Abstract
Cryptosporidium parvum is unable to synthesize fatty acids de novo, but possesses three long-chain fatty acyl-CoA synthetase (CpACS) isoforms for activating fatty acids. We have recently shown that these enzymes could be targeted to kill the parasite in vitro and in vivo. Here we demonstrated that the CpACS genes were differentially expressed during the parasite life cycle, and their proteins were localized to different subcellular structures by immunofluorescence and immuno-electron microscopies. Among them, CpACS1 displayed as an apical protein in sporozoites and merozoites, but no or little presence during the intracellular merogony until the release of merozoites, suggesting that CpACS1 probably functioned mainly during the parasite invasion and/or early stage of intracellular development. Both CpACS2 and CpACS3 proteins were present in all parasite life cycle stages, in which CpACS2 was present in the parasite and the parasitophorous vacuole membranes (PVM), whereas CpACS3 was mainly present in the parasite plasma membranes with little presence in the PVM. These observations suggest that CpACS2 and CpACS3 may participate in scavenging and transport of fatty acids across the PVM and the parasite cytoplasmic membranes, respectively.
Keywords: Apicomplexa, Enzyme, Immunofluorescence microscopy, Immuno-electron microscopy, Apical proteins, Parasitophorous vacuole membrane (PVM), Fatty acid activation
THE apicomplexan parasite Cryptosporidium parvum infects both humans and animals, and frequently causes water-borne outbreaks around the world. In AIDS patients, Cryptosporidium causes one of the opportunistic infections (AIDS-OIs) that can be chronic and live-threatening (Chen et al. 2002, Checkley et al. 2015). In developing countries, Cryptosporidium is not only one of the most common causes of moderate to severe diarrhea, but also associated with slower growth and higher death rates in children under 5 years old (Kotloff et al. 2013). This parasite is unable to synthesize virtually any nutrients de novo, including amino acids, nucleosides and fatty acids (Abrahamsen et al. 2004, Rider and Zhu 2010). Within the fatty acid associated metabolic pathways, C. parvum lacks an apicoplast responsible for synthesizing fatty acids from acetyl-CoA, but possesses a 900 kDa type I fatty acid synthase (CpFAS1) and a 1,500 kDa type I polyketide synthase (CpPKS1). Both CpFAS1 and CpPKS1 use long-chain fatty acids as substrates, and the reductase domain of CpFAS1 involved in the release of acyl-chain acts only on very long chain fatty acids, indicating that they are responsible for the elongation of fatty acyl or polyketide chains, rather than de novo synthesis of fatty acids. Therefore, Cryptosporidium has to scavenge fatty acids from host cells or the environment.
Additionally, Cryptosporidium possesses three [AMP-binding]-long chain fatty acyl-coenzyme A (CoA) synthetases (ACSs; aka, fatty acid-CoA ligases, ACLs) [EC 6.2.1.3] to catalyze the formation of fatty acyl-CoA from a fatty acid and CoA. Reactions catalyzed by ACS are essential to all cells, as fatty acids have to be thioesterified with CoA before they can enter subsequent major metabolic pathways, such as the synthesis of complex lipids and beta-oxidation (Andersson et al. 2012, Fujino et al. 1996). We have recently characterized the biochemical features of two C. parvum ACS isoforms (CpACS1 and CpACS2), and demonstrated that the ACS inhibitor triacsin C could not only inhibit CpACS enzyme activity, but also was highly efficacious against C. parvum growth both in vitro and in vivo, indicating that ACS may serve as an effective drug target in the parasite (Guo et al. 2014).
To gain more insight into the biological role of the three ACS in the parasite, we further studied the molecular features of CpACSs, including their phylogenetic positions, gene expression patterns and protein localizations in the complex life cycle of C. parvum. We showed that CpACSs belonged to the long chain ACS family, CpACS genes were differentially expressed during the parasite life cycle, and enzymes were localized to the different subcellular structures, suggesting different roles played by the three parasite ACS isoforms.
MATERIALS AND METHODS
General procedures for manipulating C. parvum and isolating nucleotide acids
The parasite (IOWA-1 strain of C. parvum) was purchased from Grass Bunch Farm (Deary, ID) and experiments only used oocysts that were less than 3-month old since harvest. Oocysts were purified from calf feces by sucrose-gradient centrifugation protocol, followed by treatment of 10% Clorox on ice for 7 min and repeated washed with pure water by centrifugation for 5-8 times (Arrowood and Sterling 1987, Zhang et al. 2012). Oocysts might be further purified by a Percoll-gradient centrifugation procedure to remove minor debris if present. Free sporozoites were prepared by incubating oocysts in PBS containing 0.25% trypsin and 0.5% taurodeoxycholic acid at 37 °C for 60 min, followed by a Percoll-based centrifugation protocol as described (Robertson et al. 1993).
For in vitro experiments, HCT-8 cells (ATCC # CCL-225) were seeded into 24-well cell culture plates and cultured in RPMI 1640 medium containing 10% fetal bovine serum (FBS) at 37 °C supplied with 5% CO2 until they reached to ~80% confluence. Purified C. parvum oocysts were added into the cell culture at a parasite:host cell ratio of 1:2 (i.e., 105 oocysts/well). After incubation at 37 °C for 3 h that allowed sporozoites to be released by excystation and invade host cells, uninvaded parasites were removed by a medium exchange. Intracellular parasites were allowed to grow for specified times before subsequent experiments including RNA isolation for expression analysis or fixation for immunofluorescence staining and immuno-electron microscopy. For gene expression analysis, total RNA was isolated from oocysts, free sporozoites and intracellular parasites developed in host cells for specified times using RNeasy Mini Kit (Qiagen, Valencia, CA) (Zhang et al. 2012).
Molecular sequence analysis and phylogeny
Phylogenetic relationship of CpACS proteins with other orthologs within the chromalveolates was determined by maximum likelihood (ML) and Bayesian inference (BI) methods. ACS orthologs were retrieved from NCBI protein database by repeated BLAST searches using ACS protein sequences from C. parvum and other apicomplexans as queries. Multiple sequence alignments were performed using MUSCLE program (v3.8.31) (http://www.drive5.com/muscle/). Identical sequences and short partial sequences were removed, and only amino acid positions that could be unambiguously aligned were used in phylogenetic reconstructions. The final dataset consisted of 120 taxa with 281 amino acid positions. In ML analysis, bootstrapping supporting values were derived from 1000 replicated sequences using TreeFinder program (version 2008) (http://www.treefinder.de). BI analysis used MrBayes program (v3.2.1) (http://mrbayes.sourceforge.net/), in which 106 generations of searches were performed with two independent runs, each run with 4 chains running simultaneously. Posterior probability (PP) values were derived after the first 25% of trees were discarded. Both ML and BI analyses used the WAG amino acid substitution model, with the consideration of the fraction of invariance (Finv) and a 4-rate gamma distribution among varied sites (Γ[4-rate]) for among-site heterogeneity (i.e., Finv + Γ[4-rate] + WAG). Consensus trees were displayed using the FigTree (v1.3.1) program (http://tree.bio.ed.ac.uk/software/figtree/), and final annotation was performed using Adobe Illustrator CS4. Sequence logos for the conserved motifs were derived from the 120 aligned chromalveolate sequences using WebLogo3 server (http://weblogo.threeplusone.com). Potential signal peptides were analyzed at SignalP 4.1 server (http://www.cbs.dtu.dk/services/SignalP/) using a model for eukaryotes.
