Abstract
H1 linker histones are key chromatin architectural proteins facilitating the formation of higher order chromatin structures. The H1 family constitutes the most heterogeneous group of histone proteins, with eleven non-allelic H1 variants in mammals. H1 variants differ in their biochemical properties and exhibit significant sequence divergence from one another, yet most of them are highly conserved during evolution from mouse to human. H1 variants are differentially regulated during development and their cellular compositions undergo dramatic changes in embryogenesis, gametogenesis, tissue maturation and cellular differentiation. As a group, H1 histones are essential for mouse development and proper stem cell differentiation. Here we summarize our current knowledge on the expression and functions of H1 variants in mammalian development and stem cell differentiation. Their diversity, sequence conservation, complex expression and distinct functions suggest that H1s mediate chromatin reprogramming and contribute to the large variations and complexity of chromatin structure and gene expression in the mammalian genome.
Keywords: Linker histones, H1 variants, mammalian development, stem cell differentiation, chromatin, epigenetic gene regulation
1. Overview: Histone H1 and its variants in mammals
The DNA of all eukaryotic nuclei is packaged into chromatin by association with histone proteins. The basic repeating unit of chromatin is the nucleosome core particle, which consists of an octamer of four core histones (H2A, H2B, H3 and H4) wrapped by 147 bp of DNA [1, 2]. Linker histone H1 binds to nucleosome core particles and the linker DNA between nucleosomes to facilitate the folding of the “beads-on-a-string” extended chromatin fiber into higher order chromatin structures, the 30-nm fiber [3–6]. For gene transcription to occur, the chromatin template plays a dynamic role. Nucleosome core particles and posttranslational modifications of core histones, such as acetylation, methylation, ubiquitination, phosphorylation and sumoylation, have been shown to play critical roles in gene activation and repression. Much less is known about the role of linker histone H1 and its variants. Here we focus our discussion on recent studies about the function of mammalian H1s in development and stem cell differentiation.
The H1 histone family is the most divergent and heterogeneous group of histones among the evolutionarily conserved histone protein families. There are multiple nonallelic linker histone variants present in higher organisms which provide additional levels of regulation on chromatin structure and function [7, 8]. In mammals, 11 H1 variants have been identified, including seven somatic H1s (H10, H1a, H1b, H1c, H1d, H1e and H1x) and four germ cell-specific H1s (H1t, H1T2, H1LS1 and H1oo) (Table 1). Among the multiple nomenclature systems for mammalian somatic H1 variants, the alphabetic nomenclature (H1a-e, H10 and H1x) and the alternative numeric system (H1.1-1.5, H1.0 and H1.x) are most commonly used [8–12]. Table 1 summarizes their nomenclature as well as H1 variants distinct expression and genomic localization patterns.
Table 1.
Mammalian histone H1 variantsa
| Histone H1 variants (Alt. symbols) | Mouse histone H1 genes
|
Human histone H1 genes
|
Gene Intronicb | Expression Cell typese | Expression Dependence on DNA replication | mRNA 3′UTR | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| Gene name | Accession no. | Chr location | Gene name | Accession no. | Chr location | |||||
| H1a (H1.1) | Hist1h1a | NM_030609 | 13 | HIST1H1A | NM_005325 | 6 | No | Somatic | Yes | Stem-loop |
| H1b (H1.5) | Hist1h1b | NM_020034 | 13 | HIST1H1B | NM_005322 | 6 | No | Somatic | Yes | Stem-loop |
| H1c (H1.2) | Hist1h1c | NM_015786 | 13 | HIST1H1C | NM_005319 | 6 | No | Somatic | Bothg | Bothg |
| H1d (H1.3) | Hist1h1d | NM_145713 | 13 | HIST1H1D | NM_005320 | 6 | No | Somatic | Yes | Stem-loop |
| H1e (H1.4) | Hist1h1e | NM_015787 | 13 | HIST1H1E | NM_005321 | 6 | No | Somatic | Yes | Stem-loop |
| H10 (H1.0, H1f0) | H1f0 | NM_008197 | 15 | H1F0 | NM_005318 | 22 | No | Somaticf, oocyte | No | Poly-A |
| H1x (H1.x, H1fx) | H1fx | NM_198622 | 6 | H1FX | NM_006026 | 3 | Y/Nc | Somatic | No | Poly-A |
| H1oo (H1foo) | H1foo | NM_138311 | 6 | H1FOO | NM_153833 | 3 | Yes | Oocytes, zygote and 2-cell embryo | No | Poly-A |
| H1t | Hist1h1t | NM_010377 | 13 | HIST1H1T | NM_005323 | 6 | No | Spermatocytes spermatids | Yes | Stem-loop |
| H1T2 | H1fnt | NM_027304 | 15 | H1FNT | NM_181788 | 12 | No | Spermatids | No | Poly-A |
| HILS1 | Hils1 | NM_018792 | 11 | HILS1 | AY286318 | 17 | Y/Nd | Spermatids | No | Poly-A |
Gene names and accession numbers are adapted from [192];
Human H1FX gene encodes multiple transcript variants with or without introns (see refs. [55, 193]);
Human HILS1 gene consists of two exons and a small 106-bp intron and mouse Hils1 gene is intronless (ref. [27]);
H10 is enriched in differentiated cells (see ref. [22]);
The H1c gene encodes two versions of mRNAs, polyadenylated and stem-loop forms, allowing for both replication-dependent and replication-independent expression (see ref. [36]).
The expression of H1 variants is differentially regulated during mammalian development and cellular differentiation [13–19]. H1a through H1e are the main types of H1 ubiquitously expressed in somatic cells, yet their expression is tightly regulated with distinct levels in different tissues and cell types [14, 15, 20, 21]. H10 is expressed mainly in differentiated and nondividing cells [22], whereas the least characterized H1x appears to exhibit a G1 phase-dependent nucleolar accumulation in cultured human cell lines [23]. H1oo and H1t are germ cell-specific H1s with expression in oocytes and testis, respectively [24, 25]. H1T2 and H1LS1 are two H1t-related H1s specifically expressed in spermatids [26–30].
All of the mammalian H1 genes are transcribed by RNA Pol II. The genes of major somatic H1 variants (H1a, H1b, H1c, H1d and H1e) and H1t are intronless and transcribed in S phase of cell cycle in a DNA replication-dependent manner (Table 1). Their mRNAs have characteristics of DNA replication-coupled histone mRNAs, lacking a poly(A) tail but terminated by a conserved 3 stem-loop structure. This stem-loop hairpin is bound by the histone RNA hairpin-binding protein or stem-loop binding protein (SLBP) which is required for histone pre-mRNA processing and enhances translation [31–35]. The transcription of these six H1 genes involves typical cell cycle regulation of histone genes: transcription initiation, 3 end processing and mRNA stability. Interestingly, H1c gene also produces a polyadenylated form of mRNA besides the 3 stem-loop form mRNA, allowing for independent regulation of expression in dividing and nondividing cells [36]. These six H1 genes (H1a-e and H1t) are linked and clustered together with core histone genes on mouse chromosome 13 and the orthologous human chromosome 6 [14, 21, 37–40]. This genomic organization is conserved from mouse to human. The other five H1 genes, H10, H1x, H1oo, H1t2 and Hils1, however, are expressed in a DNA replication-independent manner, with mRNAs polyadenylated and lacking the 3 stem-loop structure. In addition, they are not clustered with core histones but scattered in the mammalian genomes (Table 1).
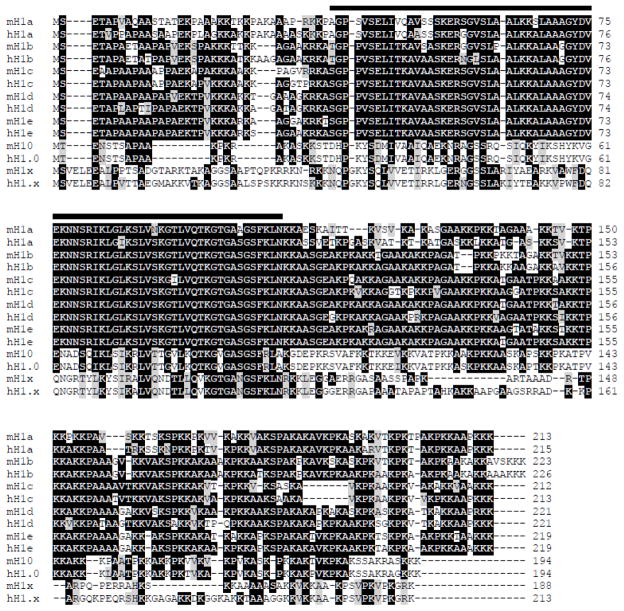
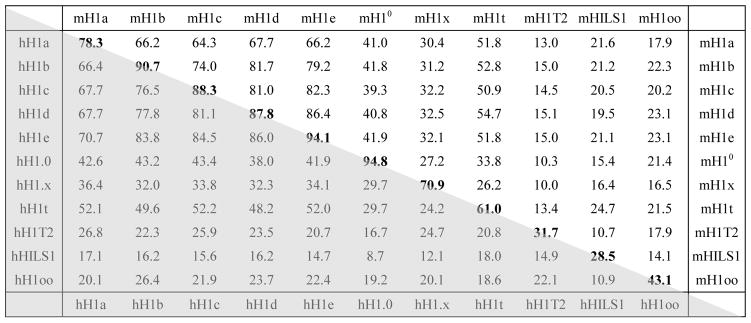
All metazoan H1s share the same tripartite domain structure that includes a short, flexible N-terminal tail domain (NTD), a central globular domain (GD) with a winged-helix motif and a long, lysine-rich C-terminal tail domain (CTD) [41–43]. The H1 globular domain is highly conserved, while the N- and C-terminal regions of H1 variants contain more sequence divergence. Comparison of the sequences of mouse and human H1 genes reveals that each mouse H1 variant gene is more similar to its human ortholog than to other mouse H1 variant genes (Fig. 1, Table 2) [44]. Table 2 summarizes the sequence comparisons among all H1 variants in mice and humans. Mouse and human H10 (H1.0) are particularly conserved with 94.8% of homology, whereas they have diverged significantly from other H1 variants with homology less than 45% (Table 2) [44]. On the other hand, H1b (H1.5), H1c (H1.2), H1d (H1.3) and H1e (H1.4) are not only highly conserved from mouse to human with over 85% sequence identity, but also share most similarities with each other, with 76–86% sequence identity. Their sequence similarity suggests that H1b, H1c, H1d and H1e may be more functionally related to each other than to other H1s. Among the 7 somatic H1s, H1x is the least conserved and the most divergent, with 70.9% identity between mouse and human H1x and less than 37% similarities with other somatic H1s. H1x also contains a lower content of basic amino acid residues [45, 46]. Germ cell-specific H1 variants are generally less conserved and more divergent than somatic H1s (Table 2). Mouse H1t shares a moderate identity of 61% with human H1t and 26–55% with somatic H1s, whereas other germ cell-specific H1s (H1oo, H1T2 and HILS1) have only 10–27% sequence identity with other H1 variants. The sequence conservation of the individual H1 variants suggests that H1 variants may have distinct roles in chromatin function and gene regulation in various cellular and developmental processes.
Fig. 1. Multiple alignment of mouse and human somatic H1 variants.
Genbank accession numbers of the protein sequences are listed in Table 1. The black line on top of amino acid sequences marks the globular domain. The conserved and the similar residues are highlighted in black and grey colors, respectively.
Table 2.
Sequence similarity among H1 variants in mouse and humana
The numbers indicate the percentage of identity between two aligned H1 variants. Sequence comparisons between mouse H1 variants are shown in the upper half of the table (non-shaded) and those between human H1 variants are shown in the lower half (shaded). The bold numbers at the diagonal represent the sequence similarity between mouse H1 variants and their human counterparts.
Consistent with the sequence heterogeneity of H1 variants, cumulative evidence suggests that H1 variants differ in their biochemical properties, affinities for chromatin, capabilities in chromatin compaction and binding partners [42, 47–62]. The globular and C-terminal domains are required for high-affinity binding of H1 to chromatin [42, 47–49]. The globular domain is suggested to bind to nucleosomes at the dyad and the linker DNA with symmetric or asymmetric models [42, 63–67], whereas the CTD is likely to bind to the linker DNA non-specifically. The globular domain of H1 variants may bind to nucleosomes in distinctive structural modes, leading to varied higher order structures [67]. Atomic force microscopy, in vitro biochemical assays and fluorescence recovery after photobleaching (FRAP) studies have suggested that H1b, H1d and H1e, the somatic H1s with longer C-terminal tails, display higher affinity for chromatin than H1a, H1c and H10, the somatic H1s with shorter C-terminal tails [51–53]. H1b and H1d are sometimes categorized into the intermediate group of chromatin binding affinity, so is H10, the most lysine-rich H1 with a short C-terminal tail [51–53]. The CTD appears to be the key determinant for the chromatin binding affinity of somatic H1s [48, 53]. The function of CTD in condensing chromatin is related to its length, the density of basic residues, the number of S/TPXK sites and its specific amino acid composition, as well as the intrinsic protein disorder in the CTD [53, 68]. The N-terminal tail appears to be dispensable for chromatin binding, nevertheless, its deletion or swapping between different H1 variants alters the binding affinity of the respective H1 variant for chromatin [47, 48, 50]. Not surprisingly, different H1 variants also display differential in vivo binding dynamics in oocytes and during ES cell nuclear transfer [69]. The binding of H1 to chromatin is also regulated by post-translational modifications [54, 55, 70] and histone chaperones [71, 72]. In addition to binding to DNA and nucleosomes, H1 variants interact with a variety of cellular proteins, which contributes to their diverse functions in various cellular processes [58–62].
Germ cell-specific H1 variants differ dramatically from somatic H1s in amino acid sequences and biochemical properties. H1t exhibits lower binding affinity for DNA and condenses chromatin to a lesser extent than somatic H1s [73–75], which may be attributed to the absence of the S/TPXK motifs, the sites for DNA binding and phosphorylation in CTD [76–78], and the single amino acid substitution of lysine observed in somatic H1s by glutamine in the H1t globular domain [79]. H1T2 is distinctive from H1t in that it is highly enriched with arginine residues and S/TPXK sites [28]. The oocyte-specific H1, H1oo, is the longest variant, with an NTD containing multiple potential phosphorylation sites and an extraordinarily long C-terminal tail rich in acidic amino acid residues [24]. Both the N-terminal and globular domains of H1oo are required for correct association with chromatin in the oocyte nucleus [69]. These intrinsic differences among H1 variants are expected to contribute to their distinct roles in chromatin compaction and gene regulation.
2. Role of histone H1 variants in mammalian development
The role of H1 in development has been interrogated in a variety of organisms. While H1 is nonessential in unicellular organisms such as yeast and Tetrahymena [80–84], H1 depletion in higher eukaryotes gives rise to more complex phenotypes [85–92], suggesting the involvement of H1 and its variants in regulation of diverse biological processes.
During mammalian development, the 11 nonallelic H1 variants are abundantly expressed and differentially regulated at all stages, from germ cells to embryos and adult tissues, suggesting their fundamental roles in development. The distinct properties of H1 variants as summarized above indicate that they are excellent candidates as mediators of chromatin reprogramming during mammalian development. In this section, we focus our discussions on the expression and functions of mammalian H1 variants in embryos, adult tissues and germ cells.
