Abstract
Many cancer types, including head and neck cancers (HNC), express programmed death ligand 1 (PD-L1). Interaction between PD-L1 and its receptor, programmed death 1 (PD-1), inhibits the function of activated T cells and results in an immunosuppressive microenvironment, but the stimuli that induce PD-L1 expression are not well characterized. Interferon gamma (IFNγ) and the epidermal growth factor receptor (EGFR) utilize Janus kinase 2 (JAK2) as a common signaling node to transmit tumor cell-mediated extrinsic or intrinsic signals, respectively. In this study, we investigated the mechanism by which these factors upregulate PD-L1 expression in HNC cells in the context of JAK/STAT pathway activation, Th1 inflammation, and HPV status. We found that wild type, overexpressed EGFR significantly correlated with JAK2 and PD-L1 expression in a large cohort of HNC specimens. Furthermore, PD-L1 expression was induced in an EGFR- and JAK2/STAT1-dependent manner, and specific JAK2 inhibition prevented PD-L1 upregulation in tumor cells and enhanced their immunogenicity. Collectively, our findings suggest a novel role for JAK2/STAT1 in EGFR-mediated immune evasion, and therapies targeting this signaling axis may be beneficial to block PD-L1 upregulation found in a large subset of HNC tumors.
Keywords: JAK2, HPV, EGFR, STAT1, PD-L1
INTRODUCTION
Cancer immunoediting implies that lymphocytes successfully suppress tumor growth (1). However, tumor cell immune escape can eventually occur by downregulating HLA class I antigen processing (2) or by providing inhibitory signals (3) (4). Programmed death-1 (PD-1) is an immune checkpoint receptor expressed by tumor infiltrating lymphocytes (TIL), which limits the function of activated T lymphocytes (3, 5). Its cognate ligand, programmed death ligand-1 (PD-L1) is expressed in many types of cancers (5). Indeed, trials targeting the PD-L1/PD-1 pathway with blocking antibodies show encouraging results where tumor PD-L1 expression enriches for clinical responders (6–10). The incidence of HPV+ tumors is rapidly increasing (11) and these tumors are more responsive to oncologic therapy, which may be in part immune mediated (12–14). A previous report suggested that PD-L1 expression contributed to immune resistance in HPV+ HNC (15). In contrast, PD-1+ CD8+ T cells with an activated phenotype may be a favorable prognostic biomarker in HPV+ patients (16). Therefore, the stimuli and signaling pathways that induce PD-L1 expression in HNC may permit more effective therapeutic approaches.
PD-L1 expression in tumor cells may be regulated by two major mechanisms. First, an “extrinsic” mechanism where an antitumor cellular immune response driven by natural killer cells (NK) and CD8+ TIL produce IFNγ, which in turn may induce PD-L1 expression on tumor cells. Second, an “intrinsic” mechanism may exist in which constitutive oncogenic signaling pathways within the tumor cell lead to PD-L1 overexpression. In glioblastoma, PTEN deletion promotes PI3K-AKT mediated PD-L1 overexpression (17), while EGFR mutant lung cancer cells have been associated with PD-L1 overexpression (18, 19). In contrast to lung cancer, in the setting of HNC, EGFR mutations are extremely rare, whereas, wild type EGFR is overexpressed in approximately 80–90% of tumors (20). We hypothesized that targeting signaling molecules involved in PD-L1 expression in HNC cells might synergize with current anti-EGFR antibody targeted immunotherapies such as cetuximab, that are known to activate NK cells and cytotoxic T lymphocytes (CTL) (21). However, because activated NK and T cells secrete IFNγ, a known stimulus for PD-L1 expression, understanding the complex signaling pathway regulating PD-L1 is crucial. Since extrinsic (IFNγ-mediated) and intrinsic (EGFR-mediated) mechanisms may cooperate to promote PD-L1 upregulation, we investigated signaling pathways that mediate both IFNγ and EGFR induced PD-L1 upregulation in tumor cells. These findings have relevance for immunotherapeutic combinations with cetuximab, which can both block EGFR signaling and stimulate IFNγ secretion via activation of NK cells and CTL (21–23).
MATERIALS AND METHODS
Tumor cell lines
HPV− HNC cell lines (JHU020, JHU022, JHU029, PCI13) and HPV+ (SCC2, SCC47, SCC90, 93VU) were cultured in IMDM complete media (Mediatech, Herndon, VA). JHU020, JHU022 and JHU029 were a gift from Dr. James Rocco in January of 2006 (Ohio State University). SCC90 and PCI13 were isolated from patients treated at the University of Pittsburgh through the explant/culture method, authenticated and validated using STTR profiling and HLA genotyping (24, 25). SCC2 and SCC47 were a kind gift from Dr. Thomas Carey in December of 2005 (University of Michigan) and 93VU a gift from Dr. Henning Bier in October of 2013 (Technische Universitat Munchen, Germany). HNC lines were tested every 6 months and were free of Mycoplasma infection.
Antibodies and treatments
The anti-PD-L1 mAb (clone 405.9A11) was previously validated (26) by Dr. Gordon J. Freeman (Dana-Farber Cancer Institute, Boston, MA), anti-pJAK2(Y1007 and Y1008) (Abcam, Cambridge, UK) were used for IHC staining. The anti-PD-L1-PE (BD Pharmingen, San Jose, CA) anti-HLA-ABC-FITC mAb (clone w6/32, E-biosciences, San Diego, CA), IgG1-PE isotype control, pSTAT1 Tyr701-PE, STAT1-PE and STAT3-PE (BD Biosciences, San Jose, CA), phospho-AKT(Thre308)-PE and primary anti-p44/42 MAPK(Erk1/2) and phospho-p44/42 MAPK(Erk1/2) (Thr202/Tyr204) and secondary anti-rabbit-PE antibodies (Cell Signaling, Danvers, MA). WB antibodies included rabbit anti-human JAK2, pJAK2 (y1007/1008), total AKT, pAKT, pERK and mouse anti-human β-actin (Cell signaling, Danvers, MA). IFNγ (R&D systems Minneapolis, MN) was used at 10IU/mL. IFNα2a (PBL Interferon Source, Piscataway, NJ) was used at 1000 IU/mL. JAK2 inhibitor BMS-911543 was characterized previously (25), provided by Bristol-Myers Squibb and used at 10uM. JAK1/3 inhibitor (ZM39923) (Tocris bioscience, Bristol, United Kingdom) was characterized previously (27, 28) and was used at 10uM. Wortmannin (Cell Signaling, Danvers, MA) was used at a 1uM. PI3Kα110 subunit inhibitor (BYL-719) was used at a 1uM and MEK1/2 inhibitor (PD0325901) was used at 5uM (Tocris Bioscience, Bristol, UK).
Flow cytometry analysis
Cell viability was determined by Zombie Aqua staining (29) (Biolegend, San Diego, CA), then incubated with fluorophore-conjugated antibodies at 1:10 dilution for 15 minutes at 4 C, then washed twice and resuspended in 2% PFA solution until analysis. Intracellular flow cytometry was performed (30) and analyzed using an LSR Fortessa cytometer (BD Biosciences), and FlowJo version 10 software (Ashland, OR). Median fluorescence intensity (MFI) fold change calculated by normalizing after subtracting the isotype control MFI.
Western Blotting
Western blotting was performed as described previously (2) using the indicated antibodies.
siRNA knockdown
Cells were transfected as described elsewhere (2). After 48h transfection, cells were incubated ±IFNγ (10IU/mL) or EGF (10ng/mL) for 48h, then harvested and analyzed by FC. siRNA STAT1: 5-CUACGAACAUGACCCUAUTT-3(s) and 5-AUAGGGUCAUGUUCGUAGGTG-3(as) siRNA STAT3: 5-GCCUCAAGAUUGACCUAGATT-3(s) and 5-UCUAGGUCAAUCUUGAGGCCT-3(as) and siRNA non-targeting 5-AGUACAGCAAACGAUACG Gtt-3 control: (s) and 5-CCGUAUCGUUUGCUGUACUtt-3(as).
Quantitative PCR (qPCR)
qPCR was performed as described previously (31). PCR probes for PD-L1 (Hs01125301_m1) and GUSB (Hs99999908_m1) (Applied Biosystems for TaqMan® Gene Expression Assay). GUSB was amplified as an internal control. Relative expression of the target genes to GUSB control gene was calculated using the ΔCT method: relative expression = 2 − ΔCT, where ΔCT = CT (target gene) − CT (GUSB).
Chromatin immunoprecipitation (ChIP) assay
Cells were serum starved for 18 h prior to incubation with IFN-γ (10 IU/ml) for 30 min at 37°C, or sequentially with cetuximab (10 ug/ml) for 30 min at 37°C. Then ChIP assay was performed as described previously (2) using the Ez-ChIP™ kit (Millipore). Purified DNA was used in quantitative RT-PCR using the EpiTect ChIP qPCR (Qiagen) SYBR-green Master Mix method using a primer for PD-L1 promoter NM_014143.2 (-)16Kb. qPCR amplification data were normalized and analyzed as %input (32) and expressed as relative enrichment to % input.
