Abstract
More than a hundred de novo single gene mutations and copy-number variants have been implicated in autism, each occurring in a small subset of cases. Mutant mouse models with syntenic mutations offer research tools to gain an understanding of the role of each gene in modulating biological and behavioral phenotypes relevant to autism. Knockout, knockin and transgenic mice incorporating risk gene mutations detected in autism spectrum disorder and comorbid neurodevelopmental disorders are now widely available. At present, autism spectrum disorder is diagnosed solely by behavioral criteria. We developed a constellation of mouse behavioral assays designed to maximize face validity to the types of social deficits and repetitive behaviors that are central to an autism diagnosis. Mouse behavioral assays for associated symptoms of autism, which include cognitive inflexibility, anxiety, hyperactivity, and unusual reactivity to sensory stimuli, are frequently included in the phenotypic analyses. Over the past 10 years, we and many other laboratories around the world have employed these and additional behavioral tests to phenotype a large number of mutant mouse models of autism. In this review, we highlight mouse models with mutations in genes that have been identified as risk genes for autism, which work through synaptic mechanisms and through the mTOR signaling pathway. Robust, replicated autism-relevant behavioral outcomes in a genetic mouse model lend credence to a causal role for specific gene contributions and downstream biological mechanisms in the etiology of autism.
Keywords: Anxiety-like, autism, cognition, genes, hyperactivity, mice, mutant models, neurodevelopmental, repetitive behavior, sensory reactivity, sociability, social behavior, vocalizations
Autism spectrum disorder (ASD) is a neurodevelopmental syndrome with a prevalence of over 1% of the population (CDC 2014; Elsabbagh et al. 2012; Kim et al. 2011). Diagnosis by the current Diagnostic and Statistical Manual of Mental Disorders (DSM-5) criteria is based on two categories of behavioral symptoms: (1) unusual reciprocal social interactions and impaired social communication; and (2) stereotyped and repetitive patterns of behaviors, with restricted interests and activities (American Psychiatric Association 2013; Lord & Bishop 2015). Associated symptoms, which are present in subsets of individuals with ASD, include intellectual disabilities, anxiety, seizures, hyperactivity, sleep disruption and unusual reactivity to sensory stimuli. Early behavioral interventions are the current standard of care and offer the best long-term outcomes at present (Lord & Jones 2013; Rogers et al. 2012). Intensive behavioral intervention is highly effective in teaching young children to improve their social skills and redirect their repetitive behaviors. However, even the best behavioral therapies do not work for all, are expensive, are labor- and time-intensive and are not available in many geographic regions. Understanding the causes of autism is the first step in the development of effective medical therapeutics to improve symptoms and reverse the disorder’s trajectory.
Hypotheses about the genetic causes of ASD originally arose from observations of a male–female bias, 4:1 or greater, and high concordance between identical twins, 50–90%, as compared to less than 10% for non-identical twins and siblings (Fombonne 2009; Hallmayer et al. 2011; Miles 2011; Nordenbaek et al. 2014; Ritvo et al. 1985; Sandin et al. 2014; Smalley et al. 1988). Intensive searches for the genes causing ASD employed genome-wide association approaches in the early 2000s, progressed to analyses of copy-number variants (CNVs), and are now proceeding with exome and whole genome sequencing in thousands of individuals. Early findings quickly revealed that ASD is not a monogenic disorder. In contrast to disorders such as Huntington’s disease and Fragile X syndrome, there is no one specific gene responsible for all cases of autism. Rather, a growing number of de novo single gene mutations and CNVs have been identified in people with autism (Alarcon et al. 2008; Bucan et al. 2009; Butler et al. 2005; Buxbaum et al. 2007; Cook & Scherer 2008; Crepel et al. 2014; Glessner et al. 2009; Iossifov et al. 2014; Krumm et al. 2015; Kumar et al. 2009; Lawson-Yuen et al. 2008; Leblond et al. 2014; Michaelson et al. 2012; Morrow 2010; Neale et al. 2012; O’Roak et al. 2011; Pinto et al. 2010; Szatmari et al. 2007; Vernes et al. 2008; Wang et al. 2009; Yuen et al. 2015). Mutations in common gene variants and de novo coding mutations may be responsible for up to 50% of ASD cases (Gaugler et al. 2014; Iossifov et al. 2014; Miles 2011). Over 100 risk genes and CNVs for ASD have been published, each one appearing in only a relatively small number of individuals (Butler et al. 2005; Coe et al. 2014; De Rubeis et al. 2014; Gaugler et al. 2014; Iossifov et al. 2014; Li et al. 2014; Parikshak et al. 2013; Pinto et al. 2014; Willsey & State 2015). Epigenetic risk factors have been implicated in ASD, including chromatin remodeling and methylation mechanisms, such as CHD8 (Bernier et al. 2014; Cotney et al. 2015; O’Roak et al. 2012; Wilkinson et al. 2015), HDAC (Foley et al. 2012; Moldrich et al. 2013) and MECP2 (Shibayama et al. 2004; Theoharides et al. 2015), Further, environmental risk factors, such as parental age (Kong et al. 2012) and atypical maternal autoantibodies (Braunschweig et al. 2013; Brimberg et al. 2013; Diamond et al. 2013; Piras et al. 2014), are associated with a higher incidence of ASD.
One of the most intriguing aspects regarding the genetics of ASD is the enigma of how these many risk factors converge to result in the same general cluster of symptoms diagnosed as ASD. One possibility is that there are underlying convergent downstream mechanisms which contribute to ASD symptomotology. No definitive biomarkers have yet been identified across all diagnosed cases. Rather, subsets of biological factors may define subgroups of individuals with ASD. Stratification by subgroup, either by behavioral category or biomarker, may offer the best strategy for focused clinical trials. Intensive searches are underway to define abnormalities in neurophysiology, neuroanatomy, brain chemistry, immune markers and other key biological systems (Ecker et al. 2013; Jeste & Geschwind 2014; Levitt & Veenstra-VanderWeele 2015). High heterogeneity of symptoms across cases suggests that autism is actually multiple disorders, analogous to the plural concept of ‘cancers’, with different genetic etiologies and biological defects, to be treated with different classes of therapeutics. The concept of ‘autisms’ is implicit in the current use of the term ASD, implemented in the 2013 edition of the DSM-5.
Readers of Genes, Brain and Behavior are well aware of methods to interrogate genetic hypotheses of human disorders by targeting the homologous mutation in another species and then explicating the consequent phenotypic outcomes. Knockout (KO) and humanized knockin mice, and more recently KO rats, have been generated for many of the single gene mutations and CNVs that were identified in ASD populations and for comorbid neurodevelopmental disorders such as Fragile X and tuberous sclerosis (TSC) (Baudouin et al. 2012; Ey et al. 2011; Silverman et al. 2010a; Zoghbi & Bear 2012). Some of these mutant mouse models are now being employed in preclinical testing of pharmacological targets to treat the core symptoms of ASD (Silverman & Crawley 2014; Spooren et al. 2012; Vorstman et al. 2014).
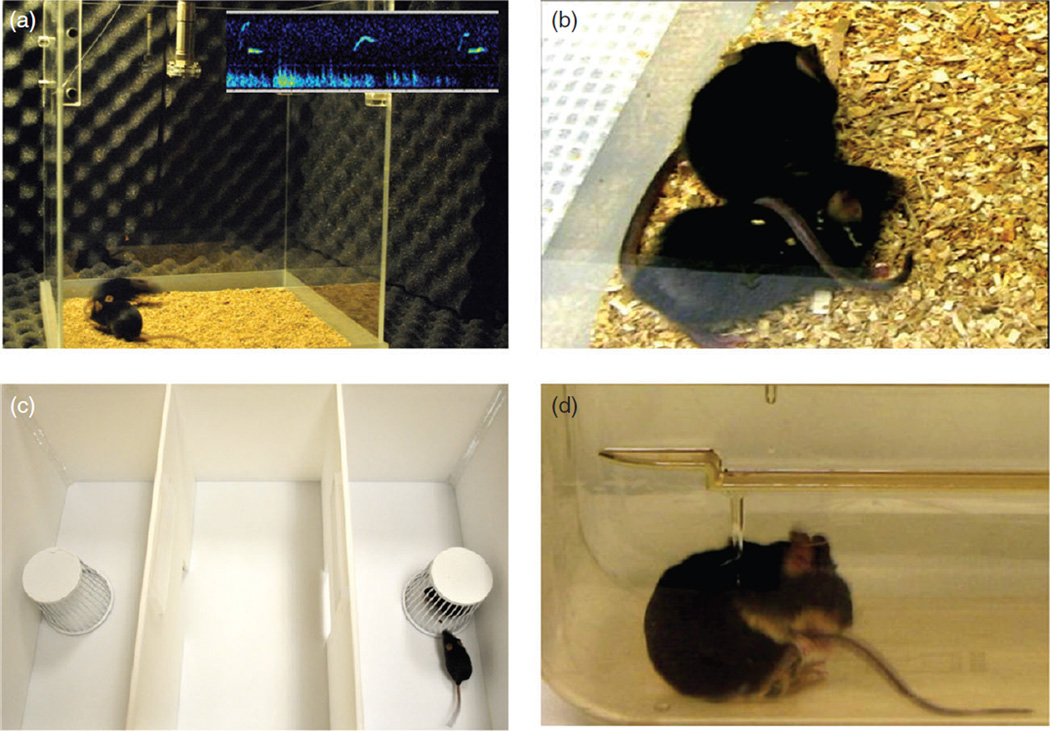
As genetic mouse models emerged, our behavioral neuroscience laboratory invested in methods development to design mouse behavioral assays with high relevance to the diagnostic symptoms of autism (Crawley 2004). Because the clinical phenotype of this uniquely human disorder is complex and heterogeneous, we initiated discussions with autism clinical experts, to understand the critical symptoms that could be most meaningfully modeled in mice. Clinical researchers, including colleagues at the University of California Davis MIND Institute, Weill Cornell Medical College, University of North Carolina, University of Washington, University College London and the National Institute of Mental Health Intramural Research Program, kindly allowed us to observe diagnostic interviews and watch videotapes of children with ASD. Knowledge gained through these sessions, and from lectures and conversations with many other generous colleagues working with children, adolescents and adults with ASD, guided our thinking in the development of mouse behavioral assays that dovetail with the natural behavioral repertoire of mice. Considering the types of social approach abnormalities and inappropriate social behaviors that are common in ASD, we developed a mouse 3-chambered social approach assay (Moy et al. 2004; Nadler et al. 2004), refined methods for scoring reciprocal social interactions in juvenile and adult mice (McFarlane et al. 2008), adapted measures for the detection of responses to social olfactory cues (Yang & Crawley 2009), and developed call categories for ultrasonic vocalizations emitted in response to social cues during reciprocal social interactions (Scattoni et al. 2008, 2011). Further, we established observational scoring methods to quantify motor stereotypies and repetitive behaviors, such as self-grooming and digging, along with assembling a set of established behavioral assays relevant to anxiety, intellectual impairment, hyperactivity and sensory reactivity (McFarlane et al. 2008; Moy et al. 2008a,b; Roullet & Crawley 2011; Silverman et al. 2010a,b, 2012, 2013, 2015; Wohr et al. 2011a; Yang et al. 2011, 2012a, 2015), which are now widely used. A small subset of these assays is illustrated in Figure 1. This review presents examples and summaries of ASD-relevant phenotypes discovered by our lab and many other excellent behavioral genetics labs, revealing the phenotypic consequences of targeting mutations in ASD risk genes.
Figure 1. Examples of rodent behavioral assays with face validity to the diagnostic symptoms of autism.
(a) Two mice interacting in a Noldus Phenotyper 3000 reciprocal social interaction chamber equipped with an Avisoft ultrasonic microphone. The inset shows representative ultrasonic vocalizations recorded during adult male–female social interaction. (b) Close-up of two mice displaying ‘crawl over and under’ during the reciprocal social interaction test session. (c) 3-Chambered social approach apparatus offers automated scoring of time spent with a novel social partner vs. time spent with a novel object. (d) BTBR mouse engaged in repetitive self-grooming. Photos by Jane Hayes, Michael Pride, Jill Silverman and Mu Yang, MIND Institute, University of California Davis School of Medicine.
A remarkable number of risk genes for ASD code for synaptic proteins. Cell adhesion proteins, including contactin-associated proteins, neuroligins and neurexins connect dendrites with axons to promote synapse formation. Postsynaptic scaffolding proteins, such as shanks and neuroligins, strengthen synapses and maintain synaptic transmission. Postsynaptic receptors, such as NMDA and metabotropic glutamate receptors, GABA receptors of varying subunit compositions, serotonin transporter and receptor subtypes, and oxytocin receptors, mediate excitatory and inhibitory synaptic signals. Sodium channels, potassium channels and downstream signaling pathways, such as the PTEN/PI3 kinase/Akt/mTOR pathway, mediate postsynaptic events and critical cellular functions. Mutations and common variants of the genes for these proteins have been identified in small numbers of individuals with autism and related disorders (Butler et al. 2005; Cheah et al. 2013; Frazier et al. 2014; Han et al. 2012; Krey et al. 2013; Rosander & Hallbook 2015; Tavassoli et al. 2014; Veenstra-VanderWeele et al. 2012; Weiss et al. 2003).
Mice with targeted mutations in many of these genes were generated by outstanding molecular genetics laboratories and generously donated to public repositories such as The Jackson Laboratory. Behavioral phenotypes have been published for some of these mutant lines. In most cases, one original publication describes the behavioral, electrophysiological, neuroanatomical, and/or biochemical phenotypes of the new mouse model of autism. In some cases, the first findings have been replicated by the same laboratory in additional publications. In a few cases, behavioral phenotypes have been replicated by other laboratories. We summarize some of the strongest findings below. Table 1 provides descriptions of gene mutations associated with human ASD. Table 2 summarizes the behavioral phenotypes in the corresponding mouse models, focusing on a subset of ASD risk genes that are involved in synaptic function and the mTOR signaling pathway. Here, we will refer to mice without any functional alleles (homozygous null) as KO mice, mice with one functional allele as heterozygous (Het) mice, mice with targeted amino acid substitutions as knockin mice, and littermate controls with both functional alleles as wildtype (WT) mice.
Table 1.
