Abstract
Background
Lung cancer results in the highest number of cancer deaths worldwide. The segmentation of lung nodules is an important task in computer systems to help physicians differentiate malignant lesions from benign lesions. However, it has already been observed that this may be a difficult task, especially when nodules are connected to an anatomical structure.
Methods
This paper proposes a method to estimate the background of the nodule area and how this estimation is used to facilitate the segmentation task.
Results
Our experiments indicate more than 99% of accuracy with less than 1% of false positive rate (FPR).
Conclusions
The proposed methods achieved better results than a state-of-the-art approach, indicating potential to be used in medical image processing systems.
Keywords: Lung nodules, segmentation, background estimation, chest CT
Introduction
Lung cancer is currently considered one of the most common and dangerous types of cancer. According to World Health Organization, in 2012 lung cancer resulted in 1.59 million deaths worldwide and caused approximately two times more deaths than liver cancer, the second leading cause of death by cancer (1). Statistics indicate 0.87 as the overall ratio of mortality to incidence of lung cancer and relative lack of variability in survival in different world regions (1). However, the cancer mortality is reduced when cases can be detected and treated in early stages. For about 20 years, computed tomography (CT) examinations have been widely used for early detection of cancer (2).
The segmentation of lung nodules is an important part of two different systems that are related to the prevention and diagnosis of lesions. The first system is the computer-aided diagnosis (CAD) system, which aims to improve the ability to detect the nodules and can help to classify the nodule as malignant or benign. The second system focuses on content-based medical image retrieval (CBMIR), where the system identifies a set of images from a database that have similar characteristics to the lung nodule that the physician is viewing. The goal of these systems is to assist the physician in differentiating between malignant and benign lesions and therefore resulting in timelier treatment and better prognosis. The segmentation is usually performed after a pre-processing step and before a feature extraction step that quantifies characteristics of the nodule. When using the CAD system, these features will be used in the classification algorithm. When using the CBMIR system, these features are the input of the similarity calculation between different cases. Since characteristics of the nodule, such as size and shape, are important for the analysis of lung nodules (3), the feature extraction step depends on a proper determination of the lesion area. Consequently, the segmentation significantly influences the system outputs, and providing accurate segmentation is an important goal.
Related work and motivation
Sluimer et al. (2) published a survey covering the literature and reviewed all relevant work related to the computer analysis of chest CT. Considering the lung nodule segmentation, they found that while thresholding-based approaches are likely the most common approaches proposed in the literature, approaches based on clustering, mathematical morphology, template matching, and other techniques have also been proposed. More recently, Farag et al. (4) reviewed the literature and also proposed approaches using level sets and 3D morphological operations. But more importantly, Farag et al. observed that the algorithms proposed in the literature frequently fail when the nodule is connected to another structure, e.g., the pleural surface. This failure is due to the assumption of a difference between the pixel intensities (in the Hounsfield scale) of the nodule and its surrounding, which is not always valid. Tan et al. (5) also observed this same issue, and as a result, proposed to combine thresholding, watershed, active contours and Markov random field in a rule-based approach. Although they obtained good results, their experiments were performed on only a small number of cases. Moreover, their proposed method is considered very complex, where the accuracy of each step is dependent on the previous step, as well as on a set of defined parameters. In contrast, Farag et al. (4) performed their experiments using a very large number of cases, collecting images from four different databases. Farag et al.’s paper presents a level set algorithm that searches for elliptical regions, based on the idea that the “head” of the nodule tends to assume a rounded shape. After the level set minimization, they used a thresholding technique as post-processing, determining the lesion area inside the elliptical region.