2.1. Embryogenesis
Mammalian development starts from a zygote, when the egg is fertilized. Fertilization induces the completion of the second meiotic division of the oocyte and the formation of two separate pronuclei within the zygote, i.e., a haploid paternal pronucleus from the sperm and a haploid maternal pronucleus from the oocyte. At this stage, the protamine-bound sperm chromatin becomes decondensed and the protamines are quickly replaced by the oocyte-specific H1, H1oo, leading to an enlarged sperm pronucleus [24, 93]. The two pronuclei subsequently fuse and cell division ensues.
Before activation of the zygotic genome at the two-cell stage, the oocyte provides the materials necessary for the first cell division. H1oo, the oocyte-specific H1 variant, remains the major H1 variant present till the late two-cell stage embryo (Fig. 2) [24, 93]. It is surmised that the incorporation of H1oo may facilitate the decondensation of paternal chromatin and nuclear reprogramming after fertilization. Evidence supporting this includes that the Xenopus homolog of H1oo, B4/H1M, is shown to be crucial for the reactivation of pluripotency genes in somatic nuclei transferred to Xenopus oocytes [94] and that ectopic expression of H1oo-GFP in moue embryonic stem cells (ESCs) leads to increased nuclease sensitivity at specific targets of H1oo-GFP [95]. In vitro experiments using Xenopus oocyte extracts showed that B4/H1M is necessary for the architecture and proper segregation of mitotic chromosomes even though it is not required for DNA replication and nuclear envelope assembly [96, 97]. Despite its presumed important roles, H1oo is not required for early embryogenesis in vivo as demonstrated by the lack of observable phenotypes of H1oo−/− mice (Table 3) [98, 99]. This result is reminiscent of the knockouts of single somatic H1s (Table 3). While the lack of obvious phenotypes in single somatic H1 knockouts is probably due to the compensation by the remaining somatic H1 variants (as discussed below), the major somatic H1 variants, H1a-e, are unlikely to compensate upon the deletion of H1oo due to the apparent lack of cross regulation of somatic H1 genes with the H1oo gene.
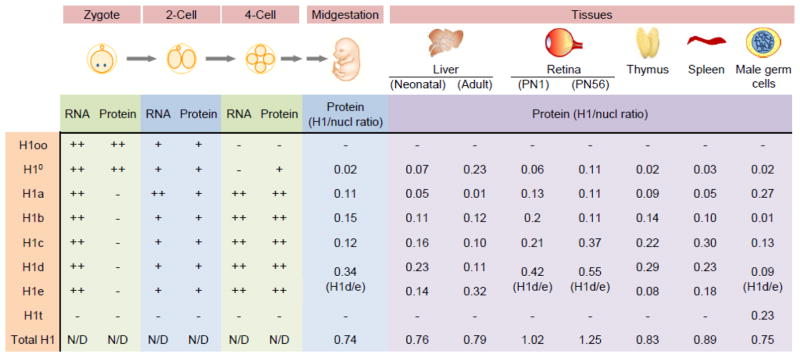
Fig. 2. The expression patterns of histone H1 variants during mouse development.
Currently available data of indicated developmental stages in embryogenesis and postnatal tissues are shown. From zygotes to four-cell stage embryos, “++”, “+” and “−” represent relative “high expression”, “low expression” and “no expression detected”, respectively, of the temporal expression change of each H1 variant, not intended for comparisons among different H1 variants. N/D: expression not determined. Data derived from refs. [24, 93, 100–102, 104]. RNA levels were measured by semi-quantitative RT-PCR, and protein levels were measured by Western blotting or immunofluorescence microscopy. The oocyte-specific H1oo is also enriched in zygotes but gradually decreased until complete disappearance at the four-cell stage [24, 93]. The maternal H10 mRNA and protein are readily detected at the zygotic stage but are degraded and replaced by zygotic H10 counterparts at the late two-cell stage [100–102]. Synthesis of H1a-e from the maternal mRNA transcripts starts at the late one-cell stage, resulting in appearance of H1a-e proteins in two-cell stage embryos [104]. The maternal mRNAs of H1a-e are degraded in late two-cell embryos concomitant with the accumulation of their zygotic transcripts [101]. Individual H1 variant- and the total H1-to-nucleosome ratios of midgestation embryos and postnatal tissues were adapted from refs. [13, 109, 117, 135]. No expression data in mice is available for H1x. As H1d and H1e are co-eluted in HPLC analysis, they were quantified together in embryos and certain tissue samples.
Table 3.
Summary of properties of mice and ESCs with altered H1 expression
| H1 manipulationa | H1/nucleosome ratiob | Phenotype | References | |
|---|---|---|---|---|
| In mice | ||||
|
| ||||
| H1 OE | H10 | N/D | Normal (10–20 fold increase in H10 mRNA, N/C in H10 protein) | [198] |
|
| ||||
| Single H1 KOs | H10−/− | N/C (adult liver: 0.68) | Normal | [103] |
| Decreased dendritic cell production; reduced response to vaccination with antigens | [118] | |||
| H1a−/− | N/C (male germ cells: 0.73) | Normal | [107, 135] | |
| H1c−/− | N/C | Normal | [108] | |
| Increased resistance to x-ray induced apoptosis in thymocytes and intestine | [120] | |||
| RD (PN56 retina: WT 1.25/KO 0.99) | Normal retina | [117] | ||
| H1d−/− | N/C | Normal | [108] | |
| H1e−/− | N/C | Normal | [108] | |
| H1oo−/− | N/D | Normal | [98] | |
| Or homeostasis | [99] | |||
| H1t−/− | N/C (male germ cells: 0.68) | Normal | [132–134] | |
| H1T2−/− | N/D | Abnormal sperms and male infertility | [28] | |
|
| ||||
| Double H1 KOs | H10−/−H1c−/− | N/C (adult liver: 0.71) | Normal | [108] |
| H10−/−H1d−/− | N/C (adult liver: 0.71) | Normal | [108] | |
| H10−/−H1e−/− | N/C (adult liver: 0.76) | Normal | [108] | |
| H1c−/−H1e−/− | N/C (adult liver: 0.72) | Mild growth retardation | [13] | |
| RD (retina) | Abnormal rod photoreceptor packing | [117] | ||
| H1a−/−H1t−/− | RD (male germ cells: WT 0.78/KO 0.59) | Normal | [135] | |
|
| ||||
| Triple H1 KOs | H1c−/−H1e−/−H10−/− | RD (adult liver: WT 0.79/KO 0.64) | Growth retardation; lower birth rate; small thymus | [13] |
| RD (retina) | Abnormal rod photoreceptor packing | [117] | ||
| H1c−/−H1d−/−H1e−/− | RD (midgestation embryo: WT 0.74/KO 0.40) | Embryonic lethality by E11.5 | [13] | |
|
| ||||
| Other compound H1 KOs with multiple H1 null alleles | H1c−/−H1d+/−H1e−/− | RD (thymus:WT0.83/KO 0.41) | Low birth rate; severe growth retardation | [13] |
| H1c−/−H1d+/−H1e−/−H10−/− | N/D | Low birth rate; neonatal lethality | [13] | |
| H1c−/−H1d−/−H1e−/−H10−/− | N/D | Embryonic lethality | [13] | |
|
| ||||
| In ESCs | ||||
|
| ||||
| Single H1 KOs (mouse) | H10−/− | N/D | Normal ESC differentiation | # |
|
| ||||
| H1c−/− | RD (WT 0.45/KO 0.38) | Normal cell morphology and proliferation; normal ESC differentiation and teratoma formation; specific expression changes of Hox genes | [154] | |
| H1d−/− | RD (WT 0.45/KO 0.35) | [154] | ||
| H1e−/− | RD (WT 0.45/KO 0.35) | [154] | ||
|
| ||||
| Triple H1 KO (mouse) | H1c−/−H1d−/−H1e−/− | RD (ESCs: WT 0.46/KO 0.25) | Reduced chromatin compaction; specific changes in gene expression and DNA methylation | [153] |
| Chromocenter clustering and de-repression of major satellite repeats | [109] | |||
| Enhanced DNA damage response | [199] | |||
| Decreased chromatin flexibility in gene rich TAD | [163] | |||
| RD (EBs: WT 0.62/KO 0.36) | ESCs more resistant to spontaneous differentiation; impaired EB and neural differentiation | [16] | ||
| [16] | ||||
| Dysregulated in specification of mesendoderm and neuroectoderm | [105] | |||
|
| ||||
| H1 KD (human) | H10 | N/D | Lack of induction of a few differentiation associated genes | [106] |
OE, overexpression; KO, knockout; KD, knockdown.
N/C, no change; RD, reduced; N/D, not determined; WT, wildtype.
Pan, Zhang, Fan unpublished observation.
Surprisingly, the other H1 variant protein abundantly present in the oocyte and the zygote is H10, the replacement H1 variant accumulated in differentiated cells (Figs 2, 3). H10 diminishes at the four-cell stage and is present at minimum levels during early embryogenesis, which is consistent with rapid DNA replication and cell divisions at this stage (Figs 2, 3) [100–102]. Similar to H1oo−/− mice, H10 null mice are fertile and develop normally, suggesting that H10 is not required for early embryogenesis either [103]. These studies suggest that, despite their unique expression patterns and properties in the oocyte and the zygote, H1oo and H10 are dispensable for zygotic genome activation and embryogenesis. Nevertheless, H1oo and H10 might help define active and inactive transcription domains in the genome and facilitate proper activation of the zygotic genome. The lack of H1oo or H10 may reduce the efficiency of chromatin reprograming in zygotes, even though the surviving H1oo−/− or H10−/− embryos and mice are not affected. It is also conceivable that the protein synthesis of other H1 variants is increased in these embryos to compensate for the loss of H1oo or H10.
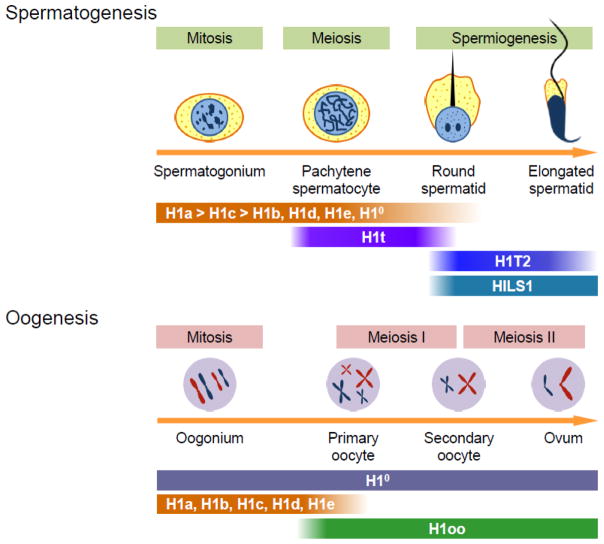
Fig. 3. The expression patterns of histone H1 variants during gametogenesis.
During spermatogenesis, male germ cells undergo unique chromatin remodeling. Somatic H1s were detected in spermatogonia, predominated by H1a and H1c, whose levels steadily decrease in meiotic spermatocytes until their disappearance in round spermatids [126, 133]. H1t is expressed in pachytene spermatocytes and persists until round spermatids [127–130]. The expression of H1T2 and HILS1 is confined to spermatids, with H1T2 in the nuclei of early spermatids and HILS1 in both early and elongated spermatids [26–29]. Similar to spermatogonia, oogonia express a complement of somatic H1s. During oogenesis, somatic H1s are gradually replaced by the oocyte-specific H1oo [24, 101]. However, H10 is retained throughout oogenesis [100–102].
Synthesis of other somatic H1s, including H1a, H1b, H1c, H1d and H1e, from the maternal mRNA transcripts starts at late one-cell stage, reaching its maximum rate in early two-cell stage embryos (Fig. 2) [104]. The maternal mRNAs of these somatic H1s are degraded in late two-cell embryos, concomitant with the accumulation of their zygotic transcripts [101]. Resumption of somatic H1 proteins occurs during the four-cell stage of embryogenesis, accompanied by the disappearance of H1oo (Fig. 2) [24, 102]. The expression patterns of individual H1 variants after the four-cell stage have not been fully defined. The incorporation of somatic H1s into chromatin is likely to facilitate proper cell differentiation and germ layer specification during early embryonic development. This is suggested by studies demonstrating important roles of H1 in ESC differentiation and in loss of mesodermal competence during midblastula to neurula transition in Xenopus [16, 90, 105, 106]. HPLC analysis of histone extracts from E10.5 mouse embryos at mid-gestation indicates a total H1-to-nucleosome ratio of 0.74 that is approximately 3 H1 molecules per 4 nucleosomes (Fig. 2) [13]. H1d, H1e and H1b are the most abundant H1s at this stage, accounting for ~67% of the total H1, whereas H10 is present at a very low level, with an H10-to-nucleosome ratio of 0.02. Given that the accumulation of H10 only occurs in differentiated and non-dividing cells, this expression pattern of H1 variants is consistent with the rapid cell division in midgestation embryos [13].
The functions of somatic H1 variants in mammalian embryonic development have been extensively interrogated by targeted gene inactivation. Although somatic H1s are abundant in embryos, mice with single deletion of H1a, H1c, H1d, or H1e produce litters with normal live pups and litter size, suggesting no obvious defects in embryogenesis in these mice (Table 3) [103, 107, 108]. Even in the context of H10 deficiency, further deletion of H1c, H1d or H1e generates mice that are fertile with no obvious phenotypes (Table 3) [108]. Intercrosses of H1c/H1e double heterozygous mice produce H1c/H1e double null mice at the expected Mendelian ratio, suggesting that compound deletion of H1c and H1e does not interrupt embryogenesis [108]. However, neonatal males of H1c/H1e double knockout appear to be smaller than wild-type male littermates. Such phenotype is not observed in neonatal H1c−/−H1e−/− females [13]. These results suggest that the loss of H1c/H1e may not be fully compatible with the rapid growth in male embryos, resulting in moderate impairment in their growth. Due to the compensatory increase of the remaining H1 variants controlled by yet unknown mechanisms, single or double H1 deletion does not significantly alter the H1 content in mouse tissues, suggesting somatic H1 variants are, at least partially, functionally redundant during embryogenesis and in adult tissues [13, 108].
H10/H1c/H1e triple homozygous knockout mice are significantly underrepresented in F2 offspring from intercrosses of H10 null mice and H1c/H1e double null mice, suggesting that the loss of H10/H1c/H1e impairs embryogenesis [13]. Currently it is not determined at which stage during embryogenesis some of the H10−/−H1c−/−H1e−/− embryos are lost. Nevertheless, the surviving H10/H1c/H1e triple knockout mice appear normal and fertile despite their mild growth retardation and smaller litter size [13].
The essentiality of histone H1 in mammalian development was demonstrated by the compound deletion of three major somatic H1 variants, H1c, H1d and H1e. Given that most of somatic H1 genes are clustered on chromosome 13 in mice (Table 1), a sequential gene targeting strategy was employed to generate cis triply targeted H1c+/−H1d+/−H1e+/− ESCs for production of H1c/H1d/H1e triple knockout mice [13]. H1c/H1d/H1e triple heterozygous (H1c+/−H1d+/−H1e+/−) mice are fertile and viable, but exhibit growth retardation. H1c/H1d/H1e triple heterozygotes are underrepresented in litters from intercrosses of H1c+/−H1d+/−H1e+/− mice and 17% of the triple heterozygotes die during embryogenesis [13]. H1c−/−H1d−/−H1e−/− homozygous mutants are underrepresented at E7.5 with increasingly fewer surviving homozygous mutant embryos at the following developmental stages. All triple knockout embryos die by E11.5, indicating embryonic lethality at midgestation. Recovered E9.5 embryos display a broad spectrum of defects, ranging from mild growth restriction and modest developmental delay to severe compromise, such as neural tube aberration, failed chorioallantoic fusion, expanded pericardia, shortened tails and regions of excess tissue [13]. H1c−/−H1d−/−H1e−/− embryos have a 50% reduction in the total H1 level at E10.5, suggesting that compensation among somatic H1 variants is disrupted upon depletion of H1c, H1d and H1e. The embryonic lethality phenotype can be partially rescued by a single wild-type H1d allele, as H1c−/−H1d+/−H1e−/− (5−) mutant mice are produced from crosses of H1c/H1e double knockout mice with H1c/H1d/H1e triple heterozygotes. However, the majority of these 5(−) mice die in late embryogenesis and only one-third survive to newborns ([13]; Zhang and Fan, unpublished observation). The surviving 5(−) mutant mice are severely retarded in growth and development [13]. These findings demonstrate that significant functional redundancy exists in the H1 family and that the total H1 content, rather than any individual H1 variants, is essential for mammalian embryogenesis.
