Abstract
The budding yeast Saccharomyces cerevisiae is a useful model for elucidating the pathways that control life span and the influence of environmental factors, such as calorie restriction (CR). For 75 years, CR has been studied for its ability to delay diseases of aging in mammals, from cancer to cardiovascular disease (McCay et al., Nutr Rev 33:241–243, 1975). In many other species, reducing calorie intake extends life span, including unicellular organisms (Jiang et al., FASEB J 14:2135–2137, 2000; Lin et al., Science 289:2126–2128, 2000), invertebrates (Rogina and Helfand, Proc Natl Acad Sci U S A 101:15998–16003, 2004), and rodents (Martín-Montalvo et al., Oncogene 30:505–520, 2011). Here we describe how to calorically restrict yeast cells, the methods used to determine the replicative life span (RLS) of budding yeast cells, how to selectively kill daughter cells using the mother enrichment program (MEP), how to measure recombination frequency at the rDNA locus, how to isolate large quantities of old cells, and how to analyze the circular forms of DNA known as extrachromosomal rDNA circles (ERCs), a cause of aging in S. cerevisiae (Petes, Cell 19:765–774, 1980; Sinclair and Guarente, Cell 91:1033–1042, 1997; Defossez et al., Mol Cell 3:447–455, 1999).
Keywords: Yeast aging, Life span, RLS, Bud scars, Mother cell enrichment program, ERC
1 Introduction
The quest to understand why we age and how to slow it has been a goal ever since humans evolved. In recent years, great strides have been made in elucidating genes that influence the pace of aging. Studying humans and even rodents is a daunting and costly task that is not amenable to genetic screening. Instead, many researchers have turned to simpler organisms with shorter life spans to identify conserved longevity genes and small molecules that might regulate it.
The simple dietary manipulation known as calorie restriction (CR) can extend life span of rodents and delay most age-related diseases. But what underlies this effect? In recent years, the budding yeast Saccharomyces cerevisiae has become one of the leading models for uncovering conserved causes of aging and understanding how CR works. Today, dozens of yeast longevity genes are known and we have a good understanding of how CR works in yeast, the best for any studied species [1, 2].
There are two ways to define yeast life span. One is the number of days a yeast culture remains viable in a hypometabolic stationary phase. This is known as the “chronological life span” [3]. Alternatively, one can determine life span for a single cell based on the number of times it divides, known as “replicative life span” or “RLS” [4].
Despite yeast being a simple eukaryote, accurately determining its replicative life span is not a simple task, even with the invention of the “mother enrichment program” or “MEP.” The MEP is an estradiol-inducible daughter killing system [5] that allows much higher purity of old cells. The MEP also allows RLS to be determined as a function of viability over time. But the system is not perfect: ~8 % of daughter cells in the MEP can still form micro-colonies due to “leakiness” [5]. To precisely quantify mean and maximum RLS of a given strain, micromanipulation is still necessary. Only with time does one become skilled at this technique, and often beginners do not obtain reliable data the first time. Even using the MEP, very pure populations of old cells require a magnetically sorting procedure to separate the mothers from daughters in a culture.
2 Materials
2.1 Preparation of Yeast for Replicative Life Span Analysis
2.2× supplemented yeast extract/peptone(YEP)media:22.2g/L Bacto Yeast extract (Becton, Dickinson and Company), 44.4 g/L Bacto Peptone (Becton, Dickinson and Company) in water and filter sterilized with a 0.22 μm vacuum filter. Supplement the media by adding 40 mL/L 0.5 % adenine solution and 20 mL/L for the remaining 1 % amino acid solutions.
Use flat wooden toothpicks that have a rounded end opposite to the pointed tip. Diamond brand is suitable. Sterilize by autoclaving in a small beaker with aluminum foil on the top and toothpicks point down. Discard any toothpicks that have a sharp protruding splinter that could damage the agar.
4 % agar: Bacto Agar (Becton, Dickinson and Company); 8 g of agar is added into 200 mL of water and autoclaved.
18 M Ω Milli-Q grade water is used for all media preparations.
Petri plates: 100 × 15 mm (Falcon).
