Abstract
Periodontal disease initiated by subgingival pathogens is linked with diminished secretion of saliva, and implies pathogenic bacteria dissemination to or affects secondary sites such as the salivary glands. MicroRNAs activated in response to bacteria may modulate immune responses against pathogens. Therefore, Sprague-Dawley rats were infected by oral lavage consisting of polymicrobial inocula, namely Porphyromonas gingivalis, Tannerella forsythia, and Treponema denticola, or sham-infected for 12 weeks (n=6). We quantified inflammatory miRNA expression levels of miRNA-132, miR-146a, and miR-155 at secondary sites to the primary infection of the gingiva, including submandibular salivary glands, lacrimal glands, and pancreas. The presence of bacteria was detected in situ at secondary sites. Infected rat gingiva showed increased relative expression of miR-155. In contrast, miRNA-155 expression was decreased in submandibular salivary glands, along with positive identification of P. gingivalis in 2/6 and T. denticola in 1/6 rat salivary glands. Furthermore, miRNA-132 and miRNA-146a were significantly decreased in the pancreas of infected rats. This study is the first to show primary periodontal infections can alter miRNA profiles in secondary sites such as the salivary gland and pancreas. Whether these alterations contribute to pathologies of salivary glands in Sjögren’s syndrome or of pancreas in diabetes warrants further investigation.
Keywords: periodontal pathogens, polymicrobial infection, microRNA, submandibular glands, experimental periodontitis
1. INTRODUCTION
Periodontitis is a chronic polymicrobial dysbiotic inflammatory disease of the periodontal tissues and results in decreased tooth support [1, 2]. Periodontal disease results from a polymicrobial synergy among Gram-negative anaerobic microorganisms in subgingival biofilm including Porphyromonas gingivalis, Tannerella forsythia, and Treponema denticola which shifts relative abundance or influence of microbial species within the community known as “dysbiosis” thereby enabling the manifestation of periodontal disease [3]. Periodontal disease has also been linked to several systemic diseases, such as atherosclerosis [4], diabetes [5], rheumatoid arthritis [6], Alzheimer’s disease [7], adverse pregnancy outcomes [8], and respiratory diseases [9]. However, exact mechanisms contributing to development of these systemic diseases are unknown. Presumably, periodontal pathogens mediate disease progression by direct and indirect mechanisms through recurrent bacteremia resulting in inflammation at distal sites [10, 11]. Oral bacteria P. gingivalis, T. denticola, T. forsythia, and Fusobacterium nucleatum are capable of dissemination from the primary site of infection in the gingiva into vascular tissues and peripheral organs, as observed through the identification of bacterial 16S ribosomal RNA (rRNA) and bacterial genomic DNA in infected mice and rats at secondary sites [12–17]. Recent metagenomic analysis of ancient human ilial bone tissues from Otzi the Iceman, a 5,300-year-old Copper Age natural ice mummy, detected T. denticola corroborating evidence for haematogenous dissemination of this periodontal pathogen [18].
During the course of periodontal disease, the presence of pathogenic bacteria or their effector molecules activates local innate immune responses that typically result in inflammation of the host’s tissues that, if unchecked, ultimately results in progressive bacterial attachment and alveolar bone resorption characterized by gingival pocket formation and recession. Understanding the molecular mechanisms that govern the magnitude of inflammatory responses is vital and the study of microRNA becomes an indispensable tool at this juncture. MicroRNAs (miRNAs) are small (~20–22 nucleotides) non-coding, endogenously expressed, RNA sequences that can post-transcriptionally inhibit protein synthesis of their targeted messenger RNAs (mRNAs) [19]. MiRNAs are involved in regulation of various biological processes within cells, tissues, and in various pathological processes in autoimmune disorders and infection [20, 21]. The monocyte/macrophage inflammatory response to infection involves the differential expression of several miRNAs including miR-155, -146, and -132, which are critical for resolution of inflammation in a timely manner to avoid damage to the host.
MiRNA (miR)-155 is involved in regulating toll-like receptor (TLR) 2/4-mediated NF-κB activation, thus limits the production of inflammatory cytokines [22, 23]. MiR-146a is another miRNA that is commonly implicated with the TLR-mediated pathways and inflammatory cytokine inhibition, and shows significant increases after stimulation with bacterial lipopolysaccharide (LPS) in monocytes [23, 24]. MiR-146a acts as a negative feedback regulator of the innate immune response by targeting two adapter proteins, TRAF6 (TNF receptor–associated factor 6) and IRAK1 (IL-1 receptor-associated kinase 1), that are crucial for proinflammatory signaling [25]. MiR-132 has been shown to regulate neuronal morphogenesis and dendritic plasticity of cultured neurons [26]. MiR-132 may also be responsible for limiting inflammation in the mouse brain by targeting acetylcholinesterase [26]. MiR-132 can also modulate inflammation induced by early-stage Kaposi’s sarcoma–associated herpesvirus (KSHV) infection. MiR-132 also shows significant increases after LPS stimulation and can also modulate antiviral immunity [23, 27].
