Abstract
Fusobacterium nucleatum is a strictly anaerobic, Gram negative bacterial species that has been associated with dental infections, pre-term labour, appendicitis, inflammatory bowel disease, and, more recently, colorectal cancer. The species is unusual in its phenotypic and genotypic heterogeneity, with some strains demonstrating a more virulent phenotype than others; however, as yet the genetic basis for these differences is not understood. Bacteriophage are known to contribute to the virulence phenotype of several bacterial species. In this work, we set out to characterize the bacteriophage associated with F. nucleatum subsp. animalis strain 7-1, a highly invasive isolate from the human gastrointestinal tract. As well, we used computational approaches to predict and compare bacteriophage signatures across available sequenced Fusobacterium nucleatum genomes.
Introduction
Recently the potential importance of the strictly anaerobic, Gram negative bacterial species, Fusobacterium nucleatum, in gastrointestinal (GI) diseases such as inflammatory bowel disease and colorectal cancer has become evident1,2,3. F. nucleatum is unusual in its heterogeneity, with a wide range of phenotypic and genotypic variation evident within the species4. For example, a high degree of serovar and ribotype heterogeneity, as well as differences in 16S rRNA gene-based DGGE profiling, have been observed among F. nucleatum strains4,5. It is also evident that there may be strain-dependent differences in virulence (e.g. invasive ability)6. As such, a greater understanding of the virulence potential of F. nucleatum is warranted, given the emergence of this species as an opportunistic pathogen.
Bacteriophages are viruses that can infect only bacteria7. They are often used as powerful tools for the study of bacterial genetics, and, given their host specificity, are useful in the identification and characterization of their host bacterial species8. The contribution of phage to the pathogenicity of their bacterial hosts has been well documented9, 10. Exotoxins are the most widely recognized virulence factor linked to phage infection, with the most common example being the cholera toxin gene located in the genome of CTXɸ, a bacteriophage from Vibrio cholerae11. Bacteriophages have also been shown to alter other host bacterial properties including bacterial adhesion, colonization, invasion, the spread through human tissues, resistance to immune defences, resistance to antibiotics and transmissibility among humans9.
Therefore, the identification and characterization of F. nucleatum bacteriophage may help to define their roles in pathogenesis. In addition, a greater understanding of bacteriophage and their associated genomes may help to elucidate the evolution of the species, as well as to delineate a method for strain typing, which for the heterogeneous F. nucleatum species is very complex4. Previous to this work, only 1 F. nucleatum bacteriophage, Fnpɸ2, has been described in detail8.
With this in mind, we successfully induced, purified and subsequently fully sequenced and analyzed two bacteriophage from the highly invasive F. nucleatum subspecies animalis strain 7-1 (also known as strain EAVG_002), and designated them ɸFunu1 (Genbank accession no. KR131710) and ɸFunu2 (Genbank accession nos. KR131711 and KR131712). We also computationally predicted phage across all sequenced Fusobacterium genomes.
Materials and Methods
Bacterial strains and media
F. nucleatum subspecies animalis strain 7-1 was examined for the presence of prophage. This highly invasive strain was isolated directly from a biopsy taken from inflamed Sigmoid colon tissue from a male Crohn's disease patient6. The F. nucleatum strain was grown in a broth culture of tryptic soy broth supplemented with hemin (5 μg/mL) and menadione (1 μg/mL).
Mitomycin C induction
Briefly, to induce prophage, an early log-phase broth culture of F. nucleatum subspecies animalis 7-1 was incubated at 37°C until its absorbance at 600 nm was between 0.2-0.3, at which time mitomycin C was added at a final concentration of 2.5 μg/mL. Absorbance at 600 nm was measured for 4 hours, at which time lysis was observed. The lysed culture was then treated with DNase and RNase (1 μg/mL), centrifuged at low speed and the supernatant was sterilized by filtration using a 0.45μm polyethersulfone membrane12, 13. Plaque assays to assess bacteriophage load were also performed on cell free-lysates12.
