Abstract
17β-estradiol (E2) modulates gene expression in the hypothalamic arcuate nucleus (ARC) to control homeostatic functions. In the ARC, estrogen receptor (ER) α is highly expressed and is an important contributor to E2’s actions, controlling gene expression through estrogen response element (ERE)-dependent and -independent mechanisms. The objective of this study was to determine if known E2-regulated genes are regulated through these mechanisms. The selected genes have been shown to regulate homeostasis and have been separated into three subsections: channels, receptors, and neuropeptides. To determine if ERE-dependent or ERE-independent mechanisms regulate gene expression, two transgenic mouse models, an ERα knock-out (ERKO) and an ERα knock-in/knock-out (KIKO), which lacks a functional ERE binding domain, were used in addition to their wild-type littermates. Females of all genotypes were ovariectomized and injected with oil or estradiol benzoate (E2B). Our results suggest that E2B regulates multiple genes through these mechanisms. Of note, Cacna1g and Kcnmb1 channel expression was increased by E2B in WT females only, suggesting an ERE-dependent regulation. Furthermore, the NKB receptor, Tac3r, was suppressed by E2B in WT and KIKO females but not ERKO females, suggesting that ERα-dependent, ERE-independent signaling is necessary for Tac3r regulation. The adrenergic receptor Adra1b was suppressed by E2B in all genotypes indicating that ERα is not the primary receptor for E2B’s actions. The neuropeptide Tac2 was suppressed by E2B through ERE-dependent mechanisms. These results indicate that E2B activates both ERα-dependent and independent signaling in the ARC through ERE-dependent and ERE-independent mechanisms to control gene expression.
Keywords: 17β-estradiol, ovariectomy, ERα, arcuate nucleus
Introduction
The steroid hormone 17β-estradiol (E2) is known to regulate gene expression throughout the brain. E2 primarily uses two classical nuclear receptors, estrogen receptor α (ERα, Esr1,) and ERβ (Esr2) to regulate gene expression [1]. In the classical ER-mediated mechanism, ligand binding to the receptor initiates receptor recognition of the estrogen response element (ERE) to regulate gene transcription. In addition to classical regulation of gene expression, E2 also functions through ERE-independent mechanisms. As reviewed in McDevitt et al. (2008), these mechanisms include ligand-independent ER signaling, rapid effects through a membrane-associated ER, and ERE-independent signaling through protein-protein interactions (AP-1, etc.) [2].
In the hypothalamus, E2 mediates numerous homeostatic functions including reproduction, energy homeostasis, core body temperature, fluid balance, motivational behaviors, and stress physiology by regulating central neural pathways. Many of these pathways originate in or pass through the arcuate nucleus (ARC) of the hypothalamus [1, 3-5]. In the ARC, ERα is highly expressed and is the primary receptor used by E2 to control many homeostatic functions [5, 6]. Few studies have examined the physiological effects of ERα-mediated, ERE-dependent, and ERE-independent signaling on hypothalamic (ARC) gene expression.
Recently, the development of an ERα knock-in/knock-out (KIKO) mouse model lacking a functional DNA-binding domain (no ERE binding) gives insight to nonclassical, ERα-mediated, ERE-independent signaling while retaining ERβ-mediated signaling and other extra-nuclear initiated pathways (GPER, Gq-mER) [7, 8]. While KIKO females are infertile due to an absence of the LH surge [9, 10], they exhibit similar body weights, feeding, activity, oxygen consumption, glucose homeostasis and hypothalamic leptin sensitivity compared to wild-type (WT) females, unlike their total ERKO counterparts [11]. However, recent data from our lab suggest that ERα-mediated ERE-independent signaling partially restores the post-ovariectomy (ovx) weight gain but is not sufficient to mediate E2’s attenuation of this weight gain [12].
E2 control of homeostatic functions occurs, in part, through regulation of important genes in the ARC. However, only a few studies have examined which signaling mechanisms E2 utilizes to control the expression of these genes. The KIKO mouse model provides an appropriate tool to increase our understanding of how homeostatic genes are regulated by E2-induced ERα-mediated mechanisms in the ARC. In the ARC, E2 is known to regulate the expression of a variety of cation channels including calcium channels and potassium channels [13-16]. The expression and activity of these cation channels are involved in regulating ARC proopiomelanocortin (POMC) and neuropeptide Y (NPY) neurons and their neuronal excitability [1, 12, 14-20]. Furthermore, E2 is known to regulate the mRNA expression of signaling molecules such as calmodulin and phosphatidylinositol-4,5-biphosphate 3-kinase (PI3K) [16, 21-23] and neurotransmitter enzymes like tyrosine hydroxylase (TH) and glutamate decarboxylase [1, 24]. Many of these E2-regulated genes are involved in reproduction, energy homeostasis, and hormone receptor signaling [1, 3, 25-27].
E2 is also known to suppress or augment the expression of a variety of neuropeptides and receptors in the hypothalamus, depending upon the experimental paradigm. In rodents, E2 increases Pomc and suppresses Npy expression in the ARC [1] and increases the expression of growth hormone [28]. It is also well known that E2 differentially regulates steroid receptors in the hypothalamus such as ERα, ERβ, and progesterone receptor (Pgr) [1, 16, 29]. Other hypothalamic hormone and neurotransmitter receptors are regulated by E2 in the hypothalamus include growth hormone secretagogue receptor (Ghsr) [30] and serotonin receptor 2C receptor (5HT2c) [31]. The mechanism underlying the regulation of all of these genes is largely unknown. Our current study focused on the regulation of ARC gene expression by E2.
E2 also regulates ARC KNDy (Kisspeptin-Neurokinin B-Dynorphin) neuronal gene expression [32]. Kisspeptin is involved in mediating negative and positive feedback of E2 on the hypothalamic-pituitary-gonadal (HPG) axis and potentially has a role in energy homeostasis and core body temperature [33]. Previous studies indicated that the Kiss1 gene is regulated by E2 through ERE-independent mechanisms in the mediobasal hypothalamus while dynorphin expression is ERE-dependent [34]. Thus, we used these genes as positive and negative controls for ERE-dependent and ERE-independent gene expression. Nothing is known about the mechanisms behind the regulation of Neurokinin B (NKB, Tac2) or the KNDy receptors (Kiss1r, Tac3r) by E2 in the ARC. Therefore, the objective of this study was to determine if E2-responsive, homeostatic genes involved in reproduction and energy homeostasis that have been identified to be E2-responsive in the hypothalamus are regulated by E2 in the ARC through ERE-dependent or ERE-independent mechanisms using ovx WT, KIKO, and ERKO females.
Experimental
Animal care
All animal procedures were in compliance with institutional guidelines based on National Institutes of Health standards and were performed with Institutional Animal Care and Use Committee approval at Rutgers University. Adult C57BL/6 mice were housed under constant photoperiod conditions (12/12 h light/dark cycle) and maintained at a controlled temperature (25°C). Animals were given low phytoestrogen chow diet (<75 isoflavone ppm, Lab Diet Advanced Protocol 5V75, St. Louis, MO, USA) and water ad libitum. Animals were weaned on postnatal day 21 (PD21). Genotype was determined by using PCR products of extracted DNA from ear clippings, using previously published protocols [9]. We used three genotypes of mice: WT, KIKO, and ERKO (provided by Dr. Ken Korach, NIEHS) [9]. Crossing heterozygous WT/KI males expressing the nonclassical ERα knock-in with WT/KO heterozygous females generated WT and KIKO females. Crossing heterozygous WT/KO males and females generated WT and ERKO females. WT females used in the experiments were littermates generated from both colonies.
Drugs
17β-estradiol benzoate (E2B) was purchased from Steraloids (Newport, RI, USA) and dissolved in ethanol (1mg/ml) prior to mixing in sesame oil (Sigma-Aldrich). Ketamine was purchased from Henry Schein Animal Health (Melville, NY, USA) and used for sedation prior to sacrifice.
Ovariectomy
Adult females (7-22 weeks and > 14 g body weight) were bilaterally ovx under isoflurane anesthesia 7 days prior to sacrifice using sterile no-touch techniques according to the NIH “Guidelines for Survival Rodent Surgery.” Animals were given a dose of analgesic [4 mg/kg carprofen (Rimadyl®)] one day following surgery for pain management. Animals typically lost 1-2 g of weight one day after surgery. Females were monitored daily and allowed to recover for 5 days prior to the first injection of E2B or oil. The active metabolite of E2B is 17β-estradiol. Females were injected in the morning at 1000 h on post-ovx days 5 and 6 and sacrificed on post-ovx day 7 in the morning at 1000 h.
Experimental design
Females of each genotype (WT, KIKO, ERKO) were ovx and separated into a control sesame oil-treated group (n=6-9 per genotype) and an E2B-treated group (n=6-9 per genotype). An E2B injection protocol was used that has been shown to alter gene expression in the hypothalamus [19]. Animals were injected subcutaneously at 1000 h on post-ovx day 5 with either 0.25 μg of E2B or sesame oil. On post-ovx day 6, a 1.5 μg dose of E2B or sesame oil was injected at 1000 h. We did not include intact females in our experimental design as neither ERKO nor KIKO females exhibit a normal estrous cycle, which makes it difficult to compare among intact WT, KIKO, and ERKO females [11]. Animals were sacrificed on post-ovx day 7 at 1000 h. Animals were sedated with ketamine (100 μl of 100 mg/ml stock, i.p.) and decapitated. Brains were removed and rinsed in ice-cold Sorensen’s Phosphate Buffer (0.2 M sodium phosphate, dibasic; and 0.2 M sodium phosphate, monobasic) for 30-60 sec. The basal hypothalamus (BH) was cut using a brain slice matrix (Ted Pella, Inc., Redding, CA, USA) into 1-mm thick coronal rostral and caudal blocks corresponding to Plates 42 to 47 and Plates 48 to 53, respectively, from The Mouse Brain in Stereotaxic Coordinates [35]. The slices were transferred to a 50/50 Pyrogard water/RNAlater® (Life Technologies, Grand Island, NE, USA) solution and fixed overnight at 4°C. The ARC tissue, found in two slices, was microdissected using a dissecting microscope, following our previous publications [1, 12, 14, 35]. The microdissected sections represent the entirety of the ARC tissue. Dissected tissue was stored at −80 °C until RNA extraction. Trunk blood was collected at sacrifice to measure plasma E2 levels. Uteri were removed and wet weight was recorded. Wet uterine weight (mg) is a sensitive measure of circulating E2 in WT mice.
Tissue extraction
RNA was extracted from ARC using Ambion RNAqueous® Micro Kits (Life Technologies, Inc., Carlsbad, CA, USA) according to the manufacturer’s protocol, followed by DNase-I treatment to remove contamination by genomic DNA. RNA samples were run on a NanoDrop™ ND-2000 spectrophotometer (ThermoFisher, Inc., Waltham, MA, USA) to assess quantity, followed by an Agilent 2100 Bioanalyzer run using the RNA 6000 Nano Kit (Agilent Technologies, Inc., Santa Clara, CA, USA) to assess quality. Samples with a RNA integrity number (RIN) > 8 were used for quantitative real-time PCR (qPCR).