Quantitative analysis of CpACS gene expression
The relative expression levels of CpACS1, CpACS2 and CpACS3 genes in oocysts, sporozoites and intracellular parasites developed in HCT-8 cells for 6 h to 72 h were evaluated by quantitative real-time RT-PCR (qRT-PCR) using the One-Step RT-PCR QuantiTect SYBR Green RT-PCR Kit (Qiagen, Valencia, CA). The levels of C. parvum 18S rRNA (Cp18S) were determined in parallel amplifications for normalization as previously described (Cai et al. 2005). Each of the qRT-PCR reactions contained 0.5 μM of primers, 1 ng of total RNA from oocysts and sporozoites or 15 ng total RNA from intracellular parasites in 20 μL volume. Real-time RT-PCR amplifications were performed in a CFX Connect Real-Time PCR detection system (Bio-Rad Labs, Hercules, CA), in which 30 min reaction at 50 °C was allowed for reverse transcription, followed by 40 thermal cycles each at 94 °C for 20 sec, 50 °C for 30 sec and 72 °C for 30 sec. The following primer pairs specific to CpACS genes and 18S rRNA were used: 5’ GCA GAA AGA CAA CAA GAT GGC 3’ and 5’ TCC CTC AAT TCT TTC TGG TGA 3’ for CpACS1 (amplicon size = 102 bp); 5’ TAT ATG GGT GTT AGG GTG CCA 3’ and 5’ CCT GGA CTG CTC CAA TAT GAA 3’ for CpACS2 (127 bp); 5’ TGG TGT CCA GGT TTC ACT TTC 3’ and 5’ TTC AAT TCA CCA AGC CTC ATC 3’ for CpACS3 (96 bp); and 5’ TTG TTC CTT ACT CCT TCA GCA C 3’ and 5’ TCC TTC CTA TGT CTG GAC CTG 3’ for Cp18S rRNA (175 bp).
Each experimental condition included at least three biological replicates (RNA samples) and two technical replicates (RT-PCR reactions). Threshold cycle (CT) values were used for computing the relative levels of expression, in which data were first normalized by computing the ΔCT values between CpACS mRNA (ΔCT[sample]) and Cp18S rRNA (ΔCT[Cp18S]) for all samples. PCR amplification efficiencies (ε) for individual genes were determined by equation:
| (1) |
in which slopes were obtained by real-time RT-PCR with serial dilutions of a pooled total RNA sample, followed by linear regression of CT values against common logarithms of relative RNA concentrations (dilutions). Two types of relative expression levels were plotted. The first one was the percent levels of mRNA of a specified gene in various samples in relative to the one with the highest level of expression, which was derived from relative fold changes (f) determined by equation:
| (2) |
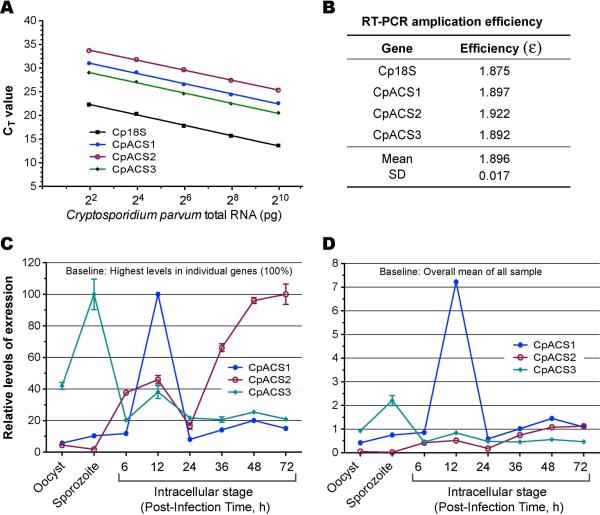
where ΔΔCT values were those between a specified sample and the highest one. Second, because the PCR amplification efficiencies for all three genes and Cp18S rRNA were nearly identical (i.e., ε = 1.896 ± 0.017) (see Fig. 2A and 2B), we were able to also use the ΔΔCT method to directly compare the relative levels of expressions of between three CpACS genes in all samples against the overall mean (see Fig. 2D).
Figure 2.
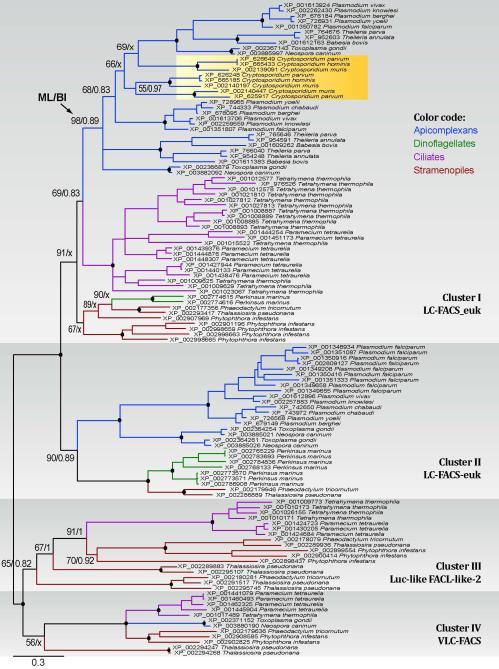
Phylogenetic tree inferred from ACS proteins among chromalveolates using Bayesian inference (BI) and maximum likelihood (ML) methods. Numbers at the nodes indicate posterior probability and bootstrapping proportion supporting values obtained in BI and ML analyses. Numbers of sequences contained in specified clusters are indicated in parentheses after the species names.
Production and affinity purification of anti-CpACS polyclonal antibodies
Polyclonal antibodies against CpACS1, CpACS2 and CpACS3 were commercially produced by Alpha Diagnostic International (San Antonio, TX). Antibodies to CpACS1 and CpACS3 were produced using recombinant proteins as antigens (i.e., amino acids from positions 99 to 254 and 86 to 288, respectively), each in two specific pathogen-free (SPF) rabbits by a standard 63-day protocol, which included 5 injections at multiple sites at 14 day interval. Pre-immune bleeds were collect prior to immunization, and two immune bleeds were collected at weeks 7 and 9, separately. Antibodies to CpACS2 were generated in two SPF hens using synthesized peptide CTAAFVYGQLGREGTKLGSN corresponding to amino acids at positions from 271-290 by a standard 63-day immunization protocol, including isolation of chicken IgY from the 6 egg yolks.
