Abstract
Methylation-mediated silencing of G0S2 has been detected in a variety of solid tumors, whereas G0S2 induction is associated with remissions in patients with acute promyelocytic leukemia, implying that G0S2 may possess tumor suppressor activity. In this study, we clearly demonstrate that G0S2 opposes oncogene-induced transformation using G0S2-null immortalized mouse embryonic fibroblasts (MEFs). G0S2-null MEFs were readily transformed with HRAS or EGFR treatment compared to wildtype MEFs. Importantly, restoration of G0S2 reversed HRAS-driven transformation. G0S2 is known to regulate fat metabolism by attenuating adipose triglyceride lipase (ATGL), but repression of oncogene-induced transformation by G0S2 was independent of ATGL inhibition. Gene expression analysis revealed that an upregulation of gene signatures associated with transformation, proliferation, and MYC targets in G0S2-null MEFs. RNAi-mediated ablation and pharmacologic inhibition of MYC abrogated oncogene-induced transformation of G0S2-null MEFs. Furthermore, we found that G0S2 was highly expressed in normal breast tissues compared to malignant tissue. Intriguingly, high levels of G0S2 were also associated with a decrease in breast cancer recurrence rates, especially in estrogen receptor-positive subtypes, and overexpression of G0S2 repressed the proliferation of breast cancer cells in vitro. Taken together, these findings indicate that G0S2 functions as a tumor suppressor in part by opposing MYC activity, prompting further investigation of the mechanisms by which G0S2 silencing mediates MYC-induced oncogenesis in other malignancies.
Keywords: G0S2, MYC, Ras, transformation, breast cancer, PNPLA2
Introduction
G0/G1 switch gene 2 (G0S2) was originally identified as being transiently induced in human peripheral blood mononuclear cells during the transition from G0 to G1 phase (1). However, a direct role in cell cycle regulation has not been determined. G0S2 only exists in vertebrates with no homologs in lower organisms and the gene encodes a small basic protein highly conserved between species. However, the lack of homology with well-characterized proteins or protein modules has made assigning functions to this protein a challenge. Recent studies support a role for G0S2 in fat homeostasis due to the interaction of the central hydrophobic domain of G0S2 with adipose triglyceride lipase (ATGL) a key lipolysis enzyme in adipose and other tissues (2–4). G0S2 can regulate lipid droplet size in adipocytes and multiple reports have demonstrated alterations in adiposity, energy balance and thermogenesis in engineered G0s2 mouse models (5–9). G0S2 has also been implicated in immune regulation based on its increased expression in peripheral blood and bone-marrow-derived mononuclear cells isolated from patients with autoimmune disease including vasculitis, acute graft-versus host disease, psoriasis, rheumatoid arthritis and lupus (10–13).
One of the first putative functions of G0S2 was as a tumor suppressor. This was based on several studies showing that G0S2 is epigenetically silenced by gene promoter methylation in head and neck and lung cancer, adrenocarcinoma, and a variety of human cancer cell lines (14–17). G0S2 has also been reported to be associated with induced terminal differentiation and cell cycle withdrawal of adipocytes, quiescence of hematopoietic stem cells and senescence of human dermal fibroblasts (18–21). Further, G0S2 was reported to decrease proliferation and growth of leukemic xenografts and to induce apoptosis in lung and colon cancer cells (22,23). While these studies imply that G0S2 has antitumor activity that potentially would oppose oncogenesis, this had not been formally demonstrated.
Previously, we have shown that G0S2 is one of the most highly induced genes during retinoic acid mediated growth arrest of human bronchial epithelial cells and induced terminal differentiation of acute promyelocytic leukemia cells, which implied an association between G0S2 and reduced tumorigenicity (24,25). In the current study, we utilized G0s2 null mouse embryonic fibroblasts (MEFs) to demonstrate that the absence of G0S2 promotes oncogene-induced cellular transformation that is closely associated with a basal increase in MYC pathway activation. In addition, we found that G0S2 was substantially repressed in human breast cancer and that G0S2 expression was associated with a low rate of breast cancer recurrence. Together these findings strongly support a tumor suppressive role for G0S2. This finding would provide a new opportunity to target breast cancers and potentially other tumors where G0S2 is silenced.
Materials and Methods
MEF generation, drugs, and cell proliferation assays
The generation of G0s2 null mice in the C57BL/6 background was previously described (6). Mouse embryonic fibroblasts (MEFs) were isolated from 12-day old embryos. In brief, embryos were detached from the amniotic sac and decapitated. Bodies were finely minced in 1× trypsin/EDTA (Invitrogen) and cultured in DMEM media (Gibco) supplemented with 10% fetal bovine serum (FBS) (Invitrogen). Embryos were genotyped during expansion. The MEF wild type (E2 and E10) and G0s2 null (E4 and E6) cell lines were generated by continuous culturing of the MEFs. Each MEF line corresponds to an individual embryo. Knockout MEF lines were confirmed to be G0s2 null by PCR assay of genomic DNA and by real-time PCR assay of total RNA as described (6) and all MEF lines were used at passage number 20 or less. T47D and BT474 breast cancer cell lines were obtained from the American Type Culture Collection (ATCC) and authenticated by the ATCC with karyotyping and short tandem repeat (STR) profiling. Cells were frozen within 1 month of purchase and used within 2 months after resuscitation. T47D was cultured in DMEM with 10% FBS and BT474 was cultured in RPMI 1640 (Cellgro) with 10% FBS. Atglistatin, a selective small molecular weight ATGL inhibitor with an IC50 of 0.7 µM, was purchased from Selleck Chemicals (26). The selective BET bromodomain inhibitor (+/−)-JQ1 was purchased from Sigma (27). All other drugs and chemicals were purchased from Sigma. For cell proliferation assays, BT474 cells were plated at 2 × 104 cells per well of a 12-well tissue culture plate. T47D cells were plated at 1 × 105 cells per well of a 6-well tissue culture plate. Cell proliferation was assessed by manually counting trypan blue viable cells with a hemocytometer.
RNAi knockdown and overexpression
Lentiviral silencing shRNAs targeting mouse ATGL (PNPLA2) (V2LMM_11149) and MYC (TRCN0000042517) were purchased from Open Biosystems along with respective controls, GIPZ non-silencing lentiviral shRNA control (RHS4346) and TRC lentiviral non-targeting shRNA control (RHS6848). The sequence of shATGL is TCTTCACACACTCTGCAAG and the sequence of shMYC is TATGCACCAGAGTTTCGAAGC. Lentiviral stocks for shATGL and sh-control were generated from 293T cells using the trans-lentiviral packaging kit with calcium phosphate transfection reagent (Open Biosystems). Lentiviral stocks for shMYC and sh-control were generated from 293T cells using HIV packaging mix (GeneCopoeia). MEFs were cultured with lentiviral stocks for 24 hours and stable pools were selected with 2.0 µg/mL puromycin. For G0S2 overexpression, lentiviral open reading frames (LentiORFs) expressing human G0S2 (PLOHS_100009070) were purchased from Dharmacon along with LentiORF RFP pLOC vector positive control. The insert sequence for G0S2 is available from GenBank (DQ894610). Lentiviral stocks for G0S2 and control were generated from 293T cells using the trans-lentiviral packaging kit with calcium phosphate transfection reagent (Open Biosystems). Stable pools were selected with 7.0 µg/mL and 15.0 µg/mL blasticidin S for BT474 and T47D, respectively.
Foci formation assay
MEFs were plated at 1 × 106 cells per 10-cm dish in DMEM with 10% FBS. The next day, indicated cell lines were transfected with 1.5 µg of either insertless vector pcDNA 3.1+ (Invitrogen) or human HRAS (V12) expression plasmid (a gift from Dr. Michael Cole, Geisel School of Medicine at Dartmouth) or wild-type EGFR expression plasmid (Addgene 11011) along with 1.0 µg of GFP expression plasmid pEGFP-C1 (Clontech) to control for transfection efficiency. Transfections were performed with EndoFectin™-Lenti (GeneCopoeia) and Opti-MEM I (1×) Reduced Serum Medium (Life Technologies) according to the manufacturers’ protocols. The day after transfection, cells were split into 10-cm dishes and GFP expression was determined with a florescent microscope the following day. Cells were allowed to grow to confluence and media was changed every 3 days. Cells were fixed with methanol and stained with Giemsa 14 days after transfection to visualize transformed foci. The number of foci was manually counted. For G0S2 co-transfection foci assay, MEFs were transfected with 1.5 µg of HRAS (V12) expression plasmid and either 7.0 µg of insertless vector pcDNA 3.1+ or human G0S2 expression plasmid pCMV-SPORT6-G0S2 (Thermo Fisher). For co-transfection of shRNA, 1.5 µg HRAS (V12) was co-transfected with 5.0 µg of shMYC, shATGL or corresponding shControl lentiviral expression plasmid.
