Pancreatic ductal adenocarcinoma (PDAC) is typically diagnosed after the disease has metastasized; it is among the most lethal forms of cancer. We recently described aberrant expression of a protein, ORF1p, encoded by Long INterspersed Element (LINE-1) retrotransposon, in PDAC 1. To test whether LINE-1 expression leads to somatic insertions of this mobile DNA, we used a targeted method to sequence LINE-1 insertion sites in matched PDAC and normal samples. We found evidence of 465 somatic LINE-1 insertions in 20 PDAC genomes absent from corresponding normal samples. In cases where matched normal tissue, primary PDAC, and metastatic disease sites were available, insertions were found in primary and metastatic in differing proportions. Two adenocarcinomas secondarily involving the pancreas but originating in the stomach and duodenum acquired insertions with a similar discordance between primary and metastatic sites. Together, our findings show that LINE-1 contributes to the genetic evolution of PDAC and suggest that somatic insertions are acquired discontinuously in gastrointestinal neoplasms.
Pancreatic ductal adenocarcinoma (PDAC) affects about 270,000 people worldwide each year and is the fourth most common cause of cancer deaths in the United States. One and five year survival rates are 25% and 6%, respectively; most cases are locally advanced or metastatic when the disease is recognized clinically. Somatically acquired mutations are central to the development of PDAC. A typical case acquires 50 mutations in protein coding sequences. These include a few causative (driver) mutations, like KRASG12D or KRASG12V, and many inconsequential (passenger) mutations 2,3. Most mutations occur while the neoplasm is localized to the pancreas, before metastasis 4.
Acquired structural alterations to PDAC genomes are less well characterized. In this study, we focused on Long INterspersed element (LINE-1, L1) retrotransposon insertions. L1 is a mobile genetic sequence comprising about 17% of our genome. Most copies are invariant and not competent to transpose. However, some L1Hs or L1PA1 sequences are a significant source of inherited genetic variation and encode endonuclease and reverse transcriptase activities that promote their propagation in genomes 5, 6, 7. We recently described expression of a LINE-1 encoded protein, open reading frame 1 protein (ORF1p), in 89% of PDACs and 27% of pancreatic intraepithelial neoplasias (PanINs) 1. In the current study, we tested whether somatic retrotransposition events would be evident in PDAC genomes.
Matched normal and tumor samples were collected postmortem. We extracted gDNA and used one-sided PCR to selectively amplify segments downstream of L1Hs insertions for sequencing, an approach we term Transposon Insertion Profiling by sequencing (TIP-seq) (Fig. 1a). Similar approaches have been previously reported 8,9. Twenty-two individuals donated samples used in this study (Supplementary Table 1). All were initially diagnosed with PDAC; this was confirmed in 20 cases. Two were reclassified as adenocarcinomas likely originating in gastric or duodenal sites and secondarily involving the pancreas.
Figure 1.
Experimental approach. (a) Normal (N); primary (P) tumor samples; and metastatic (M) samples were collected. Sections were evaluated histologically to assess viability and composition prior to DNA extraction. (b) Transposon Insertion Profiling (TIP)-seq. A full-length (6kb) LINE-1 is diagrammed (blue). TSD, target site duplication; UTR, untranslated region; ORF, open reading frame. PCR amplicons initiate from an oligonucleotide complementary to the 3' UTR and extend to a restriction enzyme site. These are sheared and sequenced. Alignment to the reference genome shows read pileups (peaks, lower left). There are no alignments 5′ of the insertion; reads align to the plus (blue) and minus (orange) strands 3' of the insertion. Peaks found in neoplastic samples and not matched normal samples indicate potential somatic insertions. Reads spanning the polyadenylated tail and unique DNA indicate the point of insertion. Insertions are validated by spanning PCR (lower right). The gel shows four lanes with a marker in the leftmost; amplicons from normal (N), primary (P) tumor, and metastasis (M) are shown. The “empty” or pre-insertion site is amplified in all samples, whereas the insertion is recovered from primary and metastatic disease sites. (c) Structure of LINE-1 insertions. LINE-1 is frequently 5' truncated (upper left). Insertions can have inverted 5′ ends (upper right). The diagram shows a sequence breakpoint (//); the 3′ LINE-1 sequence is in the regular orientation (rightward arrow), and 5′ LINE-1 sequence is in the opposing orientation (leftward arrow). Finally, 3′ transduction events incorporate DNA downstream of the parental LINE-1 at the new insertion site.
Genomic DNA was digested with restriction enzymes, ligated to vectorette oligonucleotides, and amplified using a primer specific for the 3′ UTR of L1Hs repeats (Fig. 1b). Amplicons were sheared and sequenced to an average read count of 43 million and sequences aligned to the reference genome assembly. We identified most L1Hs in the reference genome in all samples (averaging 400 elements in the top 500 peaks, range 390 – 413; Supplementary Table 2). As expected, many known inherited polymorphisms were also seen.