When performing experiments with our image database, we also observed the difficulty in segmenting a lesion that is connected to other structures. Intensity-based techniques such as thresholding and clustering may be able to generate accurate results for isolated nodules, but these techniques fail when the nodule is connected to the pleural tissue or chest wall structures and mediastinal tissues that present similar Hounsfield units (4,5). Although some segmentation approaches assume a previous segmentation of the whole lung differentiating the regions inside and outside the lung cavities, this method also may not be the solution, since nodules connected to the pleural tissue are frequently misidentified as part of the outer regions (6). Therefore, our main goal for this project is to propose an approach that is able to eliminate other structures from the image, facilitating the segmentation of the lesion.
Contributions and paper organization
First of all, it is important to observe that our method is proposed for the segmentation of lung nodules on a previously defined region of interest (ROI). This ROI must be rectangular and includes the area surrounding the lung nodule. Also, the segmentation is applied to each slice of the CT individually, and no information between slices has been used.
In this paper, the segmentation is based on a proposed background estimation method, more specifically an estimation of the ROI background. And, to estimate the ROI background, we make an important assumption: since the lung nodule is the only object to be segmented, the nodule is considered to be the only image foreground, and therefore, all other pixels (including structures that may be connected to the nodule) are consequently considered image background. We also demonstrate how this technique is able to facilitate the segmentation of a lung nodule and that a simple thresholding technique combined with morphological operations is able to accurately determine the nodule region in the chest CT slice. This paper is organized as follows. First, image sets and our proposed method for background estimation and lung nodule segmentation are described in the following section. Next, we present experimental results applying our approach on two different databases of chest CT images, including a comparison with a state-of-the-art method. Finally, conclusions about the paper and indications of possible future works are presented in the final section.
Materials and methods
We performed our experiments with two image sets: the Hamilton Health Sciences (HHS) database and the LIDC-IDRI database. The first one is a collection of 100 CTs from patients of the HHS, a network of six hospitals and a cancer center in Hamilton, Canada, with a catchment area serving more than 2.3 million people. This collection includes benign and malignant cases, with a large variability of sizes and shapes of lung nodules. The LIDC-IDRI database (7) is a publicly available database containing more than 2,000 chest CTs. Each case has been analyzed by one to four experienced radiologists, where the nodule areas were manually determined in the CT slices, i.e., the segmentation ground truths.
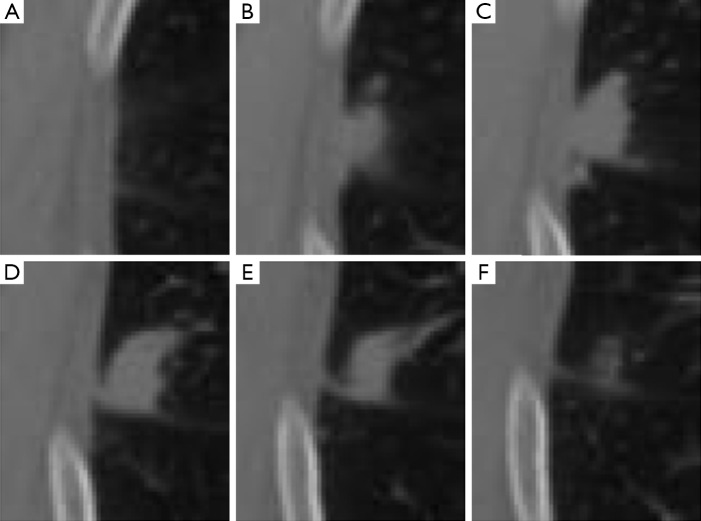
Our approach was developed to determine the lung nodule area (i.e., the foreground) based on a predefined ROI. Analyzing the ROI, we observed that the background of the defined ROI for an image (i.e., a CT slice) with a lesion is visually similar to the same region in a nearby CT slice without the presence of a lesion. We illustrate this idea in Figure 1, where we present ROIs, which are defined by an expert, for slices 55 to 60, respectively images A to F. Although nodule is visible only on images B to E (slices 56 to 59), the reader may also observe that by extracting this same region from the slices that are located before and after the lesion (i.e., slices 55 and 60, shown on images A and F), a similar anatomical structure is captured. However, it is also important to observe that although similar, the background information may present changes. Throughout the CT scanning, the visible lung area (i.e., the pleural cavity) enlarges and shrinks in size, and other structures such as vessels and other chest organs may become visible.