Immunohistochemistry (IHC) protocol
The University of Pittsburgh IRB #99-069 approved the use of clinical samples and written informed consent was obtained. Slides were deparaffinized and rehydrated. Antigen retrieval was performed using Diva Retrieval (Biocare Medical, Concord, CA) and a decloaking chamber at 124°C, 3 minutes, and cooled for 10 min. Slides were placed on an Autostainer Plus (Dako, Carpenteria, CA) using a TBST rinse buffer (Dako) and stained using: 3% H2O2 (ThermoFisher Scientific, Pittsburgh, PA) for 5 minutes, CAS Block (Invitrogen, Grand Island, NY) for 10 minutes, the primary antibody for PD-L1 (clone 405.9A11) (33) and the pJAK2 (Y1007-1008) (clone E132) used per instructions. The secondary consisted of Envision Dual Link + (Dako) polymer for 30 min, rinsed, then a TBST holding rinse was applied for 5 min. The substrate used was 3,3, Diaminobenzidine + (Dako) for 7 minutes and counterstained with hematoxylin. PD-L1 and pJAK2 staining were quantified by positive pixel count v9 algorithm (Aperio). A head and neck pathologist blinded to clinical patient data examined tumor sections. Scoring was determined by %tumor stained for PD-L1 or pJAK2, respectively. Tumors with <5% tumor cells positive staining were considered negative.
Cellular cytotoxicity assays
Cytotoxicity was determined by 51Cr release assay as described before (34). Tumor targets and NK cells were co-incubated with cetuximab or IgG1 isotype (10μg/mL) for 4h. Controls for spontaneous and maximal lysis were included. Specific lysis = (experimental lysis − spontaneous lysis)/(experimental lysis − maximal lysis) × 100.
TCGA data retrieval and analysis
RNAseq data from queried genes were downloaded from the UCSC cancer genomics browser (https://genome-cancer.ucsc.edu). The HNC gene expression profile from 500 HNC specimens was measured experimentally (35). The RSEM units to quantitate RNAseq expression data were described previously (36). Correlations from TCGA data were calculated using Pearson r test and linear regression curve fits were graphed using GraphPad PRISM software v6.
IPA Ingenuity pathway software analysis
Software was accessed via the University of Pittsburgh license. Path Explorer tool available in IPA Ingenuity was utilized for associating EGFR and IFNγ pathways with PD-L1 expression.
RESULTS
PD-L1 protein expression is higher in HPV+ HNC tumor specimens
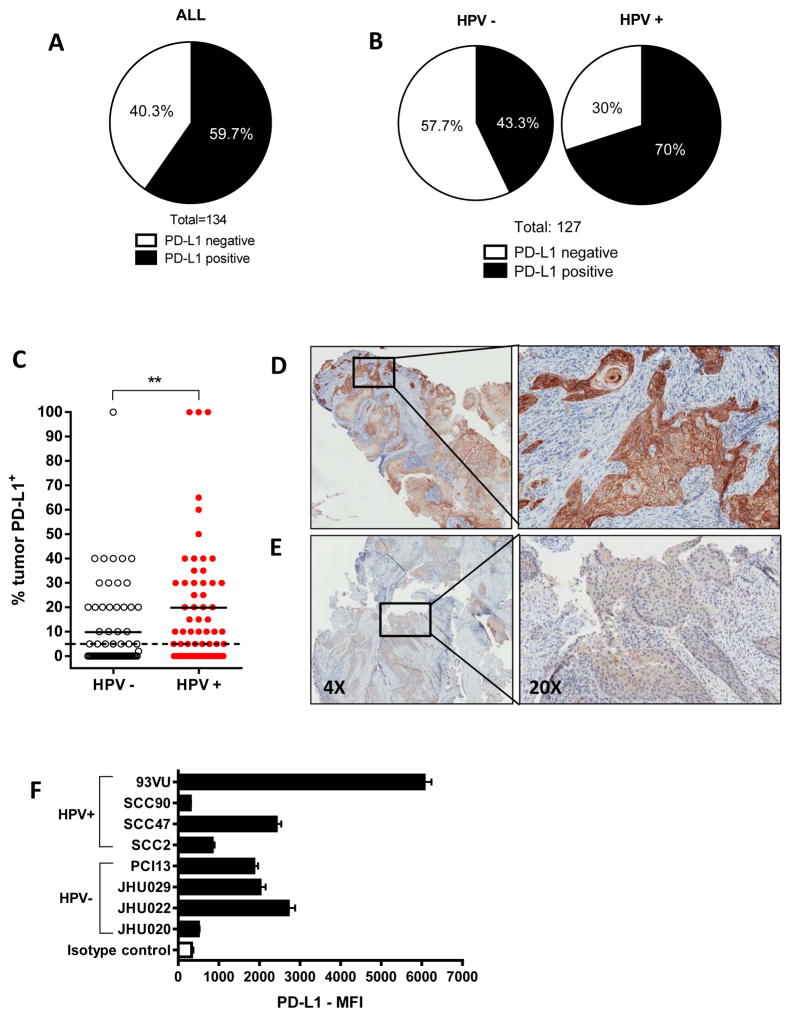
IHC staining of tumor specimens (n=134) revealed that 59.7% of HNC patients express detectable PD-L1 on the tumor cell surface, as determined by >5% positive tumor cells (Figure 1A). Furthermore, when segregated by HPV status (n=127, 63 HPV− and 64 HPV+), we noted that PD-L1 expression was more frequent in HPV+ specimens (70% vs. 43.3%, respectively, Figure 1B) and the % PD-L1 expression was also significantly higher in in HPV+ tumors (Figure 1C). Interestingly, PD-L1 expression was more intense on the cell membrane than in the cytoplasm and was heterogeneously expressed within the microenvironment, generally forming clusters of PD-L1+ tumor cells with a higher intensity at the cluster periphery (Figure 1D and E). To study the stimuli and pathways by which PD-L1 is upregulated in vitro, we analyzed a panel of HPV+ and HPV− HNC lines for PD-L1 expression, which was expressed variably (Figure 1F) similar to patient tumors by IHC.
Figure 1. PD-L1 protein expression is higher in HPV+ tumor specimens.
A. PD-L1 protein expression in HNC tumor specimens (IHC, n=134). Tumors were considered positive for PD-L1 when higher than 5% tumor staining threshold. 59.7% of tumors were PD-L1+. B. PD-L1 expression in HPV− and HPV+ tumor specimens using the same criteria as in A. 70% HPV+ vs. 43.3% HPV− specimens were PD-L1+ C. percent PD-L1+ tumor area in HPV− and HPV+ specimens. Dotted line represents the 5% tumor positive, solid lines represent the median. (Mann-Whitney, ** P<0.001). D. Representative high intensity, 100% PD-L1+, HPV+ tumor. E. Representative low intensity, 50% PD-L1+ HPV− tumor. Insets on the left represent magnification (20X) on the right. F. HNSCC cells express heterogeneous levels of PD-L1.
HPV+ tumor specimens show higher Th1 type expression profile
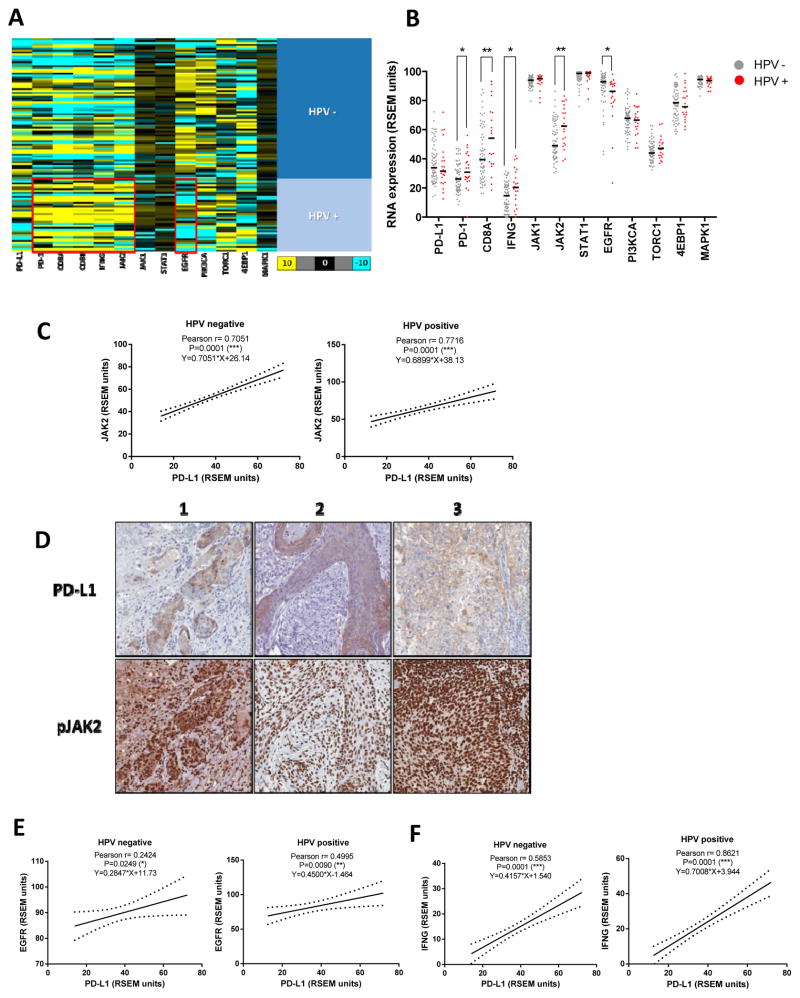
We then analyzed PD-L1 expression in a large cohort of HNC specimens for which gene expression TCGA repository data were available (35). Since PD-L1 expression has been linked with that of CD8 and IFNγ (15, 23), we investigated the Th1 mRNA expression profile of HPV+ versus HPV− specimens. Pooled data from 88 HNC specimens were plotted using a heat map, segregated by HPV status (Figure 2A, red boxes depict higher expression in HPV+ tumors). A Th1 type expression profile (PD-1, CD8A, CD8B, IFNG and JAK2) was significantly higher in HPV+ than HPV− tumors (Figure 2B) suggesting that activated immune effector cells readily infiltrate HPV+ tumors, which may be important for PD-L1 induction due to this source of IFNγ. Importantly, JAK2 expression (but not JAK1) was also higher in HPV+ tumors (Figure 2B). Therefore, JAK2 was associated with a Th1 profile and with PD-L1 expression, particularly in HPV+ tumors.