After subtracting the known germline variants, significant read pile-ups (peaks) from normal tissue TIP-seq indicated novel constitutional L1Hs insertions. We examined these locations for potential roles in cancer risk. No exonic LINE-1 could be sequence verified. No insertions were seen in the familial pancreatic cancer genes, breast cancer 2, early onset (BRCA2) 10; partner and localizer of BRCA2 (PALB2) 11; protease, Ser1 (PRSS1) 12; or cyclin-dependent kinase inhibitor 2 (CDKN2) 13.
To identify somatically acquired insertions, we compared TIP-seq profiles of matched tumor and normal gDNA samples. We found a total of 268 cancer-specific insertions in 18 primary PDAC cases (Fig. 2a, Supplementary Tables 3 and 4). There was significant variability between cases in the numbers of somatic insertions identified (range 0–65; average 15 insertions/case).
Figure 2.
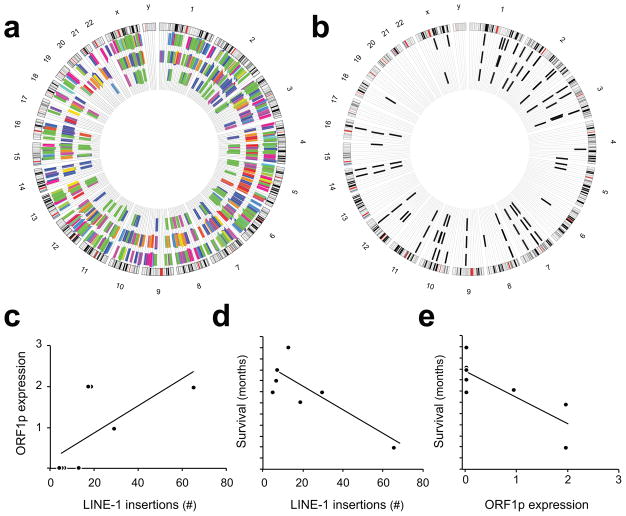
Somatic LINE-1 insertions in PDAC. (a) Genomic positions are arrayed clockwise in a Circos plot. Positions of all somatic LINE-1 insertion sites are marked with radial hashes color- coded by case. Insertions found in the primary tumor are shown in the outermost wheel; insertions found in the metastasis are found in the middle wheel. Those shared by the two sites of disease are marked innermost. (b) Image b. shows insertions from a single case. Twenty-nine (29) LINE-1 insertions were identified in the primary tumor, and 34 in the metastatic sample. Twenty-three (23) were shared between the two sites of disease. (c) LINE-1 ORF1p protein expression levels as scored by immunohistochemistry (0-3) corresponded to LINE-1 insertion events detected in primary PDAC tumors though not to statistical significance (p=0.07, ANOVA). (d) Survival after diagnosis was inversely related to somatically acquired LINE-1 insertions found in the primary tumor (p=0.025). Each hash on the y-axis indicates one month. (e) Survival after diagnosis was inversely related to LINE-1 ORF1p immunoreactivity (p=0.03).
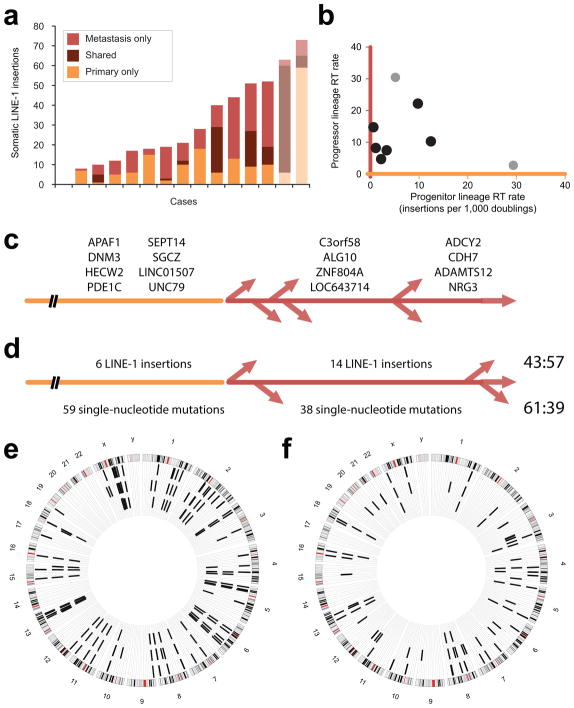
To identify somatic insertions at sites of metastatic PDAC, we assayed 15 metastases in 15 cases and found 242 insertion candidates not present in corresponding normal tissue samples (range 0–42, average 16 insertions/site). In 13 cases with both primary and metastatic cancers available, we found 45 insertions shared between the two sites of disease. One case harbored 23 of these shared insertions (Fig. 2b, Fig. 3a, Supplementary Tables 3 and 4).