Figure 1.
Example of ROI extracted for a lung nodule connected to the pleural tissue. An expert observed the nodule in CT slices 56–59 and the area defined as ROI was extracted from each slice, respectively, images shown in (B)–(E). The same area is also extracted from the previous and posterior slices to those where the nodule is visible, i.e., slices 55 and 60, respectively, (A) and (F). ROI, region of interest; CT, computed tomography.
Considering that modern CT scanners generate images with thin slice thickness, the differences in the background between a slice and its previous or following slices are usually very small. These small differences are very similar to the differences between frames of a video, i.e., the motion of the objects in a scene. In the case illustrated in Figure 1, for example, the visible pleural tissue presents a “motion” (i.e., changes) similar to a smooth shift to the left when analyzing figures (A) to (F). Based on these similarities, we propose to estimate the ”motion” of these anatomical structures in a similar way that motion is estimated in a sequence of video frames. In the next section, we review two motion estimation techniques originally proposed for videos. Then, in the following section, we detail how we used these methods to estimate the background of the chest CTs and how it is possible to obtain accurate lung nodule segmentation results using a simple thresholding-based method following motion estimation.
Motion estimation methods review
Lucas-Kanade method
This is a classic technique (8) to estimate the optical flow between two frames (i.e., images) based on the idea that an affine model can represent the small and essentially constant flow in a local neighborhood of the pixel under consideration. In other words, it assumes that the brightness of a pixel p does not change in the subsequent frame considering a motion (Vx, Vy). Based on these assumptions, and making use of Taylor series approximation, the local image flow must satisfy the following Eq. [1]:
| [1] |
qi refers to an image pixel, and Ix(qi), Iy(qi), It(qi) indicate the partial derivatives of the image I with respect to position x, y and time t, evaluated at the point qi and at the current time. Considering all n pixels inside a window (i.e., the neighborhood of the pixel p), we can define a set of equations and write them in matrix form Av = b where Eq. [2]:
| [2] |
A solution can be determined using the least squares methodology, and consequently the motion is known for pixel p. After obtaining the motion vectors for all image pixels, a motion compensated image can be reconstructed. Considering that the least squares solution may not be integer, the motion compensation step is performed using interpolation.
SubME method
Chan et al. (8) proposed to start this motion estimation technique using a block matching step, i.e., pixels inside the block (i.e., the window) under consideration are compared to pixels in a search range (a pre-defined neighborhood of pixels). Although Chan et al. indicate that any block matching algorithm can be used to determine a motion (Vx, Vy), the full search was applied (i.e., all pixels are considered) and the sum of absolute differences was used as a metric to determine the best matching block.
Considering that the motion is obtained using the block matching step, (Vx, Vy) is computed as integer values, and no interpolation is needed for the motion compensation. Moreover, after shifting the image, the difference between (Vx, Vy) and the true displacement now have a high probability of being small (an assumption made by the Lucas-Kanade method that is not always valid). Consequently, the Taylor approximation is also valid, and Eq. [2] can be used to refine the motion estimation and obtain subpixel precision to the reconstructed image.
According to experiments by Chan et al. (8), SubME tends to obtain faster motion estimation. However, considering that we are performing this task on relatively small images (the previously defined ROIs), we did not observe significant speed improvement. Moreover, we observed good results from both methods when visually analyzing the reconstructed images, and hence we considered both methods in our experiments.