Antibodies were subjected to an antigen-based affinity purification procedure using a previously described protocol with minor modification (Kurien 2009). Briefly, for CpACS1 and CpACS3 antibodies, the recombinant antigens were separated on an SDS-PAGE gel and transferred to nitrocellulose membranes. The blots were stained with Ponceau Red and the strips containing the recombinant protein were cut out for subsequent antibody purification. For CpACS2 antibody, synthetic peptide was directly blotted on nitrocellulose membranes and air-dried. All membranes were blocked with 5% BSA in TBST (Tris buffered saline containing 0.02% Tween 20) for 1 h, and then incubated separately with antisera to CpACS1 and CpACS3 (second bleeds), or with anti-CpACS2 IgY overnight at 4 °C on an orbital shaker. The blots were washed three times with TBST (10 min each), and purified antibodies were eluted in 0.2 M glycine (pH 2.2) by vigorous shaking on an orbital shaker for 2 minutes, followed by a quick neutralization with 1 M Tris-HCl (pH 7.6) buffer.
Western blot analysis
Free sporozoites were prepared by an in vitro excystation procedure, in which C. parvum oocysts were incubated in PBS buffer containing 0.5% taurodeoxycholic acid (TDC) and 0.25% trypsin for 45 minutes at 37 °C. Freshly released sporozoites were collected and washed at least three times by centrifugation with culture medium containing 10% fetal bovine serum (FBS) to neutralize trypsin. Sporozoites were lysed with Radioimmunoprecipitation assay (RIPA) lysis buffer (Sigma-Aldrich, St. Louis, MO) containing 1% Triton X-100 and protease inhibitor cocktail for eukaryotes (Sigma-Aldrich, St. Louis, MO) on ice overnight, followed by additional 1 h incubation at room temperature. After centrifugation at 500 × g for 5 min, soluble proteins in supernatants were mixed with sample buffer, heated at 100 °C for 5 min, and subjected to sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) (~4 × 107 sporozoites/lane).
Fractioned proteins were transferred onto nitrocellulose membranes, followed by washes with TBST buffer and incubation for 1 h with blocking buffer (5% fat-free milk in TBST). Membranes were incubated with affinity-purified primary antibodies against the three CpACS proteins and their corresponding pre-immune antibodies (1 h), washed 3 times (10 min each), and then incubated with horseradish peroxidase (HRP)-conjugated secondary antibodies (i.e., goat anti-rabbit IgG for CpACS1 and CpACS3, and rabbit anti-chicken IgG for CpACS2). After washes (3 times, 10 min each), the blots were treated with an ECL Plus chemiluminescent substrates for 1 minute (Pierce Biotechnology, Rockford, lL), and the chemiluminescent signals were exposed to Kodak BioMax light films (Carestream Health, Rochester, NY), followed by the film development in a Konica SRX-101A film processor (Konica Minolta Medical Imaging, Wayne, NJ). All washes were carried out using TBST buffer and all incubations were performed at room temperature or as specified.
Immunofluorescence microscopy (IFM)
Free sporozoites were prepared as described above. Type I merozoites were collected from the supernatant of cultured HCT-8 cells infected with C. parvum for 12-18 h. Different intracellular stages of parasites were obtained by infecting HCT-8 cell monolayers grown on glass coverslips for 3 to 24 h, followed by fixation of monolayers in 4% formaldehyde for 30 min at room temperature. After 3 washes in PBS, excessive formaldehyde was quenched with 50 mM NH4Cl for 15 min. Fixed cells were permeabilized with 0.2% Triton X-100 and 0.01% SDS in PBS for 5 min, washed 3 times in PBS (5 min each). Cells were then blocked with 5% FBS in PBS for 30 min, followed by incubation with affinity-purified anti-CpACS antibodies (1:500 dilution in 5% FBS-PBS) for 30 min, three washes with 5% FBS-PBS, incubation with tetramethylrhodamine (TRITC)-conjugated goat anti-rabbit or rabbit anti-chicken IgG (1:400 dilution in 5% FBS-PBS) for 30 min, and three washes with 5% FBS-PBS. After an additional wash with PBS, coverslips were mounted onto slides with a Prolong Gold Antifade reagent containing 4′,6-diamidino-2-phenylindole (DAPI) for counter-staining of nuclei (Molecular Probes/Invitrogen, Grand Island, NY). In antigen-competition IFM experiment, affinity-purified antibodies were first presoaked with their corresponding antigens (i.e., recombinant CpACS isoforms) and MBP (as negative control) that were conjugated onto the nitrocellulose membranes. These antibodies were then presoaked with corresponding antigens for 30 min and used as primary antibodies to label free sporozoites as described above. Cells labeled with fluorescent molecules were examined with an Olympus BX51 research microscope equipped with appropriate filter sets. Images were captured with a Retiga SRV CCD Digital Camera (QImaging, Surry, BC). Fluorescence images were manipulated with Adobe Photoshop CS6. To better visualize the immunofluorescence labeling, images were subjected to adjustments of signal levels and color balance that were uniformly applied to the entire images without local manipulations.
Colloidal-gold immuno-electron microscopy (IEM)
For immuno-electron microscopic labeling of CpACS proteins, free sporozoites and intracellular parasites were prepared by excystation and in vitro cell culture as described above, but cells were grown in LabTek Permanox chamber slides. Infected monolayers were washed in PBS and fixed in 4% paraformaldehyde mixed with 0.1% glutaraldehyde (Electron Microscopy Sciences, Hatfield, PA) in PBS for 15 h at 4 °C. Fixed samples were washed with 0.05 M maleate buffer containing 0.5 mM CaCl2 and 2% sucrose, incubated for 30 min at 20 °C in 0.5% uranyl acetate in maleate buffer, and then washed again in maleate buffer. Freshly excysted sporozoties were similarly fixed and washed, but enrobed with melted 1.5% agar in maleate buffer at 50 °C, followed by sedimentation by centrifugation. After cooling down to room temperature, the pellets were minced into small segments (~0.5 cubic mm) for further processing. These samples were dehydrated in a graded ethanol series, then infiltrated with LR White acrylic resin (Electron Microscopy Sciences, Hatfield, PA) and cured at 50 °C for 13 h. Selected segments of embedded samples were transferred to LR White in gelatin capsules and cured at 50 °C for an additional 14 h. Thin sections were produced using a diamond knife on a Leica EM UC6 ultramicrotome and mounted on film-coated grids.
Sections on grids were blocked sequentially in blocking buffer I (100 mM glycine in TBS) and II (2.5% BSA/0.2% cold water fish gelatin), and then incubated with primary antibodies at 4 °C overnight. After washes, samples were labeled with goat anti-rabbit IgG or rabbit anti-chicken IgY secondary antibodies conjugated with 10 nm gold beads for 2 h at room temperature, washed again, and then post-stained briefly with 2% uranyl acetate and Reynold's lead citrate. Colloidal gold labeled thin sections were examined with a Philips Morgagni 268 transmission electron microscope (FEI company, aka Field Emission Inc, Hillsboro, OR) at an accelerating voltage of 80 kV. Digital images were recorded with a MegaViewIII digital camera operated with iTEM software (Olympus Soft Imaging Systems, Münster, Germany).