Figure 2. HPV+ specimens show higher expression of a Th1 type RNA expression profile and PD-L1 expression correlates with that of JAK2, EGFR and IFNγ regardless HPV status.
A. Heat map of RNAseq expression expressed as RSEM units (as described in Materials and Methods) of PD-L1, PD-1, CD8A, CD8B, IFNγ, JAK2, JAK1, STAT1, EGFR, PIK3CA, TORC1, 4EBP1 and MAPK1 (66 HPV− and 22 HPV+) TCGA database (35), red boxes emphasize higher expression of a Th1 profile in HPV+ specimens and higher EGFR expression in HPV− counterparts (color code, yellow 10 fold higher, turquoise 10 fold lower relative expression change over black). B. HPV+ tumor specimens show significantly higher expression of a Th1 type expression profile: PD-1, CD8A, IFNγ and JAK2. EGFR expression is significantly higher in HPV− tumor specimens (Mann-Whitney * P<0.05, ** P<0.001). C. PD-L1 expression significantly correlated with JAK2 in both HPV− and HPV+ specimens (Pearson r and linear regression curve fit, *** p<0.0001). D. PD-L1 and pJAK2 IHC staining in adjacent sections and matching areas of HNC specimens. PD-L1 (top panel) was predominantly expressed on the tumor cell membrane. Phospho-JAK2 (bottom panel) exhibits strong nuclear staining with occasional weak-moderate cytoplasmic staining. PD-L1 positive tumor islands are also diffusely strongly positive for phospho-JAK2 (3 representative specimens out of 23) E. PD-L1 mRNA expression significantly correlated with EGFR in both HPV− and HPV+ specimens (Pearson r and linear regression curve fit * P<0.05 ** P<0.001). F. PD-L1 mRNA expression significantly correlated with IFNγ regardless HPV status (Pearson r and linear regression curve fit *** P<0.0001). 66 HPV− and 22 HPV+ tumor specimens collected from TCGA database.
PD-L1 expression correlates with that of JAK2, EGFR, IFNγ and a Th1 profile regardless HPV status
Given that JAK2 is a common signaling molecule downstream of the EGFR and IFNγ pathways, we found that PD-L1 and JAK2 mRNA expression were highly correlated (n=500) and persisted when the cohort was segregated by HPV status (Figure 2C). In order to further assess the relationship between JAK2 and PD-L1 in vivo at the protein level, we determined phospho-JAK2 and PD-L1 using IHC from adjacent sections of HNC specimens (n=23). Corroborating our previous findings, PD-L1 was predominantly expressed on the tumor cell membrane while phospho-JAK2 exhibited strong nuclear staining, with occasional weak-moderate cytoplasmic staining. PD-L1 positive tumor islands were found to be strongly positive for phospho-JAK2 (Figure 2D). Furthermore, we also found a significant correlation between EGFR and PD-L1 expression, which was somewhat weaker in HPV− tumors (Figure 2E). Likewise, PD-L1 expression was highly correlated with a Th1 type expression profile (IFNγ, CD8A and PD-1) regardless of HPV status (Figure 2F and Table 1A). In addition, correlation of EGFR and CD8 or JAK2 was only significant in HPV+ tumors, given that their expression level was higher than in HPV− tumors. However, this finding did not preclude the fact that JAK2 could also be important for PD-L1 expression in HPV− tumors given that they were strongly correlated regardless of HPV status.
Table 1. A. Correlation of PD-L1, EGFR and IFNG with a Th1 profile in HPV negative and HPV positive tumors.
PD-L1 mRNA expression highly correlated with that of PD-1 and CD8A regardless HPV status. In addition, EGFR expression correlated with that of CD8A or JAK2 in HPV+ but not HPV− tumors. As expected, IFNγ shows a strong correlation with that of CD8A and JAK2 regardless HPV status B. Correlation of PD-L1 and STAT3, PI3K and MAPK pathway components in HPV negative and HPV positive tumors. PD-L1 mRNA expression did not show a strong correlation with STAT3, PI3K and MAPK pathways regardless HPV status. (TCGA, 66 HPV− and 22 HPV+ tumor specimens).
| A
| ||||
|---|---|---|---|---|
| Correlation (XY) | HPV Negative | HPV Positive | ||
|
| ||||
| Pearson r | P value | Pearson r | P value | |
| PD-L1 vs PD-1 | 0.5937 | < 0.0001 (***) | 0.7538 | < 0.0001 (***) |
| PD-L1 vs CD8A | 0.5157 | < 0.0001 (***) | 0.8363 | < 0.0001 (***) |
|
| ||||
| EGFR vs CD8A | −0.01368 | 0.4566 (ns) | 0.4705 | 0.0136 (*) |
| EGFR vs JAK2 | 0.1853 | 0.0681 (ns) | 0.438 | 0.0207 (*) |
|
| ||||
| IFNγ vs CD8A | 0.7747 | < 0.0001 (***) | 0.9396 | < 0.0001 (***) |
| IFNγ vs JAK2 | 0.7264 | < 0.0001 (***) | 0.7391 | < 0.0001 (***) |
| B
| ||||
|---|---|---|---|---|
| Correlation (XY) | HPV Negative | HPV Positive | ||
|
| ||||
| Pearson r | P value | Pearson r | P value | |
| PD-L1 vs STAT3 | 0.2503 | 0.0427 (*) | 0.3867 | 0.0754 (ns) |
|
| ||||
| PD-L1 vs AKT1 | −0.2048 | 0.099 (ns) | 0.00568 | 0.98 (ns) |
| PD-L1 vs TORC1 | −0.2743 | 0.0258 (*) | −0.1973 | 0.3787 (ns) |
| PD-L1 vs 4EBP1 | −0.2488 | 0.044 (*) | −0.2206 | 0.3238 (ns) |
|
| ||||
| PD-L1 vs MAPK1 | −0.1659 | 0.1832 (ns) | 0.07862 | 0.728 (ns) |
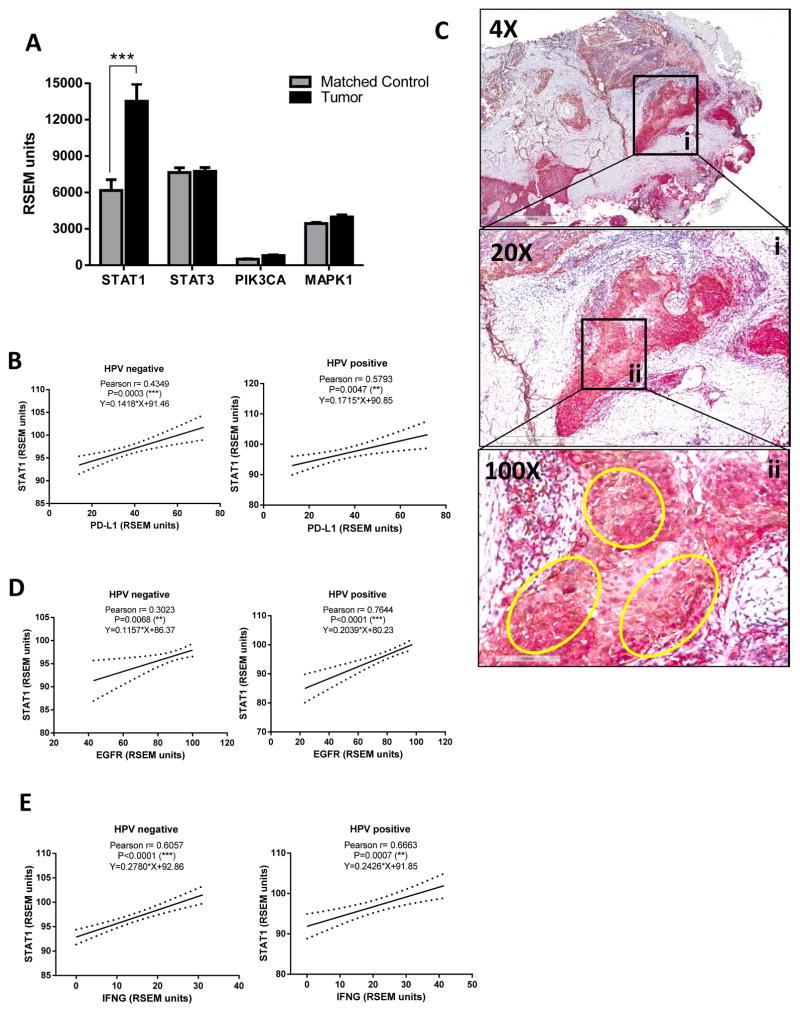
STAT1 but not STAT3, PIK3CA or MAPK1 expression is higher in tumor tissue and strongly correlates with PD-L1, EGFR and IFNγ regardless of HPV status
Since PD-L1 expression strongly correlated with a Th1 type expression profile in the tumor microenvironment, we hypothesized that PD-L1 may depend on STAT1 activation, a known Th1 type transcription factor. Indeed, STAT1 emerged as one of the highly predicted transcription factors binding to PD-L1 promoter region and common to EGFR and IFNγ pathways when utilizing previously validated software for transcription factor binding prediction (MATCH) and pathway exploration (Ingenuity IPA) (37). Given that previous reports presented STAT3, PI3K and MAPK as possibly involved in PD-L1 expression in other tumor types, we included these in our investigation. We pooled RNAseq data collected from 46 paired specimens of tumor vs. matched normal mucosa and found that STAT1 (but not STAT3, PIK3CA or MAPK1) was significantly upregulated in tumor tissue (Figure 3A). Furthermore, STAT1 expression highly correlated with that of PD-L1 in TCGA (n=500), which was preserved when segregated by HPV status (Figure 3B). Concordant with TCGA data, we found that STAT1 protein was widely expressed in HNC tumor tissues, and that PD-L1 positive tumor islands were also strongly positive for total STAT1 staining (Figure 3C, circled areas highlight co-localization, 100X inset). Interestingly, STAT1 expression also showed strong correlation with that of EGFR (Figure 3D). As expected, STAT1 tumor expression also was strongly correlated with that of IFNγ (Figure 3E). Notably, STAT3 and PI3K pathway components (AKT1, TORC1 or 4EBP1) showed no correlation with PD-L1 in HPV+ tumors and only weakly in the HPV− HNC. Likewise, MAPK1 was not correlated with PD-L1 expression in either cohort (Table 1B). Overall, our findings suggest that the JAK2/STAT1 pathway may serve as an important common mediator for both EGFR- and IFNγ-mediated PD-L1 expression in HNC tumors, regardless of HPV status.