Examples of autism risk genes identified by human genetic studies
| Gene | Identification method | Investigated population | Reference for genetic study |
|---|---|---|---|
| CNTNAP2 | |||
| Association with ASD |
Linkage study | Old Order Amish families (4 children and 6 parents) with an autosomal recessive founder (null mutation) |
Strauss et al. (2006) |
| Linkage, association and gene expression studies |
172 parent–child trios from the Autism Genetic Resource Exchange (AGRE) resource |
Alarcon et al. (2008) | |
| Genome-wide linkage study, family based-association mapping |
National Institute of Mental Health Autism Genetics Initiative Repository; Stage I: 72 muliplex families (148 affected, 292 controls); Stage II: 1295 parent–child trios (145 mulitplex families with 303 affected children) |
Arking et al. (2008) | |
| Cytogenetics | 34 year old female | Rossi et al. (2008) | |
| Cytogenetics | 11 year old male | Poot et al. (2010) | |
| Limited association with ASD |
Cytogenetics, linkage and resequencing |
635 ASD cases and 942 controls | Bakkaloglu et al. (2008) |
| Genetic association analyses; transmission disequilibrium test |
186 multiplex (408 trios) and 323 simplex families with ASD from the AGRE resource |
Sampath et al. (2013) | |
| No association with ASD |
Next generation sequencing | 2704 ASD cases, 2747 controls | Murdoch et al. (2015) |
| NLGN1 | |||
| Association with ASD |
Whole genome copy number variant study |
859 ASD cases, 1409 controls; 1336 ASD cases, 1110 controls |
Glessner et al. (2009) |
| Limited association with ASD |
Mutation analysis, linkage analysis, association analysis |
30 ASD cases; 19 families with 41 ASD cases; 100 families with 122 ASD cases |
Ylisaukko-oja et al. (2005) |
| NLGN3 | |||
| Association with ASD |
Amino acid sequencing, linkage study |
36 ASD sibling pairs, 122 ASD trios, 350 unrelated controls |
Jamain et al. (2003) |
| Limited association with ASD |
Mutation analysis, linkage analysis, association analysis |
30 ASD cases; 19 families with 41 ASD cases; 100 families with 122 ASD cases |
Ylisaukko-oja et al. (2005) |
| NLGN4 | |||
| Association with ASD |
Cytogenetic analysis, fluorescence in situ hybridization (FISH) |
8 females, 3 with ASD | Thomas et al. (1999) |
| Amino acid sequencing, linkage study |
36 ASD sibling pairs, 122 ASD trios, 350 unrelated controls |
Jamain et al. (2003) | |
| Linkage analysis and gene sequencing |
10 members of a French family with ASD and intellectual disability, 200 controls |
Laumonnier et al. (2004) | |
| Direct sequencing and mutation analysis |
148 unrelated ASD cases, 48 ADHD and bipolar cases, 288 unaffected controls |
Yan et al. (2005) | |
| Chromosome analysis | Family with 1 ASD proband, 96 controls |
Lawson-Yuen et al. (2008) | |
| Single nucleotide polymorphism microarrays and karyotyping |
427 unrelated ASD families | Marshall et al. (2008) | |
| Limited association with ASD |
Mutation analysis, linkage analysis, association analysis |
30 ASD cases; 19 families with 41 ASD cases; 100 families with 122 ASD cases |
Ylisaukko-oja et al. (2005) |
| No association with ASD |
PCR genetic screen, denaturing high performance liquid chromatography |
196 ASD cases from 183 multiplex and 13 simplex families |
Vincent et al. (2004) |
| NRXN1 | |||
| Gene scanning and sequencing | 103 Caucasian ASD cases, 61 Afro-American ASD cases, 535 Caucasian controls |
Feng et al. (2006) | |
| Linkage study, using comparative analysis of hybridization intensities |
1496 multiplex ASD families (at least 2 affected individuals) (7917 family members) |
Szatmari et al. (2007) | |
| Single nucleotide polymorphism microarrays and karyotyping |
427 unrelated ASD families | Marshall et al. (2008) | |
| Cytogenic analysis | 1 ASD case study | Zahir et al. (2008) | |
| Whole genome CNV study | 859 ASD cases, 1409 controls; 1336 ASD cases, 1110 controls |
Glessner et al. (2009) | |
| Comparative genomic hybridization microarrays |
3540 cases with developmental disorders, ASD, intellectual disability |
Ching et al. (2010) | |
| SHANK1 | |||
| Microarray | 1158 Canadian and 456 European ASD cases, 15122 controls |
Sato et al. (2012) | |
| Meta-analysis of copy-number and coding-sequence variants |
5657 ASD cases, 19163 controls; 76 0 – 214 7 ASD cases, 492–1090 controls depending on the SHANK gene |
LeBlond et al. (2014) | |
| SHANK2 | |||
| Genome-wide microarray scan; DNA sequencing |
396 ASD cases, 184 individuals with intellectual disability, 659 unaffected controls |
Berkel et al. (2010) | |
| Dense genotyping arrays | 996 European ASD cases, 1287 matched controls |
Pinto et al. (2010) | |
| Meta-analysis of copy-number and coding-sequence variants |
5657 ASD cases, 19, 163 controls; 76 0 – 214 7 ASD cases, 492–1090 controls depending on the SHANK gene |
LeBlond et al. (2014) | |
| SHANK3 | |||
| FISH analysis; direct sequencing | 226 families with at least 1 ASD child, 270 controls |
Durand et al. (2007) | |
| DNA sequencing and microarray-based comparative intensity analysis |
400 Canadian ASD cases; HapMap collection was used for comparison |
Moessner et al. (2007) | |
| Single nucleotide polymorphism microarrays and karyotyping |
427 unrelated ASD families | Marshall et al. (2008) | |
| Gene sequencing | 427 ASD cases, 190 controls | Gauthier et al. (2009) | |
| Denaturing high performance liquid chromatography, direct sequencing, multiplex ligation-dependent probe amplification, array comparative genomic hybridization |
3 cohorts: 133 American ASD cases; 88 Italian ASD cases; 104 American ASD cases; 560 American controls and 422 Italian controls |
Boccuto et al. (2013) | |
| Meta-analysis of copy-number and coding-sequence variants |
5657 ASD cases, 19, 163 controls; 76 0 – 2147 ASD cases, 492–1090 controls depending on the SHANK gene |
LeBlond et al. (2014) | |
| PTEN | |||
| Direct DNA sequencing | 18 ASD cases with macrocephaly | Butler et al. (2005) | |
| Direct DNA sequencing, multiplex ligation-dependent probe amplification |
88 ASD cases with macrocephaly | Buxbaum et al. (2007) | |
| Direct DNA sequencing | 2 ASD cases with macrocephaly | Herman et al. (2007) | |
| Direct DNA Sequencing | 114 cases of ASD, developmental delay or macrocephaly |
Varga et al. (2009) | |
| Direct DNA sequencing | 93 cases of ASD or developmental delays with macrocephaly |
McBride et al. (2010) | |
Table 2.
Examples of genetic mouse models of autism
| Mouse model | Mouse phenotype | Reference |
|---|---|---|
| Cntnap2 knockout mouse |
|
Penagarikano et al. (2011) |
| Neuroligin-1 knockout mouse |
|
Blundell et al. (2010) |
| Neuroligin-2* knockout mouse |
|
Chubykin et al. (2007), Blundell et al. (2009) and Wohr et al. (2013) |
| Neuroligin-3* knockout mouse |
|
Radyushkin et al. (2009) |
| Neuroligin-3* knockin mouse (R451C substitution) |
|
Tabuchi et al. (2007) and Etherton et al. (2011) |
| Neuroligin-3* knockin mouse (R451C substitution) |
|
Chadman et al. (2008) |
|
Nlgn4 knockout mouse (gene trap insertion downstream of exon I) |
|
Jamain et al. (2008) and El-Kordi et al. (2013) |
|
Nlgn4* knockout mouse (gene trap insertion downstream of exon I) |
|
Ey et al. (2012) |
| Neurexin 1α* knockout mouse |
|
Etherton et al. (2009) |
| Neurexin 1α* knockout mouse |
|
Grayton et al. (2013) |
|
Shank1* knockout mouse (targeted replacement of PDZ domain (exons XIV and XV)) |
|
Hung et al. (2008) |
|
Shank1* knockout mouse (targeted replacement of PDZ domain (exons XIV and XV)) |
|
Silverman et al. (2011) |
|
Shank1* knockout mouse (targeted replacement of PDZ domain (exons XIV and XV)) |
|
Wohr et al. (2011b) |
|
Shank2 knockout mouse (lacking all Shank2 isoforms) |
|
Schmeisser et al. (2012) |
|
Shank2 knockout mouse (exon VI and VII microdeletion and a frameshift for both Shank splice variants) |
|
Won et al. (2012) |
|
Shank3A* heterozygous mouse (ankyrin repeat domain deletion (exon IV-IX)) |
|
Bozdagi et al. (2010) |
|
Shank3A* heterozygous and knockout mice (ankyrin repeat domain deletion (exon IV-IX)) |
|
Yang et al. (2012b) and Drapeau et al. (2014) |
|
Shank3A knockout mouse (ankyrin repeat domain deletion (exon IV-IX)) |
|
Wang et al. (2011) |
|
Shank3A* knockout mouse (ankyrin repeat domain deletion) |
|
Peca et al. (2011) |
|
Shank3B knockout mouse (PDZ domain deletion) |
|
Peca et al. (2011) |
|
Shank3* knockout mouse (exon 21 deletion, including the Homer binding domain) |
|
Kouser et al. (2013) |
|
Nse-Cre Pten conditional knockout mouse (Pten deletion restricted to subsets of postmitotic cortical and hippocampal neurons) |
|
Kwon et al. (2006), Ogawa et al. (2007) and Zhou et al. (2009) |
| Pten heterozygous mouse |
|
Page et al. (2009); Clipperton-Allen and Page (2014) and Clipperton-Allen and Page (2015) |
The asterisk (*) following the name of a mouse model indicates that its social behaviors were normal, using assays with high face validity to the types of social deficits which characterize autism.
Mouse models of genetic risk factors for autism
The CNTNAP2 gene, located on chromosome 7, encodes contactin-associated protein-like 2 (CASPR2), a member of the neurexin superfamily of proteins, functioning as a cell adhesion molecule and receptor (Rodenas-Cuadrado et al. 2014). This protein, which contains a putative PDZ binding domain, mediates interactions of neurons and glia during central nervous system development. It also is located in myelinated axons and directs potassium channel localization within differentiating neurons (Poliak et al. 1999, 2003). CASPR2 directly binds to the transcription factor FOXP2 (forkhead box protein P2), which has been implicated in speech and language development (Fischer & Hammerschmidt 2011; Vernes et al. 2008). Several mutations in the CNTNAP2 locus, including rare, common and deletion variants, have been associated with ASD (Alarcon et al. 2008; Arking et al. 2008; Poot et al. 2010; Rossi et al. 2008; Strauss et al. 2006), although more recent studies indicate a limited contribution of CNTNAP2 dysregulation to ASD (Bakkaloglu et al. 2008; Murdoch et al. 2015; Sampath et al. 2013). Mice lacking Cntnap2 exhibited reduced juvenile ultrasonic vocalizations, reduced social interaction time and increased repetitive behaviors (Penagarikano et al. 2011). In addition to these deficits in ASD-related behaviors, Cntnap2 KO mice also displayed abnormal neuronal cortical migration, asynchronous neuronal firing in the cortex, a reduced number of inhibitory interneurons, behavioral perseveration in a cognitive task, hyperactivity and seizures (Penagarikano et al. 2011).
Neuroligins are cell adhesion molecules located at the postsynaptic side of the synapse, interacting with their presynaptic partner proteins, the neurexins (Bang & Owczarek 2013). Neuroligins contribute to synaptic neurotransmission through their influence on synaptic formation and are distributed at excitatory and inhibitory synapses in an isoform-dependent manner (Hu et al. 2015). For example, neuroligin-1 is primarily located at excitatory synapses (Budreck et al. 2013; Chih et al. 2005; Song et al. 1999), while neuroligin-2 is found at inhibitory synapses (Varoqueaux et al. 2004); neuroligin-3 can be found at both these locations (Budreck & Scheiffele 2007). At the synapse, neuroligins bind to PSD-95, a scaffolding protein important for postsynaptic strengthening and synapse organization, particularly with ion channels and receptors, such as the glutamatergic NMDA receptor (Bolliger et al. 2001; Irie et al. 1997; Kim et al. 1995, 2008; Kornau et al. 1995; Niethammer et al. 1996; Shipman & Nicoll 2012). Several studies suggest that neuroligins are involved in synapse modulation and specification rather than synapse formation (Chubykin et al. 2007; Krueger et al. 2012; Varoqueaux et al. 2006). Neuroligin proteins encoded by X-linked genes, such as NLGN3 and NLGN4 which map to Xq13 and Xp22.3, respectively, have been associated with ASD in large genome-wide scans (Auranen et al. 2002; Glessner et al. 2009; Philippe et al. 1999), but strong associations have not been found in all studies (Vincent et al. 2004; Ylisaukko-oja et al. 2005). Using amino acid sequencing in linkage and proband case studies, deletions and frameshifts in NLGN3 and NLGN4 sequences have been identified in individuals with ASD (Jamain et al. 2003; Laumonnier et al. 2004; Lawson-Yuen et al. 2008; Marshall et al. 2008; Thomas et al. 1999; Yan et al. 2005).
Neuroligin-1 KO mice exhibited minimal deficits in social behavior, but displayed increased grooming and spatial learning impairments, along with impaired hippocampal long-term potentiation (Blundell et al. 2010). Neuroligin-2 heterozygous and KO mice showed normal social interactions in social approach, but displayed increased anxiety-like behavior, decreased pain sensitivity and poor motor coordination (Blundell et al. 2009; Wohr et al. 2013). In addition, neuroligin-2 KO mice had decreased inhibitory neurotransmission, as well as decreased immunostaining of inhibitory synapse markers (Blundell et al. 2009; Chubykin et al. 2007). Neuroligin-3 knockin (R451C) mice, with an arginine to cysteine substitution at site 451, did not display robust autism-relevant behaviors, but rather had mild developmental differences, (e.g. slower righting reflexes), enhanced spatial learning acquisition and reduced acoustic startle (Chadman et al. 2008; Etherton et al. 2011; Tabuchi et al. 2007), suggesting that this ASD-related point mutation delayed development, altered learning and reduced sensitivity to stimuli. Neuroligin-3 knockin mice also displayed increased inhibitory neurotransmission in the barrel cortex, increased excitatory neurotransmission and enhanced long-term potentiation in the hippocampus, increased dendritic branching in the hippocampus, as well as increased protein levels of inhibitory synaptic markers, while KO mice did not (Etherton et al. 2011; Tabuchi et al. 2007). Neuroligin-3 KO mice displayed normal sociability, but impairments in fear conditioning and olfaction, as well as hyperactivity and decreased total brain volume (Radyushkin et al. 2009). Characterization of Nlgn4 KO mice revealed that loss of this neuroligin resulted in reduced sociability and ultrasonic vocalizations, as well as a reduction in total brain volume (El-Kordi et al. 2013; Jamain et al. 2008). However, phenotypic analysis of later generations of the same line of Nlgn4 KO did not find any genotype differences in sociability, ultrasonic vocalizations, anxiety-related behaviors or general locomotor activity (Ey et al. 2012).
Neurexins are a class of cell adhesion proteins found on the presynaptic terminal of synapses that bind to neuroligins (Bang & Owczarek 2013). Numerous association studies have identified mutations in the NRXN1 gene, located on chromosome 2, in intellectual disabilities and other syndromes, including several cases of autism (Ching et al. 2010; Feng et al. 2006; Glessner et al. 2009; Gregor et al. 2011; Marshall et al. 2008; Szatmari et al. 2007; Zahir et al. 2008). Neurexin-1α KO mice displayed increased grooming, reduced locomotor activity, reduced sensorimotor gating and increased aggression (Etherton et al. 2009; Grayton et al. 2013). Further studies are necessary to determine the exact contribution of specific neurexin and neuroligin mutations to ASD-relevant behaviors.
The SHANK family of genes located on chromosome 22q encodes scaffolding proteins that assist in the synaptic organization of excitatory glutamatergic neurons by binding to postsynaptic density proteins, signaling molecules, postsynaptic receptors and cytoskeletal proteins (Boeckers et al. 2002; Grabrucker et al. 2011; Lim et al. 1999; Naisbitt et al. 1999; Sheng & Kim 2000; Tu et al. 1999). SHANK3 can bind to neuroligins, suggesting disrupted cell adhesion may contribute to ASD (Meyer et al. 2004). Genetic studies have identified de novo and inherited mutations in SHANK1 (Sato et al. 2012), SHANK2 (Berkel et al. 2010, 2012; Pinto et al. 2010) and SHANK3 (Boccuto et al. 2013; Durand et al. 2007; Gauthier et al. 2009, 2010; Marshall et al. 2008; Moessner et al. 2007). A recent meta-analysis of SHANK mutations has suggested that ASD severity due to SHANK mutations may be related to which gene is mutated, such that SHANK3 mutations have a higher frequency and penetrance in individuals with ASD, compared to SHANK1 and SHANK2 (Leblond et al. 2014). 22q13 deletion syndrome, also known as Phelan-McDermid syndrome, is caused by a deletion on the distal part of the long arm of chromosome 22 and is associated with ASD-like behaviors, including disrupted social behavior, repetitive behaviors, motor dysfunctions, seizures and moderate to severe intellectual disability (Kolevzon et al. 2014; Phelan & McDermid 2012). SHANK3 is one of the most commonly mutated genes within the Phelan-McDermid critical region and is thought to underlie most of the neural consequences of this deletion (Phelan & McDermid 2012).
While Shank1 KO mice do not display robust autism-relevant social deficits (Silverman et al. 2011), Shank1 KO mice emitted fewer ultrasonic vocalizations as pups, exhibited reduced scent marking and abnormal vocalizations as adults and had motor impairments (Silverman et al. 2011; Wohr et al. 2011b). Shank1 KO mice also displayed dendritic spine abnormalities, including weaker basal synaptic neurotransmission (Hung et al. 2008). Shank2 KO mice had abnormalities in several ASD-relevant behaviors, including reduced sociability, as measured by fewer social contacts during same-sex interactions, and reduced preference for social novelty, as well as higher levels of repetitive behaviors (e.g. grooming and jumping) and abnormal ultrasonic vocalizations (Schmeisser et al. 2012; Won et al. 2012). In addition, Shank2 KO mice had a reduced number of hippocampal dendritic spines and reduced glutamatergic neurotransmission in the hippocampus (Schmeisser et al. 2012; Won et al. 2012).