RESULTS AND DISCUSSION
CpACS enzymes belong to the long-chain ACS subfamily
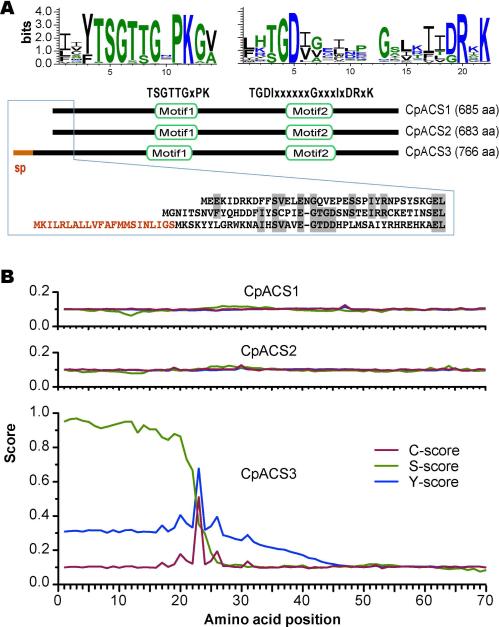
ACS enzymes can be divided into four major subfamilies based on the chain length of their preferred acyl groups: short-chain (C2 - C4) (SC)-ACS, medium-chain (C4 - C12) (MC)-ACS, long-chain (C12 - C20) (LC)-ACS, and very long-chain (C18-C26) (VLC)-ACS (Soupene and Kuypers 2008, Andersson et al. 2012). The number of LC-ACSs varies among organisms (e.g., at least 9 in Arabidopsis thaliana, 11 in Plasmodium falciparum and 5 in humans) (Schnurr et al. 2004, Soupene and Kuypers 2008, Matesanz et al. 2003). Among Cryptosporidium species, each of the three sequenced genomes (i.e., C. parvum, C. homins and C. muris) contains three LC-ACS genes (see genome sequences and annotation at http://www.CryptoDB.org). In C. parvum, CpACS1 (GenBank:AAP41029), CpACS2 (GenBank: XP_626248) and CpACS3 (GenBank: XP_625917) are comprised of 685, 683 and 766 amino acids (aa), predicting molecular weights of 78.2, 77.1 and 86.1 kDa, respectively. All three CpACS isoforms contained two conserved domains: the AMP/ATP-binding motif (TSGTTGxPK) and the fatty acyl-CoA synthetase signature motif (TGDIxxxxxxGxxxIxDRxK) (Fig. 1A). The AMP/ATP-binding sites are highly conserved not only in the ACS family, but also in many other ATP-dependent AMP-binding enzymes for adenylate formation (Melton et al. 2011, Black et al. 1997). N-terminal signal peptides were absent in both the CpACS1 and CpACS2 proteins. However, SignalP analysis indicated the presence of a signal peptide typically found in secretory proteins in CpACS3 (S-score: 0.97) with a predicted cleavage site between amino acids 22 and 23 (Fig. 1B).
Figure 1.
Structures and domains of CpACS1, CpACS2 and CpACS3 proteins. A) Domain organization and sequence logos of amino acids conserved among chromaveolates; B) Prediction of N-terminal signal peptide (sp) for secretion in CpACS3 by SignalP algorithm, in which S-score predicts signal peptide, while C-score and Y-score predict cleavage site.
Phylogenetic trees inferred from chromalveolate ACS orthologs by maximum likelihood (ML) and Bayesian inference (BI) methods divided the LC- and VLC-ACS proteins into 4 major clusters (Fig. 2): clusters I and II with top hits to eukaryotic-type LC-ACS domains in the NCBI conserved domain database (e.g., LC-ACS-euk [cd05927]); cluster III with Firefly Luc-like (cd05911) and uncharacterized FACL-like-2 (cd05917) domains; and cluster IV with VLC-ACS (cd05907) and bubblegum VLC-ACS (cd05933). Sequences from stramenopiles and ciliates were present in all 4 clusters, whereas those from a dinoflagellate (Perkinsus) and apicomplexans were present only in clusters I and II (LC-ACS) with the exception of Toxoplasma and Neospora that also possessed two sequences in cluster IV (VLC-ACS). These observations suggest that: 1) there was an ancient split between LC- and VLC-ACS genes that predated the split between alveolates and stramenopiles; 2) VLC-ACS genes were lost in most apicomplexans and at least one dinoflagellate (Perkinsus), but were retained in some coccidia (e.g., Toxoplasma and Neospora); and 3) gene expansions occurred within each group, particularly among the LC-ACS genes in clusters I and II. All three Cryptosporidium ACS proteins were predicated to be LC-ACS enzymes in cluster I (Fig. 2), which was in full agreement with our recently reported biochemical data (i.e., CpACS1 and CpACS2 preferred to using C10 - C18 fatty acids) (Guo et al. 2014).
CpACS genes were differentially expressed
We examined transcript expression during parasite growth and used immunofluorescence to examine protein localization during parasite development in vitro. Real-time qRT-PCR based analysis revealed that CpACS genes were differentially expressed at different parasite developmental stages (Fig. 3). More specifically, the CpACS1 and CpACS2 genes were expressed at much higher levels in the early and later intracellular developmental stages, respectively. On the other hand, the CpACS3 gene had much higher levels of expression in oocysts and sporozoites. These observations imply that the three CpACS enzymes might play different roles during the parasite development. This notion was also supported by immunofluorescence staining using affinity-purified polyclonal antibodies against the three CpACS isoforms as described below (Fig. 5-10).
Figure 3.
Differential expression of the 3 CpACS genes in C. parvum at different life cycle stages as determined by qRT-PCR. A) Standard curves showing linear relationship between detection threshold cycles (CT values) and the levels of CpACS transcripts using serially diluted, pooled total RNA isolated from infected HCT-8 cells. Cp18S rRNA was used as control for normalization; B) PCR amplification efficiencies Cp18S rRNA and CpACS transcripts derived from standard curves; C) Normalized levels of three CpACS transcripts in different life cycle stages in relative to the highest level for each gene (i.e., comparison between different developmental stages, but not between different genes); D) Normalized levels of three CpACS transcripts in relative to the overall mean of all three genes in all developmental stages.
Figure 5.
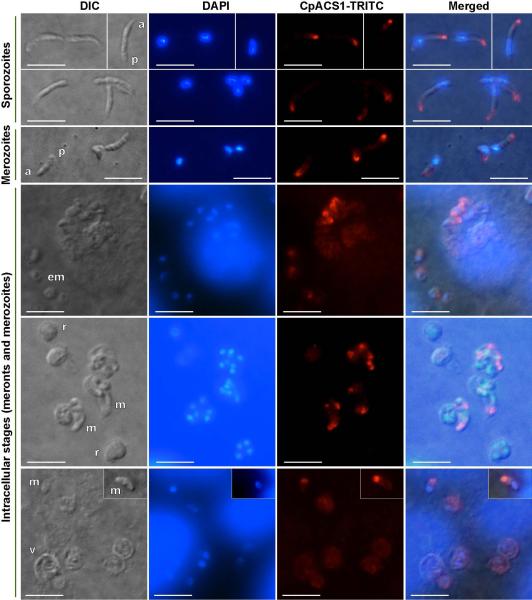
Immunofluorescence labeling showing that CpACS1 protein is an apical protein and mainly localized in the anterior region of C. parvum sporozoites, merozoites and intracellular parasites. Images were taken by differential interference contrast microscopy (DIC), fluorescence microscopy using filter sets for 4′,6-diamidino-2-phenylindole (DAPI) staining nuclei and tetramethylrhodamine (TRITC) labeling CpACS proteins, and superimposed (Merged). Abbreviations: a, anterior region of sporozoites or merozoites; p, posterior region of sporozoites or merozoites; em, early stage meronts; m, merozoites; r, meronts; v, parasitophorous vacuole membrane (PVM). Bars = 5 μm.