Figure 3. STAT1, but not STAT3, PI3KCA or MAPK1 expression is higher in tumor tissue and strongly correlates with that of PD-L1, EGFR and IFNγ regardless HPV status.
A. Expression of STAT1 but not STAT3, PIK3CA or MAPK1 was significantly higher in tumor specimens when compared with matched control mucosa (TCGA, 46 HNC tumor specimens and matched controls, Mann-Whitney, *** P<0.0001). B. STAT1 expression is strongly correlated with PD-L1 regradless HPV status (Pearson r and linear regression curve fit, ** P<0.001 *** P<0.0001) C. PD-L1+ tumor islands are also strongly positive for STAT1 protein in HNC specimens. Representative section of a HNC specimen co-stained for PD-L1 (brown chromogen) and STAT1 (red chromogen). Insets represent magnification of the tumor area, yellow circles indicate co-localization. D–E. STAT1 expression correlated with that of EGFR and IFNγ regardless HPV status. (Pearson r and linear regression curve fit, ** P<0.001 *** P<0.0001).
IFNγ-mediated PD-L1 upregulation is JAK2/STAT1 dependent
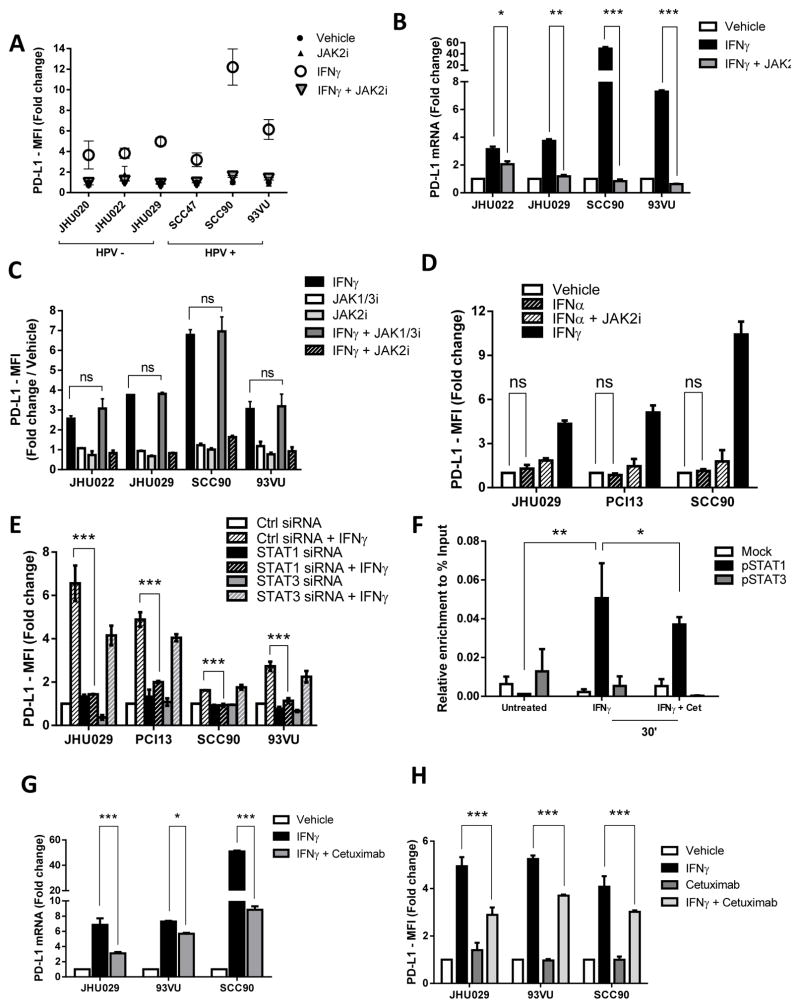
Based on our TCGA analysis and previous reports linking IFNγ with PD-L1 expression at the mRNA level, we investigated the signaling pathway by which IFNγ upregulates PD-L1 expression in vitro. Indeed, a panel of HPV+ and HPV− HNC cell lines upregulated PD-L1 expression after IFNγ treatment (Figure 4A). Given that IFNγ-mediated PD-L1 upregulation was linked with PI3K pathway activation (17), we tested whether wortmannin (pan-PI3K inhibitor) or BYL-719 (PI3Kα110 subunit specific inhibitor) could prevent IFNγ-mediated PD-L1 upregulation. PI3K pathway inhibition did not induce PD-L1 downregulation, under conditions in which these inhibitors effectively prevented AKT phosphorylation (Supplementary Figure 1 A–D). Since IFNγ signals via JAK1 and JAK2, we utilized a clinical grade, selective JAK2 inhibitor BMS-911345 (JAK2i) which was previously characterized (38), finding an abrogation of IFNγ-mediated PD-L1 upregulation in all cell lines tested, both at the mRNA and protein level (Figure 4A and 4B, Supplementary Figure 1E–F). Interestingly, specific JAK1/3 inhibition (JAK1/3i) did not show a significant downregulation of IFNγ-mediated PD-L1 protein expression (Figure 4C). We then used IFNα, which signals via JAK1 and TYK2, but not JAK2. IFNα treatment did not upregulate PD-L1 expression (Figure 4D) but still induced pSTAT1 upregulation, although to a lesser extent than IFNγ, in all cell lines tested. Moreover, JAK2 inhibition did not affect HLA-ABC upregulation (Supplementary Figure 2A–B), which suggests that the kinetics of IFNα-induced pSTAT1 binding to the PD-L1 promoter differ for HLA-ABC.
Figure 4. IFNγ mediated PD-L1 upregulation is JAK2/STAT1 dependent.