Mutant mice have been generated for each of the Shank3 isoforms, with deletions in various domains of the Shank3 gene, some of which displayed social deficits while others displayed normal sociability (Jiang & Ehlers 2013). Reduced sociability, reduced ultrasonic vocalizations and high levels of repetitive self-grooming were dependent on which isoform was deleted (Bozdagi et al. 2010; Drapeau et al. 2014; Kouser et al. 2013; Peca et al. 2011; Wang et al. 2011; Yang et al. 2012b). Reduced basal neurotransmission as well as abnormalities in neuronal morphology (e.g. neuronal hypertrophy, dendritic spine deficits) have been identified in most of these models (Bozdagi et al. 2010; Kouser et al. 2013; Peca et al. 2011; Wang et al. 2011; Yang et al. 2012b), underscoring the importance of the Shank proteins in maintaining normal synaptic function and neuronal structure. Therefore, several different ASD-relevant mutations in the SHANK gene family have been modeled in mice, and while the phenotypes differ between the specific models, it appears that complete or partial loss of Shank proteins may be detrimental to normal social behaviors and may induce high levels of repetitive behaviors in mice.
ASD risk genes in the mTOR signaling pathway
While many of the identified risk genes for ASD involve synaptic proteins, mutations in several components of the mTOR pathway are also implicated in ASD (Bourgeron 2009), suggesting that normal function of this intracellular signaling pathway is necessary for proper synaptic transmission and neuronal activity. The mTOR pathway is critical for protein synthesis, cellular proliferation and growth (Hershey et al. 2012; Laplante & Sabatini 2012). Activated tyrosine kinase receptors recruit and activate phosphoinositide 3-kinase (PI3K), which converts phosphatidylinositol (4,5)-biphosphate (PIP2) to phosphatidylinositol (3, 4, 5)-triphosphate (PIP3). PIP3 recruits many proteins to the membrane through pleckstrin homology domains, including PDK1 and the serine/threonine kinase AKT, an important downstream effector of PIP3. Phosphatase and tensin homolog located on chromosome 10 (PTEN) is a lipid and protein phosphatase that negatively regulates Akt activity by working in opposition to PI3K, converting PIP3 back to PIP2 (Maehama & Dixon 1998; Stambolic et al. 1998). Akt is fully activated following phosphorylation by PDK1 and mTOR complex 2 (mTORC2), one of two complexes that involves the evolutionarily conserved serine/threonine kinase, mammalian target of rapamycin (mTOR) (Bayascas & Alessi 2005; Sarbassov et al. 2005). Phosphorylated Akt is involved in a wide range of cellular processes, including cell proliferation, survival and growth, via a myriad of downstream signaling proteins, including TSC1 and TSC2. Activated Akt inhibits a protein complex composed of TSC1 (hamartin) and TSC2 (tuberin) through phosphorylation (Huang & Manning 2009; Tee et al. 2002). This disinhibits the GTPase Rheb and actives mTOR complex 1 (mTORC1) (Sato et al. 2008). Once activated, mTORC1 upregulates protein synthesis by phosphorylating several key proteins, including S6 kinase 1 (S6K1), which subsequently regulates Fragile X mental retardation protein (FMRP) through phosphorylation (Narayanan et al. 2008). Several key proteins, such as PTEN, TSC1, TSC2 and FMRP, are strongly implicated in subsets of ASD cases, and are discussed below. However, additional components of the mTOR pathway are also under investigation for their contribution to ASD-related deficits, such as the mTOR substrate eIF4E (Gkogkas et al. 2013).
PTEN mutations are most often associated with a variety of hamartoma syndromes, including Cowden syndrome (Eng 2003), which are characterized by benign focal malformations. PTEN was identified as an ASD candidate gene after several case and prospective studies revealed an association in individuals with ASD and macrocephaly (enlarged head size) (Butler et al. 2005; Buxbaum et al. 2007; Herman et al. 2007; McBride et al. 2010; Varga et al. 2009). Modeling PTEN mutations in mice has focused on two distinct strategies: conditional KO mice, in which distinct cell types lack the gene and protein product, and heterozygous null mice that possess constitutive haploinsufficiency. Conditional KO mice with a Pten deletion restricted to a subset of post-mitotic hippocampal and cortical neurons exhibited deficits in social interaction, sociability and preference for social novelty as well as impaired performance in the Morris water maze and macrocephaly (Kwon et al. 2006; Zhou et al. 2009). Similarly, heterozygous KO mice with a constitutive Pten mutation exhibited macrocephaly and deficits in sociability (Clipperton-Allen & Page 2014), but these sociability impairments were limited to female mice in one study (Page et al. 2009). Additionally, Allen, Page and colleagues also showed that Pten heterozygous mice exhibited elevated repetitive behaviors (i.e. digging, self-grooming). To date, the results obtained with Pten mutant mice suggest that Pten dysfunction leads to several ASD-relevant behaviors, although more extensive behavioral characterization of the various Pten conditional mouse models would further elucidate this phosphatase’s contribution to ASD.
Tuberous sclerosis is a genetic condition resulting from mutations in the TSC1 or TSC2 gene, which negatively regulate mTOR activity (Curatolo & Maria 2013). Individuals that carry mutations in either TSC gene have a higher than expected occurrence of ASD-like features (50%) or an ASD diagnosis (29%) (Curatolo et al. 2010). Tsc1 heterozygous mice exhibited deficits in social interactions, hippocampus-dependent contextual fear conditioning, and hidden platform Morris water maze tasks (Goorden et al. 2007). Social deficits were not identified in Tsc2 heterozygous mice, except when the mice were also treated prenatally with Poly I:C, a maternal immune activation model (Ehninger et al. 2012). However, Tsc2 mutations did produce learning and memory deficits in Morris water maze hidden platform performance and contextual fear conditioning (Ehninger et al. 2008). A dominant negative Tsc2 mutation that binds to Tsc1 and inhibits its activity also led to reduced social interactions and a reduced preference for social novelty, although these mice exhibited normal sociability in the 3-chambered social approach task (Chevere-Torres et al. 2012). A specific Tsc1 disruption limited to cerebellar Purkinje cells produced deficits in sociability, repetitive behavior and cognitive flexibility (Tsai et al. 2012). Similarly, Tsc2 disruption limited to cerebellar Purkinje cells replicated deficits in sociability and social preference, which were accompanied by increased repetitive behaviors (Reith et al. 2013). The study of these Tsc1 and Tsc2 mutant mice has provided knowledge as to how these single gene mutations can model some ASD-relevant phenotypes and contribute to the overall ASD phenotype.
Fragile X syndrome (FXS) is caused by a hypermethylated CGG repeat expansion in the FMR1 gene that leads to a drastic reduction in its protein product, FMRP. FMRP is an RNA binding protein that has been implicated in regulating protein expression (Bagni & Greenough 2005; Chen & Joseph 2015; Crawford et al. 2001; Kazdoba et al. 2014). Individuals with FXS are characterized by intellectual disability and a variety of physical abnormalities, and frequently display social dysfunction, anxiety and repetitive behaviors (Berry-Kravis et al. 2002; Lightbody & Reiss 2009). The diagnostic criteria for ASD commonly occur in individuals with FXS, with recent estimates ranging from 18% to 47% (Clifford et al. 2007; Demark et al. 2003; Hatton et al. 2006; Kaufmann et al. 2004; Rogers et al. 2001). Conversely, the proportion of FXS in the ASD population has been estimated at 3–8% (Cohen et al. 1991; Fombonne et al. 1997). The first and most widely tested mouse model of FXS is the Dutch-Belgian Consortium Fmr1 KO mouse, which has been maintained on multiple background strains. Recent work with the Fmr1 KO mouse has evaluated ASD-relevant phenotypes, including social deficits and repetitive behaviors. Fmr1 KO mice on a C57BL/6 (B6) background exhibited decreased sociability in some cases (Dahlhaus & El-Husseini 2010) or decreased sniffing during a social approach test when maintained on a B6/FVB hybrid background (McNaughton et al. 2008). In contrast, other studies revealed normal sociability and reduced preference for social novelty in Fmr1 KO mice maintained on FVB and B6 backgrounds (Liu & Smith 2009; Pietropaolo et al. 2011). In tests of direct social interaction, Fmr1 KO mice on a B6 background showed deficits in social interaction in some studies (Mineur et al. 2006), but as with much of the Fmr1 mouse behavioral literature, there are caveats to these behavioral impairments (Kazdoba et al. 2014). Paylor and colleagues demonstrated increased social interaction in Fmr1 KO mice on a B6 background (Spencer et al. 2005, 2008). Importantly, the Paylor team presented data interpreted as higher social anxiety in these mice. Fmr1 KO mice also exhibited elevated repetitive behaviors, another core symptom of ASD. Specifically, Fmr1 KO mice on B6 or hybrid backgrounds had elevated levels of self-grooming, but not when they were maintained on an FVB background (McNaughton et al. 2008; Pietropaolo et al. 2011). Fmr1 KO mice on B6 and B6 hybrid backgrounds also had higher levels of marble burying (Gholizadeh et al. 2014; Spencer et al. 2011; Veeraragavan et al. 2012). Intellectual disabilities which characterize FXS have been evaluated in a variety of cognitive tasks. In some reports, Fmr1 KO mice demonstrated cognitive deficits consistent with the intellectual impairments that characterize FXS. Deficits have been observed in contextual, cued and trace fear conditioning and/or context discrimination in Fmr1 KO mice on B6 and sighted FVB backgrounds (Auerbach et al. 2011; Ding et al. 2014; Paradee et al. 1999; Zhao et al. 2005). However, many other reports have described normal cognitive abilities in Fmr1 KO mice maintained on B6, FVB/129 hybrid, and albino B6 backgrounds, including fear conditioning (Baker et al. 2010; Dobkin et al. 2000; Peier et al. 2000; Uutela et al. 2012; Van Dam et al. 2000). Similarly, spatial navigation and reversal deficits were observed during Morris water maze testing in Fmr1 KO mice maintained on B6, albino B6 and FVB backgrounds in some labs (Baker et al. 2010; D’Hooge et al. 1997; Kooy et al. 1996; The Dutch-Belgian Fragile X Consortium et al. 1994) but these impairments were not seen in other studies (Paradee et al. 1999; Uutela et al. 2012; Yan et al. 2004). Thus, although the gene and its product are absent in Fmr1 mice, behavioral phenotypes relevant to the human syndrome and to ASD appear to be variable, with phenotypes potentially dependent on a variety of methodological and environmental factors. While genetic background is one potential factor, there does not appear to be a clear segregation of behavioral outcomes on the B6 vs. FVB backgrounds, either in the social and repetitive behavioral domains most relevant to autism, or in the cognitive domains most relevant to FXS. In contrast to the inconsistent behavioral literature on Fmr1 mice, investigations of neuroanatomical, electrophysiological, genetic and biochemical phenotypes in Fmr1 KO mice have allowed researchers to gain considerable insight into biological mechanisms underlying Fragile X syndrome.
Conclusions
The summary above and in Table 2 provides descriptions of behavioral and biological phenotypes in representative genetic mouse models of ASD. The small subset of genetic mouse models included herein focuses primarily on risk genes that mediate the formation and strengthening of synapses, and postsynaptic downstream signaling through the mTOR pathway. Our selection is presented for its possible usefulness in conceptualizing a cluster of genes with potentially interrelated actions through synaptic and postsynaptic intracellular mechanisms. The appeal of this convergence concept, proposed by many autism researchers (Delorme et al. 2013; Geschwind & State 2015; Silverman & Crawley 2014; Spooren et al. 2012), includes the possibility of developing pharmacological treatments for ASD that act through impaired synaptic mechanisms, perhaps using compounds repurposed from other uses involving synaptic dysfunction. Several intriguing preclinical studies with mouse models of ASD indicate improvements in social behaviors and/or reductions in repetitive behaviors and/or amelioration of cognitive deficits after pharmacological treatments. Promising results from pharmacological interventions in mouse models of ASD and FXS include mGluR5 antagonists (Michalon et al. 2012; Silverman et al. 2012; Tian et al. 2015), GABA agonists (Han et al. 2014; Henderson et al. 2012; Silverman et al. 2015), rapamycin (Burket et al. 2014; Ehninger & Silva 2011; Zhou et al. 2009), d-cycloserine (Burket et al. 2013; Yadav et al. 2012), BDNF and ampakines (Lauterborn et al. 2007; Silverman et al. 2013), IGF-1 (Bozdagi et al. 2013) and oxytocin (Huang et al. 2014; Meziane et al. 2014; Modi & Young 2012; Penagarikano et al. 2015). Descriptions of additional mouse models of ASD are available in several other recent review articles (Ey et al. 2011; Kas et al. 2014; Silverman & Crawley 2014).
Caveats abound for the methods and interpretations of mouse behavioral phenotypes relevant to the core symptoms of autism. The first is genetic background. Just as humans with the same mutation may present with different symptoms, possibly due to protective or susceptibility genes in their genetic backgrounds, mice present with different phenotypes when a mutation is bred into different inbred strains, such as C57BL/6J, FVB/NJ and substrains of 129, each of which has its own idiosyncratic behavioral traits (Crawley et al. 1997). Varying behavioral phenotypes have been reported for mutant mouse models of autism and FXS, as described above, when outcomes of the mutation were placed on divergent backgrounds and directly compared (Moy et al. 2009; Pietropaolo et al. 2011; Spencer et al. 2011). The second is experimental design. Employing large Ns and using WT littermates as controls are essential to avoid over-interpretations of phenotypes which were actually caused by environmental influences that affect mouse behaviors. The third is statistical analysis of behavioral data. Two-way ANOVAs are often required, in which treatment is one factor and genotype is the other factor, rather than simple t-tests. Stringent post-hoc tests, such as Newman-Keuls, Dunnett’s and Tukey’s, avoid the false positives that may be obtained from more forgiving post-hoc tests such as Fisher’s LSD. Statistical comparisons must match the original experimental design. For example, our 3-chambered social approach task is not sensitive enough to compare the absolute number of seconds spent with the novel mouse across genotypes, or across drug doses. Data on social approach are correctly analyzed with a simple paired t-test or equivalent, which compares time with the novel mouse vs. time with the novel object within genotype only, or within a drug dose only, to provide a yes-or-no outcome measure, i.e. sociability or absence of sociability. The fourth is corroboration within the behavioral domain. Conducting two or more assays that interrogate the same behavior (e.g. at least two social tasks, or two repetitive behavior assays, or two anxiety-related tests, or several learning and memory tasks that tap into different cognitive domains) allows for stronger interpretations and generalizability of a finding within a given domain. Including relevant controls, such as open field exploration for tasks that require locomotor activity, and pain or foot shock sensitivity for fear conditioning, will ensure that a motor or sensory artifact is not the cause of a significant effect in the behavioral domain of primary interest. Large numbers of mouse behavioral assays relevant to the diagnostic and associated symptoms of ASD are available to choose from (Crawley 2012; Silverman et al. 2010a), along with a wealth of relevant control tests (Crawley 2007).
One additional caveat, arguably the most important, is replicability. While all scientific discoveries require replication, behavioral findings require extra attention to reproducibility because of the strong influence of the various environmental factors on mouse behaviors. Stressors, such as construction noise or rough handling, can greatly affect scores on sensitive behavioral assays for anxiety-related, social and cognitive phenotypes. These types of variable findings remain primarily anecdotal, because they are difficult to document and publish, although they are common knowledge among behavioral neuroscientists. Loss of behavioral phenotypes across breeding generations may occur due to attritional loss of the mutation or drift in background genes. Basic methodological issues, such as the age of the mice at testing, composition of the chow diet, properties of the testing equipment and testing room, social testing in single vs. mixed genotype dyads, cleaning of the equipment between subject mice, and simple random chance beyond P <0.05, may create conditions that yield a positive finding that cannot be replicated. Following published procedures by expert behavioral neuroscientists can help alleviate some of these concerns. However, the best strategy to ensure high replicability is for the research team to conduct the entire experiment again in a separate cohort of mice prior to publishing. Further, to confirm the ultimate strength of a finding for the scientific community, several different laboratories should conduct essentially the same experiment with the same line of mice. When a social deficit in a genetic mouse model of ASD is replicated across cohorts and by many labs, the strength of a finding is ensured. Highly robust, well-replicated behavioral phenotypes relevant to the symptoms of ASD in a genetic mouse model can then effectively inform our understanding of the role of that gene in the symptomotology of ASD and serve as preclinical outcome measures for therapeutic discovery.