Electron microscopy of bacteriophage particles
The phage suspension was dropped onto copper grids with carbon-coated formvar film and incubated for 30 seconds. Excess solution was drained away on filter paper and washed five times with de-ionized water to remove filtrate debris. The grids were then incubated with 1% uranyl acetate for 10 seconds and the negatively stained phage particles were them viewed with a Phillips CM10 transmission electron microscope at 120kV.
Bacteriophage purification and DNA extraction
Bacteriophages in the cell-free filtrate were precipitated using polyethylene glycol (10% w/v) and sodium chloride (1M). Subsequently, the bacteriophages were purified using a cesium chloride (CsCl) gradient (1.2 g/L- 1.6 g/L) and ultracentrifugation14, 15. Phage particles forming a band at the 1.3 g/L density were collected and dialysis was performed using SM buffer12. Genomic DNA was extracted by incubating the purified bacteriophage particles in SM buffer with Proteinase K (50 μg/mL) and SDS (0.5% w/v) at 56°C for 1h followed by a phenol:chloroform (1:1) extraction and DNA precipitation by ethanol14.
Bacteriophage DNA sequencing
Sequencing was accomplished using Illumina Sequencing. Briefly, target DNA was sheared using the Covaris AFA process (Covaris, Woburn, MA), and then a sequencing library was generated using Kapa library prep and amplification kits (Kapa Biosystems, Inc., Wilmington, MA), with dual indexing using the Agilent Bravo system (Agilent Technologies, Santa Clara, CA). Library samples were quantified by qPCR using the Illumina Eco platform (Illumina, San Diego, CA), denatured and then amplified onto an 8-channel flowcell using an Illumina CBot. Paired end sequencing was performed on the Illumina HiSeq platform with dual index reads and v.3 chemistry. The de novo sequencing strategy generated 1656436 reads and attained approximately 428-fold coverage. Assembly of all the reads was performed using ALLPATHS version R45962, which gave a 35x fragment coverage. The ALLPATHS parameter ASSISTED_PATCHING=2.1 was used with a reference sequence generated from another F. nucleatum 7_1 assembly containing the phage sequences. Phage assemblies were analyzed using the GAEMR genome analysis package (http://www.broadinstitute.org/software/gaemr/) and were reviewed prior to annotation.
Bacteriophage gene annotation
Annotations of the obtained scaffolds were done using the Broad Institute's prokaryotic annotation pipeline. Briefly, the protein-coding genes were predicted with Prodigal16 and filtered to remove genes with ≥70% overlap to tRNAs or rRNAs. The tRNAs were identified by tRNAscan-SE17. The rRNA genes were predicted using RNAmmer18. The gene product names were assigned based on top BLAST hits against SwissProt protein database (≥70% identity and ≥70% query coverage), and protein family profile search against the TIGRfam hmmer equivalogs. Additional annotation analyses performed include PFAM19, TIGRfam20, KEGG21, COG22, GO23, EC24, SignalP25, and TMHMM26.
Computational phage prediction and cluster analysis
In order to compare our sequenced phage with phage from other Fusobacterium strains, we computationally predicted bacteriophage in a set of 29 genomes, using the PHAST phage prediction software27, which was able to correctly predict the ɸFunu1 and ɸFunu2 phage regions within Fusobacterium nucleatum 7_1. We analyzed the set of 26 fusobacterial genomes and one Leptotrichia buccalis outgroup from a previous fusobacterial comparative analysis dataset28, together with two additional Fusobacterium strains from Genbank: Fusobacterium nucleatum subsp. polymorphum 13-3C (Genbank accession GCA_000523555.1) and Fusobacterium nucleatum CC53 (Genbank accession GCA_000347315.1). To cluster our predicted phage, we performed all versus all pairwise alignment of predicted prophage sequences using the NUCmer program from the MUMmer suite29 (using the –maxmatch parameter). We collapsed aligned regions, computed pairwise overall coverage values, and discarded values below 80%. Using the remaining overall coverage as input, we clustered phage sequences using the MCL algorithm30 (using the mcxload parameter “--stream-mirror” and the mcl parameters “-I 1.4”).