A. JAK2 inhibitor BMS-911345 (JAK2i) abrogates IFNγ-mediated PD-L1 protein upregulation regardless HPV status. HNSCC cells were treated with vehicle control, JAK2i (10uM), IFNγ (10IU/mL) or the combination for 48h, harvested and PD-L1 expression was determined by flow cytometry (FC) B. JAK2i abrogates IFNγ-mediated PD-L1 mRNA upregulation. Cell lines were treated with vehicle control, IFNγ (10IU/mL) or JAK2i (10uM) or the combination for 24h; PD-L1 mRNA was determined by qPCR and expressed as fold change over vehicle C. JAK1/3 inhibition did not prevent IFNγ-mediated PD-L1 upregulation. Cell lines were treated JAK1/3i (10uM), JAK2i (10uM), IFNγ (10IU/mL) or the combination for 48h and PD-L1 expression was determined by FC (ANOVA, ns= not significant) D. IFNα did not upregulate PD-L1 expression. Cells were incubated with IFNα (1000 IU/mL), JAK2i (10uM) and the combination for 48 h. IFNγ (10IU/mL) was used as a positive control. PD-L1 expression was determined by FC (ANOVA, ns= not significant). E. IFNγ-mediated PD-L1 upregulation is abrogated when STAT1, but not STAT3, is silenced. Cells were incubated with STAT1 siRNA, STAT3 siRNA or control siRNA (10nM) for 48h then were left untreated or treated with IFNγ (10IU/mL) for additional 48h, harvested and PD-L1 expression was determined by FC. F. pSTAT1 but not pSTAT3 binds to the PD-L1 promoter region after IFNγ treatment as determined by ChIP assay. Cells were either left untreated or treated with IFNγ (10IU/mL) or IFNγ+cetuximab for 30 minutes, ChIP assay showed enrichment of pSTAT1 in the PD-L1 promoter (black bars). PD-L1 enrichment calculated as % input DNA (refer to Materials and Methods) (ANOVA * P<0.05, ** P<0.01). G. Cetuximab-mediated EGFR blockade downregulated IFNγ-mediated PD-L1 mRNA upregulation. Cell lines were either treated with vehicle, IFNγ (10IU/mL), cetuximab (10ug/mL) or IFNγ+cetuximab for 24 h. harvested and mRNA was quantified by qPCR and expressed as fold change over vehicle (ANOVA, * P<0.05, *** P<0.0001) H. Cetuximab-mediated EGFR blockade downregulated the IFNγ-mediated PD-L1 protein upregulation. Cell lines were either treated as in G for 48h harvested and PD-L1 protein expression was determined by FC (ANOVA, *** P<0.0001)
In order to determine whether the IFNγ-mediated PD-L1 upregulation was solely STAT1 dependent, we silenced each transcription factor using siRNA technology (80–90% knockdown efficiency, supplementary Figure 3A). STAT1 but not STAT3 knockdown potently impaired IFNγ-mediated upregulation of PD-L1 (Figure 4E). Moreover, chromatin immunoprecipitation (ChIP) assays documented that pSTAT1 but not pSTAT3 binds to the promoter region of PD-L1 after IFNγ treatment (Figure 4F). Interestingly, cetuximab mediated EGFR blockade downregulated IFNγ induced pSTAT1 binding to the PD-L1 promoter and significantly downregulated the IFNγ-mediated PD-L1 upregulation at the mRNA and protein level, respectively. (Figure 4G–H and Supplementary Figure 3B).
EGFR-mediated PD-L1 upregulation is JAK2/STAT1 dependent
Since EGFR strongly correlated with PD-L1 expression in TCGA specimens and a previous report showed that EGFR activating mutations induce PD-L1 in lung cancers (18), we hypothesized that wild type EGFR, overexpressed in 80–90% of HNC, may promote PD-L1 upregulation. EGF treatment induced upregulation of PD-L1 protein in 7 of 8 HNC lines studied, though to a lesser extent than that induced by IFNγ (Figure 5A). This effect was also seen at the mRNA level (Figure 5B). Although EGFR activates multiple downstream pathways, including PI3K, MAPK and JAK/STAT pathway, TCGA analysis yielded weak if any correlation between PD-L1 and PIK3CA or MAPK1 (Table 1A). However, a strong correlation was observed with that of JAK2 and STAT1 (Figure 2C and 3B respectively). Given that JAK2 serves as a common signaling molecule for both IFNγ and EGFR pathways, we investigated whether EGF-mediated PD-L1 upregulation was JAK2 and/or STAT1 dependent. Indeed, basal expression of PD-L1 in HNC cell lines was downregulated by JAK2 but not JAK1/3 inhibition (Figure 5C). Furthermore, EGF induced JAK2 phosphorylation (Supplementary Figure 4A) and upregulation of basal PD-L1 expression (Figure 5D). Additionally, specific JAK2, but not JAK1/3, inhibition prevented EGF induced PD-L1 upregulation (Figure 5D and Supplementary Figure 4B). Interestingly, the EGF-mediated PD-L1 upregulation was higher in cell lines with a higher EGFR expression (JHU029 and JHU022 vs 93VU and SCC90). Likewise, JAK2 inhibition more strongly downregulated basal and EGF-mediated PD-L1 expression in the EGFRhigh cell lines (Figure 5D, JHU022 and JHU029).
Figure 5. EGFR-mediated PD-L1 upregulation is JAK2/STAT1 dependent.
A. EGF upregulates PD-L1 protein expression. Cells were either left untreated or treated with EGF (10ng/mL) for 48h, IFNγ (10IU/mL) was used as a positive control. Cells were harvested and PD-L1 surface expression was determined by FC B. EGF treatment upregulates PD-L1 mRNA expression. Cells were treated as in A for 24h, harvested and PD-L1 mRNA expression was determined by qPCR and expressed as fold change over vehicle control (ANOVA, * P<0.05, ** P<0.01, *** P<0.001). C. JAK2 but not JAK1/3 inhibition downregulates baseline expression of PD-L1. Cells were treated with JAK1/3 inhibitor (JAK1/3i, 10uM) or JAK2i (10uM) for 48h and PD-L1 expression level was determined by FC (ANOVA, * P<0.05, ** P<0.001). D. JAK2 but not JAK1/3 inhibition prevents EGF-mediated PD-L1 upregulation. Cells were treated with JAK1/3i (10uM) or JAK2i (10uM) for 48h and PD-L1 expression level was determined by FC (ANOVA, ns= non significant ** P<0.001 *** P<0.0001). E. EGF induces pSTAT1y701 upregulation. Cells were treated with EGF (10ng/mL) for 1, 2, 4, 24 and 48 hours, harvested, fixed and permeabilized and pSTAT1y701 or total STAT1 expression were determined by ICF. F. STAT1 silencing prevents EGF induced PD-L1 upregulation. Cells were treated with control siRNA or STAT1 siRNA (10nM) and EGF (10ng/mL) for 48h, harvested and PD-L1 expression was determined by FC (ANOVA, *** P<0.001).
Since EGFR activates PI3K and MAPK pathways, we tested whether these mediated PD-L1 upregulation after EGFR stimulation. We found that neither wortmannin-mediated PI3K inhibition nor MEK1/2-mediated MAPK inhibition prevented EGF-induced PD-L1 upregulation (Supplementary Figure 4C–D). However, these inhibitors effectively suppressed AKT and ERK phosphorylation, respectively (Supplementary Figure 4E–F). In light of this result and the positive correlation found between EGFR and STAT1, we hypothesized that EGF may be activating STAT1 phosphorylation, mediated by JAK2. Indeed, EGF induced STAT1 (tyrosine701) phosphorylation reaching its maximum peak at 24 hours, while total STAT1 levels remained stable (Figure 5E). Furthermore, siRNA-targeted STAT1 knockdown efficiently suppressed total STAT1 levels, (Supplementary Figure 5) as well as significantly abrogating EGF induced PD-L1 upregulation (Figure 5F).
JAK2 inhibition prevents tumor PD-L1 expression and enhances cetuximab mediated NK cell cytotoxicity
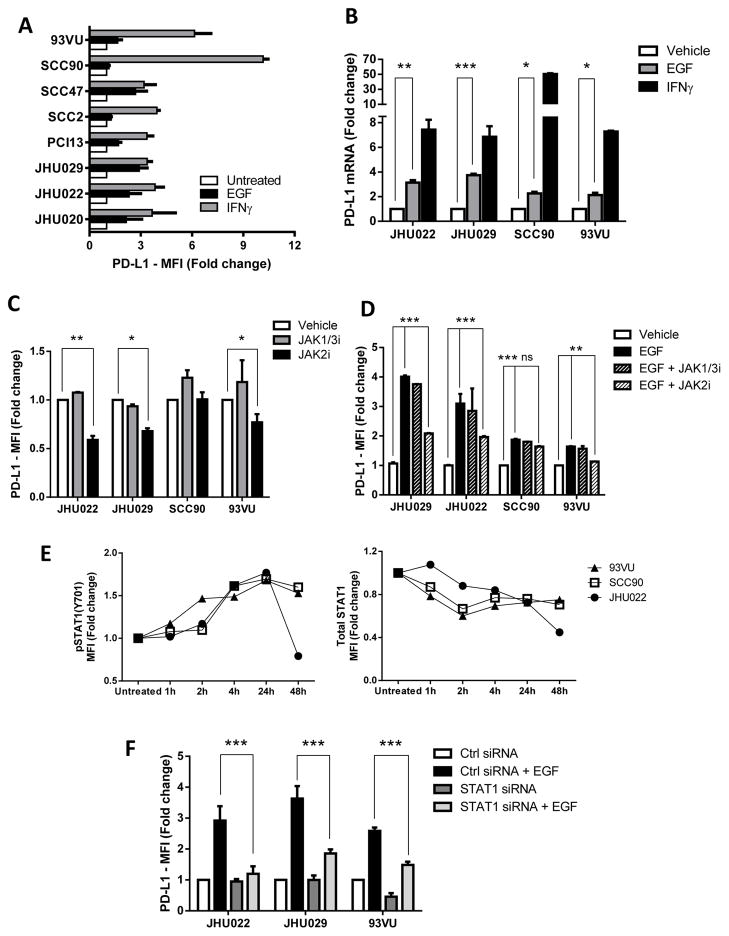
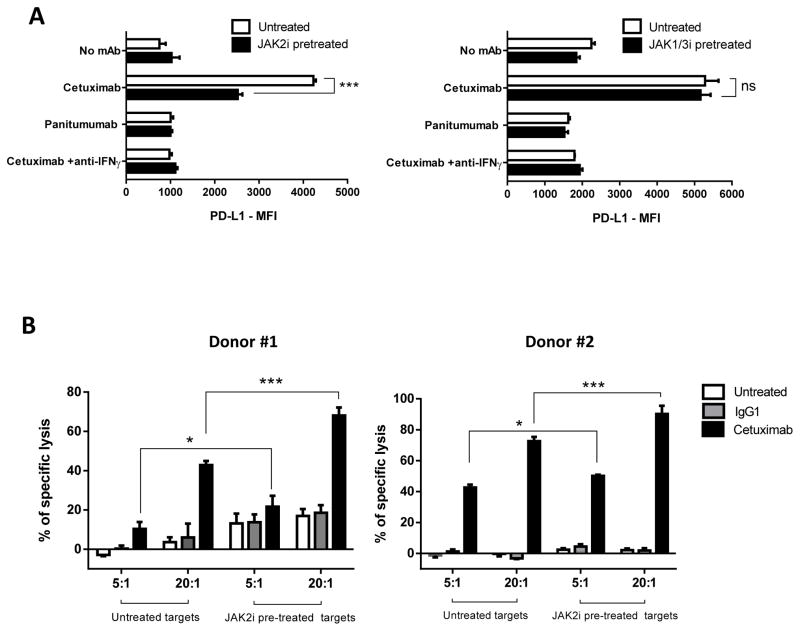
Since JAK2 represents a key player in PD-L1 upregulation in both EGFR (intrinsic) and IFNγ (extrinsic) pathways in vitro, we tested whether JAK2 inhibition enhanced NK mediated killing via antibody dependent cell cytotocixity (ADCC) (21) against PD-L1+ HNC cells. When NK cells were co-cultured with HNC targets and cetuximab, activated NK cells upregulated tumor PD-L1 expression in an IFNγ dependent fashion (Figure 6A, open bars). As a control, the EGFR specific mAb panitumumab (IgG2 isotype) which does not bind to CD16 on NK cells, did not induce PD-L1 upregulation, most likely because of a lack of NK cell activation and IFNγ secretion (39). Importantly, the IFNγ mediated PD-L1 upregulation on HNC cells was prevented when they were pre-treated with the JAK2 inhibitor (left panel, closed bars), but not with a JAK1/3 specific inhibitor (right panel, closed bars). We therefore tested the hypothesis that NK cells would more efficiently lyse JAK2 inhibitor pre-treated tumor cells, in the setting of reduced PD-L1 expression. Indeed, NK cells showed aproximately 25% higher specific lysis of HNC cells pre-treated with the JAK2 inhibitor (Figure 6B). Overall, these results confirm that JAK2 is an important regulator of PD-L1 expression in HNC tumor cells, and its inhibition reverses PD-L1 mediated tumor cell escape from cetuximab mediated ADCC.