Amino acid and adenine (Sigma-Aldrich) stock solutions (w/v): 0.5 % adenine, 1 % uridine, 1 % tryptophan, 1 % histidine, and 1 % leucine solutions are prepared with water and filter sterilized using a 0.22 μm vacuum filter. These supplement the auxotrophies that are particular to the W303AR strain of yeast. Different amino acids may be required for other strains.
1.05× YEP: 10.5 g/L Bacto Yeast extract (Becton, Dickinson and Company), 21.1 g/L Bacto Peptone (Becton, Dickinson and Company) in water and autoclaved. Supplement after autoclaving as with the preparation of 2.2× YEP media, but use half the amount of solutions.
40 % (w/v) glucose stock solution.
Dissection needle kit (Cora Styles Needles n Blocks, Cat. No. 1025, 105 Cypress Pt., Hendersonville, NC 28739, Phone# 828-629-9528). We use 10 or 25 μM needles that are made by polishing flat the ends of optic fibers.
17β-Estradiol (Sigma, St. Louis, MO), only if using the MEP.
2.2 Magnetic Sorting of Old Yeast
Phosphate-buffered saline (PBS): A 10× stock solution is prepared with 1.37 M NaCl, 27 mM KCl, 43 mM Na2H- PO4·7H2O, 14 mM KH2PO4,adjusted to pH 7.4 with HCl if required and then promptly autoclaved. A 1× working solution is made by diluting 1:10 with sterile water.
EZ-Link Sulfo-NHS-LC-Biotin, m.w. 556.59 (Thermo Scientific/Pierce).
Streptavidin-coated paramagnetic iron beads (Thermo Scientific/Pierce). Other labs have used Dynabeads streptavidin (Invitrogen/Life Technologies).
Calcofluor fluorescent brightener 28 (Sigma, St. Louis, MO).
17β-Estradiol (Sigma, St. Louis, MO), only if using the MEP.
2.3 Isolation of ERCs
1× TE buffer: 10 mM Tris–Cl, pH 8.0 and 1 mM EDTA, pH 8.0.
Sorbitol solution: 0.9 M sorbitol (Fisher), 0.1 M Tris–Cl, pH 8.0, 0.1 M EDTA are prepared with water and filter sterilized using a 0.22 μm filter.
Zymolyase solution: Dissolve 0.3 mg/mL Zymolyase (20,000 U/g; ICN Immunobiologicals) in sorbitol solution and store at 4 °C.
2-Mercaptoethanol.
10 % (w/v) sodium dodecyl sulfate (SDS).
5 M potassium acetate solution (do not adjust pH).
2.4 Analysis of ERCs
Random primed DNA labeling kit (Roche Diagnostics).
ProbeQuant™ Sephadex G-50 columns (Amersham Pharmacia).
dCTP32 (PerkinElmer).
Pre-hybridization buffer: 100 μl boiled salmon sperm DNA (5 mg/mL stock ssDNA), 1 g dextran sulfate, 1 mL 10 % SDS, 0.56 g NaCl in 10 mL water. Heat at 65 °C for 30 min to dissolve the salts into solution.
10× TBE electrophoresis stock buffer: 0.89 M Tris base, 0.89 M boric acid, 20 mM EDTA, pH 8.0. A 1× working solution is made by diluting 1:10 with water.
0.4 M NaOH solution.
0.25 M HCl solution.
20× SSC stock solution: 3 M NaCl, 0.3 M Na3citrate·2H2O. Adjust the pH to 7.0 with 1 M HCl. A 2× working solution is made by diluting 1:10 with water.
Nylon membrane (NEN).
Wash buffers: 2× SSC 0.1 % SDS (low stringency) at room temp, 1× SSC 0.1 % SDS (medium stringency) at 60 °C, 0.1× SSC 0.5 % SDS (high stringency) at 60 °C, 0.1× SSC (brief wash) at room temp.
2.5 Recombination Analysis
1.05× YEP: prepared in the same manner as previously described.
2.2× YEP: unlike 2.2× YEP that is prepared for life span assay plates, do not supplement this stock solution with adenine or histidine. The solution can also be autoclaved rather than filter sterilized. Otherwise, it is prepared in the same manner.
3 % agar: Bacto Agar (Becton, Dickinson and Company); 6 g of agar is added into 200 mL of water and autoclaved.
Large Petri plates: 150 × 15 mm (VWR).