Acknowledgments
This work was supported by MIND Institute Autism Research Training Program MH0073124 (T.K.), Autism Speaks Targeted Award #8703 (J.C.), NINDS 1R01NS085709-01 (J.C., P.L.), and the NIH-funded MIND Institute Intellectual and Developmental Disabilities Research Center (U54 HD079125). We thank Michael Pride, Jane Hayes, Jill Silverman and Mu Yang for the photographs that appear in Figure 1.
REFERENCES
- Alarcon M, Abrahams BS, Stone JL, Duvall JA, Perederiy JV, Bomar JM, Sebat J, Wigler M, Martin CL, Ledbetter DH, Nelson SF, Cantor RM, Geschwind DH. Linkage, association, and gene-expression analyses identify CNTNAP2 as an autism-susceptibility gene. Am J Hum Genet. 2008;82:150–159. doi: 10.1016/j.ajhg.2007.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- American Psychiatric Association. Diagnostic and Statistical Manual of Mental Disorders. Washington, DC: American Psychiatric Publishing; 2013. [Google Scholar]
- Arking DE, Cutler DJ, Brune CW, Teslovich TM, West K, Ikeda M, Rea A, Guy M, Lin S, Cook EH, Chakravarti A. A common genetic variant in the neurexin superfamily member CNTNAP2 increases familial risk of autism. Am J Hum Genet. 2008;82:160–164. doi: 10.1016/j.ajhg.2007.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auerbach BD, Osterweil EK, Bear MF. Mutations causing syndromic autism define an axis of synaptic pathophysiology. Nature. 2011;480:63–68. doi: 10.1038/nature10658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auranen M, Vanhala R, Varilo T, Ayers K, Kempas E, Ylisaukko-Oja T, Sinsheimer JS, Peltonen L, Jarvela I. A genomewide screen for autism-spectrum disorders: evidence for a major susceptibility locus on chromosome 3q25-27. Am J Hum Genet. 2002;71:777–790. doi: 10.1086/342720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagni C, Greenough WT. From mRNP trafficking to spine dysmorphogenesis: the roots of fragile X syndrome. Nat Rev Neurosci. 2005;6:376–387. doi: 10.1038/nrn1667. [DOI] [PubMed] [Google Scholar]
- Baker KB, Wray SP, Ritter R, Mason S, Lanthorn TH, Savelieva KV. Male and female Fmr1 knockout mice on C57 albino background exhibit spatial learning and memory impairments. Genes Brain Behav. 2010;9:562–574. doi: 10.1111/j.1601-183X.2010.00585.x. [DOI] [PubMed] [Google Scholar]
- Bakkaloglu B, O’Roak BJ, Louvi A, Gupta AR, Abelson JF, Morgan TM, Chawarska K, Klin A, Ercan-Sencicek AG, Stillman AA, Tanriover G, Abrahams BS, Duvall JA, Robbins EM, Geschwind DH, Biederer T, Gunel M, Lifton RP, State MW. Molecular cytogenetic analysis and resequencing of contactin associated protein-like 2 in autism spectrum disorders. Am J Hum Genet. 2008;82:165–173. doi: 10.1016/j.ajhg.2007.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bang ML, Owczarek S. A matter of balance: role of neurexin and neuroligin at the synapse. Neurochem Res. 2013;38:1174–1189. doi: 10.1007/s11064-013-1029-9. [DOI] [PubMed] [Google Scholar]
- Baudouin SJ, Gaudias J, Gerharz S, Hatstatt L, Zhou K, Pun-nakkal P, Tanaka KF, Spooren W, Hen R, De Zeeuw CI, Vogt K, Scheiffele P. Shared synaptic pathophysiology in syn-dromic and nonsyndromic rodent models of autism. Science. 2012;338:128–132. doi: 10.1126/science.1224159. [DOI] [PubMed] [Google Scholar]
- Bayascas JR, Alessi DR. Regulation of Akt/PKB Ser473 phosphorylation. Mol Cell. 2005;18:143–145. doi: 10.1016/j.molcel.2005.03.020. [DOI] [PubMed] [Google Scholar]
- Berkel S, Marshall CR, Weiss B, Howe J, Roeth R, Moog U, Endris V, Roberts W, Szatmari P, Pinto D, Bonin M, Riess A, Engels H, Sprengel R, Scherer SW, Rappold GA. Mutations in the SHANK2 synaptic scaffolding gene in autism spectrum disorder and mental retardation. Nat Genet. 2010;42:489–491. doi: 10.1038/ng.589. [DOI] [PubMed] [Google Scholar]
- Berkel S, Tang W, Trevino M, Vogt M, Obenhaus HA, Gass P, Scherer SW, Sprengel R, Schratt G, Rappold GA. Inherited and de novo SHANK2 variants associated with autism spectrum disorder impair neuronal morphogenesis and physiology. Hum Mol Genet. 2012;21:344–357. doi: 10.1093/hmg/ddr470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernier R, Golzio C, Xiong B, et al. Disruptive CHD8 mutations define a subtype of autism early in development. Cell. 2014;158:263–276. doi: 10.1016/j.cell.2014.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry-Kravis E, Grossman AW, Crnic LS, Greenough WT. Understanding fragile X syndrome. Curr Paediatr. 2002;12:316–324. [Google Scholar]
- Blundell J, Tabuchi K, Bolliger MF, Blaiss CA, Brose N, Liu X, Sudhof TC, Powell CM. Increased anxiety-like behavior in mice lacking the inhibitory synapse cell adhesion molecule neuroligin 2. Genes Brain Behav. 2009;8:114–126. doi: 10.1111/j.1601-183X.2008.00455.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blundell J, Blaiss CA, Etherton MR, Espinosa F, Tabuchi K, Walz C, Bolliger MF, Sudhof TC, Powell CM. Neuroligin-1 deletion results in impaired spatial memory and increased repetitive behavior. J Neurosci. 2010;30:2115–2129. doi: 10.1523/JNEUROSCI.4517-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boccuto L, Lauri M, Sarasua SM, Skinner CD, Buccella D, Dwivedi A, Orteschi D, Collins JS, Zollino M, Visconti P, Dupont B, Tiziano D, Schroer RJ, Neri G, Stevenson RE, Gurrieri F, Schwartz CE. Prevalence of SHANK3 variants in patients with different subtypes of autism spectrum disorders. Eur J Hum Genet. 2013;21:310–316. doi: 10.1038/ejhg.2012.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeckers TM, Bockmann J, Kreutz MR, Gundelfinger ED. ProSAP/Shank proteins - a family of higher order organizing molecules of the postsynaptic density with an emerging role in human neurological disease. J Neurochem. 2002;81:903–910. doi: 10.1046/j.1471-4159.2002.00931.x. [DOI] [PubMed] [Google Scholar]
- Bolliger MF, Frei K, Winterhalter KH, Gloor SM. Identification of a novel neuroligin in humans which binds to PSD-95 and has a widespread expression. Biochem J. 2001;356:581–588. doi: 10.1042/0264-6021:3560581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourgeron T. A synaptic trek to autism. Curr Opin Neurobiol. 2009;19:231–234. doi: 10.1016/j.conb.2009.06.003. [DOI] [PubMed] [Google Scholar]
- Bozdagi O, Sakurai T, Papapetrou D, Wang X, Dickstein DL, Takahashi N, Kajiwara Y, Yang M, Katz AM, Scattoni ML, Harris MJ, Saxena R, Silverman JL, Crawley JN, Zhou Q, Hof PR, Buxbaum JD. Haploinsufficiency of the autism-associated Shank3 gene leads to deficits in synaptic function, social interaction, and social communication. Mol Autism. 2010;1:15. doi: 10.1186/2040-2392-1-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozdagi O, Tavassoli T, Buxbaum JD. Insulin-like growth factor-1 rescues synaptic and motor deficits in a mouse model of autism and developmental delay. Mol Autism. 2013;4:9. doi: 10.1186/2040-2392-4-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braunschweig D, Krakowiak P, Duncanson P, Boyce R, Hansen RL, Ashwood P, Hertz-Picciotto I, Pessah IN, Van de Water J. Autism-specific maternal autoantibodies recognize critical proteins in developing brain. Transl Psychiatry. 2013;3:e277. doi: 10.1038/tp.2013.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brimberg L, Sadiq A, Gregersen PK, Diamond B. Brain-reactive IgG correlates with autoimmunity in mothers of a child with an autism spectrum disorder. Mol Psychiatry. 2013;18:1171–1177. doi: 10.1038/mp.2013.101. [DOI] [PubMed] [Google Scholar]
- Bucan M, Abrahams BS, Wang K, et al. Genome-wide analyses of exonic copy number variants in a family-based study point to novel autism susceptibility genes. PLoS Genet. 2009;5:e1000536. doi: 10.1371/journal.pgen.1000536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budreck EC, Scheiffele P. Neuroligin-3 is a neuronal adhesion protein at GABAergic and glutamatergic synapses. Eur J Neurosci. 2007;26:1738–1748. doi: 10.1111/j.1460-9568.2007.05842.x. [DOI] [PubMed] [Google Scholar]
- Budreck EC, Kwon OB, Jung JH, Baudouin S, Thommen A, Kim HS, Fukazawa Y, Harada H, Tabuchi K, Shigemoto R, Scheiffele P, Kim JH. Neuroligin-1 controls synaptic abundance of NMDA-type glutamate receptors through extracellular coupling. Proc Natl Acad Sci USA. 2013;110:725–730. doi: 10.1073/pnas.1214718110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burket JA, Benson AD, Tang AH, Deutsch SI. D-Cycloserine improves sociability in the BTBR T+ Itpr3tf/J mouse model of autism spectrum disorders with altered Ras/Raf/ERK1/2 signaling. Brain Res Bull. 2013;96:62–70. doi: 10.1016/j.brainresbull.2013.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burket JA, Benson AD, Tang AH, Deutsch SI. Rapamycin improves sociability in the BTBR T(+)Itpr3(tf)/J mouse model of autism spectrum disorders. Brain Res Bull. 2014;100:70–75. doi: 10.1016/j.brainresbull.2013.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler MG, Dasouki MJ, Zhou XP, Talebizadeh Z, Brown M, Takahashi TN, Miles JH, Wang CH, Stratton R, Pilarski R, Eng C. Subset of individuals with autism spectrum disorders and extreme macrocephaly associated with germline PTEN tumour suppressor gene mutations. J Med Genet. 2005;42:318–321. doi: 10.1136/jmg.2004.024646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buxbaum JD, Cai G, Chaste P, Nygren G, Goldsmith J, Reichert J, Anckarsater H, Rastam M, Smith CJ, Silverman JM, Hollander E, Leboyer M, Gillberg C, Verloes A, Betancur C. Mutation screening of the PTEN gene in patients with autism spectrum disorders and macrocephaly. Am J Med Genet B Neuropsychiatr Genet. 2007;144B:484–491. doi: 10.1002/ajmg.b.30493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CDC (Centers for Disease Control and Prevention) & Developmental Disabilities Monitoring Network Surveillance Year 2010 Principal Investigators. Prevalence of autism spectrum disorder among children aged 8 years - autism and developmental disabilities monitoring network, 11 sites, United States, 2010. Morb Mortal Wkly Rep Surveill Summ. 2014;63:1–21. [PubMed] [Google Scholar]
- Chadman KK, Gong S, Scattoni ML, Boltuck SE, Gandhy SU, Heintz N, Crawley JN. Minimal aberrant behavioral phenotypes of neuroligin-3 R451C knockin mice. Autism Res. 2008;1:147–158. doi: 10.1002/aur.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheah CS, Westenbroek RE, Roden WH, Kalume F, Oakley JC, Jansen LA, Catterall WA. Correlations in timing of sodium channel expression, epilepsy, and sudden death in Dravet syndrome. Channels (Austin) 2013;7:468–472. doi: 10.4161/chan.26023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen E, Joseph S. Fragile X mental retardation protein: a paradigm for translational control by RNA-binding proteins. Biochimie. 2015;114:147–154. doi: 10.1016/j.biochi.2015.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevere-Torres I, Maki JM, Santini E, Klann E. Impaired social interactions and motor learning skills in tuberous sclerosis complex model mice expressing a dominant/negative form of tuberin. Neurobiol Dis. 2012;45:156–164. doi: 10.1016/j.nbd.2011.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chih B, Engelman H, Scheiffele P. Control of excitatory and inhibitory synapse formation by neuroligins. Science. 2005;307:1324–1328. doi: 10.1126/science.1107470. [DOI] [PubMed] [Google Scholar]
- Ching MS, Shen Y, Tan WH, et al. Deletions of NRXN1 (neurexin-1) predispose to a wide spectrum of developmental disorders. Am J Med Genet B Neuropsychiatr Genet. 2010;153B:937–947. doi: 10.1002/ajmg.b.31063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chubykin AA, Atasoy D, Etherton MR, Brose N, Kavalali ET, Gibson JR, Sudhof TC. Activity-dependent validation of excitatory versus inhibitory synapses by neuroligin-1 versus neuroligin-2. Neuron. 2007;54:919–931. doi: 10.1016/j.neuron.2007.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clifford S, Dissanayake C, Bui QM, Huggins R, Taylor AK, Loesch DZ. Autism spectrum phenotype in males and females with fragile X full mutation and premutation. J Autism Dev Disord. 2007;37:738–747. doi: 10.1007/s10803-006-0205-z. [DOI] [PubMed] [Google Scholar]
- Clipperton-Allen AE, Page DT. Pten haploinsufficient mice show broad brain overgrowth but selective impairments in autism-relevant behavioral tests. Hum Mol Genet. 2014;23:3490–3505. doi: 10.1093/hmg/ddu057. [DOI] [PubMed] [Google Scholar]
- Clipperton-Allen AE, Page DT. Decreased aggression and increased repetitive behavior in Pten haploinsufficient mice. Genes Brain Behav. 2015;14:145–157. doi: 10.1111/gbb.12192. [DOI] [PubMed] [Google Scholar]
- Coe BP, Witherspoon K, Rosenfeld JA, et al. Refining analyses of copy number variation identifies specific genes associated with developmental delay. Nat Genet. 2014;46:1063–1071. doi: 10.1038/ng.3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen IL, Sudhalter V, Pfadt A, Jenkins EC, Brown WT, Vietze PM. Why are autism and the fragile-X syndrome associated? Conceptual and methodological issues. Am J Hum Genet. 1991;48:195–202. [PMC free article] [PubMed] [Google Scholar]
- Cook EH, Jr, Scherer SW. Copy-number variations associated with neuropsychiatric conditions. Nature. 2008;455:919–923. doi: 10.1038/nature07458. [DOI] [PubMed] [Google Scholar]
- Cotney J, Muhle RA, Sanders SJ, Liu L, Willsey AJ, Niu W, Liu W, Klei L, Lei J, Yin J, Reilly SK, Tebbenkamp AT, Bichsel C, Pletikos M, Sestan N, Roeder K, State MW, Devlin B, Noonan JP. The autism-associated chromatin modifier CHD8 regulates other autism risk genes during human neurodevelopment. Nat Commun. 2015;6:6404. doi: 10.1038/ncomms7404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford DC, Acuna JM, Sherman SL. FMR1 and the fragile X syndrome: Human genome epidemiology review. Genet Med. 2001;3:359–371. doi: 10.1097/00125817-200109000-00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawley JN. Designing mouse behavioral tasks relevant to autistic-like behaviors. Ment Retard Dev Disabil Res Rev. 2004;10:248–258. doi: 10.1002/mrdd.20039. [DOI] [PubMed] [Google Scholar]
- Crawley JN. What’s Wrong With My Mouse?: Behavioral Phenotyping of Transgenic and Knockout Mice. Hoboken, NJ: John Wiley & Sons, Inc; 2007. [Google Scholar]
- Crawley JN. Translational animal models of autism and neu-rodevelopmental disorders. Dialogues Clin Neurosci. 2012;14:293–305. doi: 10.31887/DCNS.2012.14.3/jcrawley. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawley JN, Belknap JK, Collins A, Crabbe JC, Frankel W, Henderson N, Hitzemann RJ, Maxson SC, Miner LL, Silva AJ, Wehner JM, Wynshaw-Boris A, Paylor R. Behavioral phenotypes of inbred mouse strains: implications and recommendations for molecular studies. Psychopharmacology (Berl) 1997;132:107–124. doi: 10.1007/s002130050327. [DOI] [PubMed] [Google Scholar]