Results
Mitomycin C induction
Mitomycin C successfully induced lysis for F. nucleatum subspecies animalis strain 7-1. Two bacteriophages were obtained from this strain and were designated ɸFunu1 and ɸFunu2. Phage titers in induced lysates were undetermined as both ɸFunu1 and ɸFunu2 were unable to produce plaques using a standard plaque assay and a variety of different F. nucleatum including the originating strain and isolates representative of the animalis, vincentii and polymorphum subspecies (data not shown).
Sequence and analysis of Fusobacterium phage ɸFunu1
The genome of ɸFunu1 consisted of linear double-stranded DNA (dsDNA), with one scaffold of length of 39921 bp and GC content of 27%. ɸFunu1 mapped to a co-linear stretch of the 7_1 genome from positions 810,600 to 854,000. Annotation of ɸFunu1 revealed 66 coding DNA sequences. Further genome analysis revealed that 71.2% of the ɸFunu1 genes (47 of 66) encoded unique proteins with no reliable identity to database entries. Annotation also revealed that ɸFunu1 had zero tRNA genes, one integrase gene, one capsid gene and seven genes associated with DNA replication, recombination and repair. According to Phast (http://phast.wishartlab.com/) ɸFunu1 was most similar to BcepMu, a Mu-like myoviridiae phage from Burkholderia cenocepacia31. Further verification from Virfam (http://biodev.cea.fr/virfam/Default.aspx) confirmed ɸFunu1 is most related to viruses from the family myoviridiae.
Sequence and analysis of Fusobacterium phage ɸFunu2
The genome of the ɸFunu2 consisted of linear dsDNA comprised of two scaffolds (lengths 38,801 and 1043 bp), for a total length of 39,844 bp, with GC content of 27.7%. The ɸFunu2 genome mapped to a co-linear stretch of the 7_1 genome from position 2,205,500 to 2,244,400. Annotation by Prodigal revealed 71 coding sequences. Of these 71 genes, 56 (78.9%) encoded hypothetical proteins with no reliable identity to database entries. Genome analysis also revealed ɸFunu2 had no tRNA genes, one integrase gene, one envelope (coat) gene and nine genes associated with DNA replication, recombination and repair. No significant homology to known bacterial virulence genes were detected through comparison of sequences to the Virulence Factors of Pathogenic Bacteria (VFPB) database (http://www.mgc.ac.cn/VFs/). Interestingly, the ɸFunu2 genome includes a toxin secretion/bacteriophage lysis holin gene, yet does not contain a known endolysin gene, and we were unable to produce plaques in a plaque assay with purified phage on soft agar. Both PHAST and VIRFAM showed that ɸFunu2 is most similar to SboM-AG3, a phage that infects Shigella boydii that also belongs to the myoviridiae family32. Using the NUCmer program to align the DNA sequences of ɸFunu1 and ɸFunu2, we observed a short region of sequence similarity between them.