Our previous study showed miR-146a was significantly increased in response to periodontal infections in the ApoE−/− mouse model of atherosclerosis in the periodontium and systemically at the spleen, suggesting that miRNAs associated with inflammation can be altered as a result of periodontal pathogens [28]. The mucus coat serves as a defense barrier protecting soft gingival tissue as well as oral mucosa from bacterial insults [28, 29]. And the development of bacterial infection can result from diminished saliva secretion [30]. Because periodontal disease development and inhibition of salivary gland function tend to be associated [29, 31–33], our current study continued the work of Rivera et al. on rat polymicrobial periodontal disease model [14] and evaluated the expression levels of miR-155, miR-146a, and miR-132 at sites peripheral or distant to periodontal infection - the submandibular salivary glands, lacrimal glands, and pancreas.
2. MATERIALS AND METHODS
2.1 Bacterial Strains and Inoculum Preparation
P. gingivalis FDC 381, T. denticola ATCC 35404, and T. forsythia ATCC 43037 were grown in an anaerobic chamber at 37°C as described previously [14]. The mixed bacterial inoculum was prepared as described in Rivera et al. [14]. In brief, equal cell concentrations of P. gingivalis, T. denticola, and T. forsythia were sequentially suspended and mixed at 5 minute intervals in reduced transport fluid. The suspension was then mixed with an equal volume of 8% sterile carboxy methylcellulose (CMC) in phosphate-buffered saline (PBS).
2.2 Oral Infection and Sampling
This study was carried out in accordance with the recommendations from the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. The protocol was approved by the University of Florida Institutional Animal Care and Use Committee (Protocol 201004367) [14]. Female Sprague-Dawley rats were obtained from Harlan laboratories (Harlan, Indianapolis, IN, USA) and fed powdered normal chow and water ad libitum. All rats were administered sulfamethoxazole (0.87 mg per mL) and trimethoprim (0.17 mg per ml) daily for 10 days in the drinking water to reduce endogenous oral flora and rat mouths were swabbed with 0.12% chlorhexidine gluconate mouth rinse to enhance subsequent colonization of periodontal bacteria [14]. Rats were randomly sorted into two groups (n = 6 per group): polymicrobial infected and sham-infected groups. The infected group was given polymicrobial inocula, as described above, of 1 × 109 cells via oral lavage every other week for 12 weeks to establish a stable infection as described elsewhere [14]. The control was sham-infected with sterile 8% CMC following the same schedule [14]. Following 12 weeks of infection, rats were euthanized gingival tissues; submandibular salivary glands, lacrimal glands, and pancreas were collected and stored at −80°C until further usage.
2.3 Quantitative reverse transciptase PCR (qRT-PCR)
Total RNA including miRNAs were isolated from each tissue and individually processed using a miRNA isolation kit (Ambion mirVana miRNA Isolation Kit, Life Technologies, Carlsbad, CA, USA) following the manufacturer’s protocol. The quality and concentration of the RNA were assessed using a spectrophotometer (NanoDrop ND-1000 spectrophotometer, NanoDrop Technology Inc., Wilmington, DE, USA). Step-wise dilutions were made to standardize concentrations between samples. Subsequently, quantitative stem-loop reverse transcription and quantitative real-time PCR for miRNA-132, -155, and -146a (Life Technologies, Carlsbad, CA, USA) were used following the manufacturer’s protocol to quantify our miRNAs of interest. The relative expression levels of miRNAs were determined following the 2−ΔΔ CT method as described by Livak and Schmittgen [34].
2.4 Fluorescence in situ hybridization (FISH) for Bacterial Localization
Modified FISH protocol [13] was used to identify periodontal bacteria that were metabolically active within tissues using probes specific for bacterial 16S rRNA [13, 35]. Submandibular salivary glands and lacrimal glands were fixed in 4% paraformaldehyde, paraffin embedded, cut sequentially, and mounted with two sections per slide. One section was used as a control and the other for bacterial detection. Sections were de-paraffinized by a series of xylene and ethanol washes of increasing dilution. Slides were blocked for 30 minutes at 37°C with Denhardt’s reagent (Fisher Scientific, Waltham, MA, USA). Two hybridization solutions (900 mM NaCl, 20 mM Tris-HCl pH 7.5, 0.01% SDS, 20% formamide) were made, one control and one mixed with 5 µg/mL of Alexafluor-568 3’-labeled oligonucleotide probe specific for 16S rRNA (Invitrogen, Carlsbad, CA, USA) of either P. gingivalis, T. denticola, or T. forsythia. Sections were covered with the respective hybridization buffers and placed in a dark, moist environment at 48°C for 3 hours. Slides were rinsed with pre-warmed washing buffer (20 mM Tris-HCl, pH7.5, 5 mM EDTA, 0.01% SDS, 0.225 M NaCl) and then incubated with washing buffer for 25 min in the dark. All solutions contained protectRNA (Sigma-Aldrich, St. Louis, MO, USA) to prevent RNA degradation. Slides were mounted and counterstained for fluorescence screening.
2.5 Image Acquisition
Hybridized slides were viewed with the Zeiss Axiovert 200M microscope fitted with a Zeiss AxioCam MRm camera (Carl Zeiss Microscopy, Thornwood, NY, USA). The Alexafluor-568 was visualized in the Cy3 channel. Unlabeled sections were used as background controls for Alexafluor-568. Background Cy3 noise was estimated by the control tissue unstained by Alexaflour-568 [13, 15].
2.6 Statistical Analyses
Two-tailed Student’s t-test was used to compare miRNA expression between sham and infected groups for each miRNA in each tissue examined. In addition, a two-tailed nonparametric Mann-Whitney test was used to compare miRNA expression between sham and infected groups for miR-155 in the salivary gland. For all statistical analyses, P < 0.05 was considered significant.