- Crepel A, De Wolf V, Brison N, Ceulemans B, Walleghem D, Peuteman G, Lambrechts D, Steyaert J, Noens I, Devriendt K, Peeters H. Association of CDH11 with non-syndromic ASD. Am J Med Genet B Neuropsychiatr Genet. 2014;165B:391–398. doi: 10.1002/ajmg.b.32243. [DOI] [PubMed] [Google Scholar]
- Curatolo P, Maria BL. Tuberous sclerosis. Handb Clin Neurol. 2013;111:323–331. doi: 10.1016/B978-0-444-52891-9.00038-5. [DOI] [PubMed] [Google Scholar]
- Curatolo P, Napolioni V, Moavero R. Autism spectrum disorders in tuberous sclerosis: pathogenetic pathways and implications for treatment. J Child Neurol. 2010;25:873–880. doi: 10.1177/0883073810361789. [DOI] [PubMed] [Google Scholar]
- Dahlhaus R, El-Husseini A. Altered neuroligin expression is involved in social deficits in a mouse model of the fragile X syndrome. Behav Brain Res. 2010;208:96–105. doi: 10.1016/j.bbr.2009.11.019. [DOI] [PubMed] [Google Scholar]
- De Rubeis S, He X, Goldberg AP, et al. Synaptic, transcriptional and chromatin genes disrupted in autism. Nature. 2014;515:209–215. doi: 10.1038/nature13772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delorme R, Ey E, Toro R, Leboyer M, Gillberg C, Bourgeron T. Progress toward treatments for synaptic defects in autism. Nat Med. 2013;19:685–694. doi: 10.1038/nm.3193. [DOI] [PubMed] [Google Scholar]
- Demark JL, Feldman MA, Holden JJ. Behavioral relationship between autism and fragile x syndrome. Am J Ment Retard. 2003;108:314–326. doi: 10.1352/0895-8017(2003)108<314:BRBAAF>2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- D’Hooge R, Nagels G, Franck F, Bakker CE, Reyniers E, Storm K, Kooy RF, Oostra BA, Willems PJ, De Deyn PP. Mildly impaired water maze performance in male Fmr1 knockout mice. Neuroscience. 1997;76:367–376. doi: 10.1016/s0306-4522(96)00224-2. [DOI] [PubMed] [Google Scholar]
- Diamond B, Honig G, Mader S, Brimberg L, Volpe BT. Brain-reactive antibodies and disease. Annu Rev Immunol. 2013;31:345–385. doi: 10.1146/annurev-immunol-020711-075041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Q, Sethna F, Wang H. Behavioral analysis of male and female Fmr1 knockout mice on C57BL/6 background. Behav Brain Res. 2014;271:72–78. doi: 10.1016/j.bbr.2014.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobkin C, Rabe A, Dumas R, El Idrissi A, Haubenstock H, Brown WT. Fmr1 knockout mouse has a distinctive strain-specific learning impairment. Neuroscience10. 2000;0:423–429. doi: 10.1016/s0306-4522(00)00292-x. [DOI] [PubMed] [Google Scholar]
- Drapeau E, Dorr NP, Elder GA, Buxbaum JD. Absence of strong strain effects in behavioral analyses of Shank3-deficient mice. Dis Model Mech. 2014;7:667–681. doi: 10.1242/dmm.013821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand CM, Betancur C, Boeckers TM, et al. Mutations in the gene encoding the synaptic scaffolding protein SHANK3 are associated with autism spectrum disorders. Nat Genet. 2007;39:25–27. doi: 10.1038/ng1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ecker C, Spooren W, Murphy D. Developing new pharmacotherapies for autism. J Intern Med. 2013;274:308–320. doi: 10.1111/joim.12113. [DOI] [PubMed] [Google Scholar]
- Ehninger D, Silva AJ. Rapamycin for treating tuberous sclerosis and autism spectrum disorders. Trends Mol Med. 2011;17:78–87. doi: 10.1016/j.molmed.2010.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehninger D, Han S, Shilyansky C, Zhou Y, Li W, Kwiatkowski DJ, Ramesh V, Silva AJ. Reversal of learning deficits in a Tsc2+/− mouse model of tuberous sclerosis. Nat Med. 2008;14:843–848. doi: 10.1038/nm1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehninger D, Sano Y, de Vries PJ, Dies K, Franz D, Geschwind DH, Kaur M, Lee YS, Li W, Lowe JK, Nakagawa JA, Sahin M, Smith K, Whittemore V, Silva AJ. Gestational immune activation and Tsc2 haploinsufficiency cooperate to disrupt fetal survival and may perturb social behavior in adult mice. Mol Psychiatry. 2012;17:62–70. doi: 10.1038/mp.2010.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Kordi A, Winkler D, Hammerschmidt K, Kastner A, Krueger D, Ronnenberg A, Ritter C, Jatho J, Radyushkin K, Bourgeron T, Fischer J, Brose N, Ehrenreich H. Development of an autism severity score for mice using Nlgn4 null mutants as a construct-valid model of heritable monogenic autism. Behav Brain Res. 2013;251:41–49. doi: 10.1016/j.bbr.2012.11.016. [DOI] [PubMed] [Google Scholar]
- Elsabbagh M, Divan G, Koh YJ, Kim YS, Kauchali S, Marcin C, Montiel-Nava C, Patel V, Paula CS, Wang C, Yasamy MT, Fombonne E. Global prevalence of autism and other pervasive developmental disorders. Autism Res. 2012;5:160–179. doi: 10.1002/aur.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eng C. PTEN: one gene, many syndromes. Hum Mutat. 2003;22:183–198. doi: 10.1002/humu.10257. [DOI] [PubMed] [Google Scholar]
- Etherton MR, Blaiss CA, Powell CM, Sudhof TC. Mouse neurexin-1alpha deletion causes correlated electrophysiological and behavioral changes consistent with cognitive impairments. Proc Natl Acad Sci USA. 2009;106:17998–18003. doi: 10.1073/pnas.0910297106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etherton M, Foldy C, Sharma M, Tabuchi K, Liu X, Shamloo M, Malenka RC, Sudhof TC. Autism-linked neuroligin-3 R451C mutation differentially alters hippocampal and cortical synaptic function. Proc Natl Acad Sci USA. 2011;108:13764–13769. doi: 10.1073/pnas.1111093108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ey E, Leblond CS, Bourgeron T. Behavioral profiles of mouse models for autism spectrum disorders. Autism Res. 2011;4:5–16. doi: 10.1002/aur.175. [DOI] [PubMed] [Google Scholar]
- Ey E, Yang M, Katz AM, Woldeyohannes L, Silverman JL, Leblond CS, Faure P, Torquet N, Le Sourd AM, Bourgeron T, Crawley JN. Absence of deficits in social behaviors and ultrasonic vocalizations in later generations of mice lacking neuroligin4. Genes Brain Behav. 2012;11:928–941. doi: 10.1111/j.1601-183X.2012.00849.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J, Schroer R, Yan J, Song W, Yang C, Bockholt A, Cook EH, Jr, Skinner C, Schwartz CE, Sommer SS. High frequency of neurexin 1beta signal peptide structural variants in patients with autism. Neurosci Lett. 2006;409:10–13. doi: 10.1016/j.neulet.2006.08.017. [DOI] [PubMed] [Google Scholar]
- Fischer J, Hammerschmidt K. Ultrasonic vocalizations in mouse models for speech and socio-cognitive disorders: insights into the evolution of vocal communication. Genes Brain Behav. 2011;10:17–27. doi: 10.1111/j.1601-183X.2010.00610.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foley AG, Gannon S, Rombach-Mullan N, Prendergast A, Barry C, Cassidy AW, Regan CM. Class I histone deacetylase inhibition ameliorates social cognition and cell adhesion molecule plasticity deficits in a rodent model of autism spectrum disorder. Neuropharmacology. 2012;63:750–760. doi: 10.1016/j.neuropharm.2012.05.042. [DOI] [PubMed] [Google Scholar]
- Fombonne E. Epidemiology of pervasive developmental disorders. Pediatr Res. 2009;65:591–598. doi: 10.1203/PDR.0b013e31819e7203. [DOI] [PubMed] [Google Scholar]
- Fombonne E, Du Mazaubrun C, Cans C, Grandjean H. Autism and associated medical disorders in a French epidemiological survey. J Am Acad Child Adolesc Psychiatry. 1997;36:1561–1569. doi: 10.1016/S0890-8567(09)66566-7. [DOI] [PubMed] [Google Scholar]
- Frazier TW, Embacher R, Tilot AK, Koenig K, Mester J, Eng C. Molecular and phenotypic abnormalities in individuals with germline heterozygous PTEN mutations and autism. Mol Psychiatry. 2014;20:1132–1138. doi: 10.1038/mp.2014.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaugler T, Klei L, Sanders SJ, Bodea CA, Goldberg AP, Lee AB, Mahajan M, Manaa D, Pawitan Y, Reichert J, Ripke S, Sandin S, Sklar P, Svantesson O, Reichenberg A, Hultman CM, Devlin B, Roeder K, Buxbaum JD. Most genetic risk for autism resides with common variation. Nat Genet. 2014;46:881–885. doi: 10.1038/ng.3039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauthier J, Spiegelman D, Piton A, Lafreniere RG, Laurent S, St-Onge J, Lapointe L, Hamdan FF, Cossette P, Mot-tron L, Fombonne E, Joober R, Marineau C, Drapeau P, Rouleau GA. Novel de novo SHANK3 mutation in autistic patients. Am J Med Genet B Neuropsychiatr Genet. 2009;150B:421–424. doi: 10.1002/ajmg.b.30822. [DOI] [PubMed] [Google Scholar]
- Gauthier J, Champagne N, Lafreniere RG, et al. De novo mutations in the gene encoding the synaptic scaffolding protein SHANK3 in patients ascertained for schizophrenia. Proc Natl Acad Sci USA. 2010;107:7863–7868. doi: 10.1073/pnas.0906232107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geschwind DH, State MW. Gene hunting in autism spectrum disorder: on the path to precision medicine. Lancet Neurol. 2015 doi: 10.1016/S1474-4422(15)00044-7. http://dx.doi.org/10.1016/S1474-4422(15)00044-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gholizadeh S, Arsenault J, Xuan IC, Pacey LK, Hampson DR. Reduced phenotypic severity following adeno-associated virus-mediated Fmr1 gene delivery in fragile X mice. Neuropsychopharmacology. 2014;39:3100–3111. doi: 10.1038/npp.2014.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gkogkas CG, Khoutorsky A, Ran I, Rampakakis E, Nevarko T, Weatherill DB, Vasuta C, Yee S, Truitt M, Dallaire P, Major F, Lasko P, Ruggero D, Nader K, Lacaille JC, Sonenberg N. Autism-related deficits via dysregulated eIF4E-dependent translational control. Nature. 2013;493:371–377. doi: 10.1038/nature11628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glessner JT, Wang K, Cai G, et al. Autism genome-wide copy number variation reveals ubiquitin and neuronal genes. Nature. 2009;459:569–573. doi: 10.1038/nature07953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goorden SM, van Woerden GM, van der Weerd L, Cheadle JP, Elgersma Y. Cognitive deficits in Tsc1+/− mice in the absence of cerebral lesions and seizures. Ann Neurol. 2007;62:648–655. doi: 10.1002/ana.21317. [DOI] [PubMed] [Google Scholar]
- Grabrucker AM, Schmeisser MJ, Schoen M, Boeckers TM. Postsynaptic ProSAP/Shank scaffolds in the cross-hair of synaptopathies. Trends Cell Biol. 2011;21:594–603. doi: 10.1016/j.tcb.2011.07.003. [DOI] [PubMed] [Google Scholar]
- Grayton HM, Missler M, Collier DA, Fernandes C. Altered social behaviours in neurexin 1alpha knockout mice resemble core symptoms in neurodevelopmental disorders. PLoS One. 2013;8:e67114. doi: 10.1371/journal.pone.0067114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregor A, Albrecht B, Bader I, et al. Expanding the clinical spectrum associated with defects in CNTNAP2 and NRXN1. BMC Med Genet. 2011;12:106. doi: 10.1186/1471-2350-12-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallmayer J, Cleveland S, Torres A, Phillips J, Cohen B, Torigoe T, Miller J, Fedele A, Collins J, Smith K, Lotspeich L, Croen LA, Ozonoff S, Lajonchere C, Grether JK, Risch N. Genetic heritability and shared environmental factors among twin pairs with autism. Arch Gen Psychiatr y. 2011;68:1095–1102. doi: 10.1001/archgenpsychiatry.2011.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han S, Tai C, Westenbroek RE, Yu FH, Cheah CS, Potter GB, Rubenstein JL, Scheuer T, de la Iglesia HO, Catter-all WA. Autistic-like behaviour in Scn1a+/− mice and rescue by enhanced GABA-mediated neurotransmission. Nature. 2012;489:385–390. doi: 10.1038/nature11356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han S, Tai C, Jones CJ, Scheuer T, Catterall WA. Enhancement of inhibitory neurotransmission by GABAA receptors having alpha2,3-subunits ameliorates behavioral deficits in a mouse model of autism. Neuron. 2014;81:1282–1289. doi: 10.1016/j.neuron.2014.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatton DD, Sideris J, Skinner M, Mankowski J, Bailey DB, Jr, Roberts J, Mirrett P. Autistic behavior in children with fragile X syndrome: prevalence, stability, and the impact of FMRP. Am J Med Genet A. 2006;140a:1804–1813. doi: 10.1002/ajmg.a.31286. [DOI] [PubMed] [Google Scholar]
- Henderson C, Wijetunge L, Kinoshita MN, Shumway M, Hammond RS, Postma FR, Brynczka C, Rush R, Thomas A, Pay-lor R, Warren ST, Vanderklish PW, Kind PC, Carpenter RL, Bear MF, Healy AM. Reversal of disease-related pathologies in the fragile X mouse model by selective activation of GABAB receptors with arbaclofen. Sci Transl Med. 2012;4:152ra128. doi: 10.1126/scitranslmed.3004218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman GE, Butter E, Enrile B, Pastore M, Prior TW, Som-mer A. Increasing knowledge of PTEN germline mutations: two additional patients with autism and macrocephaly. Am J Med Genet A. 2007;143a:589–593. doi: 10.1002/ajmg.a.31619. [DOI] [PubMed] [Google Scholar]
- Hershey JW, Sonenberg N, Mathews MB. Principles of translational control: an overview. Cold Spring Harb Perspect Biol. 2012;4:a011528. doi: 10.1101/cshperspect.a011528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu X, Luo JH, Xu J. The interplay between synaptic activity and neuroligin function in the CNS. Biomed Res Int. 2015;2015:498957. doi: 10.1155/2015/498957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Manning BD. A complex interplay between Akt, TSC2, and the two mTOR complexes. Biochem Soc Trans. 2009;37:217–222. doi: 10.1042/BST0370217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H, Michetti C, Busnelli M, Manago F, Sannino S, Scheg-gia D, Giancardo L, Sona D, Murino V, Chini B, Scattoni ML, Papaleo F. Chronic and acute intranasal oxytocin produce divergent social effects in mice. Neuropsychopharmacology. 2014;39:1102–1114. doi: 10.1038/npp.2013.310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung AY, Futai K, Sala C, Valtschanoff JG, Ryu J, Woodworth MA, Kidd FL, Sung CC, Miyakawa T, Bear MF, Weinberg RJ, Sheng M. Smaller dendritic spines, weaker synaptic transmission, but enhanced spatial learning in mice lacking Shank1. J Neurosci. 2008;28:1697–1708. doi: 10.1523/JNEUROSCI.3032-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iossifov I, O’Roak BJ, Sanders SJ, et al. The contribution of de novo coding mutations to autism spectrum disorder. Nature. 2014;515:216–221. doi: 10.1038/nature13908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irie M, Hata Y, Takeuchi M, Ichtchenko K, Toyoda A, Hirao K, Takai Y, Rosahl TW, Sudhof TC. Binding of neuroligins to PSD-95. Science. 1997;277:1511–1515. doi: 10.1126/science.277.5331.1511. [DOI] [PubMed] [Google Scholar]