Blood preparation
Whole trunk blood was centrifuged (1300 rpm at 4 °C for 30 min). The supernatant was subjected to an additional 15 minutes of centrifugation (4 °C at 1300 rpm), then the plasma supernatant was removed and stored at −20 °C until E2 analysis. E2 concentration of plasma was analyzed using Mouse Calbiotech ELISA at the Ligand Assay and Analysis Core of the University of Virginia’s Center for Research in Reproduction [36]. The Calbiotech ELISA is specific to E2 detection.
Quantitative real-time PCR
cDNA was synthesized from 200 ng of total RNA using Superscript III reverse transcriptase (Life Technologies, Inc.), 4 μl 5x buffer, 25 mM MgCl2, 10 mM dNTP (Clontech Laboratories, Inc., Mountain View, CA, USA), 100 ng random hexamer primers (Promega Corporation, Madison, WI, USA), 40 U/μl Rnasin (Promega), and 100 mM DTT in DEPC-treated water (Gene Mate, Bioexpress, Inc., Kaysville, UT, USA) in a total volume of 20 μl. Reverse transcription was conducted using the following protocol: 5 min at 25 °C, 60 min at 50 °C, 15 min at 70 °C. A 1:20 dilution of the cDNA was produced using nuclease-free water (Gene Mate) for a final cDNA concentration of 0.5 ng/μl and stored at −20 °C. BH tissue RNA, which contains the ARC, from a male mouse was used as a positive control. A negative tissue control (BH with no reverse transcriptase) was also used.
A Taqman® Low Density Array (TLDA) (Life Technologies, Inc.) was used to analyze ARC gene expression in WT females. This array consisted of Taqman® expression assays of genes known to be E2-regulated and/or involved in reproduction and energy homeostasis (see Table 1 for a listing of genes analyzed). Data presented graphically only include genes significantly regulated by E2B, with the exception of Gpr30. The TLDA was designed so each expression assay was run in triplicate, including the reference gene, Actb, and the internal control, 18S. On each TLDA plate, one WT experimental female sample was run in duplicate and a calibrator sample (male BH) was run on the remaining wells. The same calibrator sample was run on each plate. Taqman® primers were ordered for all genes, and KIKO and ERKO samples were analyzed. Positive and negative tissue control samples and master mix (nuclease-free water) controls were added to each run. In KIKO and ERKO qPCR plates, an additional sample (termed “pool”) was analyzed that included all the control oil samples from each respective genotype, to account for inter-plate variation (data from Cq values analyzed across plates). qPCR was performed on a StepOnePlus™ Real-Time PCR System (Life Technologies, Inc.) using Taqman® Gene Expression Master Mix. For qPCR, we used 4 μg of cDNA (equivalent to 2 ng of total RNA). The amplification protocol for all genes was as follows: a holding stage consisted of 2 min at 50 °C and 95 °C for 10 min, followed by a cycling stage of 95 °C for 15 sec (denaturing) and at 60 °C (annealing) for 1 min for 40 cycles.
Table 1.
List of genes analyzed in the Taqman® Low Density Array (TLDA)
| Gene Name | Gene Abbreviation | Taqman Assay # |
|---|---|---|
| ATP-binding cassette, subfamily C | Abcc8 | Mm00803450_m1 |
| B cell leukemia/lymphoma 2 | Bcl2 | Mm00477631_m1 |
| Cav3.1 | Cacna1g | Mm00486572_m1 |
| Cav3.2 | Cacna1h | Mm00445382_m1 |
| Calmodulin 1 | Calm1 | Mm01336281_g1 |
| Estrogen receptor α | Esr1 | Mm00433149_m1 |
| Estrogen receptor β | Esr2 | Mm00599821_m1 |
| Glutamate decarboxylase 1 | Gad1 | Mm04207432_g1 |
| Glutamate decarboxylase 2 | Gad2 | Mm00484623_m1 |
| G protein-coupled receptor 30 | Gpr30 | Mm01194815_m1 |
| Growth hormone | Gh | Mm00433590_g1 |
| Serotonin receptor 2C | Htr2c | Mm00434127_m1 |
| Calcium-activated potassium channel subunit β1 | Kcnmb1 | Mm00466621_m1 |
| Kir6.2 | Kcnj11 | Mm00440050_s1 |
| Calcium-activated potassium channel subunit β4 | Kcnmb4 | Mm00465684_m1 |
| KCNQ5 (Kv7.5) | Kcnq5 | Mm01226041_m1 |
| Kisspeptin 1 | Kiss1 | Mm03058560_m1 |
| Kisspeptin receptor | Kiss1r | Mm00475046_m1 |
| Neuropeptide Y | Npy | Mm03048253_m1 |
| Prodynorphin | Pdyn | Mm00457573_m1 |
| Progesterone receptor | Pgr | Mm00435628_m1 |
| PI3K p55γ | Pik3r3 | Mm00725026_m1 |
| Proopiomelanocortin | Pomc | Mm00435874_m1 |
| Tachykinin 2 | Tac2 | Mm00436885_m1 |
| Tachykinin 3 receptor | Tac3r | Mm00445346_m1 |
| Tyrosine hydroxylase | Th | Mm00447557_m1 |
| Nuclear receptor coactivator 1 | Ncoa | Mm01318933_m1 |
| Mammalian target of rapamycin | Mtor | Mm00444968_m1 |
| Sirtuin 1 | Sirt1 | Mm00490758_m1 |
| Beta actin | Actb | Mm01205647_g1 |
| 18s ribosomal RNA | 18S | Hs99999901_s1 |
Genes that were analyzed using a TLDA and their respective Taqman® assay #s are listed. For KIKO and ERKO analysis, identical Taqman® assays were used.
In addition to the genes analyzed in the TLDA, we also analyzed the mRNA expression of four additional genes: Adra1b, Cart, Ghsr and Chrm1, which were found to be E2-regulated in our preliminary investigations or from the literature [37, 38]. Primers for these genes were designed to span exon-exon junctions and were synthesized by Life Technologies, Inc., using Clone Manager 5 software (Sci Ed Software, Cary, NC, USA). See Table 2 for a listing of synthesized primers used for qPCR. For qPCR of these four genes, we used 4 μg of cDNA (equivalent to 2 ng of total RNA) amplified with either PowerSYBR® Green Master Mix (Adra1b, Chrm1; Life Technologies, Inc.) or SsoAdvanced™ SYBR Green (Cart, Ghsr; BioRad, Inc., Hercules, CA, USA) on CFX-Connect Real-time PCR Instrument (BioRad, Inc.). A standard curve was generated for each primer pair using serial dilutions of BH cDNA in triplicate. Efficiencies were calculated as a percent efficiency, listed in Table 2. Amplification protocol for Table 2 genes was as follows: initial denaturing 95 °C for 10 min (PowerSYBR®) or 3 min (SsoAdvanced™) followed by 40 cycles of amplification at 94 °C for 10 sec (denaturing), 60 °C for 45 sec (annealing), and completed with a dissociation step for melting point analysis with 60 cycles of 95 °C for 10 sec, 65 °C to 95 °C (in increments of 0.5 °C) for 5 sec and 95 °C for 5 sec. The reference genes used were β-actin (PowerSYBR®) and Gapdh (SsoAdvanced™). Positive, negative, and water blank controls were included in the qPCR plate design.
Table 2.
List of genes using designed primers
| Gene Name |
Product length |
% Eff | Primer sequence | Base pair # | Accession # |
|---|---|---|---|---|---|
| Adra1b | 84 | 100 | F: CTTCATCGCTCTCCCACTTG | 1174-1193 | NM_007416 |
| R: TAGCCCAGCCAGAACACT | 1240-1257 | ||||
| β-actin | 63 | 100 | F: GCCCTGAGGCTCTTTTCCA | 849-867 | NM_007393.3 |
| R: TAGTTTCATGGATGCCACAGGA | 911-990 | ||||
| Cart | 169 | 93 | F: GCTCAAGAGTAAACGCATTCC | 277-297 | NM_013732 |
| R: GTCCCTTCACAAGCACTTCAA | 425-445 | ||||
| Chrm1 | 272 | 110 | F: AGCAGCTCAGAGAGGTCACAGCCA | 1331-1354 | NM_001112697 |
| R: GGGCCTCTTGACTGTATTTGGGGA | 1580-1603 | ||||
| Gapdh | 98 | 93 | F: TGACGTGCCGCCTGGAGAAA | 852-875 | NM_008084 |
| R: AGTGTAGCCCAAGATGCCCTTCAG | 106-125 | ||||
| Ghsr | 122 | 110 | F: CAGGGACCAGAACCACAAAC | 1003-1022 | NM_177330 |
| R: AGCCAGGCTCGAAAGACT | 1107-1124 |
The genes listed are those that were not included in the TLDA. Sense primer is listed first with the antisense primer below. Primer pairs were ordered from Life Technologies and designed to span exon-exon junctions using Clone 5 software. Adra1b, alpha-1B adrenergic receptor; β-actin, beta actin; Cart, cocaine- and amphetamine- regulated transcript; Chrm1, cholinergic muscarinic 1 receptor; Gapdh, glyceraldehyde-3-phosphate dehydrogenase; Ghsr, growth hormone secretagogue receptor.
Analysis of qPCR was done using the comparative Cq method using a 1:20 diluted BH cDNA (equivalent to 2 ng of RNA) sample from a male as the calibrator [39, 40]. All values were normalized to oil controls and are expressed as relative mRNA expression. In all plates, we maintained a consistent threshold level, set at the lowest but steepest slope of the exponential curve. We calculated the linear quantity of target genes using the formula 2−ΔΔCq. Data are expressed as n-fold difference from the calibrator, normalized to oil controls. The n-fold difference was used for statistical analysis.
Statistical analyses
qPCR data from the TLDA in WT females were initially analyzed using Data Assist® software (Life Technologies, Inc.) to determine significant differences between oil- and E2B-treated samples using a t-test with a false discovery rate (Benjamini-Hochberg method) set to a p < 0.05. All further statistical analyses were performed using GraphPad® Prism software (GraphPad Software, Inc., La Jolla, CA, USA). Data were expressed as mean ± SEM and analyzed by a two-way ANOVA (genotype × treatment), followed by a post-hoc Bonferroni’s multiple comparison test between oil- and E2B-treated groups within each genotype (WT, KIKO, ERKO). Uterine weights were analyzed using a two-way ANOVA (genotype × treatment) followed by Bonferroni’s multiple comparison test between oil- and E2B-treated groups, within each genotype (WT, KIKO, ERKO). Plasma E2 levels of all three genotypes were pooled together for oil vs. E2B treatment analysis, as there was no genotype effect (data not shown), and analyzed using a Student’s t-test. In all experiments, a p < 0.05 was considered to be significant. To determine relative gene expression among genotypes, we compared oil-treated WT, KIKO, and ERKO females. These data were analyzed using a one-way ANOVA followed by a post-hoc Bonferroni’s multiple comparison test.
Results
Uterine weights and plasma E2 levels
Following sacrifice, we dissected out the uterus of each female to confirm the hypertrophic actions of E2, an ERα-mediated process [41]. Past studies in our lab have suggested that E2B (250 ng/dose) replacement every other day for 4 weeks significantly increased the uterine weight in WT females [12]. E2B significantly increased the uterine weight in WT females (Table 3; ANOVA: F(2,24) = 34.75, p < 0.0001), but did not increase uterine weight in KIKO and ERKO females. There was no significant difference in body weight at sacrifice for E2B-treated females in all genotypes. The age of sacrifice of females was as follows: WT: 12-23 weeks (average: 18.2 ± 1.0 weeks); KIKO: 7-23 weeks (average: 17.2 ± 1.6 weeks); ERKO: 7-23 weeks (average: 13.9 ± 1.8 weeks). There were no age-specific effects when we analyzed the 2ΔΔCq -values for all regulated genes across genotypes (data not shown).