Glass beads for spreading can be obtained from Amazon.com from Carolina Biological Supply Company. They are sterilized by autoclaving.
3 Methods
3.1 Preparation of Yeast for Replicative Life Span (RLS) Analysis with and Without MEP
The day prior to starting the experiment, the yeast should be streaked onto agar plates containing media identical to that used for the life span assay. The yeast should be as fresh as possible. If taken directly from the freezer, allow at least 2 days of consecutive streaking for recovery. Use yeast only from a plate that has been struck within 24 h, i.e., as fresh and actively growing as possible. See Note 1 for tips on handling the yeast and plates.
Using a sterile toothpick, transfer a tiny amount (barely visible to the eye) from the previous nights’ streak onto a fresh plate that will be used for the life span analysis. The yeast should be a barely visible dot on the plate. Allow the transferred cells to grow for an additional 3–5 h at 30 °C.
After attaching the inverted plate on to the stage of the microscope, use the tip of the fiber-optic needle to drag a bunch of cells, i.e., however many will fit under the needle, approximately ≥ 1 cm away from the plated dot of cells. You may need to repeat this several times to move enough cells.
Find cells that are relatively small and healthy looking; i.e., round and exhibiting no aberrant morphology. Use the tip of the needle to array 40 cells, in sets of 5, planting a single column. Allow enough spacing between each cell so as to see no more than 3–5 cells within the field of view using a 10× objective. Each group of 5 cells can be marked by stabbing the needle into the agar, i.e., use 1 stab, 2 stabs, 3, 4, and a right angle, any pattern that will help you to find and keep track of the column of cells on the plate. The same pattern can then be marked in a notebook with columns divided into groups of 5.
After arraying the cells, remove the prepared life span assay plate from the microscope stage and incubate at 30 °C for 1–2 h, after first sealing the plate with Parafilm to prevent the loss of moisture. After returning the plate to the microscope, the yeast will have undergone about 1–2 divisions. If a cell died before the first division, we excluded it from the data set assuming the transfer was deleterious. Most wild-type strains divide at least five times before senescing.
With the needle, pull apart the cells and leave behind the smaller budded cell (Fig. 1). This starting “virgin” cell will henceforth be the mother cell during the assay. Discard the rest of the divided cells by moving them to the left, corresponding to about 1/3 of a field of view away from the virgin mother cell. Not all cells will be ready for separation at the same time. Thus, you may need to come back after another 30 min to 1 h after the others were separated. Importantly, mother cells are dragged carefully in the opposite direction to the daughters every 2–5 divisions about ¼ of a field of view to avoid running out of nutrients. If using the MEP, first grow the mother cells on YPD medium with no estradiol. Move the founding virgin cells to agar with 1 μM estradiol [5] (Fig. 2).
Incubate the sealed plate at 30 °C for 1–2 h. After transferring the plate back to the microscope, pick off and, from now onwards, discard the smaller daughter buds from your starting virgin mother cell and record the number of divisions for each arrayed mother cell.
Repeat step 7 throughout the day. The mother cells get enlarged after progressive divisions and eventually become much easier to tell apart from the daughter cells. Their rate of division will also slow down, with some old cells taking up to 4 h to divide. If a cell does not divide after 8 h we considered it dead because we did not see any cell produce a viable daughter after this time. Old cells may try to bud, and the attached bud may bud, but we do not consider these true offspring because the cells did not fully separate. Unless you are using the MEP, you can store plates in the cold room until the next day but it is preferable to let the cells divide as many times as possible during the day.
When all of the cells have died (~10 days), you are now ready to graph their replicative life spans. Each row pertaining to each specific cell’s division number is tallied, and the numbers are plotted as the % cells that are still alive during each round of division. For example, at the start of the assay, 100 % of the cells are alive after 0 divisions have occurred, whereas 0 % of the cells are alive at the amount of divisions that the last remaining mother cell has undergone.