3. RESULTS
3.1 MiRNAs are differentially expressed among tissues
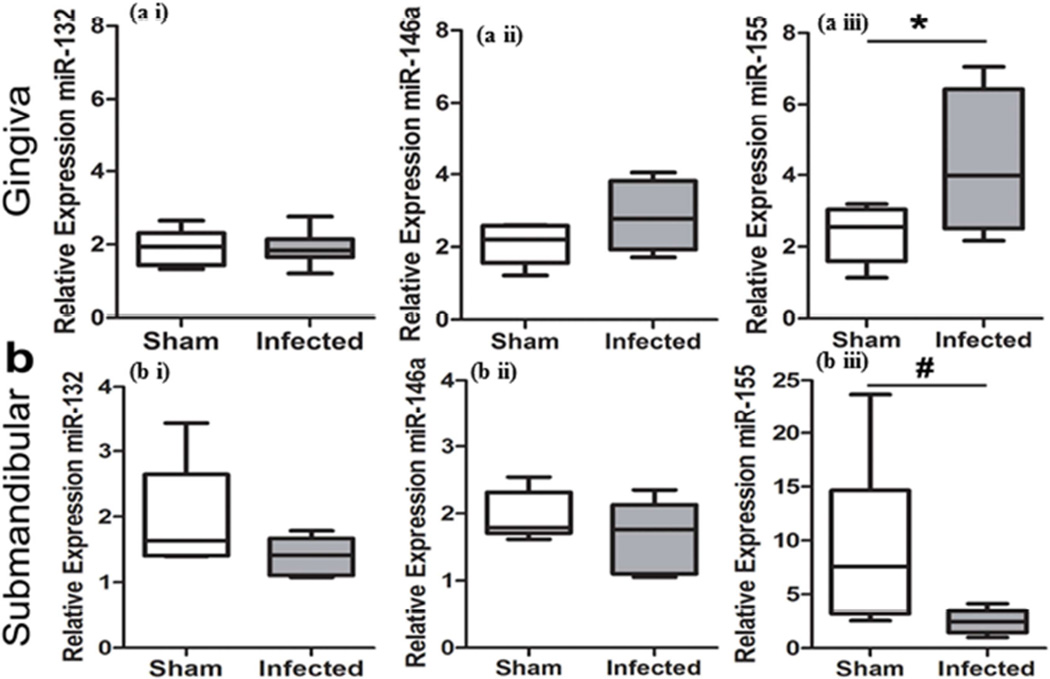
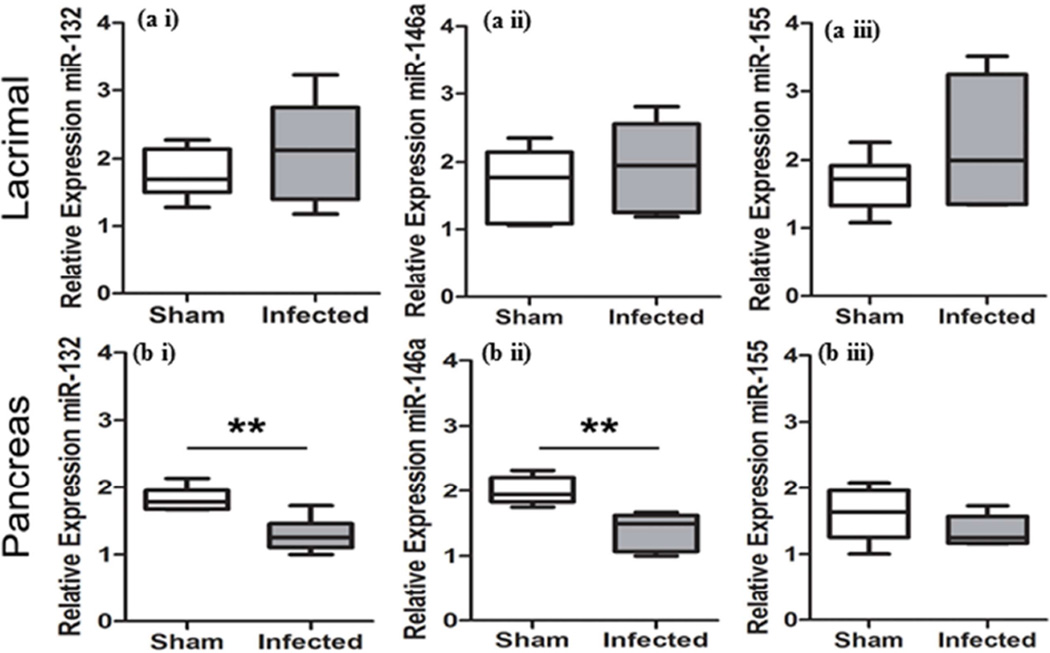
Relative expressions of miRNAs were evaluated in gingival tissues, submandibular glands, lacrimal glands, and pancreas to determine the local and systemic alterations in miRNA expression following oral polymicrobial infection. Gingival and lacrimal gland tissue showed a general trend for elevated miR-132 and miR-146a in the infected group, which were not statistically significant (Figure 1ai, 1aii and Figure 2ai, 2aii). Of note, miR-155 was significantly elevated in the gingiva (Figure 1aiii). In contrast, submandibular glands exhibited a general decrease in miR-132, miR-146a, and miR-155 in the infected group (Figure 1b). Initial analysis of miR-155 in the submandibular salivary glands using a Student’s t-test showed P-value of 0.0577. However, because variances were significantly different between the groups (F test, P = 0.0006), a two-tailed Mann-Whitney t-test was used, which resulted in P-value of 0.026 (Figure 1biii). Furthermore, pancreas tissues from the infected group exhibited a significant reduction in both miR-132 and miR-146a relative expression (Figure 2bi and 2bii).