Table 3.
Body and uterine weights
| Genotype | Treatment | Body weight (g) |
Uterine weight (mg) |
|---|---|---|---|
| WT | Oil | 24.0 ± 0.6 | 27.5 ± 0.9 |
| E2B | 23.6 ± 0.7 | 98.3 ± 2.1**** | |
| KIKO | Oil | 22.1 ± 1.6 | 38.8 ± 5.3 |
| E2B | 22.4 ± 0.9 | 43.0 ± 5.1 | |
| ERKO | Oil | 19.5 ± 0.3 | 10.0 ± 1.3 |
| E2B | 21.0 ± 0.9 | 13.2 ± 2.4 |
Uterine weights are expressed relative to body weight. Data were analyzed with a two-way ANOVA with Bonferroni’s multiple comparison tests
p < 0.0001
We pooled plasma E2 data across genotypes (WT, KIKO, ERKO) because there was no genotype effect observed (data not shown). Plasma E2 concentrations were as follows: oil-treated: 5.2 ± 0.4 pg/ml; E2B-treated: 28.82 ± 7.0 pg/ml. There was a significant increase in plasma E2 levels 24 h post-injection between oil- and E2B-treated groups (p < 0.001).
E2B dose regulates ARC gene expression
E2B-treatment in ovx WT females significantly regulated the mRNA expression of 14 genes in the ARC (Table 4). These genes include those that function as cation channels, receptors for hormones and neurotransmitters, and neuropeptides. E2 replacement significantly suppressed the mRNA expression of Adra1b, Cart, Chrm1, Esr1, Kiss1, Pdyn, NKB (Tac2), and Tac3r and significantly augmented expression of Cacna1g, Esr2, Ghsr, Kcnmb1, Kiss1r, and Pgr. The remaining genes in Table 4 were not regulated by E2B in WT females.
Table 4.
TLDA Gene Expression Analysis
| Gene Name | Functional Name (Protein) | Oil | E2B | p |
|---|---|---|---|---|
| Abcc8 | ATP-binding cassette, subfamily C member 8 | 1.02 ± 0.10 | 0.97 ± 0.11 | - |
| Adra1b | Alpha-1B adrenergic receptor | 1.02 ± 0.04 | 0.67 ± 0.03 | **** |
| Bcl2 | Apoptosis regulator Bcl-2 | 1.00 ± 0.03 | 0.99 ± 0.06 | - |
| Cacna1g | Voltage-dependent T-type calcium channel subunit alpha-1G |
1.01 ± 0.08 | 1.61 ± 0.17 | *** |
| Cacna1h | Voltage-dependent T-type calcium channel subunit alpha-1H |
1.01 ± 0.05 | 1.03 ± 0.06 | - |
| Calm1 | Calmodulin | 1.01 ± 0.07 | 1.13 ± 0.05 | - |
| Cart | Cocaine- and amphetamine-regulated transcript protein |
1.08 ± 0.08 | 0.54 ± 0.02 | * |
| Chrm1 | Muscarinic acetylcholine receptor M1 | 1.08 ± 0.03 | 0.33 ± 0.04 | **** |
| Esr1 | Estrogen receptor α | 1.01 ± 0.06 | 0.66 ± 0.08 | * |
| Esr2 | Estrogen receptor β | 1.03 ± 0.13 | 1.60 ± 0.12 | * |
| Gad1 | Glutamate decarboxylase 1 | 1.01 ± 0.06 | 0.94 ± 0.07 | - |
| Gad2 | Glutamate decarboxylase 2 | 1.01 ± 0.05 | 1.03 ± 0.06 | - |
| Gh | Growth Hormone (Somatotropin) | 1.03 ± 0.11 | 1.01 ± 0.11 | - |
| Ghsr | Growth hormone secretagogue receptor type 1 | 1.16 ± 0.09 | 2.47 ± 0.15 | **** |
| Gpr30 | G-protein coupled estrogen receptor 1 | 1.03 ± 0.12 | 0.84 ± 0.04 | - |
| Htr2c | 5-hydroxytryptamine receptor 2C | 1.01 ± 0.09 | 1.20 ± 0.14 | - |
| Kcnj11 | ATP-sensitive inward rectifier potassium channel 11 |
1.01 ± 0.07 | 0.92 ± 0.05 | - |
| Kcnmb1 | Calcium-activated potassium channel subunit beta-1 |
1.16 ± 0.25 | 2.45 ± 0.41 | ** |
| Kcnmb4 | Calcium-activated potassium channel subunit beta-4 |
1.03 ± 0.11 | 1.30 ± 0.07 | - |
| Kcnq5 | Potassium voltage-gated channel subfamily KQT member 5 |
1.00 ± 0.04 | 1.12 ± 0.11 | - |
| Kiss1 | Kisspeptin | 1.02 ± 0.11 | 0.12 ± 0.02 | *** |
| Kiss1r | Kisspeptin receptor | 1.00 ± 0.02 | 1.29 ± 0.05 | ** |
| Mtor | Serine/threonine-protein kinase mammalian target of rapamycin |
1.00 ± 0.04 | 1.08 ± 0.06 | - |
| Ncoa | Nuclear receptor coactivator 1 | 1.00 ± 0.05 | 1.07 ± 0.04 | - |
| Npy | Neuropeptide Y | 1.13 ± 0.25 | 0.99 ± 0.19 | - |
| Pdyn | Prodynorphin | 1.04 ± 0.15 | 0.55 ± 0.03 | ** |
| Pgr | Progesterone receptor | 1.00 ± 0.04 | 2.12 ± 0.20 | **** |
| Pik3r3 | Phosphatidylinositol-4,5 bisphosphate 3-kinase regulatory subunit gamma |
1.00 ± 0.03 | 1.12 ± 0.06 | - |
| Pomc | Pro-opiomelanocortin | 1.01 ± 0.08 | 0..94 ± 0.10 | - |
| Sirt1 | NAD-dependent protein deacetylase sirtuin-1 | 1.00 ± 0.04 | 1.06 ± 0.03 | - |
| Tac2 | Tachykinin 2 (Neurokinin B) | 1.04 ± 0.16 | 0.19 ± 0.03 | **** |
| Tac3r | Tachykinin 3 receptor | 1.04 ± 0.15 | 0.35 ± 0.06 | **** |
| Th | Tyrosine hydroxylase | 1.04 ± 0.13 | 1.01 ± 0.05 | - |
Data were analyzed by a Students t-test for each gene
p < 0.05;
p < 0.01;
p < 0.001;
p < 0.0001
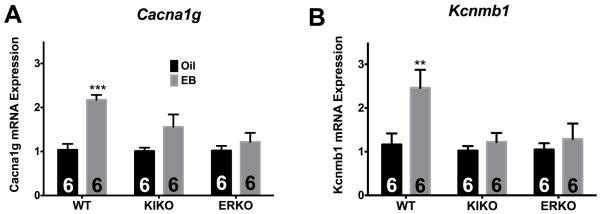
After studying WT ARC mRNA expression, we then analyzed all the genes in ARC tissue from KIKO and ERKO females injected with either sesame oil or E2B using individual Taqman® assays or custom primers. In comparing the genotypes, we found that the mRNA expression of two cation channels, which are involved in neuronal excitability, were upregulated by E2B in WT females only: Cav3.1 (Cacna1g), a subunit of the T-type Ca2+ channels, (Figure 1A; ANOVA: F(2,30) = 4.004, p < 0.05); and Kcnmb1, the β1 regulatory subunit for Ca2+-activated potassium channel (Figure 1B; ANOVA: F(1,27) = 6.221, p < 0.05). The mRNA expression of both genes was increased more than two-fold in WT females injected with E2B.
Figure 1. E2B regulates channel gene expression in the ARC.
(A) Cacna1g and (B) Kcnmb1. Results of qPCR analyses represent gene expression of oil- (black bars) and E2B- (gray bars) treated females in WT, KIKO, and ERKO genotypes. The number of animals in each treatment group is listed within each bar. Genes are expressed as relative n-fold changes, normalized to oil controls, within each genotype (WT, KIKO, ERKO). A two-way ANOVA (genotype × treatment) followed by post-hoc Bonferroni’s multiple comparison test was used to determine significant differences between treatments, within genotype. **p < 0.01; ***p < 0.001.
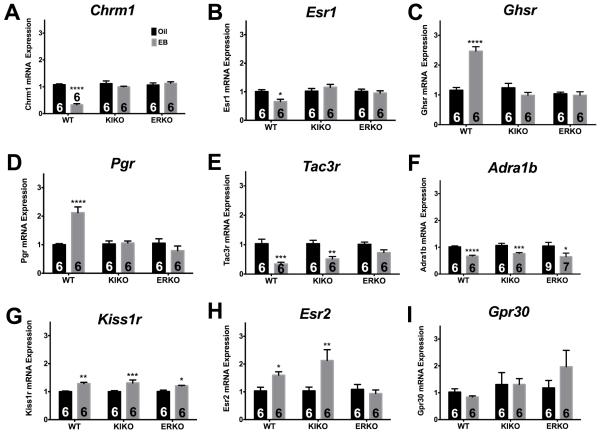
In addition, multiple receptors involved in reproduction and energy balance showed a change in gene expression with E2B treatment. The mRNA expression of the cholinergic muscarinic receptor 1 (Chrm1) was suppressed threefold (Figure 2A; ANOVA: F(2,30) = 25.64; p < 0.0001), and ERα (Esr1) by ~35% (Figure 2B; ANOVA: F(2,29) = 4.143; p < 0.05) in WT females. E2B increased expression of growth hormone secretagogue receptor (Ghsr) (Figure 2C; ANOVA: F(2, 30) = 28.45; p < 0.0001) and progesterone receptor (Pgr) (Figure 2D; ANOVA: F(2,29) = 13.43; p < 0.0001) in WT females. Expression of Tac3r, the NKB receptor, was suppressed twofold in both WT and KIKO females by E2B (Figure 2E; ANOVA: F(1,29) = 40.58, p < 0.0001). Expression of adrenergic receptor, Adra1b, was suppressed by E2B across all genotypes (Figure 2F; ANOVA: F(1,34) = 17.64, p < 0.001). Expression of the kisspeptin receptor, Kiss1r, was increased by E2B across all genotypes (Figure 2G; ANOVA: F(1,29) = 35.49, p < 0.0001). Finally, ERβ (Esr2) expression was augmented by E2B in both WT and KIKO females (Figure 2H; ANOVA: F(2,28) = 4.452, p < 0.05). GPER/GPR30 expression was not regulated by E2 in the ARC of any genotype (Figure 2I).
Figure 2. E2B regulates receptor gene expression in the ARC.
