Abstract
Background
Studies of mitochondrial morphology vary in techniques. Most use one morphological parameter while others describe mitochondria qualitatively. Because mitochondria are so dynamic, a single parameter does not capture the true state of the network and may lead to erroneous conclusions. Thus, a gestalt method of analysis is warranted.
New Method
This work describes a method combining immunofluorescence assays with computerized image analysis to measure the mitochondrial morphology within neuritic projections of a specific population of neurons. Six parameters of mitochondrial morphology were examined utilizing ImageJ to analyze colocalized signals.
Results
Using primary neuronal cultures from Drosophila, we tested mitochondrial morphology in neurites of dopaminergic (DA) neurons. We validate our model using mutants with known defects in mitochondrial morphology. Furthermore, we show a difference in mitochondrial morphology between cells treated as control or with a neurotoxin inducing PD (Parkinson's Disease in humans)-like pathology. We also show interactions between morphological parameters and experimental treatment.
Comparison with Existing Methods
Our method is a significant improvement of previously described methods. Six morphometric parameters are quantified, providing a gestalt analysis of mitochondrial morphology. Also it can target specific populations of mitochondria using immunofluorescence assay and image analysis.
Conclusions
We found that our method adequately detects differences in mitochondrial morphology between treatment groups. We conclude that some parameters may be unique to a mutation or a disease state, and the relationship between parameters is altered by experimental treatment. We suggest at least four variables should be considered when using mitochondrial structure as an experimental endpoint.
Keywords: Drosophila, mitochondria, drp1, opa1, Parkinson
Introduction
Mitochondria are a dynamic collective of organelles that, in addition to supplying to the cell with energy, keenly respond to cellular stress. They have important functional relationships with apoptosis, metabolism, and other diseases (Rugarli and Langer, 2012). However, there is seemingly no agreement on how to best describe or report the morphology of these organelles. Some studies use individual mitochondria (Wang et al., 2011), while others examine all of the mitochondria in a cell (Koopman et al., 2006; Dagda, 2009; Leonard et al., 2015). Also important is which cellular compartment is best to measure mitochondria. In neurons, the mitochondria in the soma are likely not as critical as mitochondria in the dendrites and/or axons for synaptic transmission (Cheng et al., 2010; Court and Coleman, 2012). Thus, measuring one parameter from individual mitochondria is not capturing the totality of the state of the network in terms of morphology.
The range of possible morphologies for mitochondria is wide and dependent on cell-type, particularly for neurons and especially for the dopaminergic (DA) neurons implicated in Parkinson's disease (PD). Mitochondria in these highly-specialized neurons are smaller and have a lower total mass compared to mitochondria in other neurons (Liang et al., 2007; Perier and Vila, 2012), and are selectively vulnerable to oxidative stress (Surmeir et al., 2013; Wiemerslage et al., 2013). In general, damaged mitochondria are fragmented, round, and swollen (Chang and Reynolds, 2006; Berman et al., 2008; Perier and Vila, 2012), and such changes in mitochondria are implicated in Parkinson's disease (PD) (Dawson et al., 2010; Nakamura et al., 2011; Saxena and Caroni, 2011; Botella et al., 2011). A difficulty in studying neurodegenerative disorders such as PD is that many of the neurons implicated in the disease are already dead by the time symptoms appear (Jellinger, 2012; Smith et al., 2012). However, mitochondria could serve as an early biomarker to screen for neuroprotective therapies by trying to reverse changes in mitochondrial morphology caused by neurodegenerative processes.
In this work, we utilize our previous cell culture model of PD-like degeneration (Wiemerslage et al., 2013) and adapt an ImageJ macro written by Dagda et al., 2009 to quantify the mitochondrial morphology specifically in DA neurons. First, we quantify different parameters of morphology using mutants (e.g. drp1, opa1) with known morphological defects in their mitochondria. Then we compare mitochondrial morphology between control and PD-like treatment. Lastly, we examine the specific parameters measured to determine how many parameters are needed for a comprehensive assessment of mitochondrial morphology.
Methods
Fly stocks
All lines were obtained from the Bloomington Drosophila Stock Center (BDSC) unless otherwise noted. Flies were kept at 25°C and raised on standard cornmeal agar diet. A ‘Cantonized’ white eye stock w118 served as wild-type. Morphometric control experiments used drp1/CyO-GFP and opa1/CyO-GFP (gifts from Dr. Leo Pallanck, University of Washington).
Drosophila primary neuronal cultures
Cultures were prepared as previously described in Park and Lee (2006). Briefly, mid-gastrula embryos at developmental stage 7 were harvested in a laminar-flow hood and plated onto round, glass coverslips (Bellco Glass, Inc., Vineland NJ, USA). Cultures were incubated in 4-5% CO2 at 24-25° C for up to 9 days in vitro (DIV). Culture medium (DDM1) was a mixture of high glucose Hams's F-12/Delbecco's medium (Irvine Scientific, Santa Ana, CA), L-glutamine (2.5mM; Irvine Scientific), HEPES (20 mM), and four supplements: putrescine (100 μM), progesterone (20 ng/mL), transferrin (100 μg/mL), and insulin (50 μg/mL). At 3 DIV, all cultures had 50% of the culture medium replaced with new medium.
Pharmacological treatments
All drugs were added to cultures at 3 DIV after baseline images were acquired, except for experiments with delayed use of rescue therapy. Drug remained in the dish once treated (i.e. was never washed out). Cultures were handled and treated in a laminar flow hood (Forma Scientific, model 1849). Drugs dissolved in ddH2O were sterilized by filtration through a 0.2 μm cellulose acetate filter before use/storage. 1-methyl-4-phenylpyridinium (MPP+) iodide (Sigma) was prepared as a 40mM stock solution dissolved in ddH2O and stored in darkness at -70° C. MPP+ was handled according to guidelines reviewed in Przedborski et al. (2001).
Immunofluorescence assay
Cultures were fixed with 4% paraformaldehyde for 40 minutes on ice, and then washed 3 times, 10 minutes for each wash. Wash solution was 10mM phosphate buffered saline containing 0.5% bovine serum albumin. All washes were at room temperature (≈25°C). Blocking and permeabilization was performed using 0.1% Triton X-100 and 5% normal goat serum (Sigma) for 30 minutes on ice. After 1 more wash for 10 minutes, permeabilized cultures were incubated overnight (≈16 hours) at 4° C with a 1:1,000 ratio of primary antibody (mouse anti-tyrosine hydroxylase, ImmunoStar) diluted in wash. The next day, primary antibody was removed and cultures were washed 3 times, 10 minutes for each wash. Cultures were next treated with a 1:2,000 ratio of secondary antibody (FITC or TRITC labeled goat anti-mouse, Invitrogen) diluted in wash and placed on ice for 1 hour. Secondary antibody was then removed and cultures were again washed 3 times, 10 minutes for each wash. Coverslips were next mounted onto glass slides upon rows of fluorogel with Tris buffer (Electron Microscopy Sciences), covered with an extra drop of fluorogel, then topped with coverglass (Electron Microscopy Sciences), and edges sealed with clear fingernail polish.