Electron Microscopy of the bacteriophage particles
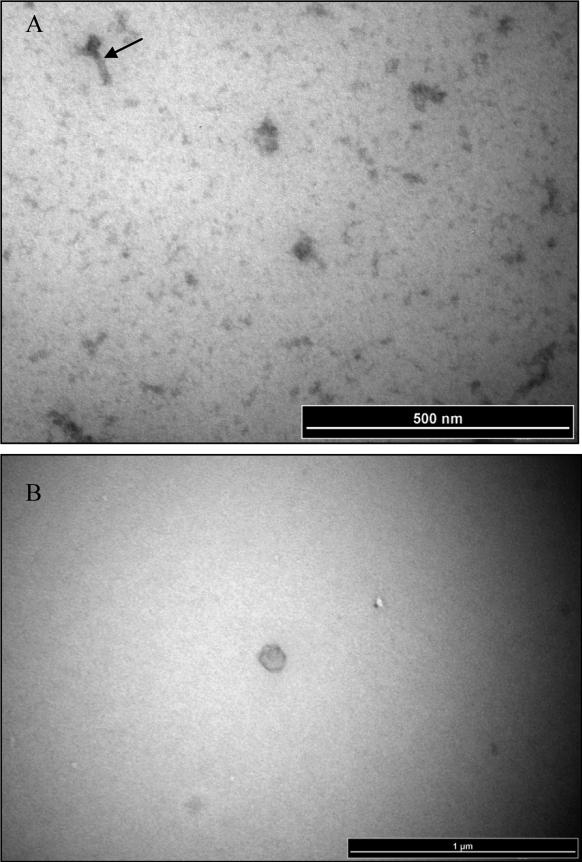
Two distinct phage morphologies were observed, representing both ɸFunu1 and ɸFunu2 (see Figure 2A and B). Both virion morphologies have a rough pentagonal outline indicating an icosahedral nature, as is the case with most virions from the myoviridae family. The virion head size ranges from 30-40 nm in diameter for ɸFunu1 and 50-60 nm for ɸFunu2. As suggested by the annotation of the ɸFunu1 genome indicating tail proteins, ɸFunu1 is assumed to be the phage with a tail visible in the electron micrographs. The tail region appears to be 60-120 nm in length and includes a distinct neck region.
Figure 2.
Representative electron micrograph images of phage particles isolated and purified from F.nucleatum strain 7_1. Panels A) ɸFunu1; and B) ɸFunu2.
Arrow in panel A indicates a putative phage tail structure. Note the absence of a tail structure for the representative virion head imaged in panel B.
Predicted phage in other Fusobacterial strains
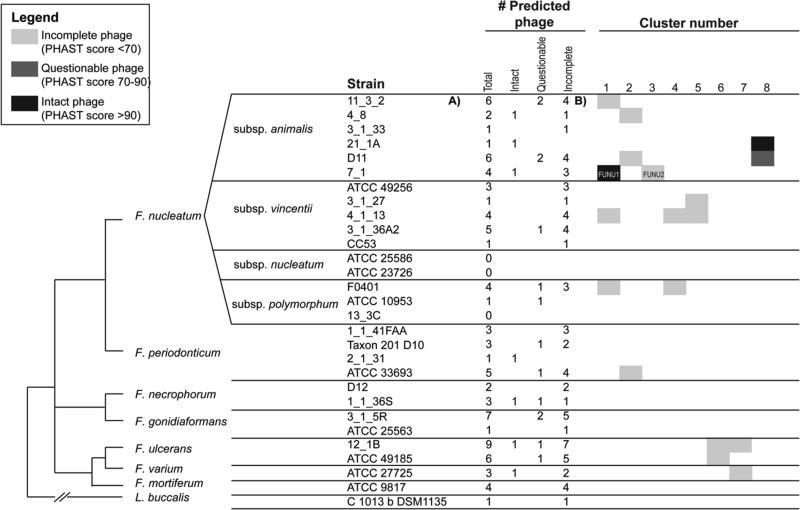
In order to compare phage content of Fusobacterium nucleatum subsp animalis 7_1 with phage in other sequenced Fusobacterium strains, we used the PHAST tool27 to computationally predict phage in 28 Fusobacterium strains and one related Leptotrichia strain (see Materials and Methods). PHAST was able to correctly predict the presence of two prophage (ɸFunu1 and ɸFunu2) in Fusobacterium nucleatum subsp animalis 7_1. Based on our phage predictions in the other strains, we believe that many strains of F. nucleatum harbour temperate bacteriophage within their genomes, and that there is a high level of phage diversity within the Fusobacterium genus. Across the 29 genomes, we observed a total of 87 predicted phage (7 “intact”, 14 “questionable”, and 66 “incomplete”) (see Figure 1).
Figure 1.
Results of clustering analysis carried out for predicted phage sequences. Strains are arranged according to previously published phylogeny (28). F. nucleatum strains 11_3_2, 4_1_13 and F0401 all contain similar phage sequences that cluster with ɸFunu1. ɸFunu2 is unique among the strains examined.
