particular note, a Forkhead (FKH) binding motif was among the top five enriched motifs for all six possible transitions (Figure 1F and Table S2). FOXD3, a FKH TF previously found to be essential for ESC self-renewal, was particularly striking as its binding motif showed the strongest enrichment in four of the six transitions (Table S2, Figure 1F) (Hanna et al., 2002; Liu and Labosky, 2008). Therefore, we focused our attention on FOXD3.
FOXD3 Localizes to Distinct Regions of Chromatin during the ESC to EpiC Transition preceding Maximum Expression of Proximal Genes
FOXD3 is a known regulator of pluripotency both in vitro and in vivo (Hanna et al., 2002). Furthermore, Foxd3 KO mouse ESCs have a self-renewal defect and show aberrant expression of a large number of genes (Liu and Labosky, 2008; Plank et al., 2014). Foxd3 is expressed at approximately equal levels in ESCs and EpiCs (Figure S3A). In order to determine the genomic localization of FOXD3 in ESCs and EpiCs, we knocked in a 3X-FLAG tag at the C-terminal end of the endogenous Foxd3 locus (Figure S3B). The tagged protein was expressed and nuclear as expected (Figures S3C and S3D). Despite ESCs and EpiCs being highly related pluripotent cell types, ChIP-seq for FOXD3-3X-FLAG revealed that FOXD3 bound largely mutually exclusive sites in ESCs (7,000 sites) and EpiCs (8,387 sites), with only a small number (1,650) of overlapping loci (Figures 2A and S3E and Table S2). De novo motif finding and FIMO analysis with the FOXD3 position weight matrix (PWM) confirmed highly significant enrichment for the FKH binding motifs at these sites (Figures 3A and 3B, Table S2, Data S2, and Data S3). These results suggested that FOXD3 regulates different genes in ESCs and EpiCs by binding the same motif, but at distinct sites in the genome in the two cell states.
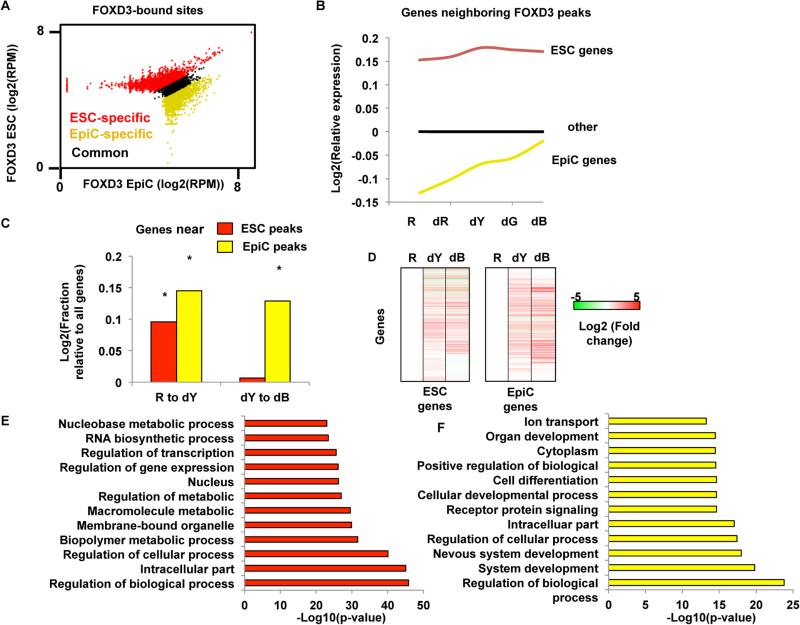
Figure 2. FOXD3 Is Mobilized during the Transition from the ESC to EpiC State, and Its Departure Is Associated with Increased Gene Expression.
(A) Scatterplot of reads per million (RPM) at the union of all peaks identified by MACS (Zhang et al., 2008) in both ESCs and EpiCs. Peaks were re-characterized as ESC-specific (red), EpiC-specific (yellow), and common peaks (black) based on their distance from the x = y diagonal (threshold set at ± 0.1) to obtain high-confidence specific peaks (q < 0.01). ESC only peaks = 5,350. EpiC only peaks = 6,737. Common peaks = 1,650. Samples were prepared in triplicate.
(B) mRNA expression (from Illumina microarray data) for genes closest to FOXD3 peaks in ESC (red line) and EpiC (yellow line), as well as all other genes on the array (blue line). Microarray experiments were performed in R (ESC), dR, dY (EpiC), dG, and dB cells. dG and dB (differentiated Black) represent further sequential differentiation following EpiCs during continued culture in the absence of LIF and 2i. For this experiment, all populations were collected by FACS. Data shown are all relative to all genes in the ESC state.
(C) Fraction of genes with increased expression from ESCs (R) to EpiCs (dY), as well as from EpiCs (dY) to dG, for genes near FOXD3 peaks compared with all other genes. Genes associated with ESC FOXD3 peaks increase with transition to EpiC but then level off. In contrast, genes associated with EpiC FOXD3 peaks continue to go up after EpiC. Asterisk (*) denotes Z scores > 2, p < 0.05 relative to genes without FOXD3 peaks.
(D) Heatmap showing hierarchical clustering (single linkage) of all genes shown in (F) and (G) for the transition from ESC (R) to EpiC (dY) to dB. Data are shown as a log2 transformed fold change relative to ESCs.
(E) Top 12 GO terms identified by gene ontology analysis using DAVID for genes near FOXD3 peaks in ESCs.
(F) Same as in (D) for genes near FOXD3 peaks in EpiCs.
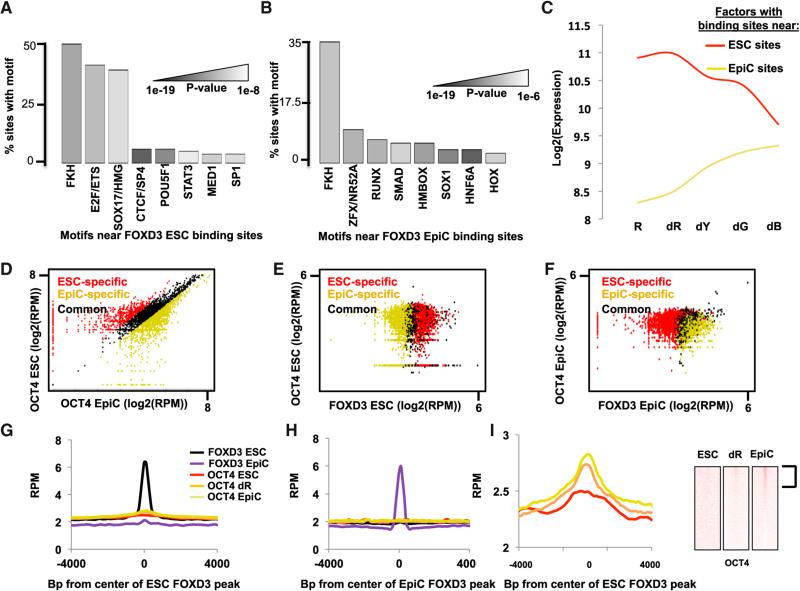
Figure 3. FOXD3 Precedes Binding of Other TFs, Such as OCT4, at Enhancers.
(A) Motifs enriched within a 200 bp window surrounding FOXD3 peaks in ESCs cells, shown as a percentage of peaks containing the motif. Shades of gray represent p values as shown (full data in Table S2).
(B) Same as (A) but for FOXD3 peaks in EpiCs (full data in Table S2).
(C) Average expression of TFs whose motifs were found to be neighboring FOXD3 in either ESCs or EpiCs. For motifs with multiple potential binding TFs, an average of all family members was used. The red line is for TFs to motifs associated with FOXD3 sites in ESCs and the yellow line is for TFs to motifs associated with Foxd3 sites in EpiCs.
(D) Scatterplot of RPM in ESCs versus EpiCs at all OCT4 peaks. ESC-specific OCT4 peaks are in red, EpiC-specific peaks are in yellow, and common peaks are in black. Similar to FOXD3, OCT4 shows very distinct binding sites in ESCs and EpiCs, even though sites are distinct from FOXD3. Peaks were identified similarly to FOXD3 peaks in Figure 3A.