Soft agar assay
Wild-type (WT) E2, WT E2 stably overexpressing HRAS (V12), G0S2 −/− E6, and G0S2 −/− E6 stably overexpressing HRAS (V12) were used in the soft agar assay. MEFs were plated at 2 × 104 cells per well of a 6-well plate in DMEM containing 20% FBS and 3.2% SeaKEM ME Agarose (Cambrex Bioscience) in an 8:1 ratio. The bottom layer was made up of DMEM containing 20% FBS and 3.2% agarose in a 2:1 ratio. Cells were cultured every 4 days with fresh media. After 2 weeks of culture, colonies were stained with 3-[4,5-Dimethylthiazol-2-yl]-2,5-diphenyltetrazolium bromide (MTT) Reagent (Sigma) in phosphate buffered saline (PBS).
Real-time PCR and Immunoblot analyses
Generation of cDNA was performed with the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems). Real-time polymerase chain reaction (PCR) assays were performed with iTaq Universal SYBR Green Supermix (Bio-Rad Laboratories) and the ddCt method was employed with normalization to GAPDH. Primer sequences are available upon request. For immunoblot analysis, cells were lysed in radio-immunoprecipitation assay buffer with protease inhibitor cocktail tablets (Roche), separated by SDS-PAGE and transferred to nitrocellulose membranes. Antibodies to actin (sc-1615, Santa Cruz), ATGL (2138, Cell Signaling Technology), MYC (sc-764, Santa Cruz) were used.
Gene expression microarray analysis
RNA was extracted with Trizol reagent. Expression analysis was performed with MouseRef-8 bead chip arrays (Illumina) and scanned on the BeadArray Reader (Illumia) according to the manufacturer’s instructions. Raw data were normalized (quantile) and analyzed with Genome Studio software (Illumina). Data were imported in GeneSifter (vizX labs) for pairwise statistical analyses using the t-test and the Benjamini–Hochberg correction. GSEA software was downloaded from the Broad website (http://www.broadinstitute.org/gsea/index.jsp). The number of permutations was 1,000 and the permutation type was gene_set. Gene expression microarray data has been submitted to the NCBI GEO repository as GSE74696.
In silico and survival analysis
G0S2 expression from TCGA Breast (http://tcga-data.nci.nih.gov/tcga/) (28,29) was obtained from Oncomine (https://www.oncomine.com). Kaplan-Meier log-rank tests were performed using default parameters in SurvExpress (30). Patients’ data were divided into low or high expression groups at the median expression value in all cases. Univariant Cox proportional analysis was performed using the above datasets and the default parameters in SurvExpress (30).
Statistics
When a value for statistical significance is provided, a two-sample, two-tailed t-test assuming unequal variance was performed.
Results
G0S2 inhibits oncogene-induced transformation in an ATGL independent manner
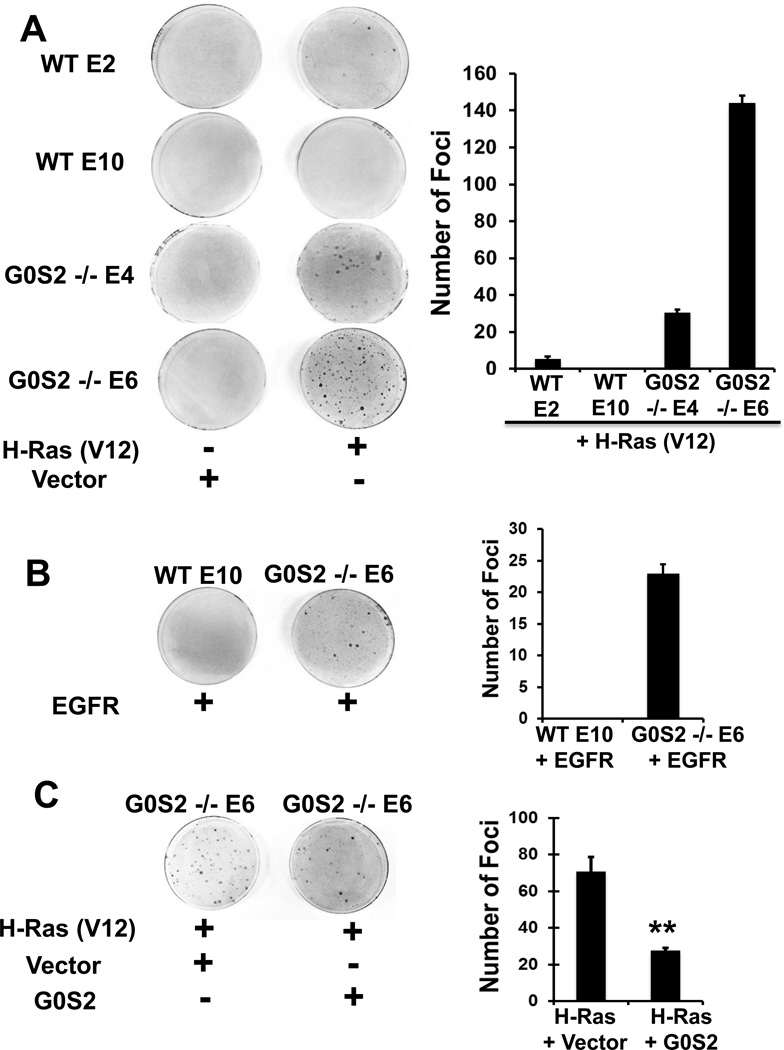
To first assess potential tumor suppressor activity of G0S2, we assessed the transformation rate of wild-type to G0s2 null MEFs in response to HRAS (V12). MEFs were transfected with either insertless control vector or HRAS (V12) expression plasmid along with a GFP expression plasmid. Transfection efficiency and plasmid integration were comparable between the cell lines as assessed by GFP fluorescence (data not shown). When transfected with HRAS (V12), G0s2 null MEFs formed a substantially higher number of foci as compared to wild-type MEFs (Fig. 1A), indicating a higher efficiency of HRAS in inducing cellular transformation in the absence of G0S2. This is despite the similar basal proliferation rate between G0s2 null and wild-type MEFs. That these cells were truly transformed by HRAS (V12) was confirmed by soft agar assay (data not shown). Similar results were observed when cells were transfected with an upstream member of the Ras pathway, EGFR (Fig. 1B). To confirm that the observed transforming phenotype was due to G0S2 deficiency, G0S2 was re-introduced into G0S2 null MEFs by co-transfecting HRAS (V12) with either a G0S2 expression plasmid or insertless control vector. G0S2 reconstitution significantly diminished (p < 0.01) the transformation efficiency in response to HRAS (Fig. 1C). These data suggest that G0S2 acts as a barrier to oncogenic transformation.
Figure 1. G0s2 null cells have an increased susceptibility to undergo oncogene induced transformation.
A, Wild-type (WT-E2, WT-E10) and G0s2 null (G0S2−/− E4, G0S2−/− E6) immortalized MEFs were transfected with either an insertless control vector or an HRAS (V12) expression plasmid. Cells were stained 14 days after transfection with Giemsa. Left are representative plates. Right is the average of two biological replicates where error bars indicate the range in values. The experiment was repeated four times with similar results. B, WT-E10 and G0S2 −/− E6 cells were transfected with either an insertless control vector or an EGFR expression plasmid. Cells were stained 14 days after transfection with Giemsa. Left are representative plates. Right is the average of two biological replicates where error bars indicate the range in values. The experiment was repeated two times with similar results. C, G0S2 −/− E6 cells were transfected with an HRAS (V12) expression plasmid and either an insertless control vector or G0S2 expression plasmid. Cells were stained 14 days after transfection with Giemsa. Left are representative plates. Right is the average of biological triplicate determinations and error bars are standard deviation. ** = P < 0.01. The experiment was repeated three times with similar results.