Figure 1.
Polymicrobial infection with periodontal pathogens P. gingivalis, T. denticola, and T. forsythia alters miRNA expression levels in the gingiva and submandibular salivary glands following 12 weeks of infection in rats. Total RNAs from a) gingiva, b) submandibular salivary glands, were evaluated by qRT-PCR to determine the relative miRNA expression levels of miR-132 (a i and b i), -146a (a ii and b ii), and -155(a iii and b iii) between infected and sham-infected rats (n = 6). Middle line indicates mean, box indicates the upper and lower quartiles, and whiskers indicate minimum and maximum values. *P < 0.05 by Student’s two-tailed t-test, #P < 0.05 Mann-Whitney two-tailed t-test.
Figure 2.
Polymicrobial infection with periodontal pathogens P. gingivalis, T. denticola, and T. forsythia alters miRNA expression levels in lacrimal glands and pancreas following 12 weeks of infection in rats a) lacrimal glands, and b) pancreas were evaluated by qRT-PCR to determine the relative miRNA expression levels of miR-132 (ai and bi), -146a (aii and bii), and -155 (aiii and biii) between infected and sham-infected rats (n = 6). Middle line indicates mean, box indicates the upper and lower quartiles, and whiskers indicate minimum and maximum values. **P < 0.01 by Student’s two-tailed t-test.
3.2 Presence of P. gingivalis and T. denticola within the submandibular salivary glands
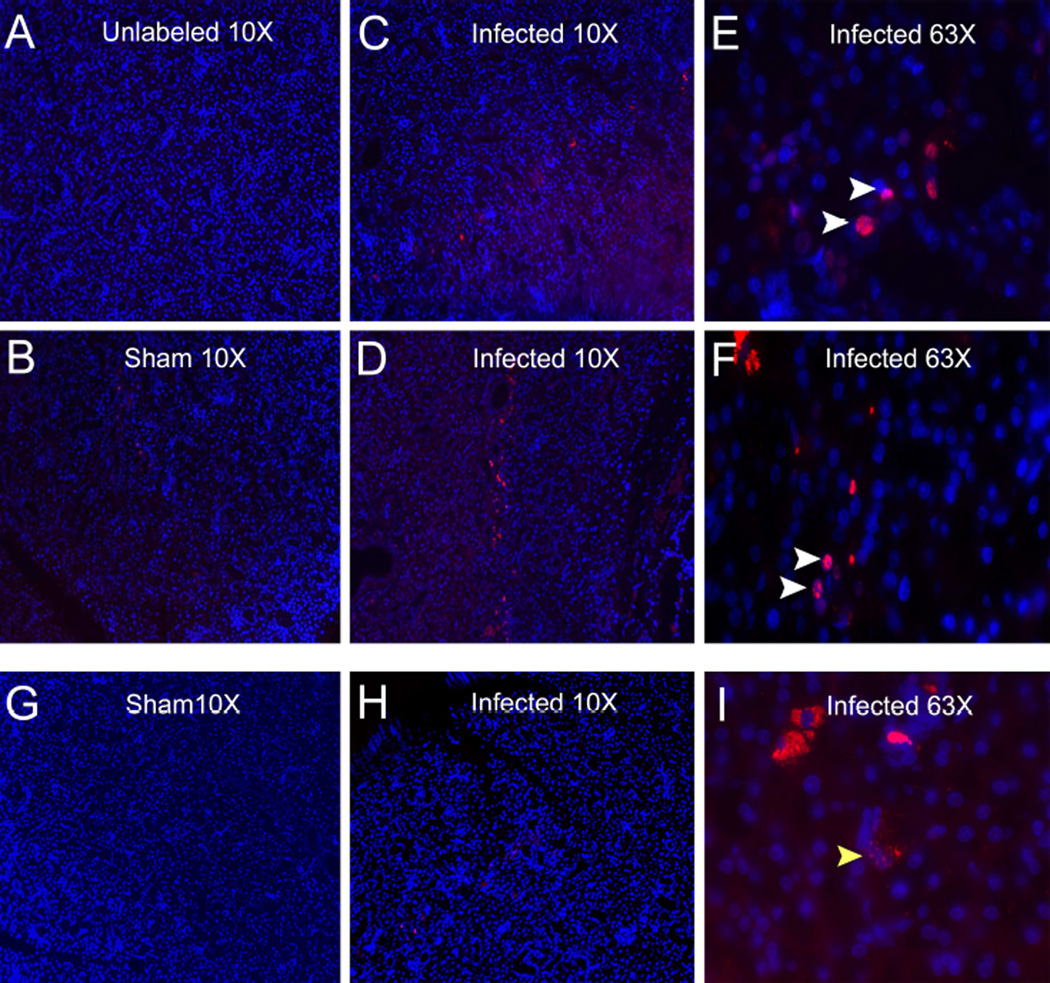
To further understand miRNA alterations of periodontal infection on the submandibular glands, bacterial presence was tested using species-specific probes that utilized complementary nucleotide sequences for the detection of metabolically active bacterial 16S rRNA. P. gingivalis was identified within the submandibular salivary glands of 2 out of 6 infected rats (Figure 3C–3F). The positive P. gingivalis labeling was localized in the perinuclear region of salivary gland cells as demonstrated by significant co-localization with the DAPI nuclear staining. In addition, T. denticola was also identified in submandibular salivary glands of one rat also positive for P. gingivalis (Figure 3H and 3I). The sham-infected rats showed no positive labeling for bacteria (Figure 3B and 3G). Furthermore, evaluation of the lacrimal glands of infected and sham-infected rats also showed no positive labeling for bacteria (data not shown).