(A) Chrm1, (B) Esr1, (C) Ghsr, (D) Pgr, (E) Tac3r, (F) Adra1b, (G) Kiss1r, (H) Esr2, and (I) Gpr30. Results of qPCR analyses represent gene expression of oil- (black bars) and E2B- (gray bars) treated females in WT, KIKO, and ERKO genotypes. The number of animals in each treatment group is listed within each bar. Genes are expressed as relative n-fold changes, normalized to oil controls, within each genotype (WT, KIKO, ERKO). A two-way ANOVA (genotype × treatment) followed by post-hoc Bonferroni’s multiple comparison test was used to determine significant differences between treatments, within genotype. *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001.
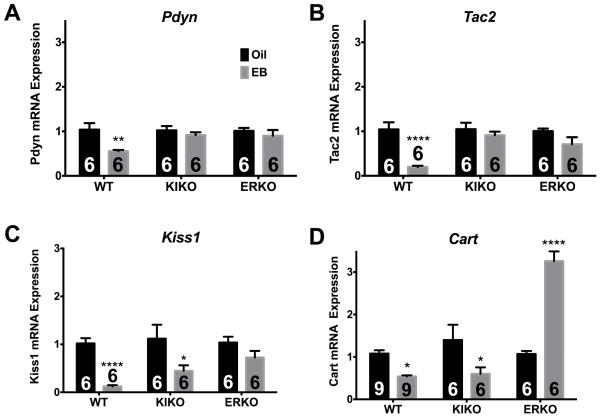
E2B also regulated several important neuropeptide genes in the ARC. In WT females only, prodynorphin (Pdyn) and neurokinin B (Tac2) expression were suppressed by E2B twofold (Figure 3A: ANOVA: F(1, 29) = 9.322, p < 0.01) and fivefold (Figure 3B; ANOVA: F(2, 29) = 5.183, p < 0.05), respectively. Kisspeptin (Kiss1) gene expression was suppressed fivefold in WT females and twofold in KIKO females (Figure 3C; ANOVA: F(1,28) = 27.63, p < 0.0001). Interestingly, Cart expression was suppressed by E2B in WT and KIKO females, but was increased by E2B in ERKO females (Figure 3D; ANOVA: F(2,42) = 7.719, p < 0.01). Surprisingly, we did not find a significant change in other E2-regulated arcuate genes including POMC and TH, which may be due to differences in treatment paradigms or rodent models [1, 42, 43].
Figure 3. E2B regulates neuropeptide gene expression in the ARC.
(A) Pdyn, (B) Tac2, (C) Kiss1, and (D) Cart. Results of qPCR analyses represent gene expression of oil- (black bars) and E2B- (gray bars) treated females in WT, KIKO, and ERKO genotypes. The number of animals in each treatment group is listed within each bar. Genes are expressed as relative n- fold changes, normalized to oil controls, within each genotype (WT, KIKO, ERKO). A two-way ANOVA (genotype × treatment) followed by post-hoc Bonferroni’s multiple comparison test was used to determine significant differences between treatments, within genotype. *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001.
ARC genes are differentially expressed in KIKO and ERKO ovx oil-treated females
Differences in gene expression across these genotypes may provide insight into signaling and feedback mechanisms involved in ERE-dependent, ERE-independent and ERα-independent signaling in the ARC. To determine if mRNA expression of the selected genes was different between the genotypes, we calculated the relative mRNA expression for each gene by normalizing the KIKO and ERKO oil groups to the WT oil group. Data were analyzed using a one-way ANOVA followed by post-hoc Bonferroni’s multiple comparison tests between WT vs. KIKO, WT vs. ERKO, and KIKO vs. ERKO females. See Table 5 for results.
Table 5.
Comparison of gene expression between genotypes in oil-treated females
| Gene List | WT | KIKO | ERKO |
|---|---|---|---|
| Abcc8 | 1.02 ± 0.10 | 0.32 ± 0.14 b | 0.21 ± 0.08 c |
| Adra1b | 1.03 ± 0.12 | 3.97 ± 0.64 b | 2.64 ± 0.29 |
| Bcl2 | 1.00 ± 0.03 | 0.24 ± 0.07 d | 0.02 ± 0.01 d D |
| Cacna1g | 1.01 ± 0.08 | 1.97 ± 0.15 b | 2.36 ± 0.24 c |
| Cacna1h | 1.01 ± 0.05 | 0.82 ± 0.21 | 0.09 ± 0.02 c B |
| Calm1 | 1.01 ± 0.07 | 1.07 ± 0.35 | 0.18 ± 0.05 A |
| Cart | 1.02 ± 0.10 | 0.19 ± 0.05 d | 0.77 ± 0.12 B |
| Chrm1 | 1.10 ± 0.31 | 0.34 ± 0.07 A | 0.34 ± 0.06 A |
| Esr1 | 1.01 ± 0.06 | 0.97 ± 0.09 | 0.44 ± 0.03 c C |
| Esr2 | 1.03 ± 0.13 | 1.80 ± 0.45 | 2.00 ± 0.32 |
| Gad1 | 1.01 ± 0.06 | 0.66 ± 0.16 | 0.16 ± 0.06 c A |
| Gad2 | 1.01 ± 0.05 | 0.15 ± 0.03 d | 0.09 ± 0.02 A |
| Ghsr | 1.01 ± 0.07 | 1.58 ± 0.42 | 1.11 ± 0.14 |
| Gpr30 | 1.03 ± 0.12 | 0.61 ± 0.21 | 0.12 ± 0.03 b |
| Htr2c | 1.01 ± 0.09 | 0.26 ± 0.09 d | 0.18 ± 0.07 d |
| Kcnj11 | 1.01 ± 0.07 | 0.83 ± 0.15 | 0.13 ± 0.06 c C |
| Kcnmb1 | 1.16 ± 0.25 | 6.87 ± 1.45 | 14.04 ± 1.91 d A |
| Kcnmb4 | 1.03 ± 0.11 | 0.50 ± 0.20 | 0.13 ± 0.06 b |
| Kcnq5 | 1.00 ± 0.04 | 0.85 ± 0.33 | 0.20 ± 0.06 |
| Kiss1 | 0.92 ± 0.27 | 0.02 ± 0.01 b | 0.17 ± 0.05 A |
| Kiss1r | 1.48 ± 0.53 | 0.45 ± 0.11 | 6.84 ± 1.66 b C |
| Mtor | 1.00 ± 0.04 | 0.54 ± 0.18 | 0.23 ± 0.06 b |
| Ncoa | 1.00 ± 0.05 | 0.67 ± 0.16 | 0.18 ± 0.04 c A |
| Npy | 1.13 ± 0.25 | 0.87 ± 0.49 | 0.45 ± 0.23 |
| Pdyn | 1.04 ± 0.15 | 2.39 ± 0.23 c | 1.50 ± 0.10 B |
| Pgr | 1.00 ± 0.04 | 3.42 ± 0.35 c | 2.59 ± 0.38 A |
| Pik3r3 | 1.00 ± 0.03 | 1.01 ± 0.28 | 0.24 ± 0.09 a A |
| Pomc | 1.01 ± 0.08 | 0.44 ± 0.17 A | 0.17 ± 0.07 c |
| Sirt1 | 1.00 ± 0.04 | 0.59 ± 0.15 A | 0.18 ± 0.02 c A |
| Tac2 | 1.08 ± 0.18 | 0.01 ± 0.01 d | 0.13 ± 0.05 c |
| Tac3r | 1.38 ± 0.83 | 0.05 ± 0.02 | 0.12 ± 0.03 |
| Th | 1.04 ± 0.13 | 0.23 ± 0.05 d | 0.05 ± 0.01 d |
Data were normalized to WT oil and analyzed by a one-way ANOVA followed by post-hoc Bonferroni’s multiple comparison test, within gene. Lowercase letters denote changes observed between WT and KIKO or ERKO
=p < 0.05;
=p < 0.01;
=p < 0.001;
=p < 0.0001
Uppercase letters sign denotes changes observed between KIKO and ERKO females
=p < 0.05;
=p < 0.01;
=p < 0.001;
=p < 0.0001
The mRNA levels of multiple genes were lower in KIKO and ERKO oil-treated females as compared to WT females. Interestingly, Cart expression was lower in KIKO females but not ERKO females compared to WT females. Cart expression in KIKO females was lower than in ERKO females (ANOVA: F(2, 14) = 19.43, p < 0.0001). For eight genes, mRNA expression in KIKO and ERKO females was lower compared to WT females, with no difference observed between KIKO and ERKO females. These genes include: Abcc8 (the regulatory subunit for KATP channels; ANOVA: F(2,14) = 15.63, p < 0.001), Chrm1 (ANOVA: F(2,14) = 5.657, p < 0.05), Gad2 (glutamate decarboxylase 2; ANOVA: F(2,14) = 181.8, p < 0.0001), Htr2c (the 5HT2c serotonin receptor, ANOVA: F(2,14) = 30.72, p < 0.0001), Kiss1 (ANOVA: F(2,14) = 10.43, p < 0.01), Pomc (ANOVA: F(2,14) = 12.48, p < 0.001), Tac2 (ANOVA: F(2,14) = 26.99, p < 0.0001), and Th (ANOVA: F(2,14) = 47.44, p < 0.0001). For another two genes, mRNA expression was lower in KIKO and ERKO females compared to WT females, with mRNA expression also lower in ERKO females than in KIKO females. These genes were Bcl2 (the anti-apoptotic gene B cell leukemia/lymphoma 2; ANOVA: F(2, 14) = 124.8, p < 0.0001), and Sirt1 (Sirtuin 1 or NAD-dependent deactylase; ANOVA: F(2, 14) = 16.58, p < 0.001).
The mRNA expression of three other genes was lower in ERKO females, but not KIKO females, compared to WT females. These genes include Gpr30 (ANOVA: F(2, 14) = 10.05, p < 0.01), Kcnmb4 (Ca2+-activated potassium channel subunit β4; ANOVA: (2, 14) = 10.04, p 0.01), and Mtor (mammalian target of rapamycin; ANOVA: F(2, 14) = 10.19, p < 0.01). Additionally, six genes were expressed at lower levels in ERKO females compared to both WT and KIKO females. These genes were Cacna1h (a subunit of the T-type Ca2+ channels; ANOVA: F(2, 14) = 10.92, p < 0.001), Esr1 (ANOVA: F(2,14) = 22.96, p < 0.0001), Gad1 (glutamate decarboxylase 1; ANOVA: F(2,14) = 15.48, p < 0.001), Kcnj11 (Kir6.2, the channel subunit for KATP channels; ANOVA: F(2,14) = 20.18, p < 0.0001), Ncoa (a coactivator of transcription; ANOVA: F(2,14) = 15.17; p < 0.001), and Pik3r3 (PI3K p55γ subunit; ANOVA: F(2,14) = 6.116, p < 0.05). Interestingly, expression of calmodulin (Calm1) was lower in ERKO females compared to KIKO females, but not WT females (ANOVA: F(2,14) = 5.163, p < 0.05).