(E) Scatterplots of RPM for peaks of OCT4 (y axis) versus RPM for peaks of FOXD3 (x axis) in ESCs. ESC-specific FOXD3 peaks are in red, EpiC-specific peaks are in yellow, and common peaks are in black.
(F) Same as (E), but in EpiCs.
(G) Metagene analysis summarizing OCT4 and FOXD3 binding for regions surrounding FOXD3 peaks in ESCs. Shown are the data for ESCs in a 4,000 bp region surrounding the FOXD3 peaks using a moving window average of 500 bp at 100 bp steps.
(H) Same as (G), but in EpiCs.
(I) Magnification of OCT4 binding data shown in (G) along with heatmap showing binding of OCT4 in dR and EpiCs, at sites bound by FOXD3 in ESCs.
In ESCs, FOXD3 peaks were in proximity to genes that are expressed at levels greater than average genome-wide expression, but whose expression increased further upon differentiation to EpiCs before reaching a plateau in the later stages (Figures 2B–2D). These genes were enriched for gene ontology categories consistent with the high rate of cellular metabolism associated with the rapid proliferation of epiblast cells in vivo (Figure 2E, Table S2) (Snow, 1977). In contrast, FOXD3 peaks in EpiCs were in proximity to genes that were expressed to a lesser extent in ESCs, showed increased expression in the transition from ESC to EpiC, and increased even further with the transition to dG and dB (Figures 2B–2D, Table S2). EpiC-specific peaks were enriched for a number of categories associated with gastrulation and organogenesis, representing genes that need to be expressed shortly after the epiblast stage (Figure 2F). This trend was consistent across a majority of the genes in the respective transitions (Figures 2C and 2D). Together, these findings show that FOXD3 relocalizes during the ESC to EpiC transition and is bound near functionally distinct genes in these two cell states, preceding their maximal expression.
FOXD3 Precedes Binding of Other Regulatory TFs
Sequence analysis around the FOXD3-bound sites uncovered a number of motifs that were distinct between ESCs and EpiCs (Figures 3A and 3B). ESC-specific FOXD3 sites were enriched for motifs for housekeeping and pluripotency factors such as E2F1 and POU5F1/OCT4, respectively. In contrast, EpiC-specific sites were enriched for motifs for a number of developmental regulators including HNF, ZFX, and HOX. Therefore, FOXD3 appears to move from sites generally important in pluripotent stem cells to sites associated with developmental processes. Interestingly, the average expression of the TFs predicted to bind the uncovered motifs in ESCs and EpiCs followed distinct patterns. Expression of TFs predicted to bind motifs enriched at ESC sites remained high with differentiation to dR but then decreased as cells transitioned to the EpiC state, while those at EpiC sites increased with differentiation even beyond the EpiC state (Figure 3C).
An enrichment of the POU5F1/OCT4 motif in ESC-specific FOXD3 binding sites was especially intriguing as recent work by Wysocka and colleagues showed that OCT4, like FOXD3, relocalizes during early ESC differentiation (Buecker et al., 2014). This link prompted the question of whether FOXD3 and OCT4 colocalize during the ESC to EpiC transition. Therefore, we performed ChIP-seq for OCT4 in ESCs, dR, and EpiCs (Table S1). As previously shown, OCT4 relocalized during the ESC to EpiC transition (Figure 3D, Table S3). However, there was no correlation between FOXD3-bound and OCT4-bound sites in ESCs or EpiCs (Figures 3E–3H). To determine the basis for enrichment of the OCT4 motif at ESC-specific FOXD3 sites, we evaluated OCT4 binding at these sites in ESCs, dR, and EpiCs. Surprisingly, OCT4 binding at the ESC FOXD3 bound sites was associated with differentiation to dR and EpiCs (Figure 3I), suggesting that OCT4 follows FOXD3 binding. Analysis of individual loci confirmed this order of events (Data S4). Therefore, FOXD3 precedes the binding of other key regulatory factors such as OCT4, consistent with binding of FOXD3 preparing the chromatin for binding of other TFs as cells transition.
FOXD3 Binding Initiates Cell-State-Specific Enhancers by Promoting Nucleosome Removal
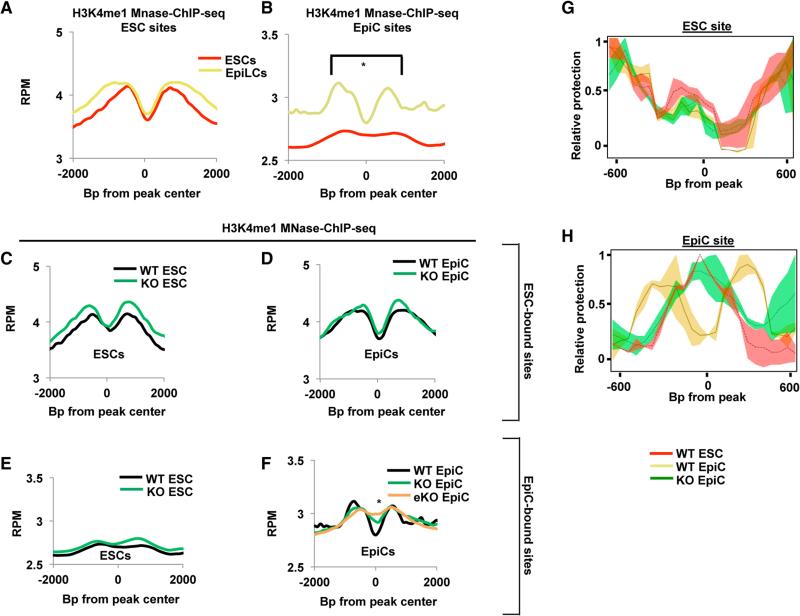
To ask whether the binding of FOXD3 activates enhancers in the two pluripotent cell types, we analyzed recently published FAIRE-seq data in ESCs and EpiCs (Buecker et al., 2014). Sites bound by FOXD3 in ESCs also had FAIRE-seq peaks in both ESCs and EpiCs, consistent with nucleosome depletion in both states (Figure S4A). Conversely, sites bound by FOXD3 in EpiCs showed a much higher FAIRE-seq peak in EpiCs, consistent with removal of nucleosomes from these sites as cells transitioned to the EpiC state (Figure S4A). Similar to the FAIRE-seq data, DNaseI hypersensitivity (HS)-seq showed increased chromatin accessibility at FOXD3 ESC sites relative to EpiC sites in ESCs (Figure S4B) (Stamatoyannopoulos et al., 2012). Furthermore, high-resolution micrococcal nuclease (MNase)-ChIP-seq for H3K4me1 in ESCs and EpiCs showed both increased H3K4me1 and formation of a dip with binding of FOXD3 (Figures 4A and 4B). Together, these data show a strong association between FOXD3 binding and nucleosome removal.
Figure 4. FOXD3 Binds Nucleosome Occupied Sites and Establishes Primed Enhancers.
(A) Metagene analysis of H3K4me1 MNase-ChIP-seq data for ESCs (red line) and EpiCs (yellow line) in a 4,000 bp window surrounding FOXD3-bound sites in ESCs.
(B) Same as (A) but for EpiC-bound sites. Asterisk (*) denotes paired Student's t test, p < 0.05.
(C) Metagene analysis of H3K4me1 MNase-ChIP-seq data from WT and Foxd3 KO ESCs using a 4,000 bp window surrounding FOXD3-bound sites in ESCs. Data are shown for WT (black line) and Foxd3 KO (green line) cells.
(D) Same as (C) but in EpiCs. In addition, data are shown for KO of Foxd3 16 hr prior to differentiation (orange line; KO-l stands for KO long).
(E) Metagene analysis of H3K4me1 MNase-ChIP-seq data from WT and Foxd3 KO ESCs using a 4,000 bp window surrounding FOXD3-bound sites in EpiCs. Data are shown for WT (black line) and Foxd3 KO (green line) cells.
(F) Same as (E) but in EpiCs. In addition, data are shown for KO of Foxd3 16 hr prior to differentiation (orange line; eKO stands for early KO).
(G) MNase tiling qPCR at an ESC enhancer (see Supplemental Experimental Procedures for primers) in WT ESCs, WT EpiCs, and early Foxd3 KO EpiCs. The graph represents an average of n = 2–3 experiments for each primer pair. Unpaired Student's t tests were conducted for WT and Foxd3 KO EpiCs, and the heatmap for the p values for this test is shown below the graph. ANOVA for a three-way comparison of WT ESCs, WT EpiCs, and Foxd3 KO EpiCs was also performed (p = 0.97964, not shown on graph).