Figure 6. JAK2 inhibition prevents NK mediated PD-L1 upregulation on tumor cells and enhances cetuximab mediated NK cell cytotoxicity.
A. PD-L1 expression is upregulated on tumor cells when co-cultured with cetuximab-activated NK cells in IFNγ dependent fashion (left panel, open bars). JAK2i pre-treatment of tumor targets prevented PD-L1 upregulation (left panel, closed bars). In contrast, JAK1/3 inhibition did not prevent PD-L1 upregulation under the same conditions (right panel, closed bars). Tumor target cells were incubated in media alone or JAK2i (10uM) supplemented media for 48 hours then co-cultured with NK cells for 24 hours either alone or in the presence of cetuximab (10ug/mL), panitumumab (10ug/mL) or cetuximab+anti-IFNγ blocking antibody (50ug/mL), harvested and PD-L1 expression on tumor cells was determiend by FC. Data representattive of two independent experiments with similar results B. Higher NK-cetuximab mediated lysis of JAK2i pre-treated targets (closed black bars, 5:1 and 20:1 effector:target ratio). Tumor cells were pre-treated with JAK2i (10uM) for 48 hours then labeled with 51Cr and co-cultured with purified NK cells plus media, IgG1 control (10ug/mL) or cetuximab (10ug/mL) for 4h (ANOVA, * P<0.05, *** P<0.0001).
DISCUSSION
Given the known importance of HPV infection in the etiology of HNC, several studies have sought to correlate PD-L1 expression with HPV status. Recent reports have shown higher PD-L1 expression in HPV+ tumors (15, 16, 40, 41). Lyford-Pike et al. (15) reported only 29% PD-L1 positivity among a small cohort (n=9) of HPV− HNC patients while Malm et al. reported 80% (41) making the association of PD-L1 expression with HPV status controversial. In our large series of 134 patients, the majority (approximately 60%) was PD-L1+. Most importantly, we found HPV+ tumors to be more frequently PD-L1 positive (70%, n=64) and have a significant higher percent area and intensity of PD-L1. Importantly, a previous study showed that PD-L1 co-localized with CD3 in 56% of tumors while 44% showed a diffuse pattern with no co-localization noted (41), raising the question of how PD-L1 is regulated in those tumors. Thus, PD-L1 expression could be “extrinsically” induced by IFNγ secreting TILs (where co-localization was found), particularly in HPV+ tumors and “intrinsically” induced via endogenous EGFR signaling (where no co-localization was found), particularly in HPV− tumors.
Since HPV+ tumors showed higher PD-L1 protein expression in vivo and to extend findings reported previously, we took advantage of the RNAseq expression data available in TCGA (n=500). HPV+ tumors have significantly higher expression of a Th1 type profile including CD8A, PD-1, IFNG and JAK2 (Figure 2A–B). Our findings are concordant with those of a previous report where HPV+ HNC tumors showed more PD-1+ CD8+ T cell infiltration, which correlated with favorable clinical outcome (16). These data suggest that PD-1 expressing cells are biologically relevant and may play a crucial role in HPV+ disease and PD-L1 induction. Importantly, we report that PD-L1 expression highly correlated with that of JAK2 at the mRNA (TCGA) and protein level in vivo (IHC). Additionally, we found that pJAK2 staining was significantly higher in HPV+ than HPV− specimens (data not shown). Interestingly, PD-L1 was also strongly correlated with a Th1 type profile regardless of HPV status suggesting that the PD-L1/PD-1 axis represents an important mechanism of immune evasion in both HPV negative and positive tumors, such that HPV− tumors may rely more on a tumor intrinsic (EGFR-driven) PD-L1 expression, while HPV+ tumors rely more on a tumor extrinsic IFNγ-mediated Th1 like response.
STAT1 was upregulated in HNC tumors when compared with paired autologous normal mucosa and that was significantly correlated with PD-L1 expression regardless of HPV status. Notably, components of other signaling pathways such as PI3K and MAPK, which have been previously associated with PD-L1 expression in other types of cancer (17, 28) did not show significant correlation with PD-L1 expression in HPV+ tumors or induce PD-L1. Indeed, the unique biology, mutational landscape and predominant signaling pathways in HNC may explain the discrepancy in PD-L1 expression. It has been recently reported that PTEN loss-of-function mutations are frequent in glioblastoma (31.9% of specimens, TCGA data) (42). Furthermore, PD-L1 was upregulated after PTEN loss/PI3K activation in glioblastoma lines, suggesting that gliomas may rely more on this signaling pathway (17). Likewise, in the setting of NSCLC, PD-L1 protein was upregulated after EGFR/RAS/MAPK pathway activating mutations. Indeed, HRAS and EGFR mutations in NSCLC are far more frequent than in HNC (2–5%) (43, 44). Therefore, mutant EGFR may induce a stronger MAPK pathway activation than wild type EGFR. On the other hand, PTEN or PIK3CA mutations are rather infrequent in HNC, 7% and 8% respectively (44). Hence, in the setting of HNC the intrinsic oncogenic signaling mostly depends on overexpressed wild type EGFR stimulation and presents as a unique feature of this type of cancer, in which the JAK/STAT3 oncogenic pathway is best characterized (45). Importantly, in our series, STAT3 showed no significant correlation with PD-L1 expression in HPV+ tumors and only a weak correlation in HPV− tumors, most likely because of higher EGFR expression in these tumors versus HPV+ ones.
Concordant with other types of cancer, IFNγ induced PD-L1 upregulation in all of the HNC cell lines tested in our study, however its upregulation was not PI3K dependent as reported for glioma, lymphoma or lung cancer (28, 46, 47). We believe this is the first report that specific JAK2 inhibition completely abrogates IFNγ-mediated PD-L1 upregulation at the mRNA and protein level. Interestingly, IFNα, which does not signal via JAK2, did not upregulate PD-L1 expression, confirming the specific role of JAK2 upregulating PD-L1. Likewise, we should emphasize that IFNα not only induces STAT1 phosphorylation but also STAT2, and complexes with IRF9 in the transcription factor assembly cascade, forming the ISGF3 transcription complex, where IRF9 is the main DNA binding domain (48). In contrast IFNγ mainly induces the formation of pSTAT1 dimers that directly bind to the promoter region of the target gene, which explains why IFNα may not upregulate PD-L1 although it still upregulates pSTAT1. In addition, and corroborating our TCGA findings, in vitro knockdown experiments show that STAT1 but not STAT3 mediates IFNγ induced PD-L1 upregulation, further supported by ChIP assay, providing additional evidence that pSTAT1 but not pSTAT3 binds to the PD-L1 promoter region as early as 30 minutes after IFNγ treatment. Our findings complement that of a previous report performed in lung cancer that shows IRF-1 binding to the PD-L1 promoter after IFNγ treatment (49). Interestingly, we are the first to report that cetuximab-mediated EGFR blockade significantly downregulates IFNγ-induced PD-L1 expression, suggesting cross-talk between the IFNγ and EGFR pathways in regulating PD-L1 expression mediated through STAT1 modulation.