Based on our clustering analysis (see Materials and Methods), F. nucleatum strains 11_3_2, 4_1_13 and F0401 all contain similar phage sequences that cluster with ɸFunu1 (Figure 1). These were all predicted by PHAST as “incomplete” phage predictions (PHAST score <70). Sequences similar to ɸFunu2 could not be detected in the sequenced genomes of all other F. nucleatum strains sequenced to date.
Discussion
Bacteriophage are often analyzed to help give clues to the pathogenicity of bacteria since the discovery of the cholera toxin within ɸCTX11. Due to the genotypic, phenotypic, phylogenetic and biochemical heterogeneity observed within the F. nucleatum genus4 and the uncertainty about Fusobacterium spp. influence on various inflammatory diseases3, we hoped to find clues about this opportunistic pathogen's virulence potential within the genome of its harboured bacteriophage. Although both ɸFunu1 and ɸFunu2 genome sequences did not definitively identify with virulence associated genes, many of their genes coded for hypothetical proteins of unknown function, and it is possible that these predicted genes are involved in as-yet undetermined virulence mechanisms. Regardless, we have still obtained valuable information concerning the predicted gene content of the two F. nucleatum bacteriophages, and in addition, through our phage sequencing efforts, the phage sequence clustering algorithm designed at The Broad Institute, as well as PHAST, may be further evaluated for prediction of phage genomes within the Fusobacterium species.
Both ɸFunu1 and ɸFunu2 possess some interesting characteristics. This includes the inability of the phage lysates to induce plaque formation in a range of different F. nucleatum isolates using a simple plaque assay. Although ɸFunu2 does not appear to have any predicted proteins related to a phage tail, which may provide answers as to why it cannot bind to F. nucleatum and induce active lysis, ɸFunu1 has multiple proteins believed to be involved in tail generation. These tails were observed in the electron micrographs (see Figure 2A). However, there may be a defect in these tail proteins in terms of binding capabilities. Perhaps during the isolation and purification of the phage, pivotal phage tail proteins, responsible for bacterial attachment, may have been sheared off, rendering the phage unable to propagate by integration into another fusobacterial cell. Interestingly, ɸFunu2 does appear to have a holin gene, suggesting its ability to form pores in the bacterial cell membranes exposing the peptidoglycan, there is no suggestion of an endolysin gene which would allow the bacteriophage to fully degrade the bacterial cell membrane's peptidoglycan33. This gene loss may be responsible for the inability of ɸFunu2 to induce plaque formation. It also may also explain the improper formation of the full phage particle, as seen in some of the electron micrographs (see Figure 2B). Since the release of the bacteriophage from Fusobacterium nucleatum subsp animalis 7_1 does not appear to be associated with bacterial cell rupture, it is more difficult to determine the infectivity of these two bacteriophage. It is possible that F. nucleatum bacteriophages are able to encase themselves in the F. nucleatum bacterial cell outer membrane and bleb out of the bacteria without causing lysis. The phenomenon of a budding bacteriophage has been previously reported for certain mycoplasmaviruses34.
In addition to the induction, purification and sequencing of the two bacteriophages from Fusobacterium nucleatum subsp animalis 7_1, we also tested the phage sequence clustering algorithm, designed at the Broad Institute. This algorithm and the PHAST phage prediction software were used to predict other phages located in the genomes of 28 other fully sequenced Fusobacterium strains isolated from patients with IBD or undergoing colorectal cancer screening. This was done for two reasons. The first was with the sequencing of ɸFunu1 and ɸFunu2 we could determine the validity of the prediction software. The second reasoning was to see if the heterogeneity observed in the phage isolates from the 7-1strain was also observed in other Fusobacterium strains. As with any genomic analysis, there are shortcomings to the computational analysis. Because we also predicted two additional “incomplete” phage sequences within Fusobacterium nucleatum subsp animalis 7_1, which we did not identify in our experimental phage extraction, it is possible that our computational phage prediction is identifying a substantial number of false positives. In addition, the diverse assembly qualities of the genomes in our dataset will impact our ability to identify phage using computational methods. It is more difficult to identify phage in genomes with a large number of scaffolds, such as F. nucleatum CC53 (only one phage identified). However, we saw substantial diversity in the number of predicted phage even within the finished genomes in our dataset: some of our finished genomes had zero or only one predicted phage (including F. nucleatum subsp. polymorphum 10953, F. nucleatum subsp. nucleatum 25586, F. nucleatum subsp. vincentii 3_1_27), while some of our finished genomes had higher numbers of predicted phage (F. nucleatum subsp. vincentii 3_1_36A2). In addition, some of the F. nucleatum phage clusters did not correlate with phylogeny, indicating the possibility of horizontal transfer of mobile phage elements across different F. nucleatum subspecies. For instance, phage cluster 1 includes members from the animalis, vincentii, and polymorphum subspecies, and cluster 2 includes members from the vincentii subspecies, as well as from F. periodonticum. In contrast, cluster 4 appears to be specific to the vincentii subspecies, and cluster 5 appears to be specific to F. ulcerans.