Figure 10.
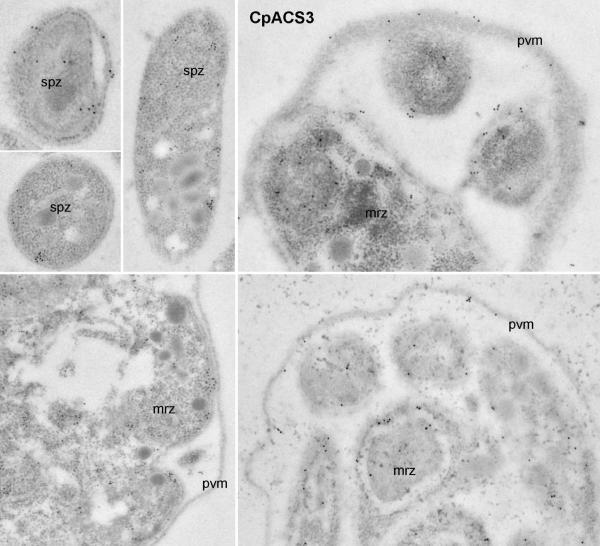
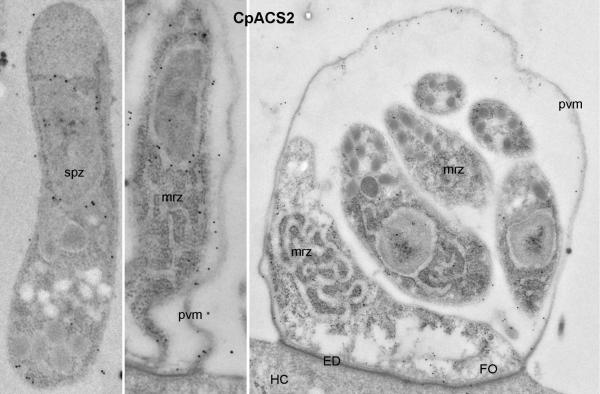
Colloidal-gold immuno-electron microscopic labeling of CpACS3 protein in sporozoites (spz) and meronts. God particles were present on the parasite cytoplasmic membranes and cytosol. However, unlike CpACS2 protein, few particles were present in the parasitophorous vacuole membranes (pvm). See figure 6 legend for abbreviations.
Western blot analysis
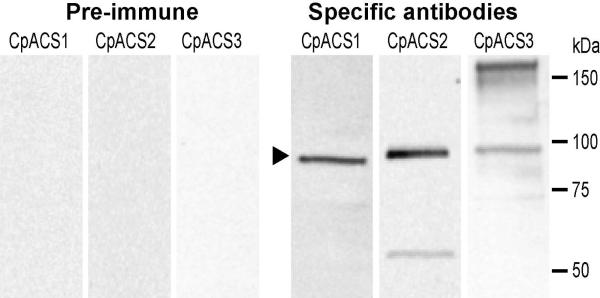
Western blot analysis using affinity-purified antibodies recognized three CpACS proteins slightly below 100 kDa (Fig. 4), which appeared to be slightly larger than their predicted molecular weights (i.e., 78.1 kDa for CpACS1, 77.1 kDa for CpACS2, and 86.1 or 83.6 kDa for intact or SP-cleaved CpACS3). This was possibly due to the combined effects of high isoelectric points of these proteins (i.e., at 9.24, 8.94 and 9.72) and a small amount of detergents present in samples, causing protein samples migrating slightly slower than the molecular standards. Pre-immune serums and IgY subjected to the same affinity-purification did not label any proteins (Fig. 4). However, antibody to CpACS2 labeled an additional minor band at ~55 kDa, while that to CpACS3 recognized a major band at ~160 kDa (Fig. 4). Based on the sizes, it was possible that CpACS2 was more sensitive to certain protease degradation, whereas CpACS3 might form protein dimers that could not be fully resolved by the SDS-PAGE fractionation, similar to the human ACSL6 isoforms observed by other investigators (Soupene et al. 2010).
Figure 4.
Western blot analysis of the three CpACS proteins. Crude protein extracts were prepared from free C. parvum sporozoites by in vitro excystation. Primary antibodies were affinity-purified. Antibodies from pre-immune animals were also affinity-purified and used as control. For clarity and easy comparison, images from the three membrane strips were digitally scaled according to positions of protein standards. The region corresponding to the CpACS bands were indicated by an arrowhead.
CpACS proteins were localized to different subcellular structures
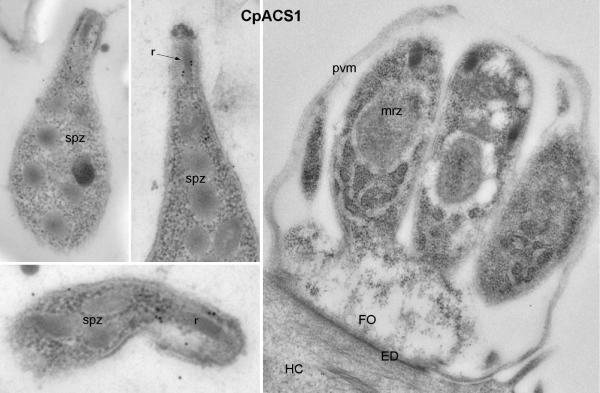
Indirect immunofluorescence microscopy (IFM) and colloidal-gold immuno-electron microscopy (IEM) using affinity-purified antibodies indicated that CpACS proteins were differentially localized in the parasite. No signals were produced using pre-immune sera that were affinity-purified in parallel with anti-serums and IgY (data not shown). Among the three CpACS proteins, CpACS1 was highly concentrated in the anterior region of free sporozoites and merozoites (Fig. 5). It was briefly retained in the anterior region after sporozoites and merozoites entered host cells (i.e., single-nuclear early stage of merogony), fully dissolved in the mid-stage of multi-nuclear meronts with no or weak fluorescence signal, and then reappeared in the anterior region of mature merozoites which were free or being released from the host cells (Fig. 5). In agreement with IFM, colloidal-gold IEM also labeled CpACS1 to the apical location of CpACS1 in sporozoites, but failed to label it to any structure in the intracellular meronts (Fig. 6). The parasite anterior region contains the unique apical complex comprised of various cytoskeletal structures and secretory organelles (i.e., micronemes and rhoptries). Our data indicate that CpACS1 is an apical protein, likely associated with the rhoptries. It is also indicative that CpACS1 may mainly participate in the activation of fatty acids required during the invasion and/or early stage of intracellular development, and/or required for the homeostasis of the secretory organelles.