Microscopic detection of mitochondria in dopaminergic neurons
To visualize mitochondria from DA neurons, cultures were stained with both MitoTracker Orange (Invitrogen) and anti-TH antibody (ImmunoStar). At 7 DIV but before staining, cultures were treated with 50 nM MitoTracker Orange (Invitrogen) in the original culture medium for 1 hour at 25° C prior to immunostaining. Culture medium was then removed. Neuronal cultures were fixed and stained as usual (described above) with anti-TH primary antibody and FITC-labeled secondary antibody. The colocalization of signals between MitoTracker and anti-TH staining specifically identified mitochondria in DA neurons for analysis.
Image acquisition
Prepared/stained cultures were viewed under a fluorescence microscope (Olympus IX71 with 100W Mercury lamp). Mitochondria were observed with a LUCPLFLN 40× lens (NA, 0.60). Two fluorescence filter sets were also used as following: Chroma 41017 (FRITC) and 41002 (TRITC). Images were taken using Spot CCD digital camera (2 megapixels, Diagnostic Instruments, Sterline Heights, MI). For a 12-bit image, the intensity ranges from 0 (no signal) to 4095 (maximum saturation). To determine the best threshold for our analysis of mitochondria, the raw MitoTracker signal of several images of mitochondria was compared to its “ideal” threshold level. The ideal threshold level was considered the one where a manual count of the mitochondria from the raw image gave the same results as the Analyze Particles function in ImageJ. The mean ideal threshold level was then set as the threshold to be applied for all future analysis of mitochondria.
Analysis of mitochondrial morphology in dopamine neurons
After images were acquired, they were analyzed using a macro developed for ImageJ (Rasband 1997-2006) software (See Appendix A for code, installation, and use of macro). ImageJ allows for users to write programs (typically macros) in a “Java-like” language. This macro combines a plugin created by Dagda et al. (2009) with the colocalization highlighter plugin from the WCIF version of ImageJ (http://www.uhnresearch.ca/facilities/wcif/imagej/). Analysis of mitochondrial morphology is performed on a merged image between images of the MitoTracker and anti-TH signals. Before the merge, however, both channels are subjected to a threshold to make them binary images. Then an overlap of those two binary images is made. This overlapped image is then used for analysis. The signals are painted by the threshold function in ImageJ based on signal intensity. For mitochondria, this threshold was determined to be 20% of the maximum intensity, which is about 4× the typical background intensity. For the anti-TH signal, we chose a threshold (also around 20% of the maximum intensity) which completely saturated the cell with signal so that all mitochondria within would colocalize.
Colocalized signals were measured using a macro adapted from the plugin created by Dagda et al. (2009). Major differences in the macro from Dagda's original, were changing the threshold to 20% of the maximum intensity and the measured particle range to 5-500 pixels. This macro quantifies mitochondrial morphometrics using four parameters: 1) number of mitochondria, 2) size, 3) interconnectivity, and 4) elongation. These four parameters taken together provide a gestalt “snapshot” of the mitochondrial phenotype.
The macro only considers signals within a size range. With our microscope settings and image size, the mean size for mitochondria is around 40 pixels2 (1.4 μm2), matching what is commonly reported across species (0.75 to 3 μm2; Bereiter-Hahn, 1990; Rafelski and Wallace, 2008). Thus, the lower and upper limits of the size range had to be decreased from Dagda's original macro. If the lower limit was too low, it would include tiny artifacts as signals, which do not provide meaningful parameter values. The upper limit is less of a concern. In healthy neurons, many of the mitochondria are interconnected, combining to a pixel area much greater than the average mitochondria size, which is the phenomenon that is measured by the interconnectivity score. However, because the soma is typically completely saturated in our images by both the anti-tyrosine hydroxylase antibody and MitoTracker signals, it is not included in analysis. This is easily accomplished by simply drawing around the soma with the selection tool in ImageJ (Figure 1). An upper limit of 500 pixels2 was used to exclude what are probable artifacts (e.g. the occasional piece of broken glass added during culturing). Thus, a particle size range of 5-500 pixels2 was chosen for non-somatic mitochondria.
Figure 1. Visualizing mitochondria and quantifying their morphology in a specific cell type.
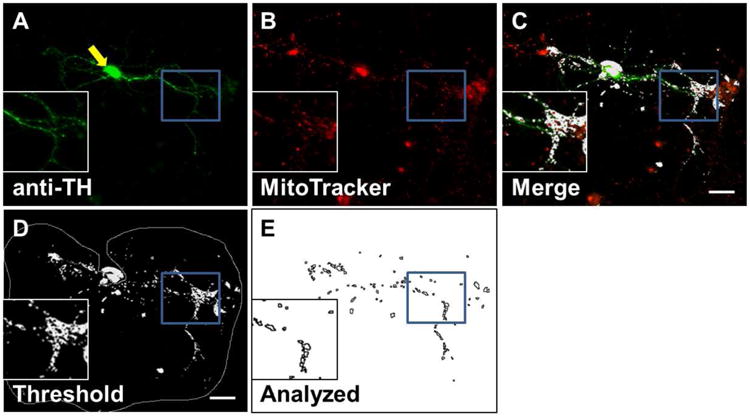
Example images from immunocytochemistry method of quantifying mitochondria in neurites of dopaminergic (DA) neurons. A) DA neurons (green) are identified by antibody to tyrosine hydroxylase (anti-TH). B) Mitochondria (red) are identified by MitoTracker staining. C) Colocalized signals (white) from anti-TH and MitoTracker show as white and represent mitochondria specifically from DA soma and neurites. D) Threshold painting of the mitochondrial signal. The soma is indicated by a yellow arrow. The grey line indicates the selected area to be analyzed (soma excluded). E) Resulting analyzed image from D. Soma and particles outside of specified size range are excluded. Outlines of remaining mitochondrial signals are drawn in black. Image taken at 7 days in vitro (DIV), scale bar = 20 μm.
Determining the four parameters of mitochondrial morphology requires computation. Counting the number of mitochondria and determining the mean size is a straightforward task; the ImageJ software simply counts each signal and simultaneously determines the pixel area, perimeter, and circularity for each signal. From these measurements, the ImageJ macro calculates a score for interconnectivity and elongation as the following:
| (1) |
| (2) |
Interconnectivity describes the network of the mitochondria, and is calculated by dividing the mean area by the mean perimeter of all the particles analyzed. Higher scores for interconnectivity signify that mitochondria have more physical connections, while lower scores signify that the mitochondria are more fragmented. Elongation is best thought of as the shape of mitochondria. Higher values are more abstract shapes, while a value of 1 would be considered a perfect circle.
Statistics
All statistics are reported as mean ± 1 SEM. Analysis is performed using either ANOVA with pairwise comparisons using Tukey's Honest Significant Difference correction, or Student's t-test. Significance scores are: * for p < 0.05, ** for p < 0.01, and *** for p < 0.001. All distributions are tested for normality and homogeneity of variance before testing. Principal component analysis was performed with R statistical software using the FactoMineR package (Lê et al., 2008). Results were considered significant if the percentage of inertia summing from the first two eigenvalues exceeded values listed in a significance table based on 10,000 analyses with similar numbers of individuals and independent variables (Lê et al., 2008).
Results
Targeting Mitochondria in a Specific Cell Type and Compartment
Our method to quantify mitochondria builds on previous work (Dagda et al., 2009) that analyzes their morphology using four parameters: number, size, interconnectivity, and elongation. Previous studies may measure only one parameter and ignore other potential factors in the mitochondrial network, whereas this analysis gives a comprehensive assessment of the mitochondrial morphology. We improve upon this method to allow for the analysis of mitochondrial morphology between different cell types as well as cell compartments.