Figure 3.
Polymicrobial infection with periodontal pathogens P. gingivalis, T. denticola, and T. forsythia results in hematogenous dissemination from the oral cavity into the salivary glands of rats. Fluorescence in situ hybridization of rat submandibular salivary glands identifies presence of 16S rRNA for oral bacteria (red) counter-stained with DAPI (blue). A) Control unlabeled salivary gland, B) representative sham-infected rat salivary gland, C) Representative salivary gland tissue sections of polymicrobial infected rat #2 showing P. gingivalis at lower magnification 10× and E) at higher magnification 63×. D) Representative salivary gland tissue sections of polymicrobial infected rat #3 showing presence of P. gingivalis (10× magnification). and F) at higher magnification 63×. White arrow heads indicate perinuclear localization of P. gingivalis. G) Representative sham-infected salivary gland and H) Representative salivary gland tissue sections of polymicrobial infected rat #3 showing the presence of T. denticola. (10× lower magnification). I) Under higher magnification (63×). Yellow arrow head indicate localization of T. denticola.
4. DISCUSSION
Chronic periodontal disease fosters bacteria that can hematogenously disseminate and trigger significant systemic disease processes in non-oral sites. Periodontal lesions have been shown to be reservoirs for chronic recurrent systemic bacteremia [12, 13, 15, 36]. The current study was designed to explore the effects of chronic periodontal disease with potential bacteremia and translocation of bacteria on the immune response, as quantified in terms of miRNA expression profiles, at secondary sites of infection. Our previous study, Rivera et al. showed that periodontal disease as well as the systemic immune responses to periodontal pathogens were evidenced in the infected group of rats by significant alveolar bone resorption, inflammation of the epithelium, and increased specific antibodies against P. gingivalis, T. forsythia, and T. denticola in polymicrobial infected rats [14]. Our current findings indicate increased inflammatory miR-155 in the gingiva of infected rats. Interestingly, decreased miR-155 was evident in the submandibular salivary glands and miR-132 and miR-146a were both reduced in the pancreas of infected rats.
In vivo and in vitro studies together demonstrate the complexity of oral bacterial involvement in miRNA expression patterns. Gram-negative bacterial cell walls contain LPS endotoxins, which are well known to increase miR-146a and miR-155 in RAW264.7 macrophages and miR-146a, miR-132, and miR-155 in THP-1 monocytes in vitro [23, 37]. A previous study of human periodontal disease gingiva found miR-146a increased but miR-155 reduced relative to healthy tissues [38]. Interestingly, similar to our current infection model, another recent study showed miR-155 increased in human periodontal disease gingiva in comparison to healthy tissues [39]. Conversely, oral pathogens P. gingivalis, T. denticola, and T. forsythia monoinfections or polymicrobial infections in human THP-1 monocytes resulted in distinct differential miRNA expression patterns in vitro. Interestingly, live T. denticola alone inhibited miR-146a expression and live P. gingivalis or T. denticola alone inhibited miR-132 expression when compared with heat-killed bacteria. Of note, both live and heat-killed P. gingivalis, T. denticola, and T. forsythia failed to increase miR-155 expression in vitro [28]. Similarly, we showed miR-132 and miR-146a were reduced in pancreas and miR-155 reduced in submandibular salivary glands in the periodontitis rat model. Therefore, these results support a possible mechanism for oral Gram-negative bacteria to modulate miRNA responses during infection in vivo.
Altered miRNA expression may be due to dissemination of oral pathogens, although the exact mechanisms are still unknown. In our study, metabolically active P. gingivalis and T. denticola was identified in infected rat submandibular salivary glands. It is unlikely that these bacteria were present before oral inoculation in these rats, as they are not found in their natural oral flora. In addition, an antibiotic course was given prior to infection to these rat and sham-infected rats showed no presence of these bacteria. Therefore, bacterial presence in submandibular salivary glands provides strong evidence of haematogenous or possible retrograde dissemination of these bacteria from the site of the oral cavity. As such, it is possible to speculate that oral bacterial dissemination may act directly to inhibit inflammatory miR-155 expression in the salivary glands. miRNA expression level may be increased in infected animals at the beginning of infection course and then diminish during subsequent infections.
Surprisingly, the pancreas of infected rats showed significant reductions in miR-132 and miR-146a. Although the pancreas were not evaluated for presence of oral periodontal pathogens, it is possible that periodontal pathogens may also alter pancreatic inflammatory miRNA expression patterns. For instance, diabetes, a disease in which blood sugar levels are dysregulated is an important risk factor for more severe and progressive periodontitis, and periodontitis in turn can also exacerbate the progression of diabetes [40]. This bidirectional association between periodontal disease and diabetes implicates the potential role of periodontal pathogens on modulation of inflammation through miRNAs.