- Jamain S, Quach H, Betancur C, Rastam M, Colineaux C, Gillberg IC, Soderstrom H, Giros B, Leboyer M, Gillberg C, Bourgeron T. Mutations of the X-linked genes encoding neuroligins NLGN3 and NLGN4 are associated with autism. Nat Genet. 2003;34:27–29. doi: 10.1038/ng1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamain S, Radyushkin K, Hammerschmidt K, Granon S, Boretius S, Varoqueaux F, Ramanantsoa N, Gallego J, Ronnenberg A, Winter D, Frahm J, Fischer J, Bourgeron T, Ehrenreich H, Brose N. Reduced social interaction and ultrasonic communication in a mouse model of monogenic heritable autism. Proc Natl Acad Sci USA. 2008;105:1710–1715. doi: 10.1073/pnas.0711555105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeste SS, Geschwind DH. Disentangling the heterogeneity of autism spectrum disorder through genetic findings. Nat Rev Neurol. 2014;10:74–81. doi: 10.1038/nrneurol.2013.278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang YH, Ehlers MD. Modeling autism by SHANK gene mutations in mice. Neuron. 2013;78:8–27. doi: 10.1016/j.neuron.2013.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kas MJ, Glennon JC, Buitelaar J, Ey E, Biemans B, Crawley J, Ring RH, Lajonchere C, Esclassan F, Talpos J, Noldus LP, Burbach JP, Steckler T. Assessing behavioural and cognitive domains of autism spectrum disorders in rodents: current status and future perspectives. Psychopharmacology (Berl) 2014;231:1125–1146. doi: 10.1007/s00213-013-3268-5. [DOI] [PubMed] [Google Scholar]
- Kaufmann WE, Cortell R, Kau AS, Bukelis I, Tierney E, Gray RM, Cox C, Capone GT, Stanard P. Autism spectrum disorder in fragile X syndrome: communication, social interaction, and specific behaviors. Am J Med Genet A. 2004;129a:225–234. doi: 10.1002/ajmg.a.30229. [DOI] [PubMed] [Google Scholar]
- Kazdoba TM, Leach PT, Silverman JL, Crawley JN. Modeling fragile X syndrome in the Fmr1 knockout mouse. Intractable Rare Dis Res. 2014;3:118–133. doi: 10.5582/irdr.2014.01024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E, Niethammer M, Rothschild A, Jan YN, Sheng M. Clustering of Shaker-type K+ channels by interaction with a family of membrane-associated guanylate kinases. Nature. 1995;378:85–88. doi: 10.1038/378085a0. [DOI] [PubMed] [Google Scholar]
- Kim J, Jung SY, Lee YK, Park S, Choi JS, Lee CJ, Kim HS, Choi YB, Scheiffele P, Bailey CH, Kandel ER, Kim JH. Neuroligin-1 is required for normal expression of LTP and associative fear memory in the amygdala of adult animals. Proc Natl Acad Sci USA. 2008;105:9087–9092. doi: 10.1073/pnas.0803448105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YS, Leventhal BL, Koh YJ, Fombonne E, Laska E, Lim EC, Cheon KA, Kim SJ, Kim YK, Lee H, Song DH, Grinker RR. Prevalence of autism spectrum disorders in a total population sample. Am J Psychiatry. 2011;168:904–912. doi: 10.1176/appi.ajp.2011.10101532. [DOI] [PubMed] [Google Scholar]
- Kolevzon A, Angarita B, Bush L, Wang AT, Frank Y, Yang A, Rapaport R, Saland J, Srivastava S, Farrell C, Edelmann LJ, Buxbaum JD. Phelan-McDermid syndrome: a review of the literature and practice parameters for medical assessment and monitoring. J Neurodev Disord. 2014;6:39. doi: 10.1186/1866-1955-6-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong A, Frigge ML, Masson G, Besenbacher S, Sulem P, Magnusson G, Gudjonsson SA, Sigurdsson A, Jonasdottir A, Wong WS, Sigurdsson G, Walters GB, Steinberg S, Helgason H, Thorleifsson G, Gudbjartsson DF, Helgason A, Magnusson OT, Thorsteinsdottir U, Stefansson K. Rate of de novo mutations and the importance of father’s age to disease risk. Nature. 2012;488:471–475. doi: 10.1038/nature11396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kooy RF, D’Hooge R, Reyniers E, Bakker CE, Nagels G, De Boulle K, Storm K, Clincke G, De Deyn PP, Oostra BA, Willems PJ. Transgenic mouse model for the fragile X syndrome. Am J Med Genet. 1996;64:241–245. doi: 10.1002/(SICI)1096-8628(19960809)64:2<241::AID-AJMG1>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- Kornau HC, Schenker LT, Kennedy MB, Seeburg PH. Domain interaction between NMDA receptor subunits and the postsynaptic density protein PSD-95. Science. 1995;269:1737–1740. doi: 10.1126/science.7569905. [DOI] [PubMed] [Google Scholar]
- Kouser M, Speed HE, Dewey CM, Reimers JM, Widman AJ, Gupta N, Liu S, Jaramillo TC, Bangash M, Xiao B, Worley PF, Powell CM. Loss of predominant Shank3 isoforms results in hippocampus-dependent impairments in behavior and synaptic transmission. J Neurosci. 2013;33:18448–18468. doi: 10.1523/JNEUROSCI.3017-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krey JF, Pasca SP, Shcheglovitov A, Yazawa M, Schwemberger R, Rasmusson R, Dolmetsch RE. Timothy syndrome is associated with activity-dependent dendritic retraction in rodent and human neurons. Nat Neurosci. 2013;16:201–209. doi: 10.1038/nn.3307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krueger DD, Tuffy LP, Papadopoulos T, Brose N. The role of neurexins and neuroligins in the formation, maturation, and function of vertebrate synapses. Curr Opin Neurobiol. 2012;22:412–422. doi: 10.1016/j.conb.2012.02.012. [DOI] [PubMed] [Google Scholar]
- Krumm N, Turner TN, Baker C, Vives L, Mohajeri K, Wither-spoon K, Raja A, Coe BP, Stessman HA, He ZX, Leal SM, Bernier R, Eichler EE. Excess of rare, inherited truncating mutations in autism. Nat Genet. 2015;47:582–588. doi: 10.1038/ng.3303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar RA, Marshall CR, Badner JA, Babatz TD, Mukamel Z, Aldinger KA, Sudi J, Brune CW, Goh G, Karamohamed S, Sutcliffe JS, Cook EH, Geschwind DH, Dobyns WB, Scherer SW, Christian SL. Association and mutation analyses of 16p11.2 autism candidate genes. PLoS One. 2009;4:e4582. doi: 10.1371/journal.pone.0004582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon CH, Luikart BW, Powell CM, Zhou J, Matheny SA, Zhang W, Li Y, Baker SJ, Parada LF. Pten regulates neuronal arborization and social interaction in mice. Neuron. 2006;50:377–388. doi: 10.1016/j.neuron.2006.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laplante M, Sabatini DM. mTOR signaling in growth control and disease. Cell. 2012;149:274–293. doi: 10.1016/j.cell.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laumonnier F, Bonnet-Brilhault F, Gomot M, Blanc R, David A, Moizard MP, Raynaud M, Ronce N, Lemonnier E, Calvas P, Laudier B, Chelly J, Fryns JP, Ropers HH, Hamel BC, Andres C, Barthelemy C, Moraine C, Briault S. X-linked mental retardation and autism are associated with a mutation in the NLGN4 gene, a member of the neuroligin family. Am J Hum Genet. 2004;74:552–557. doi: 10.1086/382137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauterborn JC, Rex CS, Kramar E, Chen LY, Pandyarajan V, Lynch G, Gall CM. Brain-derived neurotrophic factor rescues synaptic plasticity in a mouse model of fragile X syndrome. J Neurosci. 2007;27:10685–10694. doi: 10.1523/JNEUROSCI.2624-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson-Yuen A, Saldivar JS, Sommer S, Picker J. Familial deletion within NLGN4 associated with autism and Tourette syndrome. Eur J Hum Genet. 2008;16:614–618. doi: 10.1038/sj.ejhg.5202006. [DOI] [PubMed] [Google Scholar]
- Leblond CS, Nava C, Polge A, et al. Meta-analysis of SHANK Mutations in Autism Spectrum Disorders: a gradient of severity in cognitive impairments. PLoS Genet. 2014;10:e1004580. doi: 10.1371/journal.pgen.1004580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levitt P, Veenstra-VanderWeele J. Neurodevelopment and the origins of brain disorders. Neuropsychopharmacology. 2015;40:1–3. doi: 10.1038/npp.2014.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Shi M, Ma Z, Zhao S, Euskirchen G, Ziskin J, Urban A, Hallmayer J, Snyder M. Integrated systems analysis reveals a molecular network underlying autism spectrum disorders. Mol Syst Biol. 2014;10:774. doi: 10.15252/msb.20145487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lightbody AA, Reiss AL. Gene, brain, and behavior relationships in fragile X syndrome: evidence from neuroimaging studies. Dev Disabil Res Rev. 2009;15:343–352. doi: 10.1002/ddrr.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim S, Naisbitt S, Yoon J, Hwang JI, Suh PG, Sheng M, Kim E. Characterization of the Shank family of synaptic proteins. Multiple genes, alternative splicing, and differential expression in brain and development. J Biol Chem. 1999;274:29510–29518. doi: 10.1074/jbc.274.41.29510. [DOI] [PubMed] [Google Scholar]
- Liu ZH, Smith CB. Dissociation of social and nonsocial anxiety in a mouse model of fragile X syndrome. Neurosci Lett. 2009;454:62–66. doi: 10.1016/j.neulet.2009.02.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lord C, Bishop SL. Recent advances in autism research as reflected in DSM-5 criteria for autism spectrum disorder. Annu Rev Clin Psychol. 2015;11:53–70. doi: 10.1146/annurev-clinpsy-032814-112745. [DOI] [PubMed] [Google Scholar]
- Lord C, Jones RM. New strategies and findings for behavioral interventions in autism spectrum disorders. Ann N Y Acad Sci. 2013;1304:70–76. doi: 10.1111/nyas.12311. [DOI] [PubMed] [Google Scholar]
- Maehama T, Dixon JE. The tumor suppressor, PTEN/MMAC1, dephosphorylates the lipid second messenger, phosphatidylinositol 3,4,5-trisphosphate. J Biol Chem. 1998;273:13375–13378. doi: 10.1074/jbc.273.22.13375. [DOI] [PubMed] [Google Scholar]
- Marshall CR, Noor A, Vincent JB, et al. Structural variation of chromosomes in autism spectrum disorder. Am J Hum Genet. 2008;82:477–488. doi: 10.1016/j.ajhg.2007.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride KL, Varga EA, Pastore MT, Prior TW, Manickam K, Atkin JF, Herman GE. Confirmation study of PTEN mutations among individuals with autism or developmental delays/mental retardation and macrocephaly. Autism Res. 2010;3:137–141. doi: 10.1002/aur.132. [DOI] [PubMed] [Google Scholar]
- McFarlane HG, Kusek GK, Yang M, Phoenix JL, Bolivar VJ, Crawley JN. Autism-like behavioral phenotypes in BTBR T+tf/J mice. Genes Brain Behav. 2008;7:152–163. doi: 10.1111/j.1601-183X.2007.00330.x. [DOI] [PubMed] [Google Scholar]
- McNaughton CH, Moon J, Strawderman MS, Maclean KN, Evans J, Strupp BJ. Evidence for social anxiety and impaired social cognition in a mouse model of fragile X syndrome. Behav Neurosci. 2008;122:293–300. doi: 10.1037/0735-7044.122.2.293. [DOI] [PubMed] [Google Scholar]
- Meyer G, Varoqueaux F, Neeb A, Oschlies M, Brose N. The complexity of PDZ domain-mediated interactions at gluta-matergic synapses: a case study on neuroligin. Neuropharmacol-ogy. 2004;47:724–733. doi: 10.1016/j.neuropharm.2004.06.023. [DOI] [PubMed] [Google Scholar]
- Meziane H, Schaller F, Bauer S, Villard C, Matarazzo V, Riet F, Guillon G, Lafitte D, Desarmenien MG, Tauber M, Mus-catelli F. An early postnatal oxytocin treatment prevents social and learning deficits in adult mice deficient for Magel2, a gene involved in Prader-Willi syndrome and autism. Biol Psychiatry. 2014;78:85–94. doi: 10.1016/j.biopsych.2014.11.010. [DOI] [PubMed] [Google Scholar]
- Michaelson JJ, Shi Y, Gujral M, et al. Whole-genome sequencing in autism identifies hot spots for de novo germline mutation. Cell. 2012;151:1431–1442. doi: 10.1016/j.cell.2012.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalon A, Sidorov M, Ballard TM, Ozmen L, Spooren W, Wettstein JG, Jaeschke G, Bear MF, Lindemann L. Chronic pharmacological mGlu5 inhibition corrects fragile X in adult mice. Neuron. 2012;74:49–56. doi: 10.1016/j.neuron.2012.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miles JH. Autism spectrum disorders--a genetics review. Genet Med. 2011;13:278–294. doi: 10.1097/GIM.0b013e3181ff67ba. [DOI] [PubMed] [Google Scholar]
- Mineur YS, Huynh LX, Crusio WE. Social behavior deficits in the Fmr1 mutant mouse. Behav Brain Res. 2006;168:172–175. doi: 10.1016/j.bbr.2005.11.004. [DOI] [PubMed] [Google Scholar]
- Modi ME, Young LJ. The oxytocin system in drug discovery for autism: animal models and novel therapeutic strategies. Horm Behav. 2012;61:340–350. doi: 10.1016/j.yhbeh.2011.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moessner R, Marshall CR, Sutcliffe JS, Skaug J, Pinto D, Vincent J, Zwaigenbaum L, Fernandez B, Roberts W, Szatmari P, Scherer SW. Contribution of SHANK3 mutations to autism spectrum disorder. Am J Hum Genet. 2007;81:1289–1297. doi: 10.1086/522590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moldrich RX, Leanage G, She D, Dolan-Evans E, Nelson M, Reza N, Reutens DC. Inhibition of histone deacetylase in utero causes sociability deficits in postnatal mice. Behav Brain Res. 2013;257:253–264. doi: 10.1016/j.bbr.2013.09.049. [DOI] [PubMed] [Google Scholar]
- Morrow EM. Genomic copy number variation in disorders of cognitive development. J Am Acad Child Adolesc Psychiatry. 2010;49:1091–1104. doi: 10.1016/j.jaac.2010.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moy SS, Nadler JJ, Perez A, Barbaro RP, Johns JM, Mag-nuson TR, Piven J, Crawley JN. Sociability and preference for social novelty in five inbred strains: an approach to assess autistic-like behavior in mice. Genes Brain Behav. 2004;3:287–302. doi: 10.1111/j.1601-1848.2004.00076.x. [DOI] [PubMed] [Google Scholar]
- Moy SS, Nadler JJ, Poe MD, Nonneman RJ, Young NB, Koller BH, Crawley JN, Duncan GE, Bodfish JW. Development of a mouse test for repetitive, restricted behaviors: relevance to autism. Behav Brain Res. 2008a;188:178–194. doi: 10.1016/j.bbr.2007.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moy SS, Nadler JJ, Young NB, Nonneman RJ, Segall SK, Andrade GM, Crawley JN, Magnuson TR. Social approach and repetitive behavior in eleven inbred mouse strains. Behav Brain Res. 2008b;191:118–129. doi: 10.1016/j.bbr.2008.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moy SS, Nadler JJ, Young NB, Nonneman RJ, Grossman AW, Murphy DL, D’Ercole AJ, Crawley JN, Magnuson TR, Lauder JM. Social approach in genetically engineered mouse lines relevant to autism. Genes Brain Behav. 2009;8:129–142. doi: 10.1111/j.1601-183X.2008.00452.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murdoch JD, Gupta AR, Sanders SJ, Walker MF, Keaney J, Fernandez TV, Murtha MT, Anyanwu S, Ober GT, Raubeson MJ, DiLullo NM, Villa N, Waqar Z, Sullivan C, Gonzalez L, Willsey AJ, Choe SY, Neale BM, Daly MJ, State MW. No evidence for association of autism with rare heterozygous point mutations in contactin-associated protein-like 2 (CNTNAP2), or in other contactin-associated proteins or contactins. PLoS Genet. 2015;11:e1004852. doi: 10.1371/journal.pgen.1004852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadler JJ, Moy SS, Dold G, Trang D, Simmons N, Perez A, Young NB, Barbaro RP, Piven J, Magnuson TR, Crawley JN. Automated apparatus for quantitation of social approach behaviors in mice. Genes Brain Behav. 2004;3:303–314. doi: 10.1111/j.1601-183X.2004.00071.x. [DOI] [PubMed] [Google Scholar]