We used immunofluorescence assay to target non-somatic mitochondria in specific cell populations for analysis, in this case dopaminergic (DA) neurons. We stained Drosophila primary neuronal cultures with both MitoTracker Orange and an antibody to tyrosine hydroxylase (TH). MitoTracker stains mitochondria in all cells, while the antibody to TH stains only DA neurons. We then used the overlapping image of the two signals to identify mitochondria in the neurites of DA neurons, specifically (Figure 1).
Quantifying Mitochondrial Morphology in Drosophila Dopaminergic Neurons using Mutants with Known Morphological Defects
Mitochondria vary their morphology via fission and fusion. In general, fusion processes appear neuroprotective, while fission processes are implicated in cell death (Berman et al., 2008; Perier and Vila, 2012). We suspected that loss-of-function mutants for both of these processes would adequately benchmark the detectable range of our method. Thus, we validated our method using mutant strains with known defects/phenotypes in mitochondrial fusion and fission: dynamin-related protein 1 (drp1) mutant, which prevents mitochondrial fission, and optic atrophy 1 (opa1) mutant, which prevents mitochondrial fusion (Okamoto and Shaw, 2005).
Flies homozygous for drp1 and opa1 loss-of-function mutations are embryonic lethal, so the mutations are maintained over a balancer chromosome: CyO-GFP. Using the balanced fly lines (drp1/CyO-GFP and opa1/CyO-GFP), single-embryonic cultures were prepared as previously described (Darya et al., 2009). Briefly, from either strain, the genotype of any single-embryonic culture is one of the following three: homozygous mutant (-/-), heterozygous GFP/-, or homozygous GFP/GFP. Homozygous mutant cultures were selected by intensity of GFP signal. Coverslips homozygous for the drp1 or opa1 mutation had no GFP signa, while coverslips homozygous for the GFP balancer had an intense live-GFP signal (10× more luminous than background) and were used as controls. After 7 days in vitro (DIV), cultures were stained with MitoTracker and processed for immunocytochemistry staining with anti-TH.
Importantly, expression of GFP in the balancer chromosome (e.g. CyO) is driven by an actin promotor (Act) and limited to non-neuronal tissue (refer to Figure 2A in Darya et al., 2009). Thus, DA neurons showing green signal of the secondary FITC-tagged antibody for the primary anti-TH, were clearly distinguished from the non-neuronal Act-GFP+ cells. Furthermore, no Act-GFP signal was found in the neurites. Thus, the colocalization signal from the MitoTracker and anti-TH antibody identifies mitochondria specifically in DA neurons for analysis.
Figure 2. Mitochondrial morphology in fission and fusion mutants.
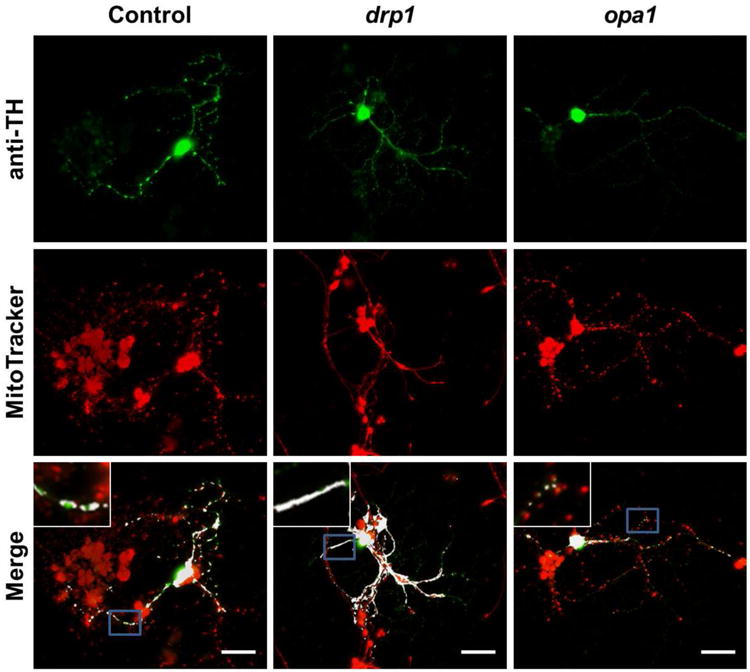
Quantification of mitochondrial morphology in control and mutant neuronal cultures. Top row: dopaminergic (DA) cells for control, drp1 mutants, and opa1 mutants. Signal is from antibody to tyrosine hydroxylase (TH), selectively marking soma and neurites of DA neuron. Middle row: images of MitoTracker staining for control, drp1 and opa1. Mitochondria from all cell types are stained. Bottom row: overlapped image of anti-TH and MitoTracker signals. Colocalized signals are shown in white, i.e. are mitochondria from DA neurons. Single-embryo cell cultures on individual glass coverslips were prepared from drp1/CyO-GFP and opa1/CyO-GFP fly lines. At 3 DIV, coverslips were selected by the presence/intensity of the live-GFP signal. Coverslips without a live-GFP signal were homozygous for drp1 or opa1. Coverslips with the brightest live-GFP signals were used as controls. At 7 DIV, coverslips were stained with anti-TH and MitoTracker and images were taken for analysis (scale bar = 20 μm, insets are of areas outlined in blue).
We detected differences in mitochondrial morphology using both mutant strains. Cultures from drp1 mutants had elongated and widely connected mitochondria, while opa1 mutants had isolated and round mitochondria (Figure 3A). Both drp1 and opa1 reduced the number of mitochondria (per DA neuron) from 96 ± 7 (control, see below) to 41 ± 11 (57% decrease) and 58±5 (43% decrease), respectively (Figure 2). A decreased number of mitochondria in drp1 mutants was expected because mitochondria are highly interconnected in the drp1 mutants.
Figure 3. Cells with decreased mitochondrial fission or fusion have changes in all measured parameters.
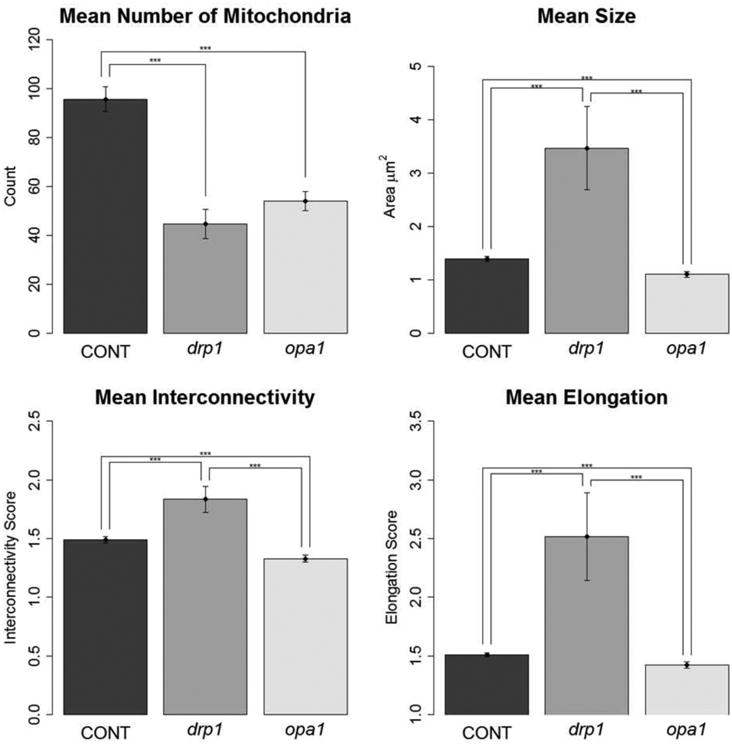
Quantification of mitochondrial morphology in control and mutant neuronal cultures. Graphs showing morphological characteristics of mitochondria: number, size, interconnectivity, and elongation. drp1 mutants had larger, more interconnected, and elongated mitochondria compared to control and opa1, while opa1 had smaller, less interconnected, and less elongated mitochondria compared to drp1 and control. Both drp1 and opa1 had fewer mitochondria than control – likely because in drp1 cells, the mitochondria are all connected and thus not counted as separate, and in opa1 cells because the fragmented mitochondria are translocated to the soma where they are not included in analysis. At 7 DIV, images were taken for analysis (Tukey's HSD test; number of experiments: Control, n = 7; drp1, n = 3; opa1 n = 3; *** p < 0.001).