2.2. Tissue-specific functions
A plethora of studies have shown that the expression of histone H1 variants varies massively across various tissues and even at different developmental stages of the same tissue (Fig. 2) [13, 20, 108–110]. Given the presence of both replication-dependent and replication-independent H1 variant genes and the multitude levels of transcriptional and translational regulation distinct for each H1 gene, the relative abundance of mRNA levels of different H1 variants often does not correlate with the protein composition of H1 variants in specific cell types or tissues. In any specific tissue, the protein composition of H1 variants is a result of the combinatory effects of the relative proportions of dividing and nondividing cells as well as the complex regulatory mechanisms controlling transcription, mRNA processing and the stability of mRNA and proteins for each H1 variant [14, 20, 22, 111–113].
Earlier studies using gel electrophoresis, such as two-dimensional polyacrylamide gel electrophoresis (PAGE), acid/urea PAGE and SDS PAGE, to analyze H1 histones extracted from mouse or rat tissues found that H1a through H1e are present in almost all tissues examined, consisting with the ubiquitous expression features of somatic H1s. More recent HPLC analyses of histone extracts allow the separation and quantitative measurement of different histone fractions. Using H2B as an internal control, this method provides an accurate assessment of the ratios of H1-to-nucleosome, including ratios of total H1-to-nucleosome and individual H1 variants-to-nucleosome, and standardizes the cross comparisons of the amounts of H1 variants in different cell types and tissues. Figure 2 summarizes available data on the H1 to nucleosome ratios in adult tissues.
Combining data sets from gel electrophoresis and HPLC analyses, we could note the dramatic increase of H10 and H1e accompanied by the decline of H1a, H1c and H1d during tissue maturation and reduction of cell division. This trend is prominent in liver, lung, kidney and cerebral cortex during development (Fig. 2) [13, 110, 114–116]. For example, H10 and H1e account for 9.5% and 19% of the total H1, respectively, in neonatal mouse liver, but their compositions reach 29% and 40% of the total H1 in adult liver (Fig. 2) [13]. Likewise, in differentiating cortical neurons, H1e reaches the highest proportion of the total H1 accompanied by an escalation of H10 [110, 114–116]. During retinal maturation, however, H1c is markedly upregulated (Fig. 2) [117]. Consistent with the accumulation of H10 upon terminal cell division, H10 is at minimal levels in tissues maintaining active cell proliferation, such as thymus and spleen. H1c and H1d are abundant in most tissues, including adult thymus and spleen, while the level of H1a is rather low in most adult tissues. The distantly related variant H1x is expressed in all tissues examined [45], however, it is not expressed at a comparable level as other somatic H1s that can be clearly detected by HPLC analysis of histone extracts (Fig. 2).
The distinct regulation of H1 variants expression and the specific H1 composition in each tissue during mammalian development may suggest that the abundance and relative proportion of each H1 variant are important for maintaining proper development and tissue maturation. However, mice deficient of any one of the somatic H1 variants, including those of high abundance in specific tissues, do not display obvious defects in particular tissue functions (Table 3) [13, 103, 107, 108, 117]. For example, H1c reaches 30% of the total H1 in rod photoreceptors of adult mice, yet H1c−/− mice have normal rod photoreceptor packing and global heterochromatin condensation in retina [117]. Similarly, H1 proteins in adult liver are largely composed of H1e and H10 which account for 42% and 27% of the total H1, respectively. The loss of either H1e or H10 has no effects on the size or maturation of mouse liver [13, 103].
Functional redundancy among H1 family members and compensation of the lost H1 variants by the remaining H1s are thought to account for the lack of phenotypes in single knockouts of H1 variants. Indeed, even when two major somatic H1 variants are depleted, mice appear to be normal (Table 3). HPLC analysis of histone extracts from mouse tissues indicates that the total H1 levels in these single and double knockouts remain unchanged, suggesting compensation of the deleted H1s by upregulation of the remaining somatic H1s. For example, H10 and H1e together account for 70% of the total H1 in adult liver, yet H10−/−H1e−/− double mutant mice appear to be normal with a typical total H1-to-nucleosome ratio of 0.76 in mature liver [108]. The compensatory effects are most prominent within the group of H10, H1c, H1d and H1e, indicating potential cross regulatory mechanisms among these H1s [13, 103, 107, 108, 117]. These results suggest a large capacity of increased expression and synthesis of the remaining H1 variants that is sufficient to compensate for the loss of up to two abundant H1 variants.
Disruption of compensation by compound knockout of more than two major H1 variants leads to severe phenotypes, ranging from mild growth retardation to embryonic lethality as summarized in Table 3. In addition, H10−/−H1c−/−H1e−/− and H1c−/−H1d+/−H1e−/− mice have smaller thymuses than those of wild-type littermates. HPLC analysis showed a 40–50% reduction in H1 content in thymus of these mutant mice, suggesting that a low H1 content is not compatible with the rapid cell division in thymus. These studies on compound H1 knockout mice indicate that the total H1 level is critical for normal tissue development and functions [13].
Despite the compensation among H1 variants and the lack of general defects in single and double knockouts of H1 variants, emerging evidence suggests that, the loss of single somatic H1 variants can lead to specific defects. For example, H10 is upregulated during cellular differentiation in all cell types, including dendritic cells [22, 118]. The deletion of H10 does not lead to general defects in differentiation or cell division in most tissues examined, but specifically impairs production and function of dendritic cells in mutant mice [103, 118]. This apparent lack of compensation of H10 by the remaining somatic H1s could be due to a potential failure in upregulation of the remaining H1s or may be attributed to specific cellular functions of H10 in dendritic cells not directly connected to its general role in chromatin condensation, such as H10-specific interactions with other proteins [119]. Similarly, H1c−/− mice display increased resistance to x-ray induced apoptosis in thymocytes and intestines, which is connected to H1c being a signal transducer in apoptosis induced by DNA double-strand breaks (DSB) [120, 121]. DSB induces the translocation of H1c from the nucleus to the cytoplasm which destabilizes mitochondrial membranes and leads to the release of cytochrome c and apoptosis [120–122].
2.3. Gametogenesis
Gametogenesis is the process of producing haploid gametes from diploid germ cells by meiotic cell division in gonads. It includes spermatogenesis and oogenesis in males and females, respectively. Mammalian gametogenesis is characterized by distinct chromatin remodeling and condensation processes where histone H1 plays a fundamental role [123].
Spermatogenesis has three phases: 1) spermatogonia renewal and proliferation, in which spermatogonial stem cells undergo multiplication to form preleptotene primary spermatocytes; 2) meiosis, in which preleptotene primary spermatocytes enter meiosis to form round spermatids; 3) spermiogenesis, in which spermatids undergo dramatic morphological changes to form elongated spermatozoa [124]. Chromatin condensation during spermatogenesis is mediated by a unique set of male germ cell specific proteins, including testis-specific linker histones, transition proteins and protamines [123]. During spermatogenesis, somatic H1 variants are progressively replaced by testis-specific H1 variants, followed by the replacement of most histones with transition proteins and protamines, resulting in dramatic chromatin condensation culminating in mature spermatozoa.
Spermatogonia express a complement of somatic H1 variants, mainly H1a and H1c [25, 125]. H1a is the predominant H1 variant in prepachytene spermatocytes, comprising approximately 70% of the total H1 [25], but its level steadily decreases in meiotic spermatocytes and postmeiotic spermatids until it fully disappears (Fig. 3) [126]. H1t, the testis-specific H1 variant, starts to accumulate in pachytene spermatocytes and persists until round spermatids, constituting up to 55% of the total H1 (Figs 2, 3) [127–130]. Both H1a and H1t have relatively low chromatin binding affinity and are weak chromatin condensers [51–53, 73–75], which presumably contributes to the adoption of loosely compacted chromatin in early spermatocytes followed by a relatively open chromatin in pachytene spermatocytes and facilitates the genetic recombination in pachytene spermatocytes and/or the replacement of histones with transition proteins in early spermatids [131]. However, despite their unique expression patterns and abundance during these developmental stages, H1a and H1t appear to be dispensable for spermatogenesis. Mice lacking H1a or H1t are fertile and show normal spermatogenesis [107, 132–134]. HPLC analysis indicates that the total amount of H1 proteins in H1a−/− and H1t−/− mice remain unchanged. Interestingly, the loss of H1a is mainly compensated by increased expression of other somatic H1 variants, H1c, H1d and H1e, whereas the loss of H1t is compensated by upregulation of H1a and H1c [133, 135]. Depletion of both H1a and H1t, which together account for nearly 70% of the total H1 in male germ cells, disrupts the biochemical compensation and leads to a 25% decrease in the total H1 level. Such depletion causes specific changes in gene expression in germ cells without overt phenotypes in H1a−/−H1t−/− mice, which are fertile and have normal spermatogenesis [135]. No defects in homologous chromosome pairing and recombination or differences in chromatin condensation are observed in male germ cells of H1a/H1t double knockout mice, further demonstrating that H1a and H1t are dispensable for chromatin condensation and structural changes during spermatogenesis [135]. In H1a−/−H1t−/− male germ cells, major somatic H1 variants, H1c, H1d and H1e, are markedly upregulated, suggesting potential roles of somatic H1s in spermatogenesis and in compensating for the loss of H1s specific for or enriched in spermatogenesis.
The other two testis-specific H1 variants, H1T2 and HILS1, are most distantly related and least conserved H1 variants (Table 2). The expression of H1T2 and HILS1 is confined to spermatids (Fig. 3) [26–29]. H1T2 is present in the nuclei of early spermatids and has a particular polarized localization in round and elongating spermatids, associated with chromatin at the apical pole of the spermatid nucleus [26]. The loss of H1T2 in mice severely impairs chromatin condensation and the morphological transformation of round spermatids [26, 28]. These results demonstrate that H1T2 is required for normal spermiogenesis and plays a critical role in chromatin condensation and in establishing cell polarity of elongating spermatids [26, 28]. H1T2 has been found to interact with protamines and its elimination leads to a decrease in protamine content, suggesting a functional role of H1T2 in histone-to-protamine transition [28]. H1T2 and the histone methyltransferase Ezh2 have been found to interact and colocalize at the apical poles of round spermatids, which may suggest a role of H1T2 in chromatin remodeling through regulating histone H3K27 methylation during spermatid elongation [136].
HILS1 is expressed in early and elongated spermatids, and its nuclear localization largely overlaps with that of transition proteins and protamines (Fig. 3) [27]. HILS1 has a higher DNA binding affinity than H1T2, which may contribute to the increased chromatin condensation in late spermatids [28]. No knockout study has been reported for HILS1. Nonetheless, decreased expression of HILS1 in sperms was found to be associated with reduced sperm mobility in human males, offering a potential link between HILS1 and male fertility [137].
Similar to spermatogenesis, the maternal germline stem cells, oogonia, go through mitotic and meiotic phases to produce fertilizable eggs. However, oogenesis does not involve a spectacular chromatin condensation process as observed in spermatogenesis. Instead, oocytes sustain a chromatin structure much like somatic cells. Oogonia express a complement of somatic H1s, including H1a-e and H10. Starting from the meiotic phase, H1a-e are gradually replaced by the oocyte-specific H1oo (Fig. 2) [24, 101]. The aforementioned unique expression pattern and properties of H1oo suggest that it may have a special role in organizing chromatin structure during oocyte maturation. Indeed, knockdown of H1oo expression by microinjection of antisense morpholino oligonucleotides against H1oo into mouse oocytes or injection of H1oo siRNA into bovine oocytes dramatically impairs first polar body extrusion and meiotic maturation of germinal vesicle stage oocytes in vitro [138, 139]. Such defects can be rescued by H1oo, but not somatic H1s, indicating the requirement of H1oo for proper meiotic progression during oogenesis [138, 139]. However, H1oo−/− mice do not exhibit obvious phenotypes [98, 99], demonstrating that the loss of H1oo can be tolerated in vivo during oogenesis (Table 3). Further examination of chromatin and meiosis maturation of oocytes from H1oo−/− mice will help elucidate processes and regulatory mechanisms of chromatin remodeling during mammalian oogenesis.
Although H10 is retained throughout oogenesis [100–102], the role of H10 in this process is not clear. H10 null mice are fertile [103], suggesting that H10 is dispensable. As discussed above for other developmental stages, the lack of phenotypes in single H1 knockouts is likely due to the compensation by the remaining somatic H1 variants. The mRNA transcripts of somatic H1s are present in meiotic oocytes and persist until newly fertilized embryos [100, 101], serving as a reservoir for H1 synthesis before zygotic genome activation in early embryos.
In summary, gametogenesis involves a series of chromatin and morphological reorganization steps to produce fertilizable germ cells. The replacement of somatic H1s with germ cell-specific counterparts suggests that germ cell-specific H1s play a critical role in sperm and oocyte production. Given their distinct biochemical properties from somatic H1s, germ cell-specific H1 variants are likely to mediate or participate in the dramatic chromatin remodeling processes occurring in gametogenesis. However, their exact roles in these processes and early embryogenesis remain underexplored and warrant further investigation.
3. Role of linker histone H1 in stem cell differentiation
Stem cells have the remarkable potential to give rise to different cell types in mammalian tissues. According to their differentiation capacity, stem cells can be categorized into two major groups: pluripotent stem cells such as embryonic stem cells (ESCs) derived from the inner cell mass of a blastocyst, which have the potential to differentiate into all cell types in the body, and adult stem cells, which exist in specific tissues and are capable to generate certain cell types in the tissues. Stem cells are fundamental to embryonic development and tissue generation and can serve as great in vitro systems for studying the regulatory mechanisms controlling these processes. Progenitor cells, the early descendants of adult stem cells, have limited capacity in self-renewal and differentiation. Recent studies have uncovered linker histones as important regulators for differentiation of embryonic stem cells and progenitor cells.
Pluripotent stem cells have an open chromatin state with globally hyperactive transcription and undergo chromatin condensation upon differentiation, rendering the chromatin less permissive to transcription [140–142]. Differentiation of pluripotent stem cells involves dramatic molecular changes, including silencing of pluripotency genes and activation of lineage-specific genes [143], as well as global chromatin remodeling and epigenetic reprogramming [144, 145]. FRAP studies have shown that ESCs are characterized by hyperdynamic binding of chromatin proteins, such as linker histones and heterochromatin protein 1 (HP1), and that ESC differentiation is accompanied by a reduction in the mobility of these proteins [141, 146]. The rapid exchange of H1 proteins appears to be required for ESC differentiation to proceed, as overexpression of an H10 protein with a tandem duplication of CTD for restricting its mobility perturbed ESC differentiation [141]. It is surmised that hyperdynamic binding of H1 and other chromatin proteins provides a “breathing” window rendering the chromatin more accessible to remodeling factors in response to differentiation signals [141]. The high H1 mobility in ESCs may be achieved through citrullination, which neutralizes positive charges and results in a reduced binding affinity for chromatin. Peptidylarginine deiminase 4 (PADI4) catalyzes citrullination on arginine 54 within the globular domain of H1c (H1.2), H1d (H1.3) and H1e (H1.4). PADI4 expression is restricted to pluripotent stem cells and necessary to maintain pluripotency transcriptional network [147]. These findings suggest that H1 dynamics plays a fundamental role in regulating chromatin plasticity during ESC differentiation.