We observed other gene-expression differences between genotypes. Adra1b expression was higher in KIKO females compared to WT females (ANOVA: F(2,14) = 10.82, p < 0.01). Additionally, expression of Cacna1g (ANOVA: F(2,14) = 14.64, p < 0.001) and Pgr (ANOVA: F(2,14) = 14.04, p < 0.001) was higher in both KIKO and ERKO females compared to WT females. Expression of Kiss1r (ANOVA: F(2,12) = 16.29, p < 0.001) and Kcnmb1 (ANOVA: F(2,14) = 18.41, p < 0.001) was higher in ERKO females compared to both WT and KIKO females. Interestingly, expression of Pdyn was higher in KIKO females compared to WT females and lower in ERKO compared to KIKO females (ANOVA: F(2,14) = 16.41, p < 0.001). Finally, there was no change observed in Esr2, Ghsr, Npy, Tac3r, and Kcnq5 (a subunit of the potassium channel that produces the M-current) in the ARC across oil-treated females in WT, KIKO, and ERKO genotypes.
Discussion
Previous studies have demonstrated that E2 regulates ARC gene expression to control homeostatic functions through ERα-mediated mechanisms. In the present study, to distinguish between ERE-dependent and ERE-independent transcriptional mechanisms, we compared gene expression in WT, ERα KO and an ERα KIKO mouse models. Previously, this KIKO model, which lacks ERα-mediated ERE-dependent signaling, was has been used to delineate such ERα-mediated signaling in the uterus [44] and the HPG axis [34, 45]. Studies on osteoblasts suggested that E2 regulation of gene transcription occurs through both ERE-dependent and ERE-independent mechanisms (including non-genomic mechanisms) [46]. The classical ERα signaling pathway is genomic ERE-dependent gene transcription in which E2 binds to ERα in the nucleus and then ERα binds to ERE to regulate expression of multiple genes. In addition to the classical ERα regulation of gene expression, there are ERE-independent mechanisms that include ERα/β-mediated, non-genomic second messenger pathways and ERα/β-independent signaling through membrane estrogen receptors [7, 8]. ERE-independent signaling includes PI3K and mitogen-activated protein kinase second messenger signaling cascades from membrane-associated ERα/β, protein-protein interactions with other transcription factors and ligand-independent mechanisms [2].
Recent evidence suggests that E2 signals through multiple membrane ERs including GPER (GPR30) [1, 3]. However, there are few studies that determine how E2 signals through ERE-dependent and ERE-independent mechanisms to regulate gene expression in the hypothalamus. This present study compared gene expression in WT, KIKO, and ERKO mouse models to characterize mechanisms of E2 regulation of ARC gene expression. We determined that regulation of ARC gene expression by E2B, with E2 being the active hormone, occurs through both ERE-dependent and ERE-independent mechanisms. The genes regulated by ERα-mediated, ERE-dependent and ERE-independent mechanisms in this study include cation channels, receptors, and neuropeptides associated with reproduction, energy balance, stress, and other homeostatic functions. However, it is necessary to note that our ERE-independent KIKO mouse model is nonselective to other hormone response element (HRE) motifs and may bind to other HREs to regulate transcription [47]. The development of an “EAAE” ERα mouse that lacks ERE- and HRE-dependent signaling would be useful in future studies to distinguish between those two types of signaling [47].
In the current study, E2 (or treatment with E2B) increased channel expression of Cacna1g and Kcnmb1 in WT females only, a finding supported by previous studies, but not in KIKO or ERKO [16, 17, 19]. Voltage-dependent T-type calcium channel subunit alpha-1G (Cacna1g, also known as Cav3.1) is one subunit of low voltage-activated (T-type) calcium channels that is important in burst firing and neurotransmitter release. The elevated expression of Cacna1g expression in KIKO and ERKO females suggests that ERα-mediated, ERE-mediated signaling independent of ligand differentially regulates Cacna1g expression. Past studies have suggested that E2 regulates expression of the Cav3 subunits [13, 15]. For example, in GnRH neurons of the preoptic area (POA), a high E2B dose increased Cacna1g expression transiently [13]. Increased expression of Cacna1g in the ARC led to increased neuronal excitability and burst firing to regulate hypothalamic neurons involved in reproduction and energy homeostasis [13, 15].
E2 has been found to regulate mRNA expression of a range of potassium channels in the guinea pig [16]. In the current study, we found that expression of Kcnmb1, a calcium-activated potassium channel (a MaxiK channel), was increased by E2B treatment in WT females only. Collectively, these studies emphasize the role of E2 regulation of channel expression in the ARC, which may translate to regulation of electrophysiological properties of ARC neurons [48]. Future studies in our lab will investigate the neurophysiological effects of the E2-induced gene expression. Furthermore, the results of our study indicate that in the mouse ARC, Kcnmb1 MaxiK channel expression is regulated through ERE-dependent mechanisms. Elevated expression of Kcnmb1 in oil-treated ERKO females also indicates that the loss of ERα positively affects ARC Kcnmb1 gene expression.
In addition to the above findings related to channel expression regulation, the results of this study indicate that, through multiple ER-mediated mechanisms, E2B regulates several hormone and neurotransmitter receptors in the ARC. These receptors are involved in a number of functions including reproduction, neuronal excitability, and energy balance. Collectively, our data suggest that E2B regulates the expression of several receptors involved in feeding, stress, and neurotransmission through multiple receptor-mediated mechanisms. Chrm1 is a muscarinic acetylcholine receptor involved in a number of functions including memory consolidation, neuronal excitability and signal transduction [49]. Only a few studies to date have examined Chrm1 expression in the ARC. In the present study, E2B suppressed Chrm1 expression in the ARC through ERE-dependent signaling. Past studies in the rat hippocampus also suggest that Chrm1 is decreased in response to immediate E2 replacement following ovx [50]. The current study used a different E2 replacement paradigm, yet it would be informative to examine Chrm1 expression at different time points after E2B administration in the ARC to determine the time-dependent regulation of Chrm1 expression. We also found suppressed Chrm1 expression in KIKO and ERKO females compared to WT. This difference may result in a decrease in muscarinic signaling in the ARC of KIKO and ERKO females, which can be examined in future experiments using electrophysiology to assess muscarinic activity in ARC neurons. Furthermore, the role of muscarinic receptors in the control of homeostatic functions in the ARC has not been previously investigated and the effects of the suppression by E2 in these functions are unknown.
Expression of another neurotransmitter receptor, Adra1b, was suppressed by E2B treatment in WT, KIKO and ERKO females. Alpha-1B adrenergic receptor (Adra1b) is a receptor for catecholamines (norepinephrine/epinephrine) involved in arousal, feeding behaviors, cell growth, and proliferation. Previous studies in rats report that E2 increases Adra1b expression in the hypothalamus and POA [51-53]. E2 was administered via subcutaneous injection 24- and 48-hours prior to sacrifice, similar to the present study. The difference between those studies and the current study indicate the differential effects of E2 on adrenergic signaling between rodent species and between discrete hypothalamic nuclei. Furthermore, previous reports suggest that E2 inhibits catecholamine secretion in vitro [54, 55]. It should be noted that Adra1b suppression occurs in ERKO animals, suggesting that this is an ERα-independent mechanism. A decrease of Adra1b expression in the ARC would suppress, in part, the actions of noradrenergic/adrenergic signals from the hindbrain that control hypothalamic functions including arousal, feeding behavior, and energy expenditure [56, 57]. Genotype-specific analysis indicates that Adra1b expression and activity is different between KIKO and WT females, which may be a mechanism behind the differences in feeding behavior between the genotypes [12]. Future studies should examine if the difference in expression correlates with changes in noradrenergic signaling in ARC neurons, specifically NPY neurons.
The growth hormone secretagogue receptor, GHSR, is a GPCR involved in ghrelin signaling. E2B treatment has been shown to increase Ghsr expression in the ARC in mice [30]. We determined that the increase in ARC Ghsr expression by E2, which is regulated by ERE-dependent mechanisms, as E2B-treatment did not affect Ghsr expression in KIKO or ERKO females. Ghrelin signaling in the hypothalamus illuminates the relationship of E2 on feeding behavior control by ARC neurons. Ghsr activation by ghrelin excites NPY neurons [58] and initiates a signaling cascade that increases transcription of NPY and AgRP [59]. Activation of NPY neurons is associated with an increase in food intake, which is opposed by the actions of E2 [60]. However, Ghsr is expressed throughout the heterogeneous population of ARC neurons including POMC, TH, and KNDy neurons [30, 61, 62], and the increase in ARC Ghsr expression by E2 is most likely involved in other homeostatic functions of these neurons. Cell-type specific analysis will be necessary to address this contradictory information of E2’s regulation of GHSR signaling in the ARC.
Our results also indicate that steroid receptors associated with reproduction are regulated by E2 through both ERE-dependent and ERE-independent mechanisms. The classical role of E2 in the control of reproduction is through feedback mechanisms of the HPG axis and regulation of steroid receptor expression. The results of this study agree with past studies on E2 regulation of the estrogen receptor, ERα (Esr1). In the hypothalamus, E2 has been shown to decrease expression of Esr1 [63]. This is supported by our study, which suggests that the E2-mediated Esr1 suppression occurs through ERE-dependent mechanisms. There are no previous studies that identify the mechanism for this E2-mediated Esr1 decrease. Furthermore, E2B treatment augmented the expression of ERβ (Esr2) in WT and KIKO females. These data indicate that, unlike with ERα, ERβ is regulated in the ARC by ERE-independent, ERα-mediated signaling. GPER/GPR30, a membrane estrogen receptor, was not regulated by E2 in any genotype. ERα (Esr1) expression was lower in ERKO compared to WT and KIKO females. GPER (Gpr30) expression was lower in ERKO compared to WT females. Lastly, there was no difference in expression of ERβ (Esr2) between all genotypes.
In addition, E2B treatment increased expression of Pgr in WT females only. These results are consistent with multiple studies that suggest E2B increases Pgr in the hypothalamus, priming the brain for progesterone’s reproductive and behavioral actions [64, 65], but also suggest that Pgr is regulated through ERE-dependent, ERα-mediated signaling pathway. Pgr expression is two- to threefold higher in both KIKO and ERKO females compared to WT females. Interestingly, the relative expression of Pgr in oil-treated KIKO and ERKO females is equal to the relative levels of Pgr in E2B-treated, WT females. Higher expression of Pgr in the ARC may play a role in the lack of normal estrous cycles in KIKO and ERKO females.
In the present study, E2B treatment regulated multiple neuropeptides that are coexpressed in KNDy neurons of the ARC and their receptors. Kisspeptin-expressing neurons are expressed in two main regions in the rodent hypothalamus [32]. The first region is the anteroventral periventricular (AVPV) nucleus, which is referred to as the surge center for its role in the LH surge in female rodents [32]. The second region is the ARC, which contains kisspeptin neurons that coexpress Kisspeptin (Kiss1), Neurokinin B (Tac2) and Dynorphin (Dyn, Pdyn) [66]. These KNDy neurons are known for their contribution to negative feedback of E2 on the HPG axis and are hypothesized to be the pulse generator for the secretion of GnRH into the median eminence [67, 68]. Additional studies suggest that kisspeptin neurons directly contact GnRH neurons in the median eminence to control GnRH excitability and pulsatility [68]. Since then, KNDy neurons have also been shown to integrate feeding signals through input from peptides such as ghrelin, leptin, and insulin [69, 70]. Our results corroborate previous data demonstrating that Kiss1 regulation by E2 is nonclassically mediated [34]. Kiss1 expression also was lower in both KIKO and ERKO females than WT females. Because ARC Kiss1 is involved in negative feedback, perhaps the lower expression indicates that negative feedback is disrupted in these genotypes, in part, due to lower expression of Kiss1 [67, 68]..