For size, interconnectivity, and elongation, the trend for each of the three treatment groups was the same: drp1 had the highest value, opa1 the lowest, and controls fell in the middle. drp1 mitochondria had a mean area of 2.8 ± 0.2 μm2, 131% larger than control at 1.2 ± 0.1 μm2. opa1 mitochondria were 37% smaller than control at 0.8 ± 0.1 μm2. drp1 mitochondria were 40% more interconnected than control, with an interconnectivity score of 1.85 ± 0.07 compared to 1.33 ± 0.03. opa1 mitochondria were 15% less interconnected compared to control with an interconnectivity score of 1.12 ± 0.07. Lastly, drp1 mitochondria were 40% more elongated than control, with an elongation score of 1.85 ± 0.07 compared to 1.33 ± 0.03. opa1 mitochondria were 8% less elongated compared to control with an elongation score of 1.12 ± 0.07 (Figure 3). Thus, the method successfully detected a difference in each parameter of mitochondrial morphology in the neurites of DA cells with altered mitochondrial fission and fusion.
Degeneration of Mitochondria in Dopaminergic Neurons by a Parkinson's disease-like Neurotoxin
After validation of our two methods, we tested mitochondrial morphology in our previously reported in vitro model of Parkinson's disease (PD) (Wiemerslage et al., 2013). Our method specifically tested non-somatic, mitochondria (i.e. mitochondria in the neurites) of DA neurons. Non-somatic mitochondria are likely more important for synaptic signaling and functionality than somatic mitochondria (Cheng et al., 2010; Court and Coleman, 2012). Indeed, axons are thought to possess a unique, genetically-controlled self-destruction program which functions somewhat in parallel to apoptosis (Barrientos et al., 2011), and mitochondria are a major part of the mechanisms controlling axonal degeneration (Court and Coleman, 2012). Moreover, mitochondria are implicated in the death of DA cells, the major pathogenic event in PD (Cicchetti et al., 2009; Perier and Vila, 2012). Thus, we wanted to examine the changes in mitochondrial morphology of these cells in response to PD-like treatment. We used the neurotoxin 1-methyl-4-phenylpyridinium (MPP+), as it models PD both in terms of symptoms/behaviors produced as well as molecular events (Manning-Boğ et al., 2007; Ghosh et al., 2012). Moreover, it specifically inhibits complex-I of the mitochondria (Levy et al., 2009).
PD-like treatment decreased all the parameters of mitochondrial morphology compared to control (Figure 4A). 40 μM of MPP+ decreased all the measured parameters compared to control: the number of mitochondria decreased 53% from 88 ± 8 to 41 ± 3, the size of mitochondria decreased 47% from 1.5 ± 0.1 to 0.8 ± 0.1 μm2, interconnectivity decreased 27% from 1.56 ± 0.04 to 1.14 ± 0.03, and elongation decreased 7% from 1.50 ± 0.02 to 1.40 ± 0.02 (Figure 4B). Thus, our results show that MPP+ fragments mitochondria: decreasing the number, shrinking them, breaking down the network, and making them rounder.
Figure 4. MPP+ treatment causes fragmented mitochondrial morphology.
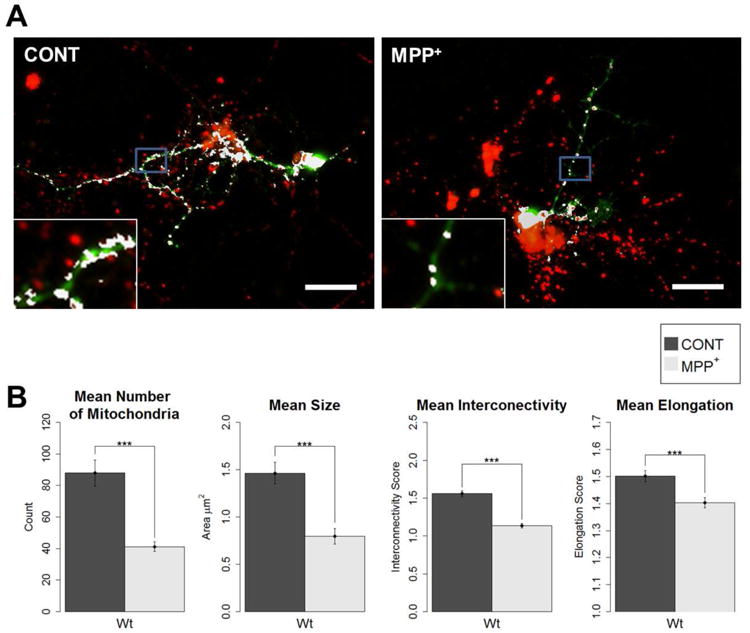
Primary neuronal cell cultures were prepared from wild-type fly embryos. At 3 DIV, cultures were treated with 40 μM MPP+, or as control. At 7DIV, cells were stained with anti-TH and MitoTracker for the colocalization analysis of mitochondria in DA neurons. A) Example images of control and MPP+-treated dopaminergic (DA) neurons from immunocytochemistry colocalization method (anti-TH and MitoTracker). Mitochondria of dopaminergic neurons are shown in white (scale bar = 20 μm, insets are of areas outlined in blue). B) All four parameters showed a significant decrease from control when treated with MPP+. Primary neuronal cell cultures were prepared from wild-type fly embryos. At 3 DIV, cultures were treated with 40 μM MPP+, or as control. At 7DIV, cells were stained with MitoTracker and anti-TH for the colocalization analysis, and images were taken for analysis (Student's t-test; *** p < 0.001); number of DA neurons analyzed: Control = 29, MPP+ = 51).
Justification of Parameters
Dagda et al. 2009 used four parameters for his analysis of somatic mitochondria in SH-SY5Y cells: the number of mitochondria, the mean size, the interconnectivity score, and the elongation score. We wondered if other parameters may improve the analysis. We included two more parameters: total area, and perimeter. Respectively, they are the sum of all pixels measured from each mitochondrion for a given cell, and the mean perimeter of all mitochondria. Moreover, we wondered if some parameters could be consolidated into one. Some studies report only one parameter of mitochondria to make conclusions regarding morphology. Thus, the independence of these parameters from each other is important. Researchers may be justified in measuring only the size or shape if all the parameters signify the same phenomena – health of the mitochondrial network. To answer these questions, we examined the correlations of all the above parameters between each other. This analysis was performed from 52 individual control cells, not individual mitochondria, i.e., the number of mitochondria, size, interconnectivity, elongation, total area, and perimeter were each determined for a given cell and the correlation coefficient was determined between the resulting measurements of the 52 cells (i.e., n = 52 for all tests).