Figure 6.
Colloidal-gold immuno-electron microscopic labeling of CpACS1 protein in sporozoites (spz) and a mature meront containing well-formed merozoites (mrz). Gold particles were only present in the sporozoite apical region associated with rhoptries (r), but absent in other regions and in the meront. ED, electron-dense membrane structure at the host cell-parasite interface; FO, feeder organelle (a structure at the base of meronts unique to Cryptosporidium); HC, host cell.
A number of proteins have been previously localized to the apical region in Cryptosporidium, including two P-type ATPases (Ca2+ transporter CpATPase1 and CpATPase3 with unknown substrate) (LaGier et al. 2002, Zhu and Keithly 1997), thrombospondin-related proteins (TRAP-C1 and CpTSP8) (Putignani et al. 2008, Spano et al. 1998), ATP-binding cassette proteins (ABC1 and ABC2) (Zapata et al. 2002), a serine protease (SUB1) (Wanyiri et al. 2009), a modular protein with ricin B and LCCL domains (Cpa135) (Tosini et al. 2004), and a galactose/N-acetylgalactosamine-specific lectin (p30) (Bhat et al. 2007). The present study expands the Cryptosporidium apical proteins to include an enzyme involved in fatty acid metabolism.
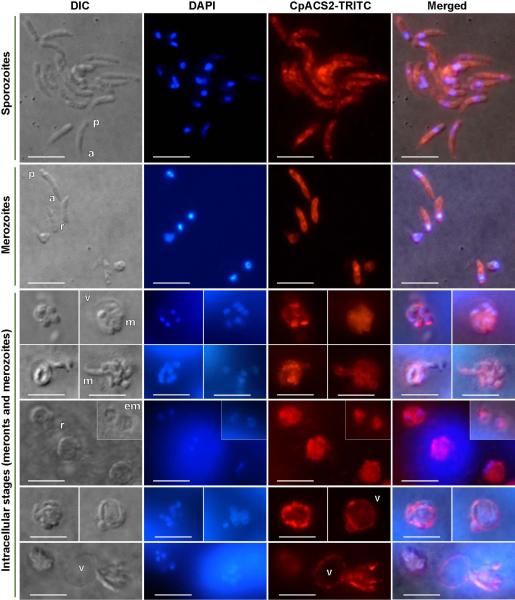
Unlike CpACS1, CpACS2 protein was distributed throughout the entire parasite cells in virtually all parasite developmental stages (Fig. 7 and 8), implying that CpACS2 might play a more general housekeeping role in the parasite. It appeared to be not homogenously distributed in the parasite, with stronger fluorescence signal from some sporozoites or merozoites (Fig. 7). IEM indicated that CpACS2 was generally concentrated along with the cytoplasmic and intracellular membranes (Fig. 8). Moreover, CpACS2 was also present in the parasitophorous vacuole membrane (PVM), a host cell-derived membrane structure separating the intracellular parasites from the intestinal lumen environment (Fig. 7 and 8). Since ACSs can function as fatty acid transporters (Weimar et al. 2002, Zou et al. 2003), we hypothesize that CpACS2 in PVM may also be involved in the scavenging of fatty acids from host cells and/or the environment. We have previously reported that at least three other enzymes related to fatty acid metabolism were localized to the PVM, including a long-type fatty acyl-CoA binding protein (CpACBP1) (Zeng et al. 2006), one of the two oxysterol-binding protein-related proteins (CpORP1) (Zeng and Zhu 2006), and a long chain fatty acid elongase (CpLCE1) (Fritzler et al. 2007). In other apicomplexans, a few ACS isoform were biochemically characterized, in which the Plasmodium falciparum ACS1 (PfACS1) was also found to be present in vesicle-like structures and distributed to the PVM (Matesanz et al. 1999). The presence of CpACS2 in PVM further supports the involvement of this unique organelle in fatty acid metabolism in the Cryptosporidium, ranging from fatty acid/lipid-sensing (CpORP1), activation/transport of fatty acids (CpACS2), elongation of fatty acyl-CoA (CpLCE1), to the storage/pool-forming of fatty acyl-CoA thioesters (CpACBP1).
Figure 7.
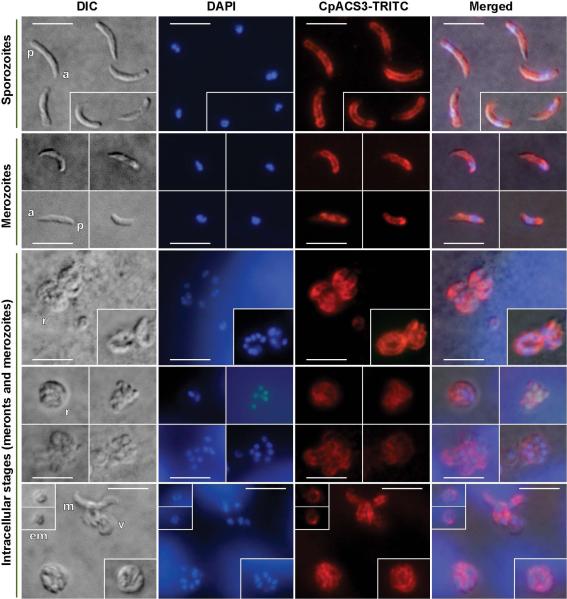
Immunofluorescence labeling showing that CpACS2 protein is localized in the cytosol and cytoplasmic membranes of all tested parasite development stages, as well as on the parasitophorous vacuole membrane. Bars = 5 μm. See figure 5 legend for abbreviations.
Figure 8.
Colloidal-gold immuno-electron microscopic labeling of CpACS2 protein in sporozoites (spz) and a mature meront. Gold particles were mainly present along with intracellular membrane structures in sporozoites (spz) and merozoites (mrz), as well as on the parasitophorous vacuole membrane (pvm). See figure 6 legend for other abbreviations.
Similar to CpACS2, CpACS3 was distributed in the entire free sporozoites, merozoites and the intracellular parasites from early to late developmental stages (Fig. 9 and 10), suggesting that CpACS3 might also function as a housekeeping protein in the parasite. However, unlike CpACS2, CpACS3 protein appeared to be more concentrated on the cytoplasmic membranes of the parasite, and with little presence on PVM. These observations suggest that CpACS3 is a parasite surface protein, probably responsible for the synthesis of fatty acyl-CoA thioesters on and/or transport of fatty acids across the parasite cytoplasmic membranes coupled with activation of long-chain fatty acids.
Figure 9.
Immunofluorescence labeling showing that CpACS3 protein is a surface protein mainly localized on the cytoplasmic membranes of all tested parasite developmental stages. See figure 4 legend for abbreviations. Bars = 5 μm. See figure 5 legend for abbreviations.
Our bioinformatics analysis indicated that CpACS3 contained an N-terminal signal peptide (Fig. 1B), which was probably responsible for targeting the protein to the cytoplasmic membranes. It was noticeable that N-terminal signal peptides were not present in CpACS2 as well as other PVM proteins reported so far (e.g., CpACBP1, CpORP1 and CpLCE1) (Fritzler et al. 2007, Zeng et al. 2006, Zeng and Zhu 2006), suggesting that signal peptides were not required for targeting proteins to the Cryptosporidium PVM, or if required, they were not present at the N-termini of proteins.