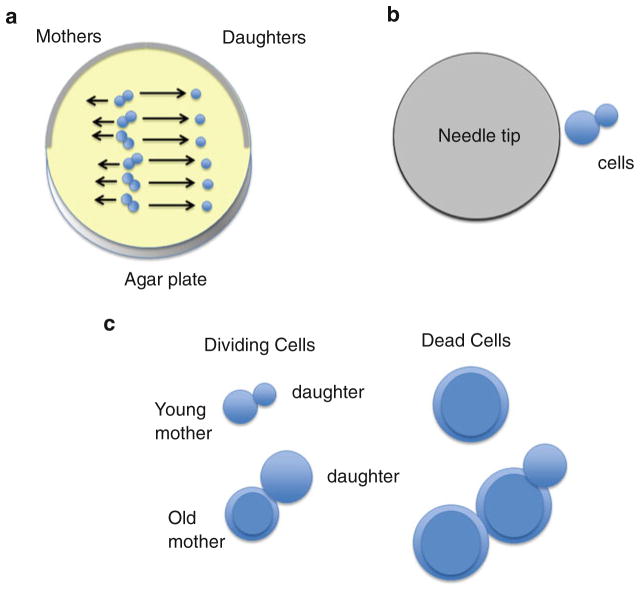
Fig. 1.
The yeast replicative life span (RLS) assay. Individual yeast cells are planted in a single column on agar plates, using a 25 or 50 μm fiber-optic needle. If using the MEP, then the virgin cells are transferred from an island of YPD (yeast extract/Bacto Peptone/glucose) or the test medium to the same medium with 1 μM estradiol (see Fig. 2). Micromanipulation of yeast is typically performed using a 10× objective. After the placement of “virgin” mother cells in an array on the plate, daughter cells that bud off are removed using the needle and are swept away, at least one field of view, from the mother cells so as not to compete with them for nutrients. The amount of divisions each mother cell has undergone during each micromanipulation session is then recorded. (a) Move daughter cells at least one field of view away from the mothers, which can be moved in the opposite direction if daughter colonies grow too large. (b) Relative sizes of the 25 μM needle and young yeast cells. (c) Appearance of young mothers, old mothers, and dead cells. Cells at the end of life are always large, with a thick cell wall, and may have cells permanently attached. Their final morphology depends on the strain and the medium
Fig. 2.
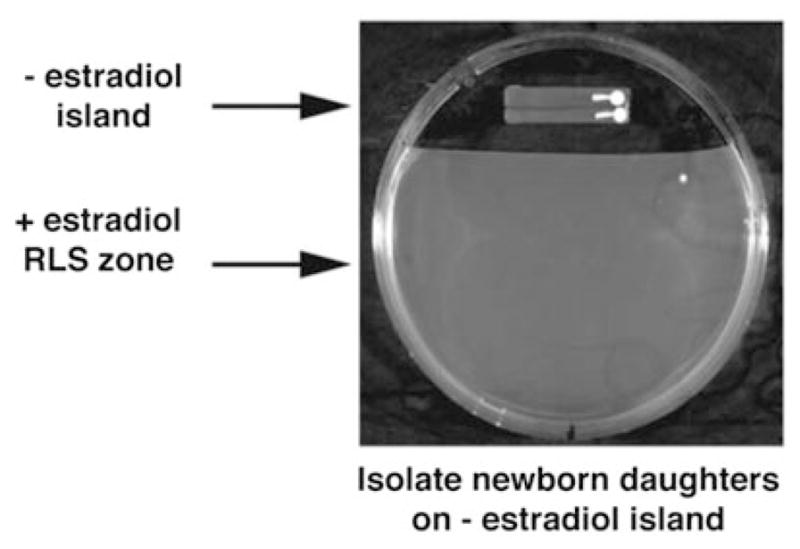
Use of the mother enrichment program (MEP) in RLS assays (from ref. 5). After incubation on YPD (or the test medium) for at least two divisions, virgin cells are transferred to the estradiol-containing portion of the plate for life span analysis. Life spans are then performed, as described in Fig. 1. Image reproduced from ref. 5
3.2 Magnetic Sorting of Old Yeast Cells with and Without MEP
Inoculate 5 mL of 1× YPD with yeast and grow overnight at 30°C while shaking at 200 rpm.
Next morning, resuspend enough cells into 50 mL 1× YPD to reach an optical density at 600 nm (O.D. 600) of approx. 1.0 (2 × 107 haploid cells/mL) later that day. This usually entails diluting ~1 mL of overnight culture into 50 mL 1× YPD. The starting O.D. 600 should be about ~0.15–0.20.