(H) Same as (F) but for an EpiC enhancer, with t test p values shown as a heatmap. ANOVA p = 1.111e–07, not shown on graph.
To determine whether FOXD3 is required for nucleosomal removal at these enhancers, we used a conditional KO model and a Tamoxifen-inducible cre recombinase drivenoff a ubiquitous promoter (Mundell and Labosky, 2011). Addition of Tamoxifen resulted in loss of FOXD3 protein within 24-48 hr (Figure S3F). We devised a strategy to knock out FOXD3 in both self-renewing and differentiating ESCs (see Figure S3G and Experimental Procedures). Of important note, the later condition, which includes the addition of Tamoxifen 16 hr after initiation of differentiation, resulted in diminishing levels of FOXD3 during, rather than before, differentiation into EpiCs. ChIP-seq and MNase-ChIP-seq for H3K4me1 in KO ESCs and EpiCs were compared to that of corresponding WT cells. Surprisingly, the loss of FOXD3 had little effect on H3K4me1 at any FOXD3 sites in either cell state (Figures 4C–4F, S4C, and S4D). Furthermore, it had no effect on the size of the dip at the FOXD3 ESC sites in either cell state (Figures 4C–4D and S4C). However, there was a partial, yet significant decrease in the dip at EpiC sites when FOXD3 was removed during differentiation, suggesting that FOXD3 is needed for the establishment of nucleosome-depleted regions (Figures 4F and S4D). The remaining dip may be due to residual FOXD3, especially during the early stages of differentiation. Therefore, we repeated this experiment with the addition of Tamoxifen 32 hr earlier so FOXD3 protein would be gone sooner during differentiation. The earlier deletion of Foxd3 led to an almost complete loss of the dip, once again with no change in H3K4me1 levels (Figure 4F). MNase-qPCR across an ESC and an EpiC FOXD3 binding site validated that the dips represented changes in nucleosome occupancy and not the less likely scenario of a very focal loss of methylation of H3K4 within an otherwise strong H3K4me1 peak (Figures 4G and 4H). Together, these data show that FOXD3 initiates enhancer activitybyremoving nucleosomes atsites of H3K4me1,and itdoes so atnew enhancer sites asthe cells transition from ESCs to EpiCs, priming them for future activation.
FOXD3 Suppresses Enhancer Activity by Inhibiting Acetylation
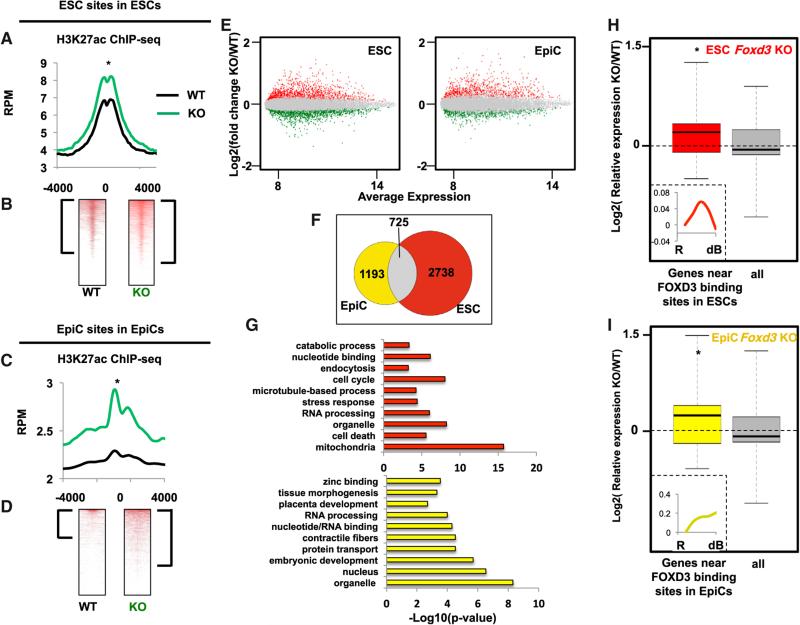
FOXD3's role in nucleosome depletion and association with H3K4me1 suggested a role for FOXD3 in promoting enhancer activation and thus neighboring gene activation. Indeed, FOXD3 binding at EpiC sites during differentiation was associated with increased gene expression. However, expression of these genes increased further as FOXD3 was lost both at ESC and EpiC sites (Figures 2B–2D). Also, the recruitment of FOXD3 to EpiC sites was not associated with increased H3K27ac, a mark of enhancer activation (Figure S4E). Therefore, we asked how FOXD3 may be influencing histone acetylation. We removed Foxd3 in ESCs and 16 hr into differentiation toward EpiCs as described (Figure S3G). In the later condition, FOXD3 would still have the opportunity to bind and initiate many of the EpiC enhancers prior to its loss (see Figure 4F, green line). ChIP-seq under these conditions showed a significant increase in H3K27ac in both ESCs and EpiCs at corresponding FOXD3 sites with FOXD3 loss (Figures 5A–5D). In contrast, there was no increase in H3K27ac at FOXD3 EpiC sites in the context of ESCs, sites where FOXD3 had yet to bind (Figure S4E). These data suggest that once FOXD3 binds, it actively maintains a locally H3K27 hypoacetylated state, which in turn represses neighboring gene activation.
Figure 5. FOXD3 Maintains Lower Levels of Histone Acetylation at Enhancers.
(A) Metagene analysis of H3K27ac ChIP-seq data from WT and Foxd3 KO ESCs using a 4,000 bp window surrounding FOXD3-bound sites in EpiCs. Data are shown for WT (black line) and Foxd3 KO (green line) cells. Asterisk (*) denotes paired Student's t test, p < 0.05.
(B) Data in (A) shown as heatmaps. Brackets represent genes with significant peaks of H3K27ac (signed-rank test, p < 0.05).
(C) Same as (A) but for H3K27ac ChIP-seq at EpiC sites. Asterisk (*) denotes paired Student's t test, p < 0.05.
(D) Data in (C) shown as heatmaps. Brackets represent genes with significant peaks of H3K27ac (signed-rank test, p < 0.05).
(E) Scatterplot showing average gene expression on the x axis and fold change between WT and Foxd3 KO cells on the y axis. Data are shown on the left for ESCs and the right for EpiCs. Points in red are significantly upregulated (adj. p < 0.05) and points in green are significantly downregulated (adj. p < 0.05).
(F) Venn diagram showing overlap of upregulated and downregulated genes upon Foxd3 KO in ESCs and EpiCs (adj. p < 0.05).
(G) Top ten GO terms identified by gene ontology analysis using DAVID for genes affected by Foxd3 KO in ESCs (top) and EpiCs (bottom).
(H) Microarray expression data for WT and Foxd3 KO ESCs. Plotted are all genes that are significantly changed upon Foxd3 KO (“all”) versus genes near FOXD3 binding sites. (Asterisk denotes p < 0.05 and Z > 2 in comparison between two populations). All n = 2142 and FOXD3 n = 511. Inset: expression of FOXD3-bound and Foxd3 KO-affected genes (n = 511) during differentiation.
(I) Same as (G), but for EpiCs. All n = 1,800 and FOXD3 n = 412. Inset: expression of FOXD3-bound and Foxd3 KO-affected genes (n = 412) during differentiation. ChIP-seq samples were performed in duplicate.
Expression microarray analysis on WT and Foxd3 KO cells under these conditions showed hundreds of genes upregulated and downregulated, representing both direct and indirect effects of FOXD3 regulation. These genes were largely non-overlapping between the ESCs and EpiCs (Figures 5E and 5F). Gene ontology showed an enrichment for cellular metabolism in ESCs and development processes in EpiCs, reiterating the distinct biological processes regulated by FOXD3 in these two cell types (Figure 5G). Importantly, genes that are proximal to FOXD3 binding sites, and thus likely directly regulated by FOXD3, showed a strong bias toward increased expression upon FOXD3 loss (Figures 5H and 5I). These same genes normally increase in expression at a subsequent stage in differentiation (Figures 5H and 5I insets). Therefore, FOXD3 maintenance of a hypoacetylated state maintains transcriptional repression of neighboring genes, thereby tightly regulating the timing of their expression.