- Naisbitt S, Kim E, Tu JC, Xiao B, Sala C, Valtschanoff J, Weinberg RJ, Worley PF, Sheng M. Shank, a novel family of postsynaptic density proteins that binds to the NMDA receptor/PSD-95/GKAP complex and cortactin. Neuron. 1999;23:569–582. doi: 10.1016/s0896-6273(00)80809-0. [DOI] [PubMed] [Google Scholar]
- Narayanan U, Nalavadi V, Nakamoto M, Thomas G, Ceman S, Bassell GJ, Warren ST. S6K1 phosphorylates and regulates fragile X mental retardation protein (FMRP) with the neuronal protein synthesis-dependent mammalian target of rapamycin (mTOR) signaling cascade. J Biol Chem. 2008;283:18478–18482. doi: 10.1074/jbc.C800055200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neale BM, Kou Y, Liu L, et al. Patterns and rates of exonic de novo mutations in autism spectrum disorders. Nature. 2012;485:242–245. doi: 10.1038/nature11011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niethammer M, Kim E, Sheng M. Interaction between the C terminus of NMDA receptor subunits and multiple members of the PSD-95 family of membrane-associated guanylate kinases. J Neurosci. 1996;16:2157–2163. doi: 10.1523/JNEUROSCI.16-07-02157.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordenbaek C, Jorgensen M, Kyvik KO, Bilenberg N. A Danish population-based twin study on autism spectrum disorders. Eur Child Adolesc Psychiatry. 2014;23:35–43. doi: 10.1007/s00787-013-0419-5. [DOI] [PubMed] [Google Scholar]
- Ogawa S, Kwon CH, Zhou J, Koovakkattu D, Parada LF, Sinton CM. A seizure-prone phenotype is associated with altered free-running rhythm in Pten mutant mice. Brain Res. 2007;1168:112–123. doi: 10.1016/j.brainres.2007.06.074. [DOI] [PubMed] [Google Scholar]
- O’Roak BJ, Deriziotis P, Lee C, Vives L, Schwartz JJ, Girirajan S, Karakoc E, Mackenzie AP, Ng SB, Baker C, Rieder MJ, Nickerson DA, Bernier R, Fisher SE, Shendure J, Eichler EE. Exome sequencing in sporadic autism spectrum disorders identifies severe de novo mutations. Nat Genet. 2011;43:585–589. doi: 10.1038/ng.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Roak BJ, Vives L, Fu W, et al. Multiplex targeted sequencing identifies recurrently mutated genes in autism spectrum disorders. Science. 2012;338:1619–1622. doi: 10.1126/science.1227764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page DT, Kuti OJ, Prestia C, Sur M. Haploinsufficiency for Pten and Serotonin transporter cooperatively influences brain size and social behavior. Proc Natl Acad Sci USA. 2009;106:1989–1994. doi: 10.1073/pnas.0804428106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paradee W, Melikian HE, Rasmussen DL, Kenneson A, Conn PJ, Warren ST. Fragile X mouse: strain effects of knockout phenotype and evidence suggesting deficient amygdala function. Neuroscience. 1999;94:185–192. doi: 10.1016/s0306-4522(99)00285-7. [DOI] [PubMed] [Google Scholar]
- Parikshak NN, Luo R, Zhang A, Won H, Lowe JK, Chandran V, Horvath S, Geschwind DH. Integrative functional genomic analyses implicate specific molecular pathways and circuits in autism. Cell. 2013;155:1008–1021. doi: 10.1016/j.cell.2013.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peca J, Feliciano C, Ting JT, Wang W, Wells MF, Venkatraman TN, Lascola CD, Fu Z, Feng G. Shank3 mutant mice display autistic-like behaviours and striatal dysfunction. Nature. 2011;472:437–442. doi: 10.1038/nature09965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peier AM, McIlwain KL, Kenneson A, Warren ST, Paylor R, Nelson DL. (Over)correction of FMR1 deficiency with YAC transgenics: behavioral and physical features. Hum Mol Genet. 2000;9:1145–1159. doi: 10.1093/hmg/9.8.1145. [DOI] [PubMed] [Google Scholar]
- Penagarikano O, Abrahams BS, Herman EI, Winden KD, Gdalyahu A, Dong H, Sonnenblick LI, Gruver R, Almajano J, Bragin A, Golshani P, Trachtenberg JT, Peles E, Geschwind DH. Absence of CNTNAP2 leads to epilepsy, neuronal migration abnormalities, and core autism-related deficits. Cell. 2011;147:235–246. doi: 10.1016/j.cell.2011.08.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penagarikano O, Lazaro MT, Lu XH, Gordon A, Dong H, Lam HA, Peles E, Maidment NT, Murphy NP, Yang XW, Golshani P, Geschwind DH. Exogenous and evoked oxytocin restores social behavior in the Cntnap2 mouse model of autism. Sci Transl Med. 2015;7:271ra278. doi: 10.1126/scitranslmed.3010257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phelan K, McDermid HE. The 22q13.3 deletion syndrome (Phelan-McDermid Syndrome) Mol Syndromol. 2012;2:186–201. doi: 10.1159/000334260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippe A, Martinez M, Guilloud-Bataille M, Gillberg C, Rastam M, Sponheim E, Coleman M, Zappella M, Aschauer H, Van Maldergem L, Penet C, Feingold J, Brice A, Leboyer M. Genome-wide scan for autism susceptibility genes. Paris Autism Research International Sibpair Study. Hum Mol Genet. 1999;8:805–812. doi: 10.1093/hmg/8.5.805. [DOI] [PubMed] [Google Scholar]
- Pietropaolo S, Guilleminot A, Martin B, D’Amato FR, Cru-sio WE. Genetic-background modulation of core and variable autistic-like symptoms in Fmr1 knock-out mice. PLoS One. 2011;6:e17073. doi: 10.1371/journal.pone.0017073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto D, Pagnamenta AT, Klei L, et al. Functional impact of global rare copy number variation in autism spectrum disorders. Nature. 2010;466:368–372. doi: 10.1038/nature09146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto D, Delaby E, Merico D, et al. Convergence of genes and cellular pathways dysregulated in autism spectrum disorders. Am J Hum Genet. 2014;94:677–694. doi: 10.1016/j.ajhg.2014.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piras IS, Haapanen L, Napolioni V, Sacco R, Van de Water J, Persico AM. Anti-brain antibodies are associated with more severe cognitive and behavioral profiles in Italian children with Autism Spectrum Disorder. Brain Behav Immun. 2014;38:91–99. doi: 10.1016/j.bbi.2013.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poliak S, Gollan L, Martinez R, Custer A, Einheber S, Salzer JL, Trimmer JS, Shrager P, Peles E. Caspr2, a new member of the neurexin superfamily, is localized at the juxtaparanodes of myelinated axons and associates with K+ channels. Neuron. 1999;24:1037–1047. doi: 10.1016/s0896-6273(00)81049-1. [DOI] [PubMed] [Google Scholar]
- Poliak S, Salomon D, Elhanany H, Sabanay H, Kiernan B, Pevny L, Stewart CL, Xu X, Chiu SY, Shrager P, Furley AJ, Peles E. Juxtaparanodal clustering of Shaker-like K+ channels in myelinated axons depends on Caspr2 and TAG-1. J Cell Biol. 2003;162:1149–1160. doi: 10.1083/jcb.200305018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poot M, Beyer V, Schwaab I, Damatova N, Van’t Slot R, Prothero J, Holder SE, Haaf T. Disruption of CNTNAP2 and additional structural genome changes in a boy with speech delay and autism spectrum disorder. Neurogenetics. 2010;11:81–89. doi: 10.1007/s10048-009-0205-1. [DOI] [PubMed] [Google Scholar]
- Radyushkin K, Hammerschmidt K, Boretius S, Varoqueaux F, El-Kordi A, Ronnenberg A, Winter D, Frahm J, Fischer J, Brose N, Ehrenreich H. Neuroligin-3-deficient mice: model of a monogenic heritable form of autism with an olfactory deficit. Genes Brain Behav. 2009;8:416–425. doi: 10.1111/j.1601-183X.2009.00487.x. [DOI] [PubMed] [Google Scholar]
- Reith RM, McKenna J, Wu H, Hashmi SS, Cho SH, Dash PK, Gambello MJ. Loss of Tsc2 in Purkinje cells is associated with autistic-like behavior in a mouse model of tuberous sclerosis complex. Neurobiol Dis. 2013;51:93–103. doi: 10.1016/j.nbd.2012.10.014. [DOI] [PubMed] [Google Scholar]
- Ritvo ER, Freeman BJ, Mason-Brothers A, Mo A, Ritvo AM. Concordance for the syndrome of autism in 40 pairs of afflicted twins. Am J Psychiatry. 1985;142:74–77. doi: 10.1176/ajp.142.1.74. [DOI] [PubMed] [Google Scholar]
- Rodenas-Cuadrado P, Ho J, Vernes SC. Shining a light on CNTNAP2: complex functions to complex disorders. Eur J Hum Genet. 2014;22:171–178. doi: 10.1038/ejhg.2013.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers SJ, Wehner DE, Hagerman R. The behavioral phenotype in fragile X: symptoms of autism in very young children with fragile X syndrome, idiopathic autism, and other developmental disorders. J Dev Behav Pediatr. 2001;22:409–417. doi: 10.1097/00004703-200112000-00008. [DOI] [PubMed] [Google Scholar]
- Rogers SJ, Estes A, Lord C, Vismara L, Winter J, Fitzpatrick A, Guo M, Dawson G. Effects of a brief Early Start Denver model (ESDM)-based parent intervention on toddlers at risk for autism spectrum disorders: a randomized controlled trial. J Am Acad Child Adolesc Psychiatry. 2012;51:1052–1065. doi: 10.1016/j.jaac.2012.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosander C, Hallbook T. Dravet syndrome in Sweden: a population-based study. Dev Med Child Neurol. 2015;57:628–633. doi: 10.1111/dmcn.12709. [DOI] [PubMed] [Google Scholar]
- Rossi E, Verri AP, Patricelli MG, Destefani V, Ricca I, Vetro A, Ciccone R, Giorda R, Toniolo D, Maraschio P, Zuffardi O. A 12 Mb deletion at 7q33-q35 associated with autism spectrum disorders and primary amenorrhea. Eur J Med Genet. 2008;51:631–638. doi: 10.1016/j.ejmg.2008.06.010. [DOI] [PubMed] [Google Scholar]
- Roullet FI, Crawley JN. Mouse models of autism: testing hypotheses about molecular mechanisms. Curr Top Behav Neurosci. 2011;7:187–212. doi: 10.1007/7854_2010_113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampath S, Bhat S, Gupta S, O’Connor A, West AB, Arking DE, Chakravarti A. Defining the contribution of CNTNAP2 to autism susceptibility. PLoS One. 2013;8:e77906. doi: 10.1371/journal.pone.0077906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandin S, Lichtenstein P, Kuja-Halkola R, Larsson H, Hultman CM, Reichenberg A. The familial risk of autism. JAMA. 2014;311:1770–1777. doi: 10.1001/jama.2014.4144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarbassov DD, Guertin DA, Ali SM, Sabatini DM. Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science. 2005;307:1098–1101. doi: 10.1126/science.1106148. [DOI] [PubMed] [Google Scholar]
- Sato T, Umetsu A, Tamanoi F. Characterization of the Rheb-mTOR signaling pathway in mammalian cells: constitutive active mutants of Rheb and mTOR. Methods Enzymol. 2008;438:307–320. doi: 10.1016/S0076-6879(07)38021-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato D, Lionel AC, Leblond CS, et al. SHANK1 deletions in males with autism spectrum disorder. Am J Hum Genet. 2012;90:879–887. doi: 10.1016/j.ajhg.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scattoni ML, Gandhy SU, Ricceri L, Crawley JN. Unusual repertoire of vocalizations in the BTBR T+tf/J mouse model of autism. PLoS One. 2008;3:e3067. doi: 10.1371/journal.pone.0003067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scattoni ML, Ricceri L, Crawley JN. Unusual repertoire of vocalizations in adult BTBR T+tf/J mice during three types of social encounters. Genes Brain Behav. 2011;10:44–56. doi: 10.1111/j.1601-183X.2010.00623.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmeisser MJ, Ey E, Wegener S, et al. Autistic-like behaviours and hyperactivity in mice lacking ProSAP1/Shank2. Nature. 2012;486:256–260. doi: 10.1038/nature11015. [DOI] [PubMed] [Google Scholar]
- Sheng M, Kim E. The Shank family of scaffold proteins. J Cell Sci. 2000;113(Pt 11):1851–1856. doi: 10.1242/jcs.113.11.1851. [DOI] [PubMed] [Google Scholar]
- Shibayama A, Cook EH, Jr, Feng J, Glanzmann C, Yan J, Craddock N, Jones IR, Goldman D, Heston LL, Sommer SS. MECP2 structural and 3′-UTR variants in schizophrenia, autism and other psychiatric diseases: a possible association with autism. Am J Med Genet B Neuropsychiatr Genet. 2004;128B:50–53. doi: 10.1002/ajmg.b.30016. [DOI] [PubMed] [Google Scholar]
- Shipman SL, Nicoll RA. Dimerization of postsynaptic neuroligin drives synaptic assembly via transsynaptic clustering of neurexin. Proc Natl Acad Sci USA. 2012;109:19432–19437. doi: 10.1073/pnas.1217633109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman JL, Crawley JN. The promising trajectory of autism therapeutics discovery. Drug Discov Today. 2014;19:838–844. doi: 10.1016/j.drudis.2013.12.007. [DOI] [PubMed] [Google Scholar]
- Silverman JL, Yang M, Lord C, Crawley JN. Behavioural phenotyping assays for mouse models of autism. Nat Rev Neurosci. 2010a;11:490–502. doi: 10.1038/nrn2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman JL, Yang M, Turner SM, Katz AM, Bell DB, Koenig JI, Crawley JN. Low stress reactivity and neuroendocrine factors in the BTBR T+tf/J mouse model of autism. Neu-roscience. 2010b;171:1197–1208. doi: 10.1016/j.neuroscience.2010.09.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman JL, Turner SM, Barkan CL, Tolu SS, Saxena R, Hung AY, Sheng M, Crawley JN. Sociability and motor functions in Shank1 mutant mice. Brain Res. 2011;1380:120–137. doi: 10.1016/j.brainres.2010.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman JL, Smith DG, Rizzo SJ, Karras MN, Turner SM, Tolu SS, Bryce DK, Smith DL, Fonseca K, Ring RH, Crawley JN. Negative allosteric modulation of the mGluR5 receptor reduces repetitive behaviors and rescues social deficits in mouse models of autism. Sci Transl Med. 2012;4:131ra151. doi: 10.1126/scitranslmed.3003501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman JL, Oliver CF, Karras MN, Gastrell PT, Crawley JN. AMPAKINE enhancement of social interaction in the BTBR mouse model of autism. Neuropharmacology. 2013;64:268–282. doi: 10.1016/j.neuropharm.2012.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman JL, Pride MC, Hayes JE, Puhger KR, Butler-Struben HM, Baker S, Crawley JN. GABA receptor agonist R-baclofen reverses social deficits and reduces repetitive behavior in two mouse models of autism. Neuropsychopharmacology. 2015;40:2228–2239. doi: 10.1038/npp.2015.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smalley SL, Asarnow RF, Spence MA. Autism and genetics. A decade of research. Arch Gen Psychiatr y. 1988;45:953–961. doi: 10.1001/archpsyc.1988.01800340081013. [DOI] [PubMed] [Google Scholar]