Amongst the other two KNDy neuropeptides, Neurokinin B (Tac2) expression is suppressed by E2 in WT females only. Our data suggest that Tac2 expression is primarily controlled by ERE-dependent signaling much like Pdyn [29, 41]. As with Kiss1, Tac2 expression is lower in both KIKO and ERKO females. The suppressed expression of Tac2 in these genotypes also supports the hypothesis that lower expression of both Kiss1 and Tac2 play a role in the disruption of negative feedback in these genotypes. Because neurokinin B (NKB, Tac2) is involved in GnRH pulse generation, the lower expression in KIKO and ERKO contributes to the dysregulation of the GnRH actions on the gonadotropes in these genotypes [67,68]. Finally, Pdyn expression, the other KNDy neuropeptide, is suppressed by E2B through ERE-dependent transcription. These data support previous studies and was used in this study as a “negative” control for ERE-independent signaling [34]. Interestingly, Pdyn expression was higher in KIKO females compared to WT females unlike Kiss1 and Tac2 expression. Elevated Pdyn may further disrupt the pulse generator in this genotype as this neuropeptide is considered a negative regulator of the pulse generator [67,68]. Because Kiss1 is the only KNDy gene that is regulated by ERa ERE-independent mechanisms, the KNDy pulse generator (NKB, Dyn) for GnRH secretion is primarily controlled by ERE-dependent mechanisms.
KNDy-associated receptors are also regulated by E2. Tac3r, the NKB receptor, is suppressed in WT and KIKO females by E2B treatment, suggesting that, like Kiss1, Tac3r suppression is through an ERα-mediated, ERE-independent pathway [71]. These results suggest that both ERE-dependent and ERE-independent mechanisms are involved in the control of KNDy neuropeptides and their receptors and are necessary to maintain a functional HPG axis. In fact, current evidence suggests that mutations in Tac3r lead to problems in reproductive development including hypogonadotrophic hypogonadism, similar to mutations in Kiss1r [72, 73]. The decrease in Tac3r and Tac2 associated with E2B treatment may both be involved in KNDy-mediated control of negative feedback and the GnRH pulse generator.
Kiss1r, the receptor for Kiss1, is increased by E2B treatment in all three genotypes. This upregulation suggests that E2 regulation of Kiss1r expression is ERα-independent and potentially mediated by ERβ, GPER, or perhaps the putative STX-responsive, Gq-coupled mER (Gq-mER) [1, 16, 74]. Alternatively, the increase in Kiss1r expression may be due, in part, to a decrease in the desensitization of the receptor, as Kiss1 peptide and gene expression is highly reduced by E2 in the ARC [25, 32, 75]. Future studies should examine E2 regulation of Kiss1r peptide expression selectively in the median eminence, where KNDy neurons contact GnRH neurons expressing the kisspeptin receptor [19, 66]. Kiss1r expression was higher in ERKO females compared to their WT and KIKO females, which correlated with a decrease in the ligand expression in the ERKO. However, the effect of E2 across the genotypes was similar, suggesting that the E2-mediated increase is robust and not dependent on relative baseline expression levels.
Lastly, Cart gene expression, which produces a neuropeptide involved in reward, stress, and feeding behavior [76, 77], is differentially regulated by E2 in the three genotypes. CART activity has been shown to suppress feeding much like E2 in rodents and primates [76, 77]. However, in this experiment, E2B treatment suppressed Cart in WT and KIKO females. These results are supported by previous studies that have reported the suppression of Cart by E2 in the rodent ARC [38] and suggest that the suppression of the Cart gene in the ARC is a compensatory homeostatic response for the increase in CART protein activity in the paraventricular nucleus to suppress feeding [78]. Interestingly, E2B augmented Cart expression in ERKO females. The mechanism behind this effect of E2B is unknown, but certainly involves the other ER in the ARC. To date, there have been no studies that examine which ER-mediated mechanism regulates Cart expression or their contribution to the physiological effects of CART in the hypothalamus.
There are a number of additional receptors that can be involved in ERα-independent, E2 signaling and may be involved in the control of Kiss1r, Adra1b, and Cart. In addition to ERα, the classical receptor ERβ can also regulate E2-responsive genes through ERE-dependent and ERE-independent signaling. Although ERα is more highly expressed in the ARC compared to ERβ, studies suggest that ERβ signaling is still present [27] and may function as a compensatory mechanism for gene regulation by E2 in the ERKO since Esr2 is expressed in ARC during development [79]. In addition, non-nuclear ER-mediated E2 signaling can also regulate gene expression through G-protein coupled membrane estrogen receptors (GPER) and the putative Gq-mER [80]. Evidence suggests that GPER is activated by E2 and is important for rapid cell signaling and homeostatic functions, although is it not regulated by E2 in the ARC [81, 82]. Gq-mER activates PLC-PKC signal transduction pathway and functions in energy homeostasis, bone remodeling, and core body temperature including activation of hypothalamic neurons [7] as well as control of ARC gene expression [1].
We found that there were a number of genes that were not regulated by E2B but did show differences in expression amongst genotypes. These genes include Abcc8, Bcl2, Cacna1h, Calm1, Gad1, Gad2, Gpr30, Htr2c, Kcnj11, Kcnmb4, Mtor, Ncoa1, Pik3r3, Pomc, Sirt1, and Th. The range of expression differences amongst the genotypes differs for each gene. While there is no easily discernable pattern of expression differences, these changes may result in deleterious effects of ERα loss (knockout) on reproductive and energy homeostasis. Furthermore, it is important to note that the present study only examined differences in gene expression among oil-treated genotypes, which may not produce functional differences in ARC neurons or in the protein of these enzymes, signaling molecules, receptors, and cation channels. Future experiments will be needed to confirm these potential effects and if these differences are due to a developmental role of ERα signaling, both ERE-dependent and ERE-independent, in ARC gene expression. Nonetheless, comparison of gene expression across genotypes is an important tool for enhancing our understanding of ERα’s role in ARC gene expression and homeostatic functions.
While past studies have examined regulation of gene expression by E2 in the ARC, few studies to date have used the ERα KIKO transgenic mouse model as a tool to identify molecular mechanisms of E2 regulation. The results of our study indicate that genes involved in hypothalamic functions such as reproduction, energy homeostasis, and neuronal excitability are regulated by E2 through multiple ERα-mediated and ERα-independent pathways as has been previously suggested [1]. It is important to note that gene expression in our study is a measurement of the steady-state mRNA expression. Unfortunately, due to the small size of the ARC nucleus in mice, it is not possible to conduct immunoblotting of protein expression within a single animal. Because mRNA and protein levels seldom correlate when comparing gene expression and immunohistochemistry [25], future studies will examine the expression and activity of many of these genes, especially the cation channels and GPCRs, using electrophysiology. These experiments would further characterize the role of ERα-mediated, ERE-dependent and -independent signaling on ARC gene expression and neuronal functions. Determining these signaling pathways is key to understanding the physiological effects of estrogens during the reproductive cycle and in hormone replacement therapies.
Highlights.
WT, ERKO, and KIKO females were used to study ERE-dependent and ERE-independent signaling.
Gene regulation by E2B occurs through ERE-dependent and ERE-independent mechanisms.
Gene expression varies greatly in oil-treated females between the genotypes.
Channels, receptors, and neurotransmitters are regulated in the ARC.
Acknowledgements
The authors wish to thank Dr. Michael Pierce and the Rutgers SEBS Core Facility for the use of the StepOne Plus™ Real-Time PCR System. The University of Virginia Center for Research in Reproduction Ligand Assay and Analysis Core is supported by the Eunice Kennedy Shriver NICHD/NIH (SCCPIR) Grant U54-HD298934. This research is supported by National Institutes of Health Grant R00DK083457 (T.A.R.) and NJ06107 (USDA-NIFA) (T.A.R.).
Abbreviations
- Abcc8
ATP-binding cassette, subfamily C
- Actb
beta actin
- Adra1b
alpha-1B adrenergic receptor
- AgRP
agouti-related protein
- ARC
arcuate nucleus
- AVPV
anteroventral periventricular nucleus
- Bcl2
B cell leukemia/lymphoma 2
- BH
basal hypothalamus
- Cacna1g
T-type, voltage-dependent, calcium channel alpha 1G subunit
- Cart
cocaine- and amphetamine-regulated transcript
- Chrm1
cholinergic muscarinic 1 receptor
- E2
17β-estradiol
- E2B
17β-estradiol benzoate
- ER
estrogen receptor
- ERE
estrogen response element
- ERKO
ERα knock-out
- ERα
estrogen receptor alpha
- ERβ
estrogen receptor beta
- Esr1
estrogen receptor alpha
- Esr2
estrogen receptor beta
- Gapdh
glyceraldehyde-3-phosphate dehydrogenase
- Ghsr
growth hormone secretagogue receptor
- GnRH
gonadotropin releasing hormone
- GPCR
g-protein coupled receptor
- GPER
G protein-coupled estrogen receptor 1
- HPG
hypothalamic-pituitary-gonadal
- HRE
hormone response element
- Kcnmb1
calcium-activated potassium channel subunit β1
- KIKO
knock-in/knock-out
- Kiss1
kisspeptin
- Kiss1r
kisspeptin receptor
- KNDy
Kisspeptin-Neurokinin B-Dynorphin
- Mtor
mammalian target of rapamycin
- NPY
neuropeptide Y
- ovx
ovariectomized
- Pdyn
prodynorphin
- Pgr
progesterone receptor
- POMC
proopiomelanocortin
- qPCR
quantitative real-time polymerase chain reaction
- Tac2
tachykinin 2
- Tac3r
tachykinin 3 receptor
- TH
tyrosine hydroxylase
- TLDA
Taqman Low Density Array
- WT
wild-type
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosure statement: The authors have no conflicts of interest to disclose.