FOXD3 Directly Recruits the Nucleosome Remodeler BRG1 to Enhancers
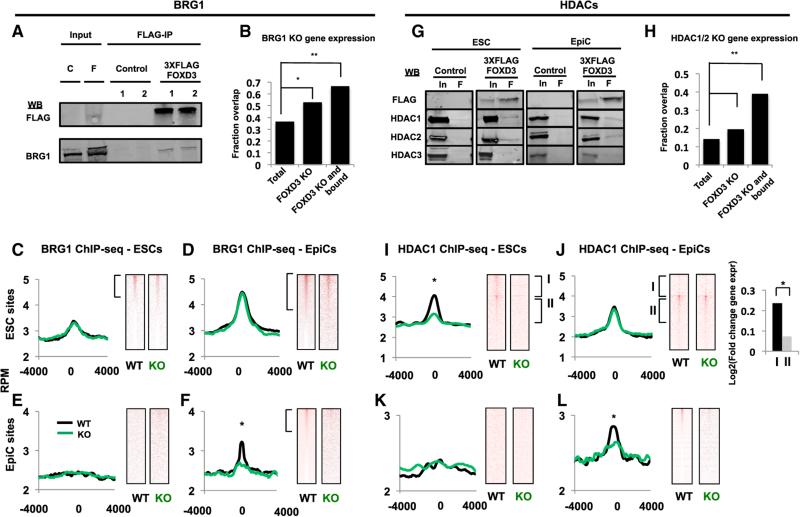
To determine whether FOXD3 directly regulates both initiation and suppression of enhancer activity, we asked whether FOXD3 simultaneously recruits enzymes that could produce both outcomes. First we asked if FOXD3 is associated with BRG1, a key component of the SWI/SNF nucleosome remodeling complex. This complex removes nucleosomes in an ATP-dependent fashion and plays a critical role in ESC self-renewal and differentiation (Ho et al., 2009; Takebayashi et al., 2013). Immunoprecipitation (IP) of FOXD3 showed a physical interaction with BRG1 (Figure 6A). Furthermore, analysis of published expression changes in Brg1 KO relative to Foxd3 KO ESCs showed a significant overlap that was further increased when we focused on genes next to FOXD3-bound enhancers (Figure 6B) (Takebayashi et al., 2013).
Figure 6. FOXD3 Recruits the Chromatin Remodeling Swi/Snf Complex and the Histone Deacetylases HDAC1/2 to Target Enhancers.
(A) Immunoprecipitation using M2 anti-FLAG resin (FLAG-IP) on extract from WT (C, Control) and FOXD3-3X-FLAG targeted (F, 3XFLAG FOXD3) cell lines (1, 2). Western blots for FLAG and BRG1 are shown. Inset: 10% of input. Representative blot for n = 3 is shown.
(B) Overlap of mRNA expression changes in Brg1 KO and Foxd3 KO relative to WT mouse ESCs. The proportion of genes affected by Brg1 KO is shown as a total of all genes, genes altered by Foxd3 loss, and genes both bound by FOXD3 and altered by Foxd3 loss. The latter two categories are significantly enriched for genes whose expression is changed by Brg1 deletion. Results of a Chi-square test are shown. *χ2 = 125, p < 2.2e–16. **χ2 = 346, p < 2.2e–16.
(C) Metagene analysis (left) of BRG1 ChIP-seq data from WT and Foxd3 KO ESCs using a 4,000 bp window surrounding FOXD3-bound sites in ESCs. WT (black line) and Foxd3 KO (green line) cells are shown. Data are visualized in heatmaps on the right.
(D) Same as (C), but in EpiCs.
(E) Same as (C), but for FOXD3 EpiC bound sites.
(F) Same as (D), but for FOXD3 EpiC bound sites. Asterisk (*) denotes paired Student's t test, p < 0.05.
(G) Immunoprecipitation using M2 anti-FLAG resin (F) on extracts from WT (Control) and FOXD3-3X-FLAG targeted (3XFLAG FOXD3) cell lines. Western blots for FLAG, HDAC1, HDAC2, and HDAC3 are shown. Inset: 10% input. Representative blots for n = 3 are shown.
(H) Overlap of mRNA expression changes in HDAC1/2 and Foxd3 KO relative to WT mouse ESCs. The proportion of genes affected by HDAC1/2 KO is shown as a total of all genes, genes altered by Foxd3 loss, and genes both bound by FOXD3 and altered by Foxd3 loss. The latter two categories are significantly enriched for genes changed by HDAC1/2 deletion. Results of a Chi-square test are shown. *χ2 = 49, p = 2.5e–12. **χ2 = 127, p < 2.2e–16.
(I) Metagene analysis (left) of HDAC1 ChIP-seq data from WT and Foxd3 KO ESCs using a 4,000 bp window surrounding FOXD3-bound sites in ESCs. Data are shown for WT (black line) and Foxd3 KO (green line) cells. Asterisk (*) denotes paired Student's t test, p < 0.05. Data are visualized in heatmaps on the right. Top bracket represents genes that have significant peaks of HDAC1 in ESCs that are lost in EpiCs (signed-rank test, p < 0.05), and bottom bracket represents genes that have significant peaks of HDAC1 in ESCs that are maintained in EpiCs (signed-rank test, p < 0.05).
(J) Same as (I) but in EpiCs (same brackets on heatmaps as C). Expr, log2 fold change in expression of associated genes from ESC to EpiC, showing a greater fold increase in expression of genes that lose HDAC1. The groups are significantly different, as per an unpaired Student's t test (p < 0.05).
(K) Same as (I) but for FOXD3 EpiC bound sites.
(L) Same as (I) but for FOXD3 EpiC bound sites. Asterisk (*) denotes paired Student's t test, p < 0.05. ChIP-seq samples were performed in duplicate.
We next asked whether depletion of FOXD3 affects recruitment of BRG1 to chromatin. We performed ChIP-seq for BRG1 in WT and Foxd3 KO ESCs and EpiCs, and determined occupancy at ESC and EpiC FOXD3 binding sites. In ESCs, BRG1 was bound at FOXD3 ESC sites, but KO of Foxd3 had no effect on BRG1 binding (Figures 6C and 6E). With differentiation to EpiCs, the ESC sites acquired additional BRG1, consistent with the increase in gene expression of the cognate genes (Figure 6D). However, with differentiation, BRG1 was recruited along with FOXD3 to the FOXD3 EpiC sites, and the deletion of Foxd3 led to a significant loss of BRG1 binding (Figures 6E and 6F, Data S5). These results were verified at six ESC sites and six EpiC sites using ChIP-qPCR (Figure S5C). Together, these findings show that FOXD3 recruits, but is not required for the maintenance of, BRG1 at enhancers.
FOXD3 Also Recruits and Maintains HDAC1/2 at Enhancers
Next, we asked whether FOXD3 is also directly regulating his-tone acetylation. Histone acetylation is regulated by a balance of the activities of the acetyltransferases and deacetylases. H3K27ac is primarily catalyzed by the acetyltransferase P300 (Creyghton et al., 2010; Hamed et al., 2013; Pasini et al., 2010; Francetic et al., 2012; Rada-Iglesias et al., 2011), but we observed that KO of Foxd3 slightly reduced the levels of P300 in ESCs and had no effect in EpiCs, suggesting that the increased acetylation could not be explained by P300 levels (Figure S5B). Therefore, we hypothesized that FOXD3 recruits HDACs to counteract HAT activity. HDACs1–3 regulate H3K27ac and have been shown to play important roles in early development and ESC differentiation (Haberland et al., 2009). IP of FOXD3 showed that FOXD3 protein interacts with HDAC1/2, but not HDAC3, in both ESCs and EpiCs cells (Figure 6G). As was the case with Brg1, evaluation of published expression data for HDAC1/2 double KO ESCs showed significant overlap in expression changes with Foxd3 KO cells, which was further enriched when considering only genes neighboring the FOXD3 enhancers (Figure 6H) (Jamaladdin et al., 2014).