EGFR and PD-L1 showed a higher correlation in HPV+ tumors that corresponds to the higher correlation seen with CD8A, JAK2 and STAT1 as well. These otherwise counterintuitive results may be explained by the fact that PD-L1 expression might be more dependent on the strength of EGFR/JAK2 pathway activation than EGFR higher expression in HPV− tumors, which may induce an increased STAT1 activation and induction of PD-L1 expression. Alternatively, other immune cells infiltrating the tumor microenvironment may also express PD-L1, such as dendritic cells, macrophages, monocytes, B cells as well as tumor associated fibroblasts and stromal cells (26, 50). PD-L1 expression on these cells may confound the strength of correlation with that of EGFR given that the expression of the latter is mostly on tumor cells, given that the TCGA RNAseq values represent unfractionated tumor.
Thus, JAK2/STAT1 signaling is a major common regulator for PD-L1 transcription driven by IFNγ and EGFR pathways. Since EGFR mutations are very rare in HNC (2%) (43), EGFR pathway overactivation, rather than activating mutations, are more important for PD-L1 upregulation in HNC. We are the first to report that wild type EGFR pathway induces PD-L1 upregulation at the mRNA and protein level, and that specific JAK2 inhibition significantly downregulated baseline and EGF-induced PD-L1 upregulation, though not completely, suggesting that other alternative pathways also contribute to PD-L1 expression in HNC. Noteworthy, JAK2 inhibition more effectively downregulated basal and EGF-mediated PD-L1 expression in EGFRhigh expressing cell lines (JHU029 and JHU022). In addition, we are the first to report that EGFR stimulation induces phosphorylation of STAT1, which in turn mediates PD-L1 upregulation, since its silencing completely abrogated this effect, EGFR and JAK2 inhibition may synergize downregulating the “intrinsic” PD-L1 expression. Most important, however, is the speculation of potential added benefit of JAK2 inhibition with the simultaneous blockade of the “extrinsic” IFNγ-mediated PD-L1 upregulation, which seems to be more important in HPV+ tumors. An added benefit of using combined JAK2 inhibition and cetuximab mediated EGFR blockade would be that the JAK2 mediated downregulation of PD-L1 expression on tumor cells would ultimately enhance the effector properties of PD-1+ NK cells activated in the tumor microenvironment. Indeed, we show that JAK2 inhibition in tumor targets enhances cetuximab mediated ADCC to a significant extent, therefore reversing PD-L1 mediated immunoescape of tumor cells to NK cell killing. Moreover, JAK2 inhibition might synergize with mAbs targeting PD-1 and/or CTLA-4 by enhancing ADCC of PD-1+CTL4+ lymphoid or myeloid suppressor immune infiltrates in the tumor microenvironment as well as reducing PD-L1 expression.
Supplementary Material
Acknowledgments
Financial support: National Institute of Health grants R01 DE19727, P50 CA097190, CA110249, University of Pittsburgh Cancer Center Support Grant P30CA047904 and P50CA101942 (GF).
Footnotes
Conflict of interest: Robert L. Ferris has received research grants from Bristol Myers Squibb and AZ/Medimmune and has been a consultant/advisory board member for Celgene, Merck, BMS and AZ/Medimmune. Gordon J. Freeman reports having ownership interest (including patents) for Merck, Bristol Myers Squibb, Roche, EMD Serono, Amplimmune, Boehringer Ingelheim and Novartis as well as a consultant/advisor for Novartis, Roche, BMS, Eli Lilly and Surface Oncology.
References
- 1.Dunn GP, Old LJ, Schreiber RD. The three Es of cancer immunoediting. Annual review of immunology. 2004;22:329–60. doi: 10.1146/annurev.immunol.22.012703.104803. [DOI] [PubMed] [Google Scholar]
- 2.Leibowitz MS, Andrade Filho PA, Ferrone S, Ferris RL. Deficiency of activated STAT1 in head and neck cancer cells mediates TAP1-dependent escape from cytotoxic T lymphocytes. Cancer immunology, immunotherapy : CII. 2011;60:525–35. doi: 10.1007/s00262-010-0961-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jie HB, Gildener-Leapman N, Li J, Srivastava RM, Gibson SP, Whiteside TL, et al. Intratumoral regulatory T cells upregulate immunosuppressive molecules in head and neck cancer patients. British journal of cancer. 2013;109:2629–35. doi: 10.1038/bjc.2013.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schreiber RD, Old LJ, Smyth MJ. Cancer immunoediting: integrating immunity’s roles in cancer suppression and promotion. Science. 2011;331:1565–70. doi: 10.1126/science.1203486. [DOI] [PubMed] [Google Scholar]
- 5.Pardoll DM. The blockade of immune checkpoints in cancer immunotherapy. Nature reviews Cancer. 2012;12:252–64. doi: 10.1038/nrc3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Topalian SL, Hodi FS, Brahmer JR, Gettinger SN, Smith DC, McDermott DF, et al. Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. The New England journal of medicine. 2012;366:2443–54. doi: 10.1056/NEJMoa1200690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brahmer JR, Tykodi SS, Chow LQ, Hwu WJ, Topalian SL, Hwu P, et al. Safety and activity of anti-PD-L1 antibody in patients with advanced cancer. The New England journal of medicine. 2012;366:2455–65. doi: 10.1056/NEJMoa1200694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Topalian SL, Drake CG, Pardoll DM. Targeting the PD-1/B7-H1(PD-L1) pathway to activate anti-tumor immunity. Current opinion in immunology. 2012;24:207–12. doi: 10.1016/j.coi.2011.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Taube JM, Klein A, Brahmer JR, Xu H, Pan X, Kim JH, et al. Association of PD-1, PD-1 ligands, and other features of the tumor immune microenvironment with response to anti-PD-1 therapy. Clinical cancer research : an official journal of the American Association for Cancer Research. 2014;20:5064–74. doi: 10.1158/1078-0432.CCR-13-3271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Seiwert TY, Burtness B, Weiss J, Gluck I, Eder JP, Pai SI, et al. A phase Ib study of MK-3475 in patients with human papillomavirus (HPV)-associated and non-HPV-associated head and neck (H/N) cancer. ASCO Meeting Abstracts. 2014;32:6011. [Google Scholar]
- 11.Ramqvist T, Dalianis T. Oropharyngeal cancer epidemic and human papillomavirus. Emerging infectious diseases. 2010;16:1671–7. doi: 10.3201/eid1611.100452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chung CH, Gillison ML. Human papillomavirus in head and neck cancer: its role in pathogenesis and clinical implications. Clinical cancer research : an official journal of the American Association for Cancer Research. 2009;15:6758–62. doi: 10.1158/1078-0432.CCR-09-0784. [DOI] [PubMed] [Google Scholar]
- 13.Fischer CA, Zlobec I, Green E, Probst S, Storck C, Lugli A, et al. Is the improved prognosis of p16 positive oropharyngeal squamous cell carcinoma dependent of the treatment modality? International journal of cancer Journal international du cancer. 2010;126:1256–62. doi: 10.1002/ijc.24842. [DOI] [PubMed] [Google Scholar]
- 14.Spanos WC, Nowicki P, Lee DW, Hoover A, Hostager B, Gupta A, et al. Immune response during therapy with cisplatin or radiation for human papillomavirus-related head and neck cancer. Archives of otolaryngology--head & neck surgery. 2009;135:1137–46. doi: 10.1001/archoto.2009.159. [DOI] [PubMed] [Google Scholar]
- 15.Lyford-Pike S, Peng S, Young GD, Taube JM, Westra WH, Akpeng B, et al. Evidence for a role of the PD-1:PD-L1 pathway in immune resistance of HPV-associated head and neck squamous cell carcinoma. Cancer research. 2013;73:1733–41. doi: 10.1158/0008-5472.CAN-12-2384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Badoual C, Hans S, Merillon N, Van Ryswick C, Ravel P, Benhamouda N, et al. PD-1-expressing tumor-infiltrating T cells are a favorable prognostic biomarker in HPV-associated head and neck cancer. Cancer research. 2013;73:128–38. doi: 10.1158/0008-5472.CAN-12-2606. [DOI] [PubMed] [Google Scholar]
- 17.Parsa AT, Waldron JS, Panner A, Crane CA, Parney IF, Barry JJ, et al. Loss of tumor suppressor PTEN function increases B7-H1 expression and immunoresistance in glioma. Nature medicine. 2007;13:84–8. doi: 10.1038/nm1517. [DOI] [PubMed] [Google Scholar]
- 18.Azuma K, Ota K, Kawahara A, Hattori S, Iwama E, Harada T, et al. Association of PD-L1 overexpression with activating EGFR mutations in surgically resected nonsmall-cell lung cancer. Annals of oncology : official journal of the European Society for Medical Oncology / ESMO. 2014;25:1935–40. doi: 10.1093/annonc/mdu242. [DOI] [PubMed] [Google Scholar]