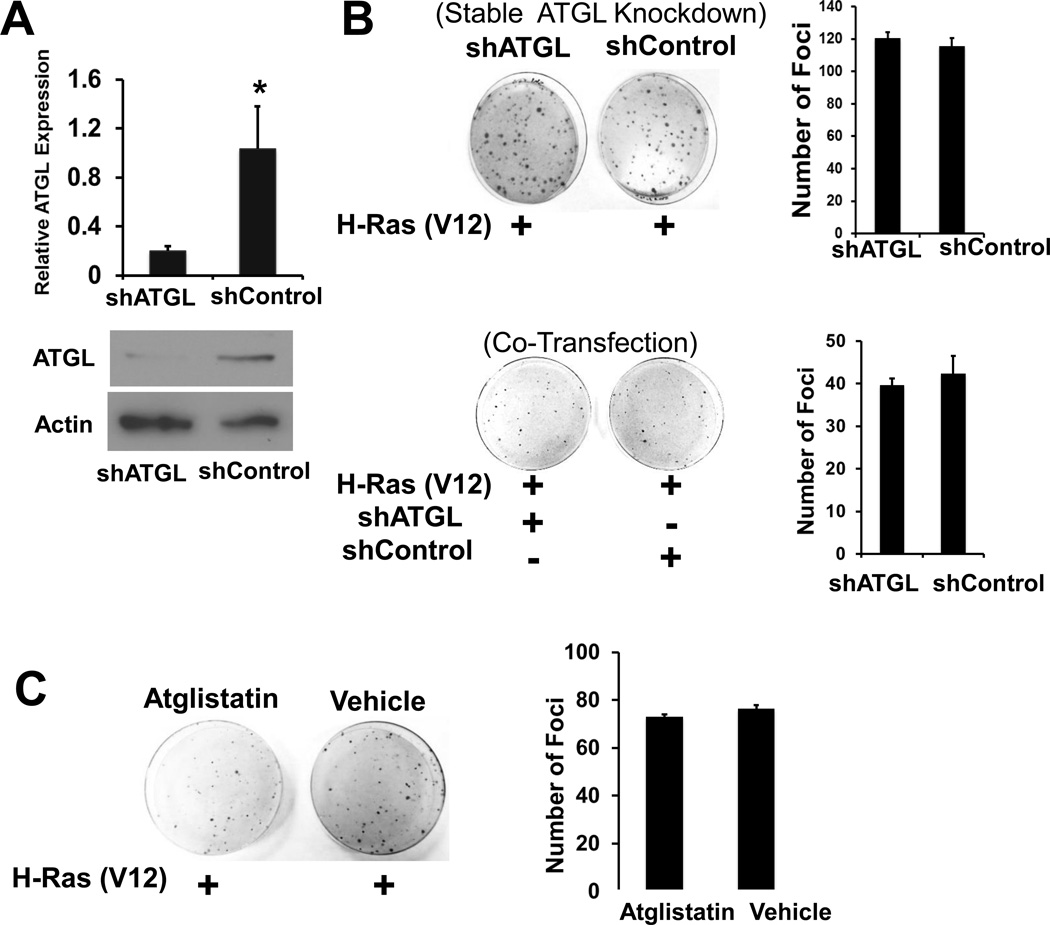
Since G0S2 is known to be a negative regulator of ATGL, it was determined whether the increased transformation potential of G0s2 null MEFs was due to the release of ATGL inhibition (2). Stable shRNA-mediated knockdown of ATGL in G0s2 null MEFs was achieved at both the mRNA and protein levels (Fig. 2A). Stable knockdown of ATGL (shATGL) had no effect on subsequent HRAS (V12)-mediated transformation of G0s2 null MEFs as compared to control cells (shControl) (Fig. 2B). Similar results were obtained with two additional, independent ATGL shRNAs (data not shown). In addition, no effects on transformation were seen when the HRAS (V12) expression plasmid was co-transfected with individual ATGL-targeting shRNA plasmids (Fig. 2B). As a third approach, atglistatin, a competitive small molecule inhibitor targeting ATGL was utilized at a dose known to achieve complete inhibition of ATGL (26). G0s2 null MEFs continually treated with atglistatin had a similar rate of HRAS (V12)-mediated transformation as compared to vehicle-treated cells (Fig. 2C). Collectively, these results indicate that G0S2 inhibits oncogene-induced transformation independent of ATGL.
Figure 2. Atgl knockdown does not affect transformation of G0s2 null cells.
A, Atgl was effectively knocked down with ATGL-targeting lentiviral shRNA. Top, real-time PCR assay, bottom, Immunoblot. Bars are the average of biological triplicates and error bars are standard deviation. * P < 0.05. B, Left are representative plates from foci formation assays comparing the ability of HRAS expression plasmid to transform G0S2 null cells stably transduced with either shATGL or shControl lentivirus (top) or co-transfected with HRAS and either shATGL or shControl expression plasmids (bottom). Right, is the average of biological triplicate determinations and error bars are standard deviation. The experiments were repeated three times with similar results. C, G0S2 null MEFs were transfected with HRAS and the following day treated with either 40 µM of ATGL inhibitor atglistatin or DMSO (Vehicle) every 2 days. Left are representative plates. Right is the average of biological triplicate determinations and error bars are standard deviation. The experiment was repeated two times with similar results.
G0s2 null MEFs have gene signatures associated with transformation, proliferation and MYC transcriptional responses
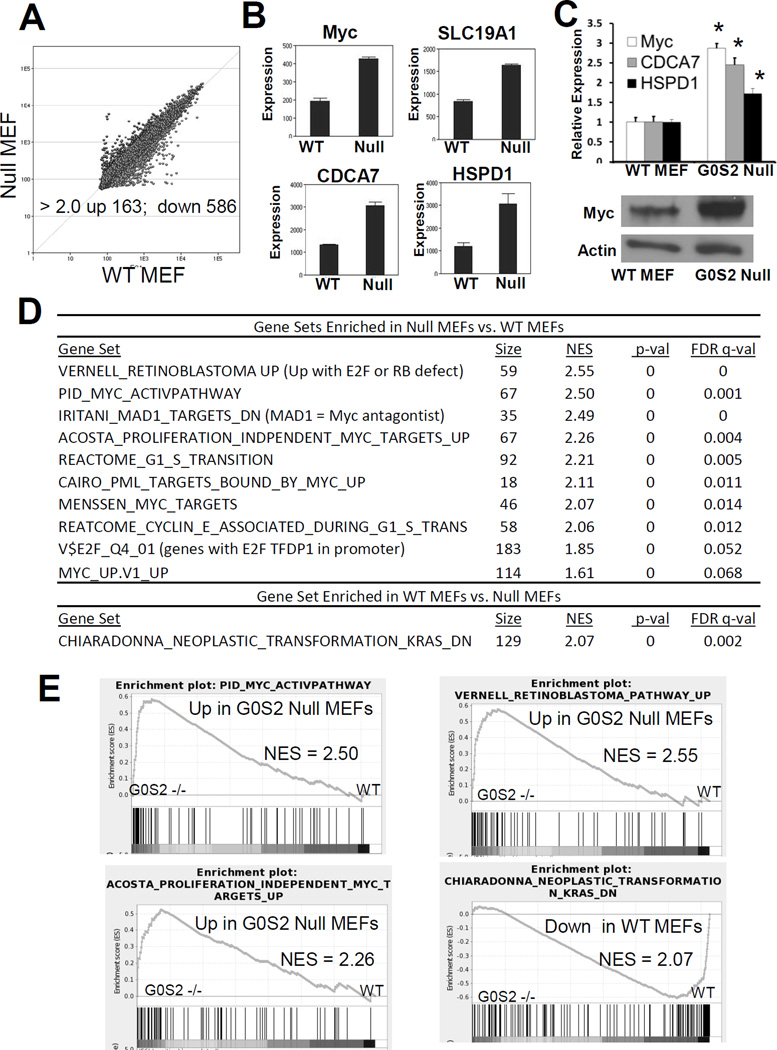
We conducted microarray-based gene expression analysis to compare gene expression changes between wild-type and G0s2 null MEFs. There was a robust reprogramming of gene expression in G0s2 null MEFs with 163 genes upregulated and 568 downregulated at least 2-fold as compared to wild-type MEFs (Fig. 3A and supplemental Table S1). Gene Set Enrichment Analysis (GSEA) indicated that G0s2 null MEFs were highly enriched in the expression of gene sets corresponding to transformation and proliferation (Fig. 3D and Fig. 3E). Interestingly, among those gene sets with the highest normalized enrichment scores (NES), many were gene signatures associated with MYC transcriptional responses, implicating an association between G0S2 and MYC (Fig. 3D and Fig. 3E). Indeed, MYC and several direct MYC target genes including SLC19A1, CDCA7 and HSPD1 were found to be upregulated in the microarray studies (Fig. 3B). MYC and MYC target genes were confirmed to be enhanced in G0s2 null cells in independent samples and MYC protein expression was also augmented in G0s2 null cells (Fig. 3C). These analyses indicate that G0S2 ablation alone results in upregulation of oncogenic transcriptional pathways associated with transformation.