ChIP-seq for HDAC1 in WT and Foxd3 KO ESCs and EpiCs showed that HDAC1 colocalized with FOXD3 and deletion of Foxd3 led to loss of HDAC1 at these sites (Figures 6I–6L, Data S5). ChIP-qPCR at the same six ESC and six EpiC enhancers done for BRG1 validated these results and extended them to HDAC2, which is commonly found in a complex with HDAC1 (Lai and Wade, 2011; Jamaladdin et al., 2014) (Figures S5D and S5E). Consistent with a role for FOXD3 in recruiting as well as maintaining HDAC1 at the FOXD3-bound enhancer sites, HDAC1 was not found at the EpiC enhancers prior to FOXD3 binding (compare Figure 6K to 6L). Interestingly, though, HDAC1 did remain following the departure of FOXD3 at a subset of the ESC sites (compare Figure 6I to 6J). Indeed, these sites could be divided into two subsets. At one subset HDAC1 departed along with FOXD3 (Figure 6I and 6J, group 1, top brackets on heatmaps). At the other subset, HDAC1 remained and became FOXD3 independent (Figure 6I and 6J, group 2, bottom brackets). Consistent with these differences, genes associated with HDAC departure showed a much greater increase in expression than genes where HDAC remained. (Figure 6J, right panel). Thus, FOXD3 primes genes that can then be either activated or repressed depending on future molecular signals.
FOXD3 Can Simultaneously Recruit BRG1 and HDACs to Individual Enhancer Sites
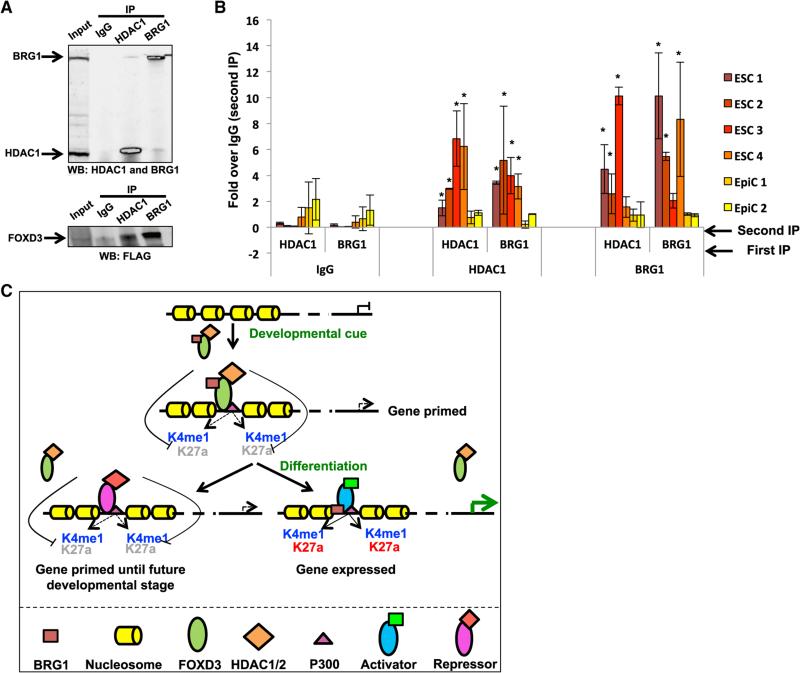
To confirm that BRG1 and HDACs were simultaneously recruited to FOXD3-bound sites, different combinations of IP-western blot and sequential ChIP were performed. Co-IP showed that BRG1 and HDAC1 interacted with each other as well as FOXD3 (Figure 7A). Furthermore, sequential ChIP for HDAC1 and BRG1 in ESCs confirmed the simultaneous presence of both enzymes at four FOXD3 ESC binding sites, but not two EpiC site controls (Figure 7B). Finally, sequential IP for FOXD3 followed by IP for BRG1 and then western blot for all three proteins showed that FOXD3, BRG1, and HDAC1 can be found in a single complex (Figure S6B). These findings confirm that FOXD3 can recruit both BRG1 and HDACs to the same locus, establishing an open enhancer whose activity is simultaneously attenuated by suppression of H3K27ac (Figure 7C).
Figure 7. FOXD3 Recruits HDAC1/2 and the SWI/SNF Complex to Sites to Both Initiate and Attenuate Enhancers.
(A) Top: immunoprecipitation using IgG, anti-HDAC1, or anti-BRG1 antibodies on extracts from ESCs. Western blots for HDAC1 and BRG1 are shown. Bottom: same as top, but with western blot for FLAG (i.e., FOXD3). Inset: 10% input. Representative blot for n = 3 is shown.
(B) Sequential ChIP for HDAC1 and BRG1 at four ESC FOXD3-bound sites and two EpiC FOXD3-bound sites in ESCs. Shown are all combinations of sequential ChIP with IgG, HDAC1, and BRG1 antibodies. In all cases, the results are expressed relative to IgG in second IP control. Background is shown as IgG used in first IP, which is used for all significance calculations. Error bars = SD (n = 3–4). Significance calculations were performed using a pairwise t test. p < 0.05.
(C) Model of FOXD3 function at developmental enhancers. FOXD3 primes enhancers for future gene expression. Upon FOXD3 departure, these genes can be directly activated or repressed by alternative TFs.
DISCUSSION
The data presented here uncover a role for the TF FOXD3 as both an initiator and a suppressor of enhancer function in pluripotent stem cells. This function allows FOXD3 to serve as a priming factor for developmental enhancers during the differentiation of pluripotent cells toward specialized cell types. Numerous pieces of evidence support this interpretation. First, while binding of FOXD3 leads to partial upregulation of neighboring gene expression, expression is further increased at many of these genes with ongoing differentiation and departure of FOXD3 from the associated enhancers. Second, movement of FOXD3 from ESC-specific to EpiC-specific sites is associated with the removal of nucleosomes at the newly bound enhancers, and deletion of Foxd3 just prior to the transition blocks nucleosomal depletion from the EpiC sites. Third, deletion of Foxd3 in steady-state ESCs or during differentiation to EpiCs does not lead to reoccupation by nucleosomes, but instead to increased H3K27 acetylation. Fourth, FOXD3, BRG1, and HDAC1/2 can be found as a single complex and bound together at the same enhancer site. Therefore, we propose a model wherein FOXD3 plays a dual role during ESC differentiation, initiating enhancer function by promoting nucleosome depletion while simultaneously repressing maximal activation by suppressing H3K27 acetylation (Figure 7C). Together, this dual function primes enhancers not for immediate, but rather future, tissue-specific expression of neighboring genes.
FOXD3 is a member of the FKH family of proteins, a group of highly conserved TFs that contain a wing-helix DNA binding domain conferring the potential to bind nucleosome-occluded DNA (Benayoun et al., 2011; Lalmansingh et al., 2012; Lam et al., 2013). Thus FKH family members have been described as pioneer TFs (Lupien et al., 2008; Sekiya et al., 2009; Shim et al., 1998; Xu et al., 2009). Similar to FOXD3, the FKH TF FOXA2 displaces nucleosomes in conjunction with chromatin remodeling factors during ESC differentiation (Li et al., 2012). However, none of these factors have previously been shown to simultaneously recruit BRG1 and HDACs.
Our studies were initiated by genome-wide analysis of enhancer marks during the ESC to EpiC transition. One of the most striking events uncovered in this analysis was a dramatic shift in H3K27ac withlittle changein H3K4me1 (states 5 and 6, Figure 1C). This is consistent with a number of reports showing that transitioning between primed and active enhancers is a predominant gene regulatory mechanism in early development (Buecker et al., 2014; McKay et al., 2014). The highly dynamic nature of FOXD3 binding itself was quite surprising. A recent study showed that the central pluripotency factor OCT4 also moved to distinct enhancer sites during the ESC to EpiC transition (Buecker et al., 2014). CTCF and OTX2 binding correlated with the distinct binding patterns of OCT4 in ESCs and EpiCs, respectively. In contrast, we found no correlation of the FOXD3 movements with OCT4 movements. Furthermore, we found no enrichment of the OTX2 motif at the FOXD3 sites in EpiCs. Interestingly, though, ESC-specific FOXD3 sites showed a slight enrichment for the OCT4 motif. However, OCT4 bound these sites in the EpiC state. Therefore, FOXD3 precedes rather than colocalizes with OCT4, and it is likely to precede the binding of numerous other pluripotency and developmental TFs during this cell state transition.