Background estimation and segmentation of lung nodules
The first step of our segmentation algorithm is to compute the background of the input image/slice I to be segmented. To perform this task, we must find the reference image Iref, the nearest slice without indication of lesion. Considering that a ROI has been previously defined to the set of slices to be segmented (i.e., all slices with the presence of a nodule), there are only two possible reference images/slices Iref: (I) the contiguous slice superior to this set of slices; and (II) the slice inferior to the same set. For example, lets consider again the previously presented case, where a nodule is present from slice 56 to slice 59, with ROI expertly defined in each of these slices (as can be seen in Figure 1). When performing the segmentation procedure for this case, slice 55 will be defined as Iref if slice 56 or 57 is segmented. Meanwhile, slice 60 will be defined as Iref for slices 58 and 59.
Then, based on the ROI defined for each image I, we crop the ROI R. In other words, the image R is a sub-image of I extracting only the rectangular region containing the lung nodule. Using the same pixel coordinates, we also crop Rref from Iref, i.e., the exact same rectangular region in a slice without the presence of lesion. Considering again the example in Figure 1, Figure 1A is defined as Rref if the ROI R to be segmented is Figure 1B,C, and Figure 1F is defined as Rref if the ROI R to be segmented is Figure 1D,E. However, it is important to observe that the ROI R can be defined differently for each slice considering differences in size or shape between the visible nodule areas in each slice. Consequently, although the same Iref can be used as reference image for two or more CT slices, the Rref is defined specifically for each input slice I since it is dependent of the pixel coordinates of the predefined ROI R.
After the ROI extraction, we estimate the motion between Rref and R using one of the methods presented in section II-A. Then, based on the computed motion vectors, pixels of Rref are moved/shifted, and we obtain a reconstructed image Rbkg. This new image presents the same background information (without the visible nodule) as Rref, but moves the visible structures in the direction of the “motion” existent between Rref and R. Consequently, Rbkg estimates the background of R.
After obtaining Rbkg, we can use this image to facilitate the lesion segmentation of R. We calculate Rsub = R − Rbkg. This subtraction enhances the lesion area and decreases the intensities of other image areas. Before we apply a thresholding algorithm to obtain the lung nodule segmentation, we should refine Rsub. If the subtraction results in pixels lower than zero, this indicates that a structure (e.g., a vessel), originally in the slice, used as reference is not visible/present in I. To eliminate this structure, all pixels lower than zero are truncated to zero. We then divide all pixel intensities by the maximum value obtained from the subtraction. In this way, we obtain an image scaled to the range [0, 1]. Mean filtering (with a 3×3 kernel) is used to smooth possible noise. Next, we compute the Otsu’s threshold (9), and all pixels with intensities above the threshold provide us a binary mask which is our first estimative of lesion area. If thresholding results in more than one connected region, we eliminate all regions except the one closest to the ROI center.
Morphological operations are the final steps of our proposed segmentation method: a closing, a dilation, and hole filling. The closing operation (using a disk with two pixels of radius as a structuring element) aims to obtain a more rounded shape, what is typical of lung nodules. The dilation (using a disk with five pixels of radius as structuring element) is responsible for generating a slightly larger area. This step is necessary because experts (i.e., medical doctors), when defining ground truths, tend to determine the borders as slightly wider than the lesion itself, to guarantee that no lesion portion is missed. Finally, the last step will obviously fill possible remaining holes inside the segmented area.
In Figure 2, we present a workflow with all the steps of our approach, and the reader may also see an example illustrating each of these steps in Figure 3. Based on a predefined ROI to a nodule connected to the pleural surface, Figure 3 presents the image resulting from each step of our method until obtaining the nodule segmentation.
Figure 2.