It is important to note that the lacrimal glands did not exhibit any significant alterations in the inflammatory miRNA profiles in this study; therefore we hypothesize a possible supporting role for a direct effect of bacteria on the miRNA expression profiles in the salivary glands rather than a general systemic response. Our study has a limitation in that we used only one time point of infection i.e. 12 weeks, and in the small number of rats used. Further studies with multiple time points of infection would be critical for determining the timing and specific cell populations involved in infection-associated miRNA expression. Hence, there is an urgent need to expand our current study to further understand the mechanisms by which periodontal pathogens may directly affect the differential expression of miRNAs in the local and distant sites in vivo.
5. CONCLUSIONS
It is postulated that heavy microbial growth, which may occur in the setting of poor oral hygiene as well as resulting from decreased salivary flow, play complimentary roles in the pathogenesis of acute bacterial infections of the salivary glands [41, 42]. However, the role of periodontal disease in Sjögren’s syndrome, a chronic autoimmune disease of the exocrine glands, still remains controversial. Earlier studies suggest Sjögren’s syndrome was not associated with increased periodontal disease [43–45]. However, more recent studies indicate Sjögren’s syndrome is associated with higher gingival inflammation and increased periodontal disease [46–49]. Interestingly, periodontal disease development is also associated with inhibition of salivary gland function [29, 31–33]. The role of miRNA in modulating the development of diseases such as Sjögren’s syndrome is a new avenue offering plausible mechanisms for observed associations with periodontal disease. The implications of periodontal disease pathogens affecting miRNA expression of the salivary glands have not been previously explored. Our study is the first to identify the alterations of miRNAs in salivary glands of rats with chronic periodontal disease, potentially as a result of translocation of pathogenic bacteria from the primary site of infection. Thus, our study highlights these intricate associations between periodontal disease, diabetes, and Sjögren’s syndrome and emphasizes the need to initiate further investigations to better understand these conditions.
HIGHLIGHTS.
We show that the relative expression of miRNA-155 was increased in periodontal bacteria-infected rat gingiva.
We observed decreased miRNA-155 expression in submandibular salivary glands, along with positive identification of P. gingivalis and T. denticola in rat salivary glands.
We found miRNA-132 and miRNA-146a expression to significantly decrease in the pancreas of infected rats.
We report for the first time that primary periodontal infections can alter miRNA profiles in secondary sites such as the salivary gland and pancreas.
Acknowledgments
This work was supported in part by National Institute of Health/National Institute of Dental and Craniofacial Research grant DE019862 (Shannon Holliday), DE019644 (S.C.), and T90 DE021990 (A.G.). In addition, we would like to acknowledge Dr. Kaleb M. Pauley and Mercedes F. Rivera for collecting tissue samples and assisting with experimental procedures, respectively.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
CONFLICT OF INTEREST
The authors declare no conflict of interests.
REFERENCES
- 1.Socransky SS, Haffajee AD, Cugini MA, Smith C, Kent RL., Jr Microbial complexes in subgingival plaque. J Clin Periodontol. 1998;25:134–144. doi: 10.1111/j.1600-051x.1998.tb02419.x. [DOI] [PubMed] [Google Scholar]
- 2.Socransky SS, Haffajee AD. Periodontal microbial ecology. Periodontol. 2005;38:135–187. doi: 10.1111/j.1600-0757.2005.00107.x. 2000. [DOI] [PubMed] [Google Scholar]
- 3.Hajishengallis G. Periodontitis: from microbial immune subversion to systemic inflammation. Nat Rev Immunol. 2015;15:30–44. doi: 10.1038/nri3785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lockhart PB, Bolger AF, Papapanou PN, Osinbowale O, Trevisan M, Levison ME, et al. Periodontal disease and atherosclerotic vascular disease: does the evidence support an independent association?: a scientific statement from the American Heart Association. Circulation. 2012;125:2520–2244. doi: 10.1161/CIR.0b013e31825719f3. [DOI] [PubMed] [Google Scholar]
- 5.Preshaw PM, Alba AL, Herrera D, Jepsen S, Konstantinidis A, Makrilakis K, et al. Periodontitis and diabetes: a two-way relationship. Diabetologia. 2012;55:21–31. doi: 10.1007/s00125-011-2342-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bingham CO, 3rd, Moni M. Periodontal disease and rheumatoid arthritis: the evidence accumulates for complex pathobiologic interactions. Curr Opin Rheumatol. 2013;25:345–353. doi: 10.1097/BOR.0b013e32835fb8ec. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Singhrao SK, Harding A, Simmons T, Robinson S, Kesavalu L, Crean S. Oral inflammation, tooth loss, risk factors, and association with progression of Alzheimer's disease. J Alzheimers Dis. 2014;42:723–737. doi: 10.3233/JAD-140387. [DOI] [PubMed] [Google Scholar]