Figure 3.
Retrotransposition (RT) events. (a) Somatic LINE-1 insertions. Stacked bars show insertions found in only the primary tumor (orange, bottom); those shared by the primary and metastatic samples (middle); and those only in the metastasis (red). Confirmed PDAC cases are shown in colors indicated by the legend; two other gastrointestinal adenocarcinomas are shown in lighter hues. (b) RT rate was calculated for each case during two phases of clonal evolution. The progenitor lineage rate (orange, x-axis) is proportional to the number of LINE-1 insertions acquired within a cellular lineage in the primary tumor antedating its seeding a metastatic site. These are shared by the primary and metastatic sites. The progressor lineage rate (red, y-axis) reflects insertions found only in the metastatic sample. PDAC cases are indicated with black dots; other adenocarcinomas are in grey. (c) Somatically acquired LINE-1 in PDAC. Twenty insertions were detected in the primary and metastatic samples for this case and not in normal DNA. Gene names are listed for 8 intronic insertions. These occurred in the progenitor lineage of the primary tumor (leftmost, orange); they were PCR amplified from all metastases. Eight insertions were found by TIP-seq at a metastatic site and not in the primary tumor; nearest gene names are given for intergenic insertions. Four were present in two additional metastases and occurred in a progressor clone of the primary tumor (middle, red). The remaining four were unique to the metastasis profiled by TIP-seq (rightmost) and occurred later in the primary tumor or at that metastatic site. (d) Somatically acquired LINE-1 and single-nucleotide mutations in PDAC. Numbers of events occurring in the progenitor (orange) and progressor (red) lineages are shown; ratios are on the right. (e) LINE-1 insertions in a gastric adenocarcinoma. Sixty insertions were detected in the primary tumor; most (54) were shared with the metastatic site. (f) LINE-1 insertions in a duodenal adenocarcinoma. Twenty-seven insertions were identified in the primary, and 42 in the metastasis. Eighteen were shared between the two sites.
LINE-1 ORF1p expression level scored by immunohistochemistry correlated with numbers of somatically acquired LINE-1 insertions albeit not to statistical significance (p=0.07). Survival after diagnosis was inversely correlated with somatically acquired LINE-1 insertions found in the primary tumor and with LINE-1 ORF1p immunoreactivity (p=0.025 and p=0.03, respectively)(Fig. 2c–e).
Two cases of adenocarcinoma that infiltrated the pancreas but were later recognized to have originated in nearby stomach and small bowel were also included. We found evidence for 51 and 63 somatic LINE-1 insertions in these PDAC mimics.
TIP-seq reads cover only the 3′ end of L1Hs insertions. To validate and sequence the entirety of somatically acquired insertions, we designed PCR primers flanking predicted insertion sites. We Sanger sequenced amplicons larger than those corresponding to the preinsertion or ‘empty’ site present in tumor and absent from normal. In total, 117 loci were considered; 92 were in a region allowing primer design and had short-read sequences consistent with a LINE-1 insertion by manual review. Of these, 81 (88%) could be PCR validated; for 75 (81%), we recovered Sanger reads of the insertion. Validation rates were similar regardless of tissue of origin.
Structures of sequenced insertions are provided (Supplementary Table 5). These show expected hallmarks of somatic retrotransposition (Fig. 1c) 14. All insertions are 5′ truncated with an average size approximating 1 kilobase; the average target site duplication (TSD) is 11bp. Inverted or reversed orientation of the 5′ L1Hs segment was commonly seen 15 (Supplementary Figure 1, Supplementary Table 6). Transduction events of sufficient size to identify the templating element 16 were recovered in two instances. Both were found in the same PDAC case and reflect activity of a known active element on 14q23.1 17, which copied a 324bp segment 3′ of its insertion site to positions on chromosomes 9 and 16 during development of the primary tumor (Supplementary Table 5).
We next used somatic LINE-1 insertion numbers to infer rates of retrotransposition during the development and dissemination of PDAC. For this, we applied a model described for this disease relating mutation rate to the rate of cell division, numbers of acquired neutral mutations (N), and time 4. We expect insertions shared between primary and metastatic samples of a case arose in the progenitor lineage of the primary tumor. These likely occurred after a genetic growth advantage began the primary tumor, but before a cell existed in the primary with all of the insertions that would be carried to the metastatic site. Six cases, all of which had somatic LINE-1 insertions shared in primary and metastatic tissues, were considered. In aggregate, 259 independent insertions were found in these cases, with 45 LINE-1 found in both the primary tumor and the sampled metastatic site. Numbers of shared insertions (N) ranged from 1 to 23 (average, 7.5), reflecting retrotransposition rates in progenitor lineages ranging from 0.5 to 12.4 insertions per 1,000 cell doublings (average, 4.0).