When the O.D.600 reaches ~1.0, spin down cells with a clinical centrifuge at 1,000 × g for 5 min. Wash cells once in 1 mL of sterile 1× PBS. Resuspend the cells in 1 mL 1× PBS and transfer into a microfuge tube.
The cells are now ready to be biotinylated. Remove the EZ-Link Biotin from the freezer and thaw at room temperature. It is VERY sensitive to moisture. When thawed, add 8 mg of sterile EZ-link biotin per 1 × 108 cells. Gently shake the cells for 15 min at room temperature.
Wash the cells 7 times using 1 mL 1× PBS to remove the excess biotin label.
Resuspend cells in 1 mL YPD. Determine the cell density using a hemacytometer. We use a Bright-Line hemacytometer (Hausser Scientific, Horsham, PA). Add 2 × 108 cells to 4 L of freshly made 1× YPD (autoclaved for 20 min). If using the MEP, 17β-estradiol is added to the culture after 2 h growth at 30° at a final concentration of 1 μm. Shake at 30 °C overnight or for up to 120 h if using the MEP. Viability can be assessed during this time if using the MEP, which serves as an indicator of RLS. Samples are taken and pelleted by centrifugation at 800 × g. All but 100 μL of the supernatant is removed, washed once with 1 mL of fresh YPD, and pelleted again. Cells are then resuspended in 500 μL YPD and plated on YPD. Colonies are counted after 3 days incubation at 30 °C. Viability is calculated as CFUs/mL and expressed as percentage of viability compared to CFUs/mL at the 0 or 4 h time points [5] 1,000 × g and thoroughly aspirate off as much medium as possible. Resuspend the cells in 20 mL cold 1× PBS and place on ice.
Wash the magnetic beads twice with 1× PBS. Add 250 μL of the washed magnetic bead slurry per 1 × 108 cells. Incubate on ice with occasional swirling for 2 h in order for the biotinylated cells to bind to the magnetic beads.
The sorting procedure is performed in a 4 °C room. Add equal volumes of the resuspended magnetic bead-coated cells into 16 × 150 mm glass culture tubes and insert into a magnetic tube rack. We use a BioMag tube rack (Polysciences Inc., Warrington, PA) for the magnetic separation. Wait 15–20 min for the magnetic bead-coated older cells to move towards the side of the tube facing the magnet. Remove young cells that are still in the solution gently with a 10 mL pipette and save the solution. Add the same volume of cold 1× YPD into each tube so that the meniscus reaches the top of the magnet and wait 15–20 min again for magnetic separation to occur.
Repeat step 6, using cold 1× YPD each time and vortexing gently to resuspend the bead-coated cells. Repeat this step eight times. Check the yield using a hemacytometer. The recovery is typically 50 % of the starting amount of cells.
Count bud scars by first adding 20 μL of cells into 100 μL 1× PBS. Add a few grains of calcofluor fluorescence brightener and incubate for 5 min at room temperature. Add 1 mL 1× PBS and centrifuge at 1,000 × g in a microfuge for 20 s. Wash cells once in 1 mL 1× PBS, spin down again, and resuspend in 100 μL of 1× PBS. With a single sorting most cells will have 4–9 bud scars.
Place approximately 10 μL of cells onto a glass slide and cover with a coverslip. Observe the stained bud scars using UV fluorescence microscopy. If your cell sorting has been successful, your old population of cells should contain on average ~5–12 more bud scars per cell than the young population of cells (Fig. 3).
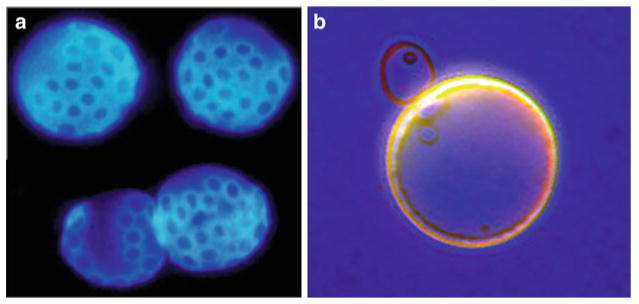
Fig. 3.