References
- [1].Roepke TA, Xue C, Bosch MA, Scanlan TS, Kelly MJ, Ronnekleiv OK. Genes associated with membrane-initiated signaling of estrogen and energy homeostasis. Endocrinology. 2008;149:6113–24. doi: 10.1210/en.2008-0769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].McDevitt MA, Glidewell-Kenney C, Jimenez MA, Ahearn PC, Weiss J, Jameson JL, et al. New insights into the classical and non-classical actions of estrogen: Evidence from estrogen receptor knock-out and knock-in mice. Mol Cell Endocrinol. 2008;290:24–30. doi: 10.1016/j.mce.2008.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Roepke TA, Bosch MA, Rick EA, Lee B, Wagner EJ, Seidlova-Wuttke D, et al. Contribution of a membrane estrogen receptor to the estrogenic regulation of body temperature and energy homeostasis. Endocrinology. 2010;151:4926–37. doi: 10.1210/en.2010-0573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Santollo J, Torregrossa AM, Eckel LA. Estradiol acts in the medial preoptic area, arcuate nucleus, and dorsal raphe nucleus to reduce food intake in ovariectomized rats. Horm Behav. 2011;60:86–93. doi: 10.1016/j.yhbeh.2011.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Shughrue PJ, Lane MV, Merchenthaler I. Comparative distribution of estrogen receptor-alpha and -beta mRNA in the rat central nervous system. J Comp Neurol. 1997;388:507–25. doi: 10.1002/(sici)1096-9861(19971201)388:4<507::aid-cne1>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- [6].Mitra SW, Hoskin E, Yudkovitz J, Pear L, Wilkinson HA, Hayashi S, et al. Immunolocalization of estrogen receptor beta in the mouse brain: Comparison with estrogen receptor alpha. Endocrinology. 2003;144:2055–67. doi: 10.1210/en.2002-221069. [DOI] [PubMed] [Google Scholar]
- [7].Qiu J, Bosch MA, Tobias SC, Grandy DK, Scanlan TS, Ronnekleiv OK, et al. Rapid signaling of estrogen in hypothalamic neurons involves a novel G-protein-coupled estrogen receptor that activates protein kinase C. J Neurosci. 2003;23:9529–40. doi: 10.1523/JNEUROSCI.23-29-09529.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Qiu J, Bosch MA, Tobias SC, Krust A, Graham SM, Murphy SJ, et al. A G-protein-coupled estrogen receptor is involved in hypothalamic control of energy homeostasis. J Neurosci. 2006;26:5649–55. doi: 10.1523/JNEUROSCI.0327-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Hewitt SC, O'Brien JE, Jameson JL, Kissling GE, Korach KS. Selective disruption of ERα DNA-binding activity alters uterine responsiveness to estradiol. Mol Endocrinol. 2009;23:2111–6. doi: 10.1210/me.2009-0356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Jakacka M, Ito M, Martinson F, Ishikawa T, Lee EJ, Jameson JL. An estrogen receptor ERα deoxyribonucleic acid-binding domain knock-in mutation provides evidence for nonclassical ER pathway signaling in vivo. Mol Endocrinol. 2002;16:2188–201. doi: 10.1210/me.2001-0174. [DOI] [PubMed] [Google Scholar]
- [11].Park CJ, Zhao Z, Glidewell-Kenney C, Lazic M, Chambon P, Krust A, et al. Genetic rescue of nonclassical ERα signaling normalizes energy balance in obese ERα-null mutant mice. J Clin Invest. 2011;121:604–12. doi: 10.1172/JCI41702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Mamounis KJ, Yang JA, Yasrebi A, Roepke TA. Estrogen response element-independent signaling partially restores post-ovariectomy body weight gain but is not sufficient for 17β-estradiol's control of energy homeostasis. Steroids. 2013;81:88–98. doi: 10.1016/j.steroids.2013.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Zhang C, Bosch MA, Rick EA, Kelly MJ, Ronnekleiv OK. 17β-estradiol regulation of T-type calcium channels in gonadotropin-releasing hormone neurons. J Neurosci. 2009;29:10552–62. doi: 10.1523/JNEUROSCI.2962-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Bosch MA, Hou J, Fang Y, Kelly MJ, Ronnekleiv OK. 17β-estradiol regulation of the mRNA expression of T-type calcium channel subunits: Role of estrogen receptor alpha and estrogen receptor beta. J Comp Neurol. 2009;512:347–58. doi: 10.1002/cne.21901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Qiu J, Bosch MA, Jamali K, Xue C, Kelly MJ, Ronnekleiv OK. Estrogen upregulates T-type calcium channels in the hypothalamus and pituitary. J Neurosci. 2006;26:11072–82. doi: 10.1523/JNEUROSCI.3229-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Roepke TA, Malyala A, Bosch MA, Kelly MJ, Ronnekleiv OK. Estrogen regulation of genes important for K+ channel signaling in the arcuate nucleus. Endocrinology. 2007;148:4937–51. doi: 10.1210/en.2007-0605. [DOI] [PubMed] [Google Scholar]
- [17].Roepke TA, Qiu J, Smith AW, Ronnekleiv OK, Kelly MJ. Fasting and 17β-estradiol differentially modulate the M-current in neuropeptide Y neurons. J Neurosci. 2011;31:11825–35. doi: 10.1523/JNEUROSCI.1395-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Bosch MA, Kelly MJ, Ronnekleiv OK. Distribution, neuronal colocalization, and 17β-E2 modulation of small conductance calcium-activated K+ channel (SK3) mRNA in the guinea pig brain. Endocrinology. 2002;143:1097–107. doi: 10.1210/endo.143.3.8708. [DOI] [PubMed] [Google Scholar]
- [19].Bosch MA, Tonsfeldt KJ, Ronnekleiv OK. mRNA expression of ion channels in GnRH neurons: Subtype-specific regulation by 17β-estradiol. Mol Cell Endocrinol. 2013;367:85–97. doi: 10.1016/j.mce.2012.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Ibrahim N, Bosch MA, Smart JL, Qiu J, Rubinstein M, Ronnekleiv OK, et al. Hypothalamic proopiomelanocortin neurons are glucose responsive and express K(ATP) channels. Endocrinology. 2003;144:1331–40. doi: 10.1210/en.2002-221033. [DOI] [PubMed] [Google Scholar]
- [21].Malyala A, Kelly MJ, Ronnekleiv OK. Estrogen modulation of hypothalamic neurons: Activation of multiple signaling pathways and gene expression changes. Steroids. 2005;70:397–406. doi: 10.1016/j.steroids.2005.03.004. [DOI] [PubMed] [Google Scholar]
- [22].Malyala A, Pattee P, Nagalla SR, Kelly MJ, Ronnekleiv OK. Suppression subtractive hybridization and microarray identification of estrogen-regulated hypothalamic genes. Neurochem Res. 2004;29:1189–200. doi: 10.1023/b:nere.0000023606.13670.1d. [DOI] [PubMed] [Google Scholar]
- [23].Malyala A, Zhang C, Bryant DN, Kelly MJ, Ronnekleiv OK. PI3K signaling effects in hypothalamic neurons mediated by estrogen. J Comp Neurol. 2008;506:895–911. doi: 10.1002/cne.21584. [DOI] [PubMed] [Google Scholar]
- [24].Curran-Rauhut MA, Petersen SL. Regulation of glutamic acid decarboxylase 65 and 67 gene expression by ovarian steroids: Identification of two functionally distinct populations of GABA neurones in the preoptic area. J Neuroendocrinol. 2002;14:310–7. doi: 10.1046/j.1365-2826.2002.00780.x. [DOI] [PubMed] [Google Scholar]
- [25].Bosch MA, Xue C, Ronnekleiv OK. Kisspeptin expression in guinea pig hypothalamus: Effects of 17β-estradiol. J Comp Neurol. 2012;520:2143–62. doi: 10.1002/cne.23032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Jamali K, Naylor BR, Kelly MJ, Ronnekleiv OK. Effect of 17β-estradiol on mRNA expression of large-conductance, voltage-dependent, and calcium-activated potassium channel alpha and beta subunits in guinea pig. Endocrine. 2003;20:227–37. doi: 10.1385/ENDO:20:3:227. [DOI] [PubMed] [Google Scholar]
- [27].Roepke TA. Oestrogen modulates hypothalamic control of energy homeostasis through multiple mechanisms. J Neuroendocrinol. 2009;21:141–50. doi: 10.1111/j.1365-2826.2008.01814.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Addison ML, Rissman EF. Sexual dimorphism of growth hormone in the hypothalamus: Regulation by estradiol. Endocrinology. 2012;153:1898–907. doi: 10.1210/en.2011-1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Zuloaga DG, Yahn SL, Pang Y, Quihuis AM, Oyola MG, Reyna A, et al. Distribution and estrogen regulation of membrane progesterone receptor-beta in the female rat brain. Endocrinology. 2012;153:4432–43. doi: 10.1210/en.2012-1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Frazao R, Lemko HM, da Silva RP, Ratra DV, Lee CE, Williams KW, et al. Estradiol modulates Kiss1 neuronal response to ghrelin. Am J Physiol Endocrinol Metab. 2014;306:E606–14. doi: 10.1152/ajpendo.00211.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Mirkes SJ, Bethea CL. Oestrogen, progesterone and serotonin converge on GABAergic neurones in the monkey hypothalamus. J Neuroendocrinol. 2001;13:182–92. doi: 10.1046/j.1365-2826.2001.00612.x. [DOI] [PubMed] [Google Scholar]
- [32].Brock O, Bakker J. The two kisspeptin neuronal populations are differentially organized and activated by estradiol in mice. Endocrinology. 2013;154:2739–49. doi: 10.1210/en.2013-1120. [DOI] [PubMed] [Google Scholar]
- [33].Mittelman-Smith MA, Williams H, Krajewski-Hall SJ, McMullen NT, Rance NE. Role for kisspeptin/neurokinin B/dynorphin (KNDy) neurons in cutaneous vasodilatation and the estrogen modulation of body temperature. Proc Natl Acad Sci USA. 2012;109:19846–51. doi: 10.1073/pnas.1211517109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Gottsch ML, Navarro VM, Zhao Z, Glidewell-Kenney C, Weiss J, Jameson JL, et al. Regulation of Kiss1 and dynorphin gene expression in the murine brain by classical and nonclassical estrogen receptor pathways. J Neurosci. 2009;29:9390–5. doi: 10.1523/JNEUROSCI.0763-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Franklin KBJ, Paxinos G. The mouse brain in stereotaxic coordinates. Third. Academic Press; San Diego: 2008. 3rd. [Google Scholar]
- [36].Haisenleder DJ, Schoenfelder AH, Marcinko ES, Geddis LM, Marshall JC. Estimation of estradiol in mouse serum samples: Evaluation of commercial estradiol immunoassays. Endocrinology. 2011;152:4443–7. doi: 10.1210/en.2011-1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Nedungadi TP, Briski KP. Effects of estradiol on acute and recurrent insulin-induced hypoglycemia-associated patterns of arcuate neuropeptide Y, proopiomelanocortin, and cocaine- and amphetamine-related transcript gene expression in the ovariectomized rat. Neuroendocrinology. 2007;86:270–6. doi: 10.1159/000109678. [DOI] [PubMed] [Google Scholar]
- [38].Silva LE, Castro M, Amaral FC, Antunes-Rodrigues J, Elias LL. Estradiol-induced hypophagia is associated with the differential mRNA expression of hypothalamic neuropeptides. Braz J Med Biol Res. 2010;43:759–66. doi: 10.1590/s0100-879x2010007500059. [DOI] [PubMed] [Google Scholar]
- [39].Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt method. Methods. 2001;25:402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- [40].Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Kurita T, Lee K, Saunders PT, Cooke PS, Taylor JA, Lubahn DB, et al. Regulation of progesterone receptors and decidualization in uterine stroma of the estrogen receptor-alpha knockout mouse. Biol Reprod. 2001;64:272–83. doi: 10.1095/biolreprod64.1.272. [DOI] [PubMed] [Google Scholar]