Figure 3. G0s2 null cells have upregulated signatures associated with transformation and MYC.
A, Scatter plot of microarray analysis (Illumina MouseRef-8 v2) from differential gene expression profiles of G0s2 null MEFs vs. WT MEFs. Each point represents the average of 3 biological replicates. Points above the diagonal line represent genes upregulated and points below the diagonal line repressed in G0s2 null MEFs. The number of altered genes above a 2-fold threshold was changed with P < 0.01. B, Microarray results for MYC and MYC targets, SLC19A1, CDCA7, and HSPD1. C, Top, Real-time PCR assay confirmation of induction of MYC and MYC targets CDCA7 and HSPD1 in G0s2 null MEFs as compared to wild-type MEFs. Bottom, MYC protein is expressed at a high level in G0s2 null cells as compared to wild-type MEFs. Bars are average of biological triplicates. Error bars are standard deviation. * = P <0.05. D–E, Gene Set Enrichment Analyses (GSEA) of G0s2 null MEFs as compared to control MEFs indicating that G0s2 null MEFs are enriched for gene signatures associated with MYC activation, proliferation and transformation.
Repression of MYC opposes transformation of G0s2 null cells
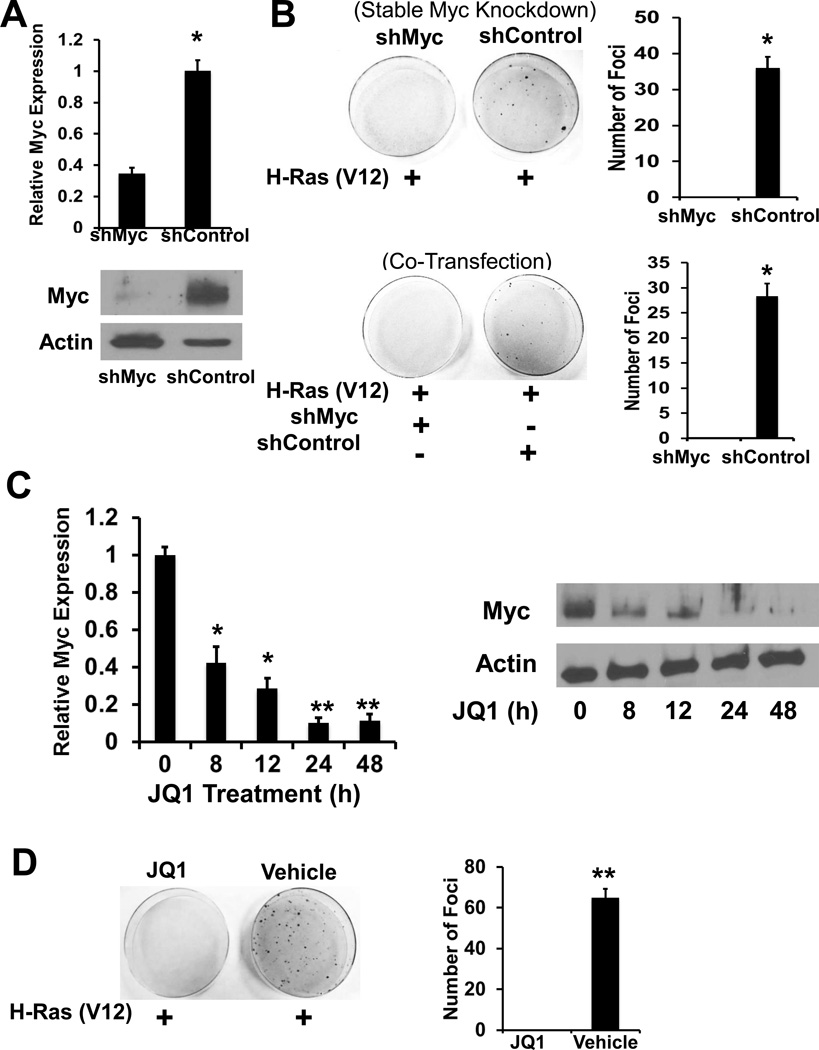
To examine the role of MYC in the susceptibility of G0s2 null MEFs to transform, these cells were transduced with shRNA specific to MYC (shMYC) or control shRNA (shControl) (Fig. 4A). Both stable and co-transfected introduction of MYC-targeting shRNA abrogated the ability of G0s2 null MEFs to be transformed with HRAS (Fig. 4B). To corroborate these findings, a small molecule inhibitor of the BRD family of BET proteins, (+/−)-JQ1, was utilized. As noted in prior studies (27), 500nM JQ1 decreased MYC expression in G0s2 null MEFs (Fig. 4C). G0s2 null MEFs treated with JQ1 were quite resistant to HRAS induced transformation (Fig. 4D).
Figure 4. Repression of MYC opposes transformation of G0s2 null MEFs.
A, MYC is effectively knocked down with MYC targeted lentiviral shRNA. Top, Real-time PCR assay, bottom, Immunoblot. Bars are the average of biological triplicates and error bars are standard deviation, * = P < 0.05. B, Left are representative plates from foci formation assays comparing the ability of HRAS expression plasmid to transform G0s2 null cells stably transduced with either shMYC or shControl lentivirus (top) or transiently transfected with shMYC or shControl expression plasmids (bottom). Right, is the average of biological triplicate determinations and error bars are standard deviation, * = P < 0.001. The experiments were repeated three times with similar results. C, BRD4 inhibitor JQ1 inhibits MYC expression in G0s2 null cells. G0s2 null cells were treated with 500 nM JQ1 for the indicated times and MYC expression was assessed by real-time PCR assay (left) and Immunoblot (right). Bars are the average of biological triplicates and error bars are standard deviation, * = P <0.05, ** = P < 0.001. D, G0s2 null MEFs were transfected with HRAS and the following day treated with either 500 nM JQ1 or DMSO (vehicle) every 2 days. Left are representative plates. Right is the average of biological triplicate determinations and error bars are standard deviation, ** = P < .001. The experiment was repeated two times with similar results.