The original four parameters used by Dagda et al. 2009 (number of mitochondria, size, interconnectivity, and elongation) were almost completely independent. There was only one strong correlation between these four parameters, which was between size and interconnectivity. They had a positive correlation of 0.91 (p < 0.001), while the other correlations between the other original four parameters were within a -0.06 and 0.13 correlation. The added parameters (total area and perimeter) had strong correlations to other parameters, and hence are not independent (Table 1). From these results, it 3 parameters are completely independent of each other: the number of mitochondria, the mean size, and the elongation score.
Table 1.
Independence of parameters is dependent upon experimental condition.
| Control | drp1 | opa1 | MPP+ | ||
|---|---|---|---|---|---|
| Number vs. | Size | -0.04 | -0.22 | 0.23 | -0.04 |
|
|
|||||
| Interconn. | 0.04 | 0.15 | 0.26* | 0.16 | |
|
|
|||||
| Elongation | -0.06 | -0.33 | 0.05 | 0.12 | |
|
|
|||||
| Area | 0.78* | 0.66*** | 0.73*** | 0.74*** | |
|
|
|||||
| Perimeter | -0.09 | -0.29 | 0.11 | -0.12 | |
|
| |||||
| Size vs. | Interconn. | 0.91** | 0.51** | 0.90*** | -0.01 |
|
|
|||||
| Elongation | 0.1 | 0.92*** | 0.39** | 0.01 | |
|
|
|||||
| Area | 0.52*** | 0.22 | 0.76*** | 0.53*** | |
|
|
|||||
| Perimeter | 0.93*** | 0.98*** | 0.89*** | 0.90*** | |
|
| |||||
| Interconn. vs. | Elongation | 0.13 | 0.2 | 0.22 | -0.01 |
|
|
|||||
| Area | 0.57** | 0.73*** | 0.76*** | -0.39** | |
|
|
|||||
| Perimeter | 0.73*** | 0.33 | 0.63*** | -0.39** | |
|
| |||||
| Elongation vs. | Area | 0.04 | 0 | 0.29** | 0.14 |
|
|
|||||
| Perimeter | 0.32* | 0.97*** | 0.53*** | -0.04 | |
|
| |||||
| Area vs. | Perimeter | 0.44** | 0.08 | 0.53*** | 0.39*** |
Table shows notable comparisons and their respective correlational coefficients between parameters for a given experimental condition (Pearson's r;
p < 0.05,
p < 0.01,
p < 0.001; Control n = 52, drp1 n = 26, opa1 n = 64, MPP+ n = 51).
We further explored the independence of the parameters by comparing relationships within the mitochondrial mutant strains and PD-like treatment group. There were notable differences in the relationships between parameters depending on the four conditions: control, drp1 strain, opa1 strain, and PD-like treatment group (MPP+). The mutant strains had positive correlations between size and elongation, while the control and PD group had no relationship. drp1 also had no relationship between interconnectivity and perimeter, while the control and opa1 groups had positive correlations and the PD group had a negative relationship. The PD group also did not show the same relationship between size and interconnectivity or interconnectivity and total area as in the other 3 groups. Most comparisons show the same trend in all groups (Table 1). Thus, the experimental condition can affect the relationship between parameters.
Specific Parameters Correlate with Treatment Groups
Lastly, we performed a principal component analysis (PCA) to further describe the relationship of the parameters to each other between the different treatment groups. Here we found that different treatment groups correlate well with parameters unique to their phenotype. PCA accounted for 84.1% of the variability (considered significant based on criteria listed in methods under statistical analysis subheading, 85.1 > 44.9). Interestingly, the different treatment groups had centers of gravity separate from each other (Figure 5): the control group was placed in the opposite quadrant of both the MPP+ and opa1 groups, while the drp1 group fell into an adjacent quadrant by itself. Thus, these parameters can be uniquely specific for treatment groups with different mitochondrial morphology.
Figure 5. Parameters of mitochondrial morphology correlate with specific treatment groups.
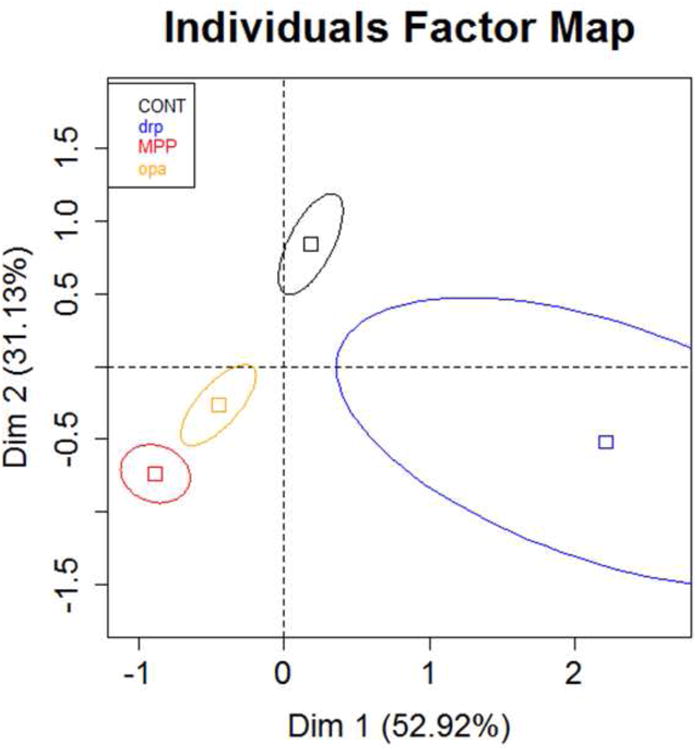
Principal component analysis was used to compare the relationships of the 6 mitochondrial morphology parameters in each of the 4 treatment groups. The different treatment groups had a center of gravity in separate quadrants, except for the MPP+ and opa1 treatments, which clustered together - expected as both had similar phenotypes of mitochondrial morphology. The control group appeared to have opposite correlations with the parameters compared to the MPP+ and opa1 treatments, while the drp1 treatment differed from all three of the other groups. Individuals factor map shows squares for the center of gravity for each of the 4 treatment groups surrounded by 95% confidence ellipses (CONT n = 52, drp1 n = 26, opa1 n = 64, MPP+ n = 51).
Discussion
Our work establishes a method for measuring specific populations of mitochondria, based on cell type and cellular location. It will be a useful tool in studies examining cell death, development, synaptogenesis, and bioenergetics. In all of these fields, mitochondria are key organelles whose morphology could serve as a useful endpoint between experimental groups. Specifically in our study, we show that mitochondrial morphology in dopaminergic (DA) cells changes in response to a Parkinson's disease (PD)-like treatment. We quantify mitochondrial morphology using four parameters: number, size, interconnectivity and elongation. Moreover, we validate our method using mutants with known defects in mitochondrial morphology: drp1 and opa1. Lastly, we determine how many parameters are needed for a comprehensive examination of mitochondrial morphology.