- 8.Madianos PN, Bobetsis YA, Offenbacher S. Adverse pregnancy outcomes (APOs) and periodontal disease: pathogenic mechanisms. J Periodontol. 2013;84:S170–S180. doi: 10.1902/jop.2013.1340015. [DOI] [PubMed] [Google Scholar]
- 9.Saini R, Saini S, Sharma S. Periodontitis: A risk factor to respiratory diseases. Lung India. 2010;27:189. doi: 10.4103/0970-2113.68313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Otomo-Corgel J, Pucher JJ, Rethman MP, Reynolds MA. State of the science: chronic periodontitis and systemic health. J Evid Based Dent Pract. 2012;12:20–28. doi: 10.1016/S1532-3382(12)70006-4. [DOI] [PubMed] [Google Scholar]
- 11.Han YW, Wang X. Mobile microbiome: oral bacteria in extra-oral infections and inflammation. J Dent Res. 2013;92:485–491. doi: 10.1177/0022034513487559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rivera MF, Lee JY, Aneja M, Goswami V, Liu L, Velsko IM, et al. Polymicrobial infection with major periodontal pathogens induced periodontal disease and aortic atherosclerosis in hyperlipidemic ApoE(null) mice. PLoS One. 2013;8:e57178. doi: 10.1371/journal.pone.0057178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Velsko IM, Chukkapalli SS, Rivera MF, Lee JY, Chen H, Zheng D, et al. Active invasion of oral and aortic tissues by Porphyromonas gingivalis in mice causally links periodontitis and atherosclerosis. PLoS One. 2014;9:e97811. doi: 10.1371/journal.pone.0097811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rivera MF, Chukkapalli SS, Velsko IM, Lee JY, Bhattacharyya I, Dolce C, et al. Bis-enoxacin blocks rat alveolar bone resorption from experimental periodontitis. PLoS One. 2014;9:e92119. doi: 10.1371/journal.pone.0092119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chukkapalli SS, Rivera MF, Velsko IM, Lee JY, Chen H, Zheng D, et al. Invasion of oral and aortic tissues by oral spirochete Treponema denticola in ApoE(−/−) mice causally links periodontal disease and atherosclerosis. Infect Immun. 2014;82:1959–1967. doi: 10.1128/IAI.01511-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chukkapalli SS, Rivera-Kweh MF, Velsko IM, Chen H, Zheng D, Bhattacharyya I, et al. Chronic oral infection with major periodontal bacteria Tannerella forsythia modulates systemic atherosclerosis risk factors and inflammatory markers. Pathog Dis. 2015;73 doi: 10.1093/femspd/ftv009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Velsko IM, Chukkapalli SS, Rivera-Kweh MF, Chen H, Zheng D, Bhattacharyya I, et al. Fusobacterium nucleatum Alters Atherosclerosis Risk Factors and Enhances Inflammatory Markers with an Atheroprotective Immune Response in ApoE(null) Mice. PLoS ONE. 2015;10:e0129795. doi: 10.1371/journal.pone.0129795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maixner F, Thomma A, Cipollini G, Widder S, Rattei T, Zink A. Metagenomic analysis reveals presence of Treponema denticola in a tissue biopsy of the Iceman. PLoS One. 2014;9:e99994. doi: 10.1371/journal.pone.0099994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Winter J, Jung S, Keller S, Gregory RI, Diederichs S. Many roads to maturity: microRNA biogenesis pathways and their regulation. Nat Cell Biol. 2009;11:228–234. doi: 10.1038/ncb0309-228. [DOI] [PubMed] [Google Scholar]
- 20.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guarnieri DJ, DiLeone RJ. MicroRNAs: a new class of gene regulators. Ann Med. 2008;40:197–208. doi: 10.1080/07853890701771823. [DOI] [PubMed] [Google Scholar]
- 22.Ma X, Becker Buscaglia LE, Barker JR, Li Y. MicroRNAs in NF-kappaB signaling. J Mol Cell Biol. 2011;3:159–166. doi: 10.1093/jmcb/mjr007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Taganov KD, Boldin MP, Chang KJ, Baltimore D. NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc Natl Acad Sci U S A. 2006;103:12481–12486. doi: 10.1073/pnas.0605298103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ceribelli A, Satoh M, Chan EK. MicroRNAs and autoimmunity. Curr Opin Immunol. 2012;24:686–691. doi: 10.1016/j.coi.2012.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Boldin MP, Taganov KD, Rao DS, Yang L, Zhao JL, Kalwani M, et al. miR-146a is a significant brake on autoimmunity, myeloproliferation, and cancer in mice. J Exp Med. 2011;208:1189–1201. doi: 10.1084/jem.20101823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wanet A, Tacheny A, Arnould T, Renard P. miR-212/132 expression and functions: within and beyond the neuronal compartment. Nucleic Acids Res. 2012;40:4742–4753. doi: 10.1093/nar/gks151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lagos D, Pollara G, Henderson S, Gratrix F, Fabani M, Milne RS, et al. miR-132 regulates antiviral innate immunity through suppression of the p300 transcriptional co352 activator. Nat Cell Biol. 2010;12:513–519. doi: 10.1038/ncb2054. [DOI] [PubMed] [Google Scholar]