Following FOXD3 binding and priming of enhancers, there are appears to be two alternative paths that can be taken with FOXD3 departure (Figure 7C). At a subset of enhancers, HDACs depart, flanking histones show increased H3K27ac, and neighboring genes become activated. In a second set, HDAC remains and the neighboring gene remains suppressed. This decision is likely driven by the subsequent recruitment of additional TFs that replace FOXD3 at these sites with transcriptional activators promoting HDAC exit or transcriptional repressors retaining HDACs (Figure 7C). Such a mechanism has been proposed for two enhancer targets of FOXD3 in different settings—an Albumin enhancer during hepatocyte specification and a Vpreb enhancer in pre-B cell specification (Liber et al., 2010; Xu et al., 2007, 2009). Our data are consistent with and potentially broaden this model to many TFs.
Here, we focus on the role of FOXD3 in pluripotent stem cells, extending from ESCs to EpiCs. Consistent with a critical role for FOXD3 in this stage of development, KO embryos arrest prior to gastrulation (Hanna et al., 2002). Furthermore, Foxd3 KO mouse ESCs have a self-renewal defect and show aberrant expression of a large number of genes (Liu and Labosky, 2008; Plank et al., 2014; and this study). Interestingly, both overexpression and knockdown of FOXD3 in human ESC causes premature differentiation, with the former skewing cells toward paraxial mesoderm and the latter skewing cells toward mesendoderm (Arduini and Brivanlou, 2012). Thus, FOXD3 levels must be tightly regulated for normal development, likely limiting the number of target sites that FOXD3 binds in a specific cell context. Of note, FOXD3 regulates specification of neural crest cells and is expressed in both the precursor and specified states, akin to its role during the ESC to EpiC transition (Chang and Kessler, 2010; Hochgreb-Hägele and Bronner, 2013; Nitzan et al., 2013; Teng et al., 2008; Thomas and Erickson, 2009; Wang et al., 2011). It will be important to determine whether the FOXD3 bound sites are analogously bound at distinct sites in the neural crest precursor and specified states. Interestingly, recent work in melanoma, which arises from a neural crest derivative, showed that gene regulation by FOXD3 is responsible for the acquisition of drug resistance (Abel et al., 2013; Basile et al., 2012).
The dual role described here is unlikely to be unique to FOXD3, but instead common to other TFs, especially those with important roles in stem and progenitor cells where future expression programs need to be poised for activation with further differentiation. Interestingly, the NuRD complex contains both HDAC and chromatin remodeling activity (Denslow and Wade, 2007; Hu and Wade, 2012; Wong et al., 1998). Furthermore, NuRD and BRG1 physically interact and depletion of BRG1 leads to loss of NuRD chromatin localization. However, the association of NuRD and BRG1 had not been previously linked to a sequence-specific binding factor, but rather, a chromatin mark (Yildirim et al., 2011). In addition to normal development, dual-function TFs are likely to be important players in disease, especially when the differentiation state of a cell is altered as it is in cancer, such as FOXD3 in melanoma. Therefore, the identification of these TFs should greatly advance our understanding of how cells retain specific developmental potentials and how they can be harnessed to manipulate cell fate transitions in development and disease.
EXPERIMENTAL PROCEDURES
Cell Lines and Cell Culture
All ESCs were maintained in Knockout DMEM (Invitrogen) supplemented with 15% FBS, LIF, and 2i (PD0325901 and CHIR99021), and differentiations were performed in the absence of LIF and 2i. A detailed protocol is available in the Supplemental Information.
IP and ChIP
IP, ChIP, sequential ChIP, and sequential IP were performed as per standard protocols, with 5 to 10 million cells per experiment for ChIP, 30 million cells for IP, 100 million cells for sequential ChIP, and 2 billion cells for sequential IP. For ChIP, sequential ChIP, and sequential IP, the chromatin was cross-linked. All IPs were incubated overnight. A detailed protocol is available in the Supplemental Information.
ChIP-Seq
ChIP-seq libraries were made as per the Tru-Seq Illumina protocol with minor modifications. Samples were run on the Illumina HiSeq 2500. A detailed protocol is available in the Supplemental Information. All sequencing data can be found at GEO: GSE58408.
MNase-qPCR
This assay was adapted from a previously published protocol (Krishnakumar and Kraus, 2010). A detailed protocol is available in the Supplemental Information.
ChIP-Seq Data Analysis
Fastq files were mapped to mm10 using Bowtie 2. All samples were deduplicated to eliminate PCR bias. ChromHMM was used to identify chromatin states (Ernst and Kellis, 2012). To find nucleosome-depleted regions, the H3K27ac data was used as described previously (Wamstad et al., 2012). Two programs were used for motif finding, RSAT-tool and HOMER, both with default settings (Heinz et al., 2010; Thomas-Chollier et al., 2011). MACSv1.4 was used for FOXD3 and OCT4 peak finding (Zhang et al., 2008). A detailed protocol is available in the Supplemental Information.
Microarray Analysis
Total RNA was prepared from WT and FOXD3 KO (1mM Tamoxifen for 36 hr) using the standard Trizol protocol, reverse-transcribed, and hybridized to the Illumina Mouse Ref 8 v 2.0 Beadchip. More details on protocol and data analysis are available in the Supplemental Information. All array data can be found at GEO: GSE58408.
Supplementary Material
Highlights.
Differentially regulated enhancers in ESCs and EpiCs harbor FOXD3 binding sites
FOXD3 binding precedes other transcription factors and gene activation
FOXD3 recruits BRG1 and promotes nucleosome removal
FOXD3 also recruits HDACs to maintain deacetylation and restrict expression
ACKNOWLEDGMENTS
We thank Jennifer Plank and Scott Oakes for technical advice and Glencijoy David, Brandon Chu, Greg Bronevetsky, and Richard Lao for technical support. We acknowledge Indiana University for access to their Mason cluster of computers. We thank the following people for critical reading of the manuscript: Marco Conti, Diana Laird, Barbara Panning, Brian DeVeale, Jacob Freimer, Jennifer Plank, Archana Shenoy, Mayya Shveygert, John Vincent, and Julia Ye. This project was funded by NIH (R01 GM101180, U54 HD055764) and CIRM (RN2-00906-1) to R.B. and an AP Gianinni Postdoctoral Award to R.K.
Footnotes
AUTHOR CONTRIBUTIONS
R.K. designed and performed the experiments and data analysis. A.F.C. contributed to Figures S1A and S1B and S5C–S5E; M.G.P. contributed to Figures 6A, 6G, and 7A; and M.D. contributed to Figures 4G–4H and 6G. R.J.P. generated the dual-reporter ESCs and P.A.L. provided the 3XFLAG-FOXD3 targeting construct. R.K. and R.B. conceived of the project and wrote the manuscript.
ACCESSION NUMBERS
The accession number for the data reported in this paper is GEO: GSE58408.
SUPPLEMENTAL INFORMATION
Supplemental Information for this article includes six figures, Supplemental Experimental Procedures, three tables, and five data files and can be found with this article online at http://dx.doi.org/10.1016/j.stem.2015.10.003.