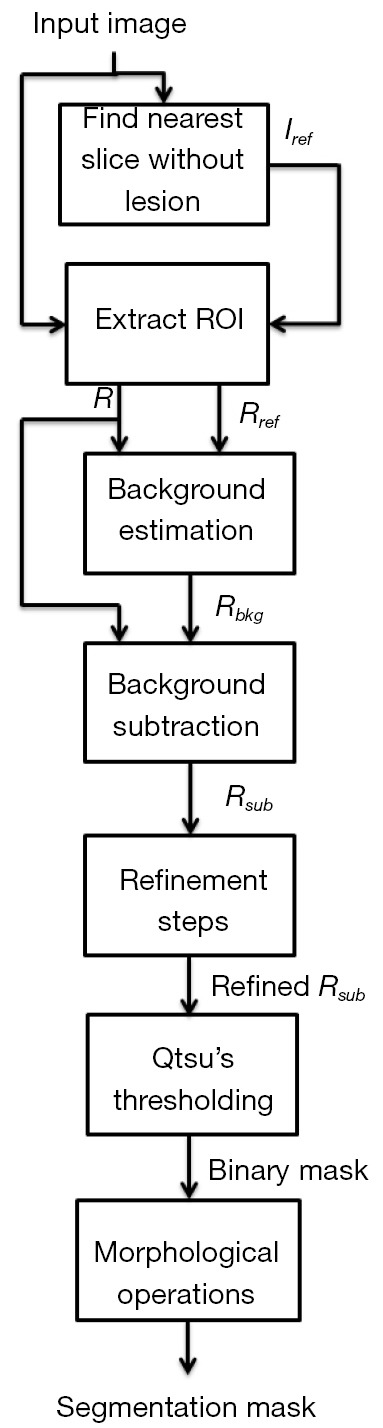
Workflow of our approach for the background estimation and segmentation of lung nodules. ROI, region of interest.
Figure 3.
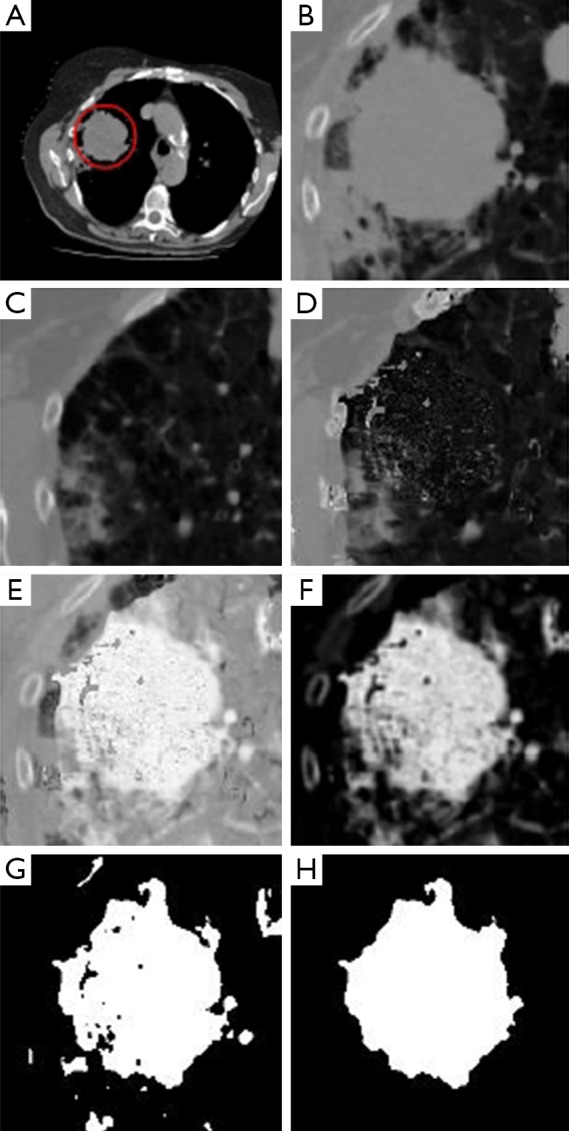
An example illustrating all the steps of the proposed approach: (A) Input image and ROI defined in red by an expert; (B) ROI extracted from the input image; (C) ROI extracted from the reference image; (D) background estimation result; (E) result of background subtraction; (F) result after refinement of image; (G) result of applying Otsu’s threshold; (H) final segmentation result after morphological operations. ROI, region of interest.