The role of H1 dynamics in modulating differentiation is not exclusive to pluripotent stem cells, but has also been shown in murine erythroleukemia (MEL) cells, an excellent model for studying terminal cell differentiation [148]. Concomitant with MEL differentiation is the dramatic increase in H10 and H1c levels [149] as well as the increased chromatin binding affinity of all histone H1 variants [150]. H1 phosphorylation by cyclin-dependent kinase (CDK) destabilizes H1-chromatin interactions and reduces its binding to chromatin [48, 151, 152]. Overexpression of H10 mutants mimicking the constitutively phosphorylated H1 sufficiently inhibits MEL differentiation, demonstrating that dephosphorylation of H1 is necessary for erythroid differentiation. It is proposed that H1 phosphorylation in MEL cells may cause an alteration in local chromatin structure without the displacement of H1 from chromatin as observed for H1 citrullination in ESCs [150].
During ESC differentiation, not only is the mobility of H1 reduced, but also the level of total H1 is progressively increased. ESCs have a total H1-to-nucleosome ratio of around 0.46, compared with that above 0.7 in differentiated cells [13, 15, 153]. During embryoid body (EB) differentiation of ESCs, the H1-to-nucleosome ratio increases to 0.62 (Table 3) [16]. The increase in the total H1 level during ESC differentiation is due to the elevation in protein levels of H1c, H1d, H1e and H10 [16], which likely contributes to chromatin condensation. The dynamic changes in H1 expression suggest histone H1 and its variants as important modulators of cellular differentiation. However, deletion of H1c, H1d, H1e, or H10, despite a slight decrease in the total H1 level in mouse ESCs, does not impair ESC differentiation (Table 3) ([154] and Pan, Zhang, Fan unpublished observations). This is consistent with the findings from single H1 knockout studies that individual H1 variants are not required for mammalian development (see Section 2.1). Nevertheless, H10 (H1.0) knockdown in human ESCs impairs the induction of HNF4, Sox17 and FoxA2, genes associated with differentiation [106], suggesting a potential role of H10 in regulating specific genes during human ESC differentiation.
The important role of H1 in ESC differentiation has been demonstrated in H1c/H1d/H1e triple null ESCs. These ESCs, derived from H1c/H1d/H1e triple knockout (H1 TKO) blastocysts, have 50% reduction in the total H1 level, offering a useful system to test the necessity of H1 in ESC differentiation [16, 105, 153]. The loss of H1c, H1d and H1e does not appear to affect self-renewal or expression of pluripotency factors in undifferentiated ESCs, and H1 TKO ESCs display growth rate, cell morphology and overall transcriptome characteristic of WT ESCs [16, 153]. However, H1 TKO ESCs are more resistant to spontaneous differentiation, and H1 TKO EBs are impaired in producing cells and morphologies of three primitive germ layers (Table 3) [16]. When induced for neural differentiation, putative H1 TKO EBs display severe defects in neurite outgrowth and the induction of neural lineage specific genes. A common theme in the various differentiation schemes is the failure in effective repression of pluripotency genes such as Oct4 and Nanog in H1 TKOs, which also occurs in vivo in H1 TKO embryos [16]. H1 TKO ESCs are dysregulated in specification of mesendoderm and neuroectoderm [105], further suggesting specific effects of H1 in these lineages. The fact that H1 occupancy at the promoter region of the pluripotency gene Oct4 increases during ESC differentiation and that the loss of H1 impairs DNA methylation and changes of histone marks at this region necessary for Oct4 silencing suggests H1 as a critical regulator for epigenetic silencing of Oct4 [16, 106]. Thus, the effects of H1 on the silencing of pluripotency genes are likely to be direct.
Other evidence establishes specific H1 variants as important regulators for differentiation of progenitor cells. During terminal differentiation of retinal cells in mice, the expression of H1c, along with H10 and H1e, is dramatically increased, contributing to the highly compact heterochromatin and nuclear architecture in mouse rod cells [117]. Although the loss of H1c does not lead to abnormalities in rod cell differentiation, H1c/H1e double knockouts and H10/H1c/H1e triple knockouts displayed defects in rod development and rod cell packing, presumably due to the impairment in the terminal differentiation of rod photoreceptors.
While aforementioned studies suggest the necessity of a sufficient total H1 level for proper differentiation, H1b has been found to participate in the inhibition of myogenesis during myoblast differentiation [90, 155]. H1b is specifically recruited by the homeoprotein Msx1 to the core enhancer region of the MyoD gene, which establishes a repressive chromatin state at the MyoD gene locus and inhibits the expression of MyoD and myogenic differentiation of myoblast C2C12 cells [155]. Knockdown of H1b by shRNA abolishes the Msx1-mediated repression of myogenic differentiation, and coexpression of Msx1 and H1b in Xenopus embryos synergistically inhibits Xenopus MyoD [155]. These results are reminiscent of the previous findings in Xenopus embryos where inhibition of the accumulation of Xenopus H1c (xH1c), the ortholog of mouse H1b, by ribozymes, leads to the failure in loss of mesodermal competence and the lack of MyoD silencing [90]. The effects appear to be specific to H1b, as injection of H1e into Xenopus embryos does not inhibit MyoD expression [155].
4. Potential mechanisms of histone H1 in the regulation of mammalian development and stem cell differentiation
The broad defects in mammalian development and stem cell differentiation uncovered through H1 manipulation as discussed above suggest that H1 plays crucial and versatile roles in these processes. While the exact mechanisms underlying the regulatory roles of H1 in these processes remain to be explored, a few potential mechanisms can be gleaned from the studies of functions of H1 in chromatin structure and gene expression.
Chromatin structure undergoes dramatic changes during development and cellular differentiation. As a key architectural chromatin protein, H1 modulates the bulk chromatin structure in various aspects, such as chromatin condensation, nucleosome repeat length (NRL), histone modifications and chromatin dynamicity. The H1 stoichiometry is positively correlated with nucleosome repeat length (NRL) (reviewed in ref. [15]), both of which are key determinants of the degree of chromatin compaction [156]. Higher H1 content leads to a longer NRL and more compact chromatin fibers, while H1 depletion causes reduction in NRL and chromatin decondensation [13, 109, 117, 153, 157]. The H1 content progressively increases during stem cell differentiation or terminal cell differentiation, as shown in the total H1-to-nucleosome ratio changing from 0.45 in ESCs to 0.62 in differentiating EBs, or from 1.02 in new born retina to 1.25 in adult retinal cells (Fig. 2, Table 3) [16, 117]. H1 stoichiometry regulates histone modifications in bulk chromatin as shown for specific histone methylations and histone acetylation [153, 158, 159]. The H1 level is also pivotal for promoting heterochromatin formation, such as the establishment and proper function of pericentric heterochromatin and chromocenter clustering [86, 109, 159–162].
The amount and modifications of H1 histones can regulate chromatin dynamicity. Chromatin in pluripotent stem cells is found to be hyperdynamic [141]. The proper level of chromatin flexibility and dynamicity seems to be necessary for chromatin remodeling during development and cellular differentiation, as both the increase of H1 binding affinity in ESCs (by overexpression of the H10 mutant with two C-terminal domains) and the failure to increase H1 binding by dephosphorylation during MEL differentiation (by overexpression of a H10 mutant mimicking the constitutively phosphorylated H10) inhibit cellular differentiation in respective mouse ESCs and erythroleukemia cells [141, 146, 150]. In addition, H1 depletion reduces chromatin flexibility in gene-rich topologically associating domains (TADs) [163].
It is not determined if any or all of the aspects of bulk chromatin structure per se are responsible for the defects in development and cell differentiation displayed in compound H1 knockouts. The lack of overall phenotype in vivo in single knockouts, combined with increasingly more severe phenotypes caused by more dramatic depletion of H1 in compound knockouts, establishes a compensatory effect among the H1 family and a common role of H1 variants in development and cellular differentiation. These results, together with the studies showing the necessity of H1 in chromatin architecture and assembly of mitotic chromosomes as well as the roles of H1 in pericentromeric heterochromatin function and proper chromocenter clustering [86, 97, 109, 164], indicate that H1 s regulatory roles in bulk chromatin structure could contribute to general cell functionality required for robust development and proper cell differentiation.
In contrast to its global effects on bulk chromatin, H1 plays rather specific roles in gene regulation in vivo. Even though early studies with in vitro constituted chromatin indicate a general repressive role of H1 for transcription by all three types of RNA polymerases (reviewed in [165, 166]), depletion of H1 in a variety of organisms and cells shows limited gene expression changes in vivo [81, 84, 135, 153, 167]. rRNA genes, transgenes, major satellites and genes at pericentromeric regions, imprinted genes, sex chromosome genes and pluripotency genes, as well as developmental genes important for embryogenesis and gametogenesis, have been characterized as target genes of H1 in various systems [16, 85, 88, 90, 91, 109, 153–155, 168–171]. Interestingly, these genes are often subjected to epigenetic regulation.
Indeed, recent studies have established H1 as an important epigenetic factor for gene regulation. Genes affected by triple deletion of H1c/H1d/H1e in ESCs are enriched in genes normally regulated by DNA methylation, and H1 depletion leads to hypomethylation at the regulatory regions of these H1-regulated genes, such as the imprinting control regions (ICR) of H19/Igf2 and Gtl2/Dlk loci in ESCs [153]. These results reveal a novel link between H1 and DNA methylation, which was also strengthened by studies on the immunoglobulin heavy chain locus [172], the Rhox gene cluster [170] and the pluripotency gene Oct4 locus [154]. The identification of DNA methyltransferases DNMT1 and DNMT3B as H1 interacting partners further provides a mechanism for H1 regulating DNA methylation through direct recruitment of DNMTs [173, 174]. Likewise, H1 has been shown in various studies to regulate histone modifications by interaction with histone modifying enzymes, including PRC2-Ezh2 polycomb complex, which catalyzes H3K27 di- and tri- methylation [175–178], SirT1, a histone deacetylase preferentially targeting to H3K9Ac and H4K16Ac [179, 180], and Cul4A, an E3 ubiquitin ligase responsible for H4K31 ubiquitination [62]. H1 occupancy can also prevent the binding of certain histone modifying enzymes to specific regulatory loci [174]. Nevertheless, given the general non-DNA-sequence-dependent nature for the binding of H1, DNMTs, and histone modifying enzymes to DNA, how the specificity is achieved at these target gene loci remains elusive. It is noteworthy that many of these H1-regulated genes, such as imprinted genes, are particularly sensitive to local changes in chromatin structure and epigenetic modifications, with a lower threshold for eliciting changes in gene expression and epigenetic states.
H1 variants may also participate in specific gene regulation in a variant-specific manner. As mentioned earlier, H1 variants have distinct expression patterns during development and cell differentiation. They differ from each other in amino acid sequence, biochemical property and binding affinity for chromatin, which may confer specificity in chromatin compaction, protein interactions and gene regulation (See Section 1). While genome-wide mapping studies of H1 variants categorize H1 binding as a repressive epigenetic mark by its positive correlation with repressive histone marks, such as H3K9me3, and negative correlation with active histone marks, such as H3K4me3, different H1 variants do have distinct enriched regions despite their overall co-localization and common binding patterns at the genome-wide scale [109, 181, 182]. Indeed, it has been shown that in vivo occupancy of H1a (H1.1) and H1d (H1.3) differ dramatically at the regulatory region of H19 gene [171], suggesting that distinct binding by different H1 variants could contribute to the establishment and maintenance of epigenetic patterns and gene regulation at specific loci.
H1 variants may interact with transcription factors, resulting in specific gene regulation of the target genes of those regulatory proteins. The best examples come from studies on H1c (H1.2) and H1b (H1.5). H1c was found to repress p53 target genes in HeLa cells through its interaction with p53, a group of cofactors, and ribosomal proteins [61]. During early embryogenesis, H1c forms a complex with CHD8 to suppress p53-mediated transcription and apoptosis [169]. H1b interacts with the homeoprotein Msx1 to repress myoD expression, thus inhibiting myogenic differentiation in myoblast C2C12 cells and mesodermal competence in Xenopus animal caps [90, 155]. On the other hand, H1b was found to bind forkhead box transcription factor FoxP3 in human regulatory T (Treg) cells to regulate the expression of FoxP3 target genes, such as IL2 and CTL4, thus modulating Treg function [183]. In the human promonocytic THP-1 cells, H1b binds to NF-κB factor RelB and forms a repressor complex at the promoters of TNF-α and IL-1β, silencing these proinflammatory genes [184]. These results suggest that H1 variants regulate specific gene expression in a context dependent manner.
A variety of other specific roles of H1 variants in cellular processes in mammalian cells have been identified. For instance, H10 (H1.0) was found to preferentially interact with a group of proteins in the nucleolus, indicating a specific role of H10 in rRNA biogenesis and mRNA metabolism [119]. Other than the aforementioned role of H1 in apoptosis through regulating p53 and CHD8 target genes, H1c was shown to translocate from the nucleus to the cytoplasm and stimulate cytochrome c release from mitochondria upon DSB damage, thus acting as a signal transducer in apoptosis induced by DSB [120]. More recently, H1c and H1x have been found to be ubiquitinated at K63 and that H1 is a key target for E3 ubiquitin ligase RNF8 and E2 ubiquitin-conjugating enzyme UBC13 in DSB signaling [185]. In addition, H1 may stimulate non-homologous end-joining and homologous recombination mediated repair after DNA damage [186, 187].
Most of the mechanistic studies mentioned in this section were carried out in cell lines, which are more amiable for genetic manipulation and environmental stimuli. It is not yet clear whether any or all of the specific roles of H1 variants discussed here are responsible for the phenotypes observed in mammalian development and stem cell differentiation in compound H1 knockouts. Future studies in in vivo setting to identify the key regulatory pathways that are responsible for the defects in compound H1 knockouts should provide new insights.
5. Concluding remarks and future directions
During the past decades, we have seen an enormous amount of advances in the understanding of chromatin structure and function, especially in connection with chromatin modifying activities and chromatin remodeling complexes. Compared with core histones, linker histones have not drawn adequate attention and thus remain underexplored [57]. Investigating the role of H1 variants in mammalian development and stem cell differentiation in vivo as well as the underlying mechanisms has been challenging partly due to the complexity/heterogeneity and the compensatory effects of mammalian H1 variants and the lack of antibodies of high affinity and specificity for H1 variants and their post-translational modifications. However, a plethora of recent studies as summarized in this review have shed significant new insights on the role of H1 in these processes.
We have focused our discussion on the pivotal roles of H1 in mammalian development, including embryogenesis, tissue maturation and gametogenesis, and also reviewed its role in stem cell differentiation with connection to development. These studies establish that the total H1 level, rather than individual H1s, is critical for viability and robust development and required for proper ESC differentiation. There is significant redundancy and compensation among somatic H1 variants during development. While the regulatory circuit governing the compensation among somatic H1s remains poorly understood, such redundancy suggest commonality in their structural and functional roles at the genome-wide scale, which is also supported by their largely overlapping genomic localization in multiple cell types [109, 181, 182]. On the other hand, the high sequence conservation and the exquisite differential expression patterns of H1 variants are indicative of an amazing evolutionary adaption. The presence of multiple H1 variants may also provide a powerful cushion for potential perturbations in chromatin milieu.