In contrast, we expected the 82 insertions identified at metastatic sites and not the corresponding primary tumors occurred later. To test this in one case, we attempted to amplify 8 of these insertions in 6 additional metastatic lesions. Four insertions (4 of 8) were uniquely amplified in the single metastasis (Fig. 3c). Four were found in two additional metastases (totaling 3 of 7 lesions). The latter were acquired in the primary tumor in a subclone that would seed multiple metastases, though as a comparably late event (i.e., subsequent to the birth of the progenitor clone). None were detected in the primary tumor. Overall, these findings validate using these insertions to estimate retrotransposition rates in what is termed the progressor clone 4. This ranges from 4.6 to 30 insertions per 1,000 cell doublings (average, 12.7).
Interestingly, within each PDAC case, we found no concordance between retrotransposition rates for these two phases of disease (Fig. 3b). This is also true of primary and metastatic diseases originating in the tubular gastrointestinal tract if a similar pace of disease spread is assumed. The gastric adenocarcinoma case had many shared insertions (Fig. 3e) and disproportionately acquired these in the progenitor lineage of the primary tumor; its metastatic site had little evidence of continued retrotransposition. Conversely, the duodenal tumor accrued few somatic LINE-1 insertions in the progenitor lineage but produced a progressor clone that acquired many new insertions (Fig. 3f). These data suggest to us that retrotransposition occurs discontinuously during tumor evolution.
To test for concordance between acquired single nucleotide changes and LINE-1 insertions, we compared proportions of shared (i.e., primary and metastasis) and ‘metastasis- only’ alterations in a PDAC case. Single-nucleotide mutations were detected by exome sequencing. For single-nucleotide mutations, we found a ratio of shared to ‘metastasis-only’ mutations of approximately 60:40 (61:39). This is similar to ratios previously reported 4. The ratio is essentially reversed for LINE-1 insertions in the same case (43:57)(Fig. 3d). The discrepancy in the proportions of single-nucleotide mutations and LINE-1 insertions found is additional evidence of asynchrony in the rates of these two mutational mechanisms.
Somatic LINE-1 insertions accumulate throughout PDAC genomes. Although loosely aggregated on individual chromosomes in some cases, hotspots for integration were not readily appreciated when all sites were considered (Fig. 4a). No exonic insertions were found. Intronic insertions were seen in 202 genes. Gene set enrichment analysis do not clearly implicate pathways required for tumor development. Together, these data indicate that LINE-1 insertions are not commonly canonical ‘driver’ mutations, and that any one insertion is likely dispensable for tumorigenesis.
Figure 4.
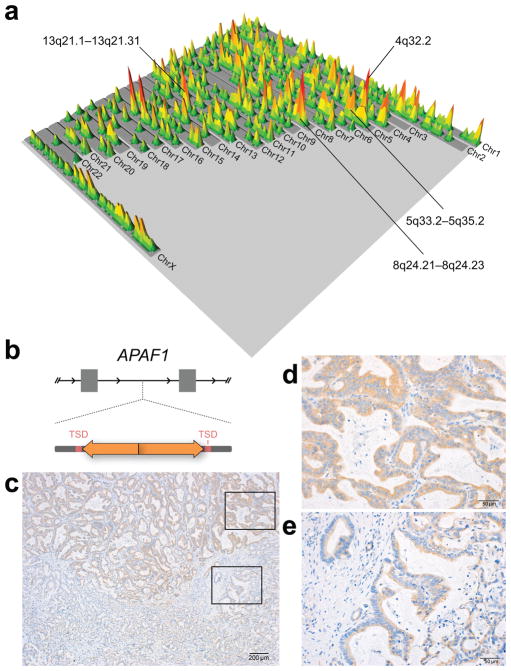
Effects of somatic LINE-1 insertions in PDAC. (a) A landscape plot showing the human genome. Insertion sites are indicated as raised peaks; areas accumulating more independent insertions are reflected by peak height and color. The pattern is distributed; highly recurrently mutated loci do not stand out. Areas accruing the most LINE-1 insertions are marked. Somatic LINE-1 insertions occurred over a 17MB region on 4q32.2 in 6 cases. The nearest gene is follistatin-like 5 isoform (FSTL5), a putative tumor suppressor gene infrequently mutated in pancreatic cancer. None of the insertions were within the FSTL5 transcription unit. A 19MB interval on 5q33.2-5q35.2 showed somatic L1 insertions in 11 cases. An 11MB interval spanning 8q24.21-8q24.23 showed insertions in 6 cases with some cases showing multiple independent insertions. Seven cases had evidence for insertions over a 6MB interval on 13q21.1-13q21.31. Insertions were broadly distributed within each of these regions with no single gene locus recurrently involved. Peaks at 16p11.2 were precisely superimposed but in satellite DNA, could not be PCR validated, and are likely artifact. (b) The schematic illustrates an intronic LINE-1 in apoptotic protease activating factor 1 (APAF1). The L1 is a 443bp insert with an inverted and truncated 5′ end; it is flanked by 16bp TSDs. (c) An immunohistochemistry slide (20x) shows two regions of the primary tumor with differential expression of APAF1 protein. Immunoreactivity is indicated in brown; the counterstain is blue. The tumor shows higher APAF1 expression in one sector (upper right inset, (d) 100x magnification) and reduced immunoreactivity in a less differentiated region (lower right inset, (e) ). Other tumors showed uniform APAF1 expression.