Overall, the most interesting discovery, although not unexpected, was the heterogeneity observed in the two sequenced and annotated phages, ɸFunu1 and ɸFunu2, as well the heterogeneity observed when looking at the predicted Fusobacterium phages using the cluster analysis and phage prediction software. These data further suggest that F. nucleatum heterogeneity may be related in part to phage acquisition. The roles of ɸFunu1 and ɸFunu2 in F. nucleatum subspecies animalis strain 7-1 virulence are as-yet undefined, but since F. nucleatum strains are known to differ both in virulence (e.g. invasive ability) as well as phage complement, further investigation of the roles of genes encoded by these phage genomes is warranted.
Highlights.
Two phage particles were induced, purified and analyzed from an invasive F.nucleatum subspecies animalis strain, and designated ɸFunu1 and ɸFunu2.
Electron microscopy, genome sequencing and protein annotation were performed on ɸFunu1 and ɸFunu2 in an effort to better understand the role these bacteriophage may have in F. nucleatum virulence.
ɸFunu1 and ɸFunu2 are myoviridae of genome lengths 43,921 bp and 39,844 bp respectively.
PHAST: A Fast Phage Search Tool, was assessed for its ability to properly predict bacteriophage within various fusobacterial genomes.
Acknowledgements
The authors acknowledge a Crohn's and Colitis Canada Grant in Aid of Research, awarded to E.A.-V. This project was also funded in part by the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services, under contract no. HHSN272200900018C (A.M.-M. and A.M.E.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Literature Cited
- 1.Castellarin M, Warren RL, Freeman JD, Warren L, Dreolini L, Krzywinski M, Holt RA. Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Genome Research. 2012:299–306. doi: 10.1101/gr.126516.111. doi:10.1101/gr.126516.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kostic AD, Gevers D, Pedamallu CS, Michaud M, Duke F, Earl AM, Meyerson M. Genomic analysis identifies association of Fusobacterium with colorectal carcinoma. Genome Research. 2012:292–298. doi: 10.1101/gr.126573.111. doi:10.1101/gr.126573.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Allen-Vercoe E, Strauss J, Chadee K. Fusobacterium nucleatum: An emerging gut pathogen? Gut Microbes. 2011;2(5):294–298. doi: 10.4161/gmic.2.5.18603. doi:10.4161/gmic.2.5.18603. [DOI] [PubMed] [Google Scholar]
- 4.Thurnheer T, Guggenheim B, Gruica B, Gmur R. Infinite serovar and ribotype heterogeneity among oral Fusobacterium nucleatum strains? Anaerobe. 1999;5:79–92. doi:10.1006/anae.1999.0188. [Google Scholar]
- 5.Strauss J, White A, Ambrose C, McDonald J, Allen-Vercoe E. Phenotypic and genotypic analyses of clinical Fusobacterium nucleatum and Fusobacterium periodonticum isolates from the human gut. Anaerobe. 2008;14(6):301–309. doi: 10.1016/j.anaerobe.2008.12.003. doi:10.1016/j.anaerobe.2008.12.003. [DOI] [PubMed] [Google Scholar]
- 6.Strauss J, Kaplan GG, Beck PL, Rioux K, Panaccione R, Devinney R, Allen-Vercoe E. Invasive potential of gut mucosa-derived Fusobacterium nucleatum positively correlates with IBD status of the host. Inflammatory Bowel Diseases. 2011;17(9):1971–1978. doi: 10.1002/ibd.21606. doi:10.1002/ibd.21606. [DOI] [PubMed] [Google Scholar]
- 7.Fong J. Bacteriophage. University of California; Berkeley, California, U.S.A.: 1941. [Google Scholar]