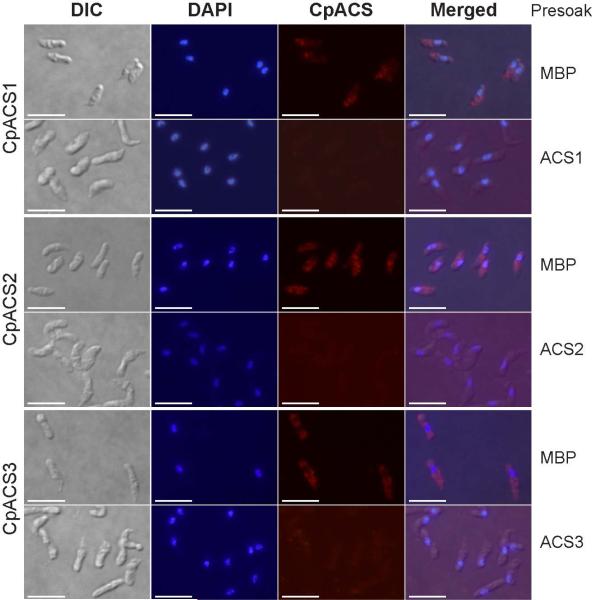
In summary, we have observed differential gene expressions and protein distribution patterns of the three CpACS isoforms during the parasite life cycle by IFM and IEM (Fig. 5-10). The specificity of the antibodies was validated by western blot analysis using affinity-purified antibodies derived from pre- and post-immune animals (Fig. 4), as well as by an antigen-competition IFM assay, in which pre-soaking of antibodies with their corresponding CpACS proteins, but not with MBP control, was able to eliminate the immunofluorescence labeling in sporozoites (Fig. 11). These observations enable us to hypothesize that CpACS1 is mainly responsible for the fatty acyl-CoA synthesis during the parasite invasion and/or early stage of intracellular development. CpACS2 may function throughout the entire parasite life cycle in scavenging fatty acids across the PVM coupled with fatty acid activation and fatty acyl-CoA synthesis within the parasite. CpACS3 mainly as a surface protein is involved in the fatty acyl-CoA synthesis on and/or transport of fatty acids across the parasite cytoplasmic membranes.
Figure 11.
Antigen-competition immunofluorescence microscopy. In this assay, specific fluorescence labeling of ACS proteins in the parasite sporozoites was eliminated by presoaking CpACS antibodies with their corresponding antigens, but not by presoaking them with maltose-binding protein (MBP). Bars = 5 μm. See figure 5 legend for abbreviations.
ACKNOWLEDGMENTS
Research reported in this study was supported by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health under award number R01AI44594 (to G.Z.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
LITERATURE CITED
- Abrahamsen MS, Templeton TJ, Enomoto S, Abrahante JE, Zhu G, Lancto CA, Deng M, Liu C, Widmer G, Tzipori S, Buck GA, Xu P, Bankier AT, Dear PH, Konfortov BA, Spriggs HF, Iyer L, Anantharaman V, Aravind L, Kapur V. Complete genome sequence of the apicomplexan, Cryptosporidium parvum. Science. 2004;304:441–5. doi: 10.1126/science.1094786. [DOI] [PubMed] [Google Scholar]
- Andersson CS, Lundgren CA, Magnusdottir A, Ge C, Wieslander A, Martinez Molina D, Hogbom M. The Mycobacterium tuberculosis very-long-chain fatty acyl-CoA synthetase: structural basis for housing lipid substrates longer than the enzyme. Structure. 2012;20:1062–70. doi: 10.1016/j.str.2012.03.012. [DOI] [PubMed] [Google Scholar]
- Arrowood MJ, Sterling CR. Isolation of Cryptosporidium Oocysts and Sporozoites Using Discontinuous Sucrose and Isopycnic Percoll Gradients. J. Parasitol. 1987;73:314–319. [PubMed] [Google Scholar]
- Bhat N, Joe A, PereiraPerrin M, Ward HD. Cryptosporidium p30, a galactose/N-acetylgalactosamine-specific lectin, mediates infection in vitro. J. Biol. Chem. 2007;282:34877–87. doi: 10.1074/jbc.M706950200. [DOI] [PubMed] [Google Scholar]
- Black PN, Zhang Q, Weimar JD, DiRusso CC. Mutational analysis of a fatty acyl-coenzyme A synthetase signature motif identifies seven amino acid residues that modulate fatty acid substrate specificity. J. Biol. Chem. 1997;272:4896–903. doi: 10.1074/jbc.272.8.4896. [DOI] [PubMed] [Google Scholar]
- Cai X, Woods KM, Upton SJ, Zhu G. Application of quantitative real-time reverse transcription-PCR in assessing drug efficacy against the intracellular pathogen Cryptosporidium parvum in vitro. Antimicrob. Agents Chemother. 2005;49:4437–42. doi: 10.1128/AAC.49.11.4437-4442.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Checkley W, White AC, Jr., Jaganath D, Arrowood MJ, Chalmers RM, Chen XM, Fayer R, Griffiths JK, Guerrant RL, Hedstrom L, Huston CD, Kotloff KL, Kang G, Mead JR, Miller M, Petri WA, Jr., Priest JW, Roos DS, Striepen B, Thompson RC, Ward HD, Van Voorhis WA, Xiao L, Zhu G, Houpt ER. A review of the global burden, novel diagnostics, therapeutics, and vaccine targets for cryptosporidium. Lancet Infect. Dis. 2015;15:85–94. doi: 10.1016/S1473-3099(14)70772-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen XM, Keithly JS, Paya CV, LaRusso NF. Cryptosporidiosis. N Engl J Med. 2002;346:1723–31. doi: 10.1056/NEJMra013170. [DOI] [PubMed] [Google Scholar]
- Fritzler JM, Millership JJ, Zhu G. Cryptosporidium parvum long-chain fatty acid elongase. Eukaryot. Cell. 2007;6:2018–28. doi: 10.1128/EC.00210-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujino T, Kang MJ, Suzuki H, Iijima H, Yamamoto T. Molecular characterization and expression of rat acyl-CoA synthetase 3. J. Biol. Chem. 1996;271:16748–52. doi: 10.1074/jbc.271.28.16748. [DOI] [PubMed] [Google Scholar]
- Guo F, Zhang H, Fritzler JM, Rider SD, Jr., Xiang L, McNair NN, Mead JR, Zhu G. Amelioration of Cryptosporidium parvum infection in vitro and in vivo by targeting parasite fatty acyl-coenzyme A synthetases. J. Infect. Dis. 2014;209:1279–87. doi: 10.1093/infdis/jit645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotloff KL, Nataro JP, Blackwelder WC, Nasrin D, Farag TH, Panchalingam S, Wu Y, Sow SO, Sur D, Breiman RF, Faruque AS, Zaidi AK, Saha D, Alonso PL, Tamboura B, Sanogo D, Onwuchekwa U, Manna B, Ramamurthy T, Kanungo S, Ochieng JB, Omore R, Oundo JO, Hossain A, Das SK, Ahmed S, Qureshi S, Quadri F, Adegbola RA, Antonio M, Hossain MJ, Akinsola A, Mandomando I, Nhampossa T, Acacio S, Biswas K, O'Reilly CE, Mintz ED, Berkeley LY, Muhsen K, Sommerfelt H, Robins-Browne RM, Levine MM. Burden and aetiology of diarrhoeal disease in infants and young children in developing countries (the Global Enteric Multicenter Study, GEMS): a prospective, case-control study. Lancet. 2013;382:209–222. doi: 10.1016/S0140-6736(13)60844-2. [DOI] [PubMed] [Google Scholar]