G0S2 expression in human breast cancer
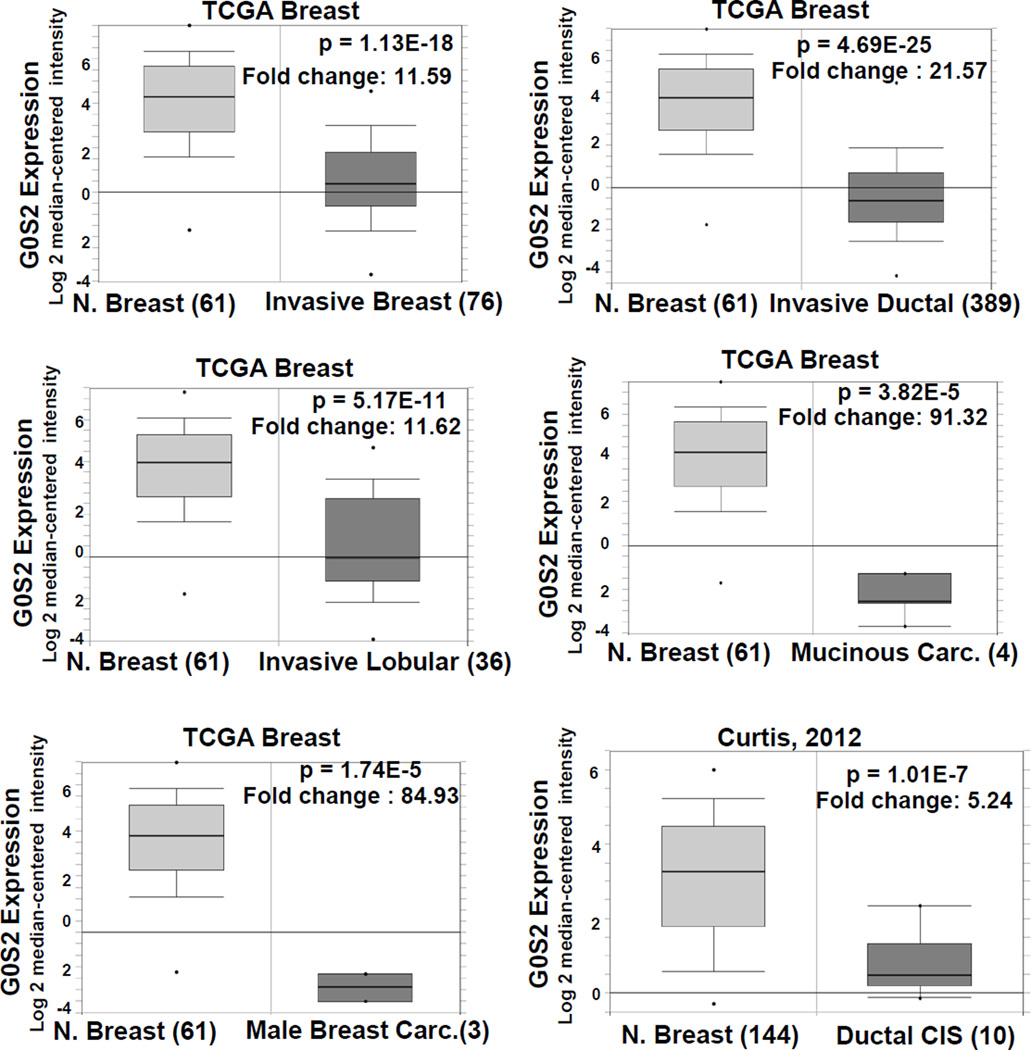
Based on the above evidence that G0S2 possesses tumor suppressor activity, in silico analysis was performed to determine whether G0S2 is deregulated in human cancers. A wide spectrum of cancer types from the Oncomine database was assessed for G0S2 deregulation (31). Unexpectedly, the most prominent alteration found was an appreciable loss of G0S2 expression in breast cancers as compared to normal controls. In the Oncomine (31) database, 17 of 43 analyses demonstrated a greater than 4-fold decline in G0S2 expression in breast cancer vs. normal with a p-value < 0.0001 (Fig. 5 and supplemental Table S2). This includes large and highly significant decreases in G0S2 expression in a wide spectrum of breast cancer classifications, for example, from the TCGA breast database, mucinous breast cancer vs. normal (p = 3.825E-5, 91.3 fold), invasive breast cancer vs. normal (p = 1.13E-18, 11.59 fold), male breast cancer (p = 1.74E-5, 84.93 fold), invasive lobular breast cancer vs. normal (p = 5.17E-11, 11.62 fold), and invasive ductal breast carcinoma vs. normal (p = 4.69E-25, 21.57 fold). The study by Radvanyi et al. (28) confirms the result for invasive ductal carcinoma and the study of Curtis et al. (29) found highly significant declines in G0S2 expression in mucinous breast carcinoma, invasive ductal and lobular breast carcinoma, medullary breast carcinoma, tubular breast carcinoma, ductal breast carcinoma in situ and breast phyllodes tumor (Supplemental Table S2). In contrast, using the same cutoffs no analysis in the Oncomine database showed a significant increase in G0S2 expression in breast cancer vs. normal breast tissue. Other tumor types from Oncomine with significantly decreased expression of G0S2 as compared to normal were leukemia (9 out of 23 analyses), sarcoma (5 out of 20 analyses), kidney cancer (3 out of 20 analyses) and lung cancer (4 out of 27 analyses).
Figure 5. G0S2 expression is repressed in breast cancer.
Data from TCGA Breast (https://tcga-data.nci.nih.gov/tcga/) and (29) comparing microarray-based G0S2 expression from non-malignant breast tissue and malignant breast tissue. Data were obtained through the Oncomine database (31). For details of all breast cancer datasets with decreased G0S2 expression, see Table S2.
G0S2 expression is associated with reduced breast cancer proliferation and recurrence
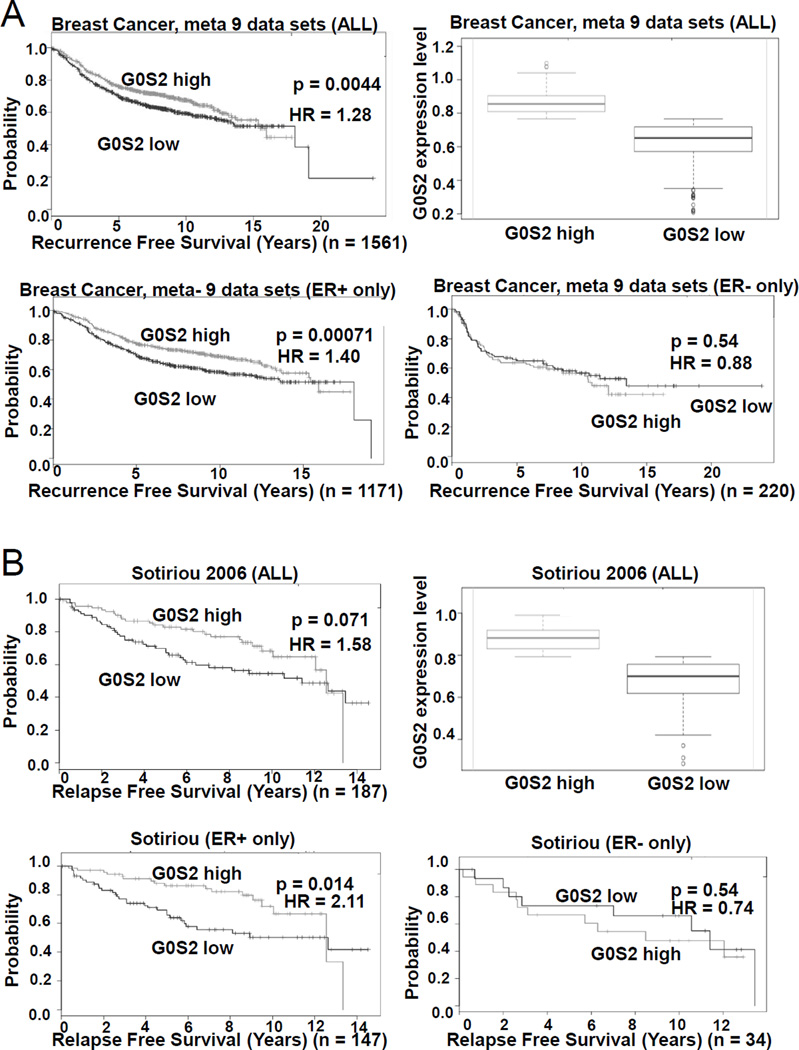
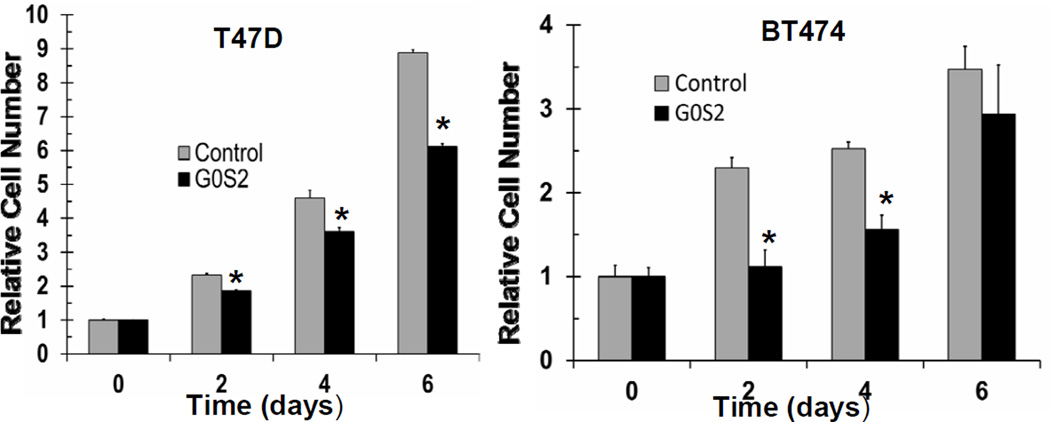
To probe the clinical relevance of G0S2 expression in breast cancer, we utilized the SurvExpress web resource to conduct univariant Cox survival analysis (30). From a total of 31 breast cancer datasets, six distinct studies demonstrated that when divided at the median, those primary tumors with high G0S2 expression had a lower rate of recurrence as compared to those with low levels of G0S2 (Fig. 6 and supplemental Table S3) (30, 32–35). Notably, the association between high G0S2 and low breast cancer recurrence was more apparent in estrogen receptor (ER)-positive as compared to ER-negative patients (Fig. 6 and supplemental Table S3). In addition, associations between high G0S2 expression and positive clinical outcomes for ER-positive breast cancer were also noted across several studies in the KMPlot, GOBO, bc-GeneExMiner and PrognoScan databases (data not shown) (36–39). To further investigate a potential role for G0S2 in breast cancer, the ER-positive breast cancer cell lines T47D and BT474 were engineered to stably express G0S2. Engineered G0S2 over-expression resulted in a consistent decrease in cell proliferation (Fig. 7). This result is consistent with the clinical observations noted above supporting a growth suppressive role for G0S2 in human breast cancer.