Results
In the following sections, we detail experiments that have been performed to evaluate the accuracy of our proposed approach for background estimation and segmentation of lung nodules in chest CT.
HHS database
Each case of the HHS database has been analyzed by an expert radiologist, and a geometrical form (e.g., an ellipse or a polygon) was drawn around each identified nodule to determine the ROI. In order to extract the ROI to be used as input to our method, we computed the bounding box of the drawn ROI (i.e., converted it to a rectangle), and then enlarged this region to 15 pixels in each direction (i.e., 30 horizontally and 30 vertically) to guarantee that all nodule pixels were within the ROI.
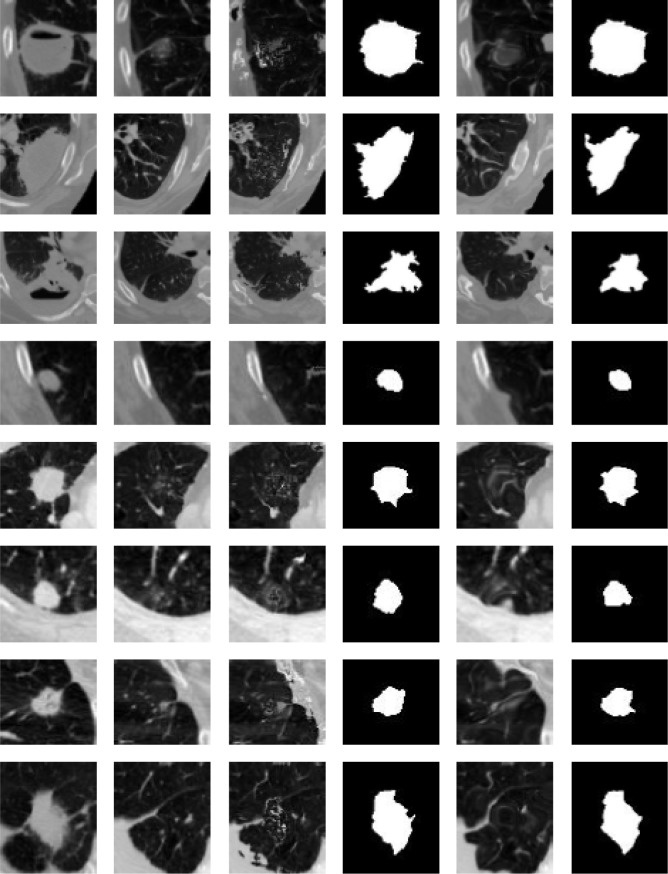
In Figure 4, we present the results for our background estimation approach based on the Lucas-Kanade and the SubME method. Readers may also observe the posterior lung nodule segmentation masks for each presented case. The results generated by the two approaches usually tend to be very similar and accurate. However, when carefully analyzing the results, the reader may also observe two possible issues of our approach. Specifically the seventh and eighth rows (bottom two rows) present examples with significant background difference between the image to be segmented and the reference image. Interestingly, the “motion” between these two images is not small as assumed by the Lucas-Kanade method. Considering the SubME method, it is possible to see artifacts in some of the reconstructed images (e.g., fourth and sixth rows), where the estimated background images distorted the pleural structure. These artefacts are a consequence of the block-matching step, which mistakenly matches a pleural region with a nodule region of similar intensities. However, these issues do not significantly influence the final segmentation result.
Figure 4.
Examples of results of our proposed approach. In the first column, images to be segmented. In the second column, the reference images. In the third and fourth columns, the estimated background images and the final segmentation results based on the Lucas-Kanade method. In the fifth and sixth columns, the estimated background images and the final segmentation results based on the SubME method.
LIDC-IDRI database
Very recently, Farag et al. (4) proposed their segmentation approach based on level sets, and as part of their experiments, they collected 315 cases from the LIDC-IDRI database (10). When comparing their segmentation results with the ground truths, they reported a 95% true positive rate (TPR), i.e., on average, 95% of the nodule pixels were correctly segmented using their approach; and 10% of false positive rate (FPR), i.e., on average, 10% of the background pixels were mistakenly segmented as nodule pixels. Moreover, they indicated an accuracy of 81% when their segmentation achieved a 1% FPR.