- Song JY, Ichtchenko K, Sudhof TC, Brose N. Neuroligin 1 is a postsynaptic cell-adhesion molecule of excitatory synapses. Proc Natl Acad Sci USA. 1999;96:1100–1105. doi: 10.1073/pnas.96.3.1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer CM, Alekseyenko O, Serysheva E, Yuva-Paylor LA, Paylor R. Altered anxiety-related and social behaviors in the Fmr1 knockout mouse model of fragile X syndrome. Genes Brain Behav. 2005;4:420–430. doi: 10.1111/j.1601-183X.2005.00123.x. [DOI] [PubMed] [Google Scholar]
- Spencer CM, Graham DF, Yuva-Paylor LA, Nelson DL, Paylor R. Social behavior in Fmr1 knockout mice carrying a human FMR1 transgene. Behav Neurosci. 2008;122:710–715. doi: 10.1037/0735-7044.122.3.710. [DOI] [PubMed] [Google Scholar]
- Spencer CM, Alekseyenko O, Hamilton SM, Thomas AM, Serysheva E, Yuva-Paylor LA, Paylor R. Modifying behavioral phenotypes in Fmr1KO mice: genetic background differences reveal autistic-like responses. Autism Res. 2011;4:40–56. doi: 10.1002/aur.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spooren W, Lindemann L, Ghosh A, Santarelli L. Synapse dysfunction in autism: a molecular medicine approach to drug discovery in neurodevelopmental disorders. Trends Pharmacol Sci. 2012;33:669–684. doi: 10.1016/j.tips.2012.09.004. [DOI] [PubMed] [Google Scholar]
- Stambolic V, Suzuki A, de la Pompa JL, Brothers GM, Mirtsos C, Sasaki T, Ruland J, Penninger JM, Siderovski DP, Mak TW. Negative regulation of PKB/Akt-dependent cell survival by the tumor suppressor PTEN. Cell. 1998;95:29–39. doi: 10.1016/s0092-8674(00)81780-8. [DOI] [PubMed] [Google Scholar]
- Strauss KA, Puffenberger EG, Huentelman MJ, Gottlieb S, Dobrin SE, Parod JM, Stephan DA, Morton DH. Recessive symptomatic focal epilepsy and mutant contactin-associated protein-like 2. N Engl J Med. 2006;354:1370–1377. doi: 10.1056/NEJMoa052773. [DOI] [PubMed] [Google Scholar]
- Szatmari P, Paterson AD, Zwaigenbaum L, et al. Mapping autism risk loci using genetic linkage and chromosomal rearrangements. Nat Genet. 2007;39:319–328. doi: 10.1038/ng1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabuchi K, Blundell J, Etherton MR, Hammer RE, Liu X, Powell CM, Sudhof TC. A neuroligin-3 mutation implicated in autism increases inhibitory synaptic transmission in mice. Science. 2007;318:71–76. doi: 10.1126/science.1146221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavassoli T, Kolevzon A, Wang AT, Curchack-Lichtin J, Halpern D, Schwartz L, Soffes S, Bush L, Grodberg D, Cai G, Buxbaum JD. De novo SCN2A splice site mutation in a boy with Autism spectrum disorder. BMC Med Genet. 2014;15:35. doi: 10.1186/1471-2350-15-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tee AR, Fingar DC, Manning BD, Kwiatkowski DJ, Cantley LC, Blenis J. Tuberous sclerosis complex-1 and −2 gene products function together to inhibit mammalian target of rapamycin (mTOR)-mediated downstream signaling. Proc Natl Acad Sci USA. 2002;99:13571–13576. doi: 10.1073/pnas.202476899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Dutch-Belgian Fragile X Consortium. Bakker CE, Verheij C, Willemsen R, van der Helm R, Oerlemans F, Vermey M, Bygrave A, Hoogeveen A, Oostra BA, Reyniers E, De Boule K, D’Hooge R, Cras P, van Velzen D, Nagels G, Martin J-J, De Deyn PP, Darby JK, Willems PJ. Fmr1 knockout mice: a model to study fragile X mental retardation. Cell. 1994;78:23–33. [PubMed] [Google Scholar]
- Theoharides TC, Athanassiou M, Panagiotidou S, Doyle R. Dysregulated brain immunity and neurotrophin signaling in Rett syndrome and autism spectrum disorders. J Neuroimmunol. 2015;279:33–38. doi: 10.1016/j.jneuroim.2014.12.003. [DOI] [PubMed] [Google Scholar]
- Thomas NS, Sharp AJ, Browne CE, Skuse D, Hardie C, Dennis NR. Xp deletions associated with autism in three females. Hum Genet. 1999;104:43–48. doi: 10.1007/s004390050908. [DOI] [PubMed] [Google Scholar]
- Tian D, Stoppel LJ, Heynen AJ, Lindemann L, Jaeschke G, Mills AA, Bear MF. Contribution of mGluR5 to pathophysiology in a mouse model of human chromosome 16p11.2 microdeletion. Nat Neurosci. 2015;18:182–184. doi: 10.1038/nn.3911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai PT, Hull C, Chu Y, Greene-Colozzi E, Sadowski AR, Leech JM, Steinberg J, Crawley JN, Regehr WG, Sahin M. Autistic-like behaviour and cerebellar dysfunction in Purkinje cell Tsc1 mutant mice. Nature. 2012;488:647–651. doi: 10.1038/nature11310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu JC, Xiao B, Naisbitt S, Yuan JP, Petralia RS, Brakeman P, Doan A, Aakalu VK, Lanahan AA, Sheng M, Worley PF. Coupling of mGluR/Homer and PSD-95 complexes by the Shank family of postsynaptic density proteins. Neuron. 1999;23:583–592. doi: 10.1016/s0896-6273(00)80810-7. [DOI] [PubMed] [Google Scholar]
- Uutela M, Lindholm J, Louhivuori V, Wei H, Louhivuori LM, Pertovaara A, Akerman K, Castren E, Castren ML. Reduction of BDNF expression in Fmr1 knockout mice worsens cognitive deficits but improves hyperactivity and sensorimotor deficits. Genes Brain Behav. 2012;11:513–523. doi: 10.1111/j.1601-183X.2012.00784.x. [DOI] [PubMed] [Google Scholar]
- Van Dam D, D’Hooge R, Hauben E, Reyniers E, Gantois I, Bakker CE, Oostra BA, Kooy RF, De Deyn PP. Spatial learning, contextual fear conditioning and conditioned emotional response in Fmr1 knockout mice. Behav Brain Res. 2000;117:127–136. doi: 10.1016/s0166-4328(00)00296-5. [DOI] [PubMed] [Google Scholar]
- Varga EA, Pastore M, Prior T, Herman GE, McBride KL. The prevalence of PTEN mutations in a clinical pediatric cohort with autism spectrum disorders, developmental delay, and macrocephaly. Genet Med. 2009;11:111–117. doi: 10.1097/GIM.0b013e31818fd762. [DOI] [PubMed] [Google Scholar]
- Varoqueaux F, Jamain S, Brose N. Neuroligin 2 is exclusively localized to inhibitory synapses. Eur J Cell Biol. 2004;83:449–456. doi: 10.1078/0171-9335-00410. [DOI] [PubMed] [Google Scholar]
- Varoqueaux F, Aramuni G, Rawson RL, Mohrmann R, Missler M, Gottmann K, Zhang W, Sudhof TC, Brose N. Neuroligins determine synapse maturation and function. Neuron. 2006;51:741–754. doi: 10.1016/j.neuron.2006.09.003. [DOI] [PubMed] [Google Scholar]
- Veenstra-VanderWeele J, Muller CL, Iwamoto H, Sauer JE, Owens WA, Shah CR, Cohen J, Mannangatti P, Jessen T, Thompson BJ, Ye R, Kerr TM, Carneiro AM, Craw-ley JN, Sanders-Bush E, McMahon DG, Ramamoorthy S, Daws LC, Sutcliffe JS, Blakely RD. Autism gene variant causes hyperserotonemia, serotonin receptor hypersensitivity, social impairment and repetitive behavior. Proc Natl Acad Sci USA. 2012;109:5469–5474. doi: 10.1073/pnas.1112345109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veeraragavan S, Graham D, Bui N, Yuva-Paylor LA, Wess J, Paylor R. Genetic reduction of muscarinic M4 receptor modulates analgesic response and acoustic startle response in a mouse model of fragile X syndrome (FXS) Behav Brain Res. 2012;228:1–8. doi: 10.1016/j.bbr.2011.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernes SC, Newbury DF, Abrahams BS, Winchester L, Nicod J, Groszer M, Alarcon M, Oliver PL, Davies KE, Geschwind DH, Monaco AP, Fisher SE. A functional genetic link between distinct developmental language disorders. N Engl J Med. 2008;359:2337–2345. doi: 10.1056/NEJMoa0802828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent JB, Kolozsvari D, Roberts WS, Bolton PF, Gurling HM, Scherer SW. Mutation screening of X-chromosomal neuroligin genes: no mutations in 196 autism probands. Am J Med Genet B Neuropsychiatr Genet. 2004;129B:82–84. doi: 10.1002/ajmg.b.30069. [DOI] [PubMed] [Google Scholar]
- Vorstman JA, Spooren W, Persico AM, Collier DA, Aigner S, Jagasia R, Glennon JC, Buitelaar JK. Using genetic findings in autism for the development of new pharmaceutical compounds. Psychopharmacology (Berl) 2014;231:1063–1078. doi: 10.1007/s00213-013-3334-z. [DOI] [PubMed] [Google Scholar]
- Wang K, Zhang H, Ma D, et al. Common genetic variants on 5p14.1 associate with autism spectrum disorders. Nature. 2009;459:528–533. doi: 10.1038/nature07999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, McCoy PA, Rodriguiz RM, Pan Y, Je HS, Roberts AC, Kim CJ, Berrios J, Colvin JS, Bousquet-Moore D, Lorenzo I, Wu G, Weinberg RJ, Ehlers MD, Philpot BD, Beaudet AL, Wetsel WC, Jiang YH. Synaptic dysfunction and abnormal behaviors in mice lacking major isoforms of Shank3. Hum Mol Genet. 2011;20:3093–3108. doi: 10.1093/hmg/ddr212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss LA, Escayg A, Kearney JA, Trudeau M, MacDonald BT, Mori M, Reichert J, Buxbaum JD, Meisler MH. Sodium channels SCN1A, SCN2A and SCN3A in familial autism. Mol Psychiatry. 2003;8:186–194. doi: 10.1038/sj.mp.4001241. [DOI] [PubMed] [Google Scholar]
- Wilkinson B, Grepo N, Thompson BL, Kim J, Wang K, Evgrafov OV, Lu W, Knowles JA, Campbell DB. The autism-associated gene chromodomain helicase DNA-binding protein 8 (CHD8) regulates noncoding RNAs and autism-related genes. Transl Psychiatry. 2015;5:e568. doi: 10.1038/tp.2015.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willsey AJ, State MW. Autism spectrum disorders: from genes to neurobiology. Curr Opin Neurobiol. 2015;30:92–99. doi: 10.1016/j.conb.2014.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wohr M, Roullet FI, Crawley JN. Reduced scent marking and ultrasonic vocalizations in the BTBR T+tf/J mouse model of autism. Genes Brain Behav. 2011a;10:35–43. doi: 10.1111/j.1601-183X.2010.00582.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wohr M, Roullet FI, Hung AY, Sheng M, Crawley JN. Communication impairments in mice lacking Shank1: reduced levels of ultrasonic vocalizations and scent marking behavior. PLoS One. 2011b;6:e20631. doi: 10.1371/journal.pone.0020631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wohr M, Silverman JL, Scattoni ML, Turner SM, Harris MJ, Saxena R, Crawley JN. Developmental delays and reduced pup ultrasonic vocalizations but normal sociability in mice lacking the postsynaptic cell adhesion protein neuroligin2. Behav Brain Res. 2013;251:50–64. doi: 10.1016/j.bbr.2012.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Won H, Lee HR, Gee HY, Mah W, Kim JI, Lee J, Ha S, Chung C, Jung ES, Cho YS, Park SG, Lee JS, Lee K, Kim D, Bae YC, Kaang BK, Lee MG, Kim E. Autistic-like social behaviour in Shank2-mutant mice improved by restoring NMDA receptor function. Nature. 2012;486:261–265. doi: 10.1038/nature11208. [DOI] [PubMed] [Google Scholar]
- Yadav R, Gupta SC, Hillman BG, Bhatt JM, Stairs DJ, Dravid SM. Deletion of glutamate delta-1 receptor in mouse leads to aberrant emotional and social behaviors. PLoS One. 2012;7:e32969. doi: 10.1371/journal.pone.0032969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan QJ, Asafo-Adjei PK, Arnold HM, Brown RE, Bauchwitz RP. A phenotypic and molecular characterization of the fmr1-tm1Cgr fragile X mouse. Genes Brain Behav. 2004;3:337–359. doi: 10.1111/j.1601-183X.2004.00087.x. [DOI] [PubMed] [Google Scholar]
- Yan J, Oliveira G, Coutinho A, Yang C, Feng J, Katz C, Sram J, Bockholt A, Jones IR, Craddock N, Cook EH, Jr, Vicente A, Sommer SS. Analysis of the neuroligin 3 and 4 genes in autism and other neuropsychiatric patients. Mol Psychiatry. 2005;10:329–332. doi: 10.1038/sj.mp.4001629. [DOI] [PubMed] [Google Scholar]
- Yang M, Crawley JN. Simple behavioral assessment of mouse olfaction. Current Protocols in Neuroscience. 2009:8–24. doi: 10.1002/0471142301.ns0824s48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M, Silverman JL, Crawley JN. Automated three-chambered social approach task for mice. Current Protocols in Neuroscience. 2011:8–26. doi: 10.1002/0471142301.ns0826s56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M, Abrams DN, Zhang JY, Weber MD, Katz AM, Clarke AM, Silverman JL, Crawley JN. Low sociability in BTBR T+tf/J mice is independent of partner strain. Physiol Behav. 2012a;107:649–662. doi: 10.1016/j.physbeh.2011.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M, Bozdagi O, Scattoni ML, Wohr M, Roullet FI, Katz AM, Abrams DN, Kalikhman D, Simon H, Woldeyohannes L, Zhang JY, Harris MJ, Saxena R, Silverman JL, Buxbaum JD, Crawley JN. Reduced excitatory neurotransmission and mild autism-relevant phenotypes in adolescent Shank3 null mutant mice. J Neurosci. 2012b;32:6525–6541. doi: 10.1523/JNEUROSCI.6107-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M, Mahrt EJ, Lewis F, Foley G, Portmann T, Dolmetsch RE, Portfors CV, Crawley JN. 16p11.2 deletion syndrome mice display sensory and ultrasonic vocalization deficits during social interactions. Autism Res. 2015 doi: 10.1002/aur.1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ylisaukko-oja T, Rehnstrom K, Auranen M, Vanhala R, Alen R, Kempas E, Ellonen P, Turunen JA, Makkonen I, Riikonen R, Nieminen-von Wendt T, von Wendt L, Peltonen L, Jarvela I. Analysis of four neuroligin genes as candidates for autism. Eur J Hum Genet. 2005;13:1285–1292. doi: 10.1038/sj.ejhg.5201474. [DOI] [PubMed] [Google Scholar]
- Yuen RK, Thiruvahindrapuram B, Merico D, et al. Whole-genome sequencing of quartet families with autism spectrum disorder. Nat Med. 2015;21:185–191. doi: 10.1038/nm.3792. [DOI] [PubMed] [Google Scholar]
- Zahir FR, Baross A, Delaney AD, Eydoux P, Fernandes ND, Pugh T, Marra MA, Friedman JM. A patient with vertebral, cognitive and behavioural abnormalities and a de novo deletion of NRXN1alpha. J Med Genet. 2008;45:239–243. doi: 10.1136/jmg.2007.054437. [DOI] [PubMed] [Google Scholar]
- Zhao MG, Toyoda H, Ko SW, Ding HK, Wu LJ, Zhuo M. Deficits in trace fear memory and long-term potentiation in a mouse model for fragile X syndrome. J Neurosci. 2005;25:7385–7392. doi: 10.1523/JNEUROSCI.1520-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Blundell J, Ogawa S, Kwon CH, Zhang W, Sinton C, Powell CM, Parada LF. Pharmacological inhibition of mTORC1 suppresses anatomical, cellular, and behavioral abnormalities in neural-specific Pten knock-out mice. J Neurosci. 2009;29:1773–1783. doi: 10.1523/JNEUROSCI.5685-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoghbi HY, Bear MF. Synaptic dysfunction in neurodevelopmental disorders associated with autism and intellectual disabilities. Cold Spring Harb Perspect Biol. 2012;4:a009886. doi: 10.1101/cshperspect.a009886. [DOI] [PMC free article] [PubMed] [Google Scholar]