Our method improves upon a previous method originally reported by Dagda et al. (2009). This method measures four parameters of mitochondrial morphology: number, size, interconnectivity, and elongation. Whereas Dagda et al. (2009) only examined somatic mitochondria in a putatively DA, cancer cell-line, while we test non-somatic (i.e., neuritic) mitochondria within a specific cell population (DA neurons) using primary neuronal cultures from Drosophila. We validate the quantification of non-somatic, mitochondria in DA neurons by using fly strains with mutated drp1 or opa1. drp1 mutants have mitochondria which are unable to divide and we found them to have fewer, but larger, more interconnected, and more elongated mitochondria compared to control. opa1 mutants, which have decreased mitochondrial fusion, also had fewer mitochondria but were smaller, less interconnected, and less elongated as control. Thus, we successfully limited our analysis both spatially (measuring mitochondria only in dendritic/axonal projections) and specifically to a cell type, and were able to detect differences in all four parameters.
An example of the importance of measuring multiple parameters for mitochondrial morphology is illustrated by our results for drp1 mutants. Cells with mutant drp1 had an increase in size and interconnectivity compared to control, but the number of mitochondria was lower than in control. This decrease in the number of mitochondria is not because of a loss in mitochondrial biomass, but because most of them are connected to each other and counted as one, large mitochondria, as indicated by the high interconnectivity score. Thus, basing the cell's health only on the number of mitochondria is incomplete: healthy cells have higher number of mitochondria, are larger, more interconnected, and are less round compared to unhealthy cells (Dagda et al., 2009; Perier and Vila, 2012), i.e., an unhealthy cell may still have a large number of mitochondria, but those mitochondria may otherwise have a phenotype indicating they are damaged (i.e. round, fragmented). Thus, measuring only one parameter may incorrectly assess a cell's mitochondrial network. We therefore recommend a comprehensive assessment of mitochondrial morphology and health using multiple parameters.
As a proof of principle, our method found damaged mitochondria in the neurites of DA neurons exposed to PD-like treatment. The neurotoxin 1-methyl-4-phenylpyridinium (MPP+) decreased all parameters of mitochondrial morphology compared to control. Mitochondria are implicated in PD pathology (Dawson et al., 2010; Nakamura et al., 2011; Saxena and Caroni, 2011; Botella et al., 2011), and moreover, their demise may be an early event in the pathogenesis of PD (Martinou and Youle, 2011; Wang et al., 2011). Thus, our method provides a powerful tool not only for studying altered mitochondria morphology related to PD pathology, but also for screening neuroprotective therapies in PD. Future experiments could test if the changes in mitochondrial morphology induced by MPP+ are reversible with neuroprotective drugs.
Several parameters of mitochondrial morphology can be quantified, but many of these are likely redundant. Previous work by Koopman et al. (2006) produced a list of 13 different features describing mitochondrial morphology, but concluded that 7 of these features were redundant with the same measurement used for elongation in this work. Area, perimeter, and aspect ratio (the ratio between the major and minor axis of the ellipse equivalent of the mitochondrion) were considered independent (Koopman et al., 2006). Another study utilizing a machine-learning algorithm classified mitochondria into 4 separate categories, and then quantified the total area for each category, as well as a general count of the number of mitochondria in each category (Leonard et al., 2015). The categories used were based on similar elements employed by Dagda et al., (2009): interconnectedness, size, and shape. However, this analysis was also limited to the soma, so it is unknown if it could detect differences in neuritic mitochondria. It is possible that mitochondria in the neurites will not match the same classifications or have the same distributions as in the soma. Thus, a raw measurement, rather than a classification, may be a more appropriate quantification of mitochondrial morphology. Furthermore, testing both cellular compartments (soma and neurites) is probably warranted.
Regarding the independence of the parameters, we found the number of mitochondria, the mean area (size), and the elongation score to be independent of each other. However, our two added parameters, perimeter and total area, showed strong positive correlations with other parameters and thus, are not necessary. We consider perimeter to be a unique measurement worth capturing, but its consolidation into interconnectivity is warranted given that perimeter will increase with size. Indeed size and perimeter had a near-perfect, positive correlation.
The interconnectivity score should remain in the analysis in our opinion. Although there were strong, positive correlations between area and interconnectivity in the control, drp1, and opa1 experimental conditions, there was no relationship in the PD-like treatment group. This discrepancy justifies the inclusion of interconnectivity, as size and interconnectivity measurements may capture two separate morphological phenomena in the disease state. Also, only in the PD-like treatment group, there was a negative relationship between interconnectivity and perimeter, and no relationship between interconnectivity and total area. These differences highlight the ability of the method to discriminate between experimental conditions, and suggest differential morphological processes at work in the PD-like treatment group. The negative correlation between interconnectivity and perimeter is expected, as we found decreased interconnectivity in the PD-like treatment group compared to control and perimeter is in the denominator of the interconnectivity measurement, so mathematically interconnectivity decreases as perimeter increases. This may also explain why we did not see a positive correlation between interconnectivity and perimeter in the drp1 condition. Size was likely increasing much faster than perimeter compared to control. Thus, while there was still a positive correlation of 0.33, the result was not significant.
The drp1 and opa1 conditions also had notable correlations between parameters. Both had positive correlations between size and elongation while there was no correlation in the control or PD-like treatment group. Both of these results make sense, as drp1 mutants have large mitochondria with abstract shapes and opa1 mitochondria are smaller and rounder. There was also a trending, though not significant, result between the number of mitochondria and size, with drp1 mutants having a slightly negative correlation and opa1 mutants having a slightly positive correlation. Again, this makes sense, as the number of mitochondria in the drp1 mutants decreases because they are all conjoining and increasing size, while the opposite happens in opa1 mutants, mitochondria split, thus increasing the number and decreasing the average size. Overall, the parameters adequately represent the phenomena they are meant to measure, and are mostly independent of each other as well for multiple experimental conditions.
Interestingly, the relationships between parameters changed for different experimental treatments. For example, size and interconnectivity had a strong, positive correlation in the control group, but no relationship in the PD-like treatment group. We believe this highlights the importance of measuring multiple parameters, and also may provide ideas for novel analysis in the future. Because we see an “uncoupling” of these two parameters, it could mean the mitochondria are assembling themselves by different molecular processes due to the PD-like treatment. Thus, one could screen for cellular machinery which uncouples the parameters of mitochondrial morphology which we describe here. Indeed, here we identify drp1 and opa1 as two proteins which relate morphological parameters differently from control neurons in the example of size versus elongation.
A principal component analysis (PCA) further supported the idea that each parameter could specifically describe a treatment group, accurately separating the control group from all three other treatments, as well as grouping the PD-like and opa1 treatments together. Moreover, the correlations described for each treatment in the PCA matched the observed phenotypes. For example, the control group had a positive correlation with the number of mitochondria and this group indeed had the highest number of mitochondria compared to all three treatment groups. Moreover, the PD-like and opa1 treatments had a negative correlation with the number of mitochondria, which again matched what was found in direct comparisons for the number of mitochondria. Based on this analysis, we are confident that parameters could accurately describe treatment groups with different mitochondrial morphology.