- Kurien BT. Affinity purification of autoantibodies from an antigen strip excised from a nitrocellulose protein blot. Methods Mol Biol. 2009;536:201–11. doi: 10.1007/978-1-59745-542-8_22. [DOI] [PubMed] [Google Scholar]
- LaGier MJ, Keithly JS, Zhu G. Characterisation of a novel transporter from Cryptosporidium parvum. Int. J. Parasitol. 2002;32:877–87. doi: 10.1016/s0020-7519(02)00018-8. [DOI] [PubMed] [Google Scholar]
- Matesanz F, Duran-Chica I, Alcina A. The cloning and expression of Pfacs1, a Plasmodium falciparum fatty acyl coenzyme A synthetase-1 targeted to the host erythrocyte cytoplasm. J. Mol. Biol. 1999;291:59–70. doi: 10.1006/jmbi.1999.2964. [DOI] [PubMed] [Google Scholar]
- Matesanz F, Tellez MM, Alcina A. The Plasmodium falciparum fatty acyl-CoA synthetase family (PfACS) and differential stage-specific expression in infected erythrocytes. Mol. Biochem. Parasitol. 2003;126:109–12. doi: 10.1016/s0166-6851(02)00242-6. [DOI] [PubMed] [Google Scholar]
- Melton EM, Cerny RL, Watkins PA, DiRusso CC, Black PN. Human fatty acid transport protein 2a/very long chain acyl-CoA synthetase 1 (FATP2a/Acsvl1) has a preference in mediating the channeling of exogenous n-3 fatty acids into phosphatidylinositol. J. Biol. Chem. 2011;286:30670–9. doi: 10.1074/jbc.M111.226316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putignani L, Possenti A, Cherchi S, Pozio E, Crisanti A, Spano F. The thrombospondin-related protein CpMIC1 (CpTSP8) belongs to the repertoire of micronemal proteins of Cryptosporidium parvum. Mol. Biochem. Parasitol. 2008;157:98–101. doi: 10.1016/j.molbiopara.2007.09.004. [DOI] [PubMed] [Google Scholar]
- Rider SD, Jr., Zhu G. Cryptosporidium: genomic and biochemical features. Exp. Parasitol. 2010;124:2–9. doi: 10.1016/j.exppara.2008.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson LJ, Campbell AT, Smith HV. In vitro excystation of Cryptosporidium parvum. Parasitology. 1993;106(Pt 1):13–9. doi: 10.1017/s003118200007476x. [DOI] [PubMed] [Google Scholar]
- Schnurr J, Shockey J, Browse J. The acyl-CoA synthetase encoded by LACS2 is essential for normal cuticle development in Arabidopsis. The Plant cell. 2004;16:629–42. doi: 10.1105/tpc.017608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soupene E, Kuypers FA. Mammalian long-chain acyl-CoA synthetases. Exp. Biol. Med. 2008;233:507–21. doi: 10.3181/0710-MR-287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soupene E, Dinh NP, Siliakus M, Kuypers FA. Activity of the acyl-CoA synthetase ACSL6 isoforms: role of the fatty acid Gate-domains. BMC Biochem. 2010;11:18. doi: 10.1186/1471-2091-11-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spano F, Putignani L, Naitza S, Puri C, Wright S, Crisanti A. Molecular cloning and expression analysis of a Cryptosporidium parvum gene encoding a new member of the thrombospondin family. Mol. Biochem. Parasitol. 1998;92:147–62. doi: 10.1016/s0166-6851(97)00243-0. [DOI] [PubMed] [Google Scholar]
- Tosini F, Agnoli A, Mele R, Gomez Morales MA, Pozio E. A new modular protein of Cryptosporidium parvum, with ricin B and LCCL domains, expressed in the sporozoite invasive stage. Mol. Biochem. Parasitol. 2004;134:137–47. doi: 10.1016/j.molbiopara.2003.11.014. [DOI] [PubMed] [Google Scholar]
- Wanyiri JW, Techasintana P, O'Connor RM, Blackman MJ, Kim K, Ward HD. Role of CpSUB1, a subtilisin-like protease, in Cryptosporidium parvum infection in vitro. Eukaryot. Cell. 2009;8:470–7. doi: 10.1128/EC.00306-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weimar JD, DiRusso CC, Delio R, Black PN. Functional role of fatty acylcoenzyme A synthetase in the transmembrane movement and activation of exogenous long-chain fatty acids. Amino acid residues within the ATP/AMP signature motif of Escherichia coli FadD are required for enzyme activity and fatty acid transport. J. Biol. Chem. 2002;277:29369–76. doi: 10.1074/jbc.M107022200. [DOI] [PubMed] [Google Scholar]
- Zapata F, Perkins ME, Riojas YA, Wu TW, Le Blancq SM. The Cryptosporidium parvum ABC protein family. Mol. Biochem. Parasitol. 2002;120:157–61. doi: 10.1016/s0166-6851(01)00445-5. [DOI] [PubMed] [Google Scholar]
- Zeng B, Zhu G. Two distinct oxysterol binding protein-related proteins in the parasitic protist Cryptosporidium parvum (Apicomplexa). Biochem. Biophys. Res. Commun. 2006;346:591–99. doi: 10.1016/j.bbrc.2006.05.165. [DOI] [PubMed] [Google Scholar]
- Zeng B, Cai X, Zhu G. Functional characterization of a fatty acyl-CoA-binding protein (ACBP) from the apicomplexan Cryptosporidium parvum. Microbiology. 2006;152:2355–63. doi: 10.1099/mic.0.28944-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Guo F, Zhu G. Involvement of Host Cell Integrin alpha2 in Cryptosporidium parvum Infection. Infect. Immun. 2012;80:1753–8. doi: 10.1128/IAI.05862-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu G, Keithly JS. Molecular analysis of a P-type ATPase from Cryptosporidium parvum. Mol. Biochem. Parasitol. 1997;90:307–16. doi: 10.1016/s0166-6851(97)00168-0. [DOI] [PubMed] [Google Scholar]
- Zou Z, Tong F, Faergeman NJ, Borsting C, Black PN, DiRusso CC. Vectorial acylation in Saccharomyces cerevisiae. Fat1p and fatty acyl-CoA synthetase are interacting components of a fatty acid import complex. J. Biol. Chem. 2003;278:16414–22. doi: 10.1074/jbc.M210557200. [DOI] [PubMed] [Google Scholar]