To explore the possibility that insertions influence gene expression, we prioritized insertions in close proximity of exons or potential regulatory sites as indicated by ENCODE project annotations. One example is a somatic LINE-1 detected in an intron of apoptotic protease activating factor 1 (APAF1); it is a 443bp insertion with an inverted and truncated 5′ end. The L1 is located in a small intron (1.6kb) of the APAF1 gene adjacent to a DNAse hypersensitive region that can be bound by transcription factors. We used PCR to detect the insertion in the primary tumor and in all seven metastatic disease sites sampled in the same case. Immunohistochemistry to evaluate APAF1 protein showed a variegated pattern of expression in the primary tumor sample in this case (Fig. 4b–e), while other tumors displayed uniform APAF1 immunoreactivity. Repeated attempts to amplify the insertion from microdissected sectors of the tissue were unsuccessful. Our findings are consistent with an acquired event altering APAF-1 expression in this case. However, they do not exclude other underlying mechanisms nor imply that this is a common feature in pancreatic cancers. To date, targeted studies have demonstrated LINE-1 retrotransposition in lung 8, colon 18, and hepatocellular 19 carcinoma, and whole genome sequencing from The Cancer Genome Atlas (TCGA) 20,21 and the International Cancer Genome Consortium (ICGC) 22 has shown more broadly that human malignancies of various origins are characterized by different levels of L1 activity. Our work adds pancreatic cancer to this picture. We show that PDAC is highly active for retrotransposition, and moreover, that single lineages acquire multiple LINE-1 integrations over their evolution.
The variability in numbers of acquired insertions between PDAC cases is similar to that previously described for other types of tumors. We have ascribed this to differences in each person’s inherited complement of active LINE-1 or to tumor-specific idiosyncrasies of retrotransposon control, but we expected new insertions to be acquired steadily during the course of disease. Our discovery of discordant overall rates of retrotransposition in matched progenitor and progressor subclones challenges this assumption. Similarly, where single-nucleotide mutation data are available, we find a lack of concordance between proportions of single-nucleotide mutations and LINE-1 insertions occurring in different phases of disease. Our findings suggest that LINE-1 insertions in gastrointestinal tract cancers occur discontinuously. This is consistent with variable activity of individual LINE-1 elements previously reported in prostate cancer and lung cancer 22. Cellular mechanisms restricting somatic retrotransposition are not well understood, and perhaps the discontinuity we see in the timing of insertions is owed to discrete breaches in host defenses. Tumors with many insertions may have acquired these in series of breaches or even in isolated catastrophic episodes that can be likened to chromothripsis in cancer genomes 23.
Although many LINE-1 insertions can occur in metastatic progenitor lineages, they appear dispensable for PDAC. Some cases had no evidence of retrotransposition, and we have no evidence of recurrent insertion sites or exonic insertions. While we cannot definitively assign function to acquired LINE-1 insertions, our findings leave open the possibility that somatically acquired LINE-1 influence local gene regulation and contribute to cancer cell phenotypes. Emerging technologies such as single cell LINE-1 insertion site sequencing 24 and phenotyping should yield unprecedented opportunities to understand the timing of LINE-1 insertion events and how they contribute to genetic and phenotypic heterogeneity in cancer.
Methods
Tissue samples
All participants were consented as part of the Gastrointestinal Cancer Rapid Medical Donation Program (GICRMDP) at the Johns Hopkins Hospital. The study was approved by the Johns Hopkins School of Medicine Institutional Review Board (IRB); protocol numbers are J03103 and NA_00036610. Each specimen was examined as a frozen section by a trained pathologist to ensure that (i.) no cancer cells were identified in grossly normal tissue fragments, and (ii.) that viable cancer cells were present in the cancer specimens. K-ras mutation and immunohistochemistry (CDX2, CK7, CK20) were used to identify two of the 22 cases that are more appropriately classified as adenocarcinomas originating in the nearby tubular gastrointestinal tract and secondarily involving the pancreas. Age at diagnosis for confirmed PDAC patients ranged from 36–85 years; average age was 65.