In summary, this work describes a method for quantifying mitochondrial morphology in a specific population of neurons. We validate our model using mutants with known defects in mitochondrial morphology, and we show a difference between treatments as a proof of principle. We also explore the independence of the parameters measured in the method. We conclude that at least four variables should be considered when measuring mitochondrial structure as an experimental endpoint: the number of mitochondria, the size, the interconnectivity, and elongation (shape). At this time, however, it is unknown which parameter is the most important in regards to the health of the cell. Also unknown is which cellular compartment offers the best description of mitochondrial changes in response to treatment. Thus, for future analysis we suggest two separate analyses: one for mitochondria in the soma and another for mitochondria in the neurites, as we would expect each cellular compartment to show different values for the testing parameters. The currently provided method can accomplish analysis of both compartments.
Highlights.
Mitochondrial morphology of dopaminergic neurites can be quantified by at least 4 independent variables.
A gestalt method of analysis measuring multiple parameters is required to adequately quantify mitochondrial morphology.
Mutations with known effects on mitochondrial morphology change the relationship between morphological parameters of mitochondria.
The method successfully differentiates control mitochondria from those damaged by Parkinson's disease-like treatment and genetic manipulations affecting mitochondrial fusion/fission.
Acknowledgments
This work was partially supported by NIH grant (NS050260) and Korea Institute of Science & Technology (Brain Science Institute), Seoul, Korea.
Abbreviations
- DA
dopaminergic
- MPP+
1-methyl-4-phenylpyridinium
- PD
Parkinson's disease
- TH
tyrosine hydroxylase
Appendix A
ImageJ code for analysis of mitochondrial morphology (immunocytochemistry method)
| // Displays the colocalization of two images and then measures the morphometrics of the |
| //overlapped signal |
| // Installation notes: |
| // Download and Install WCIF ImageJ package from |
| // http://www.uhnresearch.ca/facilities/wcif/fdownload.html |
| // Save this code as .txt file and move file to >Program Files>ImageJ>macros |
| // Make sure it's the correct ImageJ Folder (the WCIF version) if you have multiple versions |
| // open WCIF ImageJ and install the macro by selecting from the ImageJ toolbar: |
| // Plugins>Macros>Install… and then select the .txt file saved earlier. |
| // Push F8, a window will appear to select a file. |
| // First select the red image |
| // another window will appear, now select the green image |
| // ImageJ will then merge the two images and present a threshold image of the colocalized |
| // points. There will also be the original images as well as merge between the two with |
| // colocalized points as white. The image to be analyzed will be autoselected. |
| // Draw around the area of the image to be analyzed. Only points within this area |
| // will be measured |
| // Push F10. ImageJ will perform the measurements and report the results in a new window. |
| // After copying the results, push F11 to close all windows. |
| // Push F9 and repeat |
| // Code below |
| macro “Close All Windows [F11] ” //F11 used as a shortcut to close all active windows |
| { |
| while (nImages>0) { |
| selectImage(nImages); |
| close(); |
| } |
| } |
| macro “Open Images for colocalization [F8] ” //F8 used as shortcut |
| { |
| open(); //select red image |
| run(“8-bit”); //makes image 8-bit |
| open(); //select green image |
| run(“8-bit”); //makes images 8-bit |
| run(“Colocalization Highligter”, “ratio=50 threshold_channel_1=52 threshold_channel_2=52 display=255 colocalized”); //Colocalization Highlighter is a tool from the WCIF version of |
| //ImageJ, adjust threshold here if necessary |
| //user may wish to use this image for making figures |
| setTool(3); //activates freehand selection tool for drawing around region of interest |
| selectWindow(“Colocalized points (8-bit) ”); //selects image containing only colocalized |
| //signals |
| setThreshold(15, 255); //threshold setting not important here |
| } |
| macro “Measure Morphometrics [F10] ” //F10 used as shortcut |
| { |
| run(“Clear Results”); |
| run(“Set Measurements… ”, “area perimeter circularity redirect=None decimal=2”); |
| run(“Analyze Particles… ”, “minimum=5 maximum=500 bins=100 show=Outlines display summarize”); //set size range of particles here |
| for (i=0; i<nResults; i++){ |
| MP += getResult(‘Perim.’,i); |
| MA +=getResult(‘Area’, i); |
| MC +=abs(getResult(‘Circ.’, i)); //old version of ImageJ reports circularities of -1 if the |
| //calculated circularity > 1, so absolute value needed here |
| }//loop to assign variables from results |
| { |
| AMP= (MP/i); //calculates mean perimeter |
| AMA= (MA/i); //calculates mean area |
| AMC= (MC/i); //calculates mean circularity |
| Rmorph= (AMA/AMP); //measures the area perimeter ratio, a.k.a. interconnectivity |
| print(getTitle()); |
| print(“Count:” +i); |
| print(“Total Area:” +MA); |
| print(“Avg. Perimeter:” +AMP); |
| print (“Avg. Area:” + AMA); |
| print (“Avg. Circularity:”+ AMC); |
| print (“Area/Perim:”+ Rmorph); |
| selectWindow(“Results”); |
| selectWindow(“Log”); |
| } |
| } |
Footnotes
Conflicts Of Interest: The authors declare no conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Baens M, Noels H, Broeckx V, Hagens S, Fevery S, Billiau AD, Vankelecom H, Marynen P. The dark side of EGFP: defective polyubiquitination. PLoS One. 2006;1:e54. doi: 10.1371/journal.pone.0000054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrientos SA, Martinez NW, Yoo S, Jara JS, Zamorano S, Hetz C, Twiss JL, Alvarez J, Court FA. Axonal degeneration is mediated by the mitochondrial permeability transition pore. Neurobiology of Disease. 2011;31(3):966–78. doi: 10.1523/JNEUROSCI.4065-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bereiter-Hahn J. Behavior of mitochondria in the living cell. International Review of Cytology. 1990;122:1–63. doi: 10.1016/s0074-7696(08)61205-x. [DOI] [PubMed] [Google Scholar]
- Berman SB, Pineda FJ, Hardwick JM. Mitochondrial fission and fusion dynamics: the long and short of it. Cell Death and Differentiation. 2008;15:1147–52. doi: 10.1038/cdd.2008.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botella JA, Bayersdorfer F, Gmeiner F, Schneuwly S. Modelling Parkinson's disease in Drosophila. NeuroMolecular Medicine. 2011;11:268–80. doi: 10.1007/s12017-009-8098-6. [DOI] [PubMed] [Google Scholar]
- Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–15. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- Chang DTW, Reynolds IJ. Mitochondrial trafficking and morphology in healthy and injured neurons. Progress in Neurobiology. 2006;80:241–68. doi: 10.1016/j.pneurobio.2006.09.003. [DOI] [PubMed] [Google Scholar]