To compare the results from Farag et al. approach with our proposed method, we collected the first 350 cases of the LIDC-IDRI database. We then extracted the ROI for each case based on the ground truths available in the database. Considering all ground truths (up to four for each slice), we computed the bounding box of all pixels that have been delimited as nodule area by at least one of the experts. To diminish the influence of the manual delimitations, we enlarged the bounding box to 30 pixels in each direction (i.e., 60 horizontally and 60 vertically). We chose a slightly higher number of cases than Farag et al. for two reasons: (I) they did not provide details on how they selected cases from the LIDC-IDRI database, and we hoped that by selecting a higher number, we would include all cases that they used; (II) they used an automated method to extract the ROI from each slice (11) but eliminated cases in which this pre-processing step failed. Although we did not use an automated method, we believe that the ROIs we extracted are very similar to those used in their experiments based on the results presented by Farag et al. (4).
Comparing our binary segmentation masks with the respective ground truths, we obtained very promising results. Using the SubME method for background estimation, the final segmentation resulted in 93.53% for TPR, 0.89% for FPR, and 99.11% for accuracy. Compared with the results from Farag et al., the TPR is slightly lower, but since the number of background pixels wrongly segmented is significantly lower, we obtained more accurate results. Moreover, when applying the Lucas-Kanade method for background estimation, the final segmentation resulted in 95.66% for TPR, 0.98% for FPR, and 99.02% for accuracy. Furthermore, when comparing these results with the approach proposed by Farag et al. (4) we observed that the main advantage of our proposed method is that there is a significantly lower FPR. This is a result of our background estimation step, which enables the elimination of all present structures besides the nodule and makes it possible for the posterior segmentation step not to confuse background pixels as lesion.
When the LIDC-IDRI database was proposed, it has been observed that experts may interpret the boundaries of lesions in different ways, and consequently the manually determined ground truths, for a specific slice, may present variability (7). As a result, we also measured the variability of our proposed lesion segmentation approach by comparing it to the different (up to four) ground truths of each image. Considering only the ground truths which our approach resulted in higher accuracy for each slice, and using the SubME method, our approach resulted in an average of 94.78% for TPR, 0.86% for FPR, and 99.13% for accuracy. Considering the ground truths that resulted in lower accuracy, we obtained 93.99% for TPR, 0.89% for FPR, and 99.11% for accuracy on average. Using the Lucas-Kanade method, the computed average was 98.04% for TPR, 0.94% for FPR, and 99.06% for accuracy for the higher accuracy situations; and for the lower accuracy situations, we obtained 93.26% for TPR, 0.97% for FPR, and 99.03% for accuracy. These results indicate that although experts may not completely agree when interpreting lung nodules boundaries, our approach can obtain consistent segmentation results.
Conclusions
We proposed a novel technique to estimate the background of nodules in chest CTs and demonstrated how a simple thresholding-based algorithm can take advantage of this pre-processing step and achieve accurate nodule segmentation results. Considering all performed experiments, we understand that the Lucas-Kanade method achieves better background estimations for this application. Although the FPR and accuracy levels are slightly better using the SubME method, it is important to observe the higher TPR using the Lucas-Kanade method. Higher TPR levels indicate that larger portions of the nodules are correctly segmented, and this characteristic will positively influence the posterior processing step of a CAD or CBMIR system, which is typically the feature extraction.
According to our preliminary results using a publicly available database, our approach demonstrated better results than a state-of-the-art method. Furthermore, we believe that our approach can be adapted and may be also useful for other medical applications and diseases. We intend to further use the proposed background estimation and segmentation approach in a complete CBMIR system for lung nodules.