We incubated 30–40 mg of frozen tissue in a lysis buffer with RNAse A (Life Technologies; Grand Island, NY) prior to an overnight proteinase K (NEB; Ipswich, MA) digestion. We extracted high molecular weight genomic DNA using phenol-chloroform and precipitated the DNA with ethanol. Quality was verified by spectrophotometry and agarose gel electrophoresis.
TIP-seq
Transposon Insertion Profiling (TIP-seq) uses a ligation mediated PCR to amplify genomic DNA 3′ of LINE-1 insertions 1. For each sample, we split 10μg of genomic DNA into six parallel restrictions enzymes digests: AseI, BspHI, BstYI, HindIII, NcoI, PstI (NEB). We ligated vectorette oligonucleotide adapters designed with sticky-ends to match each restriction enzyme cut site to the digested DNA fragments. Touchdown PCR was run using ExTaq HS polymerase (Takara Bio; Shiga, Japan). An L1PA1-specific primer (5′-AGATATACCTAATGCTAGATGACACA-3′) initiates extension through the LINE-1 3′ UTR and polyA tail into adjacent DNA. The 3′ most ACA trinucleotide is selective for the youngest, active L1 subfamily (L1PA1).
We combined six PCR reactions for each sample and purified the DNA for sequencing library preparation. Using a Covaris E210 (Covaris; Woburn, MA) we sheared amplicons to an average size of 300bp. We then performed end-repair, dA-tailing, and index-specific adapter ligation steps according to Illumina’s TruSeq DNA Sample Prep v2 kit protocol (Illumina; San Diego, CA). Using 2% Size-Select E-gels (Life Technologies; Carlsbad, CA) we size-selected our adapter-ligated DNA at approximately 450bp before performing a final PCR amplification. After purifying the PCR amplified libraries we submitted them for quality control and Illumina HiSeq 100bp paired-end sequencing at HudsonAlpha Institute for Biotechnology (HudsonAlpha; Huntsville, AL). Similar methods for LINE-1 insertion site amplification for next generation sequencing have been described previously 2, 3.
Peak Calling and Filtering
We aligned demultiplexed sequencing reads to the human reference genome assembly (March 2006 human reference sequence NCBI Build 36.1, hg18) using BOWTIE 4 and called peaks using CisGenome 5. Consensus sequences of the insertion sites were created by multi-sequence alignment of initially unaligned reads which when trimmed to 35bp became alignable to a peak interval. We used Galaxy to identify differences in putative insertion sites across samples. We required peaks to rank in the top 1000; insertions predicted by lower ranking peaks were less commonly validated in subsequent PCRs. Known L1Hs (including polymorphic L1Hs) and L1PA2 insertions were removed. Consensus sequences were required to have an ‘A’ or ‘T’ homopolymer and sequence unambiguously alignable to the reference assembly.
PCR Validations
To validate candidate somatic insertions we manually reviewed insertion site consensus sequences and their alignment to the reference assembly by BLAT. We designed primers around the insertion site coordinate using Primer3. Long-range spanning PCR was performed using LATaq (Takara Bio; Shiga, Japan) on both normal and tumor DNA and amplicons run on agarose gels. When we observed an amplicon larger than the ‘empty’ allele present only in tumor DNA we excised the band and extracted the DNA for Sanger sequencing. LINE-1 length, target site duplications (TSD), and 5′ inverted structures are reported from the Sanger reads.
Retrotransposition rate estimates
We used a previously described model for clonal evolution in PDAC 6. This model assumes a constant mutation rate summarized by the following relationship :
where r is the mutation rate per generation, Tgen is the cellular doubling time, which we took as 2.3 days 7, T1 is the time interval for the development of the metastatic progenitor cell in the primary tumor, and N1 is the number of LINE-1 insertions detected that were shared by the primary tumor and the metastatic sample. An analogous relationship was used to estimate retrotransposition rates in the progressor subclone, with T2 reflecting time for continued retrotransposition, and N2 signifying LINE-1 insertions detected uniquely in the metastatic sample. T1 and T2 were were taken as 11.7 and 6.8 years, respectively 6.
Gene expression and gene set enrichment analysis
We overlapped peak intervals corresponding to somatic LINE-1 insertions +/− 1 kb with RefSeq transcription units using Galaxy. This identified 248 unique annotations, 202 of which were Entrez genes. We performed gene set enrichment studies using the Broad Institute website 8, 9.