REFERENCES
- Abel EV, Basile KJ, Kugel CH, 3rd, Witkiewicz AK, Le K, Amaravadi RK, Karakousis GC, Xu X, Xu W, Schuchter LM, et al. Melanoma adapts to RAF/MEK inhibitors through FOXD3-mediated upregulation of ERBB3. J. Clin. Invest. 2013;123:2155–2168. doi: 10.1172/JCI65780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arduini BL, Brivanlou AH. Modulation of FOXD3 activity in human embryonic stem cells directs pluripotency and paraxial mesoderm fates. Stem Cells. 2012;30:2188–2198. doi: 10.1002/stem.1200. [DOI] [PubMed] [Google Scholar]
- Arnold SJ, Robertson EJ. Making a commitment: cell lineage allocation and axis patterning in the early mouse embryo. Nat. Rev. Mol. Cell Biol. 2009;10:91–103. doi: 10.1038/nrm2618. [DOI] [PubMed] [Google Scholar]
- Basile KJ, Abel EV, Aplin AE. Adaptive upregulation of FOXD3 and resistance to PLX4032/4720-induced cell death in mutant B-RAF melanoma cells. Oncogene. 2012;31:2471–2479. doi: 10.1038/onc.2011.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benayoun BA, Caburet S, Veitia RA. Forkhead transcription factors: key players in health and disease. Trends Genet. 2011;27:224–232. doi: 10.1016/j.tig.2011.03.003. [DOI] [PubMed] [Google Scholar]
- Bradley A, Evans M, Kaufman MH, Robertson E. Formation of germ-line chimaeras from embryo-derived teratocarcinoma cell lines. Nature. 1984;309:255–256. doi: 10.1038/309255a0. [DOI] [PubMed] [Google Scholar]
- Buecker C, Srinivasan R, Wu Z, Calo E, Acampora D, Faial T, Simeone A, Tan M, Swigut T, Wysocka J. Reorganization of enhancer patterns in transition from naive to primed pluripotency. Cell Stem Cell. 2014;14:838–853. doi: 10.1016/j.stem.2014.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulger M, Groudine M. Functional and mechanistic diversity of distal transcription enhancers. Cell. 2011;144:327–339. doi: 10.1016/j.cell.2011.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calo E, Wysocka J. Modification of enhancer chromatin: what, how, and why? Mol. Cell. 2013;49:825–837. doi: 10.1016/j.molcel.2013.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang LL, Kessler DS. Foxd3 is an essential Nodal-dependent regulator of zebrafish dorsal mesoderm development. Dev. Biol. 2010;342:39–50. doi: 10.1016/j.ydbio.2010.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chenoweth JG, McKay RDG, Tesar PJ. Epiblast stem cells contribute new insight into pluripotency and gastrulation. Dev. Growth Differ. 2010;52:293–301. doi: 10.1111/j.1440-169X.2010.01171.x. [DOI] [PubMed] [Google Scholar]
- Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ, Hanna J, Lodato MA, Frampton GM, Sharp PA, et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc. Natl. Acad. Sci. USA. 2010;107:21931–21936. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denslow SA, Wade PA. The human Mi-2/NuRD complex and gene regulation. Oncogene. 2007;26:5433–5438. doi: 10.1038/sj.onc.1210611. [DOI] [PubMed] [Google Scholar]
- ENCODE Project Consortium An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst J, Kellis M. ChromHMM: automating chromatin-state discovery and characterization. Nat. Methods. 2012;9:215–216. doi: 10.1038/nmeth.1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francetic T, Le May M, Hamed M, Mach H, Meyers D, Cole PA, Chen J, Li Q. Regulation of Myf5 Early Enhancer by Histone Acetyltransferase p300 during Stem Cell Differentiation. J. Mol. Biol. 2012 doi: 10.4172/2168-9547.1000103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haberland M, Montgomery RL, Olson EN. The many roles of histone deacetylases in development and physiology: implications for disease and therapy. Nat. Rev. Genet. 2009;10:32–42. doi: 10.1038/nrg2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamed M, Khilji S, Chen J, Li Q. Stepwise acetyltransferase association and histone acetylation at the Myod1 locus during myogenic differentiation. Sci. Rep. 2013;3:2390. doi: 10.1038/srep02390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanna LAL, Foreman RKR, Tarasenko IAI, Kessler DSD, Labosky PAP. Requirement for Foxd3 in maintaining pluripotent cells of the early mouse embryo. Genes Dev. 2002;16:2650–2661. doi: 10.1101/gad.1020502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkins RD, Hon GC, Yang C, Antosiewicz-Bourget JE, Lee LK, Ngo Q-M, Klugman S, Ching KA, Edsall LE, Ye Z, et al. Dynamic chromatin states in human ES cells reveal potential regulatory sequences and genes involved in pluripotency. Cell Res. 2011;21:1393–1409. doi: 10.1038/cr.2011.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi K, Ohta H, Kurimoto K, Aramaki S, Saitou M. Reconstitution of the mouse germ cell specification pathway in culture by pluripotent stem cells. Cell. 2011;146:519–532. doi: 10.1016/j.cell.2011.06.052. [DOI] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC. Simple Combinations of Lineage-Determining Transcription Factors Prime cis-Regulatory Elements Required for Macrophage and B Cell Identities. Mol. Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho L, Ronan JL, Wu J, Staahl BT, Chen L, Kuo A, Lessard J, Nesvizhskii AI, Ranish J, Crabtree GR. An embryonic stem cell chromatin remodeling complex, esBAF, is essential for embryonic stem cell self-renewal and pluripotency. Proc. Natl. Acad. Sci. USA. 2009;106:5181–5186. doi: 10.1073/pnas.0812889106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochgreb-Hägele T, Bronner ME. A novel FoxD3 gene trap line reveals neural crest precursor movement and a role for FoxD3 in their specification. Dev. Biol. 2013;374:1–11. doi: 10.1016/j.ydbio.2012.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu G, Wade PA. NuRD and pluripotency: a complex balancing act. Cell Stem Cell. 2012;10:497–503. doi: 10.1016/j.stem.2012.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamaladdin S, Kelly RDW, O'Regan L, Dovey OM, Hodson GE, Millard CJ, Portolano N, Fry AM, Schwabe JWR, Cowley SM. Histone deacetylase (HDAC) 1 and 2 are essential for accurate cell division and the pluripotency of embryonic stem cells. Proc. Natl. Acad. Sci. USA. 2014;111:9840–9845. doi: 10.1073/pnas.1321330111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jouneau A, Ciaudo C, Sismeiro O, Brochard V, Jouneau L, Vandormael-Pournin S, Coppée J-Y, Zhou Q, Heard E, Antoniewski C, Cohen-Tannoudji M. Naive and primed murine pluripotent stem cells have distinct miRNA expression profiles. RNA. 2012;18:253–264. doi: 10.1261/rna.028878.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller G. Embryonic stem cell differentiation: emergence of a new era in biology and medicine. Genes Dev. 2005;19:1129–1155. doi: 10.1101/gad.1303605. [DOI] [PubMed] [Google Scholar]
- Kojima Y, Kaufman-Francis K, Studdert JB, Steiner KA, Power MD, Loebel DAF, Jones V, Hor A, de Alencastro G, Logan GJ, et al. The transcriptional and functional properties of mouse epiblast stem cells resemble the anterior primitive streak. Cell Stem Cell. 2014;14:107–120. doi: 10.1016/j.stem.2013.09.014. [DOI] [PubMed] [Google Scholar]
- Krishnakumar R, Kraus WL. PARP-1 regulates chromatin structure and transcription through a KDM5B-dependent pathway. Mol. Cell. 2010;39:736–749. doi: 10.1016/j.molcel.2010.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai AY, Wade PA. Cancer biology and NuRD: a multifaceted chromatin remodelling complex. Nat. Rev. Cancer. 2011;11:588–596. doi: 10.1038/nrc3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalmansingh AS, Karmakar S, Jin Y. Multiple modes of chromatin remodeling by Forkhead box proteins. Biochim. Biophys. Acta. 2012;1819:707–715. doi: 10.1016/j.bbagrm.2012.02.018. [DOI] [PubMed] [Google Scholar]
- Lam EW-F, Brosens JJ, Gomes AR, Koo C-Y. Forkhead box proteins: tuning forks for transcriptional harmony. Nat. Rev. Cancer. 2013;13:482–495. doi: 10.1038/nrc3539. [DOI] [PubMed] [Google Scholar]
- Lanner F, Rossant J. The role of FGF/Erk signaling in pluripotent cells. Development. 2010;137:3351–3360. doi: 10.1242/dev.050146. [DOI] [PubMed] [Google Scholar]
- Li Z, Gadue P, Chen K, Jiao Y, Tuteja G, Schug J, Li W, Kaestner KH. Foxa2 and H2A.Z mediate nucleosome depletion during embryonic stem cell differentiation. Cell. 2012;151:1608–1616. doi: 10.1016/j.cell.2012.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liber D, Domaschenz R, Holmqvist P-H, Mazzarella L, Georgiou A, Leleu M, Fisher AG, Labosky PA, Dillon N. Epigenetic priming of a pre-B cell-specific enhancer through binding of Sox2 and Foxd3 at the ESC stage. Cell Stem Cell. 2010;7:114–126. doi: 10.1016/j.stem.2010.05.020. [DOI] [PubMed] [Google Scholar]