- 19.Akbay EA, Koyama S, Carretero J, Altabef A, Tchaicha JH, Christensen CL, et al. Activation of the PD-1 pathway contributes to immune escape in EGFR-driven lung tumors. Cancer discovery. 2013;3:1355–63. doi: 10.1158/2159-8290.CD-13-0310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maiti GP, Mondal P, Mukherjee N, Ghosh A, Ghosh S, Dey S, et al. Overexpression of EGFR in head and neck squamous cell carcinoma is associated with inactivation of SH3GL2 and CDC25A genes. PloS one. 2013;8:e63440. doi: 10.1371/journal.pone.0063440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee SC, Srivastava RM, Lopez-Albaitero A, Ferrone S, Ferris RL. Natural killer (NK): dendritic cell (DC) cross talk induced by therapeutic monoclonal antibody triggers tumor antigen-specific T cell immunity. Immunologic research. 2011;50:248–54. doi: 10.1007/s12026-011-8231-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Srivastava RM, Lee SC, Andrade Filho PA, Lord CA, Jie HB, Davidson HC, et al. Cetuximab-activated natural killer and dendritic cells collaborate to trigger tumor antigen-specific T-cell immunity in head and neck cancer patients. Clinical cancer research : an official journal of the American Association for Cancer Research. 2013;19:1858–72. doi: 10.1158/1078-0432.CCR-12-2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li J, Jie HB, Lei Y, Gildener-Leapman N, Trivedi S, Green T, et al. PD-1/SHP-2 inhibits Tc1/Th1 phenotypic responses and the activation of T cells in the tumor microenvironment. Cancer research. 2015;75:508–18. doi: 10.1158/0008-5472.CAN-14-1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhao M, Sano D, Pickering CR, Jasser SA, Henderson YC, Clayman GL, et al. Assembly and initial characterization of a panel of 85 genomically validated cell lines from diverse head and neck tumor sites. Clinical cancer research : an official journal of the American Association for Cancer Research. 2011;17:7248–64. doi: 10.1158/1078-0432.CCR-11-0690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Heo DS, Snyderman C, Gollin SM, Pan S, Walker E, Deka R, et al. Biology, cytogenetics, and sensitivity to immunological effector cells of new head and neck squamous cell carcinoma lines. Cancer research. 1989;49:5167–75. [PubMed] [Google Scholar]
- 26.Curiel TJ, Wei S, Dong H, Alvarez X, Cheng P, Mottram P, et al. Blockade of B7-H1 improves myeloid dendritic cell-mediated antitumor immunity. Nature medicine. 2003;9:562–7. doi: 10.1038/nm863. [DOI] [PubMed] [Google Scholar]
- 27.Chikamatsu K, Sakakura K, Toyoda M, Takahashi K, Yamamoto T, Masuyama K. Immunosuppressive activity of CD14+ HLA-DR- cells in squamous cell carcinoma of the head and neck. Cancer science. 2012;103:976–83. doi: 10.1111/j.1349-7006.2012.02248.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Han SJ, Ahn BJ, Waldron JS, Yang I, Fang S, Crane CA, et al. Gamma interferon-mediated superinduction of B7-H1 in PTEN-deficient glioblastoma: a paradoxical mechanism of immune evasion. Neuroreport. 2009;20:1597–602. doi: 10.1097/WNR.0b013e32833188f7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schmitt NC, Trivedi S, Ferris RL. STAT1 Activation Is Enhanced by Cisplatin and Variably Affected by EGFR Inhibition in HNSCC Cells. Mol Cancer Ther. 2015;14:2103–11. doi: 10.1158/1535-7163.MCT-15-0305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Krutzik PO, Nolan GP. Intracellular phospho-protein staining techniques for flow cytometry: monitoring single cell signaling events. Cytometry Part A : the journal of the International Society for Analytical Cytology. 2003;55:61–70. doi: 10.1002/cyto.a.10072. [DOI] [PubMed] [Google Scholar]
- 31.Leibowitz MS, Srivastava RM, Andrade Filho PA, Egloff AM, Wang L, Seethala RR, et al. SHP2 is overexpressed and inhibits pSTAT1-mediated APM component expression, T-cell attracting chemokine secretion, and CTL recognition in head and neck cancer cells. Clinical cancer research : an official journal of the American Association for Cancer Research. 2013;19:798–808. doi: 10.1158/1078-0432.CCR-12-1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Haring M, Offermann S, Danker T, Horst I, Peterhansel C, Stam M. Chromatin immunoprecipitation: optimization, quantitative analysis and data normalization. Plant Methods. 2007;3:11. doi: 10.1186/1746-4811-3-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen BJ, Chapuy B, Ouyang J, Sun HH, Roemer MG, Xu ML, et al. PD-L1 expression is characteristic of a subset of aggressive B-cell lymphomas and virus-associated malignancies. Clinical cancer research : an official journal of the American Association for Cancer Research. 2013;19:3462–73. doi: 10.1158/1078-0432.CCR-13-0855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jie HB, Schuler PJ, Lee SC, Srivastava RM, Argiris A, Ferrone S, et al. CTLA-4(+) Regulatory T Cells Increased in Cetuximab-Treated Head and Neck Cancer Patients Suppress NK Cell Cytotoxicity and Correlate with Poor Prognosis. Cancer research. 2015;75:2200–10. doi: 10.1158/0008-5472.CAN-14-2788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cancer Genome Atlas N. Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature. 2015;517:576–82. doi: 10.1038/nature14129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kel AE, Gossling E, Reuter I, Cheremushkin E, Kel-Margoulis OV, Wingender E. MATCH: A tool for searching transcription factor binding sites in DNA sequences. Nucleic acids research. 2003;31:3576–9. doi: 10.1093/nar/gkg585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Purandare AV, McDevitt TM, Wan H, You D, Penhallow B, Han X, et al. Characterization of BMS-911543, a functionally selective small-molecule inhibitor of JAK2. Leukemia. 2012;26:280–8. doi: 10.1038/leu.2011.292. [DOI] [PubMed] [Google Scholar]
- 39.Lopez-Albaitero A, Ferris RL. Immune activation by epidermal growth factor receptor specific monoclonal antibody therapy for head and neck cancer. Archives of otolaryngology--head & neck surgery. 2007;133:1277–81. doi: 10.1001/archotol.133.12.1277. [DOI] [PubMed] [Google Scholar]
- 40.Ukpo OC, Thorstad WL, Lewis JS., Jr B7-H1 expression model for immune evasion in human papillomavirus-related oropharyngeal squamous cell carcinoma. Head and neck pathology. 2013;7:113–21. doi: 10.1007/s12105-012-0406-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Malm IJ, Bruno TC, Fu J, Zeng Q, Taube JM, Westra W, et al. Expression profile and in vitro blockade of programmed death-1 in human papillomavirus-negative head and neck squamous cell carcinoma. Head & neck. 2014 doi: 10.1002/hed.23706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ramkissoon SH, Bi WL, Schumacher SE, Ramkissoon LA, Haidar S, Knoff D, et al. Clinical implementation of integrated whole-genome copy number and mutation profiling for glioblastoma. Neuro Oncol. 2015;17:1344–55. doi: 10.1093/neuonc/nov015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McBride SM, Rothenberg SM, Faquin WC, Chan AW, Clark JR, Ellisen LW, et al. Mutation frequency in 15 common cancer genes in high-risk head and neck squamous cell carcinoma. Head & neck. 2014;36:1181–8. doi: 10.1002/hed.23430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stransky N, Egloff AM, Tward AD, Kostic AD, Cibulskis K, Sivachenko A, et al. The mutational landscape of head and neck squamous cell carcinoma. Science. 2011;333:1157–60. doi: 10.1126/science.1208130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sen M, Pollock NI, Black J, DeGrave KA, Wheeler S, Freilino ML, et al. JAK kinase inhibition abrogates STAT3 activation and head and neck squamous cell carcinoma tumor growth. Neoplasia. 2015;17:256–64. doi: 10.1016/j.neo.2015.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yamamoto R, Nishikori M, Tashima M, Sakai T, Ichinohe T, Takaori-Kondo A, et al. B7-H1 expression is regulated by MEK/ERK signaling pathway in anaplastic large cell lymphoma and Hodgkin lymphoma. Cancer science. 2009;100:2093–100. doi: 10.1111/j.1349-7006.2009.01302.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ota K, Azuma K, Kawahara A, Hattori S, Iwama E, Tanizaki J, et al. Induction of PD-L1 Expression by the EML4-ALK Oncoprotein and Downstream Signaling Pathways in Non-Small Cell Lung Cancer. Clinical cancer research : an official journal of the American Association for Cancer Research. 2015;21:4014–21. doi: 10.1158/1078-0432.CCR-15-0016. [DOI] [PubMed] [Google Scholar]
- 48.Sadzak I, Schiff M, Gattermeier I, Glinitzer R, Sauer I, Saalmuller A, et al. Recruitment of Stat1 to chromatin is required for interferon-induced serine phosphorylation of Stat1 transactivation domain. Proc Natl Acad Sci U S A. 2008;105:8944–9. doi: 10.1073/pnas.0801794105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lee SJ, Jang BC, Lee SW, Yang YI, Suh SI, Park YM, et al. Interferon regulatory factor-1 is prerequisite to the constitutive expression and IFN-gamma-induced upregulation of B7-H1 (CD274) FEBS letters. 2006;580:755–62. doi: 10.1016/j.febslet.2005.12.093. [DOI] [PubMed] [Google Scholar]
- 50.Freeman GJ, Long AJ, Iwai Y, Bourque K, Chernova T, Nishimura H, et al. Engagement of the PD-1 immunoinhibitory receptor by a novel B7 family member leads to negative regulation of lymphocyte activation. The Journal of experimental medicine. 2000;192:1027–34. doi: 10.1084/jem.192.7.1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.