Immunohistochemistry
APAF1 Slides were deparaffinized and heated at 98°C for 20 minutes citrate buffer (pH 6) for antigen retrieval. Endogenous peroxidase was blocked using 3% hydrogen peroxide for 5 minutes at room temperature. A rabbit polyclonal antibody (Cat # HPA031373, Sigma-Aldrich, St. Louis, MO) was used as the primary antibody at 1:60 for 30 minutes. The reaction was developed with a biotin-free Bond polymer detection system on a Bond-Leica autostainer (Leica Microsystems, Bannockburn, IL); the 3,3-Diaminobenzidine chromogen-substrate was used for detection. Slides were counterstained with hematoxylin, dehydrated, and coverslipped. LINE-1 ORF1p Slides were deparaffinized and heated at 98°C for 20 minutes citrate buffer (pH 6) for antigen retrieval. After blocking, we incubated with a 1:1000 dilution of a monoclonal LINE-1 ORF1p antibody targeting amino acids 35 to 44 of LINE-1 ORF1p 10 overnight in a 4°C humid chamber. Slides were then processed using the Histostain-SP Broad Spectrum kit according to the manufacturer’s instructions (Cat # 95-9643, Life Technologies).
Supplementary Material
Twin priming events in somatic LINE-1 insertions. Different (independent) insertion events are stacked vertically. Each was Sanger sequenced in its entirety to identify the segments shown. The x-axis indicates the length of the insertion. Insertions are drawn to scale and centered on the breakpoint between 5′ to 3′ oriented LINE-1 sequence (i.e., the 3′ LINE-1 end in regular orientation, right facing arrow) and sequence in the reverse orientation (i.e., the 5′ opposing orientation, left facing arrow). The lengths of target site duplications (TSDs) are indicated by red shading at the extreme ends of each schematic.
Acknowledgments
This work was started by funding from the Sol Goldman Pancreatic Cancer Research Center (KHB and NR) and supported also by the Fred and Janet Sanfilippo Fund in the Department of Pathology at the Johns Hopkins University School of Medicine (NR); a Burroughs Wellcome Fund Career Award for Biomedical Scientists Program (KHB); and NIH awards F31CA180682 (AM-M), R01CA163705 (KHB), R01GM103999 (KHB), P50CA62924 (RHH, CI-D), R01CA179991 (CI-D), as well as the NIGMS Center for Systems Biology of Retrotransposition P50GM107632 (KHB and JDB). Computational resources were provided through the National Science Foundation funded MRI-R2 project #DBI-0959894. The authors would like to thank Haig Kazazian, Szilvia Solyom, and Adam Ewing for helpful discussions. This work is dedicated to Dr. Frank Kretzer.
Footnotes
Author Contributions
N. Rodic , R. Hruban, C. Iacobuzio-Donahue, J. Boeke, and K. Burns conceived of the project; N. Rodic, A.M.-M., and C. Iacobuzio-Donahue. obtained tissues and reviewed histology; J. Steranka, A. Moyer, P. Shen, and P. Mita designed and performed molecular biology assays; N. Rodic, R. Sharma, M. Taylor, and N. Barker performed and reviewed immunostains; Z. Kohutek, C. R. Huang, and D. Ahn designed and performed sequence analysis; N. Rodic, J.Steranka, J. Boeke, and K. Burns interpreted data; J. Steranka summarized data for the Supplementary Tables; N. Rodic and K.Burns wrote the manuscript. All authors contributed edits and approved of the final manuscript.
References
- 1.Rodić N, et al. Long Interspersed Element-1 Protein Expression Is a Hallmark of Many Human Cancers. The American journal of pathology. 2014 doi: 10.1016/j.ajpath.2014.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vogelstein B, et al. Cancer genome landscapes. Science. 2013;339:1546–1558. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jones S, et al. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science. 2008;321:1801–1806. doi: 10.1126/science.1164368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yachida S, et al. Distant metastasis occurs late during the genetic evolution of pancreatic cancer. Nature. 2010;467:1114–1117. doi: 10.1038/nature09515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burns KH, Boeke JD. Human transposon tectonics. Cell. 2012;149:740–752. doi: 10.1016/j.cell.2012.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beck CR, Garcia-Perez JL, Badge RM, Moran JV. LINE-1 elements in structural variation and disease. Annu Rev Genomics Hum Genet. 2011;12:187–215. doi: 10.1146/annurev-genom-082509-141802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hancks DC, Kazazian HH., Jr Active human retrotransposons: variation and disease. Curr Opin Genet Dev. 2012;22:191–203. doi: 10.1016/j.gde.2012.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Iskow RC, et al. Natural mutagenesis of human genomes by endogenous retrotransposons. Cell. 2010;141:1253–1261. doi: 10.1016/j.cell.2010.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ewing AD, Kazazian HH., Jr High-throughput sequencing reveals extensive variation in human-specific L1 content in individual human genomes. Genome Res. 2010 doi: 10.1101/gr.106419.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Murphy KM, et al. Evaluation of candidate genes MAP2K4, MADH4, ACVR1B, and BRCA2 in familial pancreatic cancer: deleterious BRCA2 mutations in 17% Cancer Res. 2002;62:3789–3793. [PubMed] [Google Scholar]