- 8.Machuca P, Daille L, Vinés E, Berrocal L, Bittner M. Isolation of a novel bacteriophage specific for the periodontal pathogen Fusobacterium nucleatum. Applied and Environmental Microbiology. 2010;76(21):7243–7250. doi: 10.1128/AEM.01135-10. doi:10.1128/AEM.01135-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wagner PL, Waldor MK. Bacteriophage Control of Bacterial Virulence. Infection and Immnunity. 2002;70(8):3985–3993. doi: 10.1128/IAI.70.8.3985-3993.2002. doi:10.1128/IAI.70.8.3985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Preus HR, Olsen I, Gjermo P. Bacteriophage infection--a possible mechanism for increased virulence of bacteria associated with rapidly destructive periodontitis. Acta Odontologica Scandinavica. 1987;45(1):49–54. doi: 10.3109/00016358709094353. doi:10.3109/00016358709094353. [DOI] [PubMed] [Google Scholar]
- 11.McLeod SM, Kimsey HH, Davis BM, Waldor MK. CTX and Vibrio cholerae: Exploring a newly recognized type of phage-host cell relationship. Molecular Microbiology. 2005;57(2):347–356. doi: 10.1111/j.1365-2958.2005.04676.x. doi:10.1111/j.1365-2958.2005.04676.x. [DOI] [PubMed] [Google Scholar]
- 12.Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, N.Y.: 1989. [Google Scholar]
- 13.Timme T, Brennan P. Induction of bacteriophage from members of the Mycobacterium avium, Mycobacterium intracellulare, Mycobacterium scrofulaceum Serocomplex. Journal of General Microbiology. 1984;130:2059–2066. doi: 10.1099/00221287-130-8-2059. [DOI] [PubMed] [Google Scholar]
- 14.Sambrook J, Russel DW. Molecular cloning: A laboratory manual. Cold Spring Harbour Laboratory Press; Cold Spring Harbour, N.Y.: 2001. [Google Scholar]
- 15.Humphrey SB, Stanton TB, Jensen NS, Zuerner RL. Purification and characterization of VSH-1, a generalized transducing bacteriophage of Serpulina hyodysenteriae. Journal of Bacteriology. 1997;179(2):323–329. doi: 10.1128/jb.179.2.323-329.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hyatt D, Chen G-L, Locascio PF, Land ML, Larimer FW, Hauser LJ. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics. 2010;11:119. doi: 10.1186/1471-2105-11-119. doi:10.1186/1471-2105-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lowe TM, Eddy SR. tRNAscan-SE: A program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Research. 1997;25(5):955–964. doi: 10.1093/nar/25.5.955. doi:10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lagesen K, Hallin P, Rødland EA, Stærfeldt HH, Rognes T, Ussery DW. RNAmmer: Consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Research. 2007;35(9):3100–3108. doi: 10.1093/nar/gkm160. doi:10.1093/nar/gkm160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Finn RD, Tate J, Mistry J, Coggill PC, Sammut SJ, Hotz H-R, Bateman A. The Pfam protein families database. Nucleic Acids Research, 36(Database) 2007:D281–D288. doi: 10.1093/nar/gkm960. doi:10.1093/nar/gkm960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haft DH, Loftus BJ, Richardson DL, Yang F, Eisen JA, Paulsen IT, White O. TIGRFAMs: a protein family resource for the functional identification of proteins. Nucleic Acids Research. 2001;29(1):41–43. doi: 10.1093/nar/29.1.41. doi:10.1093/nar/29.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ogata H, Goto S, Sato K, Fujibuchi W, Bono H, Kanehisa M. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Research. 1999;27(1):29–34. doi: 10.1093/nar/27.1.29. doi:10.1093/nar/27.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tatusov RL, Koonin EV, Lipman DJ. A genomic perspective on protein families. Science (New York, N.Y.) 1997;278(5338):631–637. doi: 10.1126/science.278.5338.631. doi:10.1126/science.278.5338.631. [DOI] [PubMed] [Google Scholar]