- [42].Priest CA, Roberts JL. Estrogen and tamoxifen differentially regulate beta-endorphin and cFos expression and neuronal colocalization in the arcuate nucleus of the rat. Neuroendocrinology. 2000;72:293–305. doi: 10.1159/000054598. [DOI] [PubMed] [Google Scholar]
- [43].Blum M, McEwen BS, Roberts JL. Transcriptional analysis of tyrosine hydroxylase gene expression in the tuberoinfundibular dopaminergic neurons of the rat arcuate nucleus after estrogen treatment. J Biol Chem. 1987;262:817–21. [PubMed] [Google Scholar]
- [44].Hewitt SC, Korach KS. Estrogenic activity of bisphenol a and 2,2-bis(p-hydroxyphenyl)-1,1,1-trichloroethane (HPTE) demonstrated in mouse uterine gene profiles. Environ Health Perspect. 2011;119:63–70. doi: 10.1289/ehp.1002347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Glidewell-Kenney C, Hurley LA, Pfaff L, Weiss J, Levine JE, Jameson JL. Nonclassical estrogen receptor alpha signaling mediates negative feedback in the female mouse reproductive axis. Proc Natl Acad Sci USA. 2007;104:8173–7. doi: 10.1073/pnas.0611514104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Roforth MM, Atkinson EJ, Levin ER, Khosla S, Monroe DG. Dissection of estrogen receptor alpha signaling pathways in osteoblasts using RNA-sequencing. PLoS One. 2014;9:e95987. doi: 10.1371/journal.pone.0095987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Hewitt SC, Li L, Grimm SA, Winuthayanon W, Hamilton KJ, Pockette B, et al. Novel DNA motif binding activity observed in vivo with an estrogen receptor alpha mutant mouse. Mol Endocrinol. 2014;28:899–911. doi: 10.1210/me.2014-1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Roepke TA, Ronnekleiv OK, Kelly MJ. Physiological consequences of membrane-initiated estrogen signaling in the brain. Front Biosci (Landmark Ed) 2011;16:1560–73. doi: 10.2741/3805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Nathanson NM. A multiplicity of muscarinic mechanisms. Enough signaling pathways to take your breath away. Proc Natl Acad Sci USA. 2000;97:6245–7. doi: 10.1073/pnas.97.12.6245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Cardoso CC, Ricardo VP, Frussa-Filho R, Porto CS, Abdalla FM. Effects of 17β-estradiol on expression of muscarinic acetylcholine receptor subtypes and estrogen receptor alpha in rat hippocampus. Eur J Pharmacol. 2010;634:192–200. doi: 10.1016/j.ejphar.2010.02.032. [DOI] [PubMed] [Google Scholar]
- [51].Karkanias GB, Ansonoff MA, Etgen AM. Estradiol regulation of alpha 1B-adrenoceptor mRNA in female rat hypothalamus-preoptic area. J Neuroendocrino. 1996;8:449–55. doi: 10.1046/j.1365-2826.1996.04716.x. [DOI] [PubMed] [Google Scholar]
- [52].Petitti N, Karkanias GB, Etgen AM. Estradiol selectively regulates alpha 1B-noradrenergic receptors in the hypothalamus and preoptic area. J Neurosc. 1992;12:3869–76. doi: 10.1523/JNEUROSCI.12-10-03869.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Quesada A, Etgen AM. Functional interactions between estrogen and insulin-like growth factor-I in the regulation of alpha 1B-adrenoceptors and female reproductive function. J Neurosci. 2002;22:2401–8. doi: 10.1523/JNEUROSCI.22-06-02401.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Machado JD, Alonso C, Morales A, Gomez JF, Borges R. Nongenomic regulation of the kinetics of exocytosis by estrogens. J Pharmacol Exp Ther. 2002;301:631–7. doi: 10.1124/jpet.301.2.631. [DOI] [PubMed] [Google Scholar]
- [55].Kim YJ, Hur EM, Park TJ, Kim KT. Nongenomic inhibition of catecholamine secretion by 17β-estradiol in PC12 cells. J Neurochem. 2000;74:2490–6. doi: 10.1046/j.1471-4159.2000.0742490.x. [DOI] [PubMed] [Google Scholar]
- [56].Bienkowski MS, Rinaman L. Noradrenergic inputs to the paraventricular hypothalamus contribute to hypothalamic-pituitary-adrenal axis and central Fos activation in rats after acute systemic endotoxin exposure. Neuroscience. 2008;156:1093–102. doi: 10.1016/j.neuroscience.2008.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Rinaman L. Hindbrain noradrenergic lesions attenuate anorexia and alter central cFos expression in rats after gastric viscerosensory stimulation. J Neurosci. 2003;23:10084–92. doi: 10.1523/JNEUROSCI.23-31-10084.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Cowley MA, Smith RG, Diano S, Tschop M, Pronchuk N, Grove KL, et al. The distribution and mechanism of action of ghrelin in the CNS demonstrates a novel hypothalamic circuit regulating energy homeostasis. Neuron. 2003;37:649–61. doi: 10.1016/s0896-6273(03)00063-1. [DOI] [PubMed] [Google Scholar]
- [59].Nogueiras R, Williams LM, Dieguez C. Ghrelin: New molecular pathways modulating appetite and adiposity. Obes Facts. 2010;3:285–92. doi: 10.1159/000321265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Hillebrand JJ, de Wied D, Adan RA. Neuropeptides, food intake and body weight regulation: A hypothalamic focus. Peptides. 2002;23:2283–306. doi: 10.1016/s0196-9781(02)00269-3. [DOI] [PubMed] [Google Scholar]
- [61].Guan JL, Okuda H, Takenoya F, Kintaka Y, Yagi M, Wang L, et al. Synaptic relationships between proopiomelanocortin- and ghrelin-containing neurons in the rat arcuate nucleus. Regul Pept. 2008;145:128–32. doi: 10.1016/j.regpep.2007.09.028. [DOI] [PubMed] [Google Scholar]
- [62].Pirnik Z, Majercikova Z, Holubova M, Pirnik R, Zelezna B, Maletinska L, et al. Effect of ghrelin receptor agonist and antagonist on the activity of arcuate nucleus tyrosine hydroxylase containing neurons in C57BL/6 male mice exposed to normal or high fat diet. J Physiol Pharmacol. 2014;65:477–86. [PubMed] [Google Scholar]
- [63].Pinzone JJ, Stevenson H, Strobl JS, Berg PE. Molecular and cellular determinants of estrogen receptor alpha expression. Mol Cell Biol. 2004;24:4605–12. doi: 10.1128/MCB.24.11.4605-4612.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Diotel N, Servili A, Gueguen MM, Mironov S, Pellegrini E, Vaillant C, et al. Nuclear progesterone receptors are up-regulated by estrogens in neurons and radial glial progenitors in the brain of zebrafish. PLoS One. 2011;6:e28375. doi: 10.1371/journal.pone.0028375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Moffatt CA, Rissman EF, Shupnik MA, Blaustein JD. Induction of progestin receptors by estradiol in the forebrain of estrogen receptor-alpha gene-disrupted mice. J Neurosci. 1998;18:9556–63. doi: 10.1523/JNEUROSCI.18-22-09556.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Oakley AE, Clifton DK, Steiner RA. Kisspeptin signaling in the brain. Endocr Rev. 2009;30:713–43. doi: 10.1210/er.2009-0005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Navarro VM, Tena-Sempere M. Kisspeptins and the neuroendocrine control of reproduction. Front Biosci. 2011;3:267–75. doi: 10.2741/s150. [DOI] [PubMed] [Google Scholar]
- [68].Lehman MN, Coolen LM, Goodman RL. Minireview: Kisspeptin/neurokinin B/dynorphin (KNDy) cells of the arcuate nucleus: A central node in the control of gonadotropin-releasing hormone secretion. Endocrinology. 2010;151:3479–89. doi: 10.1210/en.2010-0022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].De Bond JA, Smith J. Kisspeptin and energy balance in reproduction. Reproduction. 2013;147:R53–63. doi: 10.1530/REP-13-0509. [DOI] [PubMed] [Google Scholar]
- [70].Fernandez-Fernandez R, Martini AC, Navarro VM, Castellano JM, Dieguez C, Aguilar E, et al. Novel signals for the integration of energy balance and reproduction. Mol Cell Endocrinol. 2006;254-255:127–32. doi: 10.1016/j.mce.2006.04.026. [DOI] [PubMed] [Google Scholar]
- [71].Sinkevicius KW, Burdette JE, Woloszyn K, Hewitt SC, Hamilton K, Sugg SL, et al. An estrogen receptor-alpha knock-in mutation provides evidence of ligand-independent signaling and allows modulation of ligand-induced pathways in vivo. Endocrinology. 2008;149:2970–9. doi: 10.1210/en.2007-1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Topaloglu AK, Reimann F, Guclu M, Yalin AS, Kotan LD, Porter KM, et al. Tac3 and Tacr3 mutations in familial hypogonadotropic hypogonadism reveal a key role for neurokinin B in the central control of reproduction. Nat Genet. 2009;41:354–8. doi: 10.1038/ng.306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Gianetti E, Tusset C, Noel SD, Au MG, Dwyer AA, Hughes VA, et al. Tac3/Tacr3 mutations reveal preferential activation of gonadotropin-releasing hormone release by neurokinin B in neonatal life followed by reversal in adulthood. J Clin Endocrinol Metab. 2010;95:2857–67. doi: 10.1210/jc.2009-2320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Hammond R, Gibbs RB. GPR30 is positioned to mediate estrogen effects on basal forebrain cholinergic neurons and cognitive performance. Brain Res. 2011;1379:53–60. doi: 10.1016/j.brainres.2010.11.098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Eghlidi DH, Haley GE, Noriega NC, Kohama SG, Urbanski HF. Influence of age and 17β-estradiol on kisspeptin, neurokinin B, and prodynorphin gene expression in the arcuate-median eminence of female rhesus macaques. Endocrinology. 2010;151:3783–94. doi: 10.1210/en.2010-0198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Hill JW, Elmquist JK, Elias CF. Hypothalamic pathways linking energy balance and reproduction. Am J Physiol Endocrinol Metab. 2008;294:E827–32. doi: 10.1152/ajpendo.00670.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Koylu EO, Balkan B, Kuhar MJ, Pogun S. Cocaine and amphetamine regulated transcript (CART) and the stress response. Peptides. 2006;27:1956–69. doi: 10.1016/j.peptides.2006.03.032. [DOI] [PubMed] [Google Scholar]
- [78].Dandekar MP, Nakhate KT, Kokare DM, Subhedar NK. Involvement of CART in estradiol-induced anorexia. Physiol Behav. 2012;105:460–9. doi: 10.1016/j.physbeh.2011.09.001. [DOI] [PubMed] [Google Scholar]
- [79].Wilson ME, Rosewell KL, Kashon ML, Shughrue PJ, Merchenthaler I, Wise PM. Age differentially influences estrogen receptor-alpha (ERα) and estrogen receptor-beta (ERβ) gene expression in specific regions of the rat brain. Mech Ageing Dev. 2002;123:593–601. doi: 10.1016/s0047-6374(01)00406-7. [DOI] [PubMed] [Google Scholar]
- [80].Qiu J, Ronnekleiv OK, Kelly MJ. Modulation of hypothalamic neuronal activity through a novel G-protein-coupled estrogen membrane receptor. Steroids. 2008;73:985–91. doi: 10.1016/j.steroids.2007.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Revankar CM, Cimino DF, Sklar LA, Arterburn JB, Prossnitz ER. A transmembrane intracellular estrogen receptor mediates rapid cell signaling. Science. 2005;307:1625–30. doi: 10.1126/science.1106943. [DOI] [PubMed] [Google Scholar]
- [82].Davis KE, Carstens EJ, Irani BG, Gent LM, Hahner LM, Clegg DJ. Sexually dimorphic role of G protein-coupled estrogen receptor (GPER) in modulating energy homeostasis. Horm Behav. 2014;66:196–207. doi: 10.1016/j.yhbeh.2014.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]