Disruption of the compensation among somatic H1s results in global effects on chromatin structure but specific gene expression changes [13, 16, 103, 107, 108, 132–135, 154]. The mechanisms underlying the global vs. specific effects regulated by H1 are important avenues to pursue. New links between H1 and other epigenetic regulatory mechanisms are likely to emerge from future studies. In addition, identification of key H1 target genes and the signaling pathways involved in physiological and pathological contexts should lead to new insights into the role of H1.
Given their differences in biochemical properties, interaction partners, chromatin binding affinity and spatial temporal expression patterns, H1 variants are bound to have distinct functions, probably in a specific and context-dependent manner, in development and cellular differentiation. These differential properties of H1 variants offer a means to fine-tune the regulation of chromatin structures and gene expression, culminated in optimal physiological performance and response to environmental stress. Indeed, such variant-specific functions may be especially important in diseases, such as cancers and immunological diseases, as nicely illustrated by the prominent effects arising from manipulation of H1 variants in cancer cells and the identification of H1 variant mutations as driver mutations in tumors [167, 171, 188–191]. Future research aimed at H1 variant-specific functions in the context of various diseases should provide novel insights into the role of H1 variants in mammalian development and cellular differentiation.
Highlights.
The properties and expression patterns of mammalian H1 variants are reviewed.
H1 plays fundamental roles in embryogenesis, tissue maturation and gametogenesis.
H1 is a key regulator for stem cell differentiation.
Potential mechanisms of H1 in regulating development and cell differentiation are discussed.
The role of H1 in epigenetic gene regulation is highlighted.
Acknowledgments
We apologize to all colleagues whose work could not be directly cited here due to restrictions on references and space limitations. This work is in part supported by NIH grant GM085261 (Y. Fan), Georgia Research Alliance Distinguished Cancer Scientists Award (Y. Fan), Chen Fellowship, NSF Science and Technology Center Emergent Behaviors of Integrated Cellular Systems (EBICS; CBET0939511), and Georgia Institute of Technology.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Wolffe AP, Kurumizaka H. The nucleosome: a powerful regulator of transcription. Progress in nucleic acid research and molecular biology. 1998;61:379–422. doi: 10.1016/s0079-6603(08)60832-6. [DOI] [PubMed] [Google Scholar]
- 2.Davey CA, Sargent DF, Luger K, Maeder AW, Richmond TJ. Solvent mediated interactions in the structure of the nucleosome core particle at 1.9 a resolution. J Mol Biol. 2002;319:1097–1113. doi: 10.1016/S0022-2836(02)00386-8. [DOI] [PubMed] [Google Scholar]
- 3.Finch JT, Klug A. Solenoidal model for superstructure in chromatin. Proc Natl Acad Sci U S A. 1976;73:1897–1901. doi: 10.1073/pnas.73.6.1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thoma F, Koller T, Klug A. Involvement of histone H1 in the organization of the nucleosome and of the salt-dependent superstructures of chromatin. J Cell Biol. 1979;83:403–427. doi: 10.1083/jcb.83.2.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wolffe AP. Histone H1. Int J Biochem Cell Biol. 1997;29:1463–1466. doi: 10.1016/s1357-2725(97)00026-5. [DOI] [PubMed] [Google Scholar]
- 6.Olins AL, Olins DE. Spheroid chromatin units (v bodies) Science. 1974;183:330–332. doi: 10.1126/science.183.4122.330. [DOI] [PubMed] [Google Scholar]
- 7.Brown DT. Histone variants: are they functionally heterogeneous? Genome Biol. 2001;2:REVIEWS0006. doi: 10.1186/gb-2001-2-7-reviews0006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Parseghian MH, Hamkalo BA. A compendium of the histone H1 family of somatic subtypes: an elusive cast of characters and their characteristics. Biochem Cell Biol. 2001;79:289–304. [PubMed] [Google Scholar]
- 9.Seyedin SM, Kistler WS. H1 histone subfractions of mammalian testes. 1. Organ specificity in the rat. Biochemistry. 1979;18:1371–1375. doi: 10.1021/bi00574a038. [DOI] [PubMed] [Google Scholar]
- 10.Albig W, Meergans T, Doenecke D. Characterization of the H1.5 gene completes the set of human H1 subtype genes. Gene. 1997;184:141–148. doi: 10.1016/s0378-1119(96)00582-3. [DOI] [PubMed] [Google Scholar]
- 11.Parseghian MH, Henschen AH, Krieglstein KG, Hamkalo BA. A proposal for a coherent mammalian histone H1 nomenclature correlated with amino acid sequences. Protein science: a publication of the Protein Society. 1994;3:575–587. doi: 10.1002/pro.5560030406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Talbert PB, Ahmad K, Almouzni G, Ausio J, Berger F, Bhalla PL, Bonner WM, Cande WZ, Chadwick BP, Chan SW, Cross GA, Cui L, Dimitrov SI, Doenecke D, Eirin-Lopez JM, Gorovsky MA, Hake SB, Hamkalo BA, Holec S, Jacobsen SE, Kamieniarz K, Khochbin S, Ladurner AG, Landsman D, Latham JA, Loppin B, Malik HS, Marzluff WF, Pehrson JR, Postberg J, Schneider R, Singh MB, Smith MM, Thompson E, Torres-Padilla ME, Tremethick DJ, Turner BM, Waterborg JH, Wollmann H, Yelagandula R, Zhu B, Henikoff S. A unified phylogeny-based nomenclature for histone variants. Epigenetics & chromatin. 2012;5:7. doi: 10.1186/1756-8935-5-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fan Y, Nikitina T, Morin-Kensicki EM, Zhao J, Magnuson TR, Woodcock CL, Skoultchi AI. H1 linker histones are essential for mouse development and affect nucleosome spacing in vivo. Mol Cell Biol. 2003;23:4559–4572. doi: 10.1128/MCB.23.13.4559-4572.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang ZF, Sirotkin AM, Buchold GM, Skoultchi AI, Marzluff WF. The mouse histone H1 genes: gene organization and differential regulation. J Mol Biol. 1997;271:124–138. doi: 10.1006/jmbi.1997.1166. [DOI] [PubMed] [Google Scholar]
- 15.Woodcock CL, Skoultchi AI, Fan Y. Role of linker histone in chromatin structure and function: H1 stoichiometry and nucleosome repeat length. Chromosome Res. 2006;14:17–25. doi: 10.1007/s10577-005-1024-3. [DOI] [PubMed] [Google Scholar]
- 16.Zhang Y, Cooke M, Panjwani S, Cao K, Krauth B, Ho PY, Medrzycki M, Berhe DT, Pan C, McDevitt TC, Fan Y. Histone h1 depletion impairs embryonic stem cell differentiation. PLoS Genet. 2012;8:e1002691. doi: 10.1371/journal.pgen.1002691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Khochbin S. Histone H1 diversity: bridging regulatory signals to linker histone function. Gene. 2001;271:1–12. doi: 10.1016/s0378-1119(01)00495-4. [DOI] [PubMed] [Google Scholar]
- 18.Godde JS, Ura K. Dynamic alterations of linker histone variants during development. Int J Dev Biol. 2009;53:215–224. doi: 10.1387/ijdb.082644jg. [DOI] [PubMed] [Google Scholar]
- 19.Perez-Montero S, Carbonell A, Azorin F. Germline-specific H1 variants: the “sexy” linker histones. Chromosoma. 2015 doi: 10.1007/s00412-015-0517-x. [DOI] [PubMed] [Google Scholar]
- 20.Lennox RW, Cohen LH. The histone H1 complements of dividing and nondividing cells of the mouse. J Biol Chem. 1983;258:262–268. [PubMed] [Google Scholar]
- 21.Doenecke D, Albig W, Bouterfa H, Drabent B. Organization and expression of H1 histone and H1 replacement histone genes. Journal of cellular biochemistry. 1994;54:423–431. doi: 10.1002/jcb.240540409. [DOI] [PubMed] [Google Scholar]
- 22.Zlatanova J, Doenecke D. Histone H1 zero: a major player in cell differentiation? FASEB J. 1994;8:1260–1268. doi: 10.1096/fasebj.8.15.8001738. [DOI] [PubMed] [Google Scholar]
- 23.Stoldt S, Wenzel D, Schulze E, Doenecke D, Happel N. G1 phase-dependent nucleolar accumulation of human histone H1x. Biology of the cell/under the auspices of the European Cell Biology Organization. 2007;99:541–552. doi: 10.1042/bc20060117. [DOI] [PubMed] [Google Scholar]
- 24.Tanaka M, Hennebold JD, Macfarlane J, Adashi EY. A mammalian oocyte-specific linker histone gene H1oo: homology with the genes for the oocyte-specific cleavage stage histone (cs-H1) of sea urchin and the B4/H1M histone of the frog. Development. 2001;128:655–664. doi: 10.1242/dev.128.5.655. [DOI] [PubMed] [Google Scholar]
- 25.Lennox RW, Cohen LH. The alterations in H1 histone complement during mouse spermatogenesis and their significance for H1 subtype function. Dev Biol. 1984;103:80–84. doi: 10.1016/0012-1606(84)90009-5. [DOI] [PubMed] [Google Scholar]
- 26.Martianov I, Brancorsini S, Catena R, Gansmuller A, Kotaja N, Parvinen M, Sassone-Corsi P, Davidson I. Polar nuclear localization of H1T2, a histone H1 variant, required for spermatid elongation and DNA condensation during spermiogenesis. Proc Natl Acad Sci U S A. 2005;102:2808–2813. doi: 10.1073/pnas.0406060102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yan W, Ma L, Burns KH, Matzuk MM. HILS1 is a spermatid-specific linker histone H1-like protein implicated in chromatin remodeling during mammalian spermiogenesis. Proc Natl Acad Sci U S A. 2003;100:10546–10551. doi: 10.1073/pnas.1837812100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tanaka H, Iguchi N, Isotani A, Kitamura K, Toyama Y, Matsuoka Y, Onishi M, Masai K, Maekawa M, Toshimori K, Okabe M, Nishimune Y. HANP1/H1T2, a novel histone H1-like protein involved in nuclear formation and sperm fertility. Mol Cell Biol. 2005;25:7107–7119. doi: 10.1128/MCB.25.16.7107-7119.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Iguchi N, Tanaka H, Yomogida K, Nishimune Y. Isolation and characterization of a novel cDNA encoding a DNA-binding protein (Hils1) specifically expressed in testicular haploid germ cells. International journal of andrology. 2003;26:354–365. doi: 10.1046/j.0105-6263.2003.00449.x. [DOI] [PubMed] [Google Scholar]
- 30.Iguchi N, Tanaka H, Yamada S, Nishimura H, Nishimune Y. Control of mouse hils1 gene expression during spermatogenesis: identification of regulatory element by transgenic mouse. Biol Reprod. 2004;70:1239–1245. doi: 10.1095/biolreprod.103.024760. [DOI] [PubMed] [Google Scholar]
- 31.Whitfield ML, Zheng LX, Baldwin A, Ohta T, Hurt MM, Marzluff WF. Stem-loop binding protein, the protein that binds the 3′ end of histone mRNA, is cell cycle regulated by both translational and posttranslational mechanisms. Mol Cell Biol. 2000;20:4188–4198. doi: 10.1128/mcb.20.12.4188-4198.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marzluff WF, Gongidi P, Woods KR, Jin J, Maltais LJ. The human and mouse replication-dependent histone genes. Genomics. 2002;80:487–498. [PubMed] [Google Scholar]
- 33.Ling J, Morley SJ, Pain VM, Marzluff WF, Gallie DR. The histone 3′-terminal stem-loop-binding protein enhances translation through a functional and physical interaction with eukaryotic initiation factor 4G (eIF4G) and eIF3. Mol Cell Biol. 2002;22:7853–7867. doi: 10.1128/MCB.22.22.7853-7867.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gorgoni B, Andrews S, Schaller A, Schumperli D, Gray NK, Muller B. The stem-loop binding protein stimulates histone translation at an early step in the initiation pathway. RNA. 2005;11:1030–1042. doi: 10.1261/rna.7281305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Allard P, Yang Q, Marzluff WF, Clarke HJ. The stem-loop binding protein regulates translation of histone mRNA during mammalian oogenesis. Dev Biol. 2005;286:195–206. doi: 10.1016/j.ydbio.2005.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cheng GH, Nandi A, Clerk S, Skoultchi AI. Different 3′-end processing produces two independently regulated mRNAs from a single H1 histone gene. Proc Natl Acad Sci U S A. 1989;86:7002–7006. doi: 10.1073/pnas.86.18.7002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang ZF, Krasikov T, Frey MR, Wang J, Matera AG, Marzluff WF. Characterization of the mouse histone gene cluster on chromosome 13: 45 histone genes in three patches spread over 1Mb. Genome Res. 1996;6:688–701. doi: 10.1101/gr.6.8.688. [DOI] [PubMed] [Google Scholar]
- 38.Albig W, Doenecke D. The human histone gene cluster at the D6S105 locus. Human genetics. 1997;101:284–294. doi: 10.1007/s004390050630. [DOI] [PubMed] [Google Scholar]
- 39.Albig W, Kioschis P, Poustka A, Meergans K, Doenecke D. Human histone gene organization: nonregular arrangement within a large cluster. Genomics. 1997;40:314–322. doi: 10.1006/geno.1996.4592. [DOI] [PubMed] [Google Scholar]
- 40.Volz A, Albig W, Doenecke D, Ziegler A. Physical mapping of two histone gene clusters on human chromosome 6p22.1-22.2. DNA sequence: the journal of DNA sequencing and mapping. 1997;8:173–179. doi: 10.3109/10425179709034070. [DOI] [PubMed] [Google Scholar]
- 41.Chapman GE, Hartman PG, Bradbury EM. Studies on the role and mode of operation of the very-lysine-rich histone H1 in eukaryote chromatin. The isolation of the globular and non-globular regions of the histone H1 molecule. Eur J Biochem. 1976;61:69–75. doi: 10.1111/j.1432-1033.1976.tb09998.x. [DOI] [PubMed] [Google Scholar]
- 42.Allan J, Hartman PG, Crane-Robinson C, Aviles FX. The structure of histone H1 and its location in chromatin. Nature. 1980;288:675–679. doi: 10.1038/288675a0. [DOI] [PubMed] [Google Scholar]
- 43.Ramakrishnan V, Finch JT, Graziano V, Lee PL, Sweet RM. Crystal structure of globular domain of histone H5 and its implications for nucleosome binding. Nature. 1993;362:219–223. doi: 10.1038/362219a0. [DOI] [PubMed] [Google Scholar]
- 44.Drabent B, Franke K, Bode C, Kosciessa U, Bouterfa H, Hameister H, Doenecke D. Isolation of two murine H1 histone genes and chromosomal mapping of the H1 gene complement. Mammalian genome: official journal of the International Mammalian Genome Society. 1995;6:505–511. doi: 10.1007/BF00356166. [DOI] [PubMed] [Google Scholar]