Distinguishing old yeast cells from young. (a) Old yeast cells that have been sorted using magnetic sorting and stained with calcofluor dye. Each blue stained ring is a “bud scar,” a deposit of chitin that remains on the surface of the cell wall at each site of division (photo credit James Claus). (b) Young and old yeast cells stained with bromophenol blue, showing the larger size and thicker cell wall of the old mother cell
3.3 Isolation of ERCs
Resuspend cells in 500 μl sorbitol solution. Add 25 μl Zymolyase and 50 μl of 0.28 M 2-mercaptoethanol (20 μl 2-mercaptoethanol in 980 μl water). Depending on the activity of the Zymolyase, incubate at 30 °C for 15–45 min. You can monitor the efficiency of spheroplasting (i.e., digestion of the cell wall) by adding ~2 μl of 10 % SDS into a 10 μl aliquot of cells. If 85–95 % cells appear lysed when viewed under a microscope, then they are ready for the next step (see Note 2).
Add 80 μl of 10 % SDS, invert several times to mix and incubate for 20 min at 65 °C. The solution should become viscous.
Add 200 μl of 5 M potassium acetate solution. Invert several times and leave on ice at least 30 min. A white precipitate should form.
Centrifuge for 3 min at 16,000–20,000 × g (or maximum speed) in a microcentrifuge for 10 min. Remove supernatant and add 1 mL 100 % ethanol. Mix well by inverting several times and centrifuge at 12,000 rpm (or maximum speed) for 10–20 min.
Resuspend the DNA pellet in 300 μl TE. Add 1 μl of 10 mg/mL of RNase and incubate for 30 min at 37 °C.
Precipitate the DNA by adding 1 mL of ice cold 100 % ethanol and centrifuging at 12,000 rpm (or maximum speed) in a microfuge for 10 min. After removing the ethanol, dry the pellet in a speed vac and resuspend in 50 μl of TE. The resuspended DNA pellet can now be stored frozen at −80 °C.
3.4 Analysis of ERCs
The analysis of ERCs essentially involves performing a Southern blot and probing for ERCs with a radio-labeled rDNA fragment. Begin by casting a large 1 % agarose gel using 1× TBE. Load samples, along with molecular weight markers and electrophorese overnight at 30 V.
Soak the gel for 30 min in 500 mL water containing 25 μl of 10 mg/mL ethidium bromide solution and photograph with a ruler using UV light.
Wash the gel for 30 s with water and then soak the gel for 30 min in 0.25 M HCl, with gentle rocking.
Rinse the gel in water again and soak in 1 L of 0.4 M NaOH for 20 min, along with gentle rock in, to denature the DNA.
Set up a transfer apparatus, using 0.4 M NaOH as the transfer buffer. The transfer apparatus consists of a shallow glass tray that is filled two thirds of the way to the top with transfer buffer. A glass plate is then used to partially cover the top of the tray, leaving a 1–2 in. gap on either side. A rectangular piece of 3M Whatman paper is then cut, wetted in transfer buffer, and placed over the top of the glass plate, with the ends being sufficiently long enough to soak in the transfer buffer through the 1–2 in. gaps. This will act as a wick for the transfer. The wick should also be approximately an inch wider, on either side, than the nylon transfer membrane.
Drain the gel and place carefully on top of the wick, using a glass pipette to gently roll away any air bubbles that may be trapped under the gel.
Cut a piece of nylon transfer membrane, corresponding to the size of the gel. Wet the membrane in transfer buffer and lay on top of the gel, using the pipette to smooth away any air bubbles.
Cut three pieces of Whatman 3M paper corresponding to the same shape as the nylon membrane, only a few millimeters smaller at each edge. Soak the piece in transfer buffer and lay each of them on top of the membrane and gel, again using the glass pipette to carefully smooth away any air bubbles after each piece is laid.
Place a stack of paper towels, 3–4 in. high, on top the gel setup; lay a piece of glass plate over the stack and add a light weight on top to stabilize the plate. Use a bubble level to ensure the plate is perfectly level.
Wrap the outer part of the transfer apparatus, i.e., the exposed wick, with Saran wrap to prevent evaporation of the transfer buffer. The transfer apparatus is now left to transfer for 12 h.
Rinse the membrane with 2× SSC to remove any agarose residue. The wet membrane is then irradiated with UV light (254 nM) at an intensity of 1,200 J/cm2. This corresponds to a setting of 1,200 using a Stratagene UV crosslinker.