Acknowledgements
The authors would like to thank the National Cancer Institute and the Foundation for the National Institutes of Health and their critical role in the creation of the free publicly available LIDC-IDRI database. The authors also thank CAPES—Brazilian Ministry of Education—for financial support.
Footnotes
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- 1.World Health Organization. Cancer. Fact sheet Fact sheet N°297. Available: http://www.who.int/mediacentre/factsheets/fs297/en/, 2014.
- 2.Sluimer I, Schilham A, Prokop M, van Ginneken B. Computer analysis of computed tomography scans of the lung: a survey. IEEE Trans Med Imaging 2006;25:385-405. [DOI] [PubMed] [Google Scholar]
- 3.Kligerman S, Abbott G. A radiologic review of the new TNM classification for lung cancer. AJR Am J Roentgenol 2010;194:562-73. [DOI] [PubMed] [Google Scholar]
- 4.Farag AA, El Munim HE, Graham JH, Farag AA. A novel approach for lung nodules segmentation in chest CT using level sets. IEEE Trans Image Process 2013;22:5202-13. [DOI] [PubMed] [Google Scholar]
- 5.Tan Y, Schwartz LH, Zhao B. Segmentation of lung lesions on CT scans using watershed, active contours, and Markov random field. Med Phys 2013;40:043502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kawata Y, Niki N, Ohmatsu H, Kusumoto M, Kakinuma R, Yamada K, Mori K, Nishiyama H, Eguchi K, Kaneko M, Moriyama N. Pulmonary Nodule Classification Based on Nodule Retrieval from 3-D Thoracic CT Image Database. In: Christian Barillot, David R. Haynor, Pierre Hellier, editors. Medical Image Computing and Computer-Assisted Intervention – MICCAI 2004. Berlin: Springer Berlin Heidelberg, 2004;3217:838-46. Available online: http://link.springer.com/chapter/10.1007%2F978-3-540-30136-3_102
- 7.Lucas BD, Kanade T. An iterative image registration technique with an application to stereo vision. Proceedings of Imaging Understanding Workshop 1981:121-130. [Google Scholar]
- 8.Chan SH, Vo DT, Nguyen TQ. Subpixel motion estimation without interpolation. Acoustics Speech and Signal Processing (ICASSP), 2010 IEEE International Conference on IEEE 2010:722-5. [Google Scholar]
- 9.Otsu N. A threshold selection method from graylevel histograms. IEEE Transactions on Systems, Man, and Cybernetics 1979;9:62-6. [Google Scholar]
- 10.Armato SG, 3rd, McLennan G, Bidaut L, McNitt-Gray MF, Meyer CR, Reeves AP, Zhao B, Aberle DR, Henschke CI, Hoffman EA, Kazerooni EA, MacMahon H, Van Beeke EJ, Yankelevitz D, Biancardi AM, Bland PH, Brown MS, Engelmann RM, Laderach GE, Max D, Pais RC, Qing DP, Roberts RY, Smith AR, Starkey A, Batrah P, Caligiuri P, Farooqi A, Gladish GW, Jude CM, Munden RF, Petkovska I, Quint LE, Schwartz LH, Sundaram B, Dodd LE, Fenimore C, Gur D, Petrick N, Freymann J, Kirby J, Hughes B, Casteele AV, Gupte S, Sallamm M, Heath MD, Kuhn MH, Dharaiya E, Burns R, Fryd DS, Salganicoff M, Anand V, Shreter U, Vastagh S, Croft BY, Clarke LP. The Lung Image Database Consortium (LIDC) and Image Database Resource Initiative (IDRI): a completed reference database of lung nodules on CT scans. Med Phys 2011;38:915-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Farag AA, Graham J, Farag AA, Elshazly S, Falk R. Parametric and non-parametric nodule models: design and evaluation. Proc. of Third International Workshop on Pulmonary Image Processing in conjunction with MICCAI-10 2010:151-62. Available online: http://www.lungworkshop.org/2010/proc2010/farag.pdf