Figure 6. G0S2 expression in breast cancer is associated with decreased recurrence.
Kaplan-Meier log-rank test and univariant Cox proportion analysis was performed using the SurvExpress database (30) with cases divided into two groups above and below the median G0S2 expression value. A, Results from a 9 data set meta-analysis constructed by SurvExpress. Depicted are the Kaplan-Meier analysis for all patients, ER-positive only, and ER-negative only patients;respectively. Expression levels of G0S2 for all patients in the G0S2 high and G0S2 low groups are also provided (Whisker plot). B, Results from (33) utilizing SurvExpress. For additional details and a list of other studies supporting an association between recurrence free survival and G0S2 level, see Table S3.
Figure 7. G0S2 overexpression results in deceased breast cancer cell proliferation.
T47D and BT474 breast cancer cells stably expressing G0S2 lentivirus or control lentivirus were assessed for changes in cell proliferation. Data points are the average of biological triplicates, * = P < 0.05. Experiments were repeated three times and with similar results obtained. Overexpression of G0S2 in T47D and BT474 cells was confirmed with real-time PCR and Immunoblot analyses (data not shown).
Discussion
Various reports indicate that G0S2 has distinct functions mediated by distinct protein-protein interactions in different tissues, including roles in regulating energy utilization, fat metabolism and immune function (40–42). A tumor suppressive role for G0S2 has also been proposed based on the frequent epigenetic silencing of the G0S2 promoter in a variety of cancers (14–17). Here we provide formal evidence that G0S2 possesses anti-tumor and anti-oncogenic activity consistent with tumor suppression. This includes an increased susceptibility of G0s2 null cells to undergo cellular transformation. Unbiased, genome-wide expression analysis and molecular knockdown and pharmacologic inhibitor studies indicated that the increased rate of transformation is likely due, at least in part, to enhanced activation of MYC signaling pathways. We also uncovered a potential role for G0S2 specifically in human breast cancer that has clinical relevance for predicting outcomes and providing additional molecular targets to combat this disease. Our work lends further evidence for a role for G0S2 beyond adipose metabolism and provides a strong rationale to further investigate and more fully understand G0S2 in cancer, especially as an anti-neoplastic target.
The G0S2 gene has been identified as frequently methylated in de novo genome-wide studies in head and neck cancer, non-small cell lung cancer and adrenocortical carcinomas (14–17). In addition, re-expression of G0S2 was induced in a number of cell lines after treatment with DNA methylation inhibitors (14,16,17,22). Yamada et al. demonstrated that treatment of K567 leukemia cells with 5-azacytidine resulted in a 24-fold increase in G0S2 expression and a reduction in cell proliferation that could be restored with G0S2 shRNA (22). Further, engineered expression of G0S2 inhibited the proliferation of leukemic cells and xenografts (22). Welch et al., demonstrated that engineered expression of G0S2 in a lung and a colon cancer cell line induced apoptosis through G0S2 binding to Bcl-2 (23). Further evidence associating G0S2 with anti-proliferation comes from G0S2 overexpression and knockdown studies showing that G0S2 can growth arrest 3T3-L1 and non-small cell lung cancer cells and also promote quiescence of hematopoietic stem cells (18–21,43). Our data on G0s2 null MEFs and breast cancer cells engineered to express G0S2 are consistent with these studies and support an overall tumor suppressive activity of G0S2. Of note, we did not see evidence of apoptosis when G0S2 was expressed in breast cancer cells but rather a consistent decrease in cell proliferation.
The current work demonstrates that G0s2 deletion increases the susceptibility of cells to undergo oncogene-induced transformation. This effect on transformation appears to be independent of the known role of G0S2 as an inhibitor of ATGL, as genetic and pharmacologic inhibition of ATGL did not have a discernable effect on foci formation (2–4). Rather, an unbiased approach found a basal upregulation of MYC and MYC target gene signatures upon G0s2 deletion. Although MYC alone is known to have only weak transforming activity on fibroblasts, it is well established that MYC can synergize with other oncogenes to cause efficient transformation of primary and immortalized fibroblast cells (44). Thus, MYC alteration is a likely mechanism accounting for G0S2 opposition of transformation. Indeed, oncogenic transformation of G0s2 null cells was abolished upon MYC inhibition. Precisely how G0S2 interacts with MYC is unclear. G0S2 is a small protein devoid of homology with any other proteins and has no known protein motifs. It is possible that the interaction of G0S2 with MYC may be indirect and non-specific; for example if G0S2 generally interacts with and sequesters mRNAs or inhibits the general transcriptional machinery in a manner that preferentially alters the MYC transcriptional signature during cell cycle progression. However, we note that G0S2 alters MYC levels itself and this could account for the alterations seen in MYC targets upon G0S2 depletion. Although beyond the scope of the current work, it will be of interest to dissect the cross-talk between G0S2 and MYC pathway activation.
Prior database mining suggested that G0S2 expression could be decreased in cancer (23). We provide evidence that G0S2 is prominently decreased in breast cancer as compared to normal breast tissue across multiple studies and disease states. Further, we provide evidence that low levels of G0S2 are associated with negative outcomes in breast cancer patients, particularly those of ER-positive status. Although the basis for the association with ER is not yet known, it is accepted that MYC can be a driver of breast cancer and is a well-established ER target gene (45,46). Further, overexpression of MYC is known to favor tamoxifen resistance (45,46). Interestingly, MYC activation is a known driver of cell cycle progression closely associated with increased CDK4/6 activity. CDK4/6 inhibitors like palbociclib have recently been shown to have promising clinical activity specifically in ER+ breast cancer (47). It is tempting to speculate that the association of G0S2 with the increased survival of ER+ breast cancer and the ability of G0S2 to repress MYC signaling may be related and have clinical implications. For example, G0S2 levels could potentially be a predictor of palbociclib or tamoxifen response and altering G0S2 levels may be a strategy to increase the efficacy of endocrine and CDK4/6 inhibitor therapies in ER+ breast cancers.
Although our work highlights a mechanism involving MYC, it does not rule out an ATGL-dependent tumor suppressor role for G0S2 in other contexts. It is possible that G0S2 downregulation in cancer could lead to increased ATGL-mediated lipolysis to meet the fatty acid building block and metabolic demands of rapidly proliferating cancer cells (48). In apparent conflict with this model, targeted disruption of G0S2 prevents diet-induced weight gain and insulin resistance in mice (5–9). Obese postmenopausal women have an increased risk of aggressive ER+ breast cancer, presumably due to elevated estrogen production by adipose tissue (49). Obesity-associated insulin resistant type 2 diabetes is also associated with increased risk and aggressiveness of ER+ breast cancer due in part to induction of estrogen production (49). Hence it appears that there may be a dichotomy of action concerning G0S2. However, it is tempting to speculate that women who are obese despite high G0S2 levels could be partially protected from the pro-estrogenic effects of obesity due to limitation in liberation of free fatty acids that supports production of energy and building blocks required for rapid cell division. In contract, overweight women with low levels of G0S2 could have both increased estrogen and increased lipolysis to support cell division.