Add 10 mL pre-hybridization buffer into a 200 mL capacity cylindrical glass hybridization bottle (Boekel Scientific). Place the transferred membrane into the bottle and incubate for 1 h at 65 °C while rolling.
The radio-labeled probe is prepared as per kit instructions. After the reaction is complete, bring the probe reaction volume up to 50 μl with 1× TE. Prepare a Sephadex G-50 column by spinning for 1 min at 550 × g, in order to compact the bead matrix. Pipette the 50 μl probe reaction volume into the prepared Sephadex G-50 column and spin for 2 min at 550 × g.
Boil the radio-labeled probe DNA for 5 min and carefully add into the bottle containing your membrane and pre-hybridization solution. Incubate overnight at 65 °C.
The membrane is washed with the wash buffers until counts are reduced to only severalfold over background. The exact washing times must be empirically determined by the user. The bound probe can now be analyzed using either a Phosphorimager cassette or X-ray film.
3.5 Recombination Analysis
Cells are first inoculated in the appropriate media, i.e., containing 2 % glucose for controls and either 0.1 or 0.5 % glucose for calorie-restricted cells, and preincubated overnight at 30 °C while shaking (see Note 3). The following day, cells are reinoculated into fresh media at an O.D. 600 of ~0.15–0.20 and allowed to grow until log phase has been reached (O.D. 600 of 0.8–1.0).
Prepare serial dilutions of cells in 1× PBS until a final concentration of 3–4 × 103 cells/mL is reached. Pipette 500 μl of this final dilution onto large plates and spread evenly using sterile glass beads along with a gentle shaking motion.
After the plates are dry, incubate at 30 °C until colonies are large enough to be easily counted by eye. This usually takes about 2 days of growth at 30 °C. Plates are then transferred into 4 °C for at least 2–3 days or until red pigmentation becomes evident, following a marker loss event.
Half-sectored colonies are counted by eye on each plate. Colonies that are completely red are subtracted from the total colony number on each plate. A half-sectored colony represents a marker loss event that has occurred during the first cell division. The wild-type W303AR strain, when grown in media containing 2 % glucose, has a recombination frequency of approximately 1 × 10−3, whereas deleting SIR2 elevates the frequency of recombination approximately tenfold, to approximately 1 × 10−2.
Acknowledgments
This work was supported by grants from the NIA/NIH, The Ellison Medical Foundation, and The Glenn Foundation for Medical Research.
Footnotes
- Since the procedure takes approximately 10–14 days to complete, it is imperative that the agar plates do not overly dehydrate. Plates should always be carefully wrapped in Saran wrap or Parafilm when in the incubator or cold room. Plates should also be poured with more agar than usual to take into account dehydration and shrinkage of the agar. We usually pour the plates with the agar level reaching two thirds to three quarters of the way to the top of the plate.
- The technique takes practice to manipulate cells and to differentiate the old mother from daughter cells, especially in the first two days when there is little size difference. The researcher should not let the yeast divide more than once when starting out. In this case, two researchers may work as a team to check on the cells every hour. The more experienced researcher can also help the novice distinguish mother from daughter.
- If the agar eventually shrinks to a point at which it is difficult for the dissection needle to reach the surface, part of the plate’s edge may be then carefully cut away using a hot scalpel to allow for more freedom of motion. However, it is best to reserve this until absolutely necessary since the larger opening will now permit faster dehydration to occur. Also, when cutting the plate, it should be tilted upwards to prevent any toxic fumes from the molten plastic from coming into contact with the cells.
- Plates should be poured a day before using so that they are given ample time to evaporate away excess moisture; otherwise dissection becomes difficult. If you are testing the effect of a labile compound (e.g., resveratrol), however, you should make the compound freshly and use the plates within 24 h and complete the experiment as fast as possible, allowing cells to grow for 10–15 h per day. We took a microscope home so we could dissect late at night and first thing in the morning. Plates can be air-dried upside down in an incubator or a laminar low hood. If the plates change color (e.g., resveratrol plates turn brown), this is a sign that the compound has been compromised.