- 23.Conesa A, Götz S, García-Gómez JM, Terol J, Talón M, Robles M. Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics. 2005;21(18):3674–3676. doi: 10.1093/bioinformatics/bti610. doi:10.1093/bioinformatics/bti610. [DOI] [PubMed] [Google Scholar]
- 24.Tian W, Arakaki AK, Skolnick J. EFICAz: A comprehensive approach for accurate genome-scale enzyme function inference. Nucleic Acids Research. 2004;32(21):6226–6239. doi: 10.1093/nar/gkh956. doi:10.1093/nar/gkh956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Petersen TN, Brunak S, von Heijne G, Nielsen H. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nature Methods. 2011;8(10):785–786. doi: 10.1038/nmeth.1701. doi:10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- 26.Krogh A, Larsson B, von Heijne G, Sonnhammer EL. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. Journal of Molecular Biology. 2001;305(3):567–580. doi: 10.1006/jmbi.2000.4315. doi:10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 27.Zhou Y, Liang Y, Lynch KH, Dennis JJ, Wishart DS. PHAST: A Fast Phage Search Tool. Nucleic Acids Research. 2011;39(SUPPL. 2):347–352. doi: 10.1093/nar/gkr485. doi:10.1093/nar/gkr485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McGuire A, Cochrane K, Griggs AD, Haas BJ, Abeel T, Zeng Q, Nice JB, MacDonald H, Birren BW, Berger BW, Allen-Vercoe E, Earl AM. Evolution of Invasion in a Diverse Set of Fusobacterium Species. Microbiome. 2014;5(6):1–11. doi: 10.1128/mBio.01864-14. doi:10.1128/mBio.01864-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kurtz S, Phillippy A, Delcher AL, Smoot M, Shumway M, Antonescu C, Salzberg SL. Versatile and open software for comparing large genomes. Genome Biology. 2004;5(2):R12. doi: 10.1186/gb-2004-5-2-r12. doi:10.1186/gb-2004-5-2-r12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Enright A,J, Dongen SV, Ouzounis CA. An efficient algorithm for large-scale detection of protein families. Nucleic Acids Research. 2002;30(7):1575–1584. doi: 10.1093/nar/30.7.1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Summer EJ, Gonzalez CF, Carlisle T, Mebane LM, Cass AM, Savva CG, Young R. Burkholderia cenocepacia phage BcepMu and a family of Mu-like phages encoding potential pathogenesis factors. Journal of Molecular Biology. 2004;340(1):49–65. doi: 10.1016/j.jmb.2004.04.053. doi:10.1016/j.jmb.2004.04.053. [DOI] [PubMed] [Google Scholar]
- 32.Anany H, Lingohr EJ, Villegas A, Ackermann H-W, She Y-M, Griffiths MW, Kropinski AM. A Shigella boydii bacteriophage which resembles Salmonella phage ViI. Virology Journal. 2011;8(1):242. doi: 10.1186/1743-422X-8-242. doi:10.1186/1743-422X-8-242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang IN, Smith DL, Young R. Holins: the protein clocks of bacteriophage infections. Annual Review of Microbiology. 2000;54:799–825. doi: 10.1146/annurev.micro.54.1.799. doi:10.1146/annurev.micro.54.1.799. [DOI] [PubMed] [Google Scholar]
- 34.Maniloff J, Cadden SP, Putzrath RM. Maturation of an enveloped budding phage: mycoplasmavirus L2. Progress in Clinical Biological Research. 1981;64:503–13. [PubMed] [Google Scholar]