- 11.Jones S, et al. Exomic sequencing identifies PALB2 as a pancreatic cancer susceptibility gene. Science. 2009;324:217. doi: 10.1126/science.1171202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Whitcomb DC, et al. Hereditary pancreatitis is caused by a mutation in the cationic trypsinogen gene. Nat Genet. 1996;14:141–145. doi: 10.1038/ng1096-141. [DOI] [PubMed] [Google Scholar]
- 13.Whelan AJ, Bartsch D, Goodfellow PJ. Brief report: a familial syndrome of pancreatic cancer and melanoma with a mutation in the CDKN2 tumor-suppressor gene. N Engl J Med. 1995;333:975–977. doi: 10.1056/NEJM199510123331505. [DOI] [PubMed] [Google Scholar]
- 14.Ostertag EM, Kazazian HH., Jr Biology of mammalian L1 retrotransposons. Annu Rev Genet. 2001;35:501–538. doi: 10.1146/annurev.genet.35.102401.091032. [DOI] [PubMed] [Google Scholar]
- 15.Ostertag EM, Kazazian HH., Jr Twin priming: a proposed mechanism for the creation of inversions in L1 retrotransposition. Genome Res. 2001;11:2059–2065. doi: 10.1101/gr.205701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goodier JL, Ostertag EM, Kazazian HH., Jr Transduction of 3'-flanking sequences is common in L1 retrotransposition. Hum Mol Genet. 2000;9:653–657. doi: 10.1093/hmg/9.4.653. [DOI] [PubMed] [Google Scholar]
- 17.Beck CR, et al. LINE-1 retrotransposition activity in human genomes. Cell. 2010;141:1159–1170. doi: 10.1016/j.cell.2010.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Solyom S, et al. Extensive somatic L1 retrotransposition in colorectal tumors. Genome Res. 2012;22:2328–2338. doi: 10.1101/gr.145235.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shukla R, et al. Endogenous retrotransposition activates oncogenic pathways in hepatocellular carcinoma. Cell. 2013;153:101–111. doi: 10.1016/j.cell.2013.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee E, et al. Landscape of Somatic Retrotransposition in Human Cancers. Science. 2012;337:967–971. doi: 10.1126/science.1222077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Helman E, et al. Somatic retrotransposition in human cancer revealed by whole-genome and exome sequencing. Genome Res. 2014;24:1053–1063. doi: 10.1101/gr.163659.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tubio JM, et al. Mobile DNA in cancer. Extensive transduction of nonrepetitive DNA mediated by L1 retrotransposition in cancer genomes. Science. 2014;345:1251343. doi: 10.1126/science.1251343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stephens PJ, et al. Massive genomic rearrangement acquired in a single catastrophic event during cancer development. Cell. 2011;144:27–40. doi: 10.1016/j.cell.2010.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Evrony GD, et al. Single-neuron sequencing analysis of L1 retrotransposition and somatic mutation in the human brain. Cell. 2012;151:483–496. doi: 10.1016/j.cell.2012.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
Methods References
- 1.Huang CR, et al. Mobile interspersed repeats are major structural variants in the human genome. Cell. 2010;141:1171–1182. doi: 10.1016/j.cell.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Iskow RC, et al. Natural mutagenesis of human genomes by endogenous retrotransposons. Cell. 2010;141:1253–1261. doi: 10.1016/j.cell.2010.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ewing AD, Kazazian HH., Jr High-throughput sequencing reveals extensive variation in human-specific L1 content in individual human genomes. Genome Res. 2010 doi: 10.1101/gr.106419.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ji H, et al. An integrated software system for analyzing ChIP-chip and ChIP-seq data. Nat Biotechnol. 2008;26:1293–1300. doi: 10.1038/nbt.1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yachida S, et al. Distant metastasis occurs late during the genetic evolution of pancreatic cancer. Nature. 2010;467:1114–1117. doi: 10.1038/nature09515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Amikura K, Kobari M, Matsuno S. The time of occurrence of liver metastasis in carcinoma of the pancreas. International journal of pancreatology : official journal of the International Association of Pancreatology. 1995;17:139–146. doi: 10.1007/BF02788531. [DOI] [PubMed] [Google Scholar]
- 8.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mootha VK, et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34:267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 10.Rodić N, et al. Long Interspersed Element-1 Protein Expression Is a Hallmark of Many Human Cancers. The American journal of pathology. 2014 doi: 10.1016/j.ajpath.2014.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Twin priming events in somatic LINE-1 insertions. Different (independent) insertion events are stacked vertically. Each was Sanger sequenced in its entirety to identify the segments shown. The x-axis indicates the length of the insertion. Insertions are drawn to scale and centered on the breakpoint between 5′ to 3′ oriented LINE-1 sequence (i.e., the 3′ LINE-1 end in regular orientation, right facing arrow) and sequence in the reverse orientation (i.e., the 5′ opposing orientation, left facing arrow). The lengths of target site duplications (TSDs) are indicated by red shading at the extreme ends of each schematic.