The relationship between adiposity and breast cancer is complex and G0S2 likely functions as a tumor suppressor via multiple mechanisms (50). The current studies were conducted in replete lipid conditions, which are distinct from the microenvironment of human tumors. It is likely that the defined activity of G0S2 as a major regulator of lipolysis influences adiposity and energy metabolism of both the normal and malignant breast. Further work is required to better understand the precise relationship between G0S2, MYC and lipid metabolism in breast and other cancers.
Supplementary Material
Acknowledgements
The authors would like to thank Dr. Craig Tomlinson, Dr. Joanna Hamilton and Heidi Trask from the Geisel School of Medicine at Dartmouth, Genomics Shared Resource for performing expression array analysis. We thank Todd Miller and Manabu Kurokawa from the Department of Pharmacology and Toxicology, Geisel School of Medicine at Dartmouth for providing T47D and BT474 cell lines and Michael Cole from Department of Pharmacology and Toxicology, Geisel School of Medicine at Dartmouth for the HRAS expression plasmid.
Grant Support
This work was supported by a United States Department of Defense Investigator-Initiated Research Award PR093629 (MJS) and a Reach grant from the Alex’s Lemonade Stand Foundation (MJS) as well as by National Institutes of Health (NIH) and National Cancer Institute grants R21-CA177781 (MJS), R01-CA190722 (ED), R01-CA087546 (ED), R01-CA062275 (ED) and by a Waxman Cancer Research Foundation grant (ED) and a UT-STARS award (ED).
Footnotes
Disclosure of Conflicts of Interest
The authors declare no conflicts of interest.
REFERENCES
- 1.Siderovski DP, Blum S, Forsdyke RE, Forsdyke DR. A set of human putative lymphocyte G0/G1 switch genes includes genes homologous to rodent cytokine and zinc finger protein-encoding genes. DNA Cell Biol. 1990;9:579–587. doi: 10.1089/dna.1990.9.579. [DOI] [PubMed] [Google Scholar]
- 2.Yang X, Lu X, Lombès M, Rha GB, Chi YI, Guerin TM, et al. The G(0)/G(1) switch gene 2 regulates adipose lipolysis through association with adipose triglyceride lipase. Cell Metab. 2010;11:194–205. doi: 10.1016/j.cmet.2010.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cornaciu I, Boeszoermenyi A, Lindermuth H, Nagy HM, Cerk IK, Ebner C, et al. The minimal domain of adipose triglyceride lipase (ATGL) ranges until leucine 254 and can be activated and inhibited by CGI-58 and G0S2, respectively. PLoS One. 2011;6:e26349. doi: 10.1371/journal.pone.0026349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schweiger M, Paar M, Eder C, Brandis J, Moser E, Gorkiewicz G, et al. G0/G1 switch gene-2 regulates human adipocyte lipolysis by affecting activity and localization of adipose triglyceride lipase. J Lipid Res. 2012;53:2307–2317. doi: 10.1194/jlr.M027409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang X, Xie X, Heckmann BL, Saarinen AM, Czyzyk TA, Liu J. Targeted disruption of G0/G1 switch gene 2 enhances adipose lipolysis, alters hepatic energy balance, and alleviates high-fat diet-induced liver steatosis. Diabetes. 2014;63:934–946. doi: 10.2337/db13-1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ma T, Lopez-Aguiar AG, Li A, Lu Y, Sekula D, Nattie EE, et al. Mice lacking G0S2 are lean and cold-tolerant. Cancer Biol Ther. 2014;15:643–650. doi: 10.4161/cbt.28251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.El-Assaad W, El-Kouhen K, Mohammad AH, Yang J, Morita M, Gamache I, et al. Deletion of the gene encoding G0/G 1 switch protein 2 (G0s2) alleviates high-fat-diet-induced weight gain and insulin resistance, and promotes browning of white adipose tissue in mice. Diabetologia. 2015;58:149–157. doi: 10.1007/s00125-014-3429-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang Y, Zhang Y, Qian H, Lu J, Zhang Z, Min X, et al. The G0/G1 switch gene 2 is an important regulator of hepatic triglyceride metabolism. PLoS One. 2013;8:e72315. doi: 10.1371/journal.pone.0072315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Heckmann BL, Zhang X, Xie X, Saarinen A, Lu X, Yang X, et al. Defective adipose lipolysis and altered global energy metabolism in mice with adipose overexpression of the lipolytic inhibitor G0/G1 switch gene 2 (G0S2) J Biol Chem. 2014;289:1905–1916. doi: 10.1074/jbc.M113.522011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kobayashi S, Ito A, Okuzaki D, Onda H, Yabuta N, Nagamori I, et al. Expression profiling of PBMC-based diagnostic gene markers isolated from vasculitis patients. DNA Res. 2008;15:253–265. doi: 10.1093/dnares/dsn014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Verner J, Kabathova J, Tomancova A, Pavlova S, Tichy B, Mraz M, et al. Gene expression profiling of acute graft-vs-host disease after hematopoietic stem cell transplantation. Exp Hematol. 2012;40:899–905. doi: 10.1016/j.exphem.2012.06.011. [DOI] [PubMed] [Google Scholar]
- 12.Nakamura N, Shimaoka Y, Tougan T, Onda H, Okuzaki D, Zhao H, et al. Isolation and expression profiling of genes upregulated in bone marrow-derived mononuclear cells of rheumatoid arthritis patients. DNA Res. 2006;13:169–183. doi: 10.1093/dnares/dsl006. [DOI] [PubMed] [Google Scholar]
- 13.Ishii T, Onda H, Tanigawa A, Ohshima S, Fujiwara H, Mima T, et al. Isolation and expression profiling of genes upregulated in the peripheral blood cells of systemic lupus erythematosus patients. DNA Res. 2005;1(2):429–439. doi: 10.1093/dnares/dsi020. [DOI] [PubMed] [Google Scholar]
- 14.Kusakabe M, Kutomi T, Watanabe K, Emoto N, Aki N, Kage H, et al. Identification of G0S2 as a gene frequently methylated in squamous lung cancer by combination of in silico and experimental approaches. Int J Cancer. 2010;126:1895–1902. doi: 10.1002/ijc.24947. [DOI] [PubMed] [Google Scholar]
- 15.Barreau O, Assié G, Wilmot-Roussel H, Ragazzon B, Baudry C, Perlemoine K, et al. Identification of a CpG island methylator phenotype in adrenocortical carcinomas. J Clin Endocrinol Metab. 2013;98:E174–E184. doi: 10.1210/jc.2012-2993. [DOI] [PubMed] [Google Scholar]
- 16.Chang X, Monitto CL, Demokan S, Kim MS, Chang SS, Zhong X, et al. Identification of hypermethylated genes associated with cisplatin resistance in human cancers. Cancer Res. 2010;70:2870–2879. doi: 10.1158/0008-5472.CAN-09-3427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kusakabe M, Watanabe K, Emoto N, Aki N, Kage H, Nagase T, et al. Impact of DNA demethylation of the G0S2 gene on the transcription of G0S2 in squamous lung cancer cell lines with or without nuclear receptor agonists. Biochem Biophys Res Commun. 2009;390:1283–1287. doi: 10.1016/j.bbrc.2009.10.137. [DOI] [PubMed] [Google Scholar]