- Liu Y, Labosky PA. Regulation of embryonic stem cell self-renewal and pluripotency by Foxd3. Stem Cells. 2008;26:2475–2484. doi: 10.1634/stemcells.2008-0269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupien M, Eeckhoute J, Meyer CA, Wang Q, Zhang Y, Li W, Carroll JS, Liu XS, Brown M. FoxA1 translates epigenetic signatures into enhancer-driven lineage-specific transcription. Cell. 2008;132:958–970. doi: 10.1016/j.cell.2008.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marks H, Kalkan T, Menafra R, Denissov S, Jones K, Hofemeister H, Nichols J, Kranz A, Stewart AF, Smith A, Stunnenberg HG. The transcriptional and epigenomic foundations of ground state pluripotency. Cell. 2012;149:590–604. doi: 10.1016/j.cell.2012.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKay RD, Crawford GE, Scacheri PC, Tesar PJ. Epigenomic Comparison Reveals Activation of “Seed” Enhancers during Transition from Naive to Primed Pluripotency. Cell Stem Cell. 2014;14:854–863. doi: 10.1016/j.stem.2014.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mundell NA, Labosky PA. Neural crest stem cell multipotency requires Foxd3 to maintain neural potential and repress mesenchymal fates. Development. 2011;138:641–652. doi: 10.1242/dev.054718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy G, Dániel B, Jónás D, Nagy L, Barta E. A novel method to predict regulatory regions based on histone mark landscapes in macrophages. Immunobiology. 2013;218:1416–1427. doi: 10.1016/j.imbio.2013.07.006. [DOI] [PubMed] [Google Scholar]
- Najm FJ, Chenoweth JG, Anderson PD, Nadeau JH, Redline RW, McKay RDG, Tesar PJ. Isolation of epiblast stem cells from preimplantation mouse embryos. Cell Stem Cell. 2011;8:318–325. doi: 10.1016/j.stem.2011.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols J, Smith A. The origin and identity of embryonic stem cells. Development. 2011;138:3–8. doi: 10.1242/dev.050831. [DOI] [PubMed] [Google Scholar]
- Nitzan E, Pfaltzgraff ER, Labosky PA, Kalcheim C. Neural crest and Schwann cell progenitor-derived melanocytes are two spatially segregated populations similarly regulated by Foxd3. Proc. Natl. Acad. Sci. USA. 2013;110:12709–12714. doi: 10.1073/pnas.1306287110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parchem RJ, Ye J, Judson RL, LaRussa MF, Krishnakumar R, Blelloch A, Oldham MC, Blelloch R. Two miRNA clusters reveal alternative paths in late-stage reprogramming. Cell Stem Cell. 2014;14:617–631. doi: 10.1016/j.stem.2014.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasini D, Malatesta M, Jung HR, Walfridsson J, Willer A, Olsson L, Skotte J, Wutz A, Porse B, Jensen ON, Helin K. Characterization of an antagonistic switch between histone H3 lysine 27 methylation and acetylation in the transcriptional regulation of Polycomb group target genes. Nucleic Acids Res. 2010;38:4958–4969. doi: 10.1093/nar/gkq244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plank JL, Suflita MT, Galindo CL, Labosky PA. Transcriptional targets of Foxd3 in murine ES cells. Stem Cell Res. (Amst.) 2014;12:233–240. doi: 10.1016/j.scr.2013.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rada-Iglesias A, Bajpai R, Swigut T, Brugmann SA, Flynn RA, Wysocka J. A unique chromatin signature uncovers early developmental enhancers in humans. Nature. 2011;470:279–283. doi: 10.1038/nature09692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossant J, Tam PPL. Blastocyst lineage formation, early embryonic asymmetries and axis patterning in the mouse. Development. 2009;136:701–713. doi: 10.1242/dev.017178. [DOI] [PubMed] [Google Scholar]
- Sekiya T, Muthurajan UM, Luger K, Tulin AV, Zaret KS. Nucleosome-binding affinity as a primary determinant of the nuclear mobility of the pioneer transcription factor FoxA. Genes Dev. 2009;23:804–809. doi: 10.1101/gad.1775509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shim EY, Woodcock C, Zaret KS. Nucleosome positioning by the winged helix transcription factor HNF3. Genes Dev. 1998;12:5–10. doi: 10.1101/gad.12.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shlyueva D, Stampfel G, Stark A. Transcriptional enhancers: from properties to genome-wide predictions. Nat. Rev. Genet. 2014;15:272–286. doi: 10.1038/nrg3682. [DOI] [PubMed] [Google Scholar]
- Smith AG. Embryo-derived stem cells: of mice and men. Annu. Rev. Cell Dev. Biol. 2001;17:435–462. doi: 10.1146/annurev.cellbio.17.1.435. [DOI] [PubMed] [Google Scholar]
- Snow MHL. Gastrulation in the mouse: Growth and regionalization of the epiblast. Development. 1977;42:293–303. [Google Scholar]
- Stamatoyannopoulos JA, Snyder M, Hardison R, Ren B, Gingeras T, Gilbert DM, Groudine M, Bender M, Kaul R, Canfield T, et al. Mouse ENCODE Consortium An encyclopedia of mouse DNA elements (Mouse ENCODE). Genome Biol. 2012;13:418. doi: 10.1186/gb-2012-13-8-418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takebayashi S, Lei I, Ryba T, Sasaki T, Dileep V, Battaglia D, Gao X, Fang P, Fan Y, Esteban MA, et al. Murine esBAF chromatin remodeling complex subunits BAF250a and Brg1 are necessary to maintain and reprogram pluripotency-specific replication timing of select replication domains. Epigenetics Chromatin. 2013;6:42. doi: 10.1186/1756-8935-6-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng L, Mundell NA, Frist AY, Wang Q, Labosky PA. Requirement for Foxd3 in the maintenance of neural crest progenitors. Development. 2008;135:1615–1624. doi: 10.1242/dev.012179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas AJ, Erickson CA. FOXD3 regulates the lineage switch between neural crest-derived glial cells and pigment cells by repressing MITF through a non-canonical mechanism. Development. 2009;136:1849–1858. doi: 10.1242/dev.031989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas-Chollier M, Defrance M, Medina-Rivera A, Sand O, Herrmann C, Thieffry D, van Helden J. RSAT 2011: regulatory sequence analysis tools. Nucleic Acids Res. 2011;39:W86–W91. doi: 10.1093/nar/gkr377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wamstad JA, Alexander JM, Truty RM, Shrikumar A, Li F, Eilertson KE, Ding H, Wylie JN, Pico AR, Capra JA, et al. Dynamic and coordinated epigenetic regulation of developmental transitions in the cardiac lineage. Cell. 2012;151:206–220. doi: 10.1016/j.cell.2012.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W-D, Melville DB, Montero-Balaguer M, Hatzopoulos AK, Knapik EW. Tfap2a and Foxd3 regulate early steps in the development of the neural crest progenitor population. Dev. Biol. 2011;360:173–185. doi: 10.1016/j.ydbio.2011.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whyte WA, Bilodeau S, Orlando DA, Hoke HA, Frampton GM, Foster CT, Cowley SM, Young RA. Enhancer decommissioning by LSD1 during embryonic stem cell differentiation. Nature. 2012;482:221–225. doi: 10.1038/nature10805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong J, Moreno GT, Young MK, Côté J, Wang W. NURD, a novel complex with both ATP-dependent chromatin-remodeling and histone deacetylase activities. Mol. Cell. 1998;2:851–861. doi: 10.1016/s1097-2765(00)80299-3. [DOI] [PubMed] [Google Scholar]
- Xu J, Pope SD, Jazirehi AR, Attema JL, Papathanasiou P, Watts JA, Zaret KS, Weissman IL, Smale ST. Pioneer factor interactions and unmethylated CpG dinucleotides mark silent tissue-specific enhancers in embryonic stem cells. Proc. Natl. Acad. Sci. USA. 2007;104:12377–12382. doi: 10.1073/pnas.0704579104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Watts JA, Pope SD, Gadue P, Kamps M, Plath K, Zaret KS, Smale ST. Transcriptional competence and the active marking of tissue-specific enhancers by defined transcription factors in embryonic and induced pluripotent stem cells. Genes Dev. 2009;23:2824–2838. doi: 10.1101/gad.1861209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yildirim O, Li R, Hung JH, Chen PB, Dong X, Ee LS. Mbd3/ NURD complex regulates expression of 5-hydroxymethylcytosine marked genes in embryonic stem cells. Cell. 2011;147:1498–1510. doi: 10.1016/j.cell.2011.11.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, Liu XS. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.