- Cheng A, Hou Y, Mattson MP. Mitochondria and neuroplasticity. ASN Neuro. 2010;2(5):243–56. doi: 10.1042/AN20100019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cicchetti F, Drouin-Ouellet J, Gross RE. Environmental toxins and Parkinson's diseases: what have we learned from pesticide-induced animal models? Trends in Pharmacological Sciences. 2009;30(9):475–83. doi: 10.1016/j.tips.2009.06.005. [DOI] [PubMed] [Google Scholar]
- Court FA, Coleman MP. Mitochondria as a central sensor for axonal degenerative stimuli. Trends in Neurosciences Article. 2012 doi: 10.1016/j.tins.2012.04.001. 2012. [DOI] [PubMed] [Google Scholar]
- Dagda RK, Cherra SJ, III, Kulich SM, Tandon A, Park D, Chu CT. Loss of PINK1 function promotes mitophagy through effects on oxidative stress and mitochondrial fission. Journal of Biological Chemistry. 2009;284(20):13843–55. doi: 10.1074/jbc.M808515200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darya K, Ganguly A, Lee D. Quantitative analysis of synaptic boutons in cultured Drosophila neurons. Brain Research. 2009;1280:1–12. doi: 10.1016/j.brainres.2009.05.026. [DOI] [PubMed] [Google Scholar]
- Dawson TM, Ko HS, Dawson VL. Genetic animal models of Parkinson's disease. Neuron. 2010;66:646–61. doi: 10.1016/j.neuron.2010.04.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh A, Chandran K, Kalivendi SV, Joseph J, Antholine WE, Hillard CJ, Kanthasamy Ar, Kanthasamy An, Kalyanaraman B. Neuroprotection by a mitochondria-targeted drug in a Parkinson's disease model. Free Radical Biology & Medicine. 2010;49:1674–1684. doi: 10.1016/j.freeradbiomed.2010.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman S, Roy N. Reply to “Human neural progenitor cells: better blue than green? Nature Medicine. 2000;6:483–4. doi: 10.1038/74920. [DOI] [PubMed] [Google Scholar]
- Jellenger KA. Neuropathology of sporadic Parkinson's disease: evaluation and changes of concepts. Movement Disorders. 2012;27(1):8–30. doi: 10.1002/mds.23795. [DOI] [PubMed] [Google Scholar]
- Koopman WJH, Visch HJ, Smeitink JAM, Willems PHGM. Simultaneous quantitative measurement and automated analysis of mitochondrial morphology, mass, potential, and motility in living human skin fibroblasts. Cytometry Part A. 2006;69A(1):1–12. doi: 10.1002/cyto.a.20198. [DOI] [PubMed] [Google Scholar]
- Lê S, Rennes A, Josse J, Husson F. FactoMineR: an R package for multivariate analysis. Journal of Statistical Software. 2008:1–18. [Google Scholar]
- Leonard AP, Cameron RB, Speiser JL, Wolf BJ, Peterson YK, Schnellmann RG, et al. Rohrer B. Quantitative analysis of mitochondrial morphology and membrane potential in living cells using high-content imaging, machine learning, and morphological binning. Biochimica et Biophysica Acta (BBA) - Molecular Cell Research. 2015;1853(2):348–360. doi: 10.1016/j.bbamcr.2014.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy OA, Malagelada C, Greene LA. Cell death pathways in Parkinson's disease: proximal triggers, distal effectors, and final steps. Apoptosis. 2009;14:478–500. doi: 10.1007/s10495-008-0309-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang CL, Wang TT, Luby-Phelps K, German DC. Mitochondrial mass is low in mouse substantia nigra dopamine neurons: implications for Parkinson's disease. Experimental Neurology. 2007;203:370–80. doi: 10.1016/j.expneurol.2006.08.015. [DOI] [PubMed] [Google Scholar]
- Liu HS, Jan MS, Chou CK, Chen PH, Ke NJ. Is Green Fluorescent Protein Toxic to the Living Cells? Biochemical and Biophysical Research Communications. 1999;260:712–7. doi: 10.1006/bbrc.1999.0954. [DOI] [PubMed] [Google Scholar]
- Manning-Boğ AB, Langston JW. Model fusion: the next phase in developing animal models for Parkinson's disease. Neurotoxicity Research. 2007;11(3,4):219–40. doi: 10.1007/BF03033569. [DOI] [PubMed] [Google Scholar]
- Martinou JC, Youle RJ. Mitochondria in Apoptosis: Bcl-2 family Members and Mitochondrial Dynamics. Developmental Cell. 2011;21(1):92–101. doi: 10.1016/j.devcel.2011.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura K, Nemani VM, Azarbal F, Skibinski G, Levy JM, Egami K, Munishkina L, Zhang J, Gardner B, Wakabayashi J, Sesaki H, Cheng Y, Finkbeiner S, Nussbaum RL, Masliah E, Edwards RH. Direct membrane association drives mitochondrial fission by the Parkinson disease-associated protein alpha-synuclein. Journal of Biological Chemistry. 2011;286(23):20710–26. doi: 10.1074/jbc.M110.213538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto K, Shaw JM. Mitochondrial morphology and dynamics in yeast and multicellular eukaryotes. Annual Review of Genetics. 2005;39:503–36. doi: 10.1146/annurev.genet.38.072902.093019. [DOI] [PubMed] [Google Scholar]
- Perier C, Vila M. Mitochondrial biology and Parkinson's disease. Cold Spring Harbor Perspectives in Medicine. 2012;4:a009332. doi: 10.1101/cshperspect.a009332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Przedborski S, Jackson-Lewis V, Naini AB, Jakowec M, Petzinger G, Miller R, Akam M. The Parkinsonian toxin 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine (MPTP): a technical review of its utility and safety. Journal of Neurochemistry. 2001;76:1265–74. doi: 10.1046/j.1471-4159.2001.00183.x. [DOI] [PubMed] [Google Scholar]
- Rafelski SM, Marshall WF. Building the cell: design principles of cellular architecture. Nature Reviews Molecular Cell Biology. 2008;9:593–602. doi: 10.1038/nrm2460. [DOI] [PubMed] [Google Scholar]
- Rasband WS. US Natl Inst Health; Bethesda, Maryland, USA: 1997-2006. ImageJ. http://rsb.info.nih.gov/ij/ [Google Scholar]
- Rugarli EI, Langer T. Mitochondrial quality control: a matter of life and death for neurons. European Molecular Biology Organization Journal. 2012;31:1336–49. doi: 10.1038/emboj.2012.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxena S, Caroni P. Selective neuronal vulnerability in neurodegenerative diseases: from stressor thresholds to degeneration. Neuron. 2011;71:35–48. doi: 10.1016/j.neuron.2011.06.031. [DOI] [PubMed] [Google Scholar]
- Smith Y, Wichmann T, Factor SA, DeLong MR. Parkinson's disease therapeutics: new developments and challenges since the introduction of levodopa. Neuropsychopharmacology. 2012;37:213–46. doi: 10.1038/npp.2011.212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surmeier DJ, Schumacker PT. Calcium, bioenergetics, and neuronal vulnerability in Parkinson's disease. Journal of Biological Chemistry. 2013;288(15):10736–41. doi: 10.1074/jbc.R112.410530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Su B, Liu W, He X, Gao Y, Castellani RJ, Perry G, Smith MA, Zhu X. DLP1-dependent mitochondrial fragmentation mediates 1-methyl-4-phyenylpyridinium toxicity in neurons: implications for Parkinson's disease. Aging Cell. 2011;10:807–23. doi: 10.1111/j.1474-9726.2011.00721.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiemerslage L, Schultz BJ, Ganguly A, Lee D. Selective degeneration of dopaminergic neurons by MPP+ and its rescue by D2 autoreceptors in Drosophila primary culture. Journal of Neurochemistry. 2013;126:529–40. doi: 10.1111/jnc.12228. [DOI] [PMC free article] [PubMed] [Google Scholar]