- 45.Yamamoto T, Horikoshi M. Cloning of the cDNA encoding a novel subtype of histone H1. Gene. 1996;173:281–285. doi: 10.1016/0378-1119(96)00020-0. [DOI] [PubMed] [Google Scholar]
- 46.Happel N, Schulze E, Doenecke D. Characterisation of human histone H1x. Biol Chem. 2005;386:541–551. doi: 10.1515/BC.2005.064. [DOI] [PubMed] [Google Scholar]
- 47.Allan J, Mitchell T, Harborne N, Bohm L, Crane-Robinson C. Roles of H1 domains in determining higher order chromatin structure and H1 location. J Mol Biol. 1986;187:591–601. doi: 10.1016/0022-2836(86)90337-2. [DOI] [PubMed] [Google Scholar]
- 48.Hendzel MJ, Lever MA, Crawford E, Th’ng JP. The C-terminal domain is the primary determinant of histone H1 binding to chromatin in vivo. J Biol Chem. 2004;279:20028–20034. doi: 10.1074/jbc.M400070200. [DOI] [PubMed] [Google Scholar]
- 49.Luque A, Collepardo-Guevara R, Grigoryev S, Schlick T. Dynamic condensation of linker histone C-terminal domain regulates chromatin structure. Nucleic Acids Res. 2014;42:7553–7560. doi: 10.1093/nar/gku491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vyas P, Brown DT. N- and C-terminal domains determine differential nucleosomal binding geometry and affinity of linker histone isotypes H1(0) and H1c. J Biol Chem. 2012;287:11778–11787. doi: 10.1074/jbc.M111.312819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Clausell J, Happel N, Hale TK, Doenecke D, Beato M. Histone H1 subtypes differentially modulate chromatin condensation without preventing ATP-dependent remodeling by SWI/SNF or NURF. PLoS One. 2009;4:e0007243. doi: 10.1371/journal.pone.0007243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Orrego M, Ponte I, Roque A, Buschati N, Mora X, Suau P. Differential affinity of mammalian histone H1 somatic subtypes for DNA and chromatin. BMC biology. 2007;5:22. doi: 10.1186/1741-7007-5-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Th’ng JP, Sung R, Ye M, Hendzel MJ. H1 family histones in the nucleus. Control of binding and localization by the C-terminal domain. J Biol Chem. 2005;280:27809–27814. doi: 10.1074/jbc.M501627200. [DOI] [PubMed] [Google Scholar]
- 54.Catez F, Ueda T, Bustin M. Determinants of histone H1 mobility and chromatin binding in living cells. Nat Struct Mol Biol. 2006;13:305–310. doi: 10.1038/nsmb1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Happel N, Doenecke D. Histone H1 and its isoforms: contribution to chromatin structure and function. Gene. 2009;431:1–12. doi: 10.1016/j.gene.2008.11.003. [DOI] [PubMed] [Google Scholar]
- 56.Izzo A, Kamieniarz K, Schneider R. The histone H1 family: specific members, specific functions? Biol Chem. 2008;389:333–343. doi: 10.1515/BC.2008.037. [DOI] [PubMed] [Google Scholar]
- 57.Harshman SW, Young NL, Parthun MR, Freitas MA. H1 histones: current perspectives and challenges. Nucleic Acids Res. 2013;41:9593–9609. doi: 10.1093/nar/gkt700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.McBryant SJ, Lu X, Hansen JC. Multifunctionality of the linker histones: an emerging role for protein-protein interactions. Cell research. 2010;20:519–528. doi: 10.1038/cr.2010.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Szerlong HJ, Herman JA, Krause CM, DeLuca JG, Skoultchi A, Winger QA, Prenni JE, Hansen JC. Proteomic characterization of the nucleolar linker histone H1 interaction network. J Mol Biol. 2015;427:2056–2071. doi: 10.1016/j.jmb.2015.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ni JQ, Liu LP, Hess D, Rietdorf J, Sun FL. Drosophila ribosomal proteins are associated with linker histone H1 and suppress gene transcription. Genes Dev. 2006;20:1959–1973. doi: 10.1101/gad.390106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kim K, Choi J, Heo K, Kim H, Levens D, Kohno K, Johnson EM, Brock HW, An W. Isolation and characterization of a novel H1.2 complex that acts as a repressor of p53-mediated transcription. J Biol Chem. 2008;283:9113–9126. doi: 10.1074/jbc.M708205200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kim K, Lee B, Kim J, Choi J, Kim JM, Xiong Y, Roeder RG, An W. Linker Histone H1.2 cooperates with Cul4A and PAF1 to drive H4K31 ubiquitylation-mediated transactivation. Cell reports. 2013;5:1690–1703. doi: 10.1016/j.celrep.2013.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.An W, Leuba SH, van Holde K, Zlatanova J. Linker histone protects linker DNA on only one side of the core particle and in a sequence-dependent manner. Proc Natl Acad Sci U S A. 1998;95:3396–3401. doi: 10.1073/pnas.95.7.3396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhou YB, Gerchman SE, Ramakrishnan V, Travers A, Muyldermans S. Position and orientation of the globular domain of linker histone H5 on the nucleosome. Nature. 1998;395:402–405. doi: 10.1038/26521. [DOI] [PubMed] [Google Scholar]
- 65.Brown DT, Izard T, Misteli T. Mapping the interaction surface of linker histone H1(0) with the nucleosome of native chromatin in vivo. Nat Struct Mol Biol. 2006;13:250–255. doi: 10.1038/nsmb1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Fan L, Roberts VA. Complex of linker histone H5 with the nucleosome and its implications for chromatin packing. Proc Natl Acad Sci U S A. 2006;103:8384–8389. doi: 10.1073/pnas.0508951103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhou BR, Jiang J, Feng H, Ghirlando R, Xiao TS, Bai Y. Structural Mechanisms of Nucleosome Recognition by Linker Histones. Mol Cell. 2015;59:628–638. doi: 10.1016/j.molcel.2015.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lu X, Hamkalo B, Parseghian MH, Hansen JC. Chromatin condensing functions of the linker histone C-terminal domain are mediated by specific amino acid composition and intrinsic protein disorder. Biochemistry. 2009;48:164–172. doi: 10.1021/bi801636y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Becker M, Becker A, Miyara F, Han Z, Kihara M, Brown DT, Hager GL, Latham K, Adashi EY, Misteli T. Differential in vivo binding dynamics of somatic and oocyte-specific linker histones in oocytes and during ES cell nuclear transfer. Molecular biology of the cell. 2005;16:3887–3895. doi: 10.1091/mbc.E05-04-0350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wood C, Snijders A, Williamson J, Reynolds C, Baldwin J, Dickman M. Post-translational modifications of the linker histone variants and their association with cell mechanisms. The FEBS journal. 2009;276:3685–3697. doi: 10.1111/j.1742-4658.2009.07079.x. [DOI] [PubMed] [Google Scholar]
- 71.Kepert JF, Mazurkiewicz J, Heuvelman GL, Toth KF, Rippe K. NAP1 modulates binding of linker histone H1 to chromatin and induces an extended chromatin fiber conformation. J Biol Chem. 2005;280:34063–34072. doi: 10.1074/jbc.M507322200. [DOI] [PubMed] [Google Scholar]
- 72.Lusser A, Urwin DL, Kadonaga JT. Distinct activities of CHD1 and ACF in ATP-dependent chromatin assembly. Nat Struct Mol Biol. 2005;12:160–166. doi: 10.1038/nsmb884. [DOI] [PubMed] [Google Scholar]
- 73.De Lucia F, Faraone-Mennella MR, D’Erme M, Quesada P, Caiafa P, Farina B. Histone-induced condensation of rat testis chromatin: testis-specific H1t versus somatic H1 variants. Biochem Biophys Res Commun. 1994;198:32–39. doi: 10.1006/bbrc.1994.1005. [DOI] [PubMed] [Google Scholar]
- 74.Khadake JR, Rao MR. DNA- and chromatin-condensing properties of rat testes H1a and H1t compared to those of rat liver H1bdec; H1t is a poor condenser of chromatin. Biochemistry. 1995;34:15792–15801. doi: 10.1021/bi00048a025. [DOI] [PubMed] [Google Scholar]
- 75.Talasz H, Sapojnikova N, Helliger W, Lindner H, Puschendorf B. In vitro binding of H1 histone subtypes to nucleosomal organized mouse mammary tumor virus long terminal repeat promotor. J Biol Chem. 1998;273:32236–32243. doi: 10.1074/jbc.273.48.32236. [DOI] [PubMed] [Google Scholar]
- 76.Bharath MM, Chandra NR, Rao MR. Prediction of an HMG-box fold in the C-terminal domain of histone H1: insights into its role in DNA condensation. Proteins. 2002;49:71–81. doi: 10.1002/prot.10204. [DOI] [PubMed] [Google Scholar]
- 77.Caterino TL, Hayes JJ. Structure of the H1 C-terminal domain and function in chromatin condensation. Biochem Cell Biol. 2011;89:35–44. doi: 10.1139/O10-024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Pradeepa MM, Rao MR. Chromatin remodeling during mammalian spermatogenesis: role of testis specific histone variants and transition proteins. Society of Reproduction and Fertility supplement. 2007;63:1–10. [PubMed] [Google Scholar]
- 79.Ramesh S, Bharath MM, Chandra NR, Rao MR. A K52Q substitution in the globular domain of histone H1t modulates its nucleosome binding properties. FEBS Lett. 2006;580:5999–6006. doi: 10.1016/j.febslet.2006.09.061. [DOI] [PubMed] [Google Scholar]
- 80.Patterton HG, Landel CC, Landsman D, Peterson CL, Simpson RT. The biochemical and phenotypic characterization of Hho1p, the putative linker histone H1 of Saccharomyces cerevisiae. J Biol Chem. 1998;273:7268–7276. doi: 10.1074/jbc.273.13.7268. [DOI] [PubMed] [Google Scholar]
- 81.Hellauer K, Sirard E, Turcotte B. Decreased expression of specific genes in yeast cells lacking histone H1. J Biol Chem. 2001;276:13587–13592. doi: 10.1074/jbc.M011196200. [DOI] [PubMed] [Google Scholar]
- 82.Downs JA, Kosmidou E, Morgan A, Jackson SP. Suppression of homologous recombination by the Saccharomyces cerevisiae linker histone. Mol Cell. 2003;11:1685–1692. doi: 10.1016/s1097-2765(03)00197-7. [DOI] [PubMed] [Google Scholar]
- 83.Shen X, Yu L, Weir JW, Gorovsky MA. Linker histones are not essential and affect chromatin condensation in vivo. Cell. 1995;82:47–56. doi: 10.1016/0092-8674(95)90051-9. [DOI] [PubMed] [Google Scholar]
- 84.Shen X, Gorovsky MA. Linker histone H1 regulates specific gene expression but not global transcription in vivo. Cell. 1996;86:475–483. doi: 10.1016/s0092-8674(00)80120-8. [DOI] [PubMed] [Google Scholar]
- 85.Jedrusik MA, Schulze E. A single histone H1 isoform (H1.1) is essential for chromatin silencing and germline development in Caenorhabditis elegans. Development. 2001;128:1069–1080. doi: 10.1242/dev.128.7.1069. [DOI] [PubMed] [Google Scholar]
- 86.Lu X, Wontakal SN, Emelyanov AV, Morcillo P, Konev AY, Fyodorov DV, Skoultchi AI. Linker histone H1 is essential for Drosophila development, the establishment of pericentric heterochromatin, and a normal polytene chromosome structure. Genes Dev. 2009;23:452–465. doi: 10.1101/gad.1749309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Perez-Montero S, Carbonell A, Moran T, Vaquero A, Azorin F. The embryonic linker histone H1 variant of Drosophila, dBigH1, regulates zygotic genome activation. Developmental cell. 2013;26:578–590. doi: 10.1016/j.devcel.2013.08.011. [DOI] [PubMed] [Google Scholar]
- 88.Bouvet P, Dimitrov S, Wolffe AP. Specific regulation of Xenopus chromosomal 5S rRNA gene transcription in vivo by histone H1. Genes Dev. 1994;8:1147–1159. doi: 10.1101/gad.8.10.1147. [DOI] [PubMed] [Google Scholar]
- 89.Kandolf H. The H1A histone variant is an in vivo repressor of oocyte-type 5S gene transcription in Xenopus laevis embryos. Proc Natl Acad Sci U S A. 1994;91:7257–7261. doi: 10.1073/pnas.91.15.7257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Steinbach OC, Wolffe AP, Rupp RA. Somatic linker histones cause loss of mesodermal competence in Xenopus. Nature. 1997;389:395–399. doi: 10.1038/38755. [DOI] [PubMed] [Google Scholar]
- 91.Prymakowska-Bosak M, Przewloka MR, Slusarczyk J, Kuras M, Lichota J, Kilianczyk B, Jerzmanowski A. Linker histones play a role in male meiosis and the development of pollen grains in tobacco. The Plant cell. 1999;11:2317–2329. doi: 10.1105/tpc.11.12.2317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Przewloka MR, Wierzbicki AT, Slusarczyk J, Kuras M, Grasser KD, Stemmer C, Jerzmanowski A. The “drought-inducible” histone H1s of tobacco play no role in male sterility linked to alterations in H1 variants. Planta. 2002;215:371–379. doi: 10.1007/s00425-002-0758-9. [DOI] [PubMed] [Google Scholar]
- 93.Gao S, Chung YG, Parseghian MH, King GJ, Adashi EY, Latham KE. Rapid H1 linker histone transitions following fertilization or somatic cell nuclear transfer: evidence for a uniform developmental program in mice. Dev Biol. 2004;266:62–75. doi: 10.1016/j.ydbio.2003.10.003. [DOI] [PubMed] [Google Scholar]
- 94.Jullien J, Astrand C, Halley-Stott RP, Garrett N, Gurdon JB. Characterization of somatic cell nuclear reprogramming by oocytes in which a linker histone is required for pluripotency gene reactivation. Proc Natl Acad Sci U S A. 2010;107:5483–5488. doi: 10.1073/pnas.1000599107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hayakawa K, Ohgane J, Tanaka S, Yagi S, Shiota K. Oocyte-specific linker histone H1foo is an epigenomic modulator that decondenses chromatin and impairs pluripotency. Epigenetics. 2012;7:1029–1036. doi: 10.4161/epi.21492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Dasso M, Dimitrov S, Wolffe AP. Nuclear assembly is independent of linker histones. Proc Natl Acad Sci U S A. 1994;91:12477–12481. doi: 10.1073/pnas.91.26.12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Maresca TJ, Freedman BS, Heald R. Histone H1 is essential for mitotic chromosome architecture and segregation in Xenopus laevis egg extracts. J Cell Biol. 2005;169:859–869. doi: 10.1083/jcb.200503031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kihara M, Kirkman N, Adashi EY, King G. Oocytes from H1foo knockout mice develop normally. MGI Direct Data Submission. 2005 http://www.informatics.jax.org/reference/J:101326.
- 99.W.T.S. Institute. Alleles produced for the KOMP project by the Wellcome Trust Sanger Institute. MGI Direct Data Submission. 2009 http://www.informatics.jax.org/reference/J:148605.