- 28.Nahid MA, Rivera M, Lucas A, Chan EK, Kesavalu L. Polymicrobial infection with periodontal pathogens specifically enhances microRNA miR-146a in ApoE−/− mice during experimental periodontal disease. Infect Immun. 2011;79:1597–1605. doi: 10.1128/IAI.01062-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Slomiany BL, Slomiany A. Porphyromonas gingivalis lipopolysaccharide interferes with salivary mucin synthesis through inducible nitric oxide synthase activation by ERK and p38 kinase. Biochem Biophys Res Commun. 2002;297:1149–1153. doi: 10.1016/s0006-291x(02)02354-9. [DOI] [PubMed] [Google Scholar]
- 30.Bradley PJ. Microbiology and Management of Sialadenitis. Curr Infect Dis Rep. 2002;4:217–224. doi: 10.1007/s11908-002-0082-3. [DOI] [PubMed] [Google Scholar]
- 31.Slomiany BL, Slomiany A. Porphyromonas gingivalis lipopolysaccharide-induced up-regulation in endothelin-1 interferes with salivary mucin synthesis via epidermal growth factor receptor transactivation. IUBMB Life. 2004;56:601–607. doi: 10.1080/15216540400020361. [DOI] [PubMed] [Google Scholar]
- 32.Matto J, Saarela M, Alaluusua S, Oja V, Jousimies-Somer H, Asikainen S. Detection of Porphyromonas gingivalis from saliva by PCR by using a simple sample-processing method. J Clin Microbiol. 1998;36:157–160. doi: 10.1128/jcm.36.1.157-160.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sanchez GA, Miozza V, Delgado A, Busch L. Determination of salivary levels of mucin and amylase in chronic periodontitis patients. J Periodontal Res. 2011;46:221–227. doi: 10.1111/j.1600-0765.2010.01332.x. [DOI] [PubMed] [Google Scholar]
- 34.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 35.Moter A, Gobel UB. Fluorescence in situ hybridization (FISH) for direct visualization of microorganisms. J Microbiol Methods. 2000;41:85–112. doi: 10.1016/s0167-7012(00)00152-4. [DOI] [PubMed] [Google Scholar]
- 36.Pinho MM, Faria-Almeida R, Azevedo E, Manso MC, Martins L. Periodontitis and atherosclerosis: an observational study. J Periodontal Res. 2013;48:452–457. doi: 10.1111/jre.12026. [DOI] [PubMed] [Google Scholar]
- 37.Cheng Y, Kuang W, Hao Y, Zhang D, Lei M, Du L, et al. Downregulation of miR-27a* and miR-532-5p and upregulation of miR-146a and miR-155 in LPS-induced RAW264.7 macrophage cells. Inflammation. 2012;35:1308–1313. doi: 10.1007/s10753-012-9443-8. [DOI] [PubMed] [Google Scholar]
- 38.Xie YF, Shu R, Jiang SY, Liu DL, Zhang XL. Comparison of microRNA profiles of human periodontal diseased and healthy gingival tissues. Int J Oral Sci. 2011;3:125–134. doi: 10.4248/IJOS11046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stoecklin-Wasmer C, Guarnieri P, Celenti R, Demmer RT, Kebschull M, Papapanou PN. MicroRNAs and their target genes in gingival tissues. J Dent Res. 2012;91:934–940. doi: 10.1177/0022034512456551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Southerland JHT, W G, Offenbacher S. Diabetes and Periodontal Infection: Making the Connection. Clinical Diabetes. 2005;23:171–178. [Google Scholar]
- 41.Bradley PJ. Microbiology and Management of Sialadenitis. Curr Infect Dis Rep. 2002;4:217–224. doi: 10.1007/s11908-002-0082-3. [DOI] [PubMed] [Google Scholar]
- 42.Raad II, Sabbagh MF, Caranasos GJ. Acute bacterial sialadenitis: a study of 29 cases and review. Rev Infect Dis. 1990;12:591–601. doi: 10.1093/clinids/12.4.591. [DOI] [PubMed] [Google Scholar]
- 43.Boutsi EA, Paikos S, Dafni UG, Moutsopoulos HM, Skopouli FN. Dental and periodontal status of Sjogren's syndrome. J Clin Periodontol. 2000;27:231–235. doi: 10.1034/j.1600-051x.2000.027004231.x. [DOI] [PubMed] [Google Scholar]
- 44.Kuru B, McCullough MJ, Yilmaz S, Porter SR. Clinical and microbiological studies of periodontal disease in Sjogren syndrome patients. J Clin Periodontol. 2002;29:92–102. doi: 10.1034/j.1600-051x.2002.290202.x. [DOI] [PubMed] [Google Scholar]
- 45.Jorkjend L, Johansson A, Johansson AK, Bergenholtz A. Periodontitis, caries and salivary factors in Sjogren's syndrome patients compared to sex- and age-matched controls. J Oral Rehabil. 2003;30:369–378. doi: 10.1046/j.1365-2842.2003.01088.x. [DOI] [PubMed] [Google Scholar]
- 46.Najera MP, al-Hashimi I, Plemons JM, Rivera-Hidalgo F, Rees TD, Haghighat N, et al. Prevalence of periodontal disease in patients with Sjogren's syndrome. Oral Surg Oral Med Oral Pathol Oral Radiol Endod. 1997;83:453–457. doi: 10.1016/s1079-2104(97)90144-x. [DOI] [PubMed] [Google Scholar]
- 47.Pers JO, d'Arbonneau F, Devauchelle-Pensec V, Saraux A, Pennec YL, Youinou P. Is periodontal disease mediated by salivary BAFF in Sjogren's syndrome? Arthritis Rheum. 2005;52:2411–2414. doi: 10.1002/art.21205. [DOI] [PubMed] [Google Scholar]
- 48.Antoniazzi RP, Miranda LA, Zanatta FB, Islabao AG, Gustafsson A, Chiapinotto GA, et al. Periodontal conditions of individuals with Sjogren's syndrome. J Periodontol. 2009;80:429–435. doi: 10.1902/jop.2009.080350. [DOI] [PubMed] [Google Scholar]
- 49.Scardina GA, Ruggieri A, Messina P. Periodontal disease and sjogren syndrome: a possible correlation? Angiology. 2010;61:289–293. doi: 10.1177/0003319709344576. [DOI] [PubMed] [Google Scholar]