- With the plates being continually opened and closed during this time period, it is important to avoid contamination. If a small fungal or bacterial growth occurs far from the plated yeast, it can be successfully removed by cutting away the contaminated section with a sterile scalpel blade. Another troublesome source of contamination may be the presence of fruit flies which given the chance, will walk all over the surface of the plate, usually ruining the experiment.
- When a cell is picked up on the needle, the plates should be moved gently since a sudden jarring motion may cause a cell to drop off.
- Old mother cells are especially fragile and must not be overly “bashed” with the tip of the dissection needle to dislodge recalcitrant daughter cells. At this point they have a higher tendency to “pop” under pressure. A good technique is to move the needle tip close to the daughters and allow them to move gently to the tip due to surface tension. Never lift a cell off the agar unless absolutely necessary and then only for 20 s or less, to avoid the cells drying out.
- Obviously, mother cells should not be allowed to overgrow since it then becomes next to impossible to keep score, but they should also not be dissected too often since we have found that this tends to shorten their life span, possibly due to mechanical damage. Two to three divisions between each dissection session for W303 are about right.
- Try to dissect the cells every day. Keeping the plates in the cold room for an entire day is acceptable, but longer periods, i.e., over an entire weekend, should be avoided because life span can be reduced. The rate at which the cells are dividing will determine how many times they can be dissected during the day.
- Someone just beginning should limit themselves to no more than two columns of cells to dissect at once, i.e., one experiment and one control. Later on, as skill level and rapidity increases, 3–9 columns of cells may be attempted. We would discourage placing more than three columns of cells onto one plate, since this increases nutrient depletion and allows the plate to dehydrate faster since each individual plate is kept open longer during dissections.
- For life span dissections, we have found it best not to autoclave the 2× YEP medium but to instead filter though a 0.22 μm filter prior to use. We also supplement the medium with additional amino acids, corresponding to the auxotrophies that are unique to a strain. If a compound is added to the medium, be aware that it could be sensitive to light or oxygen. Such is the case for many polyphenols, e.g., resveratrol.
- It is best that the cells be dragged across the surface of the agar with the needle, rather than being lifted off of the surface. This ensures that the cells are continuously in contact with the media.
- The more dissections you can perform per day, especially during the first week, the better results you will obtain, i.e., longer overall life spans.
- We use Petri plates that have been removed from the plastic wrapper and left out on the bench overnight, with the covers closed. This allows any potentially volatile plasticizers to evaporate from the plates.
- When the yeast cell wall is digested, the spheroplasts will have a “ghostly” or clear outline when viewed under a light microscope. In contrast, yeast with intact cell walls will appear to be much more refractive.
- Large plates for the recombination analysis should contain 2 % glucose, regardless of the starting glucose concentration during liquid culture; otherwise the red coloration will not develop.
- It helps to store the plates for several days at 4 °C after colonies arise. Over time, the red coloration becomes more intense, allowing for easier detection of half-sectored colonies.
- The omission of histidine from the large plates allows for a more intense red coloration to develop, which allows for easier detection of sectored colonies.
References
- 1.Longo VD, Shadel GS, Kaeberlein M, Kennedy B. Replicative and chronological aging in Saccharomyces cerevisiae. Cell Metab. 2012;16:18–31. doi: 10.1016/j.cmet.2012.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Medvedik O, Lamming DW, Kim KD, Sinclair DA. MSN2 and MSN4 link calorie restriction and TOR to sirtuin-mediated lifespan extension in Saccharomyces cerevisiae. PLoS Biol. 2007;5:e261. doi: 10.1371/journal.pbio.0050261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fabrizio P, Longo VD. The chronological life span of Saccharomyces cerevisiae. Methods Mol Biol. 2007;371:89–95. doi: 10.1007/978-1-59745-361-5_8. [DOI] [PubMed] [Google Scholar]
- 4.Barton AA. Some aspects of cell division in Saccharomyces cerevisiae. J Gen Microbiol. 1950;4:84–86. doi: 10.1099/00221287-4-1-84. [DOI] [PubMed] [Google Scholar]
- 5.Lindstrom DL, Gottschling DE. The mother enrichment program: a genetic system for facile replicative life span analysis in Saccharomyces cerevisiae. Genetics. 2009;183:413–422. doi: 10.1534/genetics.109.106229. [DOI] [PMC free article] [PubMed] [Google Scholar]