- 100.Clarke HJ, Bustin M, Oblin C. Chromatin modifications during oogenesis in the mouse: removal of somatic subtypes of histone H1 from oocyte chromatin occurs post-natally through a post-transcriptional mechanism. J Cell Sci. 1997;110( Pt 4):477–487. doi: 10.1242/jcs.110.4.477. [DOI] [PubMed] [Google Scholar]
- 101.Clarke HJ, McLay DW, Mohamed OA. Linker histone transitions during mammalian oogenesis and embryogenesis. Developmental genetics. 1998;22:17–30. doi: 10.1002/(SICI)1520-6408(1998)22:1<17::AID-DVG3>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 102.Fu G, Ghadam P, Sirotkin A, Khochbin S, Skoultchi AI, Clarke HJ. Mouse oocytes and early embryos express multiple histone H1 subtypes. Biol Reprod. 2003;68:1569–1576. doi: 10.1095/biolreprod.102.012336. [DOI] [PubMed] [Google Scholar]
- 103.Sirotkin AM, Edelmann W, Cheng G, Klein-Szanto A, Kucherlapati R, Skoultchi AI. Mice develop normally without the H1(0) linker histone. Proc Natl Acad Sci U S A. 1995;92:6434–6438. doi: 10.1073/pnas.92.14.6434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Wiekowski M, Miranda M, Nothias JY, DePamphilis ML. Changes in histone synthesis and modification at the beginning of mouse development correlate with the establishment of chromatin mediated repression of transcription. J Cell Sci. 1997;110( Pt 10):1147–1158. doi: 10.1242/jcs.110.10.1147. [DOI] [PubMed] [Google Scholar]
- 105.Nguyen GD, Gokhan S, Molero AE, Yang SM, Kim BJ, Skoultchi AI, Mehler MF. The role of H1 linker histone subtypes in preserving the fidelity of elaboration of mesendodermal and neuroectodermal lineages during embryonic development. PLoS One. 2014;9:e96858. doi: 10.1371/journal.pone.0096858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Terme JM, Sese B, Millan-Arino L, Mayor R, Belmonte JC, Barrero MJ, Jordan A. Histone H1 variants are differentially expressed and incorporated into chromatin during differentiation and reprogramming to pluripotency. J Biol Chem. 2011;286:35347–35357. doi: 10.1074/jbc.M111.281923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Rabini S, Franke K, Saftig P, Bode C, Doenecke D, Drabent B. Spermatogenesis in mice is not affected by histone H1.1 deficiency. Experimental cell research. 2000;255:114–124. doi: 10.1006/excr.1999.4767. [DOI] [PubMed] [Google Scholar]
- 108.Fan Y, Sirotkin A, Russell RG, Ayala J, Skoultchi AI. Individual somatic H1 subtypes are dispensable for mouse development even in mice lacking the H1(0) replacement subtype. Mol Cell Biol. 2001;21:7933–7943. doi: 10.1128/MCB.21.23.7933-7943.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Cao K, Lailler N, Zhang Y, Kumar A, Uppal K, Liu Z, Lee EK, Wu H, Medrzycki M, Pan C, Ho PY, Cooper GP, Jr, Dong X, Bock C, Bouhassira EE, Fan Y. High-resolution mapping of h1 linker histone variants in embryonic stem cells. PLoS Genet. 2013;9:e1003417. doi: 10.1371/journal.pgen.1003417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Dominguez V, Pina B, Suau P. Histone H1 subtype synthesis in neurons and neuroblasts. Development. 1992;115:181–185. doi: 10.1242/dev.115.1.181. [DOI] [PubMed] [Google Scholar]
- 111.Franke K, Drabent B, Doenecke D. Expression of murine H1 histone genes during postnatal development. Biochimica et biophysica acta. 1998;1398:232–242. doi: 10.1016/s0167-4781(98)00062-1. [DOI] [PubMed] [Google Scholar]
- 112.Pehrson JR, Cole RD. Histone H1 subfractions and H10 turnover at different rates in nondividing cells. Biochemistry. 1982;21:456–460. doi: 10.1021/bi00532a006. [DOI] [PubMed] [Google Scholar]
- 113.Khochbin S, Gorka C, Lawrence JJ. Multiple control level governing H10 mRNA and protein accumulation. FEBS Lett. 1991;283:65–67. doi: 10.1016/0014-5793(91)80554-g. [DOI] [PubMed] [Google Scholar]
- 114.Pina B, Martinez P, Simon L, Suau P. Differential kinetics of histone H1(0) accumulation in neuronal and glial cells from rat cerebral cortex during postnatal development. Biochem Biophys Res Commun. 1984;123:697–702. doi: 10.1016/0006-291x(84)90285-7. [DOI] [PubMed] [Google Scholar]
- 115.Pina B, Martinez P, Suau P. Changes in H1 complement in differentiating rat-brain cortical neurons. Eur J Biochem. 1987;164:71–76. doi: 10.1111/j.1432-1033.1987.tb10994.x. [DOI] [PubMed] [Google Scholar]
- 116.Garcia-Segura LM, Luquin S, Martinez P, Casas MT, Suau P. Differential expression and gonadal hormone regulation of histone H1(0) in the developing and adult rat brain. Brain research. Developmental brain research. 1993;73:63–70. doi: 10.1016/0165-3806(93)90046-d. [DOI] [PubMed] [Google Scholar]
- 117.Popova EY, Grigoryev SA, Fan Y, Skoultchi AI, Zhang SS, Barnstable CJ. Developmentally regulated linker histone H1c promotes heterochromatin condensation and mediates structural integrity of rod photoreceptors in mouse retina. J Biol Chem. 2013;288:17895–17907. doi: 10.1074/jbc.M113.452144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Gabrilovich DI, Cheng P, Fan Y, Yu B, Nikitina E, Sirotkin A, Shurin M, Oyama T, Adachi Y, Nadaf S, Carbone DP, Skoultchi AI. H1(0) histone and differentiation of dendritic cells. A molecular target for tumor-derived factors. J Leukoc Biol. 2002;72:285–296. [PubMed] [Google Scholar]
- 119.Kalashnikova AA, Winkler DD, McBryant SJ, Henderson RK, Herman JA, DeLuca JG, Luger K, Prenni JE, Hansen JC. Linker histone H1.0 interacts with an extensive network of proteins found in the nucleolus. Nucleic Acids Res. 2013;41:4026–4035. doi: 10.1093/nar/gkt104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Konishi A, Shimizu S, Hirota J, Takao T, Fan Y, Matsuoka Y, Zhang L, Yoneda Y, Fujii Y, Skoultchi AI, Tsujimoto Y. Involvement of histone H1.2 in apoptosis induced by DNA double-strand breaks. Cell. 2003;114:673–688. doi: 10.1016/s0092-8674(03)00719-0. [DOI] [PubMed] [Google Scholar]
- 121.Cascone A, Bruelle C, Lindholm D, Bernardi P, Eriksson O. Destabilization of the outer and inner mitochondrial membranes by core and linker histones. PLoS One. 2012;7:e35357. doi: 10.1371/journal.pone.0035357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Okamura H, Yoshida K, Amorim BR, Haneji T. Histone H1.2 is translocated to mitochondria and associates with Bak in bleomycin-induced apoptotic cells. Journal of cellular biochemistry. 2008;103:1488–1496. doi: 10.1002/jcb.21537. [DOI] [PubMed] [Google Scholar]
- 123.Kimmins S, Sassone-Corsi P. Chromatin remodelling and epigenetic features of germ cells. Nature. 2005;434:583–589. doi: 10.1038/nature03368. [DOI] [PubMed] [Google Scholar]
- 124.Clermont Y. Kinetics of spermatogenesis in mammals: seminiferous epithelium cycle and spermatogonial renewal. Physiological reviews. 1972;52:198–236. doi: 10.1152/physrev.1972.52.1.198. [DOI] [PubMed] [Google Scholar]
- 125.Bucci LR, Brock WA, Meistrich ML. Distribution and synthesis of histone 1 subfractions during spermatogenesis in the rat. Experimental cell research. 1982;140:111–118. doi: 10.1016/0014-4827(82)90162-8. [DOI] [PubMed] [Google Scholar]
- 126.Franke K, Drabent B, Doenecke D. Testicular expression of the mouse histone H1.1 gene. Histochem Cell Biol. 1998;109:383–390. doi: 10.1007/s004180050239. [DOI] [PubMed] [Google Scholar]
- 127.Meistrich ML, Bucci LR, Trostle-Weige PK, Brock WA. Histone variants in rat spermatogonia and primary spermatocytes. Dev Biol. 1985;112:230–240. doi: 10.1016/0012-1606(85)90137-x. [DOI] [PubMed] [Google Scholar]
- 128.Steger K, Klonisch T, Gavenis K, Drabent B, Doenecke D, Bergmann M. Expression of mRNA and protein of nucleoproteins during human spermiogenesis. Molecular human reproduction. 1998;4:939–945. doi: 10.1093/molehr/4.10.939. [DOI] [PubMed] [Google Scholar]
- 129.Drabent B, Bode C, Bramlage B, Doenecke D. Expression of the mouse testicular histone gene H1t during spermatogenesis. Histochem Cell Biol. 1996;106:247–251. doi: 10.1007/BF02484408. [DOI] [PubMed] [Google Scholar]
- 130.Drabent B, Bode C, Miosge N, Herken R, Doenecke D. Expression of the mouse histone gene H1t begins at premeiotic stages of spermatogenesis. Cell and tissue research. 1998;291:127–132. doi: 10.1007/s004410050986. [DOI] [PubMed] [Google Scholar]
- 131.Churikov D, Zalenskaya IA, Zalensky AO. Male germline-specific histones in mouse and man. Cytogenetic and genome research. 2004;105:203–214. doi: 10.1159/000078190. [DOI] [PubMed] [Google Scholar]
- 132.Drabent B, Saftig P, Bode C, Doenecke D. Spermatogenesis proceeds normally in mice without linker histone H1t. Histochem Cell Biol. 2000;113:433–442. doi: 10.1007/s004180000146. [DOI] [PubMed] [Google Scholar]
- 133.Lin Q, Sirotkin A, Skoultchi AI. Normal spermatogenesis in mice lacking the testis-specific linker histone H1t. Mol Cell Biol. 2000;20:2122–2128. doi: 10.1128/mcb.20.6.2122-2128.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Fantz DA, Hatfield WR, Horvath G, Kistler MK, Kistler WS. Mice with a targeted disruption of the H1t gene are fertile and undergo normal changes in structural chromosomal proteins during spermiogenesis. Biol Reprod. 2001;64:425–431. doi: 10.1095/biolreprod64.2.425. [DOI] [PubMed] [Google Scholar]
- 135.Lin Q, Inselman A, Han X, Xu H, Zhang W, Handel MA, Skoultchi AI. Reductions in linker histone levels are tolerated in developing spermatocytes but cause changes in specific gene expression. J Biol Chem. 2004;279:23525–23535. doi: 10.1074/jbc.M400925200. [DOI] [PubMed] [Google Scholar]
- 136.Lambrot R, Jones S, Saint-Phar S, Kimmins S. Specialized distribution of the histone methyltransferase Ezh2 in the nuclear apical region of round spermatids and its interaction with the histone variant H1t2. Journal of andrology. 2012;33:1058–1066. doi: 10.2164/jandrol.111.013870. [DOI] [PubMed] [Google Scholar]
- 137.Jedrzejczak P, Kempisty B, Bryja A, Mostowska M, Depa-Martynow M, Pawelczyk L, Jagodzinski PP. Quantitative assessment of transition proteins 1, 2 spermatid-specific linker histone H1-like protein transcripts in spermatozoa from normozoospermic and asthenozoospermic men. Archives of andrology. 2007;53:199–205. doi: 10.1080/01485010701426430. [DOI] [PubMed] [Google Scholar]
- 138.Furuya M, Tanaka M, Teranishi T, Matsumoto K, Hosoi Y, Saeki K, Ishimoto H, Minegishi K, Iritani A, Yoshimura Y. H1foo is indispensable for meiotic maturation of the mouse oocyte. The Journal of reproduction and development. 2007;53:895–902. doi: 10.1262/jrd.19008. [DOI] [PubMed] [Google Scholar]
- 139.Yun Y, An P, Ning J, Zhao GM, Yang WL, Lei AM. H1foo is essential for in vitro meiotic maturation of bovine oocytes. Zygote. 2015;23:416–425. doi: 10.1017/S0967199414000021. [DOI] [PubMed] [Google Scholar]
- 140.Efroni S, Duttagupta R, Cheng J, Dehghani H, Hoeppner DJ, Dash C, Bazett-Jones DP, Le Grice S, McKay RD, Buetow KH, Gingeras TR, Misteli T, Meshorer E. Global transcription in pluripotent embryonic stem cells. Cell stem cell. 2008;2:437–447. doi: 10.1016/j.stem.2008.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Meshorer E, Yellajoshula D, George E, Scambler PJ, Brown DT, Misteli T. Hyperdynamic plasticity of chromatin proteins in pluripotent embryonic stem cells. Developmental cell. 2006;10:105–116. doi: 10.1016/j.devcel.2005.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Gaspar-Maia A, Alajem A, Meshorer E, Ramalho-Santos M. Open chromatin in pluripotency and reprogramming. Nat Rev Mol Cell Biol. 2011;12:36–47. doi: 10.1038/nrm3036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Loebel DA, Watson CM, De Young RA, Tam PP. Lineage choice and differentiation in mouse embryos and embryonic stem cells. Dev Biol. 2003;264:1–14. doi: 10.1016/s0012-1606(03)00390-7. [DOI] [PubMed] [Google Scholar]
- 144.Jaenisch R, Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat Genet. 2003;33(Suppl):245–254. doi: 10.1038/ng1089. [DOI] [PubMed] [Google Scholar]
- 145.Rasmussen TP. Embryonic stem cell differentiation: a chromatin perspective. Reproductive biology and endocrinology: RB&E. 2003;1:100. doi: 10.1186/1477-7827-1-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Melcer S, Hezroni H, Rand E, Nissim-Rafinia M, Skoultchi A, Stewart CL, Bustin M, Meshorer E. Histone modifications and lamin A regulate chromatin protein dynamics in early embryonic stem cell differentiation. Nature communications. 2012;3:910. doi: 10.1038/ncomms1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Christophorou MA, Castelo-Branco G, Halley-Stott RP, Oliveira CS, Loos R, Radzisheuskaya A, Mowen KA, Bertone P, Silva JC, Zernicka-Goetz M, Nielsen ML, Gurdon JB, Kouzarides T. Citrullination regulates pluripotency and histone H1 binding to chromatin. Nature. 2014;507:104–108. doi: 10.1038/nature12942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Tsiftsoglou AS, Pappas IS, Vizirianakis IS. Mechanisms involved in the induced differentiation of leukemia cells. Pharmacology & therapeutics. 2003;100:257–290. doi: 10.1016/j.pharmthera.2003.09.002. [DOI] [PubMed] [Google Scholar]
- 149.Helliger W, Lindner H, Grubl-Knosp O, Puschendorf B. Alteration in proportions of histone H1 variants during the differentiation of murine erythroleukaemic cells. The Biochemical journal. 1992;288( Pt 3):747–751. doi: 10.1042/bj2880747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Yellajoshyula D, Brown DT. Global modulation of chromatin dynamics mediated by dephosphorylation of linker histone H1 is necessary for erythroid differentiation. Proc Natl Acad Sci U S A. 2006;103:18568–18573. doi: 10.1073/pnas.0606478103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Contreras A, Hale TK, Stenoien DL, Rosen JM, Mancini MA, Herrera RE. The dynamic mobility of histone H1 is regulated by cyclin/CDK phosphorylation. Mol Cell Biol. 2003;23:8626–8636. doi: 10.1128/MCB.23.23.8626-8636.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Hill CS, Rimmer JM, Green BN, Finch JT, Thomas JO. Histone-DNA interactions and their modulation by phosphorylation of -Ser-Pro-X-Lys/Arg- motifs. EMBO J. 1991;10:1939–1948. doi: 10.1002/j.1460-2075.1991.tb07720.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Fan Y, Nikitina T, Zhao J, Fleury TJ, Bhattacharyya R, Bouhassira EE, Stein A, Woodcock CL, Skoultchi AI. Histone H1 depletion in mammals alters global chromatin structure but causes specific changes in gene regulation. Cell. 2005;123:1199–1212. doi: 10.1016/j.cell.2005.10.028. [DOI] [PubMed] [Google Scholar]