- 18.Zandbergen F, Mandard S, Escher P, Tan NS, Patsouris D, Jatkoe T, et al. The G0/G1 switch gene 2 is a novel PPAR target gene. Biochem J. 2005;392:313–324. doi: 10.1042/BJ20050636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Choi H, Lee H, Kim TH, Kim HJ, Lee YJ, Lee SJ, et al. G0/G1 switch gene 2 has a critical role in adipocyte differentiation. Cell Death Differ. 2014;21:1071–1080. doi: 10.1038/cdd.2014.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yoon IK, Kim HK, Kim YK, Song IH, Kim W, Kim S, et al. Exploration of replicative senescence-associated genes in human dermal fibroblasts by cDNA microarray technology. Exp Gerontol. 2004;39:1369–1378. doi: 10.1016/j.exger.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 21.Yamada T, Park CS, Burns A, Nakada D, Lacorazza HD. The cytosolic protein G0S2 maintains quiescence in hematopoietic stem cells. PLoS One. 2012;7:e38280. doi: 10.1371/journal.pone.0038280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yamada T, Park CS, Shen Y, Rabin KR, Lacorazza HD. G0S2 inhibits the proliferation of K562 cells by interacting with nucleolin in the cytosol. Leuk Res. 2014;38:210–217. doi: 10.1016/j.leukres.2013.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Welch C, Santra MK, El-Assaad W, Zhu X, Huber WE, Keys RA, et al. Identification of a protein, G0S2, that lacks Bcl-2 homology domains and interacts with and antagonizes Bcl-2. Cancer Res. 2009;69:6782–6789. doi: 10.1158/0008-5472.CAN-09-0128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ma Y, Koza-Taylor PH, DiMattia DA, Hames L, Fu H, Dragnev KH, et al. Microarray analysis uncovers retinoid targets in human bronchial epithelial cells. Oncogene. 2003;22:4924–4932. doi: 10.1038/sj.onc.1206728. [DOI] [PubMed] [Google Scholar]
- 25.Kitareewan S, Blumen S, Sekula D, Bissonnette RP, Lamph WW, et al. G0S2 is an all-trans-retinoic acid target gene. Int J Oncol. 2008;33:397–404. [PMC free article] [PubMed] [Google Scholar]
- 26.Mayer N, Schweiger M, Romauch M, Grabner GF, Eichmann TO, Fuchs E, et al. Development of small-molecule inhibitors targeting adipose triglyceride lipase. Nat Chem Biol. 2013;9:785–787. doi: 10.1038/nchembio.1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Delmore JE, Issa GC, Lemieux ME, Rahl PB, Shi J, Jacobs HM, et al. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell. 2011;146:904–917. doi: 10.1016/j.cell.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Radvanyi L, Singh-Sandhu D, Gallichan S, Lovitt C, Pedyczak A, Mallo G, et al. The gene associated with trichorhinophalangeal syndrome in humans is overexpressed in breast cancer. Proc Natl Acad Sci U S A. 2005;102:11005–11010. doi: 10.1073/pnas.0500904102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Curtis C, Shah SP, Chin SF, Turashvili G, Rueda OM, Dunning MJ, et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature. 2012;18(486):346–352. doi: 10.1038/nature10983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Aguirre-Gamboa R, Gomez-Rueda H, Martìnez-Ledesma E, Martínez-Torteya A, Chacolla-Huaringa R, Rodriguez-Barrientos A, et al. SurvExpress: an online biomarker validation tool and database for cancer gene expression data using survival analysis. PLoS One. 2013;8:e74250. doi: 10.1371/journal.pone.0074250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rhodes DR, Kalyana-Sundaram S, Mahavisno V, Varambally R, Yu J, Briggs BB, Barrette TR, et al. Oncomine 3.0: genes, pathways, and networks in a collection of 18,000 cancer gene expression profiles Neoplasia. 2007;9:166–180. doi: 10.1593/neo.07112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang Y, Klijn JG, Zhang Y, Sieuwerts AM, Look MP, Yang F, et al. Gene-expression profiles to predict distant metastasis of lymph-node-negative primary breast cancer. Lancet. 2005;365:671–679. doi: 10.1016/S0140-6736(05)17947-1. [DOI] [PubMed] [Google Scholar]
- 33.Sotiriou C, Wirapati P, Loi S, Harris A, Fox S, Smeds J, et al. Gene expression profiling in breast cancer: understanding the molecular basis of histologic grade to improve prognosis. J Natl Cancer Inst. 2006;98(4):262–272. doi: 10.1093/jnci/djj052. [DOI] [PubMed] [Google Scholar]
- 34.Ivshina AV, George J, Senko O, Mow B, Putti TC, Smeds J, et al. Genetic reclassification of histologic grade delineates new clinical subtypes of breast cancer. Cancer Res. 2006;66:10292–10301. doi: 10.1158/0008-5472.CAN-05-4414. [DOI] [PubMed] [Google Scholar]
- 35.Zhang Y, Sieuwerts AM, McGreevy M, Casey G, Cufer T, Paradiso A, et al. The 76-gene signature defines high-risk patients that benefit from adjuvant tamoxifen therapy. Breast Cancer Res Treat. 2009;116:303–309. doi: 10.1007/s10549-008-0183-2. [DOI] [PubMed] [Google Scholar]
- 36.Györffy B, Lanczky A, Eklund AC, Denkert C, Budczies J, Li Q, et al. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Res Treat. 2010;123:725–731. doi: 10.1007/s10549-009-0674-9. [DOI] [PubMed] [Google Scholar]
- 37.Ringnér M, Fredlund E, Häkkinen J, Borg Å, Staaf J. GOBO: gene expression-based outcome for breast cancer online. PLoS One. 2011;6:e17911. doi: 10.1371/journal.pone.0017911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jézéquel P, Campone M, Gouraud W, Guérin-Charbonnel C, Leux C, Ricolleau G, et al. bc-GenExMiner: an easy-to-use online platform for gene prognostic analyses in breast cancer. Breast Cancer Res Treat. 2012;131:765–775. doi: 10.1007/s10549-011-1457-7. [DOI] [PubMed] [Google Scholar]
- 39.Mizuno H, Kitada K, Nakai K, Sarai A. PrognoScan: a new database for meta-analysis of the prognostic value of genes. BMC Med Genomics. 2009;2:18. doi: 10.1186/1755-8794-2-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Heckmann BL, Zhang X, Xie X, Liu J. The G0/G1 switch gene 2 (G0S2): regulating metabolism and beyond. Biochim Biophys Acta. 2013;1831:276–281. doi: 10.1016/j.bbalip.2012.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nielsen TS, Müller N. Adipose triglyceride lipase and G0/G1 switch gene 2: approaching proof of concept. Diabetes. 2014;63:847–849. doi: 10.2337/db13-1838. [DOI] [PubMed] [Google Scholar]
- 42.Kioka H, Kato H, Fujikawa M, Tsukamoto O, Suzuki T, Imamura H, et al. Evaluation of intramitochondrial ATP levels identifies G0/G1 switch gene 2 as a positive regulator of oxidative phosphorylation. Proc Natl Acad Sci U S A. 2014;111:273–278. doi: 10.1073/pnas.1318547111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zagani R, El-Assaad W, Gamache I, Teodoro JG. Inhibition of adipose triglyceride lipase (ATGL) by the putative tumors suppressor G0S2 or a small molecule inhibitor attenuates the growth of cancer cells. Oncotarget. 2015;6:28282–28295. doi: 10.18632/oncotarget.5061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Prochownik EV. c-Myc: linking transformation and genomic instability. Curr Mol Med. 2008;8:446–458. doi: 10.2174/156652408785747988. [DOI] [PubMed] [Google Scholar]
- 45.Butt AJ, McNeil CM, Musgrove EA, Sutherland RL. Downstream targets of growth factor and oestrogen signalling and endocrine resistance: the potential roles of c-Myc, cyclin D1 and cyclin E. Endocr Relat Cancer. 2005;(Suppl 1):S47–S59. doi: 10.1677/erc.1.00993. [DOI] [PubMed] [Google Scholar]
- 46.Cowling VH, Cole MD. Turning the tables: Myc activates Wnt in breast cancer. Cell Cycle. 2007;6:2625–2627. doi: 10.4161/cc.6.21.4880. [DOI] [PubMed] [Google Scholar]
- 47.Mangini NS, Wesolowski R, Ramaswamy B, Lustberg MB, Berger MJ. Palbociclib: A novel cyclin-dependent kinase inhibitor for hormone receptor-positive advanced breast cancer. Ann Pharmacother. 2015;49:1252–1260. doi: 10.1177/1060028015602273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Currie E, Schulze A, Zechner R, Walther TC, Farese RV., Jr Cellular fatty acid metabolism and cancer. Cell Metab. 2013;18:153–161. doi: 10.1016/j.cmet.2013.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rose DP, Gracheck PJ, Vona-Davis L. The interactions of obesity, inflammation and insulin resistance in breast cancer. Cancers. 2015;7:2147–2168. doi: 10.3390/cancers7040883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dirat B, Bochet L, Escourrou G, Valet P, Muller C. Unraveling the obesity and breast cancer links: a role for cancer-associated adipocytes? Endocr Dev. 2010;19:45–52. doi: 10.1159/000316896. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.