- 154.Zhang Y, Liu Z, Medrzycki M, Cao K, Fan Y. Reduction of Hox gene expression by histone H1 depletion. PLoS One. 2012;7:e38829. doi: 10.1371/journal.pone.0038829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Lee H, Habas R, Abate-Shen C. MSX1 cooperates with histone H1b for inhibition of transcription and myogenesis. Science. 2004;304:1675–1678. doi: 10.1126/science.1098096. [DOI] [PubMed] [Google Scholar]
- 156.Routh A, Sandin S, Rhodes D. Nucleosome repeat length and linker histone stoichiometry determine chromatin fiber structure. Proc Natl Acad Sci U S A. 2008;105:8872–8877. doi: 10.1073/pnas.0802336105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Gunjan A, Brown DT. Overproduction of histone H1 variants in vivo increases basal and induced activity of the mouse mammary tumor virus promoter. Nucleic Acids Res. 1999;27:3355–3363. doi: 10.1093/nar/27.16.3355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Gunjan A, Sittman DB, Brown DT. Core histone acetylation is regulated by linker histone stoichiometry in vivo. J Biol Chem. 2001;276:3635–3640. doi: 10.1074/jbc.M007590200. [DOI] [PubMed] [Google Scholar]
- 159.Lu X, Wontakal SN, Kavi H, Kim BJ, Guzzardo PM, Emelyanov AV, Xu N, Hannon GJ, Zavadil J, Fyodorov DV, Skoultchi AI. Drosophila H1 regulates the genetic activity of heterochromatin by recruitment of Su(var)3-9. Science. 2013;340:78–81. doi: 10.1126/science.1234654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Nielsen AL, Oulad-Abdelghani M, Ortiz JA, Remboutsika E, Chambon P, Losson R. Heterochromatin formation in mammalian cells: interaction between histones and HP1 proteins. Mol Cell. 2001;7:729–739. doi: 10.1016/s1097-2765(01)00218-0. [DOI] [PubMed] [Google Scholar]
- 161.Daujat S, Zeissler U, Waldmann T, Happel N, Schneider R. HP1 binds specifically to Lys26-methylated histone H1.4, whereas simultaneous Ser27 phosphorylation blocks HP1 binding. J Biol Chem. 2005;280:38090–38095. doi: 10.1074/jbc.C500229200. [DOI] [PubMed] [Google Scholar]
- 162.Hale TK, Contreras A, Morrison AJ, Herrera RE. Phosphorylation of the linker histone H1 by CDK regulates its binding to HP1alpha. Mol Cell. 2006;22:693–699. doi: 10.1016/j.molcel.2006.04.016. [DOI] [PubMed] [Google Scholar]
- 163.Ea V, Sexton T, Gostan T, Herviou L, Baudement MO, Zhang Y, Berlivet S, Le Lay-Taha MN, Cathala G, Lesne A, Victor JM, Fan Y, Cavalli G, Forne T. Distinct polymer physics principles govern chromatin dynamics in mouse and Drosophila topological domains. BMC genomics. 2015;16:607. doi: 10.1186/s12864-015-1786-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Hashimoto H, Takami Y, Sonoda E, Iwasaki T, Iwano H, Tachibana M, Takeda S, Nakayama T, Kimura H, Shinkai Y. Histone H1 null vertebrate cells exhibit altered nucleosome architecture. Nucleic Acids Res. 2010;38:3533–3545. doi: 10.1093/nar/gkq076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Wolffe A. Chromatin: Structure and Function. Elsevier Science & Technology Books; 1999. [Google Scholar]
- 166.Vignali M, Workman JL. Location and function of linker histones. Nature structural biology. 1998;5:1025–1028. doi: 10.1038/4133. [DOI] [PubMed] [Google Scholar]
- 167.Sancho M, Diani E, Beato M, Jordan A. Depletion of human histone H1 variants uncovers specific roles in gene expression and cell growth. PLoS Genet. 2008;4:e1000227. doi: 10.1371/journal.pgen.1000227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Alami R, Fan Y, Pack S, Sonbuchner TM, Besse A, Lin Q, Greally JM, Skoultchi AI, Bouhassira EE. Mammalian linker-histone subtypes differentially affect gene expression in vivo. Proc Natl Acad Sci U S A. 2003;100:5920–5925. doi: 10.1073/pnas.0736105100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Nishiyama M, Oshikawa K, Tsukada Y, Nakagawa T, Iemura S, Natsume T, Fan Y, Kikuchi A, Skoultchi AI, Nakayama KI. CHD8 suppresses p53-mediated apoptosis through histone H1 recruitment during early embryogenesis. Nat Cell Biol. 2009;11:172–182. doi: 10.1038/ncb1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Maclean JA, Bettegowda A, Kim BJ, Lou CH, Yang SM, Bhardwaj A, Shanker S, Hu Z, Fan Y, Eckardt S, McLaughlin KJ, Skoultchi AI, Wilkinson MF. The rhox homeobox gene cluster is imprinted and selectively targeted for regulation by histone h1 and DNA methylation. Mol Cell Biol. 2011;31:1275–1287. doi: 10.1128/MCB.00734-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Medrzycki M, Zhang Y, Zhang W, Cao K, Pan C, Lailler N, McDonald JF, Bouhassira EE, Fan Y. Histone h1.3 suppresses h19 noncoding RNA expression and cell growth of ovarian cancer cells. Cancer Res. 2014;74:6463–6473. doi: 10.1158/0008-5472.CAN-13-2922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Giambra V, Volpi S, Emelyanov AV, Pflugh D, Bothwell AL, Norio P, Fan Y, Ju Z, Skoultchi AI, Hardy RR, Frezza D, Birshtein BK. Pax5 and linker histone H1 coordinate DNA methylation and histone modifications in the 3′ regulatory region of the immunoglobulin heavy chain locus. Mol Cell Biol. 2008;28:6123–6133. doi: 10.1128/MCB.00233-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Kashiwagi K, Nimura K, Ura K, Kaneda Y. DNA methyltransferase 3b preferentially associates with condensed chromatin. Nucleic Acids Res. 2011;39:874–888. doi: 10.1093/nar/gkq870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Yang SM, Kim BJ, Norwood Toro L, Skoultchi AI. H1 linker histone promotes epigenetic silencing by regulating both DNA methylation and histone H3 methylation. Proc Natl Acad Sci U S A. 2013;110:1708–1713. doi: 10.1073/pnas.1213266110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Martin C, Cao R, Zhang Y. Substrate preferences of the EZH2 histone methyltransferase complex. J Biol Chem. 2006;281:8365–8370. doi: 10.1074/jbc.M513425200. [DOI] [PubMed] [Google Scholar]
- 176.Cao R, Wang L, Wang H, Xia L, Erdjument-Bromage H, Tempst P, Jones RS, Zhang Y. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science. 2002;298:1039–1043. doi: 10.1126/science.1076997. [DOI] [PubMed] [Google Scholar]
- 177.Margueron R, Li G, Sarma K, Blais A, Zavadil J, Woodcock CL, Dynlacht BD, Reinberg D. Ezh1 and Ezh2 maintain repressive chromatin through different mechanisms. Mol Cell. 2008;32:503–518. doi: 10.1016/j.molcel.2008.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Shen X, Liu Y, Hsu YJ, Fujiwara Y, Kim J, Mao X, Yuan GC, Orkin SH. EZH1 mediates methylation on histone H3 lysine 27 and complements EZH2 in maintaining stem cell identity and executing pluripotency. Mol Cell. 2008;32:491–502. doi: 10.1016/j.molcel.2008.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Vaquero A, Scher M, Lee D, Erdjument-Bromage H, Tempst P, Reinberg D. Human SirT1 interacts with histone H1 and promotes formation of facultative heterochromatin. Mol Cell. 2004;16:93–105. doi: 10.1016/j.molcel.2004.08.031. [DOI] [PubMed] [Google Scholar]
- 180.Li JY, Patterson M, Mikkola HK, Lowry WE, Kurdistani SK. Dynamic distribution of linker histone H1.5 in cellular differentiation. PLoS Genet. 2012;8:e1002879. doi: 10.1371/journal.pgen.1002879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Izzo A, Kamieniarz-Gdula K, Ramirez F, Noureen N, Kind J, Manke T, van Steensel B, Schneider R. The genomic landscape of the somatic linker histone subtypes H1.1 to H1.5 in human cells. Cell reports. 2013;3:2142–2154. doi: 10.1016/j.celrep.2013.05.003. [DOI] [PubMed] [Google Scholar]
- 182.Millan-Arino L, Islam AB, Izquierdo-Bouldstridge A, Mayor R, Terme JM, Luque N, Sancho M, Lopez-Bigas N, Jordan A. Mapping of six somatic linker histone H1 variants in human breast cancer cells uncovers specific features of H1.2. Nucleic Acids Res. 2014;42:4474–4493. doi: 10.1093/nar/gku079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Mackey-Cushman SL, Gao J, Holmes DA, Nunoya JI, Wang R, Unutmaz D, Su L. FoxP3 interacts with linker histone H1.5 to modulate gene expression and program Treg cell activity. Genes and immunity. 2011;12:559–567. doi: 10.1038/gene.2011.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.El Gazzar M, Yoza BK, Chen X, Garcia BA, Young NL, McCall CE. Chromatin-specific remodeling by HMGB1 and linker histone H1 silences proinflammatory genes during endotoxin tolerance. Mol Cell Biol. 2009;29:1959–1971. doi: 10.1128/MCB.01862-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Thorslund T, Ripplinger A, Hoffmann S, Wild T, Uckelmann M, Villumsen B, Narita T, Sixma TK, Choudhary C, Bekker-Jensen S, Mailand N. Histone H1 couples initiation and amplification of ubiquitin signalling after DNA damage. Nature. 2015 doi: 10.1038/nature15401. [DOI] [PubMed] [Google Scholar]
- 186.Hashimoto H, Sonoda E, Takami Y, Kimura H, Nakayama T, Tachibana M, Takeda S, Shinkai Y. Histone H1 variant, H1R is involved in DNA damage response. DNA Repair (Amst) 2007;6:1584–1595. doi: 10.1016/j.dnarep.2007.05.003. [DOI] [PubMed] [Google Scholar]
- 187.Rosidi B, Wang M, Wu W, Sharma A, Wang H, Iliakis G. Histone H1 functions as a stimulatory factor in backup pathways of NHEJ. Nucleic Acids Res. 2008;36:1610–1623. doi: 10.1093/nar/gkn013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Sjoblom T, Jones S, Wood LD, Parsons DW, Lin J, Barber TD, Mandelker D, Leary RJ, Ptak J, Silliman N, Szabo S, Buckhaults P, Farrell C, Meeh P, Markowitz SD, Willis J, Dawson D, Willson JK, Gazdar AF, Hartigan J, Wu L, Liu C, Parmigiani G, Park BH, Bachman KE, Papadopoulos N, Vogelstein B, Kinzler KW, Velculescu VE. The consensus coding sequences of human breast and colorectal cancers. Science. 2006;314:268–274. doi: 10.1126/science.1133427. [DOI] [PubMed] [Google Scholar]
- 189.Wood LD, Parsons DW, Jones S, Lin J, Sjoblom T, Leary RJ, Shen D, Boca SM, Barber T, Ptak J, Silliman N, Szabo S, Dezso Z, Ustyanksky V, Nikolskaya T, Nikolsky Y, Karchin R, Wilson PA, Kaminker JS, Zhang Z, Croshaw R, Willis J, Dawson D, Shipitsin M, Willson JK, Sukumar S, Polyak K, Park BH, Pethiyagoda CL, Pant PV, Ballinger DG, Sparks AB, Hartigan J, Smith DR, Suh E, Papadopoulos N, Buckhaults P, Markowitz SD, Parmigiani G, Kinzler KW, Velculescu VE, Vogelstein B. The genomic landscapes of human breast and colorectal cancers. Science. 2007;318:1108–1113. doi: 10.1126/science.1145720. [DOI] [PubMed] [Google Scholar]
- 190.Li H, Kaminski MS, Li Y, Yildiz M, Ouillette P, Jones S, Fox H, Jacobi K, Saiya-Cork K, Bixby D, Lebovic D, Roulston D, Shedden K, Sabel M, Marentette L, Cimmino V, Chang AE, Malek SN. Mutations in linker histone genes HIST1H1 B, C, D, and E; OCT2 (POU2F2); IRF8; and ARID1A underlying the pathogenesis of follicular lymphoma. Blood. 2014;123:1487–1498. doi: 10.1182/blood-2013-05-500264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Okosun J, Bodor C, Wang J, Araf S, Yang CY, Pan C, Boller S, Cittaro D, Bozek M, Iqbal S, Matthews J, Wrench D, Marzec J, Tawana K, Popov N, O’Riain C, O’Shea D, Carlotti E, Davies A, Lawrie CH, Matolcsy A, Calaminici M, Norton A, Byers RJ, Mein C, Stupka E, Lister TA, Lenz G, Montoto S, Gribben JG, Fan Y, Grosschedl R, Chelala C, Fitzgibbon J. Integrated genomic analysis identifies recurrent mutations and evolution patterns driving the initiation and progression of follicular lymphoma. Nat Genet. 2014;46:176–181. doi: 10.1038/ng.2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Medrzycki M, Zhang Y, Cao K, Fan Y. Expression analysis of mammalian linker-histone subtypes. Journal of visualized experiments: JoVE. 2012 doi: 10.3791/3577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Thierry-Mieg D, Thierry-Mieg J. AceView: a comprehensive cDNA-supported gene and transcripts annotation. Genome Biol. 2006;7(Suppl 1):S12 11–14. doi: 10.1186/gb-2006-7-s1-s12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Drabent B, Kardalinou E, Doenecke D. Structure and expression of the human gene encoding testicular H1 histone (H1t) Gene. 1991;103:263–268. doi: 10.1016/0378-1119(91)90284-i. [DOI] [PubMed] [Google Scholar]
- 195.Panyim S, Chalkley R. A new histone found only in mammalian tissues with little cell division. Biochem Biophys Res Commun. 1969;37:1042–1049. doi: 10.1016/0006-291x(69)90237-x. [DOI] [PubMed] [Google Scholar]
- 196.Seyedin SM, Kistler WS. Isolation and characterization of rat testis H1t. An H1 histone variant associated with spermatogenesis. J Biol Chem. 1980;255:5949–5954. [PubMed] [Google Scholar]
- 197.Lea MA. Relationship of H1(0) histone to differentiation and cancer. Cancer biochemistry biophysics. 1987;9:199–209. [PubMed] [Google Scholar]
- 198.Tonjes RR, Paul D, Doenecke D. Transgenic mice transcribing the human H1 zero histone gene exhibit a normal phenotype. Eur J Biochem. 1997;245:97–102. doi: 10.1111/j.1432-1033.1997.00097.x. [DOI] [PubMed] [Google Scholar]
- 199.Murga M, Jaco I, Fan Y, Soria R, Martinez-Pastor B, Cuadrado M, Yang SM, Blasco MA, Skoultchi AI, Fernandez-Capetillo O. Global chromatin compaction limits the strength of the DNA damage response. J Cell Biol. 2007;178:1101–1108. doi: 10.1083/jcb.200704140. [DOI] [PMC free article] [PubMed] [Google Scholar]